Abstract
DNA double-strand breaks (DSBs) are linked to neurodegeneration and senescence. However, it is not clear how DSB-bearing neurons influence neuroinflammation associated with neurodegeneration. Here, we characterize DSB-bearing neurons from the CK-p25 mouse model of neurodegeneration using single-nucleus, bulk, and spatial transcriptomic techniques. DSB-bearing neurons enter a late-stage DNA damage response marked by nuclear factor κB (NFκB)–activated senescent and antiviral immune pathways. In humans, Alzheimer’s disease pathology is closely associated with immune activation in excitatory neurons. Spatial transcriptomics reveal that regions of CK-p25 brain tissue dense with DSB-bearing neurons harbor signatures of inflammatory microglia, which is ameliorated by NFκB knockdown in neurons. Inhibition of NFκB in DSB-bearing neurons also reduces microglia activation in organotypic mouse brain slice culture. In conclusion, DSBs activate immune pathways in neurons, which in turn adopt a senescence-associated secretory phenotype to elicit microglia activation. These findings highlight a previously unidentified role for neurons in the mechanism of disease-associated neuroinflammation.
Neurons with DNA damage trigger microglia activation and neurodegeneration through antiviral-like signaling.
INTRODUCTION
Loss of genomic integrity is linked to aging and neurodegeneration (1, 2). DNA damage repair pathways are transcriptionally prominent in the aging brain, and many age-associated neurodegenerative diseases exhibit both accumulation of DNA lesions and reduced DNA repair efficiency (3–5). The most toxic of these lesions, the DNA double-strand break (DSB), can drive many phenotypes of aging including senescence, mutation, and cell death. Postmitotic neurons are particularly susceptible to these threats due to their long life span, high metabolic activity, and limited DSB repair capacity. While DSB accumulation in neurons is a well-documented feature of aging and neurodegeneration, the transcriptional profile adopted by such population of neurons remains largely unknown.
The accumulation of DSBs is an early feature of Alzheimer’s disease (AD), suggesting that they may act as an initiating lesion of toxicity (6). Multiple mouse models of neurodegeneration phenocopy increased DSBs at early pathological stages, including the Tau P301S and P301L tauopathy models, the inducible CK-p25 model, and the hAPP-J20 amyloid pathology model (7–9). Recent studies characterizing DNA strand breaks in postmitotic neurons reveal that break location may underlie the functional decline of the brain in aging and neurodegenerative disease (10, 11). However, regardless of location, the downstream biological effects of DSB burden in neurons are unclear.
Here, we sought to characterize the biological consequences of DSB accumulation in neurons. We also investigated how this affects mechanisms of neuroinflammation in age-associated neurodegenerative disease. We used fluorescence-activated nuclei sorting (FANS) followed by bulk and single-nucleus RNA sequencing (snRNA-seq) to transcriptionally characterize neurons burdened with DSBs in the CK-p25 mouse model of neurodegeneration. We found that DSB-bearing neurons activate innate immune signaling pathways reminiscent of those expressed by senescent cells and neurons infected by virus (12–14). This is accompanied by degradation of neuronal identity. The gene expression patterns of DSB-burdened neurons were enriched in excitatory neurons from AD postmortem human brain. Spatial transcriptomics of the CK-p25 forebrain revealed that signatures of microglial inflammation were proximally associated with DSB-bearing neurons. Correspondingly, suppression of the nuclear factor κB (NFκB) transcription factor in neurons reduced microglial proliferation and activation at both early and late stages of disease and also rescued synapse loss. Together, these data establish a previously unidentified signaling relationship between neurons burdened with DSBs and microglia in age-associated neurodegenerative disease.
RESULTS
Identification of DSB-bearing neurons at early stages of disease in a mouse model of neurodegeneration
We used the CK-p25 mouse model of inducible neurodegeneration to understand how DSB-bearing neurons contribute to disease development. In these mice, the CamkII promoter drives the expression of the neurotoxic protein fragment p25 through a doxycycline (dox)–off system (15). P25 is the calpain-cleaved product of p35, an activator of cyclin-dependent kinase 5 (CDK5). Previously, we determined that the first pathologies observed in these mice are increased DSBs in neurons (7) and activation of microglia (16). These pathologies occur 1 to 2 weeks after the onset of p25 expression when mice are taken off dox. Intracellular amyloid-β accumulation also occurs as early as 2 weeks after induction (17). Neuronal loss, learning deficits, and tau hyperphosphorylation are also observed in the following 4 to 12 weeks (15, 17, 18). These pathological events are inducible and occur in a highly concerted and predictable manner.
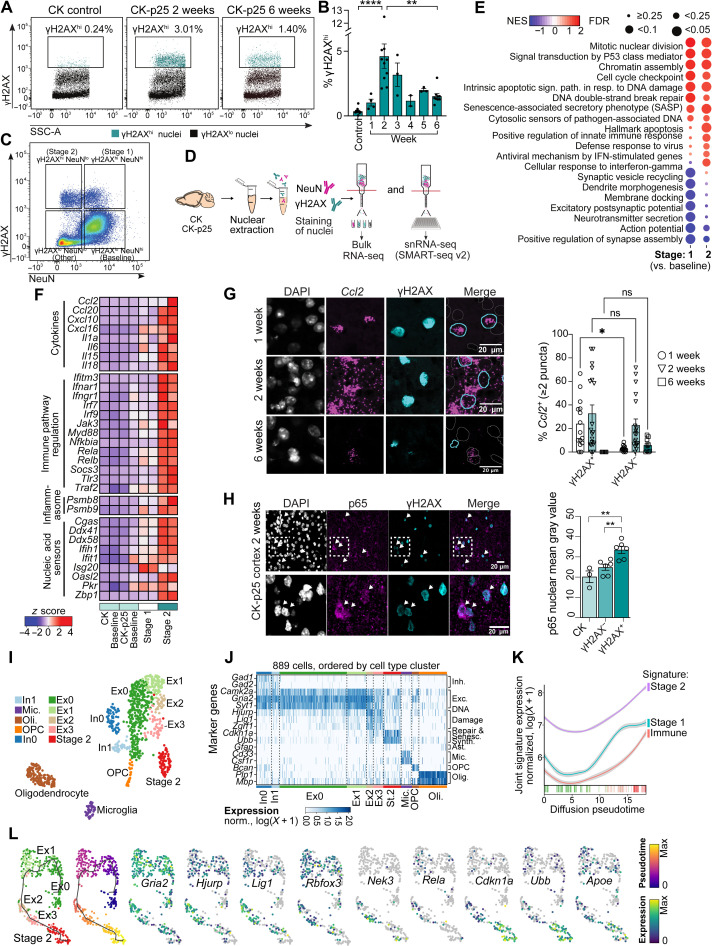
The phosphorylation of the histone variant H2A.X (γH2AX) by ataxia-telangiectasia mutated (ATM) kinase occurs rapidly after DSB detection. This posttranslational histone modification is essential for efficient DSB recognition and repair and is a robust DSB biomarker. Using flow cytometry, we identified a distinct population of nuclei expressing high levels of γH2AX (γH2AXhi) at the 2-week time point in the CK-p25 cortex, but not in the CK control cortex (Fig. 1A). CK mice express the CamkII-driven tetracycline transactivator, but not p25. We performed a timeline analysis to determine when γH2AXhi nuclei begin to accumulate in the CK-p25 cortex. γH2AXhi nuclei were detectable as early as 1 week after induction (1.038 ± 0.2627% population), peaked at 2 weeks (4.614 ± 0.9416% population), and gradually decreased thereafter (Fig. 1B). The significant reduction in γH2AXhi nuclei at 6 weeks corresponds with previous observations of neuronal loss in this model, suggesting that γH2AXhi cells degenerate by 6 weeks (15, 18). We were able to observe similar γH2AXhi population dynamics using immunofluorescent microscopy (fig. S1, A and B). We did not detect γH2AXhi nuclei after only 4 days off dox, suggesting that 1 week is the earliest time point at which this population appears.
Fig. 1. Neurons marked by DNA DSBs activate inflammatory signaling at early stages of neurodegeneration.
(A) Flow cytometry dot plots of γH2AXhi nuclei (turquoise) from CK and CK-p25 cortex. (B) Quantification of percent γH2AXhi for 1 to 6 weeks. Each data point represents percent γH2AXhi nuclei from one mouse. (C) Representative dot plot of γH2AX and NeuN immunoreactivity in 2-week CK-p25 cortex. (D) RNA-seq workflow. Bulk RNA-seq: n = 2 per genotype for bulk RNA-seq, 2-week time point. snRNA-seq: n = 3 per genotype at 1- and 2-week time points. (E) Differential Gene Ontology terms from bulk RNA-seq. FDR, false discovery rate; IFN, interferon; NES, normalized enrichment score. (F) Heatmap of differentially expressed genes associated with inflammation from bulk RNA-seq. (G) Left: Representative images of Ccl2 γH2AX staining for 1-, 2-, and 6-week CK-p25 cortex. Right: Quantification of number γH2AX+ and γH2AX− cells with ≥2 Ccl2 puncta. Data points represent one image from one mouse. Three to four images were taken per mouse (1 week, n = 4; 2 weeks, n = 5; 6 weeks, n = 3). DAPI, 4′,6-diamidino-2-phenylindole. (H) Top: Representative image of p65 in 2-week CK-p25. Bottom: Quantification of p65 mean intensity for γH2AX+ and γH2AX− nuclei. Data points represent 20 to 60 nuclei from one mouse. (I) UMAP (Uniform Manifold Approximation and Projection) of gated populations from CK and CK-p25 cortex at 1- and 2-week time points. (J) Marker gene expression for each cell type cluster. (K and L) Trajectory analysis of Ex0, Ex1, Ex2, Ex3, and stage 2 neurons. Smoothened gene signature expression across pseudotime. Immune = stage 2 immune genes. Padj < 0.05, log2 fold change ≥ 1.0 (K). Individual genes across trajectory (L). Error bars represent SEM; ****P < 0.0001, **P < 0.01, and *P < 0.05; ns, not significant. One-way analysis of variance (ANOVA) with Tukey’s test for multiple comparisons (B and H). Two-way ANOVA followed by Sidak’s test for multiple comparisons (G). Data are pooled from four independent experiments (B). Data are representative of two independent experiments (G and H).
We next sought to identify the cell type composition of γH2AXhi nuclei. Through immunofluorescent microscopy, we found that while most γH2AX+ nuclei were immunoreactive for NeuN, a general neuron marker, 21.51 ± 2.36% did not have strong NeuN immunoreactivity (fig. S1, C and D). Furthermore, none of the γH2AX+ nuclei overlapped with markers of microglia (Iba1), astrocytes [glial fibrillary acidic protein (GFAP)], or oligodendrocytes and oligodendrocyte precursor cells (Olig2), indicating that the 21.51% with low NeuN immunoreactivity were not likely of glial origin (fig. S1, C and D). We found that 74.17 ± 1.27% of γH2AX+ cells with low NeuN immunoreactivity were labeled with Neurod1, a neuronal transcription factor (fig. S1, E and F). γH2AX+ cells (88.98 ± 2.16%) with high NeuN immunoreactivity were labeled with Neurod1 as well (fig. S1, E and F). In addition, we observed that 85.22 ± 4.52% of γH2AX+ cells with low NeuN immunoreactivity were labeled with green fluorescent protein (GFP), which is fused to the p25 transgene and driven by the CamKII promoter, an excitatory neuron-specific gene (fig. S1, G and H). γH2AX+ cells (89.15 ± 3.55%) with high NeuN expression were also labeled with GFP (fig. S1, G and H). These immunostaining analyses indicated that γH2AX+ nuclei with low NeuN immunoreactivity were likely excitatory neurons with some level of cell identity degradation. Degradation of neuronal identity is observed within aging and AD patients (19), and reduced NeuN expression has been proposed as an indicator of declining neuronal health (20). Reduced NeuN expression or reduced NeuN immunoreactivity in neurons is an established feature of neuronal damage due to irradiation, ischemia, and axotomy (21–23).
To follow up on this observation, we performed additional flow cytometry analyses of γH2AXhi nuclei, staining for both CamKIIa and NeuN. In support of our immunostaining analyses, we found that all γH2AXhi nuclei were also labeled by CamKIIa (CamKIIahi), even those with low NeuN immunoreactivity (NeuNlo) (fig. S1, I and J). Last, to validate that DNA damage response pathways were active in γH2AXhi nuclei, we confirmed colabeling with the active form of ATM kinase (p-ATM S1981) (fig. S1, K and L).
DSB-bearing neurons express antiviral and senescence genes
To formally analyze the population of γH2AXhi cells, we classified nuclei into four distinct populations: γH2AXlo NeuNhi (“baseline” neurons), γH2AXhi NeuNhi (stage 1 neurons), γH2AXhi NeuNlo (stage 2 neurons), and γH2AXlo NeuNlo (putative nonneuronal cells, which we refer to as “other”) (Fig. 1C). We used FANS followed by bulk RNA-seq to transcriptionally profile each population (Fig. 1D). All nuclei were collected for sequencing at the 2-week time point, which was when we observed the peak density of γH2AXhi nuclei.
We performed three differential expression analyses to characterize gene expression changes in these populations: stage 1 versus CK-p25 baseline, stage 2 versus CK-p25 baseline, and stage 2 versus stage 1. Large transcriptional changes were observed in the first two comparisons. A total of 3031 up-regulated and 717 down-regulated transcripts were identified in stage 1, and 5055 up-regulated and 3792 down-regulated transcripts were identified in stage 2 (log2 fold change ≥ |1.0|, adjusted P < 0.05) (fig. S2, A to C). Comparatively, few transcripts were found differentially expressed between stage 2 and stage 1 (186 up-regulated and 42 down-regulated transcripts in stage 2). Gene set enrichment analysis (GSEA) (24) revealed that both stage 1 and stage 2 neurons displayed a significant enrichment for genes implicated in DSB repair, apoptotic signaling, and cell cycle reentry, and a significant reduction in synaptic processes (Fig. 1E). These pathways were previously found to be dysregulated in both the CK-p25 mouse and AD human brain tissue (7, 25).
We also found that a number of innate immune pathways were enriched in stage 1 and stage 2 neurons. This included Gene Ontology terms “Senescence-associated secretory phenotype (SASP)”, “Cytosolic sensors of pathogen-associated DNA,” and “Positive regulation of innate immune response” (Fig. 1E). Upon closer inspection, we observed an enrichment of genes linked to DSB-mediated activation of SASP signaling, particularly in stage 2 neurons. This included nucleic acid sensors such as Cgas and Zbp1; NFκB subcomponents Rela and Relb; SASP factors Il6, Il15, Ccl2, and Cxcl10; and interferon (IFN)–stimulated genes Isg15 and Ifitm3 (Fig. 1F). Notably, a number of these genes are expressed in neurons following viral infection (14, 26). These genes are also classic markers of senescence, suggesting that DSB accumulation elicits senescent and antiviral-like pathways in neurons.
Because the CK-p25 model is characterized by the development of type I IFN–responsive microglia specifically by 2-week induction (16), we wanted to determine whether DSB-bearing neurons express cytokines before microglia. To do this, we performed multiplexed fluorescent RNAscope in situ hybridization in the CK-p25 cortex after 1 and 2 weeks of induction. We focused our profiling on Cxcl10 and Ccl2 because these pro-inflammatory chemotactic molecules were highly expressed in stage 2 neurons and are known to be secreted by neurons upon viral infection, downstream of type I IFN (14, 27, 28). First, we performed RNAscope for the excitatory neuron marker Camk2a to again confirm whether γH2AX+ nuclei were of neuronal origin. Consistent with our RNA-seq and flow cytometry data, 98.81 ± 1.19% of γH2AX+ nuclei also expressed Camk2a (fig. S2D). This high positivity rate confirms that γH2AX+ nuclei are nearly exclusively excitatory neurons. RNAscope analysis also revealed that γH2AX+ nuclei were the only cells to express Cxcl10 and Ccl2 at the 1-week time point. In contrast, Cxcl10 and Ccl2 gene expression was highly enriched in γH2AX-immunonegative nuclei at the 2-week time point (Fig. 1G and fig. S2E). Both γH2AX+ and γH2AX− nuclei had reduced Cxcl10 and Ccl2 expression by 6 weeks (Fig. 1G and fig. S2E). Analysis of the cell type markers associated with Cxcl10 expression revealed that most γH2AX+ nuclei expressing Cxcl10 were astrocytes and microglia (fig. S2F). Together, these data indicate that expression of Cxcl10 and Ccl2 in γH2AX+ cells precedes their expression in glial cells and that cytokine secretion from DSB-bearing neurons may be an early mechanism of glial cell recruitment and activation in the CK-p25 brain.
To identify the master regulators of DSB-associated neuronal immune signaling, we performed transcription factor enrichment analysis using Enrichr (29). A subset of immune pathway genes was extracted from the significantly up-regulated genes in stage 2 neurons for analysis. The overlap between this immune gene module and transcription factor target genes was then calculated. Multiple subunits of the NFκB complex were consistently enriched across Enrichr transcription factor libraries, including Rela, Nfkb1, and Relb (table S1). Notably, the NFκB transcription factor plays a well-established role in SASP activation and the DSB response (30, 31). We chose to focus on Rela, also known as p65, because it is a core member of the canonical NFκB complex, and it was most frequently enriched in the Enrichr analysis. We stained for p65 in 1-, 2-, and 6-week CK and CK-p25 cortices. NFκB is normally sequestered in the cytosol but translocates to the nucleus to form an active complex upon cellular insult. Nuclear p65 intensity was significantly higher in γH2AX+ cells compared to other γH2AX− cells at the 1- and 2-week time points (Fig. 1H and fig. S3A), supporting evidence of increased NFκB transcriptional activity. By 6 weeks, there was no significant difference between γH2AX+ and γH2AX− nuclei in nuclear p65 intensity (fig. S3A).
Single-cell RNA-seq analysis in DSB-bearing neurons
In our bulk RNA-seq analysis, inflammatory gene expression was lower in stage 1 neurons compared to stage 2 neurons (Fig. 1F). We also performed a flow cytometry analysis of stage 1 and stage 2 populations over 1 to 6 weeks and found that 1 week after induction, stage 1 neurons made up 75% of all γH2AXhi neurons, while stage 2 neurons made up the remaining 25%. By 2 weeks, however, the total population of γH2AXhi neurons was approximately 50% stage 1 and 50% stage 2 (fig. S3, B and C). This suggested that the stage 2 population may develop after stage 1, which aligned with our initial prediction when defining these distinct cellular states based on NeuN expression. To better understand the relationship between stage 1 and stage 2 neurons, we performed snRNA-seq on each FANS-gated population at both 1- and 2-week time points (Fig. 1D). A total of 1357 single-nucleus libraries were prepared using SMART-seq2 chemistry. Following quality control measures (see Materials and Methods), 889 libraries remained for downstream analysis (Fig. 1I and fig. S3D). Cells were classified into major cell type clusters based on marker gene expression, resulting in the identification of 521 excitatory neurons, 71 inhibitory neurons, 131 oligodendrocytes, 33 oligodendrocyte precursor cells (OPCs), and 50 microglia. We did not detect any astrocytes.
Most nuclei sorted under the stage 2 gate formed their own cell type cluster (Fig. 1I), accounting for the remaining 83 cells. While we did identify 13 stage 2–gated nuclei in microglia and oligodendrocyte clusters, all of the nuclei in question came from one CK-p25 mouse (fig. S3, E and F). None of the other five CK-p25 mice had stage 2–gated nuclei fall into glial clusters. Combined with our previous cell type immunostaining analysis (fig. S1, C and D), we concluded that the immune signature identified in the stage 2 population was not likely to be driven by contamination from microglia or oligodendrocytes. Compared to other neuronal clusters, the in silico stage 2 cluster was significantly enriched for the FANS-gated stage 1 and stage 2 gene signatures (fig. S3G). The stage 2 cluster expressed only moderate levels of the excitatory neuron cluster markers Camk2a, Gria1, and Syt2 and lacked marker genes of other canonical cell types such as inhibitory neurons (Gad1, Gad2), astrocytes (Gfap), microglia (Cd33, Csf1r), oligodendrocytes (Plp1, Mbp), and OPCs (Bcan). Instead, the stage 2 cluster expressed marker genes indicative of senescence, including Cdkn1a and Ubb (Fig. 1J). These distinctive cell type markers further suggested to us that the stage 2 cluster represented a population of DSB-bearing neurons engaged in a senescence-like inflammatory response.
To further examine stage 1 and stage 2 cell type heterogeneity, we subclustered all neuronal cells. This resulted in the identification of four excitatory neuron subclusters (Ex0 to Ex3) and two inhibitory neuron subclusters (In0 and In1) (Fig. 1I). Stage 1 cells were enriched in subclusters Ex2 and Ex3 compared to Ex0 and Ex1 (fig. S3F). In addition to excitatory neuron markers, subclusters Ex2 and Ex3 were also marked by genes associated with DNA repair and DNA synthesis, such as Holliday junction–recognizing protein (Hjurp), the helicase Zgrf1, and ligase I (Lig1). Both Hjurp and Zgrf1 are involved in homologous recombination, a form of DSB repair that requires entry into S phase of the cell cycle. Expression of these genes by postmitotic neurons may reflect erroneous cell cycle reentry in an effort to repair DSBs (32). LigI is involved in both DNA replication and numerous DNA repair pathways, including alternative end-joining of DSBs. These genes indicate that Ex2 and Ex3 cells were likely engaged in a DSB repair response and cell cycle reentry (Fig. 1J).
Hjurp, Zgrf1, and LigI were not expressed in the stage 2 cluster. Combined with the enrichment for the classical senescence marker Cdkn1a, and the fact that these cells are still γH2AXhi, we hypothesized that stage 2 cells may be engaged in an even later stage of response to DSBs. This motivated us to perform trajectory analysis. The single-cell analysis package Monocle 3 (33) was used to order cells from subclusters Ex0, Ex1, Ex2, Ex3, and stage 2 along a pseudotime, which was generated through a learned trajectory of subcluster gene expression differences (Fig. 1K). Both CK and CK-p25 cells were used for this analysis. Next, we identified genes that changed as a function of pseudotime. We observed sequential enrichment of stage 1 and stage 2 gene signatures along pseudotime (Fig. 1K). The expression of stage 2 signature genes specifically involved in immune response (Immune) was also enriched at the end of pseudotime (Fig. 1K). We found that genes associated with neuronal identity such as Gria2 and Kalrn were most highly expressed at the beginning of the trajectory (Fig. 1L). Meanwhile, genes associated with DSB repair and DNA synthesis like Hjurp and Lig1 peaked along the middle, and Cdkn1a expression peaked at the end of the trajectory. We also observed late-stage expression of Apoe, supporting previous observations that Apoe expression increases in neurons following injury (34). While expression of the gene encoding NeuN (Rbfox3) increased significantly in Ex1 compared to Ex0, we did not observe a significant difference in expression between Ex0 and stage 2 (fig. S3H). This indicates that reduction of NeuN in stage 2 neurons likely occurs at the posttranscriptional level and agrees with previous observations that reduced NeuN antigenicity or protein levels are observed in damaged neurons (21, 23). We also observed a significant increase in Rela expression in Ex1 and Ex2 neurons compared to Ex0, but we did not observe a significant difference in expression between Ex0 and stage 2 (fig. S3H).
DSBs elicit innate immune signaling in neurons
We next wanted to determine whether the gene signatures identified in CK-p25 mice could be recapitulated independent of p25 expression. We also wanted to test whether DSB induction is sufficient to trigger inflammatory gene expression in neurons. To accomplish this, we first treated murine primary neuron cultures with etoposide (ETP). ETP produces DSBs by inhibiting topoisomerase II function, thus preventing the religation of cleaved DNA-TopII complexes (Fig. 2A). The dose of ETP used was consistent with previous studies using ETP to investigate DSB-elicited immune activity (35, 36). Neuron cell type purity was also confirmed in primary neuronal cultures (see Materials and Methods and fig. S4A).
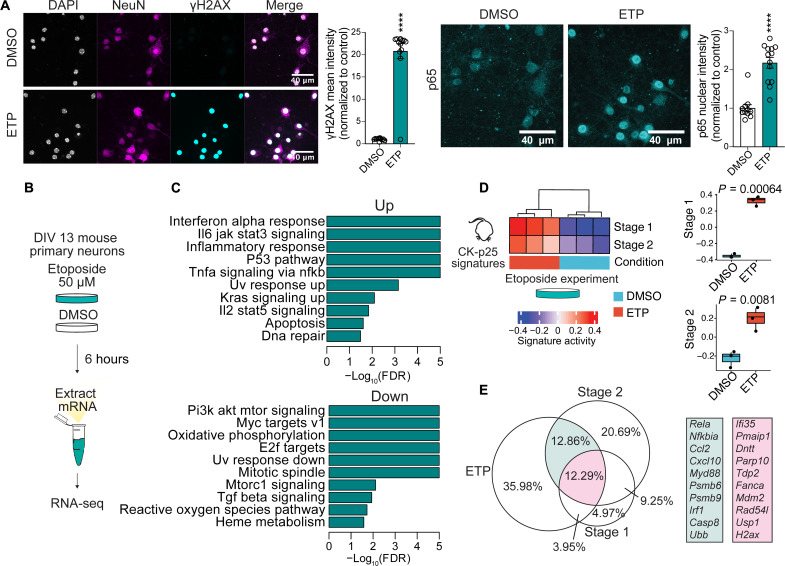
Fig. 2. Induction of DNA DSBs is sufficient to elicit immune pathway signaling in neuron primary culture.
(A) Left: Representative images of NeuN and γH2AX immunostaining in etoposide (ETP) and vehicle-treated primary cultures. Each data point represents γH2AX mean gray value for one nucleus. Right: Representative images of p65 immunostaining in ETP- and vehicle-treated primary cultures. Each data point represents p65 mean gray value for one nucleus. (B) Schematic of ETP treatment. DIV13 neuron primary cultures were treated with either 50 μM ETP or vehicle control (DMSO) for 6 hours. (C) Differential Gene Ontology (biological pathway) terms identified through GSEA from ETP versus DMSO contrast. (D) Left: Heatmap of stage 1 and stage 2 signature enrichment in DMSO- and ETP-treated neurons. Right: Quantification of stage 1 and stage 2 signature enrichment in DMSO- and ETP-treated neurons. Each data point represents one biological replicate. (E) Venn diagram of significantly up-regulated protein-coding genes from ETP-treated neurons and stage 1 and stage 2 gene signatures. Percentages are in reference to the total number of unique genes from all three gene sets. Genes overlapping in ETP and stage 2 are in turquoise. Genes overlapping in ETP, stage 2, and stage 1 are in magenta. The area of the circles is in proportion to the size of the gene sets. Error bars represent SEM; ****P < 0.0001. Student’s t test (A). Wilcoxon test (D). Data are representative of three independent experiments (A).
RNA-seq was performed to profile the transcriptional changes occurring in primary neurons after ETP treatment (Fig. 2B). There was a significant up-regulation of 7532 transcripts and down-regulation of 7106 transcripts compared to vehicle-treated control neurons [dimethyl sulfoxide (DMSO)] (adjusted P < 0.05) (fig. S4, B to D). Notably, GSEA revealed that biological pathways mediating senescent and antiviral activity such as “Interferon alpha response” and “Il6 jak stat3 signaling” were enriched in ETP-treated cultures (Fig. 2C). Furthermore, many of the cytokines up-regulated in stage 2 neurons were also up-regulated in ETP-treated neurons, including Ccl2, Cxcl10, Cxcl16, and Il6 (table 2). Stage 1 and stage 2 gene signatures were both significantly enriched in differentially expressed genes from ETP-treated neurons (P = 0.00064 and P = 0.0081, respectively) (Fig. 2D). Venn diagram analysis indicated that 12.29% of all protein-coding differentially expressed genes (DEGs) (total protein-coding DEGs from all datasets) were shared between ETP, stage 1, and stage 2 conditions (Fig. 2E). This overlap was enriched for DNA damage repair genes such as Parp10, Tdp2, Fanca, and Rad54l. A further 12.86% of all DEGs were shared between ETP and stage 2 conditions. This overlap was enriched for immune genes such as Rela, Nfkbia, and Irf1 (Fig. 2E). p65 nuclear intensity was also significantly increased in ETP-treated neurons (Fig. 2A). We also used x-ray irradiation as an independent method to induce DSBs. Exposing primary neuronal cultures to 10-Gy irradiation was also able to increase nuclear p65 intensity in neurons, as was treatment with a lower dose of ETP (25 μM) (fig. S4, E and F). Furthermore, both x-ray irradiation and 25 μM ETP significantly increased many of the genes shared by the higher-dose ETP treatment and stage 1 or stage 2 (fig. S4G). Together, these data indicated that stage 1 and stage 2 gene signatures can be recapitulated in primary neuron cultures independent of p25 expression.
A DSB-associated immune signature is conserved in AD human neurons
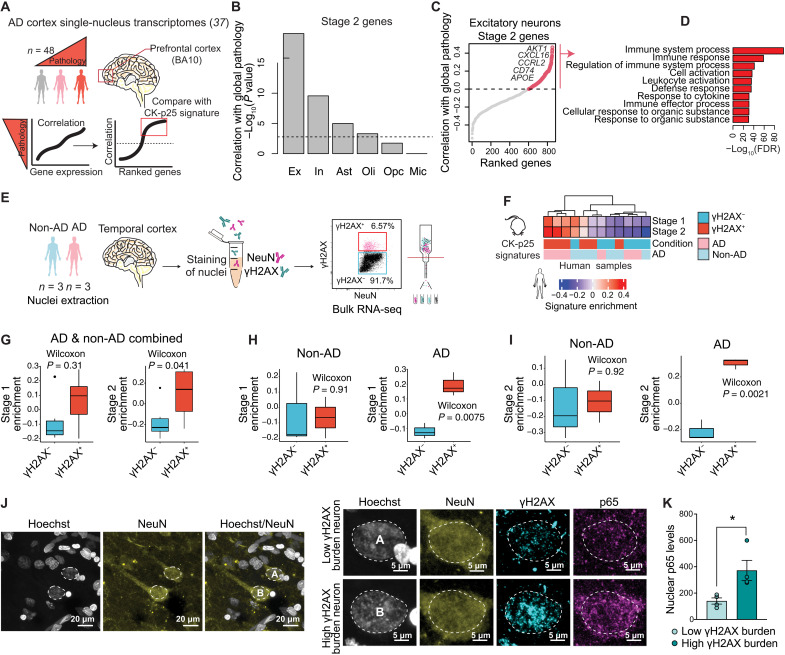
We next sought to determine whether signatures of DSB-bearing neurons could also be detected in the human brain. First, we used the stage 2 gene signature to identify neurons with DSB-associated immune activation in a previously published snRNA-seq dataset of individuals with and without AD pathology (Fig. 3A) (37). We assessed stage 2 gene expression in all major brain cell types: excitatory neurons, inhibitory neurons, astrocytes, oligodendrocytes, OPCs, and microglia. For each cell type, genes were ranked on the basis of their expression and correlation to “global pathology,” a variable quantifying three major AD pathologies: neuritic plaques, diffuse plaques, and neurofibrillary tangles (38). Genes that positively correlated with global pathology for each cell type were then sampled for the enrichment of stage 2 genes (Fig. 3A). The strongest enrichment of stage 2 genes was identified in excitatory neurons (P = 8.7 × 10−19) and inhibitory neurons (P = 7.7 × 10−7). Astrocytes and oligodendrocytes were also found to have a significant enrichment of stage 2 genes correlating with global pathology, albeit to a much lesser extent (Fig. 3B) (Bonferroni adjusted P < 0.01). Therefore, while we cannot exclude the possibility that human astrocytes and oligodendrocytes may carry some disease-associated enrichment for stage 2 genes, the stage 2 signature is largely driven by neurons in this dataset. Furthermore, when assessing the stage 2 genes positively correlated with global pathology in excitatory neurons, we again observed an enrichment of immune-related biological pathways, including the genes Ccl2-like receptor (CCRL2), CD74, CXCL16, and APOE (Fig. 3, C and D).
Fig. 3. Inflammatory signaling in DSB-bearing neurons is positively correlated with Alzheimer’s disease pathology.
(A) Schematic of the stage 2 signature analysis in the AD snRNA-seq dataset (37). (B) Quantification of stage 2 signature correlation with global pathology in cell type clusters (37). Stage 2 genes were tested for significant and positive correlation with the global pathology metric for each major cell type. The −log10 P value for these tests is shown in the histogram. The dashed line indicates a P value of 0.01 after Bonferroni correction for multiple testing. Ex, excitatory neurons; In, inhibitory neurons; Ast, astrocytes; Oli, oligodendrocytes; Opc, oligodendrocyte precursor cells; Mic, microglia. (C) Stage 2 signature genes ranked by their correlation with global pathology in excitatory neurons. Example stage 2 genes are shown with red circles. (D) Gene Ontology of stage 2 signature genes positively correlated with global pathology. (E) Schematic of γH2AXhi nuclei sorting from AD and non-AD brain tissue. (F) Heatmap of stage 1 and stage 2 signature enrichment in γH2AXhi and γH2AXlo human NeuNhi nuclei. (G to I) Quantification of stage 1 and stage 2 signature enrichment in γH2AXhi and γH2AXlo human NeuNhi nuclei samples by FANS gate (G), as well as FANS gate and disease status (H and I). (J) Representative image of γH2AX, p65, and NeuN in the AD brain. Left: Two NeuN+ nuclei are outlined (white dashed line). Right: Magnification of the two outlined nuclei. Top nucleus (A) represents low DSB burden; bottom nucleus (B) represents high DSB burden. (K) Quantification of p65 nuclear enrichment in low and high DSB-burdened neurons. Each dot represents the average of 23 to 41 NeuN+ nuclei per individual. Error bars represent SEM; *P < 0.05. Student’s t test (K). Wilcoxon test (G to I).
To validate the enrichment of DSB-bearing neurons identified in our snRNA-seq analysis, we sorted γH2AXhi and γH2AXlo NeuNhi and NeuNlo nuclei from the postmortem temporal cortex of six individuals and then performed bulk RNA-seq (Fig. 3E and fig. S5, A to C). These individuals were part of the Massachusetts Alzheimer’s Disease Research Center (MADRC) cohort and were divided by Braak score as either AD (Braak 4 to 6) or non-AD (Braak 1 to 3). Age, sex, and other biological variables for each individual are available in table S3. To assess the cell type specificity of our sorted populations, we examined the expression of several cell type marker genes (fig. S5, D to F). Unlike the stage 2 nuclei from the CK-p25 mice, γH2AXhi NeuNlo nuclei from the human brain were enriched for glial markers (fig. S5, E and F). This indicated to us that human glial cells also accumulate DSBs, as has been previously reported (6), and that γH2AXhi nuclei with low NeuN expression are likely to contain a heterogeneous population of many different cell types. Thus, to ensure that we were analyzing neuronal gene expression, we focused our analysis on γH2AXhi NeuNhi nuclei.
We assessed stage 1 and stage 2 gene signature activity in NeuNhi nuclei from the human brain (Fig. 3F). γH2AXhi neurons displayed modest enrichment for the CK-p25 stage 1 and stage 2 genes (P = 0.31 and P = 0.04, respectively) (Fig. 3G). Comparing samples by AD status and γH2AX status further revealed that stage 1 and stage 2 gene activity was markedly enriched in γH2AXhi neurons from AD individuals (P = 0.0075 and P = 0.0021, respectively) (Fig. 3, H and I). Furthermore, when contrasting all γH2AXhi neurons against all γH2AXlo neurons, we found that the average log2 fold change values for both stage 1 and stage 2 gene sets were positive and significantly higher than the average log2 fold change values for all other genes (fig. S5G). We also assessed the transcriptional similarity between human γH2AXhi neurons and ETP-treated primary neurons. Compared to γH2AXlo neurons, we found that gene expression in γH2AXhi neurons tended to be more similar to gene expression in ETP-treated neurons (fig. S5H).
To further verify the enrichment of immune activation in DSB-bearing neurons from brains with AD pathology, we assessed nuclear p65 expression in NeuN-expressing neurons with high versus low burdens of DSBs in the AD postmortem brain. We acquired four prefrontal cortex tissue samples from individuals belonging to the Religious Orders Study cohort. All individuals analyzed exhibited AD pathology, described in table S4. To bin neurons by DSB burden, we analyzed γH2AX expression in NeuN-expressing nuclei. The median γH2AX expression was calculated for each brain and used to split neurons into the high or low DSB condition (fig. S6A). Neurons with high DSB burden consistently expressed higher levels of nuclear p65 compared to neurons with low DSB burden (Fig. 3, J and K, and fig. S6B). Furthermore, we observed a strong positive correlation between raw γH2AX values and raw p65 values when grouping together all neurons from all individuals (P < 0.0001) (fig. S6C).
In summary, through the analysis of an independent snRNA-seq dataset, FANS RNA-seq, and immunofluorescent microscopy, we found that stage 1 and stage 2 gene signatures were active in human DSB-bearing neurons. Furthermore, this neuronal immune signature was further amplified in the context of AD pathology, suggesting that it may serve a functional role in disease-associated neuroinflammation.
DSB-bearing neurons stimulate glial activation
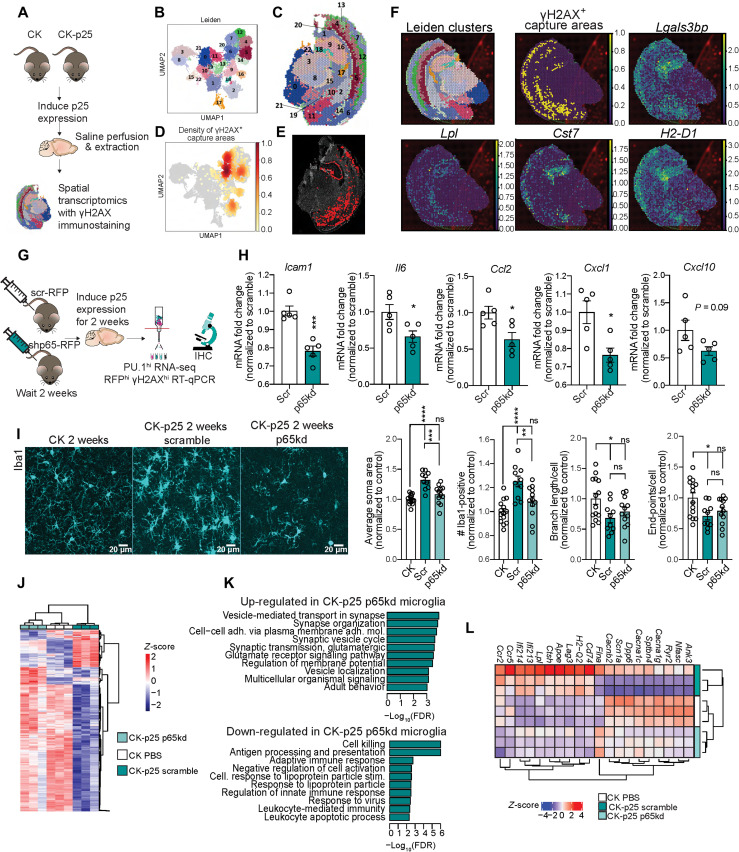
We have previously published an in-depth characterization of reactive microglia in CK-p25 mice using snRNA-seq (16). We wondered if these microglia were responding to immune signaling from DSB-bearing neurons. To address this, we performed γH2AX immunostaining followed by spatial transcriptomics on 2-week induced CK-p25 brains (Fig. 4A). The capture areas of 10X Visium spatial transcriptomics slides are large enough to encompass the transcriptional profile of three to five cells in mouse brain tissue. Therefore, we hypothesized that capture areas with γH2AX signal would also contain transcripts from nearby microglia. First, we identified 23 spatial clusters across three CK and four CK-p25 coronal sections (Fig. 4, B and C, and fig. S7, A and B). Next, we used a binary classification system to identify capture areas with γH2AX signal past a given threshold (i.e., γH2AX+ capture areas) (see Materials and Methods, Fig. 4, D and E, and fig. S7C). To determine whether the reactive microglia were enriched in γH2AX+ capture areas (16), we performed a differential comparison between all γH2AX+ capture areas and all γH2AX− capture areas. Using GSEA, we identified a robust enrichment of the microglia signature (1383 significantly up-regulated genes in reactive microglia compared to homeostatic microglia) within γH2AX+ capture areas (fig. S8A). This indicated that, overall, the reactive microglia signature was significantly associated with γH2AX+ neurons.
Fig. 4. Immune signaling in neurons recruits and activates microglia.
(A) Schematic of spatial transcriptomics experiment. CK (n = 3) and CK-p25 (n = 4) were induced for 2 weeks. Coronal brain sections were stained and imaged for γH2AX and then sequenced. (B) UMAP of capture areas from all samples. Leiden clusters are indicated by color and number. Each dot represents one capture area. (C) Leiden clusters superimposed onto a CK-p25 brain slice used for spatial transcriptomics. (D) UMAP indicating the density of γH2AX+ capture areas. (E) γH2AX+ capture areas identified in one CK-p25 brain slice. (F) Spatial clusters, γH2AX+ capture areas, and reactive microglia signature gene expression in one CK-p25 sample. (G) Schematic of neuronal p65 knockdown experiment. CK-p25 mice received retro-orbital injections of scramble shRNA–red fluorescent protein (RFP) adeno-associated virus (AAV) or shp65-RFP AAV. CK mice received retro-orbital injections of phosphate-buffered saline (PBS). Mice recovered for 2 weeks before being taken off dox. Brains were collected at the 2-week time point. (H) Reverse transcription quantitative polymerase chain reaction of immune genes in sorted RFPhi γH2AXhi nuclei. (I) Representative images of Iba1 in CK, scramble, and p65kd cortex. (J) Quantification of (left to right) Iba1+ soma area, number Iba1+ per image, average branch length per Iba1+ cell, and number end points per Iba1+ cell. Each data point represents one image. Two images were taken per mouse. (K) Heatmap of differentially expressed genes from p65 versus scramble contrast. (L) Up-regulated and down-regulated Gene Ontology (biological pathway) terms identified through GSEA of p65kd versus scramble contrast. (L) Heatmap of significantly up-regulated and down-regulated genes in PU.1hi nuclei from p65kd cortex compared to PU.1hi nuclei from scramble cortex. Error bars represent SEM; ****P < 0.0001, ***P < 0.001, **P < 0.01, and *P < 0.05. Student’s t test (H). One-way ANOVA followed by Holm-Sidak’s test for multiple comparisons (I). (H) Scramble (n = 5), p65kd (n = 5). (I) CK (n = 7), scramble (n = 5), p65kd (n = 6).
By mapping the density of γH2AX+ capture areas across all spatial clusters, we further prioritized distinct spatial clusters enriched for γH2AX+ capture areas (Fig. 4D and fig. S8B). We focused on spatial clusters with γH2AX+ capture areas comprising 20% or more of total capture areas (fig. S8B). This included clusters 16 and 18, which corresponded to CA1, CA2, and CA3 and the dentate gyrus, respectively, and clusters 2, 5, 6, and 7, which were primarily localized to the cortex. Differential comparisons of γH2AX+ capture areas and γH2AX− capture areas within each cluster of interest revealed a significant enrichment of the reactive microglia signature (fig. S8A). Notably, many of the marker genes for reactive microglia displayed expression patterns that visually correlated with γH2AX+ capture areas, including the galectin gene Lgals3bp, lipase Lpl, cystatin Cst7, and major histocompatibility complex (MHC) class I gene H2-d1 (Fig. 4F).
To corroborate this finding, we also assessed the spatial relationship between microglia and DSB-bearing neurons through immunofluorescent microscopy (fig. S8C). By analyzing the density of γH2AX+ neurons for each cortical layer, we were able to identify a strong enrichment of DSB-bearing neurons in layers 2/3 and 6 (fig. S8D). Analyzing the number and size of Iba1+ microglia for each cortical layer revealed an increased density and size of microglia particularly in layers 2/3 (fig. S8, E and F), suggesting that regions with high densities of DSB-bearing neurons also have higher densities of microglia with larger soma area. The density of DSB-bearing neurons by cortical layer also seemed to associate with Cxcl10 and Ifitm3 expression levels and γH2AX+ capture areas identified through the Visium analysis (fig. S9, A to E). Collectively, these data indicated that reactive microglia are closely associated with γH2AX+ neurons, providing evidence for neuroimmune communication between damaged neurons and microglia.
To determine whether microglia responded to DSB-mediated immune signaling, we sought to inhibit a master regulator of immune gene expression in neurons. Our data revealed that NFκB transcription factor activity is increased in DSB-bearing neurons, as are the genes downstream of NFκB activation. Therefore, we focused on inhibiting NFκB transcriptional activity. To achieve this in the CK-p25 mouse, we opted for brain-wide viral delivery of short hairpin RNAs (shRNAs) targeting p65 (39). We performed retro-orbital injections of PHP.eb adeno-associated virus (AAV) shp65–red fluorescent protein (RFP) or scramble-RFP into CK-p25 mice. CK control mice were injected with phosphate-buffered saline (PBS). Two weeks after injections, mice were taken off dox and induced for 2 weeks (Fig. 4G).
Most RFP colocalized with NeuN and γH2AX, indicating that neurons were the primary targets of shRNA expression, and confirming the previously reported tropism of PHP.eb for neuronal infectivity (fig. S10, A and B) (39). The reduced expression of p65 in mice was confirmed via quantitative polymerase chain reaction (qPCR) and immunofluorescent staining (fig. S10, C and D). The number of γH2AX+ neurons remained the same between p65 knockdown (p65kd) and scramble mice (fig. S10E). Next, we sorted γH2AXhi RFPhi neurons from both p65kd and scramble mice to assess changes in immune gene expression (fig. S10F). Reverse transcription qPCR (RT-qPCR) analysis revealed that a number of immune genes were down-regulated in DSB-bearing neurons from p65kd mice compared to scramble, including Ccl2, Icam1, Il6, and Cxcl1 (Fig. 4H). Down-regulation of Ccl2 in DSB-bearing neurons was also confirmed via RNAscope (fig. S10G). This indicated that knockdown of p65kd reduces neuron immune gene signatures.
To determine whether suppressing neuron immune genes attenuates microglial activation, we performed immunofluorescent staining with the microglia marker Iba1. We found that microglia proliferation was suppressed in p65kd mice (Fig. 4I), which was confirmed through flow cytometry analysis of PU.1hi nuclei from the entire cortex (fig. S10H). We also found that microglia soma area was significantly reduced, but differences in branch length and end points were not statistically significant (Fig. 4I). These data show that suppressing neuron immune signaling significantly alters the number and morphology of microglia.
To determine whether changes in microglia number and morphology were accompanied with changes in gene expression, we performed RNA-seq on sorted PU.1hi nuclei from CK control, CK-p25-scramble, and CK-p25-p65kd cortex (Fig. 4G and fig. S10, H and I). Differential analysis comparing microglia from p65kd and scramble cortices revealed the up-regulation of 627 transcripts and down-regulation of 160 transcripts (Fig. 4J and fig. S10J). Genes that were down-regulated in microglia from p65kd mice were involved in cell killing and antigen processing, such as H2-Q2, Lag3, Ccr5, and Ccr2. Notably, the Ccr2 gene encodes the receptor for CCL2, further implying an alteration of the CCL2-CCR2 axis compared to scramble mice (Fig. 4, K and L). We also observed a down-regulation of genes related to lipoprotein processing (Fig. 4, K and L). Pathways related to synaptic activity and membrane organization were significantly up-regulated in microglia from p65kd mice. This included calcium channels Cacna1g, Cacna1c, and Cacnb2, and sodium channel Scn1a, suggesting that p65kd microglia may return to a more homeostatic state embodied by CK microglia (Fig. 4J). Together, these results suggest that neuron immune signaling disrupts microglia homeostatic activity and activate cell killing and antigen processing mechanisms in microglia.
To gain a better understanding of how microglia respond to neuronal p65kd over time, we generated a mir30-based shRNA driven by an hSyn promoter packaged in AAV PHP.eB to suppress p65 exclusively in neurons (fig. S11A). CK-p25 brains injected with mir30-based shp65 were collected for analysis at 1, 2, and 6 weeks after p25 induction (fig. S11B). After confirming suppression of p65 expression (fig. S11, C and D), we assessed microglia number and morphology at each time point (fig. S11E). We found that neuronal p65kd was able to suppress microglial proliferation for 1 to 6 weeks of p25 induction (fig. S11F). Neuronal p65kd was also able to ameliorate branch shortening and end-point reduction at 1 and 2 weeks, but not at 6 weeks (fig. S11G). While we did not observe any rescue in microglia morphology at the 6-week time point, we did observe a significant reduction in the percentage of microglia that were MHCII+ (fig. S11H). MHCII expression is a hallmark feature of late-responding microglia in CK-p25 (16). While neuronal p65kd did not affect levels of endogenous amyloid-β, phosphorylated tau, or neuronal loss, we observed a marked rescue of synaptic density as measured by vGlut1 and synaptophysin (fig. S12, A to D). We also observed a corresponding decrease in the complement protein C1q, which is implicated in synaptic loss in other models of neurodegeneration (40, 41). Combined, these data suggest that neuronal NFκB promotes microglial proliferation and synaptic degradation in the CK-p25 mouse.
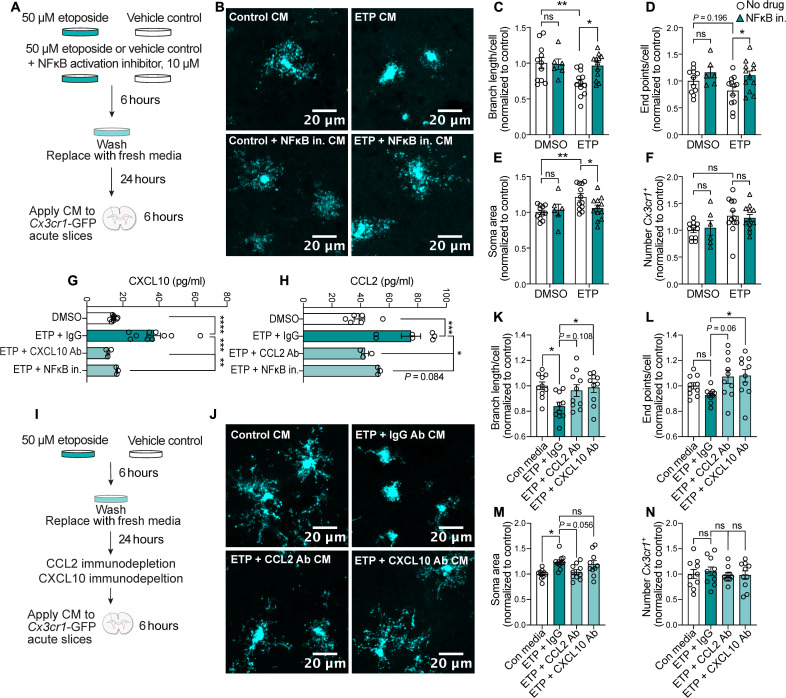
To determine whether specific cytokines expressed and secreted by DSB-bearing neurons play a role in microglia activation, we transitioned to our in vitro ETP model of DNA damage. First, we wanted to understand how conditioned medium from ETP-treated neurons affects microglia morphology. To do this, we generated organotypic acute brain slice cultures from Cx3cr1-GFP mice, which express GFP in all microglia. After ETP treatment and washout with PBS, neurons recovered in fresh medium for 24 hours. This conditioned medium was then collected and applied to Cx3cr1-GFP brain slices for 6 hours (Fig. 5A). The conditioned medium from ETP-treated neurons reduced microglia branch length (Fig. 5, B and C) but did not significantly affect the number of end points per microglia (Fig. 5D). We also observed increased soma area (Fig. 5E). Treating primary neurons with an NFκB activation inhibitor (10 μM NFκB Activation Inhibitor VI) before and during ETP treatment significantly reduced the soma area of Cx3cr1-GFP–expressing microglia and increased branch length and end points (Fig. 5, B to E). Conditioned medium from ETP-treated neurons did not elicit a robust difference in the number of microglia analyzed per image (Fig. 5F). These data further demonstrate NFκB activity as a significant mediator of immune signaling in DSB-bearing neurons.
Fig. 5. CCL2 and CXCL10 are secreted by DSB-bearing neurons to activate microglia.
(A) Schematic for treating acute Cx3cr1-GFP slices with conditioned medium (CM) from ETP-treated primary neurons. Cultures were treated with 50 μM ETP or vehicle control (DMSO) ± 10 μM NFκB Activation Inhibitor VI (IKK2 inhibitor) for 6 hours. Cultures were washed with PBS, and the medium was replaced. After 24 hours, the medium was applied to acute Cx3cr1-GFP slices for 6 hours. (B) Representative images of GFP in Cx3cr1-GFP acute slices treated with CM. (C to F) Quantification of branch length (C), end points (D), soma area per microglia (E), and number of microglia per image (F). Each data point represents the average of two images in one acute slice. (G and H) Quantification of CXCL10 (G) and CCL2 (H) from CM from control and ETP-treated primary neurons. Each data point represents one biological replicate. (I) Schematic of ETP CM experiment. Primary neurons were treated with either ETP or DMSO for 6 hours and washed with PBS, and then the medium was replaced. Cultures recovered for 24 hours before CM was collected. Immunoglobulin G (IgG), CCL2, or CXCL10 antibodies were used to immunodeplete CM before they were applied to Cx3cr1-GFP acute slices for 6 hours. (J) Representative images of microglia from acute slices treated with different CM. (K to N) Quantification of (K) branch length (L) end points, soma area per microglia (M), and number of microglia per image (N). Each data point represents the average of two images in one acute slice. Error bars represent SEM; ****P < 0.0001, ***P < 0.001, **P < 0.01, and *P < 0.05. Two-way ANOVA followed by Sidak’s test for multiple comparisons (C to F). One-way ANOVA followed by Tukey’s test for multiple comparisons (K to N). Data are combined from two independent experiments (C to F and K to N). Data are combined from three independent experiments (G and H).
Notably, conditioned medium from ETP-treated neurons was significantly enriched for CXCL10 and CCL2 (Fig. 5, G and H). Furthermore, the NFκB activation inhibitor reduced the concentration of these cytokines in conditioned medium (Fig. 5, G and H). Previously, we found that DSB-bearing neurons are the first cells to express CXCL10 and CCL2 in the CK-p25 cortex (Fig. 1F and fig. S2E). These cytokines induce the migration of macrophages and T cells to sites of viral infection and traumatic brain injury (14, 28, 42), suggesting that they may be primary constituents of immune signaling in DSB-bearing neurons. To determine whether they played a role in DSB-mediated microglia activation, we immunodepleted CCL2 or CXCL10 from conditioned medium (Fig. 5I). Both CCL2 and CXCL10 depletion increased branch length and end points per microglia compared to immunoglobulin G (IgG) control (Fig. 5, J to L). CCL2 depletion reduced microglia soma area, but CXCL10 depletion did not (Fig. 5M). Neither immunodepletion had an effect on microglia number (Fig. 5N). Combined, this suggests that while both CCL2 and CXCL10 elicit morphological changes in microglia, they may play differential roles in microglia recruitment and activation. Notably, both CCL2 and CXCL10 are up-regulated in aging and AD pathogenesis (42, 43), suggesting that their signaling activity via DSB-bearing neurons could play significant roles in neuroinflammation. Together, our data indicate that CCL2 and CXCL10 play important roles in recruiting and activating microglia to neurons burdened with DSBs. This establishes a previously unknown role for neuronal communication with microglia in the context of age-associated neurodegenerative disease and uncovers a new facet of DSB toxicity in mature postmitotic neurons.
DISCUSSION
Substantial evidence supports a causal role for microglia and neuroinflammation in AD pathogenesis (25, 44). For example, many AD risk genes and genomic loci are most active in microglia (45), and reactive microglia are thought to create a cytotoxic environment for neurons and exacerbate neurodegeneration (46, 47). Here, we provide evidence that neurons participate in this inflammatory signaling as well. Specifically, upon the accumulation of DSBs, neurons are able to engage microglia through the secretion of chemotactic and pro-inflammatory factors, thus facilitating neuroinflammation and disease progression. This is a previously undefined role for neurons in the context of neurodegenerative disease and provides a mechanistic link between genomic fragility and senescence in neurons and microglia activation.
The activation of inflammatory signaling in DSB-bearing neurons corresponds with a progressive erosion of cell identity. Using snRNA-seq, we found that DSB-bearing neurons engage DNA repair and DNA synthesis pathways first (stage 1) but transition to immune gene expression at later stages of genome toxicity (stage 2). These stage 2 cells are most enriched for senescent markers, such as Cdkn1a and Ubb, rather than neuronal markers. The age-dependent erosion of neuronal identity has been observed in both sporadic and familial AD patients, and coincides with the activation of a number of biological pathways identified in our study, including DNA damage response, cell cycle reentry, and NFκB signaling (19). Interpreted in the context of the present study, these observations suggest that the age-associated accumulation of DSBs may degrade chromatin integrity in neurons, ultimately resulting in the activation of immune signaling that engage microglia.
While we initially identified the stage 2 gene signature in CK-p25 mice by virtue of reduced NeuN immunoreactivity in a subset of DSB-bearing neurons, we also identified stage 2 characteristics in DSB-bearing neurons with NeuN expression, including neurons from the human postmortem brain and primary culture. These observations indicate that there are likely high levels of heterogeneity in the damaged neuron population and that a simple stage 1 versus stage 2 dichotomy may not fully encompass all transcriptional states. However, the stage 1 and stage 2 populations are able to serve as proof of principle that a heterogeneous response to DSBs in neurons does exist and that transitions between these transcriptional states may occur during the progression of neurodegenerative disease.
The SASP and canonical antiviral response are activated through the detection of nonendogenous nucleic acids within the cytosol. Thus, it is likely that loss of nuclear and genomic integrity may lead to the release of DNA fragments in DSB-bearing neurons. We observed increased expression of nucleic acid sensors in DSB-bearing neurons, including cGAS, which plays a significant role in innate immune activation associated with senescence (12). Nevertheless, many other mechanisms may be at play, including derepression of transposable elements (48), epigenetic drift, and ATM-mediated NFκB transcription (30). There is also evidence that nucleic acids released from mitochondria play roles in neuron immune gene expression (49). It remains to be addressed whether DSB-induced immune gene expression is a product of age-associated decline in DNA repair or whether multiple cellular functions that are also known to decline with age engage immune signaling as a convergent mechanism of neuronal distress. Therefore, while the current study does not address the mechanism by which DSBs in the CK-p25 mouse activate immune gene expression, a rich foundation of evidence for DSBs activating immune gene expression through multiple pathways exists for future studies.
Our data do not exclude the possibility that other cell types in the brain are capable of initiating a SASP in response to DNA damage. cGAS-STING–mediated senescence has also been described in ATM knockout microglia (50). Additional senescent phenotypes described in microglia (51), astrocytes (52), and OPCs (53) have also been shown to play a functional role in mouse models of neurodegenerative disease. However, despite the fact that neurons are the brain cell type most vulnerable to DSB accumulation, few studies have examined the relationship between DNA damage and senescence in neurons. Observations of senescent neurons have been observed in other studies (54), indicating that an investigation of the effects of senescent neurons in neurodegenerative disease is merited. We build upon these foundational observations by providing a detailed transcriptional analysis of such a population of neurons, linking them to a late-stage DSB response, and mechanistically establishing their role in microglia recruitment and activation in neurodegeneration.
We identified NFκB as a major regulator of immune gene expression in DSB-bearing neurons from both CK-p25 mice, primary neurons, and postmortem human brain. In addition, we found that suppression of NFκB activity through p65 knockdown or small-molecule inhibition in neurons was sufficient to reduce activated microglia morphology and gene expression. NFκB has been previously identified as a therapeutic target for AD, albeit in the context of microglia and astrocytes (55). Our results now indicate that this transcription factor plays a pivotal role in DSB-bearing neurons as well. In rodent models of learning and memory, NFκB activity in neurons is neuroprotective. Suppression of NFκB in forebrain excitatory neurons impairs spatial learning and neuronal plasticity (56) and exacerbates cell death following exposure to neurotoxic stimuli (57). Therefore, the primary role of neuronal NFκB may be to regulate synaptic processes, but it can also orchestrate immune activation in response to cell stressors. It is possible that NFκB and inflammatory activity in DSB-bearing neurons may initially function as a neuroprotective process. While we observed a notable reduction in synaptic gene expression in stage 1 neurons, this down-regulation was virtually absent in stage 2 neurons (Fig. 1E). However, we also observed a notable rescue of synaptic loss in p65kd mice at the 6-week time point, indicating that there is a complex relationship at play between the neuroprotective effects of NFκB and the synaptic pruning activity it may promote in microglia.
Last, we identify CCL2 and CXCL10 as primary signaling molecules secreted from DSB-bearing neurons to recruit and activate microglia. In the CK-p25 model, DSB-bearing neurons are the first cell type to express CXCL10 and CCL2. Mainly astrocytes and microglia express these chemokines at a later time point, presumably in response to DSB-bearing neurons. Furthermore, spatial transcriptomics revealed that signatures of reactive microglia are closely associated with γH2AX+ neurons, suggesting that DSB-bearing neurons are hubs for neuroimmune communication. Last, immunodepletion of CCL2 or CXCL10 in conditioned medium from ETP-treated neurons was able to prevent branch shortening and end-point reduction in microglia in acute slice culture. Notably, increased levels of both of these cytokines are implicated in the pathogenesis of AD and affect blood-brain barrier permeability to aid the infiltration of peripheral monocytes (28, 58). However, manipulation of either signaling axis seems to have varying effects on AD pathology. For example, CCR2 deficiency in murine models of AD aggravate amyloid pathology and cognitive decline, but CCL2 overexpression seems to also increase amyloid deposition (59–61). Another study demonstrates that deficiency of the CXCL10 receptor CXCR3 reduces amyloid deposition and behavioral deficits (62). These findings suggest that CCL2 and CXCL10 are crucial for effective microglia recruitment and activation, but imbalance in these signaling axes have detrimental effects on cognition and pathology clearance.
In summary, we leveraged bulk, single-nuclei, and spatial transcriptomic techniques in parallel with in vitro and in vivo manipulations of NFκB signaling to characterize DSB-bearing neurons and their relationship with microglia in the context of age-associated neurodegenerative disease. We demonstrate that DSB accumulation elicits senescent and antiviral-like signaling in neurons, which recruits and activates microglia in an NFκB-dependent manner. Our data posit that neurons play meaningful roles in neuroinflammation, which historically has been thought to be driven largely by glial cells. Crucially, this axis of neuron-microglia communication is mediated by DNA damage accumulation in neurons, revealing that two hallmarks of AD, genome fragility and neuroinflammation, are mechanistically linked.
MATERIALS AND METHODS
Fluorescence-activated nuclei sorting
Frozen cortices were disrupted with a handheld homogenizer in ice-cold PBS with protease inhibitors (catalog no. 11836170001, Roche, Basel, Switzerland) and ribonuclease inhibitors (catalog no. EO0382, Thermo Fisher Scientific, Waltham, MA). Samples were fixed with 1% paraformaldehyde for 10 min at room temperature and then quenched with 2.5 M glycine for 5 min. Nuclei were isolated through dounce homogenization followed by filtration with a 70-μm cell strainer (catalog no. 21008-952, VWR, Radnor, PA). The following antibodies were used to tag nuclei: anti-H2A.X–phosphorylated (Ser139) antibody conjugated to allophycocyanin (APC) (catalog no. 613416, BioLegend, San Diego, CA), anti-ATM–phosphorylated (Ser1981) antibody conjugated to phycoerythrin (PE) (catalog no. 651203, BioLegend, San Diego, CA), anti-NeuN antibody conjugated to Alexa Fluor 488 (catalog no. MAB377X, EMD Millipore, Burlington, MA), anti-PU.1 antibody conjugated to Alexa Fluor 647 (catalog no. 2240S, Cell Signaling Technology, Danvers, MA), anti-RFP antibody (catalog no. 600-401-379, Rockland Antibodies and Assays, Gilbertsville, PA), and anti–CaMKII-α (6G9) antibody (catalog no. 50049S, Cell signaling Technology, Danvers, MA). Antibodies were incubated with nuclei in 1% bovine serum albumin (BSA)/PBS at 4°C for 1 hour or overnight. For nonconjugated antibodies, samples were washed once with 1% BSA/PBS and then resuspended with 1:1000 Alexa Fluor secondary antibody (Thermo Fisher Scientific, Waltham, MA) for 1 hour at 4°C. Samples were passed through a 40-μm filter (21008-949, VWR) and stained with 4′,6-diamidino-2-phenylindole (DAPI) (catalog no. D9542, Sigma-Aldrich, St. Louis, MO) before sorting. Sorting was performed on FACSAria at the Koch Institute Flow Cytometry Core (BD Biosciences, USA). At least 50,000 nuclei of each cell type were collected for RNA-seq. Nuclei were sorted into 1% BSA/PBS and then spun at 2000g for 15 min in a cooled centrifuge (catalog no. 97058-916, Eppendorf, North America) for downstream analysis. For snRNA-seq, nuclei were not fixed. Single nuclei were sorted into a 96-well plate and then transported immediately to the Massachusetts Institute of Technology (MIT) BioMicro Center for library preparation. Forty-eight nuclei were sorted for each gated CK-p25 population per mouse, and 32 nuclei were sorted for each gated CK population per mouse. Flow cytometry analysis was performed using FlowJo software (Ashland, OR).
Bulk RNA-seq
For bulk RNA-seq of sorted nuclei, samples were treated for 15 min with proteinase K at 50°C and then for 13 min at 80°C. RNA was extracted using a Direct-zol RNA MicroPrep kit according to the manufacturer’s instructions (catalog no. R2062, Zymo Research, Irvine, CA). Purified RNA samples underwent fragment analysis at the MIT BioMicro Center. Libraries were generated from samples passing quality control (DV200 < 50%). Library generation was performed using SMARTer Stranded Total RNA-Seq Kit v2 - Pico Input Mammalian (catalog no. 634412, Takara Bio Inc.) and then submitted to the MIT BioMicro Center for quality control (fragment analysis and qPCR), followed by sequencing. Paired-end sequencing was performed using the Illumina NextSeq500 platform according to standard operating procedures.
For bulk RNA-seq of ETP-treated primary neurons, cultures were collected in TRIzol LS (catalog no. 10296028, Thermo Fisher Scientific, Waltham, MA). mRNA was purified and extracted using a Direct-zol RNA MicroPrep kit according to the manufacturer’s instructions. RNA was submitted to the MIT BioMicro Center for library preparation and sequencing. Libraries were prepared using the NEBNext Ultra II RNA Library Prep Kit for Illumina (catalog no. E7770, New England Biolabs, Ipswich, MA). Single-end sequencing was performed using the Illumina NextSeq500 platform according to standard operating procedures.
Mouse bulk RNA-seq read cleaning and bulk RNA-seq pipeline
For the paired-end fastq files, the first three nucleotides were trimmed off the second sequencing read using cutadapt version 1.16. TrimGalore version 0.4.5 was used in paired mode to trim adapters and low-quality portions of reads. Reads were aligned using Salmon version 0.12.0 to mouse genome version GRCm38.94 and human genome version GRCh38.94. For downstream analysis, TPM files from Salmon were imported into R version 3.6.1 using tximport version 1.14.0 with the option to generate estimated counts using abundance estimates scaled up to library size and additionally scaled using the average transcript length over samples and the library size (countsFromAbundance = lengthScaledTPM).
Mouse bulk RNA-seq differential analysis
We performed differential analysis using R version 3.6.1 and DESeq2 version 1.26.0. For each corresponding cell type and condition, we performed differential expression using only the corresponding samples. Genes were categorized as significant if they met the cutoff threshold (log2 fold change ≥ |1.0|, adjusted P < 0.05).
Mouse bulk RNA-seq GSEA
We used the complete set of RNA-seq results for each differential analysis for downstream GSEA processing. Genes for which adjusted P value could not be calculated were excluded. We ran GSEA version 3.0 in ranked list mode with default settings. The Gene Ontology biological pathway gene sets from Molecular Signatures Database v7.3 (MSigDB) were used for analysis. Genes were ranked by the sign of the fold change times the negative base 10 log of the adjusted P value.
Enrichr
Significantly up-regulated genes from the stage 2 versus baseline contrast were filtered on the basis of their occupation in MSigDB Gene Ontology biological processes containing the keyword “immune.” This resulted in a stage 2 immune module consisting of 940 genes. The corresponding Entrez gene symbols were then entered into the Enrichr website: https://maayanlab.cloud/Enrichr/. The resulting transcription factor enrichment data were downloaded as tables from the following transcription factor libraries: ChEA 2016, ENCODE and ChEA Consensus TFs from ChIP-X, TRRUST Transcription Factors 2019, Enrichr Submission TF-Gene Coocurrence, TRANSFAC and JASPAR PWMs, and ENCODE TF ChIP-seq 2015. These data are available in table S1. More information about the Enrichr libraries can be accessed on the Enrichr website.
Smart-seq pipeline
SMART-seq2
Single-nuclei library preparation was performed by the MIT BioMicro Center using the SMART-Seq v4 Ultra Low Input RNA Kit for Sequencing (Takara Bio Inc.) according to the manufacturer’s instructions. Libraries were sequenced on a MiSeq Illumina sequencer according to standard operating procedures.
Read processing
We aligned 40–base pair (bp) paired-end reads to the mm10 genome for each of 1357 single nuclei from 12 mice (64 cells each for six CK controls and an average of 162 cells in each of six CK-p25 mice) using Burrows-Wheeler Alignment–maximal exact match (options: -k 15 -M) and filtered out improperly aligned reads and secondary alignments with samtools (options: -f 3 -F 1280). We ran HTSeq-count10 on each cell’s filtered bam file to compute the cell’s transcriptomic coverage over each gene’s exons in vM25 GENCODE gene annotation.
Transgene detection
We first aligned 40-bp single-end reads from each sequenced single nucleus separately with the STAR aligner6 against the b37 genome with decoy contigs using a two-pass alignment (options: --outFilterMultimapNmax 20 --alignSJoverhangMin 8 --alignSJDBoverhangMin 1 --alignIntronMin 20 --alignIntronMax 1000000). We then used Picard tools to revert and merge the alignment with unaligned reads and marked duplicates on the merged bam file. We identified and removed alignments on decoy contigs, sorted and fixed NM, MD, and UQ tags with Picard tools, filtered duplicates, unmapped, and nonprimary alignment reads, and split reads by Ns in their CIGAR string.
Cell type annotation
Cell identities
We used SCANPY11 to process and cluster the expression profiles and infer cell identities. We kept only 21,859 protein-coding genes detected in at least 3 cells and filtered out 349 cells with less than 100 expressed genes, leaving 1008 cells over the 12 mice. We used the filtered dataset to calculate the low-dimensional embedding of the cells [Uniform Manifold Approximation and Projection (UMAP)] from the log1p matrix principal components analysis (PCA) with k = 50 and nearest neighbor graph with N = 20 (UMAP default parameters, min_dist = 0.2) and clustered it with Leiden clustering (resolution = 2), giving 17 preliminary clusters. We then manually assigned clusters based on the following two to three major marker genes per class: excitatory neurons: Camk2a, Gria2, and Syt1; inhibitory neurons: Gad1 and Gad2; astrocytes: Gfap; microglia: Cd33 and Csf1r; oligodendrocytes: Plp1 and Mbp; OPCs: Bcan; stage 2: Cdkn1a and Ubb. We merged clusters sharing marker genes to obtain seven final clusters, defining two broad neuronal subtypes (484 excitatory and 108 inhibitory cells), three glial clusters (50 microglia, 131 oligodendrocytes, and 33 OPCs), 179 stage 2 cells, and 23 cells with both high read counts and broad, nonspecific marker gene expression, likely due to doublets or other sorting artifacts. The doublet cluster was removed from downstream analyses. We also identified a cluster of cells that expressed fewer than 500 genes and had high expression of mitochondrial genes. Because of the questionable quality of these cells, this cluster was also removed from downstream analysis. We further sub-annotated neuronal subtypes from neuronal clusters with distinctive expression in the original Leiden clustering, resulting in four excitatory subclusters (314 Ex0, 91 Ex1, 32 Ex2, and 47 Ex3 neurons) and two inhibitory subclusters (71 In0 and 37 In1 neurons).
Signature analysis on single cells
For each signature, a joint expression value was calculated as the number of reads per 10,000 reads in each cell coming from all of the signature’s genes (genes with adjusted P < 0.05). Signature expression was transformed by log1p and averaged across all cells in a given neuronal subtype and mouse to obtain average signature values for plotting. We used two-sided t tests to compare the signature expression levels of pairs of excitatory neuronal subtypes (fig. S3G).
Trajectory analysis
Pseudotime
We performed pseudotime analysis using Monocle3 (v0.2.3.0) on the in silico Ex0, Ex1, Ex2, Ex3, and stage 2 neuronal populations. We normalized total counts per cell in the read count matrix to the median number of counts, log1p transformed the matrix, and regressed out the counts per gene using Monocle. We clustered the subsetted data in a new UMAP, clustered cells, learned a trajectory graph, and ordered cells by choosing the initial node as the Ex0 end of the graph to get a pseudotime across the graph. We plotted the log1p read counts across the gene set as well as the generalized additive model smoothed fits of each signature’s per-cell expression values across pseudotime (Fig. 1K).
Human snRNA-seq analysis
Single-nucleus transcriptomic sequencing data from postmortem cortical samples (prefrontal cortex, Brodmann area 10) of 48 subjects with varying levels of AD pathology were obtained from a previous publication (37). Individual-level cell type expression profiles were computed by averaging for each individual the normalized gene expression profiles across cells of the same cell type. Average profiles were subsequently mean-centered and scaled to compute gene-wise correlation coefficients of gene expression versus individual-level measures of global AD pathology burden reported as part of the Religious Orders Study and Memory and Aging Project (ROSMAP) cohort. Briefly, global burden of AD pathology is a quantitative summary of AD pathology derived from counts of three AD pathologies: neuritic plaques (n), diffuse plaques (d), and neurofibrillary tangles (nft), as determined by microscopic examination of silver-stained slides (37).
Global consistency between gene signatures observed in CK-p25 mice and a neuronal-specific association between gene expression and AD pathology in human tissue was assessed statistically using a nonparametric resampling test. To test whether cell type–specific expression of CK-p25 stage 2 signature genes tends to correlate with pathology in the human brain, a z-score statistic was computed to quantify the deviation of their correlation coefficient rank scores, relative to random expectation. Expected scores were estimated by randomly sampling same-sized gene sets (n = 1000 replicates). This analysis was performed for excitatory neurons, inhibitory neurons, and microglia cells independently.
Stage 1 and 2 signature generation
Stage 1 and stage 2 gene signatures were curated by performing differential expression analysis on the CK-p25 bulk RNA-seq dataset. Stage 1 versus baseline provided the genes for the stage 1 signature, and stage 2 versus baseline provided the genes for the stage 2 signature. Only genes that met the cutoff threshold (log2 fold change ≥ |1.0|, adjusted P < 0.05) were retained.
Visium spatial transcriptomics library generation
Mice were transcardially perfused with ice-cold saline, and then brains were dissected and flash-frozen in Optimal Cutting Temperature (OCT) Compound. A cryostat was used to generate 10-μm coronal sections of the hippocampus. These sections were applied to 10X Visium Spatial Gene Expression slides. Sections were immunostained with γH2AX and DAPI following the manufacturer’s instructions. Sectioning and staining was performed at the MIT Hope Babette Tang (1983) Histology Core Facility. Sections were imaged immediately after staining using the Olympus FV1200 Laser Scanning Confocal Microscope at the MIT Microscopy Core Facility. Sections were then used to generate 10X Visium Spatial Gene Expression Libraries according to the manufacturer’s instructions at the MIT BioMicro Center. Libraries were sequenced on a NovaSeq6000 Illumina sequencer according to standard operating procedures.
Visium spatial transcriptomics data processing
Samples were processed using Scanpy 1.7.2. The seven sample data matrices were merged into one matrix, which was then processed. The sample id and location of each capture area of the resulting matrix were saved and used for visualization. Counts were normalized (total count of 10,000 per capture area) and logarithmized (using Scanpy’s log1p function). The resulting counts matrix, called raw normalized, was used for expression visualization and differential expression analysis. For dimension resolution purposes, the raw normalized matrix was further processed: Genes that were not characterized as highly variable enough were filtered out (minimum mean of 0.0125, max mean of 3, minimum dispersion of 0.5), and linear regression was performed to eliminate the effect of covariates (total_counts and mitochondrial gene percentage). The data were then scaled (standard scaling, max value of 10). Afterward, PCA was performed, as well as sample-level batch correction, using Harmony. Then, a knn network was constructed for the creation of a UMAP embedding. Clusters were found using the Leiden algorithm.
Visium immunohistochemistry image processing
Immunostaining images were first processed as shades of gray pictures. The largest autofluorescence artifacts were removed manually. The signal was then amplified and cleaned using an 85% contrast increase on each image.
As the immunochemistry images of the tissue align perfectly to the pictures taken for 10X Visium purposes, calibration was performed to reconstruct the capture areas grid on the immunostaining images. In that image, for each capture area, the mean signal within a circle maximizing image coverage is calculated and recorded as the DNA damage signal. After standard scaling of this variable, a threshold of 0.4 SD was set to assign capture areas as positive or negative for DNA damage.
Visium differential expression and reactive microglia signature analysis
Differential expression analysis was performed using Wilcoxon rank sum test, and the resulting P values were corrected using Benjamini-Hochberg false discovery rate correction. For GSEA, the package gseapy was used, with 100 permutation and the signal-to-noise method. As input, the raw normalized matrix was used, though only containing the genes considered as highly variable in the dataset.
Reverse transcription qPCR
RNA was extracted from primary tissue cultures using the RNeasy Plus Mini Kit (catalog no. 74136, Qiagen, Hilden, Germany). Reverse transcription was performed using Invitrogen SuperScript IV First Strand Synthesis System with Oligo dT primers according to the manufacturer’s protocol (catalog no. 18091050, Thermo Fisher Scientific, Waltham, MA). Complementary DNA (cDNA) was quantified with a NanoDrop spectrophotometer. qPCR was performed using a Bio-Rad CFX-96 quantitative thermal cycler (catalog no. 1855195, Bio-Rad, Hercules, CA) and SsoFast EvaGreen Supermix (catalog no. 1725202, Bio-Rad, Hercules, CA). Relative changes in gene expression were determined using the 2−ΔΔCt method. Cycle numbers for the gene Gapdh or Rpl11 were used for housekeeping Ct values.
Immunofluorescent microscopy
Mice were transcardially perfused with ice-cold PBS and then fixed with ice-cold 4% paraformaldehyde in PBS. Dissected brains were drop-fixed overnight in 4% paraformaldehyde in PBS at 4°C. Forebrains were sectioned with a vibrating microtome (Leica BioSystems, Wetzlar Germany) to generate 40-μm coronal slices. Slices were blocked for 2 hours at room temperature in blocking buffer (10% normal goat serum, 0.3% Triton X-100, and PBS) and then incubated with primary antibody overnight at 4°C. Slices were washed 3 × 10 min with PBS, and Alexa Fluor secondary antibodies (Thermo Fisher Scientific) were added at a 1:1000 dilution for 1 hour at room temperature. Slices were washed again 3 × 10 min with PBS, then stained with DAPI (catalog no. D9542, Sigma-Aldrich, St. Louis, MO), and mounted onto Fisherbrand Superfrost Plus Microscope Slides (catalog no. 12-550-15, Thermo Fisher Scientific, Waltham, MA) with Fluoromount-G slide mounting medium (catalog no. 100502-406, VWR, Radnor, PA).
Primary neurons cultured on cover glass (catalog no. 194310012A, VWR, Radnor, PA) were washed once with PBS and then fixed with 4% paraformaldehyde/PBS for 15 min at room temperature. Immunostaining proceeded as described.
Free-floating 40-μm sections from postmortem human brain were blocked for 1 hour at room temperature in blocking buffer before being incubated in primary antibodies for 72 hours at 4°C with gentle rocking in blocking buffer (1:500, anti-NeuN, catalog no. 266 004, Synaptic Systems; 1:100, anti–phospho-histone H2A.X, catalog no. 05-636, Millipore; 1:300, anti-NFκB p65, catalog no. 51-0500, Invitrogen). Samples were then rinsed three times in 1× PBS for 5 min and incubated in secondary antibodies (1:1000, Alexa Fluor 488, 594, or 647; Thermo Fisher Scientific) for 2 hours at room temperature. After 1× PBS rinse, samples were incubated in 1:10,000 Hoechst in PBS (catalog no. H3569, Invitrogen) followed by 2 min in TrueBlack Lipofuscin Autofluorescence Quencher (catalog no. 23007, Biotium) with three subsequent 1× PBS rinses before mounting and imaging.
Mounted samples were imaged with a Zeiss LSM 710 confocal microscope. Images were quantified using ImageJ (NIH Image Analysis) and Imaris (Oxford Instruments). Image analysis was blinded for images from Figs. 4 and 5 and figs. S4 and S10 to S12. The number of slices and images taken for each experiment is described in figure legends. For PHP.eb AAV shp65-RFP cell type analysis, 40-μm coronal brain sections were stained for RFP and either NeuN, Iba1, GFAP, or Olig2. First, RFP-positive cells were identified in each image. Then, the percent of RFP-positive cells that also had positive immunoreactivity for a given cell type marker was quantified. Amyloid-β and pTau181 intensity were calculated using Imaris software (Oxford Instruments, UK).
Antibodies for immunofluorescent microscopy in mouse
The following antibodies were used: anti–phospho-histone H2A.X (Ser139) antibody, clone JBW301, EMD Millipore, 05-636, 1:500; NeuN, Synaptic Systems, 266 004, 1:1000; CaMKII-α (6G9) mouse monoclonal antibody (mAb), Cell Signaling Technology, 50049S, 1:200; NEUROD1 polyclonal antibody, Proteintech, 12081-1-AP, 1:200; anti-GFP antibody, Abcam, ab13970, 1:500; Iba1, Synaptic Systems, 234 004, 1:1000; anti-GFAP antibody, Abcam, ab53554, 1:500; recombinant anti-Olig2 antibody (EPR2673), Abcam, ab109186, 1:500; RFP antibody preadsorbed, Fisher Scientific, 600-401-379, 1:200; NFκB p65 (D14E12) XP rabbit mAb, Cell Signaling Technology, 8242S, 1:500; NFκB p65 polyclonal antibody, Invitrogen, 51-0500, 1:100; anti–MHC class II (I-A/I-E), clone M5/114, EMD Millipore, MABF33, 1:500; anti-nestin antibody (rat 401), Abcam, ab6142, 1:1000; anti-C1q antibody (4.8), Abcam, ab182451, 1:500; synapsin 2 antibody (guinea pig), Synaptic Systems, 106 004, 1:500; VGlut1 (rabbit), Synaptic Systems, 135 303, 1:500; purified anti-mouse/rat β-amyloid antibody, BioLegend, 805801, 1:500; phospho-Tau (Thr181) (D9F4G) rabbit mAb, Cell Signaling Technology, 12885S, 1:500.
Primary neuron culture
Cortices were dissected from E15 Swiss-Webster embryos in ice-cold Hanks’ balanced salt solution (HBSS) (catalog no. 14175103, Thermo Fisher Scientific, Waltham, MA) and dissociated with papain (catalog no. LS003126, Worthington Biochemical Corp., Lakewood, NJ) and deoxyribonuclease I (catalog no. 10104159001, Roche, Basel, Switzerland). Cells were resuspended in plating medium [neurobasal medium (catalog no. 21103049, Thermo Fisher Scientific, Waltham, MA), 1% penicillin/streptomycin solution (catalog no. 400-109, Gemini Bio-Products, Sacramento, CA), and 10% fetal bovine serum] and filtered through a 100-μm cell strainer (catalog no. 21008-950, VWR, Radnor, PA). Cell density was quantified using the Countess II Automated Cell Counter (catalog no. AMQAX1000, Thermo Fisher Scientific, Waltham, MA) and then plated on poly-d-lysine–coated 12-well culture dishes, 0.5 × 106. Cultures were maintained in 5% CO2 at 37°C in a cell culture incubator. After allowing 4 hours for the cells to adhere to the plate, the medium was replaced and maintained with neurobasal medium supplemented with B-27 (catalog no. 17504-044, Invitrogen, Carlsbad, CA), 1% penicillin/streptomycin, and 1% GlutaMAX supplement (catalog no. 35050-079, Thermo Fisher Scientific, Waltham, MA).
We also performed cell type immunostaining analysis to assess the cell type composition of our primary cultures. In total, 82.87% of cells were identified as mature neurons (NeuN expression), and another 9.31% were neural precursor cells (nestin expression) (fig. S4A). A trace number of cells were also identified as astrocytes (GFAP expression, 2.46%) and oligodendroglia (Olig2 expression without nestin expression, 1.35%) (fig. S4A).
Primary neuron treatments
Primary cortical neuron cultures (DIV11 to DIV13) were treated with 50 μM ETP prepared from 20 mM stock (catalog no. E1383-250MG, Sigma-Aldrich, St. Louis MO). Cultures were treated for 6 hours before collection for downstream experiments. RT-qPCR was used to measure gene expression. We were also able to observe increased expression of nuclear p65 and immune genes in primary neuron cultures treated with 25 μM ETP.
X-ray irradiation
Primary cortical neurons (DIV11 to DIV13) were exposed to 10- or 0-Gy x-ray irradiation using X-Rad320 from Precision X-Ray. Cultures were given 24 hours to recover before proceeding with experiments.
NFκB activation inhibitor
Primary neuron cultures were treated with 10 μM NFκB Activation Inhibitor VI, a benzoxathiole compound from Abcam (catalog no. ab145954). Cultures were pretreated for at least 30 min before ETP exposure.
RNAscope in situ hybridization
Fluorescent in situ hybridization was performed using RNAscope Multiplex Fluorescent Reagent Kit v2 according to the manufacturer’s instructions (catalog no. 323100, Advanced Cell Diagnostics, Newark, CA). Probes targeting murine Ccl2 (catalog no. 311791), Cxcl10 (catalog no. 408921), Aif1-C2 (catalog no. 319141-C2), Olig2-C3 (catalog no. 447091-C3), GFAP-C3 (catalog no. 313211-C3), and Rbfox3-C2 (catalog no. 313311-C2) were purchased from Advanced Cell Diagnostics. Following the RNAscope protocol, slices were stained for γH2AX (catalog no. 05-636, EMD Millipore, Burlington, MA). Samples were imaged with a Zeiss LSM 710 confocal microscope at 40× objective. Images were analyzed with ImageJ.
RNAscope analysis
ImageJ version 2.1.0 thresholding was used to identify γH2AX+ nuclei and generate regions of interest (ROIs). Process -> “Find Maxima” was used to count mRNA puncta within ROIs. Prominence > 100.00 with “Strict” setting. The number of γH2AX+ nuclei with ≥2 maxima, as well as the number of maxima per γH2AX+ nucleus, was quantified. γH2AX− nuclei were identified by thresholding for individual nuclei using the DAPI channel and then excluding ROIs that overlapped with γH2AX+ ROIs. Individual nuclei were identified following the Nuclei Watershed Separation process described on the ImageJ website (https://imagej.net/imaging/watershed).
Brain tissue samples
MADRC brain tissue samples
Fresh-frozen postmortem brain samples were provided by the MADRC. These samples were used for bulk RNA-seq. Individuals were selected on the basis of clinical diagnosis and Braak score. The three samples labeled as AD all had a clinical diagnosis of AD and a Braak score of VI. The three samples labeled as non-AD did not have a clinical diagnosis of AD and had Braak scores of I, II, and II. Sample metadata is available in table S3.
ROSMAP brain tissue samples
Fixed-frozen postmortem brain samples were chosen from the ROSMAP cohort. ROSMAP is a longitudinal cohort study of aging and dementia in elderly nuns, brothers, and priests. Sample metadata is available in table S4. In-depth description of metadata variables is available at the Rush Alzheimer’s Disease Center website (www.radc.rush.edu/docs/var/variables.htm).
p65 knockdown
Custom p65 and scramble shRNA oligos cloned into an AAV backbone (pAV-U6-RFP) were purchased from ViGene Biosciences (Rockville, MD). p65 shRNA sequences were as follows: GATCCGGCAGGCTATCAGTCAGCGCATTGTGCTTATGCGCTGACTGATAGCCTGCTTTTTA, GATCCGCGGATTGAGGAGAAACGTAAATGTGCTTTTTACGTTTCTCCTCAATCCGTTTTTA, GATCCGCACCATCAACTATGATGAGTTTGTGCTTAACTCATCATAGTTGATGGTGTTTTTA, GATCCGCCTGAGGCTATAACTCGCCTATGTGCTTTAGGCGAGTTATAGCCTCAGGTTTTTA, and shRNA scramble: GATCCGCAACAAGATGAAGAGCACCAACTCGAGTTGGTGCTCTTCATCTTGTTGTTTTTA.
p65 knockdown was confirmed via RT-qPCR in-house. PHP.eB AAV was generated by Janelia Viral Services. PHP.eB AAV shRNA or PBS was delivered retro-orbitally to anesthetized CK-p25 mice, 2 × 1011 IU/ml. For neuron-specific knockdown of p65, the plasmid SIN40C.SFFV.dTomato.miR30n (a gift from D. Heckl; Addgene, plasmid #169277) was modified with an hSyn, a neuron-specific promoter, and then cloned into an AAV backbone for generation of PHP.eB AAV.
Mir30-based p65 shRNA: AGCGAGCCTCATCCACATGAACTTGTTAGTGAAGCCACAGATGTAACAAGTTCATGTGGATGAGGCC.
Mir30-based scramble: AGCGCCCTAAGGTTAAGTCGCCCTCGTAGTGAAGCCACAGATGTACGAGGGCGACTTAACCTTAGGA.
Mice received bilateral intracranial injections [−2.00 Anterior-Posterior (AP), 1.5 Medial-Lateral (ML), −0.5 Dorsal-Ventral (DV)] of 500 nl of AAV-PHP.eb-hSyn-RFP-mir30-shp65 or AAV-PHP.eb-hSyn-RFP-mir30-scramble at a rate of 100 nl/min (both 2 × 1011 IU/ml). Two weeks after injection, mice were induced by removing dox diet. Following induction, mice were transcardially perfused with ice-cold PBS. One hemisphere was drop-fixed overnight in 4% paraformaldehyde/PBS at 4°C for immunostaining. The other hemisphere was flash-frozen in liquid nitrogen and stored at −80°C for RNA-seq.
CK-p25 mice
All mouse experiments were approved by the Committee for Animal Care of the Division of Comparative Medicine at the MIT, as well as the MIT Institutional Animal Care and Use Committee. All animal experiments were carried out at MIT. CK-p25 double transgenic mice were raised and maintained on a dox diet. All mice were induced by removing dox from their diet to drive the expression of p25-GFP in forebrain excitatory neurons. All mice were induced at 3 to 4 months old.
Conditioned medium and immunodepletion
Following ETP treatment, cultures were washed once with PBS. Cultures then recovered in fresh medium for 24 hours. The medium was collected and spun at 2,000g for 10 min to remove cellular debris. The medium was stored at −80°C for future experiments. Conditioned medium from ETP-treated neurons was incubated with IgG, Ccl2, or Cxcl10 antibodies (all 40 μg/ml) for 4 hours at 4°C. Dynabeads Protein G were added to pull down the antibody complex.
Organotypic brain slice culture
Eight- to 12-week-old male Cx3cr1-GFP mice were anesthetized with isoflurane and transcardially perfused, dissected, and sliced in ice-cold N-methyl-d-glucamine (NMDG) cutting solution containing 2.5 mM KCl, 0.5 mM CaCl2, 10 mM MgSO4, 1.25 mM NaH2PO4, 20 mM Hepes, 2 mM thiourea, 5 mM sodium ascorbate, 3 mM sodium pyruvate, 92 mM NMDG, 30 mM NaHCO3, and 25 mM d-glucose (pH 7.3 to 7.4) with HCl. The slicing chamber was bubbled with 95% O2/5% CO2, and coronal slices were cut at 250-μm thickness using a vibratome (Leica, VT1000s). After the last slice was collected, slices were transferred to a well plate containing fresh artificial cerebrospinal fluid (ACSF) and placed in an incubator set at 37°C and 95% O2/5% CO2 for 30 min. The ACSF solution contained 125 mM NaCl, 2.5 mM KCl, 1.2 mM NaH2PO4, 1.2 mM MgCl2, 2.4 mM CaCl2, 26 mM NaHCO3, and 11 mM d-glucose. Afterward, 100% of ACSF was removed and a 1:1 mixture of fresh ACSF and conditioned medium was added to the slices and placed back into the incubator for 6 hours. At the completion of the experiment, slices were fixed overnight at 4°C with 4% paraformaldehyde. Slices were then incubated in 30% glucose overnight at 4°C. Slices were subsectioned into 25-μm slices using a cryostat and then coverslipped for imaging.
Enzyme-linked immunosorbent assay
The Mouse MCP1 ELISA Kit and Mouse IP-10 ELISA Kit from Abcam (catalog nos. ab208979 and ab214563, respectively) were used to quantify CCL2 and CXCL10 in conditioned medium from primary neurons. Assays were quantified on a plate reader, and protein concentration was calculated according to manual instructions.
Microglia morphological analysis
Microglia morphology from CK-p25 and acute slice cultures was analyzed according to the protocol described by Young and Morrison (63) with minor alterations. Gray Scale Attribute Filtering (default settings, connectivity: 8) from MorphoLibJ plug-in version 1.4.1 was used to reduce background noise when thresholding images for skeleton analysis. Following skeleton analysis, Morphological Filters (Operation: Opening, Element: Octagon, Radius (in pixels): 2) from MorphoLibJ was used to quantify soma area. This analysis was performed for both Iba1 and Cx3cr1-GFP imaging experiments.
Statistical methods and reproducibility
All imaging, RT-qPCR, and enzyme-linked immunosorbent assay (ELISA) data were analyzed using either Student’s t test, one-way analysis of variance (ANOVA), two-way ANOVA, or simple linear regression. Data were plotted and statistical tests were performed on GraphPad Prism 9. Detailed statistical procedures for sequencing analysis are described in the sequencing sections of Materials and Methods.
Acknowledgments
We thank M. Saturno-Condon, M. Jennings, M. Griffin, and G. Paradis from the Swanson Biotechnology Center Flow Cytometry Facility for assistance with FANS. We thank S. Levine, N. Kamelamela, and A. Hendricks from the MIT BioMicro Center for assistance with RNA sequencing and library preparation. We thank K. Cormier from the MIT Hope Babetter Tang (1983) Histology Core Facility and J. Khun from the MIT Microscopy Core Facility for assistance with Visium Spatial Gene Expression sample preparation.
Funding: This work was supported by NIH grants AG054012 (to M.K. and L.-H.T.), AG058002 (to M.K. and L.-H.T.), AG062377 (to M.K. and L.-H.T.), NS110453 (to M.K. and L.-H.T.), NS115064 (to M.K. and L.-H.T.), AG067151 (to M.K. and L.-H.T.), AG062335 (to M.K. and L.-H.T.), MH109978 (to M.K. and L.-H.T.), MH119509 (to M.K. and L.-H.T.), HG008155 (to M.K. and L.-H.T.), and 5 R01 NS102730-03 (to L.-H.T.); CureAlz CIRCUITS (to M.K. and L.-H.T.); Glenn Foundation for Medical Research (to L.-H.T.); NIH fellowship 5F31NS113464-02 (to G.M.W.); Alzheimer’s Association grant AARF-19-618751 (to V.D.); Howard Hughes Medical Institute Hanna H. Gray Fellowship (to M.B.V.); and NIH training grant GM087237 (to C.A.B.).
Author contributions: Conceptualization: G.M.W. and L.-H.T. Methodology: G.M.W., C.A.B., E.S., J.D.-V., J.D.C., P.L.B., L.-H.T., and M.K. Investigation: G.M.W., C.A.B., E.S., J.D.-V., P.L.B., M.B.V., A.L., Q.S., N.S.L., P.L.B., and J.D.C. Visualization: G.M.W., C.A.B., E.S., J.D.-V., and M.B.V. Supervision: L.-H.T., M.K., and A.R.P. Writing—original draft: G.M.W. Writing—review and editing: G.M.W., L.-H.T., M.B.V., V.D., and P.L.B.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. All mouse sequencing data are available through GEO accession number GSE174265 (www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE174265). Human bulk RNA-seq data generated for this manuscript are available at Synapse (www.synapse.org/#!Synapse:syn33212353) under controlled use conditions set by human privacy regulations. snRNA-seq data of prefrontal cortex samples from patients with AD (37) are available at Synapse (www.synapse.org/#!Synapse:syn18485175) under controlled use conditions set by human privacy regulations. ROSMAP metadata can be accessed at www.synapse.org/#!Synapse:syn3157322.
Supplementary Materials
This PDF file includes:
Figs. S1 to S12
Other Supplementary Material for this manuscript includes the following:
Tables S1 to S4
REFERENCES AND NOTES
- 1.Madabhushi R., Pan L., Tsai L.-H., DNA damage and its links to neurodegeneration. Neuron 83, 266–282 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Welch G., Tsai L., Mechanisms of DNA damage-mediated neurotoxicity in neurodegenerative disease. EMBO Rep. 23, e54217 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lu T., Pan Y., Kao S.-Y., Li C., Kohane I., Chan J., Yankner B. A., Gene regulation and DNA damage in the ageing human brain. Nature 429, 883–891 (2004). [DOI] [PubMed] [Google Scholar]
- 4.Lovell M. A., Markesbery W. R., Oxidative DNA damage in mild cognitive impairment and late-stage Alzheimer’s disease. Nucleic Acids Res. 35, 7497–7504 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jacobsen E., Beach T., Shen Y., Li R., Chang Y., Deficiency of the Mre11 DNA repair complex in Alzheimer’s disease brains. Mol. Brain Res. 128, 1–7 (2004). [DOI] [PubMed] [Google Scholar]
- 6.Shanbhag N. M., Evans M. D., Mao W., Nana A. L., Seeley W. W., Adame A., Rissman R. A., Masliah E., Mucke L., Early neuronal accumulation of DNA double strand breaks in Alzheimer’s disease. Acta Neuropathol. Commun. 7, 77 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim D., Frank C. L., Dobbin M. M., Tsunemoto R. K., Tu W., Peng P. L., Guan J.-S., Lee B.-H., Moy L. Y., Giusti P., Broodie N., Mazitschek R., Delalle I., Haggarty S. J., Neve R. L., Lu Y., Tsai L.-H., Deregulation of HDAC1 by p25/Cdk5 in neurotoxicity. Neuron 60, 803–817 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sykora P., Misiak M., Wang Y., Ghosh S., Leandro G. S., Liu D., Tian J., Baptiste B. A., Cong W.-N., Brenerman B. M., Fang E., Becker K. G., Hamilton R. J., Chigurupati S., Zhang Y., Egan J. M., Croteau D. L., Wilson D. M. III, Mattson M. P., Bohr V. A., DNA polymerase β deficiency leads to neurodegeneration and exacerbates Alzheimer disease phenotypes. Nucleic Acids Res. 43, 943–959 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suberbielle E., Sanchez P. E., Kravitz A. V., Wang X., Ho K., Eilertson K., Devidze N., Kreitzer A. C., Mucke L., Physiologic brain activity causes DNA double-strand breaks in neurons, with exacerbation by amyloid-β. Nat. Neurosci. 16, 613–621 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu W., Hill S. E., Nathan W. J., Paiano J., Callen E., Wang D., Shinoda K., van Wietmarschen N., Colón-Mercado J. M., Zong D., De Pace R., Shih H.-Y., Coon S., Parsadanian M., Pavani R., Hanzlikova H., Park S., Jung S. K., McHugh P. J., Canela A., Chen C., Casellas R., Caldecott K. W., Ward M. E., Nussenzweig A., Neuronal enhancers are hotspots for DNA single-strand break repair. Nature 593, 440–444 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reid D. A., Reed P. J., Schlachetzki J. C. M., Nitulescu I. I., Chou G., Tsui E. C., Jones J. R., Chandran S., Lu A. T., McClain C. A., Ooi J. H., Wang T.-W., Lana A. J., Linker S. B., Ricciardulli A. S., Lau S., Schafer S. T., Horvath S., Dixon J. R., Hah N., Glass C. K., Gage F. H., Incorporation of a nucleoside analog maps genome repair sites in postmitotic human neurons. Science 372, 91–94 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dou Z., Ghosh K., Vizioli M. G., Zhu J., Sen P., Wangensteen K. J., Simithy J., Lan Y., Lin Y., Zhou Z., Capell B. C., Xu C., Xu M., Kieckhaefer J. E., Jiang T., Shoshkes-Carmel M., Tanim K. M. A. A., Barber G. N., Seykora J. T., Millar S. E., Kaestner K. H., Garcia B. A., Adams P. D., Berger S. L., Cytoplasmic chromatin triggers inflammation in senescence and cancer. Nature 550, 402–406 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rodier F., Coppé J.-P., Patil C. K., Hoeijmakers W. A. M., Muñoz D. P., Raza S. R., Freund A., Campeau E., Davalos A. R., Campisi J., Persistent DNA damage signalling triggers senescence-associated inflammatory cytokine secretion. Nat. Cell Biol. 11, 973–979 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klein R. S., Lin E., Zhang B., Luster A. D., Tollett J., Samuel M. A., Engle M., Diamond M. S., Neuronal CXCL10 directs CD8+ T-cell recruitment and control of west nile virus encephalitis. J. Virol. 79, 11457–11466 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cruz J. C., Tseng H.-C., Goldman J. A., Shih H., Tsai L.-H., Aberrant Cdk5 activation by p25 triggers pathological events leading to neurodegeneration and neurofibrillary tangles. Neuron 40, 471–483 (2003). [DOI] [PubMed] [Google Scholar]
- 16.Mathys H., Adaikkan C., Gao F., Young J. Z., Manet E., Hemberg M., De Jager P. L., Ransohoff R. M., Regev A., Tsai L.-H., Temporal tracking of microglia activation in neurodegeneration at single-cell resolution. Cell Rep. 21, 366–380 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cruz J. C., Kim D., Moy L. Y., Dobbin M. M., Sun X., Bronson R. T., Tsai L.-H., p25/cyclin-dependent kinase 5 induces production and intraneuronal accumulation of amyloid beta in vivo. J. Neurosci. 26, 10536–10541 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fischer A., Sananbenesi F., Pang P. T., Lu B., Tsai L.-H., Opposing roles of transient and prolonged expression of p25 in synaptic plasticity and hippocampus-dependent memory. Neuron 48, 825–838 (2005). [DOI] [PubMed] [Google Scholar]
- 19.Mertens J., Herdy J. R., Traxler L., Schafer S. T., Schlachetzki J. C. M., Böhnke L., Reid D. A., Lee H., Zangwill D., Fernandes D. P., Agarwal R. K., Lucciola R., Zhou-Yang L., Karbacher L., Edenhofer F., Stern S., Horvath S., Paquola A. C. M., Glass C. K., Yuan S. H., Ku M., Szücs A., Goldstein L. S. B., Galasko D., Gage F. H., Age-dependent instability of mature neuronal fate in induced neurons from Alzheimer’s patients. Cell Stem Cell 28, 1533–1548 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yousef A., Robinson J. L., Irwin D. J., Byrne M. D., Kwong L. K., Lee E. B., Xu Y., Xie S. X., Rennert L., Suh E., Van Deerlin V. M., Grossman M., Lee V. M.-Y., Trojanowski J. Q., Neuron loss and degeneration in the progression of TDP-43 in frontotemporal lobar degeneration. Acta Neuropathol. Commun. 5, 68 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ünal-Çevik I., Kılınç M., Gürsoy-Özdemir Y., Gurer G., Dalkara T., Loss of NeuN immunoreactivity after cerebral ischemia does not indicate neuronal cell loss: A cautionary note. Brain Res. 1015, 169–174 (2004). [DOI] [PubMed] [Google Scholar]
- 22.McPhail L. T., McBride C. B., McGraw J., Steeves J. D., Tetzlaff W., Axotomy abolishes NeuN expression in facial but not rubrospinal neurons. Exp. Neurol. 185, 182–190 (2004). [DOI] [PubMed] [Google Scholar]
- 23.Wu K.-L., Li Y.-Q., Tabassum A., Lu W.-Y., Aubert I., Wong C. S., Loss of neuronal protein expression in mouse hippocampus after irradiation. J. Neuropathol. Exp. Neurol. 69, 272–280 (2010). [DOI] [PubMed] [Google Scholar]
- 24.Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S., Mesirov J. P., Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gjoneska E., Pfenning A. R., Mathys H., Quon G., Kundaje A., Tsai L.-H., Kellis M., Conserved epigenomic signals in mice and humans reveal immune basis of Alzheimer’s disease. Nature 518, 365–369 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Préhaud C., Mégret F., Lafage M., Lafon M., Virus infection switches TLR-3-positive human neurons to become strong producers of beta interferon. J. Virol. 79, 12893–12904 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sokolova A., Hill M. D., Rahimi F., Warden L. A., Halliday G. M., Shepherd C. E., Monocyte chemoattractant protein-1 plays a dominant role in the chronic inflammation observed in Alzheimer’s disease. Brain Pathol. 19, 392–398 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chai Q., She R., Huang Y., Fu Z. F., Expression of neuronal CXCL10 induced by rabies virus infection initiates infiltration of inflammatory cells, production of chemokines and cytokines, and enhancement of blood-brain barrier permeability. J. Virol. 89, 870–876 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xie Z., Bailey A., Kuleshov M. V., Clarke D. J. B., Evangelista J. E., Jenkins S. L., Lachmann A., Wojciechowicz M. L., Kropiwnicki E., Jagodnik K. M., Jeon M., Ma’ayan A., Gene set knowledge discovery with Enrichr. Curr. Protoc. 1, e90 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McCool K. W., Miyamoto S., DNA damage-dependent NF-κB activation: NEMO turns nuclear signaling inside out. Immunol. Rev. 246, 311–326 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chien Y., Scuoppo C., Wang X., Fang X., Balgley B., Bolden J. E., Premsrirut P., Luo W., Chicas A., Lee C. S., Kogan S. C., Lowe S. W., Control of the senescence-associated secretory phenotype by NF-κB promotes senescence and enhances chemosensitivity. Genes Dev. 25, 2125–2136 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Leeuwen L. A. G., Hoozemans J. J. M., Physiological and pathophysiological functions of cell cycle proteins in post-mitotic neurons: Implications for Alzheimer’s disease. Acta Neuropathol. 129, 511–525 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cao J., Spielmann M., Qiu X., Huang X., Ibrahim D. M., Hill A. J., Zhang F., Mundlos S., Christiansen L., Steemers F. J., Trapnell C., Shendure J., The single-cell transcriptional landscape of mammalian organogenesis. Nature 566, 496–502 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang C., Najm R., Xu Q., Jeong D., Walker D., Balestra M. E., Yoon S. Y., Yuan H., Li G., Miller Z. A., Miller B. L., Malloy M. J., Huang Y., Gain of toxic apolipoprotein E4 effects in human iPSC-derived neurons is ameliorated by a small-molecule structure corrector. Nat. Med. 24, 647–657 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dunphy G., Flannery S. M., Almine J. F., Connolly D. J., Paulus C., Jønsson K. L., Jakobsen M. R., Nevels M. M., Bowie A. G., Unterholzner L., Non-canonical activation of the DNA sensing adaptor STING by ATM and IFI16 mediates NF-κB signaling after nuclear DNA damage. Mol. Cell 71, 745–760.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sriram G., Milling L. E., Chen J.-K., Kong Y. W., Joughin B. A., Abraham W., Swartwout S., Handly E. D., Irvine D. J., Yaffe M. B., The injury response to DNA damage in live tumor cells promotes antitumor immunity. Sci. Signal. 14, eabc4764 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mathys H., Davila-Velderrain J., Peng Z., Gao F., Mohammadi S., Young J. Z., Menon M., He L., Abdurrob F., Jiang X., Martorell A. J., Ransohoff R. M., Hafler B. P., Bennett D. A., Kellis M., Tsai L.-H., Single-cell transcriptomic analysis of Alzheimer’s disease. Nature 570, 332–337 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Variable Details | RADC; www.radc.rush.edu/docs/var/detail.htm?category=Pathology&subcategory=Alzheimer%27s+disease&variable=gpathpath.
- 39.Chan K. Y., Jang M. J., Yoo B. B., Greenbaum A., Ravi N., Wu W.-L., Sánchez-Guardado L., Lois C., Mazmanian S. K., Deverman B. E., Gradinaru V., Engineered AAVs for efficient noninvasive gene delivery to the central and peripheral nervous systems. Nat. Neurosci. 20, 1172–1179 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hong S., Beja-Glasser V. F., Nfonoyim B. M., Frouin A., Li S., Ramakrishnan S., Merry K. M., Shi Q., Rosenthal A., Barres B. A., Lemere C. A., Selkoe D. J., Stevens B., Complement and microglia mediate early synapse loss in Alzheimer mouse models. Science 352, 712–716 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dejanovic B., Huntley M. A., De Mazière A., Meilandt W. J., Wu T., Srinivasan K., Jiang Z., Gandham V., Friedman B. A., Ngu H., Foreman O., Carano R. A. D., Chih B., Klumperman J., Bakalarski C., Hanson J. E., Sheng M., Changes in the synaptic proteome in Tauopathy and rescue of tau-induced synapse loss by C1q antibodies. Neuron 100, 1322–1336.e7 (2018). [DOI] [PubMed] [Google Scholar]
- 42.Conductier G., Blondeau N., Guyon A., Nahon J.-L., Rovère C., The role of monocyte chemoattractant protein MCP1/CCL2 in neuroinflammatory diseases. J. Neuroimmunol. 224, 93–100 (2010). [DOI] [PubMed] [Google Scholar]
- 43.Bradburn S., McPhee J., Bagley L., Carroll M., Slevin M., Al-Shanti N., Barnouin Y., Hogrel J.-Y., Pääsuke M., Gapeyeva H., Maier A., Sipilä S., Narici M., Robinson A., Mann D., Payton A., Pendleton N., Butler-Browne G., Murgatroyd C., Dysregulation of C-X-C motif ligand 10 during aging and association with cognitive performance. Neurobiol. Aging 63, 54–64 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Keren-Shaul H., Spinrad A., Weiner A., Matcovitch-Natan O., Dvir-Szternfeld R., Ulland T. K., David E., Baruch K., Lara-Astaiso D., Toth B., Itzkovitz S., Colonna M., Schwartz M., Amit I., A unique microglia type associated with restricting development of Alzheimer’s disease. Cell 169, 1276–1290 (2017). [DOI] [PubMed] [Google Scholar]
- 45.Nott A., Holtman I. R., Coufal N. G., Schlachetzki J. C. M., Yu M., Hu R., Han C. Z., Pena M., Xiao J., Wu Y., Keulen Z., Pasillas M. P., O’Connor C., Nickl C. K., Schafer S. T., Shen Z., Rissman R. A., Brewer J. B., Gosselin D., Gonda D. D., Levy M. L., Rosenfeld M. G., McVicker G., Gage F. H., Ren B., Glass C. K., Brain cell type–specific enhancer–promoter interactome maps and disease - risk association. Science 366, 1134–1139 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spangenberg E. E., Lee R. J., Najafi A. R., Rice R. A., Elmore M. R. P., Blurton-Jones M., West B. L., Green K. N., Eliminating microglia in Alzheimer’s mice prevents neuronal loss without modulating amyloid-β pathology. Brain 139, 1265–1281 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Song W. M., Colonna M., The identity and function of microglia in neurodegeneration. Nat. Immunol. 19, 1048–1058 (2018). [DOI] [PubMed] [Google Scholar]
- 48.Simon M., Van Meter M., Ablaeva J., Ke Z., Gonzalez R. S., Taguchi T., De Cecco M., Leonova K. I., Kogan V., Helfand S. L., Neretti N., Roichman A., Cohen H. Y., Meer M. V., Gladyshev V. N., Antoch M. P., Gudkov A. V., Sedivy J. M., Seluanov A., Gorbunova V., LINE1 derepression in aged wild-type and SIRT6-deficient mice drives inflammation. Cell Metab. 29, 871–885.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee H., Fenster R. J., Pineda S. S., Gibbs W. S., Mohammadi S., Davila-Velderrain J., Garcia F. J., Therrien M., Novis H. S., Gao F., Wilkinson H., Vogt T., Kellis M., LaVoie M. J., Heiman M., Cell type-specific transcriptomics reveals that mutant huntingtin leads to mitochondrial RNA release and neuronal innate immune activation. Neuron 107, 891–908.e8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Song X., Ma F., Herrup K., Accumulation of cytoplasmic DNA due to ATM deficiency activates the microglial viral response system with neurotoxic consequences. J. Neurosci. 39, 6378–6394 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bussian T. J., Aziz A., Meyer C. F., Swenson B. L., van Deursen J. M., Baker D. J., Clearance of senescent glial cells prevents tau-dependent pathology and cognitive decline. Nature 562, 578–582 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chinta S. J., Woods G., Demaria M., Rane A., Zou Y., McQuade A., Rajagopalan S., Limbad C., Madden D. T., Campisi J., Andersen J. K., Cellular senescence is induced by the environmental neurotoxin paraquat and contributes to neuropathology linked to Parkinson’s disease. Cell Rep. 22, 930–940 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang P., Kishimoto Y., Grammatikakis I., Gottimukkala K., Cutler R. G., Zhang S., Abdelmohsen K., Bohr V. A., Misra Sen J., Gorospe M., Mattson M. P., Senolytic therapy alleviates Aβ-associated oligodendrocyte progenitor cell senescence and cognitive deficits in an Alzheimer’s disease model. Nat. Neurosci. 22, 719–728 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sah E., Krishnamurthy S., Ahmidouch M. Y., Gillispie G. J., Milligan C., Orr M. E., The cellular senescence stress response in post-mitotic brain cells: Cell survival at the expense of tissue degeneration. Life 11, 229 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wyss-Coray T., Rogers J., Inflammation in Alzheimer disease—A brief review of the basic science and clinical literature. Cold Spring Harb. Perspect. Med. 2, a006346 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kaltschmidt B., Ndiaye D., Korte M., Pothion S., Arbibe L., Prüllage M., Pfeiffer J., Lindecke A., Staiger V., Israël A., Kaltschmidt C., Mémet S., NF-κB regulates spatial memory formation and synaptic plasticity through protein kinase A/CREB signaling. Mol. Cell. Biol. 26, 2936–2946 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fridmacher V., Kaltschmidt B., Goudeau B., Ndiaye D., Rossi F. M., Pfeiffer J., Kaltschmidt C., Israël A., Mémet S., Forebrain-specific neuronal inhibition of nuclear factor-κB activity leads to loss of neuroprotection. J. Neurosci. 23, 9403–9408 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dimitrijevic O. B., Stamatovic S. M., Keep R. F., Andjelkovic A. V., Effects of the chemokine CCL2 on blood–brain barrier permeability during ischemia–reperfusion injury. J. Cereb. Blood Flow Metab. 26, 797–810 (2006). [DOI] [PubMed] [Google Scholar]
- 59.El Khoury J., Toft M., Hickman S. E., Means T. K., Terada K., Geula C., Luster A. D., Ccr2 deficiency impairs microglial accumulation and accelerates progression of Alzheimer-like disease. Nat. Med. 13, 432–438 (2007). [DOI] [PubMed] [Google Scholar]
- 60.Naert G., Rivest S., CC Chemokine receptor 2 deficiency aggravates cognitive impairments and amyloid pathology in a transgenic mouse model of Alzheimer’s disease. J. Neurosci. 31, 6208–6220 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yamamoto M., Horiba M., Buescher J. L., Huang D., Gendelman H. E., Ransohoff R. M., Ikezu T., Overexpression of monocyte chemotactic protein-1/CCL2 in β-amyloid precursor protein transgenic mice show accelerated diffuse β-amyloid deposition. Am. J. Pathol. 166, 1475–1485 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Krauthausen M., Kummer M. P., Zimmermann J., Reyes-Irisarri E., Terwel D., Bulic B., Heneka M. T., Müller M., CXCR3 promotes plaque formation and behavioral deficits in an Alzheimer’s disease model. J. Clin. Invest. 125, 365–378 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Young K., Morrison H., Quantifying microglia morphology from photomicrographs of immunohistochemistry prepared tissue using ImageJ. J. Vis. Exp. 57648 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S12
Tables S1 to S4