Abstract
Disrupted circadian rhythmicity is a prominent feature of modern society and has been designated as a probable carcinogen by the World Health Organization. However, the biological mechanisms that connect circadian disruption and cancer risk remain largely undefined. We demonstrate that exposure to chronic circadian disruption [chronic jetlag (CJL)] increases tumor burden in a mouse model of KRAS-driven lung cancer. Molecular characterization of tumors and tumor-bearing lung tissues revealed that CJL enhances the expression of heat shock factor 1 (HSF1) target genes. Consistently, exposure to CJL disrupted the highly rhythmic nuclear trafficking of HSF1 in the lung, resulting in an enhanced accumulation of HSF1 in the nucleus. HSF1 has been shown to promote tumorigenesis in other systems, and we find that pharmacological or genetic inhibition of HSF1 reduces the growth of KRAS-mutant human lung cancer cells. These findings implicate HSF1 as a molecular link between circadian disruption and enhanced tumorigenesis.
Circadian disruption enhances HSF1 signaling, which could contribute to increased cancer risk in shift workers.
INTRODUCTION
In 2015, the National Health Interview Survey revealed that 12 to 35% of the workforce in various U.S. industries work irregular schedules, including night and rotating shifts (1). Several human and animal studies have demonstrated that disruption of circadian rhythms, either by genetic or environmental means, enhances cancer risk (2), including the risk of lung adenocarcinoma (LUAD) (3–7). Lung cancer is the leading cause of cancer deaths in men and women worldwide (8, 9). The LUAD subtype of non–small cell lung cancer (NSCLC) is the most prevalent form of lung cancer, and Kirsten rat sarcoma (KRAS) is the most frequently mutated oncogene in human LUAD (10). Despite extensive characterization of genetic events that contribute to lung cancer, there has been relatively little research addressing the impact of environmental circadian disruption on lung tumorigenesis in humans. The lung is under tight circadian control, as evidenced by robust 24-hour rhythms in intrinsic defense mechanisms and lung physiology indices such as lung resistance and peak expiratory flow (11). Selective ablation of bronchiolar epithelial cells results in the loss of circadian clock oscillations in mouse lung slices, demonstrating that airway epithelial cells are key circadian oscillators within the lung (12). NSCLC is a cancer of epithelial origin (13, 14), so this reinforces the hypothesis that disruption of the circadian machinery could trigger harmful events due to dysregulation of homeostasis, resulting in increased risk of lung tumor development.
The mammalian circadian machinery consists of an autoregulatory transcription-translation feedback loop. Its positive arm—heterodimers of circadian locomotor output cycle kaput (CLOCK) and brain and muscle aryl hydrocarbon receptor nuclear translocator (ARNT)-like protein 1 (BMAL1)—drives the transcription of two inhibitory arms, periods (PERs), and cryptochromes (CRYs) on one hand and nuclear receptor subfamily 1 group D members 1 and 2 (NR1D1/NR1D2; also called REV-ERBα/REV-ERBβ) on the other. While PERs and CRYs inhibit the BMAL1-CLOCK heterodimer transactivation function, REV-ERBs repress BMAL1 expression. These transcription-translation feedback loops drive 24-hour periodic expression of gene products, leading to rhythmic physiologic functions. Accumulating evidence demonstrates that circadian clock components play critical roles in regulating several hallmarks of cancer, including control of cell proliferation, cell death, DNA repair, and metabolic alteration (15–17). However, the precise mechanisms underlying the cooperation between circadian clock disruption and tumorigenesis remain poorly understood. By manipulating lighting schedules to mimic the circadian disturbance that humans encounter during rotating shift work or frequent eastbound transmeridian flights [chronic jetlag (CJL)], we show that this environmental light disruption alters gene expression in the liver and lungs of mice. Further, we show that KrasLSL-G12D/+ (K) mice, a genetically engineered mouse model (GEMM) of NSCLC (18), developed many more tumors when housed in CJL compared to normal light conditions [12-hour light:12-hour darkness (12:12LD)]. Unbiased RNA sequencing and gene expression analyses revealed a profound disruption of the circadian clock machinery and chronic elevation of the heat shock response in the lungs of mice exposed to CJL, indicating that light-induced circadian disruption perturbs homeostatic regulation of heat shock factor 1 (HSF1). Given the strong and growing evidence that HSF1 can facilitate tumorigenesis (19–22), these findings suggest that chronic elevation of HSF1 signaling could be a key molecular link between circadian disruption and increased cancer risk.
RESULTS
Experimental CJL disrupts peripheral clocks
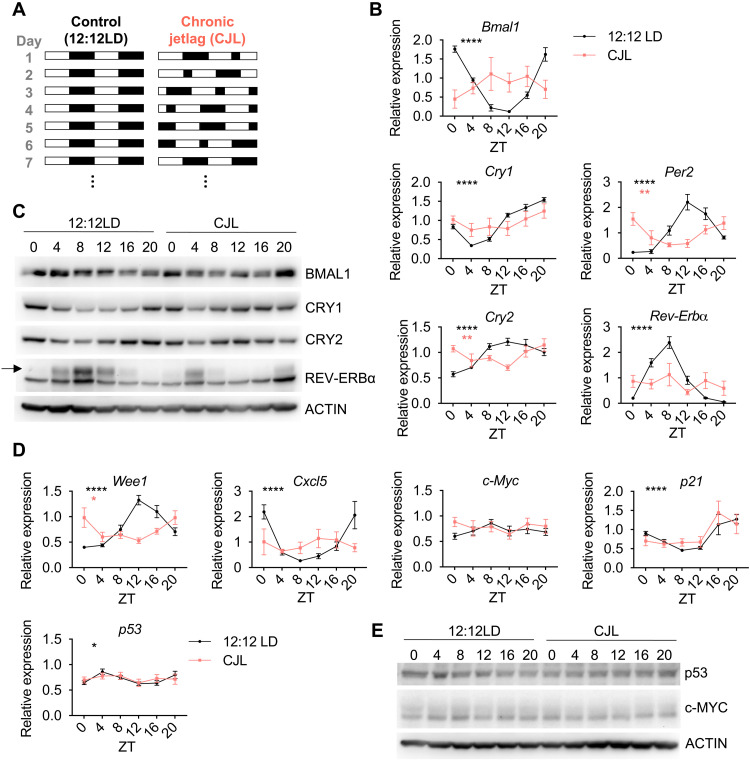
To mimic chronic disruption of a functional circadian timing system, we used a CJL protocol consisting of an 8-hour light-phase advance repeated every 2 or 3 days (Fig. 1A). This altered light/dark scheme mimics the circadian disturbance that humans face during rotating shift work (23, 24) and has been shown to increase tumor development upon Glasgow osteosarcoma inoculation (25), in chemically induced or spontaneous mouse liver cancer models (26, 27), and in a mouse model of lung cancer similar to the one used here (3). To first assess the magnitude of the disturbance of this protocol on the clock machinery and other cancer-related pathways, male and female C57BL/6J mice were housed in either normal light (12:12LD) or CJL conditions for 8 weeks before tissues were collected every 4 hours over a 24-hour period. The timing of light-dark transitions on the day of collection (day 1 in Fig. 1A) were the same for at least 24 hours before the collection (day 7 in Fig. 1A). As expected, CJL greatly affected the expression of core clock genes in the lung, liver, and, to a lesser extent, spleen, with loss of periodic expression (PJTKcycle < 0.05) for most of the genes, including Bmal1, Cry1, and Rev-Erba (Fig. 1B and fig. S1). Similarly, the components of the clock machinery were profoundly disrupted at the protein level (Fig. 1C and fig. S1C). While these data are consistent with a model in which circadian rhythms of gene expression are suppressed or disrupted by CJL, we cannot exclude the possibility that the loss of observed rhythmicity for some genes is caused by a lack of synchrony between animals or an inability to properly assign individual animals to a specific circadian “phase” rather than loss of rhythmicity within individual mice based on this analysis. Nonetheless, the observation that Per2 appears to remain rhythmic in mice exposed to CJL, while Rev-Erbα, for example, does not, suggests a desynchronization of the core clock components relative to each other. CJL exposure affected daily rhythms of core clock genes somewhat differently in male versus female mice (fig. S1). These disparities are consistent with previous studies suggesting sexual dimorphism in circadian clock mechanism and physiology (28).
Fig. 1. CJL severely impairs rhythmicity and magnitude of core clock and clock-controlled genes in the lung.
(A) Schematic representation of the CJL protocol. White and black rectangles represent periods of light and dark, respectively. Each row represents two consecutive days starting with the numbered day shown at left. (B to E) C57BL/6J male and female mice were housed in 12:12LD or CJL for 8 weeks. Lung tissues were collected at the indicated times (ZT0: light on; ZT12: light off) on day 1 of the schedule shown in (A). (B and D) Gene expression normalized to U36b4 measured by quantitative real-time PCR. Data represent means ± SEM for three males and three females per time point and light condition. Rhythmicity was determined by JTK_Cycle analyses; *PJTKcycle < 0.05, **PJTKcycle < 0.01, and ****PJTKcycle < 0.0001. (C and E) Proteins detected by immunoblot. Each lane on the Western blot represents a sample prepared from a unique animal. Representative images were taken from n = 6 biological replicates.
The expression of clock-controlled genes was also affected by CJL.Wee1 is a critical regulator of the G2-M transition of the cell cycle that is under circadian control via BMAL1-CLOCK activation that targets E-box elements in the Wee1 gene promoter (29). Wee1 expression was strongly influenced by the time of day in healthy lungs and was robustly affected by CJL (Fig. 1D). CJL also profoundly altered the diurnal expression of Cxcl5 (Fig. 1D), a key chemokine for recruiting neutrophils to the lung upon bacterial and viral exposures (30–32) that is regulated by REV-ERBα and REV-ERBβ (33). However, expression of the cell cycle regulators p53 and p21 and oncogenic transcription factor c-Myc, each of which can be regulated by circadian clock factors (34–36), were not affected by CJL in lung tissue (Fig. 1D). Although p53 and c-MYC are regulated posttranslationally by clock components (35, 36) and exhibit rhythmic expression that is affected by exposure to altered lighting conditions in mouse thymus (23), we did not observe any clear effect of CJL on the levels of these proteins in the lungs of healthy mice (Fig. 1E).
In an earlier study of mice with mammary tumors in the FVB genetic background, chronic disruption of light exposures markedly increased weight gain (37). In contrast, we found that CJL had no impact on body weight in healthy C57BL/6J mice (fig. S2A). We observed CJL-induced differences in rhythmic corticosterone levels in serum of female mice only, suggesting a more rapid light entrainment for corticosterone secretion in males and sexual dimorphism in hormonal responses (fig. S2B). In a separate group of mice that were housed first in 12:12LD for 2 weeks and then in CJL for 13 weeks with access to a running wheel, we confirmed the disruption of rhythmic behavior upon CJL; while the locomotor activity was mostly consolidated within the dark phases, the pattern and amplitude of activity during the dark hours markedly changed after only 1 week of CJL (fig. S3, A to E).
Experimental CJL increases KrasG12D-driven lung tumor burden
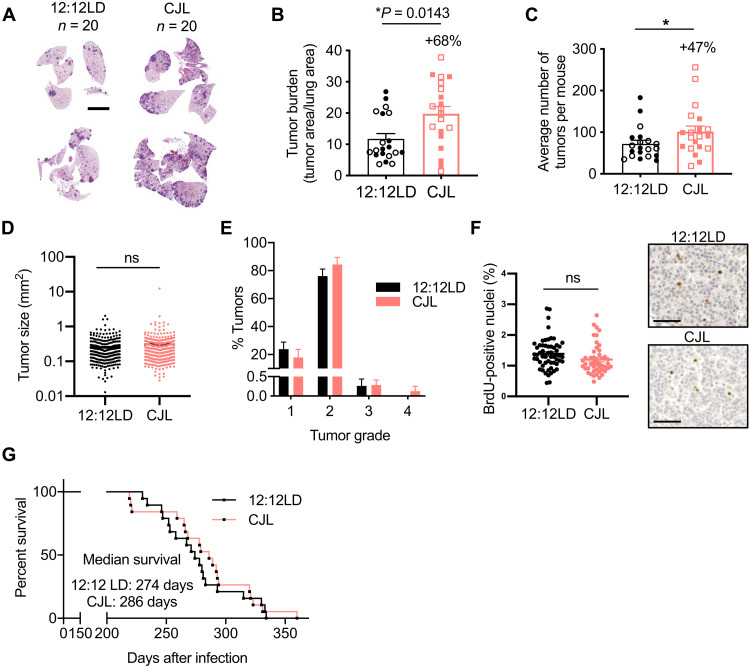
To investigate molecular mechanisms related to light-induced circadian disruption in lung tumorigenesis, we used a GEMM of NSCLC in which tumor formation is initiated by expression of oncogenic KrasG12D in a small number of lung cells (KrasLSL-G12D/+ mice, also known as K mice)—a model established to recapitulate many of the clinical features of naturally occurring KRAS-driven lung cancer (38). Male and female K mice were first infected intratracheally with lentivirus expressing CRE recombinase under the control of the Ubc promoter to induce tumorigenesis and, 5 weeks later, were placed in 12:12LD or CJL conditions. At 25 weeks after infection (20 weeks in CJL), we observed a notable 68% increase in tumor burden in K mice housed in CJL conditions compared to those that had remained in 12:12LD (Fig. 2, A and B). This was attributed to an increase in the number (Fig. 2C) and not the size (Fig. 2D) of tumors, suggesting that CJL affects early events in tumor progression in this model. Moreover, there was no difference in the spectrum of tumor grades assessed by histopathology (38) between the two groups, with most of the tumors being grade 2 adenomas (Fig. 2E). Further, CJL exposure had no statistically significant effect on 5-bromo-2′-deoxyuridine (BrdU) or Ki67 positivity in tumors (Fig. 2F and fig. S4), suggesting that altered light exposure does not affect the proliferation of tumor cells in this model. There was no impact of CJL on overall survival (Fig. 2G), indicating that additional factors precipitating death in K mice appeared stochastically in both light conditions. An earlier study demonstrated that CJL increased tumor burden in KrasLSL-G12D/+;p53flox/flox (KP) mice but to a much lesser extent than we observed in K mice (3). Consistent with this, we did not observe any effects of CJL on lung tumor burden, numbers, grading, or overall survival in KP mice (fig. S5), which harbor simultaneous activation of oncogenic KRAS and deletion of tumor suppressor protein p53, and thus experience more rapid and severe tumor progression. Technical differences such as use of a different virus to initiate the onset of the disease (lentivirus versus adenovirus), different mouse genetic background (C57BL/6J versus 129SvJ), and different animal facilities (distinct light intensity exposure, microbiome, etc.) may explain why Papagiannakopoulos et al. (3) observed an effect of CJL on tumor burden where we did not.
Fig. 2. CJL causes an increase in tumor burden in K mice but has no impact on survival.
Five weeks after infection with lentivirus-Cre, K mice were placed in either 12:12LD or CJL for 20 weeks (A to F) or until signs of distress (G). (A) Representative H&E-stained sections at end point. Scale bars, 5000 μm. Tumor burden (B), numbers (C), size (D), and grade (E) were assessed from H&E sections. Column data represent means ± SEM. Values for individual animals (B, C, and E) or tumors (D) are plotted. (B and C) Clear and filled symbols represent males and females, respectively. *P < 0.05 by Mann-Whitney test. (F) Representative BrdU IHC images in tumors from K mice euthanized at ZT5 and quantitation of stained positive nuclei. Scale bar, 50 μm. (G) Kaplan-Meier survival analysis for K mice placed in 12:12LD (n = 19) or CJL (n = 19) conditions.
c-MYC levels in K mice do not explain increased tumor burden upon CJL
The c-MYC oncoprotein has been shown to play a crucial role in the growth of KRAS-driven lung tumors (39), and c-MYC accumulation in mouse thymus exhibits a daily rhythm and is robustly elevated at all times of day after exposure to a single shift of light exposure (23). Furthermore, genetic deletion of Bmal1 or loss of function of period circadian regulator 2 (PER2) enhanced c-MYC accumulation in KrasG12D-driven lung tumors (3). The circadian transcriptional repressor cryptochrome 2 (CRY2) can recruit phosphorylated substrates, including c-MYC (36), to the SCFFBXL3 ubiquitin ligase, thereby promoting their ubiquitination and proteasomal degradation. We thus expected that CJL could promote tumorigenesis by perturbing the expression of CRY2, resulting in aberrant accumulation of c-MYC. Unexpectedly, CJL resulted in significantly reduced accumulation of c-MYC in KrasG12D-driven lung tumors as assessed by immunohistochemistry and Western blotting (fig. S6). Thus, while aberrant stabilization of c-MYC may contribute to enhanced cell growth and transformation caused by deletion or suppression of circadian clock components (3, 36), it does not appear to play a major role in the enhanced tumorigenesis caused by circadian disruption of environmental light exposure in the context of KRAS-driven lung cancer.
CJL further disrupts an already dysregulated clock machinery in tumors from K mice
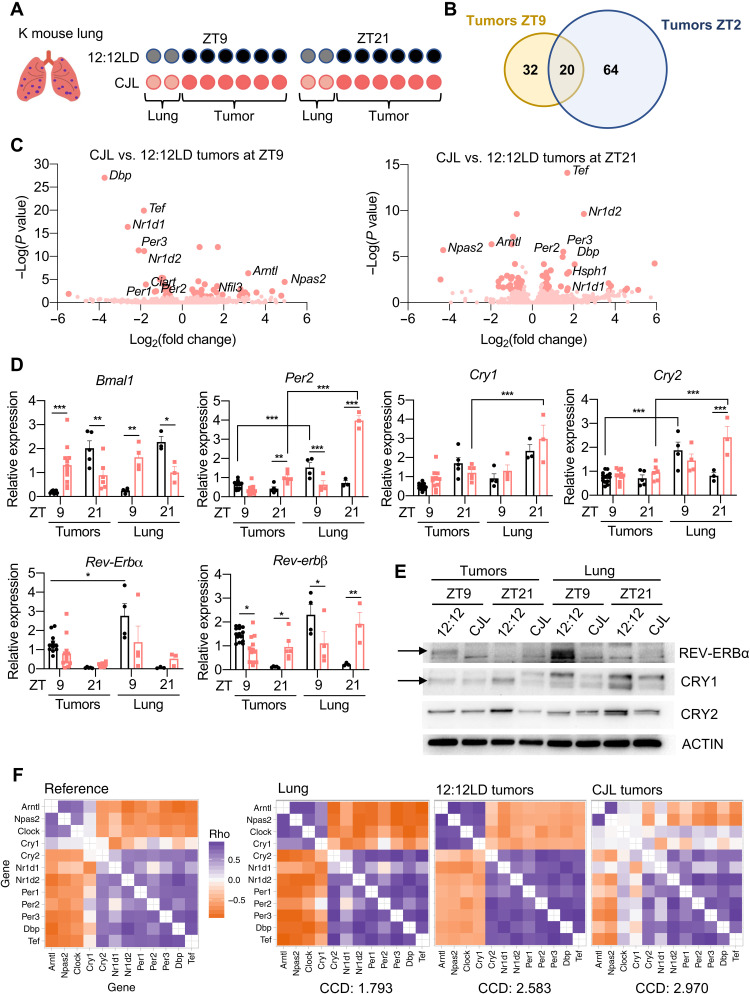
To identify mechanisms potentially underlying the protumorigenic effect of CJL in K mice, we compared the transcriptional programs in tumors and total lung from K mice housed in normal or CJL conditions. Up to 12 individual tumors were collected from each animal at either zeitgeber time (ZT, hours after lights on) ZT9 or ZT21, and the remaining lung tissue was also collected for subsequent analyses. We chose these two time points as they represent the trough and peak, respectively, of expression of the core clock component Bmal1, in healthy lung under normal light conditions. We sequenced RNA prepared from three tumors per animal and two mice per time point and lighting condition (Fig. 3A). To gain unbiased insight into transcriptional networks perturbed by CJL, we used differential expression analysis (DESeq2) (40). The “whole lung” samples also contain several tumors, but the tumor tissue represents a smaller fraction of the whole compared to the tumor samples. Pathway enrichment analysis confirmed up-regulation of KRAS signaling in resected tumors compared to whole lung samples for all conditions combined (fig. S7A). When looking at each time point individually for tumors, we identified 53 and 85 genes differentially expressed between 12:12LD and CJL at ZT9 and ZT21, respectively, with 20 genes differentially expressed at both time points (Fig. 3, B and C, and fig. S7B). The expression changes for these 20 genes were inverted between ZT9 and ZT21 (fig. S7B), suggesting that their expression is under circadian control. Core clock genes and highly rhythmic transcription factors Tef and Dbp were among the most differentially expressed genes in all groups, and expectedly, Database for Annotation, Visualization, and Integrated Discovery (DAVID) analyses highlighted biological circadian rhythms and rhythmic processes in the cluster with the highest enrichment score for tumor groups at each time point (fig. S7C). Transcript analyses by quantitative polymerase chain reeaction (qPCR) of different tumors and additional lung samples validated these findings, demonstrating significant variations in Bmal1, Per2, and Rev-erbβ mRNA between tumors from K mice housed in 12:12LD and CJL conditions (Fig. 3D). Notably, rhythmic expression of core clock genes was retained in tumors from mice housed in the 12:12LD standard light condition, but the amplitude of some clock gene expression such as Per2, Cry2, and Rev-erbα was reduced in tumor samples compared to whole lungs (Fig. 3D). This is consistent with several studies demonstrating dampening of circadian rhythms in tumors, mediated by oncogenic factors such as c-MYC or RAS (41, 42). Similar results were observed at the protein level, with a particularly marked reduction in REV-ERBα at the peak of its normal expression (ZT9) in tumors compared to total lung from K mice housed in 12:12LD (Fig. 3E). REV-ERBα was not detected at the trough of its expression (ZT21) in either tumors or total lung, indicating that the rhythmicity of REV-ERBα was probably retained in tumors but with a greatly diminished amplitude. Moreover, REV-ERBα protein levels were very low under CJL conditions in both tumors and total lung samples at these two time points, but given its very high amplitude of expression, we cannot exclude the possibility that CJL caused a shift in the phase of REV-ERBα.
Fig. 3. CJL further disrupts an already dysregulated clockwork in tumors from K mice.
Five weeks after infection with lentivirus-Cre, K mice were placed in either 12:12LD or CJL for 20 weeks. (A) For RNA sequencing, six tumors per animal and two mice per time point and light conditions were used. (B) Plots indicating the numbers of differentially expressed genes between 12:12LD and CJL by DESeq2 analyses for each condition, with adujusted P value < 0.05 and fold change > ±1.4 cutoffs. (C) Volcano plots of differentially expressed genes between 12:12LD and CJL by DESeq2 analyses for tumors collected at ZT9 and ZT21. (D) Gene expression normalized to U36b4 measured by quantitative real-time PCR. Data represent means ± SEM; n = 14 and 12 for tumors collected at ZT9 in 12:12LD and CJL, respectively; n = 5 and 6 for tumors collected at ZT21 in 12:12LD and CJL, respectively; and n = 3 to 4 for lung samples. *P < 0.05, **P < 0.01, and ***P < 0.001 by two-way ANOVA post hoc Bonferroni test. (E) Proteins detected by immunoblot. Tumors and lungs for each light condition and time point on the blot were from the same animal. Representative images were taken from n = 3 biological replicates. (F) Heatmaps of Spearman correlation between each pair of the 12 clock genes and corresponding CCD (relative to the mouse reference) in murine healthy lung from previously published dataset GSE54651 or in tumors from 12:12LD or CJL conditions.
To further assess whether CJL disrupts the clockwork in tumors from K mice, we used the clock correlation distance (CCD) algorithm, which infers the regularity of circadian clock progression in a group of samples based on the correlated coexpression of 12 clock genes (43). A higher CCD score indicates a more profound disruption of circadian rhythmicity. Originally, the CCD method was designed to evaluate circadian clocks in human cancer and revealed that clock gene coexpression in tumors is consistently perturbed compared to matched healthy tissue. Because our whole lung samples contain both healthy lung tissue and tumors, we compared the CCD score from our 12:12LD tumors (n = 12)—calculated using our data acquired by sequencing RNA—to that calculated using published data from murine healthy lung (44). This analysis revealed that the CCD score for healthy mouse lung was lower than the CCD score for tumors from K mice housed in 12:12LD. Moreover, we found that the CCD score for tumors from CJL-housed mice was higher than the CCD score for tumors collected from control animals (Fig. 3F), although the difference between groups is not statistically significant, likely due to limited numbers of samples. Together, these results strongly suggest a dysregulation of circadian clock progression in tumors from K mice compared to healthy lung tissue under normal light condition, and that exposure to CJL further perturbs an already disrupted clock within KRASG12D-driven lung tumors.
HSF1 signaling is up-regulated in response to CJL in K mice
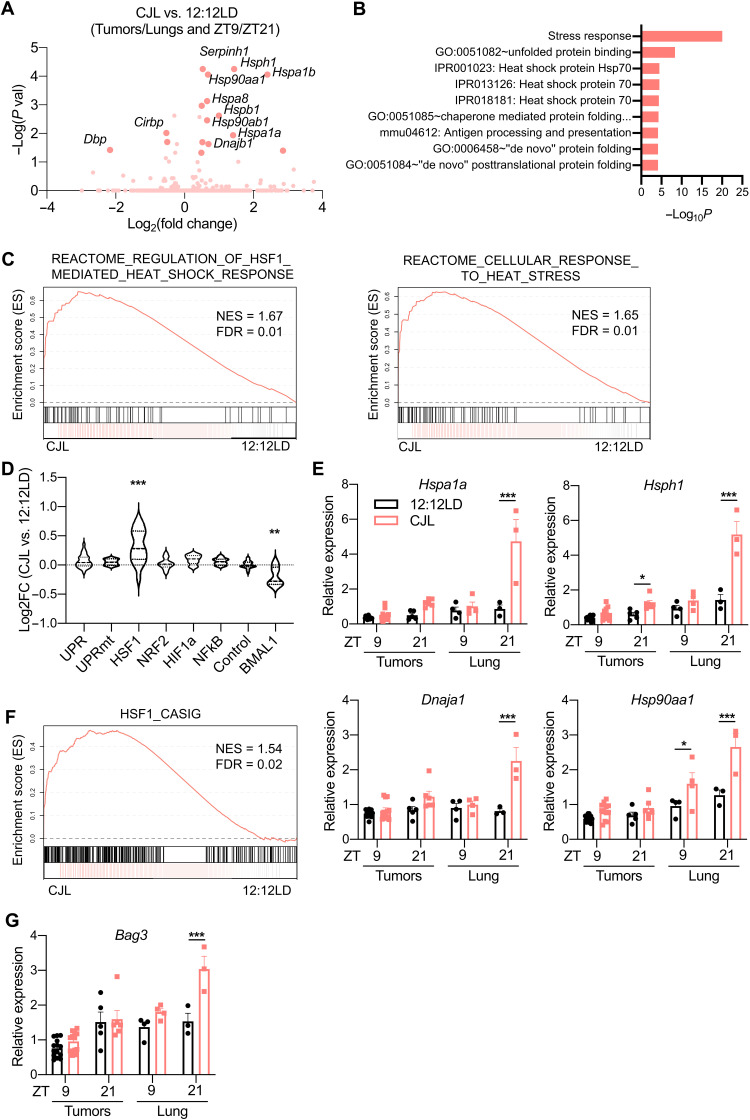
Because we were primarily interested in identifying gene networks that were consistently affected by circadian disruption within both tumors and lungs without time of sample collection as confounding factor, we searched for transcripts that were differentially expressed between all the samples (tumors + lungs) collected from mice housed in control 12:12LD lighting conditions compared to those housed in CJL lighting conditions. This comparison revealed that genes encoding various heat shock proteins (HSPs) are up-regulated in samples from CJL-exposed K mice (Fig. 4A). The same analysis including only tumor samples gave very similar results (fig. S8A). The expression of HSPs is primarily activated by the heat shock response–associated transcription factor HSF1 in response to pathologic insults that disrupt cytosolic proteostasis, including modest changes in temperature and oxidative stress (45, 46). HSPs function to enhance proteostasis capacity of the cell and prevent the pathologic accumulation of potentially toxic protein aggregates (47). Accordingly, DAVID analysis pointed to stress response and response to unfolded protein in the cluster with the highest enrichment score (Fig. 4B and fig. S8B). Strikingly, all of the transcripts that are significantly elevated in samples from mice exposed to CJL are known transcriptional targets of HSF1 (48). Gene Set Enrichment Analysis (GSEA) provided further support for the idea that CJL leads to an elevated HSF1-mediated heat shock response upon CJL in these samples (Fig. 4C). Moreover, this CJL-driven activation of HSF1 in K mice appeared to be selective, as we did not observe increased expression of genes regulated by other stress-responsive signaling pathways (Fig. 4D) (49). Consistent with dysregulation of clock gene expression measured by qPCR (Fig. 3D), we also observed a significant decrease in expression of a set of previously defined BMAL1 target genes (Fig. 4D, right) (43). Transcript analyses by qPCR from different tumors and additional lung samples validated these findings and highlighted a more pronounced up-regulation of HSF1 target gene expression at ZT21 in whole lungs from mice exposed to CJL compared to normal light conditions (Fig. 4E).
Fig. 4. CJL enhances expression of the HSF1-mediated heat shock response and cancer signature in KrasG12D-driven lung tumor model.
Five weeks after infection with lentivirus-Cre, K mice were placed in either 12:12LD or CJL for 20 weeks. (A) Volcano plots of differentially expressed genes between 12:12LD and CJL by DESeq2 analyses for all samples (tumors + lungs), taking time of collection (ZT9/21) as confounding factor. (B) DAVID analyses on the differentially expressed genes by DESeq2 in all samples between 12:12LD and CJL, taking time of collection as confounding factor. Only terms with false discovery rate (FDR) < 0.25 are shown. (C) GSEA plots for the cellular response to heat stress and HSF1-mediated heat shock response reactome gene sets applied to samples (lungs + tumors) from CJL versus 12:12LD-housed K mice. NES, normalized enrichment score. (D) Activation of stress response pathways by CJL in lungs and tumors, independently of collection time, revealed by grouped fold change for transcripts established as selective targets of each stress pathways (49) or “BMAL1 pathway” (43). **P < 0.01 and ***P < 0.001 by one-way ANOVA with Dunnett’s multiple comparison test. (E and G) Gene expression normalized to U36b4 measured by quantitative real-time PCR; T, tumors; L, lungs. Data represent means ± SEM. n = 14 and 12 for tumors collected at ZT9 in 12:12LD and CJL, respectively; n = 5 and 6 for tumors collected at ZT21 in 12:12LD and CJL, respectively; and n = 3 to 4 for lung samples. *P < 0.05, **P < 0.01, and ***P < 0.001 by two-way ANOVA post hoc Bonferroni test. (F) GSEA plot for the gene set representing the HSF1-CaSig network applied to samples (lungs + tumors) from CJL versus 12:12LD-housed K mice.
HSF1 broadly influences tumor biology (21). Notably, HSF1 activates a distinct transcriptional program in malignant cells, dubbed the HSF1 cancer signature, or HSF1-CaSig (50). We created a gene matrix based on the HSF1-CaSig defined in (50) and used GSEA to show that samples from CJL-exposed mice also exhibit robustly enriched expression of the HSF1-CaSig network compared to lungs and tumors from control mice (Fig. 4F). BCL2-associated athanogene 3, Bag3, is a molecular chaperone and HSF1 target gene that is part of the HSF1-CaSig network and is involved in apoptosis evasion (51). qPCR revealed that Bag3 is significantly up-regulated in whole lung collected at ZT21 from K mice exposed to CJL compared to normal light conditions (Fig. 4G), mirroring changes measured for other HSF1 target genes, such as Hspa1a (Fig. 4E).
Applying GSEA to a previously published gene expression profiling dataset of KrasG12V-driven lung hyperplasia and normal murine lung cells (52) showed that both the HSF1-mediated heat shock response and HSF1-CaSig gene sets were significantly enriched in hyperplastic lesions compared to normal lung cells (fig. S8C). However, these gene sets were not significantly enriched in frank adenocarcinoma compared to hyperplastic lesions in the same dataset (fig. S8D). This suggests that activation of HSF1 signaling occurs at early stages of KRAS-driven lung cancer development.
To glean cellular compositions of tumors and lung tissue of K mice and determine the source of variation of HSF1 target gene expression measured in our analyses, we used a recently developed machine learning tool, CIBERSORTx, to deconvolute our bulk RNA sequencing data (53, 54). Using a signature matrix (sigmatrix_NSCLC) derived from RNA sequencing libraries of four major subpopulations purified by fluorescence-activated cell sorting (FACS)—epithelial [epithelial cell adhesion molecule positive (EpCAM+)], immune (CD45+), endothelial (CD31+), stromal (CD10+)—generated from surgically resected primary NSCLC tumor biopsies, we determined the fractional presence of these four cell populations in whole lung and tumor samples collected from K mice exposed to either CJL or control lighting conditions. We observed increased epithelial cell content and decreased endothelial cells and leukocytes in tumors compared to whole lung (fig. S9A), consistent with the epithelial origin of LUAD (55). No significant differences in cellular composition were detected between samples collected from different lighting conditions. To complement this analysis and quantitate the presence of several immune cell populations, we performed flow cytometry on single-cell suspensions generated from whole lung tissues collected at two different times of day, ZT9 and ZT21 from K mice housed in control or CJL conditions for the prior 20 weeks. Consistent with the outcome of CIBERSORTx analysis, there was no significant impact of CJL on the presence of any of 11 cell populations detected, including B cells, CD4+ cells, and CD8+ T cells (fig. S10). We detected a significant effect of time of the day on neutrophil numbers under normal light conditions, which was lost upon CJL. Together, these data indicate that CJL does not have a major impact on broad cellular composition of late-stage tumors and tumor-bearing lungs.
CIBERSORTx further enables the imputation of gene expression in broadly defined cell types from bulk RNA sequencing data. Using the same signature matrix (sigmatrix_NSCLC) and a control “groundtruth” dataset from RNA sequencing performed in FACS-sorted samples from human NSCLC biopsies (53), we compared the imputed expression of HSF1 stress pathway regulated genes between all samples collected from mice exposed to CJL versus all samples from 12:12LD-exposed mice. Although several genes could not be reliably imputed in the different cell types, this analysis showed an up-regulation of HSF1 target gene expression in epithelial cells and, to a lesser extent, in endothelial cells, from samples collected from CJL-exposed mice (fig. S9, B and C). In contrast, imputed expression of genes regulated by other stress pathways was not affected by CJL exposure in epithelial cells (fig. S9D).
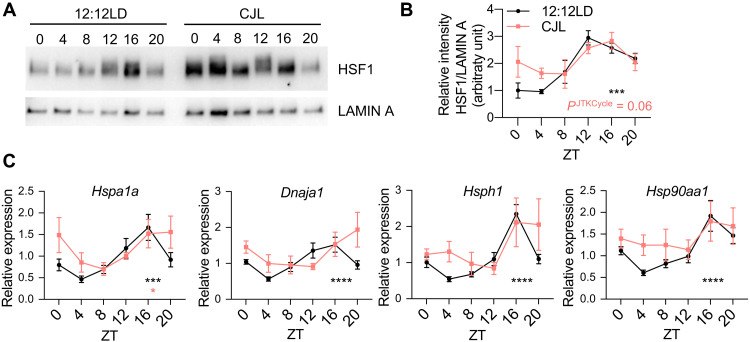
Rhythmic HSF1 nuclear accumulation and transcriptional activity are perturbed by CJL
Previous studies show that nuclear HSF1 levels fluctuate daily in the liver of mice and chipmunks in phase with body temperature rhythms and that HSF1 acts as a circadian transcription factor (56, 57). To determine whether this was also the case in the lung tissue and to assess the impact of CJL exposure on this regulation, we measured HSF1 protein levels in lung nuclear extracts over 24 hours from C57BL/6J mice housed in 12:12LD or CJL conditions (Fig. 5, A and B). Lung nuclear HSF1 protein levels exhibited robust diurnal oscillations, peaking during the dark phase, under normal light conditions. Upon CJL exposure, the amplitude of this rhythm was dampened, but the phase was retained (or only slightly shifted), leading to enhanced accumulation of nuclear HSF1 at the beginning of the light phase. In contrast, total HSF1 protein levels exhibited a moderate rhythm with lower amplitude and different phase than nuclear HSF1 levels (fig. S11, A to C). The expression profiles of HSF1 target genes from a different cohort of mice mirrored diurnal HSF1 nuclear localization (Fig. 5C), with enhanced expression upon CJL from the end of dark phase through the beginning of the light phase. These results were also consistent with increased expression of HSF1 target genes that we observed at ZT21 in K mice exposed to CJL (Fig. 4E). The turnover of nuclear HSF1 and therefore its activity are affected by extracellular signal–regulated kinase (ERK)–mediated phosphorylation of HSF1 leading to recruitment of the E3 ubiquitin ligase F-box and WD repeat domain containing 7 (FBXW7) and subsequent ubiquitination and degradation of HSF1 (58). We measured phosphorylated levels of ERK1/2 in lungs over 24 hours from C57BL/6J mice housed in 12:12LD or CJL conditions, and while there was a tendency for phospho-ERK1/2 (p-ERK1/2) to be decreased at ZT0 in the lungs of mice exposed to CJL, we did not measure any statistically significant effect of CJL on p-ERK levels (fig. S11, D and E). Similarly, we did not detect any impact of CJL exposure on p-ERK levels from tumors and lung tissues from K mice (fig. S11, F and G), indicating that the effect of CJL on HSF1 nuclear accumulation and transcriptional activity is not due to differences in posttranslational regulation of HSF1 by ERK. Together, our findings reveal that CJL perturbs homeostatic regulation of HSF1 transcriptional activity in the lung, which could lead to enhanced tumor initiation in combination with other oncogenic factors.
Fig. 5. Time-regulated HSF1 nuclear accumulation and transcriptional activity are perturbed upon CJL exposure.
C57BL/6J mice were housed in 12:12LD or CJL for 8 to 12 weeks. Lung tissues were collected at the indicated times (hours after lights on) on day 1 of the schedule shown in Fig. 1A. (A) Proteins from lung nuclear extracts detected by immunoblot. Each lane on the Western blot represents a sample prepared from a unique animal. Representative images were taken from n = 3 biological replicates. (B) Quantitation of (A). Rhythmicity was determined by JTK_Cycle analyses. ***PJTKCycle < 0.001. (C) Gene expression normalized to U36b4 measured by quantitative real-time PCR. Data represent means ± SEM for three males and three females per time point and light condition. Rhythmicity was determined by JTK_Cycle analyses. *PJTKcycle < 0.05, ***PJTKcycle < 0.001, and ****PJTKcycle < 0.0001.
HSF1 signaling affects human KRAS-mutant lung cancer
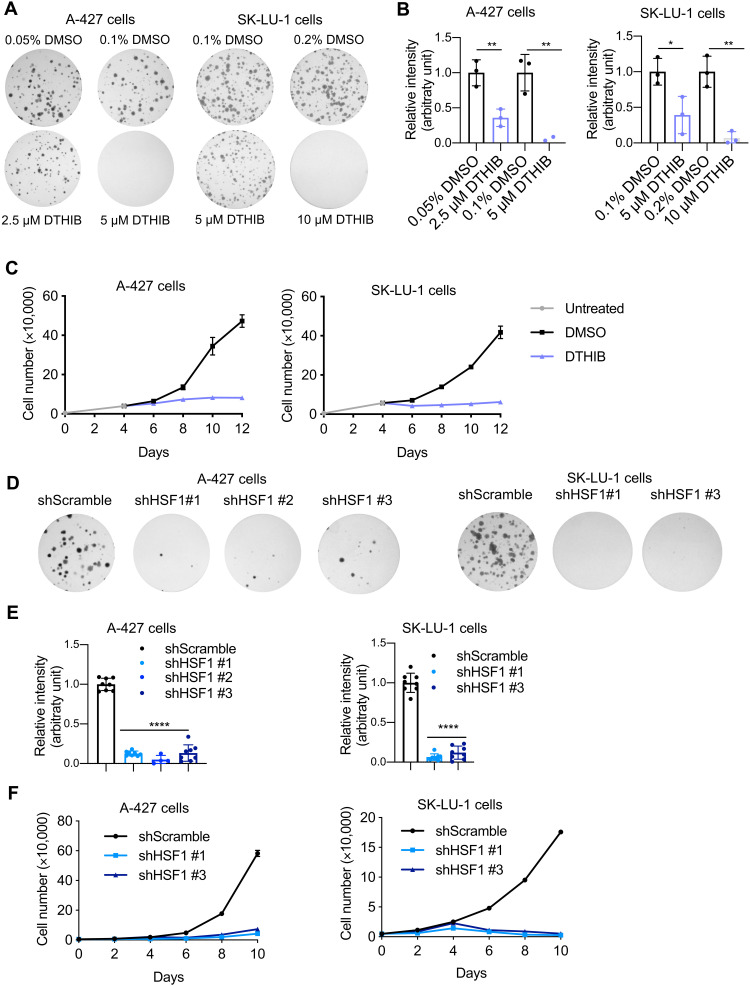
To gain insight into the potential for HSF1 to influence human LUAD, we initially treated human lung cancer cell lines with a novel direct targeted HSF1 inhibitor (DTHIB) that has been shown to stimulate degradation of nuclear HSF1 and suppress the growth of prostate cancer xenografts (59). We confirmed the potency of DTHIB for suppressing HSF1 transcriptional activity in human embryonic kidney (HEK) 293T cells expressing a heat shock element (HSE)–luciferase reporter in which HSF1 is stimulated with the activating ligand A3 (fig. S12A) (60). We found that inhibiting HSF1 with DTHIB slowed the growth of two human LUAD cell lines harboring heterozygous KRASG12D mutations (A-427 and SK-LU-1 cells) in a dose-dependent manner (Fig. 6, A and B, and fig. S12, B and C, for later treatment). We also measured decreased proliferation in these lung cancer cells treated with DTHIB (Fig. 6C). We confirmed significant reduction in the protein levels of HSF1 and downstream chaperone DNAJB1 in A-427 cells treated with 5 μM DTHIB for 48 hours, confirming compound activity in this model (fig. S12, D and E). We did not detect a significant change in KRASG12D protein level upon DTHIB treatment. However, two downstream effectors of RAS signaling (phosphorylation of ERK1/2 and accumulation of c-MYC) were significantly decreased (fig. S12, D and E), suggesting impairment of RAS signaling in these cells upon pharmacologic inhibition of HSF1. Given the potential off-target activity of DTHIB, we next wanted to determine the impact of genetic HSF1 inhibition on human lung cancer cell growth. We confirmed reduction in protein levels of HSF1 and downstream chaperones in A-427 cells subjected to three different short haipin RNAs (shRNAs) targeting HSF1, but we could not detect any consistent changes in phosphorylation of ERK1/2 and c-MYC levels in this context of genetic HSF1 knockdown (fig. S13, A and B). As observed with DTHIB, genetic depletion of HSF1 decreased the cell growth of both A-427 and SK-LU-1 human lung cancer cells, confirming their sensitivity to reduced HSF1 activity (Fig. 6, D to F). Together, these results indicate that HSF1 is an important contributor to cellular proliferation in two human cell models of KRAS-driven LUAD.
Fig. 6. Activation of HSF1 signaling plays a role in human KRAS-mutant LUAD cell growth.
(A) Representative images of crystal violet stained colonies formed by A-427 or SK-LU-1 cells treated with DTHIB or vehicle dimethyl sulfoxide (DMSO) 2 days after seeding, for 14 days. (B) Quantification of (A) from three biological replicates. Each condition was compared to controls that were plated in wells on the same plates. Bars represent means ± SD. *P < 0.05, **P < 0.01 by Student’s t test. (C) Proliferation of A-427 and SK-LU-1 cells treated with DTHIB or vehicle DMSO from day 4 after seeding. Representative of three independent biological replicates. (D) Representative images of crystal violet stained colonies formed by A-427 or SK-LU-1 cells upon genetic (shRNA) inhibition of HSF1. (E) Quantification of (D) from three biological replicates. Each condition was compared to shScramble controls that were plated in wells on the same plates. Bars represent means ± SD. ****P < 0.0001 by one-way ANOVA post hoc Bonferroni test. (F) Proliferation of A-427 and SK-LU-1 cells upon genetic (shRNA) inhibition of HSF1. Representative of three independent biological replicates.
DISCUSSION
Several hypotheses have been proposed to explain the increased cancer risk associated with circadian disruption. Studying the impact of circadian disruption in GEMMs of cancer enables us to identify molecular changes that occur in response to CJL and investigate their contributions to enhanced cancer risk in a controlled environment. Here, we show that circadian disruption promotes lung tumorigenesis in K mice, a GEMM in which tumor formation is initiated by expression of oncogenic KrasG12D in a small number of lung cells that recapitulates many of the clinical features of naturally occurring KRAS-driven lung cancer (38, 61). This provides additional evidence of the negative impact of circadian disruption on lung cancer as previously reported in a related but more severe lung cancer mouse model (3). We find that the CJL protocol used affects the number of tumors formed, but not their size, indicating that circadian disruption likely affects early events in KrasG12D-driven tumor formation rather than the growth of established tumors. We further demonstrated that HSF1 signaling is significantly elevated in mice exposed to altered light/dark cycles, revealing a previously unknown mechanism of action that likely contributes to increased tumor formation in response to circadian disruption.
The CJL protocol used in our studies, consisting of repeated light advances and mimicking the effects of rotating shift work or frequent eastbound transmeridian flights, has been previously shown to cause severe perturbations in rest-activity cycles and body temperature (3, 25, 26, 62). By housing mice in constant darkness for 2 days after 10 days of this CJL protocol exposure (to avoid any masking effect of light on circadian rhythmic patterns), previous work has demonstrated that this protocol caused dysregulation of circadian rhythms of gene expression in the suprachiasmatic nucleus and peripheral organs (25, 26, 63). In our studies, mice were kept in their respective light schedule, and tissues were sampled when all mice experienced the same light exposures for at least 24 hours before sample collection. In that way, we aimed to assess the effects of CJL in a normal light exposure context, which is more representative of what humans experience after shift work or frequent eastbound flights. As expected, we found that peripheral clocks cannot adjust their timing rapidly enough to maintain synchrony with the shifting of the environment. It appeared that some clock genes, including Per2, remained rhythmic after CJL exposure, while others (e.g., Bmal1) did not, consistent with prior work indicating that Per2 is more sensitive to entrainment signals in peripheral tissues than other core clock genes (64).
Our analysis of gene expression in tumors and tumor-bearing lung tissues from animals housed in standard or CJL conditions demonstrated that the clock machinery was highly disrupted by CJL in both tumors and lung tissue. Accumulating evidence reveals that circadian clock components play critical roles in several hallmarks of cancer, including cell proliferation, DNA damage and repair, and cell death (15), suggesting that disruption of cellular circadian rhythms within tumors could contribute to the detrimental impact of irregular light exposure in cancer. In support of this idea, previous work established that K mice, the same KrasG12D-driven NSCLC mouse model that we used, develop a greater tumor burden when the core circadian clock component Bmal1 is deleted specifically within tumors (3). Conversely, previous studies have shown that circadian functions in cancer cells are compromised or deregulated (43), in some cases due to high expression of oncogenic c-MYC (41) or RAS (42). However, circadian disruption does not seem to be a universal feature of cancer, because some cancer cells—such as melanoma, acute myeloid leukemia cells, and patient-derived cancer stem cells (CSCs) of glioblastoma—harbor an intact circadian clock despite their highly tumorigenic and metastatic potential (65–67).
Here, we demonstrate that chronic circadian disruption in vivo enhances the expression of HSF1 target genes. HSF1 promotes expression of HSPs to protect the proteome, allowing cells to survive diverse proteotoxic stresses. Over the last decade, it has become clear that HSF1 activity is exploited by cancer cells to overcome diverse stresses and intrinsic and extrinsic demands (68). High levels of HSF1 and HSF1-regulated HSPs have been measured in different types of cancers and are negatively correlated with prognosis in patients (58, 69), including those with NSCLC (70). Accordingly, in various human cancer cell lines and murine cancer models, deletion of HSF1 markedly reduces growth, survival, and metastatic potential (21, 50, 71–74), whereas its overexpression enhances the malignant phenotypes of xenografted human melanoma cells in vivo (75). Several mechanisms have been proposed to contribute to the role of HSF1 in supporting malignancy, including regulation of HSP expression and regulation of a unique transcriptional program activated by HSF1 in cancer cells dubbed as the HSF1-CaSig (50). Notably, the HSF1-CaSig was up-regulated in tumor-bearing K mouse lung upon CJL exposure (Fig. 5F). Regulation of cancer cell proteostasis by HSPs is an important feature for cancer cell survival and proliferation (76), and recently, HSPs have drawn increased attention as potential targets in cancer, especially given the role of these stress proteins in resistance to conventional therapies (77). Specifically, the growing evidence of correlation between Hsps expression profile and degree of differentiation and staging of lung tumors suggest that these proteins could be considered as therapeutic targets and biomarkers for lung cancer patient management (78, 79). Mutant oncoproteins, such as KRASG12D, may depend on HSF1-dependent regulation of HSPs to enable folding and to maintain full activity. Although we did not detect a significant change in KRASG12D protein level upon pharmacological inhibition of HSF1, its activity appeared to be impaired as measured by significant down-regulation of the phosphorylation levels of its downstream effectors ERK1/2. These findings are consistent with previous observations, in other contexts, that HSF1 supports transformation and tumorigenesis via activation of oncogenic RAS signaling (73, 80).
Our work also adds to a growing body of evidence indicating robust circadian regulation of HSF1 activity, both in healthy and tumor-bearing lung tissue. As in the liver (56), HSF1 nuclear accumulation in the lung is dependent on the time of day, peaking at night under normal light conditions. Daily rhythmic nuclear accumulation of HSF1 is associated with rhythmic expression of Hsps, which peak in the middle of the dark phase, around ZT16. Exposure to CJL disrupted these rhythms mainly by preventing the reduction of their expression that is seen between ZT20 and ZT4 under normal conditions. Deconvolution of our RNA sequencing data suggests an activation of HSF1 signaling in epithelial cells upon CJL. Since LUADs are tumors of epithelial origin (55), it is possible that epithelial cells exhibit oscillations in HSF1 activity and would be more prone to CJL effects, leading to enhanced tumorigenesis. In turn, HSF1 may be particularly sensitive to stress-induced activation in KRAS-mutant cells as reports have shown that HSF1 is activated by mitogen-activated protein kinase kinase–induced phosphorylation downstream of active RAS (80, 81). Other studies have shown that HSF1 can be activated in T lymphocytes at fever temperatures (82, 83), and KRAS-mediated lung cancer models have been documented to generate inflammatory responses (84), including T lymphocyte recruitment into the adenocarcinoma microenvironment (85). Further, several studies have documented important roles for HSF1 in cancer-associated fibroblasts in promoting disease progression (71, 86–88). Although we did not observe any increase in HSF1 target gene expression in CD10+ stroma by deconvolution of bulk RNA sequencing, this analysis lacks sufficient statistical power to make a robust conclusion regarding gene expression in cancer-associated fibroblasts or other stromal cell types. Hence, a more detailed characterization of gene expression across cell types would be required to assess the impact of CJL on HSF1 activity in tumor-initiating cells and the surrounding tumor microenvironment.
Activation of HSF1 occurs in response to a wide variety of environmental and (patho)physiological stress conditions, including heat and cold stress, reactive oxygen species, toxic chemicals, infectious agents, metabolic imbalance, and other proteotoxic stressors (89). Fluctuations in body temperature have been shown to act as a major entrainment factor for rhythmic HSF1 activity (56), and compared to day-shift nurses, night-shift nurses exhibited significant differences in peripheral skin temperature, with notably higher minimum temperature but unchanged maximum temperature (90, 91). Therefore, activation of HSF1 due to abnormal changes in body temperature could be a key component in the connection between shift work and cancer risk. The relation between the circadian clock system and HSF1 signaling is bidirectional as previous work have reported a cooperative interaction among BMAL1 and HSF1 to synchronize the cellular clock in response to heat shock, oxidative stress, and ultraviolet irradiation (92–94). Thus, disruption of cell autonomous molecular clocks by CJL could enhance HSF1 independently of body temperature. Further investigation is needed to determine why CJL enhances HSF1 activity.
Numerous cell autonomous and systemic mechanisms are susceptible to alteration upon circadian disruption and can influence tumorigenesis. In this work, we revealed that circadian disruption affects early events in tumor formation in a KRAS-driven mouse model of LUAD. Further, we demonstrated that HSF1 signaling in the lung and lung tumors is dysregulated by exposure to altered environmental lighting schedules. HSF1 has been shown to support tumorigenesis in myriad ways (95), suggesting that the enhanced HSF1 activity that we observed in response to circadian disruption could play an important role in increased tumor formation. Additional investigation is needed to determine whether the chronic elevation of HSF1 signaling that we measured in the lungs in response to circadian disruption occurs early in the disease process, whether it is present in other anatomical locations, and whether HSF1 is required for increased tumorigenesis in response to circadian disruption. We demonstrated that genetic or pharmacological inhibition of HSF1 using compound DTHIB, which has previously been shown to potently attenuate tumor progression in therapy-resistant prostate cancer models (59), reduced the growth of two different human KRAS-driven lung cancer cell lines. To further define the determinants of susceptibility to growth inhibition by DTHIB and genetic manipulation of HSF1, it will be necessary to examine additional cell lines and to investigate the impact of KRAS and other factors on growth inhibition. Of particular relevance for connecting HSF1-related therapeutic opportunities to circadian disruption, an HSP90 inhibitor reduced the growth of mouse melanoma in a time-of-day specific manner, which depended on an intact core clock system in the tumors (96). Our findings described here demonstrate that HSF1 is a potential therapeutic target for mitigating cancer risk among populations exposed to chronic circadian disruption, such as shift workers.
MATERIALS AND METHODS
Mouse models
C57BL/6J mice were purchased from the Scripps Research breeding colony at 6 weeks of age. They were group-housed except when given voluntary access to running wheels, in which case they were singly housed in running wheel cages. GEMMs, KrasLSL-G12D/+ (K) and KrasLSL-G12D/+;p53flox/flox (KP), all in pure C57BL/6J background, were obtained from the Jackson Laboratory and have been previously described (38, 61). When they were between 8 and 10 weeks old, mice were infected intratracheally with lentivirus containing Cre recombinase, lenti-Cre (PGK-Cre, gift from T. Jacks), at a viral titer of 5 × 105 plaque-forming units per mouse according to the previously established protocol (38). All experiments used both female and male mice. They were given ad libitum access to normal mouse chow and water. Sacrifices during the dark phase were carried out under red light. All animal care and treatments were in accordance with Scripps Research guidelines for the care and use of animals and approved by the Scripps Research Institutional Animal Care and Use Committee under protocol #10-0019.
CJL conditions
Mice were randomly placed into standard light conditions (12:12LD) or CJL consisting of an 8-hour light-phase advance repeated every 2 or 3 days (3, 23, 63). For GEMM studies, mice were housed in these light conditions 5 weeks after infection with lenti-Cre for 10 or 20 weeks for KP and K mice, respectively, or until showing signs of distress for the survival studies.
Running wheel activity analysis
C57BL/6J mice were singly housed and given access to running wheels with ad libitum access to food and water for several weeks, under specific light conditions as indicated in the figure legends. Voluntary running wheel activity was analyzed with ClockLab (Actimetrics) using digital recordings of wheel rotations.
Histology and tumor burden analyses
Mice were euthanized by carbon dioxide asphyxiation. Lungs were inflated through the trachea with 4% paraformaldehyde, fixed overnight, transferred to 70% ethanol, and sent to the Rodent Histopathology Core facility at Harvard Medical School for subsequent paraffin embedding and sectioning at a thickness of 5 μm. Sections were stained with hemotoxylin and eosin (H&E) for pathological examination. Histopathological grading of tumors and quantification of tumor numbers were performed with the assistance of R. Bronson, histopathologist at Harvard Medical School. The tumor size of each individual tumor was measured from H&E-stained sections using morphometric analysis in Panoramic viewer software (PerkinElmer). Tumor burden, calculated as a percentage of tumor area per total lung area per mouse, was quantified from H&E-stained sections using a Nuance automated spectral imaging system (Inform v2.1 software, Cambridge Research and Instrumentation). Briefly, the trainable tissue segmentation method was trained to identify tumor, normal lung, vessel, and space. This program was then applied to all H&E images, and each of the resulting mapped images was then screened to verify that accurate tissue segmentation had occurred.
BrdU injection
BrdU (Sigma-Aldrich, B5002) was intraperitoneally injected at 100 mg/kg. Mice were euuthanized 4 hours after injection, and lungs were inflated for paraffin embedding and subsequent immunohistochemistry.
Immunohistochemistry
Slides were deparaffinized and rehydrated. Antigen retrieval was performed at high heat (95°C) in citrate buffer (pH 6.0) for 10 min for all antibodies except BrdU, for which antigen retrieval was performed for 30 min. Endogenous peroxidase activity was quenched with Bloxall (Vector Laboratories) for 10 min. Slides were blocked for 1 hour using 10% normal goat serum (Invitrogen) and incubated overnight with the following primary antibodies: BrdU (1:200; Abcam, ab6326), Ki67 [1:200; Cell Signaling Technology (CST), 12202], and c-MYC (1:100; Abcam, ab32072). After 1-hour incubation with secondary antibody, VectaElite (VectorLabs) was applied on the sections for 30 min. Staining was visualized using the DAB Peroxidase Substrate Kit (Vector Labs, SK-4100). Slides were counterstained with hematoxylin, dehydrated, and mounted with refrax mounting medium. Immunostained slides were scanned using a PerkinElmer Slide Scanner (Panoramic MIDI Digital SlideScanner). Inform v2.1 image analysis software (Cambridge Research and Instrumentation) was used as a nonbiased method to quantitate staining as previously described (97). Quantitation of BrdU-, Ki67-, and c-MYC–positive nuclei was performed using tumors of the same histological grade.
Flow cytometry
Tumor-bearing lungs were harvested and manually dissociated to single-cell suspensions in FACS buffer [phosphate-buffered saline (PBS) supplemented with 2% fetal bovine serum and 2 mM EDTA]. Briefly, lungs were diced with scissors, incubated for 30 min in CDTI (collagenase, deoxyribonuclease, and trypsin inhibitor) buffer, and processed through a 70-μm cell strainer. After red blood cell lysis, leukocytes were enriched using Percoll gradient centrifugation. Cells were counted, and 16 × 106 cells per sample were stained with the following antibodies purchased from BioLegend. Samples were analyzed using the Cytek Aurora spectral flow cytometer. The following antibodies and fluorophores were used: LiveDead eFluor 780, CD45.2-phycoerythrin (PE)/Dazzle 594, CD4-BV605, CD8-AF700, B220-BV785, Thy1.2-fluorescein isothiocyanate, CD11c-BV650, F4/80-BV421, CD11b-PerCP/Cy5.5, γ/δ T cell receptor–PE, Ly6G-PE/Cy7, Ly6C-allophycocyanin, and NK1.1-BV711.
RNA sequencing
RNA from lung tumors and remaining lung tissues was isolated using QIAzol reagent using standard protocols (QIAGEN, catalog no. 799306). RNA purity was assessed by Agilent 2100 Bioanalyzer. Total RNA samples were sent to BGI Group, Beijing, China, for library preparation and sequencing. Reads (single-end 50 base pairs at a sequencing depth of 20 million reads per sample) were generated by BGISEQ-500.
RNA sequencing analysis
Kallisto (https://pachterlab.github.io/kallisto/) was used to align to the reference transcriptome (ftp://ftp.ensembl.org/pub/current_fasta/mus_musculus/cdna/) and estimate transcript abundance. Differential gene expression analysis (DESeq2) was carried out using R (www.r-project.org/). Differentially expressed genes were defined as having an adjusted P value < 0.05 and fold change > ±0.5. Gene Ontology analysis was conducted on selected genes using the DAVID (https://david.ncifcrf.gov/) program. GSEA (www.gsea-msigdb.org/gsea/index.jsp) was generated with transcripts per million reads (TPM) values from the above experiment using the Java GSEA package.
Lung nuclear extracts
Freshly collected lungs were mechanically homogenized in sucrose solution, and nuclei were isolated by ultracentrifugation through a denser layer of sucrose. Briefly, the whole lung was placed into a large (15 ml) dounce homogenizer on ice containing about 4 ml of ice-cold PBS and 4 ml of ice-cold homogenization solution [2.2 M sucrose with protease inhibitors, dithiothreitol (DTT), and phenylmethylsulfonyl fluoride (PMSF)]. Tissue was disrupted by pressing piston up/down 6× with loose piston and then 4× with tight piston. Homogenized tissue was added to an additional volume of ice-cold homogenization solution for a total volume of about 33 ml, which was then slowly poured on top of 10 ml of cushion solution (2.05 M sucrose with protease inhibitors, DTT, and PMSF) in the ultracentrifugation tube (Beckman polyallomer, #326823). After a 45-min spin at 24,600 rpm in prechilled SW32Ti rotor at 4°C, supernatants were carefully aspirated (the white pellets contain the nuclei). Nuclei were resuspended in 500 μl of nuclear resuspension buffer [5 mM Hepes (pH 7.6), 50 mM KCl and EDTA with DTT, protease inhibitors, and PMSF] and transferred into a small (2 ml) dounce homogenizer on ice. Nuclei pellets were further resuspended by pressing piston up/down 3× with loose piston and then 2× with tight piston. Nuclei were then transferred to fresh 1.5-ml tubes, and 500 μl of 2× NUN (NP40, urea, and NaCl) buffer (+ protease inhibitors and PMSF) was added while gently vortexing. After 20-min incubation on ice, lysates were centrifuged for 20 min in ultracentrifuge at 38,000 rpm in prechilled 70.1Ti rotor with delrin adapters at 4°C. Supernatants were then transferred to clean tubes before protein quantification by bicinchoninic acid (BCA) assay.
Cell culture
A-427 and SK-LU-1 cells were purchased from the American Type Culture Collection, cultured in Dulbecco’s modified Eagle’s medium + 10% fetal bovine serum (Thermo Fisher Scientific) and 1% penicillin-streptomycin (Gibco), and maintained in an atmosphere containing 5% CO2 at 37°C.
Generation of cell lines expressing shRNA-targeting HSF1
Lentiviral shRNAs (Sigma-Aldrich, TRCN0000007480-1-2) were produced by transient transfection in 293T cells. Beginning 48 hours after viral transduction, infected cells were cultured in selection media for 3 to 5 days.
Proliferation and colony formation assays
For proliferation assay, cells were plated into six-well plates at 5000 cells per well and counted every 2 days for 10 days. For colony formation assay, cells were plated into six-well plates at 500 cells per well, and medium was changed every 2 or 3 days with medium containing DTHIB (MedChemExpress, HY-138280) or corresponding concentration of dimethyl sulfoxide (DMSO). After indicated days of treatment, cells were washed in PBS, fixed for 10 min with 100% methanol, and stained with 0.05% crystal violet for 20 min. Plates were rinsed in deionized H2O, imaged, and quantified using the ChemiDoc XRS+ System (Bio-Rad).
HSE-Luc activity
The pGL4.41[luc2P/HSE/Hygro] plasmid (HSE-Luc; Promega) was transfected into HEK293T cells, and a stable clone expressing HSE-Luc was selected with hygromycin (200 mg/ml). For activity measurements, cells were plated in a flat, white, clear-bottom 96-well plate at a concentration of 50,000 cells per well. After 6 hours, cells were pretreated with DTHIB (5 μM) and/or vehicle overnight. Compound A3 (10 μM; a gift from R. Morimoto, Northwestern University), MG132 (10 μM; Sigma-Aldrich), or DMSO was added for an additional 6 hours. Plates were then equilibrated to room temperature and lysed by the addition of Bright-Glo reagent (100 μl; Promega) to each well. After a 10-min incubation to stabilize the signal, luminescence was then measured using an Infinite F200 PRO plate reader (Tecan) and corrected for background signal.
RNA extraction and quantitative RT-PCR
RNA was extracted from frozen tissues with Qiazol reagent using standard protocols (QIAGEN, catalog no. 799306). cDNA was prepared using QScript cDNA Supermix (VWR, catalog no. 101414-106) and analyzed for gene expression using quantitative real-time PCR with iQ SYBR Green Supermix (Bio-Rad, catalog no.1708885).
Western blots
Tissues or cells were lysed in radioimmunoprecipitation assay buffer supplemented with protease and phosphatase inhibitors. Protein lysates were separated by SDS–polyacrylamide gel electrophoresis and transferred to polyvinylidine difluoride membranes. Proteins were detected by standard Western blotting procedures. Antibodies were diluted 1:1000 for BMAL1 (Abcam, ab93806), CRY1 and CRY2 (98), c-MYC (Abcam, ab32072), HSF1 (CST-12972), KRASG12D (CST-14429), p-ERK1/2 (CST-4370), and ERK1/2 (CST-4695); 1:2000 for REV-ERBα (33), DNAJB1 (Enzo, ADI-SPA-400), and HSP90AA1 (GTX109753); 1:10,000 for LAMIN A (Sigma-Aldrich, L1293); 1:50,000 for ACTIN (Sigma-Aldrich, A1978). Imaging and band quantification were carried out using ChemiDoc XRS+ System (Bio-Rad).
Web-based analysis tools
Pathway analysis was performed with Enrichr (http://amp.pharm.mssm.edu/Enrichr). Quantifying circadian clock function using clock gene coexpression (CCD method) was carried using the web application available at https://hugheylab.shinyapps.io/deltaccd. Heatmap was generated by clustering using the Cluster 3.0 program (log2 transform data, center genes on mean, Hierarchical clustering with average linkage) (99) and then visualized with Java TreeView version 1.1.6r4 (100). The decovolution of RNA sequencing data using CIBERSORTx was carried out using the web application available at https://cibersortx.stanford.edu. For imputed gene expression, high-resolution mode analysis was performed using signature matrix “sigmatrix_NSCLC” and the control groundtruth dataset “groundtruth_NSCLCsubsets.”
Statistical analysis
Statistical analyses were performed using GraphPad Prism 8 software. Unless otherwise indicated, analysis of variance (ANOVA) was used to determine significance with a threshold of 0.05 acceptable false positive (P < 0.05). Rhythmicity was determined by JTK_Cycle analyses (101).
Acknowledgments
We thank L. Eichner and R. Svensson for helpful discussions and gifting critical reagents and expertise. We thank R. Bronson for histopathological analysis of tumors; N. Madrazo for performing HSF1 luciferase assay; V. Vartabedian for the help with flow cytometry experiments; T. Thomas, J. Valecko, and Y. Slivers for administrative assistance; and X. Jing for technical assistance.
Funding: This work was funded by NIH grant CA211187 (to K.A.L.), Brown Foundation for Cancer Research (to K.A.L.), NIH grant DK107604 (to R.L.W.), NIH grant R00CA204593 (to B.J.A.), and NSF/DBI-1759544 (to E.I.).
Author contributions: Conceptualization: M.P. and K.A.L. Methodology: M.P., K.A.L., R.J.S., R.L.W., and B.J.A. Investigation: M.P., E.I., R.M., A.B.C., and L.H.I. Visualization: M.P., K.A.L., L.H.I., and B.J.A. Supervision: K.A.L., R.L.W., R.J.S., and M.J.B. Writing—original draft: M.P. and K.A.L Writing—review and editing: M.P., K.A.L., R.J.S., R.L.W., M.J.B., and B.J.A.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. RNA sequencing data are deposited in Gene Expression Omnibus (GEO) with accession number GSE194097.
Supplementary Materials
This PDF file includes:
Figs. S1 to S13
REFERENCES AND NOTES
- 1.N. O. H. S. (NHIS-OHS, 2015); wwwn.cdc.gov/Niosh-whc/chart/ohs-workorg?OU=WORKSCHD_RCD&T=I&V=R2.
- 2.Pariollaud M., Lamia K. A., Cancer in the fourth dimension: What is the impact of circadian disruption? Cancer Discov. 10, 1455–1464 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Papagiannakopoulos T., Bauer M. R., Davidson S. M., Heimann M., Subbaraj L., Bhutkar A., Bartlebaugh J., Vander Heiden M. G., Jacks T., Circadian rhythm disruption promotes lung tumorigenesis. Cell Metab. 24, 324–331 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gu F., Xu S., Devesa S. S., Zhang F., Klerman E. B., Graubard B. I., Caporaso N. E., Longitude position in a time zone and cancer risk in the United States. Cancer Epidemiol. Biomarkers Prev. 26, 1306–1311 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ye Y., Xiang Y., Ozguc F. M., Kim Y., Liu C.-J., Park P. K., Hu Q., Diao L., Lou Y., Lin C., Guo A.-Y., Zhou B., Wang L., Chen Z., Takahashi J. S., Mills G. B., Yoo S.-H., Han L., The genomic landscape and pharmacogenomic interactions of clock genes in cancer chronotherapy. Cell Syst 6, 314–328.e2 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schernhammer E. S., Feskanich D., Liang G., Han J., Rotating night-shift work and lung cancer risk among female nurses in the United States. Am. J. Epidemiol. 178, 1434–1441 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sephton S. E., Lush E., Dedert E. A., Floyd A. R., Rebholz W. N., Dhabhar F. S., Spiegel D., Salmon P., Diurnal cortisol rhythm as a predictor of lung cancer survival. Brain Behav. Immun. 30, S163–S170 (2013). [DOI] [PubMed] [Google Scholar]
- 8.Siegel R. L., Miller K. D., Jemal A., Cancer statistics, 2020. CA Cancer J. Clin. 70, 7–30 (2020). [DOI] [PubMed] [Google Scholar]
- 9.Barta J. A., Powell C. A., Wisnivesky J. P., Global epidemiology of lung cancer. Ann. Glob. Health 85, 8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.The Cancer Genome Atlas Research Network , Comprehensive molecular profiling of lung adenocarcinoma. Nature 511, 543–550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ince L. M., Pariollaud M., Gibbs J. E., Lung physiology and defense. Curr. Opin. Physio. 5, 9–15 (2018). [Google Scholar]
- 12.Gibbs J. E., Beesley S., Plumb J., Singh D., Farrow S., Ray D. W., Loudon A. S. I., Circadian timing in the lung; a specific role for bronchiolar epithelial cells. Endocrinology 150, 268–276 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferone G., Lee M. C., Sage J., Berns A., Cells of origin of lung cancers: Lessons from mouse studies. Genes Dev. 34, 1017–1032 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spella M., Lilis I., Pepe M. A., Chen Y., Armaka M., Lamort A.-S., Zazara D. E., Roumelioti F., Vreka M., Kanellakis N. I., Wagner D. E., Giannou A. D., Armenis V., Arendt K. A., Klotz L. V., Toumpanakis D., Karavana V., Zakynthinos S. G., Giopanou I., Marazioti A., Aidinis V., Sotillo R., Stathopoulos G. T., Club cells form lung adenocarcinomas and maintain the alveoli of adult mice. eLife 8, e45571 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shafi A. A., Knudsen K. E., Cancer and the circadian clock. Cancer Res. 79, 3806–3814 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Masri S., Sassone-Corsi P., The emerging link between cancer, metabolism, and circadian rhythms. Nat. Med. 24, 1795–1803 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sulli G., Lam M. T. Y., Panda S., Interplay between circadian clock and cancer: New frontiers for cancer treatment. Trends Cancer 5, 475–494 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jackson E. L., Willis N., Mercer K., Bronson R. T., Crowley D., Montoya R., Jacks T., Tuveson D. A., Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 15, 3243–3248 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Whitesell L., Santagata S., Mendillo M. L., Lin N. U., Proia D. A., Lindquist S., HSP90 empowers evolution of resistance to hormonal therapy in human breast cancer models. Proc. Natl. Acad. Sci. U.S.A. 111, 18297–18302 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Santagata S., Hu R., Lin N. U., Mendillo M. L., Collins L. C., Hankinson S. E., Schnitt S. J., Whitesell L., Tamimi R. M., Lindquist S., Ince T. A., High levels of nuclear heat-shock factor 1 (HSF1) are associated with poor prognosis in breast cancer. Proc. Natl. Acad. Sci. U.S.A. 108, 18378–18383 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dai C., Whitesell L., Rogers A. B., Lindquist S., Heat shock factor 1 is a powerful multifaceted modifier of carcinogenesis. Cell 130, 1005–1018 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qian W., Chen K., Qin T., Xiao Y., Li J., Yue Y., Zhou C., Ma J., Duan W., Lei J., Han L., Li L., Shen X., Wu Z., Ma Q., Wang Z., The EGFR-HSF1 axis accelerates the tumorigenesis of pancreatic cancer. J. Exp. Clin. Cancer Res. 40, 25–25 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee S., Donehower L. A., Herron A. J., Moore D. D., Fu L., Disrupting circadian homeostasis of sympathetic signaling promotes tumor development in mice. PLOS ONE 5, e10995 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thaiss C. A., Zeevi D., Levy M., Zilberman-Schapira G., Suez J., Tengeler A. C., Abramson L., Katz M. N., Korem T., Zmora N., Kuperman Y., Biton I., Gilad S., Harmelin A., Shapiro H., Halpern Z., Segal E., Elinav E., Transkingdom control of microbiota diurnal oscillations promotes metabolic homeostasis. Cell 159, 514–529 (2014). [DOI] [PubMed] [Google Scholar]
- 25.Filipski E., Delaunay F., King V. M., Wu M.-W., Claustrat B., Gréchez-Cassiau A., Guettier C., Hastings M. H., Francis L., Effects of chronic jet lag on tumor progression in mice. Cancer Res. 64, 7879–7885 (2004). [DOI] [PubMed] [Google Scholar]
- 26.Filipski E., Subramanian P., Carrière J., Guettier C., Barbason H., Lévi F., Circadian disruption accelerates liver carcinogenesis in mice. Mutat. Res. 680, 95–105 (2009). [DOI] [PubMed] [Google Scholar]
- 27.Kettner N. M., Voicu H., Finegold M. J., Coarfa C., Sreekumar A., Putluri N., Katchy C. A., Lee C., Moore D. D., Fu L., Circadian homeostasis of liver metabolism suppresses hepatocarcinogenesis. Cancer Cell 30, 909–924 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anderson S. T., FitzGerald G. A., Sexual dimorphism in body clocks. Science 369, 1164–1165 (2020). [DOI] [PubMed] [Google Scholar]
- 29.Matsuo T., Yamaguchi S., Mitsui S., Emi A., Shimoda F., Okamura H., Control mechanism of the circadian clock for timing of cell division in vivo. Science 302, 255–259 (2003). [DOI] [PubMed] [Google Scholar]
- 30.Mei J., Liu Y., Dai N., Favara M., Greene T., Jeyaseelan S., Poncz M., Lee J. S., Worthen G. S., CXCL5 regulates chemokine scavenging and pulmonary host defense to bacterial infection. Immunity 33, 106–117 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nouailles G., Dorhoi A., Koch M., Zerrahn J., Weiner J. III, Faé K. C., Arrey F., Kuhlmann S., Bandermann S., Loewe D., Mollenkopf H.-J., Vogelzang A., Meyer-Schwesinger C., Mittrücker H.-W., McEwen G., Kaufmann S. H. E., CXCL5-secreting pulmonary epithelial cells drive destructive neutrophilic inflammation in tuberculosis. J. Clin. Invest. 124, 1268–1282 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gibbs J., Ince L., Matthews L., Mei J., Bell T., Yang N., Saer B., Begley N., Poolman T., Pariollaud M., Farrow S., Demayo F., Hussell T., Worthen G. S., Ray D., Loudon A., An epithelial circadian clock controls pulmonary inflammation and glucocorticoid action. Nat. Med. 20, 919–926 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pariollaud M., Gibbs J. E., Hopwood T. W., Brown S., Begley N., Vonslow R., Poolman T., Guo B., Saer B., Jones D. H., Tellam J. P., Bresciani S., Tomkinson N. C. O., Wojno-Picon J., Cooper A. W. J., Daniels D. A., Trump R. P., Grant D., Zuercher W., Willson T. M., MacDonald A. S., Bolognese B., Podolin P. L., Sanchez Y., Loudon A. S. I., Ray D. W., Circadian clock component REV-ERBα controls homeostatic regulation of pulmonary inflammation. J. Clin. Intest. 128, 2281–2296 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gréchez-Cassiau A., Rayet B., Guillaumond F., Teboul M., Delaunay F., The circadian clock component BMAL1 is a critical regulator of p21WAF1/CIP1 expression and hepatocyte proliferation. J. Biol. Chem. 283, 4535–4542 (2008). [DOI] [PubMed] [Google Scholar]
- 35.Gotoh T., Vila-Caballer M., Santos C. S., Liu J., Yang J., Finkielstein C. V., The circadian factor Period 2 modulates p53 stability and transcriptional activity in unstressed cells. Mol. Biol. Cell 25, 3081–3093 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huber A.-L., Papp S. J., Chan A. B., Henriksson E., Jordan S. D., Kriebs A., Nguyen M., Wallace M., Li Z., Metallo C. M., Lamia K. A., CRY2 and FBXL3 cooperatively degrade c-MYC. Mol. Cell 64, 774–789 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van Dycke K. C. G., Rodenburg W., van Oostrom C. T. M., van Kerkhof L. W. M., Pennings J. L. A., Roenneberg T., van Steeg H., van der Hors G. T. J., Chronically alternating light cycles increase breast cancer risk in mice. Curr. Biol. 25, 1932–1937 (2015). [DOI] [PubMed] [Google Scholar]
- 38.DuPage M., Dooley A. L., Jacks T., Conditional mouse lung cancer models using adenoviral or lentiviral delivery of Cre recombinase. Nat. Protocols 4, 1064–1072 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Soucek L., Whitfield J. R., Sodir N. M., Massó-Vallés D., Serrano E., Karnezis A. N., Swigart L. B., Evan G. I., Inhibition of Myc family proteins eradicates KRas-driven lung cancer in mice. Genes Dev. 27, 504–513 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550–550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Altman B. J., Hsieh A. L., Sengupta A., Krishnanaiah S. Y., Stine Z. E., Walton Z. E., Gouw A. M., Venkataraman A., Li B., Goraksha-Hicks P., Diskin S. J., Bellovin D. I., Simon M. C., Rathmell J. C., Lazar M. A., Maris J. M., Felsher D. W., Hogenesch J. B., Weljie A. M., Dang C. V., MYC disrupts the circadian clock and metabolism in cancer cells. Cell Metab. 22, 1009–1019 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Relógio A., Thomas P., Medina-Pérez P., Reischl S., Bervoets S., Gloc E., Riemer P., Mang-Fatehi S., Maier B., Schäfer R., Leser U., Herzel H., Kramer A., Sers C., Ras-mediated deregulation of the circadian clock in cancer. PLOS Genet. 10, e1004338 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shilts J., Chen G., Hughey J. J., Evidence for widespread dysregulation of circadian clock progression in human cancer. PeerJ 6, e4327 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang R., Lahens N. F., Ballance H. I., Hughes M. E., Hogenesch J. B., A circadian gene expression atlas in mammals: Implications for biology and medicine. Proc. Natl. Acad. Sci. 111, 16219–16224 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shamovsky I., Nudler E., New insights into the mechanism of heat shock response activation. Cell Mol. Life Sci. 65, 855–861 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Akerfelt M., Morimoto R. I., Sistonen L., Heat shock factors: Integrators of cell stress, development and lifespan. Nat. Rev. Mol. Cell Biol. 11, 545–555 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lindquist S., The heat-shock response. Annu. Rev. Biochem. 55, 1151–1191 (1986). [DOI] [PubMed] [Google Scholar]
- 48.Kovács D., Sigmond T., Hotzi B., Bohár B., Fazekas D., Deák V., Vellai T., Barna J., HSF1Base: A comprehensive database of HSF1 (heat shock factor 1) target genes. Int. J. Mol. Sci. 20, 5815 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grandjean J. M. D., Plate L., Morimoto R. I., Bollong M. J., Powers E. T., Wiseman R. L., Deconvoluting stress-responsive proteostasis signaling pathways for pharmacologic activation using targeted RNA sequencing. ACS Chem. Biol. 14, 784–795 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mendillo M. L., Santagata S., Koeva M., Bell G. W., Hu R., Tamimi R. M., Fraenkel E., Ince T. A., Whitesell L., Lindquist S., HSF1 drives a transcriptional program distinct from heat shock to support highly malignant human cancers. Cell 150, 549–562 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Behl C., Breaking BAG: The co-chaperone BAG3 in health and disease. Trends Pharmacol. Sci. 37, 672–688 (2016). [DOI] [PubMed] [Google Scholar]
- 52.Ambrogio C., Gómez-López G., Falcone M., Vidal A., Nadal E., Crosetto N., Blasco R. B., Fernández-Marcos P. J., Sánchez-Céspedes M., Ren X., Wang Z., Ding K., Hidalgo M., Serrano M., Villanueva A., Santamaría D., Barbacid M., Combined inhibition of DDR1 and Notch signaling is a therapeutic strategy for KRAS-driven lung adenocarcinoma. Nat. Med. 22, 270–277 (2016). [DOI] [PubMed] [Google Scholar]
- 53.Newman A. M., Steen C. B., Liu C. L., Gentles A. J., Chaudhuri A. A., Scherer F., Khodadoust M. S., Esfahani M. S., Luca B. A., Steiner D., Diehn M., Alizadeh A. A., Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat. Biotechnol. 37, 773–782 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Steen C. B., Liu C. L., Alizadeh A. A., Newman A. M., Profiling Cell type abundance and expression in bulk tissues with CIBERSORTx. Methods Mol. Biol. 2117, 135–157 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sainz de Aja J., Dost A. F. M., Kim C. F., Alveolar progenitor cells and the origin of lung cancer. J. Intern. Med. 289, 629–635 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reinke H., Saini C., Fleury-Olela F., Dibner C., Benjamin I. J., Schibler U., Differential display of DNA-binding proteins reveals heat-shock factor 1 as a circadian transcription factor. Genes Dev. 22, 331–345 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tsukamoto D., Hasegawa T., Hirose S.-i., Sakurai Y., Ito M., Takamatsu N., Circadian transcription factor HSF1 regulates differential HSP70 gene transcription during the arousal-torpor cycle in mammalian hibernation. Sci. Rep. 9, 832 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kourtis N., Moubarak R. S., Aranda-Orgilles B., Lui K., Aydin I. T., Trimarchi T., Darvishian F., Salvaggio C., Zhong J., Bhatt K., Chen E. I., Celebi J. T., Lazaris C., Tsirigos A., Osman I., Hernando E., Aifantis I., FBXW7 modulates cellular stress response and metastatic potential through HSF1 post-translational modification. Nat. Cell Biol. 17, 322–332 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dong B., Jaeger A. M., Hughes P. F., Loiselle D. R., Hauck J. S., Fu Y., Haystead T. A., Huang J., Thiele D. J., Targeting therapy-resistant prostate cancer via a direct inhibitor of the human heat shock transcription factor 1. Sci. Transl. Med. 12, eabb5647 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Calamini B., Silva M. C., Madoux F., Hutt D. M., Khanna S., Chalfant M. A., Saldanha S. A., Hodder P., Tait B. D., Garza D., Balch W. E., Morimoto R. I., Small-molecule proteostasis regulators for protein conformational diseases. Nat. Chem. Biol. 8, 185–196 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jackson E. L., Olive K. P., Tuveson D. A., Bronson R., Crowley D., Brown M., Jacks T., The differential effects of mutant p53 alleles on advanced murine lung cancer. Cancer Res. 65, 10280–10288 (2005). [DOI] [PubMed] [Google Scholar]
- 62.Hadadi E., Taylor W., Li X.-M., Aslan Y., Villote M., Rivière J., Duvallet G., Auriau C., Dulong S., Raymond-Letron I., Provot S., Bennaceur-Griscelli A., Acloque H., Chronic circadian disruption modulates breast cancer stemness and immune microenvironment to drive metastasis in mice. Nat. Commun. 11, 3193 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Filipski E., Innominato P. F., Wu M., Li X.-M., Iacobelli S., Xian L.-J., Lévi F., Effects of light and food schedules on liver and tumor molecular clocks in mice. J. Natl. Cancer Inst. 97, 507–517 (2005). [DOI] [PubMed] [Google Scholar]
- 64.Kornmann B., Schaad O., Bujard H., Takahashi J. S., Schibler U., System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLOS Biol. 5, e34 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee Y., Lahens N. F., Zhang S., Bedont J., Field J. M., Sehgal A., G1/S cell cycle regulators mediate effects of circadian dysregulation on tumor growth and provide targets for timed anticancer treatment. PLOS Biol. 17, e3000228 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Puram R. V., Kowalczyk M. S., de Boer C. G., Schneider R. K., Miller P. G., Conkey M. M., Tothova Z., Tejero H., Heckl D., Järås M., Chen M. C., Li H., Tamayo A., Cowley G. S., Rozenblatt-Rosen O., Al-Shahrour F., Regev A., Ebert B. L., Core circadian clock genes regulate leukemia stem cells in AML. Cell 165, 303–316 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dong Z., Zhang G., Qu M., Gimple R. C., Wu Q., Qiu Z., Prager B. C., Wang X., Kim L. J. Y., Morton A. R., Dixit D., Zhou W., Huang H., Li B., Zhu Z., Bao S., Mack S. C., Chavez L., Kay S. A., Rich J. N., Targeting glioblastoma stem cells through disruption of the circadian clock. Cancer Discov. 9, 1556–1573 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Carpenter R. L., Gökmen-Polar Y., HSF1 as a cancer biomarker and therapeutic target. Curr. Cancer Drug Targets 19, 515–524 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhou Z., Li Y., Jia Q., Wang Z., Wang X., Hu J., Xiao J., Heat shock transcription factor 1 promotes the proliferation, migration and invasion of osteosarcoma cells. Cell Prolif. 50, e12346 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cui J., Tian H., Chen G., Upregulation of nuclear heat shock factor 1 contributes to tumor angiogenesis and poor survival in patients with non–small cell lung cancer. Ann. Thorac. Surg. 100, 465–472 (2015). [DOI] [PubMed] [Google Scholar]
- 71.Scherz-Shouval R., Santagata S., Mendillo M. L., Sholl L. M., Ben-Aharon I., Beck A. H., Dias-Santagata D., Koeva M., Stemmer S. M., Whitesell L., Lindquist S., The reprogramming of tumor stroma by HSF1 is a potent enabler of malignancy. Cell 158, 564–578 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xi C., Hu Y., Buckhaults P., Moskophidis D., Mivechi N. F., Heat shock factor Hsf1 cooperates with ErbB2 (Her2/Neu) protein to promote mammary tumorigenesis and metastasis. J. Biol. Chem. 287, 35646–35657 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Min J. N., Huang L., Zimonjic D. B., Moskophidis D., Mivechi N. F., Selective suppression of lymphomas by functional loss of Hsf1 in a p53-deficient mouse model for spontaneous tumors. Oncogene 26, 5086–5097 (2007). [DOI] [PubMed] [Google Scholar]
- 74.Gabai V. L., Meng L., Kim G., Mills T. A., Benjamin I. J., Sherman M. Y., Heat shock transcription factor Hsf1 is involved in tumor progression via regulation of hypoxia-inducible factor 1 and RNA-binding protein HuR. Mol. Cell. Biol. 32, 929–940 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Toma-Jonik A., Widlak W., Korfanty J., Cichon T., Smolarczyk R., Gogler-Piglowska A., Widlak P., Vydra N., Active heat shock transcription factor 1 supports migration of the melanoma cells via vinculin down-regulation. Cell. Signal. 27, 394–401 (2015). [DOI] [PubMed] [Google Scholar]
- 76.Chatterjee S., Burns T. F., Targeting heat shock proteins in cancer: A promising therapeutic approach. Int. J. Mol. Sci. 18, 1978 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lang B. J., Guerrero-Giménez M. E., Prince T. L., Ackerman A., Bonorino C., Calderwood S. K., Heat shock proteins are essential components in transformation and tumor progression: Cancer cell intrinsic pathways and beyond. Int. J. Mol. Sci. 20, 4507 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mittal S., Rajala M. S., Heat shock proteins as biomarkers of lung cancer. Cancer Biol. Ther. 21, 477–485 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chatterjee S., Bhattacharya S., Socinski M. A., Burns T. F., HSP90 inhibitors in lung cancer: Promise still unfulfilled. Clin. Adv. Hematol. Oncol. 14, 346–356 (2016). [PubMed] [Google Scholar]
- 80.Dai C., Santagata S., Tang Z., Shi J., Cao J., Kwon H., Bronson R. T., Whitesell L., Lindquist S., Loss of tumor suppressor NF1 activates HSF1 to promote carcinogenesis. J. Clin. Invest. 122, 3742–3754 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tang Z., Dai S., He Y., Doty R. A., Shultz L. D., Sampson S. B., Dai C., MEK guards proteome stability and inhibits tumor-suppressive amyloidogenesis via HSF1. Cell 160, 729–744 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Murapa P., Gandhapudi S., Skaggs H. S., Sarge K. D., Woodward J. G., Physiological fever temperature induces a protective stress response in T lymphocytes mediated by heat shock factor-1 (HSF1). J. Immunol. 179, 8305–8312 (2007). [DOI] [PubMed] [Google Scholar]
- 83.Gandhapudi S. K., Murapa P., Threlkeld Z. D., Ward M., Sarge K. D., Snow C., Woodward J. G., Heat shock transcription factor 1 is activated as a consequence of lymphocyte activation and regulates a major proteostasis network in T cells critical for cell division during stress. J. Immunol. 191, 4068–4079 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ji H., Houghton A. M., Mariani T. J., Perera S., Kim C. B., Padera R., Tonon G., McNamara K., Marconcini L. A., Hezel A., El-Bardeesy N., Bronson R. T., Sugarbaker D., Maser R. S., Shapiro S. D., Wong K. K., K-ras activation generates an inflammatory response in lung tumors. Oncogene 25, 2105–2112 (2006). [DOI] [PubMed] [Google Scholar]
- 85.Busch S. E., Hanke M. L., Kargl J., Metz H. E., MacPherson D., Houghton A. M., Lung cancer subtypes generate unique immune responses. J. Immunol. 197, 4493–4503 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Grunberg N., Pevsner-Fischer M., Goshen-Lago T., Diment J., Stein Y., Lavon H., Mayer S., Levi-Galibov O., Friedman G., Ofir-Birin Y., Syu L. J., Migliore C., Shimoni E., Stemmer S. M., Brenner B., Dlugosz A. A., Lyden D., Regev-Rudzki N., Ben-Aharon I., Scherz-Shouval R., Cancer-associated fibroblasts promote aggressive gastric cancer phenotypes via heat shock factor 1-mediated secretion of extracellular vesicles. Cancer Res. 81, 1639–1653 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liao Y., Xue Y., Zhang L., Feng X., Liu W., Zhang G., Higher heat shock factor 1 expression in tumor stroma predicts poor prognosis in esophageal squamous cell carcinoma patients. J. Transl. Med. 13, 338 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shi Y., Sun L., Zhang R., Hu Y., Wu Y., Dong X., Dong D., Chen C., Geng Z., Li E., Fan Y., Thrombospondin 4/integrin α2/HSF1 axis promotes proliferation and cancer stem-like traits of gallbladder cancer by enhancing reciprocal crosstalk between cancer-associated fibroblasts and tumor cells. J. Exp. Clin. Cancer Res. 40, 14 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kmiecik S. W., Mayer M. P., Molecular mechanisms of heat shock factor 1 regulation. Trends Biochem. Sci. 47, 218–234 (2022). [DOI] [PubMed] [Google Scholar]
- 90.Bracci M., Ciarapica V., Copertaro A., Barbaresi M., Manzella N., Tomasetti M., Gaetani S., Monaco F., Amati M., Valentino M., Rapisarda V., Santarelli L., Peripheral skin temperature and circadian biological clock in shift nurses after a day off. Int. J. Mol. Sci. 17, 623 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Molzof H. E., Prapanjaroensin A., Patel V. H., Mokashi M. V., Gamble K. L., Patrician P. A., Misaligned core body temperature rhythms impact cognitive performance of hospital shift work nurses. Neurobiol. Learn. Mem. 160, 151–159 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tamaru T., Hattori M., Ninomiya Y., Kawamura G., Varès G., Honda K., Mishra D. P., Wang B., Benjamin I., Sassone-Corsi P., Ozawa T., Takamatsu K., ROS stress resets circadian clocks to coordinate pro-survival signals. PLOS ONE 8, e82006 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tamaru T., Hattori M., Honda K., Benjamin I., Ozawa T., Takamatsu K., Synchronization of circadian Per2 rhythms and HSF1-BMAL1:CLOCK interaction in mouse fibroblasts after short-term heat shock pulse. PLOS ONE 6, e24521 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kawamura G., Hattori M., Takamatsu K., Tsukada T., Ninomiya Y., Benjamin I., Sassone-Corsi P., Ozawa T., Tamaru T., Cooperative interaction among BMAL1, HSF1, and p53 protects mammalian cells from UV stress. Commun. Biol. 1, 204 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang G., Cao P., Fan Y., Tan K., Emerging roles of HSF1 in cancer: Cellular and molecular episodes. Biochim. Biophys. Acta Rev. Cancer 1874, 188390 (2020). [DOI] [PubMed] [Google Scholar]
- 96.Lee Y., Fong S. Y., Shon J., Zhang S. L., Brooks R., Lahens N. F., Chen D., Dang C. V., Field J. M., Sehgal A., Time-of-day specificity of anticancer drugs may be mediated by circadian regulation of the cell cycle. Sci. Adv. 7, eabd2645 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Eichner L. J., Brun S. N., Herzig S., Young N. P., Curtis S. D., Shackelford D. B., Shokhirev M. N., Leblanc M., Vera L. I., Hutchins A., Ross D. S., Shaw R. J., Svensson R. U., Genetic analysis reveals AMPK is required to support tumor growth in murine kras-dependent lung cancer models. Cell Metab. 29, 285–302.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lamia K. A., Papp S. J., Yu R. T., Barish G. D., Uhlenhaut N. H., Jonker J. W., Downes M., Evans R. M., Cryptochromes mediate rhythmic repression of the glucocorticoid receptor. Nature 480, 552–556 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.de Hoon M. J. L., Imoto S., Nolan J., Miyano S., Open source clustering software. Bioinformatics 20, 1453–1454 (2004). [DOI] [PubMed] [Google Scholar]
- 100.Saldanha A. J., Java Treeview--Extensible visualization of microarray data. Bioinformatics 20, 3246–3248 (2004). [DOI] [PubMed] [Google Scholar]
- 101.Hughes M. E., Hogenesch J. B., Kornacker K., JTK_CYCLE: An efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. J. Biol. Rhythms 25, 372–380 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S13