Abstract
The connections between metabolic state and therapy resistance in multiple myeloma (MM) are poorly understood. We previously reported that electron transport chain (ETC) suppression promotes sensitivity to the BCL-2 antagonist venetoclax. Here, we show that ETC suppression promotes resistance to proteasome inhibitors (PIs). Interrogation of ETC-suppressed MM reveals integrated stress response–dependent suppression of protein translation and ubiquitination, leading to PI resistance. ETC and protein translation gene expression signatures from the CoMMpass trial are down-regulated in patients with poor outcome and relapse, corroborating our in vitro findings. ETC-suppressed MM exhibits up-regulation of the cystine-glutamate antiporter SLC7A11, and analysis of patient single-cell RNA-seq shows that clusters with low ETC gene expression correlate with higher SLC7A11 expression. Furthermore, erastin or venetoclax treatment diminishes mitochondrial stress–induced PI resistance. In sum, our work demonstrates that mitochondrial stress promotes PI resistance and underscores the need for implementing combinatorial regimens in MM cognizant of mitochondrial metabolic state.
Inhibition of the electron transport chain/OXPHOS promotes resistance to proteasome inhibitors in multiple myeloma.
INTRODUCTION
Multiple myeloma (MM) is a plasma cell malignancy accounting for 12,000 deaths and 32,000 diagnoses in 2021 in the United States (1). Despite administration of proteasome inhibitors (PIs), immunomodulatory agents, and monoclonal antibody–based combinatorial regimens, MM remains incurable with a 5-year survival rate of approximately 54% (1), underscoring the need to address potential resistance mechanisms at the outset. Altered cellular metabolism is now recognized as a hallmark of cancer, leading to increased investigation into the role of metabolic alterations and heterogeneity in MM progression, therapy efficacy, and resistance mechanisms. Mitochondria are critical sources of adenosine triphosphate (ATP), reducing equivalents, reactive oxygen species (ROS), oncometabolites, and biomolecules that regulate protein synthesis, gene expression, and kinase activity (2). Last, mitochondria are also the site of cytochrome c release in the apoptosis cascade. While mitochondria are considered important targets for cancer therapy, with multiple drugs such as metformin, phenformin, VLX600, BCL-2 (B-cell lymphoma-2) family antagonists, and antibiotics like tigecycline targeting mitochondrial function (3, 4), the negative effects of targeting mitochondria on sensitivity to other therapies are relatively underexplored.
PIs are backbone MM therapeutics targeting plasma cell biology. MM cells retain their ability to produce antibodies after malignant transformation, resulting in greater dependence on cellular proteostasis machinery and higher sensitivity to proteasome inhibition (5–7). Despite being an effective first-line therapy, resistance to PIs eventually develops. Several mechanisms of PI resistance have been investigated such as deregulation of components of the ubiquitin proteasome system, mutations in the proteasome subunit β type 5 (PSMB5) of the 20S proteasome, autophagy, up-regulation of the antioxidant response, and down-regulation of protein synthesis (8–11). Previous studies investigating the role of metabolism have implicated oxidative phosphorylation (OXPHOS), glycolysis, antioxidant defense, and serine metabolism in promoting PI resistance (12–18). Nevertheless, the cellular and molecular mechanisms that connect mitochondrial metabolic states or electron transport chain (ETC) activity to the proteostasis machinery are unclear.
The sensitivity of MM cells to proteasome inhibition is determined by a balance between proteasomal “load” versus “capacity” (6, 7, 19, 20). Proteasomal load is directly related to the number of proteins tagged with ubiquitin for proteasomal degradation, and capacity is determined by the expression and activity of the proteasome. Furthermore, the ubiquitin-proteasome system and the protein translation machinery are ATP dependent, and their activity is affected by ROS levels (21, 22). Therefore, as key sources of ATP and ROS, mitochondria can directly or indirectly modulate changes in proteasomal load or capacity. We have previously shown that ETC inhibition sensitizes MM cells to BCL-2 antagonist venetoclax. Here, we show that acute mitochondrial stress upon ETC inhibition up-regulates the integrated stress response (ISR) leading to down-regulation of protein synthesis and reduced proteasomal load, resulting in resistance to proteasome inhibition. Investigation of patient sample data reveals down-regulation of ETC/OXPHOS, tricarboxylic acid (TCA) cycle, and protein translation–related pathways in patients with poor progression-free survival (PFS) and relapsed-refractory (RR) disease compared to those with better PFS and newly diagnosed (ND) MM, respectively, supporting our in vitro observations. We identify key metabolic and transcriptional changes in venetoclax-sensitive and PI-resistant ETC-inhibited MM cells and provide evidence of the existence of metabolic heterogeneity and importance of mitochondrial function in regulating therapy sensitivity in MM.
RESULTS
ETC suppression induces the ISR in MM
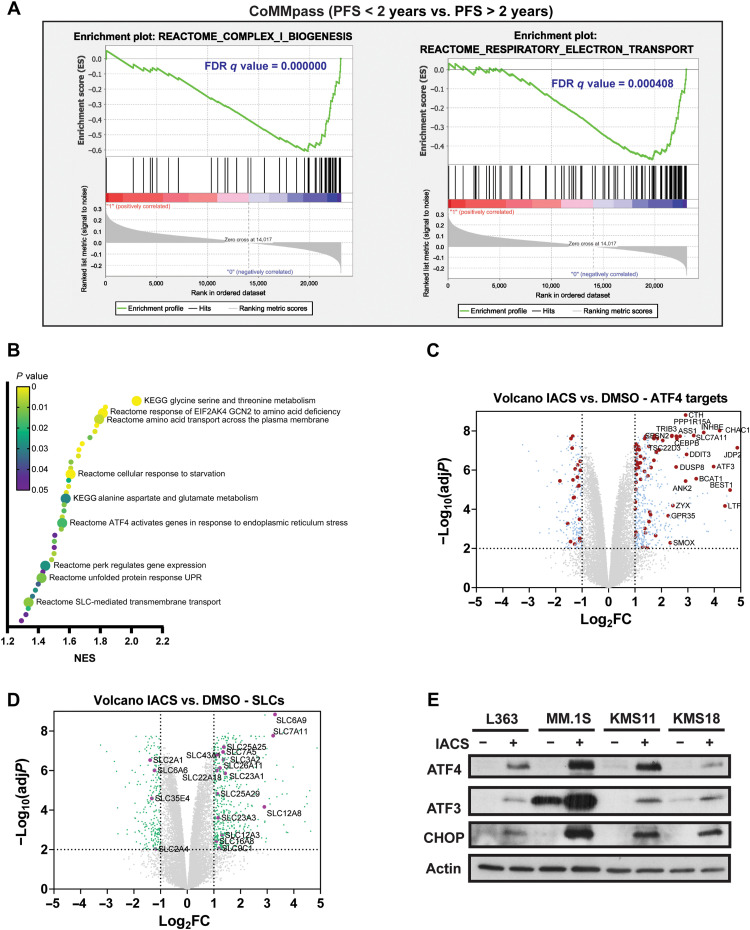
We previously demonstrated that reduced ETC activity correlates with venetoclax sensitivity, and ETC suppression increases BCL-2 dependence and venetoclax sensitivity in MM (23). Given the role of mitochondrial metabolism in determining venetoclax sensitivity (23–28), we inquired whether ETC activity correlates with patient prognosis and queried the CoMMpass (Clinical Outcomes in Multiple Myeloma to Personal Assessment of Genetic Profile) MM trial (NCT0145429, IA15) RNA sequencing (RNA-seq) data where 93.4% of patients were initially treated with PI-containing regimens. Patients were divided into two cohorts on the basis of a PFS cutoff of 2 years. Gene set enrichment analysis (GSEA) analysis of patients with poor PFS (<2 years) versus better PFS (>2 years) showed down-regulation of complex I biogenesis, TCA cycle, and OXPHOS-related pathways in poor survival patients (Fig. 1A and fig. S1), suggesting that mitochondrial metabolism plays a role in governing therapy sensitivity in MM. To identify inducible transcriptional changes occurring upon ETC suppression, we performed RNA-seq analysis of the L363 MM cell line treated with the ETC complex I inhibitor, IACS-010759 (IACS) (29). GSEA analysis of IACS- versus vehicle-treated [dimethyl sulfoxide (DMSO)] cells showed up-regulation of pathways related to the ISR and amino acid metabolism and transport (Fig. 1B). ISR induction results in phosphorylation of the eukaryotic translation initiation factor 2 subunit alpha (eIF2α), leading to inhibition of global protein translation and induction of activating transcription factor 4 (ATF4), which controls several metabolic and oxidative stress response genes (30, 31). We detected up-regulation of several ATF4 targets and solute carrier family (SLC) members with IACS treatment, including amino acid transporters (Fig. 1, C and D). Furthermore, Western blot analysis of key ISR proteins revealed induction of ATF4, ATF3, and C/EBP (CCAAT/enhancer binding protein) homologous protein (CHOP) confirming up-regulation of the ISR (Fig. 1E).
Fig. 1. ETC suppression induces the integrated stress response.
(A) Key mitochondrial pathways down-regulated in GSEA performed in patients with MM from the CoMMpass trial (NCT0145429) with poor prognosis (PFS < 2 years, n = 426) versus better prognosis (PFS > 2 years, n = 341). Gene sets for shown pathways are derived from the Reactome database. FDR, false discovery rate. (B) Pathways up-regulated in GSEA of RNA-seq data from L363 cells treated with 25 nM IACS for 9 hours compared to vehicle control (n = 3). KEGG, Kyoto Encyclopedia of Genes and Genomes. (C) Volcano plot showing differentially expressed ATF4 targets. ATF4 target genes with |log2FC| ≥ 1 and adjusted P < 0.01 are shown in red circles, and non-ATF4 target genes with |log2FC| ≥ 1 and adjusted P < 0.01 are shown in blue circles. ATF4 target genes with |log2FC| ≥ 2 and adjusted P < 0.01 are labeled. (D) Volcano plot showing differentially expressed SLCs with |log2FC| ≥ 1 and adjusted P < 0.01 labeled and shown in purple circles and other genes with |log2FC| ≥ 1 and adjusted P < 0.01 shown in green circles. (E) Expression levels of ATF4, ATF3, and CHOP evaluated in L363, MM.1S, KMS11, and KMS18 cells treated with 25 nM IACS for 24 hours by immunoblotting using actin as loading control.
ETC suppression promotes glutamine reductive carboxylation and elevates glutathione synthesis in MM
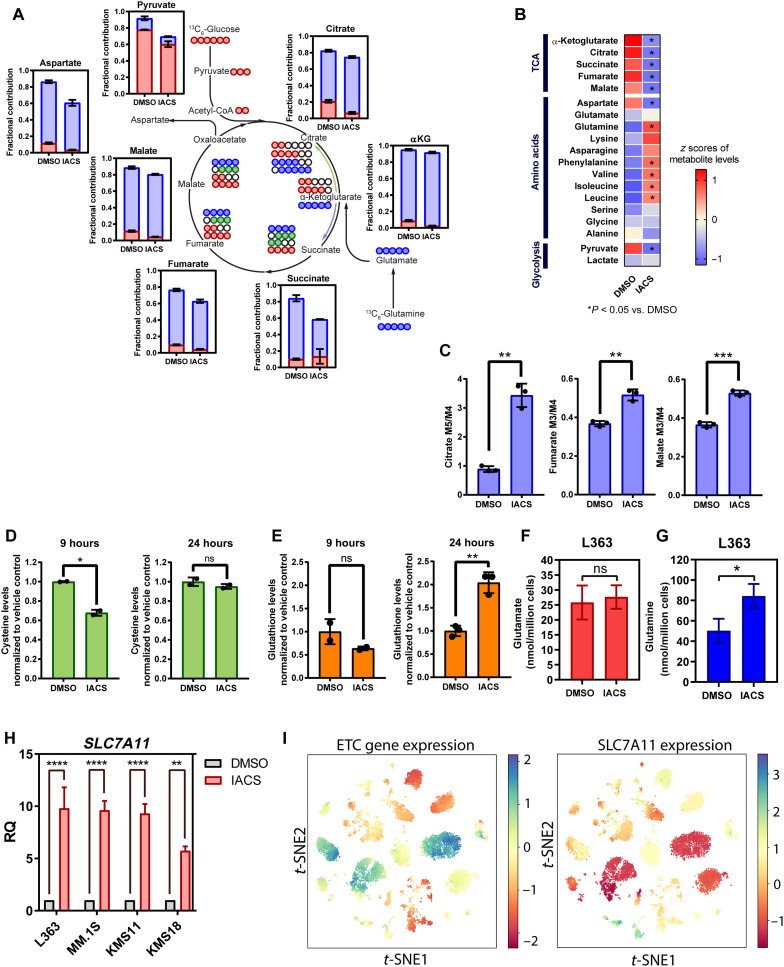
To investigate the effect of ETC inhibition on cellular metabolism, we performed a Mito Stress assay and stable carbon isotope tracing with uniformly labeled U13C-glucose or U13C-glutamine in L363 cells treated with IACS. As expected, we found reduction in basal and maximal oxygen consumption rates (OCRs) in IACS-treated L363 cells along with an increase in extracellular acidification rate (ECAR), suggesting an increase in glycolysis (fig. S2, A and B). Furthermore, stable carbon isotope tracing revealed reduction in intracellular levels of TCA cycle intermediates and pyruvate (Fig. 2A). Inspection of glucose and glutamine carbon contribution showed a reduction in glucose carbon incorporation in TCA cycle intermediates (Fig. 2A and fig. S2C). We also observed a substantial reduction in glutamine carbon contribution to succinate (Fig. 2A and fig. S2D). Congruent with the observed increase in amino acid transporter expression (Fig. 1D), we detected an increase in intracellular levels of most amino acids (Fig. 2B). Furthermore, elevated citrate M5/M4, fumarate M3/M4, and malate M3/M4 isotopomer ratios suggest a redirection of glutamine carbons toward reductive carboxylation upon IACS treatment (Fig. 2C). Mitochondrial dysfunction has been previously reported to result in up-regulation of glutamine reductive carboxylation (29, 32–34).
Fig. 2. ETC suppression promotes glutamine reductive carboxylation and elevates GSH synthesis in MM assessed in L363 cells ± treated with IACS for 9 hours.
(A) Isotopolog labeling profiles of TCA cycle–related metabolites in DMSO-treated and IACS-treated (25 nM) cells supplemented with U-13C-glucose or U-13C-glutamine for 9 hours (n = 3). Data are presented as mean values ± SD. (B) TCA cycle, glycolysis, and amino acid levels were measured in L363 cells treated with IACS for 9 hours by gas chromatography–mass spectrometry (GC-MS) (n = 3). (C) Citrate M5/M4, fumarate M3/M4, and malate M3/M4 isotopomer ratios in DMSO- and IACS-treated cells, supplemented with U-13C-glucose or U-13C-glutamine for 9 hours (n = 3). Data are presented as mean values ± SD. P values are calculated using two-tailed t tests with Welch’s correction. (D) Intracellular cysteine levels were evaluated in L363 cells treated with IACS for 9 and 24 hours (n = 2). Data are presented as mean values ± SD. P values are calculated using two-tailed t tests with Welch’s correction. (E) Intracellular GSH levels were evaluated in L363 cells treated with IACS for 9 and 24 hours (n = 2). Data are presented as mean values ± SD. P values are calculated using two-tailed t tests with Welch’s correction. (F) Intracellular glutamate and (G) glutamine levels in L363 cells treated with IACS for 9 hours. Total levels of glutamine and glutamate were measured from the GC-MS by summing up signals of all labeled and unlabeled isotopologs for each metabolite. Signals were converted to concentrations using external calibration curves (H) SLC7A11 mRNA expression in MM lines treated with IACS for 24 hours. Data are represented as means ± SEM (n = 3 independent experiments done in duplicates). (I) t-distributed stochastic neighbor embedding (t-SNE) of average expression of ETC genes (data S1) and SLC7A11 in plasma cells derived from patients with MM. ns, not significant. *P 0.01 to 0.05, **P 0.001 to 0.1, ***P 0.0001 to 0.001, and ****P < 0.0001.
In addition, the two most up-regulated amino acid transporters detected upon IACS treatment were the glycine transporter, SLC6A9, and the cystine glutamate antiporter, SLC7A11 (xCT) (L363 ± IACS RNA-seq data; Fig. 1, C and D). We observed maintenance of glycine levels despite reduction in the glucose mean enrichment (fig. S2C) in glycine, suggesting increased uptake of glycine on complex I inhibition. In addition to SLC7A11, the expression of two genes involved in cysteine metabolism, i.e., cystathionine gamma-lyase (CTH), which catalyzes the synthesis of cysteine via the trans-sulfuration pathway, and glutathione (GSH)–specific gamma-glutamylcyclotransferase 1 (CHAC1), which catalyzes cysteine synthesis via GSH cycling, was also significantly up-regulated upon ETC inhibition (Fig. 1C). A separate evaluation of intracellular cysteine levels on IACS treatment at 9 hours revealed that total cysteine levels were reduced at 9 hours (Fig. 2D). Given the up-regulation of SLC7A11, we questioned whether cysteine levels are eventually restored at a later time point. As anticipated, we found that cysteine levels were restored to the level of the vehicle control at 24 hours (Fig. 2D). Since cysteine is required for GSH synthesis, we measured GSH levels and found that GSH levels were slightly reduced at 9 hours but elevated at 24 hours following the pattern of amino acid import of cystine (Fig. 2E). Notably, SLC7A11 exports glutamate to import cystine, and increase in SLC7A11 expression can lead to a reduction in glutamate levels; however, we found no change in glutamate levels on IACS treatment (Fig. 2F). Furthermore, we found that intracellular glutamine, which can be converted to glutamate, and the expression of the primary glutamine transporter ASCT2 (Alanine-Serine-Cysteine transporter 2) were increased on IACS treatment in L363 cells (Fig. 2G and fig. S2E, respectively). The increased entry of glutamine is indirectly responsible for both cystine uptake via the xCT antiporter system by sustaining glutamate export and fueling reductive carboxylation where glutamine-derived α-KG (α-ketoglutarate) is reductively carboxylated by isocitrate dehydrogenase 2 (IDH2) to generate citrate for fatty acid synthesis. Given the elevation in reductive carboxylation in complex I–inhibited cells, we tested whether IACS treatment sensitized MM to the inhibition of IDH2. Mutant IDH2 inhibitors have been shown to inhibit the WT (wild type) protein at higher concentrations. We found that IACS and enasidenib [a mutant IDH2 inhibitor (35)] synergized with each other (fig. S2F), suggestive of increased reliance of ETC-suppressed MM on reductive carboxylation.
Next, we confirmed that complex I inhibition induced SLC7A11 expression in a panel of MM cell lines (Fig. 2H). We also analyzed single-cell RNA-seq data [Ledergor et al. (36), GSE117156] from plasma cells isolated from 14 patients with MM for expression of SLC7A11 and ETC genes to determine whether SLC7A11 expression correlates with ETC gene expression in patients with MM. Clusters with low ETC gene expression were found to correlate with higher SLC7A11 expression (Fig. 2I). Furthermore, examination of ETC gene expression showed inter- and intratumor heterogeneity in MM ETC gene expression (fig. S3A). In addition, investigation of CoMMpass data revealed that higher SLC7A11 expression correlated with poor PFS and overall survival (fig. S3B). These results in sum show that ETC suppression induces notable changes in amino acid synthesis and homeostasis in MM.
ETC suppression antagonizes proteasome inhibition–induced cytotoxicity
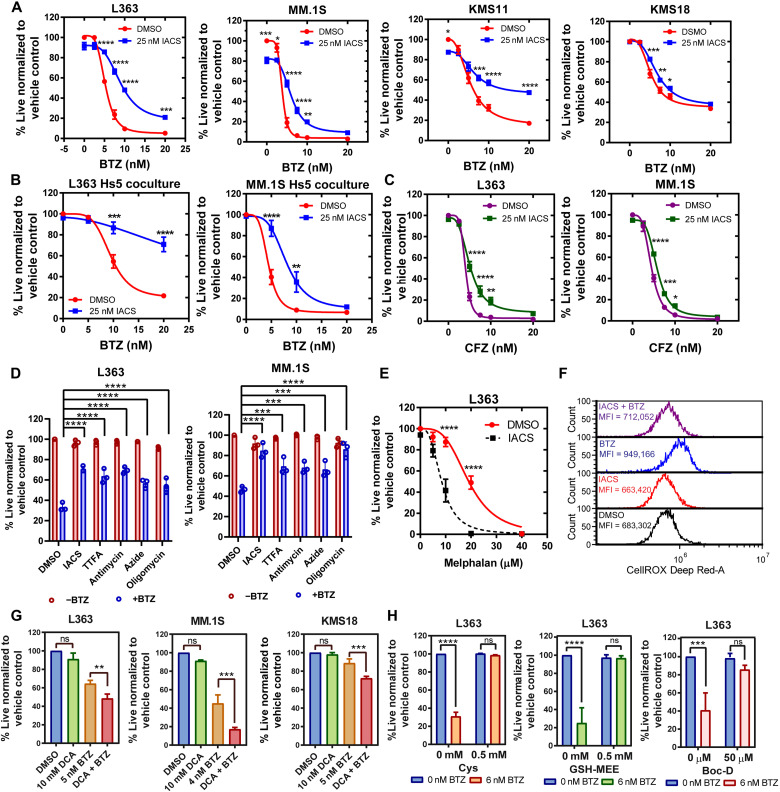
We have previously shown that ETC suppression synergizes with BCL-2 antagonist venetoclax (23). Subsequently, given the profound metabolic rewiring detected upon IACS treatment, we questioned whether ETC suppression influences the efficacy of other MM therapies. Since PIs are backbone MM therapy, we first tested the effect of complex I inhibition on PI sensitivity. We found that IACS treatment antagonized cell death induced upon treatment with PI bortezomib (BTZ; Fig. 3A). To explore the physiological relevance of the observed effect of ETC inhibition promoting resistance to BTZ, we cocultured MM cells with Hs5 stromal cells and tested the impact of IACS on MM cell sensitivity to a dose range of BTZ. While BTZ sensitivity was maintained in the MM lines cocultured with Hs5 stromal cells, IACS continued to promote resistance to BTZ in the MM cell lines cocultured with the Hs5 stromal cells (Fig. 3B).
Fig. 3. ETC suppression antagonizes proteasome inhibition–induced cell death.
(A) Dose-response curves for cotreatment of indicated lines with 25 nM IACS and increasing doses of BTZ for 24 hours. (B) Dose-response curves for indicated MM lines cocultured with Hs5 cells cotreated with 25 nM IACS and increasing doses of BTZ for 24 hours. (C) Dose-response curves for cotreatment of indicated lines with 25 nM IACS and increasing doses of CFZ for 24 hours. (D) L363 cells ± 6 nM BTZ and MM.1S cells ± 4 nM BTZ cotreated with DMSO, IACS, TTFA, antimycin A, sodium azide, and oligomycin for 24 hours. Data are represented as means ± SD (n = 3). Adjusted P values are calculated using a two-way analysis of variance (ANOVA) with post hoc Dunnett’s multiple comparisons test. (E) Dose-response curves for cotreatment of L363 with 25 nM IACS and increasing doses of melphalan for 24 hours. (F) Cellular ROS levels measured in L363 cells treated with 25 nM IACS ± 6 nM BTZ for 24 hours using CellROX Deep Red. One representative experiment is shown. (G) Viability of indicated MM lines pretreated with 10 mM DCA (24 hours) and cotreated with BTZ for an additional 24 hours. Data are presented as mean values ± SD. P values are calculated using two-tailed t tests with Welch’s correction. (H) MM lines treated with 6 nM BTZ and Cys, GSH-MEE, or Boc-D for 24 hours. Cellular viability for (A) to (E), (G), and (H) was assessed by annexin V/DAPI (4′,6-diamidino-2-phenylindole) flow cytometric staining, unless indicated. Data are represented as means ± SEM (n = 3). Adjusted P values are calculated using a two-way ANOVA with Sidak’s multiple comparisons test, unless indicated. *P 0.01 to 0.05, **P 0.001 to 0.1, ***P 0.0001 to 0.001, and ****P < 0.0001.
In addition, complex I inhibition antagonized the irreversible PI carfilzomib (CFZ), and inhibition of other ETC complexes—complex II by thenoyltrifluoroacetone (TTFA), complex III by antimycin a, complex IV by sodium azide, and complex V by oligomycin—uniformly promoted resistance to BTZ (Fig. 3, C and D). However, similar to our previous observation of ETC inhibition increasing sensitivity to venetoclax (23), we found that IACS treatment increased the sensitivity of L363 cells to the DNA alkylating agent melphalan, another MM therapeutic (Fig. 3E), suggesting that ETC inhibition selectively promotes resistance to PI. On the contrary, OXPHOS activation by dichloroacetate (DCA) pretreatment [which inhibits PDHK (pyruvate dehydrogenase kinase) to activate PDH (37)] increased sensitivity to BTZ (Fig. 3G). Evaluation of ROS levels in IACS- and/or BTZ-cotreated cells showed that BTZ elevated cytosolic ROS while IACS elevated mitochondrial ROS, both of which were reduced in the cotreated samples correlating with reduced cell death in MM treated with BTZ + IACS versus BTZ alone (Fig. 3F and fig. S4A). Last, BTZ-induced cell death was rescued by supplementation with GSH-MEE (monoethyl ester)/cysteine or the pan-caspase apoptosis inhibitor Boc-D [Boc-D-FMK, (Boc-Asp(OMe)-Fluoromethyl Ketone] (Fig. 3H), suggesting the role of ROS in BTZ-induced apoptosis, as also shown by others (38).
BTZ is a reversible inhibitor of the chymotrypsin-like (CT-L) activity of the 26S proteasome. To eliminate the possibility of IACS interfering with BTZ uptake, binding, or inhibition of the proteasome, we measured CT-L activity of the proteasome and found that it was unaffected by the addition of IACS- to BTZ-treated cells (fig. S4B). Furthermore, evaluation of the expression of the PSMB5, which is the target of BTZ, showed no change in the expression of this subunit on IACS and BTZ treatment (fig. S4C).
Since glutamine is a key carbon source for the anabolic replenishment of TCA cycle intermediates and consequently supports ETC activity, we tested the effects of glutamine deprivation on BTZ sensitivity. Notably, glutamine deprivation promoted resistance to BTZ only in the L363 and KMS18 cells, which was not detected in the MM.1S line (fig. S5A). We also found that glucose deprivation promoted resistance to BTZ in KMS18 cells but not in L363 and MM.1S cells (fig. S5B). Furthermore, treatment with the glutamine antagonist 6-diazo-5-oxo-norleucine (DON) mimicked ETC inhibition–induced resistance to PI (fig. S5C). In sum, both glutamine deprivation and glucose deprivation variably promote BTZ resistance in a cell context–specific manner, unlike direct inhibition of the ETC, which consistently promotes BTZ resistance across multiple cell lines (Fig. 3, A to D). Next, we assessed the BTZ sensitivity of a limited panel of MM cell lines and found that those with high ETC activity [reported by us in (23)] exhibited higher BTZ sensitivity and vice versa (fig. S6).
To discern the mechanistic basis of reduced apoptosis in cells cotreated with IACS and BTZ, we evaluated the expression of BCL-2 family members (fig. S7A). We found increased expression of antiapoptotic protein MCL-1 (myeloid cell leukemia-1) and proapoptotic sensitizer protein NOXA on BTZ treatment, which were reduced in IACS + BTZ–cotreated cells. MCL-1 down-regulation has been previously linked to PI sensitivity in MM, and MCL-1 accumulation on PI treatment has been shown to reduce the proapoptotic effects of proteasome inhibition (38–41). On the other hand, NOXA up-regulation has been shown to play a key role in PI sensitivity in MM and other cancers (41, 42). Therefore, to query the role of NOXA in ETC suppression–induced resistance to BTZ, we treated KMS18 NOXA knockout (KO) cells with IACS ± BTZ to determine their BTZ sensitivity and changes in the expression of BCL-2 family members. As expected, NOXA KO cells had lower BTZ sensitivity, but IACS still induced further resistance to BTZ in NOXA KO MM cells (fig. S7B). We did not detect notable changes in the expression of the BCL-2 family proteins in the KMS18 NOXA KO cells compared to vector control cells (fig. S7, C and D). The induction of apoptosis by the mitochondrial pathway is dictated both by the expression of the BCL-2 family members and the sequestration/binding properties of the various pro- and antiapoptotic BCL-2 family proteins. While we detected a reduction of NOXA on treatment with IACS + BTZ compared to BTZ alone, IACS still promoted resistance to BTZ in the NOXA KO cells with no additional compensatory changes in evaluated BCL-2 family proteins, suggesting that IACS-induced resistance to BTZ could be due to changes in the binding properties of the proapoptotic/sensitizer BCL-2 proteins.
ETC suppression promotes resistance to proteasome inhibition by decreasing proteasomal load
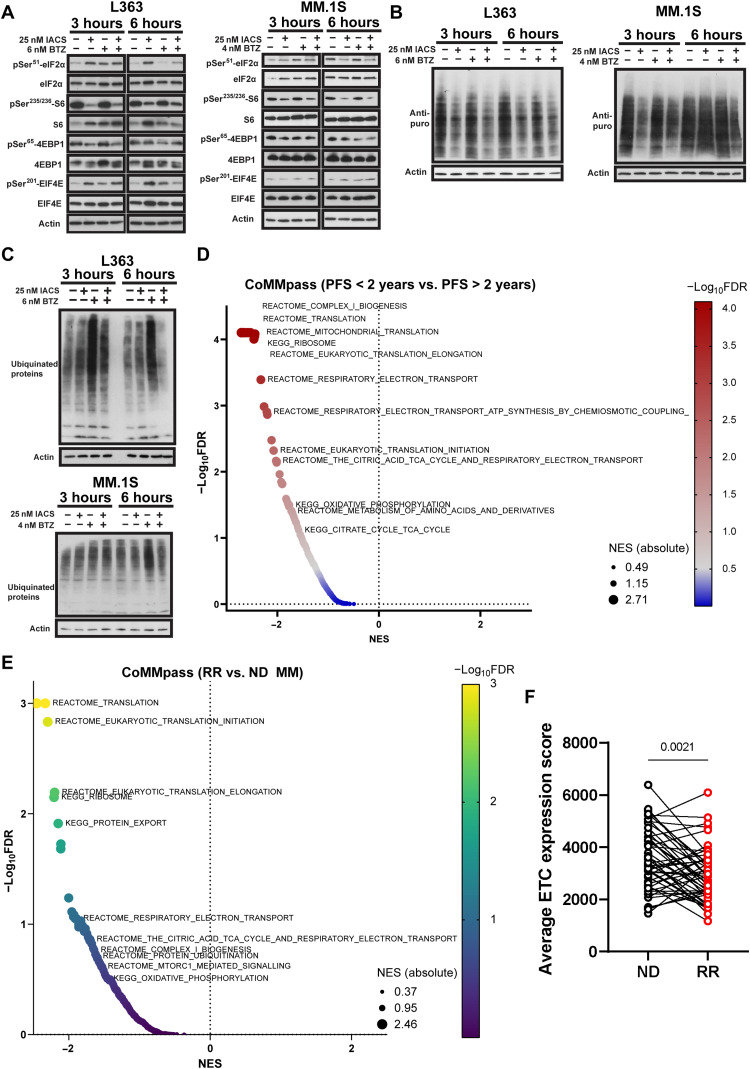
Since PI sensitivity is dependent on protein synthesis, we performed Western blot analysis of IACS ± BTZ–treated cells to assess proteins governing translation. Ribosomal protein S6 is a component of the 40S subunit of the ribosome, and reduction in phosphorylation of S6 has been previously reported to correlate with reduced protein synthesis. Hyperphosphorylation of eukaryotic translation initiation factor 4E binding protein 1 (4EBP1) releases eIF4E from 4EBP1, allowing translation of capped mRNAs, and phosphorylation of eIF4E has been shown to attenuate its affinity for capped mRNA (43, 44). IACS treatment increased pSer51-eIF2α and pSer209-eIF4E with reduction in pSer65-4EBP1 and pSer235/236-S6, indicating down-regulation of global protein translation and translation initiation at the eIF4F complex (Fig. 4A and fig. S8A). Reduction in pSer235/236-S6 was not observed in BTZ-treated cells (Fig. 4A).
Fig. 4. ETC suppression promotes resistance to proteasome inhibition by reducing proteasomal load.
(A) Expression and phosphorylation status of eIF2α, S6, 4EBP1, and eIF4E evaluated in L363 and MM.1S cells treated with IACS ± BTZ for 3 and 6 hours by immunoblotting using actin as loading control. (B) Puromycin incorporation evaluation by immunoblotting in L363 and MM.1S cells treated with IACS ± BTZ for 3 and 6 hours followed by incubation with puromycin (1 μg/ml) for 1 hour. Actin was evaluated as loading control. (C) Expression of ubiquitinated proteins in L363 and MM.1S cells treated with IACS ± BTZ for 3 and 6 hours was evaluated by immunoblotting using actin as loading control. (D) GSEA was performed to compare differential expression in metabolic and functional pathways in patients with MM from the CoMMpass trial (NCT0145429) with poor prognosis (PFS < 2 years, n = 426) versus those with better prognosis (PFS > 2 years, n = 341). Gene sets for compared pathways are derived from the KEGG and Reactome databases. (E) GSEA was performed to compare differential expression in metabolic and functional pathways in serial samples from 50 patients with MM at diagnosis and relapse. Gene sets for compared pathways are derived from the KEGG and Reactome databases. (F) Average ETC gene (data S1) expression score of ND versus RR samples. P value is calculated by Wilcoxon matched pairs signed-rank test.
Consistent with these changes, we detected a notable reduction in protein synthesis on IACS treatment using a puromycin incorporation assay (Fig. 4B) (45). Evaluation of immunoglobulin levels also revealed a substantial reduction in immunoglobulin secreted by IACS-treated cells (fig. S8B). In addition, we found a marked reduction in global protein ubiquitination levels in IACS + BTZ–treated cells compared to BTZ-treated cells suggesting a reduction in proteasomal load upon coinhibition of complex I and the proteasome (Fig. 4C). As ubiquitination is an ATP-dependent process, the observed reduction in ubiquitination could be due to a decrease in ATP levels. Therefore, we measured ATP levels on IACS ± BTZ treatment. We found a modest reduction in ATP levels in cells treated with IACS ± BTZ (fig. S8C) which, in addition to the reduction in protein synthesis on IACS treatment, could account for the observed reduction in ubiquitination levels in IACS + BTZ–treated cells compared to BTZ-treated cells. Similarly, evaluation of ubiquitinated proteins in BTZ-treated L363 and KMS18 cells grown with/without glutamine showed a reduction in ubiquitinated proteins in cells grown without glutamine as compared to cells grown with glutamine (fig. S8, D and E), in agreement with the observed resistance to BTZ in both cell lines on glutamine deprivation (fig. S5A). On the other hand, we observed a reduction in ubiquitinated proteins in KMS18 and not L363 BTZ-treated cells grown without glucose compared to cells grown in the presence of glucose (fig. S8, D and E), in agreement with the absence of glucose promoting resistance to BTZ only in KMS18 cells (fig. S5B).
Congruent with our in vitro results, GSEA analysis of data from 1150 patients treated primarily with combined PI regimens from the CoMMpass MM trial (NCT0145429, IA15) showed that Reactome and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways related to complex I biogenesis, OXPHOS/ETC, TCA cycle, and translation are down-regulated in patients with poor (<2 years) versus better PFS (>2 years) (Fig. 4D). Similarly, interrogation of serial samples taken from 50 patients (CoMMpass) at diagnosis and relapse shows down-regulation of translation, ETC/TCA cycle, complex I biogenesis, ubiquitin-mediated proteolysis, and mammalian target of rapamycin complex 1 (mTORC1) signaling pathways along with a reduction in the average expression of ETC genes, suggesting that mitochondrial metabolic state correlates with PI sensitivity in patients (Fig. 4, E to F). In sum, our data suggest that ETC inhibition promotes PI resistance by reducing proteasomal load and concomitant down-regulation of ETC/OXPHOS, and protein translation is observed in PI-resistant patients.
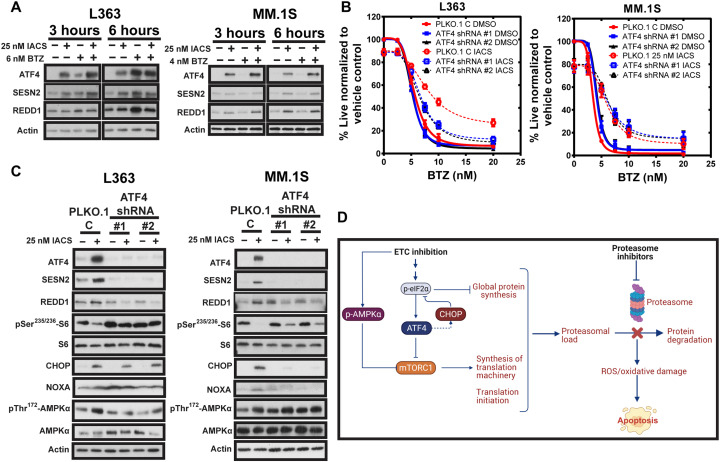
ATF4 knockdown overcomes ETC suppression–induced PI resistance in L363 cells
We next queried the role of ETC suppression–induced ATF4 induction in PI resistance. Previous studies have shown that ATF4 induction of proteasome inhibition can attenuate PI-induced cytotoxicity by inducing MCL-1 and CHOP or up-regulating autophagy and reducing proteasomal load (46–48). Furthermore, recent work by the Sabatini group (49) showed that mitochondrial stress–induced ATF4 up-regulation inhibits mTORC1 via induction of leucin sensor Sestrin2 (SESN2) and regulated in development and DNA damage responses 1 (REDD1). SESN2 and REDD1 can negatively regulate the mTORC1, a metabolic rheostat that tunes cellular protein synthesis in response to nutrient levels (50, 51). We found that IACS treatment led to early induction of ATF4, SESN2, and REDD1 and reduction in pSer235/236-S6, suggesting mTORC1 inhibition (Fig. 5A), supportive of our detection of reduced protein synthesis (Fig. 5B). We further questioned whether ATF4 knockdown (KD) could restore sensitivity to PI on ETC suppression. ATF4 KD partially restored sensitivity to BTZ in ETC-suppressed L363 cells but not in MM.1S cells (Fig. 5B). ATF4 KD efficiencies are shown in Fig. 5C. KD of ATF4 suppressed induction of SESN2 and REDD1 in both IACS-treated L363 and MM.1S, while it almost entirely restored S6 phosphorylation in L363 cells but only partially restored it in MM.1S cells (Fig. 5C). We observed a reduction in pThr172-AMPKα (AMP-activated protein kinase α) induction in IACS-treated L363 but not MM.1S cells. The Sabatini group (49) also showed that in addition to mTORC1 inhibition via the ATF4-SESN2/REDD1 axis, disruption of mitochondrial function also inhibits mTORC1 signaling at early time points through AMPKα activation. The sustained activation of AMPKα in MM.1S ATF4 KD cells correlated with partial inhibition of mTORC1 and reduction in pS6 in the absence of ATF4 induction, which could contribute to partial suppression of protein synthesis and proteasomal load. Furthermore, we observed differences in CHOP levels in IACS-treated L363 and MM.1S ATF4 KD cells. CHOP has been shown to play a proapoptotic role in PI-induced apoptosis (52). Here, we observed that CHOP was maintained in L363 ATF4 KD cells but reduced in MM.1S ATF4 KD cells on IACS treatment. The down-regulation of CHOP in MM.1S ATF4 KD cells could thus reduce cytotoxicity, contributing to continued resistance in ETC-suppressed MM.1S ATF4 KD cells in contrast to L363 ATF4 KD cells. A schematic depicting the suggested mechanisms of ETC suppression–induced resistance to PI is shown in Fig. 5D.
Fig. 5. ATF4 KD partially overcomes ETC suppression–induced PI resistance in L363 cells.
(A) Expression of ATF4, SESN2, and REDD1 evaluated in L363 and MM.1S cells treated with IACS ± BTZ for 3 and 6 hours by immunoblotting using actin as loading control. (B) Dose-response curves for cotreatment of L363 and MM.1S ATF4 KD and vector control cells with 25 nM IACS and increasing doses of BTZ for 24 hours. Cell viability was assessed by annexin V/DAPI flow cytometric staining. Data are represented as means ± SEM (n = 3). (C) L363 and MM.1S ATF4 KD and PLKO.1 vector control cells treated with IACS for 24 hours were evaluated for expression of ATF4 and its targets and p-S6 by immunoblotting using actin as loading control. (D) Schematic showing the mechanistic basis for how ETC inhibition affects the ISR, protein synthesis, proteasomal load, and efficacy of PI, created with BioRender.com.
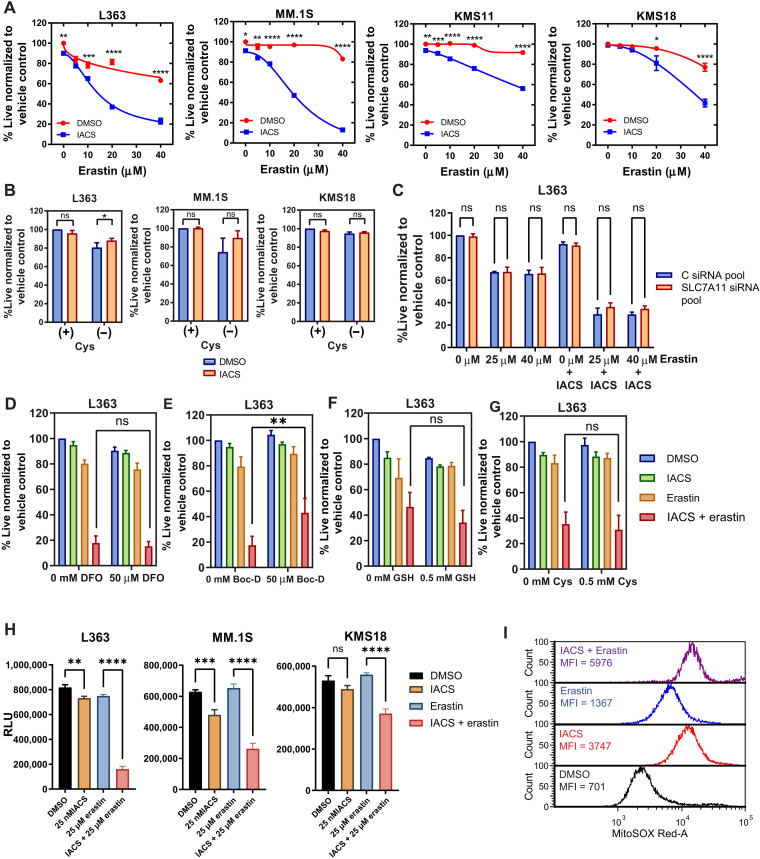
ETC suppression increases sensitivity to erastin in MM
Given the elevation in SLC7A11 expression upon ETC suppression, we tested whether perturbation of SLC7A11 or cystine deprivation induced synthetic lethality in ETC-suppressed MM. IACS treatment sensitized MM cells to the SLC7A11 inhibitor, erastin (Fig. 6A). Cystine deprivation (Fig. 6B) and SLC7A11 KD (Fig. 6C and fig. S9B; L363 KD efficiency; fig. S9A), however, had no effect on the viability of ETC-suppressed cells. Erastin’s cytotoxic effects on IACS-treated cells were maintained in SLC7A11 KD L363 and MM.1S cells (Fig. 6C and fig. S9B) and cysteine deprivation or SLC7A11 KD thus did not phenocopy the effects of erastin in IACS-treated MM.
Fig. 6. ETC suppression sensitizes MM cells to erastin.
(A) Dose-response curves for indicated lines cotreated with 25 nM IACS and erastin for 24 hours. Data are represented as means ± SEM (n = 3). (B) MM lines treated with 25 nM IACS ± 0.2 mM cysteine for 24 hours. (C) Control small interfering RNA (siRNA) or SLC7A11 siRNA pool–transfected L363 cells were treated with IACS (25 nM), erastin (25 and 40 μM), or the combination for 24 hours, and cell viability was assessed by annexin V/DAPI flow cytometric staining. n = 3 independent experiments. Data are presented as mean values ± SEM. Cells from (C) were used for RNA isolation and cDNA preparation for quantitative reverse transcription polymerase chain reaction (qRT-PCR) evaluation of SLC7A11 KD efficiency (fig. S9). (D to G) L363 cells treated with 25 nM IACS, 25 μM erastin, and the combination were treated with (D) deferoxamine (DFO), (E) Boc-D, (F) GSH, and (G) Cys for 24 hours, followed by evaluation of cell viability by annexin V/DAPI flow cytometric staining. (H) ATP levels of L363, MM.1S, and KMS18 upon IACS (25 nM), erastin (25 μM), and combination treatment evaluated by a CellTiter-Glo luminescence assay. Data are presented as relative luminescence units (RLU) compared to the control. Data are represented as means ± SEM (n = 3). Adjusted P values are calculated using Tukey’s multiple comparisons test. (I) Mitochondrial superoxide levels in L363 cells treated with 25 nM IACS ± 25 μM erastin for 24 hours using MitoSOX Red. One representative experiment is shown (n = 3). For (B) and (D) to (G), Data are represented as means ± SEM (n = 3). Adjusted P values are calculated using a two-way ANOVA with Sidak’s multiple comparisons test. *P 0.01 to 0.05, **P 0.001 to 0.1, ***P 0.0001 to 0.001, and ****P < 0.0001.
SLC7A11 inhibition by erastin has been shown to activate iron-dependent ferroptosis, which is typically accompanied by antioxidant depletion and can be reversed by treatment with iron chelators (53). The iron chelator deferoxamine did not rescue cell death induced by erastin ± IACS, while treatment with broad-spectrum caspase inhibitor Boc-D–FMK partially rescued cell death (Fig. 6, D and E), suggesting that erastin ± IACS induced apoptotic and not ferroptotic cell death. Evaluation of ROS levels on IACS and erastin cotreatment showed marginal elevation in primarily mitochondrial ROS (Fig. 6I and fig. S9C). Supportive of these observations, supplementation with GSH-MEE or cysteine did not rescue cell death of the IACS and erastin cotreated cells (Fig. 6, F and G). Furthermore, while we observed an increase in GSH levels on ETC suppression, inhibition of GSH synthesis using buthionine sulfoximine also did not increase cell death on ETC suppression (fig. S9D), suggesting that while ETC-suppressed MMs show elevated SLC7A11 and GSH levels, they do not exhibit increased reliance on cystine uptake or GSH synthesis for maintaining viability.
Erastin has previously been reported to induce RAS–RAF–mitogen-activated protein kinase kinase–dependent cell death by directly binding and inhibiting voltage-dependent anion channels (VDAC2/3) (54), altering the permeability of the mitochondrial outer membrane. VDAC regulates flux of metabolites such as ATP, adenosine diphosphate, and Pi and respiratory substrates across the mitochondrial outer membrane into the mitochondria and the release of mitochondrial ATP to the cytosol. Indirect evaluation of ATP levels using a CellTiter-Glo assay shows suppression of ATP in the IACS + erastin–treated cells (Fig. 6H). In conclusion, our data suggest that erastin’s mechanism of cytotoxicity in ETC-suppressed MM may involve profound depletion of cellular ATP.
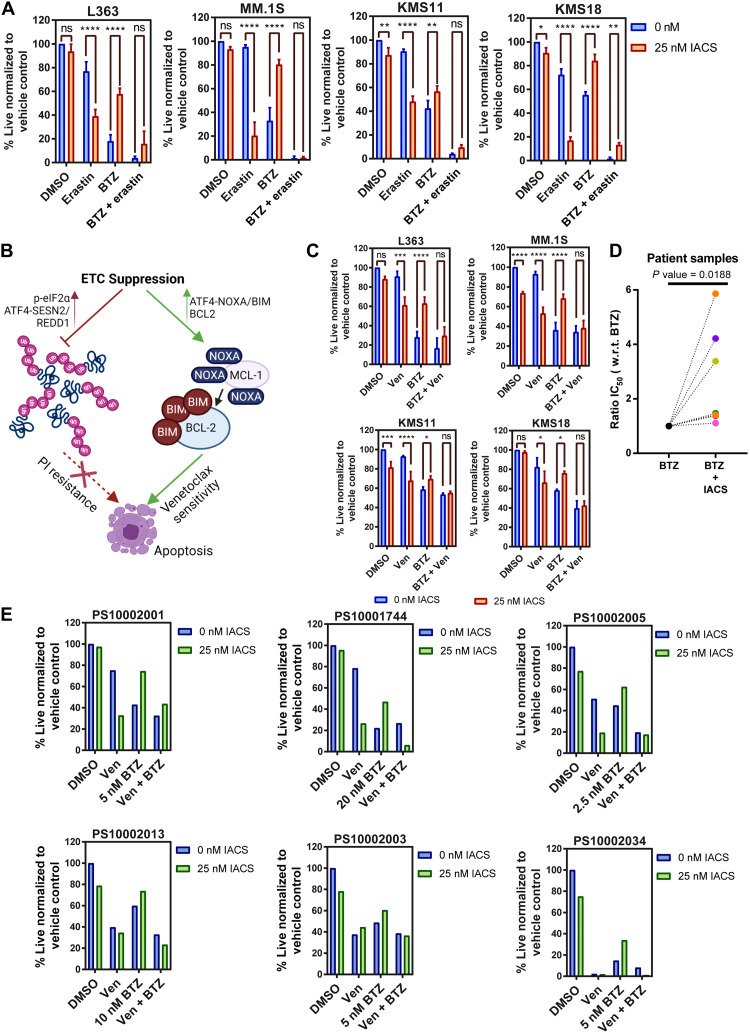
Erastin and venetoclax overcome ETC suppression–induced PI resistance
BTZ cytotoxicity is known to involve the depletion of intracellular cysteine (52). Evaluation of the expression of key genes involved in cysteine import and metabolism such as SLC7A11, CTH, CHAC1, and cystathionine beta-synthase (CBS) demonstrated SLC7A11, CBS, and CTH to be elevated in the paired BTZ RR compared to ND patients, pointing toward an elevation in cysteine sourcing and anabolism playing a role in PI resistance (fig. S10, A to E). Given the elevated sensitivity of ETC-inhibited MM to erastin, we inquired whether erastin could prevent resistance to PI on ETC suppression. We found that erastin treatment reduced ETC suppression–induced BTZ resistance in MM (Fig. 7A). Here, we observed that erastin increases sensitivity to BTZ and cysteine supplementation reverses the enhanced cytotoxicity detected with erastin and BTZ cotreatment, supportive of erastin targeting SLC7A11 and inhibiting cystine import (fig. S11). In sum, our results overall suggest that in the context of complex I inhibition, erastin provides the additional benefit of depleting ATP, creating a bioenergetic crisis to elicit cytotoxicity in BTZ-treated MM.
Fig. 7. Erastin and venetoclax combination overcomes ETC suppression–induced PI resistance.
(A) L363, MM.1S, KMS11, and KMS18 cells treated with 25 nM IACS, 6 nM BTZ (except 4 nM BTZ for MM.1S), and/or 25 μM erastin for 24 hours. (B) Schematic depicting mechanism of ETC suppression–induced resistance to PI and sensitivity to venetoclax (Ven), created with BioRender.com. (C) L363, MM.1S, KMS11, and KMS18 cells treated with 25 nM IACS, 6 nM BTZ (except 4 nM BTZ for MM.1S), and/or 0.5 μM venetoclax for 24 hours. Data are represented as means ± SD (n = 3). Adjusted P values are calculated using a two-way ANOVA with post hoc Sidak’s multiple comparisons test. (D) Ratio of BTZ median inhibitory concentration (IC50) values for seven myeloma patient bone marrow aspirates treated with vehicle control and 25 nM IACS for 24 hours. CD38-PE and CD45-APC-Cy7 were used to gate MM cells. n = 7 biologically independent samples. P value is calculated using a two-tailed ratio paired t test. (E) Patient samples treated with 25 nM IACS, BTZ (dose shown in the figure), and 0.1 μM Ven for 24 hours. CD38-PE and CD45-APC-Cy7 were used to gate MM cells. All viability is assessed by annexin V/DAPI flow cytometric staining. *P 0.01 to 0.05, **P 0.001 to 0.1, ***P 0.0001 to 0.001, and ****P < 0.0001.
We previously demonstrated that ETC suppression induces sensitivity to venetoclax via ATF4-mediated induction of NOXA (23). NOXA displaces proapoptotic BIM (Bcl-2-like protein 11) from MCL-1 to BCL-2, leading to a more venetoclax-sensitive, “BCL-2-primed” state. A model summarizing the divergent effects of ETC suppression on PI and venetoclax sensitivity in MM is presented in Fig. 7B. We next questioned whether treatment with venetoclax could diminish ETC suppression–induced resistance to BTZ. Venetoclax treatment resulted in reduced resistance to BTZ in IACS-treated cells (Fig. 7C).
As seen in MM cell lines, flow cytometric analysis of primary cells from bone marrow aspirates of patients with MM showed antagonism between IACS and BTZ (Fig. 7D). Details of patient samples and median inhibitory concentration values are summarized in table S1. Last, we tested the efficacy of venetoclax in alleviating BTZ resistance in ETC-suppressed MM patient samples. All patient samples tested showed increased sensitivity to BTZ with venetoclax in the context of ETC suppression (Fig. 7E). Cotreatment with venetoclax and BTZ had minimal impact on the viability of the non-MM cells contained within the aspirate (fig. S12).
DISCUSSION
Transformed plasma cells are in a perpetual proanabolic state and primarily reside in the hypoxic bone marrow microenvironment. The microenvironmental or pharmacological drug-induced suppression of OXPHOS can lead to profound cellular changes that influence MM therapy efficacy, a relatively underexplored area. Furthermore, complex I, the largest ETC complex, comprises 45 subunits, is prone to mutations, and is functionally suppressed in various diseases including cancer (55, 56). By using RNA-seq and stable isotope tracing, we have explored the cellular and metabolic changes that accompany the suppression of the mitochondrial ETC in MM cells. Together, the TCA cycle and ETC are important sources of ATP, ROS, precursors for amino acid/protein, and nucleotide and fatty acid synthesis. We show in MM that inhibition of the ETC results in a reduction in key metabolites and induction of a nutrient deprivation signal leading to compensatory changes in metabolism and nutrient uptake. The ensuing metabolic reprogramming results in notable alterations in amino acid levels, evident from stable carbon isotope tracing data, and GSH levels, in agreement with recently published work by the Frezza group (57).
We demonstrate here that OXPHOS suppression using a complex I inhibitor, IACS, results in up-regulation of the ISR consequent to increased eIF2a phosphorylation and ATF4 translation, in agreement with previous reports (57–59). ATF4 functions as a master transcription factor under the ISR. ATF4 can induce a complex prosurvival stress response that includes changes in metabolism and redox homeostasis genes including SESN2, REDD1, CHOP, ASS1, and ASNS. We observe a reduction of protein synthesis on ETC suppression with concordant induction of ATF4 targets (SESN2 and REDD1) that can reduce proteasomal load and collectively promote PI resistance.
We also detect up-regulation of ATF4 target genes involved in cysteine metabolism and transport. Cysteine is one of the three amino acids required for GSH synthesis, a key cellular antioxidant. Intracellular cysteine maintenance is dependent on import of cystine via SLC7A11 and subsequent reduction to cysteine. Increase in ATF4 targets CTH and CHAC1, previously implicated in poor patient outcome and disease progression, also point toward an elevated reliance of ETC-suppressed MM on cysteine synthesis via trans-sulfuration and GSH cycling (60, 61). Suraweera et al. (52) showed that PI-induced toxicity is mediated by depletion of cysteine and can be prevented by cysteine supplementation, which we have recapitulated. In addition, we show that erastin cotreatment sensitizes MM to BTZ that is reversed by cysteine supplementation. The up-regulation of SLC7A11 and the enzymes involved in cysteine anabolism (CTH and CHAC1) along with increase in GSH levels upon IACS treatment suggest that ETC-suppressed cells source additional cysteine to increase GSH synthesis. However, we show here that while cysteine supplementation reverses erastin-induced BTZ sensitivity, it does not overcome erastin’s cytotoxicity in ETC-suppressed MM nor does cysteine deprivation reverse ETC suppression–induced BTZ resistance. These results suggest that ETC suppression–induced changes in cysteine metabolism are not causal in the ETC suppression–induced resistance to BTZ. This is likely because cysteine overcomes BTZ-induced cellular proteotoxic ROS; however, here, ETC suppression largely eliminates proteotoxic stress and cellular ROS by suppressing protein translation. The relevance of SLC7A11 induction in the ETC-suppressed cells may be to maintain glutamine flux and metabolism, which we have not tested. Nevertheless, we find that ETC-suppressed MM is sensitive to the SLC7A11 inhibitor, erastin. Erastin has also been shown to inhibit mitochondrial VDAC2/3 to induce cell death (54). Erastin could thus potentially be mediating a bipronged effect by targeting SLC7A11, increasing BTZ sensitivity, and, additionally, targeting VDAC. Cell death induced by erastin in ETC-suppressed cells was not reversed by iron chelation but was reversed with a pan-caspase inhibitor, and thus, erastin likely does not induce ferroptosis in ETC-suppressed MM. VDAC opening mediated by erastin can lead to mitochondrial dysfunction, bioenergetic failure, and, ultimately, cell death. Whether erastin induces apoptotic cell death by VDAC inhibition in ETC-suppressed MM requires further investigation. Our study overall suggests that erastin can increase BTZ sensitivity and also overcome ETC suppression–induced BTZ resistance albeit by potentially different mechanisms, i.e., targeting cystine entry or creating a bioenergetic crisis, respectively.
Reductive carboxylation has been shown to support citrate formation for lipid synthesis and tumor growth under conditions of impaired mitochondrial metabolism (32). IDH2 inhibition has been shown to sensitize MM to BTZ (62). In addition, elevated ETC activity and mitochondrial respiration have recently been implicated in resistance to IDH mutant inhibitors in AML (acute myeloid leukemia) (63). Here, we show that enasidenib increases cell death in complex I–inhibited MM cells potentially because of elevated dependence of ETC-suppressed cells on IDH function to support reductive carboxylation; however, the contribution of IDH1 and/or IDH2 to glutamine-fueled reductive carboxylation requires further interrogation.
PIs are used as standard MM induction therapy and are highly efficacious in MM because of the higher dependence of myeloma cells on the proteasome to maintain proteostasis. Nevertheless, most patients relapse on PI therapy because of the development of resistance. Mechanisms of PI resistance have been extensively investigated; however, literature on the effect of alterations in mitochondrial metabolism on PI sensitivity is sparse. Previous studies have reported a contradictory role of mitochondrial metabolism in PI sensitivity. Song et al. (13) showed in a pair of MM cell lines (KMS28BM and KMS20) that mitochondrial membrane potential, OCR and ATP, and mitochondrial Ca2+ concentrations were directly correlated with BTZ resistance and combination treatment with BTZ and 2-methoxyestradiol, a superoxide dismutase inhibitor, sensitized the BTZ-resistant KMS20 cell line. Other studies have shown elevated OXPHOS in isogenic PI-resistant or Lo19S state (where a subunit of the 19S regulatory complex is down-regulated) cell lines (14, 16, 18, 64). In contrast, a study by Ling et al. (65) investigating the effect of complex I and complex III inhibition on PI sensitivity reported a decrease in ROS levels, mitochondrial membrane potential, and cytochrome c release, suggesting reduced apoptosis in human H460 non–small cell lung cancer (NSCLC) cells on coinhibition of the ETC and proteasome. Recently, metformin was found to promote resistance to BTZ in the NSCLC cell line H1299 by suppressing assembly of the proteasome subunits (66). Although metformin is not a potent complex I inhibitor, from a translational perspective, these observations and our work suggest avoiding the combination of metformin with PI therapy. Similarly, PI therapy should not be combined with drugs that may indirectly antagonize the ETC/TCA cycle activity such as DON or its prodrugs or specific antibiotics targeting ETC activity. Metformin is being currently evaluated in a phase 1 clinical trial with nelfinavir in combination with BTZ in patients with RR MM (67). Nelfinavir has been found to sensitize MM cells to BTZ in part by targeting VDAC (68), supportive of our findings with erastin. Although the mechanistic basis of the metformin-BTZ combination therapy has not been elucidated, it is possible that targeting VDAC and ETC activity using nelfinavir and metformin, respectively, may enhance PI cytotoxicity by severely depleting ATP and inhibiting protein synthesis.
PI treatment has been shown to affect ETC activity as well. Using an in vitro model of PI stress recovery in RPMI 8226 cells, mimicking typical clinical pharmacokinetics and antitumor response in patients with MM, Saavedra-García et al. (69) recently showed that cellular recovery from proteasome inhibition involved suppression of mitochondrial function. They observed down-regulation of mitochondrial proteins and OCR during stress resolution after PI treatment. Furthermore, complex I subunits including NDUFB8 were among the most suppressed during stress recovery. Consistent with this, our analysis of 50 serial samples from patients at diagnosis and relapse on PI-containing regimens from the CoMMpass trial also showed down-regulation of ETC gene expression. Other studies have also shown that early response to BTZ treatment results in reduced mitochondrial metabolism. Jaganathan et al. (70) showed that BTZ treatment results in reduced mitochondrial OCR in RPMI 8226. Tsvetkov et al. (18) also observed reduced basal OCR in control cells treated with BTZ. Mutations in complex I subunits have been detected in various diseases including cancer (55, 56, 71). Recently Hoang et al. (72) showed that mutations in genes comprising subunits of ETC complexes I and IV are frequently acquired at relapse in MM. These observations in addition to the results presented in our study suggest that down-regulation of mitochondrial metabolism in MM plays a role in response to PI-induced stress and development of resistance.
To clarify the role of mitochondrial function in PI sensitivity/resistance, we directly investigated the effect of coinhibition of the ETC and proteasome in MM. Our results show that ETC suppression (and glutamine/glucose deprivation in a cell context–specific manner and DON treatment) antagonizes PI-induced apoptosis, while elevation of OXPHOS with DCA treatment increases PI sensitivity. Furthermore, GSEA analysis of CoMMpass RNA-seq data reveals ETC/OXPHOS down-regulation to correlate with poor prognosis and therapy resistance in patients with MM. MM cells depend on the bone marrow microenvironment for proliferation and survival. Stromal cells have been shown to secrete cytokines such as interleukin-6 that promote MM survival in part by inducing antiapoptotic proteins such as MCL-1 (73), and our results show that complex I inhibition–induced resistance to BTZ is maintained in the presence of stromal cells. To the best of our knowledge, our work is the first investigation of the effect of directly modulating ETC activity on PI sensitivity in MM as opposed to indirect observations made previously. Our current findings, together with our previous work on ETC activity/glutamine metabolism and venetoclax sensitivity (23, 24), underscore the importance of mitochondrial metabolism in determining MM therapy sensitivity/resistance. In addition, intratumoral metabolic heterogeneity consequent to location, proximity to nutrients, and hypoxia is increasingly recognized (74). The existence of metabolically heterogeneous subpopulations within a tumor that responds variably to therapy can lead to survival of therapy refractory populations and emergence of resistance. We show in untreated MM, at the single-cell level, heterogeneity in ETC gene expression that inversely correlates with SLC7A11 expression, corroborating our in vitro results showing that ETC suppression elevates SLC7A11, supporting the use of SLC7A11 as a potential biomarker that may distinguish inter- and intratumor heterogeneity in ETC activity. Further work is needed to evaluate the suitability of SLC7A11 as a functional biomarker of ETC activity and PI resistance.
MATERIALS AND METHODS
Chemicals and reagents
Standard chemicals, TTFA (#T27006), rotenone (#R8875), sodium azide (#S8032), antimycin A (#A8674), GSH-MEE (#G1404), cysteine (C7352), and melphalan (#M2011) were purchased from Sigma-Aldrich. Venetoclax (#2253-1) and IACS-010759 (#B2231-1) were purchased from BioVision Inc. CFZ (#S2853) was purchased from Selleckchem. BTZ (#IC0218385905) was purchased from MP Biomedicals. Erastin (#571203-78-6) and enasidenib (#HY-18690) were purchased from MedChemExpress. Oligomycin was purchased from Merck Millipore (#495455). Polybrene (#TR-1003-G) was purchased from Millipore. Puromycin (#631306) was purchased from Clontech. Invitrogen MitoSOX Red (#M36008) and CellROX Deep Red (#C10422) were purchased from Thermo Fisher Scientific.
Cell lines
MM cell lines were obtained from the following sources: RPMI 8226 (American Type Culture Collection); L363, KMS11, and JJN3 (M. Kuehl, National Cancer Institute, MD); MM.1S (S. Rosen, City of Hope, CA); KMS18, KMS12BM, KMS12PE, and KMS27 (Japanese Collection of Research Bioresources); and KARPAS-620, OCI-MY5, and H1112 (J. Keats, TGen, a City of Hope affiliate). Human embryonic kidney (HEK) 293T cells were obtained from the American Type Culture Collection. KMS18 CRISPR control and NOXA KO were from L. Boise, Winship Cancer Institute, Emory University.
Cell culture
MM cell lines were cultured in complete RPMI 1640 (Corning, #10-040-CV) with 10% fetal bovine serum (FBS), and HEK293T cells were cultured in Dulbeccoʼs modified Eagle’s medium (Sigma-Aldrich, #D5030) with 10% FBS and penicillin (100 U/ml)/streptomycin (100 μg/ml) and maintained in a 37°C incubator with 5% CO2.
Reverse transcription quantitative polymerase chain reaction
Reagents/probes used are as listed: Thermo Scientific GeneJET RNA Purification Kit (#K0732) used for RNA isolation; cDNA synthesis, Thermo Scientific RevertAid First Strand cDNA Synthesis Kit (#K1622); Thermo Scientific DyNAmo ColorFlash probe quantitative polymerase chain reaction (PCR) kit; Bio-Rad S1000 Thermal Cycler and SLC7A11 probe (Assay_ID # hs00921938_m1) and (#F456S) with Applied Biosystems 7500 Real-Time PCR system.
Immunoblotting
Cells were harvested and washed twice with phosphate-buffered saline (PBS), and whole-cell lysates were prepared with radioimmunoprecipitation assay (RIPA) buffer supplemented with phosphatase and protease inhibitors and 1% phenylmethylsulfonyl fluoride for 20 min on ice. Cell debris was removed by centrifugation at 14,000 rpm at 4°C for 15 min. Total protein concentration was determined using Bio-Rad Protein Assay Dye Reagent Concentrate (#5000006). Samples were normalized to a total protein concentration of 20 to 30 μg in 1× Laemmli buffer containing β-mercaptoethanol and boiled for 10 min at 95°C followed by resolution by SDS–polyacrylamide gel electrophoresis (Bio-Rad, #4568096 and #4568093). Proteins were transferred to polyvinylidene fluoride (PVDF) membrane (Millipore, # IPVH00010). PVDF membranes were incubated in blocking solution [5% milk in tris-buffered saline (TBS) with Tween 20] for 1 hour, followed by washing and incubation with primary antibodies [1:1000 dilution made in BLOK Casein in TBS (G-Biosciences, #786-196)] overnight. After washing, secondary antibodies were added at a dilution of 3:10,000 in blocking solution for 1 hour.
Antibodies
ATF4 (#11815), ATF3 (#33593), p-eIF2α (#3398), eIF2α (#2103), p-S6 (#2317), S6 (#4858), p-4EBP1 (#9456), 4EBP1 (#9644), p-EIF4E (#9741), EIF4E (9742S), p-AMPK (#2535S), AMPK (#2535), SESN2 (#8487), CHOP (#5554), ubiquitin (#3936), BCL-xL (#2764), BIM (#2933), PUMA (#4976), BAX (#2772), BID (#2002), BCL-2 (#4223), p-AMPKα (#2535), AMPKα (#2532), actin (#3700), and actin (#4970) were obtained from Cell Signaling Technology. MCL-1 (#sc-819) was obtained from Santa Cruz Biotechnology. ATF4 (#ab23760) was obtained from Abcam. β-Actin (#A5441), BAK (#06-536), and NOXA (#OP180) were obtained from Sigma-Aldrich. REDD1 (#10638-1-AP) was obtained from Proteintech. Anti-puromycin (#EQ0001) was obtained from Kerafast.
Bioinformatics analysis
Unpaired analysis
GSEA was performed on DESeq2-normalized RNA-seq expression data from CoMMpass IA15a dataset using GSEA 4.10 for Windows. Samples were divided into two cohorts on the basis of the reported PFS with a cutoff of 2 years. Most recent versions of KEGG (v7.4) and Reactome (v7.4) gene sets were downloaded from MSigDB and used for the analysis. Default GSEA parameters were used with 1000 permutations over gene sets.
Paired analysis
The R package PLNSeq v1.0 was used to estimate differential gene expression across paired samples (N = 50, CoMMpass IA15a) that were acquired from the same patient before and after treatment. Differential expression was used to prerank genes for further GSEA analysis. GSEA was performed using cloud-based GSEAPreranked package (https://cloud.genepattern.org) with default parameters and 1000 permutations.
Single-cell RNA-seq analysis
The single-cell RNA-seq data by Ledergor et al. (36) were downloaded from the online repository and analyzed in Python. The data from empty wells were removed, as per the information provided by Ledergor et al. Cells with a library size smaller than 500 or greater than 10,000 were removed. Cells with less than 100 UMIs (unique molecular identifiers) for immunoglobulin genes were removed, as discussed by Ledergor et al. Cells that had greater than 4000 UMIs for mitochondrial genes were also removed. Low expression genes, defined as being nonzero in less than 100 cells, were removed. The data were normalized by library size and square root–transformed. Since the data were collected in different experimental batches, the Scanorama library was used to correct for batch effects (75). Data imputation was performed with MAGIC (76). The total expression of ETC-related genes was calculated to define the ETC score. The 20% most variable genes were selected, and the plasma cells from patients with MM were clustered via Louvain clustering of the first 50 principal components.
Cell viability/proliferation assays
A total of 0.125 × 106 and 0.250 × 106 cells/ml for cell lines and patient samples, respectively, were plated in 12-well plates and treated with the indicated concentration of drug for the duration of the experiment. For coculture experiments, MM cells that were added to Hs5 cells were preplated and incubated for 48 hours. Cells were harvested and washed twice with PBS. Cell line samples were stained with annexin V–fluorescein isothiocyanate (BD Pharmingen, #556419) and 4′,6-diamidino-2-phenylindole (DAPI). Patient samples were stained with anti–CD38-phyocerythrin and anti–CD45-allophycocyanin-Cy7 (BD Biosciences, #335808 and #557833) to identify MM cells, as previously performed (23). All samples were collected following an Emory University Institutional Review Board (IRB)–approved protocol in compliance with all relevant ethical regulations. The authors affirm that human research participants provided informed consent to participate in the study (IRB00057236). Samples were acquired on Cytek Aurora flow cytometer and analyzed with the De Novo Software FCS Express version 4.
CellTiter-Glo assay was performed using 10,000 cells per 100 microliters plated in triplicates in 96-well white flat-bottom plates (Greiner CELLSTAR, # 655083). One hundred microliters of CellTiter-Glo assay reagent (Promega, # G9248) was added to each well, and the plate was incubated for 10 min at room temperature. ATP levels were assessed using a BioTek SYNERGY H1 microplate reader following the manufacturer’s instructions.
Enzyme linked immunosorbent assay
A total of 4 × 106 cells were washed two times with PBS and resuspended in 6 ml of RPMI media with vehicle control or 25 nM IACS and incubated for 24 hours. Cell suspension was centrifuged at 1000 rpm for 4 min, and the supernatants were collected. Lysates of the cell pellets were prepared in RIPA buffer, and the lambda and kappa light chain antibodies were measured using Bethyl Human Lambda ELISA Quantitation Set (E80-116) and Bethyl Human Kappa ELISA Quantitation Set (#E80-115) following the manufacturer’s protocol.
Generation of shRNA KDs
Bacterial clones containing ATF4 short hairpin RNAs were obtained from Sigma-Aldrich (TRCN0000013573 and TRCN0000013574) and plated on LB agar plates containing ampicillin. Plasmids purified from ampicillin-resistant colonies were transfected into HEK293T to produce virus, and myeloma cells were transduced with lentiviral particles, with transduction performed in cells using jetPRIME Polyplus transfection reagent (#114-07). Medium was changed after 4 hours, and the cells were incubated for 72 hours to allow for virus production. The supernatant was collected and filtered using a 0.45-μm filter to remove cells. L363 and MM.1S cells were transduced with lentiviral particles using a standard transduction protocol. Briefly, 0.5 × 106 cells were resuspended in 500 μl of virus solution and plated per well in a 24-well plate. Polybrene Infection/Transfection Reagent (Sigma-Aldrich, #TR-1003-G) was added to a final concentration of 4 μg/ml. Spinfection was performed by centrifugation at 2500 rpm at room temperature for 1.5 hours. Cells were harvested and washed with PBS and resuspended in RPMI growth media and allowed to grow for 72 hours. L363 and MM.1S cells were selected for 7 days in media containing puromycin (0.5 and 0.05 μg/ml, respectively).
RNA sequencing
A total of 5 × 106 L363 cells were treated with vehicle control or 25 nM IACS in triplicates for 9 hours. The cells were pelleted and washed with PBS and sent to BGI Americas for DNBSEQ Eukaryotic Strand-specific Transcriptome Resequencing.
Stable isotope tracer–based flux analysis
Metabolites were extracted for the analysis of amino acids, TCA, and glycolytic intermediates. Cells were cultured in six-well plates with U-[13C6]-glucose (Cambridge Isotope Laboratories Inc., catalog no. CLM-1396-PK) and quenched after 24 hours with 800 μl of ice-cold methanol/water (1:1) solution containing 1 μg of norvaline. Cells were scraped while on ice, followed by 800 μl of high-performance liquid chromatography grade chloroform to extract lipids. Cell extracts were then vortexed vigorously for 30 min at 4°C and dried in a SpeedVac (Thermo Scientific) for 2 to 4 hours without heat and stored at −80°C. Dry extracts were derivatized with 30 μl of methoxyamine hydrochloride (Fisher Scientific, PI45950) at 30°C for 2 hours, followed by incubation with 45 μl of MBTSTFA [N-methyl-N-(tert-butyldimethylsilyl)] + 1% TBDMCS (trifluoroacetamide + 1% tert- butyldimetheylchlorosilane) (Sigma-Aldrich, M-108-5X1ML) at 55°C for 1 hour. Analysis was performed using an Agilent 7890 GC with a 30-m HP-5MSUI capillary column and Agilent 5977B MS in scan mode. The following thermal gradient was used: 100°C for 3 min, ramp of 5°C/min to 300°C, and hold at 300°C for a total run time of 48 min. The relative abundance of metabolites was estimated from the integrated peak area of ions corresponding to all potential isotopologs. Metabolite abundances were normalized to internal standard norvaline’s signal and quantified using six-point calibration with external standards. Isotopic correction of raw gas chromatography–mass spectrometry and liquid chromatography–mass spectrometry spectrum data for all reported metabolites was corrected for naturally occurring 13C using the IsoCorrectoR package (v 1.5.1) in R (CRAN 4.0.3).
Measurement of oxygen consumption and extracellular acidification rates
OCR and ECAR were measured using Seahorse bioscience extracellular flux (XFe24) analyzer. A total of 125,000 L363 cells were washed 1× with PBS and plated in triplicates in XF Base Medium supplemented with 11 mM glucose, 2 mM glutamine, and 1 mM sodium pyruvate, in Cell-Tak (Corning) precoated 24-well plates following the manufacturer’s recommendations. OCR and ECAR were evaluated over time using the Mito Stress test protocol basally and after acute injection of IACS or DMSO (final concentration, 25 nM), followed by oligomycin (final concentration, 2.5 μM), carbonyl cyanide p-trifluoromethoxyphenylhydrazone (final concentration, 0.5 μM), and antimycin plus rotenone (final concentration, 2 μM each).
Acknowledgments
We would like to thank Y.-H. T. Lin for the 50 paired-relapse specimen analysis, A. Hammond, Emory University for editorial assistance, and the Pediatric/Winship Flow Cytometry Core.
Funding: This work was supported by National Cancer Institute, NIH grant R01 CA208328, National Cancer Institute, NIH grant R01 CA247367, and Leukemia Lymphoma Society TRP Award #6573-19. The Winship Invest$ Award Research reported in this publication was supported, in part, by the Biostatistics and Bioinformatics Shared Resource of Winship Cancer Institute of Emory University and NIH/NCI under award number P30CA138292.
Author contributions: Conceptualization: A.S. and M.S. Methodology: A.S., R.N., K.B., P.G., C.L.E., and O.A. Serial sample identification: A.P.W. Metabolomics, tracing studies, RNA-seq, CoMMpass, and sc-RNA-seq bioinformatic analyses: A.A. and A.M. RNA-seq analyses: B.D. RNA-seq analyses oversight: M.B. Patient sample purification: S.M.M. and V.A.G. NOXA gene editing: S.M.M. Myeloma patient sample collection oversight: A.K.N. and S.L. Supervision: D.N. and M.S. Writing (original draft): A.S. and M.S. Writing (review and editing): A.S., M.S., L.H.B., V.A.G., B.G.B., and A.P.W.
Competing interests: A.K.N. received grant/research support from Takeda, Amgen, Janssen, BMS, GSK, Karyopharm, Pfizer, Merck, KITE, Genentech, and Arch Oncology; received grant/research support (for investigator-initiated studies) from Takeda, Amgen, GSK, Merck, and Janssen; and served on advisory boards and received honorarium from Bristol Myers Squibb, Janssen, Takeda, Amgen, Sanofi, Adaptive, GlaxoSmithKline, Oncopeptides, Karyopharm, SecuraBio, and Beyond Springs. S.L. receives research support from Takeda, Celgene, Novartis, BMS, and Janssen for trials conducted at Emory and a consultant for Takeda, Celgene, Novartis, BMS, Amgen, Janssen, GSK, and AbbVie. L.H.B. receives research funding from AstraZeneca (2019), consultancy and honoraria from AstraZeneca, and performs consultancy for Genentech (2019) and AbbVie. M.B. is an advisor for Sanofi-Genzyme. A.P.W. is a member of the scientific advisory board and holds equity stakes in Indapta Therapeutics and Protocol Intelligence LLC. Emory University has submitted a U.S. Patent Application No. 17/348,487 on 15 June 2021. Inventors on the application entitled “Uses of electron transport chain complex I or complex II inhibitors in treating cancer, combination therapies, and diagnostic methods related thereto” are A.S. and M.S. The other authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S12
Table S1
Other Supplementary Material for this manuscript includes the following:
Data S1
REFERENCES AND NOTES
- 1.American Cancer Society, Cancer Facts & Figures 2022 (American Cancer Society, 2022). [Google Scholar]
- 2.Pavlova N. N., Zhu J., Thompson C. B., The hallmarks of cancer metabolism: Still emerging. Cell Metab. 34, 355–377 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weinberg S. E., Chandel N. S., Targeting mitochondria metabolism for cancer therapy. Nat. Chem. Biol. 11, 9–15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fulda S., Galluzzi L., Kroemer G., Targeting mitochondria for cancer therapy. Nat. Rev. Drug Discov. 9, 447–464 (2010). [DOI] [PubMed] [Google Scholar]
- 5.Salmon S. E., Smith B. A., Immunoglobulin synthesis and total body tumor cell number in IgG multiple myeloma. J. Clin. Invest. 49, 1114–1121 (1970). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Obeng E. A., Carlson L. M., Gutman D. M., Harrington W. J. Jr., Lee K. P., Boise L. H., Proteasome inhibitors induce a terminal unfolded protein response in multiple myeloma cells. Blood 107, 4907–4916 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meister S., Schubert U., Neubert K., Herrmann K., Burger R., Gramatzki M., Hahn S., Schreiber S., Wilhelm S., Herrmann M., Jäck H.-M., Voll R. E., Extensive immunoglobulin production sensitizes myeloma cells for proteasome inhibition. Cancer Res. 67, 1783–1792 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Saavedra-Garcia P., Martini F., Auner H. W., Proteasome inhibition in multiple myeloma: Lessons for other cancers. Am. J. Physiol. Cell Physiol. 318, C451–C462 (2020). [DOI] [PubMed] [Google Scholar]
- 9.Lü S., Wang J., The resistance mechanisms of proteasome inhibitor bortezomib. Biomark. Res. 1, 13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Acosta-Alvear D., Cho M. Y., Wild T., Buchholz T. J., Lerner A. G., Simakova O., Hahn J., Korde N., Landgren O., Maric I., Choudhary C., Walter P., Weissman J. S., Kampmann M., Paradoxical resistance of multiple myeloma to proteasome inhibitors by decreased levels of 19S proteasomal subunits. eLife 4, e08153 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Desantis V., Saltarella I., Lamanuzzi A., Mariggiò M., Racanelli V., Vacca A., Frassanito M., Autophagy: A new mechanism of prosurvival and drug resistance in multiple myeloma. Transl. Oncol. 11, 1350–1357 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Song I.-S., Jeong Y. J., Jeong S. H., Heo H. J., Kim H. K., Lee S. R., Ko T. H., Youm J. B., Kim N., Ko K. S., Rhee B. D., Han J., Combination treatment with 2-methoxyestradiol overcomes bortezomib resistance of multiple myeloma cells. Exp. Mol. Med. 45, e50 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song I., Kim H., Lee S., Jeong S., Kim N., Ko K., Rhee B., Han J., Mitochondrial modulation decreases the bortezomib-resistance in multiple myeloma cells. Int. J. Cancer 133, 1357–1367 (2013). [DOI] [PubMed] [Google Scholar]
- 14.Thompson R. M., Dytfeld D., Reyes L., Robinson R. M., Smith B., Manevich Y., Jakubowiak A., Komarnicki M., Przybylowicz-Chalecka A., Szczepaniak T., Mitra A. K., Van Ness B. G., Luczak M., Dolloff N. G., Glutaminase inhibitor CB-839 synergizes with carfilzomib in resistant multiple myeloma cells. Oncotarget 8, 35863–35876 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zaal E. A., Wu W., Jansen G., Zweegman S., Cloos J., Berkers C. R., Bortezomib resistance in multiple myeloma is associated with increased serine synthesis. Cancer Metab. 5, 7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tibullo D., Giallongo C., Romano A., Vicario N., Barbato A., Puglisi F., Parenti R., Amorini A. M., Wissam Saab M., Tavazzi B., Mangione R., Brundo M. V., Lazzarino G., Palumbo G. A., Volti G. L., Raimondo F. D., Lazzarino G., Mitochondrial functions, energy metabolism and protein glycosylation are interconnected processes mediating resistance to bortezomib in multiple myeloma cells. Biomolecules 10, 696 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu X., Xia J., Zhang J., Zhu Y., Wu Y., Guo J., Chen S., Lei Q., Meng B., Kuang C., Feng X., He Y., Shen Y., Li X., Qiu L., Li G., Zhou W., Phosphoglycerate dehydrogenase promotes proliferation and bortezomib resistance through increasing reduced glutathione synthesis in multiple myeloma. Br. J. Haematol. 190, 52–66 (2020). [DOI] [PubMed] [Google Scholar]
- 18.Tsvetkov P., Detappe A., Cai K., Keys H. R., Brune Z., Ying W., Thiru P., Reidy M., Kugener G., Rossen J., Kocak M., Kory N., Tsherniak A., Santagata S., Whitesell L., Ghobrial I. M., Markley J. L., Lindquist S., Golub T. R., Mitochondrial metabolism promotes adaptation to proteotoxic stress. Nat. Chem. Biol. 15, 681–689 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bianchi G., Oliva L., Cascio P., Pengo N., Fontana F., Cerruti F., Orsi A., Pasqualetto E., Mezghrani A., Calbi V., Palladini G., Giuliani N., Anderson K. C., Sitia R., Cenci S., The proteasome load versus capacity balance determines apoptotic sensitivity of multiple myeloma cells to proteasome inhibition. Blood 113, 3040–3049 (2009). [DOI] [PubMed] [Google Scholar]
- 20.Cenci S., Oliva L., Cerruti F., Milan E., Bianchi G., Raule M., Mezghrani A., Pasqualetto E., Sitia R., Cascio P., Pivotal advance: Protein synthesis modulates responsiveness of differentiating and malignant plasma cells to proteasome inhibitors. J. Leukoc. Biol. 92, 921–931 (2012). [DOI] [PubMed] [Google Scholar]
- 21.Huang H., Zhang X., Li S., Liu N., Lian W., McDowell E., Zhou P., Zhao C., Guo H., Zhang C., Yang C., Wen G., Dong X., Lu L., Ma N., Dong W., Dou Q. P., Wang X., Liu J., Physiological levels of ATP negatively regulate proteasome function. Cell Res. 20, 1372–1385 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Livnat-Levanon N., Kevei E., Kleifeld O., Krutauz D., Segref A., Rinaldi T., Erpapazoglou Z., Cohen M., Reis N., Hoppe T., Glickman M. H., Reversible 26S proteasome disassembly upon mitochondrial stress. Cell Rep. 7, 1371–1380 (2014). [DOI] [PubMed] [Google Scholar]
- 23.Bajpai R., Sharma A., Achreja A., Edgar C. L., Wei C., Siddiqa A. A., Gupta V. A., Matulis S. M., McBrayer S. K., Mittal A., Rupji M., Barwick B. G., Lonial S., Nooka A. K., Boise L. H., Nagrath D., Shanmugam M., Electron transport chain activity is a predictor and target for venetoclax sensitivity in multiple myeloma. Nat. Commun. 11, 1228 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bajpai R., Matulis S. M., Wei C., Nooka A. K., Von Hollen H. E., Lonial S., Boise L. H., Shanmugam M., Targeting glutamine metabolism in multiple myeloma enhances BIM binding to BCL-2 eliciting synthetic lethality to venetoclax. Oncogene 35, 3955–3964 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pollyea D. A., Stevens B. M., Jones C. L., Winters A., Pei S., Minhajuddin M., D’Alessandro A., Culp-Hill R., Riemondy K. A., Gillen A. E., Hesselberth J. R., Abbott D., Schatz D., Gutman J. A., Purev E., Smith C., Jordan C. T., Venetoclax with azacitidine disrupts energy metabolism and targets leukemia stem cells in patients with acute myeloid leukemia. Nat. Med. 24, 1859–1866 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chan S. M., Thomas D., Corces-Zimmerman M. R., Xavy S., Rastogi S., Hong W.-J., Zhao F., Medeiros B. C., Tyvoll D. A., Majeti R., Isocitrate dehydrogenase 1 and 2 mutations induce BCL-2 dependence in acute myeloid leukemia. Nat. Med. 21, 178–184 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guièze R., Liu V. M., Rosebrock D., Jourdain A. A., Hernández-Sánchez M., Zurita A. M., Sun J., Ten Hacken E., Baranowski K., Thompson P. A., Heo J. M., Cartun Z., Aygün O., Iorgulescu J. B., Zhang W., Notarangelo G., Livitz D., Li S., Davids M. S., Biran A., Fernandes S. M., Brown J. R., Lako A., Ciantra Z. B., Lawlor M. A., Keskin D. B., Udeshi N. D., Wierda W. G., Livak K. J., Letai A. G., Neuberg D., Harper J. W., Carr S. A., Piccioni F., Ott C. J., Leshchiner I., Johannessen C. M., Doench J., Mootha V. K., Getz G., Wu C. J., Mitochondrial reprogramming underlies resistance to BCL-2 inhibition in lymphoid malignancies. Cancer Cell 36, 369–384.e13 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bosc C., Saland E., Bousard A., Gadaud N., Sabatier M., Cognet G., Farge T., Boet E., Gotanègre M., Aroua N., Mouchel P.-L., Polley N., Larrue C., Kaphan E., Picard M., Sahal A., Jarrou L., Tosolini M., Rambow F., Cabon F., Nicot N., Poillet-Perez L., Wang Y., Su X., Fovez Q., Kluza J., Argüello R. J., Mazzotti C., Avet-Loiseau H., Vergez F., Tamburini J., Fournié J.-J., Tiong I. S., Wei A. H., Kaoma T., Marine J.-C., Récher C., Stuani L., Joffre C., Sarry J.-E., Mitochondrial inhibitors circumvent adaptive resistance to venetoclax and cytarabine combination therapy in acute myeloid leukemia. Nat. Cancer 2, 1204–1223 (2021). [DOI] [PubMed] [Google Scholar]
- 29.Molina J. R., Sun Y., Protopopova M., Gera S., Bandi M., Bristow C., McAfoos T., Morlacchi P., Ackroyd J., Agip A. A., Al-Atrash G., Asara J., Bardenhagen J., Carrillo C. C., Carroll C., Chang E., Ciurea S., Cross J. B., Czako B., Deem A., Daver N., de Groot J. F., Dong J. W., Feng N., Gao G., Gay J., Do M. G., Greer J., Giuliani V., Han J., Han L., Henry V. K., Hirst J., Huang S., Jiang Y., Kang Z., Khor T., Konoplev S., Lin Y. H., Liu G., Lodi A., Lofton T., Ma H., Mahendra M., Matre P., Mullinax R., Peoples M., Petrocchi A., Rodriguez-Canale J., Serreli R., Shi T., Smith M., Tabe Y., Theroff J., Tiziani S., Xu Q., Zhang Q., Muller F., DePinho R. A., Toniatti C., Draetta G. F., Heffernan T. P., Konopleva M., Jones P., Di Francesco M. E., Marszalek J. R., An inhibitor of oxidative phosphorylation exploits cancer vulnerability. Nat. Med 24, 1036–1046 (2018). [DOI] [PubMed] [Google Scholar]
- 30.Harding H. P., Zhang Y., Zeng H., Novoa I., Lu P. D., Calfon M., Sadri N., Yun C., Popko B., Paules R., Stojdl D. F., Bell J. C., Hettmann T., Leiden J. M., Ron D., An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell 11, 619–633 (2003). [DOI] [PubMed] [Google Scholar]
- 31.Wortel I. M. N., van der MEEr L. T., Kilberg M. S., van Leeuwen F. N., Surviving stress: Modulation of ATF4-mediated stress responses in normal and malignant cells. Trends Endocrinol. Metab. 28, 794–806 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mullen A. R., Wheaton W. W., Jin E. S., Chen P. H., Sullivan L. B., Cheng T., Yang Y., Linehan W. M., Chandel N. S., DeBerardinis R. J., Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature 481, 385–388 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fendt S.-M., Bell E. L., Keibler M. A., Olenchock B. A., Mayers J. R., Wasylenko T. M., Vokes N. I., Guarente L., Heiden M. G. V., Stephanopoulos G., Reductive glutamine metabolism is a function of the α-ketoglutarate to citrate ratio in cells. Nat. Commun. 4, 2236 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gaude E., Schmidt C., Gammage P. A., Dugourd A., Blacker T., Chew S. P., Saez-Rodriguez J., O’Neill J. S., Szabadkai G., Minczuk M., Frezza C., NADH shuttling couples cytosolic reductive carboxylation of glutamine with glycolysis in cells with mitochondrial dysfunction. Mol. Cell 69, 581–593.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yen K., Travins J., Wang F., David M. D., Artin E., Straley K., Padyana A., Gross S., DeLaBarre B., Tobin E., Chen Y., Nagaraja R., Choe S., Jin L., Konteatis Z., Cianchetta G., Saunders J. O., Salituro F. G., Quivoron C., Opolon P., Bawa O., Saada V., Paci A., Broutin S., Bernard O. A., de Botton S., Marteyn B. S., Pilichowska M., Xu Y., Fang C., Jiang F., Wei W., Jin S., Silverman L., Liu W., Yang H., Dang L., Dorsch M., Penard-Lacronique V., Biller S. A., Su S.-S. M., AG-221, a first-in-class therapy targeting acute myeloid leukemia harboring oncogenic IDH2 mutations. Cancer Discov. 7, 478–493 (2017). [DOI] [PubMed] [Google Scholar]
- 36.Ledergor G., Weiner A., Zada M., Wang S.-Y., Cohen Y. C., Gatt M. E., Snir N., Magen H., Koren-Michowitz M., Herzog-Tzarfati K., Keren-Shaul H., Bornstein C., Rotkopf R., Yofe I., David E., Yellapantula V., Kay S., Salai M., Yehuda D. B., Nagler A., Shvidel L., Orr-Urtreger A., Halpern K. B., Itzkovitz S., Landgren O., San-Miguel J., Paiva B., Keats J. J., Papaemmanuil E., Avivi I., Barbash G. I., Tanay A., Amit I., Single cell dissection of plasma cell heterogeneity in symptomatic and asymptomatic myeloma. Nat. Med. 24, 1867–1876 (2018). [DOI] [PubMed] [Google Scholar]
- 37.Pratt M. L., Roche T. E., Mechanism of pyruvate inhibition of kidney pyruvate dehydrogenasea kinase and synergistic inhibition by pyruvate and ADP. J. Biol. Chem. 254, 7191–7196 (1979). [PubMed] [Google Scholar]
- 38.Pérez-Galán P., Roué G., Villamor N., Montserrat E., Campo E., Colomer D., The proteasome inhibitor bortezomib induces apoptosis in mantle-cell lymphoma through generation of ROS and NOXA activation independent of p53 status. Blood 107, 257–264 (2006). [DOI] [PubMed] [Google Scholar]
- 39.Nencioni A., Hua F., Dillon C. P., Yokoo R., Scheiermann C., Cardone M. H., Barbieri E., Rocco I., Garuti A., Wesselborg S., Belka C., Brossart P., Patrone F., Ballestrero A., Evidence for a protective role of MCL-1 in proteasome inhibitor-induced apoptosis. Blood 105, 3255–3262 (2005). [DOI] [PubMed] [Google Scholar]
- 40.Podar K., Gouill S. L., Zhang J., Opferman J. T., Zorn E., Tai Y. T., Hideshima T., Amiot M., Chauhan D., Harousseau J. L., Anderson K. C., A pivotal role for MCL-1 in Bortezomib-induced apoptosis. Oncogene 27, 721–731 (2008). [DOI] [PubMed] [Google Scholar]
- 41.Gomez-Bougie P., Wuillème-Toumi S., Ménoret E., Trichet V., Robillard N., Philippe M., Bataille R., Amiot M., NOXA up-regulation and MCL-1 cleavage are associated to apoptosis induction by bortezomib in multiple myeloma. Cancer Res. 67, 5418–5424 (2007). [DOI] [PubMed] [Google Scholar]
- 42.Qin J.-Z., Ziffra J., Stennett L., Bodner B., Bonish B. K., Chaturvedi V., Bennett F., Pollock P. M., Trent J. M., Hendrix M. J. C., Rizzo P., Miele L., Nickoloff B. J., Proteasome inhibitors trigger NOXA-mediated apoptosis in melanoma and myeloma cells. Cancer Res. 65, 6282–6293 (2005). [DOI] [PubMed] [Google Scholar]
- 43.Scheper G. C., van Kollenburg B., Hu J., Luo Y., Goss D. J., Proud C. G., Phosphorylation of eukaryotic initiation factor 4E markedly reduces its affinity for capped mRNA. J. Biol. Chem. 277, 3303–3309 (2002). [DOI] [PubMed] [Google Scholar]
- 44.Zuberek J., Wyslouch-Cieszynska A., Niedzwiecka A., Dadlez M., Stepinski J., Augustyniak W., Gingras A.-C., Zhang Z., Burley S. K., Sonenberg N., Stolarski R., Darzynkiewicz E., Phosphorylation of eIF4E attenuates its interaction with mRNA 5′ cap analogs by electrostatic repulsion: Intein-mediated protein ligation strategy to obtain phosphorylated protein. RNA 9, 52–61 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schmidt E. K., Clavarino G., Ceppi M., Pierre P., SUnSET, a nonradioactive method to monitor protein synthesis. Nat. Methods 6, 275–277 (2009). [DOI] [PubMed] [Google Scholar]
- 46.Hu J., Dang N., Menu E., De Bruyne E., Xu D., Van Camp B., Van Valckenborgh E., Vanderkerken K., Activation of ATF4 mediates unwanted MCL-1 accumulation by proteasome inhibition. Blood 119, 826–837 (2012). [DOI] [PubMed] [Google Scholar]
- 47.Milani M., Rzymski T., Mellor H. R., Pike L., Bottini A., Generali D., Harris A. L., The role of ATF4 stabilization and autophagy in resistance of breast cancer cells treated with Bortezomib. Cancer Res. 69, 4415–4423 (2009). [DOI] [PubMed] [Google Scholar]
- 48.Xian M., Cao H., Cao J., Shao X., Zhu D., Zhang N., Huang P., Li W., Yang B., Ying M., He Q., Bortezomib sensitizes human osteosarcoma cells to adriamycin-induced apoptosis through ROS-dependent activation of p-eIF2α/ATF4/CHOP axis. Int. J. Cancer 141, 1029–1041 (2017). [DOI] [PubMed] [Google Scholar]
- 49.Condon K. J., Orozco J. M., Adelmann C. H., Spinelli J. B., van der Helm P. W., Roberts J. M., Kunchok T., Sabatini D. M., Genome-wide CRISPR screens reveal multitiered mechanisms through which mTORC1 senses mitochondrial dysfunction. Proc. Natl. Acad. Sci. U.S.A. 118, e2022120118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ben-Sahra I., Manning B. D., mTORC1 signaling and the metabolic control of cell growth. Curr. Opin. Cell Biol. 45, 72–82 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Proud C. G., mTORC1 signalling and mRNA translation. Biochem. Soc. Trans. 37(Pt 1), 227–231 (2009). [DOI] [PubMed] [Google Scholar]
- 52.Suraweera A., Münch C., Hanssum A., Bertolotti A., Failure of amino acid homeostasis causes cell death following proteasome inhibition. Mol. Cell 48, 242–253 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dixon S. J., Lemberg K. M., Lamprecht M. R., Skouta R., Zaitsev E. M., Gleason C. E., Patel D. N., Bauer A. J., Cantley A. M., Yang W. S., Morrison B. III, Stockwell B. R., Ferroptosis: An iron-dependent form of nonapoptotic cell death. Cell 149, 1060–1072 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yagoda N., von Rechenberg M., Zaganjor E., Bauer A. J., Yang W. S., Fridman D. J., Wolpaw A. J., Smukste I., Peltier J. M., Boniface J. J., Smith R., Lessnick S. L., Sahasrabudhe S., Stockwell B. R., RAS–RAF–MEK-dependent oxidative cell death involving voltage-dependent anion channels. Nature 447, 865–869 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sharma L. K., Lu J., Bai Y., Mitochondrial respiratory complex I: Structure, function and implication in human diseases. Curr. Med. Chem. 16, 1266–1277 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Philley J. V., Kannan A., Qin W., Sauter E. R., Ikebe M., Hertweck K. L., Troyer D. A., Semmes O. J., Dasgupta S., Complex-I alteration and enhanced mitochondrial fusion are associated with prostate cancer progression. J. Cell. Physiol. 231, 1364–1374 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ryan D. G., Yang M., Prag H. A., Blanco G. R., Nikitopoulou E., Segarra-Mondejar M., Powell C. A., Young T., Burger N., Miljkovic J. L., Minczuk M., Murphy M. P., von Kriegsheim A., Frezza C., Disruption of the TCA cycle reveals an ATF4-dependent integration of redox and amino acid metabolism. eLife 10, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Quirós P. M., Prado M. A., Zamboni N., D’Amico D., Williams R. W., Finley D., Gygi S. P., Auwerx J., Multi-omics analysis identifies ATF4 as a key regulator of the mitochondrial stress response in mammals. J. Cell Biol. 216, 2027–2045 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guo X., Aviles G., Liu Y., Tian R., Unger B. A., Lin Y.-H. T., Wiita A. P., Xu K., Correia M. A., Kampmann M., Mitochondrial stress is relayed to the cytosol by an OMA1-DELE1-HRI pathway. Nature 579, 427–432 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang Y.-H., Huang J.-T., Chen W.-L., Wang R.-H., Kao M.-C., Pan Y.-R., Chan S.-H., Tsai K.-W., Kung H.-J., Lin K.-T., Wang L.-H., Dysregulation of cystathionine γ-lyase promotes prostate cancer progression and metastasis. EMBO Rep. 20, e45986 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li D., Liu S., Xu J., Chen L., Xu C., Chen F., Xu Z., Zhang Y., Xia S., Shao Y., Wang Y., Ferroptosis-related gene CHAC1 is a valid indicator for the poor prognosis of kidney renal clear cell carcinoma. J. Cell. Mol. Med. 25, 3610–3621 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bergaggio E., Riganti C., Garaffo G., Vitale N., Mereu E., Bandini C., Pellegrino E., Pullano V., Omedè P., Todoerti K., Cascione L., Audrito V., Riccio A., Rossi A., Bertoni F., Deaglio S., Neri A., Palumbo A., Piva R., IDH2 inhibition enhances proteasome inhibitor responsiveness in hematological malignancies. Blood 133, 156–167 (2019). [DOI] [PubMed] [Google Scholar]
- 63.Stuani L., Sabatier M., Saland E., Cognet G., Poupin N., Bosc C., Castelli F. A., Gales L., Turtoi E., Montersino C., Farge T., Boet E., Broin N., Larrue C., Baran N., Cissé M. Y., Conti M., Loric S., Kaoma T., Hucteau A., Zavoriti A., Sahal A., Mouchel P. L., Gotanègre M., Cassan C., Fernando L., Wang F., Hosseini M., Chu-Van E., Le Cam L., Carroll M., Selak M. A., Vey N., Castellano R., Fenaille F., Turtoi A., Cazals G., Bories P., Gibon Y., Nicolay B., Ronseaux S., Marszalek J. R., Takahashi K., DiNardo C. D., Konopleva M., Pancaldi V., Collette Y., Bellvert F., Jourdan F., Linares L. K., Récher C., Portais J. C., Sarry J. E., Mitochondrial metabolism supports resistance to IDH mutant inhibitors in acute myeloid leukemia. J. Exp. Med. 218, e20200924 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Besse L., Besse A., Mendez-Lopez M., Vasickova K., Sedlackova M., Vanhara P., Kraus M., Bader J., Ferreira R. B., Castellano R. K., Law B. K., Driessen C., A metabolic switch in proteasome inhibitor-resistant multiple myeloma ensures higher mitochondrial metabolism, protein folding and sphingomyelin synthesis. Haematologica 104, e415–e419 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ling Y. H., Liebes L., Zou Y., Perez-Soler R., Reactive oxygen species generation and mitochondrial dysfunction in the apoptotic response to bortezomib, a novel proteasome inhibitor, in human H460 non-small cell lung cancer cells. J. Biol. Chem. 278, 33714–33723 (2003). [DOI] [PubMed] [Google Scholar]
- 66.Schlesser C., Meul T., Stathopoulos G., Meiners S., Metformin induces resistance of cancer cells to the proteasome inhibitor bortezomib. Biomolecules 12, 756 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gonsalves W. I., Kumar S., Perez G., Saathoff S., Wilson D. M., Young A., Ailawadhi S., Trial in progress: Phase I open-label study of metformin and nelfinavir in combination with bortezomib in patients with relapsed and/or refractory multiple myeloma. Blood 138, 2735–2735 (2021). [Google Scholar]
- 68.Besse A., Besse L., Stolze S. C., Zaal E., Berkers C., Everts B., Phuyal S., Kraus M., Sobh A., Silzle T., Vulpe C., Farhan H., Overkleeft H. S., Driessen C., Nelfinavir interacts with mitochondrial VDACs and disrupts oxidative phosphorylation in proteasome inhibitor resistant myeloma. Clin. Lymphoma Myeloma Leuk. 19, e125–e126 (2019). [Google Scholar]
- 69.Saavedra-García P., Roman-Trufero M., Al-Sadah H. A., Blighe K., López-Jiménez E., Christoforou M., Penfold L., Capece D., Xiong X., Miao Y., Parzych K., Caputo V. S., Siskos A. P., Encheva V., Liu Z., Thiel D., Kaiser M. F., Piazza P., Chaidos A., Karadimitris A., Franzoso G., Snijders A. P., Keun H. C., Oyarzún D. A., Barahona M., Auner H. W., Systems level profiling of chemotherapy-induced stress resolution in cancer cells reveals druggable trade-offs. Proc. Natl. Acad. Sci. U.S.A. 118, e2018229118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jaganathan S., Malek E., Vallabhapurapu S., Vallabhapurapu S., Driscoll J. J., Bortezomib induces AMPK-dependent autophagosome formation uncoupled from apoptosis in drug resistant cells. Oncotarget 5, 12358–12370 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wallace D. C., A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: A dawn for evolutionary medicine. Annu. Rev. Genet. 39, 359–407 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hoang P. H., Cornish A. J., Chubb D., Jackson G., Kaiser M., Houlston R. S., Impact of mitochondrial DNA mutations in multiple myeloma. Blood Cancer J. 10, 46 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gupta V. A., Matulis S. M., Conage-Pough J. E., Nooka A. K., Kaufman J. L., Lonial S., Boise L. H., Bone marrow microenvironment–derived signals induce MCL-1 dependence in multiple myeloma. Blood 129, 1969–1979 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xiao Z., Dai Z., Locasale J. W., Metabolic landscape of the tumor microenvironment at single cell resolution. Nat. Commun. 10, 3763 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hie B., Bryson B., Berger B., Efficient integration of heterogeneous single-cell transcriptomes using Scanorama. Nat. Biotechnol. 37, 685–691 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.van Dijk D., Sharma R., Nainys J., Yim K., Kathail P., Carr A. J., Burdziak C., Moon K. R., Chaffer C. L., Pattabiraman D., Bierie B., Mazutis L., Wolf G., Krishnaswamy S., Pe’er D., Recovering gene interactions from single-cell data using data diffusion. Cell 174, 716–729.e27 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S12
Table S1
Data S1