Abstract
The Bacillus subtilis lacA gene, coding for β-galactosidase, has been explored as a new site able to accept DNA sequences from nonreplicating delivery vectors. Two such delivery expression vectors have been constructed and shown to be useful in obtaining regulated expression from the chromosomal location. In another experiment, it was shown that the integration of a regulatory gene at the lacA locus was able to control the expression of a transcriptional fusion at the amyE locus. These experiments demonstrate that both integration sites can be used simultaneously to obtain regulated expression of desired genes.
Several different plasmids have been used as cloning vectors in Bacillus subtilis, but many of them suffer from the disadvantage that they replicate via a single-stranded DNA intermediate (2). Several steps in the replication cycle render these plasmids highly susceptible to structural rearrangements, and these effects are often dramatically enhanced in recombinant plasmids (5). An alternative method to avoid the problem of the instability of recombinant plasmids in B. subtilis is to use integrative plasmids. Such plasmids are usually based on an Escherichia coli replicon (mostly pBR322 or one of its derivatives) and carry an antibiotic resistance marker gene that can be selected in B. subtilis and DNA sequences homologous to the B. subtilis chromosome. The most prominent and widely used systems are delivery plasmids which allow the insertion of any kind of genetic information into the bacterial chromosome. The amyE locus, coding for a nonessential α-amylase, is used in most cases for ectopic integration. This system has been developed by Shimotsu and Henner (12) and contains in its simplest form an antibiotic resistance marker and a multiple cloning site sandwiched between the two halves of the amyE gene, designated amyE-front and amyE-back. Upon transformation of B. subtilis cells, both amyE sequences will recombine at their homologous sites, thereby stably inserting the DNA sequences in between amyE-front and amyE-back into the B. subtilis chromosome via a double-crossover event (12).
In some cases, it is appropriate to have two different integration sites available, e.g., to use two different expression systems to allow the study of gene regulation. Therefore, the lacA locus has been explored as an additional site for ectopic integration of DNA sequences. The lacA gene codes for β-galactosidase (β-Gal) and is weakly expressed, if at all, in B. subtilis (1). In addition, two different expression cassettes allowing the regulatable transcription of cloned genes have been developed.
Construction of the lacA delivery expression vector pAX01.
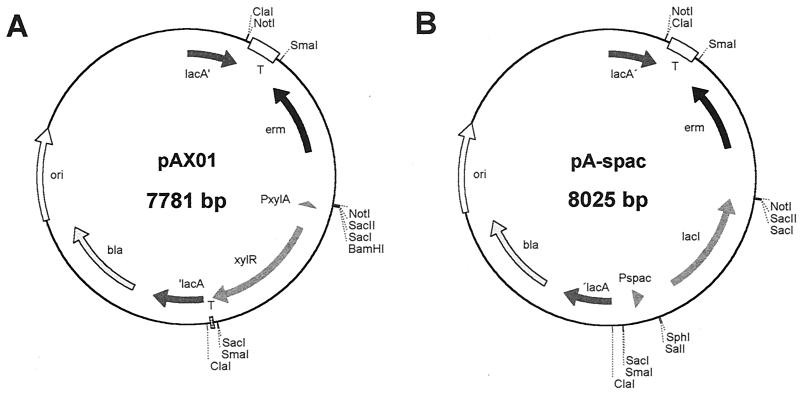
The delivery expression vector pAX01 (Fig. 1A) was assembled from different plasmids and from chromosomal DNA of B. subtilis in the following way. From pBgaB (8), the ColE1-bla backbone (3,537 bp) was PCR amplified using primers ON1 and ON2 (Table 1), which are both flanked by EcoRV restriction sites, resulting in pK1. Next, 5′ lacA (lacA-front; 500 bp; made with ON3 and ON4) and 3′ lacA (lacA-back; 500 bp; made with ON5 and ON6) were generated by PCR using chromosomal DNA of B. subtilis 1012 as a template; both fragments were flanked by SmaI and ClaI sites on their 5′ and 3′ ends, respectively. In a triple-ligation reaction, both fragments were cloned into pK1, yielding pK2.
FIG. 1.
Delivery expression vectors. (A) pAX01. (B) pA-spac. Both vectors are derivatives of pBR322 which do not replicate in B. subtilis. All the DNA sequences located between lacA′ and ′lacA allow integration into the chromosome of B. subtilis via a double-crossover event, resulting in selection for Ermr.
TABLE 1.
Oligonucleotides used in this study
| Primer | Sequence (5′-3′)a | Description |
|---|---|---|
| ON1 | GGCCATGATATC ATTCTTGAAGACGAAAGGGCCTCG | EcoRV; 5′ end of ColE1 ori-bla |
| ON2 | GGCCATGATATC CTGATATTGTCTGCATTTGCGCCG | EcoRV; 3′ end of ColE1 ori-bla |
| ON3 | GGCCATATCGAT CCTGATATTGTCTGCATTTGCGCCG | ClaI; 5′ end of lacA-front |
| ON4 | GGCCATATCGAT TCACAGTGGCAATCTCCCCCGTAT | ClaI; 3′ end of lacA-front |
| ON5 | GGCCATATCGAT TTCAAGCTATATTTGGAGTTGAGC | ClaI; 5′ end of lacA-back |
| ON6 | GGCCATCCCGGG CTAATGTGTGTTTACGACAATTCT | SmaI; 3′ end of lacA-back |
| ON7 | GGCCATCCGCGG GAGCTC GGATCCCATTTCCCCCTTTGATTT | SacII and SacI; 5′ end of xylose cassette |
| ON8 | GGCCATCCCGGG GAGCTC CTAACTTATAGGGGTAACACTTAA | SmaI and SacI; 3′ end of xylose cassette |
| ON9 | GGCCATCCCGGG GACGTTCTTGCCATTGCTGCATAAA | SmaI; 5′ end of t0 |
| ON10 | GGCCATATCGAT ATCTCTGCAGTCGCGATGATTAAT | ClaI: 3′ end of t0 |
| ON11 | GGCCATATCGAT GCGGCCGC TCTAGAGCAACGTTCTTGCCATTG | SacII and NotI; 5′ end of erm |
| ON12 | GGCCATCTCGAG GCGGCCGC ACTCTTCCTTTTTCAATATTATTG | ClaI and NotI; 3′ end of erm |
| ON13 | GGCCATATCGAT TTGACATTTTTCTTGTGGATCTGTATAATAAAGAATAATTA | ClaI and −35 and −10 regions of a hybrid promoter; 5′ end of spec |
| ON14 | GGCCATATCGAT CAATAGTTACAAATTGTTTCACTA | ClaI; 3′ end of spec |
| ON15 | GGCCATGAGCTC AGGCCTTACACAGCCCAGTCCAGACTATTCGGCA | SacI; Plac expression cassette |
| ON16 | GGCCATGAGCTC AGGCCTTAACTCACATTAATTGCGTTGCGCTCACTA | SacI; Plac expression cassette |
| ON17 | GGCCATGGATCC ATGAATGTGTTATCCTCAATTTGTTAC | BamHI; 5′ end of bgaB |
| ON18 | GGCCATGGATCC CTAAACCTTCCCGGCTTCATCATG | BamHI; 3′ end of bgaB |
| ON19 | GGCCATGTCGAC CTAAATTTTAACTTAATTTATAATTAAACG | SalI; 5′ end of bgaB |
| ON20 | GGCCATGCATGC CTAAACCTTCCCGGCTTCATCATG | SphI; 3′ end of bgaB |
| ON21 | GGCCATGCATGC ATGTTAACAAATCGTCAGCTGCT | BamHI: 5′ end of hrcA |
| ON22 | GGCCATGCATGC TGCCAAAATTCCCTTATTCATCATACAG | BamHI; 3′ end of hrcA |
G/C clamps are shown in italics; restriction sites are underlined.
In an independent series of experiments, the xylose expression cassette was fused to the transcription terminator t0 encoded by phage λ DNA (3) to avoid readthrough transcription. Using primers ON7 and ON8, the 1,440-bp xylose cassette was generated using plasmid pX (6) as a template and flanked with SacII and SacI restriction sites distal to the PxylA promoter (Fig. 1) and with SacI and SmaI sites close to the xylR repressor gene. In parallel, the 90-bp t0 terminator was amplified using oligonucleotides ON9 and ON10 and λ DNA as a template and flanked with SmaI and ClaI sites. In a last step, the xylose cassette amplicon, digested with SmaI and SacII, and the SmaI-ClaI-treated t0 terminator fragment were ligated into the ClaI-SacII-cleaved vector pBluescript II SK(+), resulting in the new plasmid pSKB.
The last cloning step in the construction of pAX01 involved assembling of the different fragments into the final expression vector. Since a selective marker gene active in B. subtilis was still missing, the erythromycin resistance gene was recovered from pMUTIN4 (13) using oligonucleotides ON11 and ON12. This amplicon was flanked by restriction sites for SacII and NotI at one end and by ClaI and NotI at the other. To assemble pAX01, three different DNA fragments were ligated: (i) pK2 linearized with ClaI and dephosphorylated, (ii) a 1,530-bp fragment carrying the xylose resistance cassette and the t0 terminator released from pSKB, and (iii) the SacII-ClaI fragment carrying the erm marker. This plasmid was constructed in a modular way, thereby allowing replacement of each part by others: (i) cleavage with ClaI releases the vector backbone, including lacA-front and lacA-back; (ii) restriction with SmaI excises the complete insert, leaving behind the two flanking terminators; (iii) application of the restriction enzyme NotI removes the erm resistance cassette; and (iv) cleavage of pAX01 with SacI liberates the xylose expression cassette. This modular structure greatly improves the versatility of this vector to adjust it to all possible applications.
Construction of a lacA::spec chromosomal insertion mutation.
Integration of pAX01 at the lacA locus can occur via a single- or a double-crossover event at the lacA locus. However, if a B. subtilis gene has been fused to the PxylA promoter, the whole vector might recombine at the site of the B. subtilis gene. Integrations into the B. subtilis chromosome will result in Ermr transformants, and experimental discrimination between these possibilities might become tedious and time-consuming. Therefore, we decided to construct a recipient strain allowing easy and fast discrimination between the various integration possibilities by screening. This was accomplished by insertion of a Specr marker within the chromosomal copy of lacA.
First, the Specr gene was amplified using ON13 and ON14, both flanked with a ClaI site, and ligated into ClaI-linearized dephosphorylated pK2, resulting in pK2-spec. Next, B. subtilis wild-type strain 1012 (10) was transformed with pK2-spec, Specr colonies were selected, and inactivation of the lacA gene was verified by PCR (data not shown). The lacA::spec recipient strain was termed IHA01. A comparable recipient strain (amyE::cat strain AM01) has already been described for the amyE locus (9). To allow the insertion of two different DNA sequences at both loci within the same strain, both antibiotic resistance marker-inactivated loci were combined into a single strain. Chromosomal DNA was prepared from IHA01 and used to transform AM01, carrying the amyE::cat marker. Chromosomal DNA from several Specr and Catr colonies was isolated, and the correct locations of the two markers were verified by PCR (data not shown); the recipient strain was designated IHA02.
Construction of the IPTG-inducible expression vector pA-spac.
The most widely used expression system in B. subtilis is based on the E. coli lac promoter, which is negatively controlled by the Lac repressor; in this system, the activity of the repressor protein is modulated by isopropyl-β-d-thiogalactopyranoside (IPTG). The E. coli-B. subtilis shuttle expression vector pREP9 is mentioned here as a paradigm (7). Using plasmid pDR66 as a template and oligonucleotides ON15 and ON16, the lac expression cassette, including the lacI gene and the lac promoter-operator region, was amplified and inserted into SacI-linearized vector pAX01, resulting in pA-spac (Fig. 1B).
The integrative expression vectors pAX01 and pA-spac express β-Gal from the lacA locus in a regulatable way.
In order to prove that both integrative expression vectors work properly, the bgaB gene, coding for heat-stable β-Gal (4), was inserted into both vectors. With pAX01, bgaB was generated from plasmid pBgaB (8) using ON17 and ON18, both flanked with BamHI sites. The BamHI-treated amplicon was then inserted into BamHI-linearized pAX01 to result in pAX01-BgaB. With pA-spac, the bgaB gene was generated from the same template using ON19 and ON20 and ligated into the SalI-SphI-cleaved vector (pA-spac-BgaB). Next, the two transcriptional fusions were recombined independently at the lacA locus using strain IHA01, and the correct integration was verified by Southern blotting (strains IHA01-Xyl-BgaB and IHA01-Spac-BgaB).
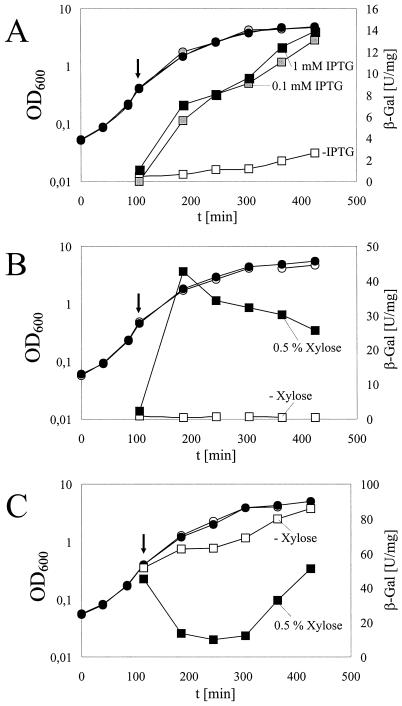
To measure the β-Gal activities of both strains, cells were grown in Luria-Bertani medium either in the absence or in the presence of an inducer for 7 h. While the addition of IPTG did not influence the growth of strain IHA01-Spac-BgaB, β-Gal activity increased steadily over time (Fig. 2A). It was found that 0.1 mM IPTG was sufficient to obtain full induction, and even the addition of 10 mM did not result in a higher induction rate (data not shown). We also observed a slow increase in β-Gal activity even in the complete absence of IPTG, demonstrating some leakiness of the system (Fig. 2A). With strain IHA01-Xyl-BgaB, no β-Gal activity was determined in the absence of the inducer xylose, while the addition of 0.5% xylose resulted in full induction after about 1 h and did not influence the rate of growth of the strain (Fig. 2B). Thereafter, β-Gal activity declined, most probably due to the consumption of the sugar xylose. Continued high levels of expression over several hours could be obtained by inducing the cells with 2% xylose, and this high concentration did not result in a higher induction rate (data not shown), as already published (6). As indicated by the data presented in Fig. 2A and B, the xylose-inducible system is superior to the IPTG-inducible system for two reasons: (i) there is no expression of the bgaB gene in the absence of an inducer, demonstrating a low level or no leakiness of the system; and (ii) it results in a higher induction rate (about 40-fold versus 15-fold with IPTG).
FIG. 2.
Induction of β-Gal activity. Bacteria were grown at 37°C in Luria-Bertani medium containing the appropriate antibiotic (erythromycin at 1 μg/ml or/and spectinomycin at 100 μg/ml) and were induced by either IPTG or xylose at the early exponential growth phase. β-Gal activities were determined at 55°C as described previously (8) using o-nitrophenyl-β-d-galactopyranoside as a substrate and are given as units per milligram of protein. All assays were repeated at least three times and yielded comparable results; the data from one experiment are shown. Growth of B. subtilis strains IHA01-Spac-BgaB (A), IHA01-Xyl-BgaB (B), and IHA13 (C) either in the absence of an inducer (●) or after the addition of an inducer (○) was recorded. β-Gal activities in the absence or presence of the inducer are as indicated, and the time point of addition of the inducer is marked by a vertical arrow. OD600, optical density at 600 nm; t, time.
The hrcA gene integrated at lacA regulates the expression of a transcriptional fusion inserted at amyE.
To test for the usage of both integration loci in a single strain, the hrcA repressor gene (11) was integrated at the lacA locus in strain AM19, where the operator recognized by HrcA was fused to bgaB at the amyE locus. hrcA was generated using oligonucleotides ON21 and ON22 as primers and B. subtilis 1012 chromosomal DNA as a template. The BamHI-flanked amplicon was ligated into BamHI-linearized and dephosphorylated vector pAX01, resulting in pAX-hrcA. The operon fusion was transferred into strain AM19 devoid of its own hrcA gene and carrying a transcriptional fusion between the promoter-operator region of the dnaK operon and bgaB, resulting in strain IHA13. β-Gal activity was then measured in the absence and in the presence of the inducer xylose. While in the absence of xylose there was considerable expression of the bgaB gene (60 to 80 U) due to the absence of an active HrcA repressor (Fig. 2C), this activity dropped to about 10 U upon the addition of 0.5% xylose (Fig. 2C). Again, upon prolonged incubation of the cells in the presence of 0.5% xylose, β-Gal activity began to rise; this effect could be prevented by using 2% xylose (data not shown). This experiment clearly demonstrates that regulated expression from one chromosomal locus will influence transcription at a distant one.
Conclusions.
(i) The lacA locus has been selected as an alternative site for ectopic integration of foreign DNA sequences. This gene codes for β-Gal and is very weakly expressed. For the easy screening of integrants, a mutated strain was constructed which carries a spectinomycin resistance cassette within lacA (lacA::spec; strain IHA01). Another strain was constructed containing, in addition, a genetically marked amyE locus (lacA::spec amyE::cat; strain IHA02). (ii) Two versatile delivery expression vectors were constructed (pAX01 and pA-spac) and tested by measuring the activity of a reporter gene fused to the regulatable promoters. (iii) To demonstrate communication between the lacA and amyE loci, the xylose-inducible hrcA repressor gene was integrated at lacA and at the promoter-operator region recognized by that repressor and fused to the bgaB reporter gene at amyE. While β-Gal activity was low in the presence of an inducer, it increased significantly in its absence. (iv) The two recipient strains and the two expression vectors can be ordered from the Bacillus Genetic Stock Center (http://bacillus.biosci.ohio-state.edu). The DNA sequences of the two expression vectors and their plasmid maps can be found at http://btbgn1.bio.uni-bayreuth.de/lsgenetik/frames.htm.
Acknowledgments
We thank John D. Quisel for providing plasmid pDR66.
This work was financed by CEC Biotech grant BI02 CT920254 and the Fonds der Chemischen Industrie.
REFERENCES
- 1.Daniel R A, Haiech J, Denizot F, Errington J. Isolation and characterization of the lacA gene encoding β-galactosidase in Bacillus subtilis and a regulator gene, lacR. J Bacteriol. 1997;179:5636–5638. doi: 10.1128/jb.179.17.5636-5638.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gruss A, Ehrlich S D. The family of highly interrelated single-stranded doexoyribonucleic acid plasmids. Microbiol Rev. 1989;53:231–241. doi: 10.1128/mr.53.2.231-241.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hayes S, Szybalski W. Control of short leftward transcripts from the immunity and ori regions in induced coliphage lambda. Mol Gen Genet. 1973;126:275–290. doi: 10.1007/BF00269438. [DOI] [PubMed] [Google Scholar]
- 4.Hirata H, Fukazawa T, Negoro S, Okada H. Structure of a β-galactosidase gene of Bacillus stearothermophilus. J Bacteriol. 1986;166:722–727. doi: 10.1128/jb.166.3.722-727.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jannière L, Bruand C, Ehrlich S D. Structurally stable Bacillus subtilis cloning vectors. Gene. 1990;87:53–61. doi: 10.1016/0378-1119(90)90495-d. [DOI] [PubMed] [Google Scholar]
- 6.Kim L, Mogk A, Schumann W. A xylose-inducible Bacillus subtilis integration vector and its application. Gene. 1996;181:71–76. doi: 10.1016/s0378-1119(96)00466-0. [DOI] [PubMed] [Google Scholar]
- 7.LeGrice S F. Regulated promoter for high-level expression of heterologous genes in Bacillus subtilis. In: Goeddel D V, editor. Gene expression technology. London, England: Academic Press Ltd.; 1990. pp. 201–214. [Google Scholar]
- 8.Mogk A, Hayward R, Schumann W. Integrative vectors for constructing single-copy transcriptional fusions between Bacillus subtilis promoters and various reporter genes encoding heat-stable enzymes. Gene. 1996;182:33–36. doi: 10.1016/s0378-1119(96)00447-7. [DOI] [PubMed] [Google Scholar]
- 9.Mogk A, Homuth G, Scholz C, Kim L, Schmid F X, Schumann W. The GroE chaperonin machine is a major modulator of the CIRCE heat shock regulon of Bacillus subtilis. EMBO J. 1997;16:4579–4590. doi: 10.1093/emboj/16.15.4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saito H, Shibata T, Ando T. Mapping of genes determining nonpermissiveness and host-specific restriction to bacteriophages in Bacillus subtilis Marburg. Mol Gen Genet. 1979;170:117–122. doi: 10.1007/BF00337785. [DOI] [PubMed] [Google Scholar]
- 11.Schulz A, Schumann W. hrcA, the first gene of the Bacillus subtilis dnaK operon, encodes a negative regulator of class I heat shock genes. J Bacteriol. 1996;178:1088–1093. doi: 10.1128/jb.178.4.1088-1093.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shimotsu H, Henner D J. Construction of a single-copy integration vector and its use in analysis of regulation of the trp operon of Bacillus subtilis. Gene. 1986;43:85–94. doi: 10.1016/0378-1119(86)90011-9. [DOI] [PubMed] [Google Scholar]
- 13.Vagner V, Dervyn E, Ehrlich S D. A vector for systematic gene inactivation in Bacillus subtilis. Microbiology. 1998;144:3097–3104. doi: 10.1099/00221287-144-11-3097. [DOI] [PubMed] [Google Scholar]