Abstract
Using the Density Functional Theory approach and in silico docking, the current study analyzes the inhibitory role of a novel α-aminophosphonate derivative against SARS-CoV-2 major protease (Mpro) and RNA dependent RNA polymerase (RdRp) of SARS-CoV-2. FT-IR, UV–Vis, and NMR (1H, 13C, 31P) approaches were used to produce and confirm the novel α-aminophosphonate derivative. The quantum chemical parameters were detremined, and the reactivity of the synthesized molecule was discussed using DFT at the B3LYP/6-31G(d,p) level. In addition, the inhibitory function of the investigated derivative for SARS-CoV-2 major protease (Mpro) and RNA dependent RNA polymerase (RdRp) was estimated using in silico docking. These discoveries could pave the way for novel SARS-CoV-2 therapies to develop and be tested.
Keywords: Mpro, RdRp, SARS–CoV-2, Synthesis, α-aminophosphonate, DFT, Molecular docking
Graphical abstract
1. Introduction
Coronaviridae virus SARS-CoV-2. This infectious disease first surfaced in the Chinese city of Wuhan in December 2019, soon spreading throughout China and then internationally, resulting in a global outbreak. In China, on November 9th [1], the COVID-19 virus had spread to over 190 countries, causing 50 million illnesses and 1 250 000 deaths [2]. Covid-19 is a respiratory illness that can be deadly in elderly or chronically ill patients. It is spread by intimate contact with sick individuals. Many doctors have been waiting for the SARS-CoV-2 coronavirus to emerge. When confronted with an unknown illness, they often had to do what they had on hand to treat their patients; this pushed them to experiment with illegal medications and therapies. Covid-19 stands for "Coronavirus Illness 2019," a disease caused by the even though a health crisis caused the COVID-19 pandemic, it infected millions of people worldwide and caused unprecedented tension and a surge in all aspects of life. COVID-19 has caused widespread misery and human suffering. It has hurt social life, health care systems, educational systems, technological, industrial, agricultural production, and financial systems worldwide, pushing people's socioeconomic well-being to the brink of collapse [3]. However, because of the pandemic's global nature, people from all over the world may band together and work together to slow and stop the spread of the disease and find new drug treatments.
In this context, and according to a study published in the journal Infectious Diseases and Therapy by the University of Pennsylvania (United States), 115 different drugs and remedies were prescribed to the first patients with Covid-19, a total of 9152 patients, and the vast majority of these treatments were administered outside of standard care protocols and without proven effectiveness, with only a small number of articles published with exploitable data on their efficacy [4]. Antivirals, particularly the Lopinavir/ritonavir combination, are among these medications, as are antibiotics and corticosteroids such as methylprednisolone. Patients have also been treated with antibodies, while others have been treated solely or with traditional medical remedies such as herbal decoctions [5]. In Algeria, for example, two medications, Hydroxychloroquine (HCQ) and Chloroquine(CQ) have been approved for the treatment of individuals with coronavirus illness 2019 (COVID-19) [6], against COVID-19 [7], [8], [9], [10], [11]. Also, some researchers are evaluating drugs approved for other vision conditions to see if they can be repositioned for treatment and prevention in animals and humans [12], [13], [14], and regulators have yet to approve any drugs for the treatment of SARS-COV-2 infection. Researchers need to understand better the dynamics of SRAS-CoV-2, the virus that causes the disease. However, due to the difficulty of experimental measurements and the rapid progression of the coronavirus, they have turned to models to better understand how the disease develops in the body, particularly in the blood and immune system [15], [16], [17], in order to model drug treatments and identify ideal delivery methods [18], [19], [20].
As a result, various research teams worldwide have already conducted an extensive study to assess the efficacy of several medications α-aminophosphonates, structural counterparts of the corresponding α-amino acids, have gotten much attention in this context, and their synthesis has gotten much attention in synthetic organic chemistry and medicinal chemistry [21]. They play an important role and exhibit various biological and biochemical properties. They effectively inhibit the growth of plasmodium falciparum, the parasite that causes malaria, in test tubes [22]. They also act as inhibitors of specific enzymes, such as HIV protease, thrombin, and human collagenase, and suppress the growth of various tumours and viruses [23,24]. Antibiotics [25], antimicrobials [26], anticancer agents 27], and antioxidants agents [28] are all examples of α-aminophosphonates. For coronaviruses, such as SARS-CoV-2 [29], [30], [31], [32], [33], enzymes such as Mpro/3CLpro and RdRp, which play significant roles in viral transcription and replication, have been regarded as interesting therapeutic targets. CoV-2 is difficult, this study sought to establish a link between some quantum descriptors and the inhibitory action of the examined α-aminophosphonates for SARS-CoV-2. The Density Functional Theory (DFT) approach derived quantum chemical parameters at the B3LYP/6-31G(d,p) level. The energy of the highest occupied molecular orbital (EHOMO), the energy of the lowest unoccupied molecular orbital (ELUMO), the energy gap (ΔEgap), global hardness (η), global softness (σ), absolute electronegativity (χ), and the electrophilicity index (ω) are some of the characteristics. This study was also used to predict the inhibitory activity of the examined α-aminophosphonate for SARS-CoV-2 in silico docking.
2. Materials and methods
2.1. Experimental
All of the chemical substances used in this research were acquired from Sigma-Aldrich and used without additional purification. The melting point of the obtained compound was determined using an open capillary method in Buchi Melting Point B-540 (60 - 900 °C). In contrast, UV–vis spectra were obtained using a JASCO V-650 UV–Vis spectrophotometer with quartz cells of 1 cm path length in Methanol between 200 and 800 nm. FT-IR spectra were obtained using a JASCO 4200 spectrometer between 4000 and 200 cm 1. Finally, the 1H, 13C, and 31P NMR spectra were obtained using a Bruker AV III 300 MHz in DMSO‑d6 with tetramethylsilane (TMS) as an internal reference.
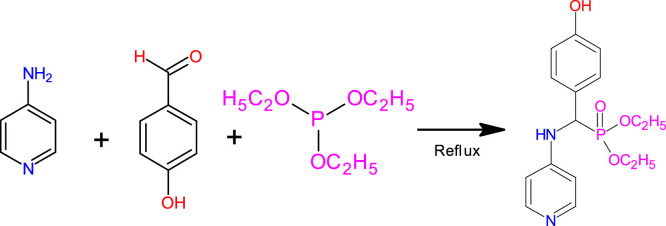
In a Kabachnik-Fields reaction [34], an equimolar amount of 4-amino pyridine (1 mmol, 0.094 mg), 4-hydroxybenzaldehyde (1 mmol, 1.22 mg), and triethyl phosphite (1 mmol, 1.66 mg) were combined to make diethyl(4-hydroxyphenyl) [(pyridine-4-yl)amino]methylphosphonate. The reaction was monitored using chromatography TLC (n-hexane/ethyl acetate) on Merck pre-coated GF254 silica plates. After that, the solid product was recrystallized in ethanol and dried at room temperature. The structure of the obtained α-aminophosphonate was confirmed by FT-IR, UV–Vis, 1H, 13C, and 31P NMR spectra (DHPAMP).
2.2. Computational details
2.2.1. Quantum chemical calculations
The Lee-Yang–Parr nonlocal correlation functional (RB3LYP) and Beck's three-parameter exchange functional (B3LYP) with the 6-31G (d, p) basis set were used in all calculations [35,36]. The Gaussian 09 program parcel [37] was used to complete the investigated structure's full optimization and vibrational study. Gauss View 5.0.8 computer program was used to visualize the results [38]. TD-SCF was used to forecast UV–Vis spectra using the B3LYP/6.31G-31G (d, p) level, while 1H and 13C chemical shift calculations were performed using the 6.31G(d,p) basis set in DMSO solution. The isotropic chemical shifts concerning tetramethylsilane were calculated using the isotropic shielding values (TMS). The dipole moment (µ), energy gap (ΔEgap), chemical hardness (η), chemical softness (σ), electronegativity (χ), and global electrophilicity index (ω) were estimated with the following formula [39], [40], [41].
| (1) |
| (2) |
| (3) |
| (4) |
| (5) |
2.2.2. Molecular docking
One of the most basic and essential strategies for drug development is molecular docking analysis [42], [43], [44]. It was carried out to determine the ligand's binding affinity for SARS-CoV-2 major protease (Mpro, also known as 3CLPro or chymotrypsin-like protease) and RNA dependent RNA polymerase (RdRp) [45]. Mpro and PLpro are proteases with the codes 6LU7 and 7BV2 [46] in the Protein Data Bank (PDB). Autodock software version 4.2.6 [47] was used to visualize the molecular docking process, including a built-in grid of 30 × 30 × 30 Ǻ3. The DHPAMP-RdRp and DHPAMP -RdRp complexes were visualized and depicted using Accelrys Discovery Studio [48].
3. Results and discussion
3.1. Experimental
3.1.1. IR spectra
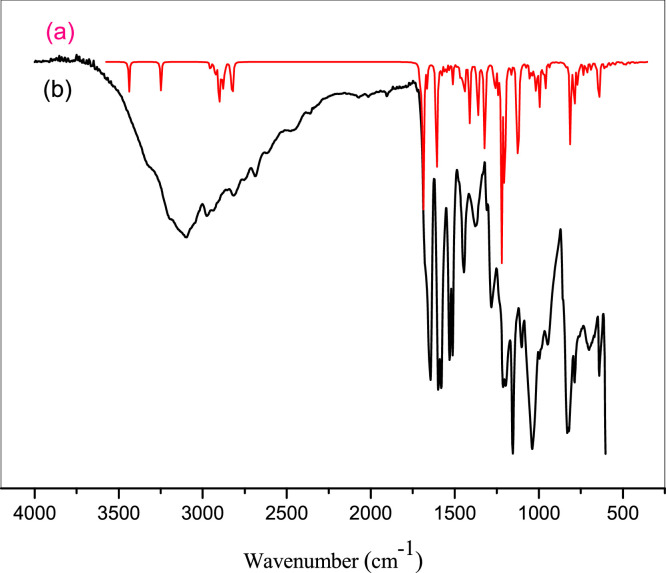
DHPAMP stands for Diethyl(4-hydroxyphenyl) [(pyridine-4-yl) amino] methyl phosphonate. Mol.Wt: 336. 322, Yield: 85% white solid, mp 90 °C. The vibrational analysis is an effective approach to locating vibrational modes linked with a compound's projected specific molecular structure. A non-linear molecule with N atoms has many potentially active observable fundamentals of three translational and three rotational degrees of freedom (3N-6). The vibrational bands of DHPAMP were allocated using the GaussView molecular visualization tool [38]. There are 126 common vibration modes for the DHPAMP molecule because it comprises 44 atoms. Fig. 2 shows the measured FT-IR spectra of DHPAMP beside the hypothesized infrared spectra. Table 1 summarizes the observed and estimated frequencies using DFT/B3LYP with the 6-31G (d, p) basis set and associated DHPAMP probable assignments. The functional group's unique peaks may be seen in the IR spectra of the α-aminophosphonate molecule: Positive evidence for the synthesis of functional groups is the elimination of the aldehyde's distinctive carbonyl peaks and the imprinting of the primary amine group on the produced molecule. νN–H (3253), νC–H(aliphatic) (2844–2999), ν(C = C), ν (C = N) aromatiques (1650–1543), ν (-P-CH2) (1381), νP= O (1215), ν (C — N) aliphatiques (1039), ν(C — C)aliphatiques (959), νP-O (764), νP–O –C (697). There is a good match between the experimental and computed frequencies (Fig. 2 ).
Fig. 1.
Synthesis procedure of DHPAMP.
Fig. 2.
(a) Calculated and (b) experimental FT-IR spectra of DHPAMP.
Table 1.
Comparison of the title compound's observed and calculated vibrational spectra.
| Experimental IR (cm−1) | Calculated(cm−1) B3LYP/6-31G(d, p) | Assignment |
|---|---|---|
| 3253 | 3526 | ν (N—H) |
| 2844–2999 | 3225–3039 | ν(C—H) arom; ν(C—H) aliph |
| 1650∼1543 | 1561–1659 | ν(C C), ν (C N) /aromatiques |
| 1381 | 1309 | ν (—P—CH2) |
| 1215 | 1211 | ν (P O) |
| 1039 | 1056 | ν (C—N) aliphatiques |
| 959 | 968 | ν(C—C)aliphatiques |
| 764 | 768 | ν(P—O) |
| 697 | 744 | ν (P—O—C) |
3.1.2. UV–vis analysis
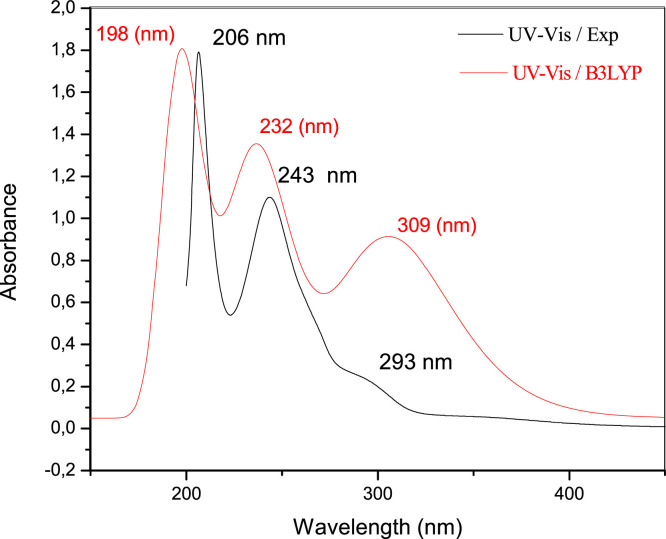
Fig. 3 shows the measured UV–vis spectra of DHPAMP and the hypothesized infrared spectra. They were carried out in methanol. The spectra of the synthesized product differ significantly from those of the starting products, with considerable displacements of the bands' maximum values and the emergence of the shoulders typical of α-aminophosphonate. DHPAMP's UV–Vis spectra are characterized by three absorption bands positioned at: A high-octane band based on λmax = 206 characterizes the molecule. In contrast, another band at λmax2 = 243 nm corresponds to the C = C group of the aromatic ring's π→π* transitions. The n→π* transitions associated with the presence of heteroatoms (O, N, and P) on the molecular structure of the examined molecule are allocated to the band that appeared λmax3 = 293 nm, while λmax1 = 198 nm, λmax2 = 232 nm, λmax3 = 309 nm are the calculated values by the (B3LYP/6-31G(d, p) technique, respectively. These findings show distinct discrepancies between the spectra of the acquired product and the spectra of the starting products, with good agreement between experimental and predicted values for the current molecule (Supplementary file).
Fig. 3.
DHPAMP's UV-visible spectrum.
3.1.3. NMR spectroscopy
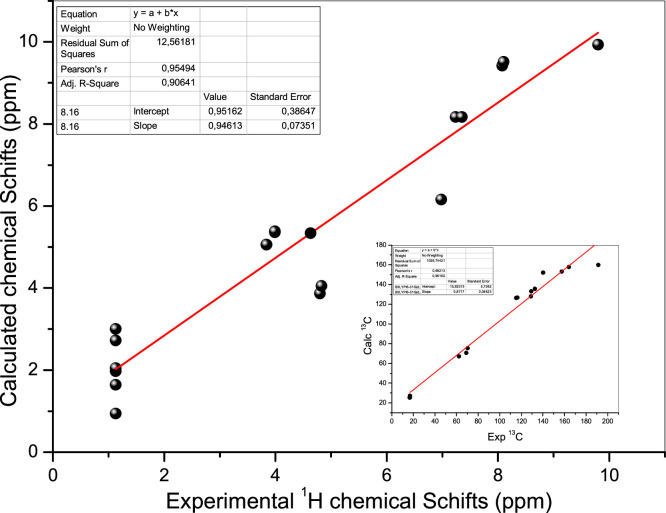
NMR spectroscopies of 1H, 13C, and 31P were also used to describe the molecule. The title compound's 1H, 13C, and 31P NMR spectra were recorded using TMS as an internal standard and DMSO as the solvent, following the numbering scheme indicated in (Supplementary file). The DFT (B3LYP) approach with the 6-31G (d, p) basis sets was used to produce GIAO 1H and 13C chemical shift values (concerning TMS), which were compared to experimental 1H and 13C chemical shift values. Table 2 summarizes the general conclusions of this calculation, along with experimental values. In addition, we present correlation graphs based on calculations in Fig. 4 to compare with experimental observations. As shown in Table 2 and Fig. 5 the experimental values are in good agreement with the DFT/B3LYP/6-31G (d,p) results.
Table 2.
Isotropic chemical shifts (ppm) for the title molecule, both experimental and theoretical (13C and 1H).
| Atom | Experimental | B3LYP/6-31G(d,p) |
|---|---|---|
| 1-C | 128.84 | 127.29 |
| 2-C | 129.14 | 128.20 |
| 3-C | 132.51 | 135.67 |
| 4-C | 129.20 | 133.14 |
| 5-C | 128.89 | 128.00 |
| 6-C | 191.30 | 159.87 |
| 11-C | 62.17 | 67.24 |
| 13-C | 163.91 | 157.64 |
| 14-C | 115.11 | 126.54 |
| 15-C | 116.34 | 126.79 |
| 16-C | 157.39 | 153.11 |
| 18-C | 140.22 | 152.19 |
| 28-C | 68.91 | 70.68 |
| 32- C | 16.68 | 25.38 |
| 37-C | 70.26 | 75.35 |
| 41- C | 16.79 | 27.34 |
| 7-H | 6.75 | 8.16 |
| 8-H | 7.75 | 6.17 |
| 9-H | 7.77 | 7.17 |
| 10-H | 6.98 | 6.16 |
| 12-H | 4.80 | 3.87 |
| 17-H | 7.24 | 8.17 |
| 19-H | 7.35 | 8.17 |
| 20-H | 8.08 | 9.42 |
| 21-H | 8.10 | 9.51 |
| 23-H | 4.83 | 4.05 |
| 26-H | 9.80 | 9.93 |
| 29-H | 3.84 | 5.06 |
| 33-H | 1.13 | 1.97 |
| 34-H | 1.13 | 1.64 |
| 35-H | 1.13 | 0.94 |
| 36-H | 3.84 | 5.06 |
| 38- H | 3.99 | 5.36 |
| 39- H | 3.99 | 5.38 |
| 42-H | 1.13 | 2.05 |
| 43-H | 1.13 | 2.72 |
| 43- H | 1.13 | 3.00 |
Fig. 4.
Experimental and theoretical NMR chemical shift values(ppm) of the DHPAMP.
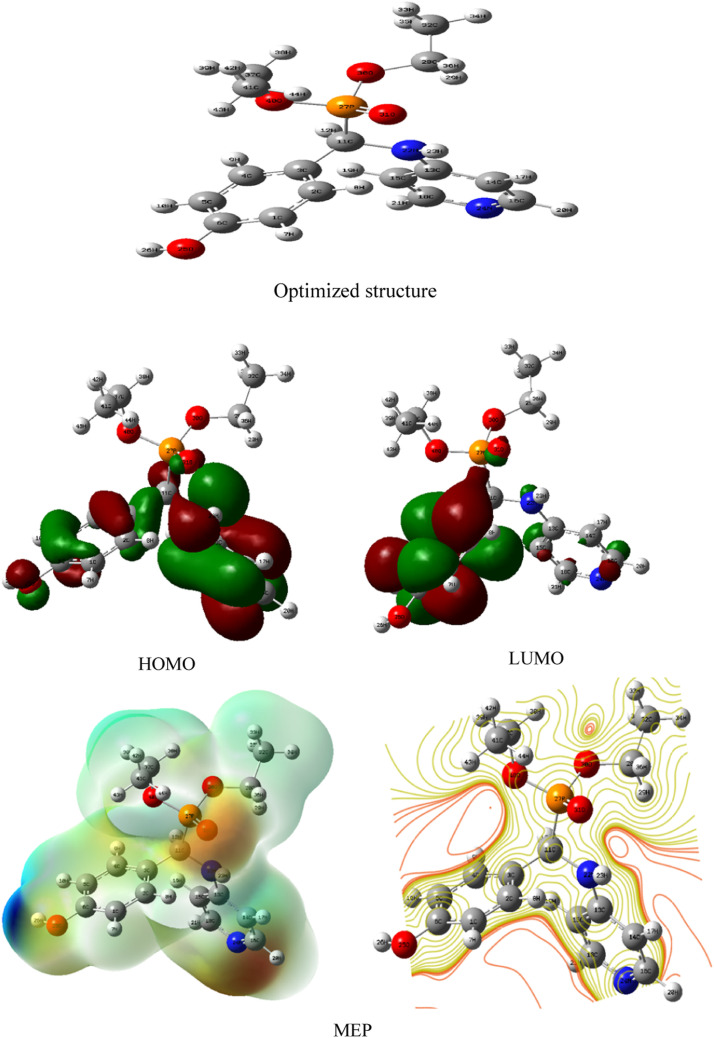
Fig. 5.
Optimized structure with atomic Mulliken charges, HOMO and LUMO frontierorbitals and MEP map of DHPAMP.
1H NMR (400 MHz, DMSO): δ (ppm): 1.13 ppm (3H, t, J = 7.0 Hz, —CH2 —CH3), 1.19 ppm (3H, t, J = 7.0 Hz, —CH2 —CH3), 3.84–3.99 ppm (m, 4H, —CH2 —CH3), 4.80 ppm (H, H—C-P), 4.83 ppm (H, -NH), 6.75 ppm (1H, arom) 6.77 ppm (1H, arom), 6.95 ppm (1H, arom), 6.98 ppm (1H, arom), 7.24 ppm (1H, arom), 7.35 ppm (1H, arom), 7.75 ppm (1H, arom), 7.77 ppm (1H, arom), 8.08 pm (1H, arom), 8.10 pm (1H, arom), 9.80 ppm (1H, OH). 13C NMR (400 MHz, DMSO): δ (ppm): 16. 68 ppm (d, J = 11.5 Hz, —CH2 —CH3),16.79 ppm (d, J = 11.8 Hz, —CH2 —CH3), 68.51- 70.26 ppm (2C, CH3 —CH2-), 62.17 ppm (1C, H—C-P), 115.11–116.34 ppm (2C, —CH arom), 128.84 ppm (1C, —CH arom), 128.89 ppm (1C, —CH arom), 129.14 ppm (1C, —CH arom), 129.20 ppm (1C, —CH arom), 132.51 ppm (1C, —CH arom), 140.22 ppm (1C, —CH arom), 157.39 ppm (1C, —CH(—CH arom), 163.91 ppm (1C, —CH(—CH arom), 191.30 ppm (1C, —CH(—CH arom). 31P NMR (400 MHz, CDCl3), δ (ppm): 22.31 ppm (C-PO (OC2H5)2).
3.2. Computational achievements
3.2.1. DFT calculations
Fig. 5 depicts the optimized structure and atom numbering of the examined molecule achieved using the B3LYP/6-31G technique. The calculated total energy (ETot), the dipole moment (µ), energy gap (ΔEgap), chemical hardness (η), chemical softness (σ), electronegativity (χ), and global electrophilicity index (ω) are all shown in Table 3 .
Table 3.
Quantum chemical parameters of DHPAMP calculated using DFT.
| Quantum chemical parameters | Values |
|---|---|
| Total energy Etot (eV) | -37,393.74736 |
| EHOMO (eV) | −5.89970 |
| ELUMO (eV) | −0.26122 |
| ΔEgap(ELUMO-EHOMO) (eV) | 5.63848 |
| Dipôle moment (µ) (debye) | 4.8495 |
| Hardness (η) | 2.81924 |
| Softness (σ) | 0.45612 |
| Electronegativity (χ) | 3.08046 |
| Electrophilicity (ω) | 1.68294 |
Frontier Molecular Orbitals (FMO) refers to two molecular orbital frontiers known as HOMO (Highest Occupied Molecular Orbital) and LUMO (Lowest Unoccupied Molecular Orbital), or LUMO is directly related to electron affinity. In contrast, HOMO is directly related to ionization potential [48,49]. These orbitals aid in understanding the molecule's chemical stability and reactivity [50,51]. Furthermore, the estimated value of ΔEgap shows the energy required to excite a molecule's electrons. If the HOMO-LUMO energy gap is small, molecules can be unstable and readily excited, whereas if the ΔEgap is big, molecules can be very stable and chemically reactive [52]. The B3LYP/6-31G (d, p) approach was used to calculate the FMOs in electronic transitions and their energies and their orbital energy gap (ΔEgap) to predict the energy characteristics and reactivity of the generated compound against the COVID-19 virus. EHOMO and ELUMO have −5.89970 eV and −0.26122 eV, respectively, and the energy separation between the HOMO and the LUMO has a value of 5.63848 eV for the most stable conformer.
The polarity of pharmacological molecules is denoted by the dipole moment (μ), which is a three-dimensional vector and is connected with the fractional electric charge distribution in these compounds [53]. As a result, its significance is demonstrated in reaction mechanisms and relates to molecules' ability to interact with other molecular species. For example, the dipole moment for the molecule DHPAMP is 4.3301 Debye, which indicates that it has a high ability to interact with the environment.
Furthermore, the hardness(η) and local softness (σ) of drug molecules can be used to assess their resistance to deformation of their electron clouds or polarization [54]. The examined compound's hardness and local softness were computed and determined to be 2.81924 and 0.45612 eV, respectively. Electronegativity (χ) is a metric that compares an atom's attraction to bonding electrons in molecules to that of other atoms. The molecule's electronegativity is calculated and found to be 3.08046 eV, showing that the ligand is an electronegative species. The electrophilicity index(ω) is used to describe a molecule's biological activity [55], with higher values indicating stronger electrophilicity and lower values indicating a weak electrophile [56]. We can divide organic molecules into three categories based on their electrophilicity values: marginal electrophiles with a value of less than 0.8 eV, moderate electrophiles between 0.8 and 1.5 eV, and strong electrophiles with a value greater than 1.5 eV [57]. The value of 1.68294 eV for the researched ligand indicates that it is a good electrophile capable of receiving an electron doublet to form bonds with another reagent, which must be a nucleophile.
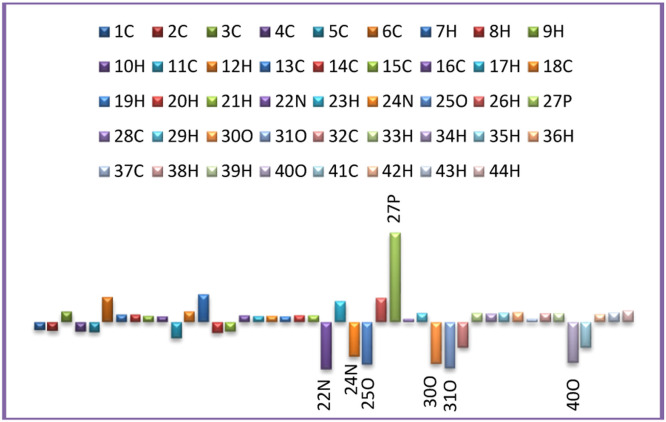
Because atomic charges affect dipole moment, molecular polarizability, electronic structure, and many other aspects of molecular systems, while Mulliken atomic charge calculation is a crucial parameter for molecular systems, medicinal compounds' atomic Mulliken charges can be utilized to prove their adsorptive locations [58]. The Mulliken atomic charges of the estimated compound were calculated using DFT with B3LYP 6-311G (d,p) as a basis set and are listed in Table 4 and are plotted in Fig. 6 . According to the data, the oxygen and nitrogen atoms have the most significant negative charges because of molecular relaxation. However, the hydrogen atoms also cover the positive charges. The majority of negative charges are concentrated on the atoms N22, N24, O25, O30, O31 and O40, which are thought to be the active region of adsorption [59]. Furthermore, most of the positive charges are carried by the P27 atom.
Table 4.
Mulliken atomic charges of the DHPAMP calculated by DFT at B3LYP/6-31G (d,p).
| Symbol | MM Charge | Symbol | MM Charge |
|---|---|---|---|
| 1C | −0.10 | 23H | 0.27 |
| 2C | −0.11 | 24N | −0.45 |
| 3C | 0.14 | 25O | −0.55 |
| 4C | −0.12 | 26H | 0.31 |
| 5C | −0.13 | 27P | 1.17 |
| 6C | 0.32 | 28C | 0.04 |
| 7H | 0.10 | 29H | 0.12 |
| 8H | 0.09 | 30O | −0.54 |
| 9H | 0.08 | 31O | −0.60 |
| 10H | 0.07 | 32C | −0.33 |
| 11C | −0.21 | 33H | 0.11 |
| 12H | 0.14 | 34H | 0.11 |
| 13C | 0.36 | 35H | 0.12 |
| 14C | −0.14 | 36H | 0.13 |
| 15C | −0.12 | 37C | 0.04 |
| 16C | 0.08 | 38H | 0.11 |
| 17H | 0.07 | 39H | 0.11 |
| 18C | 0.08 | 40O | −0.53 |
| 19H | 0.07 | 41C | −0.33 |
| 20H | 0.09 | 42H | 0.10 |
| 21H | 0.09 | 43H | 0.12 |
| 22N | −0.61 | 44H | 0.15 |
Fig. 6.
Mulliken charges charge distribution of DHPAMP compound.
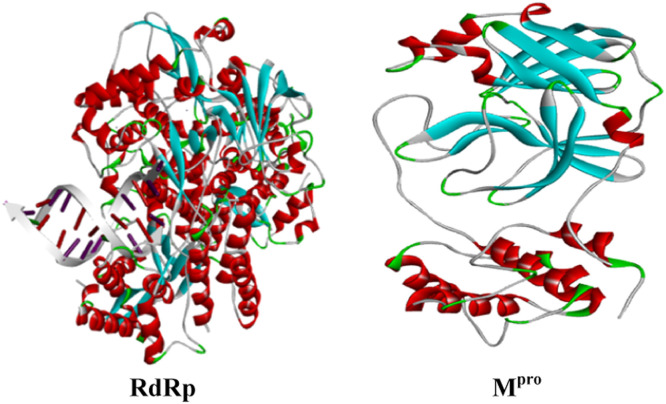
3.2.2. Structure and preparation of MPro for docking studies
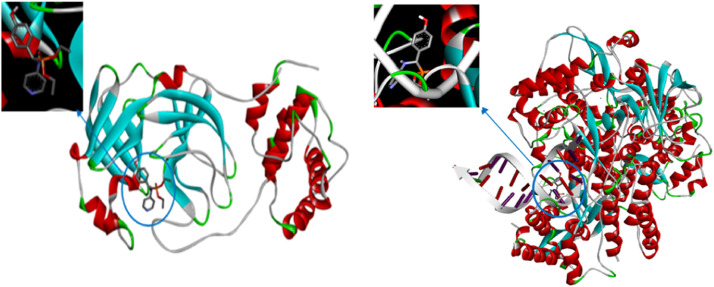
Because of its role in processing translated polyproteins, Mpro was chosen as a docking study target. In contrast, RdRp was chosen because of its significance in viral genome replication and transcription. The DHPAMP ligand was docked with receptors to determine the correct conformation of DHPAMP in the receptor and the secondary forces that result from the ligand and the receptor's active amino acids. The molecular docking of the DHPAMP ligand with the Mpro and RdRp receptors was used to determine the proper conformation of the ligand in the receptor and the secondary forces that occur from DHPAMP binding to the receptor's active amino acids. As a result, novel medication designs are developed. The non-covalent bonds, π-π* and π-σ interactions between the active amino acids of the Mpro and RdRp target receptors and the DHPAMP ligand were assessed based on the minimal binding energy. Fig. 7 depict Mpro and RdRp crystal structures, respectively, while Fig. 8 depicts the crystal structures of the best docked Mpro- DHPAMP and RdRp- DHPAMP complexes. The ligand binds to RdRp stronger than to the Main Protease, and it prefers to bind in the outer structure of Mpro, whereas it prefers to bind in the inner pocket of RdRp, as seen in Fig. 8. According to the docking studies, the ligand showed great potential for binding to the active site of COVID-19 Mpro, and RdRp could be promising therapeutic agents in COVID-19 infections.
Fig. 7.
Crystal structures of Mpro and RdRp.
Fig. 8.
Best docked model visualization with Mpro-DHPAMP and RdRp-DHPAMP complexes.
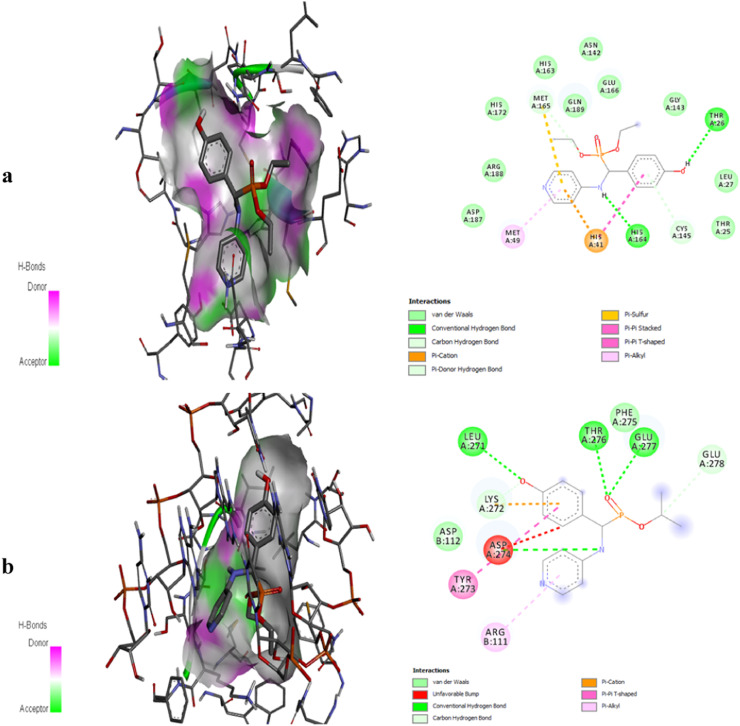
With binding affinities of −6.40 kcal/mol and −8.40, respectively, the docking of DHPAMP with SARS-CoV-2 Mpro and RdRp demonstrated significant interactions in the central pocket. For Mpro, DHPAMP created van der Waals forces with the amino acids HIS41, MET49, HIS164, MET165, GLU166 and GLN189, as well as hydrogen bonds with the amino acids SER144, HIS163, and GLU166 (Fig. 9a). While for RdRp, it created van der Waals forces with LYS272, TYR273, ASP274, PHE275, and GLU277 and hydrogen bonds with LEU271, LYS272, ASP274, THR276 and GLU277 (Fig. 9b). The lowest binding energy in molecular interactions is shown by comparing the estimated binding energies of DHPAMP and other medicines (Table 5 ). lowest bending energy in interactions with Mpro and RdRp, suggesting that it has a higher affinity for binding and that the complexes produced by Mpro-DHPAMP and RdRp-DHPAMP are more stable than those produced by the other compounds.
Fig. 9.
2D, 3D Binding-interaction diagrams of DHPAMP with SARS-CoV-2 main protease (a) and RNA dependent RNA polymerase (b).
Table 5.
Molecular docking results of DHPAMP and some drugs with Mpro and RdRp.
4. Conclusion
A novel α -aminophosphonate derivative was produced and characterized using one-pot three-component processes. The obtained chemical is a stable solid with high melting temperature, and its structure was identified using the IR, UV–Vis, 1H, 13C, and 31P NMR spectra. DFT method and molecular docking calculations were used to study derivative of α-aminophosphonate. Quantum chemical computations (DFT) B3LYP/6-31G (d, p) were used to optimize the molecular structures of this derivative, and their geometrical parameters were identified. Additionally, molecular docking research was carried out to evaluate the synthesized ligand's affinity for binding to the main protease (Mpro) of the SARS-CoV-2 and the RNA dependent RNA polymerase (RdRp). The obtained EHOMO, ELUMO, and ΔEgap values show that the ligand is a reactive and unstable species. The predicted MEP maps show potential nucleophilic and electrophilic attack sites, and the Mulliken charges show that there may be active interaction sites as well. The investigated ligand's interactions with Mpro and RdRp showed that it can bind to several bonding contacts. The derivative can be effective in treating this condition since, according to the synthesis, computational analysis, and in silico docking data, it is a potent COVID-19 viral inhibitor.
Authors' contributions
Rachida Kerkour (Doctor): Writing, review and editing, Conceptualization, Resources and Investigation, Theoretical studies and Review.
Nadjib Chafai (Doctor): Resources and Investigation, Theoretical studies.
Ouahiba Moumeni (Doctor): Supervision.
Salah Chafaa (Professor): Project management, Funding acquisition.
Declaration of Competing Interest
The authors declare no conflict of interest
Acknowledgments
Certificate of Originality
The work reported in the paper (Novel α-aminophosphonate derivates synthesis, theoretical calculation, Molecular docking, and in silico prediction of potential inhibition of SARS-CoV-2) submitted for publication in journal of molecular structure is original and has not been submitted for publication elsewhere,
All authors have seen and approved this manuscript and consent to having their name in the list of authors, thereby taking responsibility for the results contained in the manuscript.
Proper citations to the previously reported works have been given.
Funding
This research was supported by the General Directorate for Scientific Research and Technological Development (DGRSDT), Algerian Ministry of Scientific Research, Laboratory of Electrochemistry of Molecular Materials and Complex (LEMMC), Ferhat ABBAS University of Sétif.
Availability of data and material
Not applicable.
Code availability
Not applicable.
Acknowledgements
This research was supported by Algerian Ministry of Scientific Research, Laboratory of Electrochemistry of Molecular Materials and Complex (LEMMC), Ferhat ABBAS University of Sétif.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.molstruc.2022.134196.
Appendix. Supplementary materials
Data availability
Data will be made available on request.
References
- 1.Yang X., Yu Y., Xu J., Shu H., Liu H., Wu Y., Shang Y. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respirat. Med. 2020;8(5):475–481. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jackson J.K. Global economic effects of COVID-19. Congression. Res. Serv. 2021 [Google Scholar]
- 3.Ozkendir O.M., Askar M., Kocer N.E. Influence of the epidemic COVID-19: an outlook on health, Business and Scientific Studies. Lab-in-Silico. 2020;1(1):26–30. doi: 10.22034/labinsilico20011026. [DOI] [Google Scholar]
- 4.Thorlund K., Dron L., Park J., Hsu G., Forrest J.I., Mills E.J. A real-time dashboard of clinical trials for COVID-19. The Lancet Digit. Health. 2020;2(6):e286–e287. doi: 10.1016/S2589-7500(20)30086-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tai W., He L., Zhang X., Pu J., Voronin D., Jiang S., Du L. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell. Mol. Immunol. 2020;17(6):613–620. doi: 10.1038/s41423-020-0400-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30(3):269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Wit E., Feldmann F., Cronin J., Jordan R., Okumura A., Thomas T., Feldmann H. Prophylactic and therapeutic remdesivir (GS-5734) treatment in the rhesus macaque model of MERS-CoV infection. Proc. Natl. Acad. Sci. 2020;117(12):6771–6776. doi: 10.1073/pnas.1922083117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bukhari M.H., Mahmood K., Zahra S.A. Over view for the truth of COVID-19 pandemic: a guide for the Pathologists, Health care workers and community. Pak. J. Med. Sci. 2020;36:S111. doi: 10.12669/pjms.36.COVID19-S4.2519. COVID19-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Colson P., Rolain J.M., Raoult D. Chloroquine for the 2019 novel coronavirus SARS-CoV-2. Int. J. Antimicrob. Agents. 2020;55(3) doi: 10.1016/j.ijantimicag.2020.105923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gautret P., Lagier J.C., Parola P., Meddeb L., Mailhe M., Doudier B., Raoult D. Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial. Int. J. Antimicrob. Agents. 2020;56(1) doi: 10.1016/j.ijantimicag.2020.105949. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11.Khuroo M.S. Chloroquine and hydroxychloroquine in coronavirus disease 2019 (COVID-19). Facts, fiction and the hype: a critical appraisal. Int. J. Antimicrob. Agents. 2020;56(3) doi: 10.1016/j.ijantimicag.2020.106101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park S.J., Yu K.M., Kim Y.I., Kim S.M., Kim E.H., Kim S.G., Choi Y.K. Antiviral efficacies of FDA-approved drugs against SARS-CoV-2 infection in ferrets. MBio. 2020;11(3):e01114–e01120. doi: 10.1128/mBio.01114-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao J., Tian Z., Yang X. Breakthrough: chloroquine phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies. Biosci. Trends. 2020 doi: 10.5582/bst.2020.01047. [DOI] [PubMed] [Google Scholar]
- 14.Liu, C., Zhou, Q., Li, Y., Garner, L.V., Watkins, S.P., Carter, L.J., Albaiu, D. (2020). Research and development on therapeutic agents and vaccines for COVID-19 and related human coronavirus diseases. [DOI] [PMC free article] [PubMed]
- 15.Qi H., Hu Z., Yang Z., Zhang J., Wu J.J., Cheng C., Zheng L. Capacitive aptasensor coupled with microfluidic enrichment for real-time detection of trace SARS-CoV-2 nucleocapsid protein. Analytical chemistry. in order to model drug treatments and identify ideal delivery methods. 2022 doi: 10.1021/acs.analchem.1c04296. [DOI] [PubMed] [Google Scholar]
- 16.Zha T.H., Castillo O., Jahanshahi H., Yusuf A., Alassafi M.O., Alsaadi F.E., Chu Y.M. A fuzzy-based strategy to suppress the novel coronavirus (2019-NCOV) massive outbreak. Appl. Comput. Math. 2021:160–176. [Google Scholar]
- 17.Swargiary A., Wary N., Das T., Roy M.K., Boro H. Covid-19 and Fungal Co-Infection: a Literature Review with Special Reference to India. Adv. J. Sci. Eng. 2022;3(1):72–80. doi: 10.22034/advjse22031072. [DOI] [Google Scholar]
- 18.Behzadi H., Roonasi P., van der Spoel D., Manzetti S. Relationship between electronic properties and drug activity of seven quinoxaline compounds: a DFT study. J. Mol. Struct. 2015;1091:196–202. [Google Scholar]
- 19.CHAFAI N., Benbouguerra K., Chafaa S. Quantum chemical study of Hydroxychloroquine and Chloroquine drugs used as a treatment of COVID-19. Iran. J. Chem. Chem. Eng. 2020 [Google Scholar]
- 20.Benbouguerra K., Chafai N., Chafaa S., Touahria Y.I., Tlidjane H. New α-hydrazinophosphonic acid: synthesis, characterization, DFT study and in silico prediction of its potential inhibition of SARS-CoV-2 main protease. J. Mol. Struct. 2021;1239 doi: 10.1016/j.molstruc.2021.130480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kafarski P., Lejczak B. In: Synthesis of Phosphono-And Phosphinopeptides. Kukhar V.P., Hudson HR, editors. 2000. Aminophosphonic and aminophosphinic acids. chemistry and biological activity; pp. 173–205. [Google Scholar]
- 22.Palacios F., Gil M.J., de Marigorta E.M., Rodríguez M. Synthesis of 3-phosphorylated 2-aza-1, 3-dienes from imines derived from bisphosphonates. Tetrahedron Lett. 1999;40(12):2411–2414. [Google Scholar]
- 23.Oleksyszyn J., Powers J.C. Irreversible inhibition of serine proteases by peptide derivatives of (. alpha.-aminoalkyl) phosphonate diphenyl esters. Biochemistry. 1991;30(2):485–493. doi: 10.1021/bi00216a026. [DOI] [PubMed] [Google Scholar]
- 24.Green D., Patel G., Elgendy S., Baban J.A., Claeson G., Kakkar V.V., Deadman J. The facile synthesis of O, O-dialkyl 1-aminoalkanephosphonates. Tetrahedron Lett. 1993;34(43):6917–6920. [Google Scholar]
- 25.Atherton F.R., Hassall C.H., Lambert R.W. Synthesis and structure-activity relationships of antibacterial phosphonopeptides incorporating (1-aminoethyl) phosphonic acid and (aminomethyl) phosphonic acid. J. Med. Chem. 1986;29(1):29–40. doi: 10.1021/jm00151a005. [DOI] [PubMed] [Google Scholar]
- 26.Zomova A.M., Molodykh Z.V., Kudryavtseva L.A., Teplyakova L.V., Fedorov S.B., Ivanov B.E. Antimicrobial activity of O, O-diethyl N-alkylaminomethyl-phosphonates and O-ethyl N-alkylaminomethylphosphonic acids. Pharm. Chem. J. 1986;20(11):774–777. [Google Scholar]
- 27.Yang S., Gao X.W., Diao C.L., Song B.A., Jin L.H., Xu G.F., Lu P. Synthesis and antifungal activity of novel chiral α-aminophosphonates containing fluorine moiety. Chin. J. Chem. 2006;24(11):1581–1588. [Google Scholar]
- 28.Kleszczynska H., Sarapuk J. New aminophosphonates with antioxidative activity. Cell. Mol. Biol. Lett. 2001;6(1):83–92. [PubMed] [Google Scholar]
- 29.Ahn D.G., Choi J.K., Taylor D.R., Oh J.W. Biochemical characterization of a recombinant SARS coronavirus nsp12 RNA-dependent RNA polymerase capable of copying viral RNA templates. Arch. Virol. 2012;157(11):2095–2104. doi: 10.1007/s00705-012-1404-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Te Velthuis A.J., Arnold J.J., Cameron C.E., Van Den Worm S.H., Snijder E.J. The RNA polymerase activity of SARS-coronavirus nsp12 is primer dependent. Nucleic. Acids. Res. 2010;38(1):203–214. doi: 10.1093/nar/gkp904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu X., Liu Y., Weiss S., Arnold E., Sarafianos S.G., Ding J. Molecular model of SARS coronavirus polymerase: implications for biochemical functions and drug design. Nucleic. Acids. Res. 2003;31(24):7117–7130. doi: 10.1093/nar/gkg916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anand K., Palm G.J., Mesters J.R., Siddell S.G., Ziebuhr J., Hilgenfeld R. Structure of coronavirus main proteinase reveals combination of a chymotrypsin fold with an extra α-helical domain. EMBO J. 2002;21(13):3213–3224. doi: 10.1093/emboj/cdf327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang H., Yang M., Ding Y., Liu Y., Lou Z., Zhou Z., Rao Z. The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proc. Natl. Acad. Sci. 2003;100(23):13190–13195. doi: 10.1073/pnas.1835675100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fields E.K. The synthesis of esters of substituted amino phosphonic acids1a. J. Am. Chem. Soc. 1952;74(6):1528–1531. [Google Scholar]
- 35.Friesner R.A., Beachy M.D. Quantum mechanical calculations on biological systems. Curr. Opin. Struct. Biol. 1998;8(2):257–262. doi: 10.1016/s0959-440x(98)80048-1. [DOI] [PubMed] [Google Scholar]
- 36.Lee C., Yang W., Parr R.G. Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys. Rev. B. 1988;37(2):785. doi: 10.1103/physrevb.37.785. [DOI] [PubMed] [Google Scholar]
- 37.Frisch M.J., Trucks G.W., Schlegel H.B., Scuseria G.E., Robb M.A., Cheeseman J.R., Scalmani G., Barone V., Mennucci B., Petersson G.A., Nakatsuji H., Caricato M., Li X., Hratchian H.P., Izmaylov A.F., Bloino J., Zheng G., Sonnenberg J.L., Hada M., Ehara M., Toyota K., Fukuda R., Hasegawa J., Ishida M., Nakajima T., Honda Y., Kitao O., Nakai H., Vreven T., Montgomery J.A., Jr, Peralta J.E., Ogliaro F., Bearpark M., Heyd J.J., Brothers E., Kudin K.N., Staroverov V.N., Kobayashi R., Normand J., Raghavachari K., Rendell A., Burant J.C., Iyengar S.S., Tomasi J., Cossi M., Rega N., Millam J.M., Klene M., Knox J.E., Cross J.B., Bakken V., Adamo C., Jaramillo J., Gomperts R., Stratmann R.E., Yazyev O., Austin A.J., Cammi R., Pomelli C., Ochterski J.W., Martin R.L., Morokuma K., Zakrzewski V.G., Voth G.A., Salvador P., Dannenberg J.J., Dapprich S., Daniels A.D., Farkas O., Foresman J.B., Ortiz J.V., Cioslowski J., Fox D.J. Gaussian, Inc.; Wallingford, CT: 2009. Gaussian 09, Revision A.02. [Google Scholar]
- 38.GaussView V.5., Dennington R., Keith T., Millam J. Semichem Inc. 2009 [Google Scholar]
- 39.Kohn W., Sham L.J. Quantum density oscillations in an inhomogeneous electron gas. Phys. Rev. 1965;137(6A):A1697. [Google Scholar]
- 40.Feng L., Yang H., Cui X., Chen D., Li G. Experimental and theoretical investigation on corrosion inhibitive properties of steel rebar by a newly designed environmentally friendly inhibitor formula. RSC Adv. 2018;8(12):6507–6518. doi: 10.1039/c7ra13045g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rogova A.V., Gerasimova M.A., Tomilin F.N., Slyusareva E.A. XV International Conference On Pulsed Lasers and Laser Applications. SPIE; 2021. Quantum chemical study of the spectral characteristics of fluorescein dyes bound to chitosan; pp. 409–414. Vol. 12086. [Google Scholar]
- 42.Mirzaei Mirzaei M., Harismah K., Soleimani M., Mousavi S. Inhibitory effects of curcumin on aldose reductase and cyclooxygenase-2 enzymes. J. Biomol. Struct. Dyn. 2021;39(17):6424–6643. doi: 10.1080/07391102.2020.1800513. [DOI] [PubMed] [Google Scholar]
- 43.Mirzaei M., Nazemi H. In silico interactions between curcumin derivatives and monoamine oxidase-a enzyme. Biointerface Res. Appl. Chem. 2022;12:3752–3761. [Google Scholar]
- 44.Ashjaee Y., Zandi H. Molecular analysis of 5-COR derivatives of uracil and evaluating their affinity against the MPro target of COVID-19. Adv. J. Sci. Eng. 2021;2(2):79–85. [Google Scholar]
- 45.Shu B., Gong P. The uncoupling of catalysis and translocation in the viral RNA-dependent RNA polymerase. RNA Biol. 2017;14(10):1314–1319. doi: 10.1080/15476286.2017.1300221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sanders J.M., Monogue M.L., Jodlowski T.Z., Cutrell J.B. Pharmacologic treatments for coronavirus disease 2019 (COVID-19): a review. JAMA. 2020;323(18):1824–1836. doi: 10.1001/jama.2020.6019. [DOI] [PubMed] [Google Scholar]
- 47.Cosconati S., Forli S., Perryman A.L., Harris R., Goodsell D.S., Olson A.J. Virtual screening with AutoDock: theory and practice. Expert Opin. Drug Discov. 2010;5(6):597–607. doi: 10.1517/17460441.2010.484460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ribavirin Elfiky AA, Remdesivir Sofosbuvir. Galidesivir, and Tenofovir against SARS-CoV-2 RNA dependent RNA polymerase (RdRp): a molecular docking study. Life Sci. 2020;253 doi: 10.1016/j.lfs.2020.117592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xavier S., Periandy S. Spectroscopic (FT-IR, FT-Raman, UV and NMR) investigation on 1-phenyl-2-nitropropene by quantum computational calculations. Spectrochim. Acta Part A. 2015;149:216–230. doi: 10.1016/j.saa.2015.04.055. [DOI] [PubMed] [Google Scholar]
- 50.Abraham C.S., Prasana J.C., Muthu S. Quantum mechanical, spectroscopic and docking studies of 2-amino-3-bromo-5-nitropyridine by density functional method. Spectrochim. Acta Part A. 2017;181:153–163. doi: 10.1016/j.saa.2017.03.045. [DOI] [PubMed] [Google Scholar]
- 51.Asiri A.M., Karabacak M., Kurt M., Alamry K.A. Synthesis, molecular conformation, vibrational and electronic transition, isometric chemical shift, polarizability and hyperpolarizability analysis of 3-(4-Methoxy-phenyl)-2-(4-nitro-phenyl)-acrylonitrile: a combined experimental and theoretical analysis. Spectrochim. Acta Part A. 2011;82(1):444–455. doi: 10.1016/j.saa.2011.07.076. [DOI] [PubMed] [Google Scholar]
- 52.Kosar B., Albayrak C. Spectroscopic investigations and quantum chemical computational study of (E)-4-methoxy-2- [(p-tolylimino) methyl] phenol. Spectrochim. Acta Part A. 2011;78(1):160–167. doi: 10.1016/j.saa.2010.09.016. [DOI] [PubMed] [Google Scholar]
- 53.Fliszár S. Springer Science & Business Media; 2012 Dec 6. Charge Distributions and Chemical effects: a New Approach to the Electronic Structure and Energy of Molecules. [Google Scholar]
- 54.Raafat M.I., Mohamed K.A., Faten M.A. Quantum chemical studies on the inhibition of corrosion of copper surface by substituted uracils. Appl. Surf. Sci. 2008;255:2433–2441. [Google Scholar]
- 55.Hadda T.B., Berredjem M., Almalki F.A., Rastija V., Jamalis J., Emran T.B., Alqahtani A.M. How to face COVID-19: proposed treatments based on remdesivir and hydroxychloroquine in the presence of zinc sulfate. Docking/DFT/POM structural analysis. J. Biomol. Struct. Dyn. 2021:1. doi: 10.1080/07391102.2021.1930161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Noureddine O., Issaoui N., Al-Dossary O. DFT and molecular docking study of chloroquine derivatives as antiviral to coronavirus COVID-19. J. King Saud Univ. 2021;33(1) doi: 10.1016/j.jksus.2020.101248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Domingo L.R., Aurell M.J., Pérez P., Contreras R. Quantitative characterization of the global electrophilicity power of common diene/dienophile pairs in Diels–Alder reactions. Tetrahedron. 2002;58(22):4417–4423. [Google Scholar]
- 58.Kory P., Meduri G.U., Varon J., Iglesias J., Marik P.E. Review of the emerging evidence demonstrating the efficacy of ivermectin in the prophylaxis and treatment of COVID-19. Am. J. Ther. 2021 May;28(3):e299. doi: 10.1097/MJT.0000000000001377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Govindarajan M., Karabacak M., Periandy S., Tanuja D. Spectroscopic (FT-IR, FT-Raman, UV and NMR) investigation and NLO, HOMO–LUMO, nbo analysis of organic 2, 4, 5-trichloroaniline. Spectrochim. Acta Part A. 2012;97:231–245. doi: 10.1016/j.saa.2012.06.014. [DOI] [PubMed] [Google Scholar]
- 60.Vijayaraj R., Subramanian V., Chattaraj P.K. Comparison of global reactivity descriptors calculated using various density functionals: a QSAR perspective. J. Chem. Theory Comput. 2009;5(10):2744–2753. doi: 10.1021/ct900347f. [DOI] [PubMed] [Google Scholar]
- 61.Dewi Yuli Kusuma, Baiq Amelia Riyandari. Potensi Tanaman Lokal sebagai Tanaman Obat dalam Menghambat Penyebaran COVID-19. J. Pharmasci. 2020;7(2):112–128. [Google Scholar]
- 62.Hagar M., Ahmed H.A., Aljohani G., Alhaddad O.A. Investigation of some antiviral N-heterocycles as COVID 19 drug: molecular docking and DFT calculations. Int. J. Mol. Sci. 2020;21(11):3922. doi: 10.3390/ijms21113922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.EA R. Remdesivir, sofosbuvir, galidesivir, and tenofovir against SARS-CoV-2 RNA dependent RNA polymerase (RdRp): a molecular docking study. Life Sci. 2020 doi: 10.1016/j.lfs.2020.117592. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.
Not applicable.