Abstract
The direct interaction of the Escherichia coli cytotoxin RelE with its specific antidote, RelB, was demonstrated in two ways: (i) copurification of the two proteins and (ii) a positive yeast two-hybrid assay involving the relB and relE genes. In addition, the purified RelE protein exhibited ribosome-binding activity in an in vitro assay, supporting previous observations suggesting that it is an inhibitor of translation.
In Escherichia coli, a rapid accumulation of guanosine 5′-triphosphate 3-diphosphate (pppGpp) and guanosine 3′,5′-bispyrophosphate (ppGpp) occurs in response to amino acid deprivation (see reference 4 for a review). These nucleotides, collectively designated (p)ppGpp, are synthesized by a ribosome-associated enzyme, encoded by the relA gene, that is activated by amino acid deprivation. The accumulation of (p)ppGpp coincides with a global reorganization of metabolic activities known as the stringent response, which seems to be designed to promote survival of the starved bacteria. Of the many changes associated with the stringent response, the abrupt inhibition of stable RNA synthesis is perhaps the most widely studied. Strains carrying mutations in relA do not accumulate (p)ppGpp during amino acid deprivation. They exhibit what has been termed a relaxed phenotype characterized by the continued accumulation of stable RNA during starvation.
Mutations in a second gene, relB, give rise to a delayed relaxed phenotype (5, 12). Stable RNA synthesis is initially inhibited in amino acid-deprived relB mutants. However, RNA synthesis resumes about 10 min after the onset of starvation. Another characteristic of relB mutants is the unusually slow recovery from periods of starvation (5, 12–14). This lag has been attributed to a growth inhibitor that accumulates during starvation and that is thought to be a protein that inhibits translation (5, 12).
The relB gene forms an operon with two other genes, relE and relF (2). The relF gene encodes a protein that causes a rapid inhibition of growth associated with an arrest of respiration and a collapse of membrane potential (8). RelE and RelB constitute an example of a bacterial toxin-antidote system (7, 9). The overexpression of RelE results in the inhibition of bacterial growth. The coexpression of RelB neutralizes RelE toxicity. RelB also acts as a transcriptional repressor of the relBEF operon, and RelE exhibits corepressor activity. Although no direct evidence has been presented, these observations suggest that RelB directly interacts with RelE. Moreover, it appears that RelE is the growth inhibitor that accumulates during starvation of relB mutants (5, 12).
Yeast two-hybrid analysis.
The yeast two-hybrid system was employed to confirm the interaction between RelB and RelE. The Matchmaker two-hybrid system 3 (Clontech) was used for this purpose, and all protocols for the analysis were provided by Clontech. The procedures for plasmid and genomic DNA purification, restriction endonuclease digestion, DNA ligation, and PCR amplification were those described by Sambrook et al. (16). All enzymes were purchased from New England BioLabs, Inc. A 4.1-kb HindIII fragment containing the relBEF operon was first subcloned from Kohara clone 308 (11) into the low-copy-number vector pWKS30 (17), to create plasmid pJT1. The relE gene was then amplified by PCR from plasmid pJT1 using oligonucleotides 5′RelEGBK (5′GATGAAC TCATATGGCGTATTT3′) and 3′ RelEGBK (5′TGCTTTGGCTGCAGGAATGCGT3′) as primers. An NdeI site was incorporated into 5′RelEGBK and a PstI site was incorporated into 3′RelEGBK to accommodate the fusion of relE to the GAL4 DNA-binding domain in plasmid pGBKT7 (Clontech). This new construct was designated pGBKT7-E. The relB gene from plasmid pJT1 was PCR amplified using oligonucleotides 5′RelBGAD (5′AGGTGTAACATATGGGTAGCAT3′) and 3′RelBGAD (5′AATACGCCCTCGAGGTTCATCC3′) as primers. An NdeI site was incorporated into 5′RelBGAD and an XhoI site was incorporated into 3′RelBGAD to facilitate the fusion of relB to the GAL4 activation domain in the vector pGADT7 (Clontech). This recombinant plasmid was designated pGADT7-B.
The two-hybrid plasmids were analyzed in Saccharomyces cerevisiae strain AH109. AH109 carries a chromosomal lacZ reporter gene fused to the MEL1 upstream activator sequence and promoter, and this permits the use of a blue-white screen to detect two-hybrid protein interactions. Plasmids pGADT7-B and pGBKT7-E were transformed, either separately or together, into AH109, and the specific activities of β-galactosidase were quantified in the transformants in three independent experiments. The β-galactosidase specific activity of the transformant carrying pGADT7-B and pGBKT7-E was 16.6 ± 2.4 Miller units. A positive control, provided by Clontech, consisting of plasmids pGADT7-T, which encodes an activation domain–simian virus 40 large T-antigen fusion, and pGBKT7-53, which encodes a DNA-binding domain–murine p53 fusion, exhibited a similar β-galactosidase specific activity (17.2 ± 2.1 Miller units). In contrast, transformants carrying either pGADT7-B or PGBKT7-E were negative for β-galactosidase, with identical specific activities of 1.9 ± 0.5 Miller units. Collectively, these results confirm that RelB binds directly to RelE.
Cloning of relB and relE into expression vectors.
The relB gene was amplified by PCR from plasmid pJT1 using oligonucleotides RELB5X1-5A (5′CAAGAGGGGATCCACATGGGTAGC3′) and RELB5X1-3A (5′GCCATTCCTTGAATTCCCGCTCG3′). A BamHI site and a single-base change which eliminated an in-frame stop codon directly upstream of the relB gene were incorporated into RELB5X1-5A. An EcoRI site downstream of the native relB stop codon was incorporated into RELB5X1-3A. The relB PCR product was cloned into the vector pGEX-5X-1 (Pharmacia) to create plasmid pJT4. The N terminus of the RelB protein encoded on pJT4 was fused to glutathione S-transferase (GST) to facilitate its purification and detection. The relE gene was PCR amplified from pJT1 using oligonucleotides RELE30C-5A (5′GAGCTCTGATGGCGTATTTTCTGG3′) and RELE30C-3A (5′CAAGCTTTGGTTCAGAGAATGCG3′). A SacI site upstream of the ATG initiation codon was incorporated into RELE30C-5A, and a HindIII site downstream of the native relE stop codon was incorporated into RELE30C-3A. The relE gene was cloned into the vector pET-30c(+) (Novagen) to create plasmid pJT9. The expression of relE on pJT9 was dependent on phage T7 RNA polymerase, and the RelE product contained N-terminal His-tag and S-tag elements that facilitated its purification and detection. Both tags could be removed by digestion with enterokinase.
Toxicity of RelE.
The original pJT9 construct was isolated in E. coli DH5α, a strain that did not contain a copy of the T7 RNA polymerase gene and therefore could not express the cloned relE gene. However, due to the apparent cytotoxicity of the RelE protein, we were unable to transform pJT9 into E. coli strain BL21(DE3), a lysogen carrying bacteriophage λDE3 which contains a copy of the T7 RNA polymerase gene, even in the absence of isopropylthio-β-d-galactoside (IPTG). Evidently the T7 RNA polymerase gene in BL21(DE3), which is under the control of a lac promoter, exhibits a low level of expression sufficient to promote the production of enough RelE to cause cell death. It should also be noted that we were also unable to transform pJT9 into the more stringent strain, BL21(DE3) carrying plasmid pLysS (Novagen). On the other hand, we successfully cotransformed pJT9 and pJT4 into BL21(DE3). The stable maintenance of both plasmids was achieved by routinely culturing the bacterial host in medium containing kanamycin (50 μg/ml) and ampicillin (50 μg/ml), the selective agents for pJT9 and pJT4, respectively. Therefore, the toxicity of RelE was apparently neutralized by the leaky expression of RelB from pJT4.
Overexpression and copurification of RelB and RelE.
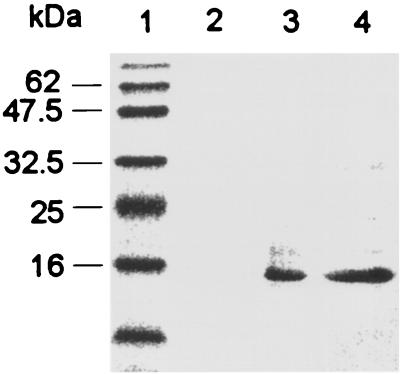
RelB and RelE were coexpressed in BL21(DE3) carrying pJT4 and pJT9 by the following procedure. Bacteria were grown in Luria-Bertani broth (Difco) containing antibiotics in a 37°C water bath shaker. When the optical density of the culture at 600 nm reached approximately 0.6, the relB and relE genes were simultaneously induced by the addition of IPTG to a final concentration of 1 mM. After 3 h of incubation, the bacteria were harvested by centrifugation. Cell extracts were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Figure 1 shows a Coomassie blue-stained gel comparing crude extracts from an uninduced culture (lane 2) and an induced culture (lane 3). IPTG induced the overexpression of a 33-kDa protein and a 16-kDa protein, and these were consistent with the expected masses of the GST fusion derivative of RelB and the His- and S-tag derivative of RelE, respectively.
FIG. 1.
Overexpression and copurification of RelB and RelE from E. coli BL211. Shown is an SDS-polyacrylamide gel stained with Coomassie blue, containing molecular mass standards (lane 1), crude extract of uninduced culture (lane 2), crude extract of induced culture (lane 3), solubilized inclusion body fraction from the induced culture which was used for affinity purification of RelE (lane 4), and RelB and RelE specifically eluted from a nickel affinity column (lane 5).
RelE was purified by nickel affinity chromatography by using the procedures outlined in the pET system manual (Novagen). Figure 1 summarizes the progress of the purification. The majority of the RelB and RelE proteins accumulated in insoluble inclusion bodies. Solubilization was achieved by the addition of 6 M urea to all buffers used in the purification procedure. Lane 4 represents a sample of the solubilized inclusion body fraction. Essentially all of the RelB and RelE proteins adsorbed to the nickel affinity column. Both RelB and RelE were specifically eluted from the column, as shown in the sample in lane 5. In a control experiment (data not shown), the 33-kDa GST-RelB fusion was purified from extracts of BL21(DE3) carrying only plasmid pJT4 by affinity chromatography on glutathione columns (Pharmacia). This purified protein did not adsorb to a nickel affinity column. Therefore, the copurification of RelB and RelE is indicative of a direct interaction between the two proteins, and this result is consistent with the results of our yeast two-hybrid analysis as well as with previous genetic analyses (9). Interestingly, the RelB-RelE interaction was stable even in the presence of 6 M urea. Moreover, the presence of the GST fusion did not interfere with the RelE-binding activity of RelB.
The identities of the RelB and RelE proteins were confirmed by Western blotting as described by Ausubel et al. (1). Protein samples were fractionated by SDS-PAGE, and the proteins were transferred to nitrocellulose (Optitran; Schleicher & Schuell). To detect RelE on this blot, we used an antibody directed specifically against the S-tag portion of our RelE derivative (anti-S protein alkaline phosphatase conjugate [Novagen]). An anti-GST antibody conjugated to alkaline phosphatase (Pharmacia) was used for the detection of RelB.
The Western blot in Fig. 2 was probed with anti-S-tag antibodies, and it confirms the overexpression and purification of HisRelE. Lane 2 contains a crude extract from an uninduced culture of strain BL21(DE3) carrying both pJT4 and pJT9, and lane 3 contains an extract from an IPTG-induced culture. The 16-kDa band in the induced extract is consistent with what was expected for the RelE protein. The absence of this band in the uninduced extract indicates that the level of apparent leaky expression, discussed above, is actually undetectable by Western blotting. This further substantiates the high degree of toxicity of RelE. Lane 4 contains a sample of the protein purified by nickel affinity chromatography.
FIG. 2.
Identification of RelE on a Western blot developed with antibodies directed against S-tag. Lanes: 1, prestained molecular mass standards which are (from top to bottom) 83, 62, 47.5, 32.5, 25, 16, and 6.5 kDa; 2, a crude extract from an uninduced culture of BL21(DE3) carrying plasmids pJT4 and pJT9; 3, crude extract from the induced culture; 4, RelB-RelE mixture eluted from the nickel affinity column.
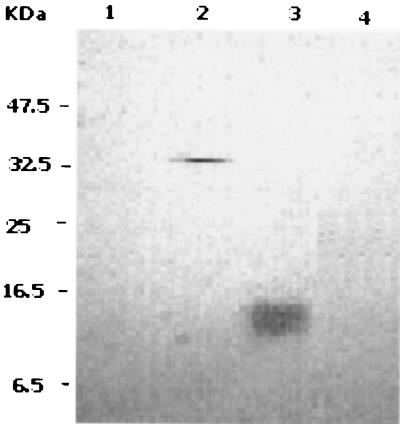
Figure 3 shows a Western blot developed with anti-GST antibodies. The 33-kDa band corresponding to the GST fusion derivative of RelB was detected only in the extract from the induced culture (compare lanes 1 and 2). Furthermore, the presence of this band in the sample eluted from the nickel affinity column clearly verified the copurification of RelB with RelE (lane 3).
FIG. 3.
Identification of RelB on a Western blot developed with antibodies directed against GST. Lanes: 1, crude extract from an uninduced culture of BL21(DE3) carrying plasmids pJT4 and pJT9; 2, crude extract from the induced culture; 3, RelB-RelE mixture eluted from the nickel affinity column.
Purification of RelB and RelE.
Several methods were tested in efforts to resolve the copurified RelB and RelE proteins. As already noted, RelB and RelE were tightly complexed even under denaturing conditions. Therefore, preparative SDS-PAGE was the most reproducible method for the purification of RelE. The RelB-RelE complex obtained by nickel affinity chromatography was resolved by SDS-PAGE, and the protein band corresponding to RelE was excised from the gel. The gel slice was manually ground up in 20 mM Tris-HCl buffer (pH 7.4) containing 0.1% SDS. The RelE protein was then eluted from the gel by incubation at 37°C for 1 h. As noted above, RelB was readily purified on a glutathione column as a GST fusion protein from extracts of BL21(DE3) expressing plasmid pJT4.
RelE binds to the ribosome.
Since RelE has been proposed to be an inhibitor of translation (9), the ability of RelE to bind to ribosomes was tested directly. Ribosomes were prepared from E. coli strain DH5α by the methods of Homann and Nierhaus (10) and of Gentry and Cashel (6) with minor modifications. Bacteria were grown to stationary phase at 37°C in 200 ml of Luria-Bertani medium (Difco), harvested, and resuspended in 5 ml of ribosome buffer (10 mM Tris-HCl [pH 7.5], 14 mM MgCl2, 1mM dithiothreitol, 10 mM potassium acetate). The cells were broken by sonication, and cell debris was removed by centrifugation at 27,000 × g for 1 h. The ribosomal fraction was collected by centrifugation of the supernatant in a Beckman Optima TLX ultracentrifuge at 200,000 × g for 3 h. The ribosomal pellet was washed with 1 ml of ribosome buffer and then centrifuged again at 200,000 × g for 3 h.
The ribosome-binding assay was performed with 1.5 ml of ribosome buffer containing 400 μg of ribosomes, as determined by the Bradford protein assay (3), and purified proteins as indicated below. Prior to use, the purified protein samples were centrifuged at 200,000 × g for 3 h to remove any insoluble material. The ribosome-binding assay mixtures were incubated at 4°C for 60 min and centrifuged at 200,000 × g for 3 h to pellet the ribosomes and any proteins bound to them. The pellets were resuspended in 20 μl of ribosome buffer. Samples of each mixture were fractionated by SDS-PAGE and analyzed by Western blotting using antibodies directed against S-tag. The results are presented in Fig. 4. Lane 1 represents a negative control composed of ribosomes alone. Lane 2 is a positive control consisting of ribosomes incubated with purified E. coli RelA (100 ng), a known ribosome-binding protein (15). The derivative of RelA used here contained S-tag and was kindly provided by X. Yang of this laboratory. The presence of the 37-kDa band corresponding to RelA in lane 2 confirmed its ribosome-binding activity. Lane 3 represents a mixture of ribosomes and RelE (100 ng). The diffuse band of about 16.5 kDa indicated that RelE was bound to the ribosome fraction. The ribosomes in lane 4 were incubated with a mixture of RelE (100 ng) and RelB (150 ng). The RelB-RelE mixture had been preincubated at 4°C overnight to facilitate the reconstitution of the RelB-RelE complex. The absence of the RelE band in this lane indicates that RelB inhibited the ribosome-binding activity of RelE.
FIG. 4.
Ribosome-binding activity of RelE in an in vitro assay. Ribosomes incubated with various purified proteins were collected by ultracentrifugation. The pellets were fractionated by SDS-PAGE and analyzed in Western blots developed with antibodies directed against S-tag. The incubation mixtures contained ribosomes and no added protein (lane 1), RelA (lane 2), RelE (lane 3), or RelB-RelE complex (lane 4).
Summary and conclusions.
Our results confirm several aspects of a working model previously proposed for RelB-RelE function (5, 9, 12). Genetic studies suggest that the specific interaction of RelE with RelB is essential for regulating the expression of the relBEF operon and for neutralizing the toxic activity of RelE. We have presented the first direct demonstration of the RelB-RelE interaction. The two proteins copurified as a tight complex. The direct binding of RelB and RelE was also confirmed by a positive yeast two-hybrid assay. The relE gene could be maintained only in the presence of a copy of relB, indicating that the RelB-RelE interaction was crucial for neutralizing the toxicity of RelE. Finally, we have demonstrated that RelE exhibits ribosome-binding activity in vitro; this is consistent with previous observations that suggest that it is an inhibitor of translation. Our current studies focus on its precise mode of action.
Acknowledgments
This study was supported by a grant from the Natural Sciences and Engineering Research Council of Canada.
We thank Xiaoming Yang for invaluable discussions and for the generous gift of RelA protein. We also thank Limei Zhang, Jessica Blaker, and David Harris for skillful technical assistance.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: Greene Publishing Associates & Wiley Interscience; 1994. [Google Scholar]
- 2.Bech F W, Jorgensen S T, Diderichsen B, Karlstrom O H. Sequence of the relB transcription unit from Escherichia coli and identification of the relB gene. EMBO J. 1985;4:1059–1066. doi: 10.1002/j.1460-2075.1985.tb03739.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 4.Cashel M, Gentry D R, Hernandez V J, Vinella D. The stringent response. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 1458–1496. [Google Scholar]
- 5.Diderichsen B, Desmarez L. Variations in phenotype of relB mutants of Escherichia coli and the effect of pus and sup mutations. Mol Gen Genet. 1980;180:429–437. doi: 10.1007/BF00425859. [DOI] [PubMed] [Google Scholar]
- 6.Gentry D R, Cashel M. Cellular localization of the Escherichia coli SpoT protein. J Bacteriol. 1995;177:3890–3893. doi: 10.1128/jb.177.13.3890-3893.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gerdes K. Toxin-antitoxin modules may regulate synthesis of macromolecules during nutritional stress. J Bacteriol. 2000;182:561–572. doi: 10.1128/jb.182.3.561-572.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gerdes K, Bech F W, Jorgensen S T, Lobner-Olesen A, Rasmussen P B, Atlung T, Boe L, Karlstrom O, Molin S, von Meyenburg K. Mechanism of postsegregational killing by the hok gene product of the parB system of plasmid R1 and its homology with the relF gene product of the E. coli relB operon. EMBO J. 1986;5:2023–2029. doi: 10.1002/j.1460-2075.1986.tb04459.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gotfredsen M, Gerdes K. The Escherichia coli relBE genes belong to a new toxin-antitoxin gene family. Mol Microbiol. 1998;29:1065–1076. doi: 10.1046/j.1365-2958.1998.00993.x. [DOI] [PubMed] [Google Scholar]
- 10.Homann H E, Nierhaus K H. Ribosomal proteins. Protein compositions of biosynthetic precursors and artificial subparticles from ribosomal subunits in Escherichia coli K 12. Eur J Biochem. 1971;20:249–257. doi: 10.1111/j.1432-1033.1971.tb01388.x. [DOI] [PubMed] [Google Scholar]
- 11.Kohara Y, Akiyama K, Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987;50:495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- 12.Lavalle R, Desmarez L, De Hauwer G. Natural messenger translation impairment in an E. coli mutant. In: Kjellgaard N O, Maaloe O, editors. Control of ribosome synthesis. Copenhagen, Denmark: Munksgaard; 1976. pp. 408–418. [Google Scholar]
- 13.Mosteller R D. Evidence that glucose starvation-sensitive mutants are altered in the relB locus. J Bacteriol. 1978;133:1034–1037. doi: 10.1128/jb.133.2.1034-1037.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mosteller R D, Kwan S F. Isolation of relaxed-control mutants of Escherichia coli K-12 which are sensitive to glucose starvation. Biochem Biophys Res Commun. 1976;69:325–332. doi: 10.1016/0006-291x(76)90525-8. [DOI] [PubMed] [Google Scholar]
- 15.Ramagopal S, Davis B D. Localization of the stringent protein of Escherichia coli on the 50S ribosomal subunit. Proc Natl Acad Sci USA. 1974;71:820–824. doi: 10.1073/pnas.71.3.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 17.Wang R F, Kushner S R. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene. 1991;100:195–199. [PubMed] [Google Scholar]