Abstract
Wastewater analysis is the most attractive alternative way for the quantification and variant profiling of SARS-CoV-2. Infection dynamics can be monitored by RT-qPCR assays while NGS can provide evidence for the presence of existing or new emerging SARS-CoV-2 variants. Herein, apart from the infection dynamic in Attica since June 1st, 2021, the monitoring of 9 mutations of the omicron and 4 mutations of the delta SARS-CoV-2 variants, utilizing both novel Nested-Seq and RT-PCR, is reported and the substitution of the delta variant (B.1.617.2) by the omicron variant (B.1.1.529) in Attica, Greece within approximately one month is highlighted. The key difference between the two methodologies is discovery power. RT-PCR can only detect known sequences cost-effectively, while NGS is a hypothesis-free approach that does not require prior knowledge to detect novel genes. Overall, the potential of wastewater genomic surveillance for the early discovery and monitoring of variants important for disease management at the community level is underlined. This is the first study, reporting the SARS-CoV-2 infection dynamic for an extended time period and the first attempt to monitor two of the most severe variants with two different methodologies in Greece.
Keywords: Wastewater, SARS-CoV-2, Omicron variant, delta variant, NGS, RT-qPCR
Graphical abstract
1. Introduction
Severe Acute Respiratory Syndrome Corona Virus 2 (SARS-CoV-2) is an enveloped respiratory virus with one of the largest RNA genomes (~30,000 nt) (Ahmed et al., 2022a; Ahmed et al., 2022b; Pipes et al., 2022). After SARS-CoV-2 prevalence since December 2019, many genome variants were observed, which are characterized by spike protein substitutions and deletions. Emerging variants of concern (VOCs) and variants of interest (VOIs) can increase transmissibility, disease severity and immune escape and may interfere with diagnostic assay performance (Ahmed et al., 2022a; Kirby et al., 2022; CDC, 2022; Jahn et al., 2021; Karthikeyan et al., 2021).
Genomic surveillance of SARS-CoV-2 basically relied on the sequencing of clinical Corona Virus Disease 2019 (COVID-19) samples. However, clinical genomic surveillance is expensive, inefficient, lacks community representation and has sampling bias due to testing of only symptomatic individuals and to systemic healthcare disparities, particularly in poor and underserved communities(Kaplan et al., 2021; Karthikeyan et al., 2021; Peccia et al., 2020; Smith et al., 2021; Wolfe et al., 2022). Variants of SARS-CoV-2 can also be tracked in community wastewater (wastewater genomic surveillance) which offers cost-effective, unbiased and real-time capture of virus spread and dynamic (Ahmed et al., 2022a; Karthikeyan et al., 2021; Michael-Kordatou et al., 2020; Smith et al., 2021). In addition, wastewater genomic surveillance tracks existing and new emerging variants, for which targeted assays do not exist as yet. These data are valuable for transmission network analysis and interpretation, as well as an emerging technology for tracing viral evolution (Karthikeyan et al., 2021; Vo et al., 2022). However, wastewater genomic surveillance remains a challenge, since low viral loads, matrix effect, heavily fragmented RNA, poor enrichment or amplification of SARS-CoV-2 genome and PCR inhibitors could lead to poor sequencing quality (Karthikeyan et al., 2021; Wolfe et al., 2022).
According to World Health Organization (WHO) and Centers for Disease Control and Prevention (CDC), Alpha (B.1.1.7 and Q lineages), Beta (B.1.351 and descendent lineages), Gamma (P.1 and descendent lineages) and Epsilon (B.1.427, B.1.429) were designated as VOCs, while Delta (B.1.617.2 and AY lineages) and Omicron (B.1.1.529 and BA lineages) are the most current VOCs identified in both clinical and wastewater samples. To date, several publications have explored SARS-CoV-2 variants in wastewater. Heijen et al. detected a single nucleotide polymorphism (SNP) (mutation N501Y) present in Beta and Alpha (Heijnen et al., 2021) while Lee et al. applied RT-qPCR assays that detect mutations present in Alpha to wastewater samples (Lee et al., 2021). Jahn et al. used wastewater genome sequencing to detect Alpha, Beta and Gamma variants (Jahn et al., 2021). Yaniv et al. developed RT-qPCR assays for Alpha, Beta, Gamma and Delta (Yaniv et al., 2021, Yaniv et al., 2021b). In addition, in our previous studies (Avgeris et al., 2021; Galani et al., 2022), we showed that wastewater-based epidemiology (WBE) can predict hospitalizations and ICP admissions, whereas using our novel Nested-Seq assay for SARS-CoV-2 mutation/variant analysis we provided real-time monitoring of SARS-CoV-2 variants and identified those strains with selective advantage to become dominant in community/population level.
From the aforementioned VOCs, Omicron variant (B.1.1.529) is more transmissible and is spreading faster than any previous variant, although causing less severe symptoms. It was detected for the first time on the 25th of November 2021 in Botswana and South Africa and was spreading quickly across South Africa and all over the world (Ahmed et al., 2022a; Ren et al., 2022). The B.1.1.59 genome included over 50 mutations, with >30 in the spike protein (Ahmed et al., 2022a). In addition, Omicron was associated with a significant increase in the risk of reinfection (2.39×), may escape the immune system's defenses and two doses of vaccination seemed to be less effective against the Omicron infection (Ahmed et al., 2022a; Andrews et al., 2022; Ren et al., 2022).
As a consequence, viral variant monitoring in wastewater will benefit from the implementation of viral Whole Genome Sequencing (WGS) but it has to overcome many challenges (Alhama et al., 2021). The main question is: NGS or PCR-based targeting of key mutations? As mentioned in the literature, during Alpha variant prevalence a S-gene “dropout” during RT-qPCR testing with certain kits indicated the presence of this mutation and the potential presence of the B.1.1.7 lineage (Ahmed et al., 2022b). In addition, there is often a lag in the availability of primers and probes and in many cases, PCR cannot distinguish between variants leading to loss of data about the range of mutations (Boudet et al., 2021; Smith et al., 2021; Yaniv et al., 2021a). On the other hand, RT-qPCR is a highly sensitive, highly specific,cost-effective, and high-throughput rapid tool which provides fast obtention and easy interpretation of the specific variants. In contrast, NGS provides massive genomic data, however is expensive, time consuming and requires experienced staff for sample and seq-data analysis (Itarte et al., 2021). For comprehensive community surveillance, both NGS and RT-qPCR are necessary but in cases where there is a lack of time and money, RT-qPCR seems to provide reliable and sufficient information.
Due to Omicron's high transmissibility, numerous research groups worldwide have already detected this VOC in wastewater. Either by using NGS technology (Agrawal et al., 2022; Rasmussen et al., 2022; Smyth et al., 2022)or by designed RT-qPCR based assays for the rapid screening of the Omicron variant followed by Sanger Sequencing (Bar-Or et al., 2022; Chassalevris et al., 2022; La Rosa et al., 2022; Oloye et al., 2022).
Nonetheless, by December 2nd, Greece reported its first Omicron case, as revealed from clinical testing, while Delta was dominant during summer of 2022 according to National Public Health Organization (NPHO) (NPHO, 2022). The aims of the current study were1) to evaluate the presence of SARS-CoV-2 VOCs in wastewater samples simultaneously by two different methods: a) our previously reported novel Nested-Seq method and b) a commercially available RT-PCR assay, 2) to compare the used methods based on actual data obtained during the study and provide the advantages and drawbacks of each method, 3) to report the total domination of Omicron variant in Attica peninsula by evaluating for the first time 4 different aspects:SARS-CoV-2 wildtype analysis, NPHO COVID-19 cases from clinical testing, novel Nested-Seq assay and RT-PCR variants assay, and 4) to correlate the results with SARS-CoV-2 infection dynamic in Attica for an extended period (from June 2021 to March 2022). A fully validated 3-step analytical protocol which includes concentration, extraction and clean-up step and RT-qPCR for the detection and quantification of N1 and N2 target genes was also used (Dimitrakopoulos et al., 2022). This study provides a comprehensive community monitoring in the Attica region and highlights the importance and effectiveness of both RT-qPCR assays and NGS.
2. Material and methods
2.1. Sampling and storage
24-hour composite flow proportional raw wastewater samples were collected from the wastewater treatment plant of Attica, the region of Greece that includes Athens metropolitan area and suburbs. The wastewater treatment plant (WWTP) that services Attica is located on Psyttaleia, an uninhabited island in the Saronic Gulf. Background and features of this WWTP that serves a large percentage of the population of Greece have been described previously (Thomaidis et al., 2016). The number of inhabitants was estimated daily as described elsewhere (Galani et al., 2021). While the WWTP facility serves 4,562,500 people, the daily estimation of the active population was taken into account for all calculations necessary in this study and this estimation is described in detail in the Supplementary Information (Tables S1 and S2). The raw wastewater samples were collected daily (from June 1st, 2021 through March 2nd, 2022) in pre-cleaned high-density polyethylene (HDPE) 2 L bottles, and transported at 4 °C to the laboratory. All samples were processed immediately upon arrival at the laboratory. Biosafety guidelines were followed during sampling, transportation and the analytical procedure.
2.2. Isolation of total nucleic acid from wastewater
The SARS-CoV-2 RNA was extracted from wastewater samples as previously described (Dimitrakopoulos et al., 2022). As a negative sample control, 40 mL of nuclease-free water were processed with each run to ensure lack of sample-to-sample carryover contamination. A quality control sample for the evaluation of the viral RNA recovery was also used in each run. Specifically, 106 gene copies from the EURM-19 synthetic single stranded RNA (European Commission, Joint Research Centre, Geel, Belgium) was spiked in the 1 mL eluate from the PureYield™ Midi Binding Column. Endogenous viral gene copies were subtracted from the recovered EURM copies in order to calculate the % recovery.
2.3. PCR reactions for the detection of N1 & N2 SARS-CoV-2 amplicons
Reverse transcription quantitative PCR (RT-qPCR) was performed in the extracted TNA by using the Water SARS-CoV-2 RT-PCR ready-to-use kit (IDEXX Laboratories, Inc., Westbrook, ME, USA), designed to target both the 2019-nCoV_N1 and 2019-nCoV_N2 target genes of the virus. Each sample was quantified by RT-qPCR in duplicate wells in a Touch CFX96™ Real-Time PCR (Bio-Rad, United States). Both positive-control and negative-control reactions were performed for quality control in each run. Five RT-qPCR standards were prepared through a ten-fold serial dilution of the EURM-19 synthetic single stranded RNA standard from 5 × 105 to 50 genome copies/well. One standard curve was prepared and analyzed in each run in triplicate. Gene copy number per PCR reaction was calculated from the standard curve according to the equation Copiesreaction = 10(Cq-a)/b, where Cq corresponds to the threshold cycle of the sample, and a and b correspond to the Y-intercept and slope of the logarithmic standard curve, respectively. In order to address the issue of PCR reaction inhibitions (Gibson et al., 2012), 4-fold and 10-fold dilutions of each TNA sample were prepared and analyzed along with the undiluted ones. Further description of the assessment of PCR inhibition and the backward calculation of the SARS-CoV-2 genome copies per liter of wastewater and per 100,000 inhabitants can be found in the Supplementary Material.
2.4. PCR reactions for the detection of SARS-CoV-2 mutations
The RT-PCR for the detection of amplicons corresponding to variant SARS-CoV-2 strains was performed in the extracted TNA by using the Wastewater SARS-CoV-2 RT-PCR Variant Panel (Promega Corp.). Specifically, daily wastewater samples were assayed for the presence of 4 different amplicons each one being able to track specific mutations associated with one or more variant strains of the SARS-CoV-2 virus. All mutations monitored were located in the spike (S) protein of the virus and corresponded to the strains mentioned in parentheses: N501Y (variants alpha [B.1.1.7], beta [B.1.351], gamma [P.1] and omicron [B.1.1.529]), Del H69-/V70- (variants alpha and omicron), Κ417Ν (variants beta and omicron) and P681R (variants delta [B.1.617.2] and kappa [B.1.617.1]). All PCR reactions were performed in a Touch CFX96™ Real-Time PCR Detection System (Bio-Rad, United States) using automatic settings for threshold and baseline. All variant amplicons were monitored in the FAM channel. In a separate channel (HEX), the multiplexed assays provided Cq values for the wild type (original Wuhan strain) equivalents of the mutant amplicons. Due to the absence of a reference material spanning the S gene-both for the wild type and the heavily mutated-an arbitrary but reasonable value of 36 cycles was set as a cut-off for each amplicon positivity.
2.5. SARS-CoV-2 mutational analysis using novel nested-Seq assay
Identification and quantification of SARS-CoV-2 variants in wastewater samples from December 2021 and January 2022 was carried out by barcoded DNA-seq targeting S gene in Ion PGM™ platform (Ion Torrent, Thermo Fisher Scientific Inc.), as previously described (Avgeris et al., 2021; Galani et al., 2022). Barcoded libraries were constructed with the Ion Xpress Plus Fragment Library Kit (Ion Torrent). Adapter ligation, nick-repair and clean-up of the ligated library were performed based on the protocol of the manufacturer, and each barcoded library was quantified with the Ion Library TaqMan Quantitation Kit (Ion Torrent) in an ABI 7500 Real-Time PCR system (Applied Biosystems). Equimolar amounts of each library were used for the downstream template preparation step on an Ion OneTouch 2 System, whereas the enrichment process was carried out on the Ion OneTouch ES instrument, using the Ion PGM Hi-Q View OT2 kit (Ion Torrent). Finally, semiconductor NGS was carried out in Ion 316TM Chip v2 using the Ion PGM™ Hi-Q™ View Sequencing kit.
Barcoded libraries were constructed from samples obtained during December 2021–January 2022, and more precisely during 1–15/12/2021 (sample 1), 16–23/12/2021 (sample 2), 27–30/12/2021 (sample 3), 01–10/01/2022 (sample 4), 16–20/01/2022 (sample 5) and 26–30/01/2022 (sample 6). S gene-related missense mutations G339D (G22578A), S371L (TC22673CT), S373P (T22679C), S375F (C22686T), K417N (G22813T), N440K (T22882G), G446S (G22898A), as well as T19R (C21618G), L452R (T22917G), D950N (G24410A) were targeted for the detection and quantification of Omicron (B.1.1529) and Delta (B.1.617.2) variants, respectively. These analyses led to the conduct of >1.5 million sequencing reads per barcode.
The in silico analysis for the mutational and variant profiling of SARS-CoV-2 included an initial alignment to the SARS-CoV-2 reference genome (NC_045512.2) with the Burrows-Wheeler Aligner (BWA-MEM) (Li and Durbin, 2009). Alignment was followed by alignment clean-up to prepare data for variant calling of SNVs and insertions/deletions, which was performed with the iVar algorithm with the recommended parameters (Grubaugh et al., 2019).
3. Results
3.1. SARS-CoV-2 variants analysis by novel nested-Seq
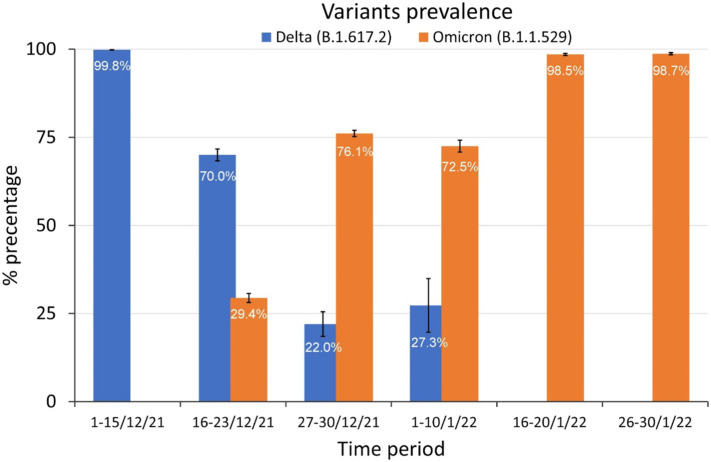
The first confirmed omicron (B.1.1529) case in Greece, was confirmed in Crete in early December 2021.In addition, Omicron variant was firstly detected in WWTP of Attica in the sampling period 16–26/12/2021 and based on the frequency of the genetic markers analyzed, in 29.4 % ± 1.3 (mean ± SE) of the total sequencing reads, compared to approximately 70.0 % ± 1.7 of the Delta variant. Interestingly, the frequencies of omicron genetic markers were immediately increased in the following sampling period, and more precisely to 76.1 % ± 0.9 and 72.5 % ± 1.7 in 27–30/12/2021 and 01–10/01/2022 samples, in combination with a corresponding reduction of Delta variant to 22.0 % ± 3.5 and 27.3 % ± 7.6, respectively. As expected, the analysis of the following libraries, 16–20/01/2022 and 26–30/01/2022, confirmed the prevalence of omicron variant (B.1.1529) in Attica, Greece in percentage 98.5 % ± 0.3 and 98.7 % ± 0.3, respectively. Despite the delta variant (B.1.617.2), specific genetic markers for the other VOC Gamma (P1) and Beta (B.1.351) were not detected at the same sampling period. The findings of these analyses are summarized in Table 1, Table 2 and illustrated in Fig. 1 .
Table 1.
Novel Nested-Seq for the detection and quantification of omicron (B.1.1.529) variant.
| Position | 1–15/12/21 | 16–23/12/21 | 27–30/12/21 | 1–10/1/22 | 16–20/1/22 | 26–30/1/22 |
|---|---|---|---|---|---|---|
| G339D (G22578A) | ND | 24.9 % | 73.98 % | 80.95 % | 99.12 % | 99.22 % |
| S371L (TC22673CT) | ND | 28.02 % | 76.22 % | 74.89 % | 98.85 % | 99.31 % |
| S373P (T22679C) | ND | 28.57 % | 76.13 % | 74.39 % | 97.59 % | 98.13 % |
| S375F (C22686T) | ND | ND | 72.26 % | 69.58 % | 97.82 % | 96.96 % |
| K417N (G22813T) | ND | 33.45 % | 78.95 % | 70.65 % | 99.42 % | 99.53 % |
| N440K (T22882G) | ND | 32.74 % | 77.79 % | 69.81 % | 98.58 % | 98.75 % |
| G446S (G22898A) | ND | 28.88 % | 77.09 % | 67.20 % | 98.24 % | 98.70 % |
| Mean ± SE | ND | 29.40 % ± 1.3 | 76.10 % ± 0.9 | 72.50 % ± 1.7 | 98.50 % ± 0.3 | 98.70 % ± 0.3 |
Table 2.
Novel Nested-Seq for the detection and quantification of delta (B.1.617.2) variant.
| Position | 1–15/12/21 | 16–23/12/21 | 27–30/12/21 | 1–10/1/22 | 16–20/1/22 | 26–30/1/22 |
|---|---|---|---|---|---|---|
| T19R (C21618G) | 99.87 % | 69.31 % | 15.99 % | 21.99 % | ND | ND |
| L452R (T22917G) | 99.90 % | 67.52 % | 21.76 % | 42.42 % | ND | ND |
| D950N (G24410A) | 99.70 % | 73.21 % | 28.19 % | 17.59 % | ND | ND |
| Mean ± SE | 99.80 % ± 0.06 | 70.00 % ± 1.7 | 22.00 % ± 3.5 | 27.30 % ± 7.6 | ND | ND |
Fig. 1.
Means of frequencies of mutations representative for delta and omicron variants for six time periods as determined by Novel Nested-Seq.
3.2. SARS-CoV-2 variants detection by RT-PCR
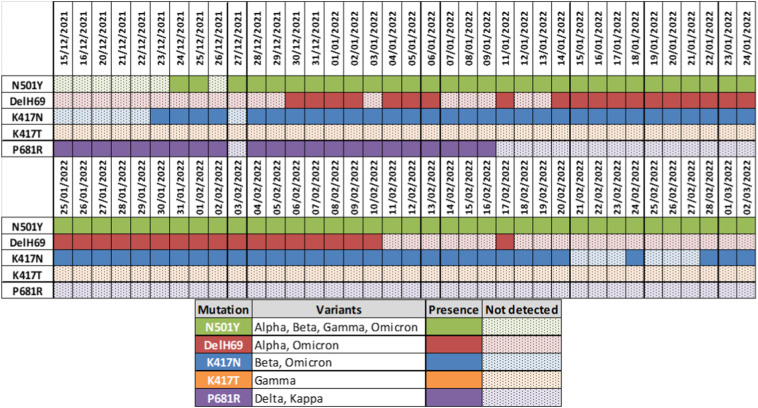
As shown in Fig. 2 , while the P681R mutation corresponding to the delta variant had been consistently detected in all samples (except the one from 27th of December) analyzed from December 15th, 2021 till January 9th 2022, two of the three “omicron” mutations (N501Y & K417N) were not detectable in Attica wastewater until December 23rd and 22nd 2021, respectively. During December 28th to January 9th, 2022, N501Y, K417N and P681R mutations were detected in wastewater indicating the parallel presence of the delta and omicron variants in Attica region during the omicron surge of the last weeks of 2021 and the first ones of 2022.
Fig. 2.
RT-PCR qualitative results for the four surrogate mutations of the SARS-CoV-2 variants omicron and delta that were circulating in the Attica region in late 2021/early 2022.
These RT-qPCR results are in general agreement with the Novel Nested-Seq results reported above. Omicron variants started appearing in the 16–23/12/21 pooled samples by sequencing and were first detected on 23/12/21 by RT-qPCR. The latter was able to detect both delta and omicron variants till 9/1/22 in complete agreement with DNA-Seqresults (lastly observed delta specific markers in the 1–10/1/22 pool of samples). One of the important inherent advantages of wastewater surveillance over conventional clinical testing of a population is that it can provide information representative of the whole population with a significant lead time before the population actually gets sick and/or tested (Karthikeyan et al., 2021). This is the case for SARS-CoV-2 as well, since viral shedding to human feces takes place early in the course of infection.
As can be seen in Fig. 3 , delta variant was present in wastewater until 12/01/2022 (P681R), while kappa variant was detected neither in wastewater nor by clinical testing. After 12/01/2022, P681R mutation strain was not detected and this phase represented the delta to omicron transition period. On the other hand, omicron variant was firstly detected on 22/12/2021 (K417N) and since 28/12/2021, omicron variant was consistently detected in all samples.
Fig. 3.
Cq values for SARS-CoV-2 mutation strains from 15/12/2021 to 2/03/2022. 40 Cq value represents no detection of mutation strains.
3.2.1. Comparison of both methods: novel nested-Seq and RT-PCR
In this study, results from both Novel Nested-Seq and RT-PCR methods for SARS-CoV-2 mutations are provided as each method is complementary to the other giving a different aspect of the presence of mutations. Each method has its own advantages and disadvantages that need to be clearly addressed and can be used for different reasons based on the laboratory's aim. NGS methods provide more precise, quantitative and detailed results about specific genetic markers and SARS-CoV-2 VOCs. However, RT-PCR assays, demonstrate the trend of VOCs and provide qualitative results about the presence of SARS-CoV-2 mutations in wastewater samples rapidly and cost-effectively. In Table 3 , the strengths and weaknesses of both methods are provided.
Table 3.
Comparison of Novel Nested-Seq and RT-PCR methods for the analysis of SARS-CoV-2 mutations.
| Novel Nested-Seq | Wastewater SARS-CoV-2 RT-PCR variant panel (Promega Corp.) | |
|---|---|---|
| Sensitivity | High | High |
| Specificity | High | Low |
| Variants/mutations | Quantitative results | Qualitative results |
| Cost | High | Low |
| Labor | 1 week | 1 day |
| Data analysis | Bioinformatics/data interpretation | Not required |
| Prior knowledge of targets | No | Required |
| Number of targets | Unlimited | Defined based on the assay |
3.3. SARS-CoV-2 infection dynamic in Attica
The results from both PCR assay and NGS were confirmed by clinical testing and NPHO reported cases. As mentioned elsewhere (Galani et al., 2022), Greece has implemented three lockdowns during 2020–2021 and the last one ended on 15th of May 2021. Since then, only local lockdowns and restrictions were announced, especially in Greek islands during summer months. SARS-CoV-2 infection dynamics in Attica was monitored from 1st of June 2021 until 2nd of March 2022, employing wastewater analysis. The viral load and the measured COVID-19 cases in Attica by NPHO are presented in Fig. 4 .
Fig. 4.
SARS-CoV-2 copies/100 K inhabitants in the wastewater from wastewater treatment plant in Athens (blue bars) and NPHO-reported COVID-19 cases (orange line) are shown for the period June 1, 2021 to March 2, 2022.
The reported COVID-19 cases can be clearly separated into 2 phases (Phase 1 and Phase 2) based on which VOC was dominant during the examined period (Fig. 2). The first phase started on 1st of June and ended on 15th of December 2021. At the beginning of this phase, viral load was the lowest of the whole study period. Since then, COVID-19 infections gradually increased due to the high tourist season (July to August) and the widespread of the highly contagious Delta variant. After schools opening on 13th of September 2021, the viral load was relatively steady with no high fluctuations. An insignificant increase was reported in November 2021. By November 29th, Greece reported its first omicron case as revealed from clinical testing, which was located in Crete. On 4th of December clinical genome sequencing revealed only two confirmed omicron variant infections in Attica which gradually led to Phase 2, when omicron Variant was spread rapidly with the greatest increase in the viral load and the number of cases in Attica from the beginning of the pandemic.
The second phase started on 16th of December and ended on 2nd of March in agreement with NGS and PCR results. The omicron variant was likely present or more widely distributed in the community than originally indicated by clinical testing alone. More specifically, on 24th of December omicron variant was detected for the first time in wastewater as resulting from PCR assay, which means that the shedding of the virus was already incredibly high. As a consequence, the Greek government enforced restrictions and measures on 30th of December due to the wide and steep spread of the omicron variant. Entertainment venues had to close at midnight and would be open only for vaccinated customers who must be seated. The measures, after a third extension, did remain in effect until January 31st. In addition, NPHO reported the highest number of confirmed COVID-19 cases on 4th of January, after New Year's Eve and Christmas Holidays, as the omicron variant seems to be more transmissible than previous VOCs. During December and January the viral load reached extremely high levels in both clinical and wastewater samples. After 17th of January viral load demonstrated a relative reduction but the observed viral levels remain a matter of concern.
4. Conclusions
In this study, two complementary methods that can untangle different aspects of the viral evolution (Novel Nested-Seqand RT-qPCR) were utilized in order to monitor the transition from delta to omicron SARS-Cov-2 virus prevalence in raw wastewater during a viral surge in Attica region, Greece in late 2021/early 2022. In addition, total viral levels were monitored using our N gene RT-qPCR validated assay. Specifically, seven omicron (G339D, S371L, S373P, S375F, K417N, N440K and G446S) and three delta (T19R, L452R and D950N) specific mutations were tracked with Novel Nested-Seq, and three omicron (N501Y, delHV69/70 and K417N) and one delta (P681R) mutations with RT-qPCR. The frequency of omicron variant in combination with delta variant was in agreement for each day or set of dates. The total percentage was almost 100 % with Novel Nested-Seq and the detection was also confirmed by RT-qPCR. The application of both methods in wastewater for the detection of SARS-CoV-2 showed that could offer either fast monitoring of known variants or the detection of variants missed by clinical testing.
Furthermore, the agreement of the two methods for the presence/absence of each variant was deemed to be very good. The viral surge during this period that was observed by the daily monitor of the total viral levels was attributed to the omicron variant that is characterized by increased transmissibility and immune evasion when compared to the delta variant mainly due to the number of mutations present in the RBD domain of the spike protein of the virus.
Overall, wastewater-based epidemiology can serve as an early alert tool that may guide decisions and policies regarding public protection measures timely before disease outbreaks take place at international, national or local level.
Funding
From the Hellenic Foundation for Research and Innovation (HFRI) and the General Secretariat for Research and Technology (GSRT).
CRediT authorship contribution statement
Aikaterini Galani: Methodology, Validation, Writing – original draft. Athina Markou: Supervision, Writing – review & editing, Project administration. Lampros Dimitrakopoulos: Methodology. Aikaterini Kontou: Validation. Marios Kostakis: Validation. Vasileios Kapes: Methodology. Marios A. Diamantopoulos: Formal analysis, Software. Panagiotis G. Adamopoulos: Formal analysis. Margaritis Avgeris: Formal analysis, Writing – review & editing. Evi Lianidou: Writing – review & editing. Andreas Scorilas: Formal analysis. Dimitrios Paraskevis: Writing – review & editing. Sotirios Tsiodras: Writing – review & editing. Meletios-Athanasios Dimopoulos: Funding acquisition, Writing – review & editing. Nikolaos Thomaidis: Conceptualization, Project administration, Visualization, Resources.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
Aikaterini Galani is a recipient of a PhD scholarship from the Hellenic Foundation for Research and Innovation (HFRI). Accordingly, part of this work has been funded by HFRI. Authors would like to acknowledge Athens Water Supply & Sewerage Company (EYDAP S.A.) and especially Mr. Konstantinos Vougiouklakis, Mr. Spyridon Dimoulas and Mr. Iraklis Karayiannis for granting permission for the collection of the wastewater samples and the Athens wastewater treatment plant operators for the collection of the samples.
Editor: Damià Barceló
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scitotenv.2022.159062.
Appendix A. Supplementary data
Supplementary material
Data availability
All data generated or analyzed during this study are included in this published article (and its supplementary information files).
References
- Agrawal S., Orschler L., Tavazzi S., Greither R., Gawlik B.M., Lackner S. Genome sequencing of wastewater confirms the arrival of the SARS-CoV-2 omicron variant at Frankfurt airport but limited spread in the city of Frankfurt, Germany, in November 2021. Microbiol. Resour. Announc. 2022;11 doi: 10.1128/MRA.01229-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bivins A., Smith W.J.M., Metcalfe S., Stephens M., Jennison A.V., et al. Detection of the Omicron (B.1.1.529) variant of SARS-CoV-2 in aircraft wastewater. Sci. Total Environ. 2022;153171 doi: 10.1016/j.scitotenv.2022.153171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Simpson S.L., Bertsch P.M., Bibby K., Bivins A., Blackall L.L., et al. Minimizing errors in RT-PCR detection and quantification of SARS-CoV-2 RNA for wastewater surveillance. Sci. Total Environ. 2022;805 doi: 10.1016/j.scitotenv.2021.149877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alhama J., Maestre J.P., Martin M.A., Michan C. Monitoring COVID-19 through SARS-CoV-2 quantification in wastewater: progress, challenges and prospects. Microb.Biotechnol. 2021;15:1719–1728. doi: 10.1111/1751-7915.13989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews N., Stowe J., Kirsebom F., Toffa S., Rickeard T., Gallagher E., et al. Covid-19 vaccine effectiveness against the Omicron (B.1.1.529) Variant. N. Engl. J. Med. 2022;386:1532–1546. doi: 10.1056/NEJMoa2119451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avgeris M., Adamopoulos P.G., Galani A., Xagorari M., Gourgiotis D., Trougakos I.P., et al. Novel nested-seq approach for SARS-CoV-2 real-time epidemiology and in-depth mutational profiling in wastewater. Int. J. Mol. Sci. 2021;22 doi: 10.3390/ijms22168498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Or I., Indenbaum V., Weil M., Elul M., Levi N., Aguvaev I., et al. National scale real-time surveillance of SARS-CoV-2 variants dynamics by wastewater monitoring in Israel. Viruses. 2022;14 doi: 10.3390/v14061229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudet A., Stephan R., Bravo S., Sasso M., Lavigne J.-P. Limitation of screening of different variants of SARS-CoV-2 by RT-PCR. Diagnostics. 2021;11:1241. doi: 10.3390/diagnostics11071241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for disease control and prevention (CDC) SARS-CoV-2 Variant Classifications and Definitions. 2022. https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-info.html#Concern
- Chassalevris T., Chaintoutis S.C., Koureas M., Petala M., Moutou E., Beta C., et al. SARS-CoV-2 wastewater monitoring using a novel PCR-based method rapidly captured the Delta-to-Omicron ΒΑ.1 transition patterns in the absence of conventional surveillance evidence. Sci. Total Environ. 2022;156932 doi: 10.1016/j.scitotenv.2022.156932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrakopoulos L., Kontou A., Strati A., Galani A., Kostakis M., Kapes V., et al. Evaluation of viral concentration and extraction methods for SARS-CoV-2 recovery from wastewater using droplet digital and quantitative RT-PCR. Case Stud. Chem. Environ. Eng. 2022;6 doi: 10.1016/j.cscee.2022.100224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galani A., Alygizakis N., Aalizadeh R., Kastritis E., Dimopoulos M.-A., Thomaidis N.S. Patterns of pharmaceuticals use during the first wave of COVID-19 pandemic in Athens, Greece as revealed by wastewater-based epidemiology. Sci. Total Environ. 2021;798 doi: 10.1016/j.scitotenv.2021.149014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galani A., Aalizadeh R., Kostakis M., Markou A., Alygizakis N., Lytras T., et al. SARS-CoV-2 wastewater surveillance data can predict hospitalizations and ICU admissions. Sci. Total Environ. 2022;804 doi: 10.1016/j.scitotenv.2021.150151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson K.E., Schwab K.J., Spencer S.K., Borchardt M.A. Measuring and mitigating inhibition during quantitative real time PCR analysis of viral nucleic acid extracts from large-volume environmental water samples. Water Res. 2012;46:4281–4291. doi: 10.1016/j.watres.2012.04.030. [DOI] [PubMed] [Google Scholar]
- Grubaugh N.D., Ladner J.T., Lemey P., Pybus O.G., Rambaut A., Holmes E.C., Andersen K.G. Tracking virus outbreaks in the twenty-first century. Nat. Microbiol. 2019;4:10–19. doi: 10.1038/s41564-018-0296-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heijnen L., Elsinga G., de Graaf M., Molenkamp R., Koopmans M.P.G., Medema G. Droplet digital RT-PCR to detect SARS-CoV-2 signature mutations of variants of concern in wastewater. Sci. Total Environ. 2021;799 doi: 10.1016/j.scitotenv.2021.149456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itarte M., Bofill-Mas S., Martínez-Puchol S., Torrell H., Ceretó A., Carrasco M., et al. Looking for a needle in a haystack. SARS-CoV-2 variant characterization in sewage. Curr. Opin. Environ. Sci. Health. 2021;24 doi: 10.1016/j.coesh.2021.100308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn K., Dreifuss D., Topolsky I., Kull A., Ganesanandamoorthy P., Fernandez-Cassi X., et al. medRxiv; 2021. Detection and Surveillance of SARS-CoV-2 Genomic Variants in Wastewater. 2021.01.08.21249379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan E.H., Wang D., Wang M., Malik A.A., Zulli A., Peccia J. Aligning SARS-CoV-2 indicators via an epidemic model: application to hospital admissions and RNA detection in sewage sludge. Health Care Manag.Sci. 2021;24:320–329. doi: 10.1007/s10729-020-09525-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karthikeyan S., Levy J.I., De Hoff P., Humphrey G., Birmingham A., Jepsen K., et al. medRxiv; 2021. Wastewater Sequencing Uncovers Early, Cryptic SARS-CoV-2 Variant Transmission. 2021.12.21.21268143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby Amy E., Welsh Rory M., Marsh Zachary A., Yu Alexander T., Vugia Duc J., Boehm Alexandria B., et al. Notes from the field: early evidence of the SARS-CoV-2 B.1.1.529 (Omicron) variant in community wastewater — United States, November–December 2021. MMWR Morb. Mortal Wkly. Rep. 2022;2022:103–105. doi: 10.15585/mmwr.mm7103a5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa G., Iaconelli M., Veneri C., Mancini P., Bonanno Ferraro G., Brandtner D., et al. The rapid spread of SARS-COV-2 Omicron variant in Italy reflected early through wastewater surveillance. Sci. Total Environ. 2022;837 doi: 10.1016/j.scitotenv.2022.155767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W.L., Imakaev M., Armas F., McElroy K.A., Gu X., Duvallet C., et al. Quantitative SARS-CoV-2 alpha variant B.1.1.7 tracking in wastewater by allele-specific RT-qPCR. Environ. Sci. Technol. Lett. 2021;8:675–682. [Google Scholar]
- Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael-Kordatou I., Karaolia P., Fatta-Kassinos D. Sewage analysis as a tool for the COVID-19 pandemic response and management: the urgent need for optimised protocols for SARS-CoV-2 detection and quantification. J. Environ. Chem. Eng. 2020;8 doi: 10.1016/j.jece.2020.104306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NPHO Hellenic National Public Health Organization. 2022. https://eody.gov.gr/en/npho/ 2021.12.21.21268143.
- Oloye F.F., Xie Y., Asadi M., Cantin J., Challis J.K., Brinkmann M., et al. Rapid transition between SARS-CoV-2 variants of concern Delta and Omicron detected by monitoring municipal wastewater from three Canadian cities. Sci. Total Environ. 2022;841 doi: 10.1016/j.scitotenv.2022.156741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peccia J., Zulli A., Brackney D.E., Grubaugh N.D., Kaplan E.H., Casanovas-Massana A., et al. Measurement of SARS-CoV-2 RNA in wastewater tracks community infection dynamics. Nat. Biotechnol. 2020;38:1164–1167. doi: 10.1038/s41587-020-0684-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pipes L., Chen Z., Afanaseva S., Nielsen R. medRxiv; 2022. Estimating the Relative Proportions of SARS-CoV-2 Strains From Wastewater Samples. 2022.01.13.22269236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen L.D., Richter S.R., Midgley S.E., Franck K.T. Detecting SARS-CoV-2 Omicron B.1.1.529 variant in wastewater samples by using nanopore sequencing. Emerg. Infect. Dis. 2022;28:1296–1298. doi: 10.3201/eid2806.220194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren S.Y., Wang W.B., Gao R.D., Zhou A.M. Omicron variant (B.1.1.529) of SARS-CoV-2: mutation, infectivity, transmission, and vaccine resistance. World J. Clin. Cases. 2022;10:1–11. doi: 10.12998/wjcc.v10.i1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith T., Holm R.H., Yeager R., Moore J.B., Rouchka E.C., Sokoloski K.J., et al. medRxiv; 2021. Combining Community Wastewater Genomic Surveillance With State Clinical Surveillance: A Framework for SARS-CoV-2 Public Health Practice. 2021.12.06.21267150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth D.S., Trujillo M., Gregory D.A., Cheung K., Gao A., Graham M., et al. Tracking cryptic SARS-CoV-2 lineages detected in NYC wastewater. Nat. Commun. 2022;13:635. doi: 10.1038/s41467-022-28246-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomaidis N.S., Gago-Ferrero P., Ort C., Maragou N.C., Alygizakis N.A., Borova V.L., et al. Reflection of socioeconomic changes in wastewater: licit and illicit drug use patterns. Environ. Sci. Technol. 2016;50:10065–10072. doi: 10.1021/acs.est.6b02417. [DOI] [PubMed] [Google Scholar]
- Vo V., Tillett R.L., Chang C.L., Gerrity D., Betancourt W.Q., Oh E.C. SARS-CoV-2 variant detection at a university dormitory using wastewater genomic tools. Sci. Total Environ. 2022;805 doi: 10.1016/j.scitotenv.2021.149930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe M., Hughes B., Duong D., Chan-Herur V., Wigginton K.R., White B.J., et al. medRxiv; 2022. Detection of SARS-CoV-2 Variant Mu, Beta, Gamma, Lambda, Delta, Alpha, and Omicron in Wastewater Settled Solids Using Mutation-specific Assays Is Associated With Regional Detection of Variants in Clinical Samples. 2022.01.17.22269439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaniv K., Ozer E., Kushmaro A. medRxiv; 2021. SARS-CoV-2 Variants of Concern, Gamma (P.1) and Delta (B.1.617), Sensitive Detection and Quantification in Wastewater Employing Direct RT-qPCR. 2021.07.14.21260495. [Google Scholar]
- Yaniv K., Ozer E., Shagan M., Lakkakula S., Plotkin N., Bhandarkar N.S., et al. Direct RT-qPCR assay for SARS-CoV-2 variants of concern (Alpha, B.1.1.7 and Beta, B.1.351) detection and quantification in wastewater. Environ. Res. 2021;201 doi: 10.1016/j.envres.2021.111653. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Data Availability Statement
All data generated or analyzed during this study are included in this published article (and its supplementary information files).