Abstract
Backgrounds
SARS-CoV-2 infection results in a broad spectrum of clinical outcomes, ranging from asymptomatic to severe symptoms and death. Most COVID-19 pathogenesis is associated with hyperinflammatory conditions driven primarily by myeloid cell lineages. The long-term effects of SARS-CoV-2 infection post recovery include various symptoms.
Methods
We performed a longitudinal study of the innate immune profiles 1 and 3 months after recovery in the Thai cohort by comparing patients with mild, moderate, and severe clinical symptoms using peripheral blood mononuclear cells (n = 62).
Results
Significant increases in the frequencies of monocytes compared to controls and NK cells compared to mild and moderate patients were observed in severe patients 1–3 months post recovery. Increased polymorphonuclear myeloid-derived suppressor cells (PMN-MDSCs) were observed in all recovered patients, even after 3 months. Increased IL-6 and TNFα levels in monocytes were observed 1 month after recovery in response to lipopolysaccharide (LPS) stimulation, while decreased CD86 and HLA-DR levels were observed regardless of stimulation. A multiplex analysis of serum cytokines performed at 1 month revealed that most innate cytokines, except for TNFα, IL4/IL-13 (Th2) and IFNγ (Th1), were elevated in recovered patients in a severity-dependent manner. Finally, the myelopoiesis cytokines G-CSF and GM-CSF were higher in all patient groups. Increased monocytes and IL-6- and TNFα-producing cells were significantly associated with long COVID-19 symptoms.
Conclusions
These results reveal that COVID-19 infection influences the frequencies and functions of innate immune cells for up to 3 months after recovery, which may potentially lead to some of the long COVID symptoms.
Keywords: COVID-19, Innate immune cells, Lipopolysaccharide, Post recovery
Introduction
The COVID-19 pandemic has affected the human population worldwide, with more than four hundred million confirmed cases and more than six million deaths (as of April 2022, Johns Hopkins University Coronavirus Resource Center). The clinical symptoms of COVID-19 present over a wide spectrum, ranging from asymptomatic or mild to severe and death, primarily depending on the host factors.1 COVID-19 pathogenesis is associated with hyperimmune activation, which leads to cytokine storm-like symptoms, acute respiratory distress syndrome (ARDS) and multiple organ failure.2
Both innate and adaptive immune responses are essential for controlling SARS-CoV-2 infection during the acute phase of infection. Many studies have investigated the immune profiles during acute infection and have identified various key molecular and cellular drivers of the disease.3, 4, 5 Immune profiling has revealed an increase in inflammatory innate immune cell infiltration of the affected lungs with a reduction in lymphocytes resulting in increased proinflammatory cytokines and chemokines.3 Furthermore, myeloid linage cells have been identified in many studies to play a critical role in exaggerated immune responses and hyperinflammation.4 , 5 Monocytes have been identified as culprits in causing atypical cytokine storms with the involvement of neutrophils in severe COVID-19 patients.6 Counterintuitively, in severe COVID-19 patients, a few studies have shown expansion of immune suppressor cells, such as myeloid-derived suppressor cells (MDSCs).7 In a Japanese cohort, expansion of PMN-MDSCs was associated with better disease outcomes.8 MAIT cells are the main subpopulation of airway T cells that comprises up to 10% and involved in immune response to viral infection, including SARS-CoV-2 infection.9 Reduction in MAIT cells but increase activated markers in MAIT cells in severe COVID-19 patients were reported.10 , 11 Decreased CCR6+CCR7+ MAIT cells11 implied the migration into the inflammatory site of infection during SARS-CoV-2. The frequency of MAIT cells in convalescent COVID-19 patients still persistently decreased compared healthy controls.10
After recovery from the disease, there are reports that some patients experience persistent symptoms that can last for months, a condition known as long COVID.12 This condition affects many organs after recovery from acute COVID-19, but the underlying causes of this persistence are currently unknown. Because symptomatic COVID-19 patients experience an exaggerated inflammatory response during the acute phase, this can potentially impact immune cell profiles and functions, even after recovery from the disease. However, the longitudinal immune cell profiles and functions after recovery are not well documented or characterized.
In this study, we investigated the innate immune cell profiles, as well as some functions of innate immune cells, in recovered COVID-19 patients at 1 and 3 months after hospital discharge in the Thai cohort. Furthermore, the biological functions of immune cells and serum cytokine profiles were measured. More understanding on such these immune responses may provide a basis for understanding long COVID. Furthermore, these results may identify predictive markers for future health problems and immunomodulatory strategies to prevent long-term health impacts in COVID-19 patients.
Materials and methods
Subjects
Recovered COVID-19 patients (n = 62) in April 2020 with mild (n = 28), moderate (n = 20), and severe (n = 14) clinical symptoms, who were diagnosed and treated at the King Chulalongkorn Memorial Hospital at 1 month (April 2020) and 3 months (June 2020) sequentially after recovery, were recruited for blood collection. COVID-19 patients were confirmed by RT–PCR positivity for SARS-CoV-2 from nasopharyngeal swabs. The classification of COVID-19 severity are shown in Supplementary Table 1, according to the COVID-19 management guideline of the Thai Ministry of Public Health.13 Some of these patients post recovery were also recruited for a post-COVID-19 symptoms review. All methods were carried out in accordance with guidelines/regulations issued and approved by Institutional Review Board of the Faculty of Medicine (IRB number 114/64).
For 40 healthy controls (18–60 years of age) with negative for SARS-CoV-2 as determined by an IgG test kit, blood was collected from the Thai Red Cross Society (IRB approval #426/63). All patients and the healthy controls in this study have not received any SARS-CoV-2 vaccination at the time of disease onset and/or blood collection.
PBMC stimulation to study innate cell profiles
One million isolated PBMCs were plated on 24-well plates and cultured with or without 100 ng/ml E. coli LPS (Sigma Aldrich) for 6 h. To perform intracellular cytokine staining, after 2 h of LPS treatment, monensin (Biolegend) was added to the culture and further incubated for 4 h.
Cell surface and intracellular cytokine staining (ICS) for flow cytometry
To determined innate immune cell profiles, treated PBMCs as indicated were stained with surface markers before fixation and permeabilization using BD Cytofix/Cytoperm™ (BD Biosciences) following staining with antibodies to cytokines. List of antibodies used for flow cytometry was shown in Supplementary Table 2. Foxp3/Transcription Factor Staining Buffer Set (eBioscience) was used for Treg panel. Flow cytometry was performed using a CytoFLEX flow cytometer (Beckman Coulter Life Sciences). All data were analyzed using FlowJo X software (Tree Star)..
Cytokine measurement using Luminex assay
Bio-Plex Pro Human Cytokine 27-plex Assay (BioRad) was performed to investigate cytokine and chemokine levels in the serum of healthy individuals and recovered COVID-19 patients. Fifty microliters of diluted sera were used for the assay following the manufacturer's instructions. Samples were measured by Bio-Plex 200 Systems (BioRad).
Post COVID-19 infection questionnaires
A post-COVID-19 symptoms questionnaire was conducted by phone. Lists of 55 symptoms based on WHO Post-COVID-19 Case Report Form (Supplementary Table 3) were asked as absent or present after 4 weeks of post-infection.
Statistical analysis
All statistical analyses were performed using GraphPad Prism 7 software. The results represented the mean ± SEM of the sample group. Group comparisons between recovered COVID-19 patients (mild, moderate, severe) and healthy controls were analyzed using Kruskal–Wallis multiple comparisons analysis. p-values less than 0.05 indicated statistical significance; p < 0.05 (∗), p < 0.01 (∗∗), p < 0.001 (∗∗∗).
Results
Demographics and clinical characteristics
Demographic information of 62 COVID-19 patients was shown in the Supplementary Table 4. The overall median age was 36 years, and age exhibited a positive correlation with disease severity. Severe patients showed a higher median age compared to other groups (Supplementary Table 4). The most frequent comorbidities were coronary artery disease/cardiac allograft vasculopathy, diabetes, hypertension and dyslipidemia. For hospital admission laboratory parameters, the severe group displayed higher WBC count, ALT, AST and CRP, which are markers of cell damage and inflammation, when compared to the mild and moderate groups.
Natural killer (NK) cell profiles in recovered COVID-19 patients
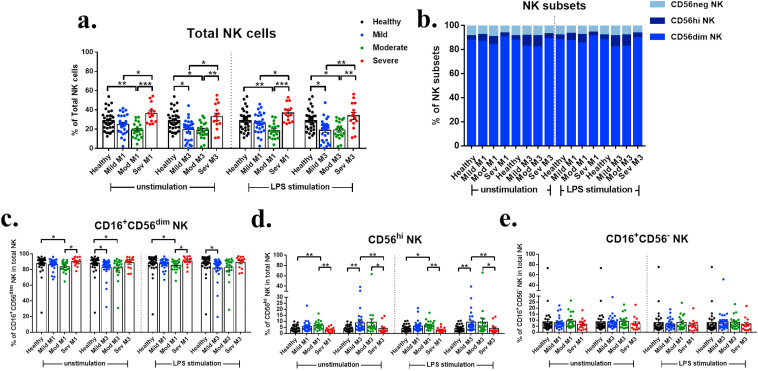
NK cell profiles of recovered patients were first examined because they play a crucial role in controlling viral infection primarily by cell-mediated cytotoxicity and secretion of IFN-γ.14 The gating strategy for flow cytometry used to identify NK population is shown in Supplementary Figs. 1–2. Severe group exhibited significantly increased frequencies of total NK cells in PBMCs compared to mild and moderate groups (Fig. 1 a). Severe group showed an increase of CD56dim subset and a decrease of CD56hi subset compared to other groups (Fig. 1b–e). The increased levels of pro-inflammatory cytokines, especially IL-6, impair the expansion and functions of NK cells15 which may be one possible mechanism of decreasing CD56hi NK subset in severe patients. To compare between 1 month and 3 months after recovery in each severity group, mild group exhibited significantly decreased total NK and CD56dim NK at 3 months (Supplementary Fig. 4). The results suggested that NK cells were transiently decreased in the mild and moderate groups and increased in the severe group at least 3 months after recovery.
Fig. 1.
NK cell profiles in COVID-19 patients after recovery. a. The percentage of total NK cells in lymphocyte population. b. Bar chart showing the percentage of NK cell subsets within the total NK cells population. c. The percentage of CD16+CD56dim among NK cells. d. The percentage of CD56hi NK among NK cells. e. The percentage of CD16+CD56- among NK cells.
Monocytes and MDSC profiles of recovered COVID-19 patients
We next investigated the monocyte profiles, as they were identified previously to play a key role in cytokine storms in COVID-19.6 Severe patients at 1 month exhibited significantly increased frequencies of total monocytes compared to healthy controls in both unstimulated and LPS-stimulated condition (Fig. 2 a). The mild group exhibited significantly increased frequencies of the classical monocyte (CD14++CD16-) and decreased the intermediate (CD14++CD16+) and the non-classical subsets (CD14+CD16+) at 1 month compared to healthy controls. LPS stimulation did not show any changes in the frequencies of monocyte subsets in any of the samples (Fig. 2c–e). In comparing profiles at 1 and 3 months within the groups, the mild group exhibited significantly decreased total monocytes at 3 months compared to 1 month (Supplementary Fig. 4).
Fig. 2.
Monocyte profiles in COVID-19 patients after recovery. a. The percentage of total monocytes in the monocyte population. b. Bar chart showing the percentage of monocyte subsets within the total monocyte population. c. The percentage of classical monocytes among total monocytes. d. The percentage of intermediate monocytes among total monocytes. e. The percentage of nonclassical monocytes among total monocytes.
There is no difference in the frequencies of MDSCs (Fig. 3 a). However, the percentage of PMN-MDSCs was significantly increased in all COVID-19 groups compared to healthy controls up to 3 months (Fig. 3b). Severe group exhibited significantly decreased of PMN-MDSCs at 3 months compared to 1 month (Supplementary Fig. 4). Previous study reported the correlation between the increase of PMN-MDSCs and the elevation of IL-1β, IL-6, IL-8 and TNF-α levels in serum, especially in severe COVID-19. The increase in the frequencies of PMN-MDSCs leads to the inhibition of IFN-γ production in T cells.16 For M-MDSCs, all COVID-19 groups displayed significantly increased frequencies at both 1 and 3 months upon LPS stimulation (Fig. 3c). Taken together, while no major changes were observed in the monocyte subsets among recovered patients, increased PMN-MDSCs at least up to 3 months after recovery were observed in patients with clinical symptoms.
Fig. 3.
MDSC profiles in COVID-19 patients after recovery. a. The percentage of total MDSCs in the monocyte population. b. The percentage of PMN-MDSCs among total MDSCs. c. The percentage of M-MDSCs among total MDSCs.
Profiles of IL-6, TNF-α producing and antigen presenting molecule expressing cells
We assessed the production of two major proinflammatory cytokines, IL-6 and TNF-α and the levels of costimulatory molecules HLA-DR and CD86 expression in LPS-stimulated monocytes. As shown in Fig. 4 , monocytes from the mild and severe groups at 1 month showed a significant increase in the percentages of IL-6- and TNF-α-producing monocytes and the level of TNF-α as shown by the MFI (Fig. 4a–d). Mild group exhibited significantly decreased MFI of IL-6 at 3 months compared to 1 month (Supplementary Fig. 4).
Fig. 4.
IL-6 and TNF-α production in monocytes of COVID-19 patients after recovery. a. The percentage of IL-6+ monocytes. b. The percentage of TNF-α+ monocytes. c. MFI of IL-6+ monocytes. d. MFI of TNF-α+ monocytes.
Moreover, in unstimulated condition, significantly decreased percentages and MFI of HLA-DR-expressing monocytes in the severe groups up to 3 months compared to healthy controls were observed (Fig. 5 a, c), and significantly decreased MFI of CD86 in all severity groups at 3 months compared to the healthy controls (Fig. 5d). These data demonstrated that SARS-CoV-2 infection resulting in clinical symptoms has a lasting impact on the responses of monocytes to stimulus by enhancing proinflammatory cytokine production and decreasing costimulatory molecule expression at least up to 3 months after recovery.
Fig. 5.
HLA-DR and CD86 in monocytes of COVID-19 patients after recovery. a. The percentage of HLA-DR + monocytes. b. The percentage of CD86+ monocytes. c. MFI of HLA-DR + monocytes. d. MFI of CD86+ monocytes.
Profiles of T lymphocytes in recovered COVID-19 patients
Innate immune cells crosstalk with adaptive immune cells to mount optimal immune responses for viral clearance. We determined the profiles of conventional T cells and MAIT cells at 1 and 3 months after recovery. A severe group at 1 month presented with significantly decreased frequencies of CD3+ T cells compared to the other groups, and this reduction in severe group, but not in mild and moderate group, persisted until 3 months after recovery (Fig. 6 a).
Fig. 6.
T cell profiles in COVID-19 patients after recovery. a. The percentage of CD3+, CD4+ and CD8+ T cells. b. The percentage of Tregs and MFI of Foxp3+ Tregs among CD3+ T cells in PBMCs.
Next, we examined the frequencies of Tregs and found a significant decrease in moderate group at 1 month but a significant decrease in mild group at 3 months compared to healthy controls (Fig. 6b). In addition, MFI of Foxp3+ in mild and moderate groups showed a significantly decreased compared to healthy controls, while a significant increase was displayed in severe group compared to mild group at 1 month. At 3 months, only mild group sustained a significant decrease of MFI of Foxp3+.
Because the roles of mucosal immune response mediated by MAIT cells were reported during SARS-CoV-2 infection, the frequencies of this subset of T cells as defined by CD161+TCRVα7.2+ T cells (Supplementary Fig. 3b) were investigated. The results showed no significant difference in their frequencies among the recovered patients. We found that the CCR6+ MAIT cells and MFI of CCR6 among all MAIT cells in all recovered groups tended to decrease but not reach statistical significance (Supplementary Fig. 5). Furthermore, it showed significant decrease of MAIT cells and increase of CCR6+ MAIT cells in mild and moderate group, respectively at 3 month (Supplementary Fig. 6).
Cytokine profiles in the serum of COVID-19 patients at 1 month after recovery
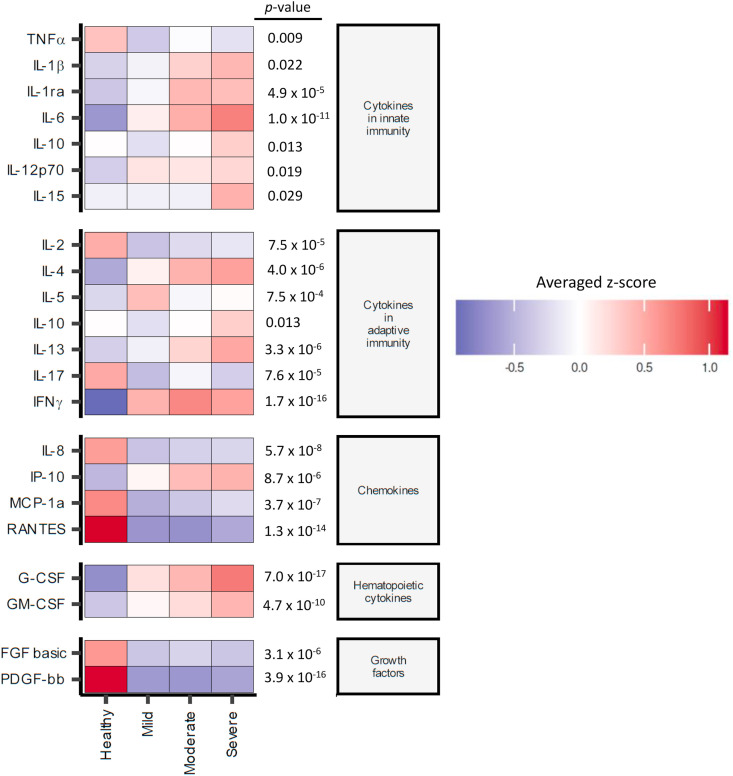
The profiles of 27 cytokines and chemokines belonging to 5 groups of different functions, including innate cytokines, adaptive cytokines, chemokines, hematopoietic cytokines and growth factors were investigated in the serum of patients 1 month after recovery. Heatmaps revealed twenty-one statistically significant differences in the levels of cytokines and chemokines among healthy controls, mild, moderate and severe COVID-19 patients (Fig. 7 ). All innate cytokines, except for TNF-α, were higher in the severe group, and the patterns displayed a correlation between disease severity and cytokine levels. For adaptive cytokines, an increase in the cytokines of mixed Th1/Th2 profiles (IL-4, IL-13 and IFNγ) was observed in patients in the moderate and severe groups, while IL-17 was lower in all patient groups. The chemokine IP-10, G-CSF and GM-CSF was higher in all patient groups regardless of severity, indicating active myelopoiesis in convalescent patients. The levels of growth factors (FGF basic and PDGF-bb) and chemokines (IL-8, MCP-1a and RANTES) were lower in recovered patients. The bar graphs in Supplementary Fig. 8 showed the comparison of cytokine/chemokine levels in each group. This result strongly indicates the lasting impact of SARS-CoV-2 infection at the systemic level of immune functions regardless of disease severity.
Fig. 7.
Serum cytokine profiles in COVID-19 patients 1 month after recovery. The serum after 1 month of recovery in COVID-19 patients were measured cytokine and chemokine profiles using a Bio-Plex multiplex immunoassay. The relative levels of 21 cytokines with statistical difference by Kruskal–Wallis multiple comparisons analysis (p-value < 0.05) were shown in the heatmap.
Association between immune profiles with long COVID symptoms
To investigate the association of the immune profiles at 1 and 3 months post recovery and long COVID symptoms in the patients, we acquired questionnaire data from the phone interview post-COVID. We obtained the data from 25 out of 62 recovered patients in our cohort. The top-ten most recognized long-term effects of COVID-1917 are fatigue, headache, attention disorder, hair loss, dyspnea, ageusia, anosmia, post-activity polypnea, joint pain and cough. We analyzed the association of the immune parameters and the 3 groups of patients, i.e. those with no long COVID symptoms, (N = 3); patients with 1–3 symptoms (N = 12); patients with >3 symptoms (N = 10). Despite a limited sample size, increased monocytes and IL-6- and TNF-α-producing cells at 1 month post recovery were significantly associated with patients with long COVID symptoms (Fig. 8 ). Furthermore, decreased MFI of CD86 and HLA-DR at 3 months post recovery were evidently observed in patients with long COVID symptoms. PMN-MDSCs and M-MDSCs were increased in patients with long COVID even 3 months post recovery. For NK cells, total NK cells were significantly decreased in LPS-stimulated PBMCs in patients with long COVID of >3 symptoms at 3 months post recovery. Moreover, there is a trend of decreased Treg cells at 1 month post recovery in patients with long COVID symptoms, although it is not statistically significance.
Fig. 8.
Long-term effects in COVID-19 patients. The 25 from 62 recovered patients were followed up with their ten common long COVID symptoms. The comparisons of the immune cell profiles among patients with 0, 1–3, and >3 long-term symptoms of COVID-19 were shown. Kruskal–Wallis multiple comparisons analysis, p-values less than 0.05 indicated statistical significance; p < 0.05 (∗), p < 0.01 (∗∗), p < 0.001 (∗∗∗).
Discussion
The exaggerated inflammatory response in COVID-19 patients potentially influences innate immune cells after recovery. Long COVID is described as a condition experienced by recovered patients that includes persistent symptoms with delayed or long-term complications beyond 4 weeks from the onset of symptoms.12 , 18 Therefore, in this study, we focused on the profiles of innate immune cells, i.e., NK, monocytes, MDSC and MAIT cells after recovery from COVID-19 up to 3 months.
During acute COVID-19, adaptive-like NK cell (CD56dim) expansion and dysfunction were reported to be associated with severe symptoms.19 , 20 Furthermore, expansion of the unconventional CD56dimCD16neg NK cell population was observed in PBMCs from patients with COVID-19 with decreased NK cell cytotoxicity.21 This abnormality was prolonged in severe patients up to 30 days after recovery.22 In our study, an increase in the frequency of NK cells without any changes in the subset compositions was observed in the severe group up to 3 months post recovery. The increase in the overall NK population only in the severe group may indicate a compensatory mechanism that drives the expansion of NK cells. Interestingly, the severe group exhibited decreased frequency of the CD56hi NK cell subset, compared with mild/moderate patients. Several studies showed the dysregulation of NK cell subset at the cell numbers and functions which is partially dependent on IL-6.23 , 24 In serum at 1 month after the disease onset, increased IL-6 was found to be correlated with disease severity.
Various studies have indicated monocytes to be the primary cell types that drive hyperinflammation, the functions of which are associated with severe clinical symptoms.4 , 6 , 8 Monocytes are also the part of trained immunity that is responsible for enhanced innate immune response after repeated exposure to stimuli such as beta-glucan and LPS.25 Increased total monocytes were observed at 1 month, and subtle changes in all monocyte subsets were observed at 1 and 3 months in the severe group.
More importantly, increased IL-6- and TNF-α-producing cells and increased expression levels were observed in mild and severe COVID-19 patients in response to LPS stimulation at 1 month but not 3 months. These enhanced responses by monocytes disappeared after 3 months, suggesting a short-lived phenomenon. These data seem indicated an enhanced response to LPS after experiencing SARS-CoV-2 infection. Whether this enhanced response is the result of innate immune memory needs further investigation. In the mild and severe groups, the percentages of IL-6- and TNF-α-producing cells were higher than those in the moderate group.
In contrast to the enhanced proinflammatory cytokines, the levels of HLA-DR, as indicated by the MFI of HLA-DR, were also lower in the moderate and severe groups, indicating reduced antigen presenting capacity. Our finding is consistent with the recent in a single-cell RNA-sequencing study showing downregulated HLA class II on monocytes of severe/critical recovered patients.26 During acute SARS-CoV-2 infection, reduced HLA-DR in myeloid cells was also reported.5 Therefore, suppressed antigen-presenting functions may be a feature in post-COVID-19 patients and may influence adaptive immune responses.
We detected drastically increase in MDSCs in recovered patients that persisted for at least 3 months. This increase was independent of LPS stimulation for PMN-MDSCs, while increased percentages of M-MDSCs required LPS stimulation. Expansion of MDSCs has been reported during various conditions, including cancer and inflammation, including SARS-CoV-2 infection.7 , 27 Our results were consistent with the results observed in the Japanese COVID-19 patients.8 , 28 MDSCs negatively regulate the immune response, through various mechanisms, including direct cell–cell contact and mediator production, for example, ROS and nitrogen species.27 Interestingly, in the serum of recovered severe patients, we detected significantly increased IL-13 and IL-4, which are involved in the induction of MDSC activation27 and increased IL-1β which are involved in MDSC recruitment and induction.29 PMN-MDSC functions to suppress antigen-specific T cell response via the production of mediators such as reactive oxygen species, Arg1, and prostaglandin E2.30 In acute SARS-CoV-2 infection, increased PMN-MDSC was pronounced in patients with ARDS and may play a role in prolong viral clearance.31 Early expansion of PMN-MDSCs suppressed IFN-γ production in SARS-CoV-2-specific T cells and strongly associated with fatal COVID-19 outcomes.32 Whether the increase in MDSCs has any impact on immune response in COVID-19 patients post recovery remains unknown.
In serum samples 1 month post recovery, most innate cytokines were higher in the moderate and severe groups in a severity-dependent manner. This result strongly agrees with previous studies that showed increased innate cytokines in COVID-19 patients during and post recovery.3 , 33 Increased Th1/Th2 signature cytokines were observed, while IL-17 was lower in post recovery patients. Among the detected chemokines, IP-10 (CXCL10) was higher in convalescent serum in a severity-dependent manner, highlighting the importance of this chemokine in COVID-19.34 Various studies identified IP-10 as a good biomarker for the prediction of COVID-19 progression and is related to the risk of death in COVID-19 patients.35 This chemokine is induced in response to IFN-γ and acts as a chemotactic factor for T cells, NK cells, monocytes/macrophages and DC. Thus, it is possible that persistent serum IP-10 during recovery at 1 month may delay the resolution of inflammatory responses even after viral clearance. Whether IP-10 plays any role in long COVID remains unanswered. Myelopoiesis cytokines were also higher in all recovered patients, indicating active recovery of the myeloid compartment in these patients. This is consistent with a report of the dysregulated myeloid compartment in severe COVID-19 groups.36
Previous studies reported the association between age and comorbidities with COVID-19 severe outcome by a defective immunological response to SARS-CoV-2 infection.37 , 38 In addition, in the study of immune response after ChAdOx1nCoV-19 vaccination, aging population showed defective innate immune responses to vaccine that resulted in weak and/or delayed innate immune activation, compared with young people.39 In this study, we found that age is one of the significant risk factors among patients. Immune profiles and functions of COVID-19 patient with combination of diabetes, hypertension and coronary artery disease (n = 5) were showed in Supplementary Fig. 9. Increase in the frequencies of M-MDSCs in unstimulated PBMCs and decrease in the MFI of CD86-expressing monocytes in LPS-stimulated PBMCs were showed in patients with comorbidities at 1 month. These suggest that age and comorbidities also affect the severity of patients and may result in impaired immune function in severe COVID-19 patients.
Based on results from our long COVID questionnaire, we observed an association between aberrant innate immune response, particularly with overactivity of monocytes at 1 month after recovery with long-term complications. Interestingly, we also observed a negative association between adaptive immunity and long COVID symptoms which may underscore the importance of adaptive immunity. Several studies indicated the persistent activation of innate immune cells after recovery from SARS-CoV-2 infections and the potential roles of epigenetic memory of innate immune cells and their progenitors in the post-acute COVID-19 sequelae.40, 41, 42 Our data are also consistent with this proposed mechanism of long COVID where hyperactivation of innate immune cells and decreasing suppressive or regulatory cells were found in patients with more than 3 long COVID symptoms.
The strength of this study is that our data provides strong evidence that immune response to acute SARS-CoV-2 infection has lasting impact on both innate and adaptive immune cells even after 3 months post recovery. We further demonstrated that this impact could influence the functions of innate immune cells upon responding to unrelated dangers. Some of these immune parameters were also correlated with the presence of the long COVID symptoms. How acute COVID-19 infection affects the immune response post recovery remains unanswered. In addition, the small sample size for following up on subjects with long COVID limits our interpretation in relationship with long COVID, which increasingly becomes public health concerns.
Taken together, we uncovered a lasting impact of COVID-19 infection on innate immune cells up to 3 months after recovery at both the cellular and cytokine levels.
Author contributions
VR and PS were responsible for sample collection, data analysis and manuscript preparation. PK analyzed serum cytokine data. WB and SK were responsible for sample collection. RR was involved in MAIT cells experiment. CT, KM, NM and KL were responsible for the design and co-ordinate the interview of post-COVID-19 symptoms. PT, LP, and OP were responsible for patient enrollment in the Biobank for biospecimen collection. TP was involved in the overall experimental design, data analysis and manuscript preparation. NH was responsible for patient enrollment, securing funding, experimental design and manuscript preparation.
Declaration of competing interest
The authors declare no competing interests.
Acknowledgments
This study was supported by the e-ASIA Joint Research Program (e-ASIA JRP) from the National Science and Technology Development Agency; Ratchadapisek Somphot Fund, Faculty of Medicine, Chulalongkorn University, grant number RA(PO)005/63; and by the National Research Council of Thailand (grant number 811/2563). VR is supported by the Postdoctoral Fellowship, Ratchadapisek Somphot Fund, Chulalongkorn University. Biospecimen collection was supported by Biobank, Faculty of Medicine, Chulalongkorn University. NM is supported by the Pink Diamond Project, Faculty of Medicine, Chulalongkorn University. We would like to thank Jiratchaya Sophonphan for some statistical analysis. We gratefully acknowledge the assistance of the study team with interview post-COVID-19 symptoms; Abhichaya Tungwongkitsiri, Chanya Mittrakulkij, Farsai Chiewbangyang, Janista Kaewsrihawong, Jirayu Sanpakit, Kanokphet Kulkiatprasert, Khemmachat Munkong, Nanthida Keawthawon, Natchanon Wattanakul, Natdanai Limchanachon, Natthapat Roopsuwankun, Natthasini Chaosuwannakij, Pasin Larpanekanan, Pawit Pitakkitnukun, Pongpon Homswad, Samapitch Ratanapraisorn, Sarunyapong Atchariyapakorn, Sasathamon Vongphanich, Sirapat Jessadapornchai, Teton Avihingsanon and Thanatorn Piyasathapornpong.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jmii.2022.09.001.
Contributor Information
study team:
Abhichaya Tungwongkitsiri, Chanya Mittrakulkij, Farsai Chiewbangyang, Janista Kaewsrihawong, Jirayu Sanpakit, Kanokphet Kulkiatprasert, Khemmachat Munkong, Nanthida Keawthawon, Natchanon Wattanakul, Natdanai Limchanachon, Natthapat Roopsuwankun, Natthasini Chaosuwannakij, Pasin Larpanekanan, Pawit Pitakkitnukun, Pongpon Homswad, Samapitch Ratanapraisorn, Sarunyapong Atchariyapakorn, Sasathamon Vongphanich, Sirapat Jessadapornchai, Teton Avihingsanon, and Thanatorn Piyasathapornpong
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Jordan R.E., Adab P., Cheng K.K. Covid-19: risk factors for severe disease and death. BMJ. 2020;368:m1198. doi: 10.1136/bmj.m1198. [DOI] [PubMed] [Google Scholar]
- 2.Ragab D., Salah Eldin H., Taeimah M., Khattab R., Salem R. The COVID-19 cytokine storm; what we know so far. Front Immunol. 2020;11:1446. doi: 10.3389/fimmu.2020.01446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lucas C., Wong P., Klein J., Castro T.B.R., Silva J., Sundaram M., et al. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature. 2020;584:463–469. doi: 10.1038/s41586-020-2588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mann E.R., Menon M., Knight S.B., Konkel J.E., Jagger C., Shaw T.N., et al. Longitudinal immune profiling reveals key myeloid signatures associated with COVID-19. Sci Immunol. 2020;5 doi: 10.1126/sciimmunol.abd6197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arunachalam P.S., Wimmers F., Mok C.K.P., Perera R., Scott M., Hagan T., et al. Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans. Science. 2020;369:1210–1220. doi: 10.1126/science.abc6261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vanderbeke L., Van Mol P., Van Herck Y., De Smet F., Humblet-Baron S., Martinod K., et al. Monocyte-driven atypical cytokine storm and aberrant neutrophil activation as key mediators of COVID-19 disease severity. Nat Commun. 2021;12:4117. doi: 10.1038/s41467-021-24360-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Agrati C., Sacchi A., Bordoni V., Cimini E., Notari S., Grassi G., et al. Expansion of myeloid-derived suppressor cells in patients with severe coronavirus disease (COVID-19) Cell Death Differ. 2020;27:3196–3207. doi: 10.1038/s41418-020-0572-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takano T., Matsumura T., Adachi Y., Terahara K., Moriyama S., Onodera T., et al. Myeloid cell dynamics correlating with clinical outcomes of severe COVID-19 in Japan. Int Immunol. 2021;33:241–247. doi: 10.1093/intimm/dxab005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lo Presti E., De Gaetano A., Pioggia G., Gangemi S. Comprehensive analysis of the ILCs and unconventional T cells in virus infection: profiling and dynamics associated with COVID-19 disease for a future monitoring system and therapeutic opportunities. Cells. 2022:11. doi: 10.3390/cells11030542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deschler S., Kager J., Erber J., Fricke L., Koyumdzhieva P., Georgieva A., et al. Mucosal-associated invariant T (MAIT) cells are highly activated and functionally impaired in COVID-19 patients. Viruses. 2021:13. doi: 10.3390/v13020241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flament H., Rouland M., Beaudoin L., Toubal A., Bertrand L., Lebourgeois S., et al. Outcome of SARS-CoV-2 infection is linked to MAIT cell activation and cytotoxicity. Nat Immunol. 2021;22:322–335. doi: 10.1038/s41590-021-00870-z. [DOI] [PubMed] [Google Scholar]
- 12.Nalbandian A., Sehgal K., Gupta A., Madhavan M.V., McGroder C., Stevens J.S., et al. Post-acute COVID-19 syndrome. Nat Med. 2021;27:601–615. doi: 10.1038/s41591-021-01283-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kowitdamrong E., Puthanakit T., Jantarabenjakul W., Prompetchara E., Suchartlikitwong P., Putcharoen O., et al. Antibody responses to SARS-CoV-2 in patients with differing severities of coronavirus disease 2019. PLoS One. 2020;15 doi: 10.1371/journal.pone.0240502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.García M., Kokkinou E., Carrasco García A., Parrot T., Palma Medina L.M., Maleki K.T., et al. Innate lymphoid cell composition associates with COVID-19 disease severity. Clin Transl Immunology. 2020;9:e1224–e. doi: 10.1002/cti2.1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Osman M., Faridi R.M., Sligl W., Shabani-Rad M.-T., Dharmani-Khan P., Parker A., et al. Impaired natural killer cell counts and cytolytic activity in patients with severe COVID-19. Blood Advances. 2020;4:5035–5039. doi: 10.1182/bloodadvances.2020002650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sacchi A., Grassi G., Bordoni V., Lorenzini P., Cimini E., Casetti R., et al. Early expansion of myeloid-derived suppressor cells inhibits SARS-CoV-2 specific T-cell response and may predict fatal COVID-19 outcome. Cell Death Dis. 2020;11:921. doi: 10.1038/s41419-020-03125-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lopez-Leon S., Wegman-Ostrosky T., Perelman C., Sepulveda R., Rebolledo P.A., Cuapio A., et al. More than 50 long-term effects of COVID-19: a systematic review and meta-analysis. Sci Rep. 2021;11 doi: 10.1038/s41598-021-95565-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Montani D., Savale L., Noel N., Meyrignac O., Colle R., Gasnier M., et al. Post-acute COVID-19 syndrome. Eur Respir Rev. 2022:31. doi: 10.1183/16000617.0185-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bjorkstrom N.K., Ponzetta A. Natural killer cells and unconventional T cells in COVID-19. Curr Opin Virol. 2021;49:176–182. doi: 10.1016/j.coviro.2021.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bjorkstrom N.K., Strunz B., Ljunggren H.G. Natural killer cells in antiviral immunity. Nat Rev Immunol. 2022;22:112–123. doi: 10.1038/s41577-021-00558-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bi J. NK cell dysfunction in patients with COVID-19. Cell Mol Immunol. 2022;19:127–129. doi: 10.1038/s41423-021-00825-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leem G., Cheon S., Lee H., Choi S.J., Jeong S., Kim E.S., et al. Abnormality in the NK-cell population is prolonged in severe COVID-19 patients. J Allergy Clin Immunol. 2021;148:996–1006 e18. doi: 10.1016/j.jaci.2021.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mazzoni A., Salvati L., Maggi L., Capone M., Vanni A., Spinicci M., et al. Impaired immune cell cytotoxicity in severe COVID-19 is IL-6 dependent. J Clin Invest. 2020;130:4694–4703. doi: 10.1172/JCI138554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Osman M., Faridi R.M., Sligl W., Shabani-Rad M.T., Dharmani-Khan P., Parker A., et al. Impaired natural killer cell counts and cytolytic activity in patients with severe COVID-19. Blood Adv. 2020;4:5035–5039. doi: 10.1182/bloodadvances.2020002650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bekkering S., Dominguez-Andres J., Joosten L.A.B., Riksen N.P., Netea M.G. Trained immunity: reprogramming innate immunity in health and disease. Annu Rev Immunol. 2021;39:667–693. doi: 10.1146/annurev-immunol-102119-073855. [DOI] [PubMed] [Google Scholar]
- 26.Li X., Garg M., Jia T., Liao Q., Yuan L., Li M., et al. Single-cell analysis reveals the immune characteristics of myeloid cells and memory T cells in recovered COVID-19 patients with different severities. Front Immunol. 2021;12 doi: 10.3389/fimmu.2021.781432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gabrilovich D.I., Nagaraj S. Myeloid-derived suppressor cells as regulators of the immune system. Nat Rev Immunol. 2009;9:162–174. doi: 10.1038/nri2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park S.J., Nam D.E., Seong H.C., Hahn Y.S. New discovery of myeloid-derived suppressor cell's tale on viral infection and COVID-19. Front Immunol. 2022;13 doi: 10.3389/fimmu.2022.842535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Elkabets M., Ribeiro V.S., Dinarello C.A., Ostrand-Rosenberg S., Di Santo J.P., Apte R.N., et al. IL-1beta regulates a novel myeloid-derived suppressor cell subset that impairs NK cell development and function. Eur J Immunol. 2010;40:3347–3357. doi: 10.1002/eji.201041037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perfilyeva Y.V., Ostapchuk Y.O., Tleulieva R., Kali A., Abdolla N., Krasnoshtanov V.K., et al. Myeloid-derived suppressor cells in COVID-19: a review. Clin Immunol. 2022;238 doi: 10.1016/j.clim.2022.109024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coudereau R., Waeckel L., Cour M., Rimmele T., Pescarmona R., Fabri A., et al. Emergence of immunosuppressive LOX-1+ PMN-MDSC in septic shock and severe COVID-19 patients with acute respiratory distress syndrome. J Leukoc Biol. 2022;111:489–496. doi: 10.1002/JLB.4COVBCR0321-129R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sacchi A., Grassi G., Bordoni V., Lorenzini P., Cimini E., Casetti R., et al. Early expansion of myeloid-derived suppressor cells inhibits SARS-CoV-2 specific T-cell response and may predict fatal COVID-19 outcome. Cell Death Dis. 2020;11:921. doi: 10.1038/s41419-020-03125-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hasichaolu, Zhang X., Li X., Li X., Li D. Circulating cytokines and lymphocyte subsets in patients who have recovered from COVID-19. BioMed Res Int. 2020;2020 doi: 10.1155/2020/7570981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang Y., Shen C., Li J., Yuan J., Wei J., Huang F., et al. Plasma IP-10 and MCP-3 levels are highly associated with disease severity and predict the progression of COVID-19. J Allergy Clin Immunol. 2020;146:119–127 e4. doi: 10.1016/j.jaci.2020.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen Y., Wang J., Liu C., Su L., Zhang D., Fan J., et al. IP-10 and MCP-1 as biomarkers associated with disease severity of COVID-19. Mol Med. 2020;26:97. doi: 10.1186/s10020-020-00230-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schulte-Schrepping J., Reusch N., Paclik D., Bassler K., Schlickeiser S., Zhang B., et al. Severe COVID-19 is marked by a dysregulated myeloid cell compartment. Cell. 2020;182:1419–1440 e23. doi: 10.1016/j.cell.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moreno Fernández-Ayala D.J., Navas P., López-Lluch G. Age-related mitochondrial dysfunction as a key factor in COVID-19 disease. Exp Gerontol. 2020;142 doi: 10.1016/j.exger.2020.111147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sanyaolu A., Okorie C., Marinkovic A., Patidar R., Younis K., Desai P., et al. Comorbidity and its impact on patients with COVID-19. SN Comprehensive Clinical Medicine. 2020;2:1069–1076. doi: 10.1007/s42399-020-00363-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shen C.F., Yen C.L., Fu Y.C., Cheng C.M., Shen T.C., Chang P.D., et al. Innate immune responses of vaccinees determine early neutralizing antibody production after ChAdOx1nCoV-19 vaccination. Front Immunol. 2022;13 doi: 10.3389/fimmu.2022.807454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hirschenberger M., Hunszinger V., Sparrer K.M.J. Implications of innate immunity in post-acute sequelae of non-persistent viral infections. Cells. 2021:10. doi: 10.3390/cells10082134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Phetsouphanh C., Darley D.R., Wilson D.B., Howe A., Munier C.M.L., Patel S.K., et al. Immunological dysfunction persists for 8 months following initial mild-to-moderate SARS-CoV-2 infection. Nat Immunol. 2022;23:210–216. doi: 10.1038/s41590-021-01113-x. [DOI] [PubMed] [Google Scholar]
- 42.Cheong J.-G., Ravishankar A., Sharma S., Parkhurst C.N., Nehar-Belaid D., Ma S., et al. Epigenetic memory of COVID-19 in innate immune cells and their progenitors. bioRxiv. 2022:2022. 02.09.479588. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.