Summary
“Pan-coronavirus” antivirals targeting conserved viral components can be designed. Here, we show that the rationally engineered H84T-banana lectin (H84T-BanLec), which specifically recognizes high mannose found on viral proteins but seldom on healthy human cells, potently inhibits Middle East respiratory syndrome coronavirus (MERS-CoV), severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (including Omicron), and other human-pathogenic coronaviruses at nanomolar concentrations. H84T-BanLec protects against MERS-CoV and SARS-CoV-2 infection in vivo. Importantly, intranasally and intraperitoneally administered H84T-BanLec are comparably effective. Mechanistic assays show that H84T-BanLec targets virus entry. High-speed atomic force microscopy depicts real-time multimolecular associations of H84T-BanLec dimers with the SARS-CoV-2 spike trimer. Single-molecule force spectroscopy demonstrates binding of H84T-BanLec to multiple SARS-CoV-2 spike mannose sites with high affinity and that H84T-BanLec competes with SARS-CoV-2 spike for binding to cellular ACE2. Modeling experiments identify distinct high-mannose glycans in spike recognized by H84T-BanLec. The multiple H84T-BanLec binding sites on spike likely account for the drug compound’s broad-spectrum antiviral activity and the lack of resistant mutants.
Keywords: antiviral, atomic force microscopy, banana, coronavirus, lectin, MERS-CoV, SARS-CoV-2, single-molecule force spectroscopy, spike, treatment
Graphical abstract
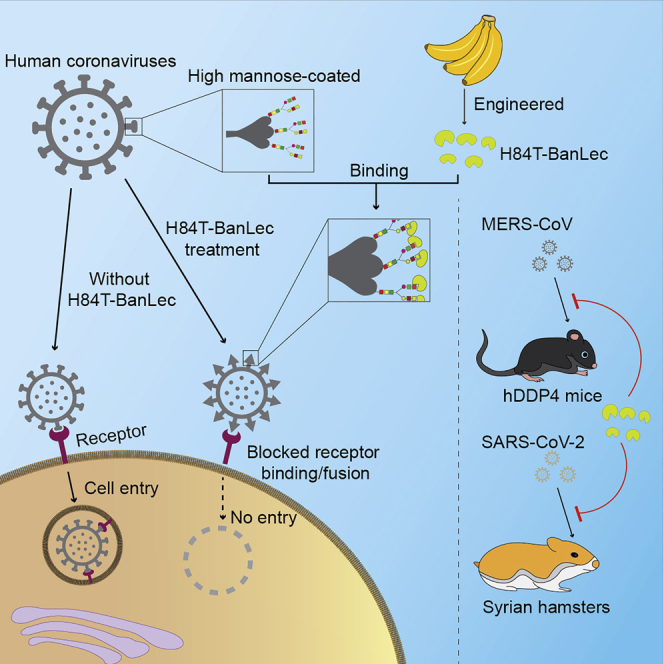
Highlights
-
•
H84T-BanLec inhibits human-pathogenic coronaviruses through inhibition of viral entry
-
•
H84T-BanLec protects against MERS-CoV and SARS-CoV-2 infections in vivo
-
•
H84T-BanLec binds to multiple SARS-CoV-2 spike high-mannose sites with high affinity
-
•
This unique mechanism likely accounts for H84T-BanLec’s broad-spectrum activity
Chan et al. investigate the antiviral activity and mechanism of H84T-BanLec against human-pathogenic coronaviruses. H84T-BanLec possesses pan-coronaviral antiviral activity through inhibition of viral entry and protects against MERS-CoV and SARS-CoV-2 infections in vivo. Binding of H84T-BanLec to multiple high-mannose glycans in spike likely accounts for its lack of resistant mutants.
Introduction
Coronaviruses are enveloped, positive-sense, single-stranded RNA viruses that have repeatedly crossed species barriers from animals to humans.1 Of the four genera in the genetically diverse Coronaviridae family (α, β, γ, and δ coronaviruses), three novel coronaviruses have emerged to cause epidemics or pandemics in humans in recent years: severe acute respiratory syndrome coronavirus (SARS-CoV) in 2002–2003, Middle East respiratory syndrome coronavirus (MERS-CoV) since 2012, and SARS-CoV-2 since 2019.2, 3, 4, 5 SARS and MERS frequently manifest as severe disease with high case fatality rates of around 10% and 35%, respectively, and coronavirus disease 2019 (COVID-19) caused by SARS-CoV-2 has caused nearly 6.4 million deaths worldwide as of August 3, 2022.6, 7, 8 Moreover, circulating common cold human coronaviruses (HCoVs) including HCoV-OC43, HCoV-229E, HCoV-NL63, and HCoV-HKU1 contribute notably to morbidity, especially in the elderly and immunocompromised.9
Despite the clinical and public health importance of these coronaviruses, effective anti-coronaviral treatment options remain scarce. Broad-spectrum antivirals such as remdesivir, interferons, nirmatrelvir/ritonavir, molnupiravir, ribavirin, lopinavir, and clofazimine have shown variable antiviral activities against one or more of these coronaviruses in vitro and/or in vivo, with remdesivir, nirmatrelvir/ritonavir, and, to a lesser degree, molnupiravir being particularly effective in early disease.10, 11, 12, 13, 14, 15 Monoclonal or single-domain antibodies are highly active in vitro and/or in vivo but are generally virus- or even strain-specific and may be ineffective against newly emerged variants such as the SARS-CoV-2 B.1.1.529 (Omicron) variant.16, 17, 18, 19, 20 Convalescent plasma may be useful in severe disease but was recently shown not to improve the overall mortality of patients with severe COVID-19 when started 8 days after symptom onset.21 Importantly, the development of novel specific antiviral agents and preparation of convalescent plasma are time consuming and inevitably lag behind highly transmissible pandemics such as COVID-19.9 Therapeutic and prophylactic options for the existing coronaviruses and emerging novel coronaviruses should be expanded by finding broad-spectrum agents that have “pan-coronaviral” antiviral activities.
Coronaviruses possess four major structural proteins, namely the spike, envelope, membrane, and nucleocapsid proteins.22 The membrane-anchored spike glycoprotein is crucial for virus-cell membrane fusion and cell entry.22 Lectins are carbohydrate-binding proteins widely found in nature, and some have potential antiviral activities.23 Clinical use of lectins, however, was limited in part by potential side effects mediated by lectin-induced mitogenicity.23 Banana lectin (BanLec) is a jacalin-related lectin isolated from the fruit of bananas (Musa acuminata).24 We have previously engineered a BanLec that has preserved broad-spectrum antiviral potency with significantly reduced mitogenicity by introducing a single amino acid substitution to replace histidine 84 with a threonine (H84T-BanLec).23 H84T-BanLec exhibits potent broad-spectrum in vitro and in vivo activities against viruses with high-mannose N-glycans, including human immunodeficiency viruses 1 and 2, influenza viruses A and B, Ebola virus, and hepatitis C virus, by binding to highly glycosylated viral proteins to block attachment, entry, membrane fusion, and/or additional steps in the viral replication cycle.23, 24, 25, 26 In this study, we report the pan-coronaviral antiviral activity of H84T-BanLec against human-pathogenic coronaviruses, including its in vivo protection against MERS-CoV and SARS-CoV-2, through binding with the glycosylated viral spike protein to inhibit virus entry into cells. Further, we used atomic force microscopy and other modalities to capture and model the strong and diverse molecular interactions of H84T-BanLec with the SARS-CoV-2 spike protein, interactions that help to explain why the molecularly engineered lectin is highly active against all SARS-CoV-2 variants tested.
Results
Pan-coronaviral antiviral activity of H84T-BanLec
To investigate whether H84T-BanLec possesses pan-coronaviral antiviral activity, we included the highly virulent MERS-CoV (lineage C betacoronavirus), the pandemic SARS-CoV-2 (lineage B betacoronavirus), SARS-CoV (lineage B betacoronavirus), HCoV-OC43 (lineage A betacoronavirus), and HCoV-229E (alphacoronavirus) to represent all the subgroups of human-pathogenic coronaviruses. H84T-BanLec significantly reduced MERS-CoV (Figure 1A) and SARS-CoV-2 (Figure 1B) RNA load in a dose-dependent manner in vitro. The mean viral load of MERS-CoV was reduced by 3.30 (Huh-7) to 7.11 (Calu-3) log10copies/mL and that of SARS-CoV-2 was reduced by 3.46 (Caco-2) log10copies/mL, respectively, at 90 nM H84T-BanLec. At ≥10 nM, H84T-BanLec completely inhibited infectious plaque formation of MERS-CoV (Figure 1C) and SARS-CoV-2 (Figure 1D). The half-maximal effective concentrations (EC50s) of H84T-BanLec against MERS-CoV and SARS-CoV-2 were 4.44 nM and 6.02 nM with selectivity indices (50% cytotoxic concentration [CC50]/EC50) of ≥2,252.3 and ≥1,661.1, respectively (Figure S1). In the cytopathic effect (CPE) inhibition assay, dose-dependent reduction of CPE could be consistently observed for both MERS-CoV (>80% in Vero and 100% in Calu-3 cells with ≥30 nM H84T-BanLec) (Figure 1E) and SARS-CoV-2 (>80% in VeroE6 cells) (Figure 1F) at 3 days post-infection (dpi). Similarly, H84T-BanLec exhibited potent antiviral activity against other human-pathogenic coronaviruses with dose-dependent reduction of mean viral RNA loads of 5.86, 3.55, and 2.87 log10copies/mL for SARS-CoV, HCoV-OC43, and HCoV-229E, respectively, at 90 nM H84T-BanLec (Figure 1G). At ≥10 nM, H84T-BanLec completely inhibited infectious SARS-CoV plaque formation with an EC50 of 5.49 nM (Figure S2A). CPE was minimally observed in cells with ≥10 (SARS-CoV) and ≥30 nM (HCoV-229E) of H84T-BanLec (Figure S2B). Importantly, H84T-BanLec was similarly active against SARS-CoV-2 variants, including the B.1.1.529 (Omicron), B.1.617.2 (Delta), B.1.1.7 (Alpha), and P.3 (Theta) variants with EC50 at low nanomolar concentrations (Figure S3). No resistant mutant was detected after eight serial passages of MERS-CoV and SARS-CoV-2 with H84T-BanLec in VeroE6 cells. These findings highlighted the pan-coronaviral antiviral activity and lack of resistance of H84T-BanLec against epidemic and circulating human-pathogenic coronaviruses, as well as emerging SARS-CoV-2 variants.
Figure 1.
Broad-spectrum in vitro antiviral activity of H84T-BanLec against human-pathogenic coronaviruses
(A–F) The antiviral activity of H84T-BanLec against MERS-CoV and SARS-CoV-2 (HKU-001a strain) was evaluated using (A and B) viral load reduction assay, (C and D) plaque reduction assay, and (E and F) CPE inhibition assay.
(G) The antiviral activity of H84T-BanLec against other human-pathogenic coronaviruses in vitro was determined by viral load reduction assay for SARS-CoV (Vero cells, ATCC CCL-81), HCoV-OC43 (BS-C-1 cells, ATCC CCL-26), and HCoV-229E (human embryonic lung fibroblasts HFL cells, in-house development). Data are mean ± SD, n = 3 biological replicates. One-way ANOVA. ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05.
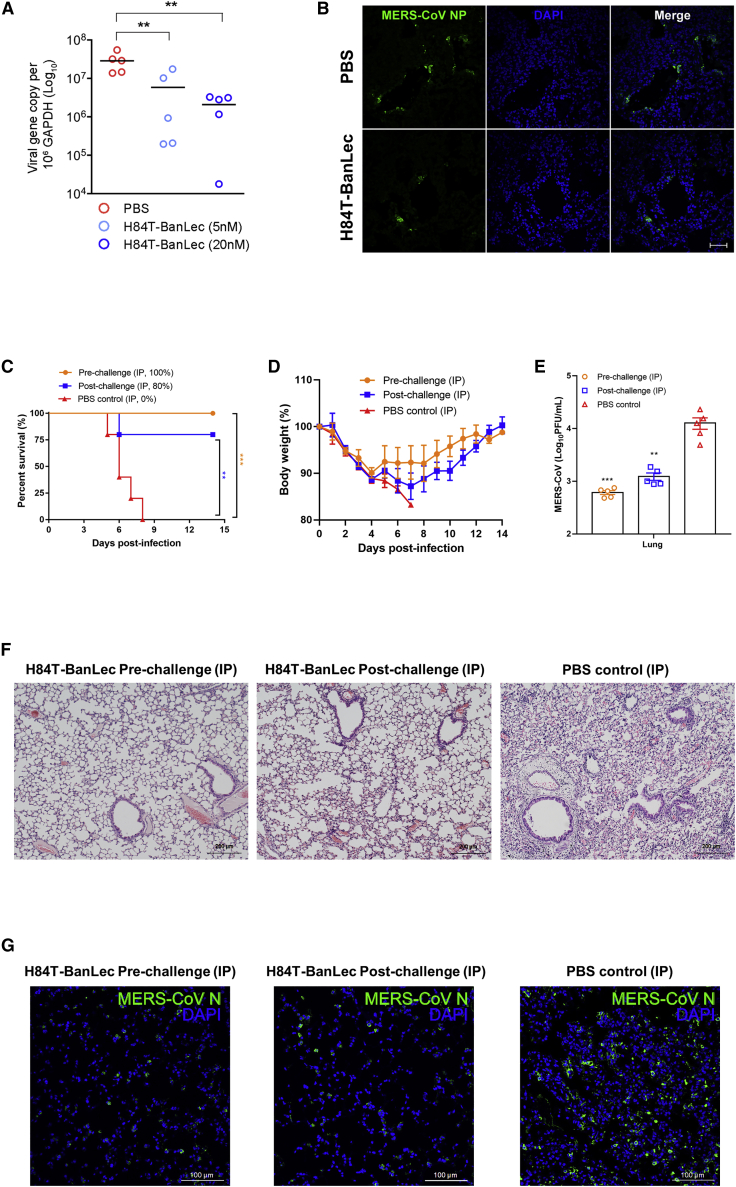
Potent ex vivo and in vivo activity of H84T-BanLec against MERS-CoV
High MERS-CoV RNA load and abundant viral nucleocapsid protein expression were detected in DMSO-treated ex vivo human lung tissue infected with MERS-CoV.27,28 Treatment with 5–20 nM H84T-BanLec significantly (p < 0.01) reduced MERS-CoV RNA load in a dose-dependent manner (Figure 2A), and there was minimal viral nucleocapsid protein expression detected in human lung tissue explant treated with 20 nM H84T-BanLec (Figure 2B). Next, we assessed the effect of H84T-BanLec in a human dipeptidylpeptidase 4-knockin (hDPP4-KI) mouse model for MERS-CoV infection.29,30 Intraperitoneal administration of H84T-BanLec resulted in significantly higher survival (100% versus 0%, p < 0.001 for pre-challenge group; 80% versus 0%, p < 0.01 for post-challenge group) (Figure 2C) and less body weight loss (Figure 2D) than the control mice receiving PBS. The mean infectious virus titer (p < 0.01) (Figure 2E) was significantly reduced in the H84T-treated mice. The PBS-treated control mouse lung sections showed evidence of diffuse interstitial pneumonia, with peribronchiolar inflammatory infiltrates and thickened alveolar walls that were only found occasionally in focal areas of the lungs of the H84T-BanLec-treated mice (Figure 2F). Immunofluorescence staining of the PBS-treated mouse lung sections showed abundant MERS-CoV nucleocapsid protein expression (green), while scarce viral nucleocapsid protein expression was observed in the lung sections of the H84T-BanLec-treated mice (Figure 2G). Together, these ex vivo and in vivo findings demonstrated the potent anti-MERS-CoV activity of H84T-BanLec.
Figure 2.
H84T-BanLec is highly potent against MERS-CoV in ex vivo human lung tissue organ culture and human dipeptidylpeptidase 4-knockin (hDPP4-KI) mice
(A) Dose-dependent reduction of MERS-CoV viral load in ex vivo human lung organ culture with increasing concentrations of H84T-BanLec treatment at 24 hours post-infection (hpi).
(B) Markedly reduced MERS-CoV nucleocapsid protein (NP) expression in ex vivo human lung organ culture in H84T-BanLec-treated samples compared with PBS control-treated samples at 24 hpi (scale bar, 50 μm). Data are mean ± SD, n = 5 biological replicates. One-way ANOVA was used to determine significant differences in viral loads. ∗∗p < 0.01.
(C–E) The hDPP4-KI mice were treated with intraperitoneal (i.p.; 15 mg/kg/animal/day every 24 h) H84T-BanLec starting at 6 h before (pre-challenge) or 24 h after MERS-CoV challenge (post-challenge) until 3 days post-infection (dpi) (n = 10 per group). Five animals per group were sacrificed at 4 dpi for viral load and histopathological studies. The survival rates and body weight changes of the remaining 5 animals per group were observed until 14 dpi or death. Survival rates (C), body weight changes (D), and viral titers (E) by plaque assay of the lungs from H84T-BanLec-treated and PBS-treated mice. Data are mean ± SD. Kaplan-Meier survival curves were analyzed by the log-rank test. Student’s t test was used to determine significant differences in viral loads and titers. ∗∗p < 0.01, ∗p < 0.05.
(F and G) Representative images of (F) haematoxylin and eosin (H&E)-stained (scale bars, 200 μm) and (G) immunofluorescence-stained (scale bars, 100 μm) lung sections at 4 dpi of mice that received H84T-BanLec or PBS. The PBS-treated control mouse lung sections showed evidence of interstitial pneumonia with peribronchiolar inflammatory infiltrates and thickened alveolar walls. In contrast, the H84T-BanLec-treated mouse lung sections demonstrated normal bronchiolar and alveolar architectures. Immunofluorescence staining of the PBS-treated mouse lung sections showed abundant MERS-CoV NP expression (green), while scarce viral NP expression was observed in the lung sections of the H84T-BanLec-treated mice.
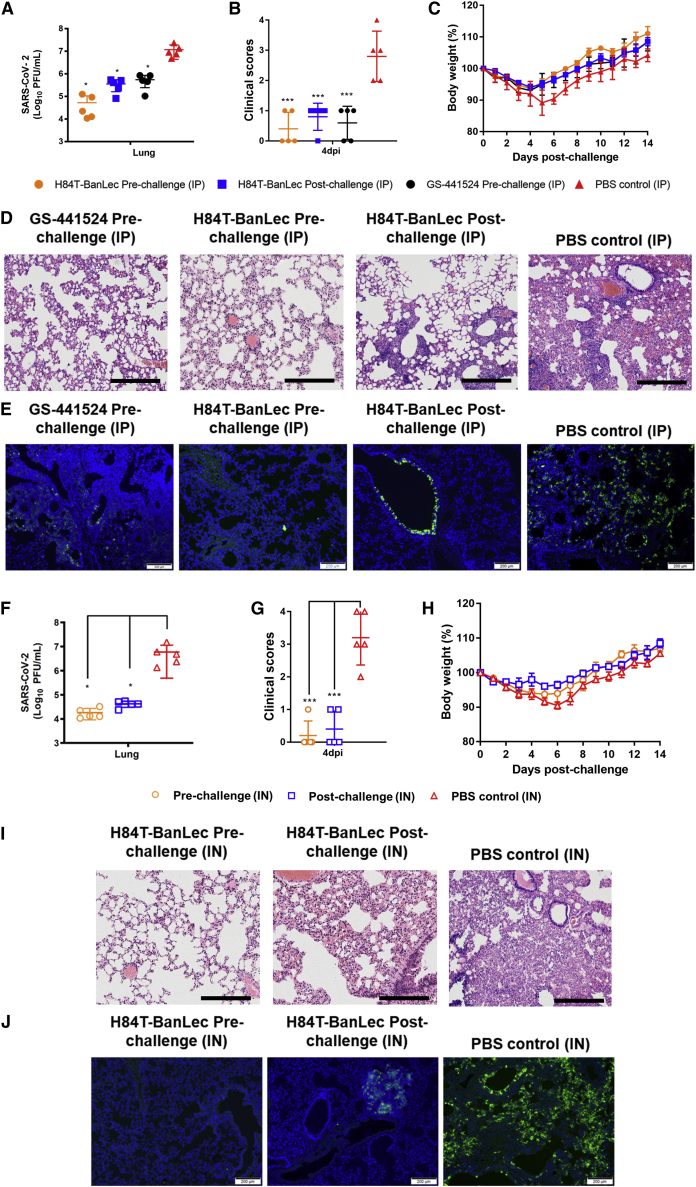
Prophylactic and therapeutic effects of systemic and intranasal H84T-BanLec against SARS-CoV-2 in golden Syrian hamsters
To thoroughly characterize the in vivo prophylactic and therapeutic effects of H84T-BanLec administered by different routes against the pandemic SARS-CoV-2 in our established golden Syrian hamster model, we dosed the animals with either intraperitoneal or intranasal H84T-BanLec, with the first dose starting 6 h before or 24 h after virus challenge.31 The hamsters that received either pre-challenge prophylactic or post-challenge therapeutic intraperitoneal H84T-BanLec had significantly lower infectious virus titers (Figure 3A) in the lung, as well as significantly lower clinical scores (Figure 3B) and less body weight loss (Figure 3C), than did the PBS-treated control hamsters. Either pre-challenge prophylactic or post-challenge therapeutic H84T-BanLec resulted in near-complete protection from lung damage except for small focal areas of mild peribronchiolar and perivascular inflammatory infiltrates (Figure 3D) and scarce SARS-CoV-2 nucleocapsid protein expression (Figure 3E) compared with PBS-treated control hamsters. Importantly, the improvement in the body weight loss, viral burden, and histopathological damages achieved with intraperitoneal (i.p.) prophylactic H84T-BanLec was comparable with that achieved with GS-441524 (active metabolite of remdesivir), further illustrating H84T-BanLec’s potent in vivo anti-SARS-CoV-2 effect. Moreover, the hamsters that received either pre-challenge prophylactic or post-challenge therapeutic intranasal H84T-BanLec also demonstrated significantly improved virological (Figure 3F), clinical (Figures 3G and 3H), and histopathological (Figures 3I and 3J) parameters when compared with the PBS-treated control hamsters. While both prophylactic and therapeutic H84T-BanLec significantly ameliorated SARS-CoV-2 infection in the hamsters, prophylactic administration conferred higher protection than therapeutic administration with generally lower viral burden and clinical scores, as well as less histopathological change. Importantly, intranasal delivery of H84T-BanLec results in good local concentration in rodents, with the highest level of H84T-BanLec in the nasal area occurring at 1 h post-administration (up to 5×108 of the average radiant efficiency) and gradually decreasing over 24 h (Figure S4). These findings demonstrated the potential of both systemic and intranasal H84T-BanLec as effective prophylactic and therapeutic anti-SARS-CoV-2 options.
Figure 3.
Intranasal or intraperitoneal H84T-BanLec reduces virus replication and alleviates histopathological changes in SARS-CoV-2-infected golden Syrian hamsters
The H84T-BanLec group hamsters were treated with (A–E) intraperitoneal (i.p.) (15 mg/kg/animal/day every 24h) or (F–J) intranasal (IN; 1.5 mg/kg/animal/day every 24 h) H84T-BanLec starting at 6 h before (pre-challenge) or 24 h after SARS-CoV-2 challenge (post-challenge) until 3 days post-infection (dpi) (n = 10 per group). The control hamsters were treated with (A–E) i.p. or (F–J) IN PBS starting at 6 h before virus challenge daily until 3 dpi. i.p. GS-441524 (25 mg/kg every 24 h as described previously)32 was used to treat an additional control group of mice for comparison. Five animals per group were sacrificed at 4 dpi for viral load and histopathological studies, and the survival rates and body weight changes of the remaining 5 animals per group were observed until 14 dpi or death. (A and F) Quantitation of virus titer by plaque assay in the lung tissues of the hamsters. (B and G) Clinical scores and (C and H) body weight changes of the hamsters. Data are mean ± SD. One-way ANOVA was used to determine significant differences in viral loads and titers. ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05. Representative images of (D) H&E-stained (scale bars, 100 μm) and (E) immunofluorescence-stained (scale bars, 200 μm) hamster lung sections at 4 dpi with or without i.p. H84T-BanLec or GS-441524. The i.p. PBS-treated control hamster lung sections showed prominent bronchiolar epithelial cell death and diffuse alveolar inflammatory infiltrates, with abundant SARS-CoV-2 nucleocapsid protein (NP) expression (green) being observed in immunofluorescence-stained lung sections. Pre-challenge i.p. administration of H84T-BanLec led to near-complete protection from lung damage and minimal SARS-CoV-2 NP expression except for small focal areas of mild peribronchiolar and perivascular inflammatory infiltrates, which were comparable with the histopathological changes observed in the lungs of hamsters treated with i.p. GS-441524. Post-challenge i.p. administration of H84T-BanLec also led to improved lung histopathological changes with only focal areas of bronchiolar cell death with viral NP expression. Representative images of (I) H&E-stained and (J) immunofluorescence-stained hamster lung sections at 4 dpi with or without IN H84T-BanLec. The IN PBS-treated control hamster lung sections showed bronchiolar epithelial cell death and diffuse lung alveolar exudation and inflammatory infiltrates. Immunofluorescence staining showed abundant and diffusely distributed SARS-CoV-2 NP expression (green). In stark contrast, pre-challenge IN administration of H84T-BanLec resulted in preserved lung architecture in the H&E-stained lung sections and absence of SARS-CoV-2 NP expression in immunofluorescence-stained sections. Post-challenge IN administration of H84T-BanLec similarly improved the histopathological changes in the hamster lung sections, although focal areas of mild epithelial cell death and alveolar inflammatory infiltrates that were viral NP-positive could occasionally be observed.
Effects on viral replication cycle and H84T-BanLec/SARS-CoV-2 spike co-localization in the lung of COVID-19 patient
To determine the steps of the SARS-CoV-2 replication cycle targeted by H84T-BanLec, we performed a time-of-drug-addition assay in a single replication cycle.33 Pre-incubation of H84T-BanLec with VeroE6 cells did not inhibit SARS-CoV-2 replication (i.e., −1 to 0 h), indicating the absence of host receptor blockage. Similarly, maintenance of H84T-BanLec in the post-virus-entry stage (i.e., 1 to 8 h) did not decrease intracellular viral load. In contrast, either mixing SARS-CoV-2 with H84T-BanLec pre-infection or adding H84T-BanLec during virus absorption (i.e., 0 to 1 h) significantly reduced viral load (p < 0.01), indicating that H84T-BanLec interfered with SARS-CoV-2 entry (Figure S5). To provide direct evidence of reduced virus uptake in cells, flow cytometry was performed to detect SARS-CoV-2 nucleocapsid protein expression with or without H84T-BanLec at as early as 2 h post-virus entry. Our results showed that the fluorescence intensity of infected cells after H84T-BanLec treatment decreased by about 55%–60%, which was comparable with the anti-SARS-CoV-2 entry effect of chloroquine at 2 hours post-infection (hpi) (Figure S6). These findings are compatible with one of the potentially expected mechanisms of action of H84T-BanLec, which has been found to inhibit the replication of other RNA viruses by binding to the high-mannose glycans found on viral proteins and inhibiting virus entry.23, 24, 25, 26 For coronaviruses, the highly glycosylated spike protein, which mediates virus entry with host cells, is the likely target of H84T-BanLec. To validate the interaction between H84T-BanLec and SARS-CoV-2 spike protein in humans, we performed co-localization experiments using the autopsied lung sections of a deceased COVID-19 patient. As shown in Figure S7, H84T-BanLec indeed co-localized with the majority of the SARS-CoV-2 spike proteins found in the autopsied human lung tissue.
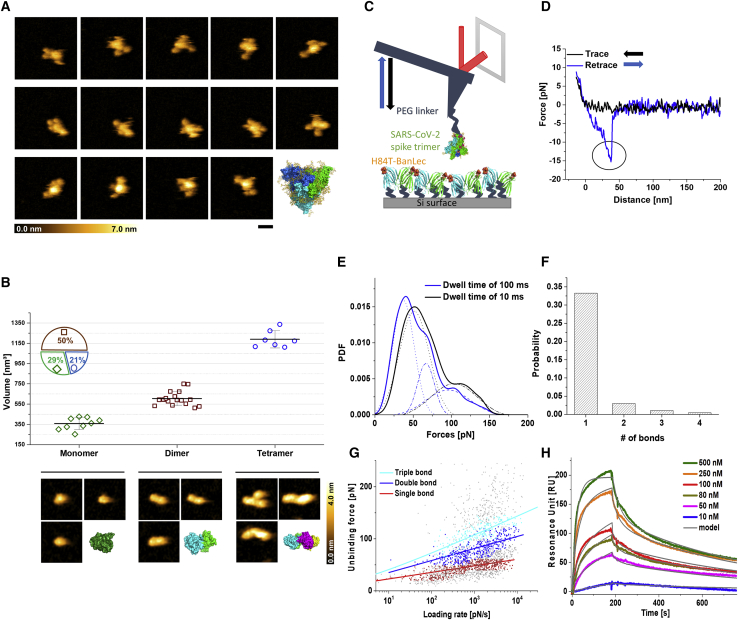
Real-time structural analysis of H84T-BanLec/SARS-CoV-2 spike binding interactions
To better understand the mechanisms underlying the broad-spectrum anti-coronavirus activity of H84T-BanLec, we investigated the binding interactions between H84T-BanLec and the SARS-CoV-2 spike at the atomic and molecular level. We first applied high-speed atomic force microscopy (HS-AFM) to visualize the isolated SARS-CoV-2 full-length spike protein and its highly dynamic movements.34, 35, 36 Snapshots captured from an HS-AFM movie revealed that three arms are waggling around a central core, in accordance with a 3-dimensional crystal structure in which three monomeric units of the spike (S) protein are entangled to form a trimeric assembly (Figure 4A). We also mapped isolated H84T-BanLec molecules to identify different oligomers and measured their respective mean volumes using AFM scans, which showed that our preparation contained 29% monomers (343 ± 73 nm3), 50% dimers (604 ± 71 nm³), and 21% tetramers (1,190 ± 85 nm3). This finding elucidates previous disagreements as to whether H84T-BanLec exists primarily as a dimer or as a tetramer.23,37 The topographical images matched well with 3-dimensional (3D) structures of each oligomeric state (Figure 4B). Next, we employed AFM-derived single-molecule force spectroscopy (SMFS) to directly measure the dissociation forces between H84T-BanLec and the full-length SARS-CoV-2 spike protein by mechanically pulling on bonds formed by molecular recognition from the two cognate molecules (Figure 4C).38,39 The SARS-CoV-2 spike trimer was conjugated to the AFM tip using a polyhistidine tag via a 6 nm-long flexible PEG linker to ensure sufficient motional freedom and unconstrained orientation for specific binding. Consecutive force-distance (F-D) measurement cycles, during which an AFM tip carrying the SARS-CoV-2 spike was brought into contact with a dense layer of H84T-BanLec molecules immobilized to a surface to allow H84T-BanLec/SARS-CoV-2 spike bonds to form, were performed. Thereafter, the AFM cantilever tip was retracted to measure the unbinding forces required to break the bonds. A typical F-D cycle initially shows nonlinear stretching behavior resulting from the flexibility of the PEG linker, then finally bond breakage occurred at a critical unbinding force (Figure 4D). The detected forces were used to construct the experimental probability density functions (PDFs) of unbinding forces resulting in dissociation of the H84T-BanLec/SARS-CoV-2 spike bond (Figure 4E). Specificity of interactions was verified in a blocking experiment and by examining spike binding to a surface covered only with PEG linker without H84T-BanLec (Figure S8A). A characteristic force distribution recorded when the spike-adorned AFM tip was allowed to rest for 10 ms on the surface containing H84T-BanLec (dwell time) shows a bimodal distribution, with the main peak at about 50 pN and a second peak at about 110 pN. The first and the second peaks represent the simultaneous rupture of 1 and 2 H84T-BanLec/SARS-CoV-2 spike bonds, respectively. Increasing the dwell time enhances the probability of multiple bond formation, as evidenced by the multimodal force histogram recorded at a dwell time of 100 ms, which verifies simultaneous unbinding of 1, 2, and 3 H84T-BanLec molecules from one homotrimeric SARS-CoV-2 spike.39 Additionally, we observed sequential bond rupture behaviors in our F-D curves arising from the breaking of up to 4 bonds (Figure 4F). These results indicated that up to 4 H84T-BanLec molecules may bind to each SARS-CoV-2 spike trimer.
Figure 4.
SARS-CoV-2 spike protein and H84T-BanLec structures and real-time observation of binding strength and affinity
(A) High-speed atomic force microscopy (HS-AFM) images of single SARS-CoV-2 spike proteins. The images in the three rows were selected from movies of three independent SARS-CoV-2 spike protein molecules. The right image in the bottom row shows a fully glycosylated (yellow) 3D model of the SARS-CoV-2 trimer (green, cyan, and blue). Scale bar, 25 nm.
(B) (Top) Volume measurements of H84T-BanLec, representing a monomer, dimer, and tetramer, depicted from an 886 frame movie. The inset pie chart displays the frequency distribution of oligomeric states. (Bottom) Representative HS-AFM images taken for the volume measurements and 3D surface representations of H84T-BanLec of each oligomeric state, taken from the X-ray structure (PDB: 4PIK). Scale bar, 5 nm. All images were captured at a scan rate of 303 ms per frame.
(C) Schematic design of single-molecule force spectroscopy (SMFS) experiments between full-length SARS-CoV-2 trimer coupled to an AFM cantilever tip and surface-immobilized H84T-BanLec.
(D) The typical force-distance curve obtained using SMFS, showing the signature of a single-bond rupture upon unbinding of the SARS-CoV-2 spike trimer from H84T-BanLec (indicated by the circle in the retrace recording).
(E) Experimental probability density functions (PDFs) of unbinding forces constructed from summing up unbinding forces, each of which is represented by a Gaussian of unitary area with the width of its measurement uncertainty. As such, they contain solely original data and can be viewed as the equivalent of continuous force histograms. PDFs for two different tip-surface interaction times, 100 (blue line) and 10 ms (black line), were fitted by a multi-Gaussian function. Maxima reveal rupture of a single or simultaneous rupture of 2 and 3 bonds, respectively (indicated by the numbers).
(F) Probability for the formation of one, two, three, or four bonds upon SARS-CoV-2 spike trimer binding to H84T-BanLec.
(G) Plot of unbinding force versus loading rate for SARS-CoV-2 spike trimer dissociating from H84T-BanLec. A Markov binding model computed the behavior of the double- (blue) and triple-bond (cyan) interactions, using parameters derived from the Bell and Evan’s single-barrier model fit of the single bond (red). Colored data points were extracted from PDFs within ± 2σ of maxima.
(H) Surface plasmon resonance measurements. H84T-BanLec was injected (in the presence of Ca2+) at the indicated concentrations to surfaces containing immobilized full-length trimeric spike to allow for binding (ascending parts), followed by wash out using buffer (descending parts). Data were fitted using the bivalent binding model, consistent with H84T-BanLec being predominantly a dimer.
To decipher the dynamic features of the H84T-BanLec/SARS-CoV-2 spike bonds, we extended our SMFS studies to dynamic force spectroscopy (DFS) experiments and varied the pulling speed. Individual dissociation forces were plotted versus their force loading rate (pulling speed times effective spring constants of molecules and AFM cantilever). H84T-BanLec/SARS-CoV-2 spike interaction forces arising from single bond breakages (first maxima ± 2σ of PDFs; Figure 4G, red dots) were well fitted by the widely used Bell-Evans model (Figure 4G, red line), yielding a kinetic dissociation rate constant of koff = 0.046 ± 0.02s−1.40,41 The kinetic on-rate constant (kon), approximated from computing the binding probability P(t) as a function of the dwell time t, according to P(t) = A(1-exp(-(t-to)/τ)), and calculated using kon = 1/τ∗Ceff (effective concentration of molecules coupled to the AFM tip), yielded kon = (9.98 ± 5.18) × 1041/Ms (Figure S8B; Table S1).39,42 The equilibrium dissociation constant (KD), determined from the kinetic rate constants (KD = koff/kon), was 0.46 μM. In addition, spontaneous thermodynamic association and dissociation were measured with surface plasmon resonance (SPR) (Figure S9A).43 H84T-BanLec was injected at different concentrations into chambers containing either the isolated SARS-CoV-2 spike receptor-binding domain (RBD) or complete SARS-CoV-2 spike trimers coupled to chip surfaces and was subsequently removed with buffer. The SARS-CoV-2 spike RBD alone failed to bind to H84T-BanLec (Figure S9B). For the SARS-CoV-2 spike trimer, experimental binding curves (Figure 4H, colored lines) recorded with SPR were fitted with the “bivalent analyte model” (Figure 4H, gray lines), which assumes two-step binding of H84T-BanLec to adjacent immobilized spike trimer binding sites. A very good match of the fit with the data confirms that H84T-BanLec is mainly present in its dimeric form. Kinetic rates and the KD value determined from spontaneous thermodynamic association and dissociation with SPR were similar to those from force probing in DFS (Table S1), providing a consistent picture for the dynamics and equilibrium of H84T-BanLec/SARS-CoV-2 spike interactions.
Interaction dynamics and structural flexibility of H84T-BanLec/SARS-CoV-2 spike complex binding sites
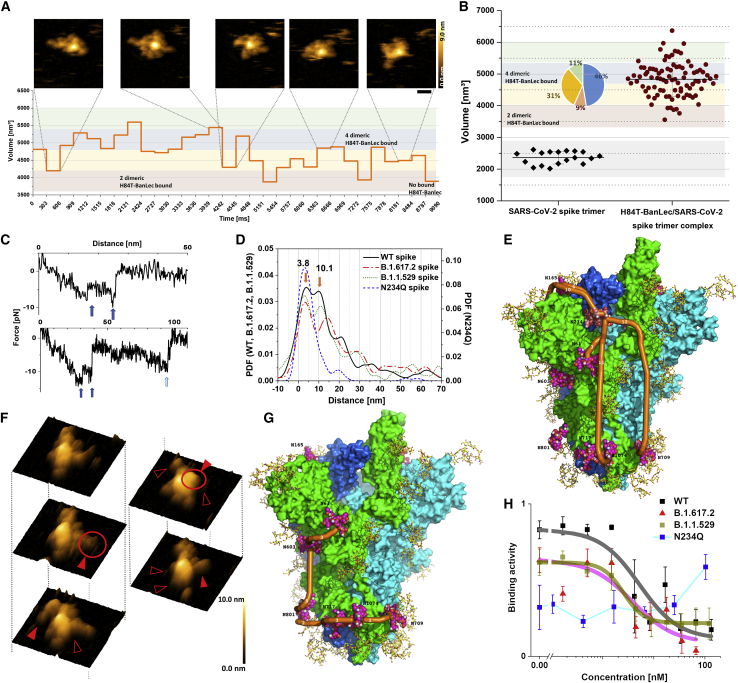
To depict the dynamics of the H84T-BanLec/SARS-CoV-2 spike interactions, we filmed the complexes using real-time HS-AFM under physiological conditions (Video 1). H84T-BanLec interacted with SARS-CoV-2 spike at the periphery around the core of the trimeric structure (Figure 5A). A sequence of 30 frames from movie recordings of H84T-BanLec/SARS-CoV-2 spike complexes that showed continuous association and dissociation was selected to quantify the complex volume in each of the frames. By comparing the volumes of the complexes with that of H84T-BanLec dimers and isolated SARS-CoV-2 spike trimers, we found that on average each SARS-CoV-2 spike trimer bound to 4.1 ± 0.7 H84T-BanLec dimers. Volume distributions showed that 46% of the SARS-CoV-2 spike trimers bound to about 4 dimers each (Figure 5B). A similar number of multimeric interactions was also observed in SMFS traces, as was evident from the sequential bond breakages (Figure 5C).
Figure 5.
Structure and dynamics of multiple bonds formed between SARS-CoV-2 spike protein and H84T-BanLec
(A) HS-AFM frames of a single SARS-CoV-2 spike trimer complexed with H84T-BanLec. Scale bar, 25 nm. All images were captured at a scan rate of 303 ms per frame. The graph in the bottom part displays the variation of the volume over time. Volume ranges indicative of the number of dimeric H84T-BanLec molecules bound to the SARS-CoV-2 spike protein are marked by colors in the background.
(B) Volume measurements of isolated SARS-CoV-2 spike protein and SARS-CoV-2 spike protein complexed with H84T-BanLec. The volumes of isolated SARS-CoV-2 spike protein were determined from 18 frames taken from three different movies. Volumes of SARS-CoV-2 spike protein associated with H84T-BanLec were taken from 3 different individual H84T-BanLec/SARS-CoV-2 spike complexes of 30 frames each. The colors in the background of the graph indicate the number of dimeric H84T-BanLec molecules bound, and the inset pie chart displays their frequency distribution (same color code as background).
(C) Examples of SMFS force-distance curves containing two or three sequential bond ruptures, respectively, upon unbinding of SARS-CoV-2 spike protein from H84T-BanLec (indicated with blue arrows).
(D) Distribution of distances of sequential bond ruptures (as exemplified in C), represented by an experimental probability density function of distances constructed from distance values measured by SMFS (main maxima of wild-type spike at 3.8 + 0.5 and 10.1 + 0.8 nm).
(E) 3D structural model of SARS-CoV-2 spike trimer with monomers in green, cyan, and blue. Complex-type glycans in yellow; high-mannose glycan (Man9) on N234 space filling with salmon carbons, oligo-mannose glycans (Man5) space filling with carbons in magenta. The shortest curved paths are indicated in orange (Man9 on N234 to Man9 on N234 [second monomer], Man5 on N61, Man5 on N709, and Man5 on N1074).
(F) Structures of the H84T-BanLec/SARS-CoV-2 spike complex shown with 3D representations of HS-AFM topographical images. The filled red arrows and empty arrows indicate association and dissociation of H84T-BanLec with the SARS-CoV-2 spike protein, respectively.
(G) 3D structural model of SARS-CoV-2 N234Q spike mutant, with the shortest curved paths between Man5 at N61, N603, N709, N717, N801, and N1074 in orange.
(H) Binding activity of SARS-CoV-2 spike protein or mutants to the surface of Vero E6 cells in the presence of H84T-BanLec, probed at the indicated concentrations in SMFS experiments.
The SARS-CoV-2 spike is known to have multiple sites that can be decorated with high mannose, and we thus investigated which of them are crucial for the interaction with H84T-BanLec.36,44,45 Following a previously described approach to localizing molecular binding sites, we charted H84T-BanLec association on the SARS-CoV-2 spike trimer surface by determining the distances between the bond ruptures.46 From our recordings, we constructed an experimental PDF. The PDF distribution of the rupture distances revealed 3 distinct peaks with maxima located at 3.8, 10.1, and 21.1 nm, respectively (Figure 5D). These findings indicated that a H84T-BanLec dimer can bind to several binding sites at distinct distances on the surface of the SARS-CoV-2 spike trimer. The B.1.617.2 (Delta) variant spike carrying the mutations L452R, T478K, and P681R and the B.1.1.529 (Omicron) variant spike carrying the mutations A67V, HV69-70del, T95I, G142D, VYY143-145del, N211del, L212I, ins214EPE, G339D, S371L, S373P, S375F, K417N, N440K, G446S, S477N, T478K, E484A, Q493R, G496S, Q498R, N501Y, Y505H, T547K, D614G, H655Y, N679K, P681H, N764K, D796Y, N856K, Q954H, N969K, and L981F showed very similar binding features to H84T-BanLec when compared with wild-type spike, with comparable kon, koff, and affinity KD in both SMFS and SPR experiments (Figures S8 and S9C; Table S1). They also showed multiple bond ruptures of up to four bonds, with distances revealing two distinct maxima in the distance rupture histogram at 3.1 and 14.0 nm for B.1.617.2 and at 3.0 and 11.1 nm for B.1.1.529 (Figure 5D), and demonstrated similar complexation features with H84T-BanLec as wild-type spike in real-time HS-AFM movies (Videos 2 and 3).
The N234 glycosylation site is the only site that exclusively carries up to 9 mannose residues on the SARS-CoV-2 spike, and this specific glycan has been suggested to control the ability of spike to assume the proper conformation for binding to ACE2.44 To depict the multimeric interaction pattern, we modeled the 9-mannose site at N234 as well as the 5-mannose sites according to the data of Watanabe et al. and calculated possible distances between the high-mannose residues.45 Note that the glycan at N165 was identified by Watanabe et al. as almost exclusively of complex type, while Zhao et al. assigned about 15% of oligomannosidic glycans to this site.47 Analyses on our own samples suggested up to 29% of oligomannosidic glycans at N165, such that we have included this site in our analysis.36 Distinct multiple binding paths along oligo (5)-mannose glycan sites within the SARS-CoV-2 spike trimer are shown with orange lines in the 3D model structure, indicating 3.2 nm for the glycan at N61, 4.3 nm for the glycan at N165, and 10–12 nm for adjacent monomers of spike or for glycans at the stalk (N709 and N1074) (Figure 5E). Indeed, the estimated distances between oligo-mannose glycans in the 3D model are in agreement with distances derived from SMFS bond dissociation ruptures. Consistent with the position of the first peak at 3.8 nm, the short path correlates with a distance between the 9-mannose site N234 and a 5-mannose site at N61 or at N165. Similar calculations for other paths, such as from N234 to other 5-mannose sites or to N234 at another spike monomer, correlated with the second peak of the length distribution at 10.1 nm, which might be slightly influenced by mechanical twists along the pulling direction in SMFS experiments. The length distributions of H84T-BanLec/SARS-CoV-2 spike dissociation revealed multimeric interactions between the H84T-BanLec dimer and different glycosylation sites on the SARS-CoV-2 spike. This was further evidenced by the 3D AFM topographical time frames, which provided direct visualization of the formation of bonds to various SARS-CoV-2 spike high-mannose sites in real time (Figure 5F).
To directly test whether N234 is crucial to the interaction between H84T-BanLec and the spike protein, we examined whether H84T-BanLec could bind as effectively to spike with a mutation at position N234 as it does to wild-type spike. The binding strength, kinetic binding rates, and affinity of the N234Q mutant for H84T-BanLec were determined with SMFS and SPR measurements (Figures S8 and S9A; Table S1). Similar to wild-type spike, the N234Q mutant also formed multiple bonds with H84T-BanLec (Figure S8C). The PDF distribution of the rupture distances, however, was strikingly different and revealed only one distinct maximum at 3.1 nm (Figure 5D), which possibly corresponded to the distances between the 5-mannose at N61 and N603 or between the oligomannosidic glycans at N709, N717, N801, and N1074 (Figure 5G). Interestingly, single bonds established between H84T-BanLec and the N234Q mutant were less stable when compared with the wild-type spike and showed a significantly higher (∼5 times) koff in SMFS experiments (Figure S8; Table S1), most likely because of the ablation of high mannose at position 234. In contrast, H84T-BanLec binds slightly more favorably to the N234Q mutant in SPR experiments, with a slightly higher kon value and a significantly lower koff rate, as well as a higher binding saturation, which could arise from reversible binding to N234Q at neighboring oligomannosidic glycans (Figure S9A; Table S1). We conclude that H84T-BanLec does not exclusively bind to the high-mannose glycan at N234 but that binding to the other indicated oligomannosidic glycans is as relevant, in agreement with our analyses of the interglycan distances.
We also assessed whether H84T-BanLec could interfere with SARS-CoV-2 spike binding to the surface of VeroE6 cells, which is a frequently used model for examining the interaction between SARS-CoV-2 and cellular receptors, especially ACE2.48 We found that H84T-BanLec in solution functionally impedes spike binding to host cell membranes and significantly reduces the binding activity of tip-coupled spike trimer to ACE2 expressed on VeroE6 cells (Figure 5H). To evaluate the inhibitory effect in detail, we titrated H84T-BanLec in the measurement solution and determined the IC50 (Figure 5H). Due to the dimeric structure of the soluble H84T-BanLec molecules used in our study, they act as bivalent effective inhibitors with IC50s lower than the affinity constants derived with SMFS for single bonds. This enhanced affinity may be a cooperative effect from binding of soluble dimeric H84T-BanLec to trimeric spike molecules. The IC50 showed a very strong affinity in the nM range, with 5.2 ± 1.1 nM for wild-type spike, 3.9 ± 0.8 nM for the B.1.617.2 (Delta) variant spike, and 2.6 ± 0.5 nM for the B.1.1.529 (Omicron) variant spike. N234Q showed relatively low binding activity for VeroE6 cells, with almost no change seen with increasing concentrations of H84T-BanLec.
Discussion
Novel coronaviruses will likely continue to emerge as important human pathogens. As evidenced by the ongoing COVID-19 pandemic, the traditional “one virus, one drug” antiviral discovery strategy is inefficient for the control of rapidly disseminating outbreaks. Broad-spectrum drug compounds with pan-coronaviral antiviral activities that target conserved viral components utilized by all coronaviruses for replication, such as the spike, are urgently needed for the current and future coronavirus pandemics. Lectins such as BanLec, griffithsin, and Flt3 receptor interacting lectin (FRIL) bind to carbohydrates to exert broad-spectrum antiviral activities but have not advanced into clinical use as antivirals, in part due to their potential mitogenicities (save griffithsin).23,49,50 To overcome this bottleneck, we have previously engineered H84T-BanLec, which exhibits broad-spectrum antiviral activities with significantly reduced mitogenicity and broad-spectrum antiviral activities against all influenza viruses tested.23,25,26 Further, it is highly effective in vivo against influenza whether given intranasally or systemically.23,25
In this study, we report the pan-coronaviral antiviral activities of H84T-BanLec against known human-pathogenic coronaviruses, including the highly virulent MERS-CoV (overall mortality rate >30%), the pandemic SARS-CoV-2, including the Delta and Omicron variants, SARS-CoV, and other circulating HCoVs. The potent antiviral activity of H84T-BanLec against MERS-CoV was well evident in the ex vivo human lung organ tissue culture and hDPP4-KI mouse models. Importantly, the in vivo anti-SARS-CoV-2 effect of H84T-BanLec was comparable with GS-441524, the active metabolite of remdesivir. Moreover, by administering H84T-BanLec via different routes and starting at different times in relation to virus challenge, we showed that this broad-spectrum lectin is highly effective against SARS-CoV-2 as an intranasal or systemic prophylaxis or therapy. For the control of pandemics caused by highly transmissible viruses like SARS-CoV-2, which causes mild or even asymptomatic infection in a significant proportion of patients, it is especially important to develop effective agents that can be administered topically (intranasally) in the outpatient setting either as prophylaxis or as early treatment to reduce the burden on healthcare and quarantine facilities. Importantly, our current study confirms that H84T-BanLec primarily exists as a dimer, which makes it highly stable and thus ideally suitable for development as a topical agent.23 H84T-BanLec also holds promise for systemic use, as it has a very long half-life (>35 h), so it likely would have to be administered only one time in the outpatient and inpatient settings.25,26 The total amount of protein required would be similar to that of anti-SARS-CoV-2 monoclonal antibodies,17,25,26 and systemic H84T-BanLec is very well tolerated in mice, hamsters, and rats (unpublished data).25,26 Unlike monoclonal antibodies, to which resistant viral mutants might be expected to develop, the high number of available binding sites of high-mannose glycans on spikes of coronaviruses will render resistance to H84T-BanLec much less likely. Indeed, we did not observe the emergence of resistant mutants after serial passages of MERS-CoV and SARS-CoV-2 with H84T-BanLec. Moreover, H84T-BanLec may be useful as part of combinatorial treatment regimens that include antivirals targeting other viral components (e.g., viral enzyme inhibitors) or host-targeting therapeutics. In the case of influenza virus, we have previously shown that H84T-BanLec is synergistic with the neuraminidase inhibitor oseltamivir.25
Regarding H84T-BanLec’s anti-coronaviral mechanism of action, we demonstrated that H84T-BanLec binds to the glycosylated spike protein in the lung tissues of a deceased patient with COVID-19. Time-of-drug-addition and flow cytometry studies showed that virus entry into cells is a major step in the SARS-CoV-2 replication cycle that is perturbed by the lectin. Further corroborating these findings, our experiments showed that H84T-BanLec can compete for binding between the ACE2 cellular receptor and the spike protein, thus inhibiting virus entry. The point mutant N234Q indeed shows less binding to VeroE6 cells, as would be predicted by the previous literature, and its response to H84T-BanLec is not really dose dependent. Of course, binding in this assay and infection do not totally correlate on a numerical basis. We would predict that the N234Q mutant will replicate relatively poorly, as N234 is crucial to the ability of the spike protein to switch to the structure necessary for fusion, and replication of the mutant will still be impeded by H84T-BanLec, although perhaps less dramatically, as H84T-BanLec will not have its “first among equals” binding site.
Extensive single-molecular structural and biophysical studies using HS-AFM and SMFS demonstrate the interaction dynamics and binding capability between H84T-BanLec and SARS-CoV-2 spike with thus far unprecedented resolution. As the SARS-CoV-2 spike is quite flexible, strong and diverse bonds accomplish maximal binding with H84T-BanLec and the resulting antiviral effects. Our data demonstrated the presence of strong interactions and depicted the formation of multiple bonds between H84T-BanLec and SARS-CoV-2 spike, which formed across two distinct spike monomers. While N234 is decorated with 9-mannose molecules and has been suggested to control the position of the RBD, our data showed that this is not the only site on spike recognized by H84T-BanLec.44 In fact, modeling and experimental data suggested a network of oligomannosidic glycans to which H84T-BanLec binds. However, H84T-BanLec does not bind to the isolated RBD, in agreement with the fact that no mannosidic glycans were determined on the RBD. An intriguing glycosylation site is found at position N165. Not all experimental glycan analyses identified this glycan as oligomannosidic, but it is placed right next to the RBD, and the compaction of the linker domains of SARS-CoV-2 observed upon binding of H84T-BanLec most likely affects the orientation of the RBD and the interaction between the RBD and ACE2. Taken together, our structural data, demonstrating strong bonds and independent interactions between H84T-BanLec and multiple high-mannose residues on the spike, sometimes on different monomers of the trimer, help explain why all strains of SARS-CoV-2 thus far tested, including Delta and Omicron, are sensitive to H84T-BanLec.
In summary, the present study reports the potent activity of H84T-BanLec against a broad range of human-pathogenic coronaviruses. The promising in vivo data and structural analysis demonstrating the high stability of H84T-BanLec and its strong binding interactions with SARS-CoV-2 spike highlight its potential to be applied clinically as both an intranasal and systemic agent for the ongoing COVID-19 pandemic and future coronavirus outbreaks. The excellent in vivo efficacy of H84T-BanLec against diverse strains of influenza further suggests its usefulness in future respiratory virus epidemics.
Limitations of the study
This study has some limitations. First, the Syrian hamster model is a nonlethal model of COVID-19. Evaluation of the effect of H84T-BanLec in additional lethal animal models such as the K18-hACE2 mouse model would provide additional information on changes in survival rate. Nevertheless, we have demonstrated the ability of H84T-BanLec to improve the survival rate of MERS-CoV-infected hDPP4-KI mice. Second, additional experiments using different dosages and time points may help to further optimize the dosing regimen of H84T-BanLec against MERS-CoV and SARS-CoV-2.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit H84T-BanLec | In house | N/A |
| Rabbit anti-SARS-CoV-2 N | In house | N/A |
| Rabbit anti-BanLec polyclonal antibody | In house | N/A |
| Goat anti-Rabbit IgG(H+L) Secondary Antibody,HRP | Thermo Fisher | Cat#: 31460 |
| Guinea pig anti-MERS-CoV N | In house | N/A |
| Goat anti-Guinea Pig IgG (H+L) Highly Cross-Adsorbed Secondary Antibody | Thermo Fisher | Cat#: A-11073 |
| Bacterial and virus strains | ||
| HCoV-229E | Chu H et al., 2022 | N/A |
| HCoV-OC43 | Chu H et al., 2022 | N/A |
| MERS-CoV (EMC/2012) | Chan JF et al., 2013 | GenBank: JX869059 |
| SARS-CoV-1 GZ50 | Chu et al., 2020 | GenBank: AY304495 |
| SARS-CoV-2 HKU001a | Chu et al., 2020 | GenBank: MT230904 |
| SARS-CoV-2 B.1.1.7 (Alpha) | Shuai et al. 2022 | GenBank: OM212469 |
| SARS-CoV-2 B.1.617.2 (Delta) | Shuai et al., 2022 | GenBank: OM212471 |
| SARS-CoV-2 P.3 (Theta) | Shuai et al. 2021 | GISAID: EPI_ISL_1660475 |
| SARS-CoV-2 BA.1.529.1 (Omicron BA.1) | Shuai et al., 2022 | GenBank: OM212472 |
| Biological samples | ||
| Human lung tissues from patients with lung tumors who underwent surgical operations | Queen Mary Hospital, Hong Kong SAR | N/A |
| Autopsied lung sections of a deceased COVID-19 patient | Department of Pathology, University of Michigan | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| SARS-CoV-2 Spike Trimer (B.1.617.2/Delta) | Acrobiosystems | Cat# SPN-C52He |
| SARS-CoV-2 Spike Trimer (B.1.1.529/Omicron) | Acrobiosystems | Cat# SPN-C52Hz |
| SARS-CoV-2 Spike protein Trimer (Wild type) | Hoffmann et.al. 2021 | N/A |
| SARS-CoV-2 Spike protein Trimer (N234Q) | Provided by M.K, BOKU This paper |
N/A |
| (3-Aminopropyl)triethoxysilane | Sigma-Aldrich | Cat# 440140 |
| Chloroform | VWR Chemicals | Cat# 22711.324 |
| Triethylamine | Sigma-Aldrich | Cat# 90335 |
| maleimide-PEG-NHS | Polypure | Cat# 21138-2790 |
| thiol-trisNTA | Provided by R.T, Institute of Biochemistry, Goethe-Universität Frankfurt Patent # US 2008/0038750 A1 |
N/A |
| tris(carboxyethyl)phosphine (TCEP) hydrochloride | Sigma-Aldrich | Cat# C4706 |
| Ethylenediaminetetraacetic acid disodium salt dihydrate | VWR | Cat# 20302.236 |
| HEPES(4-(2-Hydroxyethyl)piperazin-1-ylethanesulphonic acid) | VWR | Cat# 441485H |
| TWEEN-20 | Sigma-Aldrich | Cat# P1379 |
| Dulbecco’s Modified Eagle’s Medium with Glucose, with sodium pyruvate, with stable glutamine | Merck-Biochrom | Cat# FG0445 |
| Fetal Bovine Serum | Thermo Fisher Scientific | Cat# 10500-064 |
| streptomycin | Thermo Fisher Scientific | Cat# 15140122 |
| L-glutamine | Thermo Fisher Scientific | Cat# 25030-024 |
| Pluronic F-68 | Thermo Fisher Scientific | Cat# 24040-032 |
| G-418 | Thermo Fisher Scientific | Cat# 10131035 |
| 25-kDa linear PEI | Polysciences | Cat# 23966 |
| Tryptone N1 | Organotechnie | Cat# 19553 |
| D (+)-glucose | Carl Roth | Cat# 6887.1 |
| Critical commercial assays | ||
| CellTiter-Glo luminescent cell viability assay kit | Promega | Cat#: G7572 |
| QIAsymphony RNA Kit | Qiagen | Cat#: 931636 |
| RNeasy Mini kit | Qiagen | Cat#: 74106 |
| QuantiNova Probe RT-PCR Kit | Qiagen | Cat#: 208354 |
| Experimental models: Cell lines | ||
| BSC-1 | ATCC | CCL-26 |
| Calu-3 | ATCC | HTB-55 |
| HEK293-6E | National Research Council, NRC, Ottawa, Canada | RRID# CVCL_HF20 |
| HFL | Chu H et al. 2020 | In house |
| Huh-7 | JCRB cell bank of Okayama University | JCRB0403 |
| Vero | ATCC | CCL-81 |
| VeroE6 | ATCC | CRL-1586 |
| Experimental models: Organisms/strains | ||
| BALB/c mice | The Jackson Laboratory | Strain #:000651 |
| hDPP4-knockin mice | Chu H et al., 2021 | N/A |
| Golden Syrian hamsters | Chan JF et al., 2020 | N/A |
| Recombinant DNA | ||
| Plasmid: pCAGGS-S | Amanat et al. 2020 | N/A |
| Plasmid: pTT28 | National Research Council, NRC, Ottawa, Canada | N/A |
| Software and algorithms | ||
| Origin | Origin Lab | 2021b |
| Matlab | Mathworks | Version R2013a |
| Gwyddion | http://gwyddion.net/ | Version 2.55 |
| ProtParam | Expasy | RRID# SCR_018087 |
| NovoExpress | Agilent | Version 1.5.6 |
| GraphPad Prism | GraphPad Software, Inc. | Version 9 |
| Other | ||
| FreeStyle F17 Expression Medium | Thermo Fisher Scientific | Cat# A13835-02 |
| Pellicon XL-Ultrafiltrationsmodule Biomax, 10 kDa | Merck Millipore | Cat# PXB010A50 |
| HisTrap FF 5-mL | Cytiva | Cat# 17525501 |
| Amicon Ultra-15 | Merck Millipore | Cat# UFC910024 |
| Äkta Pure | Cytiva | N/A |
| Labscale TFF System | Merck Millipore | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to the lead contact, David M. Markovitz (dmarkov@umich.edu).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Chemicals, viruses, H84T-BanLec antibody, and cells
H84T-BanLec and D133G-BanLec were prepared in baculovirus as previously described and the H84T-BanLec antibody, which also recognizes the wild-type molecule and D133G-BanLec, was raised in a rabbit in house.23 MERS-CoV (EMC/2012) was kindly provided by Ron Fouchier (Erasmus Medical Center, the Netherlands).3 SARS-CoV-2 wild-type and variants were isolated from the respiratory tract specimens of COVID-19 patients in Hong Kong.51, 52, 53 Archived clinical strains of SARS-CoV, HCoV-OC43, and HCoV-229E were obtained from the Department of Microbiology, The University of Hong Kong (HKU).51,54 The cell lines used in this study were available in our laboratories as previously described.51,55,56 All experimental protocols involving live MERS-CoV, SARS-CoV-2, and SARS-CoV followed the approved standard operating procedures of HKU Biosafety Level 3 facility.
Animals
The use of animals at HKU was approved by the HKU Committee on the Use of Live Animals in Teaching and Research. hDPP4-KI mice (aged 5–8 weeks, male and female) and Syrian hamsters (aged 6–10 weeks, male and female) were obtained from the HKU Centre for Comparative Medicine Research. The animals were kept in cages with individual ventilation with 65% humidity and ambient temperature ranging between 21-23°C with 12-hour-interval day/night cycle for housing and husbandry before transferral to the Biosafety Level 3 facility. BALB/c mice (aged 6 weeks, female) used for the intranasal pharmacokinetics study at the University of Michigan were obtained from The Jackson Laboratory and the experiments were approved by the Institutional Animal Care and Use Committee of the University of Michigan.
Method details
Cytotoxicity and antiviral assays
The cytotoxicity of H84T-BanLec in various cell lines was determined using the CellTiter-Glo® luminescent cell viability assay (Promega Corporation, Madison, WI) according to manufacturer’s instructions and as previously described.57,58 Viral load reduction, CPE inhibition, and plaque reduction assays for coronaviruses were performed as we previously described.59,60 To test the vulnerability of H84T-BanLec to induce resistant escape mutants, a drug resistance assay was performed as we described previously.61 In brief, MERS-CoV and SARS-CoV-2 (MOI = 0.01) was passed in the presence of about 5× EC50 of H84T-BanLec (30 nM) for eight passages and in VeroE6 cells. After the eighth passage, the antiviral activities of H84T-BanLec against the wild-type and passaged viruses were compared using viral load reduction assay.
Ex vivo human lung tissue organ culture
The anti-MERS-CoV activity of H84T-BanLec was evaluated in an established ex vivo human lung tissue organ culture model as we described previously.27,28 The ex vivo human lung tissue culture experiments were approved by the Institutional Review Board of HKU/Hospital Authority Hong Kong West Cluster. The human lung tissues were obtained from patients with lung tumors who underwent surgical operations at Queen Mary Hospital, Hong Kong. Briefly, the freshly isolated human lung tissues were rinsed with the primary tissue culture medium, which contained the advanced Dulbecco’s Modified Eagle Medium (DMEM)/F12 medium supplemented with 2 mM HEPES (Gibco, Thermo Fisher Scientific, Waltham, MA, USA), 1×GlutaMAX (Gibco), 100 U/mL penicillin, 100 μg/mL streptomycin, 20 μg/mL vancomycin, 20 μg/mL ciprofloxacin, 50 μg/mL amikacin, and 50 μg/mL nystatin. Human lung tissues were cut into small pieces at comparable sizes and then infected with MERS-CoV at 1×106 plaque-forming units (PFU)/mL in a 6-well culture plate. After 2 h, the inoculum was removed and the specimens were washed with the primary tissue culture medium. The infected specimens were then transferred to inserts of 12-well transwells (Corning Life Sciences, Tewksbury, MA, USA) pre-coated with 200μL 60% Matrigel (Corning Life Sciences) diluted with the primary tissue culture medium. An additional 60% Matrigel was then added to the insert to seal the explant. The basolateral compartment was filled with 1mL of the primary tissue culture medium supplemented with 0, 5, or 20 μM H84T-BanLec. The samples were harvested at 24 hpi for confocal microscopy and qRT-PCR analysis. The virus genome copies in cell lysate samples were normalized with GAPDH copy numbers. Immunofluorescence staining of MERS-CoV nucleocapsid protein in the lung tissues was performed as previously described.27,62,63
Co-localization of H84T-BanLec and SARS-CoV-2 proteins in autopsied lung sections
Autopsied lung sections of a deceased COVID-19 patient were obtained from the Department of Pathology, University of Michigan,64 and sequentially stained with biotinylated H84T-BanLec (1:25k, Opal 620 reporter), and antibodies to SARS-CoV-2 nucleocapsid (Genetex, GTX635679, 1:1000, Opal 570 Reporter) and spike proteins (Genetex, GTX632604, 1:50, Opal 690 Reporter). After each round of antigen detection, the slides were exposed to microwave epitope retrieval in 10mM citrate buffer to remove the antibody complex, leaving behind the deposited fluorochrome. The slide was then imaged. A pseudo-coloured image was created. Original monochrome images were analyzed using the coloc 2 sub-routine in FIJI and the Pearson correlation coefficient was calculated.65 The autopsy of the deceased COVID-19 patient was exempt from Institutional Review Board approval.
hDPP4-KI mouse model for MERS-CoV infection
The in vivo anti-MERS-CoV activity of H84T-BanLec was evaluated in an established hDPP4-KI mouse model using mouse-adapted MERS-CoV (MERS-CoVMA) as described previously.29,30 Briefly, hDPP4-KI mice, aged 5–8 weeks, were divided into different groups to receive H84T-BanLec or sham (PBS) treatment. After anaesthesia, the mice were intranasally inoculated with 20 μL of virus suspension containing 2500 PFU of MERS-CoV (n = 10 per group). Each mouse in the H84T-BanLec groups was treated with intraperitoneal (15 mg/kg/animal/day every 24 h) H84T-BanLec starting at 6 h before (pre-challenge group) or 24 h after (post-challenge group) MERS-CoV challenge until 3 dpi. The negative control mice were treated with intraperitoneal PBS starting at 6h before virus challenge daily until 3 dpi. Five animals per group were sacrificed at 4 dpi for virus titer quantitation by plaque assay and histopathological studies as described previously.28,57 The survival rates and body weight changes of the remaining 5 animals per group were observed until 14 dpi or death as described previously. Immunofluorescence staining for MERS-CoV nucleocapsid protein expression was performed as we described previously.57
Golden Syrian hamster model for SARS-CoV-2 infection
The in vivo anti-SARS-CoV-2 activity of H84T-BanLec was evaluated in an established golden Syrian hamster as we described previously.31,58 Briefly, male and female golden Syrian hamsters, aged 6–10 weeks, were divided into different groups to receive H84T-BanLec or sham (PBS) treatment and their clinical scores were recorded as described previously.66 At 0 dpi, each hamster was intranasally inoculated with 100 μL of DMEM containing 105 PFU of SARS-CoV-2 (HKU-001a strain) under intraperitoneal ketamine (200 mg/kg) and xylazine (10 mg/kg) anaesthesia. The H84-BanLec group hamsters were treated with intraperitoneal (15 mg/kg/day every 24 h) or intranasal (1.5 mg/kg/animal/day every 24 h) H84T-BanLec starting at 6 h before virus challenge (pre-challenge) or 24 h after virus challenge (post-challenge) until 3 dpi (n = 10 per group). An additional group of mice received intraperitoneal GS-441524 [25 mg/kg every 24 h as described previously]32 starting at 6 h before SARS-CoV-2 challenge and continued until 3 dpi for comparison. The negative control hamsters were treated with intraperitoneal or intranasal PBS starting at 6h before virus challenge daily until 3 dpi (n = 10 per group). Five animals per group were sacrificed at 4 dpi for virus titer quantitation by plaque assay and histopathological studies as described previously.12,67 The survival rates and body weight changes of the remaining 5 animals per group were observed until 14dpi or death. Immunofluorescence staining for SARS-CoV-2 nucleocapsid protein expression was performed as we described previously.12
Time-of-drug-addition assay
Time-of-drug-addition assay was performed as we described previously to determine the phase(s) of the SARS-CoV-2 replication cycle targeted by H84T-BanLec.33 Briefly, VeroE6 cells were infected with SARS-CoV-2 (MOI = 3.0) and 20nM H84T-BanLec was added pre-infection (−1 h), at the time of infection (0 h), pre-incubated with SARS-CoV-2 pre-infection, or at 1 h post-infection. Intracellular viral genome copy numbers of the corresponding time points were determined at 8 h post-infection by qRT-PCR and compared with PBS-treated controls.
Detection of SARS-CoV-2 entry by flow cytometry
VeroE6-TMPRSS2 cells were treated with PBS or H84T-BanLec at the indicated concentrations for 1 h before infection with SARS-CoV-2 (MOI = 1.0) at 37°C for another 2 h. Afterwards, cells were dissociated into single cells using 10 mM EDTA (Invitrogen), fixed with 4% paraformaldehyde (PFA) for 15 min, permeabilized with 0.1% Triton X-100 for 5 min, and blocked using 1% BSA for 1 h. The cells were stained using an in-house anti-SARS-CoV-2 nucleoprotein antibody raised in rabbit for 1h at room temperature, followed by second antibody (goat anti-rabbit IgG secondary antibody Alexa Fluor 488 conjugate) for another 1 h. Mock cells were used for gating. The NovoCyte Advanteon BVR analyzer was used for analysis. Novoexpress software was used for data processing (Agilent).
Conjugation of Alexa fluorophore 647(AF647)-NHS (N-hydroxysuccinimide esters) to H84T-BanLec
H84T was conjugated to Alexa fluorophore 647 (AF647) by mixing 1 mg of H84T-Banlec with 25 μg of AF647-NHS (Thermo Fisher) in PBS. After 12 h of reaction at 4°C with shaking at 800 rpm, AF647-conjugated H84T-BanLec was separated by removing unconjugated AF647 with three washing steps with a centrifuge filter (Amicon® Ultra-10K centrifugal filter). AF647-conjugated H84T was resuspended in 1 mL PBS for the subsequent in vivo studies. For the in vivo intranasal pharmacokinetics study, we anesthetized BALB/c mice (female, 6 weeks old) with isoflurane and administered 20 μg of AF647-conjugated H84T in 20 μL into both nostrils. At the predetermined time points after administration, whole-body imaging was performed using an in vivo optical imaging system. The average fluorescent intensity was calculated from the nostril area.
Production of SARS-CoV-2 S-N234Q
A pCAGGS vector encoding the ectodomain of the SARS-CoV-2 S protein (GenBank: YP_009724390.1) was kindly provided by Florian Krammer (Icahn School of Medicine, Mount Sinai, New York, USA), and served as template for the generation of a pTT28 mammalian expression vector (National Research Council, NRC, Ottawa, Canada) encoding the S-N234Q spike ectodomain with a C-terminal thrombin cleavage site, T4 foldon trimerization domain and an octa-histidine tag as previously described.36,68 The sequence contains a mutated polybasic cleavage site (RRAR>A) and two proline mutations (K986P, V987P) that stabilize the protein in the pre-fusion conformation. S-N234Q was expressed in 1-L shake flasks (working volume 200 mL) using HEK293-6E cells (licensed from National Research Council, NRC, Ottawa, Canada) as previously described.69 Briefly, cells were routinely cultivated in suspension in Freestyle™ supplemented F17 medium (F17 incl. 4 mM L-glutamine, 0.1% (v/v) Pluronic F-68 and 25 μg/mL G-418, all from Thermo Fisher Scientific, Waltham, MA) in a humidified atmosphere of 8% (v/v) CO2 at 37°C shaking at 130 rpm. One μg plasmid pTT28-S-N234Q DNA and two μg linear 25-kDa linear Polyethylenimine (PEI, Polysciences, Inc., Hirschberg, DE) per mL of culture volume were combined in supplemented F17 medium (10% of total expression volume). After incubation for 15 minutes at room temperature, the mixture was added dropwise to the culture (∼2.0 × 106 cells/mL). Two- and four-days post-transfection, cells were supplemented with 0.5% (w/V) tryptone N1 (Organotechnie, La Courneuve, FR) and 0.25% (w/V) D (+)-glucose (Carl Roth, Karlsruhe, DE). Five days post-transfection the clarified supernatant (1,000 g, 10 min, RT) was further centrifuged at 10,000 g for 30 min at room temperature and 0.45-μM-filtrated. The supernatant was about five-fold concentrated using a 0,005 m2 Pellicon XL-Ultrafiltration module Biomax with a 10 kDa cut-off (# PXB010A50, Merck Millipore, Burlington, MA) on a Labscale TFF System (Merck Millipore, Burlington, MA), the buffer was exchanged to immobilized metal affinity chromatography (IMAC) binding buffer (20 mM phosphate pH 7.4, 500 mM NaCl + 0.05% (v/v) Tween-20) and further concentrated to about 8-fold in respect to the starting volume. Before downstream procedures, the concentrated and diafiltrated supernatant was supplemented with 20 mM imidazole (final concentration) and sterile-filtered. S-N234Q was purified using a HisTrap FF 5-mL column on an Äkta Pure system (both Cytiva, Marlborough, MA) and was eluted in 20 mM phosphate pH 7.4, 500 mM NaCl, 500 mM imidazole, 0.05% (v/v) Tween-20. The fractions containing the S-N234Q protein were pooled, concentrated and diafiltrated against 10 mM phosphate pH 7.4 + 0.05% (v/v) Tween-20 using an Amicon Ultra-15 centrifugal unit with a 100 kDa cutoff (# UFC910024, Merck Millipore, Burlington, MA). The protein concentration was determined by measuring the absorbance at A280 and using the calculated protein-specific extinction coefficient derived from the ProtParam tool on the ExPASy server.
Conjugation of SARS-CoV-2 spike trimer and H84T-BanLec through histidine residues of His-tagged protein
A maleimide-Poly(ethylene glycol) (PEG) linker was attached to a 3-aminopropyltriethoxysilane (APTES)-coated AFM cantilever (or silicon nitride surface for H84T-BanLec immobilization) by incubating the cantilevers for 2 h in 500 μL of chloroform containing 1mg of maleimide-PEG-N-hydroxysuccinimide (NHS) (Polypure, Oslo, Norway) and 30 μL of triethylamine. After washing 3 times with chloroform and drying with nitrogen gas, the cantilevers were immersed for 2 h in a mixture of 100 μL of 2 mM thiol-trisNTA, 2 μL of 100mM EDTA (pH 7.5), 5 μL of 1M HEPES (pH 7.5), 2 μL of 100 mM tris(carboxyethyl)phosphine (TCEP) hydrochloride, and 2.5 μL of 1M HEPES (pH 9.6) buffer, and subsequently washed with HEPES-buffer saline (HBS). Thereafter, the cantilevers were incubated for 4 h in a mixture of 4 μL of 5 mM NiCl2 and 100 μL of 0.2 μM His-tagged Spike trimer [wild-type (BOKU COVID-19 Initiative, https://portal.boku-covid19.at/signin, BOKU, Austria.); N234Q (BOKU, Austria); B.1.617.2 (SPN-C52He, Acrobiosystems, USA)]; or B.1.1.529 (SPN-C52Hz, Acrobiosystems, USA)]. After washing with HBS, the cantilevers were stored in HBS at 4°C.70
Single-molecule force spectroscopy (SMFS) measurement
Force distance measurements were performed at room temperature (∼25°C) using cantilevers with 0.01 N/m nominal spring constants (MSCT, Bruker) in TBS buffer containing 1mM CaCl2, and 0.1% Tween-20. Precise spring constant values of AFM cantilevers were determined by measuring the thermally driven mean-square bending of the cantilever using the equipartition theorem in an ambient environment.71 The deflection sensitivity was calculated from the slope of the force-distance curves recorded on a bare silicon substrate. Determined spring constants ranged from 0.008-0.015 N/m. Force-distance curves were acquired by recording at least 1000 curves with vertical sweep rates between 0.5s and 10s and at a z-range of typically 500–1000 nm, resulting in a loading rate from 10–10,000 pN/s, using a commercial AFM (Keysight Technologies, USA). The relationship between experimentally measured unbinding forces and parameters depicting the interaction potential were deciphered using the kinetic models of Bell, and Evans and Ritchie.40,41 Additionally, multiple parallel bond formation was calculated by the Williams model from the parameters derived from single-bond analysis.72
Length data analysis
Software for data analysis was written in Matlab (MathWorks, Inc.). Force curves (n = 4000, 2 different tips) were analysed for the calculation of the distance and for the construction of experimental probability density function (PDF) of distance. The distance between two unbinding events was calculated with the correction of the cantilever deflection. Thereafter, for each length difference and the corresponding standard deviation given by the pixel mash of the data recordings, single Gaussians with unitary area were constructed, summed up, and normalized to calculate the experimental probability density function. This data presentation is advantageous over conventional histograms, as the data accuracy was taken into account and binning artefacts could be avoided.
Surface plasmon resonance (SPR) measurement
SPR was used to study the kinetics of binding and dissociation of H84T-BanLec to SARS-CoV-2 spike protein in real time. SARS-CoV-2 spike protein was immobilized onto a Sensor Chip NTA (cytiva) via its His6-tag after washing the chip for at least 3 min with 350 mM EDTA and activation by applying a solution of 0.5 mM NiCl2 for 1 min. 50 nM SARS-CoV-2 spike protein was injected multiple times to generate a stable surface. For the determination of kinetic and equilibrium constants, H84T-BanLec was injected at different concentrations (10–200 nM) and subsequently removed with buffer. Running buffer was TBS, pH 7.4, containing 0.1% Tween-20, 1% BSA, and 1 mM Ca2+. The resulting experimental binding curves were fitted using the “bivalent analyte model”, assuming two-step binding of H84T-BanLec to SARS-CoV-2 spike protein.
Cell culture experiments
VeroE6 cells were grown on Petri dishes (3 mm in diameter) using DMEM containing 10% FBS, 500 unit/mL penicillin and 100 μg/mL streptomycin at 37°C with 5% CO2. For AFM measurements, the density of the cells was at about 20–80% coverage of the dish surface. The growth medium was exchanged to 2 mL physiological HEPES (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid) buffer containing 140 mM NaCl, 5 mM KCl, 1 mM MgCl2, 1 mM CaCl2, and 10 mM HEPES (pH 7.4 with NaOH). For block experiments, distinct amounts of 10 μg/mL or 500 μg/mL H84T-BanLec stock solution in HEPES buffer were added into 2 mL HEPES buffer directly on the cells in the Petri dish and thoroughly mixed.
Inhibition measurements
All force-distance curves were recorded at room temperature by using a PicoPlus 5500 AFM setup (Agilent Technologies, Chandler, AZ, USA) on living cells with the assistance of a CCD camera for localization of the cantilever tip on selected cells. The optical system of the AFM was focused on the cantilever tip, while the sample plate with the Petri dish was moved upwards by the step motor. Before the cells on the dish reached the focus, the piezo tube of the AFM was started to scan in the z-axis with a scanning range of 3μm and at a scanning frequency of 1Hz. The sample plate was moved upwards by the step motor using manual control with 1 μm per step. Due to the resistance of the liquid, a gap between the approaching curve and the retraction curve appeared, when the AFM tip was close to the sample surface. About 2 μm before the AFM tip touched the sample surface, the approaching curve was no longer parallel to the retraction curve. With this signal, the movement of the step motor was stopped. Further approaching was accomplished by gradually changing the voltage on the piezo tube. With this approaching method, the indentation force of the first contact between the AFM tip and the sample surface was controlled to be less than 30 pN. The functionalized cantilever (pegylated with SARS-CoV-2 spike protein) with a nominal spring constant of 0.01 N/m was moved downward to the cell surface and moved upward after the deflection of the cantilever reached the force limit. The deflection (z) of the cantilever was monitored by a laser beam on the cantilever surface and plotted versus the Z-position of the scanner, from which the force (F) was determined according to Hook’s law (F =kz, with k being the cantilever spring constant). When the tip tethered molecule bound to ACE2 on the cell surface, a pulling force developed during the upward movement of the cantilever causing the cantilever to bend downwards. At a critical force, i.e. the unbinding force, the tip tethered spike protein detached from ACE2, and the cantilever jumped back to its neutral position. The sweep range was fixed at 3000nm and the sweep rate was set at 1 Hz. On each cell, at least 100 force-distance cycles with 2000 data points per cycle and typical force limit of about 30 pN were recorded. The unbinding event was identified by local maximum analysis using a signal-to-noise threshold of 2. The binding activity was calculated from the fraction of curves showing unbinding events. Two-tailed Student’s t-test was performed for statistical analysis.
High-speed atomic force microscopy (HS-AFM) imaging
10 nM of purified SARS-CoV-2 spike protein was suspended in imaging buffer (10 mM Hepes, pH 7.4, 140 mM NaCl, 5 mM KCl, 1 mM CaCl, 1 mM MgCl), containing 5 μM NiCl2, of which 1.5 μL were applied to freshly cleaved mica disc (2mm diameter). After 3 min, the surface was rinsed with ∼15 μL imaging buffer (without drying) and the sample was mounted in the imaging chamber of the HS-AFM (RIBM, Japan). Similarly, H84T-BanLec stock was diluted to 2 μM in imaging buffer and 1.5 μL of the diluted solution was applied to mica for 3 min, rinsed with 15 μL imaging buffer, and imaged using HS-AFM. For following the complexation of H84T-BanLec and SARS-CoV-2 spike protein, the SARS-CoV-2 spike trimer was deposited on mic as described above, before 2 μM H84T-BanLec was added to the imaging chamber. For imaging, we used ultra-short cantilevers USC-F1.2-k0.15 (NanoWorld) with nominal spring constant 0.15N/m, resonance frequency of ∼500 kHz, and quality factor of ∼2. During image acquisition, the amplitude was set to 85–90% of the free amplitude (∼3 nm) and kept constant using a feedback loop.
HS-AFM image processing and volume measurement
Horizontal scars, which occurred due to feedback instabilities or particles sticking to the AFM tip, were selected and removed by Laplacian background substitution. A height threshold mask was used for selecting the background prior to correction of scanning artefacts. Next, Gaussian filter was applied to the images. For the volume measurement, the protein surface was selected using a height threshold mask defined from a minimum height of 0.25–0.35 nm to the maximum height of the protein structure. Image processing was done using Gwyddion 2.55.
Three-dimensional structural modelling
All structural models of the SARS-CoV-2 spike trimer were made from the fully glycosylated 3D model of the SARS-CoV-2 spike trimer, based on the cryo-EM structure of Walls et al. (PDB: 6VYB).73,74 Curved distances over the surface of the protein between glycans were calculated using the distance field algorithm.75 This algorithm determines the shortest distance between the centres of geometry of selected glycans over a grid with grid-spacing of 0.5nm, avoiding any grid points that are within 0.5 nm of any protein atom. The path was further smoothened by averaging over 5 neighbouring path points. Models of the complex of spike with H84T-BanLec were created using the structure of BanLec (PDB: 4PIK). A superposition of the bound carbohydrates of chain A of this model with the oligomannosidic glycan at N234 of the SARS-CoV-2 spike was performed to create models of the H84TBanLec/SARS-CoV-2 spike complex.
Quantification and statistical analysis
All data were analysed with GraphPad Prism software (GraphPad Software, Inc). One-way ANOVA or Student’s t test was used to determine significant differences in viral loads and titers, and Kaplan-Meier survival curves were analysed by the log rank test. p <0.05 was considered statistically significant.
Acknowledgments
This study was partly supported by funding to the University of Hong Kong: the Health and Medical Research Fund, the Food and Health Bureau, the Government of the Hong Kong Special Administrative Region (COVID190121, COVID1903010-Project 7, 20190572, and 15140762); Health@InnoHK, Innovation and Technology Commission, the Government of the Hong Kong Special Administrative Region; the General Research Fund (17119821), Collaborative Research Fund (C7060-21G), and the Theme-Based Research Scheme (T11-709/21-N) of the Research Grants Council, the Government of the Hong Kong Special Administrative Region; the Consultancy Service for Enhancing Laboratory Surveillance of Emerging Infectious Diseases and Research Capability on Antimicrobial Resistance for Department of Health of the Hong Kong Special Administrative Region Government; the National Program on Key Research Project of China (grant nos. 2020YFA0707500 and 2020YFA0707504); Sanming Project of Medicine in Shenzhen, China (grant no. SZSM201911014); the High Level-Hospital Program, Health Commission of Guangdong Province, China; the Major Science and Technology Program of Hainan Province (grant no. ZDKJ202003); the Research Project of Hainan Academician Innovation Platform (YSPTZX202004); the Hainan Talent Development Project (SRC200003); Emergency Collaborative Project (EKPG22-01) of Guangzhou Laboratory; Emergency COVID-19 Project (2021YFC0866100), Major Projects on Public Security, National Key Research and Development Program; the University of Hong Kong Outstanding Young Researcher Award; and the University of Hong Kong Research Output Prize (Li Ka Shing Faculty of Medicine); and donations from the Shaw Foundation Hong Kong, the Richard Yu and Carol Yu, Michael Seak-Kan Tong, May Tam Mak Mei Yin, Lee Wan Keung Charity Foundation Limited, the Providence Foundation Limited (in memory of the late Lui Hac Minh), Hong Kong Sanatorium & Hospital, Hui Ming, Hui Hoy and Chow Sin Lan Charity Fund Limited, the Chen Wai Wai Vivien Foundation Limited, Chan Yin Chuen Memorial Charitable Foundation, Marina Man-Wai Lee, the Hong Kong Hainan Commercial Association South China Microbiology Research Fund, the Jessie & George Ho Charitable Foundation, Perfect Shape Medical Limited, Kai Chong Tong, Tse Kam Ming Laurence, Foo Oi Foundation Limited, Betty Hing-Chu Lee, Ping Cham So, and Lo Ying Shek Chi Wai Foundation. This work was supported by the grants to D.M.M. from the Defense Threat Reduction Agency (grant no. HDTRA1-15-1-0067), the University of Michigan MTRAC (Michigan Translational Research and Commercialization Life Sciences), and the Forbes Institute of the University of Michigan Rogel Cancer Center. This work was further supported by funding from the Austrian National Foundation for Research, Technology, and Development and Research Department of the State of Upper Austria (Y.O.), from the FWF projects V584 (Y.O.), P31599 (R.Z.), P30314 (H.S., P.H.), I3183 (P.H.), the WWTF grant LS19-029 (P.H., D.C.), the ÖAW fellowship STIP13202002 (L.H.), and the European Union’s Horizon research and innovation programme (grant no. H2020-MSCA-ITN-2016) under the Marie Skolodowska-Curie grant agreement no. 721874 (P.H. and D.C.). Purified wild-type spike protein was kindly provided by the BOKU COVID-19 Initiative (https://portal.boku-covid19.at/signin). D.G.T. acknowledges funding from the Department of Pathology, University of Michigan and Allecia Wilson, and the Wayne County Medical Examiner’s office for providing access to the autopsy material. J.M.P. has received funding from the T. von Zastrow foundation, the FWF Wittgenstein award (grant no. Z 271-B19), the Austrian Academy of Sciences, the Innovative Medicines Initiative 2 Joint Undertaking (JU) under grant agreement no. 101005026, and the Canada 150 Research Chairs Program F18-01336 as well as the Canadian Institutes of Health Research COVID-19 grants F20-02343 and F20-02015. C.X. and J.J.M. were supported by NIH R01 DK125087. C.O. acknowledges funding by the Rapid Response program of the Vienna Science and Technology Fund (WWTF, Austria; grant no. COV20-015). Preliminary findings related to this work were presented as poster presentations at the 10th Asia-Pacific Congress of Medical Virology 2015 (Taipei) and Health Research Symposium 2021 (Hong Kong). The funding sources had no role in the study design, data collection, analysis, interpretation, or writing of the report. This report is dedicated to the memory of Professor Hans-Joachim Gabius.
Author contributions
J.F.-W.C., Y.O., S.Y., and H.C. contributed equally to this work. J.F.-W.C., Y.O., S.Y., H.C., P.H., K.Y.Y., and D.M.M. had roles in the study design, data collection, data analysis, data interpretation, literature search, and writing of the manuscript. J.F.-W.C., S.Y., H.C., M.-L.Y., C.C.-S.C., V.K.-M.P., C.C.-Y.C., A.J.Z., J.-P.C., Z.-W.Y., L.W., T.T.-T.Y., K.K.-H.C., H. Shuai, Y.W., Y.H., C.L., W.-M.C., Z.Q., K.K.-W.T., K.-H.C., and J.S. performed the in vitro, ex vivo, and/or animal experiments and analyzed the data. Y.O. performed force spectroscopy measurements and analyzed the data. D.C. performed HS-AFM measurements and analyzed the data. K.-Y.S., W.-K.A., and M.L. provided critical reagents and/or tissues for the experiments. R.Z. performed AFM binding activity measurements and analyzed the data. L.H. performed SPR measurement and analyzed the data. H. Seferovic developed the home-build Matlab program for distant analysis and performed HS-AFM measurements. R.T. provided thiol-trisNTA. D.G.T. performed the co-localization experiments using autopsied lung sections. M.K. produced the N234Q spike glycoprotein mutant. J.M.P. provided conceptual advice and troubleshooting. C.O. performed the molecular modeling experiments and visualized the H84T-BanLec and spike proteins. C.X. and J.J.M. designed and performed the experiments that examined the pharmacokinetics of intranasal H84T-BanLec. J.F.-W.C., P.H., K.-Y.Y., and D.M.M. oversaw the conception and supervised the study. J.F.-W.C., Y.O., D.C., R.Z., L.H., H. Seferovic, D.G.T., C.X., J.J.M., J.M.P., C.O., P.H., K.-Y.Y., and D.M.M. provided funding support.
Declaration of interests
J.F.-W.C. has received travel grants from Pfizer Corporation Hong Kong and Astellas Pharma Hong Kong Corporation Limited and was an invited speaker for Gilead Sciences Hong Kong Limited and Luminex Corporation. D.M.M. is an inventor on patents U.S. no. 8,865,867 “Lectins and Uses Thereof”; U.S. no. 9,481,717 “Lectins and Uses Thereof”; European no. 2558488 (DE, GB, FR) “Banana Lectins and Uses Thereof”; Chinese no. 103025756 “Banana Lectins and Uses Thereof”; and U.S. no. 10,450,355 “Lectins and Uses Thereof.”
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: October 18, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2022.100774.
Contributor Information
Peter Hinterdorfer, Email: peter.hinterdorfer@jku.at.
Kwok-Yung Yuen, Email: kyyuen@hku.hk.
David M. Markovitz, Email: dmarkov@umich.edu.
Supplemental information
Data and code availability
-
•
The complete sequences of MERS-CoV (EMC/2012, GenBank: JX869059), SARS-CoV (GZ50, GenBank: AY304495), SARS-CoV-2 (HKU-001a, GenBank: MT230904), SARS-CoV-2 B.1.1.7 (Alpha, GenBank: OM212469), SARS-CoV-2 B.1.617.2 (Delta, GenBank: OM212471), and SARS-CoV-2 BA.1.529.1 (Omicron BA.1, GenBank: OM212472) are available on GenBank as of the date of publication. Additional supplemental items (videos 1–3, related to Figures 4 and 5) are available from Mendeley Data at https://data.mendeley.com/datasets/gy7zt485zh/2.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon reasonable request.
References
- 1.Chan J.F.W., To K.K.W., Tse H., Jin D.Y., Yuen K.Y. Interspecies transmission and emergence of novel viruses: lessons from bats and birds. Trends Microbiol. 2013;21:544–555. doi: 10.1016/j.tim.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peiris J.S.M., Lai S.T., Poon L.L.M., Guan Y., Yam L.Y.C., Lim W., Nicholls J., Yee W.K.S., Yan W.W., Cheung M.T., et al. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D.M.E., Fouchier R.A.M. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 4.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan J.F., Yuan S., Kok K.H., To K.K., Chu H., Yang J., Xing F., Liu J., Yip C.C., Poon R.W., et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395:514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng V.C.C., Lau S.K.P., Woo P.C.Y., Yuen K.Y. Severe acute respiratory syndrome coronavirus as an agent of emerging and reemerging infection. Clin. Microbiol. Rev. 2007;20:660–694. doi: 10.1128/CMR.00023-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan J.F.W., Lau S.K.P., To K.K.W., Cheng V.C.C., Woo P.C.Y., Yuen K.Y. Middle East respiratory syndrome coronavirus: another zoonotic betacoronavirus causing SARS-like disease. Clin. Microbiol. Rev. 2015;28:465–522. doi: 10.1128/CMR.00102-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.World Health Organization Weekly Epidemiological Update on COVID-19-3 August 2022. https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19–-3-august-2022
- 9.Zumla A., Chan J.F.W., Azhar E.I., Hui D.S.C., Yuen K.Y. Coronaviruses - drug discovery and therapeutic options. Nat. Rev. Drug Discov. 2016;15:327–347. doi: 10.1038/nrd.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beigel J.H., Tomashek K.M., Dodd L.E., Mehta A.K., Zingman B.S., Kalil A.C., Hohmann E., Chu H.Y., Luetkemeyer A., Kline S., et al. Remdesivir for the treatment of Covid-19-final report. N. Engl. J. Med. 2020;383:1813–1826. doi: 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hung I.F.N., Lung K.C., Tso E.Y.K., Liu R., Chung T.W.H., Chu M.Y., Ng Y.Y., Lo J., Chan J., Tam A.R., et al. Triple combination of interferon beta-1b, lopinavir-ritonavir, and ribavirin in the treatment of patients admitted to hospital with COVID-19: an open-label, randomised, phase 2 trial. Lancet. 2020;395:1695–1704. doi: 10.1016/S0140-6736(20)31042-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yuan S., Yin X., Meng X., Chan J.F.W., Ye Z.W., Riva L., Pache L., Chan C.C.S., Lai P.M., Chan C.C.Y., et al. Clofazimine broadly inhibits coronaviruses including SARS-CoV-2. Nature. 2021;593:418–423. doi: 10.1038/s41586-021-03431-4. [DOI] [PubMed] [Google Scholar]
- 13.Sheahan T.P., Sims A.C., Zhou S., Graham R.L., Pruijssers A.J., Agostini M.L., Leist S.R., Schäfer A., Dinnon K.H., 3rd, Stevens L.J., et al. An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci. Transl. Med. 2020;12:eabb5883. doi: 10.1126/scitranslmed.abb5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.WHO Solidarity Trial Consortium. Pan H., Peto R., Hanao-Restrepo A., Preziosi M., Sathiyamoorthy V., Karim Q.A., Alejandria M.M., García G.H., Kieny M., et al. Repurposed antiviral drugs for Covid-19-interim WHO solidarity trial results. N. Engl. J. Med. 2021;384:497–511. doi: 10.1056/NEJMoa2023184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Owen D.R., Allerton C.M.N., Anderson A.S., Aschenbrenner L., Avery M., Berritt S., Boras B., Cardin R.D., Carlo A., Coffman K.J., et al. An oral SARS-CoV-2 Mpro inhibitor clinical candidate for the treatment of COVID-19. Science. 2021;374:1586–1593. doi: 10.1126/science.abl4784. [DOI] [PubMed] [Google Scholar]
- 16.Liu L., Wang P., Nair M.S., Yu J., Rapp M., Wang Q., Luo Y., Chan J.F.W., Sahi V., Figueroa A., et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature. 2020;584:450–456. doi: 10.1038/s41586-020-2571-7. [DOI] [PubMed] [Google Scholar]
- 17.Weinreich D.M., Sivapalasingam S., Norton T., Ali S., Gao H., Bhore R., Musser B.J., Soo Y., Rofail D., Im J., et al. REGN-COV2, a neutralizing antibody cocktail, in outpatients with Covid-19. N. Engl. J. Med. 2021;384:238–251. doi: 10.1056/NEJMoa2035002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu L., Iketani S., Guo Y., Chan J.F.W., Wang M., Liu L., Luo Y., Chu H., Huang Y., Nair M.S., et al. Striking antibody evasion manifested by the Omicron variant of SARS-CoV-2. Nature. 2022;602:676–681. doi: 10.1038/s41586-021-04388-0. [DOI] [PubMed] [Google Scholar]
- 19.Iketani S., Liu L., Guo Y., Liu L., Chan J.F.W., Huang Y., Wang M., Luo Y., Yu J., Chu H., et al. Antibody evasion properties of SARS-CoV-2 Omicron sublineages. Nature. 2022;604:553–556. doi: 10.1038/s41586-022-04594-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yuan S., Ye Z.W., Liang R., Tang K., Zhang A.J., Lu G., Ong C.P., Man Poon V.K., Chan C.C.S., Mok B.W.Y., et al. Pathogenicity, transmissibility, and fitness of SARS-CoV-2 Omicron in Syrian hamsters. Science. 2022;377:428–433. doi: 10.1126/science.abn8939. [DOI] [PubMed] [Google Scholar]
- 21.Simonovich V.A., Pratx L.D.B., Scibona P., Beruto M.V., Vallone M.G., Vázquez C., Savoy N., Giunta D.H., Pérez L.G., Sánchez M.D.L., et al. A randomized trial of convalescent plasma in Covid-19 severe pneumonia. N. Eng. J. Med. 2021;384:619–629. doi: 10.1056/NEJMoa2031304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chan J.F.W., Kok K.H., Zhu Z., Chu H., To K.K.W., Yuan S., Yuen K.Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microb. Infect. 2020;9:221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Swanson M.D., Boudreaux D.M., Salmon L., Chugh J., Winter H.C., Meagher J.L., André S., Murphy P.V., Oscarson S., Roy R., et al. Engineering a therapeutic lectin by uncoupling mitogenicity from antiviral activity. Cell. 2015;163:746–758. doi: 10.1016/j.cell.2015.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Swanson M.D., Winter H.C., Goldstein I.J., Markovitz D.M. A lectin isolated from bananas is a potent inhibitor of HIV replication. J. Biol. Chem. 2010;285:8646–8655. doi: 10.1074/jbc.M109.034926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coves-Datson E.M., King S.R., Legendre M., Gupta A., Chan S.M., Gitlin E., Kulkarni V.V., Pantaleón García J., et al. A molecularly engineered antiviral banana lectin inhibits fusion and is efficacious against influenza virus infection in vivo. Proc. Natl. Acad. Sci. USA. 2020;117:2122–2132. doi: 10.1073/pnas.1915152117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coves-Datson E.M., Dyall J., DeWald L.E., King S.R., Dube D., Legendre M., Nelson E., Drews K.C., Gross R., Gerhardt D.M., et al. Inhibition of Ebola virus by a molecularly engineered banana lectin. PLoS Negl. Trop. Dis. 2019;13:e0007595. doi: 10.1371/journal.pntd.0007595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou J., Chu H., Li C., Wong B.H.Y., Cheng Z.S., Poon V.K.M., Sun T., Lau C.C.Y., Wong K.K.Y., Chan J.Y.W., et al. Active replication of Middle East respiratory syndrome coronavirus and aberrant induction of inflammatory cytokines and chemokines in human macrophages: implications for pathogenesis. J. Infect. Dis. 2014;209:1331–1342. doi: 10.1093/infdis/jit504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yuan S., Chu H., Chan J.F.W., Ye Z.W., Wen L., Yan B., Lai P.M., Tee K.M., Huang J., Chen D., et al. SREBP-dependent lipidomic reprogramming as a broad-spectrum antiviral target. Nat. Commun. 2019;10:120. doi: 10.1038/s41467-018-08015-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li K., Wohlford-Lenane C.L., Channappanavar R., Park J.E., Earnest J.T., Bair T.B., Bates A.M., Brogden K.A., Flaherty H.A., Gallagher T., et al. Mouse-adapted MERS coronavirus causes lethal lung disease in human DPP4 knockin mice. Proc. Natl. Acad. Sci. USA. 2017;114:E3119–E3128. doi: 10.1073/pnas.1619109114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chu H., Shuai H., Hou Y., Zhang X., Wen L., Huang X., Hu B., Yang D., Wang Y., Yoon C., et al. Targeting highly pathogenic coronavirus-induced apoptosis reduces viral pathogenesis and disease severity. Sci. Adv. 2021;7:eabf8577. doi: 10.1126/sciadv.abf8577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chan J.F.W., Zhang A.J., Yuan S., Poon V.K.M., Chan C.C.S., Lee A.C.Y., Chan W.M., Fan Z., Tsoi H.W., Wen L., et al. Simulation of the clinical and pathological manifestations of coronavirus disease 2019 (COVID-19) in a golden Syrian hamster model: implications for disease pathogenesis and transmissibility. Clin. Infect. Dis. 2020;71:2428–2446. doi: 10.1093/cid/ciaa325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Y., Cao L., Li G., Cong F., Li Y., Sun J., Luo Y., Chen G., Li G., Wang P., et al. Remdesivir metabolite GS-441524 effectively inhibits SARS-CoV-2 infection in mouse models. J. Med. Chem. 2021;65:2785–2793. doi: 10.1021/acs.jmedchem.0c01929. [DOI] [PubMed] [Google Scholar]
- 33.Riva L., Yuan S., Yin X., Martin-Sancho L., Matsunaga N., Pache L., Burgstaller-Muehlbacher S., De Jesus P.D., Teriete P., Hull M.V., et al. Discovery of SARS-CoV-2 antiviral drugs through large-scale compound repurposing. Nature. 2020;586:113–119. doi: 10.1038/s41586-020-2577-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kodera N., Yamamoto D., Ishikawa R., Ando T. Video imaging of walking myosin V by high-speed atomic force microscopy. Nature. 2010;468:72–76. doi: 10.1038/nature09450. [DOI] [PubMed] [Google Scholar]
- 35.Preiner J., Kodera N., Tang J., Ebner A., Brameshuber M., Blaas D., Gelbmann N., Gruber H.J., Ando T., Hinterdorfer P. IgGs are made for walking on bacterial and viral surfaces. Nat. Commun. 2014;5:4394. doi: 10.1038/ncomms5394. [DOI] [PubMed] [Google Scholar]
- 36.Hoffmann D., Mereiter S., Jin Oh Y., Monteil V., Elder E., Zhu R., Canena D., Hain L., Laurent E., Grünwald-Gruber C., et al. Identification of lectin receptors for conserved SARS-CoV-2 glycosylation sites. EMBO J. 2021;40:e108375. doi: 10.15252/embj.2021108375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hopper J.T.S., Ambrose S., Grant O.C., Krumm S.A., Allison T.M., Degiacomi M.T., Tully M.D., Pritchard L.K., Ozorowski G., Ward A.B., et al. The tetrameric plant lectin BanLec neutralizes HIV through bidentate binding to specific viral glycans. Structure. 2017;25:773–782.e5. doi: 10.1016/j.str.2017.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hinterdorfer P., Baumgartner W., Gruber H.J., Schilcher K., Schindler H. Detection and localization of individual antibody-antigen recognition events by atomic force microscopy. Proc. Natl. Acad. Sci. USA. 1996;93:3477–3481. doi: 10.1073/pnas.93.8.3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rankl C., Kienberger F., Wildling L., Wruss J., Gruber H.J., Blaas D., Hinterdorfer P. Multiple receptors involved in human rhinovirus attachment to live cells. Proc. Natl. Acad. Sci. USA. 2008;105:17778–17783. doi: 10.1073/pnas.0806451105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bell G.I. Models for the specific adhesion of cells to cells. Science. 1978;200:618–627. doi: 10.1126/science.347575. [DOI] [PubMed] [Google Scholar]
- 41.Evans E., Ritchie K. Dynamic strength of molecular adhesion bonds. Biophys. J. 1997;72:1541–1555. doi: 10.1016/S0006-3495(97)78802-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Strnad M., Oh Y.J., Vancová M., Hain L., Salo J., Grubhoffer L., Nebesářová J., Hytönen J., Hinterdorfer P., Rego R.O.M. Nanomechanical mechanisms of Lyme disease spirochete motility enhancement in extracellular matrix. Commun. Biol. 2021;4:268. doi: 10.1038/s42003-021-01783-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Traxler L., Rathner P., Fahrner M., Stadlbauer M., Faschinger F., Charnavets T., Müller N., Romanin C., Hinterdorfer P., Gruber H.J. Detailed evidence for an unparalleled interaction mode between calmodulin and orai proteins. Angew. Chem. Int. Ed. Engl. 2017;56:15755–15759. doi: 10.1002/anie.201708667. [DOI] [PubMed] [Google Scholar]
- 44.Casalino L., Gaieb Z., Goldsmith J.A., Hjorth C.K., Dommer A.C., Harbison A.M., Fogarty C.A., Barros E.P., Taylor B.C., McLellan J.S., et al. Beyond shielding: the roles of glycans in the SARS-CoV-2 spike protein. ACS Cent. Sci. 2020;6:1722–1734. doi: 10.1021/acscentsci.0c01056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Watanabe Y., Allen J.D., Wrapp D., McLellan J.S., Crispin M. Site-specific glycan analysis of the SARS-CoV-2 spike. Science. 2020;369:330–333. doi: 10.1126/science.abb9983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu R., Howorka S., Pröll J., Kienberger F., Preiner J., Hesse J., Ebner A., Pastushenko V.P., Gruber H.J., Hinterdorfer P. Nanomechanical recognition measurements of individual DNA molecules reveal epigenetic methylation patterns. Nat. Nanotechnol. 2010;5:788–791. doi: 10.1038/nnano.2010.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao P., Praissman J.L., Grant O.C., Cai Y., Xiao T., Rosenbalm K.E., Aoki K., Kellman B.P., Bridger R., Barouch D.H., et al. Virus-receptor interactions of glycosylated SARS-CoV-2 spike and human ACE2 receptor. Cell Host Microbe. 2020;28:586–601.e6. doi: 10.1016/j.chom.2020.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Monteil V., Kwon H., Prado P., Hagelkrüys A., Wimmer R.A., Stahl M., Leopoldi A., Garreta E., Hurtado Del Pozo C., Prosper F., et al. Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2. Cell. 2020;181:905–913.e7. doi: 10.1016/j.cell.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.O'Keefe B.R., Giomarelli B., Barnard D.L., Shenoy S.R., Chan P.K.S., McMahon J.B., Palmer K.E., Barnett B.W., Meyerholz D.K., Wohlford-Lenane C.L., McCray P.B., Jr. Broad-spectrum in vitro activity and in vivo efficacy of the antiviral protein griffithsin against emerging viruses of the family Coronaviridae. J. Virol. 2010;84:2511–2521. doi: 10.1128/JVI.02322-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu Y.M., Shahed-Al-Mahmud M., Chen X., Chen T.H., Liao K.S., Lo J.M., Wu Y.M., Ho M.C., Wu C.Y., Wong C.H., et al. A carbohydrate-binding protein from the edible lablab beans effectively blocks the infections of influenza viruses and SARS-CoV-2. Cell Rep. 2020;32:108016. doi: 10.1016/j.celrep.2020.108016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chu H., Chan J.F.W., Yuen T.T.T., Shuai H., Yuan S., Wang Y., Hu B., Yip C.C.Y., Tsang J.O.L., Huang X., et al. Comparative tropism, replication kinetics, and cell damage profiling of SARS-CoV-2 and SARS-CoV with implications for clinical manifestations, transmissibility, and laboratory studies of COVID-19: an observational study. Lancet. Microbe. 2020;1:e14–e23. doi: 10.1016/S2666-5247(20)30004-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shuai H., Chan J.F.W., Hu B., Chai Y., Yuen T.T.T., Yin F., Huang X., Yoon C., Hu J.C., Liu H., et al. Attenuated replication and pathogenicity of SARS-CoV-2 B.1.1.529 Omicron. Nature. 2022;603:693–699. doi: 10.1038/s41586-022-04442-5. [DOI] [PubMed] [Google Scholar]
- 53.Shuai H., Chan J.F.W., Yuen T.T.T., Yoon C., Hu J.C., Wen L., Hu B., Yang D., Wang Y., Hou Y., et al. Emerging SARS-CoV-2 variants expand species tropism to murines. EBioMedicine. 2021;73:103643. doi: 10.1016/j.ebiom.2021.103643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chu H., Hou Y., Yang D., Wen L., Shuai H., Yoon C., Shi J., Chai Y., Yuen T.T.T., Hu B., et al. Coronaviruses exploit a host cysteine-aspartic protease for replication. Nature. 2022 doi: 10.1038/s41586-022-05148-4. [DOI] [PubMed] [Google Scholar]
- 55.Chan J.F.W., Chan K.H., Choi G.K.Y., To K.K.W., Tse H., Cai J.P., Yeung M.L., Cheng V.C.C., Chen H., Che X.Y., et al. Differential cell line susceptibility to the emerging novel human betacoronavirus 2c EMC/2012: implications for disease pathogenesis and clinical manifestation. J. Infect. Dis. 2013;207:1743–1752. doi: 10.1093/infdis/jit123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chan J.F.W., Yip C.C.Y., Tsang J.O.L., Tee K.M., Cai J.P., Chik K.K.H., Zhu Z., Chan C.C.S., Choi G.K.Y., Sridhar S., et al. Differential cell line susceptibility to the emerging Zika virus: implications for disease pathogenesis, non-vector-borne human transmission and animal reservoirs. Emerg. Microb. Infect. 2016;5:e93. doi: 10.1038/emi.2016.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yuan S., Chu H., Huang J., Zhao X., Ye Z.W., Lai P.M., Wen L., Cai J.P., Mo Y., Cao J., et al. Viruses harness YxxO motif to interact with host AP2M1 for replication: a vulnerable broad-spectrum antiviral target. Sci. Adv. 2020;6:eaba7910. doi: 10.1126/sciadv.aba7910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yuan S., Wang R., Chan J.F.W., Zhang A.J., Cheng T., Chik K.K.H., Ye Z.W., Wang S., Lee A.C.Y., Jin L., et al. Metallodrug ranitidine bismuth citrate suppresses SARS-CoV-2 replication and relieves virus-associated pneumonia in Syrian hamsters. Nat. Microbiol. 2020;5:1439–1448. doi: 10.1038/s41564-020-00802-x. [DOI] [PubMed] [Google Scholar]
- 59.Chan J.F.W., Chan K.H., Kao R.Y.T., To K.K.W., Zheng B.J., Li C.P.Y., Li P.T.W., Dai J., Mok F.K.Y., Chen H., et al. Broad-spectrum antivirals for the emerging Middle East respiratory syndrome coronavirus. J. Infect. 2013;67:606–616. doi: 10.1016/j.jinf.2013.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yuan S., Chan J.F.W., Chik K.K.H., Chan C.C.Y., Tsang J.O.L., Liang R., Cao J., Tang K., Chen L.L., Wen K., et al. Discovery of the FDA-approved drugs bexarotene, cetilistat, diiodohydroxyquinoline, and abiraterone as potential COVID-19 treatments with a robust two-tier screening system. Pharmacol. Res. 2020;159:104960. doi: 10.1016/j.phrs.2020.104960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhao H., To K.K.W., Sze K.H., Yung T.T.M., Bian M., Lam H., Yeung M.L., Li C., Chu H., Yuen K.Y. A broad-spectrum virus- and host-targeting peptide against respiratory viruses including influenza virus and SARS-CoV-2. Nat. Commun. 2020;11:4252. doi: 10.1038/s41467-020-17986-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chan C.M., Chu H., Wang Y., Wong B.H.Y., Zhao X., Zhou J., Yang D., Leung S.P., Chan J.F.W., Yeung M.L., et al. Carcinoembryonic antigen-related cell adhesion molecule 5 is an important surface attachment factor that facilitates entry of Middle East respiratory syndrome Coronavirus. J. Virol. 2016;90:9114–9127. doi: 10.1128/JVI.01133-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chu H., Chan C.M., Zhang X., Wang Y., Yuan S., Zhou J., Au-Yeung R.K.H., Sze K.H., Yang D., Shuai H., et al. Middle East respiratory syndrome coronavirus and bat coronavirus HKU9 both can utilize GRP78 for attachment onto host cells. J. Biol. Chem. 2018;293:11709–11726. doi: 10.1074/jbc.RA118.001897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Farkash E.A., Wilson A.M., Jentzen J.M. Ultrastructural evidence for direct renal infection with SARS-CoV-2. J. Am. Soc. Nephrol. 2020;31:1683–1687. doi: 10.1681/ASN.2020040432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lee A.C.Y., Zhang A.J., Chan J.F.W., Li C., Fan Z., Liu F., Chen Y., Liang R., Sridhar S., Cai J.P., et al. Oral SARS-CoV-2 inoculation establishes subclinical respiratory infection with virus shedding in golden Syrian hamsters. Cell Rep. Med. 2020;1:100121. doi: 10.1016/j.xcrm.2020.100121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chan J.F., Poon V.K., Chan C.C., Chik K.K., Tsang J.O., Zou Z., Chan C.C., Lee A.C., Li C., Liang R., et al. Low environmental temperature exacerbates severe acute respiratory syndrome Coronavirus 2 infection in golden Syrian hamsters. Clin. Infect. Dis. 2021:ciab817. doi: 10.1093/cid/ciab817. [DOI] [PubMed] [Google Scholar]
- 68.Amanat F., Stadlbauer D., Strohmeier S., Nguyen T.H.O., Chromikova V., McMahon M., Jiang K., Arunkumar G.A., Jurczyszak D., Polanco J., et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat. Med. 2020;26:1033–1036. doi: 10.1038/s41591-020-0913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Klausberger M., Duerkop M., Haslacher H., Wozniak-Knopp G., Cserjan-Puschmann M., Perkmann T., Lingg N., Aguilar P.P., Laurent E., De Vos J., et al. A comprehensive antigen production and characterisation study for easy-to-implement, specific and quantitative SARS-CoV-2 serotests. EBioMedicine. 2021;67:103348. doi: 10.1016/j.ebiom.2021.103348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Oh Y.J., Hubauer-Brenner M., Gruber H.J., Cui Y., Traxler L., Siligan C., Park S., Hinterdorfer P. Curli mediate bacterial adhesion to fibronectin via tensile multiple bonds. Sci. Rep. 2016;6:33909. doi: 10.1038/srep33909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hutter J.L., Bechhoefer J. Calibration of atomic-force microscope tips. Rev. Sci. Instrum. 1993;64:1868–1873. [Google Scholar]
- 72.Williams P.M. Analytical descriptions of dynamic force spectroscopy: behaviour of multiple connections. Anal. Chim. Acta. 2003;479:107–115. [Google Scholar]
- 73.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;183:1735. doi: 10.1016/j.cell.2020.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Capraz T., Kienzl N.F., Laurent E., Perthold J.W., Föderl-Höbenreich E., Grünwald-Gruber C., Maresch D., Monteil V., Niederhöfer J., Wirnsberger G., et al. Structure-guided glyco-engineering of ACE2 for improved potency as soluble SARS-CoV-2 decoy receptor. Elife. 2021;10:e73641. doi: 10.7554/eLife.73641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.de Ruiter A., Oostenbrink C. Protein-ligand binding from distancefield distances and Hamiltonian replica exchange simulations. J. Chem. Theory Comput. 2013;9:883–892. doi: 10.1021/ct300967a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The complete sequences of MERS-CoV (EMC/2012, GenBank: JX869059), SARS-CoV (GZ50, GenBank: AY304495), SARS-CoV-2 (HKU-001a, GenBank: MT230904), SARS-CoV-2 B.1.1.7 (Alpha, GenBank: OM212469), SARS-CoV-2 B.1.617.2 (Delta, GenBank: OM212471), and SARS-CoV-2 BA.1.529.1 (Omicron BA.1, GenBank: OM212472) are available on GenBank as of the date of publication. Additional supplemental items (videos 1–3, related to Figures 4 and 5) are available from Mendeley Data at https://data.mendeley.com/datasets/gy7zt485zh/2.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon reasonable request.