Summary
Background
The existing dementia risk models are limited to known risk factors and traditional statistical methods. We aimed to employ machine learning (ML) to develop a novel dementia prediction model by leveraging a rich-phenotypic variable space of 366 features covering multiple domains of health-related data.
Methods
In this longitudinal population-based cohort of the UK Biobank (UKB), 425,159 non-demented participants were enrolled from 22 recruitment centres across the UK between March 1, 2006 and October 31, 2010. We implemented a data-driven strategy to identify predictors from 366 candidate variables covering a comprehensive range of genetic and environmental factors and developed the ML model to predict incident dementia and Alzheimer's Disease (AD) within five, ten, and much longer years (median 11.9 [Interquartile range 11.2–12.5] years).
Findings
During a follow-up of 5,023,337 person-years, 5287 and 2416 participants developed dementia and AD, respectively. A novel UKB dementia risk prediction (UKB-DRP) model comprising ten predictors including age, ApoE ε4, pairs matching time, leg fat percentage, number of medications taken, reaction time, peak expiratory flow, mother's age at death, long-standing illness, and mean corpuscular volume was established. Our prediction model was internally evaluated based on five-fold cross-validation on discrimination and calibration, and it was further compared with existing prediction scales. The UKB-DRP model can achieve high discriminative accuracy in dementia (AUC 0.848 ± 0.007) and even better in AD (AUC 0.862 ± 0.015). The model was well-calibrated (Hosmer-Lemeshow goodness-of-fit p-value = 0.92), and the predictive power was solid in different incidence time groups. More importantly, our model presented an apparent superiority over existing models like Cardiovascular Risk Factors, Aging, and Incidence of Dementia Risk Score (AUC 0.705 ± 0.008), the Dementia Risk Score (AUC 0.752 ± 0.007), and the Australian National University Alzheimer's Disease Risk Index (AUC 0.584 ± 0.017). The model was internally validated in the general population of European ancestry and White ethnicity; thus, further validation with independent datasets is necessary to confirm these findings.
Interpretation
Our ML-based UKB-DRP model incorporated ten easily accessible predictors with solid predictive power for incident dementia and AD within five, ten, and much longer years, which can be used to identify individuals at high risk of dementia and AD in the general population.
Funding
This study was funded by grants from the Science and Technology Innovation 2030 Major Projects (2022ZD0211600), National Key R&D Program of China (2018YFC1312904, 2019YFA070950), National Natural Science Foundation of China (282071201, 81971032, 82071997), Shanghai Municipal Science and Technology Major Project (2018SHZDZX01), Research Start-up Fund of Huashan Hospital (2022QD002), Excellence 2025 Talent Cultivation Program at Fudan University (3030277001), Shanghai Rising-Star Program (21QA1408700), Medical Engineering Fund of Fudan University (yg2021-013), and the 111 Project (No. B18015).
Keywords: Dementia, Alzheimer's disease, Prediction model, UK biobank, Machine learning
Abbreviations: AD, Alzheimer’s disease; AUC, area under the receiver operating characteristic curve; ANU-ADRI, Australian National University Alzheimer’s disease risk index; CAIDE, cardiovascular risk factors, aging, and incidence of dementia risk score; DRS, dementia risk score; ML, machine learning; UKB-DRP, UK biobank dementia risk prediction
Research in context.
Evidence before this study
We searched PubMed without language constraints for studies about dementia prediction published between Jan. 1, 2002 and Jan 1, 2022 using the terms “(dementia OR Alzheimer's Disease) AND (prediction OR predict OR predictive) AND (longitudinal)”. Numerous dementia prediction models have been developed, most of which were score-based systems that adopted predictors varied from sociodemographic, cognitive, imaging, and biomedical, to genetic variables that were largely based on empirical knowledge, and they were primarily developed by traditional statistical methods (e.g. cox or logistic regression). Current applications of ML in dementia prediction often incorporate variables that are not easily accessible in basic clinical practice (e.g., high-level neuroimaging features and cerebrospinal fluid biomarkers), narrowing its application to research or expertise settings.
Added value of this study
We leveraged a longitudinal study cohort of UK-Biobank with richly phenotypic health-related variable space, allowing us to maximize the potential in identifying undiscovered risk factors for dementia risk predictions. The deliberately designed ML-based data-driven pipeline identified the optimal combinations of key factors, and the proposed UKB-DRP model exhibited excellent performance in discriminating future incident dementia events among a population of healthy subjects. The model is also well-generalized in predicting Alzheimer's Disease (AD). Further, our novel UKB-DRP model demonstrated an apparent superiority over existing risk prediction models.
Implications of all the available evidence
The ML-based risk prediction models can learn expressive representations from potential high-risked dementia and AD patients. Predictors leveraged by the proposed model can be rapidly available through questionnaires, physical measures, and simple blood tests. Further, several identified predictors are intervenable at an early stage, and it is worth paying more attention to their potential mechanisms to reduce or delay the development of dementia.
Alt-text: Unlabelled box
Introduction
Dementia is a group of symptoms affecting thinking, mood, and behaviour severe enough to interfere with daily life, which affects over 55 million people worldwide.1 Given the long prodromal period when neuropathological changes occur before dementia diagnosis, there is an urgent need to establish approaches to identify the population appearing normal but at high risk of developing dementia. The ability to predict dementia incidence is critically relevant for decisions of clinicians to manage patients in follow-up, and for investigators to recruit participants into clinical trials. Early precise prevention and intervention targeting the highly suspected population can effectively reduce the disease burden and save enormous medical resources from those unlikely to progress to dementia.
Dementia can be attributed to genetic and modifiable risk factors which have been incorporated into various prediction models in previous research, such as the Cardiovascular Risk Factors, Aging, and Incidence of Dementia (CAIDE) Risk Score,2 the Dementia Risk Score (DRS),3 and the Australian National University Alzheimer's Disease Risk Index (ANU-ADRI).4 Nevertheless, these models were primarily established based on predictors manually retrieved from literature like age, apolipoprotein E (ApoE), education, body mass index (BMI), physical activity, and blood pressure, while many other potential factors can be ignored. Further, the existing models were mainly conducted through traditional statistical methods, such as cox or logistic regressions, so that the predicting power and solidness can be limited.
In the present study, we employed machine learning (ML) in a large prospective cohort UK Biobank (UKB), containing 502,414 participants with over ten years of follow-up5 to establish and validate a novel UKB dementia risk prediction model, named UKB-DRP. We intend to develop a generalisable model for all-cause dementia (ACD) and its dominant subtype Alzheimer's Disease (AD) with an aim to identify a population who had a higher risk of incidence of diseases in five, ten, and much longer years. We also compared the predictive performance of the UKB-DRP to the existing prediction models mentioned above to demonstrate the superiority of the proposed ML model that was developed based on a data-driven approach. Our study performed model development and internal validation based on a population that was mainly of White ethnicity and European ancestry; thus, further independent external validation is needed to confirm our findings.
Methods
Study participants
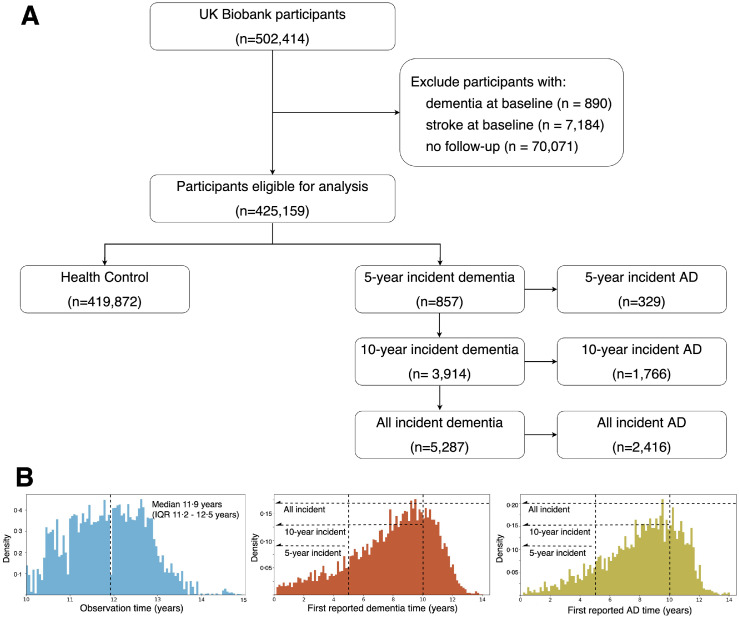
Our study adopted data from the UKB, a longitudinal cohort study with over 500,000 individuals aged 40–69 years at their baseline assessment between March 1, 2006 and October 31, 2010.5 The cohort enrolled the general population in 22 recruitment centres across the UK to undergo multiple assessments, including interviews and questionnaires covering their lifestyles and health conditions, physical measures, biological sample assays, imaging, and genotyping. As shown in Figure 1, all participants who completed the baseline assessment (n=502,414) were included except if they were: (1) with dementia at baseline (n=890), (2) with stroke at baseline (n=7184), and (3) without follow-up records (n=70,071), we finally included 425,159 non-demented participants who had at least ten years follow-up (median 11.9 years) until December 2020. The UK Biobank has research tissue bank approval from the North West Multi-centre Research Ethics Committee (https://www.ukbiobank.ac.uk/learn-more-about-uk-biobank/about-us/ethics) and provided oversight for this study. Written informed consent was obtained from all participants. Participation is voluntary, and participants are free to withdraw at any time without giving any reason.
Figure 1.
Flowchart of the participants selection.
(A) Individuals in the UK Biobank cohort were excluded if they had reported dementia at baseline, stroke at baseline, or without follow-up records. The remaining participants were classified based on their first reported years of dementia or AD after baseline.
(B) Left: distribution of observation times for the derived study population, whose median observation times were 11.9 years (IQR 11.2-12.5). Middle: distribution of first reported years of dementia after baseline. Right: distribution of first reported years of AD after baseline.
Abbreviations: AD = Alzheimer's Disease
Dementia outcomes
The primary outcome was dementia containing AD, vascular dementia, frontotemporal dementia, dementia with Lewy bodies, and dementia in other neurodegenerative or specified diseases. AD, the most common subtype of dementia, was investigated as the secondary outcome. To have an intensive survey of the incidence time, we distributed the patients into 5-year, 10-year, and all incident dementia or AD. The outcomes were ascertained and classified according to the ICD and Read codes (eTable 1, appendix p8), extracted from first occurrences data reported (field 131036-37, 130836-43), algorithmically defined (field 42018-25), death register data documented (field 40001-02), hospital inpatient data summarised (field 41270-71, 41280-81), and primary care data recorded (field 42040) dementia. Follow-up visits began from the date attending the assessment centre (field 53) to the earliest date of dementia diagnosis, death, or the last available date from the hospital or general practitioner, whichever occurred first. The diagnosis data were linked to UK electronic health records among which the dementia cases were mainly reported by professional clinicians in hospitals, family doctors in the primary care system, or staff in the death register system of the UK. However, the data of detailed diagnosis bases, e.g., lumbar puncture or imaging, which the doctors referred to was unavailable in the UKB dataset.
Candidate features
This study initially included all clinically relevant features collected at the participants’ baseline visits. A preliminary data screening step was conducted to exclude non-informative variables whose missing values were over 40% of all participants, and procedure metric variables (e.g., biological samples processing metrics, diagnosis codes, measure device IDs, etc.) that were clinical meaningless were manually cleaned by our clinicians, as well. Still, we adopted a relatively loose inclusion standard to avoid missing any potential associations, and please refer to eTable 2 (appendix pp9-10) for all candidate features for this study. Overall, a total of 366 features were adopted, including the participants’ demographic characteristics (n=3), touchscreen recorded lifestyle and health information (n=173), physical measures (n=66), cognitive function tests (n=23), and biological sample assays (n=62). Furthermore, we generated several variables not directly available from UKB (n=39), like polygenic risk score (PRS).
Predictors identification
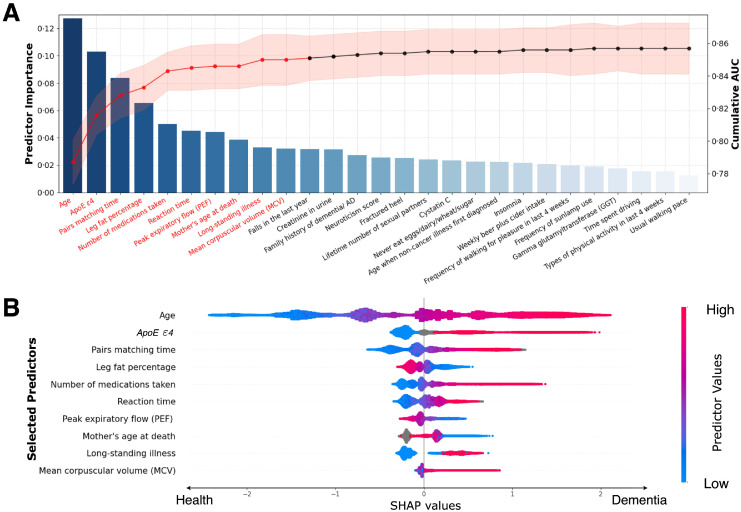
Predictors were determined mainly by two steps: candidate features ranking and sequential forward selection. In the first step, a naïve classifier called LightGBM6 was established, and each feature was ranked based on its contribution to model performance based on the information gain, which can be regarded as the predictor's ability to identify the future incidence of dementia. Based on the ranking, the top-50 features were initially chosen, and hierarchical clustering on the Spearman rank-order correlations was conducted to alleviate the multicollinearity issue (eMethods, appendix p3). Next, a sequential forward selection strategy was employed that the features within the pre-selected subset were re-ranked based on a newly developed classifier. Afterwards, consecutive classifiers were developed with sequentially added predictors based on the updated predictor importance ranking orders. The stopping point was arbitrarily determined by ourselves that we initially aimed to have an overall AUC of 0.85 (uncalibrated predicted probabilities), which seems as a prominent result. Further, no significant improvement in model performance can be observed when additional predictors come in (Figure 2A). Overall, we identified ten predictors for model development.
Figure 2.
Predictor selection and SHAP visualisation of modelling on all incident dementia population.
(A) Sequential forward selection from pre-selected candidate predictors. The bar chart represented the sorted predictor importance based on their contributions to model classifications. The line chart delineated cumulative AUCs (right axis) upon the inclusion of predictors one by each iteration. Top-10 predictors (colored in red) were finally selected for ML model building.
(B) SHAP visualisation plot of selected predictors. The width of the range of horizontal bars can be interpreted as the impact on the model prediction that the wider its range, the larger its impact. The color of the horizontal bars represented the magnitude of predictors, which was coded in a gradient from blue (low) to red (high), shown as the color bar on the right-hand side. The directions on the x-axis represented the likelihood of developing dementia (right) or being healthy (left). Readers can then infer the possibility of developing dementia given each predictor's specific value (coded in a gradient of colors).
Abbreviations: SHAP = SHapley Additive exPlanations, ApoE ε4= Apolipoprotein E (ApoE) ε4.
Model development
We implemented LightGBM6 to establish a dementia risk prediction model, named UKB-DRP, that performed the classification task of determining whether a participant falls into class 0 (predicted to remain dementia-free) or class 1 (predicted to incident dementia). The proposed model was developed based on healthy control (n=419,872) and all incident dementia (n=5287) participants from the UKB, incorporating ten identified predictors. The employed LightGBM algorithm works by starting from a weak base learner, usually a decision tree model, and sequentially training each new learner to correct the errors from the previously trained ones. In such a manner, the predictions can be added up to produce a strong overall final predictive model. The hyperparameter tuning was performed by an exhaustive selection from 1000 candidate sets of parameters and finally chosen the optimal set based on the performance measurement of the area under the Receiver Operating Characteristic (ROC) curve (AUC). Please refer to eTable 13 for detailed searching space and final adopted parameters (appendix, p21). Further, an isotonic regression7,8 served as a post-processor was performed to calibrate the raw predicted probabilities to actual risks (eMethods, appendix pp4-5). The ML algorithm was implemented with LightGBM library (v3.3.2) and scikit-learn library (v1.0.2)9 in Python (v3.9). Missing values were not imputed as the LightGBM algorithm can automatedly handle missingness in both model training and validation. In addition, we established a webpage application tool based on the Shiny package (v1.7.1) under R (v4.1.2). The source code, as well as the pre-trained models, are publicly available at https://github.com/jasonHKU0907/UKB_DRP.
The model was developed and validated using a five-fold cross-validation strategy that the validation set (one-fold of data) was kept untouched and merely used for evaluation purposes, while the hyperparameters tuning and post-calibration were performed within an inner-looped cross-validation within the training sets (four-fold of data). Please refer to appendix (eMethods, appendix p4) for detailed data partition and training process.
Statistical analysis
In the descriptive analysis of variables of interest, continuous variables were summarised by median and interquartile range, while discrete variables were summarised by frequency and percentage. Comparisons between groups (healthy control vs incident dementia/AD) were performed using the Chi-square tests for discrete variables and the Student's t-tests for continuous variables. Odds ratios were calculated using univariate analysis based on normalised data.
The model's performance was assessed in two accuracy indices, discrimination, and calibration. Discrimination was measured by the AUC, which varies between 0·5 for a non-informative model and 1 for a perfectly discriminating one. Calibration refers to the level of agreement between predicted probabilities and observed proportions of events, and it was assessed using the Hosmer-Lemeshow goodness-of-fit test10 with ten sub-groups and graphically depicted in calibration plots (eMethods, appendix pp4-5). We also reported accuracy, sensitivity, specificity, precision, and F1-score based on a cutoff defined by the achievement of the largest Youden index.11 In addition, we adopted SHapley Additive exPlanations (SHAP) plots to visualise the extent to which each predictor contributed to the target variable. All data analysis and visualisations were implemented under Python (v3.9) with packages of the scikit-learn library (v1.0.2) and Shap library (v0.40.0).12
Model deployment across dementia and AD at different incident times
To assess the generalisability of the UKB-DRP model, we deployed it to the other five target populations: 5-year/10-year incident dementia and 5-year/10-year/all incident AD. Both their AUCs and calibrations were calculated and plotted as performance measurements. Particularly, five post-processing calibrators (isotonic regressions) were performed on each of the target groups, and evaluation metrics were calculated based on those regressed outputs. Moreover, we repeated the procedures of predictors selection and model development in each of the target populations; hence, five individual ML models were established with their own optimised sets of predictors, which were used to compare with the one developed from all incident dementia.
Comparison with existing prediction scales
Existing dementia risk prediction models, including the CAIDE,2 DRS,3 and ANU-ADRI,4 were considered (please refer to eTable 5 for detailed descriptions, appendix p13). We deployed these models to the UKB and compared their performance to our model. Specifically, simple imputation (mean for continuous variables and mode for discrete variables) based on the rule of thumb was performed for the variables with missingness less than 5%, while multiple imputations for variables with missingness over 5%. We adopted Delong's test13 to assess the significance of AUCs between the UKB-DRP and the existing models; as for calibrations, we both plotted their raw predicted probabilities and regressed ones.
Role of the funding source
The funders were not involved in the study design, collection, analysis, and interpretation of data, nor did they have a role in the writing of the manuscript and decision to submit it for publication. All authors had full access to all the data in the study and accepted the responsibility to submit it for publication.
Results
Population characteristics
After filtering 78,145 participants with prevalent dementia, stroke, or missing follow-up data, we included 425,159 participants in the study (Figure 1). They were mainly comprised of white ethnicity (94.3%) with a mean age of 58 (IQR 51–64) years, and 54.4% (n = 231,187) were females and 45.6% (n=193,972) were males. During a median follow-up time of 11.9 (IQR 11.2–12.5) years, 5287 participants developed dementia, among whom 3914 were incidents within 10 years and 857 were incidents within 5 years, respectively. Specifically, for incident dementia participants, 94.9% were white ethnicity (p-value 0.053), mean age was 66 (IQR 62–68) years (p-value <0.001), and 47.4% were females (p-value <0.001). A total of 2416 patients were diagnosed with AD, among whom 1766 were incidents within 10 years and 329 were incidents within 5 years, respectively. For incident AD participants, 95.7% were white ethnicity (p-value 0.004), mean age was 66 (IQR 62–68) years (p-value<0.001), and 52.1% were females (p-value 0.024). The critical baseline predictors are presented by incident dementia and AD status in Table 1 and please refer to eTable 14 (appendix p22) for further detailed statistics on the causes of dementia.
Table 1.
The baseline characteristics of UKB participants included in the study by dementia and AD status.
| Participants Characteristics | Overall | Healthy control | Incident dementia |
Incident AD |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| (n=425,159) | (n=419,872) | (n=5287) | p-value | Odds ratio | p-value | (n=2416) | p-value | Odds ratio | p-value | |
| Age, y | 58 [51–64] | 58 [51–63] | 66 [62–68] | <0.001 | 4.57 [4.31–4.85] | <0.001 | 66 [63–68] | <0.001 | 5.03 [4.67–5.42] | <0.001 |
| Sex | ||||||||||
| female | 231,187 (54.4%) | 228,683 (54.5%) | 2504 (47.4%) | <0.001 | 0.75 [0.71–0.79] | <0.001 | 1258 (52.1%) | 0.024 | 0.91 [0.84–0.99] | 0.023 |
| male | 193,972 (45.6%) | 191,189 (45.5%) | 2783 (52.6%) | 1.33 [1.26–1.40] | <0.001 | 1158 (47.9%) | 1.10 [1.01–1.19] | 0.023 | ||
| Ethnicity (White) | 400,879 (94.3%) | 395,861 (94.3%) | 5018 (94.9%) | 0.053 | 1.13 [1.00–1.28] | 0.050 | 2317 (95.7%) | 0.004 | 1.42 [1.17–1.75] | <0.001 |
| Education, y | 11 [10–12] | 10 [11–12] | 10 [10–12] | <0.001 | 0.87 [0.86–0.89] | <0.001 | 10 [10–12] | <0.001 | 0.87 [0.86–0.89] | <0.001 |
| ApoE ε4 | ||||||||||
| Single-copy carriers | 95,618 (22.5%) | 93,684 (22.3%) | 1934 (36.6%) | <0.001 | 2.60 [2.44–2.77] | <0.001 | 1013 (41.9%) | <0.001 | 3.70 [3.37–4.07] | <0.001 |
| Double-copies carriers | 8647 (2.0%) | 8126 (1.9%) | 521 (9.9%) | 8.07 [7.31–8.90] | <0.001 | 327 (13.5%) | 13.58 [11.89–15.48] | <0.001 | ||
| Pairs matching time, s | 383 [305–497] | 382 [305–495] | 491 [376–689] | <0.001 | 1.33 [1.31–1.35] | <0.001 | 498 [381–701] | <0.001 | 1.30 [1.27–1.33] | <0.001 |
| Reaction time, ms | 547 [484–640] | 547 [484–640] | 609 [531–704] | <0.001 | 1.33 [1.31–1.36] | <0.001 | 609 [531–703] | <0.001 | 1.30 [1.27–1.34] | <0.001 |
| Long-standing illness | 141,290 (33.2%) | 138,497 (33.0%) | 2793 (52.8%) | <0.001 | 2.29 [2.17–2.42] | <0.001 | 1147 (47.5%) | <0.001 | 1.83 [1.69–1.98] | <0.001 |
| Number of medications taken | 2 [0–4] | 2 [0–4] | 4 [2–6] | <0.001 | 1.55 [1.52–1.55] | <0.001 | 3 [1–6] | <0.001 | 1.45 [1.41–1.49] | <0.001 |
| Leg fat percentage | 33.5 [22.5–41.3] | 33.5 [22.5–41.3] | 30.6 [22.0–41.2] | <0.001 | 0.94 [0.91–0.96] | <0.001 | 33.0 [22.2–41.6] | 0.498 | 0.99 [0.95–1.03] | 0.493 |
| Peak expiratory flow (PEF), L/min | 320 [225–423] | 321 [226–424] | 257 [154–360] | <0.001 | 0.66 [0.63–0.68] | <0.001 | 247 [143–351] | <0.001 | 0.62 [0.59–0.66] | <0.001 |
| Mother's age at death | 76 [66–84] | 76 [66–84] | 76 [67–84] | <0.001 | 0.94 [0.92–0.97] | <0.001 | 77 [67–84] | 0.007 | 0.95 [0.91–0.99] | 0.007 |
| Missing (still alive) | 166,362 (39.1%) | 165,656 (39.5%) | 706 (13.4%) | <0.001 | 0.24 [0.22–0.26] | <0.001 | 291 (12.0%) | <0.001 | 0.21 [0.19–0.24] | <0.001 |
| Mean corpuscular volume (MCV), fL | 91.2 [88.5–93.9] | 91.2 [88.5–93.9] | 91.8 [89.0–94.7] | <0.001 | 1.18 [1.15–1.21] | <0.001 | 91.9 [89.1–94.8] | <0.001 | 1.19 [1.14–1.24] | <0.001 |
Data are presented as median [Interquartile range] for continuous variables and number (%) for discrete variables. P-values were calculated based on the Student's t-tests for continuous variables, and Pearson's Chi-square tests for discrete variables. Odds ratios were calculated based on univariate analysis using normalised data.
Data-driven predictors selection
Among 366 candidate predictors, the initial selection procedure picked out 50 candidates, most of whom were previously discovered to be associated with dementia risk. Several predictors were highly correlated, e.g., leg fat percentage and leg fat mass, and hierarchical clustering was performed to eliminate the multicollinearity (eFigure 1A&1B, appendix p23). A set of 27 predictors was determined and sorted based on their importance to the prediction task as shown in the bar chart (Figure 2A). The sequential forward selection scheme can be demonstrated by the line chart that the model's performance (AUC on the right axis) climbed steeply when taking part in the first several predictors and gradually went to flat with gentle fluctuation when additional ones came in. Ultimately, we chose the top-10 variables as the final predictors for ML model development. Their summary statistics and unadjusted odds ratios are shown in Table 1.
We would like to elaborate on several selected predictors. The “Long-standing illness” was a self-reported variable, representing any chronic health conditions that lasted six months or longer, such as cancer, diabetes, chronic pain, heart disease, etc. Besides, the selection strategy included two time-related variables that represented participants’ cognitive functions, reaction time and pairs matching time, which were measured under two different tests of the Snap game and Pairs matching game, respectively. The Snap game was designed to test participants’ reaction time and simple processing speed. During the game, participants were shown two cards at a time on the touchscreen and instructed to press the button on the button box as quickly as possible when the symbols on the cards matched. Each pair was displayed for 2 seconds, followed by a one second gap. Reaction time (million seconds) was then recorded the button-press occurred during a gap against the previous pair. For the pairs matching game, participants were asked to memorise the position of as many matching pairs of cards as possible. The cards were then turned face down on the screen, and the participants were asked to touch as many pairs as possible in the fewest tries. The pairs matching time measured the time (seconds) consumed upon finishing the test. Please refer to eTable 4 (appendix p12) for detailed notations of all the selected predictors.
The repeated procedures were performed independently for the rest target populations, and the results are shown in eFigure 2-6 (appendix pp24-28). A summarised table of selected predictors was listed in eTable 3 (appendix p11). It can be found that age, ApoE ε4, and pairs matching time were consistently picked up in all 6 models, followed by the number of medications taken and mother's age at death in 5 models, and peak expiratory flow (PEF) in 4 models, indicating their strong predictive powers for both dementia and AD. Particularly, the reaction time extracted from the individual test and mean reaction time were crossover selected by three different models. They presented a high correlation to each other and similar contributions to the dementia prediction model. Similar scenarios were indicated in that of leg fat percentage and leg fat mass.
Model and interpretation and visualisation
To better visualise the different contributions of the predictors to the proposed ML model, we drew the SHAP plot (Figure 2B) that each participant exhibited as a data point and was coded with gradient colours representing the magnitude of the predictor. The plot can be interpreted in two aspects. On the one hand, the overall predictive power of each predictor can be visually measured by its horizontal range. Specifically, age seemed to have the widest range, indicating it had the most considerable prediction power and can significantly impact the model's output. ApoE ε4, pairs matching time, and the number of medications taken were also witnessed in relatively wide ranges, demonstrating their importance to the prediction task. On the other hand, the specific effect of each predictor can be interpreted by its value magnitude and tendency direction on the x-axis, which represents the extent of likelihood to develop dementia. Take the predictor age as an example, older participants (coloured in red) are more likely to develop dementia (right side) compared with younger ones (coloured in blue) who tend to keep healthy (left side). Similarly, for the rest predictors, participants carrying ApoE ε4, spending more time on pairs matching games, containing less leg fat, taking more medications, with longer reaction time, having lower PEF, losing mothers at a younger age, suffering long-standing illnesses, and showing the higher value of mean corpuscular volume (MCV), are prone to dementia. Their odds ratios were consistent with the explainable effects of SHAP values; please refer to eTable 11 & 12 (appendix pp19-20) for their odds ratios calculated under the univariate and multivariate analyses regarding dementia and AD at different timelines.
Model performance across different populations
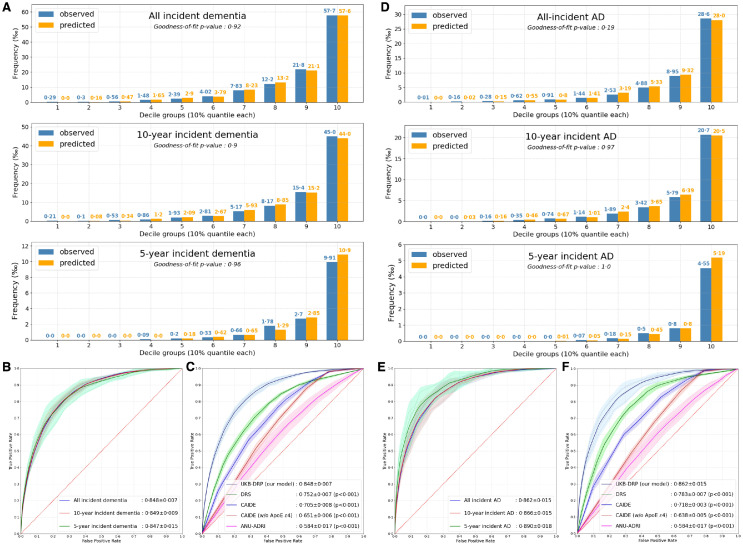
The discrimination ability of the UKB-DRP model was assessed using the AUC. According to Figure 3B, the model for all incident dementia achieved an AUC of 0.848 ± 0.007, and its deployment to the 10-year/5-year incident dementia also achieved comparable results of 0.849 ± 0.009 and 0.847 ± 0.015, respectively. All incident dementia model obtained an accuracy of 0.764 ± 0.013, sensitivity of 0.774 ± 0.024, specificity of 0.764 ± 0.013, precision of 0.040 ± 0.001, and F1-score of 0.075 ± 0.002. The deployment to the different AD population groups also observed good discrimination abilities that all/10-year/5-year/ incident AD each achieved AUCs of 0.862 ± 0.015, 0.866 ± 0.015 and 0.890 ± 0.018, respectively (Figure 3E). All incident AD model obtained an accuracy of 0.756 ± 0.013, sensitivity of 0.815 ± 0.037, specificity of 0.756 ± 0.013, precision of 0.019 ± 0.001, and F1-score of 0.037 ± 0.001. Specific metrics of 10-year/5-year dementia and AD are reported in eTable 6 (appendix p14).
Figure 3.
Performance of the UKB-DRP and existing prediction scales.
(A) Calibration plots of the UKB-DRP on dementia at different incident times.
(B) AUC plots of the UKB-DRP on dementia at different incident times.
(C) AUC plots of the UKB-DRP and existing prediction scales on all incident dementia.
(D) Calibration plots of the UKB-DRP on AD at different incident times.
(E) AUC plots of the UKB-DRP on AD at different incident times.
(F) AUC plots of the UKB-DRP and existing prediction scales on all incident AD.
The UKB-DRP model was developed based on all incident dementia population, and the results of 5-year/10-year incident dementia and all/5-year/10-year incident AD were obtained by the deployment of that model. Isotonic regressions were employed to adjust the raw predicted probabilities to the actual risks of different deployment populations, all calibration and AUC plots were drawn based on regressed probabilities, and 5-fold cross-validation strategy was conducted to calculate the results. Calibration plots of (A) and (D) was evaluated using Hosmer-Lemeshow goodness-of-fit tests on the decile groups of predicted probabilities and observed proportions of events, where a p-value greater than 0.05 indicated the goodness of fit. Delong's tests for AUCs comparison were conducted between the UKB-DRP and each of the existing prediction models on plots (C) and (F). A p-value less than 0.05 indicated the statistical significance of the difference. Notably, the CAIDE had two versions of scaling systems upon the inclusion of predictor ApoE ε4 or not, and we exhibited results derived from both versions.
Abbreviations: AUC = Area under the ROC Curve, AD = Alzheimer's Disease, w/o = without, ApoE ε4= Apolipoprotein E (ApoE) ε4, CAIDE = Cardiovascular Risk Factors, Aging, and Incidence of Dementia Risk Score, DRS = Dementia Risk Score, ANU-ADRI = Australian National University Alzheimer's Disease Risk Index, UKB-DRP = UK Biobank Dementia Risk Prediction model.
The calibration was assessed using Hosmer-Lemeshow goodness-of-fit test, where a p-value greater than 0.05 indicates sufficient goodness-of-fit. The calibration plot (Figure 3A) of all incident dementia was nicely fitted (p=0.92) that the predicted probabilities and observed actual proportions were closely matched within all decile groups. In addition, the deployment to the rest five population groups, 5-/10-year incident dementia (Figure 3A) and 5-year/10-year/all incident AD (Figure 3D), also witnessed satisfied calibrations that all had p-values greater than 0.05. Specifically, for 5-year incident dementia and AD, the predicted probabilities and observed proportions were all zeros in the first few decile groups, which were mainly resulted from insufficient target cases.
Participants with strokes at baseline were excluded from model development. This is because stroke is one of the top prevalent neurological disorders, which might influence the development and progression of dementia. Thus, we maintain a relatively healthier cerebral population to ensure the efficiency of longitudinal dementia prediction. To correct the selection bias, we performed an additional experiment that re-evaluated the UKB-DRP model after the inclusion of the participants with stroke at baseline. According to the results shown in eTable 8 (appendix, p16), no significant variations were observed, which further demonstrates the robustness of the proposed model.
Model comparison with existing prediction scales
A series of pairwise comparisons were conducted on all incident dementia and AD between the proposed UKB-DRP model and existing prediction models. Specific metrics of accuracy, sensitivity, specificity, precision, and F1-score were shown in eTable 7 (appendix p15). The DeLong's tests were performed by comparing our models to each of the existing prediction models, where all p-values less than 0.001 (p<0.05 indicates statistical significance) in both populations of all incident dementia (Figure 3C) and all incident AD (Figure 3F), demonstrating a remarkable superiority of our proposed ML model. Specifically, the CAIDE score had two versions defined by the inclusion of predictor ApoE ε4 or not, where the ApoE ε4 exhibited significant discrimination power.
Calibration plots were drawn and exhibited in eFigure 7-10 (appendix pp29-32). We naively plotted the raw predicted probabilities of each score against the observed proportions of events, and the proposed UKB-DRP model delineated a relatively good calibration (p=0.09) in all incident dementia group while all the existing models exhibited overall underestimations across all decile subgroups (eFigure 7, appendix p29). To have a fair comparison, we regressed raw probabilities from all existing models to our study cohort and plotted their calibrations (eMthods, eFigure 9-10, pp31-32). All calibrations then achieved satisfied p-values (>0.05) in both dementia and AD groups, indicating their good abilities in stratifications in general, and poor performance in raw probabilities may result from the variations in the prevalence of events within their derivation cohorts during model development (eTable 5, appendix p13).
Webpage deployment tool
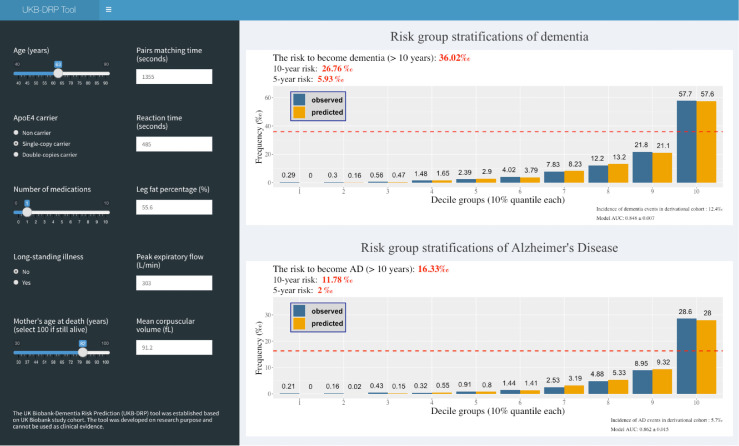
We implemented the UKB-DRP model into a web application (Figure 4) that provides risk predictions for individuals. Baseline characteristics can be inputted on the left panel and estimated risks of dementia and AD at different incident times are shown in the right panel marked in red colours. Two calibration plots are displayed to represent the stratified risk groups of all incident dementia and AD based on decile partitions. The horizontal dash line within the calibration plot gives an explicit level of risk measure. The source codes and the pre-trained weights to establish the webpage are publicly available at https://github.com/jasonHKU0907/UKB_DRP. The web application was made accessible online at https://jiayou0907.shinyapps.io/UKB-DRP-Tool/.
Figure 4.
A stylized representation of the UKB-DRP tool.
An example of a 63-year-old female participant who was enrolled in the UKB in 2008 is demonstrated on this webpage. The participant's baseline information is shown on the left panel, and UKB-DRP outputs that her risks of dementia and AD are 36.02‰ and 16.33‰, respectively. In fact, this example participant was diagnosed with Alzheimer's Disease in 2014 (6 years after) at hospital admission.
Discussion
In this study, we developed a dementia risk prediction model utilising the ML LightGBM algorithm based on large cohort data from UKB, which showed superior prognostic accuracy compared with previously published CAIDE, DRS, and ANU-ADRI. The model consisting of the ten important genetic and clinical predictors achieved AUCs of around 0.85 in predicting dementia incidence within five, ten, and much longer years. The model's performance was even better in AD predictions with all AUCs more than 0.86. In addition, the model was well-calibrated with predicted probabilities perfectly plotted against the observed proportions of events.
Compared with the models established based on variables obtained from elaborate neuropsychological tests, expensive whole genome sequencing (WGS), invasive lumbar puncture, or brain positron emission tomography (PET) imaging,14, 15, 16 our model is solely based on the easily accessible predictors which can be collected from quick questionnaires, physical measures, and simple blood tests. Therefore, this prediction model can be widely applied to medical institutions at different levels. Besides, we separately performed the feature selection and model development in six population groups, including all-time, 10-year, and 5-year incident dementia or AD. The models favoured different top features in these groups, but the dominant ones were similar, so we chose the model derived from all-time incident dementia and found remarkable generalisability into other groups. It can be concluded that our model is solid and consistent enough to predict dementia in multiple scenarios.
In our prediction model, age is the most critical factor with AUC surpassing 0.81 when combing with ApoE ε4. It is worth noting that the predicting power of PRS was highly correlated with but weaker than that of ApoE ε4. Brief cognitive tests variables, including pairs matching time and reaction time reflecting visual episodic memory and processing speed, respectively, added 1.4% to the cumulative discriminative accuracy of ROC curves, which is consistent with Calvin's study.17 The complicated long-standing illness and the number of medications taken were selected as the features predicting a higher risk of dementia, supported by the findings that comorbidity and polypharmacy were more prevalent in the dementia population18 and could predict the mortality of dementia patients.19 Except for the above familiar predictors, it is notable that many other novel factors also significantly contributed to our model. Leg fat percentage, a marker of regional body fat deposit, is first proved to be a protective factor for dementia incidence in this study. It has been proposed that leg fat percentage can reduce the risk of cardiovascular diseases20 and diabetes21 independent of BMI potentially by affecting adipose inflammation and lipid metabolism, so it is reasonable that leg fat percentage is inversely associated with dementia. Another protective factor identified in our model, PEF, is a widely adopted lung function parameter to assess and monitor airway obstruction.22 It has been extensively accepted that impaired lung function was associated with a greater risk of dementia,23,24 among which PEF was one of the strongest risk factors.25,26 The lung function contributes to the dementia process potentially through modulating the neurodegenerative pathology27 and brain structures.28,29 Furthermore, the mother's age at death is negatively related to the offspring's dementia onset, which may be explained by potential family history and psychosocial trauma.30 The last predictor MCV was observed to increase dementia risk probably due to the underlined reduction in erythrocyte lifespan resulting from increased oxidative stress and adenylate metabolism.31
This study has strengths. Our proposed UKB-DRP model was established based on a large prospective cohort containing more than 500,000 participants with at least 10 years of follow-up clinical records. Predictor determination underwent exhausted and cautious selection in a data-driven manner from a comprehensive clinical feature space, and only ten predictors were finally picked out. The UKB-DRP model was established using the LightGBM algorithm, one of the most powerful ML techniques among the other methods. It works in an ensemble strategy and perfectly fits huge datasets with large sample sizes and high feature spaces. In addition, the LightGBM algorithm can split the missingness into the optimised node; that is, it can automate handle the missingness in both training and validation, eliminating the potential bias resulting from inaccurate imputations.
Several limitations need to be considered when interpreting our findings. Firstly, the incidence of dementia especially 5-year cases was lower than that in other reported cohorts because the participants from UKB are younger, healthier, and better-educated. To ascertain potential dementia patients, we already included records from hospital inpatient data, death register data, and primary care data as suggested in the previous study.32 Secondly, our model did not identify the variables like BMI and blood pressure which were common in other models as candidate predictors. The possible explanation may be the tree-based LightGBM algorithm relies heavily on the first a few split nodes (predictors), and resting predictors were chosen on the shoulder of their pioneers; as such, those commonly used candidate predictors might not able to donate sufficient contributions, while some other more sensitive factors like leg fat percentage and long-standing illness can compensate for it. Thirdly, although we have performed hierarchical clustering to handle multicollinearity issues during predictor selection, there are still weak correlations exist within the selected predictor set (eFigure 11, appendix p33), e.g., elder individuals might suffer more comorbidities and worse cognitive functions. Thus, the potential confounding issue needs to be aware of as it might lead to inaccurate interpretations of risk factors to the prediction outcomes. Fourth, the machine learning algorithm has its own limitations as well. Although a preliminary data screening procedure was performed to include candidate predictors that are health-related and clinically meaningful (eTable 2, appendix pp9-10), the predictor selection process was merely data-driven that emphasized achieving higher performance metrics but paid less attention to the empirical claims of fundamental mechanisms, which might cause potential bias in real clinical settings.33 Besides, we tried to transparentise the employed ML model by visually interpreting with SHAP plots; however, it can still not be fully explained with the exact extent to which it can affect the model and impact the prediction outcomes, leading to misused applications under certain circumstances.34,35 Last, the model we established was based on data from UK-Biobank where the anticipated individuals were mainly of White ethnicity and European ancestry, and the proposed model has not been adequately validated in other cohorts. To somehow undermine the inadequacy of validations, two additional experiments were performed: 1) A leave-one-centre-out cross-validation by partitioning the study cohort based on the locations of 22 recruitment centres around the UK, and each time we developed a model based on the population from 21 centres and evaluated it using the rest one (eTable 10, appendix p18). 2) A subgroup validation by merely deploying the UKB-DRP model to individuals with non-British ancestries, which accounts for 11.5% (n = 49,052) of the whole population (eTable 9, appendix p17). The results of the additional two validations were consistent with our current results. Still, the validations were performed internally. Thus, our findings in populations with different backgrounds remain unclear. We believe it is worthy of performing further validation using independent external datasets as future work.
In conclusion, we identified regular and novel predictors and then translated them into a promising tool for the rapid delivery of dementia screening. In future studies, it is worthy of testing the model comprising genetic and clinical factors in real-world clinical practise for further promotion and application. What's more, studies are needed to investigate whether targeting these factors can reduce the possibility of developing dementia, which may enable precise prevention and intervention in the early stage.
Contributors
WC, YJT, and JFF conceived, designed, and supervised the project. JY implemented models’ development, validation, and statistical analysis. YRZ, HFW and MY supported the analysis and contributed to the discussion of the results. JY and YRZ drafted the manuscript, and accessed and verified the underlying data reported in the manuscript. All authors had full access to all the data in the study and accept the responsibility to submit it for publication.
Data sharing statement
All data used in this study were accessed from the publicly available UK Biobank Resource under application number 19542. These data cannot be shared with other investigators.
Declaration of interests
The authors declare no conflict of interest related to this work.
Acknowledgements
This study utilised the UK Biobank Resource under application number 19542. We would like to thank all the participants and researchers from the UK Biobank. We also thank Lingyun Chen and Tianjun Ma (who were employees of Eisai China at the time of study completion) for assisting in study design and future clinical practise. This study was funded by grants from the Science and Technology Innovation 2030 Major Projects (2022ZD0211600), National Key R&D Program of China (2018YFC1312904, 2019YFA070950), National Natural Science Foundation of China (282071201, 81971032, 82071997), Shanghai Municipal Science and Technology Major Project (2018SHZDZX01), Research Start-up Fund of Huashan Hospital (2022QD002), Excellence 2025 Talent Cultivation Program at Fudan University (3030277001), Shanghai Rising-Star Program (21QA1408700), Medical Engineering Fund of Fudan University (yg2021-013), and the 111 Project (No. B18015). Further, we would like to thank the support from the Shanghai Center for Brain Science and Brain-Inspired Technology, ZHANGJIANG LAB, Tianqiao and Chrissy Chen Institute, and the State Key Laboratory of Neurobiology and Frontiers Center for Brain Science of Ministry of Education, Fudan University.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.eclinm.2022.101665.
Contributor Information
Jian-Feng Feng, Email: jianfeng64@gmail.com.
Jin-Tai Yu, Email: Jintai_yu@fudan.edu.cn.
Wei Cheng, Email: wcheng.fdu@gmail.com.
Appendix. Supplementary materials
References
- 1.Gauthier S, Rosa-Neto P, Morais JA, Webster C. London, England: Alzheimer's Disease International; 2021. World Alzheimer report 2021: journey through the diagnosis of dementia. [Google Scholar]
- 2.Kivipelto M, Ngandu T, Laatikainen T, Winblad B, Soininen H, Tuomilehto J. Risk score for the prediction of dementia risk in 20 years among middle aged people: a longitudinal, population-based study. Lancet Neurol. 2006;5(9):735–741. doi: 10.1016/S1474-4422(06)70537-3. [DOI] [PubMed] [Google Scholar]
- 3.Walters K, Hardoon S, Petersen I, et al. Predicting dementia risk in primary care: development and validation of the Dementia Risk Score using routinely collected data. BMC Med. 2016;14:6. doi: 10.1186/s12916-016-0549-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anstey KJ, Cherbuin N, Herath PM. Development of a new method for assessing global risk of Alzheimer's disease for use in population health approaches to prevention. Prev Sci. 2013;14(4):411–421. doi: 10.1007/s11121-012-0313-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sudlow C, Gallacher J, Allen N, et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12(3) doi: 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ke G, Meng Q, Finley T, et al. Advances in neural information processing systems (NIPS 2017) Vol. 30. 2017. LightGBM: a highly efficient gradient boosting decision tree; pp. 3149–3157. [Google Scholar]
- 7.Chakravarti N. Isotonic median regression: a linear programming approach. Math Oper Res. 1989;14(2):303–308. [Google Scholar]
- 8.de Leeuw J. Correctness of Kruskal's algorithms for monotone regression with ties. Psychometrika. 1977;42:141–144. [Google Scholar]
- 9.Pedregosa F, Varoquaux G, Gramfort A, et al. Scikit-learn: machine learning in Python. J Mach Learn Res. 2011;12:2825–2830. [Google Scholar]
- 10.Hosmer DW, Jr., Lemeshow S, Sturdivant RX. 3rd ed. Wiley; Hoboken, NJ: 2013. Applied Logistic Regression. [Google Scholar]
- 11.Youden WJ. Index for rating diagnostic tests. Cancer. 1950;3(1):32–35. doi: 10.1002/1097-0142(1950)3:1<32::aid-cncr2820030106>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 12.Lundberg S, Lee SI. A unified approach to interpreting model predictions. Adv Neural Inf Process Syst. 2017;30:4765–4774. [Google Scholar]
- 13.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44(3):837–845. [PubMed] [Google Scholar]
- 14.Ota K, Oishi N, Ito K, Fukuyama H, Group S-JS. Alzheimer's disease neuroimaging I. Effects of imaging modalities, brain atlases and feature selection on prediction of Alzheimer's disease. J Neurosci Methods. 2015;256:168–183. doi: 10.1016/j.jneumeth.2015.08.020. [DOI] [PubMed] [Google Scholar]
- 15.Prestia A, Caroli A, van der Flier WM, et al. Prediction of dementia in MCI patients based on core diagnostic markers for Alzheimer disease. Neurology. 2013;80(11):1048–1056. doi: 10.1212/WNL.0b013e3182872830. [DOI] [PubMed] [Google Scholar]
- 16.Prestia A, Caroli A, Wade SK, et al. Prediction of AD dementia by biomarkers following the NIA-AA and IWG diagnostic criteria in MCI patients from three European memory clinics. Alzheimers Dement. 2015;11(10):1191–1201. doi: 10.1016/j.jalz.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 17.Calvin CM, Wilkinson T, Starr JM, et al. Predicting incident dementia 3-8 years after brief cognitive tests in the UK Biobank prospective study of 500,000 people. Alzheimers Dement. 2019;15(12):1546–1557. doi: 10.1016/j.jalz.2019.07.014. [DOI] [PubMed] [Google Scholar]
- 18.Clague F, Mercer SW, McLean G, Reynish E, Guthrie B. Comorbidity and polypharmacy in people with dementia: insights from a large, population-based cross-sectional analysis of primary care data. Age Ageing. 2017;46(1):33–39. doi: 10.1093/ageing/afw176. [DOI] [PubMed] [Google Scholar]
- 19.van de Vorst IE, Goluke NMS, Vaartjes I, Bots ML, Koek HL. A prediction model for one- and three-year mortality in dementia: results from a nationwide hospital-based cohort of 50,993 patients in the Netherlands. Age Ageing. 2020;49(3):361–367. doi: 10.1093/ageing/afaa007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen GC, Arthur R, Iyengar NM, et al. Association between regional body fat and cardiovascular disease risk among postmenopausal women with normal body mass index. Eur Heart J. 2019;40(34):2849–2855. doi: 10.1093/eurheartj/ehz391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miljkovic-Gacic I, Wang X, Kammerer CM, et al. Sex and genetic effects on upper and lower body fat and associations with diabetes in multigenerational families of African heritage. Metabolism. 2008;57(6):819–823. doi: 10.1016/j.metabol.2008.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tantucci C, Duguet A, Giampiccolo P, Similowski T, Zelter M, Derenne JP. The best peak expiratory flow is flow-limited and effort-independent in normal subjects. Am J Respir Crit Care Med. 2002;165(9):1304–1308. doi: 10.1164/rccm.2012008. [DOI] [PubMed] [Google Scholar]
- 23.Lutsey PL, Chen N, Mirabelli MC, et al. Impaired lung function, lung disease, and risk of incident dementia. Am J Respir Crit Care Med. 2019;199(11):1385–1396. doi: 10.1164/rccm.201807-1220OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Russ TC, Kivimaki M, Batty GD. Respiratory disease and lower pulmonary function as risk factors for dementia: a systematic review with meta-analysis. Chest. 2020;157(6):1538–1558. doi: 10.1016/j.chest.2019.12.012. [DOI] [PubMed] [Google Scholar]
- 25.Guo X, Waern M, Sjogren K, et al. Midlife respiratory function and Incidence of Alzheimer's disease: a 29-year longitudinal study in women. Neurobiol Aging. 2007;28(3):343–350. doi: 10.1016/j.neurobiolaging.2006.01.008. [DOI] [PubMed] [Google Scholar]
- 26.Simons LA, Simons J, McCallum J, Friedlander Y. Lifestyle factors and risk of dementia: Dubbo study of the elderly. Med J Aust. 2006;184(2):68–70. doi: 10.5694/j.1326-5377.2006.tb00120.x. [DOI] [PubMed] [Google Scholar]
- 27.Wang J, Dove A, Song R, et al. Poor pulmonary function is associated with mild cognitive impairment, its progression to dementia, and brain pathologies: a community-based cohort study. Alzheimers Dement. 2022:1–9. doi: 10.1002/alz.12625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taki Y, Kinomura S, Ebihara S, et al. Correlation between pulmonary function and brain volume in healthy elderly subjects. Neuroradiology. 2013;55(6):689–695. doi: 10.1007/s00234-013-1157-6. [DOI] [PubMed] [Google Scholar]
- 29.Yin M, Wang H, Hu X, Li X, Fei G, Yu Y. Patterns of brain structural alteration in COPD with different levels of pulmonary function impairment and its association with cognitive deficits. BMC Pulm Med. 2019;19(1):203. doi: 10.1186/s12890-019-0955-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Conde-Sala JL, Garre-Olmo J. Early parental death and psychosocial risk factors for dementia: a case-control study in Europe. Int J Geriatr Psychiatry. 2020;35(9):1051–1059. doi: 10.1002/gps.5328. [DOI] [PubMed] [Google Scholar]
- 31.Kosenko EA, Aliev G, Tikhonova LA, Li Y, Poghosyan AC, Kaminsky YG. Antioxidant status and energy state of erythrocytes in Alzheimer dementia: probing for markers. CNS Neurol Disord Drug Targets. 2012;11(7):926–932. doi: 10.2174/1871527311201070926. [DOI] [PubMed] [Google Scholar]
- 32.Wilkinson T, Schnier C, Bush K, et al. Identifying dementia outcomes in UK Biobank: a validation study of primary care, hospital admissions and mortality data. Eur J Epidemiol. 2019;34(6):557–565. doi: 10.1007/s10654-019-00499-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCradden MD, Anderson JA, Stephenson E, et al. A research ethics framework for the clinical translation of healthcare machine learning. Am J Bioethics. 2022;22(5):8–22. doi: 10.1080/15265161.2021.2013977. A. [DOI] [PubMed] [Google Scholar]
- 34.Ngiam KY, Khor W. Big data and machine learning algorithms for health-care delivery. Lancet Oncol. 2019;20(5):e262–e273. doi: 10.1016/S1470-2045(19)30149-4. [DOI] [PubMed] [Google Scholar]
- 35.Weissler EH, Naumann T, Andersson T, et al. The role of machine learning in clinical research: transforming the future of evidence generation. Trials. 2021;22(1):1–15. doi: 10.1186/s13063-021-05489-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.