Abstract
We speculate ruptured circulating tumour cells (CTC) in capillaries could release a large number of small extracellular vesicle-like vesicles, namely mechanically extruded sEV (sEVme), which can encapsulate chromosomal DNA fragments. These sEVme have similar physicochemical properties compared to small extracellular vesicles spontaneously secreted by living cells (sEVss), and thus sEVme and sEVss cannot be effectively distinguished based on their size or membrane protein markers. Meanwhile, these sEVme derived from CTC inherit oncogenic payloads, deliver cargo through the bloodstream to recipient cells, and thus may promote cancer metastasis. The validation of this speculation could facilitate our understanding of EV biogenesis and cancer pathology. The potential finding will also provide a theoretical foundation for burgeoning liquid biopsy using DNA fragments derived from harvested sEV.
Subject terms: Metastasis, Translational research
Extracellular vesicles (EV) are lipid bilayer membrane-enclosed vesicles that can mediate intercellular communication through transferring parental cell-derived proteins and nucleic acids [1]. Although the biogenesis of EV has not yet been fully understood [2], based on their mechanism of release and size EV are generally divided into three classes: exosomes (30–120 nm), microvesicles (50–1000 nm), and apoptotic bodies (500–2000 nm). According to Minimal Information for Studies of Extracellular Vesicles guidelines (MISEV2018), the term small EV (sEV) is used to describe EV that are less than 200 nm, while the term large EV as the ones that are larger than 200 nm [3]. It is noteworthy that sEV can efficiently evade phagocytosis dominated by tissue-resident macrophages and blood monocytes [4], which allows sEV to mediate long-distance intercellular communication. In comparison, large EV are more likely to encounter macrophages, patrolling monocytes, and mature dendritic cells [5], which can quickly scavenge large EV. Therefore, in recent years sEV have gained greater attention than large EV. Numerous studies demonstrated that sEV hold promise for pathophysiologic and translational discoveries. For instance, in tumour growing evidence reveals that sEV can modulate immune system, promote cancer metastasis, contribute to therapeutic resistance, enable liquid biopsy for cancer diagnostics, deliver anticancer drugs as natural nanocarriers, and treat tumours as therapeutic agents [6, 7]. In brief, sEV can efficiently evade immune clearance, successfully deliver cargo, and alter the phenotype of recipient cells. With this in mind, we used the term sEV in this perspective.
In general, proteins and RNA are described as the main cargo of sEV. It is noted that previous studies identified mtDNA and ssDNA in sEV [8, 9]. Recently, the existence of genomic dsDNA fragments and circular extrachromosomal DNA in sEV were confirmed [10–15]. We and others further found sEV-derived dsDNA represents the entire genome, reflects the mutational status, and shows identical copy number variation of parental cells [16–19]. When the evidence is taken together, one question arises as to how tumour DNA is packaged in sEV. The classic theory states exosomes and microvesicles are derived from the endolysosomal pathway and outward budding of cellular membranes, respectively. Meanwhile, a variety of RNA and proteins in the cytoplasm can be sorted and encapsulated into exosomes and microvesicles. Their forming processes are not believed to be involved with nuclear, mitochondria, or DNA fragments [20]. Although the majority of DNA fragments actually attach to the outer surface of sEV [14, 21], few studies demonstrated DNA fragments can indeed be sorted to sEV and partially disclosed the potential mechanisms [12–15]. Therefore, the fact of DNA fragments encapsulated in sEV exists but is not well understood [22–26]. Nevertheless, the presence of DNA fragments in sEV has generated interest in biogenesis, cargo selection, and the physiological significance of sEV. For example, horizontal DNA transfer to recipient cells could be achieved by sEV [25], which may inspire us to investigate and elucidate the biological roles of sEV in cancer development, metastasis, and genome evolution.
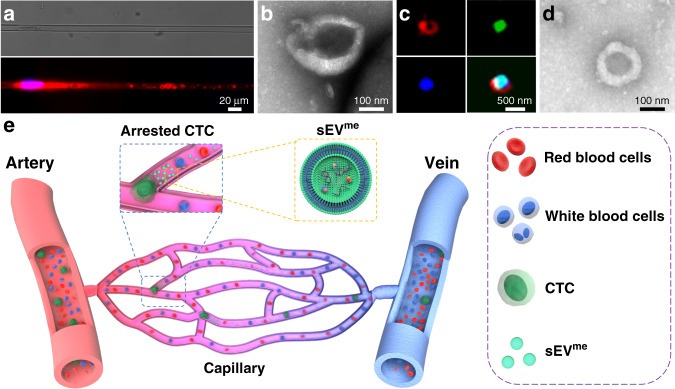
In addition to the disclosed or proposed mechanisms, based on our previous findings, we speculate that genomic DNA fragments loaded vesicles could be derived from circulating tumour cells (CTC) that are extruded in capillaries. Tumour cells that detach from the primary tumour and shed into vasculature are referred to as CTC [27]. It is believed that CTC can circulate through peripheral blood, infiltrate distant organs, and survives as disseminated seeds for eventual relapse. Due to relatively large size (12–25 µm) and stiffness, over 90% of CTC are easily arrested in the capillaries (diameter ~5 µm, length ~1 mm) of lung, liver, myocardium, and skeletal muscle during metastasis [28, 29]. Subsequently, the differential capillary blood pressure between the free ends of the arrested CTC and the external tissue pressures can rupture the membrane and traumatise arrested CTC in seconds [30]. When we investigated the mechanism of tumour cell translocation through microchannels with 4–8 µm width mimicking capillaries, we observed that under equivalent capillary blood pressure (2–3 kPa) tumour cells release EV-like vesicles (Fig. 1a). These crude EV-like vesicles have a wide range of size, ranging from tens of nanometres to micrometres. Subsequently, following the recommended protocol for sEV isolation, we harvested sEV-like vesicles, namely mechanically extruded sEV (sEVme, Fig. 1a–c). We further found mechanical extrusion of ~1 × 107 cancer cells can generate ~9 × 1010 sEVme in a few minutes [31]. The count of sEVme is much higher than that of sEV spontaneously secreted by living cells (sEVss) on the same time scale. Moreover, these sEVme contain canonical sEVss protein markers, DNA fragments, and a variety of RNA. Under electron microscopy, there is no significant difference in morphology between sEVme and sEVss (Fig. 1d). Both displayed a typical saucer-shaped morphology. Briefly, sEVme and sEVss have very similar physicochemical properties, and thus it is difficult to distinguish between the two. These findings imply that CTC under physical destruction may explosively release a large amount of sEVme into peripheral blood (Fig. 1e). On the other hand, DNA fragments could be wrapped into sEVme when CTC are squeezing through the capillary constriction. The latest studies demonstrated nuclear deformation damages the integrity of the nuclear envelope and chromosomal DNA, leading to DNA fragments may flow into cytosol [32, 33]. Afterwards, the DNA fragments can be encapsulated into sEVme (ruptured CTC) or sEVss (survived CTC) and further released to extracellular space. Taken together, we speculate that arrested CTC in capillaries can generate a mass of DNA-loaded sEVme under mechanical pressure, which could interpret why DNA fragments can be identified in these small vesicles. In the clinical setting, these CTC-derived sEVme which contain chromosomal DNA fragments may partially contribute to liquid biopsy.
Fig. 1. Proposed sEVme generation by CTC.
a Release of sEVme by arrested MDA-MB-231 breast cancer cells in microchannels mimicking capillaries (microchannel width: 10 µm; length: 600 µm; pressure: 15 mmHg). b Transmission Electron Microscopy (TEM) image of sEVme produced by arrested MDA-MB-231 cells in microchannels (microchannel width: 10 µm; length: 600 µm; pressure: 15 mmHg), which exhibits characteristic saucer-shaped morphology. c Structured Illumination Microscopy (SEM) image of sEVme produced by arrested GFP MDA-MB-231 cells in microchannels. Fluorescence: red: DID, membrane label; green: GFP, cytoplasm label; blue: Hoechst 33342, DNA label. d TEM image of sEVss harvested from cell culture supernatant of MDA-MB-231 cells, which also exhibits characteristic saucer-shaped morphology. e Illustration of releasing sEVme by arrested CTC in capillaries.
The mechanisms of sEVme generation in capillaries may involve exocytosis, shedding, and others, which depend on the external pressure exerted on the arrested CTCs. A mild-to-moderate pressure difference between the free ends of the arrested CTC may compel the plasma membrane at distal end to bulge outward and release sEVme (Fig. 1a). In brief, externally mechanical stress can influence water and ion permeation of cells [34]. The ripple effect increases membrane tension and leads to instabilities of cell shape. When the arrested CTC cannot endure external stress and fail to regulate the intracellular pressure/volume, the high membrane tension stimulates exocytosis and release sEVme, which acts to decrease membrane tension [35]. Moreover, under moderate to high-pressure sEVme can be formed as a result of the severe disruption of the cell membrane and reorganisation of lipid bilayer-forming vesicles through lipid self-assembly in seconds [36]. It is noteworthy that these two mechanisms may co-exist in sEVme generation. Nevertheless, the generation mechanisms of sEVme are not fully understood yet, and relevant research remains scarce. In this perspective, we just provide a humble remark, the exact mechanisms could be further explored in future studies.
Physical destruction of CTC in the capillary bed may causes CTC death, and thus reduces the risk of metastatic seeding of CTC. However, it is two-edged. The physically destructed CTC might still play important biological roles in metastasis. Recent studies demonstrated that sEVme derived from stem cells have similar biological functions in comparison with that of sEVss [37, 38]. Accordingly, sEVme derived from stem cells have been tested in regenerative medicine [39], such as skin rejuvenation and bone repair [40–47]. We also found the sEVme derived from highly malignant breast adenocarcinoma cell line MDA-MB-231 cells can significantly promote migration of low malignant breast adenocarcinoma cell line MCF-7 cells in a wound-healing assay, demonstrating sEVme own similar biological functions of sEVss [48]. In our recent study [49], we further compared cargo of sEVme and sEVss derived from nine cancerous cell lines using high-throughput sequencing. Protein sequencing data reveals the similarity of membrane proteins between the two groups was ~71%, while it was ~21% when pertaining to total protein cargo. Moreover, analysis of the top 1000 small RNA with RNA sequencing showed a ~65% similarity between the two groups. Our findings indicate that sEVme have biological functions in regulating cellular activities to a certain extent. The first-hand evidence supports that sEVme prepared by mechanical extrusion of stem cells can promote tissue regeneration and can be good substitutes for sEVss. On the other hand, our findings intensely imply that sEVme containing oncogenic payloads inherited from CTC can easily and extensively disseminate through the circulation system. The released sEVme have similar biofunctions in comparison with sEVss, and thus may imperceptibly influence recipient cell activities, precondition premetastatic niche, inhibit antitumor immunity, and assist CTC escaping from sequestration from capillaries to seed and propagate.
To validate our speculation, peers can generate sEVme with capillary-mimetic microchannels in vitro, measure sEVme count and morphology, and comprehensively compare cargo between sEVme and sEVss. Afterwards, engineered tumour cells bearing membrane fluorophore can be injected into mesenteric capillary bed for observation of sEVme formation in vivo with super-resolution microscopy [50–52]. The major tissue distribution of sEVme can also be tracked. To investigate biological functions of sEVme, less metastatic or normal cells can be educated with sEVme derived from highly metastatic cancer cells with a variety of assays in vitro. In the animal study, prepared sEVme can regularly be given by intravenous infusion to treat mice followed by intravenous inoculation of aggressive cancer cells. The perfusion and extravasation of inoculated cancer cells can be quantified. The potential tumour colonisation and metastasis in major organs of mice can also be enumerated [53]. Meanwhile, peers can investigate whether the number of colonisation is associated with the concentration gradient of sEVme.
On all accounts, the biogenesis of sEV in vivo might be much more complex than our current recognition, and thus it deserves further investigation. In tumour-related studies, the full understanding of sEV biogenesis would favour the understanding of metastasis and facilitate clinical therapeutics, although it might be difficult due to their nanoscale size, inadequate tumour-specific protein biomarkers [54], and lack of visualised methodologies [55].
Author contributions
YW and YX wrote the manuscript; YW and SZ edited the manuscript; all authors revised the manuscript critically; all authors approved the version to be published.
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
Not applicable.
Consent to publish
Not applicable.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Yuan Wan, Yi-Qiu Xia.
Contributor Information
Yuan Wan, Email: ywan@binghamton.edu.
Si-Yang Zheng, Email: siyangz@andrew.cmu.edu.
References
- 1.Andaloussi SE, Mäger I, Breakefield XO, Wood MJ. Extracellular vesicles: biology and emerging therapeutic opportunities. Nat Rev Drug Discov. 2013;12:347–57. doi: 10.1038/nrd3978. [DOI] [PubMed] [Google Scholar]
- 2.Théry C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol. 2002;2:569–79. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- 3.Thery C, Witwer KW, Aikawa E, Alcaraz MJ, Anderson JD, Andriantsitohaina R, et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles. 2018;7:1535750. doi: 10.1080/20013078.2018.1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Imai T, Takahashi Y, Nishikawa M, Kato K, Morishita M, Yamashita T, et al. Macrophage-dependent clearance of systemically administered B16BL6-derived exosomes from the blood circulation in mice. J Extracell Vesicles. 2015;4:26238. doi: 10.3402/jev.v4.26238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Czernek L, Chworos A, Duechler M. The uptake of extracellular vesicles is affected by the differentiation status of myeloid cells. Scand J Immunol. 2015;82:506–14. doi: 10.1111/sji.12371. [DOI] [PubMed] [Google Scholar]
- 6.Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat cell Biol. 2007;9:654–9. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 7.Abhange K, Makler A, Wen Y, Ramnauth N, Mao W, Asghar W, et al. Small extracellular vesicles in cancer. Bioact Mater. 2021;6:3705–43. doi: 10.1016/j.bioactmat.2021.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guescini M, Genedani S, Stocchi V, Agnati LF. Astrocytes and glioblastoma cells release exosomes carrying mtDNA. J Neural Transm. 2010;117:1–4. doi: 10.1007/s00702-009-0288-8. [DOI] [PubMed] [Google Scholar]
- 9.Balaj L, Lessard R, Dai L, Cho YJ, Pomeroy SL, Breakefield XO, et al. Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nat Commun. 2011;2:180. doi: 10.1038/ncomms1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Williams C, Rodriguez-Barrueco R, Silva JM, Zhang WJ, Hearn S, Elemento O, et al. Double-stranded DNA in exosomes: a novel biomarker in cancer detection. Cell Res. 2014;24:766–9. doi: 10.1038/cr.2014.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kahlert C, Melo SA, Protopopov A, Tang JB, Seth S, Koch M, et al. Identification of double-stranded genomic DNA spanning all chromosomes with mutated KRAS and p53 DNA in the serum exosomes of patients with pancreatic cancer. J Biol Chem. 2014;289:3869–75. doi: 10.1074/jbc.C113.532267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takahashi A, Okada R, Nagao K, Kawamata Y, Hanyu A, Yoshimoto S, et al. Exosomes maintain cellular homeostasis by excreting harmful DNA from cells. Nat Commun. 2017;8:15287. doi: 10.1038/ncomms15287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yokoi A, Villar-Prados A, Oliphint PA, Zhang J, Song X, De Hoff P, et al. Mechanisms of nuclear content loading to exosomes. Sci Adv. 2019;5:eaax8849. doi: 10.1126/sciadv.aax8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fischer S, Cornils K, Speiseder T, Badbaran A, Reimer R, Indenbirken D, et al. Indication of horizontal DNA gene transfer by extracellular vesicles. PLoS ONE. 2016;11:e0163665. doi: 10.1371/journal.pone.0163665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu S, Turner KM, Nguyen N, Raviram R, Erb M, Santini J, et al. Circular ecDNA promotes accessible chromatin and high oncogene expression. Nature. 2019;575:699–703. doi: 10.1038/s41586-019-1763-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Waldenström A, Gennebäck N, Hellman U, Ronquist G. Cardiomyocyte microvesicles contain DNA/RNA and convey biological messages to target cells. PLoS ONE. 2012;7:e34653. doi: 10.1371/journal.pone.0034653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kahlert C, Melo SA, Protopopov A, Tang J, Seth S, Koch M, et al. Identification of double stranded genomic DNA spanning all chromosomes with mutated KRAS and p53 DNA in the serum exosomes of patients with pancreatic cancer. J Biol Chem. 2014;289:3869–75. doi: 10.1074/jbc.C113.532267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thakur BK, Zhang H, Becker A, Matei I, Huang Y, Costa-Silva B, et al. Double-stranded DNA in exosomes: a novel biomarker in cancer detection. Cell Res. 2014;24:766–9. doi: 10.1038/cr.2014.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wan Y, Liu B, Lei H, Zhang B, Wang Y, Huang H, et al. Nanoscale extracellular vesicle-derived DNA is superior to circulating cell free DNA for mutation detection in early-stage non-small cell lung cancer. Ann Oncol. 2018;29:2379–83. doi: 10.1093/annonc/mdy458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kowal J, Tkach M, Théry C. Biogenesis and secretion of exosomes. Curr Opin Cell Biol. 2014;29:116–25. doi: 10.1016/j.ceb.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 21.Shelke GV, Jang SC, Yin Y, Lässer C, Lötvall J. Human mast cells release extracellular vesicle-associated. DNA Matters. 2016;2:e201602000034. [Google Scholar]
- 22.Jeppesen DK, Fenix AM, Franklin JL, Higginbotham JN, Zhang Q, Zimmerman LJ, et al. Reassessment of exosome composition. Cell. 2019;177:428–45. doi: 10.1016/j.cell.2019.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malkin EZ, Bratman SV. Bioactive DNA from extracellular vesicles and particles. Cell Death Dis. 2020;11:584. doi: 10.1038/s41419-020-02803-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Elzanowska J, Semira C, Costa-Silva B. DNA in extracellular vesicles: biological and clinical aspects. Mol Oncol. 2021;15:1701–14. doi: 10.1002/1878-0261.12777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kawamura Y, Yamamoto Y, Sato TA, Ochiya T. Extracellular vesicles as trans-genomic agents: Emerging roles in disease and evolution. Cancer Sci. 2017;108:824–30. doi: 10.1111/cas.13222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang L, Yang D, Yang Q, Cheng F, Huang Y. Extracellular DNA in blood products and its potential effects on transfusion. Biosci Rep. 2020;40:BSR20192770. doi: 10.1042/BSR20192770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Plaks V, Koopman CD, Werb Z. Circulating tumor cells. Science. 2013;341:1186–8. doi: 10.1126/science.1235226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chambers AF, Groom AC, MacDonald IC. Dissemination and growth of cancer cells in metastatic sites. Nat Rev Cancer. 2002;2:563. doi: 10.1038/nrc865. [DOI] [PubMed] [Google Scholar]
- 29.Xia YQ, Wan Y, Hao SJ, Nisic M, Harouaka RA, Chen YZ, et al. Nucleus of circulating tumor cell determines its translocation through biomimetic microconstrictions and its physical enrichment by microfiltration. Small. 2018;14:e1802899. doi: 10.1002/smll.201802899. [DOI] [PubMed] [Google Scholar]
- 30.Weiss L, Orr FW, Honn KV. Interactions of cancer cells with the microvasculature during metastasis. FASEB J. 1988;2:12–21. doi: 10.1096/fasebj.2.1.3275560. [DOI] [PubMed] [Google Scholar]
- 31.Wan Y, Wang L, Zhu C, Zheng Q, Wang G, Tong J, et al. Aptamer-conjugated extracellular nanovesicles for targeted drug delivery. Cancer Res. 2017;78:798–808. doi: 10.1158/0008-5472.CAN-17-2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Denais CM, Gilbert RM, Isermann P, McGregor AL, te Lindert M, Weigelin B, et al. Nuclear envelope rupture and repair during cancer cell migration. Science. 2016;352:353–8. doi: 10.1126/science.aad7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raab M, Gentili M, de Belly H, Thiam H-R, Vargas P, Jimenez AJ, et al. ESCRT III repairs nuclear envelope ruptures during cell migration to limit DNA damage and cell death. Science. 2016;352:359–62. doi: 10.1126/science.aad7611. [DOI] [PubMed] [Google Scholar]
- 34.Jiang HY, Sun SX. Cellular pressure and volume regulation and implications for cell mechanics. Biophys J. 2013;104:479a–480a. doi: 10.1016/j.bpj.2012.11.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Apodaca G. Modulation of membrane traffic by mechanical stimuli. Am J Physiol-Ren. 2002;282:F179–F190. doi: 10.1152/ajprenal.2002.282.2.F179. [DOI] [PubMed] [Google Scholar]
- 36.van Meer G, Voelker DR, Feigenson GW. Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Bio. 2008;9:112–24. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ramasubramanian L, Kumar P, Wang A. Engineering extracellular vesicles as nanotherapeutics for regenerative medicine. Biomolecules. 2019;10:48. doi: 10.3390/biom10010048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao QG, Hai B, Kelly J, Wu S, Liu F. Extracellular vesicle mimics made from iPS cell-derived mesenchymal stem cells improve the treatment of metastatic prostate cancer. Stem Cell Res Ther. 2021;12:29. doi: 10.1186/s13287-020-02097-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ilahibaks NF, Lei Z, Mol EA, Deshantri AK, Jiang L, Schiffelers RM, et al. Biofabrication of cell-derived nanovesicles: a potential alternative to extracellular vesicles for regenerative medicine. Cells. 2019;8:1509. doi: 10.3390/cells8121509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang CC, Kang M, Lu Y, Shirazi S, Diaz JI, Cooper LF, et al. Functionally engineered extracellular vesicles improve bone regeneration. Acta Biomater. 2020;109:182–94. doi: 10.1016/j.actbio.2020.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang L, Abhange KK, Wen Y, Chen Y, Xue F, Wang G, et al. Preparation of engineered extracellular vesicles derived from human umbilical cord mesenchymal stem cells with ultrasonication for skin rejuvenation. ACS Omega. 2019;4:22638–45. doi: 10.1021/acsomega.9b03561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Han C, Jeong D, Kim B, Jo W, Kang H, Cho S, et al. Mesenchymal stem cell engineered nanovesicles for accelerated skin wound closure. Acs Biomater Sci Eng. 2019;5:1534–43. doi: 10.1021/acsbiomaterials.8b01646. [DOI] [PubMed] [Google Scholar]
- 43.Wang X, Hu S, Li J, Zhu D, Wang Z, Cores J, et al. Extruded mesenchymal stem cell nanovesicles are equally potent to natural extracellular vesicles in cardiac repair. ACS Appl Mater Interfaces. 2021;13:55767–79. doi: 10.1021/acsami.1c08044. [DOI] [PubMed] [Google Scholar]
- 44.Jo W, Jeong D, Kim J, Park J. Self-Renewal of bone marrow stem cells by nanovesicles engineered from embryonic stem cells. Adv Health Mater. 2016;5:3148–56. doi: 10.1002/adhm.201600810. [DOI] [PubMed] [Google Scholar]
- 45.Qi CX, Liu XS, Zhi DK, Tai YF, Liu YF, Sun QQ, et al. Exosome-mimicking nanovesicles derived from efficacy-potentiated stem cell membrane and secretome for regeneration of injured tissue. Nano Res. 2022;15:1680–90. doi: 10.1007/s12274-021-3868-z. [DOI] [Google Scholar]
- 46.Fan JB, Lee CS, Kim S, Chen C, Aghaloo T, Lee M. Generation of small RNA-modulated exosome mimetics for bone regeneration. ACS Nano. 2020;14:11973–84. doi: 10.1021/acsnano.0c05122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim HY, Bhang SH. Stem cell-engineered nanovesicles exert proangiogenic and neuroprotective effects. Materials. 2021;14:1078. doi: 10.3390/ma14051078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xia Y-Q, Wan Y, Chen Y-Z, Zheng S-Y. Mechanical generated extracellular vesicles through capillary mimicking micro channels. Biomed Eng Annu Mtg (abstract 314) 2019.
- 49.Wen Y, Fu Q, Soliwoda A, Zhang S, Zheng M, Mao W, et al. Cell-derived nanovesicles prepared by membrane extrusion are good substitutes for natural extracellular vesicles. Extracell Vesicle. 2022;1:100004. doi: 10.1016/j.vesic.2022.100004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lai CP, Kim EY, Badr CE, Weissleder R, Mempel TR, Tannous BA, et al. Visualization and tracking of tumour extracellular vesicle delivery and RNA translation using multiplexed reporters. Nat Commun. 2015;6:7029. doi: 10.1038/ncomms8029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Verweij FJ, Revenu C, Arras G, Dingli F, Loew D, Pegtel DM, et al. Live tracking of inter-organ communication by endogenous exosomes in vivo. Dev Cell. 2019;48:573–89. doi: 10.1016/j.devcel.2019.01.004. [DOI] [PubMed] [Google Scholar]
- 52.Hyenne V, Ghoroghi S, Collot M, Bons J, Follain G, Harlepp S, et al. Studying the fate of tumor extracellular vesicles at high spatiotemporal resolution using the zebrafish embryo. Dev Cell. 2019;48:554–72. doi: 10.1016/j.devcel.2019.01.014. [DOI] [PubMed] [Google Scholar]
- 53.Plebanek MP, Angeloni NL, Vinokour E, Li J, Henkin A, Martinez-Marin D, et al. Pre-metastatic cancer exosomes induce immune surveillance by patrolling monocytes at the metastatic niche. Nat Commun. 2017;8:1–12.. doi: 10.1038/s41467-017-01433-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kowal J, Arras G, Colombo M, Jouve M, Morath JP, Primdal-Bengtson B, et al. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc Natl Acad Sci USA. 2016;113:E968–E977.. doi: 10.1073/pnas.1521230113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200:373–83. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]