Summary
Mammarenavirus are a large family of enveloped negative-strand RNA viruses that include several agents responsible for severe hemorrhagic fevers. Until now, no FDA-licensed drug has been admitted for treating an arenavirus infection, and only few effective anti-arenavirus drugs have been tested in vivo. In this work, we designed a recombinant reporter arenavirus lymphocytic choriomeningitis virus that stably expressed nanoluciferase (LCMV-Nluc). The LCMV-Nluc was proved to share similar biological properties with wild-type LCMV and the Nluc intensity reliably reflected viral replication both in vitro and in vivo. Replication of the Nluc-encoding virus in living mice can be visualized by real-time bioluminescent imaging, and bioluminescence can be detected in a variety of organs of infected mice. This work provides a novel approach that enables real-time study of the arenavirus infection and is a convenient and valuable tool for screening of compounds that are active against arenaviruses in vitro and in living mice.
Subject areas: Virology, Methodology in biological sciences
Graphical abstract

Highlights
-
•
LCMV-Nluc was constructed and shared similar biological properties with LCMV-WT
-
•
Replication of the LCMV-Nluc can be visualized by real-time bioluminescent imaging
-
•
LCMV-Nluc is a valuable tool for screening antiviral compounds in vitro
-
•
LCMV-Nluc is successfully applied for screening antiviral compounds in vivo
Virology; Methodology in biological sciences
Introduction
The family Arenaviridae can be divided into four genera: Antennavirus, Hartmanivirus, Mammarenavirus, and Reptarenavirus (Radoshitzky et al., 2015). Mammarenavirus are enveloped viruses with bisegmented, negative-stranded RNA genomes consisting of a large (L) and a small (S) segment. Mammarenavirus members are classified into Old World (OW) and New World (NW) arenaviruses, mainly according to the PAirwise Sequence Comparison (PASC) classification tool. OW arenaviruses include Lassa virus (LASV) and lymphocytic choriomeningitis virus (LCMV), whereas NW arenaviruses include Junin virus (JUNV) and Machupo virus (MACV). Although mammarenaviruses generally cause chronic and asymptomatic infections in their natural host rodents, several arenaviruses, such as LASV, JUNV, and MACV, can cause severe hemorrhagic fever in infected humans, posing serious threats to public health (Gonzalez et al., 2007; Herring et al., 2021). The lack of FDA-approved vaccines or specific drugs has limited the options to treat arenavirus infections, aside from the off-label use of ribavirin. The live-attenuated JUNV vaccine Candid#1 was developed using a conventional attenuation strategy described previously (Enria and Barrera Oro, 2002). This vaccine has been confirmed to be safe and efficacious in human volunteers (Maiztegui et al., 1998), and has been applied for reducing Argentine hemorrhagic fever (AHF) successfully (Enria and Barrera Oro, 2002). However, the vaccine has not been licensed in the United States because of concerns regarding the stability of the attenuation phenotype of Candid#1. Thus, there is an urgent need for an innovative and effective therapeutic approach to combat arenavirus infections.
A suitable animal model that faithfully represents the disease caused by pathogenic arenaviruses in human is critical for evaluating candidate drugs identified in vitro. However, this is highly challenging because of the difficulties of performing animal experimentation within Biosafety Level 4 (BSL-4) environment (Radoshitzky et al., 2012). LCMV is a neglected human pathogen that enables the antiviral studies for pathogenic arenaviruses under standard BSL-2 conditions. Conventional in vivo studies are unable to monitor real-time patterns of infection and are inadequate for large-scale primary drug screening (Caine and Osorio, 2017; Wang and Yu, 2014). Bioluminescence imaging (BLI) technique permits live visualization of virus spread in relevant tissues, enabling real-time outcome assessments for treatment regimens (Ullah et al., 2021). BLI models for many viruses, including influenza virus, respiratory syncytial virus, Zika virus, HIV and SARS-CoV-2, have been described (Pan et al., 2013; Rameix-Welti et al., 2014; Wang et al., 2020; Ullah et al., 2021; Ventura et al., 2019; Roy et al., 2021). The time course of viral infection can be easily visualized, and the relative magnitude of bioluminescence correlates well with viral replication, which can be used to evaluate antiviral therapeutic approaches in vitro and in vivo.
Genetically engineered arenaviruses expressing either green fluorescent protein (GFP) (Vazquez-Calvo et al., 2013; Iwasaki et al., 2014; Ngo et al., 2015) or firefly luciferase (Fluc) (Li et al., 2017a) genes were previously constructed; however, there has been no report on their use in vivo. Here, we rationally designed and constructed a recombinant LCMV carrying the nanoluciferase (Nluc) gene (LCMV-Nluc), which can produce glow-type luminescence (signal half-life >2 h) with a specific activity ∼150-fold greater than that of either firefly or Renilla luciferases (Hall et al., 2012). LCMV-Nluc closely mimics the wild-type virus replication kinetics and stably maintains the Nluc reporter over five generations in vitro. Further in vivo study indicated that dose-dependent bioluminescence can be detected in the abdominopelvic cavities of living mice after intraperitoneal injection with LCMV-Nluc, and the intensity of bioluminescence correlates with the viral load in the spleens and diminishes on treatment with antiviral compounds. This study provides a new approach for real-time study of arenavirus infection and evaluation of antiviral therapeutic approaches in vitro and in vivo.
Results
Development of bioluminescent LCMV reporter viruses
RNA segments of arenaviruses use an ambisense coding strategy to encode two open reading frames (ORFs) in opposite orientations and are separated by a stable hairpin structure called the intergenic region (IGR), which mediates the transcription termination (Pinschewer et al., 2005). The L segment encodes the RNA-dependent RNA polymerase L protein and zinc finger matrix protein Z, whereas the S segment encodes the NP and the envelope glycoprotein precursor (GPC). To visualize real-time LCMV replication and dynamics in vivo and to develop in vitro reporter systems, replication-competent Nluc constructs were generated by inserting the gene-encoding nanoluciferase into the N-terminal region of NP linked by a 2A autoproteolytic cleavage sequence for the parent LCMV-Armstrong strain. We then successfully rescued the Nluc reporter virus (P0) from BSR-T7 cell through a previously described reverse genetics approach (Figure 1A) (Zhu et al., 2019; Wan et al., 2021). We confirmed the rescue of rLCMV-Nluc viruses by PCR using total RNA from the supernatants of wild-type LCMV (LCMV-WT)- or LCMV-Nluc-infected BHK-21 cells using primers surrounding the Nluc fragment. As expected, the amplified bands generated from RNA extracted from LCMV-Nluc- and LCMV-WT-infected supernatants were the expected sizes of ∼1077 bp and ∼564 bp, respectively (Figure 1B). The genetic stability of LCMV-Nluc is a prerequisite for its use. Passages from P1 to P5 did not result in any apparent loss of Nluc gene, and the sequencing result of P5 LCMV Nluc did not demonstrate any mutation (Data S1), indicating that the genome of LCMV-Nluc was stable for at least five life cycles. In addition, the virus foci formation visualized by immunostaining which produced by P1-P5 LCMV-Nluc were comparable to those produced by LCMV-WT (Figure 1C). By detecting both viral RNA and titer levels, the growth kinetics of the wild-type and bioluminescent viruses were compared using a multistep growth curve. The absolute levels of viral RNA and titer were lower in LCMV-Nluc, whereas the relative levels were comparable. LCMV-WT and LCMV-Nluc both reached their peak titers at 48 hours post infection (h.p.i.) (Figures 1D–1H).
Figure 1.
In vitro characterization of bioluminescent LCMV constructs
(A) Strategy for constructing the full-length cDNA clone of LCMV-Nluc. The S segment encodes the nucleocapsid (NP) and the envelope glycoprotein precursor (GPC) of LCMV. The two open reading frames (ORFs) in opposite orientations of the S segment are separated by a stable hairpin structure called the intergenic region (IGR). The replication-competent Nluc constructs were generated by inserting the gene-encoding nanoluciferase into the N-terminal region of NP linked by a 2A autoproteolytic (P2A) cleavage sequence for the parent LCMV-Armstrong strain.
(B) Stability of P1-P5 LCMV-Nluc virus analyzed by agarose gel electrophoresis. LCMV-WT without NLuc was used as a control.
(C) The virus foci formation visualized by immunostaining of the LCMV-WT and P1-P5 LCMV-Nluc virus in BHK-21 cells.
(D–F) In vitro growth characteristics of LCMV-Nluc and LCMV-WT determined by relative mRNA (multiplicity of infection (MOI)= 0.01) in BHK-21 cell. Data represent the mean ± SD(E) In vitro relative growth characteristics of LCMV-Nluc and LCMV-WT determined by viral titers (MOI = 0.01) in BHK-21 cell. Data represent the mean ± SD(F) In vitro absolute growth characteristics of LCMV-Nluc and LCMV-WT determined by viral titers (MOI = 0.01) in BHK-21 cell.
(G–I) Data represent the mean ± SD(G) In vitro absolute growth characteristics of LCMV-Nluc and LCMV-WT determined by viral titers (MOI = 0.01) in A549 cell. Data represent the mean ± SD(H) In vitro relative growth characteristics of LCMV-Nluc and LCMV-WT determined by viral titers (MOI = 0.01) in A549 cell. Data represent the mean ± SD (I) Imaging of LCMV-Nluc in A549 cell at 48h.p.i..
(J) Luciferase avtivities of BHK-21 cell that infected with supernatants gathered from LCMV-Nluc and LCMV-WT infected BHK-21 cell at 6 h.p.i., 12 h.p.i., 24 h.p.i., 36 h.p.i., and 48 h.p.i. The luciferase signals were detected 24 h.p.i. in cell lysates.
(K) Linear correlation between viral titers and luciferase signals of the purified LCMV-Nluc virus in vitro, the Pearson correlation coefficient and two-tailed p value from a linear regression analysis are shown, Data represent the mean ± SD.
To visualize the infection of LCMV-Nluc in vitro, the LCMV-Nluc-infected-A549 cells were treated with Nano-Glo at 1:40 and imaged at 48 h.p.i. (Figure 1I). The expression of Nluc in LCMV-Nluc-infected BHK-21 cells was evaluated by the Nluc assay after cell lysis (Figure 1J). High levels of Nluc expression were detected in LCMV-Nluc-infected cells but not in LCMV-WT-infected cells. We then determined whether luminescence could serve as a direct measure of viral titer. The titer of LCMV-Nluc-infected supernatants was determined by a standard virus foci formation immunostaining assay, then the pre-seeded BHK-21 cells in 24-well plate was incubated in 200 μL of 2-fold serial dilutions of supernatants for 2 h, washed and incubated with 500 μL of fresh DMEM supplemented with 2% FBS. At 24 h.p.i., luminescence readings for serial dilutions of LCMV-Nluc-infected supernatants were taken, and paralleled with the virus titers determined by a standard immune plaque assay (read at 72 h.p.i.) Figure 1K). Viral replication levels measured by Nano-Glo assay or traditional immune plaque assay paralleled each other (r2 = 0.94), but the Nano-Glo assay could be assessed in ⅓ to ½ of the time needed for a traditional virus foci formation immunostaining assay, which favors the large-scale screening.
In vitro antiviral activity of T-705 and ribavirin against LCMV-Nluc
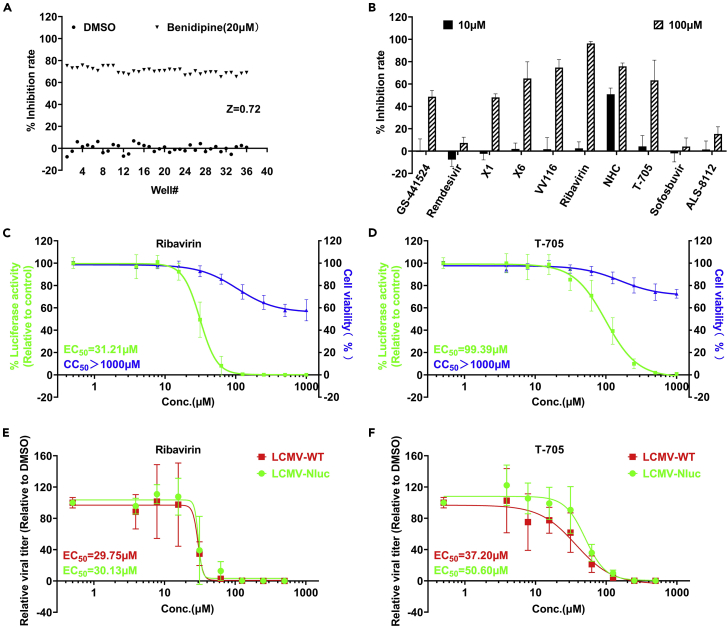
We then employed LCMV-Nluc to assess the anti-LCMV activity of nucleoside analogs in our lab. Briefly, pre-seeded Vero-E6 cells were treated with the indicated concentration of compounds or vehicle and then infected with LCMV-Nluc, and benidipine hydrochloride was chosen as the positive control (Wan et al., 2021). At 48 h.p.i., cells were lysed and Nluc was measured as the indicator of the LCMV replication level. The Z factor for the population of the 36 negative and 36 positive controls in the screen was 0.72 (Figure 2A), suggesting that our screening system was robust (Zhang et al., 1999). Among the analogs selected, T-705 and ribavirin effectively inhibited LCMV replication Figure 2B). T-705 has been reported to inhibit multiple bunyaviruses, including LASV (Rosenke et al., 2018), Crimean-Congo hemorrhagic fever virus (CCHFV) (Oestereich et al., 2014) and severe fever with thrombocytopenia syndrome virus (SFTSV) (Tani et al., 2016). Our recent study indicated that administration of T-705 can reduce case fatality in SFTS patients (Li et al., 2021). We further explored whether T-705 and ribavirin can inhibit LCMV-Nluc in a dose-dependent manner by measuring Nluc in LCMV-Nluc-infected cells, and the half maximal effective concentrations (EC50s) were 99.39 and 33.21 μM, respectively (Figures 2C and2D). By detecting viral titers in supernatant, we found EC50s were 50.60, 37.20 μM for T-705, and 30.13, 29.75 μM for ribavirin in LCMV-Nluc- and LCMV-WT-infected cells, respectively (Figures 2E and2F). More significant effect of T-705 were observed by detecting viral titer, and similar phenomenon was also observed in our previous study for SFTSV, in which T-705 induce SFTSV mutagenesis, and thus decreased viral infectivity (Li et al., 2021).
Figure 2.
In vitro application of LCMV-Nluc for anti-LCMV drug screening
(A) Screen controls. Inhibition rates of LCMV-Nluc in Vero-E6 cell are shown for all positive (Benidipine) and negative (DMSO) controls across screening plates.
(B) Screening for the anti-LCMV activities for 10 nucleoside analogs in our lab.
(C) CC50 and EC50 of ribavirin for LCMV-Nluc in Vero-E6 determined by detecting the luciferase activity.
(D) CC50 and EC50 of T-705 for LCMV-Nluc in Vero-E6 determined by detecting the luciferase activity. Experiments were performed for three times. Each time, three independent assays were performed. The plots show all three experiments performed in duplicates. Each replicate is shown. The four-parameter dose–response curve was fitted using the nonlinear regression method and EC50s were calculated in the software Prism 9.0.
(E) CC50 and EC50 of ribavirin for LCMV-Nluc in Vero-E6 determined by detecting the viral titers.
(F) CC50 and EC50 of T-705 for LCMV-Nluc in Vero-E6 determined by detecting the viral titers. The four-parameter dose–response curve was fitted using the nonlinear regression method and EC50s were calculated in the software Prism 9.0.
Characteristics of LCMV-Nluc infection in C57BL/6-Prf1tm1Sdz/J mice
We then set out to compare LCMV-WT and LCMV-Nluc in vivo and to determine whether LCMV-Nluc could be applied for visualizing real-time LCMV replication in C57BL/6-Prf1tm1Sdz/J mice. C57BL/6-Prf1tm1Sdz/J mice were chosen for their successful application as an animal model for primary hemophagocytic lymphohistiocytosis (HLH) studies in the past few years through infection with LCMV(Jordan et al., 2004). Initial mouse studies were carried out on 6- to 10-week-old C57BL/6-Prf1tm1Sdz/J mice that were mock-infected or infected with 2 × 105 PFU of LCMV-WT, 2 × 105 PFU of LCMV-Nluc or 2 × 106 PFU of LCMV-Nluc. None of the 2 × 105 PFU of LCMV-Nluc-infected C57BL/6-Prf1tm1Sdz/J mice succumbed to infection or lost body weight. However, mice infected with a higher dose (2 × 106 PFU) of LCMV-Nluc ultimately succumbed to infection or lost 20% of their initial body weight, comparable to 2 × 105 PFU of LCMV-WT-infected mice between 9 and 13 days post infection (d.p.i.). (Figures 3A and3B). Then, a rapid systemic spread of luminescence from the site of injection (i.p.) was observed in C57BL/6-Prf1tm1Sdz/J mice infected with LCMV-Nluc starting from 1 d.p.i. (Figure 3C), but not in LCMV-WT or mocked treated mice despite the same treatment of Nano-Glo. Luminescence signals were mainly seen in the abdomen of infected animals early during experimentation and the peak average radiance was obtained at 3 or 5 d.p.i. for the ventral sides (Figure 3D).
Figure 3.
In vivo luminescence of LCMV-Nluc-infected mice
(A) Temporal changes in body weight with initial body weight set to 100% for mock-infected C57BL/6-Prf1tm1Sdz/J mice and mice infected with 5 × 105 PFU of LCMV-Nluc, 5 × 106 PFU of LCMV-Nluc and 5 × 105 PFU of LCMV-WT through intraperitoneal injection (n = 6 per group). Mice were euthanized at 13 d.p.i. or once they lost 20% of their body weight.
(B and C) Survival curves of mock-infected C57BL/6-Prf1tm1Sdz/J mice and mice infected with 5 × 105 PFU of LCMV-Nluc, 5 × 106 PFU of LCMV-Nluc and 5 × 105 PFU of LCMV-WT (n = 6 per group). Logrank (Mantel-Cox) test is used for analyses (C) Bioluminescence imaging of LCMV-Nluc-infected mice was performed at 1, 3, 5, and 7 d.p.i. The ventral views of the results were shown. One of the mock-infected C57BL/6-Prf1tm1Sdz/J mouse and LCMV-WT-infected mouse was imaged as control.
(D) The average radiance of LCMV-Nluc-infected mice determined from region of interest (ROI) analysis of the ventral side.
(E) Representative BLI of tissues harvested from mock-infected- and 2 × 106 PFU of LCMV-Nluc-infected-mice.
(F) Titer of spleens dissociated from LCMV-Nluc- or LCMV-WT-infected mice that euthanized at 5 d.p.i.
(G) Titer of spleens dissociated from LCMV-Nluc- or LCMV-WT-infected mice that loss 20% of their body weight or euthanized at 13 d.p.i.
(H) Ratio of spleen weight (g) and body weight (g) for mice loss 20% of their body weight or euthanized at 13 d.p.i.
(I) HE staining of spleen from mice loss 20% of their body weight or euthanized at 13 d.p.i.
Luminescence imaging of tissues harvested from mice infected with 2 × 106 PFU of LCMV-Nluc which lost 20% of their body weight and mock infected mice (euthanized at 13 d.p.i.) is also presented in Figure 3E. Luminescence was mainly located in the spleen and some other organs in the abdominopelvic cavities, such as the liver, kidney, testis and intestine. Simultaneously, nearly no signals were detected in the brain or heart, and signals in the lung were weak, which coincided with the BLI results of live mice. In addition, the femur exhibited some signals, which might explain the occurrence of primary HLH (Figure 3E). At 5 d.p.i., the titers of spleens dissociated from LCMV-Nluc-infected mice were comparable to those from LCMV-WT-infected mice (Figure 3F). Once harvested, the spleens of mice were weighed, and obvious splenomegaly was noted in LCMV-Nluc- and LCMV-WT-infected mice (Figure 3H). Then, HE staining of the spleen showed that the boundary between the white and red pulps of the spleens harvested from LCMV-Nluc- or LCMV-WT-infected mice was indistinct as the marginal zone disappeared (Figure 3I). Taken together, these data revealed that viral propagation in vivo for LCMV-Nluc was significantly reduced as compared with LCMV-WT in C57BL/6-Prf1tm1Sdz/J mice, but we could address this shortage by increasing the amount of LCMV-Nluc given for further in vivo imaging studies, and 2 × 106 PFU was enough to reach a comparable extent of body weight loss, coinciding with the survival curve (Figures 3A and 3B). Therefore, 2 × 106 PFU of LCMV-Nluc was given for further in vivo imaging studies.
To prove the application of LCMV-Nluc-infected C57BL/6-Prf1tm1Sdz/J mice as a suitable system for future antiviral drug screening, 3 mice infected with 2 × 106 PFU of LCMV-Nluc were euthanized, and organs were harvested at 1, 3, and 5 d.p.i. to verify whether the signals detected in live mice revealed actual organ involvement. The luminescent signals of organs gradually increased from 1 to 5 d.p.i., especially for spleens (Figure S1A). Then, the virus titers in the spleen were detected and gradually increased coincident with the luminescent signals from 1 to 5 d.p.i. (Figure S1B). The spleen showed gradual enlargement (Figure S1C), and the border between the white and red pulps gradually disappeared as the virus spread from 1 to 5 d.p.i. (Figure S1D). To prove the stability of our reporter virus throughout the experimental timeline, the ratio of copy numbers of LCMV-NP to Nluc in the viral RNA of input virions and virions isolated from the spleens of mice at 1, 3, 5 d.p.i. was detected by real-time PCR analyses, and the ratio remained unchanged (Figure S1E). Thus, Nluc activity was a good surrogate for virus replication in vivo. These data demonstrate that in vivo imaging can readily be used to visualize viral spread and is a useful tool for studying the various disease outcomes of LCMV infection.
BLI application of LCMV-Nluc in the evaluation of antiviral therapeutics in living mice
We next tested the antiviral effects of ribavirin and T-705 in vivo using LCMV-Nluc. T-705 (300 mg/kg) and ribavirin (100 mg/kg) were delivered orally alone twice daily for up to 5 d.p.i. challenge with LCMV-Nluc because the peak average radiance was obtained at 3 or 5 d.p.i. for the ventral sides (Figure 4A). Temporal monitoring by BLI revealed that both ribavirin and T-705 substantially reduced LCMV-Nluc infection in the abdomen (Figures 4B and4C). The intensity of bioluminescence correlates with the viral load in ribavirin but not T-705 group (Figures 4C, E and 4F), and this may partially because T-705 increased mutation rates of viral RNA (Li et al., 2021; Arias et al., 2014), which may not affect viral RNA or protein level but reduce viral infectivity capacity significantly, however, further studies are needed to clarify the detailed mechanism of ribavirin and T-705 against LCMV. After terminal necropsy at 5 d.p.i., the livers, spleens, and intestines were harvested and imaged, and the signal was reduced in T-705- and ribavirin-treated mice (Figure S2). In addition, T-705 and ribavirin successfully protected the spleens from enlargement. The marginal zones of the spleens of treated mice were much more complete Figure 4G). These data demonstrate that our LCMV-Nluc-infected C57BL/6-Prf1tm1Sdz/J mice could be an excellent system for future anti-LCMV drug screening by real-time BLI with no need to kill the mice and dissociate the organs for virus detection.
Figure 4.
The application of LCMV-Nluc for rapid anti-LCMV drug screening
(A) Experimental design to test anti-LCMV efficacy of T-705 (300 mpk) and ribavirin (100 mpk) in vivo.
(B) BLI of LCMV-Nluc-infected mice that were mock-treated or treated with T-705 or ribavirin at 1, 3, 5 d.p.i. (Ventral Views).
(C) The average radiance determined from region of interest (ROI) analysis for the ventral side of LCMV-Nluc-infected mice that were mock-treated or treated with T-705 or ribavirin at 1, 3, 5 d.p.i. One-way ANOVA is used for analyses.
(D) Temporal changes in body weight with initial body weight set to 100% for mock-, T-705-, and ribavirin-treated mice that infected with LCMV-Nluc and LCMV-WT until 5 d.p.i. (n = 6 per group). One-way ANOVA is used for analyses.
(E) Titers of livers dissociated from LCMV-Nluc-/LCMV-WT-infected mice that were mock-treated or treated with T-705 or ribavirin at 5 d.p.i. One-way ANOVA is used for analyses.
(F) Titers of livers dissociated from LCMV-Nluc-/LCMV-WT-infected mice that were mock-treated or treated with T-705 or ribavirin at 5 d.p.i. One-way ANOVA is used for analyses.
(G) HE staining of spleen from mock-, T-705-, and ribavirin-treated mice that infected with LCMV-Nluc.
Discussion
Arenaviridae are known to have the largest number of viral species reported to cause severe viral hemorrhagic fever (VHF) (Basler, 2017; Zapata et al., 2011). However, options for treatments against arenavirus infection are limited by the lack of FDA-approved vaccines or specific drugs. Recently, multiple compounds have been identified to inhibit arenavirus effectively in vitro (Iwasaki et al., 2014; Welch et al., 2016; Saez-Ayala et al., 2018). Reverse genetics systems for highly pathogenic arenaviruses, including LASV(Albarino et al., 2011), JUNV(Albarino et al., 2009; Emonet et al., 2010) and MACV (Patterson et al., 2014) have been applied to obtain recombinant arenaviruses harboring fluorescent reporters (Welch et al., 2016; Emonet et al., 2010), which can assess viral replication levels in ⅓ to ½ of the time needed for traditional immune plaque assays. In our previous study, LCMV-GFP was rescued and applied for antiviral drug screening of an FDA-approved drug library, and five drug candidates were identified as having strong anti-LCMV effects in vitro (Wan et al., 2021). However, our in vivo evaluations of these candidates against LCMV-GFP or LCMV-mCherry were hampered by the low-resolution images produced using GFP and mCherry. BLI has been reported to offer much higher sensitivity than fluorescence imaging (approximately 500-fold more sensitive than fluorescence imaging) (Dimond et al., 2020). The ATP-independent Nluc, characterized with smaller size (19 kDa), high physical stability, and higher brightness (∼150-fold greater than that of either firefly or Renilla luciferases) (Hall et al., 2012) was eventually selected. Then, a replication-competent LCMV-Nluc was constructed and rescued successfully.
The peripheral infection with LCMV Armstrong (LCMV-Arm) in adult wild-type mice has little clinical impact and was cleared between 10 and 12 d.p.i. (Li et al., 2014). Perforin-1 (PRF1) is a pore-forming protein that plays key roles in secretory granule-dependent cell death and in defense against virus-infected cells. Recent studies and our preliminary data indicated that C57BL/6-Prf1tm1Sdz/J mice are permissive to LCMV infection (Rothman et al., 2011). In this study, we found that LCMV-WT and LCMV-Nluc replicated effectively in C57BL/6-Prf1tm1Sdz/J mice, resulting in loss of body weight and even death. Thus, our in vivo screening system was established using C57BL/6-Prf1tm1Sdz/J mice. The luminescence of LCMV-Nluc is much brighter and shows adequate sensitivity to be easily detected starting from 1 d.p.i. and increased from 1 to 5 d.p.i. This finding was in accordance with the gradual increase in titers in spleens harvested from LCMV-Nluc-infected mice. The sensitivity gives our screening system a clear advantage since the detection of bioluminescence starting from 1 d.p.i. preceded obvious weight loss or death at 9 to 13 d.p.i.
The replication, stability, and virulence must be carefully evaluated both in vitro and in vivo because the insertion of exogenous genes often results in attenuation of the recombinant viruses(Caine and Osorio, 2017; Li et al., 2017b; Ullah et al., 2021; Wang et al., 2020), as described in our study (Figures 3C and 3D). The titers of spleens harvested from LCMV-Nluc-infected mice were reduced compared with those harvested from LCMV-WT-infected mice. The defective interfering particles (DIPs), which was proposed could be a critical determinant of infection outcome for many viruses, should be considered in the future application of this model for mechanism studies (Huang and Baltimore, 1970; Ziegler and Botten, 2020). In addition, the P2A fusion of Nluc might influence the expression of NP because of ribosome fall off (Liu et al., 2017), as NP is critical for LCMV replication and suppression of host innate immunity, this could also be a possible explanation for the attenuated infection of LCMV-Nluc in animals. However, a higher dose (2 × 106 PFU) of LCMV-Nluc is adequate for mimicking the features of LCMV-WT in C57BL/6-Prf1tm1Sdz/J mice. LCMV-Nluc was proven to be genetically stable for at least five passages in vitro and for at least five days in vivo. As compared to LCMV-WT in vitro, the absolute levels of viral RNA and titer were lower in LCMV-Nluc, whereas the relative levels were comparable. LCMV-WT and LCMV-Nluc both reached their peak titers at 48 h.p.i., and the Nluc reporter intensity was highly consistent with the viral replication level in LCMV-Nluc-infected cells. This favors the application of LCMV-Nluc for large-scale antiviral drug screening in vitro and in vivo.
We then used LCMV-Nluc to evaluate the antiviral effects of multiple nucleoside analogs in Vero-E6 cells, among which T-705 and ribavirin inhibited LCMV replication in a dose-dependent manner. The Z factor in the screen was 0.72 (Figure 2A), suggesting that our screening system was robust and can be applied for large-scale screening of antiviral agents in vitro. For in vivo application, a duration of 5 days was determined to assess the anti-LCMV efficacy in vivo because the luminescent signals began to decrease from 7 d.p.i. The signals of LCMV-Nluc-infected mice treated especially with ribavirin at 5 d.p.i. were lessened compared with mock-treated mice. HE staining of spleens revealed that the border between the white and red pulps of T-705- and ribavirin-treated spleen was much clearer than that of mock-treated spleens, which confirmed the BLI results.
In addition, BLI also performed excellently for simultaneously tracking the involvement and spread of viruses in organs. The sequential spread of virus from the nasal cavity to the lungs in mice and thereafter systemically to various organs was dynamically monitored using BLI in mice (Ullah et al., 2021). In our study, rapid spread of LCMV-Nluc was recorded from the site of injection to the spleen and some other organs in the abdominopelvic cavities, such as the liver, kidney, testis, and intestine, starting from 1 d.p.i. Conventional studies of viral infection in mouse models rely on euthanizing mice at sequential time points to identify sites of infection and quantify viral titers, and an unexpected site to which virus spread might be missed once the infected tissue would not be among the standard sites collected for analysis(Luker and Luker, 2008). Nevertheless, research on both the acute and persistent infections of mice with LCMV have led to the first description of virus-induced immunopathologic disease, primary HLH and et al.(Malin et al., 2020; Weiss et al., 2018). Therefore, BLI could also provide insight into the mechanisms of LCMV infection and virus-induced immunopathologic diseases.
In summary, by performing live visualization of LCMV-Nluc via a BLI system, we can assess virus replication level, body weight and survival curves using the same set of mice, which can decrease the number of mice required for antiviral evaluation. The screening system based on LCMV-Nluc-infected C57BL/6-Prf1tm1Sdz/J mice can further enhance the information obtained using conventional measurement methods in a mouse model of LCMV. The further development of BLI-based screening systems for arenaviruses will greatly advance preclinical studies of viral infection and therapy and develop imaging biomarkers that can be translated to clinical trials and patient care for individuals infected with arenaviruses.
Limitations of the study
We found that BLI could also provide insight into the mechanisms of LCMV infection and virus-induced immunopathologic diseases, which have not been explored in this study. Besides, though the antiviral efficacy of T-705 and ribavirin has been identified in this study, the detailed mechanisms of T-705 and ribavirin needs further studies.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| LCMV-Armstrong | Zhu et al., 2019, Wan et al., 2021 | N/A |
| LCMV-Nluc | This paper | N/A |
| Critical commercial assays | ||
| KOD-Plus-Neo | TOYOBO | Cat # KOD-401 |
| In-Fusion HD cloning Kit | Takara | Cat # 639648 |
| LipofectamineTM3000 Transfection Reagent | Invitrogen | Cat #L3000015 |
| MiniBEST Viral RNA/DNA Extraction Kit | TaKaRa | Cat # 9766 |
| 2×Rapid Taq Master Mix | Vazyme | Cat # P222 |
| RNAiso Plus | Takara | Cat # 9108Q |
| Nano-Glo substrate solution | Promega | Cat # N1120 |
| TB Green® Premix Ex TaqTM II | Takara | Cat # RR820A |
| PrimeScripTMRT reagent Kit with gDNA Eraser | Takara | Cat # RR047A |
| Experimental models: Cell lines | ||
| Vero E6 cell | ATCC | CRL-1586;RRID:CVCL_0574 |
| BHK-21 cell | ATCC | CCL-10;RRID:CVCL_1915 |
| BSR-T7 cell | Buchholz et al., 1999 | N/A |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6-Prf1tm1Sdz/J mice | Jackson Laboratory | Stock No: 002407 |
| Oligonucleotides | ||
| Nluc-PCR-F: 5′-GTCTTGAGAAACCATTGAGCAACA-3′ | This paper | N/A |
| Nluc-PCR-R: 5′-TTACCACACCACTTGCACCCT-3′ | This paper | N/A |
| LCMV-NP F: 5′-GTACAAGCGCTCACAGACCT-3′ | This paper | N/A |
| LCMV-NP R: 5′-GTTACCCCCATCCAACAGGG-3′ | This paper | N/A |
| GAPDH F: 5′-ATCCCACCAACATCAAATGG-3′ | This paper | N/A |
| GAPDH R: 5′-AAGACGCCAGTAGACTCCACA-3′ | This paper | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Leike Zhang (zhangleike@wh.iov.cn).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
All animal studies were carried out in strict accordance with the National Institutes of Health guidelines under protocols approved by the Institutional Animal Care and Use Committee of Wuhan Institute of Virology. C57BL/6-Prf1tm1Sdz/J mice (Stock No: 002,407) were bought form the Jackson Laboratory and bred in a pathogen-free animal facility at the Tongji Medical College of the Huazhong University of Science and Technology. Initial experiments used groups (n = 6) of 6- to 10-week-old C57BL/6-Prf1tm1Sdz/J (perforin deficient) mice that received intraperitoneal (i.p.) injections of 2 × 105 PFU (plaque forming unit), 2 × 106 PFU of LCMV-Nluc, and 2 × 105 PFU of LCMV-WT diluted in 200 μL PBS. Mock infected mice were injected with 200 μL PBS as control. In the in vivo antiviral experiment, 6- to 10-week-old C57BL/6-Prf1tm1Sdz/J mice were infected with 2 × 106 PFU of LCMV-NLuc.
Method details
Cells and viruses
African green monkey kidney epithelial cell (Vero E6; ATCC CRL-1586), baby hamster syrian kidney cell (BHK-21; ATCC CCL-10) were obtained from ATCC, and BHK-derived cell line stably expressing T7 RNA polymerase (BSR-T7) (Buchholz et al., 1999) was provided by Dr Mingzhou Chen (Wuhan University, China). All cells were grown in Dulbecco’s Modified Eagle’s medium (DMEM; Gibco) supplemented with 10% fetal bovine serum (FBS). Plated cells were incubated at 37°C under 5% CO2. LCMV was rescued based on a T7 polymerase system and viral stocks of LCMV-Armstrong strain were prepared following previously described procedures (Zhu et al., 2019; Wan et al., 2021).
LCMV-Nluc constructs
LCMV-Nluc fusions were created by polymerase chain reaction (PCR) using KOD-Plus-Neo (TOYOBO, Cat # KOD-401) and In-Fusion HD cloning Kit (Takara, Cat # 639648). Briefly, a full-length P2A (2A Self-shearing polypeptide)-Nluc fusion construct was linked at the 3315 base of the S segment of LCMV, and the orientation of open reading frame (ORF) was same as the nucleocapsid (NP). pT7-LCMV-S-P2A-Nluc andpT7-LCMV-L plasmids were co-transfected into the BSR-T7 cell using LipofectamineTM3000 Transfection Reagent (Invitrogen, Cat #L3000015). Then P0 virus was collected by harvesting the supernatant three days later.
Stability of LCMV-Nluc constructs
The reporter viruses were continually passaged for five rounds on BHK-21 cells blindly. Every 48 h.p.i., the supernatants were harvested and the debris was removed by centrifugation at 2000 rpm/min for 10 min. The pre-seeded BHK-21 cells in 6-well plate were incubated with 400μL of the 1:1 diluted supernatant, washed with PBS for three times at 2 h.p.i., and incubated with 2 mL of fresh DMEM supplemented with 2% FBS for another 46 h. RNA extractions were performed on the viruses harvested from the culture fluids using MiniBEST Viral RNA/DNA Extraction Kit (TaKaRa, Cat # 9766). The stability of the Nluc gene was tested using a 2×Rapid Taq Master Mix (Vazyme, Cat #P222) with primers that flanked either side of the Nluc gene following the manufacturer’s instructions (Nluc-PCR-F: 5′-GTCTTGAGAAACCATTGAGCAACA-3′, Nluc-PCR-R: 5′-TTACCACACCACTTGCACCCT-3′). LCMV-WT was used as a control.
Multistep growth curve for viruses with and without the Nluc gene
When 24-well plates of BHK-21 cells reached 80 to 90% confluence, triplicate wells were infected at a MOI of 0.01 for each virus. This was considered as time point zero. At 6 h.p.i., 12h.p.i., 24 h.p.i., 36 h.p.i., 48 h.p.i., and 72 h.p.i., the total RNA of LCMV-Nluc- and LCMV-WT-infected cells was extracted using RNAiso Plus (Takara, Cat # 9108Q), reverse transcribed with PrimeScripTMRT reagent Kit with gDNA Eraser (Takara, Cat # RR047A), followed by a real-time PCR assay using TB Green Premix Ex Taq™ II (Takara, Cat # RR820A) for determining the relative mRNA of LCMV-NP using primers LCMV-NP F: 5′-GTACAAGCGCTCACAGACCT-3′, LCMV-NP R: 5′-GTTACCCCCATCCAACAGGG-3′, GAPDH F: 5′-ATCCCACCAACATCAAATGG-3′, and GAPDH R: 5′-AAGACGCCAGTAGACTCCACA-3′. Besides, LCMV-Nluc- and LCMV-WT-infected supernatants were harvested. And pre-seeded BHK-21 cells in 24-well plates were infected with 200 μL of 10-fold serial dilutions of supernatants. At 24 h.p.i., cells were washed and lysed by 1×Passive Lysis Buffer (Promega Cat #E1941). 20 μL of the diluted sample was mixed with 20 μL Nano-Glo substrate solution (Promega, Cat #N1120) following the manufacturer’s instructions and read on a GloMax Multi JR Single-tube multifunction detector (Promega). The readout was compared with the titers read by a traditional immune plaque assay (Zhu et al., 2019).
Antiviral assay
Vero-E6 cells were seeded into 96-well plates at a density of 2×104 cells/well. After 16 h incubation, the cells were pre-incubated in triplicate with indicated concentration of analogs or 2-fold serial dilutions of T-705 and ribavirin (0–1000 μM) for 50% inhibitory concentrations (IC50) 1 h before infected with LCMV-Nluc (MOI = 0.1). Supernatants were removed at 2 h.p.i., and cells were incubated with indicated concentration of compounds for another 46 h. Culture supernatants were removed at 48 h.p.i. and cells were washed and lysed by 1×Passive Lysis Buffer the luciferase signal was determined as described above. For determining the 50% cytotoxic concentration (CC50), pre-seeded Vero-E6 cells in 96-well plates were incubated in triplicate with 2-fold serial dilutions of T-705 and ribavirin (0–1000 μM) for 48 h. Culture supernatants were removed at 48 h.p.i. and cells were washed and the CC50s were determined using Cell Counting Kit-8 (Beyotime, Cat #C0039) according to the instrument.
Mouse experiments
Body weight and survival were recorded for all mice. In vivo imaging was done on LCMV-Nluc-infected mice (n = 12) every other day until 13 d.p.i., mice received PBS and LCMV-WT were imaged as control (n = 1). For imaging, Nano-Glo reagent was diluted 1:20 in 1× PBS and used immediately. 100 μL of the diluted Nano-Glo reagent was injected intraperitoneal into anesthetized mice (1% pentobarbital sodium in normal saline). In vivo imaging was performed on Living Image 4.4 (PerkinElmer). At the end of study (at 13 d.p.i. or once mice lost 20% of their body weight), mice were euthanized, and spleens were cut into half, either placed in tissue homogenization tubes for subsequent immune plaque assay or fixed in 2% paraformaldehyde for histology. Fixed tissues were sent to the Wuhan Pinuofei Biological Technology CO., LTD, where they were paraffin embedded, sectioned, and stained with hematoxylin-eosin (HE). The ratio of copy numbers of LCMV NP to Nluc in the viral RNA of input virions and virions isolated from spleens of mice was detected by real-time PCR analyses using primers Nluc-qPCR F: 5′-AGATTTCGTTGGGGACTGGC-3′, Nluc-qPCR R: 5′-CGCTCAGACCTTCATACGGG-3′, as well as LCMV-NP F and R described above.
Titration of virus from excised tissues
Spleens were homogenized in 400 μL serum free DMEM media and homogenized in 1.5mL tube using a hand-held tissue grinding machine-Coyote G50 (Coyote Bioscience, cat # 0105003) for 5000 rpm/min in 1-min interval. Homogenates were centrifuged at 2000 rpm/min for 10 min to remove tissue and cell debris. Viral loads were determined by virus foci formation in an immunostaining assay (Zhu et al., 2019).
In vivo antiviral experiment
6- to 10-week-old C57BL/6-Prf1tm1Sdz/J mice were infected with 2 × 106 PFU of LCMV-NLuc and 0.4% carboxymethyl cellulose (CMC, as control), 300 mg/kg/day T-705 or 100 mg/kg/day ribavirin were given by gavage administration twice daily. At 1, 3 and 5 d.p.i. (d.p.i.), mice were imaged and bioluminescent signals in stomach areas were quantified using Living Image software.
Quantification and statistical analysis
Data from cell and animal studies were compiled in Microsoft Excel and analyzed using Prism 9.0 software. Data are reported as mean +/− SD. The statistic significance was calculated using One-way ANOVA analyses. Significance was set to p < 0.05 (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001).
Acknowledgments
This research was supported by funds from the National Natural Science Foundation of China (No. 31970165, 31701207), innovative research grant from Wuhan Science and Technology Department (No. 2019020701011503), Chen Xiao-ping Foundation for the Development of Science and Technology of Hubei Province. We thank Xuefang An, Fan Zhang in the Core Facility and Technical Support Facility of the Wuhan Institute of Virology for their technical assistance, Dong Zhang in the Hematology Department of Beijing Friendship Hospital for providing the C57BL/6-Prf1tm1Sdz/J mice.
Author contributions
L.Z., H.C., and X.W. conceived the project and supervised this study. Y.W. and H.X. performed most of the experiments; W.W. and W.S. participated in the material preparation; J.W. providing the C57BL/6-Prf1tm1Sdz/J mice; Y.W. and L.Z. wrote the paper. H.X., R.J., F.Z., H.M., and G.X. supported and supervised the manuscript revision. All of the authors discussed the results and commented on the manuscript.
Declaration of interests
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Inclusion and diversity
We support inclusive, diversive, and equitable conduct of research.
Published: October 21, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.105090.
Contributor Information
Hongbo Chen, Email: hbchen@hust.edu.cn.
Xiaoyan Wu, Email: xwu@hust.edu.cn.
Leike Zhang, Email: zhangleike@wh.iov.cn.
Supplemental information
Data and code availability
-
•
Microscopy data, imaging data reported in this article will be shared by the lead contact on request.
-
•
This article does not report original code.
-
•
Any additional information required to reanalyze the data reported in this article is available from the lead contact on request.
References
- Albarino C.G., Bergeron E., Erickson B.R., Khristova M.L., Rollin P.E., Nichol S.T. Efficient reverse genetics generation of infectious Junin viruses differing in glycoprotein processing. J. Virol. 2009;83:5606–5614. doi: 10.1128/JVI.00276-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albarino C.G., Bird B.H., Chakrabarti A.K., Dodd K.A., Erickson B.R., Nichol S.T. Efficient rescue of recombinant Lassa virus reveals the influence of S segment noncoding regions on virus replication and virulence. J. Virol. 2011;85:4020–4024. doi: 10.1128/JVI.02556-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias A., Thorne L., Goodfellow I. Favipiravir elicits antiviral mutagenesis during virus replication in vivo. Elife. 2014;3 doi: 10.7554/eLife.03679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basler C.F. Molecular pathogenesis of viral hemorrhagic fever. Semin. Immunopathol. 2017;39:551–561. doi: 10.1007/s00281-017-0637-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchholz U.J., Finke S., Conzelmann K.K. Generation of bovine respiratory syncytial virus (Brsv) from Cdna: Brsv Ns2 is not essential for virus replication in tissue culture, and the human rsv leader region Acts as A functional Brsv genome promoter. J. Virol. 1999;73:251–259. doi: 10.1128/jvi.73.1.251-259.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caine E.A., Osorio J.E. Imaging with bioluminescent enterovirus 71 Allows for real-time visualization of tissue tropism and viral spread. J. Virol. 2017;91 doi: 10.1128/JVI.01759-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimond A., Van De Pette M., Fisher A.G. Illuminating epigenetics and inheritance in the immune system with bioluminescence. Trends Immunol. 2020:41. doi: 10.1016/j.it.2020.09.001. [DOI] [PubMed] [Google Scholar]
- Emonet S.F., Seregin A.V., Yun N.E., Poussard A.L., Walker A.G., De La Torre J.C., Paessler S. Rescue from cloned Cdnas and in vivo characterization of recombinant pathogenic romero and live-attenuated Candid #1 strains of Junin virus, the Causative agent of Argentine hemorrhagic fever disease. J. Virol. 2010;85:1473–1483. doi: 10.1128/JVI.02102-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enria D.A., Barrera Oro J.G. Junin virus vaccines. Curr. Top. Microbiol. Immunol. 2002;263:239–261. doi: 10.1007/978-3-642-56055-2_12. [DOI] [PubMed] [Google Scholar]
- Gonzalez J.P., Emonet S., De Lamballerie X., Charrel R. Arenaviruses. Curr. Top. Microbiol. Immunol. 2007;315:253–288. doi: 10.1007/978-3-540-70962-6_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall M.P., Unch J., Binkowski B.F., Valley M.P., Butler B.L., Wood M.G., Otto P., Zimmerman K., Vidugiris G., Machleidt T., et al. Engineered luciferase reporter from A deep sea shrimp utilizing A novel imidazopyrazinone substrate. ACS Chem. Biol. 2012;7:1848–1857. doi: 10.1021/cb3002478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herring S., Oda J.M., Wagoner J., Kirchmeier D., O'connor A., Nelson E.A., Huang Q., Liang Y., Dewald L.E., Johansen L.M., et al. Inhibition of arenaviruses by Combinations of orally available approved drugs. Antimicrobial Agents And Chemotherapy. 2021;65 doi: 10.1128/AAC.01146-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang A.S., Baltimore D. Defective viral particles and viral disease processes. Nature. 1970;226:325–327. doi: 10.1038/226325a0. [DOI] [PubMed] [Google Scholar]
- Iwasaki M., Ngo N., De La Torre J.C. Sodium Hydrogen exchangers Contribute to arenavirus cell entry. J. Virol. 2014;88:643–654. doi: 10.1128/JVI.02110-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan M.B., Hildeman D., Kappler J., Marrack P. An animal model of hemophagocytic lymphohistiocytosis (Hlh): Cd8+ T cells and interferon Gamma are essential for the disorder. Blood. 2004;104:735–743. doi: 10.1182/blood-2003-10-3413. [DOI] [PubMed] [Google Scholar]
- Li W., Hofer M.J., Jung S.R., Lim S.-L., Campbell I.L. Irf7-Dependent type I interferon production induces Lethal immune-Mediated disease in Stat1 Knockout mice infected with lymphocytic choriomeningitis virus. J. Virol. 2014;88:7578–7588. doi: 10.1128/JVI.03117-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Liu Q., Huang W., Wu J., Nie J., Wang M., Zhao C., Zhang L., Wang Y. An lasv gpc pseudotyped virus based reporter system enables evaluation of vaccines in mice under non-bsl-4 conditions. Vaccine. 2017 doi: 10.1016/j.vaccine.2017.07.101. [DOI] [PubMed] [Google Scholar]
- Li X.-F., Li X.-D., Deng C.-L., Dong H.-L., Zhang Q.-Y., Ye Q., Ye H.-Q., Huang X.-Y., Deng Y.-Q., Zhang B., Qin C.-F. Visualization of A neurotropic flavivirus infection in mouse reveals unique Viscerotropism Controlled by host type I interferon signaling. Theranostics. 2017;7:912–925. doi: 10.7150/thno.16615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Jiang X.M., Cui N., Yuan C., Zhang S.F., Lu Q.B., Yang Z.D., Xin Q.L., Song Y.B., Zhang X.A., et al. Clinical effect and antiviral mechanism of T-705 in treating severe fever with thrombocytopenia syndrome. Signal Transduct Target Ther. 2021;6:145. doi: 10.1038/s41392-021-00541-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Chen O., Wall J.B.J., Zheng M., Zhou Y., Wang L., Vaseghi H.R., Qian L., Liu J. Systematic Comparison of 2a peptides for cloning Multi-genes in A polycistronic Vector. Sci. Rep. 2017;7 doi: 10.1038/s41598-017-02460-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luker K.E., Luker G.D. Applications of bioluminescence imaging to antiviral research and therapy: multiple luciferase enzymes and Quantitation. Antivir. Res. 2008;78:179–187. doi: 10.1016/j.antiviral.2008.01.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiztegui J.I., Mckee K.T., Barrera Oro J.G., Harrison L.H., Gibbs P.H., Feuillade M.R., Enria D.A., Briggiler A.M., Levis S.C., Ambrosio A.M., et al. Protective efficacy of A live attenuated vaccine against Argentine hemorrhagic fever. Ahf study group. The Journal Of Infectious Diseases. 1998;177:277–283. doi: 10.1086/514211. [DOI] [PubMed] [Google Scholar]
- Malin S.G., Shavva V.S., Tarnawski L., Olofsson P.S. Functions of Acetylcholine-producing Lymphocytes in immunobiology. Curr. Opin. Neurobiol. 2020;62:115–121. doi: 10.1016/j.conb.2020.01.017. [DOI] [PubMed] [Google Scholar]
- Ngo N., Cubitt B., Iwasaki M., De La Torre J.C. Identification and mechanism of action of A novel small Molecule inhibitor of arenavirus Multiplication. J. Virol. 2015 doi: 10.1128/JVI.01587-15. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Oestereich L., Rieger T., Neumann M., Bernreuther C., Lehmann M., Krasemann S., Wurr S., Emmerich P., De Lamballerie X., Olschlager S., Gunther S. Evaluation of antiviral efficacy of ribavirin, Arbidol, and T-705 (favipiravir) in A mouse model for Crimean-Congo hemorrhagic fever. Plos. Negl. Trop Dis. 2014;8 doi: 10.1371/journal.pntd.0002804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan W., Dong Z., Li F., Meng W., Feng L., Niu X., Li C., Luo Q., Li Z., Sun C., Chen L. Visualizing influenza virus infection in living mice. Nat. Commun. 2013;4 doi: 10.1038/ncomms3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson M., Seregin A., Huang C., Kolokoltsova O., Smith J., Miller M., Yun N., Poussard A., Grant A., Tigabu B., et al. Rescue of A recombinant Machupo virus from cloned Cdnas and in vivo characterization in interferon (Alphabeta/Gamma) receptor double Knockout mice. J. Virol. 2014;88:1914–1923. doi: 10.1128/JVI.02925-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinschewer D.D., Perez M., De La Torre J.C. Dual role of the lymphocytic choriomeningitis virus intergenic region in transcription termination and virus propagation. J. Virol. 2005;79:4519–4526. doi: 10.1128/JVI.79.7.4519-4526.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radoshitzky S.R., Kuhn J.H., De Kok-Mercado F., Jahrling P.B., Bavari S. Drug discovery technologies and strategies for Machupo virus and other new World arenaviruses. Expet Opin. Drug Discov. 2012;7:613–632. doi: 10.1517/17460441.2012.687719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radoshitzky S.R., Bao Y., Buchmeier M.J., Charrel R.N., Clawson A.N., Clegg C.S., Derisi J.L., Emonet S., Gonzalez J.P., Kuhn J.H., et al. Past, present, and future of arenavirus taxonomy. Arch. Virol. 2015;160:1851–1874. doi: 10.1007/s00705-015-2418-y. [DOI] [PubMed] [Google Scholar]
- Rameix-Welti M.A., Le Goffic R., Herve P.L., Sourimant J., Remot A., Riffault S., Yu Q., Galloux M., Gault E., Eleouet J.F. Visualizing the replication of respiratory syncytial virus in cells and in living mice. Nat. Commun. 2014;5 doi: 10.1038/ncomms6104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenke K., Feldmann H., Westover J.B., Hanley P.W., Martellaro C., Feldmann F., Saturday G., Lovaglio J., Scott D.P., Furuta Y., et al. Use of favipiravir to treat Lassa virus infection in Macaques. Emerg. Infect. Dis. 2018;24:1696–1699. doi: 10.3201/eid2409.180233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman J.A., Das R., Teachey D.T., Paessler M.E., Nichols K.E. Rapamycin does not control hemophagocytic lymphohistiocytosis in Lcmv-infected perforin-deficient mice. Pediatr. Blood Cancer. 2011;57:1239–1243. doi: 10.1002/pbc.23226. [DOI] [PubMed] [Google Scholar]
- Roy C.N., Benitez Moreno M.A., Kline C., Ambrose Z. Cg dinucleotide removal in bioluminescent and fluorescent reporters improves Hiv-1 replication and reporter gene expression for dual imaging in Humanized mice. J. Virol. 2021;95 doi: 10.1128/JVI.00449-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez-Ayala M., Laban Yekwa E., Mondielli C., Roux L., Hernandez S., Bailly F., Cotelle P., Rogolino D., Canard B., Ferron F., Alvarez K. Metal Chelators for the inhibition of the lymphocytic choriomeningitis virus endonuclease domain. Antiviral Res. 2018;162:79–89. doi: 10.1016/j.antiviral.2018.12.008. [DOI] [PubMed] [Google Scholar]
- Tani H., Fukuma A., Fukushi S., Taniguchi S., Yoshikawa T., Iwata-Yoshikawa N., Sato Y., Suzuki T., Nagata N., Hasegawa H., et al. Efficacy of T-705 (favipiravir) in the treatment of infections with Lethal severe fever with thrombocytopenia syndrome virus. mSphere. 2016;1 doi: 10.1128/mSphere.00061-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah I., Prévost J., Ladinsky M.S., Stone H., Lu M., Anand S.P., Beaudoin-Bussières G., Symmes K., Benlarbi M., Ding S., et al. Live imaging of sars-Cov-2 infection in mice reveals that neutralizing Antibodies require fc function for Optimal efficacy. Immunity. 2021;54 doi: 10.1016/j.immuni.2021.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez-Calvo A., Martin-Acebes M.A., Saiz J.C., Ngo N., Sobrino F., De La Torre J.C. Inhibition of Multiplication of the prototypic arenavirus Lcmv by Valproic Acid. Antiviral Res. 2013;99:172–179. doi: 10.1016/j.antiviral.2013.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura J.D., Beloor J., Allen E., Zhang T., Haugh K.A., Uchil P.D., Ochsenbauer C., Kieffer C., Kumar P., Hope T.J., Mothes W. Longitudinal bioluminescent imaging of Hiv-1 infection during Antiretroviral therapy and treatment interruption in Humanized mice. PLoS Pathog. 2019;15 doi: 10.1371/journal.ppat.1008161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan W., Zhu S., Li S., Shang W., Zhang R., Li H., Liu W., Xiao G., Peng K., Zhang L. High-throughput screening of an fda-approved drug library identifies inhibitors against arenaviruses and sars-Cov-2. ACS Infect. Dis. 2021;7:1409–1422. doi: 10.1021/acsinfecdis.0c00486. [DOI] [PubMed] [Google Scholar]
- Wang Y.-F., Yu C.-K. Animal models of enterovirus 71 infection: Applications and Limitations. J. Biomed. Sci. 2014;21:31. doi: 10.1186/1423-0127-21-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Li P., Zhang Y., Liu Y., Tan Z., Sun J., Ke X., Miao Y., Luo D., Hu Q., et al. In vivo imaging of Zika virus reveals dynamics of viral invasion in immune-sheltered tissues and Vertical propagation during pregnancy. Theranostics. 2020;10:6430–6447. doi: 10.7150/thno.43177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss E.S., Girard-Guyonvarc'h C., Holzinger D., De Jesus A.A., Tariq Z., Picarsic J., Schiffrin E.J., Foell D., Grom A.A., Ammann S., et al. Interleukin-18 diagnostically distinguishes and pathogenically promotes human and Murine Macrophage Activation syndrome. Blood. 2018;131:1442–1455. doi: 10.1182/blood-2017-12-820852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch S.R., Guerrero L.W., Chakrabarti A.K., Mcmullan L.K., Flint M., Bluemling G.R., Painter G.R., Nichol S.T., Spiropoulou C.F., Albarino C.G. Lassa and ebola virus inhibitors identified using Minigenome and recombinant virus reporter systems. Antiviral Res. 2016;136:9–18. doi: 10.1016/j.antiviral.2016.10.007. [DOI] [PubMed] [Google Scholar]
- Zapata J.C., Pauza C.D., Djavani M.M., Rodas J.D., Moshkoff D., Bryant J., Ateh E., Garcia C., Lukashevich I.S., Salvato M.S. Lymphocytic choriomeningitis virus (Lcmv) infection of Macaques: amodel for Lassa fever. Antivir. Res. 2011;92:125–138. doi: 10.1016/j.antiviral.2011.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.H., Chung T.D., Oldenburg K.R. A simple statistical parameter for use in evaluation and Validation of high throughput screening assays. J. Biomol. Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- Zhu S., Wan W., Zhang Y., Shang W., Pan X., Zhang L.-K., Xiao G. Comprehensive interactome analysis reveals that Stt3b is required for N-Glycosylation of Lassa virus glycoprotein. J. Virol. 2019;93 doi: 10.1128/JVI.01443-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler C.M., Botten J.W. Defective interfering particles of negative-strand rna viruses. Trends Microbiol. 2020;28:554–565. doi: 10.1016/j.tim.2020.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Microscopy data, imaging data reported in this article will be shared by the lead contact on request.
-
•
This article does not report original code.
-
•
Any additional information required to reanalyze the data reported in this article is available from the lead contact on request.