Abstract
The Flavivirus genus is made up of viruses that are either mosquito-borne or tick-borne and other viruses transmitted by unknown vectors. Flaviviruses present a significant threat to global health and infect up to 400 million of people annually. As the climate continues to change throughout the world, these viruses have become prominent infections, with increasing number of infections being detected beyond tropical borders. These include dengue virus (DENV), West Nile virus (WNV), Japanese encephalitis virus (JEV), and Zika virus (ZIKV). Several highly conserved epitopes of flaviviruses had been identified and reported to interact with antibodies, which lead to cross-reactivity results. The major interest of this review paper is mainly focused on the serological cross-reactivity between DENV serotypes, ZIKV, WNV, and JEV. Direct and molecular techniques are required in the diagnosis of Flavivirus-associated human disease. In this review, the serological assays such as neutralization tests, enzyme-linked immunosorbent assay, hemagglutination-inhibition test, Western blot test, and immunofluorescence test will be discussed. Serological assays that have been developed are able to detect different immunoglobulin isotypes (IgM, IgG, and IgA); however, it is challenging when interpreting the serological results due to the broad antigenic cross-reactivity of antibodies to these viruses. However, the neutralization tests are still considered as the gold standard to differentiate these flaviviruses.
Keywords: serological cross reactivity, flaviviruses, cross-protection, antibody assays, antibodies
Introduction to Flavivirus
Characteristically, flaviviruses are RNA viruses that are enveloped and encode a positive single-stranded genome. These 11-kb viruses of the genus Flavivirus belong to the family Flaviviridae (Huang et al., 2014). Out of the 53 recognized Flavivirus spp., 40 are known to cause human diseases (Gubler et al., 2007). Flaviviruses are transmitted to vertebrates through the bites of infected mosquitoes and ticks, producing disease in animals and human (Gould & Solomon, 2008; Chong et al., 2019). The bite of an infected vector spreads the virus through the blood stream and lymphatics, and replication is seen in many organs. Flaviviruses that are pathogenic for humans include West Nile virus (WNV), yellow fever virus (YFV), Japanese encephalitis virus (JEV), dengue virus (DENV), Zika virus (ZIKV), and tick-borne encephalitis virus (TBEV) (Simmonds et al., 2017). Classification of the virus is complicated due to its extensive geographical distribution and transmission of the vectors.
As of 2015, DENV, JEV, and WNV are the most common Flavivirus infections (Daep et al., 2014). These pathogens represent a significant burden of disease, causing about 400 million cases with 100 million symptomatic cases each year with dengue alone (Bhatt et al., 2013). These viruses mainly cause hemorrhagic disease and encephalitis. For instance, the secondary infection with DENV can cause hemorrhagic fever, while infections with neurotropic Flavivirus such as JEV, TBEV, and WNV are responsible for viral encephalitis worldwide (Mathengtheng & Burt, 2014; Johnson, 2016). The clinical manifestations of Flavivirus infections can range from undifferentiated fever and mild symptoms to more severe conditions that can be fatal. Forty percent of the world’s population is affected by dengue. In the past 50 years, a 30-fold increase in dengue cases have been reported (Kyle & Harris, 2008). It has been reported that DENV infection can lead to dengue shock syndrome (DSS) or dengue hemorrhagic fever (DHF) with 20,000 fatalities every year (Webster et al., 2009). Another Flavivirus, JEV, is endemic in the Western Pacific and Southeast Asia, and it is estimated that at least 68,000 cases are reported every year in these regions and the infection may be symptomatic or asymptomatic with a 20%–30% fatality rate (Solomon, 2004).
In tropical and subtropical countries, Flavivirus infections represent a global public health problem. This has major economic, social, and individual consequences to those in these regions (Musso & Desprès, 2020). Rapid climate change, improper controlled urbanization, traveling within endemic areas, migration of populations, and extensive deforestation is associated with flaviviruses adapting to new habitats and host species. These are the main factors that contribute to the rising trend of Flavivirus infections and transmission of viruses into previously non-endemic areas (Pandit et al., 2018; Argondizzo et al., 2020). These are exemplified by ZIKV emergence in South America in 2015 and Europe in 2019 (Khawar et al., 2017; Giron et al., 2019), outbreaks of WNV in Europe and North America from the 2000s (Tsai et al., 1998; Nash et al., 2001), and YFV outbreaks in Brazil and Africa (Kraemer et al., 2017; Hamer et al., 2018).
Flavivirus genomic organization
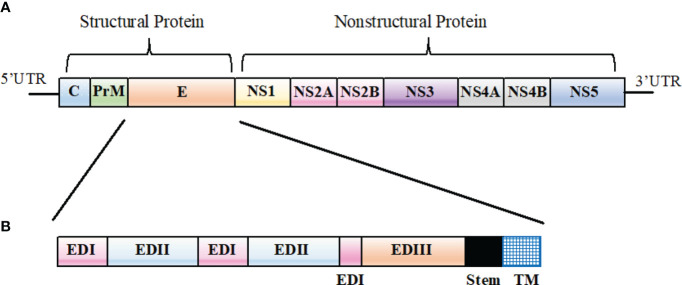
Among flaviviruses, genome organization is similar. Basically, the genome encodes three structural proteins (capsid [C], pre-membrane [prM], and envelope [E]) and seven non-structural (NS) proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) as shown in Figure 1 . The structural and non-structural proteins are essential in the formation of virus particles, viral replication, viral polyprotein processing, and cell receptor binding and entry (Mazeaud et al., 2018; Chong et al., 2019).
Figure 1.
Flavivirus genomic organization. (A) A schematic representation of Flavivirus genomic organization. (B) Primary structure of Flavivirus E protein ectodomain showing EDI (pink), EDII (light blue), and EDIII (orange). The stem region and the transmembrane (TM) domain represented in black and blue, respectively.
Cross-reactive epitopes of Flavivirus E protein
Flaviviruses enter target cells through the interaction of viral E protein with the host surface receptors. Thus, the main antigenic target for antibody responses to DENV is the E protein, which is the most exposed protein on the virus and considered as the main antigenic target for antibody responses in patients (Pierson et al., 2008; Beltramello et al., 2010; Xu et al., 2012). The E protein is made up of three domains (EDI, EDII, and EDIII), and it is EDIII that interacts with attachment factors and receptors (Pierson & Kielian, 2013). Potent neutralizing antibodies are induced by EDIII due to the presence of important antigenic epitopes with strong antigenicity. EDIII is also a common antigen used in serological diagnosis (Chávez et al., 2010; Matsui et al., 2010). On the other hand, anti-EDI and anti-EDII antibodies are less potent but show a broader cross-reactivity among different strains of flaviviruses than those against EDIII (Sun et al., 2017).
Venkatachalam and Subramaniyan (2014) performed multiple sequence alignment of E protein of the four DENV serotypes to study the conservation and homology of amino acids. Their results indicated a close relationship between DENV1 and DENV3, which showed the highest percentage of homology of up to 78.4%. DENV1 with DENV2 and DENV2 with DENV3 showed 66.1% and 66.3% amino acid identity, respectively, while DENV1 with DENV4, DENV2 with DENV4, and DENV3 with DENV4 showed 63.3%, 62.8%, and 63.4% homology, respectively, indicating that DENV4 showed a distant relationship with the three serotypes. Other similar studies using multiple sequence alignment of E protein among flaviviruses showed that the similarity between YFV and TBEV are highly related but distant from the four DENV, WNV, JEV, and ZIKV (Chakraborty, 2016). Other than that, it has been reported that the E protein of ZIKV and DENV2 share ~54% amino acid sequence identity (Priyamvada et al., 2016; Stettler et al., 2016). ZIKV also shares 57% amino acid sequence identity with WNV, 56.1% with JEV, 55.6% with DENV, and 46% with YFV (Chang et al., 2017). A phylogenetic study by Kuno et al. (1998) indicated that non-vector and vector-borne virus clusters emerged from a putative ancestor, and it is from the vector-borne cluster that tick- and mosquito-borne virus clusters emerged. Their observations suggested that the viruses of this genus evolved from non-vector group to tick-borne and then to mosquito-borne group.
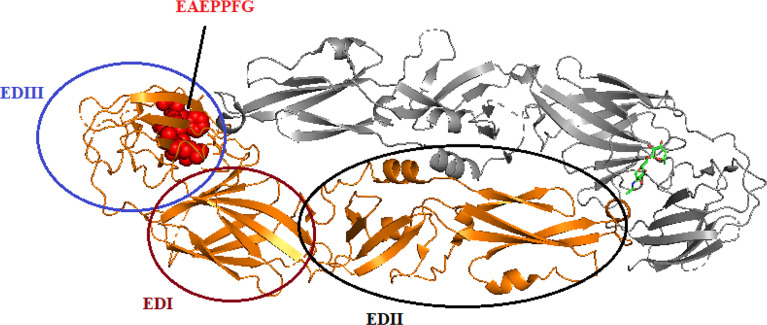
It had been reported by Li et al. (2016) that the monoclonal antibody (mAb) 3B6 binds to the EXE/DPPFG epitope in E protein domain III, which is highly conserved (approximately 85%) in DENV, WNV, ZIKV, JEV, Murray Valley encephalitis virus (MVEV), and Saint Louis encephalitis (SLEV), indicating that the function of the epitope is similar in these viruses. The cross-reactivity of DENV-, YFV-, ZIKV-, and WNV-positive sera suggested that the EXE/DPPFG epitope is an immunodominant epitope among flaviviruses. The sequences of the E protein of DENV serotypes, WNV, JEV, MVEV, and SLEV were obtained from National Center for Biotechnology Information (NCBI), and the multiple sequences alignment are shown in Table 1 . The dashes indicate identical amino acids, while the EXE/DPPFG epitope region is indicated by gray shading. Figure 2 shows the molecular structure of DENV1 E protein and the EXE/DPPFG epitope located at EDIII.
Table 1.
Sequence alignment of highly conserved flaviviruses E protein EXE/DPPFG epitope.
| Epitope | Accession No. | Sequence |
|---|---|---|
| DENV1 | NP_722460.2 | 361 KEKPVNIEAE―PPFGESYIVVGAGEKALKLS 390 |
| DENV2 | NP_739583.2 | 360 EKDSPVNIEAE―PPFGDSYIIIGVEPGQLKLN 390 |
| DENV3 | YP_001531168.2 | 358 KKEEPVNIEAE―PPFGESNIVIGIGDNALKIN 388 |
| DENV4 | NP_740317.1 | 360 NTNSVTNIELE―PPFGDSYIVIGVGNSALTLH 390 |
| WNV | YP_001527880.1 | 366 TANAKVLIELE―PPFGDSYIVVGRGEQQINH 395 |
| WNV | AEB66112.1 | 366 TANAKVLIELE―PPFGDSYIVVGRGEQQINH 395 |
| JEV | NP_059434.1 | 366 ANSKVLVEME―PPFGDSYIVVGRGDKQINH 394 |
| JEV | AAA67173.1 | 366 ANSKVLVEME―PPFGDSYIVVGRGDKQINH 394 |
| YFV | NP_740305.1 | 358 TNDDEVLIEVN―PPFGDSYIIVGRGDSRLTYQ 388 |
| YFV | QHB50138.1 | 358 TNDDEVLIEVN―PPFGDSYIIVGTGDSRLTYQ 388 |
| MVEV | NP_722531.1 | 367 ANAKVLVEIE―PPFGDSYIVVGRGDKQINHH 396 |
| SLEV | YP_009329949.1 | 367 ANNKVMIEVE―PPFGDSYIVVGRGTTQINYH 396 |
Figure 2.
The protein-dimer structure of DENV1 E protein and EAEPPFG epitope located at domain III. The brown, black, and blue circle represent the domain I, domain II, and domain III, respectively, in one E protein monomer. Another monomer is colored gray. The red spheres located in EDIII represent the EAEPPEG epitope.
Other than that, Deng et al. (2011) reported that the mAb 2A10G6, which potently neutralizes DENV serotypes, TBEV, JEV, WNV, and YFV, showed a board cross-reactivity to the 98DRXW101 motif, which is the highly conserved Flavivirus fusion loop peptide. The comparison of 2A10G6 epitope of these flaviviruses is shown in Table 2 , and the epitope region is indicated by gray shading. Figure 3 shows the structure of 2A10G6 epitope in DENV1, which is located at EDII.
Table 2.
Comparison of flaviviruses E protein sequence on the 2A10G6 epitope.
| Epitope | Accession No. | Sequence |
|---|---|---|
| DENV1 | NP_722460.2 | 90 FVCRRTFVDRGWGNGCGLFGKGSLITC 116 |
| DENV2 | NP_739583.2 | 90 FVCKHSMVDRGWGNGCGLFGKGGIVT 115 |
| DENV3 | YP_001531168.2 | 90 YVCKHTYVDRGWGNGCGLFGKGSLVT 115 |
| DENV4 | NP_740317.1 | 90 YICRRDVVDRGWGNGCGLFGKGGVVT 115 |
| WNV | YP_001527880.1 | 90 FVCRQGVVDRGWGNGCGLFGKGSIDT 115 |
| WNV | AEB66112.1 | 90 FVCRQGVVDRGWGNGCGLFGKGSIDT 115 |
| JEV | NP_059434.1 | 90 YVCKQGFTDRGWGNGCGLFGKGSIDT 115 |
| JEV | AAA67173.1 | 90 YVCKQGFTDRGWGNGCGFFGKGSDT 115 |
| YFV | NP_740305.1 | 90 NACKRTYSDRGWGNGCGLFGKGSIVA 115 |
| YFV | QHB50138.1 | 90 NACKRTYSDRGWGNGCGLFGKGSIVA 115 |
| MVEV | NP_722531.1 | 90 YLCKRGVTDRGWGNGCGLFGKGSIDT 115 |
| SLEV | YP_009329949.1 | 90 FVCKRDVVDRGWGNGCGLFGKGSIDT 115 |
Figure 3.
Structure of 2A10G6 epitope of DENV1 E protein. The 98DRXW101 motif is labeled and colored as blue, purple, yellow, and orange, respectively, according to the residue sequence.
There are several cross-reactivity epitopes on the surface of flaviviruses E protein that determine antibody binding and neutralization properties. Table 3 below summarizes some epitopes and residues that were located at different domains of E protein that show cross-reactivity among flaviviruses.
Table 3.
Flavivirus cross-reactive epitopes and residues located in E protein.
| Epitopes | Location | References |
|---|---|---|
| A1 | EDII, contains at least two independent and overlapping group-reactive epitopes, incorporating two or three of highly conserved fusion peptide residues Gly104, Gly106, and Leu107. | (Crill and Chang, 2004) |
| A5 | EDII, centers on conserved Trp231 and structurally related with neighbors Glu126 and Thr226. | |
| E111 | EDIII AB loop, EDIII residues 314–319. | (Sukupolvi-Petty et al., 2007) |
| E114 | EDIII | |
| 4E11 | EDIII, residue 307, 308, 309, 310, 311, 312, 387, 389, and 391. | (Lisova et al., 2007) |
| EDIII, residue 306, 308, 381, 387, and 389. | (Matsui et al., 2009) | |
| 4G2-1 | EDII, residue 101, 104, 106 and 107. | (Chiou et al., 2012) |
| 4G2-2 | EDIII, residue 312, 315, 331, 332, and 389. | |
| 6B3B-3, 6B6C-1 | EDIII, residue 312, 315, 329, 331, 332, and 332. | |
| 23-1, 23-2 | EDII, residue 101 and 107 | |
| 5-2 | EDI, residue 138. |
Diagnosis of Flavivirus-associated human disease
In the diagnosis of Flavivirus-associated human disease, laboratory testing is required. The tests used either directly detected the infecting agent or through the detection of antibodies targeting the infecting virus. The advantages and limitations of several laboratory techniques used for the diagnosis of Flavivirus are shown in Table 4 .
Table 4.
Methods for the diagnosis of human Flavivirus infections.
| Methods | Advantages | Limitations | References |
|---|---|---|---|
| Virus isolation |
|
|
|
| RT-PCR |
|
|
|
| Viral antigen capture |
|
|
|
| Serology |
|
|
Cross-reactivity among flaviviruses was first shown with complement fixation tests (Casals, 1957) followed by the hemagglutination-inhibition assay (Casals & Brown, 1954). Virus-neutralizing tests have strengthened this concept of cross-reactivity and enabled segregation of flaviviruses into the different arthropod-borne viruses and those with unknown arthropod vectors (Calisher et al., 1989). The antigenic similarities between flaviviruses indicate one aspect of the similarities observed among flaviviruses. Both species-specific and Flavivirus cross-reactive antibodies are produced when infection with one Flavivirus occurs. The majority of flaviviruses that are relevant to human disease were organized into eight serocomplexes that cause cross-reactive immune responses (Rathore & St. John, 2020) consistent for multiple mammalian species (Saron et al., 2018). This cross-reactivity is said to be not durable and hence not retained (Collins et al., 2017). Multiple exposures to various flaviviruses make it extremely difficult to determine the latest infection (Mansfield et al., 2011). This high degree of cross-reactivity further emphasizes the need for virological confirmation.
The detection of the viral genome using PCR-based techniques are highly specific but require clinical laboratories with advanced technology. Despite the broad antigenic cross-reactivity of anti-Flavivirus antibodies, serological assays are still widely used to detect different immunoglobulin isotypes (IgM, IgG, and IgA) (Allwinn et al., 2002; Mansfield et al., 2011; Papa et al., 2011). However, the interpretation of these serological results is challenging. The likelihood of experiencing multiple Flavivirus infections has resulted in a need to understand the effects of pre-existing immunity during a lifetime and its impact on subsequent exposures. Animal models have been used, and here, however, prior exposures were shown to be protective (Tesh et al., 2002) within the same serocomplex (Williams et al., 2001). However, for DENV, protection is seen only with the homologous serotype (Sabin, 1952). Recent studies (Saron et al., 2018) using JEV showed cross-protection against dengue viruses with increased neutralizing antibodies. With ZIKV, both cross-protection and immune-mediated pathology were noted (Rathore et al., 2019). Therefore, the major interest of this review paper is in characterizing the serological cross-reactivity of DENV serotypes, ZIKV, WNV, and JEV.
Flavivirus serological assays
Currently, there are several serological assays that are able to determine antibody levels to flaviviruses such as the Western blotting assay, dot-blot assay, neutralization tests, hemagglutination-inhibition tests, IgM/IgG antibody-capture ELISAs, immunofluorescent tests, microsphere immunoassays, high-throughput and rapid microneutralization assays, lateral flow tests, biosensors and microfluidic systems, and autologous red blood cell agglutination tests (Hobson-Peters, 2012; Chong et al., 2019).
Neutralization tests
Neutralizing anti-Flavivirus antibodies usually target highly accessible epitopes, and the detection of neutralizing anti-Flavivirus antibodies is correlated with the presence of the specific IgG in blood specimens (Roehrig et al., 2008). Neutralization tests are the most reliable serological assay and are capable of providing high specificity among flaviviruses (Russell & Nisalak, 1967). The plaque reduction neutralization test (PRNT), micro-neutralization test (MNT), and virus neutralization test (VNT) are considered as the gold standard in quantifying and detecting the levels of neutralizing antibodies against flaviviruses (Thomas et al., 2009; Guy et al., 2010; Plotkin, 2010). MNT and VNT are the alternative methods of PRNT, using 96-well microplates in the combination with enzyme-linked immunoassays, which are cost effective and widely automated (Li et al., 2017; Nurtop et al., 2018).
Calisher et al. (1989) stated that the PRNT test is the most specific serological test to differentiate the infections caused by flaviviruses in convalescent sera samples. The PRNT test can be carried out in a test tube or microtiter plate. A serial dilution of the serum sample is mixed with a standardized amount of virus. The mixture is then tested for unbound virus by testing for viral infectivity on virus-susceptible cells. Each virus that initiates a productive infection can then be measured by first restricting the spread of progeny virus by overlaying cells with a semi-solid media. This results in the formation of a localized area of infection (a plaque) that can be detected through a variety of ways. The number of plaques is calculated and compared to the initial concentration of virus to determine the percent reduction in total virus infectivity (Zannoli et al., 2018). The PRNT titer is the titer that results in a reduction of 50%–90% plaques.
However, since PRNT is a labor-intensive, time-consuming, and largely manual assay, it is challenging during the diagnosis and is expensive especially when conducting large clinical trials that might involve the testing of hundreds to thousands of samples under good clinical laboratory practice conditions (WHO, 2007; Maeda & Maeda, 2013). In addition, neutralization tests require live virus, as the assay antigen, to detect neutralizing antibodies. This has high variability between assays and between laboratories due to the differences in cell lines used, the maturation state of virus, the strain, and other variations (WHO, 2007; Thomas et al., 2009). Moreover, the laboratories that do not have access to live infectious viruses or do not meet appropriate biosafety standards are unable to perform neutralization tests.
For DENV, PRNT is the most widely accepted method in measuring the virus-neutralizing antibodies (Roehrig et al., 2008). Thomas et al. (2009) conducted a series of assays with different cell lines, virus preparation, and the presence or absence of complement to determine the variability in anti-dengue virus PRNT assays and found that modification of these conditions caused significant effects on PRNT titers, which can be impacted not only by changing a single condition but also by interaction between two or more conditions. Other than that, Rainwater-Lovett et al. (2012) had carried out a systematic review of human PRNT titers to DENV and concluded some additional factors that may influence the PRNT titers. First, the geographical areas of study population affect not only the strains that a person is exposed to but also strains against which their serum was tested. Second, based on the laboratory methods and inclusion criteria on each study, there is likely a link between the testing strains and primary or secondary DENV exposure. Lastly, the quality, duration, and magnitude of antibodies response induced by different infecting strains may differ, highlighting the differences between strains that call for further exploration.
The PRNT is useful for epidemiological or diagnostic purposes as a reduction in plaque counts of ≥80% can ensure prevention of cross-reactivity among flaviviruses in dengue endemic areas. However, the PRNT50 titer is the preferred value during the evaluation of vaccines for human use because it affords acceptable sensitivity and specificity (WHO, 2007). Timiryasova et al. (2013) had evaluated several parameters to produce reliable test performance to support the vaccine development program and validated assay reagents to ensure that they are suitable for intended use and consistently performing within the established limits. Their study showed that the dengue PRNT50 for each of the serotypes of DENV is precise, accurate, and specific, and it is suitable for detecting and measuring specific neutralizing antibodies to each DENV serotypes in human serum samples with a lower limit of quantitation of 10. However, for dengue, PRNT remains as the gold standard neutralization assay and is recommended by WHO against which any new assay will need to be validated.
Among the flaviviruses, the exposed surfaces show the highest degree of variation. The most potent neutralizing epitopes are type specific, while those that promote antibody-dependent enhancement of infection (ADE) belong to the cross-reactive group (Rey et al., 2017). Of all the structural domains of the E protein, the highly variable EDIII has the most potent neutralizing activity (Stettler et al., 2016; Zhao et al., 2016). However, these areas make up a small component of the human response against WVN and DENV (Throsby et al., 2006; Beltramello et al., 2010) and do not contribute significantly to the neutralization activity present in serum (Wahala et al., 2009). Patients infected with ZIKV who have high anti-EDIII titers have also EDIII-specific neutralizing activity, indicating the major role of these antibodies in ZIKV immunity. Some cross-reactivity with DENV from memory cells was noted in previously DENV-infected patients (Robbiani et al., 2017; Yu et al., 2017). Other than that, there are also quaternary epitopes restricted to the E–E dimer interface, and antibodies to these epitopes have also been described in WNV-, JEV-, and ZIKV-infected patients (Kaufmann et al., 2010; Hasan et al., 2017; Qiu et al., 2018). Other regions implicated include the fusion loop and the prM, but these areas are highly conserved between DENV and ZIKV (Dejnirattisai et al., 2014; Rouvinski et al., 2015), and antibody-escape mutations do not readily develop (Abbink et al., 2018). Recently, cross-reactive antibody responses were shown to promote ADE during the recent ZIKV outbreak in areas with high DENV exposure. This may have been due to the existing anti-DENV immunity and hence may also occur against a wide variety of other flaviviruses (Halstead et al., 1980; Fagbami et al., 1987). Evidence for this response was shown by passive transfer of cross-reactive antibodies isolated from ZIKV- and DENV-infected patients in AG129 mice (Stettler et al., 2016). Other studies conducted were in Stat2−/− mice after intraperitoneal administration of DENV or WNV immune sera (Bardina et al., 2017). In other human studies, it was observed that following JEV vaccination, the cross-reactive antibodies induced was found to be associated with an increased risk of symptomatic dengue illness (Anderson et al., 2011; Saito et al., 2016). However, evidence obtained from many other studies showed otherwise with no effect in ZIKV-immune macaques (George et al., 2017), whether exposed to DENV or YFV (McCracken et al., 2017). Recently, a study of pregnant women infected with ZIKV indicated that the presence of DENV antibodies did not have any significant association with congenital Zika syndrome (Halai et al., 2017).
Cross-reactivity of anti-Powassan virus (POWV) antibodies against other tick-borne flaviviruses (TBFVs) and mosquito-borne flaviviruses (MBFVs), which include TBEV, Gadgets Gully virus (GGYV), Langat virus (LGTV), SLEV, ZIKV, WNV, YFV, and Usutu virus (USUV), was shown in a study conducted by VanBlargan et al. (2021). Their results revealed that the anti-POWV antibodies were cross-reactive with other flaviviruses and cross-neutralized other TBFVs. Moreover, the studies stated that the cross-reactivity and cross-neutralization of some POWN EDIII-specific mAbs occurred because of the recognition of the lateral ridge/C–C’ loop epitope and thus were protective against TBEV and LGTV. This was similarly shown with ZIKV mAbs, which targets the epitopes on the EDIII-LR and C–C’ loop on ZIKV, which cross-reacted with DENV (Sapparapu et al., 2016; Robbiani et al., 2017; Zhao et al., 2019). Therefore, the EDIII-LR/C–C’ loop epitope represents an antigenic site with multiple TBFV and MBFV areas that can be targeted.
Enzyme-linked immunosorbent assay
The ELISA is a technique that enables many laboratories to test numerous samples simultaneously. It is used in quantifying and detecting a specific protein in a complex mixture. ELISAs are performed in a 96- or 384-well plate coated with antibodies and proteins passively bound in the well. There are several formats used for ELISAs, such as direct, indirect, or sandwich methods for capture and detection. The most important step is the attachment of the antigen to the assay microtiter plate, and this is done by either direct adsorption to the assay plate or indirectly via a capture antibody that has been attached to the plate. The antigen is then detected either directly (labeled primary antibody) or indirectly (such as labeled secondary antibody). The most widely used ELISA assay format is the sandwich ELISA assay due to its specificity and sensitivity (ThermoFisher, 2010). However, ELISA is labor-intensive to carry out, and the cost for the preparation of the antibody is high because it is a sophisticated technique that requires cell culture media to obtain a specific antibody. It is important to note that errors occur when there is insufficient blocking of the surface of microtiter plate immobilized with antigen and this can lead to high false-negative or false-positive results (Sakamoto et al., 2017).
IgM antibody capture ELISA (MAC-ELISA) can be used in the diagnosis of DENV infections by detecting specific IgM antibodies in serum samples. In the early acute phase of the disease, there is a negative window period of detection, as the antibody response has not been mounted. IgM can only be detected on days 3–5 after the onset of the disease (Shu & Huang, 2004). Other than that, during the acute phase of DENV infection or primary Flavivirus infection, IgG is undetectable. However, IgG can be detected after 3 days of the onset of the disease during secondary infection (Kit Lam et al., 2000). Therefore, the IgM : IgG ratio can be used to differentiate primary from secondary infections of the disease. Fox et al. (2011) had stated that the IgM : IgG ratio, which is equal or above 1.8, is defined as primary DENV infection, while the ratio below 1.8 is defined as secondary DENV infection. It needs to be stated that it is not advisable to use IgM alone, as it lingers in the body for more 90 days. Diagnosis based on IgM alone is usually not confirmatory.
The DENV NS1 detection ELISA was first developed in 2000. NS1 is found both as membrane bound and in soluble forms and is highly conserved (Young et al., 2000). Importantly, NS1 protein is detectable earlier during the acute phase of both primary and secondary DENV infections (Libraty et al., 2002; Duyen et al., 2011). Alcon et al. (2002) and Shu et al. (2002) reported that the NS1 protein was detectable from day 1 to 8 after the onset of illness. Therefore, NS1 detection ELISA allows for the early serological diagnosis of virus infection. The commercially available NS1 detection ELISA kits such as the Pan-E Dengue Early ELISA from Panbio (Kit Pan-E) and Platelia Dengue NS1 Ag from Bio-Rad (Kit Platelia) were evaluated by several studies (Chuansumrit et al., 2008; Guzman et al., 2010; Lima et al., 2010; Pal et al., 2014; Hunsperger et al., 2014). Guzman et al. (2010); Lima et al. (2010) and Pal et al. (2014) had reported that the overall diagnostic sensitivity and specificity of Platelia was higher than that of Pan-E, and the sensitivity was better before day 4 after the onset of disease. However, the sensitivity for serotype DENV-2 and DENV-4 were lower in both studies. The inclusion of samples from YFV and JEV infections suggested that there was no cross-reaction among flaviviruses, while the lower specificity of kit Pan-E was due to the false-positive results in patients with JEV, YFV, and acute influenza infections (Guzman et al., 2010). Several studies had reported that the sensitivity of Platelia was lower in secondary infections in comparison to primary infections (Osorio et al., 2010; Duong et al., 2011; Pal et al., 2014) due to the production of anti-NS1 antibodies, which was detected more frequently during the DENV secondary infection (Koraka et al., 2003), and the formation of immune complexes impeded the detection of free NS1 protein (Young et al., 2000; Libraty et al., 2002). Duong et al. (2011) reported that overall sensitivity increased when the NS1 antigen assay is coupled with MAC-ELISA, which allows definitive (NS1) or presumptive (IgM) diagnosis during the acute and convalescent phase of the disease and can be used as a “point of care” diagnosis.
DENV and ZIKV share genetic and antigenic determinants (Breitbach et al., 2019), which cause difficulties in serological assays. Denis et al. (2019) observed that ZIKV EDIII, which is specifically recognized by anti-ZIKV IgG, is able to distinguish ZIKV from DENV in the late phase with high sensitivity, and thus, they developed and carried out a study on ZEDIII recognition by IgG with more than 5,000 serum samples. They found out that the ZEDIII protein sequence shares 46.3% and 47.2% identity and 64.8% homology with the protein sequences of DENV-2 EDIII and DENV-4 EDIII. Then, they tested the ability of ZEDIII-immunized mouse sera to detect the recombinant EDIII to further validate the high specificity of humoral immune response towards EDIII, and their results showed that the sera of ZEDIII-positive mice did not recognize DEN-4EDIII. They found out that the purified human DEN-2EDIII-IgG or DEN-4EDIII-IgG did not cross-react with ZEDIII, and the epitopes that are recognized by ZEDIII-IgG are different. Moreover, Sapparapu et al. (2016) had reported that the ZIKV EDIII antibodies tested by ELISA did not bind to WNV-EDIII or DEN-2EDIII. Denis et al. (2019) compared their ZEDIII-based ELISA with NS1-based ELISA and found out that the sensitivity and specificity of NS1-based ELISA was higher. It has been suggested that the ZEDIII could be used a safe model for the development of vaccines.
Hemagglutination-inhibition test
Hemagglutination inhibition test (HI) has been used in the diagnosis of DENV for many years since 1950 when Sabin (1950) discovered that arboviruses are able to agglutinate certain types of red blood cells. Traditionally, HI test is used to differentiate between the primary and secondary DENV infections (WHO, 1997). Some viruses cause hemagglutination, and this property is used in the HI test. The absence of hemagglutination indicates the presence of antibodies, as these would have bound to the virus and prevented hemagglutination. HI test can be used to detect the antibodies in the case of a known hemagglutinating virus. On the other hand, for the case of unknown viruses, a panel of known antibodies can be used to identify the virus. The HI test has the advantage of being easy to perform. However, the HI test might not detect the antibodies that are not cross-reactive to the viral subtype being tested; thus, all possible subtypes of a virus should be included during the HI test to effectively detect the antibodies, which is not always feasible (Bourgeois & Oaks, 2014).
Anantapreecha et al. (2007) had conducted a study in Thailand to analyze the level of DENV cross-reactivity of HI antibodies in primary DENV infections. A total number of 101 confirmed cases of DENV 1–4 were selected, and the plasma samples were obtained twice, once during the acute phase and once during the convalescent phase. Their results showed that the HI antibodies were cross-reactive among four DENV serotypes during both acute and convalescent phase in the primary infection, which indicated that the HI test is not the best method to differentiate different serotypes of DENV due to their cross-reactivity. Other than that, several studies reported that the HI test showed a low performance in distinguishing secondary DENV infection (Graham et al., 1999; Atchareeya et al., 2006; Lukman et al., 2016). It has been reported that the HI test is unable to differentiate among the flaviviruses DENV, JEV, and WNV (Nisalak, 2015). Several studies had compared the HI test with ELISA assays and stated that the ELISA assay was more reliable than the HI test for discrimination of primary and secondary DENV infection (Vaughn et al., 1999; Matheus et al., 2005; Atchareeya et al., 2006; De Souza et al., 2007). It is important to note that HI can still be used, but the criteria for interpretation needs to be carefully considered. Any titer below <80 should be classified as primary infection, and those between 60 and 640 of both the serum pairs must be used before a classification is made. Titers above >1,280 is always considered secondary and agreed upon by many others.
Western blot test
The Western blot (WB) test was first used to identify ribosomal RNA binding proteins in 1979 (Towbin et al., 1979) and is a commonly used technique in biomedical research. There are three major steps in WB test. The proteins are first separated according to their molecular mass, charge, and structure of protein using sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) technique. The separated proteins are then transferred and immobilized on to a solid support for the reaction with antibodies, which is a process known as blotting. Finally, the primary and enzyme-conjugated secondary antibodies are used for the detection of proteins. Once detected, the target protein will be visualized as a band on a blotting membrane, X-ray film, or an imaging system (Yang & Mahmood, 2012; Burrell et al., 2017). In comparison to other techniques, WB test has advantages in detecting and semi-quantifying target proteins, allowing the detection of a single target out of a mixture of thousand proteins, obtaining the molecular weight information of the target protein, and can be used as an effective diagnostic tool (Bertoni et al., 2012; Ghosh et al., 2014). The main disadvantage of WB test is that it can only be carried out if the primary antibody against the target protein is available (Gilda et al., 2015).
Hsieh et al. (2021) used the WB test with antigens of ZIKV, WNV, and DENV 1–4 to investigate the antibody responses. Their results showed that the anti-NS1 antibodies to WNV cross-reacted to one or two DENV serotypes, and the anti-DENV NS1 antibodies were unable to differentiate the infecting DENV serotypes in the WB test. Other than that, the anti-E antibodies cross-reacted among these six flaviviruses, while the anti-prM antibodies to DENV serotypes did not cross-react to WNV or ZIKV, which suggested that the anti-DENV prM antibodies can used to distinguish between primary ZIKV infection with previous DENV infection. Moreover, they noted that during the detection of anti-NS1 antibodies, the WB test was more sensitive but less specific as compared to ELISA. This is due to the NS1 protein being present as a hexamer in the solution while NS1 protein is present as a monomer under detergent treatment in WB test, and it is likely that anti-NS1 antibodies can only recognize linear detergent-treated NS1 in ELISA but not recombinant NS1 hexamers, which then reduced the cross-reactivity in ELISA (Ocequera et al., 2007).
Immunofluorescence test
The indirect immunofluorescence assay (IFA) is a standard virological approach for the detection of antibodies based on their ability to react specifically with viral antigens produced in the infected cells, which are fixed to the wells on a glass microscope slide. Indirect IFA is a two-step assay where the primary unlabeled antibodies in the diluted serum samples will attach to the antigens followed by a fluorophore-labeled secondary anti-human antibody to detect the primary antibody, which then can be visualized using a fluorescence microscope (Sandstrom et al., 1985; Hedenskog et al., 1986; Odell & Cook, 2013). The main advantage of indirect IFA is because the secondary antibody can be used with different primary antibodies, and it is not necessary to conjugate each new antibody individually (Goding, 1996). Furthermore, by combining multiple primary antibodies with specific secondary antibodies (which are tagged with different fluorophore), specific visualization of several antigens can be done simultaneously in one sample (multicolor immunofluorescence). The limitation of the immunofluorescence assay is that the fluorescence signals from the assays are dependent on the concentration and quality of antibody, the appropriate secondary antibodies during the detection, and proper handling of the specimen (Odell & Cook, 2013). The disadvantages of indirect IFA include potential cross-reactivity and the requirement for the primary antibodies that are not generated in the same species or of distinct isotypes when performing multiple-labeling assays (Robinson et al., 2009). An immunofluorescence-based NS1 antigen determination using fluorescein isothiocyanate (FITC) conjugated to IgG antibody was developed and showed a coefficient of determination (R2) of 0.92, with a high reproducibility and stability and a low detection limit (LOD) at 15 ng ml−1 (Darwish et al., 2018). This optical immunosensor was capable of detecting NS1 analytes in plasma specimens from patients infected with the dengue virus, with low cross-reaction with plasma specimens containing JEV and Zika virus. The major limitation in this assay is that cross-reactivity regarding NS1 specificity was not conducted. The main issues with immunofluorescence assays is with regard to the degradation of fluorochromes, antigens that are not purified adequately, its usage on fixed cells, and also the cost and expertise necessary for this assay.
Currently, indirect IFA is widely used in scientific research rather than for clinical diagnostic purposes. Despite this, IFA was utilized for diagnostic purposes in the early days when ELISA kits were not yet commercially accessible by using patient serum as the primary antibody (Ryu, 2017). IFA have been used for the serodiagnosis of WNV infection (Besselaar et al., 1989), DENV infection (Boopucknavig et al., 1975), and YFV infection (Monath et al., 1981; Niedrig et al., 1999). Research on the evaluation of indirect IFA for the serological diagnosis of DENV in a population with high prevalence of arboviruses was carried out by Arai et al. (2019). They concluded that although the performance of indirect IFA was acceptable, however, for clinical diagnosis of acute infection to detect the IgM antibodies, ELISA alone is sufficient for serological diagnosis. Replacing ELISA with indirect IFA would compromise the sensitivity for IgM and might increase the number of false-negative samples for IgM.
Conclusion
Despite recent the improvements in Flavivirus-specific vaccines, the global burden of Flavivirus-associated human diseases is increasing and its area of distribution is expanding. The need for a rapid serological assay that has high sensitivity and specificity is emphasized by the fact that cross-reactive immunity influences the outcome of Flavivirus infections. The continued spread of flaviviruses worldwide has resulted in changes in the immune profile, which can change over time, further emphasizing specific diagnosis. Due to the large overlap in clinical disease, accompanied by co-circulation of different flaviviruses, cross-reactivity, and poor access to advance laboratory diagnosis tools for serological confirmation, serological diagnosis of Flavivirus infections is a great challenge. The laboratory diagnosis of flaviviruses infection has greatly improved during the last decade. However, serum samples from flaviviruses-infected patients show varying degrees of cross-reactivity to each other when conducting serological diagnosis. Identifying a single antigen with the immunoassay that provides high sensitivity and specificity is one of the difficulties during the diagnosis of Flavivirus. Neutralization test is recommended in this review paper for the correct diagnosis of Flavivirus of which DENV serotypes showed the least cross-reactivity by using PRNT, and it remains the most common assay used and recommended as the gold standard assay.
Author contributions
All authors were involved in preparation and writing of the manuscript with CR. Each provided excerpts and comments to all others. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Ministry of Higher Education (MOHE), Fundamental Research Grant Scheme (FRGS) FRGS/1/2021/SKK06/UCSI/01/1
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Abbink P., Larocca R. A., Dejnirattisai W., Peterson R., Nkolola J. P., Borducchi E. N., et al. (2018). Therapeutic and protective efficacy of a dengue antibody against zika infection in rhesus monkeys. Nat. Med. 24 (6), 721–723. doi: 10.1038/s41591-018-0056-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcon S., Talarmin A., Debruyne M., Falconar A., Deubel V., Flamand M. (2002). Enzyme-linked immunosorbent assay specific to dengue virus type 1 nonstructural protein NS1 reveals circulation of the antigen in the blood during the acute phase of disease in patients experiencing primary or secondary infections. J. Clin. Microbiol. 40 (2), 376–381. doi: 10.1128/jcm.40.02.376-381.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allwinn R., Doerr H., Emmerich P., Schmitz H., Preiser W. (2002). Cross-reactivity in flavivirus serology: new implications of an old finding? Med. Microbiol. Immunol. 190 (4), 199–202. doi: 10.1007/s00430-001-0107-9 [DOI] [PubMed] [Google Scholar]
- Anantapreecha S., A-Nuegoonpipat A., Prakrong S., Chanama S., Sa-Ngasang A., Sawanpanyalert P., et al. (2007). Dengue virus cross-reactive hemagglutination inhibition antibody responses in patients with primary dengue virus infection. Japanese J. Infect. Dis. 60 (5), 267–270. [PubMed] [Google Scholar]
- Anderson K. B., Gibbons R. V., Thomas S. J., Rothman A. L., Nisalak A., Berkelman R. L., et al. (2011). Preexisting Japanese encephalitis virus neutralizing antibodies and increased symptomatic dengue illness in a school-based cohort in Thailand. PLoS Negl. Trop. Dis. 5 (10), e1311. doi: 10.1371/journal.pntd.0001311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai K. E., Bo C. R., Silva A. P., Rodrigues S. S., Mangueira C. L. (2019). Performance evaluation of an indirect immunofluorescence kit for the serological diagnosis of dengue. Einstein (São Paulo) 18, eAO5078. doi: 10.31744/einstein_journal/2020ao5078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argondizzo A. P., Silva D., Missailidis S. (2020). Application of aptamer-based assays to the diagnosis of arboviruses important for public health in Brazil. Int. J. Mol. Sci. 22 (1), 159. doi: 10.3390/ijms22010159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atchareeya A., Songthum P., Areerat S., Sumalee C., Pathon S., Ichiro K, et al. (2006). “Comparison between haemagglutination inhibition (HI) test and IgM and IgG-capture ELISA in determination of primary and secondary dengue virus infections”. Dengue Bulletin 30, 141–145. Available at: https://apps.who.int/iris/handle/10665/170232 [Google Scholar]
- Bardina S. V., Bunduc P., Tripathi S., Duehr J., Frere J. J., Brown J. A., et al. (2017). Enhancement of zika virus pathogenesis by preexisting antiflavivirus immunity. Science 356 (6334), 175–180. doi: 10.1126/science.aal4365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltramello M., Williams K. L., Simmons C. P., Macagno A., Simonelli L., Quyen N., et al. (2010). The human immune response to dengue virus is dominated by highly cross-reactive antibodies endowed with neutralizing and enhancing activity. Cell Host Microbe 8 (3), 271–283. doi: 10.1016/j.chom.2010.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertoni T. A., Perenha-Viana M. C., Patussi E. V., Cardoso R. F., Svidzinski T. I. (2012). Western Blotting is an efficient tool for differential diagnosis of paracoccidioidomycosis and pulmonary tuberculosis. Clin. Vaccine Immunol. 19 (11), 1887–1888. doi: 10.1128/cvi.00252-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besselaar T., Blackburn N., Aldridge N. (1989). Comparison of an antibody-capture IGM enzymelinked immunosorbent assay with IGM-indirect immunofluorescence for the diagnosis of acute sindbis and West Nile infections. J. Virological Methods 25 (3), 337–345. doi: 10.1016/0166-0934(89)90060-8 [DOI] [PubMed] [Google Scholar]
- Bhatt S., Gething P., Brady O., Messina J., Farlow A., Moyes C., et al. (2013). The global distribution and burden of dengue. Nature 496 (7446), 504–507. doi: 10.1038/nature12060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonpucknavig S., Vuttivirojana O., Siripont J., Futrakul P., Nimmannitya S. (1975). Indirect fluorescent antibody technic for demonstration of serum antibody in dengue hemorrhagic fever cases. Am. J. Clin. Pathol. 64 (3), 365–371. doi: 10.1093/ajcp/64.3.365 [DOI] [PubMed] [Google Scholar]
- Bourgeois M. A., Oaks J. L. (2014). Laboratory diagnosis of viral infections. Equine Infect. Dis. 132–140.e2. doi: 10.1016/b978-1-4557-0891-8.00012-9 [DOI] [Google Scholar]
- Breitbach M. E., Newman C. M., Dudley D. M., Stewart L. M., Aliota M. T., Koenig M. R., et al. (2019). Primary infection with dengue or zika virus does not affect the severity of heterologous secondary infection in macaques. PLoS Pathog. 15 (8), e1007766. doi: 10.1371/journal.ppat.1007766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrell C. J., Howard C. R., Murphy F. A. (2017). Laboratory diagnosis of virus diseases. Fenner White’s Med. Virol. 135–154. doi: 10.1016/b978-0-12-375156-0.00010-2 [DOI] [Google Scholar]
- Calisher C. H., Karabatsos N., Dalrymple J. M., Shope R. E., Porterfield J. S., Westaway E. G., et al. (1989). Antigenic relationships between flaviviruses as determined by cross-neutralization tests with polyclonal antisera. J. Gen. Virol. 70 (1), 37–43. doi: 10.1099/0022-1317-70-1-37 [DOI] [PubMed] [Google Scholar]
- Casals J. (1957). Section of biology and division of mycology*: VIRUSES: The versatile parasites: I. @ the arthropod-borne group of animal viruses**. Trans. New York Acad. Sci. 19 (3 Series II), 219–235. doi: 10.1111/j.2164-0947.1957.tb00526.x [DOI] [PubMed] [Google Scholar]
- Casals J., Brown L. V. (1954). Hemagglutination with arthropod-borne viruses. J. Exp. Med. 99 (5), 429–449. doi: 10.1084/jem.99.5.429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty S. (2016). Computational analysis of perturbations in the post-fusion dengue virus envelope protein highlights known epitopes and conserved residues in the zika virus. F1000Research 5, 1150. doi: 10.12688/f1000research.8853.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H., Huber R. G., Bond P. J., Grad Y. H., Camerini D., Maurer-Stroh S., et al. (2017). Systematic analysis of protein identity between zika virus and other arthropod-borne viruses. Bull. World Health Organ. 95 (7), 517–525. doi: 10.2471/blt.16.182105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chávez J., Silva J., Amarilla A., Moraes Figueiredo L. (2010). Domain III peptides from flavivirus envelope protein are useful antigens for serologic diagnosis and targets for immunization. Biologicals 38 (6), 613–618. doi: 10.1016/j.biologicals.2010.07.004 [DOI] [PubMed] [Google Scholar]
- Chiou S., Fan Y., Crill W., Chang R., Chang G. (2012). Mutation analysis of the cross-reactive epitopes of Japanese encephalitis virus envelope glycoprotein. J Gen Virology 93 (6), 1185–1192. doi: 10.1099/vir.0.040238-0 [DOI] [PubMed] [Google Scholar]
- Chong H. Y., Leow C. Y., Abdul Majeed A. B., Leow C. H. (2019). Flavivirus infection–a review of immunopathogenesis, immunological response, and immunodiagnosis. Virus Res. 274, 197770. doi: 10.1016/j.virusres.2019.197770 [DOI] [PubMed] [Google Scholar]
- Chuansumrit A., Chaiyaratana W., Pongthanapisith V., Tangnararatchakit K., Lertwongrath S., Yoksan S. (2008). The use of dengue nonstructural protein 1 antigen for the early diagnosis during the febrile stage in patients with dengue infection. Pediatr. Infect. Dis. J. 27 (1), 43–48. doi: 10.1097/inf.0b013e318150666d [DOI] [PubMed] [Google Scholar]
- Collins M. H., McGowan E., Jadi R., Young E., Lopez C. A., Baric R. S., et al. (2017). Lack of durable cross-neutralizing antibodies against zika virus from dengue virus infection. Emerging Infect. Dis. 23 (5), 773–781. doi: 10.3201/eid2305.161630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crill W. D., Chang G. J. J. (2004). Localization and characterization of flavivirus envelope glycoprotein cross-reactive epitopes. J. Virology. 78 (24), 13975–13986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daep C., Muñoz-Jordán J., Eugenin E. (2014). Flaviviruses, an expanding threat in public health: focus on dengue, West Nile, and Japanese encephalitis virus. J. Neurovirol. 20 (6), 539–560. doi: 10.1007/s13365-014-0285-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejnirattisai W., Wongwiwat W., Supasa S., Zhang X., Dai X., Rouvinski A., et al. (2014). A new class of highly potent, broadly neutralizing antibodies isolated from viremic patients infected with dengue virus. Nat. Immunol. 16 (2), 170–177. doi: 10.1038/ni.3058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y., Dai J., Ji G., Jiang T., Wang H., Yang H., et al. (2011). A broadly flavivirus cross-neutralizing monoclonal antibody that recognizes a novel epitope within the fusion loop of e protein. PLoS One 6 (1), e16059. doi: 10.1371/journal.pone.0016059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denis J., Attoumani S., Gravier P., Tenebray B., Garnier A., Briolant S., et al. (2019). High specificity and sensitivity of zika EDIII-based Elisa diagnosis highlighted by a large human reference panel. PLoS Negl. Trop. Dis. 13 (9), e0007747. doi: 10.1371/journal.pntd.0007747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Souza V. A., Tateno A. F., Oliveira R. R., Domingues R. B., Araújo E. S., Kuster G. W., et al. (2007). Sensitivity and specificity of three ELISA-based assays for discriminating primary from secondary acute dengue virus infection. J. Clin. Virol. 39 (3), 230–233. doi: 10.1016/j.jcv.2007.04.005 [DOI] [PubMed] [Google Scholar]
- Duong V., Ly S., Lorn Try P., Tuiskunen A., Ong S., Chroeung N., et al. (2011). Clinical and virological factors influencing the performance of a NS1 antigen-capture assay and potential use as a marker of dengue disease severity. PLoS Negl. Trop. Dis. 5 (7), e1244. doi: 10.1371/journal.pntd.0001244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duyen H. T., Ngoc T. V., Ha D. T., Hang V. T., Kieu N. T., Young P. R., et al. (2011). Kinetics of plasma viremia and soluble nonstructural protein 1 concentrations in dengue: Differential effects according to serotype and immune status. J. Infect. Dis. 203 (9), 1292–1300. doi: 10.1093/infdis/jir014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagbami A. H., Halstead S. B., Marchette N. J., Larsen K. (1987). Cross-infection enhancement among African flaviviruses by immune mouse ascitic fluids. Cytobios 49 (196), 49–55. [PubMed] [Google Scholar]
- Fox A., Hoa L. N., Simmons C. P., Wolbers M., Wertheim H. F., Khuong P. T., et al. (2011). Immunological and viral determinants of dengue severity in hospitalized adults in ha noi, Viet nam. PLoS Negl. Trop. Dis. 5 (3), e967. doi: 10.1371/journal.pntd.0000967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- George J., Valiant W. G., Mattapallil M. J., Walker M., Huang Y. S., Vanlandingham D. L., et al. (2017). Prior exposure to zika virus significantly enhances peak dengue-2 viremia in rhesus macaques. Sci. Rep. 7 (1), 1–10. doi: 10.1038/s41598-017-10901-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh R., Gilda J. E., Gomes A. V. (2014). The necessity of and strategies for improving confidence in the accuracy of western blots. Expert Rev. Proteomics 11 (5), 549–560. doi: 10.1586/14789450.2014.939635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilda J. E., Ghosh R., Cheah J. X., West T. M., Bodine S. C., Gomes A. V. (2015). Western Blotting inaccuracies with unverified antibodies: Need for a western blotting minimal reporting standard (WBMRS). PLoS One 10 (8), e0135392. doi: 10.1371/journal.pone.0135392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giron S., Franke F., Decoppet A., Cadiou B., Travaglini T., Thirion L., et al. (2019). Vector-borne transmission of zika virus in Europe, southern France, august 2019. Eurosurveillance 24 (45), 1900655. doi: 10.2807/1560-7917.es.2019.24.45.1900655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goding J. W. (1996). Immunofluorescence. Monoclonal Antibodies, 352–399. doi: 10.1016/b978-012287023-1/50060-2 [DOI] [Google Scholar]
- Goncalves A., Peeling R. W., Chu M. C., Gubler D. J., De Silva A. M., Harris E., et al. (2017). Innovative and new approaches to laboratory diagnosis of zika and dengue: A meeting report. J. Infect. Dis. 217 (7), 1060–1068. doi: 10.1093/infdis/jix678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould E., Solomon T. (2008). Pathogenic flaviviruses. Lancet 371 (9611), 500–509. doi: 10.1016/s0140-6736(08)60238-x [DOI] [PubMed] [Google Scholar]
- Graham R. R., Laksono I., Porter K. R., Hayes C. G., Tan R., Halstead S. B. (1999). A prospective seroepidemiologic study on dengue in children four to nine years of age in yogyakarta, Indonesia i. studies in 1995-1996. Am. J. Trop. Med. Hygiene 61 (3), 412–419. doi: 10.4269/ajtmh.1999.61.412 [DOI] [PubMed] [Google Scholar]
- Gubler D. J., Kuno G., Markoff L. (2007). “Flaviviruses,” in Fields virology. 5th ed. Eds. Knipe D. M., Howley P. M., Griffin D. E., Lamb R. A., Martin M. A., Roizman B., Straus S. E. (Philadelphia, PA: Lippincott Williams & Wilkins; ), 1153–1252. [Google Scholar]
- Guy B., Guirakhoo F., Barban V., Higgs S., Monath T. P., Lang J. (2010). Preclinical and clinical development of YFV 17D-based chimeric vaccines against dengue, West Nile and Japanese encephalitis viruses. Vaccine 28 (3), 632–649. doi: 10.1016/j.vaccine.2009.09.098 [DOI] [PubMed] [Google Scholar]
- Guzman M. G., Jaenisch T., Gaczkowski R., Ty Hang V. T., Sekaran S. D., Kroeger A., et al. (2010). Multi-country evaluation of the sensitivity and specificity of two commercially-available NS1 ELISA assays for dengue diagnosis. PLoS Negl. Trop. Dis. 4 (8), e811. doi: 10.1371/journal.pntd.0000811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halai U., Nielsen-Saines K., Moreira M. L., De Sequeira P. C., Junior J. P., De Araujo Zin A., et al. (2017). Maternal zika virus disease severity, virus load, prior dengue antibodies, and their relationship to birth outcomes. Clin. Infect. Dis. 65 (6), 877–883. doi: 10.1093/cid/cix472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halstead S. B., Porterfield J. S., O’Rourke E. J. (1980). Enhancement of dengue virus infection in monocytes by flavivirus antisera *. Am. J. Trop. Med. Hygiene 29 (4), 638–642. doi: 10.4269/ajtmh.1980.29.638 [DOI] [PubMed] [Google Scholar]
- Hamer D., Angelo K., Caumes E., van Genderen P., Florescu S., Popescu C., et al. (2018). Fatal yellow fever in travelers to brazil 2018 MMWR. Morbid. Mortality Weekly Rep. 67 (11), 340–341. doi: 10.15585/mmwr.mm6711e1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan S. S., Miller A., Sapparapu G., Fernandez E., Klose T., Long F., et al. (2017). A human antibody against zika virus crosslinks the e protein to prevent infection. Nat. Commun. 8 (1), 1–6. doi: 10.1038/ncomms14722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedenskog M., Dewhurst S., Ludvigsen C., Sinangil F., Rodriguez L., Wu Y., et al. (1986). Testing for antibodies to AIDS-associated retrovirus (HTLV-III/LAV) by indirect fixed cell immunofluorescence: Specificity, sensitivity, and applications. J. Med. Virol. 19 (4), 325–334. doi: 10.1002/jmv.1890190405 [DOI] [PubMed] [Google Scholar]
- Hobson-Peters J. (2012). Approaches for the development of rapid serological assays for surveillance and diagnosis of infections caused by zoonotic flaviviruses of the Japanese encephalitis virus serocomplex. J. Biomedicine Biotechnol. 2012, 1–15. doi: 10.1155/2012/379738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh S., Tsai W., Tsai J., Stone M., Simmons G., Busch M. P., et al. (2021). Identification of anti-premembrane antibody as a serocomplex-specific marker to discriminate zika, dengue, and West Nile virus infections. J. Virol. 95 (19), e00619–21. doi: 10.1128/jvi.00619-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Higgs S., Horne K., Vanlandingham D. (2014). Flavivirus-mosquito interactions. Viruses 6 (11), 4703–4730. doi: 10.3390/v6114703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunsperger E. A., Yoksan S., Buchy P., Nguyen V. C., Sekaran S. D., Enria D. A., et al. (2014). Evaluation of commercially available diagnostic tests for the detection of dengue virus NS1 antigen and anti-dengue virus IGM antibody. PLoS Negl. Trop. Dis. 8 (10), e3171. doi: 10.1371/journal.pntd.0003171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson B. (2016). Neurotropic flaviviruses. Neurotropic Viral Infections, 229–258. doi: 10.1007/978-3-319-33133-1_9 [DOI] [Google Scholar]
- Kaufmann B., Vogt M. R., Goudsmit J., Holdaway H. A., Aksyuk A. A., Chipman P. R., et al. (2010). Neutralization of west nile virus by cross-linking of its surface proteins with fab fragments of the human monoclonal antibody CR4354. Proc. Natl. Acad. Sci. 107 (44), 18950–18955. doi: 10.1073/pnas.1011036107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khawar W., Bromberg R., Moor M., Lyubynska N., Mahmoudi H. (2017). Seven cases of zika virus infection in south Florida. Cureus 9 (3), e1099. doi: 10.7759/cureus.1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kit Lam S., Lan Ew C., Mitchell J. L., Cuzzubbo A. J., Devine P. L. (2000). Evaluation of a capture screening enzyme-linked immunosorbent assay for combined determination of immunoglobulin m and G antibodies produced during dengue infection. Clin. Diagn. Lab. Immunol. 7 (5), 850–852. doi: 10.1128/cdli.7.5.850-852.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koraka P., Burghoorn-Maas C. P., Falconar A., Setiati T. E., Djamiatun K., Groen J., et al. (2003). Detection of immune-complex-dissociated nonstructural-1 antigen in patients with acute dengue virus infections. J. Clin. Microbiol. 41 (9), 4154–4159. doi: 10.1128/jcm.41.9.4154-4159.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraemer M., Faria N., Reiner R., Golding N., Nikolay B., Stasse S., et al. (2017). Spread of yellow fever virus outbreak in Angola and the democratic republic of the Congo 2015–16: a modelling study. Lancet Infect. Dis. 17 (3), 330–338. doi: 10.1016/s1473-3099(16)30513-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuno G., Chang G. J., Tsuchiya K. R., Karabatsos N., Cropp C. B. (1998). Phylogeny of the genus flavivirus . J. Virol. 72 (1), 73–83. doi: 10.1128/jvi.72.1.73-83.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyle J., Harris E. (2008). Global spread and persistence of dengue. Annu. Rev. Microbiol. 62 (1), 71–92. doi: 10.1146/annurev.micro.62.081307.163005 [DOI] [PubMed] [Google Scholar]
- Li C., Bai X., Meng R., Shaozhou W., Zhang Q., Hua R., et al. (2016). Identification of a new broadly cross-reactive epitope within domain III of the duck tembusu virus e protein. Sci. Rep. 6 (1), 1–8. doi: 10.1038/srep36288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libraty D., Young P., Pickering D., Endy T., Kalayanarooj S., Green S., et al. (2002). High circulating levels of the dengue virus nonstructural protein NS1 early in dengue illness correlate with the development of dengue hemorrhagic fever. J. Infect. Dis. 186 (8), 1165–1168. doi: 10.1086/343813 [DOI] [PubMed] [Google Scholar]
- Li W., Cao S., Zhang Q., Li J., Zhang S., Wu W., et al. (2017). Comparison of serological assays to titrate hantaan and Seoul hantavirus-specific antibodies. Virol. J. 14 (1), 1–11.. doi: 10.1186/s12985-017-0799-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima M. D., Nogueira R. M., Schatzmayr H. G., Santos F. B. (2010). Comparison of three commercially available dengue NS1 antigen capture assays for acute diagnosis of dengue in Brazil. PLoS Negl. Trop. Dis. 4 (7), e767. doi: 10.1371/journal.pntd.0000738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisova O., Hardy F., Petit V., Bedouelle H. (2007). Mapping to completeness and transplantation of a group-specific, discontinuous, neutralizing epitope in the envelope protein of dengue virus. J Gen Virology 88 (9), 2387–2397 [DOI] [PubMed] [Google Scholar]
- Lukman N., Salim G., Kosasih H., Susanto N. H., Parwati I., Fitri S., et al. (2016). Comparison of the hemagglutination inhibition test and IGG Elisa in categorizing primary and secondary dengue infections based on the plaque reduction neutralization test. BioMed. Res. Int. 2016, 1–5. doi: 10.1155/2016/5253842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda A., Maeda J. (2013). Review of diagnostic plaque reduction neutralization tests for flavivirus infection. Veterinary J. 195 (1), 33–40. doi: 10.1016/j.tvjl.2012.08.019 [DOI] [PubMed] [Google Scholar]
- Mansfield K., Horton D., Johnson N., Li L., Barrett A., Smith D., et al. (2011). Flavivirus-induced antibody cross-reactivity. J. Gen. Virol. 92 (12), 2821–2829. doi: 10.1099/vir.0.031641-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathengtheng L., Burt F. J. (2014). Use of envelope domain III protein for detection and differentiation of flaviviruses in the free state province, south Africa. Vector-Borne Zoonotic Dis. 14 (4), 261–271. doi: 10.1089/vbz.2013.1407 [DOI] [PubMed] [Google Scholar]
- Matheus S., Deparis X., Labeau B., Lelarge J., Morvan J., Dussart P. (2005). Discrimination between primary and secondary dengue virus infection by an immunoglobulin G avidity test using a single acute-phase serum sample. J. Clin. Microbiol. 43 (6), 2793–2797. doi: 10.1128/jcm.43.6.2793-2797.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui K., Gromowski G., Li L., Schuh A., Lee J., Barrett A. (2009). Characterization of dengue complex-reactive epitopes on dengue 3 virus envelope protein domain III. Virology 384 (1), 16–20. doi: 10.1016/j.virol.2008.11.013 [DOI] [PubMed] [Google Scholar]
- Matsui K., Gromowski G., Li L., Barrett A. (2010). Characterization of a dengue type-specific epitope on dengue 3 virus envelope protein domain III. J. Gen. Virol. 91 (9), 2249–2253. doi: 10.1099/vir.0.021220-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazeaud C., Freppel W., Chatel-Chaix L. (2018). The multiples fates of the flavivirus RNA genome during pathogenesis. Front. Genet. 9. doi: 10.3389/fgene.2018.00595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken M. K., Gromowski G. D., Friberg H. L., Lin X., Abbink P., de la Barrera R., et al. (2017). Impact of prior flavivirus immunity on zika virus infection in rhesus macaques. PLoS Pathog. 13 (8), e1006487. doi: 10.1371/journal.ppat.1006487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monath T., Cropp C., Muth D., Calisher C. (1981). Indirect fluorescent antibody test for the diagnosis of yellow fever. Trans. R. Soc. Trop. Med. Hygiene 75 (2), 282–286. doi: 10.1016/0035-9203(81)90335-7 [DOI] [PubMed] [Google Scholar]
- Musso D., Desprès P. (2020). Serological diagnosis of flavivirus-associated human infections. Diagnostics 10 (5), 302. doi: 10.3390/diagnostics10050302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash D., Mostashari F., Fine A., Miller J., O’Leary D., Murray K., et al. (2001). The outbreak of West Nile virus infection in the new York city area in 1999. New Engl. J. Med. 344 (24), 1807–1814. doi: 10.1056/nejm200106143442401 [DOI] [PubMed] [Google Scholar]
- Niedrig M., Lademann M., Emmerich P., Lafrenz M. (1999). Assessment of IGG antibodies against yellow fever virus after vaccination with 17D by different assays: Neutralization test, haemagglutination inhibition test, immunofluorescence assay and Elisa. Trop. Med. Int. Health 4 (12), 867–871. doi: 10.1046/j.1365-3156.1999.00496.x [DOI] [PubMed] [Google Scholar]
- Nisalak A. (2015). LABORATORY DIAGNOSIS OF DENGUE VIRUS INFECTIONS. Southeast Asian J. Trop. Med. Public Health 46 Suppl 1, 55–76. [PubMed] [Google Scholar]
- Nurtop E., Villarroel P. M., Pastorino B., Ninove L., Drexler J. F., Roca Y., et al. (2018). Combination of elisa screening and seroneutralisation tests to expedite zika virus seroprevalence studies. Virol. J. 15 (1), 1–11. doi: 10.1186/s12985-018-1105-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oceguera L. F., 3rd, Patiris P. J., Chiles R. E., Busch M. P., Tobler L. H., Hanson C. V. (2007). Flavivirus serology by Western blot analysis. Am. J. Trop. Med. Hyg. 77 (1), 159–163. [PubMed] [Google Scholar]
- Odell I. D., Cook D. (2013). Immunofluorescence techniques. J. Invest. Dermatol. 133 (1), 1–4. doi: 10.1038/jid.2012.455 [DOI] [PubMed] [Google Scholar]
- Osorio L., Ramirez M., Bonelo A., Villar L. A., Parra B. (2010). Comparison of the diagnostic accuracy of commercial NS1-based diagnostic tests for early dengue infection. Virol. J. 7 (1), 1–10. doi: 10.1186/1743-422x-7-361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal S., Dauner A. L., Mitra I., Forshey B. M., Garcia P., Morrison A. C., et al. (2014). Evaluation of dengue NS1 antigen rapid tests and Elisa kits using clinical samples. PLoS One 9 (11), e113411. doi: 10.1371/journal.pone.0113411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandit P. S., Doyle M. M., Smart K. M., Young C. C., Drape G. W., Johnson C. K. (2018). Predicting wildlife reservoirs and global vulnerability to zoonotic flaviviruses. Nat. Commun. 9 (1), 5425. doi: 10.1038/s41467-018-07896-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papa A., Karabaxoglou D., Kansouzidou A. (2011). Acute West Nile virus neuroinvasive infections: Cross-reactivity with dengue virus and tick-borne encephalitis virus. J. Med. Virol. 83 (10), 1861–1865. doi: 10.1002/jmv.22180 [DOI] [PubMed] [Google Scholar]
- Pierson T. C., Fremont D. H., Kuhn R. J., Diamond M. S. (2008). Structural insights into the mechanisms of antibody-mediated neutralization of flavivirus infection: Implications for vaccine development. Cell Host Microbe 4 (3), 229–238. doi: 10.1016/j.chom.2008.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierson T., Kielian M. (2013). Flaviviruses: braking the entering. Curr. Opin. In Virol. 3 (1), 3–12. doi: 10.1016/j.coviro.2012.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotkin S. A. (2010). Correlates of protection induced by vaccination. Clin. Vaccine Immunol. 17 (7), 1055–1065. doi: 10.1128/cvi.00131-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priyamvada L., Quicke K., Hudson W., Onlamoon N., Sewatanon J., Edupuganti S., et al. (2016). Human antibody responses after dengue virus infection are highly cross-reactive to zika virus. Proc. Natl. Acad. Sci. 113 (28), 7852–7857. doi: 10.1073/pnas.1607931113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X., Lei Y., Yang P., Gao Q., Wang N., Cao L., et al. (2018). Structural basis for neutralization of Japanese encephalitis virus by two potent therapeutic antibodies. Nat. Microbiol. 3 (3), 287–294. doi: 10.1038/s41564-017-0099-x [DOI] [PubMed] [Google Scholar]
- Rainwater-Lovett K., Rodriguez-Barraquer I., Cummings D. A., Lessler J. (2012). Variation in dengue virus plaque reduction neutralization testing: Systematic review and pooled analysis. BMC Infect. Dis. 12 (1), 1–15. doi: 10.1186/1471-2334-12-233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathore A. P., Saron W. A., Lim T., Jahan N., St. John A. L. (2019). Maternal immunity and antibodies to dengue virus promote infection and zika virus–induced microcephaly in fetuses. Sci. Adv. 5 (2), eaav3208. doi: 10.1126/sciadv.aav3208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathore A. P., St. John A. L. (2020). Cross-reactive immunity among flaviviruses. Front. Immunol. 11. doi: 10.3389/fimmu.2020.00334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey F. A., Stiasny K., Vaney M., Dellarole M., Heinz F. X. (2017). The bright and the dark side of human antibody responses to flaviviruses: Lessons for vaccine design. EMBO Rep. 19 (2), 206–224. doi: 10.15252/embr.201745302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbiani D. F., Bozzacco L., Keeffe J. R., Khouri R., Olsen P. C., Gazumyan A., et al. (2017). Recurrent potent human neutralizing antibodies to zika virus in Brazil and Mexico. Cell 169 (4), 597–609.e11. doi: 10.1016/j.cell.2017.04.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J. P., Sturgis J., Kumar G. L. (2009). “Chapter 10 | immunofluorescence,” in IHC staining methods fifth edition, 61–65. [Google Scholar]
- Roehrig J. T., Hombach J., Barrett A. D. (2008). Guidelines for plaque-reduction neutralization testing of human antibodies to dengue viruses. Viral Immunol. 21 (2), 123–132. doi: 10.1089/vim.2008.0007 [DOI] [PubMed] [Google Scholar]
- Rouvinski A., Guardado-Calvo P., Barba-Spaeth G., Duquerroy S., Vaney M., Kikuti C. M., et al. (2015). Recognition determinants of broadly neutralizing human antibodies against dengue viruses. Nature 520 (7545), 109–113. doi: 10.1038/nature14130 [DOI] [PubMed] [Google Scholar]
- Russell P. K., Nisalak A. (1967). Dengue virus identification by the plaque reduction neutralization test. J. Immunol. (Baltimore Md.: 1950) 99 (2), 291–296. [PubMed] [Google Scholar]
- Ryu W. (2017). Diagnosis and methods. Mol. Virol. Hum. Pathogenic Viruses, 47–62. doi: 10.1016/b978-0-12-800838-6.00004-7 [DOI] [Google Scholar]
- Sabin A. B. (1950). The dengue group of viruses and its family relationships. Bacteriological Rev. 14 (3), 225–232. doi: 10.1128/br.14.3.193-258.1950 [DOI] [PubMed] [Google Scholar]
- Sabin A. B. (1952). Research on dengue during world war II 1. Am. J. Trop. Med. Hygiene 1 (1), 30–50. doi: 10.4269/ajtmh.1952.1.30 [DOI] [PubMed] [Google Scholar]
- Saito Y., Moi M. L., Takeshita N., Lim C., Shiba H., Hosono K., et al. (2016). Japanese Encephalitis vaccine-facilitated dengue virus infection-enhancement antibody in adults. BMC Infect. Dis. 16 (1), 1–11. doi: 10.1186/s12879-016-1873-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto S., Putalun W., Vimolmangkang S., Phoolcharoen W., Shoyama Y., Tanaka H., et al. (2017). Enzyme-linked immunosorbent assay for the quantitative/qualitative analysis of plant secondary metabolites. J. Natural Medicines 72 (1), 32–42. doi: 10.1007/s11418-017-1144-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel Sulca Herencia J. (2019). Laboratory tests used in the diagnostic and research of dengue virus: Present and future. Dengue Fever - Resilient Threat Face Innovation. doi: 10.5772/intechopen.80519 [DOI] [Google Scholar]
- Sandstrom E., Schooley R., Ho D., Byington R., Sarngadharan M., MacLane M., et al. (1985). Detection of human anti-HTLV-III antibodies by indirect immunofluorescence using fixed cells. Transfusion 25 (4), 308–312. doi: 10.1046/j.1537-2995.1985.25485273806.x [DOI] [PubMed] [Google Scholar]
- Sapparapu G., Fernandez E., Kose N., Cao B., Fox J. M., Bombardi R. G., et al. (2016). Neutralizing human antibodies prevent zika virus replication and fetal disease in mice. Nature 540 (7633), 443–447. doi: 10.1038/nature20564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saron W. A., Rathore A. P., Ting L., Ooi E. E., Low J., Abraham S. N., et al. (2018). Flavivirus serocomplex cross-reactive immunity is protective by activating heterologous memory CD4 T cells. Sci. Adv. 4 (7), eaar4297. doi: 10.1126/sciadv.aar4297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu P. Y., Chen L. K., Chang S. F., Yueh Y. Y., Chow L., Chien L. J, et al. (2002). Potential application of nonstructural protein NS1 serotype-specific immunoglobulin G enzyme-linked immunosorbent assay in the seroepidemiologic study of dengue virus infection: correlation of results with those of the plaque reduction neutralization test. J clin microbiol 40 (5), 1840–1844. doi: 10.1128/JCM.40.5.1840-1844.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu P., Huang J. (2004). Current advances in dengue diagnosis. Clin. Vaccine Immunol. 11 (4), 642–650. doi: 10.1128/cdli.11.4.642-650.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmonds P., Becher P., Bukh J., Gould E. A., Meyers G., Monath T., et al. (2017). ICTV virus taxonomy profile: Flaviviridae. J. Gen. Virol. 98 (1), 2–3. doi: 10.1099/jgv.0.000672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon T. (2004). Flavivirus encephalitis. New Engl. J. Med. 351 (4), 370–378. doi: 10.1056/nejmra030476 [DOI] [PubMed] [Google Scholar]
- Stettler K., Beltramello M., Espinosa D. A., Graham V., Cassotta A., Bianchi S., et al. (2016). Specificity, cross-reactivity, and function of antibodies elicited by zika virus infection. Science 353 (6301), 823–826. doi: 10.1126/science.aaf8505 [DOI] [PubMed] [Google Scholar]
- Sukupolvi-Petty S., Austin S. K., Purtha W. E., Oliphant T., Nybakken G. E. (2007). Type-and subcomplex-specific neutralizing antibodies against domain III of dengue virus type 2 envelope protein recognize adjacent epitopes. J Virology 81 (23), 12816–12826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H., Chen Q., Lai H. (2017). Development of antibody therapeutics against flaviviruses. Int. J. Mol. Sci. 19 (1), 54. doi: 10.3390/ijms19010054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesh R. B., Travassos da Rosa AP D. A., Guzman H., Araujo T. P., Shu Y. (2002). Immunization with heterologous flaviviruses protective against fatal West Nile encephalitis. Emerging Infect. Dis. 8 (3), 245–251. doi: 10.3201/eid0803.010238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ThermoFisher, S (2010) Elisa Technical guide and protocols - thermo Fisher scientific. Available at: https://assets.thermofisher.com/TFS-Assets/LSG/Application-Notes/TR0065-ELISA-guide.pdf.
- Thomas S., Jarman R., Endy T., Kalayanarooj S., Vaughn D., Nisalak A., et al. (2009). Dengue plaque reduction neutralization test (PRNT) in primary and secondary dengue virus infections: How alterations in assay conditions impact performance. Am. J. Trop. Med. Hygiene 81 (5), 825–833. doi: 10.4269/ajtmh.2009.08-0625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Throsby M., Geuijen C., Goudsmit J., Bakker A. Q., Korimbocus J., Kramer R. A., et al. (2006). Isolation and characterization of human monoclonal antibodies from individuals infected with West Nile virus. J. Virol. 80 (14), 6982–6992. doi: 10.1128/jvi.00551-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timiryasova T. M., Bonaparte M. I., Luo P., Zedar R., Hu B. T., Hildreth S. W. (2013). Optimization and validation of a plaque reduction neutralization test for the detection of neutralizing antibodies to four serotypes of dengue virus used in support of dengue vaccine development. Am. J. Trop. Med. Hygiene 88 (5), 962–970. doi: 10.4269/ajtmh.12-0461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. (1979). Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: Procedure and some applications. Proc. Natl. Acad. Sci. 76 (9), 4350–4354. doi: 10.1073/pnas.76.9.4350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai T., Popovici F., Cernescu C., Campbell G., Nedelcu N. (1998). West Nile Encephalitis epidemic in southeastern Romania. Lancet 352 (9130), 767–771. doi: 10.1016/s0140-6736(98)03538-7 [DOI] [PubMed] [Google Scholar]
- VanBlargan L. A., Errico J. M., Kafai N. M., Burgomaster K. E., Jethva P. N., Broeckel R. M., et al. (2021). Broadly neutralizing monoclonal antibodies protect against multiple tick-borne flaviviruses. J. Exp. Med. 218 (5), e20210174. doi: 10.1084/jem.20210174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughn D. W., Nguyen M. D., Kalayanarooj S., Solomon T., Nisalak A., Cuzzubbo A., et al. (1999). Rapid serologic diagnosis of dengue virus infection using a commercial capture Elisa that distinguishes primary and secondary infections. Am. J. Trop. Med. Hygiene 60 (4), 693–698. doi: 10.4269/ajtmh.1999.60.693 [DOI] [PubMed] [Google Scholar]
- Venkatachalam R., Subramaniyan V. (2014). Homology and conservation of amino acids in e-protein sequences of dengue serotypes. Asian Pacific J. Trop. Dis. 4, S573–S577. doi: 10.1016/s2222-1808(14)60681-2 [DOI] [Google Scholar]
- Wahala W., Kraus A. A., Haymore L. B., Accavitti-Loper M. A., De Silva A. M. (2009). Dengue virus neutralization by human immune sera: Role of envelope protein domain iii-reactive antibody. Virology 392 (1), 103–113. doi: 10.1016/j.virol.2009.06.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster D., Farrar J., Rowland-Jones S. (2009). Progress towards a dengue vaccine. Lancet Infect. Dis. 9 (11), 678–687. doi: 10.1016/s1473-3099(09)70254-3 [DOI] [PubMed] [Google Scholar]
- Williams D. T., Lunt R. A., Wang L. F., Daniels P. W., Newberry K. M., Mackenzie J. S. (2001). Experimental infections of pigs with Japanese encephalitis virus and closely related Australian flaviviruses. Am. J. Trop. Med. Hygiene 65 (4), 379–387. doi: 10.4269/ajtmh.2001.65.379 [DOI] [PubMed] [Google Scholar]
- World Health Organization, & World Health Organization (1997). Dengue haemorrhagic fever: Diagnosis, treatment. Prev. control 2, 12–23. [Google Scholar]
- World Health Organization (2007). Guidelines for plaque reduction neutralization testing of human antibodies to dengue viruses (Geneva: World Health Organization; ), 2007. [Google Scholar]
- Xu M., Hadinoto V., Appanna R., Joensson K., Toh Y., Balakrishnan T., et al. (2012). Plasmablasts generated during repeated dengue infection are virus glycoprotein–specific and bind to multiple virus serotypes. J. Immunol. 189 (12), 5877–5885. doi: 10.4049/jimmunol.1201688 [DOI] [PubMed] [Google Scholar]
- Yang P., Mahmood T. (2012). Western Blot: Technique, theory, and trouble shooting. North Am. J. Med. Sci. 4 (9), 429. doi: 10.4103/1947-2714.100998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young P. R., Hilditch P. A., Bletchly C., Halloran W. (2000). An antigen capture enzyme-linked immunosorbent assay reveals high levels of the dengue virus protein NS1 in the sera of infected patients. J. Clin. Microbiol. 38 (3), 1053–1057. doi: 10.1128/jcm.38.3.1053-1057.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L., Wang R., Gao F., Li M., Liu J., Wang J., et al. (2017). Delineating antibody recognition against zika virus during natural infection. JCI Insight 2 (12), e93042. doi: 10.1172/jci.insight.93042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zannoli S., Morotti M., Denicolò A., Tassinari M., Chiesa C., Pierro A., et al. (2018). Diagnostics and laboratory techniques. Chikungunya Zika Viruses, 293–315. doi: 10.1016/b978-0-12-811865-8.00009-x [DOI] [Google Scholar]
- Zhao H., Fernandez E., Dowd K. A., Speer S. D., Platt D. J., Gorman M. J., et al. (2016). Structural basis of zika virus-specific antibody protection. Cell 166 (4), 1016–1027. doi: 10.1016/j.cell.2016.07.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Xu L., Bombardi R., Nargi R., Deng Z., Errico J. M., et al. (2019). Mechanism of differential zika and dengue virus neutralization by a public antibody lineage targeting the DIII lateral ridge. J. Exp. Med. 217 (2), e20191792. doi: 10.1084/jem.20191792 [DOI] [PMC free article] [PubMed] [Google Scholar]