Abstract
A Staphylococcus aureus mutant conditionally defective in DNA ligase was identified by isolation of complementing plasmid clones that encode the S. aureus ligA gene. Orthologues of the putative S. aureus NAD+-dependent DNA ligase could be identified in the genomes of Bacillus stearothermophilus and other gram-positive bacteria and confirmed the presence of four conserved amino acid motifs, including motif I, KXDG with lysine 112, which is believed to be the proposed site of adenylation. DNA sequence comparison of the ligA genes from wild type and temperature-sensitive S. aureus strain NT64 identified a single base alteration that is predicted to result in the amino acid substitution E46G. The S. aureus ligA gene was cloned and overexpressed in Escherichia coli, and the enzyme was purified to near homogeneity. NAD+-dependent DNA ligase activity was demonstrated with the purified enzyme by measuring ligation of 32P-labeled 30-mer and 29-mer oligonucleotides annealed to a complementary strand of DNA. Limited proteolysis of purified S. aureus DNA ligase by thermolysin produced products with apparent molecular masses of 40, 22, and 21 kDa. The fragments were purified and characterized by N-terminal sequencing and mass analysis. The N-terminal fragment (40 kDa) was found to be fully adenylated. A fragment from residues 1 to 315 was expressed as a His-tagged fusion in E. coli and purified for functional analysis. Following deadenylation with nicotinamide mononucleotide, the purified fragment could self-adenylate but lacked detectable DNA binding activity. The 21- and 22-kDa C-terminal fragments, which lacked the last 76 amino acids of the DNA ligase, had no adenylation activity or DNA binding activity. The intact 30-kDa C terminus of the S. aureus LigA protein expressed in E. coli did demonstrate DNA binding activity. These observations suggest that, as in the case with the NAD+-dependent DNA ligase from B. stearothermophilus, two independent functional domains exist in S. aureus DNA ligase, consisting of separate adenylation and DNA binding activities. They also demonstrate a role for the extreme C terminus of the ligase in DNA binding. As there is much evidence to suggest that DNA ligase is essential for bacterial survival, its discovery in the important human pathogen S. aureus indicates its potential as a broad-spectrum antibacterial target for the identification of novel antibiotics.
The increasing incidence of drug resistance among bacterial pathogens, including Staphylococcus aureus, has stimulated the development of strategies targeting previously unexploited mechanisms of antibiotic action. Moreover, the emergence of vancomycin-resistant enterococci and drug-resistant Streptococcus pneumoniae has illustrated the necessity for antibacterials to combat multiply resistant gram-positive pathogens (19, 20). Attractive targets for novel antimicrobial agents can be found among genes that are essential for bacterial survival. In an effort to identify genes essential for the growth of S. aureus, a collection of temperature-sensitive mutants has been generated (13). One of the mutant strains, NT64, was found to be complemented by genes encoding an NAD+-dependent DNA ligase.
DNA ligases are essential enzymes found in all bacteria that catalyze the formation of phosphodiester bonds at single-strand breaks between adjacent 3′-OH and 5′-phosphate termini in double-stranded (ds) DNA (7, 30). This activity plays an essential role in DNA replication, repair of damaged DNA, and recombination (11, 15, 17, 18, 26). Reports describing conditional lethal mutations in the ligase gene of Escherichia coli have confirmed the essentiality of this important enzyme, since mutants are deficient in both DNA replication and repair (1, 2).
The DNA ligase family can be divided into two classes: those requiring ATP for adenylation (eukaryotic cells and phage), and those requiring NAD+ for adenylation, which include all known bacterial DNA ligases (7, 18, 21, 23, 25, 26, 29). Amino acid sequence comparisons indicate that NAD+-dependent ligases are phylogenetically unrelated to the ATP-dependent DNA ligases. Eukaryotic, bacteriophage, and viral DNA ligases show little sequence homology to DNA ligases isolated from prokaryotes, with the exception of the conserved residues within the central cofactor-binding core (28, 29). This suggests that bacterial DNA ligase may be a selective target for new antibacterials.
The first step of DNA ligation in bacteria requires adenylation by the NAD+ cofactor of an ɛ-NH2 group of lysine in the conserved KXDG motif at amino acids (aa) 112 to 115 (see Fig. 2). This first step creates an adenylated enzyme intermediate with AMP covalently bound to the enzyme and allows release of nicotinamide mononucleotide. In the second step of the reaction, the adenylate moiety is transferred from Lys-112 to the terminal 5′ phosphate at the DNA nick. A phosphodiester bond is then formed between the 5′ phosphate and the adjacent 3′ hydroxyl, producing the sealed DNA strand (16, 25).
FIG. 2.
Comparison of the conserved amino acid motifs I through IV from known and uncharacterized bacterial ligA gene sequences. Amino acids identical in all species are shown in boldface type. Motif I is the conserved region at the active site. Motifs II and III are of unknown function. Motif IV constitutes the DNA binding domain. Designations in motif IV (B1, A1, etc) refer to areas defined as beta-sheet or alpha-helix structures (4). Alignment of amino acid sequences within the C-terminal DNA binding motif IV from various DNA ligases were as follows: E. coli, aa 598 to 671; Haemophilus influenzae, aa 605 to 679; Zymomonas mobilis, aa 649 to 731; T. thermophilus, aa 594 to 676; T. filiformis, aa 594 to 667; B. stearothermophilus, aa 594 to 670; Bacillus subtilis, aa 595 to 668; Rickettsia prowazekii, aa 619 to 689; S. aureus, aa 591 to 667; Enterococcus faecalis predicted DNA ligase, aa 586 to 664; and Streptococcus pyogenes predicted DNA ligase, aa 582 to 652. The unpublished genomic sequence data used to identify the LigA orthologues were obtained from the PEDANT database (http://pedant.mips.biochem.mpg.de/).
DNA sequences of DNA ligase genes have been determined for a number of different bacterial species (8, 9, 23, 24, 25, 27), and several conserved regions have been identified and compared with those found in E. coli (4, 7). The first motif contains the active site Lys that is covalently adenylated in the intermediate reaction step. A second conserved motif, DGVVXK, and a third motif, FANPRNAAGSLRQLDPRITARRGL, starting at residue 196 in Thermus scotoductus, have unknown functions (8). A fourth motif located in the C-terminal portion of the protein is found in other DNA binding proteins (4).
A recent report described the existence of separate functional domains for the NAD+-dependent DNA ligase isolated from the moderate thermophile Bacillus stearothermophilus (28). In this species, the DNA ligase is composed of N- and C-terminal domains that are connected by a proteolytically sensitive linker region (residues 319 to 396). These separate domains were cloned and overexpressed in E. coli for detailed functional analysis. The larger, 36-kDa N-terminal domain retained full self-adenylating activity while possessing minimal DNA binding or ligation activity. The smaller, 30-kDa C-terminal domain displayed DNA binding activity comparable to that of the intact enzyme but had no adenylating activity. The separate fragments, when combined, did not function in a cooperative fashion to express ligase activity. These data taken together suggested that each domain in the B. stearothermophilus DNA ligase can function independently in vitro and cooperativity between the domains is minimal.
The independent domain functions of the B. stearothermophilus enzyme contrasts with the situation observed for ATP-dependent T7 DNA ligase, where both domains are required for self-adenylation and DNA binding. Furthermore, outside of the active site KXDG motif, the ATP- and NAD+-dependent DNA ligases share very little sequence homology; hence, questions about the structural similarities and differences between the two classes of enzyme are of great interest in terms of understanding how these DNA ligases function. X-ray crystal structures have now been reported (10, 22) for both the ATP-dependent enzyme from the T7 phage and the N-terminal adenylation domains of B. stearothermophilus and Thermus filiformis DNA ligase. Comparison of the structures revealed that, while a core fold and key nucleotide binding residues in the adenylation domains are conserved between both types of DNA ligases, sequence differences outside of this motif exist which may be specific to binding of the specific cofactor.
In the current paper, we report the cloning, overexpression in E. coli, and initial characterization of purified full-length as well as N- and C-terminal domains of S. aureus DNA ligase. This is the first report characterizing the functional DNA ligase domains from this gram-positive species, which is a significant human pathogen. Comparison with the DNA ligase characterized from B. stearothermophilus illustrates the conservation of key functional domains within this important enzyme and, in addition, demonstrates a critical role in DNA binding for the extreme C-terminal region of the DNA ligase.
The identification and characterization of the NAD+-dependent DNA ligase from S. aureus provides further insight into the potential of this enzyme as a broad-spectrum antibacterial target. In addition, our studies will facilitate future structure-function studies and the design and identification of inhibitors of DNA ligase, which could have potential as antibacterial agents.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Strains of S. aureus were grown in tryptic soy broth with erythromycin (2 μg/ml) or tetracycline (1 μg/ml) added for vector selection where appropriate. SAM23 was derived from S. aureus 8325-4 wild-type and SAM13 r− m+ 8325-4 (RN4220, from R. Novick). Plasmid pMP16 is a 6.4-kb shuttle vector derived by ligation of NarI-digested pUC19 with ClaI-digested pE194 (Apr Eryr) (13). A library of 2- to 8-kb partial Sau3AI fragments of SAM23 was prepared by ligation of genomic fragments, which were partially filled with dGTP and dATP, with SalI-cut pMP16, which had been partially filled with dCTP and dTTP. Plasmid pMP98 contained the S. aureus ligA gene on a 2.9-kb Sau3AI partial DNA fragment in pMP16.
Mutant isolation and complementation.
Temperature sensitive mutants of SAM23 were isolated following DES mutagenesis. Colonies that arose on Trypticase soy agar at 30°C were tested for the inability to grow at 43°C by replica plating. Complementing clones were isolated from a library of 2- to 8-kb S. aureus SAM23 Sau3AI-derived fragments ligated in shuttle vector pMP16, as has been described previously (13).
DNA sequence analysis.
Complementing plasmids and PCR fragments were sequenced with a PRISM dye terminator kit from Applied Biosystems, Inc. (Foster City, Calif.), and an AMI 373A automated DNA sequencer. Oligonucleotides for DNA sequencing were produced on an ABI 392 synthesizer. All sequencing reactions were performed in multiple passes from both directions. Multiple genomic PCR fragments from the NT64 mutant and the SAM23 parent were sequenced in parallel and compared to differentiate genomic mutations from possible PCR-induced artifacts. Similarity searches were performed with a BLAST program against the GenBank database. Protein alignments of putative open reading frames (ORFs) were performed using BESTFIT.
Chemicals.
[γ-32P]ATP (∼5,000 Ci/mmol) was purchased from Amersham Pharmacia Biotech, Arlington Heights, Ill. Restriction endonucleases and T4 polynucleotide kinase (10 U/ml) were purchased from Gibco/BRL, Gaithersburg, Md. T4 ligase was purchased from New England Biolabs, Inc., Beverly, Mass. Cibacron Blue 3GA type 3000-L, DEAE Sepharose CL-6B, streptomycin sulfate, and all buffer components were obtained from Sigma Chemical Co., St. Louis, Mo.
Construction of ligA expression vector.
Briefly, pMP98 was restricted with NsiI/HindIII, and a 0.4-kb NsiI fragment containing the 5′ end of the ligA coding region and a 2.2-kb NsiI/HindIII DNA fragment, which comprised the remainder of the ligA coding region and termination sequences, were isolated. The NsiI fragment was then restricted with BsmbI, and a 0.25-kb BsmbI/NsiI fragment was isolated. The 5′ end of the putative DNA ligase gene, which included the initiation ATG, was reconstructed with the ds synthetic oligonucleotide 5′-TATGGCTGATTTATCGTCTCG-3′ and 5′-CACACGAGACGATAAATCAGCCA-3′ (Genosys Biotechnologies, Inc., The Woodlands, Tex.) and isolated as a 275-bp NdeI/NsiI fragment. This fragment was ligated with the 2.2-kb NsiI/HindIII fragment to the NdeI/HindIII-cut pLEX vector. The resulting ligation products were used to transform E. coli strain GI724 by electroporation. Transformants were selected on induction base medium (Invitrogen Corp., San Diego, Calif.), which contains, per liter, 6 g of Na2HPO4, 3 g of KH2PO4, 0.5 g of NaCl, 1 g of NH4Cl, 2 g of Casamino Acids, and 0.095 g of MgCl2 and is supplemented with ampicillin (100 μg/ml). Plasmid DNA from selected transformants was analyzed by restriction enzyme digestions. Transformants containing the ligA insert were isolated and designated pLEX-ligNH. The junctions of each isolate were verified by DNA sequence analysis.
Overexpression of S. aureus ligA gene in E. coli strain GI724.
Several transformants were grown in duplicate in flasks with shaking at 30°C in the induction basal medium described above. When the cultures reached an optical density at 600 nm of 0.3 to 0.4, tryptophan was added to one flask in each set to a final concentration of 100 μg/ml, and the temperature was shifted to 37°C. Cell samples were taken at 1, 2, and 4 h postinduction. Total cell proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) to tentatively identify the DNA ligase.
Purification of S. aureus full-length DNA ligase.
The S. aureus DNA ligase was purified by a modified version of the procedure reported by Lauer et al. (9). Frozen cells of the E. coli strain GI724 (1 liter, ∼1.3 g) containing the overexpressed S. aureus DNA ligase were suspended in TKED buffer (25 mM Tris-HCl [pH 7.7], 100 mM KCl, 1 mM EDTA, and 2 mM dithiothreitol [DTT]) containing 0.5 mM phenylmethylsulfonyl fluoride. The cells were sonicated at 4°C, and the cellular debris was removed by centrifugation at 30,000 × g at 4°C for 30 min. Streptomycin sulfate (10% [wt/vol] stock) was added to the supernatant to a final concentration of 2%, and the solution was kept on ice with occasional mixing for 20 min. The resulting precipitate was removed by centrifugation (20 min at 4°C at 15,000 × g). The supernatant was loaded onto a Cibacron Blue 3GA column (1.5 by 12 cm) equilibrated with TKED buffer. The column was washed with TKED buffer and then washed with TKED buffer containing 0.2 M KCl. The DNA ligase was eluted with TKED buffer containing 0.65 M KCl The flow rate was 2.0 ml/min and 4-ml fractions were collected. The fractions containing DNA ligase were pooled and dialyzed overnight at 4°C against TKED buffer plus 10% (vol/vol) glycerol (TKEDG buffer). The dialyzed ligase fraction was loaded onto a DEAE Sepharose CL-6B column (1.5 by 10 cm) equilibrated with TKEDG buffer. The column was then washed with TKEDG buffer and ligase was eluted with a linear KCl gradient from 0.1 to 1.0 M in TKEDG. The fractions containing ligase were combined and concentrated using a Centriprep 30 unit (Amicon). The concentrate was then dialyzed against TKEDG buffer containing 15% (vol/vol) glycerol and stored at −20°C.
Expression and purification of DNA ligase N-terminal and C-terminal domains.
The N-terminal and C-terminal domains of DNA ligase were expressed as N-terminally His-tagged fusion proteins in E. coli BL21(DE3) from Novagen and BL21 Gold(DE3) from Stratagene with plasmids pET15b and pET32a(+) modified to remove the TrxTag, respectively. The N-terminal domain consisted of residues Ala1 to Lys315 of the full-length protein sequence and included the initiator methionine. The C-terminal domain consisted of residues Val391 to Ser667. Each protein was purified to >95% purity by chromatography on a Mono Q column (Amersham Pharmacia). Authenticity of the purified proteins was verified by analysis of mass by liquid chromatography (LC)-coupled mass spectroscopy (LC-MS) and N-terminal sequencing. Protein concentration was determined using the Coomassie Plus assay (Pierce Chemicals).
DNA ligase gel-based assay.
A DNA ligase assay that separates nicked substrate from ligated products by gel electrophoresis was employed using the procedure of Barker et al. (1) with slight modification. The assay is designed so that the two adjacent oligonucleotides (29-mer and 30-mer) serve as substrate DNA for DNA ligase when annealed to a 59-mer complement. S. aureus DNA ligase (0.25 μM) was added to a reaction mixture containing 50 mM Tris-HCl (pH 7.8), 5 mM MgCl2, 1 mM NH4Cl, 10 mM DTT, 27 μM NAD+, 2% polyethylene glycol 8000, and 17 μM 32P-labeled nicked oligonucleotide substrate (14). The 29-mer (5′-CCCTGTTCCAGCGTCTGCGGTGTTGCGTC-3′) and the 5′ 32P-end labeled 30-mer (5′-AGTTGTCATAGTTTGATCCTCTAGTCTGGG-3′) and the complement 59-mer (5′-CCCAGACTAGAGGATCAAACTATGACAACTGACGCAACACCGCAGACGCTGGAACAGGG-3′) come together to form a double-stranded piece of DNA with a manufactured nick that is a substrate for DNA ligase (obtained from Genosys). The assay is terminated by the addition of stop buffer containing 5.5% glycerol, 7.5 mM EDTA, 0.2% (wt/vol) SDS, and 0.015% (wt/vol) bromphenol blue. A 10% acrylamide–8 M urea running gel was used to separate product from starting material. This assay was used in all of the DNA ligase activity determinations in this study.
Band shift assay.
Binding of purified DNA ligase components to DNA was examined in buffer (50 mM Tris-HCl [pH 7.5], 10 mM EDTA, and 5 mM DTT) as previously described (28). Proteins (20 μM concentrations) were incubated with dsDNA oligonucleotide (∼0.01 μM) substrate 5′ end labeled with 32P in a total volume of 10 μl and incubated at room temperature for 60 min. One microliter of gel loading buffer (0.25% [wt/vol] bromphenol blue and 40% [wt/vol] sucrose) was then added to each reaction tube and the mixtures were analyzed on a native 6% acrylamide gel in 0.5× Tris-borate-EDTA running buffer. Gels were run at 120 V until the dye front reached three-fourths the length of the gel. Labeled bands were visualized by autoradiography.
Adenylation assay.
Adenylation assays were performed in standard DNA ligase buffer using 32P-labeled NAD+ (∼0.01 μM) and deadenylated protein (10 μM) in a total reaction volume of 10 μl as previously described (28). Reaction mixtures were incubated for 60 min at room temperature, and then reactions were terminated by the addition of SDS (3.3% final concentration) and incubation at 95°C for 3 min. Radiolabeled products were analyzed by SDS–10% PAGE, and bands were visualized by autoradiography.
Proteolysis of ligase.
Conditions for limited proteolysis of S. aureus DNA ligase with thermolysin and purification of the DNA ligase fragments are described in the legend to Fig. 3.
FIG. 3.
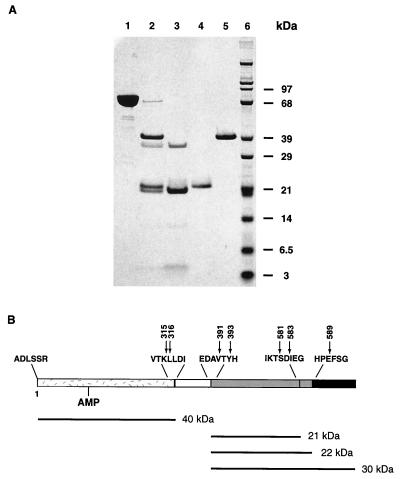
Limited proteolysis of S. aureus DNA ligase by thermolysin. (A) Purified S. aureus DNA ligase from expression in E. coli (lane 1) was incubated with thermolysin at a ratio of 10:1 (wt/wt). After 60 min, the reaction was stopped by addition of 50 mM EDTA. Products of 40, 22, and 21 kDa were produced (lane 2). Thermolysin appeared as a band of 37.5 kDa (lanes 2 and 3). The reaction mixture was chromatographed on Mono Q, and fractions containing the 21-kDa fragment and thermolysin (lane 3), the 22-kDa fragment (lane 4), and the 40-kDa fragment (lane 5) were collected and subjected to N-terminal sequencing and LC-MS analysis to identify the cleavage products. Weakly stained bands at 12 and 4 kDa (lane 3) were not identified. (B) The 40-kDa fragment represented the N-terminal domain of the DNA ligase (1 to 315 or 316) and was fully adenylated. It contained a mixture of two C termini at K315 and L316. The two smaller fragments (21 and 22 kDa) represented domains from the C terminus of the DNA ligase. The 21-kDa fragment contained the N terminus, V391, and two C termini at T581 and D583. The 22-kDa fragment had two N termini, V391 and Y393, each having a C terminus at E589. This panel summarizes the cleavages with the AMP-containing N-terminal domain shown as speckled and the C-terminal domains shown shaded gray. The missing N-C linker region is unshaded and the missing C terminus of the DNA ligase is solid black.
Nucleotide sequence accession number.
The DNA sequence corresponding to the complementing clone from pMP98 has been deposited with GenBank under accession no. AF234833.
RESULTS
Identification of the S. aureus NAD+-dependent DNA ligase gene.
The temperature-sensitive S. aureus mutant NT64 was complemented with a shuttle plasmid library of genomic fragments prepared from wild-type S. aureus strain 8325-4. Complementation of the temperature-sensitive phenotype of NT64 was observed using the plasmid clone pMP98. Sequence analysis of a 2,991-bp fragment subcloned from pMP98 revealed a large ORF. The ORF is comprised of 2,004 bp with a Shine-Dalgarno sequence, AAAGGAGG, located 9 bp from the predicted ATG start codon. The remaining sequence also revealed several other partial ORFs. A partial upstream ORF matched that of the pcrA gene of S. aureus, which encodes a DNA helicase (6). Similarities to the downstream ORF were not identified.
The translated sequence of the major ORF predicted a protein product of 667 aa with a corresponding molecular mass of 75,080 daltons. Clustal sequence alignment revealed 60% and 47% amino acid identity between the B. stearothermophilus and E. coli DNA ligases, respectively (Fig. 1). Structural alignments with orthologs from other gram-positive organisms highlighted conserved features characteristic of DNA ligases possibly involved in adenylation and DNA binding (Fig. 2). Motif I, KXDG, containing amino acid residues 112 to 115, has been shown by mutation analysis to be the site of adenylation, and Lys112 may be the AMP binding site (7, 8, 12). Motifs II and III are of unknown function, while motif IV contains the conserved C-terminal DNA binding region (4).
FIG. 1.
Clustal amino acid sequence alignment of DNA ligases from S. aureus, E. coli, and B. stearothermophilus. Regions enclosed in boxes indicate identical amino acid residues. Amino acid positions of conserved motifs (designations from references 4, 8, 9, and 26) for the S. aureus DNA ligase are as follows: motif I, 112 to 117; motif II, 278 to 283; motif III, 190 to 214; and motif IV, 591 to 667.
Identification of the temperature-sensitive mutation in NT64.
In order to identify the site of the ligase mutation present in the temperature-sensitive mutant NT64, DNA sequence analysis was carried out on PCR-amplified chromosomal DNA from the mutant and wild-type strains. A single difference was observed in the ligase gene of NT64 relative to SAM23. The mutational alteration was a G-to-A transition at nucleotide 136 of the ligA coding region, which is expected to result in the substitution of Glu for Lys46 of DNA ligase. This mutation does not reside in any of the four previously identified conserved motifs but lies in the middle of a surface-exposed alpha helix (22).
Characterization of S. aureus DNA ligase.
The S. aureus DNA ligase was overexpressed in E. coli to ∼5% of total cell protein. Purification of the enzyme was achieved by a previously published method used to obtain T. thermophilus DNA ligase (9), followed by an anion exchange chromatographic step. The protein appeared as a single band on SDS–10% PAGE and was estimated to be at least 95% pure (Fig. 3, lane 1).
Amino acid sequencing of the protein identified the N-terminal sequence as ADLS, consistent with the expected N terminus after removal of the initiator methionine. The molecular mass of enzyme determined by MS was shown to be in good agreement with the theoretical mass of adenylated protein, 75,290 Da.
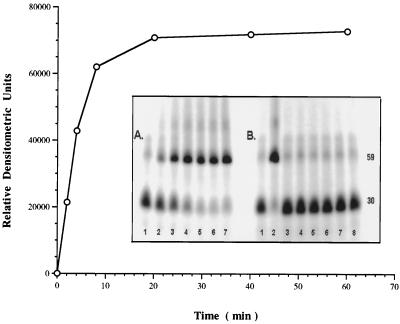
The activity of the S. aureus DNA ligase preparation was determined by a gel-based assay measuring the ligation of a 32P-labeled 30-mer with a 29-mer oligonucleotide, annealed to a complementary strand of DNA. Approximately 0.25 μM purified full-length S. aureus DNA ligase essentially completed ligation of a 17 μM concentration of the oligonucleotide mixture within 10 min under the conditions of the assay (Fig. 4A). Studies with deadenylated enzyme indicated that NAD+ was necessary for ligation activity, with maximal activity achieved at 30 μM NAD+ (data not shown). The pH optimum for the S. aureus DNA ligase was 8.5.
FIG. 4.
Ligation time course of S. aureus DNA ligase (0.25 μM) with 17 μM 32P-labeled nicked substrate (see Materials and Methods). Activity is measured in relative densitometric units of ligated product (59-mer) on a 10% acrylamide, 8 M urea gel. (A) Lanes 1 to 7 represent 0, 2, 4, 8, 18, 40, and 60 min of incubation at 30°C. (B) Gel assay comparing activity of full-length S. aureus DNA ligase with the purified N-terminal and C-terminal domains over 60 min at 30°C. Lane 1, no enzyme control; lane 2, 0.25 μM full-length ligase; lane 3, 0.25 μM 40-kDa N-terminal domain; lane 4, 0.25 μM 30-kDa C-terminal domain; and lane 5, 0.25 μM concentration of each of the N- and C-terminal domains, respectively. Lanes 6 to 8 represent the N-terminal, C-terminal, and both the N- and C-terminal domains in excess at 0.25 mM, respectively.
S. aureus DNA ligase is composed of functionally distinct N- and C-terminal domains.
A recent study by Timson and Wigley (28) indicated that the NAD+-dependent DNA ligase from B. stearothermophilus is composed of N- and C-terminal domains connected by a proteolytically sensitive linker region of 77 amino acids. In order to determine if this organization was also characteristic of the enzyme from S. aureus, proteolytic digestion was conducted with the purified DNA ligase.
Limited proteolysis of S. aureus DNA ligase with thermolysin resulted in the production of three major products with apparent molecular masses of 40, 22, and 21 kDa (Fig. 3, lane 2). Individual fragments were isolated by anion exchange chromatography (Fig. 3, lanes 3 to 5). The isolated fragments were characterized by N-terminal sequencing and mass analysis by LC-MS (Fig. 3, legend). The purified 40-kDa fragment corresponded to the N-terminal region (Fig. 3, lane 5), was fully adenylated as determined by MS, and began at residue 1 (Ala) of the mature DNA ligase. The 40-kDa fragment was composed of two distinct species terminating at K315 and L316 (Fig. 3B).
The smaller C-terminal fragments obtained after thermolysin cleavage (21 and 22 kDa) were isolated by chromatography on a Mono Q column (Fig. 3, lanes 3 and 4). The unbound fraction eluting with the void volume contained fragments of 21 kDa, from residue V391 to T581 or D583. The fraction bound to the column included two 22-kDa fragments with distinct N termini (V391 and Y393) and a common C terminus at E589. The N- and C-terminal domains obtained by treatment with thermolysin were purified and failed to demonstrate DNA ligation activity when tested individually or when combined (data not shown). Neither of the 21- or 22-kDa fragments were active in a DNA binding assay. Further inspection of the C-terminal region of the DNA ligase revealed that each of these fragments was missing the last 76 amino acids present in the full-length sequence.
In order to obtain quantities of the N- and C-terminal domains for more in-depth analysis, each was overexpressed in E. coli as the His-tagged fusion protein. Comparative ligation studies indicated that neither the 40-kDa N-terminal domain nor the 30-kDa C-terminal domains possessed ligation activity separately (Fig. 4B). The individual domains when mixed together also failed to demonstrate DNA ligase activity even when tested in an excess of 0.25 mM.
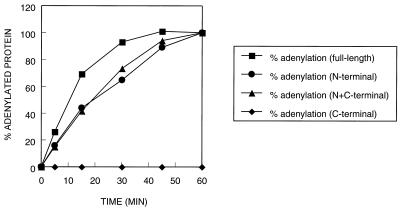
The 40-kDa N-terminal domain and the full-length DNA ligase were deadenylated in the presence of 10 mM nicotinamide mononucleotide and 10 mM magnesium chloride. Deadenylation was essentially complete as shown by mass analysis. The deadenylated N-terminal fragment as well as the full-length DNA ligase underwent rapid self-adenlyation in the presence of [32P]NAD+, while the 30-kDa C-terminal fragment was inactive (Fig. 5). The separate N-terminal fragment and the combined N- and C-terminal fragment mixture demonstrated somewhat lower adenylation activity compared with the full-length DNA ligase. Direct amino acid sequencing of the 30-kDa C-terminal domain and confirmation of mass by MS confirmed that this fragment was unadenylated at the start of the experiment.
FIG. 5.
Relative rates of adenylation of the full-length ligase and the 40-kDa N- and 30-kDa C-terminal domains. A series of assays containing a 1.0 μM concentration of protein and 0.01 μM [32P]NAD+ were incubated at room temperature and terminated at selected time points by the addition of SDS to a final concentration of 3.3%. Phosphorimaging was used to quantify the percentage of enzyme adenylated with labeled NAD+.
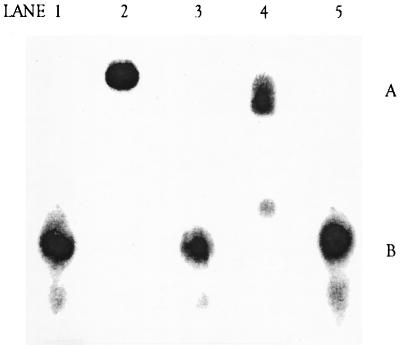
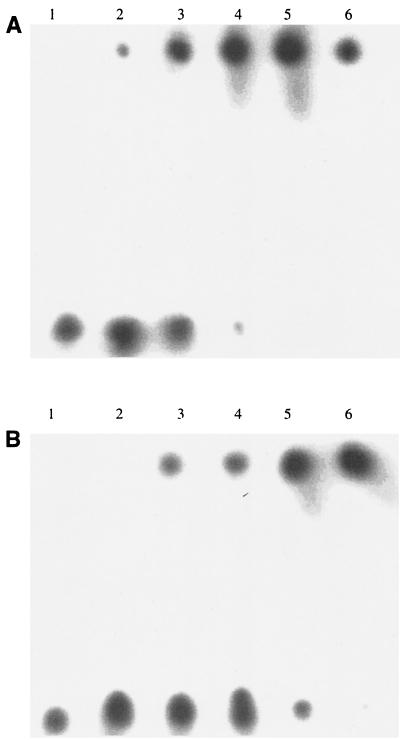
Both the C-terminal 21- and 22-kDa thermolysin-generated fragments are missing the last 76 residues of the full-length DNA ligase and were inactive in a DNA binding assay. In order to test whether the final 76 amino acids of the C terminus were required for DNA binding activity, the intact 30-kDa C terminus of the S. aureus DNA ligase (amino acids 391 to 667) expressed as the His-tagged fusion in E. coli was tested in a gel-shift DNA binding assay. The intact 30-kDa C-terminal region, unlike the versions lacking the complete C terminus, did display DNA binding activity (Fig. 6, lanes 2 and 4 compared with lane 5). The full-length DNA ligase was compared with the complete 30-kDa C-terminal fragment for DNA binding across a 100-fold protein concentration range. The full-length DNA ligase was somewhat more active than the 30-kDa fragment in the DNA binding assay (Fig. 7).
FIG. 6.
DNA band shift assay. Lane 1, 32P-labeled oligonucleotide substrate DNA only (position B); lane 2, full-length DNA ligase (creates a band-shifted oligonucleotide in position A); lane 3, 40-kDa N-terminal fragment; lane 4, complete 30-kDa C-terminal fragment; lane 5, smaller 22-kDa C-terminal fragment (minus the last 76 aa, which contain a putative DNA binding site) plus DNA. All proteins were tested at a 20 μM concentration. Each mixture was incubated at room temperature for 60 min before electrophoresis on a 6% acrylamide gel.
FIG. 7.
DNA band shift assay. (A) Full-length DNA ligase with substrate. (B) Complete 30-kDa C-terminal fragment of DNA ligase. Protein concentrations for each component were as follows: lane 1, 0 μM; lane 2, 1.0 μM; lane 3, 10 μM; lane 4, 25 μM; lane 5, 50 μM; and lane 6, 100 μM. All assays were performed with 32P-labeled oligonucleotide substrate DNA.
DISCUSSION
In this study, we have identified and characterized the NAD+-dependent DNA ligase in S. aureus that is essential for survival in vitro. Several studies have characterized the distinct differences that exist between NAD+-dependent DNA ligases and ATP-dependent DNA ligases found in bacteriophages and eukaryotes. These enzymes differ in their mass as well as energy cofactor requirement, with the bacterial DNA ligases using NAD+ while DNA ligases found in plants, animals, viruses, and bacteriophages utilize ATP (7, 8, 16, 20, 28). In addition, the NAD+-dependent DNA ligases share very little sequence homology with ATP-dependent DNA ligases outside of the active-site lysine pocket (7, 8, 22). Furthermore, recent studies of the NAD+-dependent DNA ligase from B. stearothermophilus indicate that this enzyme is comprised of two separate domains that function independently in an in vitro ligation reaction (23, 28). The presence of separate functional domains signifies a divergence from the organization of ATP-dependent DNA ligases, where the N- and C-terminal fragments function together to effect adenylation and DNA substrate binding. In order to determine whether the apparent independence of the N- and C-terminal functional domains is common to other NAD+-dependent DNA ligases from bacteria, full-length DNA ligase and truncated protein from the human pathogen S. aureus were cloned and overexpressed in E. coli.
Amino acid sequence alignment revealed that the S. aureus ligA gene product had 60% and 47% amino acid identity with the B. stearothermophilus and E. coli enzymes, respectively. Further analysis revealed the presence of conserved motifs among the DNA ligases found in these and other bacterial species. Mass determination from the deduced amino acid sequence indicated that the LigA protein from S. aureus (75,080 Da) is comparable in size to the DNA ligases from B. stearothermophilus (74,229 Da) and E. coli (73,690 Da). The full-length S. aureus DNA ligase expressed as the native protein in E. coli was essentially fully adenylated and enzymatically active when quantitated in an in vitro ligation assay using a radiolabeled oligomeric substrate. Approximately 0.25 μM purified DNA ligase essentially completed the ligation of 17 μM oligonucleotide substrate within 8 min at 30°C.
As with the DNA ligase from the moderate thermophile B. stearothermophilus, the results of our studies show that the enzyme from S. aureus is composed of two separate and functionally distinct domains. In the course of our limited proteolysis studies, we consistently obtained stable C-terminal domain fragments of 21 and 22 kDa. N-terminal sequencing and LC-MS confirmed that each of these was missing the last 76 aa of the DNA ligase; therefore, the 22-kDa fragment was isolated and tested for DNA binding activity. Unlike the 30-kDa C-terminal fragment obtained from B. stearothermophilus DNA ligase by Timson and Wigley (28), our 22-kDa fragment had no detectable DNA binding activity. In order to test whether the 76-aa truncated species was missing a critical region that is required for DNA binding, we expressed the intact S. aureus DNA ligase C-terminal domain (30 kDa), consisting of residues 391 to 667, and assessed its ability to bind DNA. In contrast to the truncated 22-kDa fragment, the complete 30-kDa construct demonstrated activity in a gel-based DNA binding assay. When compared over a 100-fold protein concentration range, the full-length DNA ligase was somewhat more active than the complete C-terminal fragment in the DNA binding assay. A conserved protein motif was proposed by Halligan (4) for several proteins known to interact with DNA. These included the bacterial (NAD+-dependent) DNA ligases and VDJP, a protein that binds the V(D)J recombinational signal sequence element (4). This motif is found at the extreme C-terminal region of all known bacterial DNA ligases and encompasses approximately 80 aa residues. This region is absent in the ATP-dependent DNA ligases (4). Our DNA binding results with the 22- and 30-kDa DNA ligase fragments are consistent with the proposal by Halligan that suggests that this motif contains an essential DNA binding region in bacterial DNA ligases. In addition, since limited proteolysis often identifies flexible interdomain regions in protein structures, our results suggest that the DNA binding region either exists as a separately folded domain or is partially flexible, as opposed to being an integral part of the larger 30-kDa C-terminal domain. Our observations with this DNA binding region of the C-terminal domain extend previous results of others (4, 10). They may also explain why the ATP-dependent DNA ligases appear to require the interaction of both subunits for DNA binding, since the C-terminal fragment alone is insufficient to bind DNA.
Additional tests with the individual domain fragments indicated that as in the case of the ligase from B. stearothermophilus, the 40-kDa N-terminal fragment of S. aureus DNA ligase possessed self-adenylation activity comparable to that of the full-length DNA ligase. This is in contrast to the ATP-dependent DNA ligase of phage T7, which requires the interaction of both N- and C-terminal domains for self-adenylation and DNA binding activities (3). Since bacterial DNA ligase has a different cofactor requirement from the eukaryotic homologue, DNA ligase is an attractive target for drug discovery compared with some other essential proteins. The current study validates the essentiality of DNA ligase in S. aureus and provides information from which to aid crystallization efforts and a structure-based approach for finding novel DNA ligase inhibitors.
Results from this study confirm that important functional regions exist within the individual domains of bacterial DNA ligases. Similarities in the activity of these subunits between the thermophilic bacterium B. stearothermophilus and the human pathogen S. aureus illustrate the conservation of these domain properties among gram-positive bacteria. Since DNA ligase provides an attractive target for the identification of novel antibiotics, further knowledge concerning the functional organization of the bacterial DNA ligases could help guide efforts to design specific antibacterial inhibitors of this enzyme. The recent structural data derived from the B. stearothermophilus DNA ligase should be relevant for gram-positive pathogens as well. Additional information concerning the structural and functional organization of the bacterial DNA ligases should further our understanding of these enzymes.
ACKNOWLEDGMENTS
We thank Jianpeng Shi for assistance in performing complementation tests and Tony Lanzetti for N-terminal sequencing data.
REFERENCES
- 1.Barker D G, Johnson A L, Johnston L H. An improved assay for DNA ligase reveals temperature-sensitive activity in cdc9 mutants of Saccharomyces cerevisiae. Mol Gen Genet. 1985;200:458–462. doi: 10.1007/BF00425731. [DOI] [PubMed] [Google Scholar]
- 2.Dermody J J, Robinson G T, Sternglanz R. Conditional-lethal deoxyribonucleic acid ligase mutant of Escherichia coli. J Bacteriol. 1979;139:701–704. doi: 10.1128/jb.139.2.701-704.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Doherty A J, Ashford S R, Wigley D B. Characterization of the proteolytic fragments of bacteriophage T7 DNA ligase. Nucleic Acids Res. 1996;24:2281–2287. doi: 10.1093/nar/24.12.2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Halligan B. A new protein motif found in DNA joining and DNA binding proteins. Nucleic Acids Res. 1993;21:5520–5521. doi: 10.1093/nar/21.23.5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Horinouchi S, Weisblum B. Nucleotide sequence and functional map of pE194, a plasmid that specifies inducible resistance to macrolide, lincosamide and streptogramin type B antibiotics. J Bacteriol. 1982;150:804–814. doi: 10.1128/jb.150.2.804-814.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iordanescu S. Characterization of the Staphylococcus aureus chromosomal gene pcrA, identified by mutations affecting plasmid replication. Mol Gen Genet. 1993;241:185–192. doi: 10.1007/BF00280216. [DOI] [PubMed] [Google Scholar]
- 7.Ishino Y, Shinagawa H, Makino K, Tsunasawa S, Sakiyama F, Nakata A. Nucleotide sequence of the lig gene and primary structure of DNA ligase of Escherichia coli. Mol Gen Genet. 1986;204:1–7. doi: 10.1007/BF00330179. [DOI] [PubMed] [Google Scholar]
- 8.Jonsson A O, Thorbjarnardottir S H, Eggertsson G, Palsdottir A. Sequence of the DNA ligase-encoding gene from Thermus scotoductus and conserved motifs in DNA ligases. Gene. 1994;151:177–180. doi: 10.1016/0378-1119(94)90652-1. [DOI] [PubMed] [Google Scholar]
- 9.Lauer G, Rudd E A, McKay D L, Ally A, Ally D, Backman K C. Cloning, nucleotide sequence, and engineered expression of Thermus thermophilus DNA ligase, a homolog of Escherichia coli DNA ligase. J Bacteriol. 1991;173:5047–5053. doi: 10.1128/jb.173.16.5047-5053.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee J Y, Chang C, Song H K, Moon J, et al. Crystal structure of NAD+-dependent DNA ligase: modular architecture and functional implications. EMBO J. 2000;19:1119–1129. doi: 10.1093/emboj/19.5.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liebert J C, Paolozzi L, Camera M G, Pedrini A M, Ghelardini P. The expression of the DNA ligase gene of Escherichia coli is stimulated by relaxation of chromosomal supercoiling. Mol Microbiol. 1989;3:269–273. doi: 10.1111/j.1365-2958.1989.tb00171.x. [DOI] [PubMed] [Google Scholar]
- 12.Luo J, Barany F. Identification of essential residues in Thermus thermophilus DNA ligase. Nucleic Acids Res. 1996;24:3079–3085. doi: 10.1093/nar/24.15.3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martin P, Li T, Sun D, Biek D P, Schmid M B. Role in cell permeability of an essential two-component system in Staphylococcus aureus. J Bacteriol. 1999;181:3666–3673. doi: 10.1128/jb.181.12.3666-3673.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maxam A M, Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci USA. 1977;74:560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Modrich P, Anraku Y, Lehman I R. Deoxyribonucleic acid ligase: isolation and physical characterization of the homogeneous enzyme from Escherichia coli. J Biol Chem. 1973;248:7495–7501. [PubMed] [Google Scholar]
- 16.Modrich P, Lehman I R. Deoxyribonucleic acid ligase: a steady state kinetic analysis of the reaction catalyzed by the enzyme from Escherichia coli. J Biol Chem. 1973;248:7502–7511. [PubMed] [Google Scholar]
- 17.Montecucco A, Pedrali-Noy G, Spadari S, Ciarrocchi G. Multiple roles of DNA ligase at the replication fork. Biochem Biophys Acta. 1988;951:330–334. doi: 10.1016/0167-4781(88)90103-0. [DOI] [PubMed] [Google Scholar]
- 18.Panasenko S M, Modrich P, Lehman I R. Modification of Escherichia coli DNA ligase by cleavage with trypsin. J Biol Chem. 1976;251:3432–3435. [PubMed] [Google Scholar]
- 19.Pfaller M A, Jones R N, Doern G V, Kugler K The Sentry Participants Group. Bacterial pathogens isolated from patients with bloodstream infection: frequencies of occurrence and antimicrobial susceptibility patterns from the SENTRY surveillance program (United States and Canada, 1997) Antimicrob Agents Chemother. 1998;42:1762–1770. doi: 10.1128/aac.42.7.1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saden H S, Jones R N, Gales A C, Winokukr P, Kugler K C, et al. Antimicrobial susceptibility patterns from pathogens isolated from patients in Latin American medical centers with a diagnosis of pneumonia: analysis of results from the SENTRY antimicrobial surveillance program (1997) Diagn Microbiol Infect Dis. 1998;32:289–301. doi: 10.1016/s0732-8893(98)00124-2. [DOI] [PubMed] [Google Scholar]
- 21.Shark K B, Conway T. Cloning and molecular characterization of the DNA ligase gene (lig) from Zymomonas mobilis. FEMS Microbiol Lett. 1992;96:19–26. doi: 10.1016/0378-1097(92)90450-3. [DOI] [PubMed] [Google Scholar]
- 22.Singleton M R, Hakansson K, Timson D J, Wigley D B. Structure of the adenylation domain of an NAD+-dependent DNA ligase. Structure. 1999;7:35–42. doi: 10.1016/s0969-2126(99)80007-0. [DOI] [PubMed] [Google Scholar]
- 23.Subramanya H S, Doherty A J, Ashford S R, Wigley D B. Crystal structure of an ATP-dependent DNA ligase from bacteriophage T7. Cell. 1996;85:607–615. doi: 10.1016/s0092-8674(00)81260-x. [DOI] [PubMed] [Google Scholar]
- 24.Takahasi M, Yamaguchi E, Uchida T. Thermophilic DNA ligase: purification and properties of the enzyme from Thermus thermophilus HB8. J Biol Chem. 1984;259:10041–10047. [PubMed] [Google Scholar]
- 25.Teraoki H, Tsukada K. Eukaryotic DNA ligase: purification and properties of the enzyme from bovine thymus, and immunochemical studies of the enzyme from animal tissues. J Biol Chem. 1982;257:4758–4763. [PubMed] [Google Scholar]
- 26.Thorbjarnardottir S H, Jonsson Z O, Andresson O S, Kristjansson J K, Eggertsson G, Palsdottir A. Cloning and sequence analysis of the DNA ligase-encoding gene of Rhodothermus marinus, and overproduction, purification and characterization of two thermophilic DNA ligases. Gene. 1995;161:1–6. doi: 10.1016/0378-1119(95)00286-f. [DOI] [PubMed] [Google Scholar]
- 27.Timson D J, Singleton M R, Wigley D B. DNA ligases in the repair and replication of DNA. Mutat Res. 2000;460:301–318. doi: 10.1016/s0921-8777(00)00033-1. [DOI] [PubMed] [Google Scholar]
- 28.Timson D J, Wigley D B. Functional domains of an NAD+-dependent DNA ligase. J Mol Biol. 1999;285:73–83. doi: 10.1006/jmbi.1998.2302. [DOI] [PubMed] [Google Scholar]
- 29.Tomkinson A E, Totty N F, Ginsburg M, Lindahl T. Location of the active site for enzyme-adenylate formation in DNA ligases. Proc Natl Acad Sci USA. 1991;88:400–404. doi: 10.1073/pnas.88.2.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zimmerman S B, Pheiffer B H. Macromolecular crowding allows blunt-end ligation by DNA ligases from rat liver or Escherichia coli. Proc Natl Acad Sci USA. 1983;80:5852–5856. doi: 10.1073/pnas.80.19.5852. [DOI] [PMC free article] [PubMed] [Google Scholar]