Abstract
Although DNA methylation has been recognised in the pathogenesis of idiopathic pulmonary fibrosis (IPF), the exact mechanisms are yet to be fully addressed. Herein, we demonstrate that lungs originated from IPF patients and mice after bleomycin (BLM)-induced pulmonary fibrosis are characterised by altered DNA methylation along with overexpression in myofibroblasts of methyl-CpG-binding domain 2 (MBD2), a reader responsible for interpreting DNA methylome-encoded information. Specifically, depletion of Mbd2 in fibroblasts or myofibroblasts protected mice from BLM-induced pulmonary fibrosis coupled with a significant reduction of fibroblast differentiation. Mechanistically, transforming growth factor (TGF)-β1 induced a positive feedback regulatory loop between TGF-β receptor I (TβRI), Smad3 and Mbd2, and erythroid differentiation regulator 1 (Erdr1). TGF-β1 induced fibroblasts to undergo a global DNA hypermethylation along with Mbd2 overexpression in a TβRI/Smad3 dependent manner, and Mbd2 selectively bound to the methylated CpG DNA within the Erdr1 promoter to repress its expression, through which it enhanced TGF-β/Smad signalling to promote differentiation of fibroblast into myofibroblast and exacerbate pulmonary fibrosis. Therefore, enhancing Erdr1 expression strikingly reversed established pulmonary fibrosis. Collectively, our data support that strategies aimed at silencing Mbd2 or increasing Erdr1 could be viable therapeutic approaches for prevention and treatment of pulmonary fibrosis in clinical settings.
Short abstract
MBD2 is upregulated in the myofibroblasts from IPF patients and facilitates the transition of myofibroblast from fibroblast via TβRI/Smad3/Mbd2/Erdr1 positive feedback regulatory loop https://bit.ly/3q0PhHA
Introduction
Idiopathic pulmonary fibrosis (IPF) is a chronic, progressive and fibrotic lung disease, usually leading to death within 3–5 years following diagnosis [1]. Despite past extensive studies, the patho-aetiology underlying IPF remains poorly understood, which has rendered its treatment largely unsuccessful [2, 3]. In general, IPF is characterised by epithelial injury and differentiation of invasive fibroblasts [4]. The differentiation of fibroblast into myofibroblast plays a critical role in fibrotic processes and contributes to the histological features of IPF lung tissues [5, 6]. Upon the stimulation of fibrotic factors such as transforming growth factor (TGF)-β and platelet-derived growth factor (PDGF), lung fibroblasts differentiate into myofibroblasts, which then produce copious amount of fibrillary extracellular matrix (ECM) proteins including type I collagen and fibronectin, predisposing to the development of matrix stiffness and pathological matrix deposition in the lung mesenchyme [7, 8].
Previous studies demonstrated that DNA methylation, one of the major epigenetic factors, is involved in the pathogenesis of IPF, but the detailed underlying mechanisms are yet to be fully elucidated [9, 10]. The information encoded by the DNA methylome is generally read by a family of methyl-CpG-binding domain (MBD) proteins (e.g. MBD1, MBD2, MBD4 and MeCP2), which selectively bind to the methylated CpG DNA, thereby inhibiting or increasing the transcription of targeted genes [11–13]. In particular, MBD2 possesses the highest binding capacity to the methylated CpG DNA [14], and therefore is involved in the pathogenesis of obesity [11], ischaemic injury [15], autoimmunity [16, 17], diabetes [18] and tumorigenesis [19]. Interestingly, MBD2 was also found to be highly expressed in the lungs of IPF patients and mice following bleomycin (BLM)-induced pulmonary fibrosis. Thus, in the present report, we generated mouse models with specific Mbd2 deficiency in fibroblasts or myofibroblasts. It was noted that loss of Mbd2 in fibroblasts or myofibroblasts significantly protected mice from BLM-induced pulmonary fibrosis along with a marked reduction of myofibroblasts by repressing fibroblast differentiation. Mechanistic studies characterised a TGF-β receptor I (TβRI)/Smad3/Mbd2/Erdr1 positive feedback regulatory loop. Specifically, TGF-β1 induces fibroblasts to undergo a global DNA hypermethylation along with Mbd2 overexpression in a TβRI/Smad3-dependent manner; while Mbd2 selectively binds to the highly methylated Erdr1 promoter to repress its expression, through which it enhances TGF-β/Smads signalling to promote fibroblast differentiation into myofibroblast. Collectively, our data support that altered MBD2 expression in fibroblasts is essential for the progression of pulmonary fibrosis, and therefore, strategies aimed at silencing Mbd2 or increasing Erdr1 could be viable treatments to attenuate fibrotic progression in clinical settings.
Materials and methods
Reagents and antibodies
Antibodies against type I collagen were purchased from EMD Millipore (Schwalbach, Germany), while antibodies against prosurfactant protein C (pro-SPC), fibronectin and α-SMA were ordered from Abcam (MA, USA). Antibodies for p-Smad2, p-Smad3, Pi3kp85, p-Pi3kp85, p-Akt, Erk1/2, mTOR, p-mTOR, p-Erk1/2, Jnk, p-Jnk, P38 and p-P38 were obtained from Cell Signaling Technology (MA, USA). Antibodies against MBD2 and glyceraldehyde 3-phosphate dehydrogenase were obtained from Santa Cruz Biotechnology (CA, USA), and Erdr1 antibody was obtained from Antibody Research Corporation (MO, USA). Murine recombinant TGF-β1 was obtained from PeproTech (London, UK), while all other reagents were purchased from Sigma (MO, USA), unless otherwise stated.
Human samples
Resected para-carcinoma lung tissues from nonsmall cell lung cancer (NSCLC) patients (n=6) and lung explant material from IPF patients (n=8) were collected in Tongji Hospital (Wuhan, China) with informed consent. IPF diagnosis was made according to the European Respiratory Society/American Thoracic Society consensus diagnostic criteria [20]. Partial lung tissues were chopped for optimal cutting temperature compound (OCT) embedding; these specimens were sliced for immunostaining. The residual lung tissues were resected into 0.5–1 cm3 and stored in liquid nitrogen for Western blotting or DNA methylation assays. The studies were approved by the human assurance committee of Tongji Hospital (TJ-IRB20210942). Clinical data are provided in supplementary table S1.
Bleomycin induction of pulmonary fibrosis
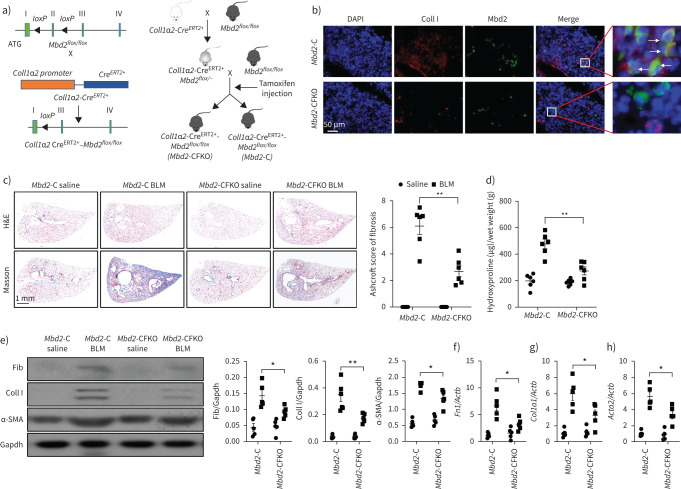
The Mbd2flox/flox mice in C57BL/6 background were generated using the clustered regularly interspaced short palindromic repeats (CRISPR)–Cas9 system by Bioray Laboratories (Shanghai, China). Two loxP sequences were inserted in the introns flanking exon 2 of Mbd2, as described in figure 4a. Coll1a2-CreERT2 and Acta2-CreERT2 transgenic mice were purchased from the Shanghai Model Organism Center (Shanghai, China). The Coll1a2-CreERT2+ and Acta2-CreERT2+ mice were crossed with Mbd2flox/flox mice to generate Coll1a2-CreERT2+Mbd2flox/flox or Acta2-CreERT2+Mbd2flox/flox mice, respectively. Mbd2 deficiency in fibroblasts or myofibroblasts was induced by intraperitoneal injection of tamoxifen (75 mg·kg−1) for five consecutive days in Coll1a2-CreERT2+Mbd2flox/flox or Acta2-CreERT2+Mbd2flox/flox 8-week-old mice to induce fibroblast-specific Mbd2 deficiency (defined as Mbd2-CFKO mice), or myofibroblast-specific Mbd2 deficiency (defined as Mbd2-CMKO mice). Littermates (Coll1a2-CreERT2–Mbd2flox/flox or Acta2-CreERT2−Mbd2flox/flox mice) administered with equal dose of tamoxifen were used as controls (defined as Mbd2-C). Conventional Mbd2 knockout mice in C57BL/6 background (defined as Mbd2−/− mice) were kindly provided by Adrian Bird (Edinburgh University, Edinburgh, UK). Wild-type C57BL/6 mice (defined as WT) were obtained from Beijing Huafukang Bioscience (Beijing, China).
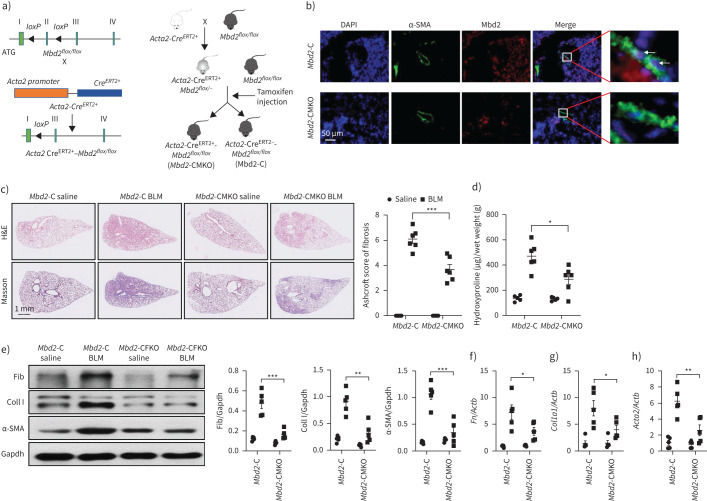
FIGURE 4.
Comparison of the severity of lung fibrosis between methyl-CpG-binding domain 2 (Mbd2)-CFKO and Mbd2-C mice after bleomycin (BLM) induction. a) Mbd2flox/flox mice were generated by inserting two loxP sequences in the same direction into the introns flanked with the exon 2 of MBD2 based on the clustered regularly interspaced short palindromic repeats (CRISPR)–Cas9 system, which could produce a nonfunctional Mbd2 protein by generating a stop codon in exon 3 after Cre-mediated gene deletion. Mbd2flox/flox then crossed with the Coll1α2-CreERT2+ transgenic mice to get the fibroblast-specific Mbd2-knockout mice following intraperitoneal injection of tamoxifen for five consecutive days. b) Representative results for co-immunostaining of Coll I and Mbd2 in lung sections from Mbd2-C and Mbd2-CFKO mice. Nuclei were stained blue using 4′,6-diamidino-2-phenylindole (DAPI), and the images were taken at an original magnification of ×400. c) Histological analysis of the severity of lung fibrosis in mice after BLM induction. Left: representative images for haematoxylin and eosin (H&E) and Masson staining. Right: quantitative mean score of the severity of fibrosis. d) Quantification of hydroxyproline contents in Mbd2-CFKO and Mbd2-C mice after BLM challenge. e) Western blot analysis of levels of fibronectin (Fib), collagen (Coll) I and α-smooth muscle actin (SMA). f–h) Results for reverse transcriptase (RT)-PCR analysis of f) Fn1, g) Coll1a1 and h) Acta2. Five to six mice were included in each study group. Gapdh: glyceraldehyde 3-phosphate dehydrogenase. Data are presented as mean±sd. *: p<0.05, **: p<0.01.
Mbd2-C, Mbd2-CFKO, Mbd2-CMKO, Mbd2−/− and WT mice were anaesthetised with 1% pentobarbital sodium (60 mg·kg−1), followed by intratracheal injection of 0.5 mg·kg−1 BLM (Nippon Kayaku, Tokyo, Japan) in 40 μL of normal saline with a high-pressure atomising needle (BJ-PW-M; Bio Jane Trading Limited, Shanghai, China). Mice administered the same volume of normal saline served as controls. Erdr1 lentiviral particles (1×107 IFU (infectious units)) were injected into the anaesthetised animals intratracheally 10 days after BLM injection. Mice administered with mock lentiviral particles were served as controls. The mice were euthanised on day 21 following BLM challenge to assess pulmonary fibrosis. The four lobes of the right lung (superior lobe, middle lobe, inferior lobe and post-caval lobe) were immediately frozen in liquid nitrogen and stored at −80°C. All mice were housed in a specified pathogen-free facility at the Tongji Hospital with a 12/12 h light/dark cycle. All experimental procedures were approved by the animal care and use committee at the Tongji Hospital (TJH-201901015).
Histological and immunohistochemical analysis
The left lungs of mice were inflated with 200 μL 4% neutral buffered paraformaldehyde, and sliced from the middle region. The left lung upper region was placed in fresh 4% neutral buffered paraformaldehyde for 24 h at room temperature. After paraffin embedding, lung from each mouse was sliced into 5-μm sections (three sections, interval of 200 μm) and subjected to haematoxylin and eosin (H&E) and Masson staining. For a single slide, five visual fields were randomly selected under 100× magnification to obtain the mean Ashcroft score by two pathologists in a blinded fashion [21]. The left lung lower region was embedded with OCT compound and stored at −80°C. For immunostaining, the OCT-embedded human and mouse lungs were sliced into 7-μm sections. The frozen sections were probed with antibodies against α-SMA, pro-SPC, Mbd2 and Erdr1, followed by staining with Alexa Fluor 594-labelled anti-mouse/rabbit or Alexa Fluor 488-conjugated anti-rabbit/mouse antibodies (Invitrogen, CA, USA), respectively. Images were obtained under a fluorescence microscope (Olympus, Shinjuku, Japan).
Culture and treatment of primary lung fibroblasts
Primary lung fibroblasts were isolated from lungs derived from mice, IPF patients and control subjects, as reported [22]. The cells were grown at 37°C and 5% carbon dioxide in DMEM supplemented with 10% fetal bovine serum (FBS) and penicillin/streptomycin. The medium was replaced every 3 days. For fibroblast differentiation, the cells were stimulated with recombinant TGF-β1 (10 ng·mL−1) at indicated time points.
Small interfering RNA transfection
Small interfering RNAs (siRNAs) specific for MBD2 and a corresponding scrambled siRNA were purchased from RiboBio (Guangzhou, China) and then transiently transfected into primary lung fibroblasts using Lipofectamine 3000 (Invitrogen, Shanghai, China), as reported [23]. Briefly, primary lung fibroblasts were seeded in 12-well plates 24 h before transfection. siRNA transfection was performed once the cells reached 60% confluence. Transfection efficiency was monitored using reverse transcriptase (RT)-PCR or Western blotting after 48 h of transfection. The following two siRNAs were used for MBD2: siRNA1 5ʹ-GCA AGA GCG AUG UCU ACU A-3ʹ and siRNA2 5ʹ-GCG AAA CGA UCC UCU CAA U-3ʹ (RiboBio). For Erdr1 siRNA transfection, Mbd2−/− lung fibroblasts cultured in DMEM supplemented with 10% FBS were transfected with an Erdr1 siRNA or control siRNA (RiboBio), as described earlier. The transfected cells were next stimulated with murine recombinant TGF-β1 (10 ng·mL−1) at indicated time points, followed by comparison of myofibroblast conversion.
Western blot analysis
The superior lobe of lung tissues from mice and human subjects and cultured cells were homogenised in RIPA lysis buffer (Beyotime, Shanghai, China) supplemented with proteinase inhibitor cocktail tablets (Roche, Wuhan, China), and the proteins were subjected to Western blotting with indicated primary antibodies, as reported previously [24]. Briefly, equal amounts of lysates were separated on 10% sodium dodecyl sulphate-polyacrylamide gel and transferred on to polyvinylidene fluoride membranes. The membranes were blocked with 5% nonfat milk for 1 h at room temperature and then incubated with primary antibodies overnight at 4°C. Next, the membranes were incubated for 1 h at room temperature with a horseradish peroxidase-conjugated secondary antibody (Servicebio, Wuhan, China), prior to the visualisation of the bands using the chemiluminescence reagents (Pierce ECL; Thermo Scientific, Ulm, Germany). The blots were recorded using X-ray film and analysed using Image Lab 6.0.1 software (Bio-Rad, Munich, Germany).
Quantitative RT-PCR analysis
Total RNA was isolated from the middle lobe of mouse lung and cultured cells using TRlzol reagent (Takara, Dalian, China). RNA quantity and quality were measured using a NanoDrop 2000 spectrophotometer (Thermo Scientific, MA, USA). Complementary DNA synthesis was performed using a Moloney murine leukaemia virus reverse transcriptase kit (Invitrogen, CA, USA). Quantitative RT-PCR was performed using the SYBR Premix Ex Taq (Takara), and the relative expression of each target gene was normalised by Actb expression, as described previously [25]. The primers used for each target gene are provided in supplementary table S2.
Hydroxyproline assays
The hydroxyproline contents in the inferior lobe of right lung were measured by using a hydroxyproline kit (Nanjing Jiancheng Institute, Nanjing, China), as described previously [26]. Briefly, the fresh lung tissues were weighed and alkaline hydrolysed for 20 min at 100°C. After adjusting pH to 6.0–6.8, the hydrolysates were refined with 25 mg active carbon and centrifuged at 1150 ×g for 10 min. The supernatants then underwent a series of chemical reactions, and optical density values were determined at 550 nm using a microplate reader (ELx800, BioTek Instruments, Winooski, VT, USA). The hydroxyproline content in lung tissue is presented as μg hydroxyproline per g lung tissue by comparing with the hydroxyproline standard provided.
RNA deep sequencing
RNAs from WT and Mbd2−/− lung fibroblasts were extracted using an RNA isolation kit (QIAGEN, Shanghai, China), respectively. RNA quality and integrity were determined using the Nanodrop 2000 Spectrophotometer and Agilent Bioanalyzer (CA, USA). RNA deep sequencing (RNA-seq) libraries were multiplexed and loaded into the Illumina HiSeq flow cell v3. All sequencing protocols were carried out as per the manufacturer's instructions using the Illumina HiSeq 1000 system and HiSeq control software.
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation (ChIP) assays were conducted using a ChIP assay kit (Beyotime) as reported previously [27]. Briefly, 1×106 lung fibroblasts were cross-linked with formaldehyde, and chromatin fragmentation was carried out according to the protocol provided. This diluted soluble chromatin solution prepared thus was then incubated with an Mbd2 antibody overnight at 4°C with rotation. Normal rabbit IgG was used to determine nonspecific bindings. Next, the mixtures were incubated with protein A+G agarose beads, and the protein–DNA complexes were eluted out after washes. The eluted DNA was subjected to ChIP-PCR with the indicated primers. The primers used for Erdr1 in the ChIP assay were 5ʹ-GGC TTT TTT AAA CTC GAT CCG-3ʹ (forward), 5ʹ-GGC AGG ACT ACA ACT CCC AG-3ʹ (reverse).
Global DNA methylation assay
Global DNA methylation was determined using a MethylFlash Methylated DNA Quantification Kit (Epigentek, NY, USA) according to the manufacturer's instructions. Briefly, the methylated DNA was detected using the capture and detection antibodies to 5-methylcytosine (5-mC) and then quantified colorimetrically by reading absorbance at 450 nm using a microplate reader (ELx800).
DNA bisulfite sequencing analysis
Genomic DNA from each preparation underwent bisulfite conversion using an EZ DNA Methylation-Direct Kit (Zymo Research, CA, USA), as reported previously [11, 13]. The resulting products were used as templates to amplify the targeted sequences, and the PCR products were directly cloned into plasmids for sequencing. The methylation state of each targeted sequence was then analysed using DNA sequencing.
Statistical analysis
Comparisons between groups were undertaken using Prism software (Prism 8.lnk; GraphPad Software, CA, USA). Two experimental groups were compared using the two-tailed t-test (data with normal distribution, homogeneity of variance) or two-tailed Mann–Whitney test (data without normal distribution). Once more than two groups were compared, one-way or two-way ANOVA with Tukey's multiple comparison test (data with normal distribution) or Kruskal–Wallis test with Dunn's post hoc tests (data without normal distribution) were used. All experiments were conducted with at least three independent replications. The data are presented as the mean±sem/sd. In all cases, p<0.05 was considered to be statistically significant.
Results
IPF features by the altered MBD2 expression and DNA methylation
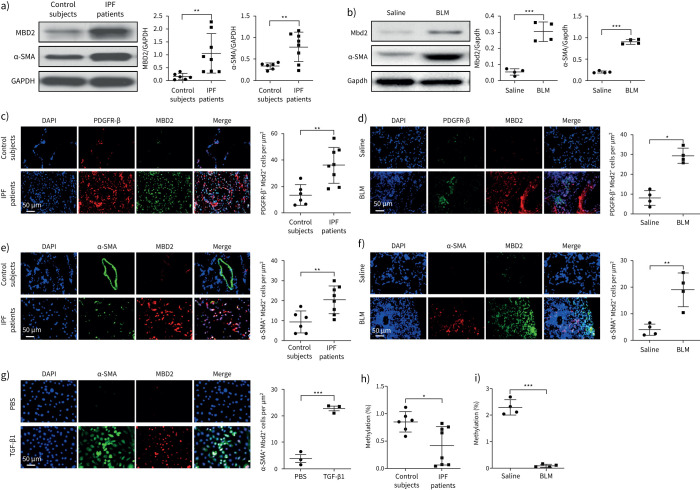
To address the role of MBD2 and DNA methylation in IPF, we first examined MBD2 expression in lungs from IPF patients and mice following BLM-induced pulmonary fibrosis. It was noted that IPF patients featured significantly higher expression of α-SMA (a marker of myofibroblasts), and a seven-fold higher MBD2 expression was noted, as compared to the lungs from control subjects (figure 1a). Similarly, lungs from mice following BLM induction also exhibited almost a three-fold higher Mbd2 expression than that of controls, along with overexpression of α-SMA in the fibrotic lungs (figure 1b). These results prompted us to characterise the cells showing altered MBD2 expression. We first checked alveolar epithelial type II cells (AECII), as their impairment and apoptosis are one of the major risk factors for initiation of IPF [28]. Compared to control subjects, co-immunostaining of MBD2 and pro-SPC, a marker of AECII, failed to detect MBD2 overexpression in pro-SPC+ AECIIs in the lung sections from IPF patients (supplementary figure S1a), and similar results were obtained in lung sections originated from BLM-induced mice (supplementary figure S1b). Consistently, no detectable difference in terms of AECII apoptosis was observed between BLM challenged WT and Mbd2−/− mice (supplementary figure S1c), which was further confirmed by the comparable expression of pro-apoptotic protein Bax (supplementary figure S1d). We then embarked on mesenchymal cells, which are a major contributor to the progression of matrix deposition and tissue distortion during the course of IPF development [29]. MBD2 was noted to be highly expressed in the lung mesenchymal cells from IPF patients, as evidenced by the co-immunostaining of PDGFR-β (figure 1c). Similarly, Mbd2 was almost undetectable in the lung sections originated from control mice, while lung sections derived from fibrotic mice were characterised by the accumulation of mesenchymal cells along with Mbd2 overexpression (figure 1d). Given that α-SMA+ and PDGFRα+ mesenchymal cells represent two separate lineages with distinct gene expression profiles in adult lungs [30], we next checked the expression pattern of MBD2 in mesenchymal cells. Indeed, increased number of α-SMA+MBD2+ and PDGFRα+MBD2+ cells were detected in the lung sections from IPF patients and pulmonary fibrotic mice (figure 1e and f, supplementary figure S2a and b). However, many more α-SMA+MBD2+ cells, and fewer PDGFRα+MBD2+ cells were observed in the lung sections derived from IPF patients and pulmonary fibrotic mice. To further confirm this observation, we induced WT primary lung fibroblast differentiation with TGF-β1, and noted that the differentiated myofibroblasts featured overexpression of Mbd2 (figure 1g).
FIGURE 1.
Analysis of methyl-CpG-binding domain 2 (MBD2) expression and DNA methylation in idiopathic pulmonary fibrosis (IPF) patients and mice with bleomycin (BLM) induction. a) Western blot analysis of MBD2 and α-smooth muscle actin (SMA) expression in the lungs of control subjects and IPF patients. b) Western blot analysis of Mbd2 and α-SMA expression in the lungs of mice following 21 days of BLM induction. c, d) Representative results for co-immunostaining of MBD2 and platelet-derived growth factor receptor (PDGFR)-β in lung sections from patients: c) control subjects and IPF patients and d) BLM-induced lung sections. e–g) Results for co-immunostaining of MBD2 and α-SMA, a myofibroblast marker, in e) lung sections from patients with IPF and control subjects, f) BLM-induced lung sections and g) transforming growth factor (TGF)-β1-induced fibroblasts. The nuclei were stained blue using 4′,6-diamidino-2-phenylindole (DAPI), and the images were taken under original magnification ×400. h) Global DNA methylation rate in the lungs of control subjects and IPF patients. i) Global DNA methylation rate in the lungs of mice following BLM induction. Eight patients with IPF and six control subjects were analysed. Five mice were analysed in each group. Data are presented as mean±sd. *: p<0.05, **: p<0.01, ***: p<0.001.
As mentioned earlier, MBD2 acts as a reader to interpret DNA methylome-encoded information; these results led us to compare the global DNA methylation levels in the lungs between IPF patients and control subjects as well as in fibrotic lungs from mice following BLM induction. Indeed, a significant global DNA demethylation was noted in the lungs from IPF patients (figure 1h), and similar results were also obtained in the fibrotic lungs from mice (figure 1i). Collectively, these data support that pulmonary fibrosis features the induction of MBD2 overexpression along with a reduction of global DNA methylation levels.
Mbd2 overexpression depends on TβRI-Smad3 signalling
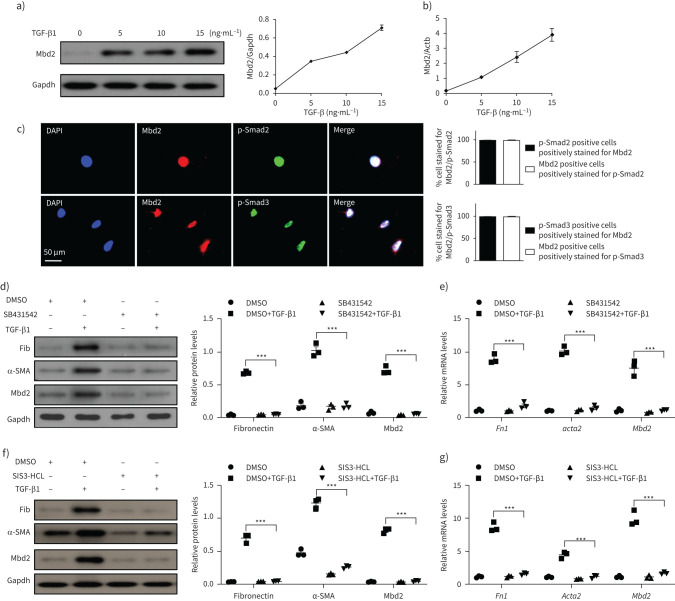
Next, we conducted Western blot and RT-PCR analysis in primary mouse lung fibroblasts following TGF-β1 stimulation, and demonstrated that TGF-β1 induces Mbd2 overexpression in a dose-dependent manner (figure 2a and b). Since canonical TGF-β signalling is crucial for the function of TGF-β1 [31], we performed co-immunostaining of Mbd2 and p-Smad2/3 in these cells following 1 h TGF-β1 stimulation. As expected, Mbd2 was colocalised with p-Smad2 and p-Smad3 in the differentiated myofibroblasts (figure 2c). These findings allowed us to assume that Mbd2 might exert its function downstream to TGF-β1-induced fibrotic signalling. To address this notion, primary mouse lung fibroblasts were treated with SB431542, a selective inhibitor of TGF-β receptor I (TβRI), and SIS3-HCl (a specific inhibitor of Smad3) followed by TGF-β1 stimulation, respectively. TGF-β1 strongly induced fibroblast differentiation as evidenced by the expression of the myofibroblast markers fibronectin and α-SMA, whereas inhibition of TβRI by SB431542 (10 nM) (figure 2d and e) or Smad3 by SIS3-HCl (5 nM) (figure 2f and g) prevented upregulation of fibronectin and α-SMA. More importantly, addition of either SB431542 (figure 2d and e) or SIS3-HCl (figure 2f and g) completely abolished TGF-β1–induced Mbd2 expression, supporting that Mbd2 overexpression was controlled by the canonical TGF-β signalling, in which TβRI and Smad3 were essential mediators.
FIGURE 2.
Transforming growth factor (TGF)-β1 induces methyl-CpG-binding domain 2 (Mbd2) in a TβRI and Smad3-dependent manner. a, b) Western blot and reverse transcriptase (RT)-PCR analysis of Mbd2 expression in the primary lung fibroblasts following different doses of TGF-β1 induction. c) Results for co-immunostaining of Mbd2 and p-Smad2 and Mbd2 and p-Smad3 in the primary lung fibroblasts following TGF-β1 induction for 1 h. The nuclei were stained blue using 4′,6-diamidino-2-phenylindole (DAPI), and the images were taken under original magnification ×400. d, e) Western blot and RT-PCR analysis of Mbd2 expression in the primary lung fibroblasts pre-treated with SB431542 following TGF-β1 induction. f, g) Western blot and RT-PCR analysis of Mbd2 expression in the primary lung fibroblasts pre-treated with SIS3-HCL following TGF-β1 induction. Data are presented as mean±sem of three independent experiments. Gapdh: glyceraldehyde 3-phosphate dehydrogenase; Fib: fibronectin; DMSO: dimethyl sulfoxide; SMA: smooth muscle actin. ***: p<0.001.
Mbd2 is required for fibroblast differentiation into myofibroblast
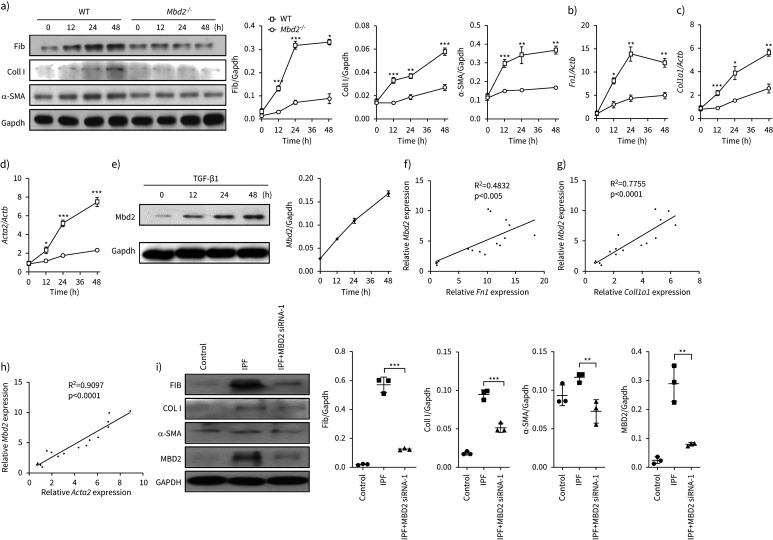
To assess the functional relevance of Mbd2 overexpression in fibroblasts, we checked the impact of Mbd2 deficiency on fibroblast differentiation. Remarkably, Mbd2 deficiency significantly repressed fibroblast differentiation, as evidenced by the attenuated expression of fibronectin, collagen I and α-SMA following TGF-β1 stimulation (figure 3a). RT-PCR analysis of Fn1 (figure 3b), Col1a1 (figure 3c) and Acta2 (figure 3d) further confirmed this observation. In line with these results, TGF-β1 time-dependently induced Mbd2 expression, and the highest expression was noted following 48 h of stimulation (figure 3e). In addition, Mbd2 overexpression was highly correlated with fibroblast differentiation, as evidenced by the detection of a positive correlation with Fn1 (R2=0.4832, p<0.005; figure 3f), Col1a1 (R2=0.7755, p<00001; figure 3g) and Acta2 expression (R2=0.9097, p<0.0001; figure 3h). More importantly, inhibition of MBD2 expression by siRNA in the lung fibroblasts derived from IPF patients and control subjects significantly attenuated the expression of fibronectin, collagen I and α-SMA (figure 3i and supplementary figure S3). Collectively, our data suggest that the induction of Mbd2 overexpression is essential for fibroblast differentiation. In support of this notion, Mbd2 deficiency did not show a significant impact on fibroblast migration (supplementary figure S4a), proliferation (supplementary figure S4b) and apoptosis (supplementary figure S4c), as determined by the Transwell assay, EdU staining and annexin V/PI staining, respectively. Together, those data indicate that Mbd2 might selectively modulate fibroblast to myofibroblast differentiation.
FIGURE 3.
Loss of methyl-CpG-binding domain 2 (Mbd2) attenuated fibroblast differentiation into myofibroblast. a) Results for time-course Western blot analysis of fibronectin (Fib), collagen I (Coll I) and α-smooth muscle actin (SMA) expression in mouse lung fibroblasts following transforming growth factor (TGF)-β1 stimulation. Left: representative Western blot; right: bar graph showing the results with three replications. b–d) Results for time-course reverse transcriptase (RT)-PCR analysis of b) Fn1, c) Coll1a1 and d) Acta2 in primary mouse lung fibroblasts following TGF-β1 treatment. e) Results for time-course Western blot analysis of Mbd2 expression in mouse lung fibroblasts following TGF-β1 stimulation. f–h) RT-PCR analysis of the correlation between Mbd2 and f) Fn1, g) Coll1a1 and h) Acta2 expression after TGF-β1 induction. i) Western blot analysis of the expression of fibronectin, collagen I, α-SMA and MBD2 in control subject and idiopathic pulmonary fibrosis (IPF) patient lung-derived fibroblasts. Gapdh: glyceraldehyde 3-phosphate dehydrogenase; WT: wild type. Data are presented as mean±sem of three independent experiments. *: p<0.05, **: p<0.01, ***: p<0.001.
Mbd2 deficiency in fibroblasts or myofibroblasts protect mice against BLM-induced lung injury and fibrosis
Since Mbd2 is required for myofibroblast polarisation, the next key question is whether Mbd2 deficiency in fibroblasts would protect mice against pulmonary fibrosis. We generated Coll1α2-CreERT2+Mbd2flox/flox mice as described, and Mbd2 deficiency in fibroblasts (Mbd2-CFKO) was induced by intraperitoneal injection of tamoxifen for five consecutive days (figure 4a) on day 12 before BLM induction. Mbd2 deficiency in collagen I+ cells was confirmed by co-immunostaining of lung sections (figure 4b, arrows), and was validated by genotyping to detect the null allele (supplementary figure S5a, box) and Coll1α2-Cre allele in DNA isolated from lung tissues (supplementary figure S5b).
The mice were sacrificed on day 21 following BLM induction. As compared to the control (Mbd2-C) mice, the Mbd2-CFKO mice displayed significantly attenuated lung injury and fibrosis as manifested by H&E and Masson's trichrome staining (figure 4c, left panel) along with much lower Ashcroft scores for the severity of pulmonary fibrosis (figure 4c, right panel). Consistently, significantly lower levels of hydroxyproline, a major component of fibrillar collagen of all types, were also detected in the Mbd2-CFKO mice (figure 4d). Western blot analysis of myofibroblast markers fibronectin, collagen I and α-SMA in the lung lysates also indicated significantly lower levels of expression in the Mbd2-CFKO mice (figure 4e), which were validated by RT-PCR analysis (figure 4f–h).
To further confirm the critical role of Mbd2 in the process of fibroblast differentiation in vivo, we further generated a myofibroblast-specific Mbd2-deficiency mouse model by crossing the Acta2-CreERT2+ mice with the Mbd2flox/flox mice as described earlier, and Mbd2 deficiency in myofibroblasts (Mbd2-CMKO) was induced by intraperitoneal injection of tamoxifen for five consecutive days on day 10 after BLM induction (figure 5a). Mbd2 deficiency in α-SMA+ cells was confirmed by co-immunostaining of lung sections (figure 5b, white arrows) and genotyping (supplementary figure S5c and d). Indeed, the Mbd2-CMKO mice exhibited strikingly alleviated lung injury and fibrosis, as evidenced by the pathologic staining and lower Ashcroft scores, compared with Mbd2-C mice, following 21 days of BLM injection (figure 5c). Additionally, remarkably lower levels of hydroxyproline were observed in the Mbd2-CMKO mice (figure 5d). Consistently, the expression of fibrotic markers was evidently blunted in Mbd2-CMKO mice as detected by Western blotting (figure 5e) and RT-PCR analysis (figure 5f–h).
FIGURE 5.
Comparison of the severity of lung fibrosis between methyl-CpG-binding domain 2 (Mbd2)-CMKO and Mbd2-C mice after bleomycin (BLM) induction. a) Schematic illustration of the generation of myofibroblast specific Mbd2 knockout mice. b) Representative results for co-immunostaining of α-smooth muscle actin (SMA) and Mbd2 in lung sections derived from Mbd2-C and Mbd2-CMKO mice. Nuclei were stained blue by 4′,6-diamidino-2-phenylindole (DAPI), and the images were taken at an original magnification of ×400. c) Histological analysis of the severity of lung fibrosis in mice after BLM induction. Left: representative images for haematoxylin and eosin (H&E) and Masson staining; right: Ashcroft scores. d) Quantification of hydroxyproline contents in Mbd2-CMKO and Mbd2-C mice after BLM challenge. e) Western blot analysis of levels of fibronectin (Fib), collagen I (CollI) and α-SMA. f–h) Results for reverse transcriptase (RT)-PCR analysis of f) Fn1, g) Coll1a1 and h) Acta2. Five to six mice were included in each study group. Data are presented as mean±sd. Gapdh: glyceraldehyde 3-phosphate dehydrogenase. *: p<0.05, **: p<0.01, ***: p<0.001.
To further validate the observed phenotype, the conventional Mbd2−/− mice were further employed for induction of pulmonary fibrosis. Similar as the Mbd2-CFKO and Mbd2-CMKO mice, the Mbd2−/− mice were remarkably resistant to BLM-induced lung injury and fibrosis (supplementary figure S6). Taken together, our data support that loss of Mbd2 in fibroblasts or myofibroblasts protects mice against BLM-induced pulmonary fibrosis.
Mbd2 represses Erdr1 expression to promote fibroblast differentiation
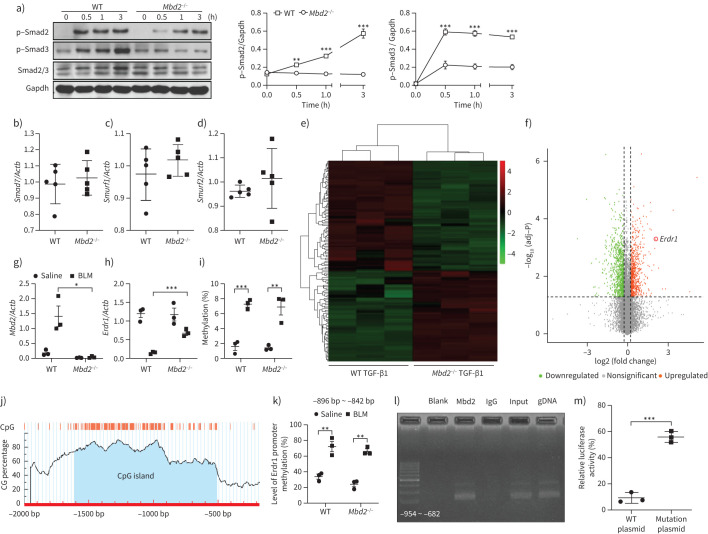
To address more detailed mechanisms by which Mbd2 deficiency attenuates fibroblast to myofibroblast conversion, we embarked on TGF-β downstream signalling molecules using the Mbd2−/− fibroblasts along with TGF-β1 stimulation. Our earlier data indicated that TGF-β1 induction of Mbd2 overexpression depends on TβRI-Smad3 signalling (figure 2d–g), and indeed, TGF-β1 stimulated a steady increase of p-Smad2/3 expression. However, it was noted that lack of Mbd2 significantly attenuated TGF-β1 induced p-Smad2/3 expression (figure 6a), but Mbd2 deficiency did not affect mitogen-activated protein kinase and phosphoinositide 3-kinase signalling as evidenced by the absence of a significant difference in terms of p-P38, p-Jnk, p-Erk1/2, p-P85, p-Akt and p-mTOR expression between TGF-β1-stimulated WT and Mbd2−/− fibroblasts (supplementary figure S7a and b). Similarly, Mbd2 deficiency did not show a perceptible effect on the expression of repressors Smad7 (figure 6b), SMAD specific E3 ubiquitin protein ligase 1 (Smurf1; figure 6c), and Smurf2 (figure 6d), for the TGF-β-Smad2/3 signalling. Collectively, our data suggest a possible positive feedback regulatory loop between TGF-β-Smads signalling and Mbd2 expression in the lung fibroblasts.
FIGURE 6.
Methyl-CpG-binding domain 2 (Mbd2) represses the expression of erythroid differentiation regulator (Erdr)1 in fibroblasts following transforming growth factor (TGF)-β1 treatment. a) Results for time-course Western blot analysis of p-Smad2, p-Smad3 and Smad2/3 expression in fibroblasts following TGF-β1 stimulation. Left: representative Western blot result for p-Smad2, p-Smad3 and Smad2/3 expression at different time points after TGF-β1 stimulation. Right: bar graph showing the results for three replicates. b–d) Reverse transcriptase (RT)-PCR results for analysis of b) Smad7, c) Smurf1 and d) Smurf2 expression in lung fibroblasts originated from wild-type (WT) and Mbd2−/− mice following TGF-β1 stimulation. e) Heat map and f) volcano plot for the differentially expressed genes identified by RNA-sequencing analysis. The colour of the heat map represents the fold enrichment in each sample. g) and h) RT-PCR analysis of g) Mbd2 and h) Erdr1 expression in lung fibroblasts from WT and Mbd2−/− following TGF-β1 stimulation. i) Global DNA methylation rate in fibroblasts after TGF-β1 treatment. j) Predicted CpG island of Erdr1 promoter. k) Results for the bisulfite DNA sequencing analysis of the Erdr1 promoter. l) Chromatin immunoprecipitation results for analysis of Mbd2 binding activity to the Erdr1 promoter. m) Relative luciferase activity in fibroblasts following TGF-β1 induction. Smurf: SMAD-specific E3 ubiquitin protein ligase. Data are presented as mean±sem of three independent experiments. Gapdh: glyceraldehyde 3-phosphate dehydrogenase. *: p<0.05, **: p<0.01, ***: p<0.001.
RNA-seq was employed to compare the expression patterns of genes related to fibroblast differentiation in WT and Mbd2−/− lung fibroblasts. Representative Mbd2-associated genes changed by more than 2.5-fold were listed in the heat map (figure 6e) and volcano plot (figure 6f). We found that 1276 mRNAs, 418 lncRNAs and one ncRNA were upregulated, while 1001 mRNAs, 185 lncRNAs and two ncRNAs were downregulated in WT lung fibroblasts as compared to that of Mbd2−/− lung fibroblasts following TGF-β1 treatment. As expected, the results of RNA-seq and RT-PCR both showed that Mbd2 was highly expressed in WT fibroblasts (figure 6g). It was noted that Erdr1, a stress-related survival factor known to inhibit synovial fibroblast migration during the progression of rheumatoid arthritis [32], was included in a cluster of genes whose expression was upregulated in the Mbd2−/− fibroblasts (figure 6f). RT-PCR analysis confirmed that TGF-β1-induced Erdr1 expression was increased 6.5-fold in Mbd2−/− lung fibroblasts compared to that in WT lung fibroblasts (figure 6h). Importantly, upon TGF-β1 stimulation WT fibroblasts manifested an eight-fold decrease of Erdr1 mRNA levels (figure 6h). Consistently, a strikingly dropping Erdr1 mRNA expression was observed in lung tissues originated from IPF patients and control subjects (supplementary figure S8).
Given that Mbd2 serves as a reader to interpret the information encoded by DNA methylation [11–13], the data prompted us to analyse DNA methylation state of genomic DNA and Erdr1 promoter by global DNA methylation assay and bisulfite DNA sequencing, respectively. Fibroblasts from the lungs of IPF patients manifested higher global genomic DNA hypermethylation than those originating from control subjects (supplementary figure S9). Remarkably, TGF-β1 induced a global genomic DNA hypermethylation in mouse fibroblasts (figure 6i), but no perceptible difference in terms of DNA methylation was noted between WT and Mbd2−/− fibroblasts (figure 6i). However, bioinformatic analysis (Laboratory of Molecular Medicine, www.urogene.org/index.html) predicted one CpG island within the Erdr1 promoter (figure 6j). Bisulfite DNA sequencing confirmed that TGF-β1 induced the Erdr1 promoter to undergo a DNA hypermethylation at regions from −896bp to −842 bp (transcription starting site as +1), which contains 12 CpG sites, and six of which showed significant methylation differences (figure 6k). Similarly, the Erdr1 promoter showed comparable methylation levels between WT and Mbd2−/− fibroblasts (figure 6k).
The next key question is whether Mbd2 selectively binds to the hypermethylated CpG DNAs within the Erdr1 promoter. ChIP assay was conducted, and analysis of the resulting ChIP-PCR products confirmed that Mbd2 bound to the Erdr1 promoter in the regions located at −954 bp to −682 bp, which included the hypermethylated CpG DNA (figure 6l). Importantly, DNA methylation-dependent luciferase reporter assays confirmed that the transcriptional activity of Erdr1 was higher in the myofibroblasts with mutant plasmid (no CpG DNA at regions from −896 bp to −842 bp) than those with the WT plasmid after TGF-β1 stimulation (figure 6m). Collectively, our data support that TGF-β1 induces Mbd2 overexpression in the lung fibroblasts by enhancing TβRI-Smad3 signalling, and Mbd2 in turn exacerbates TGF-β1 induced Smad2/3 activation; TGF-β1 also induces fibroblasts to undergo a global DNA hypermethylation, particularly for the Erdr1 promoter, and Mbd2 binds to the methylated CpG DNA to repress Erdr1 expression.
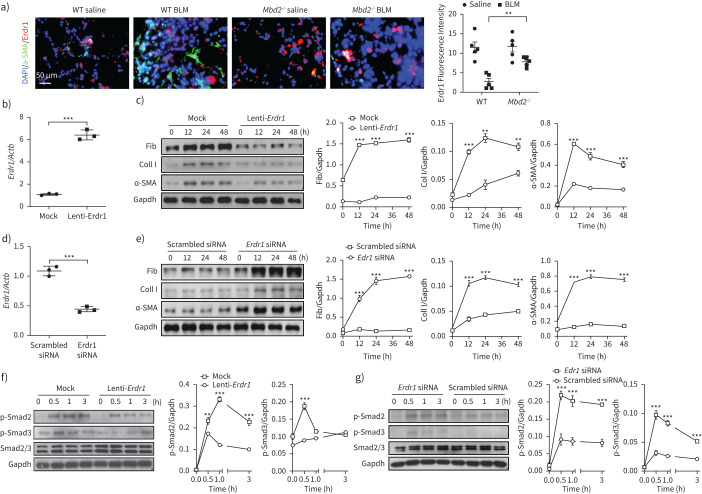
Erdr1 functions as a repressor to inhibit fibroblast differentiation
Next, we sought to address the functional relevance of Erdr1 expression in fibroblast differentiation. We first conducted immunostaining of BLM-induced lung sections. Remarkably, Mbd2−/− fibroblasts were featured by the significantly higher number of α-SMA−Erdr1+ cells, while WT fibroblasts were characterised by the markedly higher number of α-SMA+Erdr1− cells (figure 7a), suggesting that Erdr1 may suppress fibroblast differentiation. Next, WT lung fibroblasts were first transduced with mock or Erdr1 lentiviruses followed by TGF-β1 stimulation. RT-PCR confirmed a six-fold increase of Erdr1 expression in Erdr1-transduced fibroblasts as compared to that of mock-transduced fibroblasts (figure 7b). As expected, compared to mock-transduced fibroblasts, Erdr1-transduced fibroblasts featured the significantly reduced expression of fibronectin, collagen I and α-SMA during the course of TGF-β1 stimulation (figure 7c). Importantly, suppression of Erdr1 by siRNA in Mbd2−/− fibroblasts (figure 7d) restored their capability to differentiate into myofibroblast following TGF-β1 stimulation (figure 7e). To further confirm these results, we compared the expression of TGF-β downstream signalling molecules between mock and Erdr1 lentiviral transduced fibroblasts. Before TGF-β1 stimulation, both types of viral-transduced lung fibroblasts only displayed low levels of p-Smad2 and p-Smad3, while transduction of Erdr1 lentivirus almost completely abolished TGF-β1-induced Smad2 and Smad3 phosphorylation (figure 7f). Importantly, knockdown of Erdr1 by siRNA almost completely abolished the effect of Mbd2 deficiency on the suppression of fibroblast differentiation (figure 7g). Notably, Erdr1 in turn attenuated Mbd2 expression, as evidenced by the lower Mbd2 expression in Erdr1 lentiviral transduced fibroblasts (supplementary figure S10). Collectively, our data support that Erdr1 acts as a strong repressor in TGF-β-induced fibroblast differentiation.
FIGURE 7.
Erdr1 is necessary and sufficient for inhibiting fibroblast differentiation. a) Results for co-immunostaining of erythroid differentiation regulator (Erdr)1 and α-smooth muscle actin (SMA) in lung sections originated from wild-type (WT) and methyl-CpG-binding domain 2 (MBD2) knockout (Mbd2−/−) mice. b) Reverse transcriptase (RT)-PCR analysis of Erdr1 expression in WT fibroblasts following Erdr1 lentiviral transduction. c) Results for time-course Western blot analysis of fibronectin (Fib), collagen (Coll) I and α-SMA expression in transforming growth factor (TGF)-β1-induced WT fibroblasts following mock or Erdr1 lentiviral transduction. Left: representative Western blot result; right: results with three replications. d) RT-PCR results for analysis of Erdr1 expression in TGF-β1-induced Mbd2−/− fibroblasts transfected with an Erdr1 small interfering (si)RNA. e) Results for a time-course Western blot analysis of fibronectin, collagen I and α-SMA expression in TGF-β1-induced Mbd2−/− fibroblasts transfected with a scrambled or an Erdr1 siRNA. Left: representative Western blot result; right: results with three replications. f) Results for time-course Western blot analysis of p-Smad2 and p-Smad3 levels in mock or Erdr1 lentiviral transduced WT fibroblasts following TGF-β1 stimulation. g) A time-course Western blot analysis of p-Smad2 and p-Smad3 levels in scrambled or Erdr1 siRNA transfected Mbd2−/− fibroblasts following TGF-β1 stimulation. BLM: bleomycin. Data are represented as the mean±sem of three independent experiments. Gapdh: glyceraldehyde 3-phosphate dehydrogenase. **: p<0.01, ***: p<0.001.
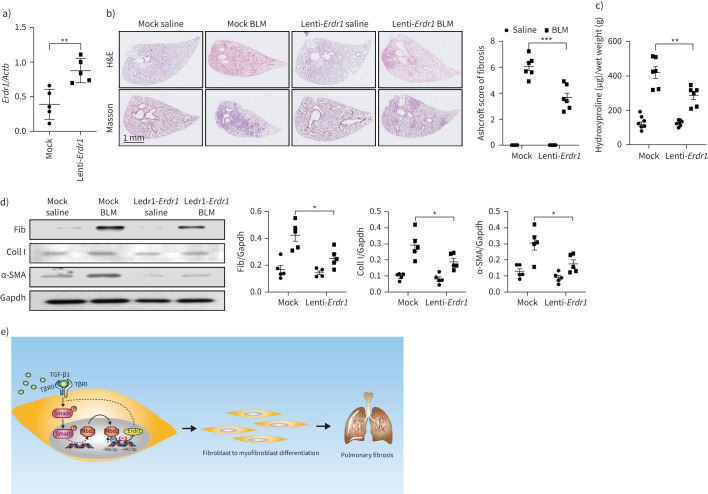
Erdr1 protects mice against BLM-induced pulmonary fibrosis
Finally, we sought to establish whether ectopic Erdr1 expression would protect mice against BLM-induced pulmonary fibrosis. We first confirmed that transduction of Erdr1 lentiviruses (1×107 IFU) by intratracheal injection resulted in a single-fold overexpression of Erdr1 in the lung (figure 8a). WT mice were next subjected to induction of pulmonary fibrosis by BLM injection, and mock or Erdr1 lentiviruses were intratracheally delivered into the lungs at day 10 after BLM induction. Indeed, transduction of Erdr1 viruses significantly alleviated BLM-induced lung fibrosis as illustrated by H&E and Masson's trichrome staining (figure 8b, left) and fibrotic scores (figure 8b, right). Consistently, Erdr1 lentiviral transduced mice manifested much lower levels of hydroxyproline in the lung (figure 8c) coupled with a significant reduction for the expression of fibronectin, collagen I and α-SMA (figure 8d). Together, our data indicate that ectopic Erdr1 expression remarkably reversed the established pulmonary fibrosis.
FIGURE 8.
Ectopic erythroid differentiation regulator (Erdr)1 expression protected mice from bleomycin (BLM)-induced lung fibrosis. a) Reverse transcriptase (RT)-PCR analysis of Erdr1 expression in BLM-induced lungs following Erdr1 lentiviral transduction. b) Lentiviral-delivered Erdr1 expression protected mice from BLM-induced pulmonary fibrosis. Left: representative results for haematoxylin and eosin (H&E) and Masson staining; right: semiquantitative Ashcroft scores relevant to the severity of fibrosis. c) Quantification of hydroxyproline contents in mice transduced with lentiviruses after BLM challenge. d) Western blot for analysis of fibronectin (Fib), collagen (Coll)I and α-smooth muscle actin (SMA) expression. e) Mechanisms underlying methyl-CpG-binding domain 2 (MBD2) regulation of fibroblast differentiation, in which transforming growth factor (TGF)-β induces Mbd2 overexpression in a TβRI/Smad3-dependent manner. Then, Mbd2 selectively binds to the regions of Erdr1 promoter to repress its expression, through which it enhances TGF-β/Smads signalling to promote fibroblast differentiating into myofibroblast. Five to six mice were included in each study group. Data are presented as mean±sd. Gapdh: glyceraldehyde 3-phosphate dehydrogenase. *: p<0.05, **: p<0.01; ***: p<0.001.
Discussion
IPF is a progressive and lethal fibrosis in the interstitium of the lung, lacking effective therapies in the clinical setting [33]. We conducted studies in patients and animals to dissect the impact of MBD2, a reader for DNA methylome-encoded information, on the development of pulmonary fibrosis. We illustrated that IPF patients and mice following BLM-induced pulmonary fibrosis were featured by the altered MBD2 expression and DNA methylation. Particularly, MBD2 was overexpressed in myofibroblasts within the lungs during the course of fibrotic processes. Therefore, loss of Mbd2 in fibroblasts or myofibroblasts provided protection for mice against pulmonary fibrosis following BLM injection by blunting the differentiation of fibroblast into myofibroblast. Mechanistic studies characterised a TβRI/Smad3/Mbd2/Erdr1 positive feedback regulatory loop. Specifically, TGF-β induces Mbd2 overexpression in a TβRI/Smad3-dependent manner, and Mbd2 selectively binds to the regions of Erdr1 promoter that contain the hypermethylated CpG DNAs to repress its expression, through which Mbd2 enhances TGF-β/Smads signalling to promote fibroblast differentiating into myofibroblast. The polarised myofibroblast then secretes copious amount of fibrillary ECM proteins to cause matrix stiffness and pathological matrix deposition in the lung mesenchyme, predisposing to the development of pulmonary fibrosis (figure 8e). These results not only provide novel insights into the understanding of the role of DNA methylation underlying pulmonary fibrosis, but also demonstrate evidence that targeting Mbd2 or Erdr1 could be a viable approach to inhibit fibroblast transition.
Although substantial effort has recently been devoted to dissect the patho-aetiology underlying pulmonary fibrosis, the exact molecular mechanisms remain poorly understood. This lack of related information has significantly hampered the development of novel and effective therapies against this devastating disorder, which renders the 5-year survival rate of IPF patients <30% after diagnosis. It is noted that the occurrence of methylation within the gene of CpG islands is closely related to fibrosis, including pulmonary fibrosis [34], liver fibrosis [35], renal fibrosis [36], cardiac fibrosis [37] and skin fibrosis [38]. Herein, we demonstrated that lungs originated from IPF patients and mice with onset of pulmonary fibrosis display altered DNA methylation coupled with MBD2 overexpression. These observations are consistent with previous studies, in which an altered DNA methylation profile was noted in the lungs of IPF patients [9, 10]. Moreover, the levels of DNA methylation-related proteins, such as DNA methyltransferase 1 (Dnmt1), Dnmt3a, Dnmt3b, MeCP2 and Mbd2 were highly elevated in the lungs of rat pneumoconiosis model, and suppression of DNA methylation by 5-aza-dC, a Dnmt inhibitor, significantly attenuates pulmonary fibrosis [39]. Together, these data support that DNA methylation is implicated in the pathogenesis of pulmonary fibrosis, but the underlying mechanisms are yet to be fully addressed.
The most exciting discovery in this report is that TGF-β1 induces lung fibroblasts to undergo a global DNA hypermethylation along with Mbd2 overexpression. The induced Mbd2 then selectively binds to the methylated CpG DNA within the Erdr1 promoter without affecting the methylation levels within the Erdr1 promoter, by which Mbd2 represses Erdr1 expression to promote the differentiation of fibroblast into myofibroblast, thereby predisposing to the development of IPF. Interestingly, Mbd2 did not seem to have a significant impact on fibroblast migration, proliferation or apoptosis. It seems contradictory in terms of global DNA hypomethylation and hypermethylation of the Erdr1 promoter in the setting of pulmonary fibrosis. In fact, regulatory mechanisms underlying DNA methylation during the progression of pulmonary fibrosis are very complicated. It is seemly that the effects of DNA methylation on pulmonary fibrosis are dependent on the hypomethylation of genes in favour of fibrosis along with the hypermethylation of critical genes against fibrosis in key types of cells (e.g. macrophages and fibroblasts), rather than the global hypomethylation or hypermethylation in DNAs derived from the lungs or cells. For example, Thy-1, a “fibrosis suppressor” gene, has been noted to be regulated by DNA methylation. Specifically, the fibroblast Thy-1 promoter region undergoes a DNA hypermethylation upon the stimulation of fibrotic factors, which blunts Thy-1 expression and causes persistent differentiation of fibroblasts during the course of pulmonary fibrosis [40].
The critical issue is the strategies to dissect the pathways relevant to Mbd2 regulation of fibroblast differentiation. Previous studies suggested compelling evidence that TGF-β/Smads signalling is a common feature of fibrotic diseases and has been shown contributing to the transition of fibroblast [5, 6, 41]. These observations prompted us to focus on the canonical TGF-β signalling in our case. Indeed, we found that Mbd2 overexpression in fibrotic condition was dependent on the canonical TGF-β signalling and characterised that TβRI and Smad3 act as the essential mediators. Additionally, loss of Mbd2 markedly inhibited TGF-β1 induced Smad2 and Smad3 phosphorylation as displayed by the significantly lower levels of p-Smad2 and p-Smad3 in Mbd2−/− fibroblasts as compared to that of WT fibroblasts. Collectively, those data provided evidence supporting that Mbd2 enhances canonical TGF-β signalling, thereby contributing to the persistent fibroblast differentiation in pulmonary fibrosis.
The last important question is how Mbd2 promotes TGF-β/Smads signalling. RNA-seq was then employed to address the underlying mechanisms. Erdr1, a stress-related survival factor, which has been found to attenuate rheumatoid arthritis by inhibition of synovial fibroblast migration [32], was identified with a 6.5-fold increase in Mbd2−/− fibroblasts compared to that of WT fibroblasts. Interestingly, under physiological conditions, the Erdr1 promoter was hypomethylated. In contrast, upon TGF-β1 stimulation, the Erdr1 promoter in fibroblasts underwent a steady DNA hypermethylation, and Mbd2 selectively bound to the methylated CpG DNA, by which it suppressed Erdr1 expression. Given the fact that Mbd2 itself does not affect DNA methylation rather by interpreting the effect of DNA methylation on the suppression of gene expression [42], and therefore, Mbd2 deficiency rendered a failure to decipher the information encoded by the change of DNA methylation in the Erdr1 promoter following TGF-β1 stimulation, although the change of DNA methylation was indeed induced. We further adopted gain and loss of function studies and confirmed that Erdr1 acts as a negative regulator to suppress fibroblast differentiation by inhibiting TGF-β/Smads signalling. In line with these results, intratracheal administration of Erdr1 lentivirus significantly attenuated BLM-induced pulmonary fibrosis. Since ectopic Erdr1 expression was administrated during the “fibrotic” phase of the model, it was more applicable and reflective to the clinical management of IPF patients.
Our study has some limitations. As no commercial antibodies for human ERDR1 could be obtained at present, we did not detect the protein level of ERDR1 in the lungs from IPF patients and control subjects, despite that we obtained consistent results by RT-PCR analysis. Although we demonstrated that Mbd2 was upregulated by TGF-β1 in a TβR1/Smad3-dependent manner, but the detailed mechanisms are yet to be fully elucidated. Furthermore, in the current study we did not address how Erdr1 as a highly conserved autocrine factor represses TGF-β/Smads signalling, which would be a major focus for our follow-up studies.
In summary, we have demonstrated that lungs originated from IPF patients and mice with onset of pulmonary fibrosis are characterised by the altered DNA methylation along with MBD2 overexpression. Therefore, loss of Mbd2 in fibroblasts or myofibroblasts protected the mice from BLM-induced lung fibrosis. Mechanistically, we have identified a positive feedback regulatory loop between TβRI, Smad3, Mbd2 and Erdr1. Specifically, the activation of TβRI/Smad3 signalling induces Mbd2 overexpression, and Mbd2 in turn further enhances TGF-β/Smads signalling through repressing the expression of Erdr1. Upon TGF-β1 stimulation, fibroblasts undergo a global DNA hypermethylation along with Mbd2 overexpression. Erdr1 acts as a negative regulator to inhibit fibroblast differentiation into myofibroblast, while CpG DNAs within the Erdr1 promoter becomes highly methylated following TGF-β1 stimulation, and Mbd2 selectively binds to those methylated CpG DNAs to repress Erdr1 expression, by which Mbd2 promotes fibroblast differentiation to exacerbate pulmonary fibrosis. Given the fact that Mbd2 itself does not involve the changes of DNA methylation and appears to be dispensable under physiological condition, our data support that strategies aimed at silencing Mbd2 or increasing Erdr1 could be a viable therapeutic approach for prevention and treatment of pulmonary fibrosis in clinical settings.
Supplementary material
Please note: supplementary material is not edited by the Editorial Office, and is uploaded as it has been supplied by the author.
Supplementary table Sl. ERJ-03697-2020.Table_S1 (37.2KB, pdf)
Supplementary table S2. ERJ-03697-2020.Table_S2 (77.3KB, pdf)
Supplementary figure S1. Representative results for coimmunostaining of MBD2 and pro-SPC, a marker of AECII in the lung sections from patients with control subjects and IPF patients (a) and BLM-induced lung sections (b). The nuclei were stained blue by DAPI, and the images were taken under original magnification ×400. c) Representative results for TUNEL assays of lung sections originated from BLM-induced WT and Mbd2−/− mice. Nuclei were stained with DAPI (blue), and images were captured at ×200 magnification. d) Western blot analysis of Bax expression in lungs originated from WT and Mbd2−/− mice following BLM induced. pro-SPC: Prosurfactant Protein C. Four to five mice were included in each study group. ERJ-03697-2020.Figure_S1 (2.8MB, tif)
Supplementary figure S2. Representative results for coimmunostaining of MBD2 and PDGFR-ɑ in the lung sections from patients with control subjects and IPF patients (a) and BLM-induced lung sections (b). The nuclei were stained blue by DAPI, and the images were taken under original magnification ×400. A total of eight patients with IPF and six control subjects were analysed. Four mice were included in each study group. The data are represented as the mean±SD. *: p<0.05; **: p<0.01. ERJ-03697-2020.Figure_S2 (1.6MB, tif)
Supplementary figure S3. a) RT-PCR analysis of the MBD2 expression after MBD2 siRNA-2 transinfected. RT-PCR analysis of the expression of FN1, COL1A1 and ACTA2 in IPF patient (b) and control subject (c) lung derived fibroblasts. The data are represented as the mean±SEM of three independent experiments. *: p<0.05; **: p<0.01; ***: p<0.001. ERJ-03697-2020.Figure_S3 (949.8KB, tif)
Supplementary figure S4. a) Transwell assay analysis of WT and Mbd2−/− fibroblast migration. b) EdU staining analysis of WT and Mbd2−/− fibroblast proliferation. The nuclei were stained blue by DAPI, and the images were taken under original magnification ×200. c) Annexin V/PI staining analysis of WT and Mbd2−/− fibroblast apoptosis. The data are represented as the mean±SEM of three independent experiments. ERJ-03697-2020.Figure_S4 (2.5MB, tif)
Supplementary figure S5. Genotyping results of Coll1ɑ2-CreERT2+Mbd2flox/flox allele (a and b). Genotyping results of Acta2-CreERT2+Mbd2flox/flox allele (c and d). ERJ-03697-2020.Figure_S5 (1.5MB, tif)
Supplementary figure S6. a) Histological analysis of the severity of lung fibrosis in mice after BLM induction. Left panel, representative images for H&E (top) and Masson staining (bottom). Right panel: A bar graph shows the quantitative mean score of the severity of fibrosis. b) Quantification of hydroxyproline contents in WT and Mbd2−/− mice after BLM challenge. c Western blot analysis of levels of fibronectin, collagen I and ɑ-SMA. Left panel: A representative Western blot result. Right panel: A bar graph shows the mean data from all mice analysed in each group. Five to six mice were included in each study group. BLM, bleomycin; Fib, Fibronectin; Col I, collagen I; ɑ-SMA, alpha-smooth muscle actin. Five to six mice were included in each study group. *: p<0.05; **: p<0.01. ERJ-03697-2020.Figure_S6 (3.2MB, tif)
Supplementary figure S7. a) A time-course analysis of the Levels of Erk1/2, p-Erk1/2, Jnk, p-Jnk, P38 and p-P38 in fibroblasts following TGF-β1 treatment. b) A time-course Western blot analysis of P85, p-P85, Akt, p-AKT, mTOR and p-mTOR expression in fibroblasts following TGF-β1 stimulation. ERJ-03697-2020.Figure_S7 (1.9MB, tif)
Supplementary figure S8. Results for RT-PCR analysis of ERDR1 expression in lung tissues from IPF patients and control subjects. *: p<0.05. ERJ-03697-2020.Figure_S8 (675KB, tif)
Supplementary figure S9. Global DNA methylation rate in fibroblasts from IPF patient and control subject lung. *: p<0.05. ERJ-03697-2020.Figure_S9 (674.7KB, tif)
Supplementary figure S10. Results for time-course Western blot analysis of Mbd2 expression in TGF-β1-induced WT fibroblasts following mock or Erdr1 lentiviral transduction. Left panel: A representative Western blot result. Right panel: A bar graph shows the results with three replications. ***: p<0.001. ERJ-03697-2020.Figure_S10 (862.7KB, tif)
Shareable PDF
Acknowledgements
We are grateful to Adrian Bird (Wellcome Trust Centre for Cell Biology, University of Edinburgh, Edinburgh, UK) for providing the conventional Mbd2 knockout mice.
Footnotes
This article has an editorial commentary: https://doi.org/10.1183/13993003.01536-2022
Author contributions: Yi Wang, Lei Zhang, Teng Huang, Guo-Rao Wu, Qing Zhou, Fa-Xi Wang, Long-Min Chen and Fei Sun performed experiments; Yongjian Xu, Jianping Zhao and Huilan Zhang provided human lung tissues and clinical data; Yongman Lv, Weikuan Gu, Jungang Xie, Fei Xiong, Shu Zhang, Qilin Yu, Ping Yang, Weining Xiong and Cong-Yi Wang designed experiments, analysed data and supported the preparation of the manuscript; Cong-Yi Wang led the investigation and wrote the manuscript.
Conflict of interest: The authors declare no competing financial interests.
Support statement: This study was supported by the National Natural Science Foundation of China (82130023, 91749207, 81920108009, 81770823, 81873656 and 81800068), the Ministry of Science and Technology (2016YFC1305002 and 2017YFC1309603), NHC Drug Discovery Program (2017ZX09304022-07), Health Center of Hubei Province (3202415), Department of Science and Technology of Hubei Province (2020DCD014), Tongji Hospital (HUST) Foundation for Excellent Young Scientist (2020YQ03), and the Innovative Funding for Translational Research from Tongji Hospital. Funding information for this article has been deposited with the Crossref Funder Registry.
References
- 1.Richeldi L, Collard HR, Jones MG. Idiopathic pulmonary fibrosis. Lancet 2017; 389: 1941–1952. doi: 10.1016/S0140-6736(17)30866-8 [DOI] [PubMed] [Google Scholar]
- 2.Yao Y, Wang Y, Zhang Z, et al. Chop deficiency protects mice against bleomycin-induced pulmonary fibrosis by attenuating M2 macrophage production. Mol Ther 2016; 24: 915–925. doi: 10.1038/mt.2016.36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kolb M, Raghu G, Wells AU, et al. Nintedanib plus sildenafil in patients with idiopathic pulmonary fibrosis. N Engl J Med 2018; 379: 1722–1731. doi: 10.1056/NEJMoa1811737 [DOI] [PubMed] [Google Scholar]
- 4.Sack C, Raghu G. Idiopathic pulmonary fibrosis: unmasking cryptogenic environmental factors. Eur Respir J 2019; 53: 1801699. doi: 10.1183/13993003.01699-2018 [DOI] [PubMed] [Google Scholar]
- 5.Boutanquoi PM, Burgy O, Beltramo G, et al. TRIM33 prevents pulmonary fibrosis by impairing TGF-β1 signalling. Eur Respir J 2020; 55: 1901346. doi: 10.1183/13993003.01346-2019 [DOI] [PubMed] [Google Scholar]
- 6.Louzada RA, Corre R, Ameziane El Hassani R, et al. NADPH oxidase DUOX1 sustains TGF-β1 signalling and promotes lung fibrosis. Eur Respir J 2021; 57: 1901949. doi: 10.1183/13993003.01949-2019 [DOI] [PubMed] [Google Scholar]
- 7.Lagares D, Ghassemi-Kakroodi P, Tremblay C, et al. ADAM10-mediated ephrin-B2 shedding promotes myofibroblast activation and organ fibrosis. Nat Med 2017; 23: 1405–1415. doi: 10.1038/nm.4419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.El Agha E, Moiseenko A, Kheirollahi V, et al. Two-way conversion between lipogenic and myogenic fibroblastic phenotypes marks the progression and resolution of lung fibrosis. Cell Stem Cell 2017; 20: 261–273. doi: 10.1016/j.stem.2016.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sanders YY, Ambalavanan N, Halloran B, et al. Altered DNA methylation profile in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med 2012; 186: 525–535. doi: 10.1164/rccm.201201-0077OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang IV, Pedersen BS, Rabinovich E, et al. Relationship of DNA methylation and gene expression in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med 2014; 190: 1263–1272. doi: 10.1164/rccm.201408-1452OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng J, Song J, He X, et al. Loss of Mbd2 protects mice against high-fat diet-induced obesity and insulin resistance by regulating the homeostasis of energy storage and expenditure. Diabetes 2016; 65: 3384–3395. doi: 10.2337/db16-0151 [DOI] [PubMed] [Google Scholar]
- 12.Zhou M, Zhou K, Cheng L, et al. MBD2 ablation impairs lymphopoiesis and impedes progression and maintenance of T-ALL. Cancer Res 2018; 78: 1632–1642. doi: 10.1158/0008-5472.CAN-17-1434 [DOI] [PubMed] [Google Scholar]
- 13.Zhong J, Yu Q, Yang P, et al. MBD2 regulates TH17 differentiation and experimental autoimmune encephalomyelitis by controlling the homeostasis of T-bet/Hlx axis. J Autoimmun 2014; 53: 95–104. doi: 10.1016/j.jaut.2014.05.006 [DOI] [PubMed] [Google Scholar]
- 14.Wang Y, Zhang L, Wu GR, et al. MBD2 serves as a viable target against pulmonary fibrosis by inhibiting macrophage M2 program. Sci Adv 2021; 7: eabb6075. doi: 10.1126/sciadv.abb6075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ge Y, Zhang R, Feng Y, et al. Mbd2 mediates retinal cell apoptosis by targeting the lncRNA Mbd2-AL1/miR-188-3p/Traf3 axis in ischemia/reperfusion injury. Mol Ther Nucleic Acids 2020; 19: 1250–1265. doi: 10.1016/j.omtn.2020.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang X, Zhao C, Zhang C, et al. Increased HERV-E clone 4-1 expression contributes to DNA hypomethylation and IL-17 release from CD4+ T cells via miR-302d/MBD2 in systemic lupus erythematosus. Cell Commun Signal 2019; 17: 94. doi: 10.1186/s12964-019-0416-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yue T, Sun F, Wang F, et al. MBD2 acts as a repressor to maintain the homeostasis of the Th1 program in type 1 diabetes by regulating the STAT1-IFN-γ axis. Cell Death Differ 2021; 29: 218–229. doi: 10.1038/s41418-021-00852-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karachanak-Yankova S, Dimova R, Nikolova D, et al. Epigenetic alterations in patients with type 2 diabetes mellitus. Balkan J Med Genet 2015; 18: 15–24. doi: 10.1515/bjmg-2015-0081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gong W, Ni M, Chen Z, et al. Expression and clinical significance of methyl-CpG binding domain protein 2 in high-grade serous ovarian cancer. Oncol Lett 2020; 20: 2749–2756. doi: 10.3892/ol.2020.11836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raghu G, Collard HR, Egan JJ, et al. An official ATS/ERS/JRS/ALAT statement: idiopathic pulmonary fibrosis: evidence-based guidelines for diagnosis and management. Am J Respir Crit Care Med 2011; 183: 788–824. doi: 10.1164/rccm.2009-040GL [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ashcroft T, Simpson JM, Timbrell V. Simple method of estimating severity of pulmonary fibrosis on a numerical scale. J Clin Pathol 1988; 41: 467–470. doi: 10.1136/jcp.41.4.467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Q, Yu J, Hu Y, et al. Indirubin alleviates bleomycin-induced pulmonary fibrosis in mice by suppressing fibroblast to myofibroblast differentiation. Biomed Pharmacother 2020; 131: 110715. doi: 10.1016/j.biopha.2020.110715 [DOI] [PubMed] [Google Scholar]
- 23.Pan T, Zhou Q, Miao K, et al. Suppressing Sart1 to modulate macrophage polarization by siRNA-loaded liposomes: a promising therapeutic strategy for pulmonary fibrosis. Theranostics 2021; 11: 1192–1206. doi: 10.7150/thno.48152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu Y, Yu J, Wang Q, et al. Tartrate-resistant acid phosphatase 5/ACP5 interacts with p53 to control the expression of SMAD3 in lung adenocarcinoma. Mol Ther Oncolytics 2020; 16: 272–288. doi: 10.1016/j.omto.2020.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Q, Liu J, Hu Y, et al. Local administration of liposomal-based Srpx2 gene therapy reverses pulmonary fibrosis by blockading fibroblast-to-myofibroblast transition. Theranostics 2021; 11: 7110–7125. doi: 10.7150/thno.61085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu M, Wang X, Xu L, et al. Chronic lung inflammation and pulmonary fibrosis after multiple intranasal instillation of PM2.5 in mice. Environ Toxicol 2021; 36: 1434–1446. doi: 10.1002/tox.23140 [DOI] [PubMed] [Google Scholar]
- 27.Wang Y, Zhu J, Zhang L, et al. Role of C/EBP homologous protein and endoplasmic reticulum stress in asthma exacerbation by regulating the IL-4/signal transducer and activator of transcription 6/transcription factor EC/IL-4 receptor α positive feedback loop in M2 macrophages. J Allergy Clin Immunol 2017; 140: 1550–1561. doi: 10.1016/j.jaci.2017.01.024 [DOI] [PubMed] [Google Scholar]
- 28.Martinez FJ, Collard HR, Pardo A, et al. Idiopathic pulmonary fibrosis. Nat Rev Dis Primers 2017; 3: 17074. doi: 10.1038/nrdp.2017.74 [DOI] [PubMed] [Google Scholar]
- 29.Haak AJ, Kostallari E, Sicard D, et al. Selective YAP/TAZ inhibition in fibroblasts via dopamine receptor D1 agonism reverses fibrosis. Sci Transl Med 2019; 11: eaau6296. doi: 10.1126/scitranslmed.aau6296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Biasin V, Crnkovic S, Sahu-Osen A, et al. PDGFRα and αSMA mark two distinct mesenchymal cell populations involved in parenchymal and vascular remodeling in pulmonary fibrosis. Am J Physiol Lung Cell Mol Physiol 2020; 318: L684–L697. doi: 10.1152/ajplung.00128.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sibinska Z, Tian X, Korfei M, et al. Amplified canonical transforming growth factor-β signalling via heat shock protein 90 in pulmonary fibrosis. Eur Respir J 2017; 49: 1501941. doi: 10.1183/13993003.01941-2015 [DOI] [PubMed] [Google Scholar]
- 32.Kim KE, Kim S, Park S, et al. Therapeutic effect of erythroid differentiation regulator 1 (Erdr1) on collagen-induced arthritis in DBA/1J mouse. Oncotarget 2016; 7: 76354–76361. doi: 10.18632/oncotarget.13047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Speca S, Dubuquoy C, Rousseaux C, et al. GED-0507 is a novel potential antifibrotic treatment option for pulmonary fibrosis. Cell Mol Immunol 2020; 17: 1272–1274. doi: 10.1038/s41423-020-0394-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee JU, Son JH, Shim EY, et al. Global DNA methylation pattern of fibroblasts in idiopathic pulmonary fibrosis. DNA Cell Biol 2019; 38: 905–914. doi: 10.1089/dna.2018.4557 [DOI] [PubMed] [Google Scholar]
- 35.Wang L, Marek GW 3rd, Hlady RA, et al. Alpha-1 antitrypsin deficiency liver disease, mutational homogeneity modulated by epigenetic heterogeneity with links to obesity. Hepatology 2019; 70: 51–66. doi: 10.1002/hep.30526 [DOI] [PubMed] [Google Scholar]
- 36.Gu Y, Chen J, Zhang H, et al. Hydrogen sulfide attenuates renal fibrosis by inducing TET-dependent DNA demethylation on Klotho promoter. FASEB J 2020; 34: 11474–11487. doi: 10.1096/fj.201902957RR [DOI] [PubMed] [Google Scholar]
- 37.Xu X, Friehs I, Zhong Hu T, et al. Endocardial fibroelastosis is caused by aberrant endothelial to mesenchymal transition. Circ Res 2015; 116: 857–866. doi: 10.1161/CIRCRESAHA.116.305629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qiu Y, Gao Y, Yu D, et al. Genome-wide analysis reveals zinc transporter ZIP9 regulated by DNA methylation promotes radiation-induced skin fibrosis via the TGF-β signaling pathway. J Invest Dermatol 2020; 140: 94–102. doi: 10.1016/j.jid.2019.04.027 [DOI] [PubMed] [Google Scholar]
- 39.Zhang N, Liu K, Wang K, et al. Dust induces lung fibrosis through dysregulated DNA methylation. Environ Toxicol 2019; 34: 728–741. doi: 10.1002/tox.22739 [DOI] [PubMed] [Google Scholar]
- 40.Sanders YY, Pardo A, Selman M, et al. Thy-1 promoter hypermethylation: a novel epigenetic pathogenic mechanism in pulmonary fibrosis. Am J Respir Cell Mol Biol 2008; 39: 610–618. doi: 10.1165/rcmb.2007-0322OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang P, Luo ML, Song E, et al. Long noncoding RNA lnc-TSI inhibits renal fibrogenesis by negatively regulating the TGF-β /Smad3 pathway. Sci Transl Med 2018; 10: eaat2039. doi: 10.1126/scitranslmed.aat2039 [DOI] [PubMed] [Google Scholar]
- 42.Rao X, Zhong J, Zhang S, et al. Loss of methyl-CpG-binding domain protein 2 enhances endothelial angiogenesis and protects mice against hind-limb ischemic injury. Circulation 2011; 123: 2964–2974. doi: 10.1161/CIRCULATIONAHA.110.966408 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please note: supplementary material is not edited by the Editorial Office, and is uploaded as it has been supplied by the author.
Supplementary table Sl. ERJ-03697-2020.Table_S1 (37.2KB, pdf)
Supplementary table S2. ERJ-03697-2020.Table_S2 (77.3KB, pdf)
Supplementary figure S1. Representative results for coimmunostaining of MBD2 and pro-SPC, a marker of AECII in the lung sections from patients with control subjects and IPF patients (a) and BLM-induced lung sections (b). The nuclei were stained blue by DAPI, and the images were taken under original magnification ×400. c) Representative results for TUNEL assays of lung sections originated from BLM-induced WT and Mbd2−/− mice. Nuclei were stained with DAPI (blue), and images were captured at ×200 magnification. d) Western blot analysis of Bax expression in lungs originated from WT and Mbd2−/− mice following BLM induced. pro-SPC: Prosurfactant Protein C. Four to five mice were included in each study group. ERJ-03697-2020.Figure_S1 (2.8MB, tif)
Supplementary figure S2. Representative results for coimmunostaining of MBD2 and PDGFR-ɑ in the lung sections from patients with control subjects and IPF patients (a) and BLM-induced lung sections (b). The nuclei were stained blue by DAPI, and the images were taken under original magnification ×400. A total of eight patients with IPF and six control subjects were analysed. Four mice were included in each study group. The data are represented as the mean±SD. *: p<0.05; **: p<0.01. ERJ-03697-2020.Figure_S2 (1.6MB, tif)
Supplementary figure S3. a) RT-PCR analysis of the MBD2 expression after MBD2 siRNA-2 transinfected. RT-PCR analysis of the expression of FN1, COL1A1 and ACTA2 in IPF patient (b) and control subject (c) lung derived fibroblasts. The data are represented as the mean±SEM of three independent experiments. *: p<0.05; **: p<0.01; ***: p<0.001. ERJ-03697-2020.Figure_S3 (949.8KB, tif)
Supplementary figure S4. a) Transwell assay analysis of WT and Mbd2−/− fibroblast migration. b) EdU staining analysis of WT and Mbd2−/− fibroblast proliferation. The nuclei were stained blue by DAPI, and the images were taken under original magnification ×200. c) Annexin V/PI staining analysis of WT and Mbd2−/− fibroblast apoptosis. The data are represented as the mean±SEM of three independent experiments. ERJ-03697-2020.Figure_S4 (2.5MB, tif)
Supplementary figure S5. Genotyping results of Coll1ɑ2-CreERT2+Mbd2flox/flox allele (a and b). Genotyping results of Acta2-CreERT2+Mbd2flox/flox allele (c and d). ERJ-03697-2020.Figure_S5 (1.5MB, tif)
Supplementary figure S6. a) Histological analysis of the severity of lung fibrosis in mice after BLM induction. Left panel, representative images for H&E (top) and Masson staining (bottom). Right panel: A bar graph shows the quantitative mean score of the severity of fibrosis. b) Quantification of hydroxyproline contents in WT and Mbd2−/− mice after BLM challenge. c Western blot analysis of levels of fibronectin, collagen I and ɑ-SMA. Left panel: A representative Western blot result. Right panel: A bar graph shows the mean data from all mice analysed in each group. Five to six mice were included in each study group. BLM, bleomycin; Fib, Fibronectin; Col I, collagen I; ɑ-SMA, alpha-smooth muscle actin. Five to six mice were included in each study group. *: p<0.05; **: p<0.01. ERJ-03697-2020.Figure_S6 (3.2MB, tif)
Supplementary figure S7. a) A time-course analysis of the Levels of Erk1/2, p-Erk1/2, Jnk, p-Jnk, P38 and p-P38 in fibroblasts following TGF-β1 treatment. b) A time-course Western blot analysis of P85, p-P85, Akt, p-AKT, mTOR and p-mTOR expression in fibroblasts following TGF-β1 stimulation. ERJ-03697-2020.Figure_S7 (1.9MB, tif)
Supplementary figure S8. Results for RT-PCR analysis of ERDR1 expression in lung tissues from IPF patients and control subjects. *: p<0.05. ERJ-03697-2020.Figure_S8 (675KB, tif)
Supplementary figure S9. Global DNA methylation rate in fibroblasts from IPF patient and control subject lung. *: p<0.05. ERJ-03697-2020.Figure_S9 (674.7KB, tif)
Supplementary figure S10. Results for time-course Western blot analysis of Mbd2 expression in TGF-β1-induced WT fibroblasts following mock or Erdr1 lentiviral transduction. Left panel: A representative Western blot result. Right panel: A bar graph shows the results with three replications. ***: p<0.001. ERJ-03697-2020.Figure_S10 (862.7KB, tif)
This one-page PDF can be shared freely online.
Shareable PDF ERJ-03697-2020.Shareable (286.3KB, pdf)