Abstract
Transcription in Archaea is initiated by association of a TATA box binding protein (TBP) with a TATA box. This interaction is stabilized by the binding of the transcription factor IIB (TFIIB) orthologue TFB. We show here that the RNA polymerase of the archaeon Methanococcus, in contrast to polymerase II, does not require hydrolysis of the β-γ bond of ATP for initiation of transcription and open complex formation on linearized DNA. Permanganate probing revealed that the archaeal open complex spanned at least the DNA region from −11 to −1 at a tRNAVal promoter. The Methanococcus TBP-TFB promoter complex protected the DNA region from −40 to −14 on the noncoding DNA strand and the DNA segment from −36 to −17 on the coding DNA strand from DNase I digestion. This DNase I footprint was extended only to the downstream end by the addition of the RNA polymerase to position +17 on the noncoding strand and to position +13 on the coding DNA strand.
Initiation of transcription in archaea is mediated by orthologues of the eucaryotic transcription factors TATA box binding protein (TBP) and transcription factor IIB (TFIIB) (11, 26, 30). These two factors, archaeal TBP (aTBP) and transcription factor B (TFB), along with RNA polymerase, are sufficient for initiation of cell-free transcription at some promoters (12, 23). This finding is in line with the data derived from analyses of archaeal genomes that indicate the absence of additional eucaryotic-like initiation factors like TFIIA, TFIIF, and TFIIH in archaea. Competition experiments using templates of different length and gel shift and footprinting experiments have shown that aTBP binds to the archaeal TATA box and that TFB stabilizes binding of aTBP to the promoter (8, 12, 23, 26, 30). This preinitiation complex seems to recruit the archaeal RNA polymerase to the promoter. The pathway of assembly of transcriptional components at the promoter, the existence of a TATA box at position −25 as a major signal directing initiation of transcription and start site selection (10, 25, 27), and the subunit structure and sequence of RNA polymerase (20) indicate a specific evolutionary relationship of the archaeal and eucaryotic RNA polymerase II (Pol II) transcriptional machinery.
In Pol II cell-free transcription systems on linear or relaxed templates, the DNA helicase activity of TFIIH is necessary for open complex formation (14). This activity requires hydrolysis of the β-γ phosphoanhydride bond of ATP in a step prior to initiation of transcription (15, 29, 31). This TFIIH requirement is bypassed by a supercoiled template at some basal Pol II promoters. ATP hydrolysis is required to drive at least two steps in the Pol II system, open complex formation and promoter clearance (25, 31). The striking similarity of the archaeal and eucaryotic Pol II systems posed the question of whether the archaeal RNA polymerase uses a similar mechanism for promoter opening.
In this study, we have used a highly purified Methanococcus cell-free system to investigate the early phase of transcription initiation. Promoter opening was analyzed by potassium permanganate footprinting, and DNA-protein interactions of the preinitiation complex were studied by DNase I footprinting.
MATERIALS AND METHODS
Reagents and enzymes.
[γ-32P]ATP and [α-32P]UTP were purchased from Hartmann Bioanalytics (Braunschweig, Germany). The modified nucleotides and the dinucleotide UpG were from Sigma (St. Louis, Mo.). Restriction endonucleases and other DNA-modifying enzymes were purchased from Fermentas (Vilnius, Lithuania) or New England Biolabs.
Templates for in vitro transcription.
The plasmid pIC-31/2 containing the promoter of the tRNAVal gene was used in standard transcription reactions (10). The plasmid pIC-31/30PRO-C25 was used in addition. It contains the wild-type tRNAVal promoter region, but all cytosine residues from positions +2 to +25 were replaced by other nucleotides (13). Templates were linearized with AlwNI, which cleaves the DNA in the vector region.
Purification of RNA polymerase.
RNA polymerase from Methanococcus thermolithotrophicus was purified as described previously (7).
Expression and purification of recombinant aTBP.
The coding region of aTBP (EMBL accession no. AJ271331) was subcloned using PCR amplification to generate the coding region with an NdeI restriction site at the 5′ end of the sequence and a BamHI site at the 3′ end. The amplified insert was then cloned between the NdeI and the BamHI restriction sites of the pET14b expression vector to generate pTBPMth.14, allowing expression of aTBP with an N-terminal six-histidine tag. BL21(DE3)(pLysS) cells containing the pTBPMth.14 plasmid were grown to an A600 of 0.8 at 25°C. Expression of the proteins was induced by addition of 1 mM isopropyl-1-β-d-thiogalactopyranoside (IPTG). Cells were harvested by centrifugation 3 h after induction, resuspended in buffer (50 mM Tris, pH 8.0, 50 mM NaCl, 20% glycerol), and disrupted by passage through a French press. The lysate was clarified by centrifugation at 4°C (100,000 × g for 20 min), and aTBP was purified by Ni-nitrilotriacetic acid agarose (Qiagen), MonoQ (Pharmacia), and Superdex 200 (Pharmacia) chromatography. The purified proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and stored at −70°C.
Expression and purification of recombinant TFB.
The coding region of TFB (EMBL accession no. AJ271467) was subcloned using PCR amplification to generate the coding region with an NdeI restriction site at the 5′ end of the sequence. The amplified insert was then cloned between the NdeI and the EcoRV restriction sites of the pET17b expression vector to generate pTFBMth.17. BL21(DE3) cells containing the pTFBMth.17 plasmid were grown to an A600 of 0.8 at 37°C. Protein expression and preparation of crude extract with buffer (50 mM Tris, pH 7.5, 300 mM NaCl) was done as described for aTBP. TFB was purified by HiTrap heparin (Pharmacia) and Superdex 200 chromatography.
In vitro transcription reactions.
In vitro transcription experiments were performed in 25-μl reaction mixtures that contained 190 fmol of template, 0.8 pmol of purified RNA polymerase, 1.7 pmol of recombinant aTBP, 1.7 pmol of recombinant TFB, and 4 μM [α-32P]UTP (370 Bq/pmol) in transcription buffer (20 mM Tris, pH 8.5, 2 mM MgCl2, 0.1 mM EDTA, 40 mM KCl, 3 mM dithiothreitol). Other ribonucleotides (Roche Diagnostics, Mannheim, Germany) used for in vitro transcription are indicated in the figure legends. After incubation for 20 min at 55°C, the reaction was terminated by adding 12.5 μl of loading buffer containing 98% formamide, 10 mM EDTA, 0.1% bromophenol blue, and 0.1% xylene cyanol. RNA products were analyzed by electrophoresis on denaturing 20% polyacrylamide gels.
Potassium permanganate footprinting.
Potassium permanganate footprinting was performed with minor modifications of the procedure described by Jiang and Gralla (18). Reaction mixtures were assembled as described above, with 20 fmol of negatively supercoiled or linearized DNA template in onefold transcription buffer. After 20 min of incubation, potassium permanganate was added to 6 mM for 3 min, followed by β-mercaptoethanol (final concentration, 700 mM) to stop the potassium permanganate reaction. EDTA to 10 mM and SDS to 0.5% were added, followed by phenol-chloroform extraction. The DNA was passed through a Sephadex G50 spin column for desalting. Potassium permanganate-hypersensitive sites were detected by asymmetric PCR using end-labeled M13 primers (located 139 bp downstream of the promoter) or M13 reverse primers (located 29 bp upstream of the promoter), 2.4 fmol of the desalted DNA, and a modified Taq DNA polymerase (Fermentas). PCR products were identified on 6% sequencing gels by using sequencing reactions with the same primer as size markers. For nonradioactive detection, a fluorescent-dye-labeled primer (DYEnamic ET primer; Amersham Pharmacia Biotech) was used instead of a radiolabeled primer, and analysis was performed on an ABI 373 or ABI PRISM 310 automated sequencer.
DNase I footprinting.
For DNase footprinting of the template strand, a 220-bp DNA fragment was generated by PCR using a biotinylated M13 and a fluorescent-dye (ABI JOE)-labeled M13 reverse primer. The fragment contains the complete insert of the plasmid pIC-31/30PRO-C25 (13). It was immobilized on streptavidin-coated paramagnetic beads (Dynabeads M280-streptavidin; Dynal, Oslo, Norway) according to the manufacturer's specifications. Reaction mixtures were formed in 20 μl of onefold transcription buffer that contained 70 fmol of DNA template pIC-31/30PRO-C25 and the components which are indicated in the figure legends. In order to get single-hit conditions, partial hydrolysis of the template DNA of each reaction mixture was performed by adding 5 μl of DNase I (Promega, Madison, Wis.) to final concentrations of 5.3, 2.6, 1.3, and 0.65 mU/μl, respectively. Cleavage was done for 1 min at 37°C in onefold transcription buffer with final concentrations of 5 mM MgCl2 and 1 mM CaCl2. DNase I digestion was terminated by addition of an equal volume of 4 M NaCl–100 mM EDTA. The nicked fragments on the beads were washed once with 100 μl of 10 mM Tris-HCl (pH 7.5)–2 M NaCl–1 mM EDTA and once with 100 μl of 10 mM Tris-HCl (pH 7.5)–1 mM EDTA. Finally, the supernatant was removed completely and the beads were mixed with 5.6 μl of loading buffer (85% formamide, 5% dextran blue, 5 mM EDTA) and 0.4 μl of X-Rodamine MapMarker (BioVentures, Inc., Murfreesboro, Tenn.). The samples were denatured for 5 min at 70°C and analyzed on an ABI 373 or ABI PRISM 310 automated sequencer.
RESULTS
Initiation of archaeal transcription does not require hydrolysis of the β-γ phosphoanhydride bond of ATP and GTP.
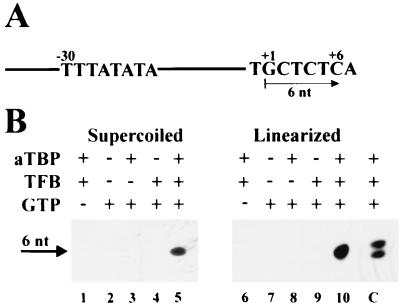
To address the question as to whether ATP is required for activation of Methanococcus transcription, a reconstituted cell-free system consisting of bacterially produced Methanococcus TBP and TFB and highly purified RNA polymerase was used. The first 6 nucleotides (nt) of RNA initiated at the Methanococcus tRNAVal wild-type promoter did not contain an A residue (Fig. 1A). Therefore, provided that hydrolysis of the β-γ bond of ATP is not required for initiation of transcription, synthesis of a hexanucleotide was expected to occur in cell-free transcription reaction mixtures containing only GTP, CTP, and UTP. Analysis of the RNA products revealed that on both supercoiled and linearized templates, a transcript of the expected size was synthesized (Fig. 1B, lanes 5 and 10). When the ATP analogue cordycepin-5′-triphosphate (3′-dATP) was added at a concentration of 50 μM in addition to GTP, CTP, and UTP, synthesis of the same product and of a slightly longer RNA product was observed (Fig. 1B, lane C). With increasing concentrations of 3′-dATP, the intensity of the upper band increased (data not shown), suggesting that the longer RNA product is a heptanucleotide carrying 3′-dAMP at its 3′ terminus. These findings supported the conclusion that the observed 6-nt transcript initiated accurately and exclusively at the G residue located 22 nt downstream of the TATA box that had been identified previously as the initiator nucleotide at this promoter (7). Control reactions showed that the synthesis of this RNA product was dependent upon the presence of two archaeal transcription factors and on the initiator nucleotide of GTP (Fig. 1B, lanes 1 to 4 and 6 to 9). These results demonstrated that a hydrolyzable β-γ phosphoanhydride bond of ATP was not necessary for initiation of transcription in Methanococcus.
FIG. 1.
Transcription in the absence of ATP. (A) Schematic drawing of the template pIC-31/2. The nucleotide sequence of the promoter region and the region around the transcription start site (+1) is shown. In the absence of ATP, synthesis of a 6-nt RNA product (+6) was expected. (B) Transcription reaction mixtures contained RNA polymerase, 4 μM UTP, 20 μM CTP, and template pIC-31/2. The presence (+) or absence (−) of aTBP, TFB, and GTP (20 μM) in the reaction mixture is indicated. For size calibration of the RNA product, the ATP analogue 3′-dATP was added to allow incorporation of the next nucleotide (lane C).
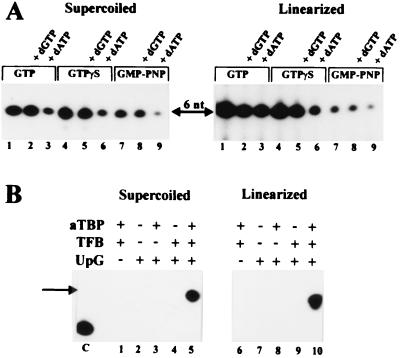
To exclude the possibility that hydrolysis of the β-γ phosphoanhydride bond of GTP catalyzed open complex formation, GTP was replaced in cell-free transcription reaction mixtures by the analogs GTPγS and GMP-PNP containing nonhydrolyzable β-γ phosphoanhydride bonds. On both linearized and supercoiled DNA as a template, GTPγS or GMP-PNP was able to replace GTP (Fig. 2A, compare lanes 1, 4, and 7). Due to ineffective incorporation of the modified nucleotide GMP-PNP into RNA, transcription was reduced in reaction mixtures containing GMP-PNP (Fig. 2A, lanes 7 to 9). However, addition of dGTP or dATP containing a hydrolyzable β-γ phosphoanhydride bond to transcription reaction mixtures did not increase the rate of hexanucleotide synthesis occurring in the presence of either GTP or of analogs of GTP (Fig. 2A, lanes 2, 3, 5, 6, 8, and 9). These findings suggested that initiation of archaeal transcription does not require hydrolysis of the β-γ phosphoanhydride bond of ATP or GTP for synthesis of the first five phosphodiester bonds (Fig. 2A, compare lanes 1 to 3, 4 to 6, and 7 to 9). In the presence of dATP, synthesis of longer RNA products occurred due to trace contaminations of dATP with ATP or to misincorporation of dAMP instead of AMP into RNA. Therefore, the intensity of the signal corresponding to the hexanucleotide was decreased in the presence of dATP (Fig. 2, lanes 3, 6, and 7), although dATP itself did not inhibit hexanucleotide synthesis.
FIG. 2.
Transcription in the presence of GTP analogs and in the presence of the dinucleotide UpG. Transcription reactions were performed as described for Fig. 1 and in Materials and Methods. (A) GTP (lanes 1 to 3) was replaced by 80 μM GTPγS (lanes 4 to 6) and 80 μM GMP-PNP (lanes 7 to 9). Addition of dGTP (20 μM) and dATP (20 μM) is indicated on top of the gel. The expected 6-nt RNA product is marked by an arrow. (B) The presence (+) or absence (−) of aTBP, TFB, and UpG (170 μM) in the reaction mixture is indicated. Note that the electrophoretic mobility of the dinucleotide-initiated transcript (labeled by an arrow) was reduced compared to that of the GTP-initiated transcript (lane C).
To provide conclusive evidence that hydrolysis of the β-γ bond of a purine nucleotide was not required for initiation of transcription, GTP was replaced in transcription assays by the dinucleotide UpG. This dinucleotide was complementary to positions −1 and +1 of the coding DNA strand at the tRNAVal promoter (Fig. 1A). Analysis of RNA products showed that a transcript was synthesized under these conditions whether a negatively supercoiled (Fig. 2B, lane 5) or linearized (Fig. 2B, lane 10) template was used. Since this transcript contained an additional uridine nucleoside at the 5′ end, the electrophoretic mobility of this transcript was reduced compared to that of the GTP-initiated hexanucleotide (Fig. 2B, lane C). These data provide evidence that archaeal transcription could be initiated by a dinucleotide and that hydrolysis of the β-γ bond of ATP or GTP was not required for initiation of transcription at this archaeal promoter.
Analysis of open complex formation.
Since ATP is required for open complex formation in the Pol II system, we have investigated DNA melting of the tRNAVal promoter in the archaeon M. thermolithotrophicus in the presence and absence of ATP. Potassium permanganate (KMnO4) was used as a chemical probe specific for thymidines in single-stranded DNA. First, we have analyzed open complex formation on linearized wild-type DNA. Transcriptional components were incubated with DNA at 55°C. Modified thymidine residues in single-stranded DNA regions were identified by an asymmetric PCR using a 32P-end-labeled primer and Taq DNA polymerase (see Materials and Methods).
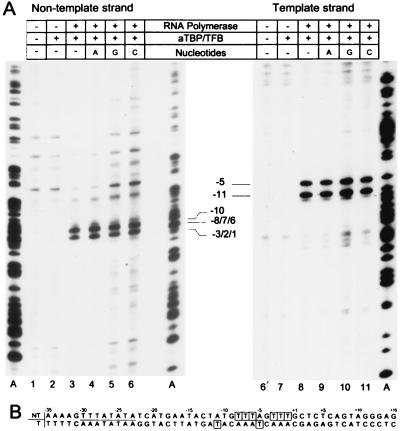
Figure 3A shows the KMnO4-detectable opening of the nontemplate and template DNA strands. Incomplete preinitiation complexes containing only aTBP and TFB showed no significant KMnO4 sensitivity on both DNA strands (lanes 2 and 7) compared with the control lanes (1 and 6). When RNA polymerase was added to the aTBP-TFB-promoter complexes, modification of thymidine residues occurred at positions −3 to −1 and −6 to −8 and also at −10 (Fig. 3A, lane 2). These findings indicate that the open region extended at least from positions −1 to −10 at the noncoding DNA strand. Addition of either ATP, GTP, or CTP had no detectable effect on the modification patterns (Fig. 3A, lanes 4 to 6 and 9 to 11). These findings provide evidence that promoter opening was catalyzed by the archaeal RNA polymerase and that this step did not require energy provided by nucleotide hydrolysis.
FIG. 3.
Open complex formation. (A) Transcription reaction mixtures for open complex formation were assembled as described in Materials and Methods. Linearized pIC-31/2 wild-type DNA was incubated with the components indicated on top of each lane for 20 min at 55°C. After treatment with KMnO4, the hyperreactive sites on the nontemplate (lanes 1 to 6) and the template strand (lanes 6′ to 11) were analyzed by asymmetric PCR using radioactive labeled primers. Positions of reactive thymidine residues were determined by comigration of a sequence ladder terminated with ddATP and are indicated in relation to the transcription start site. Compare the following: lanes 1 and 6′, without protein and nucleotides as a control; lanes 2, 3, 7, and 8, without nucleotides; lanes 4 to 6 and 9 to 11, in the presence of 20 μM ATP (A), GTP (G), or CTP (C). (B) DNA sequence of plasmid pIC-31/2 containing the promoter and the region of the transcription start site. The positions of the modified thymidine residues are boxed. Position numbers refer to the transcription start site.
When KMnO4-detectable opening of the template strand was analyzed in reaction mixtures containing aTBP, TFB, and RNA polymerase, two prominent signals corresponding to T residues at positions −5 and −11 were observed (Fig. 3A, lane 5). The analysis of the KMnO4 footprinting patterns on both DNA strands revealed that the open complex extended at this promoter at least from positions −11 to −1 (Fig. 3B, boxed T residues).
Analysis of temperature dependence of open complex formation.
M. thermolithotrophicus is a moderate thermophile that grows between 30 and 70°C with an optimum at 65°C (16), and the cell-free transcription reactions and analyses of open complex formation shown here were carried out at 55°C. To investigate the ability of the Methanococcus RNA polymerase to form open complexes at temperatures comparable to conditions allowing open complex formation in enteric bacteria and eucaryotes and to analyze the temperature dependence of open complex formation, KMnO4 sensitivity was assayed on both linearized and supercoiled DNA of the nontemplate strand containing a mutated tRNAVal promoter (pIC31/30PRO-C25; see Materials and Methods) at 40, 25, and 10°C. This template, which contains its first C residue at position 26, allows the synthesis of a 25-nt RNA product in transcription reactions carried out in the absence of CTP (13). Permanganate sensitivity was also detected by a PCR-based primer extension reaction, but a nonradiaoctive fluorescence-labeled primer was used and the modified thymidine residues causing termination of the primer extension reaction were detected by an ABI automated DNA sequencer (see Materials and Methods).
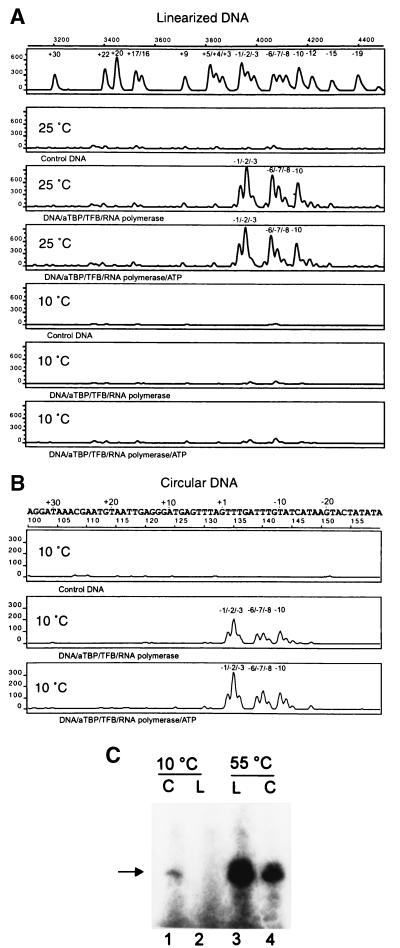
Analysis of KMnO4-detectable opening of linear DNA revealed that at 40 and 25°C, the open complex extended from position −10 to −1 (Fig. 4A and data not shown). Even at 25°C, ATP had no effect on open complex formation (Fig. 4A, compare third and fourth panels from the top).
FIG. 4.
Open complex formation and transcription at low temperatures. Transcription reactions for open complex formation were performed as described in Materials and Methods. Linearized (A) or negatively supercoiled (B) plasmid pIC-31/30PRO-C25 was incubated with the components indicated at the bottom of each panel for 20 min at the temperatures indicated. After treatment with KMnO4, the hyperreactive sites on the nontemplate strand were analyzed by asymmetric PCR using a fluorescent-dye-labeled primer. For fragment size calibration of the peaks, size markers with a different fluorescent dye were added to each probe and size correlation was done by using the global Southern method according to the instructions of the supplier. The DNA sequences of the plasmid pIC-31/30PRO-C25 are shown on top in such a way that the peaks of the chromatograms can be directly correlated to the individual base positions within the DNA sequence. The transcription start site is underlined. (C) Transcription reaction mixtures were incubated for 20 min at the temperatures indicated on top of the lanes using negatively supercoiled (C) or linearized (L) DNA.
With negatively supercoiled DNA as template, promoter opening was shown to occur even at 10°C. The T residues corresponding to position −1 to −3, −6 to −8 and −10 were clearly modified in both the presence and absence of ATP (Fig. 4B, second and third panels from the top). When ATP, GTP, and UTP were added to this reaction, strong additional permanganate signals corresponding to positions +22, +20, +17/+16, and +5 were detected and upstream signals disappeared, indicating bubble extension to the downstream DNA region (data not shown). This finding suggests that at 10°C, synthesis of a 25-nt transcript from a supercoiled template was still possible and an RNA product of this size could be also detected in cell-free transcription assays carried out at 10°C (Fig. 4C). For comparison, transcription reactions carried out at 55°C are shown in addition, with both linear and negatively supercoiled DNA as the template (Fig. 4C, lanes 3 and 4).
Analysis of DNA protein contacts in preinitiation complexes.
To couple the permanganate sensitivity assays of open regions in the DNA template to direct analysis of the RNA polymerase interaction with the promoter, DNase I footprinting experiments were carried out. The DNase I footprinting patterns of the aTBP promoter complex, the aTPB-TFB promoter complex and the aTBP-TFB-RNA polymerase promoter complex were analyzed. Footprinting reactions were carried out with DNase I by using a fluorescence-labeled DNA fragment. The nicked fragments were analyzed by an automated DNA sequencer.
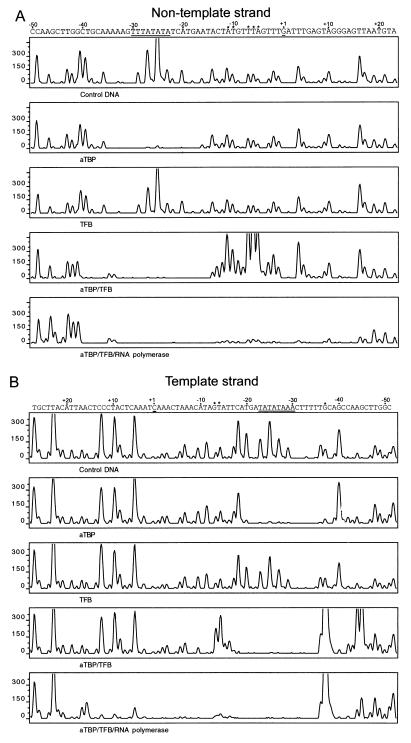
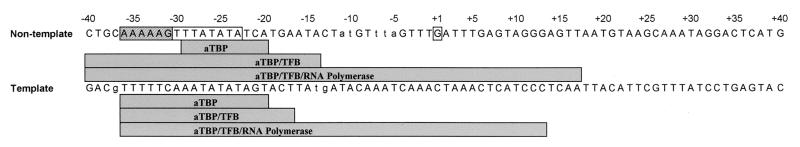
On the nontemplate strand, aTBP protected the DNA region from −29 to −20 from DNase I digestion (Fig. 5A, TFB). TFB did not produce a footprint, and the aTBP-TFB footprint extended from position −40 to −14 (Fig. 5A, TFB and aTBP/TFB). DNase I hypersensitivity sites were observed at positions −5 to −7 and at positions −10 and −11 (aTBP/TFB). When complexes containing aTBP, TFB, and RNA polymerase were analyzed, a considerable extension of the footprint to the downstream DNA region was observed. This complex protected the DNA region from −40 to +17 from DNase I digestion (Fig. 5A, bottom panel). The hypersensitivity sites disappeared in this complex.
FIG. 5.
Interaction of the transcriptional components with the tRNAVal gene. The DNA fragment was incubated with the components (25 pmol of aTBP, 8 pmol of TFB, 0.8 pmol of RNA polymerase) as indicated at the bottom of each chromatogram for 20 min at 37°C, followed by DNase I treatment. Further treatment was as described in Materials and Methods. For size calibration of the peaks of the template strand, size markers with a different fluorescent dye were added to each probe and size calling was done by using the global Southern method according to the instructions of the supplier. The calculation of the fragment length was calibrated using sequencing reactions generated with the fluorescence-labeled primer. For the analysis of the nontemplate strand, the 220-bp DNA fragment was also generated by PCR using a biotinylated M13 reverse primer and a fluorescent-dye (ABI JOE)-labeled M13 primer. The DNA sequences of the plasmid pIC-31/30PRO-C25 are shown on the tops of the chromatograms in such a way that the peaks of the chromatograms can be directly correlated to the individual base positions within the DNA sequence. The promoter and the transcription start site are underlined. *, DNase I hypersensitive sites.
On the template strand, a protection of the BRE element and the TATA box region by aTBP, from positions −36 to −20, was found (Fig. 5B, aTBP). Binding of TFB to DNA was not detected (Fig. 5B, TFB), and the aTBP-TFB footprint extended from positions −36 to −17 (Fig. 5B, aTBP/TFB). DNase I hypersensitivity sites were generated by aTBP and TFB at positions −13/−14 and −37. The complex containing RNA polymerase in addition protected the DNA region from −35 to +13 from DNase I digestion (Fig. 5B, bottom panel). The hypersensitivity sites were also on the DNA strand not observed in the complex containing the RNA polymerase in addition. The results of the footprinting experiments are summarized in Fig. 6.
FIG. 6.
Summary of the results of the DNase I footprinting experiments. The DNA sequence from −40 to +40 relative to the transcription start site is shown. The promoter and the transcription start site are boxed. The regions protected from DNase I digestion of the nontemplate and the template strand are shown as shaded rectangles below each sequence. The sequence of the TATA box is boxed and the sequence of the BRE element is hatched. DNase I hypersensitivity sites are labeled with lowercase letters.
DISCUSSION
The archaeal RNA polymerase does not require hydrolysis of the β-γ bond of ATP for initiation of transcription.
Transcription initiation by Pol II on a linearized template requires hydrolysis of the β-γ bond of ATP. Two steps during early transcription of Pol II promoters are dependent upon energy provided by ATP hydrolysis, open complex formation, and extension of the transcription bubble during RNA synthesis (31). One major finding described here is that although the archaeal transcriptional machinery strikingly resembles the Pol II machinery (see the introduction), the archaeal RNA polymerase did not require hydrolysis of the β-γ bond of ATP or GTP for initiation and thus uses a different mechanism for the early steps of transcription initiation.
Characteristics of the archaeal open complex.
Permanganate footprinting experiments have shown that aTBP- and TFB-bound promoter DNA is in the closed conformation, whereas addition of RNA polymerase results in open complex formation.
Similar to Pol II (31) and E. coli RNA polymerase (6), the archaeal enzyme is able to melt a DNA segment of 10 nt (Fig. 3) located between positions −11 and −1. In striking contrast to the Pol II system, open complex formation occurred on linear DNA in the absence of nucleotides and was not enhanced in the presence of ATP (Fig. 3 and 4). ATP-independent promoter opening of the DNA region between −1 and −12 of a Sulfolobus rRNA promoter has been shown (1). However, in this case, the template was in negatively supercoiled conformation. DNA in this conformation can be also melted by Pol II in the absence of ATP (9, 31). In the Methanococcus system, the extent of the promoter opening is independent of template topology, but negative supercoiling of DNA clearly energetically favors promoter opening as indicated by the lower temperature limit for promoter opening and RNA synthesis with negatively supercoiled DNA compared to linear DNA (Fig. 5).
DNase I footprinting analyses of an archaeal preinitiation complex.
The aTBP-TFB promoter complex has been studied in the Pyrococcus and Sulfolobus systems (12, 24). In addition, the crystal structure of the Pyrococcus preinitiation complex containing N-terminally-truncated TFB and aTBP in complex with promoter DNA was recently determined (22), but the interaction of archaeal RNA polymerase with these aTBP-TFB promoter complexes has not yet been described. Evidence for semispecific initiation of purified Sulfolobus RNA polymerase independent of a TATA box and the presence of transcription factors has been obtained (2, 17). A DNase I footprint of purified Methanococcus RNA polymerase on a homologous hisA promoter (3) and exonuclease III footprints of this enzyme on several promoters (27, 28) were reported, but Methanococcus transcription factors were not included in these studies since they were discovered later (7, 11).
A sequence of 6 nt immediately upstream of the TATA box was identified recently as the recognition element for TFB (BRE) and found to direct the polarity of archaeal transcription (2, 22). This element was also contained within the DNA region protected after addition of Methanococcus TFB. The GC base pair at position −1 of the tRNAVal promoter (relative to the TATA box) deviates from the consensus for BRE sequences RNWAAW (R = purine, W = A or T, N = any base). However, earlier mutational analyses of the tRNAVal promoter provide evidence that the major structural determinants are conserved between the BRE element of the Sulfolobus T6 and Methanococcus tRNAVal promoters. A single-point mutation of the G residue at position −1 (relative to the TATA box) to a T did not affect transcriptional activity of the tRNAVal promoter (10), suggesting that this deviation does not influence the interaction of Methanococcus TFB with BRE. By contrast, when the A residue at position −3 of the tRNAVal BRE element that has been identified as a key determinant of Sulfolobus BRE (2) was replaced by a G, the rate of transcription was reduced to 45% (10).
Similar to the Sulfolobus system (2), a DNase I hypersensitivity site 8 bp upstream of the TATA box was observed. This site was shown to correspond to a bend of the DNA in the Sulfolobus system that brings the major groove of the DNA in the BRE in close contact with a helix-turn-helix motif of TFB (22). In contrast, the DNase I hypersensitivity site downstream of the TATA box has not been observed in the Sulfolobus TPB-TFB promoter complex. This finding provides additional evidence for a significant difference in the interaction of Methanococcus and Sulfolobus TFB with promoter DNA.
Addition of RNA polymerase resulted in a large extension of the footprint to the 3′ end on both DNA strands (Fig. 5 and 6). As the footprint from position −40 to −14 can be attributed to interaction of transcription factors with DNA, the DNA region that was in contact with RNA polymerase extended at least from position −15 to +17 on the nontemplate and from −15 to +14 on the template strand. Direct contacts of the enzyme with the DNA region protected by the transcription factors could not be demonstrated even though the enzyme may also bind to this DNA region.
Comparison with Pol II and bacterial RNA polymerase holoenzyme.
The archaeal system shows similarities to both Pol II and the bacterial RNA polymerase. Like the β′βα2ς70 system, the archaeal RNA polymerase was able to melt promoter DNA without additional factors and ATP hydrolysis. By contrast, open complex formation in the Pol II system requires an additional enzymatic activity, the ATP-dependent DNA helicase of TFIIH.
The RNA polymerase of E. coli protects about 70 bp of DNA from DNase I digestion in the preinitiation complex (5, 19). In contrast to the E. coli enzyme and similar to Pol II, the archaeal enzyme bound to a preformed ternary complex of aTBP and TFB with promoter DNA, and like Pol II (4), the Methanococcus RNA polymerase extended the footprint to the DNA region downstream of the TATA box. In contrast to the Pol II system, no RNA polymerase-induced extension of the DNase I footprint to the region upstream of the TBP binding site was observed in the Methanococcus system. A further difference from the Pol II system was that archaeal TFB extended the TBP footprint on either site of the TATA box, whereas TFIIB causes an extension of the TBP footprint only to the downstream site of the TATA box (4).
Our definition of the open DNA region and of DNA protein interactions in a preinitiated complex allows an examination of the mechanism of transcription bubble and RNA polymerase translocation in isolated complexes stalled in various registers.
ACKNOWLEDGMENTS
This work was supported by the Deutsche Forschungsgemeinschaft and the Fonds der Chemischen Industrie. We appreciate the excellent technical assistance of Jutta Kock.
REFERENCES
- 1.Bell S D, Jaxel C, Nadal M, Kosa P F, Jackson S P. Temperature, template topology, and factor requirements of archaeal transcription. Proc Natl Acad Sci USA. 1998;95:15218–15222. doi: 10.1073/pnas.95.26.15218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bell S D, Kosa P L, Sigler P B, Jackson S P. Orientation of the transcription complex in archaea. Proc Natl Acad Sci USA. 1999;96:13662–13667. doi: 10.1073/pnas.96.24.13662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown J W, Thomm M, Beckler G S, Frey G, Stetter K O, Reeve J N. Archaebacterial RNA polymerase binding site and transcription of the hisA gene of Methanococcus vannielii. Nucleic Acids Res. 1988;16:135–150. doi: 10.1093/nar/16.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buratowski S, Hahn S, Guarente L, Sharp P A. Five intermediate complexes in transcription initiation by RNA polymerase II. Cell. 1989;56:549–561. doi: 10.1016/0092-8674(89)90578-3. [DOI] [PubMed] [Google Scholar]
- 5.Carpousis A J, Gralla J D. Interaction of RNA polymerase with lacUV5 promoter DNA during mRNA initiation and elongation. Footprinting, methylation, and rifampicin-sensitivity changes accompanying transcription initiation. J Mol Biol. 1985;183:165–177. doi: 10.1016/0022-2836(85)90210-4. [DOI] [PubMed] [Google Scholar]
- 6.deHaseth P L, Helmann J D. Open complex formation by Escherichia coli RNA polymerase: the mechanism of polymerase-induced strand separation of double helical DNA. Mol Microbiol. 1995;16:817–824. doi: 10.1111/j.1365-2958.1995.tb02309.x. [DOI] [PubMed] [Google Scholar]
- 7.Frey G, Thomm M, Brüdigam B, Gohl H P, Hausner W. An archaebacterial cell-free transcription system. The expression of tRNA genes from Methanococcus vannielii is mediated by a transcription factor. Nucleic Acids Res. 1990;18:1361–1367. doi: 10.1093/nar/18.6.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gohl H P, Gröndahl B, Thomm M. Promoter recognition in archaea is mediated by transcription factors: identification of aTFB from Methanococcus thermolithotrophicus as archaeal TATA-binding protein. Nucleic Acids Res. 1995;23:3837–3841. doi: 10.1093/nar/23.19.3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goodrich J A, Tjian R. TBP-TAF complexes: selectivity factors for eukaryotic transcription. Curr Opin Cell Biol. 1994;6:403–409. doi: 10.1016/0955-0674(94)90033-7. [DOI] [PubMed] [Google Scholar]
- 10.Hausner W, Frey G, Thomm M. Control regions of an archaeal gene. A TATA box and an initiator element promote cell-free transcription of the RNAVal gene of Methanococcus vannielii. J Mol Biol. 1991;222:495–508. doi: 10.1016/0022-2836(91)90492-o. [DOI] [PubMed] [Google Scholar]
- 11.Hausner W, Thomm M. Purification and characterization of a general transcription factor, aTFB, from the archaeon Methanococcus thermolithotrophicus. J Biol Chem. 1993;268:24047–24052. [PubMed] [Google Scholar]
- 12.Hausner W, Wettach J, Hethke C, Thomm M. Two transcription factors related with the eucaryal transcription factors TATA-binding protein and transcription factor IIB direct promoter recognition by an archaeal RNA polymerase. J Biol Chem. 1996;271:30144–30148. doi: 10.1074/jbc.271.47.30144. [DOI] [PubMed] [Google Scholar]
- 13.Hausner W, Lange U, Musfeld M. TFS a cleavage induction factor of the archaeal RNA polymerase. J Biol Chem. 2000;275:12393–12399. doi: 10.1074/jbc.275.17.12393. [DOI] [PubMed] [Google Scholar]
- 14.Holstege F C P, van der Vliet P C, Timmers H T M. Opening of an RNA polymerase II promoter occurs in two distinct steps and requires the basal transcription factors IIE and IIH. EMBO J. 1996;15:1666–1677. [PMC free article] [PubMed] [Google Scholar]
- 15.Holstege F C P, Fiedler U, Timmers H T M. Three transitions in the RNA polymerase II transcription complex during initiation. EMBO J. 1997;16:7468–7480. doi: 10.1093/emboj/16.24.7468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huber H, Thomm M, König H, Thies G, Stetter K O. Methanococcus thermolithotrophicus, a novel thermophilic lithotrophic methanogen. Arch Microbiol. 1982;132:47–50. [Google Scholar]
- 17.Hüdepohl U, Reiter W-D, Zillig W. In vitro transcription of two rRNA genes of the archaebacterium Sulfolobus sp. B12 indicates a factor requirement for specific initiation. Proc Natl Acad Sci USA. 1990;87:5851–5855. doi: 10.1073/pnas.87.15.5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang Y, Gralla J D. Nucleotide requirements for activated RNA polymerase II open complex formation in vitro. J Biol Chem. 1995;270:1277–1281. doi: 10.1074/jbc.270.3.1277. [DOI] [PubMed] [Google Scholar]
- 19.Krummel B, Chamberlin M J. Structural analysis of ternary complexes of Escherichia coli RNA polymerase. Deoxyribonuclease I footprinting of defined complexes. J Mol Biol. 1992;225:239–250. doi: 10.1016/0022-2836(92)90918-a. [DOI] [PubMed] [Google Scholar]
- 20.Langer D, Hain J, Thuriaux P, Zillig W. Transcription in archaea: similarity to that in eucarya. Proc Natl Acad Sci USA. 1995;92:5768–5772. doi: 10.1073/pnas.92.13.5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Linn S C, Luse D S. RNA polymerase II elongation complexes paused after the synthesis of 15- or 35-base transcripts have different structures. Mol Cell Biol. 1991;11:1508–1522. doi: 10.1128/mcb.11.3.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Littlefield O, Korkhin Y, Sigler P B. The structural basis for the oriented assembly of a TBP/TFB/promoter complex. Proc Natl Acad Sci USA. 1999;96:13668–13673. doi: 10.1073/pnas.96.24.13668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qureshi S A, Bell S D, Jackson S P. Factor requirements for transcription in the Archaeon Sulfolobus shibatae. EMBO J. 1997;16:2927–2936. doi: 10.1093/emboj/16.10.2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qureshi S A, Jackson S P. Sequence-specific DNA binding by the S. shibatae TFIIB homolog, TFB, and its effect on promoter strength. Mol Cell. 1998;1:389–400. doi: 10.1016/s1097-2765(00)80039-8. [DOI] [PubMed] [Google Scholar]
- 25.Reiter W D, Hüdepohl U, Zillig W. Mutational analysis of an archaebacterial promoter: essential role of a TATA box for transcription efficiency and start-site selection in vitro. Proc Natl Acad Sci USA. 1990;87:9509–9513. doi: 10.1073/pnas.87.24.9509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rowlands T, Baumann P, Jackson S P. The TATA-binding protein: a general transcription factor in eukaryotes and archaebacteria. Science. 1994;264:1326–1329. doi: 10.1126/science.8191287. [DOI] [PubMed] [Google Scholar]
- 27.Thomm M, Wich G. An archaebacterial promoter element for stable RNA genes with homology to the TATA box of higher eukaryotes. Nucleic Acids Res. 1988;16:151–163. doi: 10.1093/nar/16.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thomm M, Wich G, Brown J W, Frey G, Sherf B A, Beckler G S. An archaebacterial promoter sequence assigned by RNA polymerase binding experiments. Can J Microbiol. 1989;35:30–35. doi: 10.1139/m89-005. [DOI] [PubMed] [Google Scholar]
- 29.Timmers H T. Transcription initiation by RNA polymerase II does not require hydrolysis of the β-γ phosphoanhydride bond of ATP. EMBO J. 1994;13:391–399. doi: 10.1002/j.1460-2075.1994.tb06273.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wettach J, Gohl H P, Tschochner H, Thomm M. Functional interaction of yeast and human TATA-binding proteins with an archaeal RNA polymerase and promoter. Proc Natl Acad Sci USA. 1995;92:472–476. doi: 10.1073/pnas.92.2.472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yan M, Gralla J D. Multiple ATP-dependent steps in RNA polymerase II promoter melting and initiation. EMBO J. 1997;16:7457–7467. doi: 10.1093/emboj/16.24.7457. [DOI] [PMC free article] [PubMed] [Google Scholar]