Abstract
Cerulenin is a fungal mycotoxin that potently inhibits fatty acid synthesis by covalent modification of the active site thiol of the chain-elongation subtypes of β-ketoacyl-acyl carrier protein (ACP) synthases. The Bacillus subtilis fabF (yjaY) gene (fabFb) encodes an enzyme that catalyzes the condensation of malonyl-ACP with acyl-ACP to extend the growing acyl chain by two carbons. There were two mechanisms by which B. subtilis adapted to exposure to this antibiotic. First, reporter gene analysis demonstrated that transcription of the operon containing the fabF gene increased eightfold in response to a cerulenin challenge. This response was selective for the inhibition of fatty acid synthesis, since triclosan, an inhibitor of enoyl-ACP reductase, triggered an increase in fabF reporter gene expression while nalidixic acid did not. Second, spontaneous mutants arose that exhibited a 10-fold increase in the MIC of cerulenin. The mutation mapped at the B. subtilis fabF locus, and sequence analysis of the mutant fabF allele showed that a single base change resulted in the synthesis of FabFb[I108F]. The purified FabFb and FabFb[I108F] proteins had similar specific activities with myristoyl-ACP as the substrate. FabFb exhibited a 50% inhibitory concentration (IC50) of cerulenin of 0.1 μM, whereas the IC50 for FabFb[I108] was 50-fold higher (5 μM). These biochemical data explain the absence of an overt growth defect coupled with the cerulenin resistance phenotype of the mutant strain.
A universal set of genes encodes the components of the type II, or dissociated, fatty acid synthase system that is responsible for producing the multitude of fatty acid structures found in bacterial membranes (5, 36). The individual chemical transformations are carried out by separate proteins that can be purified independently from other pathway enzymes. The chain elongation steps in fatty acid biosynthesis consist of the condensation of acyl groups, which are derived from acyl-acyl carrier protein (acyl-ACP) or acyl coenzyme A (acyl-CoA), with malonyl-ACP by the β-ketoacyl-ACP synthases. These condensing enzymes are divided into two groups. The FabH class of condensing enzymes is responsible for the initiation of fatty acid elongation and utilizes acyl-CoA primers. The FabH of Escherichia coli has been extensively studied, and it selectively uses acetyl-CoA to initiate the pathway (16, 24). In contrast, Bacillus subtilis contains two FabH isozymes that differ from the E. coli enzyme in that they are selective for branched-chain acyl-CoAs (4). The FabB-FabF class of condensing enzymes together catalyze the remaining elongation steps in the pathway (5, 36). These enzymes condense malonyl-ACP with acyl-ACP to extend the acyl chain by two carbons. E. coli expresses both types of condensing enzymes and, although they have overlapping substrate specificity, FabB is responsible for a condensation reaction in unsaturated fatty acid synthesis that cannot be performed by FabF (12, 38) and FabF plays a role in the thermal regulation of fatty acid composition (9, 13). Although FabB (32), FabF (21), and FabH (7, 35) all share the same overall structure, the FabB-FabF class of enzymes possesses a Cys-His-His catalytic triad at the active site, whereas the FabH enzymes have a Cys-His-Asn configuration.
The two classes of condensing enzymes are also distinguished by their sensitivity to antibiotics. Cerulenin is a fungal epoxide that irreversibly inhibits the FabB-FabF class of elongation enzymes by covalent modification of their active-site cysteine (26, 30). Accordingly, resistance to cerulenin in E. coli is increased by the overexpression of FabB (10). Thiolactomycin is a second natural product that is known to inhibit all three condensing enzymes (23, 31, 41). The FabB-FabF class of enzymes is more sensitive to thiolactomycin than the FabH class is, and cellular resistance to thiolactomycin in E. coli is conferred by the overexpression of FabB but not FabH (44). The FabF type of condensing enzyme is more widespread in gram-positive bacteria that synthesize branched-chain saturated fatty acids. There is only a single elongation-type condensing enzyme in B. subtilis (a FabF homolog), and the inhibition of this enzyme by cerulenin would account for the sensitivity of this organism to the antibiotic. This study characterizes a spontaneous cerulenin-resistant mutant of B. subtilis and describes the transcriptional response of the fabH1-fabF operon to cerulenin.
MATERIALS AND METHODS
Materials and strains.
Sources of supplies were as follows: Amersham-Pharmacia Biotech supplied [2-14C]malonyl-CoA (specific activity, 55 Ci/mol); Sigma Chemical Co. supplied ACP, antibiotics, and microbiological media; Promega supplied molecular biology reagents; Qiagen supplied pQE32 vector, expression strains, and Ni2+-agarose resin; and Novagen supplied pET vectors and expression strains. Proteins were quantitated by the Bradford method (3). The antibodies against FabFb were raised in rabbits injected with purified FabFb. Acyl-ACP was prepared using the acyl-ACP synthetase method (16, 37). All other chemicals were reagent grade or better.
All bacterial strains and plasmids used are listed in Table 1. The B. subtilis strains were grown in either LB or Difco sporulation media (SM). E. coli strains were propagated in LB broth. β-Galactosidase assays were performed as previously described, and specific activity was expressed in Miller units (39).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| E. coli strains | ||
| DH5α | supE44 thi-1 ΔlacU169 φ80lacZΔM15 endA1 recA1 hsdR17 gyrA96 relA1 | Laboratory stock |
| DM86 | fabB(Ts) recA | 10 |
| CY288 | fabF fabB(Ts) | 10 |
| M15(pREP4) | lac ara gal mtl recA+ uvr+ F−laclq | Laboratory stock |
| BL21 (λDE3) | F−ampT hsdSB(rB mB) gal dcm | Laboratory stock |
| B. subtilis strains | Laboratory stock | |
| JH642 | trpC2 pheA1 | Laboratory stock |
| GS77 | JH642 fabF1b (cer20), cerulenin resistant | 39 |
| BS3 | trpC2 spoOA::Em | Laboratory stock |
| GS37 | JH642 amyE::pGES35 | This study |
| GS39 | BS3 amyE::pGES35 | This study |
| GS41 | GS77 amyE::pGES35 | This study |
| GS85 | JH642 Pxyl-fabFb::fabFb | This study |
| Plasmids | ||
| pJM103 | Integrational vector | 34 |
| pGES1 | pJM103 containing a DNA fragment from −341 to +1256 of fabF1b | This study |
| pGES2 | pJM103 containing a DNA fragment from −341 to +1256 of fabFb | This study |
| pGES4 | pJM103 containing a DNA fragment from −271 to +120 of fabF1b | This study |
| pGES5 | pJM103 containing a DNA fragment from +120 to +1256 of fabF1b | This study |
| pGES6 | pJM103 containing a DNA fragment from +120 to +733 of fabF1b | This study |
| pAG58 | Integrative plasmid, contains Pspac and lacI | 34 |
| pGES20 | fabFb cloned into pAG58 | This study |
| pQE32 | Expression vector | Quiagen |
| pET15b | Expression vector | Novagen |
| pGES32 | fabF1b cloned into pQE30 | This study |
| pJM116 | Integrational vector containing a promoterless lacZ gene | 6 |
| pGES35 | Putative promoter region of fabH1Fb cloned into pJM116 | This study |
| pRDC9 | Plasmid containing Pxyl and xylR | 40 |
| pGES49 | Integrational vector containing Pxyl and xylR | This study |
| pGES79 | Promoterless 5′ region of fabFb cloned under Pxyl in pGES49 | This study |
Em denotes a gene cassette conferring resistance to erythromycin. Pspac denotes an isopropyl-β-d-thiogalactopyranoside-inducible hybrid promoter containing the RNA polymerase recognition sequence from the B. subtilis phage SPO1 and the E. coli lac operator (34).
Plasmid constructions.
Plasmids pGES1 and pGES2 were constructed as follows. The oligomers 5′-TAAGGGGATCCGGTTTGGCTTGATTATG-3′ and 5′-AAATGGGAGAATTCTGCGTAAATGTCATTG-3′ (restriction sites underlined) were synthesized to amplify a chromosomal DNA fragment by PCR containing the last 290 bases of fabH1 and the complete fabFb gene from strain GS77 or JH642, respectively. The PCR products were digested with EcoRI and BamHI and cloned into pJM103, giving rise to plasmids pGES1 (containing DNA from GS77) and pGES2 (containing DNA from JH642). To obtain plasmids pGES4 and pGES5, pGES1 was digested with EcoRI and BamHI, giving a 1,577-bp fragment which was purified and further digested with HincII . The two resultant DNA fragments of 391 and 1,136 bp were purified and cloned into pJM103 to give plasmids pGES4 and pGES5, respectively. To obtain plasmid pGES6, pGES5 was digested with Sphl, and the larger DNA fragment of 4,326 bp was purified and recircularized by ligation.
Plasmid pGES20 was constructed using a DNA fragment containing the fabFb open reading frame generated by PCR using primers 5′-GTGAGGTGCACACACCATGGCTAAAAAAAG-3′ and 5′-AAATGGGAGAATTCTGCGTAAATGTCATTG-3′. The resulting fragment was digested with NcoI and BamHI and cloned into pAG58. To obtain a His-tag–FabF[I108F] fusion expression construct, the fabF1b gene was cloned into the expression vector pQE32. PCR amplification was performed using chromosomal DNA from GS77 as template and primers 5′-GTGAGGGGGATCCAGATGACTAAAAAAAG-3′ and 5′-CTACCCTTTAACTGCAGCCGGTTTGG-3′. The 1,287-bp PCR product was then digested with BamHI and EcoRI and cloned into pQE32, giving rise to pGES30. This plasmid was used to transform strain M15(pREP4). A similar strategy was used to obtain the wild-type FabF protein fusion. To this end, PCR amplification was performed using genomic DNA from the wild-type B. subtilis strain JH642 and primers 5′-GTGAGGTGCACACCATATGACTAAA-3′ and 5′-GGATCCTTGGCTTGATTATGATTGA-3′. The PCR product was first ligated into plasmid pCR2.1, and the cloned DNA was excised with NdeI and BamHI and ligated into pET15b (Novagen). This ligation mixture was used to transform E. coli strain BL21 (λDE3).
The fabH1F-lacZ transcriptional fusion was constructed by PCR amplification of chromosomal DNA using primers 5′-ATTGAACCGATTTGGTACCTAATATGCATG-3′ and 5′-AACACCAAGTATTCCAGGATCCATTAGGG-3′. The resultant DNA fragment containing the 300-bp region upstream of the putative translation start of fabH1F was digested with KpnI and BamHI and cloned into the integrational vector pJM116, generating plasmid pGES35. The plasmid was introduced by a double crossover event at the amyE locus of strains JH642, BS3, and GS77, giving rise to strains GS37, GS39, and GS41, respectively. Plasmid pGES49, containing the Pxyl promoter and the xylR repressor, was constructed as follows. Plasmid pRDC9 was digested with EcoRI and BamHI, and the fragment containing Pxyl and xylR was purified and cloned into EcoRI-BamHI-digested pAG58. To construct plasmid pGES79, PCR amplification was performed using chromosomal DNA from strain JH642 as template and primers 5′-CATTGGCATGCAAAACGGGCTTACC-3′ and 5′-GCGCCATTGCAGTCGACTGGGGCCG-3′. The 462-bp PCR product was digested with SalI and SphI and cloned into plasmid pGES49. The wild-type strain JH642 was transformed with pGES79, and colonies in which a Campbell-type recombination event at fabFb took place were screened for a xylose growth-dependent phenotype. The resultant strain was named GS85.
Enzyme purification and assay.
The proteins were expressed and purified as described previously (16). Purified His-tagged FabFb and FabF[I108F]b were dialyzed against 20 mM Tris-HCl (pH 7.6), 1 mM β-mercaptoethanol, and 1 mM dithiothreitol, concentrated with an Amicon stirred cell, and stored in 50% glycerol at −20°C.
The enzyme assay used gel electrophoresis to separate and quantify the products (17). This assay contained 50 μM ACP, 1 mM β-mercaptoethanol, 0.1 M sodium phosphate buffer (pH 7.0), 50 μM [2-14C]malonyl-CoA (specific activity, 55 Ci/mol), 12.5 μM myristoyl-ACP, FabD (0.3 μg of protein), 100 μM NADPH, and FabG (0.3 μg of protein) in a final volume of 50 μl. A mixture of ACP, 1 mM β-mercaptoethanol, and the buffer was incubated at 37°C for 30 min to ensure complete reduction of ACP, and then the remaining components (except FabFb) were added. The mixture was then aliquoted into the assay tubes and the reaction was initiated by the addition of FabFbs. The reaction mixture was incubated at 37°C for 30 min, placed in an ice slurry, gel loading buffer was added, and the entire sample was loaded onto a conformationally sensitive 15% polyacrylamide gel containing 3.5 M urea (4, 17). Electrophoresis was performed at 25°C and 32 mA/gel. The gels were dried, and product formation was quantitated by exposure in a PhosphorImager (4).
Immunoblot analysis.
Strain GS37 was grown in LB medium at 37°C. At an optical density at 525 nm (OD525) of 0.28, the culture was split and cerulenin was added at a concentration of 3.25 μg/ml to one-half of the culture. The same volume of ethanol was added to the other half of the culture. A 1-ml aliquot of each culture was harvested, centrifuged, and frozen 7 and 250 min after addition of cerulenin. The pellets were resuspended in lysis buffer (50 mM Tris-HCl [pH 8.0], 5 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, and 0.1 mM dithiothreitol), adding 180 μl of buffer per OD525 unit. A 20-μl volume of the cell resuspension was disrupted by incubating with lysozyme (500 μg/ml) for 15 min at 37°C followed by 5 min of boiling in the presence of loading buffer. Each sample was fractionated by sodium dodecyl sulfate-gel electrophoresis in a 12% acrylamide gel. Proteins were electroeluted to a nitrocellulose membrane and detected using anti-FabFb rabbit antibody and a secondary anti-rabbit immunoglobulin G conjugated to alkaline phosphatase. The blots were exposed to film, the intensity of the bands was quantified by densitometry, and the figure was generated using Adobe Photoshop.
FabFb was detected in cultures of strain GS85 grown in LB medium at 37°C to an OD525 of 0.12. The culture was split into equal parts and xylose was added to a final concentration of 0.25% (wt/vol) to one-half of the culture. The cultures were grown for 4 h, and then a 1-ml aliquot of each culture was frozen and treated as described above for strain GS37.
RESULTS
B. subtiliis yjaY gene encodes a functional FabF condensing enzyme.
B. subtilis contains three open reading frames that have homology with β-ketoacyl-ACP synthases of type II fatty acid synthesis. Two of these genes, yjaX (fabH1) and yhfB (fabH2), are related to the FabH initiating-condensing enzyme of E. coli and catalyze the initial condensation reaction in branched-chain fatty acid synthesis (4). The third gene, yjaY, had a 1,240-bp open reading frame and was predicted to encode a protein that was 54% identical and 64% similar to E. coli FabF and 43% identical and 50% similar to E. coli FabB. The genomic analysis indicated that there was only a single example of the elongation class of condensing enzymes in B. subtilis.
Complementation experiments were used to confirm the prediction that yjaY encodes a functional FabF-like condensing enzyme. E. coli fabB strains require unsaturated fatty acids for growth but synthesize saturated fatty acids normally. Strains harboring both a temperature-sensitive fabB mutation and a mutation that inactivates fabF fail to synthesize any fatty acids at the nonpermissive temperature and cannot grow even in the presence of exogenous fatty acids. Transformation of strains DM86 [fabB(Ts)] or CY288 [fabB(Ts) fabF] with plasmid pGES20, expressing the yjaY gene controlled by the Pspac promoter, showed that the E. coli fabF mutant was complemented by yjaY expression, whereas the fabB mutant was not (Table 2). Thus, based on the molecular characteristics of the yjaY gene and the ability of the yjaY to complement the fabF defect but not the fabB defect, we have designated this gene fabFb in concordance with E. coli nomenclature.
TABLE 2.
Growth of plasmid-carrying strains
| Strain | Plasmid carried | Genotype | Growth ata:
|
||
|---|---|---|---|---|---|
| 30°C | 42°C | 42°C plus oleate | |||
| DM86 | pAG58 (control) | fabB(Ts) | + | − | + |
| DM86 | pGES20 (fabFb) | fabB(Ts) | + | − | + |
| CY288 | pAG58 (control) | fabB(Ts) fabF | + | − | − |
| CY288 | pGES20 (fabFb) | fabB(Ts) fabF | + | − | + |
Growth was scored by the appearance of single colonies on solid LB medium. Oleate (18:1) when added was present as the potassium salt at a concentration of 100 μg/ml. −, no growth; +, growth.
fabFb (yjaY) is an essential gene in B. subtilis.
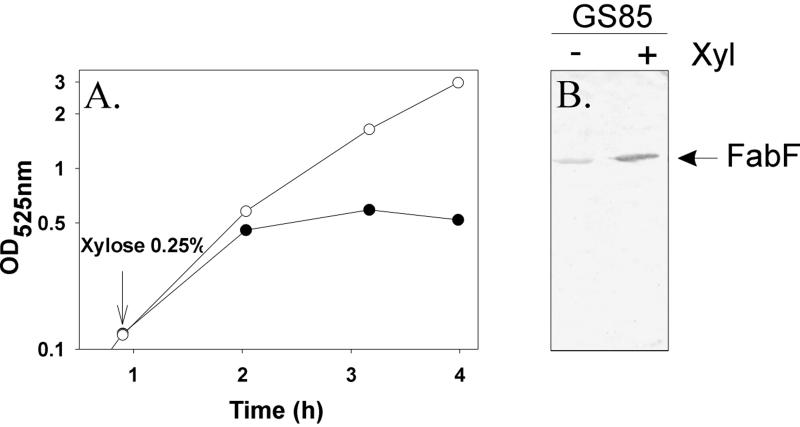
In E. coli, fabB is essential, whereas fabF is not (5). However, in B. subtilis, fabFb was the only elongation condensing enzyme detected in the genome, suggesting that this gene was essential. Several attempts to disrupt the fabFb gene by a single crossover recombination event using integrative plasmids containing internal fragments of the gene were unsuccessful. The essential nature of fabFb was verified by placing the single chromosomal copy of fabFb under control of the xyl promoter as described in Materials and Methods. The resultant strain, GS85, exhibited normal growth in the presence of xylose (Fig. 1A). The removal of xylose significantly attenuated cell growth, and growth returned to normal upon the addition of 0.25% xylose, which derepressed the expression of fabFb. The effect of xylose on the cellular FabFb content was confirmed by separation of cell extracts by gel electrophoresis followed by immunodetection (Fig. 1B). In the absence of the inducer, the FabFb content of strain GS85 was significantly lower than the levels observed in xylose-supplemented cells. These experiments demonstrated an essential role for the fabFb gene product in B. subtilis.
FIG. 1.
Effect of xylose on growth and content of FabFb of strain GS85 bearing a Pxyl-fabFb fusion. (A) Cultures of strain GS85 (Pxyl:fabFb::fabFb) were grown in nonsupplemented medium (●) or medium supplemented with 0.25% xylose (○). (B) Strain GS85 was grown either in the presence or in the absence of xylose. Cells were harvested after 4 h at 37°C (A), and the levels of FabFb were determined by immunoblotting as described in Materials and Methods.
Transcriptional regulation of the B. subtilis fabH1-fabF operon.
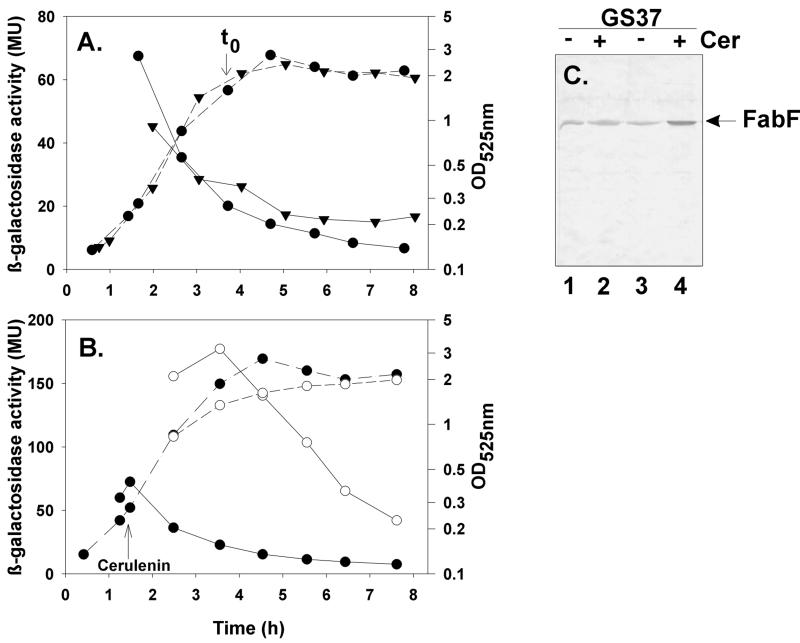
The fabFb gene is predicted to be the second gene in an operon that also contains the fabH1 gene. To study expression from the fabH1F promoter, the 300 nucleotides preceding the start codon of fabH1F were cloned in front of the promoterless lacZ gene in pJM116. The resulting plasmid (pGES35) was linearized and integrated at the nonessential amyE locus of B. subtilis, yielding strain GS37 (Table 1). The strain was grown in either SM or LB broth and assayed for β-galactosidase activity. The results indicated that the region upstream of the putative fabH1-fabF operon contained a promoter that functions primarily during vegetative growth (Fig. 2A). The promoter was active during exponential growth in SM, reaching a peak of 75 Miller units 2 h before the onset of sporulation (Fig. 2A). At this point, promoter activity decreased toward zero as the cells entered into stationary phase. β-Galactosidase production in cells grown in LB (where the cultures sporulate poorly) was similar to cells grown in SM (data not shown). These results were consistent with expression from the promoter upstream of the fabH1-fabF operon being turned off by a regulatory protein related to the cessation of growth and/or the onset of sporulation.
FIG. 2.
Expression of fabH1-fabF promoter. (A) Growth (dashed lines) and β-galactosidase specific activity (solid lines) of strains GS37 (trpC2 pheA1 amyE::pGES35) (●) and GS39 (trpC2 spoOA::Em amyE::pGES35) (▾). (B) Effect of cerulenin on growth (dashed lines) and β-galactosidase specific activity (solid lines) of strain GS37. ○, cerulenin-treated cells; ●, control cells. Note the different β-galactosidase specific activity scales between panels A and B. (C) Effect of cerulenin on FabFb content of strain GS37. Cultures were grown until early exponential phase and then treated with cerulenin as described in Materials and Methods. Cells were harvested 7 min (lane 2) or 2.5 h (lane 4) after the addition of the antibiotic. Lanes 1 and 3 contain protein extracts of untreated cultures harvested at 7 min and 2.5 h, respectively. The levels of FabFb were detected by immunoblotting and quantitated by densitometry as described in Materials and Methods to denote the onset of stationary phase.
SpoOA is the major transcription factor required for sporulation and is one candidate for a repressor of fabH1-fabF expression during the differentiation process. We tested this idea by introducing the transcription fusion into the spoOA strain B53 to yield strain GS39. The β-galactosidase activity, assayed in extracts from cells grown in SM, was similar to the pattern observed in the wild-type strain (data not shown). Therefore, SpoOA did not appear to play a role in the regulation of fabH1-fabF expression.
Regulation of fabH1-fabF expression in response to cerulenin.
Cerulenin is a potent inhibitor of FabF activity, and we used the transcriptional fusion strains to investigate whether the inhibition of lipid synthesis affected transcription of the fabH1-fabF operon. β-Galactosidase activity was assayed in strain GS37 treated with cerulenin during early exponential growth. The activity of β-galactosidase expressed from the fusion was significantly higher following the addition of cerulenin (Fig. 2B), reaching levels 8- to 10-fold higher than those observed in untreated cells. At the end of the exponential phase, β-galactosidase levels in cerulenin-treated cells began to decrease, although the activity was still significantly higher than in cells entering stationary phase without cerulenin (Fig. 2B). The transcriptional activation of the fabH1Fb promoter is not due to a side effect of cerulenin, because no effect of the antibiotic was observed on transcription of the fabH1Fb-lacZ fusion introduced into strain GS77 to obtain strain GS41 (data not shown), a cerulenin-resistant isolate described below. The levels of FabFb in cells treated or untreated with cerulenin were determined by immunodetection and quantification (Fig. 2C). Consistent with the operon fusion analysis, these experiments demonstrated that the addition of cerulenin to GS77 cultures resulted in an approximately fivefold increase of FabFb protein. We also observed an increase in PfabH1F-lacZ transcription when the levels of FabFb were decreased. These experiments were performed using a strain carrying both an isotopic pXyl-fabF fusion and an ectopic PfabH1F-lacZ fusion. In this strain, β-galactosidase activity was increased fivefold when xylose was removed from the medium. These data demonstrated that inhibition of fatty acid synthesis resulted in transcriptional induction of the genes coding for the FabH1b and FabFb condensing enzymes.
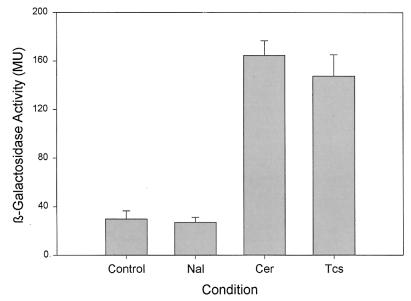
We next determined if the fabH1-fabF transcriptional response was linked to the inhibition of fatty acid synthesis or the inhibition of growth (Fig. 3). In these experiments, we used nalidixic acid, which inhibits DNA gyrase, as another inhibitor of cell growth and triclosan as a fatty acid synthesis inhibitor. Triclosan blocks the enoyl-ACP reductase (FabI) step in the fatty acid elongation cycle (18, 20, 29), although fatty acid synthesis is not the only target in gram-positive bacteria (14, 19). B. subtilis has a FabI that is potently inhibited by triclosan, but it also has another relatively triclosan-resistant enoyl-ACP reductase, FabL (19). However, the lower specific activity of FabL toward acyl-ACP substrates suggested that FabI was the principle isoform responsible for fatty acid synthesis. The addition of nalidixic acid (25 or 75 μg/ml) to cultures of the reporter strain GS37 (trpC2 pheA1 amyE::pGES35) did not trigger an increase in fabH1-fabF expression, and in fact a slight decrease in β-galactosidase activity was observed at all nalidixic acid concentrations tested. On the other hand, triclosan, either at 0.4 or 2 μg/ml, enhanced β-galactosidase transcription by approximately fivefold. These data show that the up-regulation of fabH1-fabF expression correlated with the specific inhibition of fatty acid biosynthesis and suggest that the response would be observed when any step in the pathway was blocked.
FIG. 3.
Alteration in expression from the fabH1-fabF promoter induced by antibiotics. Strain GS37 (trpC2 pheA1 amyE::pGES35) was grown in LB medium until the culture reached an OD525 of 0.3 and then was treated for 1 h with either nalidixic acid (Nal; 75 μg/ml), cerulenin (Cer; 3.3 μg/ml), or triclosan (Tcs; 0.4 μg/ml). Control cultures were untreated. Levels of β-galactosidase expression (in Miller units) were determined as described in Materials and Methods. Error bars are the standard deviation of the results from three independent experiments.
Cerulenin resistance is linked to the yjaY gene.
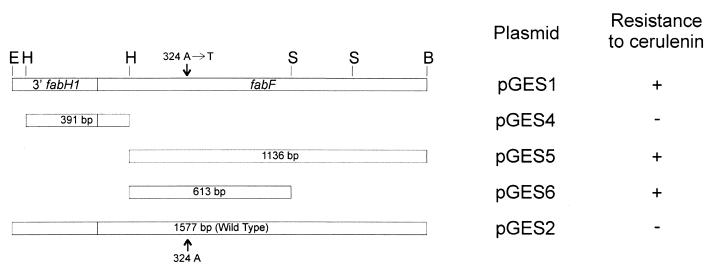
The ability of cerulenin to inhibit the growth of strains JH642 and GS77, the latter of which was selected as a spontaneous cerulenin-resistant strain (39), was compared by a conventional dilution method. The MICs were 50 μg/ml for GS77 and 5 μg/ml for JH642 after overnight cultivation. Elongation condensing enzymes in fatty acid and polyketide synthesis are uniformly inactivated by this antibiotic, with the exception of the type I synthase from Cephalosporium caerulens (42, 43). However, Saccharomyces cerevisiae acquires cerulenin resistance by a point mutation in the condensing enzyme module of the polyfunctional enzyme (22). Therefore, we reasoned that cerulenin resistance in strain GS77 was likely due to a mutation in a gene coding for a condensing enzyme involved in long-chain fatty acid synthesis. To test if a mutation in the yjaY gene was responsible for the resistance to cerulenin, we cloned a region of the chromosome containing this open reading frame from strain GS77 and from the isogenic strain JH642 into the integrative plasmid pJM103 to yield plasmids pGES1 and pGES2, respectively (Table 1 and Fig. 4). Strain JH642 was transformed with both linearized plasmids, and transformants were selected on plates containing 10 μg of cerulenin/ml. Cerulenin-resistant colonies appeared only when strain JH642 was transformed with pGES1 (Fig. 4), indicating that the mutation was linked to yjaY. Different portions of the yjaY gene from strain GS77 were subcloned into plasmid pJM103 (Fig. 4), and the resultant plasmids were transformed into strain JH642. The results of this experiment indicated that the 5′ half of the yjaY coding sequence from the resistant strain confers resistance to cerulenin when it is integrated by a double crossover event into the chromosome of the cerulenin-sensitive strain JH642.
FIG. 4.
Plasmid constructs used to localize resistance to cerulenin in strain GS77. The indicated B. subtilis DNA fragments were cloned into the integrative plasmid pJM103. The resultant linearized plasmids were used to transform strain JH642 (trpC2 pheA1), and transformants were screened for resistance to 10 μg of cerulenin/ml. DNA fragments in pGES1, -4, -5, and -6 were derived from strain GS77, and the DNA fragment in pGES2 arose from strain JH642. The transversion responsible for the resistance to the antibiotic is shown with an arrow. Restriction sites: E, EcoRI; H, HincII; S, SphI; B, BamHI.
The DNA fragments contained in plasmids pGES1 and pGES2 were sequenced. Only one difference was found: a transversion in the 240th base of the yjaY open reading frame (324 A → T) (Fig. 4). In the deduced protein sequence, this mutation is reflected by a change from Ile-108 in the wild-type protein to Phe-108 in the mutant allele (Fig. 4).
Biochemical characteristics of FabFb.
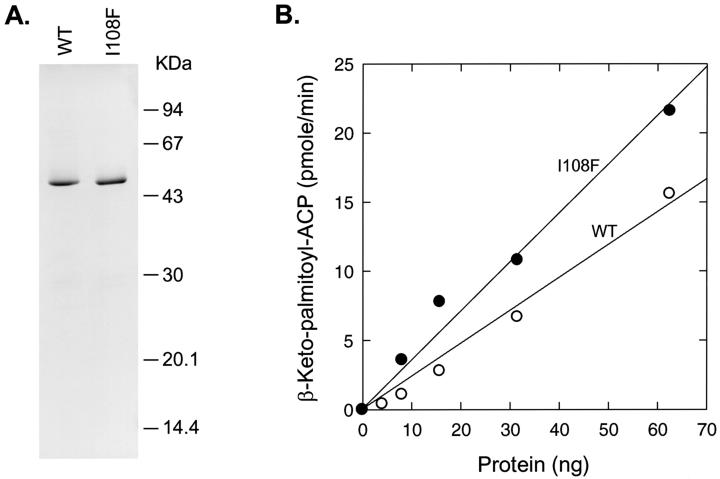
The two open reading frames coding for both the predicted FabFb and the mutant protein FabFb[I108F] were amplified by PCR and cloned into expression vectors, and the His-tagged version of the proteins was expressed and purified (Fig. 5A). The purified recombinant proteins exhibited a molecular size of 48.3 kDa, which agreed with the DNA sequence plus the His tag (Fig. 5A). The catalytic properties of FabFb and FabFb[I108F] were compared using an assay based on the incorporation of [2-14C]malonyl-CoA into myristoyl-ACP. The reaction products were separated by conformationally sensitive gel electrophoresis and quantified using PhosphorImager analysis as described in Materials and Methods. FabFb[I108F] exhibited a slightly higher specific activity (353.5 ± 16.9 pmol/min/μg) than FabFb (220.2 ± 6 pmol/min/μg) (Fig. 5B). We also examined the activity of a prototypical condensing FabB of E. coli enzyme, FabBe, under the same assay conditions. The specific activity of FabBe with C14:0-ACP was 125.3 ± 13 pmol/min/μg (data not shown). Thus, FabFb catalyzes a condensation reaction characteristic of a long-chain acyl-ACP condensing enzyme, and there was little difference between the activity of the mutant and wild-type enzymes under these in vitro assay conditions.
FIG. 5.
Purification and specific activity of FabFb and FabFb[I108F]. (A) FabFb proteins were expressed as a His-tag fusion protein and purified by Ni2+-chelate affinity chromatography as described in Materials and Methods. Samples were analyzed using sodium dodecyl sulfate-gel electrophoresis through a 12% polyacrylamide gel, and the 48.3-kDa proteins were visualized by staining with Coomassie blue. (B) Specific activities of purified FabFbs were determined using myristoyl-ACP as a substrate (○, FabFb; ●, FabFb[I108F]) using a gel electrophoresis assay as described in Materials and Methods, and the amount of the product was quantitated with a PhosphorImager and plotted as a function of the protein concentration in the assay.
Inhibition of FabFbs by cerulenin.
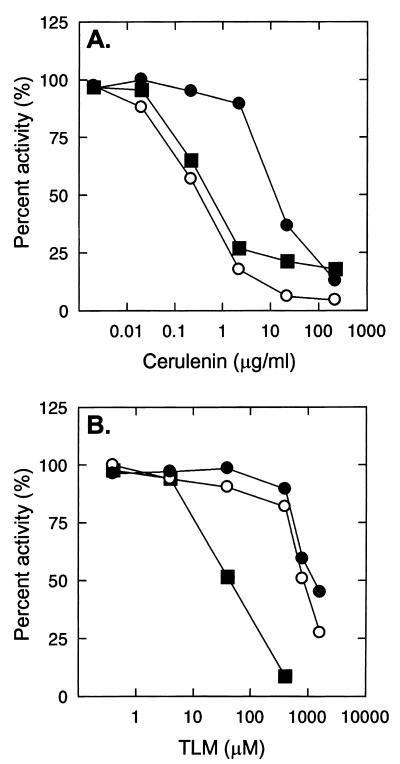
The sensitivity of FabFb and FabFb[I108F] to cerulenin and thiolactomycin was determined with the gel electrophoresis assay described above. The sensitivity of FabFb and FabBe to cerulenin was very similar, exhibiting an IC50 of 0.1 μg/ml and 0.2 μg/ml, respectively. The IC50 of cerulenin for FabFb[I108F] was 5 μg/ml, indicating that the mutant condensing enzyme is highly insensitive to the antibiotic. Notably, the magnitude of the increase in the IC50 of cerulenin between the wild-type and mutant condensing enzymes (Fig. 6A) was consistent with the differences in the cerulenin MICs for the corresponding strains. Therefore, these data demonstrate that the amino acid substitution (Ile→Phe) in the FabFb protein is responsible for the cerulenin resistance of the mutant strain GS77.
FIG. 6.
Inhibition of FabFb and FabFb[I108F] by cerulenin and thiolactomycin. (A) Inhibition of elongation condensing enzymes by cerulenin. The cerulenin IC50s for FabFb (○), FabFb[I108F] (●), and FabBe (■) were 0.1, 5, and 0.2 μg/ml, respectively. (B) Inhibition of the same group of condensing enzymes by thiolactomycin (TLM). Neither FabFb (○) nor FabFb[I108F] (●) was effectively inhibited by thiolactomycin, and the thiolactomycin IC50 for FabBe (■) was 40 μM.
Neither FabFb nor FabFb[I108F] was significantly inhibited by thiolactomycin (Fig. 6B). In a control experiment, FabBe exhibited an IC50 of thiolactomycin of 40 μM (Fig. 6B), confirming that the drug was effective against a control elongation condensing enzyme known to be a target for the antibiotic (27, 31, 44). The resistance to thiolactomycin of the FabFb condensing enzyme agreed with the previous report that this antibiotic did not inhibit the branched-chain fatty acid synthase from Bacillus species (2).
DISCUSSION
Our genetic and biochemical analysis demonstrates that FabFb, the product of the yjaY gene, is the sole elongation condensing enzyme that participates in branched-chain fatty acid biosynthesis in B. subtilis. Sequence analysis indicates that FabFb is most closely related to E. coli FabF and, accordingly, expression of FabFb complements E. coli fabF mutants but cannot complement fabB-deficient strains. Thus, FabFb is unable to catalyze the elongation of the critical intermediate(s) in unsaturated fatty acid synthesis by a type II system. B. subtilis can synthesize unsaturated fatty acids, but it utilizes a desaturase that acts on preformed phospholipid-bound fatty acids (1). FabFb is the only elongation condensing enzyme detected in the B. subtilis genome and, accordingly, genetic analysis confirmed that it is an essential enzyme. The other two condensing enzymes of B. subtilis, FabH1b and FabH2b, condense branched-chain acyl-CoA primers with malonyl-ACP to initiate fatty acid synthesis (4). One of these, FabH1b (the product of the yjaX gene), is the leading gene in the fabH1-fabF transcriptional unit. Thus, both the initiation and elongation functions of fatty acid synthesis are coregulated and coordinated with cell growth.
The elongation condensing enzymes are sensitive to two natural products, cerulenin (26, 33) and thiolactomycin (11, 25, 27, 44). The recent solution of the three-dimensional structures of the FabFe-cerulenin binary complex (30) provides the opportunity to interpret our results based on a model of the FabFb structure and its complexes with cerulenin. Cellular resistance to the effects of cerulenin occurred by the mutation in the fabFb gene to produce a FabFb[I108F] protein. Isoleucine-108 lies in the hydrophobic acyl chain-binding pocket of the FabF condensing enzymes and must rotate out of the way to accommodate the acyl chain of cerulenin (30). The I108F substitution introduces a residue into the hydrophobic channel that cannot rotate to allow the optimum interaction between FabF and cerulenin. Indeed, the FabFe[I108F] mutant is also resistant to cerulenin (45). However, properties of FabFb[I108F] (Fig. 4 and 5) are not entirely the same as those of FabFe[I108F] (45). The FabFe[I108F] mutant is essentially inactive with a 14-carbon acyl-ACP primer (45), whereas FabFb[I108F] is just as active with this substrate as the wild-type enzyme is (Fig. 5). Since fabFb is the only elongation condensing enzyme in B. subtilis and the FabFb[I108F] mutant does not have a clear growth phenotype, the FabFb[I108F] mutant protein must also catalyze the elongation of all acyl chain lengths in vivo. The reason for the apparent differences between the reactivity of the FabFe[I108F] and the FabFb[I108F] proteins with 14-carbon acyl-ACP primers is not clear. The cerulenin-producing fungus, C. caerulens, expresses a cerulenin-resistant condensing enzyme, but the molecular changes that account for this resistance are not clear (42, 43). Mutations in the gene for fatty acid synthase in S. cerevisiae result in a Gly-to-Ser replacement (22). The glycine residue corresponds to Gly-107 in FabF, which is adjacent to the isoleucine that was found to have mutated in our experiments. The fatty acid synthase of Bacillus species is remarkably insensitive to thiolactomycin (2), and our experiments show that FabFb is insensitive to thiolactomycin in vitro. The molecular differences responsible for the resistance of the Bacillus FabF to thiolactomycin await the characterization of the mode of thiolactomycin binding to other condensing enzymes.
The expression of the fabH1-fabF gene cluster is regulated by growth and the activity of fatty acid synthesis. Transcription of fabH1-fabF occurs only during exponential growth and is shut off as the cells approach stationary phase. The sporulation regulator SpoOA does not mediate this repression, but the repression may be due to the loss of an inducing signal that comes from, or is dependent on, cell growth. This finding is consistent with the requirement for constant fatty acid synthesis for membrane formation during exponential growth and cessation of this process when the cells cease to divide. An important finding in this study is the up-regulation of fabFb transcription and protein levels in response to the inhibition of the enzyme by inhibitors of fatty acid synthesis. Both the inhibition of the elongation condensing enzyme (FabF) by cerulenin and the inhibition of the enoyl-ACP reductase (FabI) with triclosan significantly increase transcription of the fabH1-fabF operon. Thus, B. subtilis has the ability to sense a decrease in the activity of the pathway and to respond by adjusting the expression of the enzymes. It will be important to test the possibility that the other genes of the type II fatty acid synthase of B. subtilis are coordinately regulated by the activity of the pathway.
How might the expression of the fabH1-fabF genes be regulated by the activity of the FabFb protein? The inactivation of the E. coli FabB and FabF enzymes triggers a dramatic increase in the accumulation of malonyl-CoA (11, 15). This finding indicates that the condensing enzymes are required for the continued utilization of malonyl-CoA, which continues to be generated by acetyl-CoA carboxylase following inhibition of fatty acid synthesis. It also seems reasonable to think that the blockade of enoyl-ACP reductase would also trigger an increase in malonyl-CoA as the pathway shuts down; however, this supposition needs to be verified. Therefore, one possibility is that fluctuations in the levels of malonyl-CoA are coupled to the expression of the fabH1-fabF transcriptional unit. The levels of malonyl-CoA are then in turn regulated in concert with fatty acid biosynthesis through feedback inhibition by the acyl-ACP end products and intermediates (8). Recently, the treatment of mice with a cerulenin analog inhibited fatty acid synthesis and reduced food intake and body weight (28). These investigators proposed that the metabolic signal mediating feeding inhibition was the accumulation of malonyl-CoA following the inhibition of fatty acid synthase. Therefore, malonyl-CoA may play a universal role signaling the status of fatty acid biosynthesis in cells.
ACKNOWLEDGMENTS
We thank Richard Heath and Suzanne Jackowski for help with preparation of the manuscript and Allen Price and Steve White for modeling of the FabF[I108F] mutant protein structure.
This work was supported by the Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), Agencia de Promoción Científica y Tecnológica (FONCYT), National Institutes of Health grant GM34496 (C.O.R.), Cancer Center (CORE) Support Grant CA 21765 (St. Jude), and the American Lebanese Syrian Associated Charities. G.S. is a fellow from CONICET and D.deM. is a Career Investigator of the same institution.
REFERENCES
- 1.Aguilar P S, Cronan J E, Jr, de Mendoza D. A Bacillus subtilis gene induced by cold shock encodes a membrane phospholipid desaturase. J Bacteriol. 1998;180:2194–2200. doi: 10.1128/jb.180.8.2194-2200.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arimyra N, Kaneda T. Type selective inhibition of microbial fatty acid synthases by thiolactomycin. Arch Microbiol. 1993;160:158–161. doi: 10.1007/BF00288719. [DOI] [PubMed] [Google Scholar]
- 3.Bradford M M. A rapid and sensitive method for quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 4.Choi K-H, Heath R J, Rock C O. β-Ketoacyl-acyl carrier protein synthase III (FabH) is a determining factor in branched-chain fatty acid biosynthesis. J Bacteriol. 2000;182:365–370. doi: 10.1128/jb.182.2.365-370.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cronan J E, Jr, Rock C O. Biosynthesis of membrane lipids. In: Neidhardt F C, Curtis R, Gross C A, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington, D.C.: American Society for Microbiology; 1996. pp. 612–636. [Google Scholar]
- 6.Dartois V, Djavakhishvili T, Hoch J A. Identification of a membrane protein involved in activation of the KinB pathway to sporulation in Bacillus subtilis. J Bacteriol. 1996;178:1178–1186. doi: 10.1128/jb.178.4.1178-1186.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davies C, Heath R J, White S W, Rock C O. The 1.8 Å cystal structure and active site architecture of β-ketoacyl-[acyl carrier protein] synthase III (FabH) from Escherichia coli. Structure. 2000;8:185–195. doi: 10.1016/s0969-2126(00)00094-0. [DOI] [PubMed] [Google Scholar]
- 8.Davis M S, Cronan J E., Jr Inhibition of Eschericia coli acetyl coenzyme A carboxylase by acyl-acyl carrier protein. J Bacteriol. 2001;183:1499–1503. doi: 10.1128/JB.183.4.1499-1503.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Mendoza D, Cronan J E., Jr Thermal regulation of membrane lipid fluidity in bacteria. Trends Biochem Sci. 1983;8:49–52. [Google Scholar]
- 10.de Mendoza D, Klages Ulrich A, Cronan J E., Jr Thermal regulation of membrane fluidity in Escherichia coli. Effects of overproduction of β-ketoacyl-acyl carrier protein synthase I. J Biol Chem. 1983;258:2098–2101. [PubMed] [Google Scholar]
- 11.Furukawa H, Tsay J-T, Jackowski S, Takamura Y, Rock C O. Thiolactomycin resistance in Escherichia coli is associated with the multidrug resistance efflux pump encoded by emrAB. J Bacteriol. 1993;175:3723–3729. doi: 10.1128/jb.175.12.3723-3729.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garwin J L, Klages A L, Cronan J E., Jr Structural, enzymatic, and genetic studies of β-ketoacyl-acyl carrier protein synthases I and II of Escherichia coli. J Biol Chem. 1980;255:11949–11956. [PubMed] [Google Scholar]
- 13.Garwin J L, Klages A L, Cronan J E., Jr β-Ketoacyl-acyl carrier protein synthase II of Escherichia coli. Evidence for function in the thermal regulation of fatty acid synthesis. J Biol Chem. 1980;255:3263–3265. [PubMed] [Google Scholar]
- 14.Heath R J, Li J, Roland G E, Rock C O. Inhibition of the Staphylococcus aureus NADPH-dependent enoyl-acyl carrier protein reductase by triclosan and hexachlorophene. J Biol Chem. 2000;275:4654–4659. doi: 10.1074/jbc.275.7.4654. [DOI] [PubMed] [Google Scholar]
- 15.Heath R J, Rock C O. Regulation of malonyl-CoA metabolism by acyl-acyl carrier protein and β-ketoacyl-acyl carrier protein synthases in Escherichia coli. J Biol Chem. 1995;270:15531–15538. doi: 10.1074/jbc.270.26.15531. [DOI] [PubMed] [Google Scholar]
- 16.Heath R J, Rock C O. Inhibition of β-ketoacyl-acyl carrier protein synthase III (FabH) by acyl-acyl carrier protein in Escherichia coli. J Biol Chem. 1996;271:10996–11000. doi: 10.1074/jbc.271.18.10996. [DOI] [PubMed] [Google Scholar]
- 17.Heath R J, Rock C O. Regulation of fatty acid elongation and initiation by acyl-acyl carrier protein in Escherichia coli. J Biol Chem. 1996;271:1833–1836. doi: 10.1074/jbc.271.4.1833. [DOI] [PubMed] [Google Scholar]
- 18.Heath R J, Rubin J R, Holland D R, Zhang E, Snow M E, Rock C O. Mechanism of triclosan inhibition of bacterial fatty acid synthesis. J Biol Chem. 1999;274:11110–11114. doi: 10.1074/jbc.274.16.11110. [DOI] [PubMed] [Google Scholar]
- 19.Heath R J, Su N, Murphy C K, Rock C O. The enoyl-[acyl-carrier-protein] reductases FabI and FabL from Bacillus subtilis. J Biol Chem. 2000;275:40128–40133. doi: 10.1074/jbc.M005611200. [DOI] [PubMed] [Google Scholar]
- 20.Heath R J, Yu Y-T, Shapiro M A, Olson E, Rock C O. Broad spectrum antimicrobial biocides target the FabI component of fatty acid synthesis. J Biol Chem. 1998;273:30316–30321. doi: 10.1074/jbc.273.46.30316. [DOI] [PubMed] [Google Scholar]
- 21.Huang W, Jia J, Edwards P, Dehesh K, Schneider G, Lindqvist Y. Crystal structure of β-ketoacyl-acyl carrier protein synthase II from E. coli reveals the molecular architecture of condensing enzymes. EMBO J. 1998;17:1183–1191. doi: 10.1093/emboj/17.5.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Inokoshi J, Tomoda H, Hashimoto H, Watanabe A, Takeshima H, Omura S. Cerulenin-resistant mutants of Saccharomyces cerevisiae with an altered fatty acid synthase gene. Mol Gen Genet. 1994;244:90–96. doi: 10.1007/BF00280191. [DOI] [PubMed] [Google Scholar]
- 23.Jackowski S, Murphy C M, Cronan J E, Jr, Rock C O. Acetoacetyl-acyl carrier protein synthase: a target for the antibiotic thiolactomycin. J Biol Chem. 1989;264:7624–7629. [PubMed] [Google Scholar]
- 24.Jackowski S, Rock C O. Acetoacetyl-acyl carrier protein synthase, a potential regulator of fatty acid biosynthesis in bacteria. J Biol Chem. 1987;262:7927–7931. [PubMed] [Google Scholar]
- 25.Jones A L, Dancer J E, Harwood J L. The effect of thiolactomycin analogues on fatty acid synthesis in peas (Pisum sativum cv. Onward) Biochem Soc Trans. 1994;22:258S. doi: 10.1042/bst022258s. [DOI] [PubMed] [Google Scholar]
- 26.Kauppinen S, Siggaard-Anderson M, van Wettstein-Knowles P. β-Ketoacyl-ACP synthase I of Escherichia coli: nucleotide sequence of the fabB gene and identification of the cerulenin binding residue. Carlsberg Res Commun. 1988;53:357–370. doi: 10.1007/BF02983311. [DOI] [PubMed] [Google Scholar]
- 27.Kremer L, Douglas J D, Baulard A R, Morehouse C, Guy M R, Alland D, Dover L G, Lakey J H, Jacobs W R, Jr, Brennan P J, Minnikin D E, Besra G S. Thiolactomycin and related analogues as novel anti-mycobacterial agents targeting KasA and KasB condensing enzymes in Mycobacterium tuberculosis. J Biol Chem. 2000;275:16857–16864. doi: 10.1074/jbc.M000569200. [DOI] [PubMed] [Google Scholar]
- 28.Loftus T M, Jaworsky D E, Frehywot G L, Townsend C A, Ronnett G V, Lane M D, Kuhajda F P. Reduced food intake and body weight in mice treated with fatty acid synthase inhibitors. Science. 2000;288:2379–2381. doi: 10.1126/science.288.5475.2379. [DOI] [PubMed] [Google Scholar]
- 29.McMurray L M, Oethinger M, Levy S. Triclosan targets lipid synthesis. Nature (London) 1998;394:531–532. doi: 10.1038/28970. [DOI] [PubMed] [Google Scholar]
- 30.Moche M, Schneider G, Edwards P, Dehesh K, Lindqvist Y. Structure of the complex between the antibiotic cerulenin and its target, β-ketoacyl-acyl carrier protein synthase. J Biol Chem. 1999;274:6031–6034. doi: 10.1074/jbc.274.10.6031. [DOI] [PubMed] [Google Scholar]
- 31.Nishida I, Kawaguchi A, Yamada M. Effect of thiolactomycin on the individual enzymes of fatty acid synthase in Escherichia coli. J Biochem (Tokyo) 1986;99:1447–1454. doi: 10.1093/oxfordjournals.jbchem.a135614. [DOI] [PubMed] [Google Scholar]
- 32.Olsen J G, Kadziola A, Wettstein-Knowles P, Siggaard-Andersen M, Lindquist Y, Larsen S. The X-ray crystal structure of β-ketoacyl [acyl carrier protein] synthase I. FEBS Lett. 1999;460:46–52. doi: 10.1016/s0014-5793(99)01303-4. [DOI] [PubMed] [Google Scholar]
- 33.Omura S. Cerulenin. Methods Enzymol. 1981;72:520–532. [PubMed] [Google Scholar]
- 34.Perego M. Integrational vectors for genetic manipulation in Bacillus subtilis. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria. Washington, D.C.: American Society for Microbiology; 1993. pp. 615–625. [Google Scholar]
- 35.Qiu X, Janson C A, Konstantinidis A K, Nwagwu S, Silverman C, Smith W W, Khandekar S, Lonsdale J, Abdel-Meguid S S. Crystal structure of β-ketoacyl-acyl carrier protein synthase III. A key condensing enzyme in bacterial fatty acid biosynthesis. J Biol Chem. 1999;274:36465–36471. doi: 10.1074/jbc.274.51.36465. [DOI] [PubMed] [Google Scholar]
- 36.Rock C O, Cronan J E., Jr Escherichia coli as a model for the regulation of dissociable (type II) fatty acid biosynthesis. Biochim Biophys Acta. 1996;1302:1–16. doi: 10.1016/0005-2760(96)00056-2. [DOI] [PubMed] [Google Scholar]
- 37.Rock C O, Garwin J L. Preparative enzymatic synthesis and hydrophobic chromatography of acyl-acyl carrier protein. J Biol Chem. 1979;254:7123–7128. [PubMed] [Google Scholar]
- 38.Rosenfeld I S, D'Agnolo G, Vagelos P R. Synthesis of unsaturated fatty acids and the lesion in fabB mutants. J Biol Chem. 1973;248:2452–2460. [PubMed] [Google Scholar]
- 39.Schujman G E, Grau R, Gramajo H C, Ornella L, de Mendoza D. De novo fatty acid synthesis is required for establishment of cell type-specific gene transcription during sporulation in Bacillus subtilis. Mol Microbiol. 1998;29:1215–1224. doi: 10.1046/j.1365-2958.1998.01004.x. [DOI] [PubMed] [Google Scholar]
- 40.Six D A, Dennis E A. The expanding superfamily of phospholipase A2 enzymes: classification and characterization. Biochim Biophys Acta. 2000;1488:1–19. doi: 10.1016/s1388-1981(00)00105-0. [DOI] [PubMed] [Google Scholar]
- 41.Slayden R A, Lee R E, Armour J W, Cooper A M, Orme I M, Brennan P J, Besra G S. Antimycobacterial action of thiolactomycin: an inhibitor of fatty acid and mycolic acid synthesis. Antimicrob Agents Chemother. 1996;40:2813–2819. doi: 10.1128/aac.40.12.2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tomoda H, Kawaguchi A, Omura S, Okuda S. Cerulenin resistance in a cerulenin-producing fungus. II. Characterization of fatty acid synthetase from Cephalosporium caerulens. J Biochem (Tokyo) 1984;95:1705–1712. doi: 10.1093/oxfordjournals.jbchem.a134784. [DOI] [PubMed] [Google Scholar]
- 43.Tomoda H, Kawaguchi A, Yasuhara T, Nakajima T, Omura S, Okuda S. Cerulenin resistance in a cerulenin-producing fungus. III. Studies on active-site peptides of fatty acid synthetase from Cephalosporium caerulens. J Biochem (Tokyo) 1984;95:1712–1723. [PubMed] [Google Scholar]
- 44.Tsay J-T, Rock C O, Jackowski S. Overproduction of β-ketoacyl-acyl carrier protein synthase I imparts thiolactomycin resistance to Escherichia coli K-12. J Bacteriol. 1992;174:508–513. doi: 10.1128/jb.174.2.508-513.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Val D, Banu G, Seshadri K, Lindqvist Y, Dehesh K. Re-engineering ketoacyl synthase specificity. Structure. 2000;8:565–566. doi: 10.1016/s0969-2126(00)00146-5. [DOI] [PubMed] [Google Scholar]