Abstract
Peptidoglycan recognition proteins (PGRPs) are important components of the innate immune system which provide the first line of defense against invading microbes. There are four members in the family of PGRPs in animals of which PGRP-S is a common domain. It is responsible for the binding to microbial cell wall molecules. In order to understand the mode of binding of PGRP-S to the components of the bacterial cell wall, the structure of the complex of camel PGRP-S (CPGRP-S) with heptanoic acid has been determined at 2.15 Å resolution. The structure determination showed the presence of four crystallographically independent protein molecules which are designated as A, B, C, and D. These four protein molecules associate in the form of two homodimers which are represented as A-B and C-D dimers. The association between molecules A and B gives rise to a shallow cleft on the surface at one end of the dimeric interface. One molecule of heptanoic acid is observed at this binding site in the A-B dimer. The association of C and D molecules results in the formation of a long zig-zag tunnel along with the C-D interface. In the cleft at the C-D interface, three molecules of hydrogen peroxide along with other non-water solvent molecules have been observed. The analysis of the several complexes of CPGRP-S with fatty acids and non-fatty acids such as peptidoglycan, lipopolysaccharide, and lipoteichoic acid shows that the fatty acids bind at the A-B site while non-fatty acids interact through C-D interface.
Keywords: Peptidoglycan recognition protein, PGRP-S, dimers, structures, ligand binding site
Introduction
Peptidoglycan recognition proteins (PGRPs) are members of the innate immune system. They bind to the unique bacterial cell wall molecules and provide the first line of defense against the invading microbes [1,2]. The first PGRP was discovered in silkworms as a protein that recognized the bacterial peptidoglycan (PGN) [3-5]. Therefore, it was named peptidoglycan recognition protein (PGRP). There are four members in this family including PGRP-short (PGRP-S), PGRP-long (PGRP-L), and PGRP-intermediates (PGRP-Iα and PGRP-Iβ) [6-9]. All the PGRPs have at least one PGRP-S domain which is responsible for the binding to bacterial cell wall molecules such as PGN, lipopolysaccharide (LPS) [10], lipoteichoic acid (LTA) [11], and mycolic acid (MA) [12]. Thus the cell wall molecules of bacteria are the targets for the recognition by PGRPs. This makes PGRPs therapeutically useful as the same cell wall molecules are absent in the eukaryotes [13]. PGRPs are widely expressed in different types of cells and tissues and their expressions are often upregulated in response to microbial infections [14-17]. Thus the potent PGRPs can provide the protection to host against microbial infections.
It is well known that the living conditions of various animals are widely different. As a result, they may be exposed to different pathogens. In view of this, their immune responses may differ greatly. Over the period, the proteins of the innate immune system including PGRPs may undergo mutations in their amino acid sequences (Figure 1) to suitably modify their structures to be able to fight the invading microbes. In view of this, it was of great interest to examine the structures of PGRP-S protein from different species. So far, structures of PGRP-S protein from Camelus dromedarius (CPGRP-S) [18], Manduca sexta (MPGRP-S) [19], Bombus terrestris (BPGRP-S) [20], Branchiostoma lanceolatum (BrPGRP-S) [21], Homo sapiens (HPGRP-S) [22] and Drosophila melanogaster (DPGRP-S) [23] are known in the unbound native forms. In the case of CPGRP-S, the structures of several complexes with various ligands are also known [10,24-26]. The structure of one complex of the human PGRP-S domain from PGRP-Iα (Iα-HPGRP-S) is also known [27]. It may be noted that the structures of the complexes of CPGRP-S with fatty acids [10,12] and non-fatty acids [10,25] showed different modes of binding. In order to establish the details of the binding sites in CPGRP-S for fatty acids and non-fatty acids, we have determined a new structure of the complex of CPGRP-S with heptanoic acid.
Figure 1.

Showing the amino acid sequence comparison of PGRP-S from camel, human, C-terminal PGN domain of human, common lancelet, Drosophila, bumblebees, and tobacco hornworm. The segments involved in the intermolecular arrangement between molecules A and B are indicated in red while those responsible for the formation of C-D dimer are shown in green. The critical residues for the formation of A-B dimer are Ser8 and Asn126 (shown in purple) while those in the formation of C-D dimer are Pro96 and Pro151 (shown in blue).
Materials and methods
Purification of PGRP-S
The fresh samples of colostrum of camels were provided by the National Research Centre on Camels, Bikaner, India. The samples were skimmed and diluted twice with 50 mM Tris-HCl, pH 7.8. The CM-Sephadex C-50 gel was pre-equilibrated with 50 mM Tris-HCl pH 7.8 and added to the diluted samples and stirred gently for about two hours using a glass rod. The gel was kept aside undisturbed for two hours. After it settled down, the solution was decanted. The gel was washed with the excess amount of 50 mM Tris-HCl pH 7.8 and then packed in a column (25×2.5 cm). It was washed with the same buffer until the absorbance was reduced to less than 0.03 at 280 nm. Thereafter, the bound basic proteins were eluted with 0.5 M NaCl in 50 mM Tris-HCl pH 7.8 and desalted by dialyzing it against triple distilled water. The desalted fractions were passed through a CM Sephadex (C-50) column (10×2.5 cm) which was also pre-equilibrated with 50 mM Tris-HCl pH 7.8 and eluted with 0.5 M NaCl in the same buffer. The eluted samples were analyzed on the sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE). The fractions corresponding to an approximate molecular weight of 20 kDa were pooled. The pooled fractions were concentrated using an Amicon ultrafiltration unit. The concentrated protein was passed through a Sephadex G-100 column (100×2 cm) using 50 mM Tris-HCl pH 7.8. This showed two peaks when the fractions were read at a wavelength of 280 nm. These were examined on SDS-PAGE which showed two bands at 40 kDa and 20 kDa indicating the presence of a dimer and a monomer in the solution of the purified protein.
Crystallization
The purified and lyophilized samples of protein were dissolved in the buffer containing 50 mM Tris-HCl pH 7.8 to a concentration of 15 mg/ml. 10 μl of protein solution was mixed with an equal volume of the reservoir solution which contained 20% glycerol, and 6% polyethylene glycol-3350 (PEG-3350) at pH 7.8. The 10 μl drops of this solution were set up in the hanging drop vapour diffusion method against the above reservoir solution. The colourless rod-shaped crystals measuring up to 0.30×0.20×0.15 mm3 were obtained within a week. The crystals were soaked for 24 hours in the reservoir solution which contained 1% heptanoic acid solution and 10% 2-methyl-2,4-pentanediol (MPD).
X-ray intensity data collection and structure determination
The soaked crystals of CPGRP-S were used for X-ray diffraction data collection at 100 K to 2.15 Å resolution with a MAR CCD-225 detector (Marresearch, Norderstedt, Germany) using synchrotron beamline BM14 at the European Synchrotron Radiation Facility (ESRF), Grenoble, France. The program, HKL-2000 [28] was used for data processing. The crystals belong to orthorhombic space group I222 with cell dimensions, a = 88.896 Å, b = 101.637 Å, c = 111.700 Å. It was clear from the unit cell dimensions and molecular weight of 19.2 kDa that there were four molecules of CPGRP-S in the asymmetric unit. The structure was determined with the molecular replacement method using the program PHASER [29]. The coordinates of the previously determined structure of CPGRP-S [18] were used as the search model. The refinement was carried out with the program REFMAC [30,31]. The manual model-building steps were carried out using programs O [32] and COOT [33]. The Fourier (2Fo-Fc) and difference Fourier (Fo-Fc) electron density maps were calculated when the value of the Rcryst factor dropped below 0.260. The maps clearly showed non-protein electron densities above 3.0 σ cut off at the contact point of molecules A and B in the A-B dimer and in the cleft at the interface of molecules C and D in the C-D dimer. Heptanoic acid (SHV) was fitted well in the electron density at the A-B site (Figure 2i) while several non-water solvent molecules including MPD, glycerol (GOL), ethylene diol (EDO), and carbonate ion (CO3 2-) were fitted well (Figure 2i). Interestingly, 9 molecules of hydrogen peroxide were also observed in the structure (Figure 2i). In addition to these, one molecule of MPD, 4 molecules of EDO, 1 carbonate, and 1 sodium ion, and 609 water molecules were also observed. The shapes and peaks of electron densities allowed proper fittings of these extra entities. The coordinates of all the extra non-protein atoms were also included in the subsequent cycles of refinement. All the steps of refinements were carried out with default values of restraints. After further cycles of refinements, the protein chains, and non-protein entities were adjusted manually using (2Fo-Fc) and (Fo-Fc) electron density maps. The final refinement cycles were carried out which gave the values of 0.220 and 0.282 for the Rcryst and Rfree factors respectively. The final electron densities for these extra entities in the structure are shown at 1.0 σ cut-offs (Figure 2ii). The overall B-factor for the main chain protein atoms was 40.9 Å2. All the four protein chains contain residues from Cys6 to Ala171 as the electron densities for residues from Glu1 to Ala5 were not observed. The overall quality of the final model was assessed with PROCHECK [34] which showed that all the residues were present in the allowed regions of the Ramachandran φ, ψ map [35]. The coordinates and other crystallographic data have been deposited in the protein data bank with accession code, 7XU8. The data collection and refinement statistics are given in Table 1.
Figure 2.
Electron density maps of various ligands with CPGRP-S. (i) The initial (Fo-Fc) map showing electron densities at 3.0 σ cut off for one heptanoic acid, nine hydrogen peroxide, one 2-methyl-2,4-pentanediol, four ethylene glycol, and one carbonate and one sodium ions and (ii) the final (2Fo-Fc) map showing electron densities at 1.0 σ cut off for the above entities.
Table 1.
Data collection and refinement statistics for the complex CPGRP-S with heptanoic acid (Values in parentheses are for the highest resolution shell)
| Data collection | |
| Beamline (ESRF, Grenoble, France) | ESRF BEAMLINE BM14 |
| Wavelength (Å) | 0.97 |
| Resolution range (Å) | 43.15-2.15 (2.20-2.15) |
| Space group | I222 |
| Unit-cell parameters (Å) | a = 89.896, b = 101.637, c = 111.700 |
| Number of molecules in the asymmetric unit | 4 |
| Vm (Å3/Da) | 2.46 |
| Solvent content (%) | 50 |
| Number of unique reflections | 38900 |
| Overall completeness (%) | 99.7 (99.7) |
| I/σ (I) | 13.1 (3.8) |
| Wilson B-factor (Å2) | 34.8 |
| Refinement statistics | |
| Rcryst | 22 |
| Rfree | 28.2 |
| Number of protein atoms | 5220 |
| Number of heptanoic acid (1) atoms | 9 |
| Number of hydrogen peroxide (9) atoms | 18 |
| Number of 2-methyl-2,4-pentanediol (1) atoms | 8 |
| Number of ethylene glycol (4) atoms | 16 |
| Number of carbonate (1) atoms | 4 |
| Sodium (1) atoms | 1 |
| Number of water oxygen atoms | 609 |
| R. M. S. deviations | |
| Bond length (Å) | 0.006 |
| Bond angles (°) | 1.425 |
| Dihedral angles (°) | 13.742 |
| Mean B-factor (Å2) | |
| Main chain | 40.9 |
| Side chain and water oxygen atoms | 45.5 |
| Overall | 43.5 |
| Ramachandran plot statistics | |
| Residues in the most favored region (%) | 89.7 |
| Residues in the additional allowed region (%) | 10.3 |
| PDB ID | 7XU8 |
Results
The structure of the complex of CPGRP-S with heptanoic acid consists of four crystallographically independent protein molecules which are designated as A, B, C, and D. These four protein molecules are associated in the form of two homodimers represented as A-B and C-D (Figure 3). The total number of atoms from four protein chains (Cys6-Ala171) is 5220. It does not include atoms from the first five residues (Glu1-Asp2-Pro3-Pro4-Ala5) as the electron densities were not observed for the N-terminal pentapeptides in all the four protein molecules in the structure. There are 9 atoms from one heptanoic acid (SHV), 18 atoms from 9 hydrogen peroxide (PEO) molecules, 8 atoms from one molecule of 2-methyl-2,4-pentanediol (MPD), 16 atoms from 4 ethylene diol (EDO) molecules, 4 atoms from one carbonate ion (CO3 2-) and 1 atom of sodium ion. There are 609 water oxygen atoms in the structure. As calculated using PROCHECK [34], there are 89.7% residues in the most favored regions of the Ramachandran’s φ, ψ map [35] whereas 10.3% residues were present in the additionally allowed regions. When the Cα traces of all the four molecules were superimposed on each other, the r.m.s. deviations were found to be less than 0.5 Å indicating that all the four molecules of CPGRP-S in the structure are conformationally identical.
Figure 3.

Structure of CPGRP-Swith four molecules, A, B, C, and D in the asymmetric unit. The bound heptanoic acid at the ligand-binding site in the A-B dimer and several solvent molecules other than the water molecules are indicated in the cleft of the C-D dimer. The numbering scheme of the heptanoic acid is shown at the bottom of the figure.
The polypeptide chain of CPGRP-S folds into a well-formed α/β globular structure (Figure 4). The overall structure includes a central β-sheet consisting of five β-strands out of which four β-strands β3 (Arg31-Thr38), β4 (Tyr71-Gly76), β6 (Ile103-Met108), and β7 (Glu142-His146) are in the parallel arrangement while the fifth strand β5 (Leu80-Arg85) is oriented in an anti-parallel sense with respect to the other four strands. The centrally located β-sheet is surrounded from three sides by three α-helices α1 (Pro46-Leu64), α2 (Pro118-Val132), and α3 (Asp157-Gln164). In addition to these structural elements, two short helices, η1 (Arg12-Trp15) and η2 (His146-Val149) are also observed.
Figure 4.
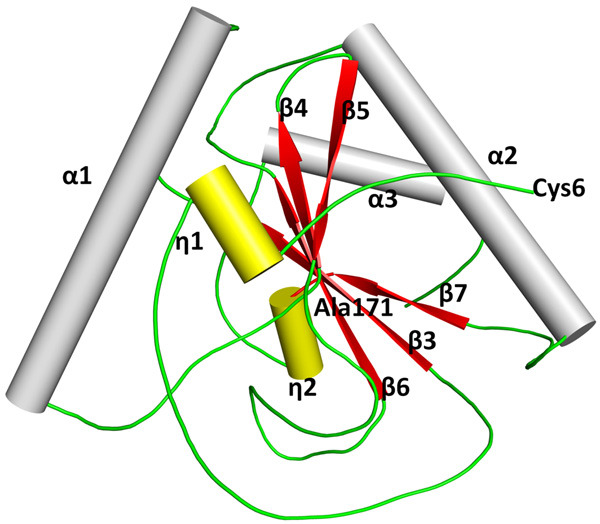
The secondary structure elements in CPGRP-S are shown. Five β-strands including β3, β4, β5, β6, and β7 are shown in red where β3, β4, β6, and β7 are in the parallel arrangement while β5 is oriented in an antiparallel sense. Three α-helices, α1, α2, and α3 are shown in grey. There are two short helices, η1 and η2 in yellow.
Discussion
One of the unique features of the structure of CPGRP-S protein pertains to its oligomerization state as it forms two types of homodimers which are designated as A-B and C-D dimers. Although the interface region in the A-B dimer is tightly packed, the association of two molecules still forms a forceps-like ligand-binding cleft at one end of the interface by orienting the α-helices, α2 from molecules A and B appropriately (Figure 5i). On the other hand, a long zig-zag cleft is formed across the interface in the C-D dimer (Figure 5ii). The two dimers are formed using nearly the opposite sides of the protein molecule leading to the formation of a polymeric arrangement with alternating A-B and C-D interfaces (Figure 5iii). The covered surface areas in A-B and C-D dimers are 640 Å2 and 484 Å2 corresponding to buried areas of nearly 10% and 7% of the total surface area of an individual molecule. This indicates that the two dimers are relatively weakly formed. However, the two dimeric states are always observed for CPGRP-S in the crystalline state [10,11,18,24-26]. On the other hand, both monomers and dimers have been observed in the solution state at concentrations of less than 0.1 mg/ml [36]. It may be mentioned here that the structures of PGRP-S protein from a few other species including MPGRP-S [19], BPGRP-S [20], BrPGRP-S [21], HPGRP-S [22], and DPGRP-S [23] are also known but none of them forms a dimeric structure similar to that of CPGRP-S. As seen from Figure 5i and 5ii, the ligand binding clefts are observed in the two dimers but both have strikingly different shapes. Since the buried surface area in the A-B dimer is relatively larger than that of the C-D dimer, the A-B dimer is more stable than the C-D dimer. Presumably, the A-B dimer is already formed before it interacts with the bacterial cell wall molecules but in the case of the C-D dimer, the interactions with the bacterial cell molecules may occur before the dimer is formed. In other words, it can be stated that the C and D molecules have a tendency to dimerize but the dimer eventually stabilizes after the interactions with cell wall molecules have occurred.
Figure 5.

Showing (i) A-B and (ii) C-D dimers with shaded ligand binding sites and (iii) showing the polymeric arrangement of A-B and C-D interfaces.
Binding of heptanoic acid to A-B dimer
A-B dimer is formed with a long interface involving residues from the N-terminus (Cys6-Glu14), loop (Cys43-Cys49) between strands β3 and helix α1, loop (Gly76-Val81) between strands β4 and β5 and residues (Pro116-Val132) from helix α2 belonging to both molecules A and B. The interface is characterized by extensive intermolecular contacts between molecules A and B. The important interactions include hydrogen bonds involving residues Ser8, Ile9, Glu14, Arg122, and Asn126 from both molecules. As seen in Figure 1, the highest sequence identity of 75% is observed between CPGRP-S and HPGRP-S but the two critical residues Ser8 and Asn126 for the dimerization in CPGRP-S are mutated to Pro11 and Gly129 in HPGRP-S. This seems to have lowered the possibility of dimer formation in HPGRP-S. As seen from Figure 3, one heptanoic acid molecule is bound to A-B dimer in the cleft. The interactions between residues of the A-B dimer and heptanoic acid are largely hydrophobic in nature. The buried surface area for the heptanoic acid in this cleft is 175 Å2 (Figure 6).
Figure 6.

Showing grasp view of the region at the A-B interface with a bound heptanoic acid in the ball and stick model.
In contrast, in the structures of DPGRP-S (PDB ID: 1XSR) and BPGRP-S (PDB ID: 5XZ4), two crystallographically independent molecules were observed but these two molecules do not associate in a manner similar to that of A-B association in the present structure (Figure 7). Thus the formation of A-B dimer in CPGRP-S is a novel feature. The comparison of the binding of heptanoic acid to the A-B dimer in the present structure with those of other fatty acids in the other complexes of CPGRP-S shows that all the fatty acids bind to CPGRP-S in the same cleft of A-B dimer with slight variations in the orientations (Figure 8i). The binding of fatty acids of different lengths to the A-B dimer indicates that the cleft has large dimensions. This suggests that the A-B dimer of CPGRP-S offers a wide scope of recognition within an overall complementarity for the fatty acids. Thus it can be summed up that CPGRP-S is capable of binding to a wide range of fatty acids through the forceps-like binding cleft at one end of the interface in the A-B dimer. Eventually, it can be extended further that CPGRP-S through A-B dimer may also recognize the molecules such as the mycolic acid of the cell wall of Mycobacterium tuberculosis.
Figure 7.
Cα-superimpositions of A-B dimer of CPGRP-S (cyan) on the Cα traces of two crystallographic independent molecules observed in (i) Drosophila (PDB ID, 1XSR, grey) and (ii) bumblebee (PDB ID, 5ZX4, orange).
Figure 8.

Superimpositions of Cα traces of (i) A-B dimer of CPGRP-S from the present complex on the Cα traces of the A-B dimer from the complexes of CPGRP-S with bound ligands at the A-B site and (ii) C-D dimer of CPGRP-S from the present complex on the Cα traces of the C-D dimer from the other complexes of CPGRP-S with bound ligands at the C-D site.
Structure of C-D dimer
The formation of the C-D dimer in CPGRP-S involves mainly three outwardly protruding loops, Tyr59-Trp66, Ala94-Trp99, and Arg147-Leu153. The most critical residues at the interface of the C-D dimer are Pro96 and Pro151. As seen in Figure 1, PGRP-S proteins from various species have residues other than proline at these two positions which seem to hamper the formations of C-D-like dimers. It may be noted that the interface in the C-D dimer is stabilized by several hydrogen bonds and several van der Waals interactions between molecules C and D. This is also a unique form of dimer as no such dimer formation is observed in the structures of PGRP-S from other species. It may be mentioned here that the two crystallographically independent molecules observed in DPGRP-S (PDB ID: 1XSR) and BPGRP-S (PDB ID: 5XZ4) did not associate to form similar dimeric structures (Figure 9). Thus it can be stated that the binding cleft at the interface similar to that of the C-D dimer is not present in the structures of PGRP-S from other species.
Figure 9.

Cα-superimpositions of C-D dimer of CPGRP-S (cyan) on the Cα traces of two crystallographic independent molecules observed in (i) Drosophila (PDB ID, 1XSR, grey) and (ii) bumblebee (PDB ID, 5ZX4, orange).
As seen in Figure 3, several solvent molecules other than water molecules such as 2-methyl-2,4-pentanediol, and ethylene glycol have been observed in the cleft. As the protein was isolated from colostrum, some molecules of hydrogen peroxide have also been observed. Previously, we have reported several other ligands in the cleft at the C-D interface [10,11,20-22]. The superimpositions of the complexes of non-fatty acids including those with LPS, LTA, and PGN show that these ligands bind to the C-D dimer at the same binding site (Figure 8ii). It clearly indicates that the cleft at the C-D interface is suitable for the binding of compounds such as PGN, LPS, LTA, and their analogues. Therefore, the binding site in the C-D dimer may be suitable for the recognition of cell wall molecules of Gram-negative and Gram-positive bacteria.
Conclusions
The present structure of the complex of camel PGRP-S with heptanoic acid shows that heptanoic acid binds to camel PGRP-S in the biding site located at one end of the interface of the A-B dimer. This structure (Figure 3) is similar to the structures of the complexes of camel PGRP-S with other fatty acids (Figure 8i). Thus, the present structure further confirms that the binding site at the interface of the A-B dimer is suitable for the binding of various fatty acids including acid. Therefore, the binding site at the A-B dimer may be the preferred site for the binding of cell wall molecules of Mycobacterium tuberculosis. On the other hand, as reported earlier, the non-fatty acids such as PGN, LPS, LTA, and their analogues bind preferably in the cleft formed at the interface of the C-D dimer (Figure 8ii). In summing up, it can be stated here that the camel PGRP-S possesses two separate binding sites which are present at the interfaces of two different dimers for the recognition of diverse cell wall molecules of bacteria. In contrast, the structures of PGRP-S from other animals are monomers and hence possess a single binding site for both fatty acids and non-fatty acids. As a result of the dimeric arrangement, the camel PGRP-S has a higher affinity for the bacterial cell wall molecules indicating a higher potency of recognition by CPGRP-S. In view of this, camel PGRP-S can be exploited for therapeutic applications against microbial infections.
Acknowledgements
The authors thank the Science and Engineering Research Board (SERB), Ministry of Science and Technology, New Delhi for the research grant under the SERB-Distinguished Fellowship program to TPS. AM and VV thank the Indian Council of Medical Research (ICMR), New Delhi for the award of fellowships. NA thanks the University Grants Commission (UGC), New Delhi, and PKS acknowledges the Department of Health Research (DHR), New Delhi for the award of fellowships.
Disclosure of conflict of interest
None.
References
- 1.Guan R, Mariuzza RA. Peptidoglycan recognition proteins of the innate immune system. Trends Microbiol. 2007;15:127–134. doi: 10.1016/j.tim.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 2.Wang Z, Zhou W, Huang B, Gao M, Li Q, Tao Y, Wang Z. Molecular and functional characterization of peptidoglycan recognition proteins of PGRP-A and of PGRP-B in ostriniafurnacalis (Lepidoptera: Crambidae) Insects. 2022;13:417. doi: 10.3390/insects13050417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dziarski R. Peptidoglycan recognition proteins (PGRPs) Mol Immunol. 2004;40:877–886. doi: 10.1016/j.molimm.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 4.Hou J, Gan Z, Chen SN, Nie P. Molecular and functional characterization of a short-type peptidoglycan recognition protein, PGRP-S in the amphibian Xenopus laevis. Dev Comp Immunol. 2019;98:13–19. doi: 10.1016/j.dci.2019.04.003. [DOI] [PubMed] [Google Scholar]
- 5.Huang L, Chen SN, Gan Z, Nie P. Molecular and functional identification of a short-type peptidoglycan recognition protein, PGRP-S, in the Chinese soft-shelled turtle Pelodiscus sinensis. Dev Comp Immunol. 2021;117:103965. doi: 10.1016/j.dci.2020.103965. [DOI] [PubMed] [Google Scholar]
- 6.Liu C, Xu Z, Gupta D, Dziarski R. Peptidoglycan recognition proteins: a novel family of four human innate immunity pattern recognition molecules. J Biol Chem. 2001;276:34686–34694. doi: 10.1074/jbc.M105566200. [DOI] [PubMed] [Google Scholar]
- 7.Hu Z, Cao X, Guo M, Li C. Identification and characterization of a novel short-type peptidoglycan recognition protein in Apostichopus japonicus. Fish Shellfish Immunol. 2020;99:257–266. doi: 10.1016/j.fsi.2020.02.013. [DOI] [PubMed] [Google Scholar]
- 8.Yashin DV, Sashchenko LP, Georgiev GP. Mechanisms of action of the PGLYRP1/Tag7 protein in innate and acquired immunity. Acta Naturae. 2021;13:91–101. doi: 10.32607/actanaturae.11102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shen D, Mei X, Guo J, Tong M, Xia D, Qiu Z, Zhao Q. Peptidoglycan recognition protein-S1 (PGRP-S1) from Diaphaniapyloalis (Walker) is involved in the agglutination and prophenoloxidase activation pathway. Gene. 2022;809:146004. doi: 10.1016/j.gene.2021.146004. [DOI] [PubMed] [Google Scholar]
- 10.Sharma P, Dube D, Sinha M, Yadav S, Kaur P, Sharma S, Singh TP. Structural insights into the dual strategy of recognition by peptidoglycan recognition protein, PGRP-S: structure of the ternary complex of PGRP-S with lipopolysaccharide and stearic acid. PLoS One. 2013;8:e53756. doi: 10.1371/journal.pone.0053756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sharma P, Dube D, Singh A, Mishra B, Singh N, Sinha M, Dey S, Kaur P, Mitra DK, Sharma S, Singh TP. Structural basis of recognition of pathogen-associated molecular patterns and inhibition of proinflammatory cytokines by camel peptidoglycan recognition protein. J Biol Chem. 2011;286:16208–16217. doi: 10.1074/jbc.M111.228163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sharma P, Yamini S, Dube D, Singh A, Mal G, Pandey N, Sinha M, Singh AK, Dey S, Kaur P, Mitra DK, Sharma S, Singh TP. Structural basis of the binding of fatty acids to peptidoglycan recognition protein, PGRP-S through second binding site. Arch Biochem Biophys. 2013;529:1–10. doi: 10.1016/j.abb.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 13.Dziarski R, Gupta D. Mammalian PGRPs: novel antibacterial proteins. Cell Microbiol. 2006;8:1059–1069. doi: 10.1111/j.1462-5822.2006.00726.x. [DOI] [PubMed] [Google Scholar]
- 14.Kong X, Liu H, Li Y, Zhang H. Two novel short peptidoglycan recognition proteins (PGRPs) from the deep sea vesicomyidae clam archivesicapackardana: identification, recombinant expression and bioactivity. Front Physiol. 2018;9:1476. doi: 10.3389/fphys.2018.01476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng W, Rus F, Hernandez A, Kang P, Goldman W, Silverman N, Tatar M. Dehydration triggers ecdysone-mediated recognition-protein priming and elevated anti-bacterial immune responses in Drosophila Malpighian tubule renal cells. BMC Biol. 2018;16:60. doi: 10.1186/s12915-018-0532-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chevée V, Sachar U, Yadav S, Heryanto C, Eleftherianos I. The peptidoglycan recognition protein PGRP-LE regulates the Drosophila immune response against the pathogen Photorhabdus. Microb Pathog. 2019;136:103664. doi: 10.1016/j.micpath.2019.103664. [DOI] [PubMed] [Google Scholar]
- 17.Jiang L, Liu W, Guo H, Dang Y, Cheng T, Yang W, Sun Q, Wang B, Wang Y, Xie E, Xia Q. Distinct functions of Bombyx mori peptidoglycan recognition protein 2 in immune responses to bacteria and viruses. Front Immunol. 2019;10:776. doi: 10.3389/fimmu.2019.00776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sharma P, Singh N, Sinha M, Sharma S, Perbandt M, Betzel C, Kaur P, Srinivasan A, Singh TP. Crystal structure of the peptidoglycan recognition protein at 1.8 A resolution reveals dual strategy to combat infection through two independent functional homodimers. J Mol Biol. 2008;378:923–932. doi: 10.1016/j.jmb.2008.03.018. [DOI] [PubMed] [Google Scholar]
- 19.Hu Y, Cao X, Li X, Wang Y, Boons GJ, Deng J, Jiang H. The three-dimensional structure and recognition mechanism of Manduca sexta peptidoglycan recognition protein-1. Insect Biochem Mol Biol. 2019;108:44–52. doi: 10.1016/j.ibmb.2019.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Y, Zhao X, Huang J, Chen M, An J. Structural insights into the preferential binding of PGRP-SAs from bumblebees and honeybees to dap-type peptidoglycans rather than lys-type peptidoglycans. J Immunol. 2019;202:249–259. doi: 10.4049/jimmunol.1800439. [DOI] [PubMed] [Google Scholar]
- 21.Wang WJ, Cheng W, Luo M, Yan Q, Yu HM, Li Q, Cao DD, Huang S, Xu A, Mariuzza RA, Chen Y, Zhou CZ. Activity augmentation of amphioxus peptidoglycan recognition protein BbtPGRP3 via fusion with a chitin binding domain. PLoS One. 2015;10:e0140953. doi: 10.1371/journal.pone.0140953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guan R, Wang Q, Sundberg EJ, Mariuzza RA. Crystal structure of human peptidoglycan recognition protein S (PGRP-S) at 1.70 Å resolution. J Mol Biol. 2005;347:683–691. doi: 10.1016/j.jmb.2005.01.070. [DOI] [PubMed] [Google Scholar]
- 23.Reiser JB, Teyton L, Wilson IA. Crystal structure of the Drosophila peptidoglycan recognition protein (PGRP)-SA at 1.56 Å resolution. J Mol Biol. 2004;340:909–917. doi: 10.1016/j.jmb.2004.04.077. [DOI] [PubMed] [Google Scholar]
- 24.Sharma P, Dube D, Singh A, Mishra B, Singh N, Sinha M, Dey S, Kaur P, Mitra DK, Sharma S, Singh TP. Structural basis of recognition of pathogen-associated molecular patterns and inhibition of proinflammatory cytokines by camel peptidoglycan recognition protein. J Biol Chem. 2011;286:16208–16217. doi: 10.1074/jbc.M111.228163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sharma P, Dube D, Sinha M, Mishra B, Dey S, Mal G, Pathak KM, Kaur P, Sharma S, Singh TP. Multiligand specificity of pathogen-associated molecular pattern-binding site in peptidoglycan recognition protein. J Biol Chem. 2011;286:31723–31730. doi: 10.1074/jbc.M111.264374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sharma P, Dube D, Sinha M, Dey S, Kaur P, Sharma S, Singh TP. Structural basis of heparin binding to camel peptidoglycan recognition protein-S. Int J Biochem Mol Biol. 2012;3:86–94. [PMC free article] [PubMed] [Google Scholar]
- 27.Guan R, Roychowdhury A, Ember B, Kumar S, Boons GJ, Mariuzza RA. Structural basis for peptidoglycan binding by peptidoglycan recognition proteins. Proc Natl Acad Sci U S A. 2004;101:17168–17173. doi: 10.1073/pnas.0407856101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 29.McCoy AJ. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr D Biol Crystallogr. 2007;63:32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 31.Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 33.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr A. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 34.Laskowski R, MacArthur M, Moss D, Thornton J. PROCHECK: a program to check stereo chemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 35.Ramachandran GN, Sasisekharan V. Conformation of polypeptides and proteins. Adv Protein Chem. 1968;23:283–438. doi: 10.1016/s0065-3233(08)60402-7. [DOI] [PubMed] [Google Scholar]
- 36.Sharma P. Structural and functional studies of peptidoglycan recognition protein and its complexes with various ligands, Ph.D. thesis. New Delhi: All India Institute of Medical Sciences; 2011. pp. 33–43. [Google Scholar]




