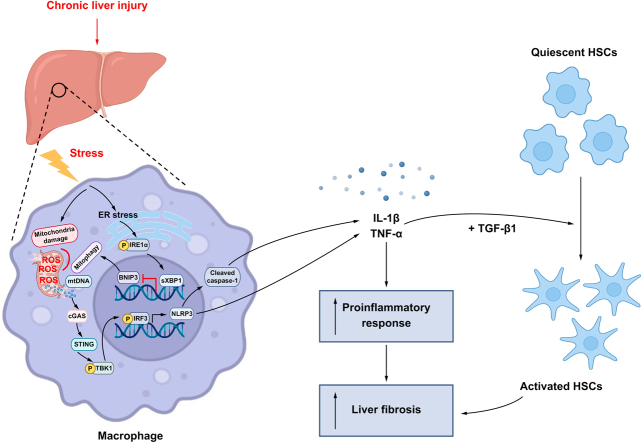
Abstract
Background & Aims
XBP1 modulates the macrophage proinflammatory response, but its function in macrophage stimulator of interferon genes (STING) activation and liver fibrosis is unknown. X-box binding protein 1 (XBP1) has been shown to promote macrophage nucleotide-binding oligomerization domain, leucine-rich repeat and pyrin domain-containing 3 (NLRP3) activation in steatohepatitis. Herein, we aimed to explore the underlying mechanism of XBP1 in the regulation of STING signalling and the subsequent NLRP3 activation during liver fibrosis.
Methods
XBP1 expression was measured in the human fibrotic liver tissue samples. Liver fibrosis was induced in myeloid-specific Xbp1-, STING-, and Nlrp3-deficient mice by carbon tetrachloride injection, bile duct ligation, or a methionine/choline-deficient diet.
Results
Although increased XBP1 expression was observed in the fibrotic liver macrophages of mice and clinical patients, myeloid-specific Xbp1 deficiency or pharmacological inhibition of XBP1 protected the liver against fibrosis. Furthermore, it inhibited macrophage NLPR3 activation in a STING/IRF3-dependent manner. Oxidative mitochondrial injury facilitated cytosolic leakage of macrophage self-mtDNA and cGAS/STING/NLRP3 signalling activation to promote liver fibrosis. Mechanistically, RNA sequencing analysis indicated a decreased mtDNA expression and an increased BCL2/adenovirus E1B interacting protein 3 (BNIP3)-mediated mitophagy activation in Xbp1-deficient macrophages. Chromatin immunoprecipitation (ChIP) assays further suggested that spliced XBP1 bound directly to the Bnip3 promoter and inhibited the transcription of Bnip3 in macrophages. Xbp1 deficiency decreased the mtDNA cytosolic release and STING/NLRP3 activation by promoting BNIP3-mediated mitophagy activation in macrophages, which was abrogated by Bnip3 knockdown. Moreover, macrophage XBP1/STING signalling contributed to the activation of hepatic stellate cells.
Conclusions
Our findings demonstrate that XBP1 controls macrophage cGAS/STING/NLRP3 activation by regulating macrophage self-mtDNA cytosolic leakage via BNIP3-mediated mitophagy modulation, thus providing a novel target against liver fibrosis.
Lay summary
Liver fibrosis is a typical progressive process of chronic liver disease, driven by inflammatory and immune responses, and is characterised by an excess of extracellular matrix in the liver. Currently, there is no effective therapeutic strategy for the treatment of liver fibrosis, resulting in high mortality worldwide. In this study, we found that myeloid-specific Xbp1 deficiency protected the liver against fibrosis in mice, while XBP1 inhibition ameliorated liver fibrosis in mice. This study concluded that targeting XBP1 signalling in macrophages may provide a novel strategy for protecting the liver against fibrosis.
Keywords: XBP1, Macrophage, Liver fibrosis, mtDNA, STING, BNIP3, Mitophagy
Abbreviations: Acta2/α-SMA, actin, alpha 2, smooth muscle, aorta; BDL, bile duct ligation; BMDMs, bone marrow-derived macrophages; BNIP3, BCL2/adenovirus E1B interacting protein 3; CCl4, carbon tetrachloride; cGAS, cyclic GMP-AMP synthase; ChIP, chromatin immunoprecipitation; CM, conditional media; Col1a1, collagen, type I, alpha 1; DMXAA, 5,6-dimethylxanthenone-4-acetic acid; ER, endoplasmic reticulum; EtBr, ethidium bromide; HSC, hepatic stellate cell; IRE1α, inositol-requiring enzyme-1α; IRF3, interferon regulatory factor 3; KEGG, Kyoto Encyclopedia of Genes and Genomes; LC3B, microtubule-associated protein 1 light chain 3 beta; LPS, lipopolysaccharide; MCD, methionine/choline-deficient diet; MnSOD, manganese superoxide dismutase; mtDNA, mitochondrial DNA; NAFLD, non-alcoholic fatty liver disease; NASH, non-alcoholic steatohepatitis; NLRP3, nucleotide-binding oligomerization domain, leucine-rich repeat and pyrin domain-containing 3; p62, sequestosome 1; PBMCs, peripheral blood mononuclear cells; ROS, reactive oxygen species; shRNAs, short hairpin RNAs; STING, stimulator of interferon genes; sXBP1, spliced XBP1; TBK1, TANK binding kinase 1; TGF-β1, transforming growth factor beta 1; Timp1, tissue inhibitor of matrix metalloproteinase 1; TLR, Toll-like receptor; TNF-α, tumour necrosis factor alpha; uXBP1, unspliced XBP1; WT, wild-type; XBP1, X-box binding protein 1
Graphical abstract
Highlights
-
•
Macrophage STING signalling can be activated by mtDNA cytosolic leakage from macrophages themselves.
-
•
Xbp1 depletion suppresses cGAS/STING/NLRP3 activation by restoring BNIP3-mediated mitophagy activation in macrophages.
-
•
XBP1 targets and inhibits the transcription of Bnip3 directly in macrophages.
-
•
Myeloid-specific Xbp1 deficiency, or STING deficiency, or Nlrp3 depletion protect livers against fibrosis in mice.
-
•
Pharmacological inhibition of XBP1 ameliorates liver fibrosis in mice.
Introduction
Liver fibrosis is a reparative response to various types of hepatic injury. Macrophages, one of the most widely studied immune cell populations in liver fibrosis, regulate both the progression and regression of liver fibrosis.1
Cyclic GMP-AMP synthase (cGAS), a cytosolic DNA sensor, and its downstream effector, stimulator of interferon genes (STING), are important for the innate immune response to intracellular DNA from the invading viral and bacterial pathogens. Recently, emerging evidence has shown the critical role of cGAS/STING in sensing endogenous DNA in autoimmune diseases and cancer.2 In the liver, STING-dependent innate immune activation of macrophages is involved in regulating non-alcoholic steatohepatitis (NASH),3,4 viral hepatitis5,6 and liver cancer.7 Given that mitochondrial DNA (mtDNA) release has been noted in stressed macrophages, we examined whether the endogenous mtDNA released from macrophages could be sensed by cGAS/STING and further induce a proinflammatory response in liver fibrosis.
X-box binding protein 1 (XBP1) is a downstream transcription factor of a transmembrane protein serine/threonine kinase, inositol-requiring enzyme-1α (IRE1α), in the endoplasmic reticulum (ER) stress signalling pathway. Stimulation of macrophages with Toll-like receptor (TLR) ligands results in IRE1α activation, which can cleave the unspliced XBP1 (uXBP1) mRNA into unconventional spliced XBP1 (sXBP1) mRNA. The sXBP1 further enhances the transcription of inflammatory cytokines. The macrophage innate immune response, which is triggered by XBP1 activation, has been described in recent studies.[8], [9], [10] Although ER stress is a major contributor to liver fibrosis,11 the role of Xbp1 in regulating macrophage STING signalling and liver fibrosis remains largely unknown.
Mitophagy is an evolutionarily conserved cellular process that removes dysfunctional and superfluous mitochondria. Although the accumulation of cytosolic mtDNA induces cGAS/STING-dependent immune activation, mitophagy affects immune homeostasis by repressing the mitochondrial injury and promoting the elimination of pathogenic mtDNA. ER stress can also regulate autophagy/mitophagy.12 Xbp1 deficiency in the nervous system protects against amyotrophic lateral sclerosis by increasing autophagy.13 Thus, herein, we hypothesised that XBP1 promotes macrophage self-mtDNA cytosolic leakage by impairing mitophagy, leading to macrophage STING activation and subsequent intrahepatic inflammation and liver fibrosis.
Materials and methods
Human liver specimens and peripheral blood mononuclear cell collection
Tissue samples (n = 54) were obtained from patients who underwent resection for liver fibrosis. The control group samples (n = 42) consisted of surgically resected specimens obtained from patients with hepatic haemangioma. In addition, peripheral blood samples were collected from 12 patients with liver fibrosis who were preparing for partial hepatectomy and 12 patients with hepatic haemangioma who underwent surgical resection (normal controls). All patients were referred to the First Affiliated Hospital of Nanjing Medical University after they provided informed consent. All research was performed in compliance with government policies and the Declaration of Helsinki. The demographic information of the study population is shown in Tables S1–S4. All procedures were approved by the Ethics Committee of the First Affiliated Hospital of Nanjing Medical University (approval number: 2020-SR-511).
Experimental animal models
Wild-type (WT), male, 6- to 8-week-old, FloxP-Xbp1 (Xbp1FL/FL), FloxP-Nlrp3 (Nlrp3FL/FL), Lyz2-Cre Xbp1-knockout (Xbp1M-KO), Lyz2-Cre Nlrp3-knockout (Nlrp3M-KO) mice on a C57BL/6 background (Nanjing Biomedical Research Institute of Nanjing University, Nanjing, China), and FloxP-Tmem173 (STING) (Tmem173FL/FL), Lyz2-Cre Tmem173-knockout (Tmem173M-KO) mice (Shanghai Model Organisms, Inc, Shanghai, China) were used in the experiments. They were fed sterilised water and food under specific pathogen-free conditions. Hepatic fibrosis models were developed using carbon tetrachloride (CCl4) injection, bile duct ligation (BDL), or a methionine/choline-deficient diet (MCD). For CCl4 models, mice were intraperitoneally injected with CCl4 (10% in olive oil, 2 ml/kg, twice per week for 8 weeks) or a vehicle (olive oil) to induce fibrosis and sacrificed 48 h after the last injection. For BDL models, the common bile duct was exposed and ligated twice with 6-0 silk sutures after a midline laparotomy. The abdomen was closed with a 6-0 suture, and the mice were allowed to wake up on a heating pad. The control mice underwent a sham operation without ligation of the common bile duct. Mice were anesthetised with isoflurane and sacrificed 2 weeks after BDL or sham surgery. For MCD models, mice were fed MCD (Research Diets, Inc., New Brunswick, CA, USA) or a normal chow diet (NCD) for 8 weeks. Liver tissue was collected for further analysis. In the CCl4 group, the BCL2/adenovirus E1B interacting protein 3 (Bnip3) siRNA or scramble siRNA (Santa Cruz, CA, USA) was mixed with mannose-conjugated polymers (Polyplus-transfection, Illkirch, France) according to the manufacturer’s instructions, and injected into the mice via the tail vein (2 mg/kg) every 3 days beginning 2 weeks after CCl4 injections. The mice were sacrificed and liver tissues were collected at the end of the treatment. In the BDL group, Bnip3 siRNA or scramble siRNA was injected into the mice every 3 days via the tail vein after BDL. In the MCD group, mice were injected with Bnip3 siRNA or scramble siRNA every 3 days via the tail vein beginning 2 weeks after the initiation of MCD feeds. The C57BL/6 background mice were intraperitoneally injected with the XBP1 inhibitor toyocamycin (0.5 mg/kg per day). In the CCl4 group, toyocamycin or PBS was injected intraperitoneally into the mice every day beginning 4 weeks after CCl4 treatment. In the BDL group, toyocamycin or PBS was injected intraperitoneally into the mice every day beginning 1 week after BDL. In the MCD group, mice were injected intraperitoneally with toyocamycin or PBS every day, beginning 4 weeks after feeding with MCD. All animal procedures were performed in strict accordance with the recommendations of the protocol (No: NMU08-092) endorsed by the Institutional Animal Care and Use Committee of Nanjing Medical University.
Statistical analysis
All data are presented as mean ± SD and were analysed using SPSS (version 22.0, IBM, Armonk, NY, USA). The Student t test and 1-way ANOVA were used to analyse the differences among the different groups. Statistical significance was set at p <0.05.
Results
Myeloid Xbp1 deficiency ameliorates experimental liver fibrosis
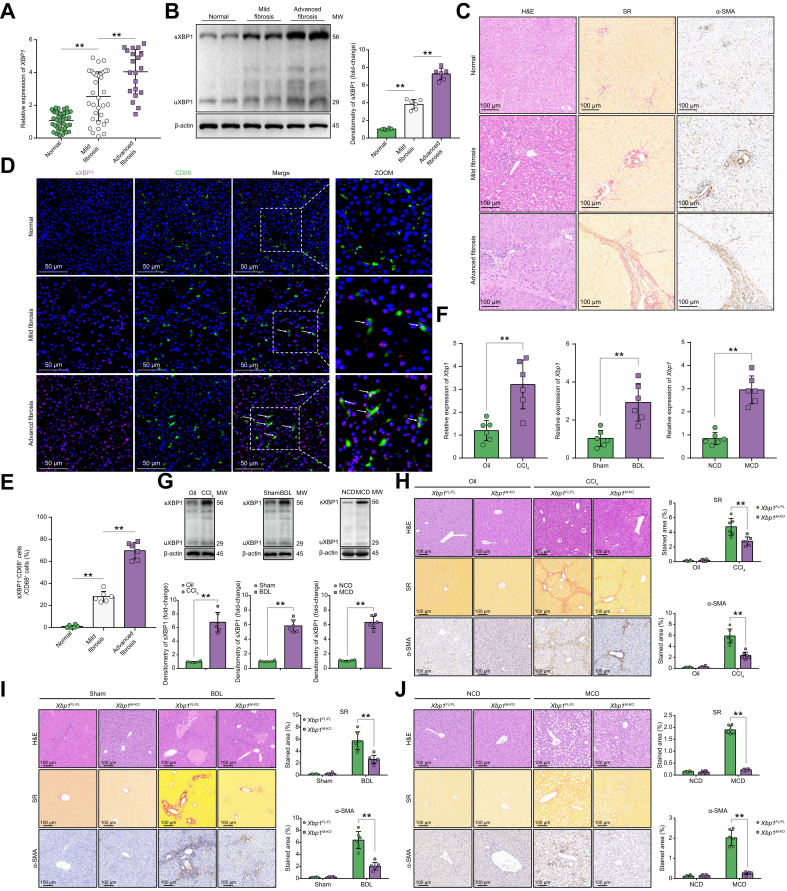
Although XBP1 activation in myofibroblasts promotes fibrotic diseases,14 the precise role of XBP1 signalling in regulating macrophage activation during liver fibrosis remains unclear. First, we compared the XBP1 expression in normal and fibrotic liver tissues from humans. Increased gene (Fig. 1A) and protein (Fig. 1B) expression of XBP1 was observed in fibrotic liver tissues. Liver fibrosis was confirmed by histopathological examination, Sirius Red staining, and immunohistochemical staining for alpha-smooth muscle actin (α-SMA) (Fig. 1C). Furthermore, enhanced sXBP1 expression was observed in macrophages from fibrotic livers (Fig. 1D) and was considerably higher in those with advanced fibrosis than in those with mild fibrosis (Fig. 1E). Similarly, increased XBP1 expression was also found in fibrotic liver tissues from different murine fibrotic models (Fig. 1F and G). In addition, the results revealed that XBP1 was expressed at higher levels in macrophages from fibrotic livers than in controls, and in macrophages than in hepatic stellate cells (HSCs) from different fibrotic models (Fig. S1A–D). Subsequently, we determined the role of myeloid Xbp1 depletion in the regulation of liver fibrosis using myeloid Xbp1-knockout (Xbp1M-KO) mice. To produce myeloid-specific knockout mice, homozygous Xbp1FL/FL mice were bred with homozygous Lyz2-Cre mice, and their heterozygous offspring were backcrossed with homozygous Xbp1FL/FL mice (Fig. S2A). The overall expression levels of XBP1 in liver tissue or hepatocytes of Xbp1FL/FL and Xbp1M-KO mice was not significantly different. Hepatic non-parenchymal cells (NPCs), hepatic macrophages, and bone marrow-derived macrophages (BMDMs) isolated from Xbp1M-KO mice exhibited significantly lower levels of XBP1 than those isolated from Xbp1FL/FL mice (Fig. S2B). Xbp1M-KO mice exhibited significant protection against liver fibrosis, as indicated by reduced Sirius Red and α-SMA staining (Fig. 1H–J), and reduced gene levels of Acta2, Col1a1, and Timp1 (Fig. S2C).
Fig. 1.
Myeloid-specific Xbp1 deficiency ameliorates experimental liver fibrosis.
(A) The gene expression of XBP1 in human liver tissues was determined by quantitative RT-PCR; 1-way ANOVA. (B) The protein levels of XBP1 in human normal and fibrotic liver tissues were analysed using Western blot technique with respect to the loading control β-actin; n = 6/group; 1-way ANOVA. (C) Normal or fibrotic liver sections from patients were subjected to H&E and Sirius Red staining and α-SMA immunohistochemical analysis (n = 6/group; original magnification 200 × ; scale bar = 100 μm). (D) Dual immunofluorescence staining showing spliced XBP1 (red) and CD68 (green) colocalization in human liver tissues. (E) The ratio of sXBP1 + CD68+/CD68+ cells; n = 6/group; 1-way ANOVA. (F) The expression levels of Xbp1 in liver tissue from mice with CCl4-, BDL-, and MCD-induced liver fibrosis were examined by quantitative real-time PCR; n = 6 mice/group; Student t test. (G) The levels of XBP1 in liver tissue from mice with CCl4-, BDL- and MCD-induced liver fibrosis were analysed using Western blot with respect to the loading control β-actin; n = 6 mice/group; Student t test. (H-J) Xbp1FL/FL and Xbp1M-KO male mice were subjected to CCl4-, BDL-, or MCD-induced experimental fibrosis, and the collected liver samples were subjected to H&E and Sirius Red staining and α-SMA immunohistochemical analysis. Scale bar = 100 μm. Representative of 6 mice/group. The proportions of the Sirius Red-positive and α-SMA-positive areas were also quantified; n = 6 mice/group; 1-way ANOVA. The values are shown as mean ± SD. Statistical significance was assessed using the Student t test or ANOVA. ∗∗p <0.01; ∗p <0.05. BDL, bile duct ligation; CCl4, carbon tetrachloride; MCD, methionine/choline deficient diet; α-SMA, alpha-smooth muscle actin; XBP1, X-box binding protein 1.
Reduced liver inflammation was observed in Xbp1M-KO mice, as evidenced by the reduced staining of intrahepatic CD11b+ macrophages and Ly6G+ neutrophils (Fig. S3A–D), the decreased TNF-α and IL-1β levels and increased IL-10 expression (Fig. S3E and F). These findings demonstrated that Xbp1 deficiency attenuated liver inflammation and fibrosis.
Xbp1 deficiency reduces macrophage STING/NLRP3 activation
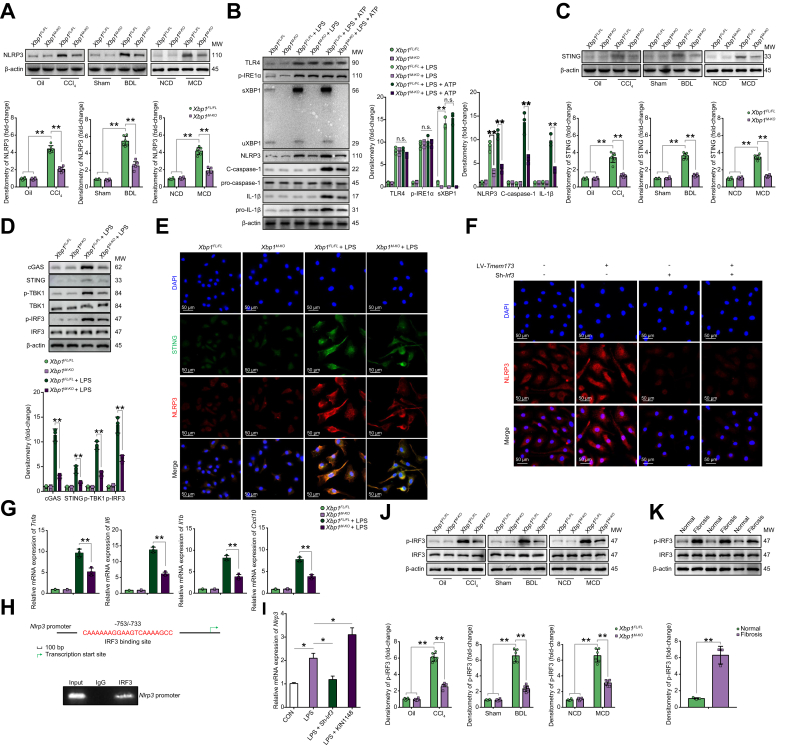
The regulatory role of nucleotide-binding oligomerization domain, leucine-rich repeat, and pyrin domain-containing 3 (NLRP3) inflammasome activation has been reported previously. Hepatocyte pyroptosis and NLRP3 inflammasome particle release induced HSC activation and liver fibrosis;15 NLRP3 inflammasome blockade reduced liver inflammation and fibrosis.16,17 We found that mice with myeloid Nlrp3 knockout developed less liver fibrosis than the WT (Nlrp3FL/FL) controls (Fig. S4A–D). To assess the effect of XBP1 on NLRP3 activation in macrophages during liver fibrosis, we analysed the hepatic NLRP3 levels of Xbp1FL/FL and Xbp1M-KO mice treated with different fibrotic models. Decreased NLRP3 activation was observed in the Xbp1M-KO (Fig. 2A). BMDMs were stimulated with lipopolysaccharide (LPS) alone or LPS and ATP, and the NLRP3 signalling pathway was analysed. TLR4, p-IRE1α, XBP1, and NLRP3 signalling pathways were activated by LPS stimulation with or without ATP, whereas those of C-caspase-1 and IL-1β, which are downstream signals in the NLRP3 pathway, were activated by LPS stimulation with ATP. Xbp1 deficiency significantly reduced NLRP3 signalling pathway activation in BMDMs in response to LPS with ATP (Fig. 2B). Moreover, 4μ8C, an antioxidant inhibitor of IRE1α, was also used to treat BMDMs; results showed that IRE1α inhibition significantly decreased the sXBP1 expression in macrophages in response to LPS with or without ATP (Fig. S4E).
Fig. 2.
XBP1 regulates macrophage activation in a STING-NLRP3-dependent manner.
(A) Western blot was performed to analyse the levels of NLRP3 in liver macrophages isolated from Xbp1FL/FL and Xbp1M-KO mice treated with CCl4, BDL, or MCD. Statistical analysis was carried out using 1-way ANOVA. (B) Western blot was performed to analyse the levels of TLR4, p-IRE1α, XBP1, NLRP3, cleaved caspase-1, pro-caspase-1, cleaved IL-1β, and pro-IL-1β in Xbp1FL/FL and Xbp1M-KO BMDMs treated with LPS or LPS + ATP. Statistical analysis was carried out using 1-way ANOVA. (C) Western blot was performed to analyse the levels of STING in liver macrophages isolated from Xbp1FL/FL and Xbp1M-KO mice treated with CCl4, BDL, or MCD. Statistical analysis was carried out using 1-way ANOVA. (D) Western blot was performed to analyse the levels of cGAS, STING, p-TBK1, TBK1, p-IRF3, and IRF3 in Xbp1FL/FL and Xbp1M-KO BMDMs treated with LPS. Statistical analysis was carried out using 1-way ANOVA. (E) Immunofluorescence staining showing STING (green) and NLRP3 (red) colocalization in Xbp1FL/FL and Xbp1M-KO BMDMs stimulated with LPS. (F) Immunofluorescence was used to detect the expression of NLRP3 in LPS-stimulated Xbp1M-KO BMDMs transfected with LV-Tmem173 or Sh-Irf3. (G) The gene levels of Tnfa, Il6, Il1b, and Cxcl10 in BMDMs; n = 3/group; 1-way ANOVA. (H) Schematic diagram of the putative IRF3 binding site within the Nlrp3 promoter. The BMDMs were subjected to ChIP assay with anti-IRF3 or IgG antibody. The representative results from 3 independent experiments are shown. (I) Relative Bnip3 mRNA expression in Xbp1FL/FL BMDMs treated with Sh-Irf3 or KIN1148 with or without LPS stimulation were determined using qPCR; n = 3 biological replicates/group; 1-way ANOVA. (J) Western blot was performed to analyse the levels of p-IRF3 in liver macrophages isolated from Xbp1FL/FL and Xbp1M-KO mice treated with CCl4, BDL, or MCD. Statistical analysis was carried out using 1-way ANOVA. (K) Western blot was performed to analyse the levels of p-IRF3 in human normal or fibrotic liver tissue; Student t test. The values are shown as the mean ± SD. Statistical significance was assessed by Student t test or ANOVA. ∗∗p <0.01; ∗p <0.05. BDL, bile duct ligation; BMDMs, bone marrow-derived macrophages; BNIP3, BCL2/adenovirus E1B interacting protein 3; CCl4, carbon tetrachloride; ChIP, chromatin immunoprecipitation; cGAS, cyclic GMP-AMP synthase; IRE1α, inositol-requiring enzyme-1α; IRF3, interferon regulatory factor 3; LPS, lipopolysaccharide; MCD, methionine/choline-deficient diet; NLRP3, nucleotide-binding oligomerization domain, leucine-rich repeat and pyrin domain-containing 3; STING, stimulator of interferon genes; TBK1, TANK binding kinase 1; TLR, toll-like receptor; TNF-α, tumour necrosis factor alpha; XBP1, X-box binding protein 1.
Macrophage STING activation is involved in liver inflammation and fibrosis,3,4 and studies have reported that NLRP3 signalling is regulated by STING activation.18,19 We next analysed the role of STING signalling in regulating the NLRP3-mediated inflammatory response in macrophages and liver fibrosis. Attenuated liver fibrosis was found in Tmem173M-KO mice (Fig. S5A–D). Xbp1M-KO mice with liver fibrosis showed decreased STING activation in liver macrophages (Fig. 2C). LPS stimulation activated STING signalling in BMDMs, as evidenced by the increased levels of cGAS, STING, p-TBK1, and p-IRF3 proteins; these effects were significantly reduced by Xbp1 depletion (Fig. 2D). As shown in Fig. 2E, reduced STING- and NLRP3-positive staining was detected in Xbp1-deficient BMDMs following LPS stimulation. Furthermore, STING overexpression by lentiviral transfection increased the NLRP3 activation in Xbp1M-KO BMDMs after LPS stimulation. Meanwhile, Irf3 blockade by short hairpin RNA (shRNA) transfection decreased the NLRP3 activation and abrogated the role of STING overexpression in its activation in Xbp1M-KO BMDMs (Fig. 2F and Fig. S5E and F). This indicates that the XBP1/STING/IRF3 axis plays a critical role in regulating NLRP3 signalling. Moreover, Xbp1 depletion in macrophages decreased proinflammatory gene expression (Fig. 2G). Bioinformatics analysis of the murine Nlrp3 promoter revealed a putative IRF3 binding site (−753/−733), which was subsequently confirmed by the chromatin immunoprecipitation (ChIP) assay (Fig. 2H). Furthermore, IRF3 activation with KIN1148, a small-molecule IRF3 agonist, enhanced Nlrp3 mRNA expression in LPS-stimulated BMDMs (Fig. 2I). The p-IRF3 levels in hepatic macrophages isolated from Xbp1FL/FL and Xbp1M-KO mice treated with CCl4, BDL, or MCD, and in human tissue samples were increased (Fig. 2J,K). Collectively, these findings suggested that Xbp1 deficiency reduced macrophage NLRP3 activation in a STING/IRF3-dependent manner.
Liver fibrosis triggers the cytosolic release of self-mtDNA in macrophages
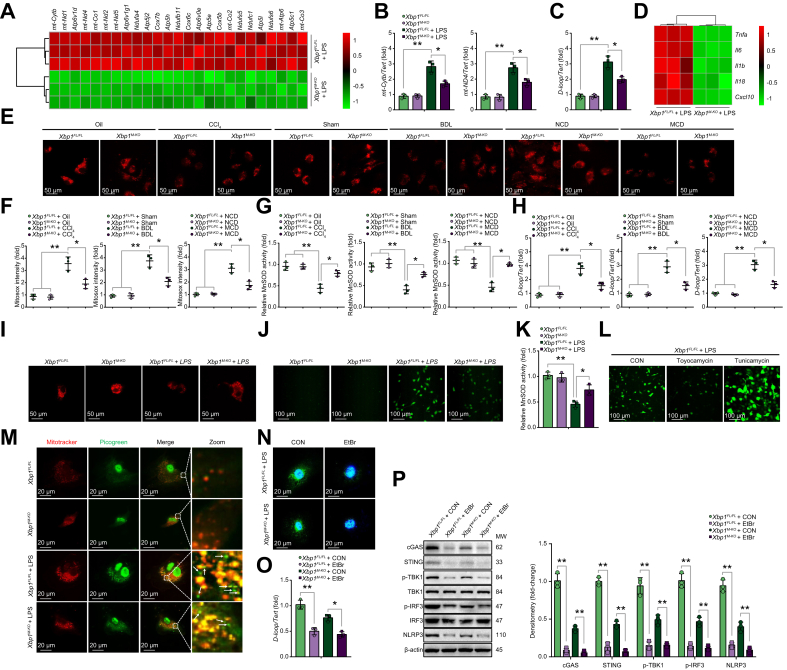
To investigate the underlying mechanism of XBP1 in regulating macrophage STING signalling, RNA sequencing analysis was conducted on Xbp1FL/FL and Xbp1M-KO BMDMs after LPS stimulation (Fig. S6A). This showed that 25 genes affecting mtDNA expression were significantly downregulated in Xbp1M-KO BMDMs compared with the Xbp1FL/FL BMDMs (Fig. 3A). The 2 representative components of mtDNA, mt-Cytb and mt-Nd4, were significantly decreased in LPS-stimulated Xbp1M-KO BMDMs (Fig. 3B). Cytosolic mtDNA is an important upstream ligand of the STING signalling pathway. Although the role of exogenous mtDNA released from other parenchymal cells in activating cGAS/cGAMP/STING has been well documented,20 it remains unclear whether endogenous mtDNA from stressed macrophages can activate STING. Our study showed that Xbp1 depletion decreased the cytosolic mtDNA accumulation in macrophages after LPS stimulation, as shown by qPCR analysis (Fig. 3C). RNA sequencing analysis demonstrated that Xbp1 depletion downregulated the expression of proinflammatory genes in LPS-stimulated macrophages (Fig. 3D). Furthermore, multiple mitochondrial abnormalities were detected in the macrophages isolated from Xbp1FL/FL mice with fibrotic liver, owing to a decreased mitochondrial membrane potential, mitochondrial superoxide accumulation, and decreased manganese superoxide dismutase (MnSOD) activity. Compared with the findings in Xbp1FL/FL mice, macrophages from Xbp1M-KO mice with fibrotic liver showed alleviated mitochondrial abnormalities and injuries (Fig. 3E–G). In particular, liver fibrosis induced mtDNA release and cytoplasmic accumulation, and this effect was significantly reduced by Xbp1 depletion (Fig. 3H). A recent study reported that LPS significantly increased the level of mtDNA in macrophages.21 We found that LPS-induced mitochondrial injury and oxidative stress in BMDMs, was mitigated by Xbp1 deficiency (Fig. 3I–K). Meanwhile, toyocamycin-induced XBP1 inhibition significantly suppressed oxidative stress; whereas, tunicamycin-induced XBP1 activation significantly promoted oxidative stress (Fig. S6B and Fig. 3L). Xbp1 depletion decreased cytosolic mtDNA accumulation in macrophages after LPS stimulation, as shown by mtDNA staining (Fig. 3M). To examine the mtDNA requirement in macrophage STING/NLRP3 activation, we depleted mtDNA using ethidium bromide (EtBr) treatment and confirmed it by staining and PCR (Fig. 3N and O). The mtDNA depletion reduced the activation of STING/NLRP3 signalling in both Xbp1FL/FL and Xbp1M-KO BMDMs after LPS stimulation (Fig. 3P). These findings suggested that liver fibrosis induced macrophagic mitochondrial injury and self-mtDNA cytosolic leakage, which subsequently activated macrophage STING/NLRP3 signalling.
Fig. 3.
Liver fibrosis triggers cytosolic release of self-mtDNA in macrophages.
(A) Heat map showing clustering of 25 downregulated genes that affect mtDNA expression in Xbp1M-KO BMDMs compared with Xbp1FL/FL BMDMs in response to LPS stimulation. (B) mtDNA was extracted as described in the Materials and methods section. Relative total mtDNA expression in Xbp1FL/FL and Xbp1M-KO BMDMs were determined using qPCR with primers specific for mtDNA (mt-Cytb, mt-Nd4) and nuclear DNA (Tert); n = 3 biological replicates/group. (C) Relative levels of total cytosolic mtDNA in Xbp1FL/FL and Xbp1M-KO BMDMs treated with LPS were determined using qPCR with primers specific for mtDNA (D-loop) and nuclear DNA (Tert); n = 3 biological replicates/group. (D) Heat map showing clustering of 5 downregulated proinflammatory genes in Xbp1M-KO BMDMs compared with Xbp1FL/FL BMDMs in response to LPS stimulation. (E) Liver macrophages were isolated as described in the Materials and methods section. The mitochondrial membrane potential of macrophages was determined by tetramethylrhodamine methyl ester (TMRM) staining. (F) Mitochondrial superoxide accumulation was determined by measuring the MitoSOX Red fluorescence intensity in macrophages isolated from the different groups according to the manufacturer’s instructions; n = 3 biological replicates/group. (G) Enzymatic activity of MnSOD in macrophages isolated from the different groups was determined according to the manufacturer’s instructions; n = 3 biological replicates/group. (H) Cytosolic mtDNA was extracted as described in the Materials and methods section. Relative amounts of total cytosolic mtDNA in macrophages isolated from the different groups were determined using qPCR with primers specific for mtDNA (D-loop) and nuclear DNA (Tert); n = 3 biological replicates/group. (I) The mitochondrial membrane potential of Xbp1FL/FL and Xbp1M-KO BMDMs treated with LPS was determined by TMRM staining. (J) The levels of ROS in BMDMs were examined using 2',7'-dichlorodihydrofluorescein diacetate (DCFH-DA) (original magnification 200 × ). (K) Enzymatic activity of MnSOD in Xbp1FL/FL and Xbp1M-KO BMDMs treated with LPS was determined according to the manufacturer’s instructions; n = 3 biological replicates/group. (L) The levels of ROS in LPS-stimulated Xbp1FL/FL BMDMs treated with or without toyocamycin or tunicamycin were examined using DCFH-DA (original magnification 200 × ). (M) mtDNA released from mitochondria in Xbp1FL/FL and Xbp1M-KO BMDMs treated with LPS, as shown by confocal microscopy. Arrowheads, mtDNA released into cytoplasm. (N) Immunofluorescence staining of cytoplasmic dsDNA confirmed mtDNA depletion by EtBr. Scale bar, 20 μm. (O) Relative total mtDNA levels in LPS-stimulated Xbp1FL/FL and Xbp1M-KO BMDMs treated with EtBr were determined using qPCR with primers specific for mtDNA (D-loop) and nuclear DNA (Tert); n = 3 biological replicates/group. (P) Western blot was performed to analyse the levels of cGAS, STING, p-TBK1, TBK1, p-IRF3, IRF3, and NLRP3 in LPS-stimulated Xbp1FL/FL and Xbp1M-KO BMDMs treated with EtBr. The values are shown as mean ± SD. Statistical significance was assessed by 1-way ANOVA. ∗∗p <0.01; ∗p <0.05. BMDMs, bone marrow-derived macrophages; cGAS, cyclic GMP-AMP synthase; EtBr, ethidium bromide; IRF3, interferon regulatory factor 3; LPS, lipopolysaccharide; MnSOD, manganese superoxide dismutase; NLRP3, nucleotide-binding oligomerization domain, leucine-rich repeat and pyrin domain-containing 3; ROS, reactive oxygen species; STING, stimulator of interferon genes; TBK1, TANK binding kinase 1.
Xbp1 deficiency promotes mitophagy activation in macrophages
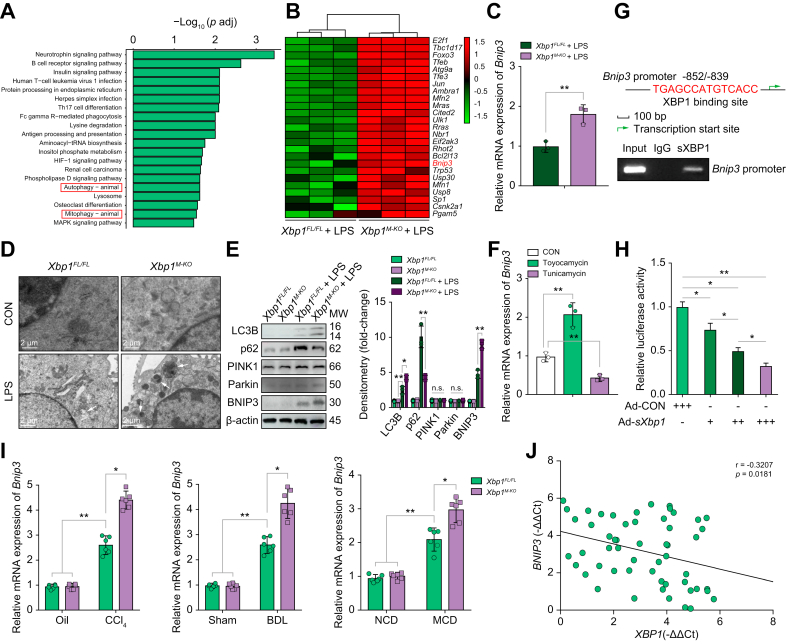
Because mitophagy has evolved to preserve mitochondrial homeostasis and promote the elimination of cytosolic pathogenic mtDNA, we examined the involvement of mitophagy in STING activation by modulating mtDNA release. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of the RNA sequencing data demonstrated that autophagy and mitophagy were significantly upregulated in LPS-stimulated Xbp1M-KO BMDMs compared with Xbp1FL/FL BMDMs (Fig. 4A). Two major mechanisms for mitophagy have been identified, 1 which is regulated by PINK1/Parkin and 1 by mitophagy receptors, including BNIP3, BCL2/adenovirus E1B interacting protein 3-like (NIX) and FUN14 domain containing 1 (FUNDC1).22 A heat map of the RNA sequencing results indicated that Bnip3 was markedly upregulated in the LPS-stimulated Xbp1M-KO BMDMs (Fig. 4B). Bnip3 mRNA expression, as detected by qPCR, showed a similar result (Fig. 4C); however, Fundc1 and Bnip3l mRNA expression was not significant (Fig. S6C). Increased mitophagy activation was observed on transmission electron microscopy in Xbp1M-KO BMDMs after LPS stimulation (Fig. 4D), and accompanied by higher microtubule-associated protein 1 light chain 3 beta (LC3B) levels and lower sequestosome 1 (p62) levels (Fig. 4E). Compared with Xbp1FL/FL BMDMs, Xbp1M-KO BMDMs showed increased BNIP3 and no significant changes in PINK1 or Parkin (Fig. 4E). Moreover, Bnip3 expression was increased when treated with toyocamycin, and decreased when treated with tunicamycin in LPS-stimulated Xbp1FL/FL BMDMs (Fig. 4F). Subsequently, we performed a bioinformatics analysis to explore the potential mechanism by which XBP1 activation regulates macrophagic BNIP3 expression. The results showed that a putative XBP1 binding site (−852/−839) was within the murine Bnip3 promoter, and was confirmed by the ChIP assay (Fig. 4G). sXBP1 stimulated the Bnip3 transcriptional activity in a concentration-dependent manner, as evidenced by the consistently decreasing Bnip3-luciferase reporter gene in LPS-stimulated BMDMs, co-transfected with pGL3-Bnip3-luciferase vector and Ad-sXbp1 (Fig. 4H). Furthermore, the gene expression levels of Bnip3 were elevated in CCl4-, BDL-, and MCD-induced fibrotic liver tissue in mice (Fig. 4I). Negative correlations between XBP1 and BNIP3 expression were also observed in human fibrotic liver tissues (Fig. 4J). These results demonstrated the following: Bnip3 is a target gene that is negatively regulated by XBP1; Xbp1 deficiency promoted mitophagy activation in macrophages; and the BNIP3 pathway plays a critical role in macrophagic mitophagy regulation.
Fig. 4.
XBP1 deficiency promotes mitophagy activation in macrophages.
(A) KEGG pathway enrichment analysis of the differentially expressed pathways in LPS-stimulated Xbp1FL/FL and Xbp1M-KO BMDMs. (B) Heat map showing the clustering of 25 upregulated genes that affect mitophagy in response to LPS according to genotype. (C) Relative Bnip3 mRNA expression in LPS-stimulated Xbp1FL/FL and Xbp1M-KO BMDMs were determined using qPCR; n = 3 biological replicates/group; Student t test. (D) Autophagic microstructures in BMDM mitochondria were examined by transmission electron microscopy, 5,000 × magnification; scale bars, 2 μm. Arrowheads, mitophagy. Representative of 3 experiments. (E) Western blot was performed to determine intracellular LC3B, p62, PINK1, Parkin, and BNIP3 protein levels in LPS-stimulated Xbp1FL/FL and Xbp1M-KO BMDMs. Statistical analysis was carried out using 1-way ANOVA. (F) Relative Bnip3 mRNA expression in LPS-stimulated Xbp1FL/FL BMDMs treated with or without toyocamycin or tunicamycin were determined using qPCR; n = 3 biological replicates/group. Statistical analysis was carried out using 1-way ANOVA. (G) Schematic diagram of the putative XBP1 binding site within the Bnip3 promoter. The BMDMs were subjected to ChIP assay with anti-XBP1 or IgG antibody. Representative results from 3 independent experiments are shown. (H) Luciferase activity in BMDMs co-transfected with Bnip3-promoter luciferase reporter plasmid and with the increasing multiplicity of infection of Ad-sXbp1 or Ad-CON. n = 3/group. Statistical analysis was carried out using 1-way ANOVA. (I) The expression levels of Bnip3 in liver tissue from mice with CCl4-, BDL-, and MCD-induced liver fibrosis were examined using quantitative real-time PCR; n = 6 mice/group; 1-way ANOVA. (J) The correlation between XBP1 levels and BNIP3 expression in human fibrotic liver tissue was assessed using Pearson’s correlation analysis; n = 54. The values are shown as the mean ± SD. Statistical significance was assessed by the Student t test or ANOVA. ∗∗p <0.01; ∗p <0.05. BDL, bile duct ligation; BMDMs, bone marrow-derived macrophages; BNIP3, BCL2/adenovirus E1B interacting protein 3; CCl4, carbon tetrachloride; ChIP, chromatin immunoprecipitation; KEGG, Kyoto Encyclopedia of Genes and Genomes; LPS, lipopolysaccharide; MCD, methionine/choline-deficient diet; XBP1, X-box binding protein 1.
BNIP3-mediated mitophagy activation is responsible for decreased mtDNA release and STING-NLRP3 activation in Xbp1-deficient macrophages
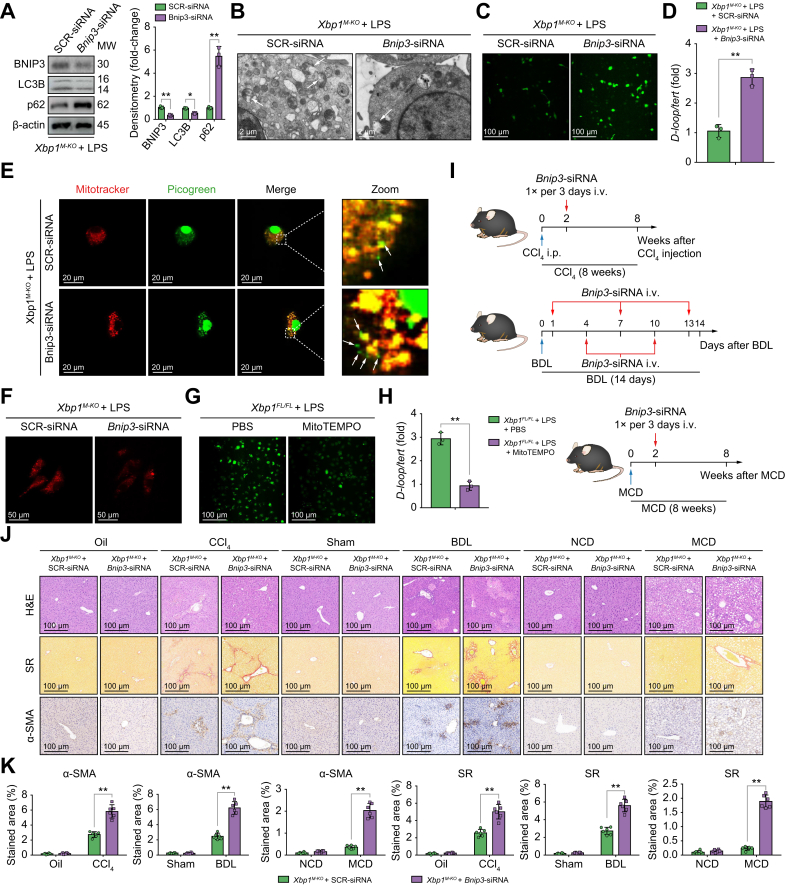
To determine the essential role of BNIP3 in mediating mitophagy in Xbp1-deficient macrophages, BNIP3 activation in Xbp1M-KO BMDMs was blocked by siRNA transfection, and was confirmed by the decreased LC3B and increased p62 protein levels (Fig. 5A) and transmission electron microscopy (Fig. 5B). Inhibition of mitophagy as a result of Bnip3 knockdown also exacerbated the oxidative stress and mitochondrial injury in Xbp1M-KO BMDMs, as indicated by increases in reactive oxygen species (ROS) production (Fig. 5C). Moreover, increased mtDNA release, cytosolic accumulation, and decreased mitochondrial membrane potential were observed in Xbp1M-KO BMDMs after Bnip3 siRNA treatment (Fig. 5D–F). Mitochondrial ROS inhibition by MitoTEMPO treatment decreased the cytosolic accumulation of mtDNA in Xbp1FL/FL BMDMs, indicating the critical role of ROS in mediating mitochondrial injury and subsequent mtDNA release (Fig. 5G and H). Mitophagy suppression by Bnip3 siRNA reversed the Xbp1 depletion-mediated reduction of cGAS/STING and NLRP3 activation (Fig. S7A–C).
Fig. 5.
BNIP3-mediated mitophagy activation decreases mtDNA release and STING-NLRP3 activation in Xbp1-deficient macrophages.
(A) Western blot was performed to examine intracellular BNIP3, LC3B, and p62 protein levels in LPS-stimulated Xbp1M-KO BMDMs transfected with SCR-siRNA or Bnip3-siRNA. Statistical analysis was carried out using the Student t test. (B) Autophagic microstructures in mitochondria in LPS-stimulated Xbp1M-KO BMDMs transfected with SCR-siRNA or Bnip3-siRNA were examined by transmission electron microscopy, 5,000 × magnification; scale bars, 2 μm. Arrowheads, mitophagy. (C) The levels of ROS in LPS-stimulated Xbp1M-KO BMDMs transfected with SCR-siRNA or Bnip3-siRNA were examined by DCFH-DA (original magnification 200 × ). (D) Relative total mtDNA amounts in LPS-stimulated Xbp1M-KO BMDMs transfected with SCR-siRNA or Bnip3-siRNA were determined using qPCR with primers specific for mtDNA (D-loop) and nuclear DNA (Tert); n = 3 biological replicates/group; Student t test. (E) mtDNA released from mitochondria in Xbp1M-KO BMDMs transfected with SCR-siRNA or Bnip3-siRNA, as shown by confocal microscopy. Arrowheads, mtDNA released into cytoplasm. (F) The mitochondrial membrane potential of Xbp1M-KO BMDMs transfected with SCR-siRNA or Bnip3-siRNA was determined by TMRM staining. (G) The levels of ROS in LPS-stimulated Xbp1FL/FL BMDMs treated with PBS or MitoTEMPO were examined using DCFH-DA (original magnification 200 × ). (H) Relative amounts of total cytosolic mtDNA in LPS-stimulated Xbp1FL/FL BMDMs treated with PBS or MitoTEMPO were determined using qPCR with primers specific for mtDNA (D-loop) and nuclear DNA (Tert); n = 3 biological replicates/group; Student t test. (I) Schematic showing the administration protocol for mannose-conjugated SCR-siRNA and Bnip3-siRNA in CCl4-, BDL-, and MCD-induced murine liver fibrosis models for the experiments shown in (J). (J) Xbp1M-KO male mice were subjected to CCl4-, BDL-, or MCD-induced experimental fibrosis and injected with SCR-siRNA or Bnip3-siRNA via the tail vein, and the collected livers were subjected to H&E and Sirius Red staining and α-SMA immunohistochemical analysis; scale bar=100 μm. Representative of 6 mice/group. (K) The proportions of the Sirius Red- and α-SMA-positive areas were quantified; n = 6 mice/group; 1-way ANOVA. The values are shown as mean ± SD. Statistical significance was assessed by Student t test or ANOVA. ∗∗p <0.01. BMDMs, bone marrow-derived macrophages; BNIP3, BCL2/adenovirus E1B interacting protein 3; LC3B, microtubule-associated protein 1 light chain 3 beta; LPS, lipopolysaccharide; NLRP3, nucleotide-binding oligomerization domain, leucine-rich repeat and pyrin domain-containing 3; p62, sequestosome 1; ROS, reactive oxygen species; STING, stimulator of interferon genes; XBP1, X-box binding protein 1.
To further test the in vivo effect of mitophagy suppression by Bnip3 knockdown, the BNIP3 activation was blocked by its siRNA in mice with CCl4-, BDL-, or MCD-induced liver fibrosis (Fig. 5I). Bnip3 siRNA abrogated the protective effect of myeloid Xbp1 depletion in alleviating liver fibrosis, as evidenced by H&E staining of hepatic tissue, increased Sirius Red and α-SMA staining of liver tissue (Fig. 5J and K), and increased gene expression of the profibrogenic markers Acta2, Col1a1, and Timp1 (Fig. S7D). These findings suggested that Xbp1 deficiency reduced mtDNA release and subsequent STING/NLRP3 activation by promoting BNIP3-mediated mitophagy in macrophages.
Macrophage XBP1/STING signalling enhances the activation of HSCs
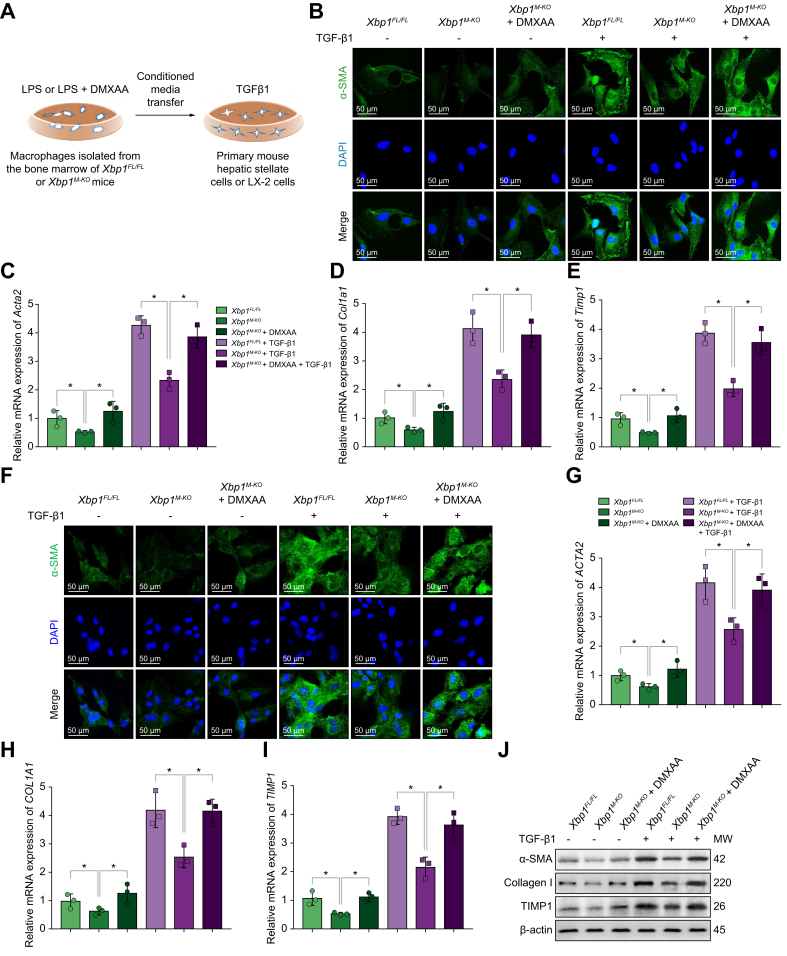
To examine the direct effects of altering STING signalling in Xbp1-deficient macrophages on HSC activation, we incubated primary mouse HSCs with conditional media (CM) of LPS-treated Xbp1FL/FL and Xbp1M-KO BMDMs in the presence or absence of transforming growth factor beta 1 (TGF-β1) (Fig. 6A). In the presence of TGF-β1, Xbp1FL/FL BMDM-CM treatment significantly enhanced α-SMA staining in primary mouse HSCs (Fig. 6B), indicating increased HSC activation. In contrast, diminished α-SMA staining was observed in primary HSCs incubated with Xbp1M-KO BMDM-CMs. Restoration of STING activation by DMXAA in Xbp1M-KO BMDMs significantly enhanced α-SMA staining in primary mouse HSCs. The expression levels of Acta2, Col1a1, and Timp1 (Fig. 6C–E) in primary mouse HSCs showed similar results. Moreover, analogous results were obtained by incubating LX-2 cells (cell lines of human hepatic stellate cells) in the absence or presence of TGF-β1 with CM from LPS-treated Xbp1FL/FL and Xbp1M-KO BMDMs (Fig. 6F–J). These results further confirmed that Xbp1 deficiency restricted the activation of HSC by macrophage STING signalling.
Fig. 6.
Macrophage XBP1/STING/NLRP3 signalling pathway mediates hepatic stellate cells activation.
(A) Schematic drawing showing that primary mouse HSCs or LX-2 cells were treated with CM of LPS-treated Xbp1FL/FL and Xbp1M-KO BMDMs and CM of LPS + DMXAA-treated Xbp1M-KO BMDMs in the absence or presence of TGF-β1 for 24 h. (B) Immunofluorescence staining for α-SMA (green) in treated primary mouse HSCs. The expression levels of Acta2 (C), Col1a1 (D), and Timp1 (E) in treated primary mouse HSCs were examined using quantitative real-time PCR; n = 3/group. (F) Immunofluorescence staining for α-SMA (green) in treated LX-2 cells. The expression levels of ACTA2 (G), COL1A1 (H), and TIMP1 (I) in treated LX-2 cells were examined using quantitative real-time PCR; n = 3/group. (J) The protein levels of α-SMA, collagen I, and TIMP1 in treated LX-2 cells were examined using Western blot. The values are shown as mean ± SD. Statistical significance was assessed by ANOVA. ∗p <0.05. Acta2/α-SMA, actin, alpha 2, smooth muscle, aorta; BMDMs, bone marrow-derived macrophages; CM, conditional media; Col1a1, collagen, type I, alpha 1; DMXAA, 5,6-dimethylxanthenone-4-acetic acid; HSC, hepatic stellate cell; LPS, lipopolysaccharide; NLRP3, nucleotide-binding oligomerization domain, leucine-rich repeat and pyrin domain-containing 3; STING, stimulator of interferon genes; TGF-β1, transforming growth factor beta 1; Timp1, tissue inhibitor of matrix metalloproteinase 1; XBP1, X-box binding protein 1.
Pharmacological inhibition of XBP1 ameliorates liver fibrosis in mice
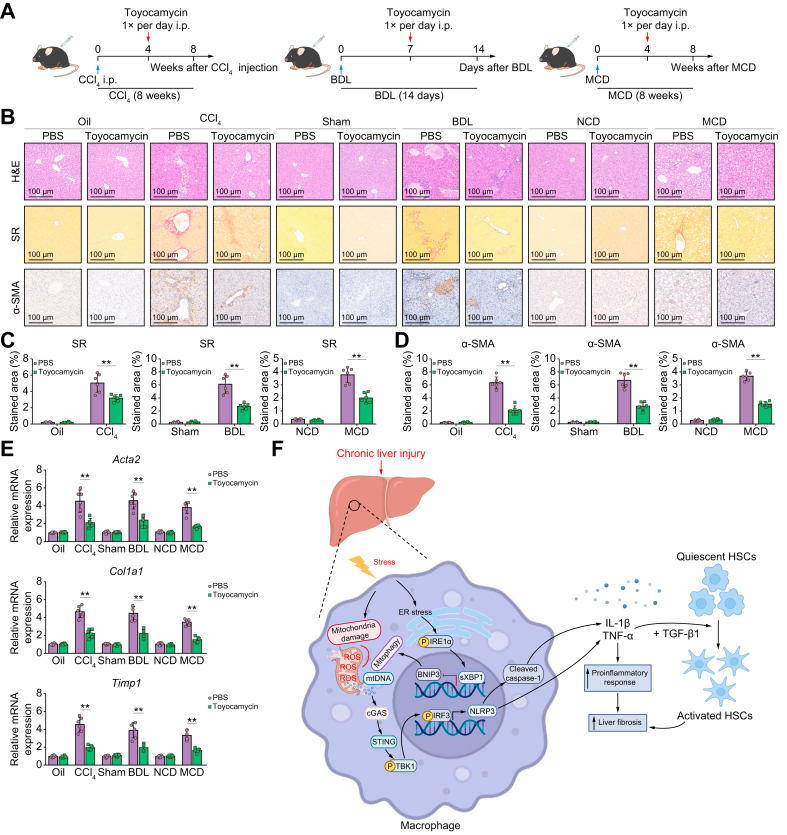
We explored the effects of XBP1 inhibition on liver fibrosis development. A selective XBP1 inhibitor (toyocamycin) was intraperitoneally injected into WT mice with CCl4-, BDL-, or MCD-induced liver fibrosis (Fig. 7A). The results indicated that pharmacological XBP1 inhibition alleviated liver fibrosis, as evidenced by hepatic tissue H&E staining, decreased Sirius Red and α-SMA staining in murine liver tissues (Fig. 7B–D), and decreased gene expression of the profibrogenic markers Acta2, Col1a1, and Timp1 (Fig. 7E). These findings show that the pharmacological inhibition of XBP1 attenuates liver fibrosis in mice. In summary, we found that macrophage STING signalling could be activated by cytosolic leakage of mtDNA from macrophages. Xbp1 depletion decreased the cGAS/STING/NLRP3 activation by restoring BNIP3-mediated mitophagy in macrophages (Fig. 7F).
Fig. 7.
Pharmacological inhibition of XBP1 ameliorates liver fibrosis in mice.
(A) Schematic showing the administration protocol for toyocamycin in CCl4-, BDL-, and MCD-induced murine liver fibrosis models for the experiments shown in (B), PBS administration was used as control. (B) WT male mice were subjected to CCl4-, BDL-, or MCD-induced experimental fibrosis and injected intraperitoneally with toyocamycin; the livers were collected and subjected to H&E and Sirius Red staining and α-SMA immunohistochemical analysis; scale bar = 100 μm. Representative of 6 mice/group. (C,D) The proportions of the Sirius Red- and α-SMA-positive areas were quantified; n = 6 mice/group. (E) The expression levels of Acta2, Col1a1, and Timp1 in liver tissue from WT mice with fibrosis treated with toyocamycin were examined using quantitative real-time PCR; n = 6 mice/group. (F) Schematic illustration of how XBP1 controls macrophage self-mtDNA cytosolic leakage and STING activation during liver fibrosis. The values are shown as the mean ± SD. Statistical significance was assessed by ANOVA. ∗∗p <0.01. Acta2/α-SMA, actin, alpha 2, smooth muscle, aorta; BDL, bile duct ligation; CCl4, carbon tetrachloride; Col1a1, collagen, type I, alpha 1; MCD, methionine/choline-deficient diet; STING, stimulator of interferon genes; Timp1, tissue inhibitor of matrix metalloproteinase 1; XBP1, X-box binding protein 1; WT, wild-type.
XBP1 modulates macrophage STING signalling activation in patients with liver fibrosis
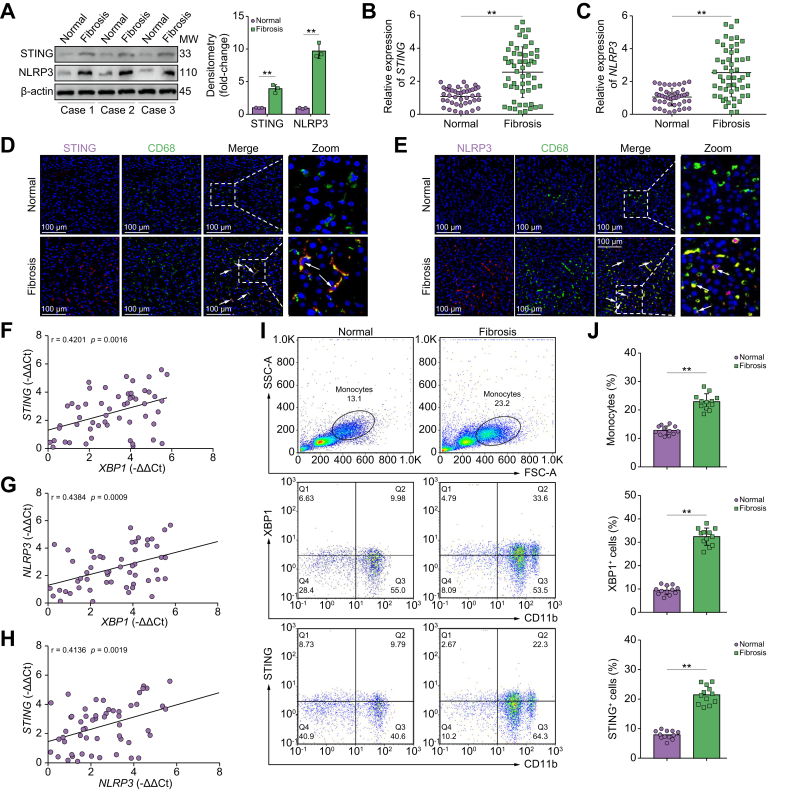
To test the clinical relevance of macrophagic STING/NLRP3 activation in liver fibrosis, we first compared the expression of STING and NLRP3 in human liver tissue with and without liver fibrosis. Increased STING and NLRP3 activation were observed, as indicated by the increased levels of protein and STING and NLRP3 mRNA (Fig. 8A–C). Additionally, there was increased co-staining of CD68 with STING/NLRP3 in human fibrotic liver tissue (Fig. 8D and E). Positive correlations between XBP1 and STING expression, XBP1 and NLRP3 expression, and STING and NLRP3 expression were observed in fibrotic liver tissue (Fig. 8F–H).
Fig. 8.
XBP1 modulates macrophage STING signalling activation in patients with liver fibrosis.
(A) The protein levels of STING and NLRP3 in human normal or fibrotic liver tissue were analysed by Western blot. Statistical analysis was carried out using the Student t test. (B) The expression level of STING in human liver tissue was analysed by quantitative RT-PCR; Student t test. (C) The expression level of NLRP3 in human liver tissue was estimated by quantitative RT-PCR; Student t test. (D) Dual immunofluorescence staining showing STING (purple) and CD68 (green) colocalization in human normal or fibrotic liver tissue. (E) Dual immunofluorescence staining showing NLRP3 (purple) and CD68 (green) colocalization in human normal or fibrotic liver tissue. (F) The correlation between XBP1 levels and STING expression in human fibrotic liver tissue was assessed using Pearson’s correlation analysis; n = 54. (G) The correlation between XBP1 levels and NLRP3 expression in human fibrotic liver tissue was assessed using Pearson’s correlation analysis; n = 54. (H) The correlation between NLRP3 levels and STING expression in human fibrotic liver tissue was assessed using Pearson’s correlation analysis; n = 54. Peripheral blood was collected and PBMCs were isolated, from patients who were clinically diagnosed with liver fibrosis or normal controls. Flow cytometry was performed to determine the expression of XBP1 and STING on CD11b-positive cells. (I) Representative flow cytometry results are shown. (J) Quantitative results are shown. The values are shown as the mean ± SD. Statistical significance was assessed by the Student t test. ∗∗p <0.01. NLRP3, nucleotide-binding oligomerization domain, leucine-rich repeat and pyrin domain-containing 3; STING, stimulator of interferon genes; XBP1, X-box binding protein 1.
Subsequently, to determine the clinical significance of XBP1 signalling in regulating macrophagic STING/NLRP3 activation, peripheral blood mononuclear cells (PBMCs) were isolated from 12 patients undergoing partial hepatectomy for liver fibrosis and normal controls. Liver fibrosis increased the monocyte proportion and enhanced XBP1/STING expression in CD11b+ PBMCs (Fig. 8I and J, and Fig. S7E). These findings indicated that macrophagic XBP1/STING/NLRP3 signalling contributed to the pathogenesis of human hepatic fibrosis.
Discussion
XBP1 modulates the macrophage proinflammatory response, but its function in macrophage STING activation and liver fibrosis is unknown. Although most studies have analysed the critical role of mtDNA release from other parenchymal cells in activating macrophage STING signalling, little is known about whether endogenous macrophagic mtDNA can activate the STING signalling pathway. Herein, we demonstrated that oxidative stress during liver fibrosis induced mitochondrial injury in macrophages, which facilitated macrophage self-mtDNA cytosolic leakage and cGAS/STING signalling activation. XBP1-mediated mitophagy suppression was identified as a key modulator of self-mtDNA leakage and subsequent cGAS/STING activation in macrophages. Thus, our findings established that XBP1 controls macrophage STING activation by regulating self-mtDNA cytosolic leakage in liver fibrosis.
STING activation in macrophages has been shown to contribute to liver fibrosis. Macrophages from patients with non-alcoholic fatty liver disease (NAFLD) showed increased STING expression.3 In the diet-induced NASH model, mice with myeloid STING knockout developed less severe hepatic steatosis, inflammation, and fibrosis than control mice.3 Similarly, hepatic cGAS/STING activation was upregulated in the CCl4-induced liver fibrosis model,23 whereas STING or Irf3 deficiency prevented liver inflammation and fibrosis.24
Accumulating evidence suggests that NLRP3 inflammasome activation is an important driver of various acute and chronic liver diseases. NLRP3 inflammasome blockade reduces liver inflammation and fibrosis in mice with experimental NASH.16 NLRP3 inflammasome particles derived from hepatocyte pyroptosis can be engulfed by HSCs to further promote liver fibrosis.15 Cytosolic DNA not only activates the cGAS-STING axis, but also directly triggers NLRP3 inflammasome activation.25,26 Furthermore, NLRP3 activation by STING signalling has been identified. Cytosolic DNA recognition by the cGAS-STING axis induces a cell death program by initiating potassium efflux upstream of NLRP3.18 We have previously reported that STING-dependent NLRP3 activation contributed to exacerbated liver ischemia and reperfusion injury in aged mice.17 We found attenuated inflammation and liver fibrosis in both Tmem173-and Nlrp3-deficient mice. In addition, NLRP3 activation was inhibited by STING deficiency in macrophages, both in vivo and in vitro, indicating the regulatory role of STING signalling in NLRP3 activation.
Exogenous mtDNA from other parenchymal cells can be engulfed by macrophages and activate STING signalling via the cGAS/cGAMP/STING pathway.20 Both in vivo and in vitro studies showed that mtDNA from stressed hepatocytes could activate macrophage STING signalling in NASH2 and liver ischemic injury18 models. A recent study showed that tumour-derived cGAMP, and not cytosolic DNA, is transferred to non-tumour cells to activate STING signalling.27 In contrast, little is known about the role of endogenous mtDNA from stressed macrophages in the regulation of STING signalling. The observations of our study were consistent with that of a recent study that LPS significantly increased the level of mtDNA in macrophages.21 Furthermore, the cytoplasmic release of mtDNA could activate the cGAS/STING pathway, leading to inflammation in amyotrophic lateral sclerosis.28 Thus, we analysed and showed that macrophage self-mtDNA cytosolic leakage activated cGAS/STING signalling in macrophages.
Mitophagy is an evolutionarily conserved cellular process that eliminates dysfunctional or superfluous mitochondria, thus fine-tuning mitochondrial numbers and preserving its homeostasis under physiological and pathological conditions. Two major mechanisms for mitophagy have been identified, involving PINK1/Parkin and mitophagy receptor signalling pathways, including BNIP3, NIX, and FUNDC1.22 Prkn- and Pink1-deficient mice show a strong inflammatory phenotype following exhaustive exercise with accumulated mtDNA mutations, and this effect is rescued by STING inhibition.29 Parkin/PINK1 mutations impair mitophagy and subsequent mtDNA release, which contributes to the pathogenesis of Parkin/PINK1-linked Parkinson’s disease.30 Failure to clear impaired mitophagy-induced cytosolic mtDNA by deletion of the autophagy-related gene Irgm1 results in STING-dependent type I interferon activation.31 BNIP3L/NIX- and FUNDC1-mediated mitophagy is required for mitochondrial network remodelling during cardiac progenitor cell differentiation and their abrogation during differentiation leads to sustained mitochondrial fission, doughnut-shaped impaired mitochondrial formation, and accumulation of mtDNA in cells.32 Therefore, specific interventions that target mitophagy to preserve and restore mitochondrial function have emerged as promising therapeutic strategies to prevent and treat inflammatory diseases.33 Our study found that XBP1 activation significantly suppressed BNIP3-mediated mitophagy and exacerbated liver fibrosis.
The XBP1 signalling effect on ER stress macrophage activation regulation has been reported recently.34 TLR4/2-activated XBP1 is required for optimal, sustained production of proinflammatory cytokines in macrophages, and Xbp1 deficiency results in a highly increased bacterial burden in mice.35 Xbp1 knockdown reduces the macrophagic LPS-induced inflammatory response in human cystic fibrosis.36 In sterile liver inflammatory injury, XBP1 is critical in mediating macrophage NLRP3 activation.9 ROS-dependent XBP1 activation in macrophages contributes to NASH progression.37 However, the effect of XBP1 signalling on macrophage STING regulation and its role in liver fibrosis remain unclear. Most of the studies on STING and XBP1 have focused on NASH models,3,38 and our current study demonstrated that the XBP1/STING/NLRP3 axis affected fibrogenesis, independent of steatosis. We found that XBP1 enhanced proinflammatory macrophage activation in a STING-dependent manner, which played an important role in promoting liver fibrosis.
Functional interplays between ER stress, mitophagy, and ROS have been identified and studied.17 The generation of ROS is a common feature of hepatic fibrogenesis, and is required for the optimal activation of multiple immune cell types.39,40 Oxidative stress in HSCs increases the unfolded protein response based on XBP1 splicing, which triggers autophagy.41 XBP1 regulates Parkinson’s disease pathophysiology by promoting PINK1-dependent mitophagy activation.42 Furthermore, ROS can induce mitophagy43 and be inhibited by mitophagy. Mitophagy is required for the clearance of damaged mitochondria, reduction of ROS production, and inhibition of inflammasome activation.44 Inhibiting mitophagy drives macrophage activation in a mitochondrial ROS-dependent manner.45,46 We analysed the crosstalk between XBP1, mitophagy, and ROS and revealed that Xbp1 knockout reduced ROS production by promoting BNIP3-mediated mitophagy activation in macrophages.
Macrophages can regulate the development of liver fibrosis depending on their heterogeneity.1 Resident activated Kupffer cells initiate the inflammatory response to liver injury. They recruit Ly6chi macrophages with proinflammatory and profibrogenic properties, ultimately promoting hepatocyte death and induce accumulation and survival of activated HSCs.[47], [48], [49] Monocyte-derived macrophages mainly function to promote inflammation and fibrosis. In a mouse model of CCl4-induced fibrosis, the Ly6chiCD11b+F4/80+ inducible, nitric oxide synthase-producing hepatic macrophage population was identified as the main profibrogenic population.50 However, the precise role of distinct types of liver macrophages in regulating liver fibrosis remains to be studied in detail. Future studies of XBP1 and mitophagy in different macrophage subpopulations is imperative to explore the specific types of macrophages responsible for liver fibrosis.
In conclusion, our findings demonstrated that XBP1 activated macrophage STING signalling by promoting self-mtDNA cytosolic leakage in liver fibrosis. These findings suggest that macrophage self-mtDNA could serve as an intrinsic trigger for macrophage cGAS/STING activation, which could be regulated by modulating XBP1/mitophagy.
Financial support
This study was supported by grants from the National Natural Science Foundation of China (Nos. 81971495, 91442117, and 82071798), CAMS Innovation Fund for Medical Sciences (No. 2019-I2M-5-035), National Science Foundation of Jiangsu Province (Nos. BRA2017533 and BK20191490), State Key Laboratory of Reproductive Medicine (No. SKLRM-K202001), and Foundation of Jiangsu Collaborative Innovation Center of Biomedical Functional Materials.
Authors’ contributions
Drafted the manuscript: QW, QB. Conducted the experiments: QW, QB, ML, RZ, JG, LL, JZ, YL, WS, ZL, MW. Participated in the research design, contributed to writing the manuscript, discussed the data, and supervised the study: ZL, LL, HZ. All authors performed data analysis and interpretation, and read and approved the final manuscript.
Data availability statement
All data relevant to the study are included in the article or are uploaded as supplementary information and protocols. The reagents, antibodies, and resources are listed in the supplementary tables.
Conflicts of interest
The authors declare no conflicts of interest.
Please refer to the accompanying ICMJE disclosure forms for further details.
Footnotes
Author names in bold designate shared co-first authorship
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jhepr.2022.100555.
Contributor Information
Zhexiong Lian, Email: zxlian1@ustc.edu.cn.
Ling Lu, Email: lvling@njmu.edu.cn.
Haoming Zhou, Email: hmzhou@njmu.edu.cn.
Supplementary data
The following are the supplementary data to this article:
References
- 1.Tacke F., Zimmermann H.W. Macrophage heterogeneity in liver injury and fibrosis. J Hepatol. 2014;60:1090–1096. doi: 10.1016/j.jhep.2013.12.025. [DOI] [PubMed] [Google Scholar]
- 2.Barber G.N. STING: infection, inflammation and cancer. Nat Rev Immunol. 2015;15:760–770. doi: 10.1038/nri3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu Y., Liu Y., An W., Song J., Zhang Y., Zhao X. STING-mediated inflammation in Kupffer cells contributes to progression of nonalcoholic steatohepatitis. J Clin Invest. 2019;129:546–555. doi: 10.1172/JCI121842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luo X., Li H., Ma L., Zhou J., Guo X., Woo S.L., et al. Expression of STING is increased in liver tissues from patients with NAFLD and promotes macrophage-mediated hepatic inflammation and fibrosis in mice. Gastroenterology. 2018;155:1971–1984. doi: 10.1053/j.gastro.2018.09.010. .e1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ding Q., Cao X., Lu J., Huang B., Liu Y.J., Kato N., et al. Hepatitis C virus NS4B blocks the interaction of STING and TBK1 to evade host innate immunity. J Hepatol. 2013;59:52–58. doi: 10.1016/j.jhep.2013.03.019. [DOI] [PubMed] [Google Scholar]
- 6.Thomsen M.K., Nandakumar R., Stadler D., Malo A., Valls R.M., Wang F., et al. Lack of immunological DNA sensing in hepatocytes facilitates hepatitis B virus infection. Hepatology. 2016;64:746–759. doi: 10.1002/hep.28685. [DOI] [PubMed] [Google Scholar]
- 7.Xu D., Tian Y., Xia Q., Ke B. The cGAS-STING pathway: novel perspectives in liver diseases. Front Immunol. 2021;12 doi: 10.3389/fimmu.2021.682736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Balce D.R., Wang Y.T., McAllaster M.R., Dunlap B.F., Orvedahl A., Hykes B.L., Jr., et al. UFMylation inhibits the proinflammatory capacity of interferon-γ-activated macrophages. Proc Natl Acad Sci U S A. 2021;118 doi: 10.1073/pnas.2011763118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yue S., Zhu J., Zhang M., Li C., Zhou X., Zhou M., et al. The myeloid heat shock transcription factor 1/β-catenin axis regulates NLR family, pyrin domain-containing 3 inflammasome activation in mouse liver ischemia/reperfusion injury. Hepatology. 2016;64:1683–1698. doi: 10.1002/hep.28739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou C.M., Luo L.M., Lin P., Pu Q., Wang B., Qin S., et al. Annexin A2 regulates unfolded protein response via IRE1-XBP1 axis in macrophages during P. aeruginosa infection. J Leukoc Biol. 2021;110:375–384. doi: 10.1002/JLB.3A1219-686RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maiers J.L., Malhi H. Endoplasmic reticulum stress in metabolic liver diseases and hepatic fibrosis. Semin Liver Dis. 2019;39:235–248. doi: 10.1055/s-0039-1681032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Senft D., Ronai Z.A. UPR, autophagy, and mitochondria crosstalk underlies the ER stress response. Trends Biochem Sci. 2015;40:141–148. doi: 10.1016/j.tibs.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hetz C., Thielen P., Matus S., Nassif M., Court F., Kiffin R., et al. XBP-1 deficiency in the nervous system protects against amyotrophic lateral sclerosis by increasing autophagy. Genes Dev. 2009;23:2294–2306. doi: 10.1101/gad.1830709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heindryckx F., Binet F., Ponticos M., Rombouts K., Lau J., Kreuger J., et al. Endoplasmic reticulum stress enhances fibrosis through IRE1α-mediated degradation of miR-150 and XBP-1 splicing. EMBO Mol Med. 2016;8:729–744. doi: 10.15252/emmm.201505925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaul S., Leszczynska A., Alegre F., Kaufmann B., Johnson C.D., Adams L.A., et al. Hepatocyte pyroptosis and release of inflammasome particles induce stellate cell activation and liver fibrosis. J Hepatol. 2021;74:156–167. doi: 10.1016/j.jhep.2020.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mridha A.R., Wree A., Robertson A.A.B., Yeh M.M., Johnson C.D., Van Rooyen D.M., et al. NLRP3 inflammasome blockade reduces liver inflammation and fibrosis in experimental NASH in mice. J Hepatol. 2017;66:1037–1046. doi: 10.1016/j.jhep.2017.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Q., Zhou H., Bu Q., Wei S., Li L., Zhou J., et al. Role of XBP1 in regulating the progression of non-alcoholic steatohepatitis. J Hepatol. 2022;77:312–325. doi: 10.1016/j.jhep.2022.02.031. [DOI] [PubMed] [Google Scholar]
- 18.Zhong W., Rao Z., Rao J., Han G., Wang P., Jiang T., et al. Aging aggravated liver ischemia and reperfusion injury by promoting STING-mediated NLRP3 activation in macrophages. Aging Cell. 2020;19 doi: 10.1111/acel.13186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gaidt M.M., Ebert T.S., Chauhan D., Ramshorn K., Pinci F., Zuber S., et al. The DNA inflammasome in human myeloid cells is initiated by a STING-cell death program upstream of NLRP3. Cell. 2017;171:1110–1124. doi: 10.1016/j.cell.2017.09.039. e1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Motwani M., Pesiridis S., Fitzgerald K.A. DNA sensing by the cGAS-STING pathway in health and disease. Nat Rev Genet. 2019;20:657–674. doi: 10.1038/s41576-019-0151-1. [DOI] [PubMed] [Google Scholar]
- 21.Ning L., Wei W., Wenyang J., Rui X., Qing G. Cytosolic DNA-STING-NLRP3 axis is involved in murine acute lung injury induced by lipopolysaccharide. Clin Transl Med. 2020;10:e228. doi: 10.1002/ctm2.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Palikaras K., Lionaki E., Tavernarakis N. Mechanisms of mitophagy in cellular homeostasis, physiology and pathology. Nat Cell Biol. 2018;20:1013–1022. doi: 10.1038/s41556-018-0176-2. [DOI] [PubMed] [Google Scholar]
- 23.Yong H., Wang S., Song F. Activation of cGAS/STING pathway upon TDP-43-mediated mitochondrial injury may be involved in the pathogenesis of liver fibrosis. Liver Int. 2021;41:1969–1971. doi: 10.1111/liv.14895. [DOI] [PubMed] [Google Scholar]
- 24.Iracheta-Vellve A., Petrasek J., Gyongyosi B., Satishchandran A., Lowe P., Kodys K., et al. Endoplasmic reticulum stress-induced hepatocellular death pathways mediate liver injury and fibrosis via stimulator of interferon genes. J Biol Chem. 2016;291:26794–26805. doi: 10.1074/jbc.M116.736991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhong Z., Liang S., Sanchez-Lopez E., He F., Shalapour S., Lin X.J., et al. New mitochondrial DNA synthesis enables NLRP3 inflammasome activation. Nature. 2018;560:198–203. doi: 10.1038/s41586-018-0372-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shimada K., Crother T.R., Karlin J., Dagvadorj J., Chiba N., Chen S., et al. Oxidized mitochondrial DNA activates the NLRP3 inflammasome during apoptosis. Immunity. 2012;36:401–414. doi: 10.1016/j.immuni.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marcus A., Mao A.J., Lensink-Vasan M., Wang L., Vance R.E., Raulet D.H. Tumor-derived cGAMP triggers a STING-mediated interferon response in non-tumor cells to activate the NK cell response. Immunity. 2018;49:754–763. doi: 10.1016/j.immuni.2018.09.016. .e754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu C.H., Davidson S., Harapas C.R., Hilton J.B., Mlodzianoski M.J., Laohamonthonkul P., et al. TDP-43 triggers mitochondrial DNA release via mPTP to activate cGAS/STING in ALS. Cell. 2020;183:636–649. doi: 10.1016/j.cell.2020.09.020. .e618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sliter D.A., Martinez J., Hao L., Chen X., Sun N., Fischer T.D., et al. Parkin and PINK1 mitigate STING-induced inflammation. Nature. 2018;561:258–262. doi: 10.1038/s41586-018-0448-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Borsche M., König I.R., Delcambre S., Petrucci S., Balck A., Brüggemann N., et al. Mitochondrial damage-associated inflammation highlights biomarkers in PRKN/PINK1 parkinsonism. Brain. 2020;143:3041–3051. doi: 10.1093/brain/awaa246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rai P., Janardhan K.S., Meacham J., Madenspacher J.H., Lin W.C., Karmaus P.W.F., et al. IRGM1 links mitochondrial quality control to autoimmunity. Nat Immunol. 2021;22:312–321. doi: 10.1038/s41590-020-00859-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lampert M.A., Orogo A.M., Najor R.H., Hammerling B.C., Leon L.J., Wang B.J., et al. BNIP3L/NIX and FUNDC1-mediated mitophagy is required for mitochondrial network remodeling during cardiac progenitor cell differentiation. Autophagy. 2019;15:1182–1198. doi: 10.1080/15548627.2019.1580095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tang C., Cai J., Yin X.M., Weinberg J.M., Venkatachalam M.A., Dong Z. Mitochondrial quality control in kidney injury and repair. Nat Rev Nephrol. 2021;17:299–318. doi: 10.1038/s41581-020-00369-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Engel A., Barton G.M. Unfolding new roles for XBP1 in immunity. Nat Immunol. 2010;11:365–367. doi: 10.1038/ni0510-365. [DOI] [PubMed] [Google Scholar]
- 35.Martinon F., Chen X., Lee A.H., Glimcher L.H. TLR activation of the transcription factor XBP1 regulates innate immune responses in macrophages. Nat Immunol. 2010;11:411–418. doi: 10.1038/ni.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lubamba B.A., Jones L.C., O'Neal W.K., Boucher R.C., Ribeiro C.M. X-box-binding protein 1 and innate immune responses of human cystic fibrosis alveolar macrophages. Am J Respir Crit Care Med. 2015;192:1449–1461. doi: 10.1164/rccm.201504-0657OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ye D., Li F.Y., Lam K.S., Li H., Jia W., Wang Y., et al. Toll-like receptor-4 mediates obesity-induced non-alcoholic steatohepatitis through activation of X-box binding protein-1 in mice. Gut. 2012;61:1058–1067. doi: 10.1136/gutjnl-2011-300269. [DOI] [PubMed] [Google Scholar]
- 38.Brenner C., Galluzzi L., Kepp O., Kroemer G. Decoding cell death signals in liver inflammation. J Hepatol. 2013;59:583–594. doi: 10.1016/j.jhep.2013.03.033. [DOI] [PubMed] [Google Scholar]
- 39.Sena L.A., Li S., Jairaman A., Prakriya M., Ezponda T., Hildeman D.A., et al. Mitochondria are required for antigen-specific T cell activation through reactive oxygen species signaling. Immunity. 2013;38:225–236. doi: 10.1016/j.immuni.2012.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mills E.L., Kelly B., O'Neill L.A.J. Mitochondria are the powerhouses of immunity. Nat Immunol. 2017;18:488–498. doi: 10.1038/ni.3704. [DOI] [PubMed] [Google Scholar]
- 41.Hernández-Gea V., Hilscher M., Rozenfeld R., Lim M.P., Nieto N., Werner S., et al. Endoplasmic reticulum stress induces fibrogenic activity in hepatic stellate cells through autophagy. J Hepatol. 2013;59:98–104. doi: 10.1016/j.jhep.2013.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.El Manaa W., Duplan E., Goiran T., Lauritzen I., Vaillant Beuchot L., Lacas-Gervais S., et al. Transcription- and phosphorylation-dependent control of a functional interplay between XBP1s and PINK1 governs mitophagy and potentially impacts Parkinson disease pathophysiology. Autophagy. 2021;17:4363–4385. doi: 10.1080/15548627.2021.1917129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.He F., Huang Y., Song Z., Zhou H.J., Zhang H., Perry R.J., et al. Mitophagy-mediated adipose inflammation contributes to type 2 diabetes with hepatic insulin resistance. J Exp Med. 2021;218 doi: 10.1084/jem.20201416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sumpter R., Jr., Sirasanagandla S., Fernández Á.F., Wei Y., Dong X., Franco L., et al. Fanconi anemia proteins function in mitophagy and immunity. Cell. 2016;165:867–881. doi: 10.1016/j.cell.2016.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Patoli D., Mignotte F., Deckert V., Dusuel A., Dumont A., Rieu A., et al. Inhibition of mitophagy drives macrophage activation and antibacterial defense during sepsis. J Clin Invest. 2020;130:5858–5874. doi: 10.1172/JCI130996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou R., Yazdi A.S., Menu P., Tschopp J. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2011;469:221–225. doi: 10.1038/nature09663. [DOI] [PubMed] [Google Scholar]
- 47.Gilgenkrantz H., Mallat A., Moreau R., Lotersztajn S. Targeting cell-intrinsic metabolism for antifibrotic therapy. J Hepatol. 2021;74:1442–1454. doi: 10.1016/j.jhep.2021.02.012. [DOI] [PubMed] [Google Scholar]
- 48.Lotersztajn S., Julien B., Teixeira-Clerc F., Grenard P., Mallat A. Hepatic fibrosis: molecular mechanisms and drug targets. Annu Rev Pharmacol Toxicol. 2005;45:605–628. doi: 10.1146/annurev.pharmtox.45.120403.095906. [DOI] [PubMed] [Google Scholar]
- 49.Mallat A., Lotersztajn S. Cellular mechanisms of tissue fibrosis. 5. Novel insights into liver fibrosis. Am J Physiol Cell Physiol. 2013;305:C789–C799. doi: 10.1152/ajpcell.00230.2013. [DOI] [PubMed] [Google Scholar]
- 50.Karlmark K.R., Weiskirchen R., Zimmermann H.W., Gassler N., Ginhoux F., Weber C., et al. Hepatic recruitment of the inflammatory Gr1+ monocyte subset upon liver injury promotes hepatic fibrosis. Hepatology. 2009;50:261–274. doi: 10.1002/hep.22950. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data relevant to the study are included in the article or are uploaded as supplementary information and protocols. The reagents, antibodies, and resources are listed in the supplementary tables.