Abstract
Objectives: Necrotizing fasciitis (NF) caused by S. aureus is a rare, aggressive and rapidly progressing superficial fascia infection with a high mortality rate. The aim of this study was to identify virulence-related genes from a complete genome sequence of a methicillin-susceptible S. aureus (MSSA) isolate recovered from a monomicrobial case of NF.
Materials and methods: The MSSA isolate UMCG579 was cultured from a pus collection from the subcutis of a patient with NF. The genome of isolate UMCG579 was sequenced using MinION (Oxford Nanopore) and MiSeq (illumina) platforms.
Results: The genome of the UMCG579 isolate was composed of a 2,741,379 bp chromosome and did not harbor any plasmids. Virulence factor profiling identified multiple pore-forming toxin genes in the UMCG579 chromosome, including the Panton-Valentine leukocidin (PVL) genes, and none of the superantigen genes. The UMCG579 isolate harbored a new sequence variant of the recently described ete gene encoding exfoliative toxin (type E). A search in the GenBank database revealed that the new sequence variant (ete2) was exclusively found among isolates (n = 115) belonging to MLST CC152. While the majority of S. aureus ete-positive isolates were recovered from animal sources, S. aureus ete2-positive isolates originated from human carriers and human infections. Comparative genome analysis revealed that the ete2 gene was located on a 8777 bp genomic island.
Conclusion: The combination of two heterogeneously distributed potent toxins, ETE2 and PVL, is likely to enhance the pathogenic ability of S. aureus isolates. Since anti-virulence therapies for the treatment of S. aureus infections continue to be explored, the understanding of specific pathogenetic mechanisms may have an important prophylactic and therapeutic value. Nevertheless, the exact contribution of ETE sequence variants to S. aureus virulence in NF infections must be determined.
Keywords: necrotizing fasciitis, Staphylococcus aureus, ETE, ETE2, PVL
Introduction
Necrotizing fasciitis (NF) is a rare but severe infection characterised by necrosis of the superficial fascia with a high mortality rate (25%–30% cases). It is a surgical diagnosis. NF can rapidly progress to life-threatening septic shock. Symptoms can be non-specific at first, but classically present with swelling and erythema (Goh et al., 2014). Moreover, intense pain disproportionate to the results of the physical examination may be observed. The progression of NF can be characterised by the development of clear bullae, crepitus and skin necrosis (Kiat et al., 2017). The presence of skin lesions (bullae) has been shown to have a positive relationship with the rate of amputation and mortality, suggesting that this symptom may be regarded as a turning point in the prognosis of the disease (Karnuta et al., 2020). As the infection progresses, toxins are released spreading into the bloodstream. It can lead to severe systemic toxicity, which can eventually result in multi-organ failure. There are a number of established risk factors for disease including an immunocompromised status, diabetes, smoking, alcohol abuse, and the use of injectable drugs. However, disease can also develop in people of all ages without underlying conditions (Burnham et al., 2016; Stevens et al., 2021). Once NF is clinically highly suspected prompt surgical intervention is needed for confirmation of diagnosis and for treatment in combination with appropriate antimicrobial treatment to reduce mortality and improve outcomes. Patients who survive often suffer from severe complications, the most common of which include; nosocomial infections, respiratory distress syndrome, acute renal failure and limb amputation.
There are two main types of NF (Stevens and Bryant, 2017). Type I is characterised by polymicrobial infections and accounts for approximately 75% of all cases. Type I NF most commonly affects elderly patients and patients with underlying medical conditions. It can be caused by a mixture of aerobic bacteria (such as Streptococcus or Enterobacteriaceae) and anaerobic bacteria (Bacteroides or Peptostreptococcus). Type II NF accounts for approximately 25% of all cases and can affect all age groups and also individuals without underlying disease. It is characteristically monomicrobial by nature. Streptococcus pyogenes is the major etiological agent of type II NF, sometimes in combination with Staphylococcus aureus. S. aureus is an uncommon causative agent of monomicrobial NF, but the number of cases caused by this pathogen is increasing (Kao et al., 2011; Thapaliya et al., 2015; Kim et al., 2019). Some experts have proposed type III NF, in which Vibrio vulnificus, Aeromonas hydrophila and possibly clostridial species are the etiological agents (Stevens and Bryant, 2017).
In NF there is a strong association between exotoxin production at the site of infection and extensive tissue necrosis and systemic toxicity (Shumba et al., 2019). S. aureus possesses an extensive arsenal of exotoxins, which can be divided into three major groups: pore-forming toxins (PFTs), exfoliative toxins (ETs), and superantigens (SAgs) (Oliveira et al., 2018). PFTs are the most common bacterial cytotoxic proteins. They disrupt host cell membranes causing cell lysis but also have additional immuno-modulatory effects. ETs are highly specific serine proteases that cleave desmoglein 1 (Amagai et al., 2000; Amagai et al., 2002), a major transmembrane component of desmosomes. This unique proteolytic attack results in a dissociation of keratinocytes in skin, which allows the bacteria to spread beneath this barrier. To date, five types of ETs have been distinguished (ETA, ETB, ETC, ETD, and ETE) with sequence identity ranging from 43% to 63% (Ullah et al., 2022). ETA, ETB, ETD, and ETE are implicated in human skin damage, while ETC is not toxic to humans and has only been recovered from a horse infection (Oliveira et al., 2018; Imanishi et al., 2019; Azarian et al., 2021; Ullah et al., 2022). SAgs constitute exotoxins that stimulate extensive T cell activation and cytokine release. There are at least 26 staphylococcal SAg toxins distinguished, including the toxic shock syndrome toxin (TSST-1), enterotoxins and staphylococcal superantigen-like toxins (Oliveira et al., 2018; Vrieling et al., 2020).
The molecular pathogenesis of NF caused by S. aureus still remains poorly understood. Each exotoxin can cause different forms of tissue damage and inflammation. NF caused by S. aureus is relatively rare and as such there are few studies determining gene content based on complete genomes. Furthermore, knowledge of S. aureus virulence factor distribution in NF is insufficient. The aim of the present study was to identify virulence-related genes from a complete genome sequence of a methicillin-susceptible S. aureus isolate recovered from a monomicrobial case of NF. Moreover, we focused on the characterization of a new sequence variant of exfoliative toxin E, which can contribute to the severity and poor clinical outcome in NF patients.
Materials and methods
Case description
A 54-year-old male patient, born in Nigeria who had been living in the Netherlands for more than 10 years, presented at the emergency department of the University Hospital Center Groningen (UMCG) with septic shock: tachypnoea (30–40/min), tachycardia (120–130 beats/min) and hypotension (80/40). He had complaints of fatigue and fever starting 2 days before admission, and an abscess under the right axilla for a few days that had been incised and drained 1 day prior by his general practitioner. Physical examination revealed a purulent wound under the right axilla. The entire right flank felt indurated, pasty, and was painful on palpation. Doubtful minimal crepitus from this area of skin was elicited. Ultrasound investigation showed the appearance of cobblestoning, which can be indicative of cellulitis. The patient was immunocompromised: his medical history included HIV (2004), diabetes mellitus type II (2016) and kidney transplantation (2016). The patient was admitted to the intensive care ward. Antibiotic treatment was immediately started with cefuroxime 1500 mg three times daily, tobramycin 7 mg/kg once daily and clindamycin 600 mg three times daily, and mycofenolate mofetil was stopped. A pus sample from the axillary wound was cultured and grew S. aureus, which was resistant to penicillin and cotrimoxazole, and susceptible to all other antibiotics tested. Blood cultures grew Brevibacterium spp from one blood culture bottle, which was considered to be contamination. The next day the erythema had spread over the entire right flank to the lateral upper leg. On the third day of admission, several large and smaller tense bullae with a clear fluid content appeared on the erythematous skin of the lower flank to upper lateral leg. This area showed pitting edema and crepitus was elicited (Figure 1). No thermal or frictional damage had occurred in this area. The Nikolsky and pseudo-Nikolsky signs on the erythematous and healthy-looking skin were negative. All mucous membranes were normal. Fluid samples taken from the bullae tested negative for herpes simplex virus (HSV)-1, HSV-2, and varicella-zoster virus. HIV PCR on plasma showed no detectable load. Histology of a skin biopsy showed dermal edema and a diffuse infiltrate with neutrophils and no eosinophils. A skin biopsy for direct immunofluorescence was negative for auto-immune blistering diseases. A diagnosis of necrotizing fasciitis was made and surgical exploration and debridement of necrotic tissue was carried out. Samples from pus collections and deep tissue were cultured from which S. aureus grew in pure cultures. Blood cultures remained negative. Intravenous antibiotic treatment was switched to flucloxacillin 12 g/day via continuous infusion and vancomycin of which the dose was based on therapeutic drug monitoring. On day 3 and 5 after admission, surgical re-explorations were performed and further debridement of necrotic tissue from the right thoracic flank and abdomen was performed. On day 7, the patient was transferred to a burn center for skin grafting. Vancomycin was stopped, and treatment with flucloxacillin was continued. After prolonged treatment on the intensive care ward in the burn center, the patient recovered and was discharged to a nursing home. The patient provided his written informed consent to participate in this case study.
FIGURE 1.
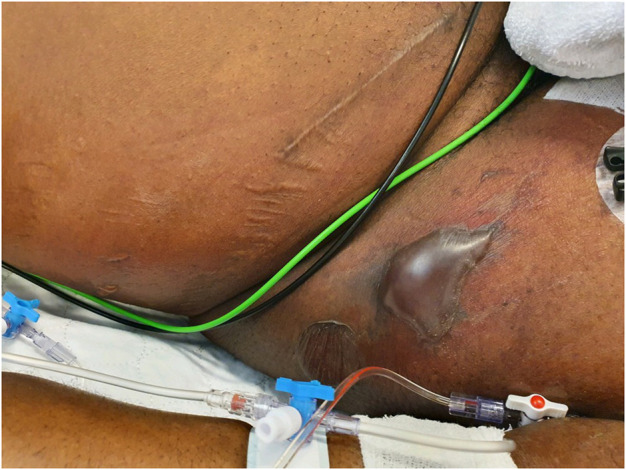
Clinical image of the right hip/inguinal region. Large superficial bullae appeared on purpuric lesions of the erythematous skin. Presumably, the bullae are caused by the systemic release of toxins by S. aureus, since no other causative pathogens were detected. A diagnosis of NF led to surgical debridement of necrotic tissue.
Bacterial isolate
The S. aureus isolate UMCG579 was cultured from a pus collection from the subcutis of the patient on December 9th 2021. The isolate was initially cultured on Columbia agar with 5% sheep blood (BA incubated for 48 h at 35°C). Species determination was performed by Maldi-TOF (Brucker, Bremen, and Germany).
Phenotypic antibiotic susceptibility testing
Automated susceptibility testing was performed by Vitek2 (bioMerieux, Marcy l'Etoile, and France). Susceptibilities were interpreted according to the current EUCAST breakpoints at the time of isolation. Susceptibility to penicillin was assessed by penicillin disk diffusion based on zone diameter and zone edge.
Extraction of genomic DNA
Total DNA was isolated using the DNeasy Blood and Tissue Kit (Qiagen). DNA was quantified using Qubit 2.0 Fluorometer (ThermoFisher Scientific) and the quality assessed by TapeStation 2200 (Agilent Technologies). A NanoDrop 2000C spectrophotometer (ThermoFisher Scientific) was used to measure the purity of the extracted DNA.
Whole genome sequencing
The genome of S. aureus isolate UMCG579 was sequenced with MinION Mk1C (Oxford Nanopore) and MiSeq (illumina) sequencers. The reads assembly was performed using SeqMan NGen version 18 (DNASTAR). Automated genome annotation was performed using the NCBI Prokaryotic Genome Annotation Pipeline (https://www.ncbi.nlm.nih.gov/genome/annotation_prok).
Data analysis
The DNA sequences were aligned using BLASTn software (https://blast.ncbi.nlm.nih.gov/Blast.cgi). The MLST sequence type (ST) was assigned through the MLST server (Larsen et al., 2012). The spa type was determined with spaTyper (Bartels et al., 2014). The search for SCCmec elements was performed using SCCmecFinder 1.2 (Kaya et al., 2018). PHASTER (PHAge Search Tool Enhanced Release) was used to analyze prophages in the genome (Arndt et al., 2016). Genomic islands in the UMCG579 genome were identified using IslandViewer 4 (Bertelli et al., 2017). The amino acid sequence alignment and phylogenetic analysis of ETs were performed with the MegAlign Pro software (DNASTAR). SNP analysis was performed using the CSI Phylogeny 1.4 server (Kaas et al., 2014). The maximum likelihood tree produced by CSI Phylogeny 1.4 server was visualized in MEGA6 software (Tamura et al., 2013).
Nucleotide sequence accession number
The sequence of the chromosome of S. aureus UMCG579 has been deposited in GenBank under accession number CP091066.
Results
Molecular characterization
The complete chromosome sequence of isolate UMCG579 consisted of 2,741,379 bp. UMCG579 did not harbor any plasmids. The chromosome showed a GC content of 32.86%, and contained 2527 coding sequences (with protein) and 83 RNAs. Based on in silico analysis, the isolate was identified as MLST ST1633, which belongs to clonal complex (CC) 152, and spa type t355. The search for SCCmec using the SCCmecFinder did not identify any SCCmec structure or its remnant DNA in this MSSA isolate. However, a sequence analysis of the UMCG579 chromosome using the IslandViewer webserver revealed five putative genomic islands (Supplementary Table S1). Among them there were two prophage regions. Analysis with the PHASTER webserver confirmed the existence of two intact prophages and additionally identified one incomplete prophage region in the genomic DNA of UMCG579 Supplementary Table S2).
Toxin gene repertoire
To better understand pathogenic mechanisms of NF infections caused by S. aureus, we investigated the content of virulence genes of UMCG579 with a focus on searching for its unique genetic features harboring potent exotoxins. Virulence factor profiling identified multiple pore-forming toxin genes in the UMCG579 chromosome and none of the superantigen genes (Table 1). However, the hla, hld and psmα genes as well as the hlg gene cluster constitute the core genome and are shared by more than 95% of S. aureus strains (Grumann et al., 2014). Moreover, the hlb, lukAB/GH and lukED genes present in the UMCG579 chromosome are widespread and can be found in the majority of strains. The lukF-PV and lukS-PV genes, encoding subunits of Panton-Valentine leukocidin (PVL), are present in a small percentage (approximately 3%) of clinical S. aureus strains and they are strongly associated with community acquired MRSA strains, particularly those isolated from skin and soft tissue infections (Vandenesch et al., 2003). In the UMCG579 chromosome the PVL genes were located on an intact phage phi2958PVL (Supplementary Table S2).
TABLE 1.
Major virulence genes in S. aureus UMCG579.
| Virulence factor gene | Position in chromosome | Protein Function |
|---|---|---|
| Adherence | ||
| clfA | 773776..777057 | clumping factor A |
| clfB | 2640621..2643311 (complement) | clumping factor B |
| Cna | 2718559..2722110 (complement) | collagen binding protein |
| ebpS | 1436472..1437932 (complement) | elastin-binding protein EbpS |
| fnbA | 2504569..2507826 (complement) | fibronectin-binding protein FnbA |
| fnbB | 2501063..2503888 (complement) | fibronectin-binding protein FnbB |
| sdrD | 532215..536324 | serine-aspartate repeat protein D |
| Pore forming toxins | ||
| Hla | 1056230..1057189 (complement) | alpha-hemolysin precursor |
| hlb | 2013235..2014227 | beta-hemolysin |
| hld | 2029162..2029242 | delta-hemolysin |
| hlgA | 2425223..2426152 | gamma-hemolysin chain II precursor |
| hlgB | 2427668..2428644 | gamma-hemolysin component B precursor |
| hlgC | 2426719..2427666 | gamma-hemolysin component C |
| lukD | 1882762..1883745 (complement) | leukocidin D component |
| lukE | 1883746..1884681 (complement) | leukocidin E component |
| lukA/lukG | 2014491..2015507 (complement) | leukocidin A/G component |
| lukB/lukH | 2015529..2016557 (complement) | leukocidin B/H component |
| lukF-PV | 1444463..1445440 (complement) | Panton-Valentine leukocidin F component |
| lukS-PV | 1445442..1446380 (complement) | Panton-Valentine leukocidin S component |
| psmα-1 | 389955..390020 (complement) | phenol-soluble modulin PSM-alpha-1 |
| psmα-2 | 389858..389923 (complement) | phenol-soluble modulin PSM-alpha-2 |
| psmα-3 | 389738..389806 (complement) | phenol-soluble modulin PSM-alpha-3 |
| psmα-4 | 389612..389674 (complement) | phenol-soluble modulin PSM-alpha-4 |
| Exoenzymes | ||
| epiP | 1874291..1875664 (complement) | lantibiotic epidermin leader peptide processing serine protease EpiP |
| sspA | 938970..939953 (complement) | Glu-specific serine endopeptidase SspA |
| sspB | 937707..938888 (complement) | cysteine protease staphopain B |
| scpA | 1984658..1985824 | cysteine protease staphopain A |
| aur | 2650383..2651912 (complement) | zinc metalloproteinase aureolysin |
| ete2 | 2223688..2224539 (complement) | exfoliative toxin E |
| sak | 1829110..1829601 (complement) | Staphylokinase |
| ednB | 2222714..2223469 | epidermal cell differentiation inhibitor B |
| hysA | 1819116..1821533 | hyaluronate lyase |
| lip1 | 2700304..2702346 (complement) | YSIRK domain-containing triacylglycerol lipase Lip1 |
| lip2 | 297349..299427 | YSIRK domain-containing triacylglycerol lipase Lip2/Geh |
| coa | 192697..194679 | Staphylocoagulase |
| nuc | 781276..781962 | Thermonuclease family protein |
| Immune modulation | ||
| ebh | 1356399..1387838 (complement) | fibronectin binding protein ebh |
| efb | 2423425..2424735 | immunoglobulin-binding protein Sbi |
| scn | 1827266..1827616 (complement) | complement inhibitor SCIN-A |
| spa | staphylococcal protein A | |
| Biofilm | ||
| icaR | 2695830..2696390 (complement) | ica operon transcriptional regulator IcaR |
| icaA | 2696554..2697792 | poly-beta-1,6 N-acetyl-D-glucosamine synthase IcaA |
| icaD | 2697756..2698061 | intracellular adhesion protein IcaD |
| icaB | 2698058..2698930 | intercellular adhesin biosynthesis polysaccharide N-deacetylase |
| icaC | 2698917..2699969 | polysaccharide intercellular adhesin biosynthesis/export protein IcaC |
A new sequence variant of exfoliative toxin E
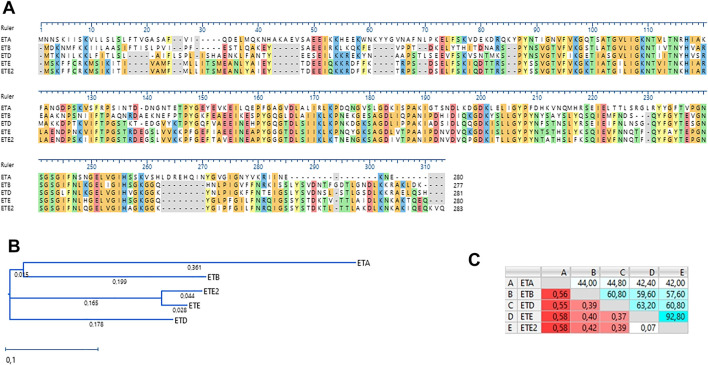
Genome sequence analysis revealed that the UMCG579 isolate harbored a sequence variant of the recently described ete gene encoding a new exfoliative toxin (type E) (Imanishi et al., 2019). We propose to name this variant ete2. Comparison of the deduced amino acid sequences of the ete and ete2 genes showed a 92.80% sequence identity (Figure 2A). Moreover, deduced sequence of the ETE2 protein was longer by three amino acids than that of ETE (283 AA versus 280 AA). The deduced amino acid sequences of the ete and ete2 genes were compared to those of other ET proteins recovered from human infections (Figure 2B). The amino acid sequence of ETE showed higher sequence identity to ETA, ETB and ETD than that of ETE2 (Figure 2C).
FIGURE 2.
Amino acid sequence alignment and phylogenetic analysis of staphylococcal ETs recovered from human infections. (A) Amino acid sequence alignment (MUSCLE). (B) A neighbor-joining tree. The scale bar indicates the evolutionary distance between the sequences determined by 0.1 substitutions per amino acid at the variable positions. (C) Distance/%Identity among analyzed amino acid sequences.
Prevalence of the ete2 gene in S. aureus genomes
A BLAST search in the GenBank database revealed that the ete2 gene was exclusively found among isolates (n = 115) belonging to MLST CC152; the majority of which were identified as ST152 (n = 112), with the remainder belonging to ST1633 (n = 2) and ST unknown (single locus variant of ST152, n = 1) (Supplementary Table S3). The isolates of CC152 possessing the ete2 gene were recovered exclusively from human carriers or human infections. The deduced amino acid sequences of ETE2 were almost always (112 isolates) identical to that of the isolate UMCG579 or differed by only a single amino acid residue (3 isolates). We also investigated if we could identify ete2-negative isolates belonging to CC152. We found six ST152 isolates that did not harbor the ete2 gene. They had a human origin and were isolated from nasal/nasopharynx swabs in Africa.
Prevalence of the ete gene in S. aureus genomes
Analysis of the ete distribution using BLAST identified the presence of this gene mostly in MLST CC130 (n = 123) and also in several genetically unrelated STs (n = 10), including ST890, ST2233, ST2867, ST2970, and ST2990 (Supplementary Table S4). Unlike ete2, the ete gene was found not only in human sources (n = 55) but also in animals (n = 71) including; European red deer (Cervus elaphus) (n = 18), domestic pig (Sus scrofa domesticus) (n = 16), domestic sheep (Ovis aries) (n = 12) European red squirrel (Sciurus vulgaris) (n = 11), domestic goat (Capra hircus) (n = 8), domestic cow (Bos taurus) (n = 3), wild goat (Capra hircus) (n = 2) and European hedgehog (Erinaceus europaeus) (n = 1). Like ete2, the ete gene was also highly conserved independently of genetic background (different MLST CCs) or animal source of isolation.
The deduced amino acid sequences of ETE were identical to that of the isolate O46 or differed by up to three amino acid residues.
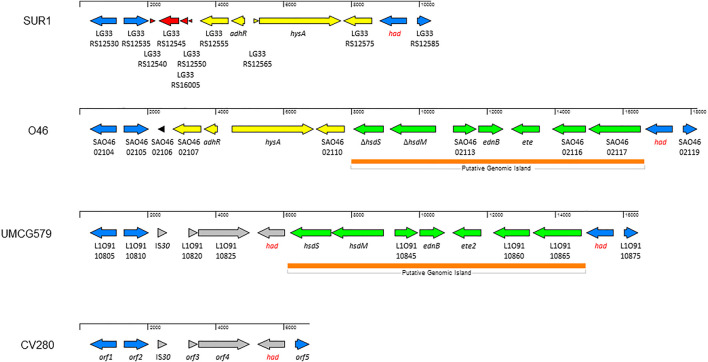
Genetic organization of genomic island harboring the ete2 gene
To determine the size and gene content of a putative genomic island harboring the ete2 gene, the genome sequence of UMCG579 was compared to that of the O46 isolate (GenBank accession number CP025395) containing a genomic island with the ete gene (Figure 3). Moreover, to determine accurate boundaries of the island, the UMCG579 genome sequence was compared to that of a genetically unrelated (MLST ST8) and exfoliative toxin negative SUR1 isolate (GenBank accession number CP009423), representative of the USA300 lineage. We searched in both UMCG579 and O46 isolates a string of genes in the vicinity of an exfoliative toxin gene that were not present in the SUR1 isolate. Comparative genome analysis revealed that the ete2 gene was located on a 8777 bp putative genomic island (Figure 3). This island contained six other genes encoding proteins associated with a restriction-modification system (hsdS and hsdM), virulence (ednB), proteolysis (L1O91_10845) and cellular activities (L1O91_10860 and L1O91_10865) (Table 2). Nucleotide alignment of the island containing the ete2 gene of UMCG579 revealed that it shared 95.67% nucleotide identity with a genomic island of the O46 isolate over 95% of its length. The most likely insertion site of the ete2-containing island in the UMCG579 chromosome was the had gene encoding HAD-IIB family hydrolase as the island was bound by two copies of this gene in direct orientation. This configuration could have arisen if the had gene in a circular molecule recombined with the had gene already present in the chromosome through homologous recombination. Two copies of the had gene (816 nucleotides in length) were not identical differing by nine nucleotides. Moreover, the copy of the had gene located upstream of ete2 possessed a mutation causing a premature stop codon, while the had copy located downstream was fully functional. Homologous recombination also generated a deletion variant, which was found in six CC152 isolates. The deletion arose through recombination between the two copies of the duplicated had gene resulting in a loss of all genes of the ete2 genomic island (Figure 3).
FIGURE 3.
Structure of the genomic islands containing the ete and ete2 genes from S. aureus isolates O46 (GenBank accession number CP025395) and UMCG579 (GenBank accession number CP091066), respectively; and comparison with genome sequences of isolates USA300_SUR1 (GenBank accession number CP009423) and CV280 (GenBank accession number JAAWZQ000000000). The arrows indicate open reading frames and their orientations. Blue arrows: genes shared by isolates SUR1, O46, and UMCG579. Yellow arrows: genes shared by isolates SUR1 and O46. Green arrows: genes shared by isolates O46 and UMCG579. Grey arrows: genes shared by isolates O46 and CV280. Red and black arrows: genes only present in isolate SUR1 or O46, respectively. ORFs are described further in Table 2.
TABLE 2.
Genes present in the genomic islands containing ete and ete2 genes.
| Isolate SUR1 gene name | Isolate O46 gene name | Isolate UMCG579 gene name | Isolate CV280 gene name | Product |
|---|---|---|---|---|
| LG33_RS12530 | SAO46_02104 | L1O91_10805 | orf1 | DeoR/GlpR family DNA-binding transcription regulator |
| LG33_RS12535 | SAO46_02105 | L1O91_10810 | orf2 | NAD-dependent protein deacylase |
| LG33_RS12540 | Absent | Absent | Absent | Hypothetical protein |
| LG33_RS12545 (frameshifted) | Absent | Absent | Absent | DUF3885 domain-containing protein |
| LG33_RS16005 (internal stop) | Absent | Absent | Absent | SAR2788 family putative toxin |
| LG33_RS12550 | Absent | Absent | Absent | Hypothetical protein |
| Absent | SAO46_02106 | Absent | Absent | Hypothetical protein |
| LG33_RS12555 | SAO46_02107 | Absent | Absent | Aldo/keto reductase |
| adhR | adhR | Absent | Absent | Transcriptional regulator, MerR family |
| LG33_RS12565 | Sequence present but not annotated | Absent | Absent | Hypothetical protein |
| hysA | hysA | Absent | Absent | Hyaluronate lyase |
| LG33_RS12575 | SAO46_02110 | Absent | Absent | M23/M37 peptidase domain protein |
| Absent | Absent | IS30 (frameshifted) | IS30 (frameshifted) | IS30 family transposase |
| Absent | Absent | L1O91_10820 | orf3 | Hypothetical protein |
| Absent | Absent | L1O91_10825 | orf4 | Glycosyltransferase |
| Absent | Absent | had (internal stop) | had (internal stop) | HAD-IIB family hydrolase |
| Absent | hsdS (frameshifted) | hsdS | Absent | Type I restriction-modification system, restriction endonuclease subunit S |
| Absent | hsdM (partial deletion) | hsdM | Absent | Type I restriction-modification system subunit M |
| Absent | SAO46_02113 | L1O91_10845 | Absent | Trypsin-like serine protease |
| Absent | ednB | ednB | Absent | Epidermal cell differentiation inhibitor |
| Absent | ete | ete2 | Absent | Exfoliative toxin type E |
| Absent | SAO46_02116 | L1O91_10860 | Absent | AAA family atpase |
| Absent | SAO46_02117 | L1O91_10865 | Absent | AAA family atpase |
| had | had | had | Absent | HAD-IIB family hydrolase |
| LG33_RS12585 | SAO46_02119 | L1O91_10875 | orf5 | MAP domain-containing protein |
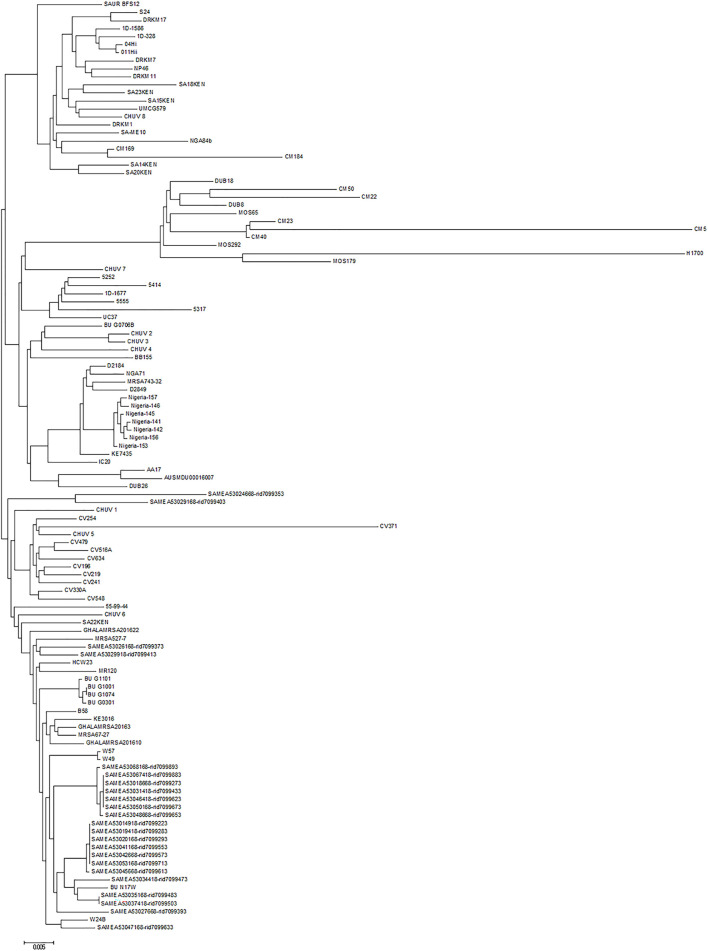
Single nucleotide polymorphism analysis
The chromosome sequences of all ete2-positive isolates (n = 116) were uploaded to the CSI Phylogeny 1.4 server (https://cge.cbs.dtu.dk/services/CSIPhylogeny) in order to investigate their single nucleotide polymorphism (SNP)-based phylogeny. Genome-wide SNP results revealed that the UMCG579 chromosome sequence was most closely related to that of CHUV 8 and SA15KEN (Figure 4), although the sequences differed by 82 and 106 SNPs, respectively. Isolates CHUV 8 and SA15KEN were recovered 6 years earlier than UMCG579, and they had different geographic origins, from Switzerland and Kenya, respectively.
FIGURE 4.
Phylogenetic relationship of S. aureus CC152 isolates containing the ete2 gene. Maximum likelihood tree was constructed on the basis of genome-wide single nucleotide polymorphism (SNP) analysis. The scale bar indicates the evolutionary distance between the sequences determined by 0.005 substitutions per nucleotide at the variable positions.
Discussion
For adequate and cost-effective infection prevention, it is important to identify S. aureus strains based on their virulence gene profiles that may be more aggressive and have a higher potential to establish a life-threatening infection. Two potent toxins that are heterogeneously distributed among S. aureus strains were identified in the UMCG579 genome, PVL and ETE2. PVL is a cytotoxin that lyses human polymorphonuclear leukocytes, especially neutrophils, and intradermal injection of purified PVL to rabbits causes edema, erythema and tissue necrosis (Cribier et al., 1992; Löffler et al., 2010). Moreover, epidemiological data demonstrates compelling evidence for PVL as an important virulence factor of S. aureus in severe skin and soft tissue infections (SSTI) and necrotizing pneumonia in humans (Gillet et al., 2007; Gillet et al., 2008; Nurjadi et al., 2015). However, several experimental studies in animal models of SSTI contradict each other. Despite evidence from a study that showed using a PVL-positive and its isogenic PVL-knockout strain that PVL plays a role in tissue pathology in the early stages of infection (Lipinska et al., 2011), other studies could not detect a contribution of this toxin in necrotizing skin infections (Li et al., 2009; Kobayashi et al., 2011).
The production of ETs in S. aureus strains is associated with superficial infections such as localized bullous impetigo and its generalized form staphylococcal scalded skin syndrome (SSSS) (Oliveira et al., 2018). All S. aureus ETs are unique glutamate-specific serine proteases that specifically cleave a peptide bond after glutamic acid residue 381, which is located in the extracellular segment of human and murine desmoglein 1 (Hanakawa et al., 2004; Bukowski et al., 2010; Imanishi et al., 2019). This cleavage causes efficient and specific abolishment of keratinocyte junctions and cell-cell adhesion in the epidermis resulting in host skin damage (Nishifuji et al., 2008). Necrotizing infections can occur after minor or major breaches of the skin (Stevens et al., 2021). Furthermore, deep-seated infections are frequently associated with S. aureus strains that produce PVL (Holmes et al., 2005). Therefore, the combined production of ETE2 and PVL may increase the risk of the occurrence of more severe symptoms during infection and be associated with poor clinical outcome in NF patients.
Conflicting results obtained in different studies regarding the role of PVL in necrotizing skin infections may result from the presence of different virulence factor profiles in the genomic DNA of S. aureus strains. S. aureus produces numerous and often functionally redundant virulence factors. Therefore, it is important to examine the full gene content from the complete genome sequences to better understand the contribution of particular S. aureus virulence determinants to NF infection. However, only a limited number of S. aureus isolates recovered from NF patients have been investigated by whole-genome sequencing (WGS). Takano and others reported that ST764 MRSA, which is a local variant of the ST5 lineage, caused invasive infections, including necrotizing fasciitis (Takano et al., 2013). Analysis of the genome sequence revealed that ST764 carried the staphylococcal enterotoxin genes seb, seg, sei, sem, sen, and seo, and was negative for the PVL genes. In another study, Bakthavatchalam and others showed genomic insights into MRSA strain of ST772 (belonging to CC1) and spa type t657 from a case of fatal necrotizing fasciitis (Bakthavatchalam et al., 2018). Analysis of the genome for virulence determinants revealed the presence of staphylococcal enterotoxin genes sea, seg, sei, sel, sem, seo, and sec3. Moreover, the genome of ST772/t657 MRSA strain possessed the prophage phiSa119 carrying an operon with the lukS/F-PV genes encoding PVL. Aswani and others analyzed the genome sequence of ST45 MSSA strain that caused severe necrotizing fasciitis in a 72-year-old diabetic male (Aswani et al., 2019). The toxin landscape of the ST45 MSSA strain consisted of the enterotoxin gene cluster (egc): seg, sen, seu, sei, sem, and seo, as well as the eta gene encoding exfoliative toxin A. The strain was lacking the genes for PVL. Furthermore, Yamagishi et al. (2020) and others presented a case of monomicrobial Fournier’s gangrene (NF of the perineal, perianal or genital regions) caused by MSSA belonging to ST8 and spa type t622. WGS and a search for virulence factors revealed that the ST8/t622 strain harbored the tst gene encoding toxic shock syndrome toxin-1 (TSST-1) and was PVL-negative.
The PVL-positive ST152 lineage seems to be a hyperepidemic community associated MSSA in Africa and associated with chronic wound infections (Wolters et al., 2020). This lineage is especially prevalent in west and central Africa (Schaumburg et al., 2014; Obasuyi et al., 2020). Recent study by Obasuyi et al. (2020) and others showed that ST152-MSSA was the most dominant MLST ST (accounting for 24.7% of all MSSA isolates) in Benin City in South-South Nigeria. This finding was in line with another study by Okon et al. (2014) and others, who found that the predominant MSSA strain recovered in tertiary-care hospitals in North-East Nigeria was ST152-MSSA. CC152 strains are not found widely outside of Africa and have been only occasionally isolated from patients in some European countries.
In this paper we showed that the UMCG579 genome contained a novel sequence variant of exfoliative toxin E (ETE2) located on a genomic island, which was acquired through homologous recombination. The amino acid sequence of ETE2 differed by 7.2% from that of ETE being also longer by 3 amino acid residues. It warrants further study to compare the biochemical properties of the two variants of this exfoliative toxin such as host-specific activity, pH optimum, thermostability or inhibition studies of the enzymes by natural and synthetic inhibitors. While the majority of S. aureus ete-positive isolates were recovered from animal sources, S. aureus ete2-positive isolates had a human origin. Most infections caused by S. aureus are due to the combined action of various factors. The combination of two potent toxins, ETE2 and PVL, is likely to enhance the virulence of S. aureus isolates.
Data availability statement
The datasets for this article are not publicly available due to concerns regarding participant/patient anonymity. Requests to access the datasets should be directed to the corresponding author.
Ethics statement
Ethical review and approval was not required for the study on human participants in accordance with the local legislation and institutional requirements. The patients/participants provided their written informed consent to participate in this study.
Author contributions
EB designed the project. AF supervised the study. AS and VA performed DNA sequencing. AS, MW-B, AR, LH, OK, and EB analyzed the data. All authors interpreted the data. AS and EB wrote the draft manuscript. All authors added comments and corrections and approved a final version of the manuscript.
Funding
This work was partly funded by the European Commission, the Dutch Ministry of Health, Welfare and Sport, the Ministry of Economy, Innovation, Digitalisation and Energy of the German Federal State of North Rhine-Westphalia and the Ministry for National and European Affairs and Regional Development of Lower Saxony within the INTERREG V A (202085) funded project EurHealth-1Health (http://www.eurhealth1health.eu).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.964358/full#supplementary-material
References
- Amagai M., Matsuyoshi N., Wang Z. H., Andl C., Stanley J. R. (2000). Toxin in bullous impetigo and staphylococcal scalded-skin syndrome targets desmoglein 1. Nat. Med. 6 (11), 1275–1277. 10.1038/81385 [DOI] [PubMed] [Google Scholar]
- Amagai M., Yamaguchi T., Hanakawa Y., Nishifuji K., Sugai M., Stanley J. R. (2002). Staphylococcal exfoliative toxin B specifically cleaves desmoglein 1. J. Invest. Dermatol. 118 (5), 845–850. 10.1046/j.1523-1747.2002.01751.x [DOI] [PubMed] [Google Scholar]
- Arndt D., Grant J. R., Marcu A., Sajed T., Pon A., Liang Y., et al. (2016). PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 44 (W1), W16–W21. 10.1093/nar/gkw387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aswani V., Najar F., Pantrangi M., Mau B., Schwan W. R., Shukla S. K. (2019). Virulence factor landscape of a Staphylococcus aureus sequence type 45 strain, MCRF184. BMC Genomics 20 (1), 123. 10.1186/s12864-018-5394-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azarian T., Cella E., Baines S. L., Shumaker M. J., Samel C., Jubair M., et al. (2021). Genomic epidemiology and global population structure of exfoliative toxin A-producing Staphylococcus aureus strains associated with staphylococcal scalded skin syndrome. Front. Microbiol. 12, 663831. 10.3389/fmicb.2021.663831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakthavatchalam Y. D., Ramaswamy B., Janakiraman R., Steve R. J., Veeraraghavan B. (2018). Genomic insights of community-acquired methicillin-resistant Staphylococcus aureus (MRSA) with reduced teicoplanin susceptibility: a case of fatal necrotizing fasciitis. J. Glob. Antimicrob. Resist. 14, 242–245. 10.1016/j.jgar.2018.05.006 [DOI] [PubMed] [Google Scholar]
- Bartels M. D., Petersen A., Worning P., Nielsen J. B., Larner-Svensson H., Johansen H. K., et al. (2014). Comparing whole-genome sequencing with Sanger sequencing for spa typing of methicillin-resistant Staphylococcus aureus . J. Clin. Microbiol. 52 (12), 4305–4308. 10.1128/jcm.01979-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertelli C., Laird M. R., Williams K. P., Lau B. Y., Hoad G., Winsor G. L., et al. (2017). IslandViewer 4: expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res. 45 (W1), W30–w35. 10.1093/nar/gkx343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukowski M., Wladyka B., Dubin G. (2010). Exfoliative toxins of Staphylococcus aureus . Toxins (Basel) 2 (5), 1148–1165. 10.3390/toxins2051148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnham J. P., Kirby J. P., Kollef M. H. (2016). Diagnosis and management of skin and soft tissue infections in the intensive care unit: a review. Intensive Care Med. 42 (12), 1899–1911. 10.1007/s00134-016-4576-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cribier B., Prévost G., Couppie P., Finck-Barbançon V., Grosshans E., Piémont Y. (1992). Staphylococcus aureus leukocidin: a new virulence factor in cutaneous infections? An epidemiological and experimental study. Dermatology 185 (3), 175–180. 10.1159/000247443 [DOI] [PubMed] [Google Scholar]
- Gillet Y., Vanhems P., Lina G., Bes M., Vandenesch F., Floret D., et al. (2007). Factors predicting mortality in necrotizing community-acquired pneumonia caused by Staphylococcus aureus containing Panton-Valentine leukocidin. Clin. Infect. Dis. 45 (3), 315–321. 10.1086/519263 [DOI] [PubMed] [Google Scholar]
- Gillet Y., Etienne J., Lina G., Vandenesch F. (2008). Association of necrotizing pneumonia with Panton-Valentine leukocidin-producing Staphylococcus aureus, regardless of methicillin resistance. Clin. Infect. Dis. 47 (7), 985–986. 10.1086/591803 [DOI] [PubMed] [Google Scholar]
- Goh T., Goh L. G., Ang C. H., Wong C. H. (2014). Early diagnosis of necrotizing fasciitis. Br. J. Surg. 101 (1), e119–125. 10.1002/bjs.9371 [DOI] [PubMed] [Google Scholar]
- Grumann D., Nübel U., Bröker B. M. (2014). Staphylococcus aureus toxins--their functions and genetics. Infect. Genet. Evol. 21, 583–592. 10.1016/j.meegid.2013.03.013 [DOI] [PubMed] [Google Scholar]
- Hanakawa Y., Schechter N. M., Lin C., Nishifuji K., Amagai M., Stanley J. R. (2004). Enzymatic and molecular characteristics of the efficiency and specificity of exfoliative toxin cleavage of desmoglein 1. J. Biol. Chem. 279 (7), 5268–5277. 10.1074/jbc.M311087200 [DOI] [PubMed] [Google Scholar]
- Holmes A., Ganner M., McGuane S., Pitt T. L., Cookson B. D., Kearns A. M. (2005). Staphylococcus aureus isolates carrying panton-valentine leucocidin genes in england and wales: frequency, characterization, and association with clinical disease. J. Clin. Microbiol. 43 (5), 2384–2390. 10.1128/jcm.43.5.2384-2390.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imanishi I., Nicolas A., Caetano A. B., Castro T. L. P., Tartaglia N. R., Mariutti R., et al. (2019). Exfoliative toxin E, a new Staphylococcus aureus virulence factor with host-specific activity. Sci. Rep. 9 (1), 16336. 10.1038/s41598-019-52777-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaas R. S., Leekitcharoenphon P., Aarestrup F. M., Lund O. (2014). Solving the problem of comparing whole bacterial genomes across different sequencing platforms. PLoS One 9 (8), e104984. 10.1371/journal.pone.0104984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao L. S., Lew D. F., Arab S. N., Todd S. R., Awad S. S., Carrick M. M., et al. (2011). Local variations in the epidemiology, microbiology, and outcome of necrotizing soft-tissue infections: a multicenter study. Am. J. Surg. 202 (2), 139–145. 10.1016/j.amjsurg.2010.07.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnuta J., Featherall J., Lawrenz J., Gordon J., Golubovsky J., Thomas J., et al. (2020). What demographic and clinical factors are associated with in-hospital mortality in patients with necrotizing fasciitis? Clin. Orthop. Relat. Res. 478 (8), 1770–1779. 10.1097/corr.0000000000001187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaya H., Hasman H., Larsen J., Stegger M., Johannesen T. B., Allesøe R. L., et al. (2018). SCCmecFinder, a web-based tool for typing of staphylococcal cassette chromosome mec in Staphylococcus aureus using whole-genome sequence data. mSphere 3 (1), e00612–17. 10.1128/mSphere.00612-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiat H. J., En Natalie Y. H., Fatimah L. (2017). Necrotizing fasciitis: how reliable are the cutaneous signs? J. Emerg. Trauma Shock 10 (4), 205–210. 10.4103/jets.Jets_42_17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T., Park S. Y., Kwak Y. G., Jung J., Kim M. C., Choi S. H., et al. (2019). Etiology, characteristics, and outcomes of community-onset necrotizing fasciitis in korea: A multicenter study. PLoS One 14 (6), e0218668. 10.1371/journal.pone.0218668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi S. D., Malachowa N., Whitney A. R., Braughton K. R., Gardner D. J., Long D., et al. (2011). Comparative analysis of USA300 virulence determinants in a rabbit model of skin and soft tissue infection. J. Infect. Dis. 204 (6), 937–941. 10.1093/infdis/jir441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen M. V., Cosentino S., Rasmussen S., Friis C., Hasman H., Marvig R. L., et al. (2012). Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 50 (4), 1355–1361. 10.1128/jcm.06094-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Diep B. A., Villaruz A. E., Braughton K. R., Jiang X., DeLeo F. R., et al. (2009). Evolution of virulence in epidemic community-associated methicillin-resistant Staphylococcus aureus . Proc. Natl. Acad. Sci. U. S. A. 106 (14), 5883–5888. 10.1073/pnas.0900743106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinska U., Hermans K., Meulemans L., Dumitrescu O., Badiou C., Duchateau L., et al. (2011). Panton-Valentine leukocidin does play a role in the early stage of Staphylococcus aureus skin infections: a rabbit model. PLoS One 6 (8), e22864. 10.1371/journal.pone.0022864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löffler B., Hussain M., Grundmeier M., Brück M., Holzinger D., Varga G., et al. (2010). Staphylococcus aureus panton-valentine leukocidin is a very potent cytotoxic factor for human neutrophils. PLoS Pathog. 6 (1), e1000715. 10.1371/journal.ppat.1000715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishifuji K., Sugai M., Amagai M. (2008). Staphylococcal exfoliative toxins: "molecular scissors" of bacteria that attack the cutaneous defense barrier in mammals. J. Dermatol. Sci. 49 (1), 21–31. 10.1016/j.jdermsci.2007.05.007 [DOI] [PubMed] [Google Scholar]
- Nurjadi D., Friedrich-Jänicke B., Schäfer J., Van Genderen P. J., Goorhuis A., Perignon A., et al. (2015). Skin and soft tissue infections in intercontinental travellers and the import of multi-resistant Staphylococcus aureus to Europe. Clin. Microbiol. Infect. 21 (6), e1–10510. 10.1016/j.cmi.2015.01.016 [DOI] [PubMed] [Google Scholar]
- Obasuyi O., McClure J., Oronsaye F. E., Akerele J. O., Conly J., Zhang K. (2020). Molecular characterization and pathogenicity of Staphylococcus aureus isolated from Benin-city, Nigeria. Microorganisms 8 (6), E912. 10.3390/microorganisms8060912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okon K. O., Shittu A. O., Kudi A. A., Umar H., Becker K., Schaumburg F. (2014). Population dynamics of Staphylococcus aureus from northeastern Nigeria in 2007 and 2012. Epidemiol. Infect. 142 (8), 1737–1740. 10.1017/s0950268813003117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira D., Borges A., Simões M. (2018). Staphylococcus aureus toxins and their molecular activity in infectious diseases. Toxins (Basel) 10 (6), E252. 10.3390/toxins10060252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaumburg F., Alabi A. S., Peters G., Becker K. (2014). New epidemiology of Staphylococcus aureus infection in Africa. Clin. Microbiol. Infect. 20 (7), 589–596. 10.1111/1469-0691.12690 [DOI] [PubMed] [Google Scholar]
- Shumba P., Mairpady Shambat S., Siemens N. (2019). The role of streptococcal and staphylococcal exotoxins and proteases in human necrotizing soft tissue infections. Toxins (Basel) 11 (6), E332. 10.3390/toxins11060332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens D. L., Bryant A. E. (2017). Necrotizing soft-tissue infections. N. Engl. J. Med. 377 (23), 2253–2265. 10.1056/NEJMra1600673 [DOI] [PubMed] [Google Scholar]
- Stevens D. L., Bryant A. E., Goldstein E. J. (2021). Necrotizing soft tissue infections. Infect. Dis. Clin. North Am. 35 (1), 135–155. 10.1016/j.idc.2020.10.004 [DOI] [PubMed] [Google Scholar]
- Takano T., Hung W. C., Shibuya M., Higuchi W., Iwao Y., Nishiyama A., et al. (2013). A new local variant (ST764) of the globally disseminated ST5 lineage of hospital-associated methicillin-resistant Staphylococcus aureus (MRSA) carrying the virulence determinants of community-associated MRSA. Antimicrob. Agents Chemother. 57 (4), 1589–1595. 10.1128/aac.01147-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30 (12), 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thapaliya D., O'Brien A. M., Wardyn S. E., Smith T. C. (2015). Epidemiology of necrotizing infection caused by Staphylococcus aureus and Streptococcus pyogenes at an Iowa hospital. J. Infect. Public Health 8 (6), 634–641. 10.1016/j.jiph.2015.06.003 [DOI] [PubMed] [Google Scholar]
- Ullah A., Khan A., Al-Harrasi A., Ullah K., Shabbir A. (2022). Three-dimensional structure characterization and inhibition study of exfoliative toxin D from Staphylococcus aureus . Front. Pharmacol. 13, 800970. 10.3389/fphar.2022.800970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenesch F., Naimi T., Enright M. C., Lina G., Nimmo G. R., Heffernan H., et al. (2003). Community-acquired methicillin-resistant Staphylococcus aureus carrying panton-valentine leukocidin genes: worldwide emergence. Emerg. Infect. Dis. 9 (8), 978–984. 10.3201/eid0908.030089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrieling M., Tuffs S. W., Yebra G., van Smoorenburg M. Y., Alves J., Pickering A. C., et al. (2020). Population analysis of Staphylococcus aureus reveals a cryptic, highly prevalent superantigen SElW that contributes to the pathogenesis of bacteremia. mBio 11 (5), e02082–20. 10.1128/mBio.02082-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolters M., Frickmann H., Christner M., Both A., Rohde H., Oppong K., et al. (2020). Molecular characterization of Staphylococcus aureus isolated from chronic infected wounds in rural Ghana. Microorganisms 8 (12), E2052. 10.3390/microorganisms8122052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi H., Kawahori K., Ohkiba N., Murooka N., Inazawa T. (2020). Monomicrobial fournier's gangrene caused by panton-valentine leukocidin-negative methicillin-susceptible Staphylococcus aureus ST8 in Japan. Intern. Med. 59 (14), 1769–1772. 10.2169/internalmedicine.4334-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets for this article are not publicly available due to concerns regarding participant/patient anonymity. Requests to access the datasets should be directed to the corresponding author.