Abstract
Escherichia coli has 12 recognized penicillin binding proteins (PBPs), four of which (PBPs 4, 5, and 6 and DacD) have dd-carboxypeptidase activity. Although the enzymology of the dd-carboxypeptidases has been studied extensively, the in vivo functions of these proteins are poorly understood. To explain why E. coli maintains four independent loci encoding enzymes of considerable sequence identity and comparable in vitro activity, it has been proposed that the dd-carboxypeptidases may substitute for one another in vivo. We tested the validity of this equivalent substitution hypothesis by investigating the effects of these proteins on the aberrant morphology of ΔdacA mutants, which produce no PBP 5. Although cloned PBP 5 complemented the morphological phenotype of a ΔdacA mutant lacking a total of seven PBPs, controlled expression of PBP 4, PBP 6, or DacD did not. Also, a truncated PBP 5 protein lacking its amphipathic C-terminal membrane binding sequence did not reverse the morphological defects and was lethal at low levels of expression, implying that membrane anchoring is essential for the proper functioning of PBP 5. By examining a set of mutants from which multiple PBP genes were deleted, we found that significant morphological aberrations required the absence of at least three different PBPs. The greatest defects were observed in cells lacking, at minimum, PBPs 5 and 6 and one of the endopeptidases (either PBP 4 or PBP 7). The results further differentiate the roles of the low-molecular-weight PBPs, suggest a functional significance for the amphipathic membrane anchor of PBP 5 and, when combined with the recently determined crystal structure of PBP 5, suggest possible mechanisms by which these PBPs may contribute to maintenance of a uniform cell shape in E. coli.
To synthesize or modify its peptidoglycan cell wall, Escherichia coli maintains 12 penicillin binding proteins (PBPs), most of which have no demonstrable physiological purpose. Although each of these enzymes mediates one or more known biochemical reactions, most are not essential because E. coli survives without them (8). Traditionally, these proteins are separated into two subfamilies, only one of which contributes to cell survival. The high-molecular-weight (HMW) PBPs (PBPs 1a, 1b, 1c, 2, and 3) are bifunctional transglycosylase/transpeptidases or monofunctional transpeptidases which synthesize and incorporate individual peptidoglycan strands into the murein sacculus (9, 10, 19, 26). Each of these HMW PBPs except PBP 1c has a defined physiological function (19, 26, 27, 29, 30). In contrast, the functions of the seven low-molecular-weight (LMW) PBPs remain enigmatic (3, 8, 13).
The LMW PBPs (PBPs 4, 5, 6, and 7, DacD, AmpC, and AmpH) are subdivided into four enzymatic classes: three monofunctional dd-carboxypeptidases, PBP 5, PBP 6, and DacD; one bifunctional dd-carboxypeptidase/dd-endopeptidase, PBP 4; one dd-endopeptidase, PBP 7; and two class C β-lactamases, AmpC and AmpH (3, 12–14, 17, 24). Among these, the dd-carboxypeptidases have been studied most extensively, but a cohesive picture of their in vivo roles has yet to be established. PBPs 4, 5, and 6 and DacD cleave the terminal d-alanine from the pentapeptide side chains of murein components (3). The considerable sequence identities and structural similarities among these proteins suggest that they all diverged from an ancient dd-carboxypeptidase, with subsequent additions and modifications of specific functional domains (16, 21).
These similarities and genetic data have led to the widespread assumption that the dd-carboxypeptidases act as functional equivalents in vivo, so that these multiple carboxypeptidases may compensate for one another in situations where one is lost or inactivated (3, 5). Unfortunately, this “equivalent substitution” hypothesis has been difficult to test because these proteins are not essential for bacterial survival. For example, a quadruple mutant grows normally even though it lacks PBPs 4, 5, and 6 and DacD (3, 8), and mutants lacking all seven LMW PBPs grow well (8). However, there is a small amount of evidence for differences among the LMW PBPs. As discussed in more detail below, morphological defects accompany the loss of PBP 5 but of no other PBP (22). Also, deletion of the dacA gene (encoding PBP 5) substantially reverses the filamentation phenotype of a temperature-sensitive ftsK allele, whereas deletion of dacB (encoding PBP 4) or dacC (encoding PBP 6) does not (4). In addition, overexpression of either PBP 5 or PBP 6, but not PBP 4, reverses the effects of a specific temperature-sensitive allele of PBP 3 (5), implying that PBPs 5 and 6 (but not 4) might perform similar functions in vivo. Finally, it has been reported that although overexpression of PBP 5 causes E. coli to become spherical and eventually lyse (18), PBP 4, PBP 6, and DacD are nonlethal (3, 17, 33), suggesting that these latter PBPs might differ from PBP 5. Because so few of these types of observations have been made, the idea of universal interchangeability among these proteins persists in the literature and minds of many workers, and none of the observations has brought us closer to understanding what these PBPs are doing. In short, the paucity of testable phenotypes has limited experimental approaches to the question of the biological function of the LMW PBPs.
Recently, we reported that PBP 5 helps maintain normal cell wall morphology and diameter in E. coli (22). By comparing isogenic mutants, we determined that no other LMW PBP could substitute for PBP 5 to correct these defects in vivo at wild-type expression levels. In this report we compare additional isogenic mutant strains to determine the minimum complement of PBP deletions necessary to generate the morphological defects. We also show that no other dd-carboxypeptidase can complement the ΔdacA phenotype in trans, nor can a PBP 5 protein lacking its carboxy-terminal amphipathic membrane anchor. Finally, we establish that overexpressing each of the dd-carboxypeptidases does lyse host cells and show that such lysis occurs only during early logarithmic growth. Collectively, the results place constraints on the equivalent substitution hypothesis for the LMW PBPs and allow us to propose a speculative mechanism by which these proteins contribute to the maintenance of uniform cell shape in E. coli.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The E. coli strains used in this work are listed in Table 1. Strains carrying the dacD insertion-deletion mutation were constructed by moving dacD::Kan536-2 from CS18-2K (8) into the indicated strains by P1 transduction (20). Plasmid vectors pBAD18-Cam and pBAD24-Amp were provided by J. Beckwith (11). Bacterial strains were maintained on Luria-Bertani (LB) broth or agar plates, supplemented when appropriate with chloramphenicol (20 μg/ml) or ampicillin (50 μg/ml) to maintain plasmids or with glucose (0.1%) to inhibit expression from pBAD promoters. Overnight cultures for morphological determinations were diluted 1:500 into fresh LB broth supplemented with 0.005 to 0.1% (wt/vol) arabinose and were incubated at 37°C with shaking until reaching an A600 of approximately 0.2. When required, aztreonam (Bristol Meyers-Squibb, New York, N.Y.) was added to a final concentration of 10 μg/ml. Other chemicals were purchased from Sigma Chemical Co. (St. Louis, Mo.) unless otherwise noted. Growth curves for determining lysis after overexpression of PBPs and for comparisons of growth rates were performed in flask assays and by growth in a Bioscreen C Microbiology Reader (Labsystems, Helsinki, Finland).
TABLE 1.
Bacterial strains and plasmids used
| Strain or plasmid | Genotype or characteristics | PBP(s) deleted | Source or reference |
|---|---|---|---|
| Strains | |||
| CS109 | W1485 rpoS rph | None | C. Schnaitman |
| CS18-2K | CS109 dacD::Kan536-2 | DacD | 8 |
| CS204-1 | CS109 dacA pbpG | 5, 7 | 8 |
| CS371-1K | CS204-1 dacD::Kan536-2 | 5, 7, DacD | This work |
| CS205-1 | CS109 dacC pbpG | 6, 7 | 8 |
| CS372-1K | CS205-1 dacD::Kan536-2 | 6, 7, DacD | This work |
| CS219-1 | CS109 dacB dacA | 4, 5 | 8 |
| CS373-1K | CS219-1 dacD::Kan536-2 | 4, 5, DacD | This work |
| CS220-1 | CS109 dacB dacC | 4, 6 | 8 |
| CS374-1K | CS220-1 dacD::Kan536-2 | 4, 6, DacD | This work |
| CS604-2 | CS109 mrcA dacB dacC pbpG ampC ampH | 1a, 4, 6, 7, AmpC, AmpH | 8 |
| CS701-1 | CS109 mrcA dacB dacA dacC pbpG ampC ampH | 1a, 4, 5, 6, 7, AmpC, AmpH | 8 |
| Plasmids | |||
| pBAD24 | Expression vector, Ampr | 11 | |
| pBAD18 | Expression vector, Camr | 11 | |
| pPJ4 | dacB cloned between the NheI and XbaI sites of pBAD18 | This work | |
| pPJ5C | dacA cloned between the NheI and HindIII sites of pBAD18 | 22 | |
| pPJ5D | C-terminally truncated dacA cloned between the NheI and HindIII of pBAD18 | This work | |
| pPJ5S | dacAS44G cloned between the NheI and XbaI sites of pBAD18 | This work | |
| pPJ6 | dacC cloned between the NheI and XbaI sites of pBAD18 | This work | |
| pPJDacD | dacD cloned between the NheI and XbaI sites of pBAD18 | This work |
Microscopic evaluation of morphological aberration.
To compare the relative morphologies of the different E. coli strains, cultures were diluted to an A600 of 0.005 and grown in LB at 37°C, and samples were removed during early logarithmic growth phase (A600 = 0.2 ± 0.02). Photographs were collected from random microscopic fields as described previously (22), and cell populations were evaluated by two to three individuals for the degree of morphological aberration. In our earlier work we could measure and quantify alterations in cell diameter (22). However, the morphological differences that we observed in this study did not lend themselves to straightforward quantification. Therefore, the results are expressed as a subjective measure of relative morphological aberration based on differences in the numbers of cells affected and the degree to which they exhibited increased diameters, uneven contours, squaring of the poles, lateral branching, and filamentation. The extent of these differences is reported on a relative scale of 0 (no observable differences from the parental strain) to 4 (numerous and extreme shape differences).
Molecular and biochemical techniques.
Plasmid preparations and DNA isolation from agarose gels were performed with commercially available kits from Qiagen (Valencia, Calif.). Competent strains were prepared and transformed by using a Bio-Rad (Hercules, Calif.) Gene Pulser electroporation apparatus according to the manufacturer's instructions. Chromosomal DNA was isolated as described previously (22). Oligonucleotides for PCR were purchased from Gibco Life Sciences (Grand Island, N.Y.). PBP expression was confirmed by using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by silver staining (25) or by labeling the PBPs with 125I-penicillin X as described previously (13). The amounts of PBPs produced were quantified by imaging SDS-PAGE gels with a Molecular Dynamics 445SI PhosphorImager (Amersham Pharmacia, Uppsala, Sweden) and measuring band intensities by using ImageQuant software (Amersham Pharmacia, Uppsala, Sweden). Enzymes and reagents were purchased from New England Biolabs (Beverly, Mass.).
Placing PBP genes under control of the arabinose promoter.
Primers B1 (5′-CTCTCTCTGAATTCTTATGCGATTTTCCAG-3′) and B2 (5′-CTCTCTCTCTAGACTTTTTGACTAATTGTTCTG-3′) were used to PCR amplify the dacB (PBP 4) gene from the CS109 chromosome. The EcoRI-XbaI fragment of this PCR product was ligated into pBAD24-Amp to create plasmid pPJ4-Amp. Afterwards, the NheI-XbaI fragment was excised from pPJ4-Amp and ligated into pBAD18-Cam, creating the PBP 4 expression vector, pPJ4. Primer pairs C1 (5′-CTCTTTTGCTAGCAGGAGGAATTCACCATGACGCAATACTCCTCTC-3′) plus C2 (5′-CTCTCTCTCTAGAAGAAGAATTAGAGAACCAGCTGC-3′) and D1 (5′-CTCTCTCGCTAGCAGGAGGAATTCACCATGCTGTTGAAACGCCGTC-3′) plus D2 (5′-CCCCCCCTCTCAGATCTGCAAAAGAAAGGTCAGGCCTTATG-3′) were used to PCR amplify the wild-type genes dacC and dacD, respectively, from the CS109 chromosome. The NheI-XbaI fragments of the dacC and dacD PCR products were ligated into pBAD18-Cam to create the PBP expression vectors pPJ6 and pPJDacD, respectively. Primers E1 (5′-CGCGGCGGCTAGCAGGAGGAATTCGTCATGAATACCATTTTTTCCGC-3′) and E2 (5′-CCAAGGATTCTAGAAGCTTTTTTTAGTTACCTTCCGGGATTTC-3′) were used to PCR amplify a truncated fragment of the dacA gene from the PBP 5 expression vector pPJ5C (22). This fragment encoded the full-length PBP 5 protein minus the 54 3′-terminal nucleotides of the dacA coding sequence, so that the C-terminal amphipathic anchor of PBP 5 was deleted. The NheI-XbaI fragment of this PCR product was ligated into pBAD18-Cam, creating plasmid pPJ5D. pPJ5S was constructed by using a Stratagene QuickChange site-directed mutagenesis kit and primers F1 (5′-CGCCGCGATCCTGCCGGCCTGACCAAAATGATG-3′) and F2 (5′-CATCATTTTGGTCAGGCCGGCAGGATCGCGGCG-3′), with pPJ5C as the template (Stratagene Co., La Jolla, Calif.). All clones were sequenced commercially at Colorado State University using the ABI Prism Terminator Cycle Sequencing technique (Fort Collins, Colo.).
RESULTS
Previously, we began to determine which PBPs were necessary to maintain the normal morphology of E. coli by comparing isogenic strains that retained only one or two active LMW PBPs, in mutants from which six to seven PBP genes had been deleted (22). Although loss of PBP 5 was the primary determinant of aberrant morphology, deletion of dacA alone had little effect on the wild-type strain (22). Thus, it was possible that an unknown combination of different PBPs could take the place of PBP 5. Therefore, to further clarify the role of PBP 5, and to determine the minimum complement of deletions necessary to yield the morphological phenotype, we examined the morphology of 41 single, double, and triple PBP mutants from our comprehensive PBP mutant set (8).
Morphological defects are associated with deletion of dacA.
In agreement with previous findings (22), no significant morphological defects were associated with loss of a single PBP (data not shown). In particular, the dacA mutant, CS12-7, showed no significant differences from the wild-type strain, CS109 (Table 2; Fig. 1A and B), and though a few aztreonam-induced filaments of CS12-7 were slightly rounded or branched, the numbers were too small to quantify (data not shown). Of special importance was that no double or triple mutant in which PBP 5 remained active exhibited significant morphological oddities (data not shown), once again implicating PBP 5 as the dominant determinant for this phenotype.
TABLE 2.
Relative morphological defects of PBP mutants
| Straina | Relative morphology scoreb | PBPsc
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1b | 1a | 4 | 5 | 6 | 7 | C | H | DacD | ||
| CS322-1 | 4 | + | + | 0 | 0 | 0 | + | + | + | + |
| CS331-1 | 4 | + | + | + | 0 | 0 | 0 | + | + | + |
| CS325-1 | 3 | + | 0 | 0 | 0 | + | + | + | + | + |
| CS345-3 | 3 | + | + | + | 0 | + | 0 | + | 0 | + |
| CS362-1 | 3 | + | 0 | + | 0 | + | + | 0 | + | + |
| CS373-1K | 3 | + | + | 0 | 0 | + | + | + | + | 0 |
| CS215-3 | 2 | + | + | + | 0 | + | + | + | 0 | + |
| CS309-1 | 2 | 0 | + | + | 0 | + | + | 0 | + | + |
| CS315-1 | 2 | + | + | 0 | 0 | + | 0 | + | + | + |
| CS326-3 | 2 | + | + | 0 | 0 | + | + | + | 0 | + |
| CS371-1K | 2 | + | + | + | 0 | + | 0 | + | + | 0 |
| CS204-1 | 1.5 | + | + | + | 0 | + | 0 | + | + | + |
| CS310-2 | 1.5 | 0 | + | 0 | 0 | + | + | + | + | + |
| CS327-1 | 1.5 | + | + | 0 | 0 | + | + | 0 | + | + |
| CS346-1 | 1.5 | + | + | + | 0 | + | 0 | 0 | + | + |
| CS349-1 | 1.5 | + | + | + | 0 | + | + | 0 | 0 | + |
| CS357-3 | 1.5 | + | 0 | + | 0 | + | + | + | 0 | + |
| CS211-2 | 1 | + | + | + | 0 | 0 | + | + | + | + |
| CS301-1 | 1 | 0 | + | + | 0 | + | 0 | + | + | + |
| CS316-1 | 1 | + | + | 0 | + | 0 | 0 | + | + | + |
| CS336-3 | 1 | + | + | + | 0 | 0 | + | + | 0 | + |
| CS337-1 | 1 | + | + | + | 0 | 0 | + | 0 | + | + |
| CS361-1 | 1 | + | 0 | 0 | + | + | + | 0 | + | + |
| CS363-1 | 1 | + | 0 | + | + | 0 | + | 0 | + | + |
| CS366A | 1 | + | 0 | + | 0 | 0 | + | + | + | + |
| CS366B | 1 | + | 0 | + | 0 | 0 | + | + | + | + |
| CS372-1K | 1 | + | + | + | + | 0 | 0 | + | + | 0 |
| CS374-1K | 1 | + | + | 0 | + | 0 | + | + | + | 0 |
| CS109 | 0 | + | + | + | + | + | + | + | + | + |
| CS12-7 | 0 | + | + | + | 0 | + | + | + | + | + |
| CS214-1 | 0 | + | 0 | + | 0 | + | + | + | + | + |
| CS216-2 | 0 | + | + | + | 0 | + | + | 0 | + | + |
| CS219-1 | 0 | + | + | 0 | 0 | + | + | + | + | + |
| CS224-2 | 0 | 0 | + | + | 0 | + | + | + | + | + |
| CS305-1 | 0 | 0 | + | + | 0 | 0 | + | + | + | + |
| CS308-3 | 0 | 0 | + | + | 0 | + | + | + | 0 | + |
| CS318-1 | 0 | + | + | 0 | + | + | 0 | 0 | + | + |
| CS319-1 | 0 | + | 0 | 0 | + | + | + | + | 0 | + |
| CS343-1 | 0 | + | + | + | + | 0 | 0 | 0 | + | + |
| CS354-3 | 0 | + | 0 | + | + | 0 | + | + | 0 | + |
| CS356-1 | 0 | + | 0 | + | 0 | + | 0 | + | + | + |
Unless otherwise described in Materials and Methods, the derivation of all other strains is as described by Denome et al. (8).
Relative degrees of morphological abnormalities were assessed visually and ranked on a scale of 0 (no difference from the parental E. coli strain, CS109) to 4 (extreme morphological alterations affecting most cells in culture).
+, present; 0, absent.
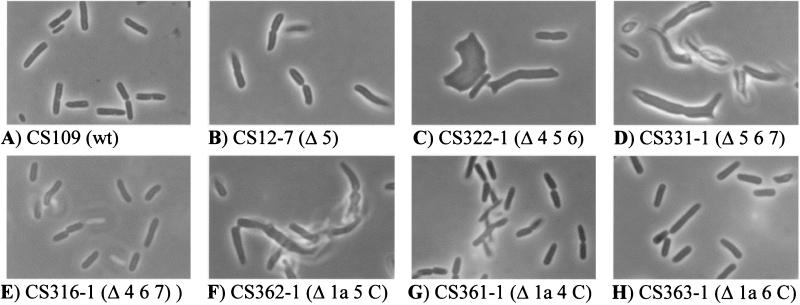
FIG. 1.
Morphology of PBP mutants of E. coli. Overnight cultures of E. coli strains were diluted 1:500 into fresh LB broth and grown at 37°C until reaching an A600 of 0.2. Cells were collected, prepared for microscopy, and photographed at a magnification of ×1,000. All photographs are displayed at equal magnification. Abbreviations: 4, PBP 4; 5, PBP 5; 6, PBP 6; 7, PBP 7; C, AmpC; wt, wild-type parent.
Although deletion of PBP 5 alone had little effect on cellular morphology, observable defects were augmented by loss of additional PBPs. The differences among mutants lacking PBP 5 were assessed subjectively by inspection of multiple photographs of cell cultures, and the strains were organized according to their relative degree of morphological aberration (Table 2). Because of their subjective nature, these rankings should be considered as a rough guide to the degree of morphological differences between strains. Nonetheless, large differences in scoring (>2 units) do depict significant differences in the frequency and/or degree of cell shape aberrations.
The mutants exhibiting the most dramatic morphological defects were those lacking PBPs 5 and 6 and either PBP 4 or PBP 7 (strains CS322-1 and CS331-1, respectively) (Table 2; Fig. 1C and D). On the other hand, the morphology of strain CS316-1, which is missing PBPs 4, 6, and 7 (described as Δ467), was nearly normal (Table 2; Fig. 1E), underscoring the importance of PBP 5 for this phenotype. This combination, loss of PBP 5 and at least one endopeptidase (PBP 4 or 7), correlated most strongly with overall morphological aberration. Eight of the 11 strains displaying abnormalities greater than 2 on the relative scale were missing PBP 5 and one of the endopeptidases, and 12 of the 17 strains scoring 1.5 or greater contained this combination of deletions. Of the 15 strains lacking PBP 5 and either PBP 4 or PBP 7, only three were of normal shape (scores below 1) (Table 2). The five strains scoring 1.5 or greater and which did not fit this pattern were missing PBP 5 and either AmpC or AmpH (or both), consistent with what we reported previously (13). Thus, two mutant combinations accounted for the most extreme morphological alterations: deletion of PBP 5 and an endopeptidase (PBP 4 or 7), which provoked the greatest deviations from normal morphology, followed in degree by deletion of PBP 5 plus one of the class C β-lactamases (AmpC or AmpH).
The data reinforced the primary role for PBP 5 in creating morphologically uniform cells. For example, strain CS362-1 (Δ1a 5 AmpC) was unable to maintain an even contour, had drastic alterations in diameter, and produced increased numbers of branched cells (Fig. 1F). In contrast, its isogenic relatives CS361-1 (Δ1a 4 AmpC) and CS363-1 (Δ1a 6 AmpC) retained near-wild-type morphology (Fig. 1G and H). In these latter two strains, PBPs 4 and 6 were deleted instead of PBP 5, confirming that the loss of neither of these proteins created the same morphological aberrations as did loss of PBP 5. Nor did the presence of PBPs 4 or 6 substitute for the absence of PBP 5 in CS362-1. Similarly, though CS346-1 (Δ5 7 AmpC) exhibited morphological defects, two isogenic relatives, CS318-1 (Δ4 7 AmpC) and CS343-1 (Δ6 7 AmpC), had wild-type morphology (Table 2). As before, deletion of PBP 5, but not PBP 4 or 6, provoked the aberrations. Comparisons of other strains yielded the same results for the activity of PBP 5 versus all other PBPs. Thus, although the absence of additional LMW PBPs augmented the morphological defects of dacA mutants, in the presence of active PBP 5, these other PBPs could not precipitate such defects when deleted individually or in combination.
Only PBP 5 complements the morphological defects of dacA mutants.
The preceding experiments addressed the question of whether wild-type levels of different LMW PBPs could substitute for PBP 5. It remained possible that these other PBPs might complement the loss of PBP 5 if they were expressed in higher quantities. Therefore, to determine which of E. coli's dd-carboxypeptidases could complement the morphological defects of dacA mutants, we cloned the dacB, dacC, and dacD genes of wild-type strain CS109 into the arabinose-inducible expression vector pBAD18-Cam (11). The PBP subclones were transformed into CS109 and into the septuple PBP mutant CS701-1, and the viability of each strain was evaluated across a range of arabinose concentrations (0.1 to 0.0001%) using a Bioscreen C Microbiology Reader to determine the most effective inducing conditions (data not shown). Final arabinose concentrations for gene induction and complementation studies were selected by comparing the growth rates of cells harboring the PBP expression vectors to those of identical strains containing the control vector pBAD18-Cam, supplemented by subjective visual assessment of morphology.
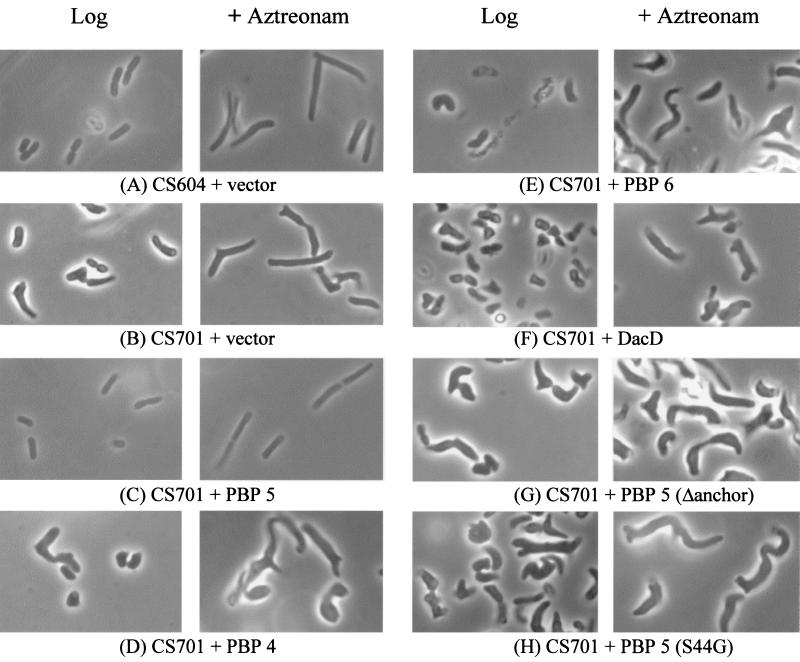
Cells were photographed during log-phase growth and again 45 min after treatment with aztreonam (10 μg/ml) to enhance any morphological aberrations by filamenting the cells. As reported previously (22), E. coli CS604-2(pBAD18-Cam), which lacks six PBPs but retains wild-type dacA, was morphologically similar to wild-type E. coli (Fig. 2A). In contrast, CS701-1(pBAD18-Cam), which is isogenic to CS604-2(pBAD18-Cam) but lacks the dacA gene, exhibited drastic alterations in morphology both before and after filamentation induced by aztreonam (Fig. 2B). Regulated expression of PBP 5 in CS701-1(pPJ5C) reversed these defects, and the complemented strain was visually indistinguishable from CS604-2(pBAD18) (Fig. 2C). In contrast, expression of cloned PBP 4 (Fig. 2D), PBP 6 (Fig. 2E), or DacD (Fig. 2F) did not complement the morphological defects of CS701-1 at any level of expression. In fact, in most cases the severity of the morphological defects in CS701-1 was exacerbated by expression of these non-PBP 5 dd-carboxypeptidases (Fig. 2D to F).
FIG. 2.
Complementation of PBP mutants by dd-carboxypeptidases. E. coli strains containing plasmids with or without cloned PBP genes were diluted 1:500 from overnight cultures into fresh LB broth supplemented with arabinose (0.005%, wt/vol) and were incubated at 37°C. When the cultures reached an A600 of 0.2, aztreonam was added to a final concentration of 10 μg/ml. Cells were photographed immediately before (Log) and 45 mins after (+ Aztreonam) aztreonam addition. All photographs are displayed at equal magnification. Abbreviations: vector, pBAD18-Cam; Δanchor, truncated PBP 5 missing the carboxy-terminal 18 amino acids; S44G, PBP 5 with serine-to-glycine substitution at amino acid 44, inactivating the active site.
Both the active site and membrane anchoring terminus of PBP 5 are required for complementation.
PBP 5 has three known domains: one containing the dd-carboxypeptidase activity, one responsible for localizing PBP 5 to the outer face of the cytoplasmic membrane, and a third domain of unknown function between these other two (7, 15, 23, 34). To determine if the enzymatic and membrane-binding activities were necessary to complement the morphological phenotype of dacA mutants, we constructed vectors to express mutant PBP 5 proteins in which one or the other of these two domains was inactivated. Plasmid pPJ5D expressed a truncated mutant protein lacking the carboxy-terminal 18 amino acids that anchor PBP 5 to the membrane. Plasmid pPJ5S expressed a full-length PBP 5 in which a Ser44-to-Gly44 mutation eliminated the protein's dd-carboxypeptidase activity (34).
Truncated PBP 5 expressed from pPJ5D was unable to complement the morphological defects of CS701-1, either before or after treatment with aztreonam (Fig. 2G). In fact, even low-level expression of the truncated protein was lethal or actually augmented the morphological abnormalities observed in these cells, indicating that membrane association of PBP 5 is important for its proper functioning in vivo. Similarly, enzymatically inactive PBP 5 encoded by pPJ5S was unable to complement the defects in CS701-1 (Fig. 2H), indicating that the dd-carboxypeptidase activity was essential for creating morphologically normal cells.
All dd-carboxypeptidases are lytic when overexpressed early in the growth phase.
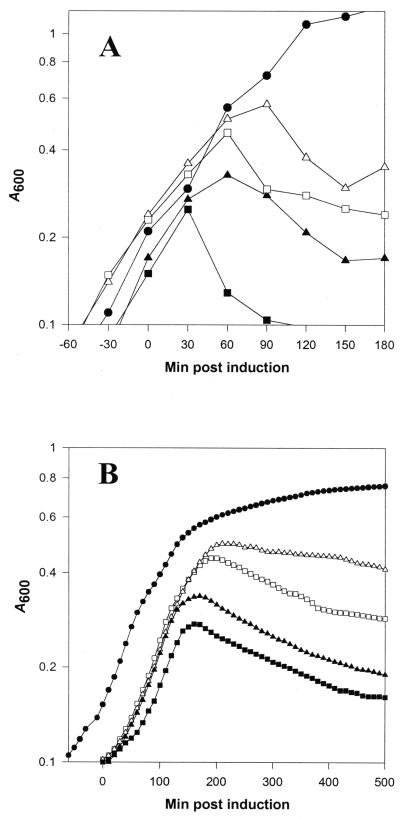
While performing complementation studies, we noted that prolonged expression of each of the dd-carboxypeptidases decreased the viability of fast-growing cultures, but that this effect diminished in slow-growing or stationary-phase cells. However, previous reports suggested that only PBP 5 induced lysis when overexpressed. Therefore, to clarify the differences between our observations and previous experiments, we overexpressed each of the dd-carboxypeptidases in E. coli CS109. All active dd-carboxypeptidases lysed CS109 within 2 h of gene induction in flask assays (Fig. 3A), and viable cell counts in these cultures dropped 2 logs within 2 h of protein induction (data not shown). The onset of lysis in cultures grown in the Bioscreen C Microbiology Reader was delayed and the rate of lysis was reduced, but the results were similar in that all of the dd-carboxypeptidases lysed E. coli (Fig. 3B). In each case, the microscopic effects of overexpressing the carboxypeptidases were similar: cells grew normally for a brief period then rapidly became spherical and lysed shortly thereafter (data not shown). This is consistent with the effects previously described for PBP 5 (28). Lysis, but not lethality, was inhibited by addition of 15% sucrose to osmotically stabilize the labile spheroplasts, and lysis did not occur if protein synthesis was interrupted by adding tetracycline (12.5 μg/ml) 30 min after gene induction (though lysis occurred normally if tetracycline was added 60 min after induction) (data not shown).
FIG. 3.
Lysis of E. coli by dd-carboxypeptidase overexpression. Overnight cultures of E. coli CS109 containing plasmids with or without cloned PBP genes were diluted 1:500 into fresh LB broth and incubated at 37°C in flasks in a shaking water bath (A) or in the Bioscreen C Microbiology Reader (B). When the A600 reached 0.2 (for flask cultures) or 0.15 (in the Bioscreen Reader), arabinose (0.1%, wt/vol) was adding to induce PBP gene expression. CS109(pBAD18), ●; CS109(pPJ4), ■; CS109(pPJ5C), ▴; CS109(pPJ6), ▵; CS109(pPJDacD), □.
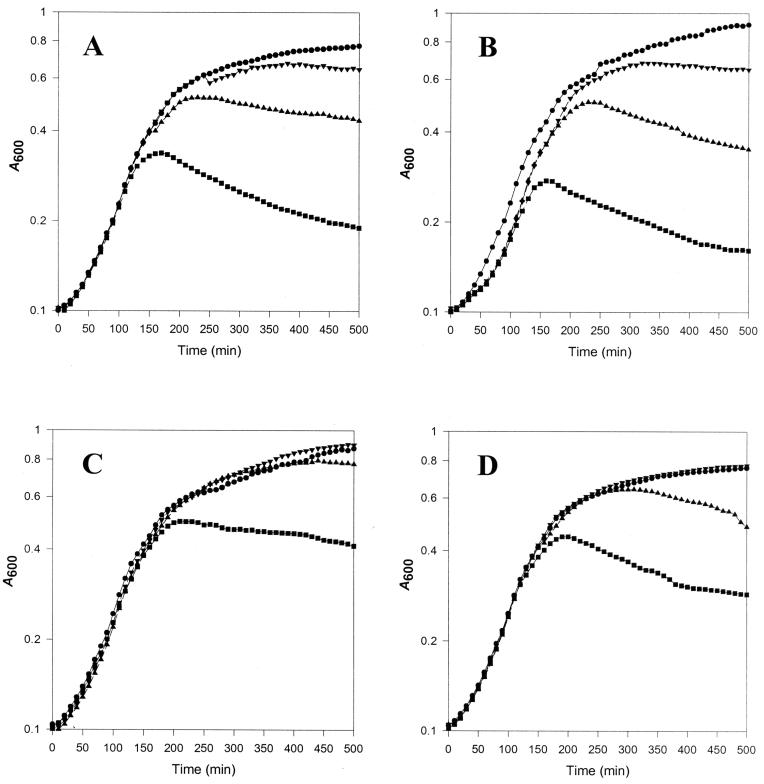
Because these lysis phenotypes were at odds with those reported previously, we examined possible reasons for the discrepancy. In the earlier studies in which dd-carboxypeptidase overexpression failed to lyse E. coli, the proteins were not expressed until cultures reached the mid or late logarithmic growth phase. Therefore, to determine if lysis sensitivity was influenced by growth phase, we induced the carboxypeptidases in cultures at different times in the growth cycle. Growth was monitored in the Bioscreen C Microbiology Reader as before, but the genes were induced by adding arabinose at time points corresponding to early, mid, and late log phases. As cells progressed from early to late log phase, induction of the various PBPs had a decreasing lytic effect (Fig. 4). This was not because PBP expression was reduced; indeed, in each case, high levels of PBPs were expressed at these times, as determined by labeling the PBPs and visualizing them by SDS-PAGE (data not shown). Thus, each of the dd-carboxypeptidases was lytic when overexpressed, but cell cultures were more sensitive to lysis during early logarithmic growth and became increasingly resistant as they approached stationary phase.
FIG. 4.
Effect of growth phase on lysis of E. coli by overexpression of dd-carboxypeptidases. E. coli CS109 was transformed with the following plasmids carrying the indicated PBPs under control of the arabinose promoter: (A) pPJ5C, PBP 5; (B) pPJ4, PBP4; (C) pPJ6, PBP 6; and (D) pPJDacD, DacD. Overnight cultures were diluted 1:500 into fresh LB broth and incubated at 37°C in a Bioscreen C Microbiology Reader. PBP expression was induced at three different times in the growth cycle by adding arabinose to a final concentration of 0.1% (wt/vol). No arabinose, ●; arabinose added at an A600 of 0.15, ■; arabinose added at an A600 of 0.30, ▴; arabinose added at an A600 of 0.60, ▾.
Removal of the C-terminal membrane anchor increases lethality of PBP 5.
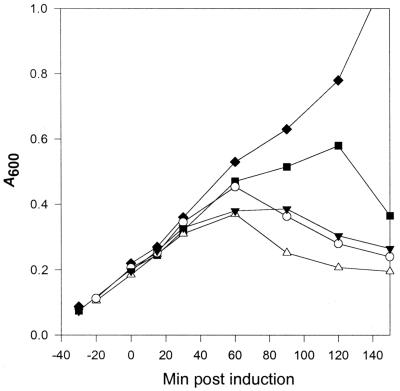
That the membrane anchoring domain of PBP 5 may serve a physiological purpose was suggested by the observation that the carboxy-truncated form of PBP 5 did not complement CS701-1 and, instead, apparently exacerbated morphological defects in the strain. In light of these results, we wished to determine if the membrane anchor affected the lytic activity of overexpressed PBP 5. Lysis of CS109 by wild-type PBP 5 (from pPJ5) and lysis by carboxy-truncated PBP 5 (from pPJ5D) were compared in flask assays (data not shown) and in the Bioscreen C Microbiology Reader, using a gradient of arabinose concentrations to induce different amounts of each cloned protein (Fig. 5). Although overexpression of truncated PBP 5 did not appreciably reduce the time of lysis onset, lysis required a substantially lower concentration of truncated PBP 5 than wild-type PBP 5. A 7-fold overexpression of truncated PBP 5 lysed CS109 with kinetics that were comparable to a 65-fold overexpression of the full-length protein (Fig. 5). Even a fourfold overexpression of truncated PBP 5 was rapidly lethal (Fig. 5). Roughly speaking, compared to the overexpression of wild-type PBP 5, only 1/10 of the amount of truncated PBP 5 was required to induce a comparable amount and extent of lysis.
FIG. 5.
Lysis of E. coli by overexpression of wild-type or carboxy-terminal truncated PBP 5. Overnight cultures of E. coli CS109 containing either pPJ5 (to express wild-type PBP 5) or pPJ5D (to express PBP 5 lacking the carboxy-terminal membrane anchor) were diluted 1:500 into fresh LB broth and incubated with shaking in flasks at 37°C to an A600 of 0.2. At that point, arabinose was added at different final concentrations to induce different levels of PBP expression. Samples for PBP quantification were collected before and 30 mins after induction of protein expression, and the amounts of wild-type and truncated PBP 5 expressed from equal numbers of cells were quantified by comparing the intensities of PBP bands after separation by SDS-PAGE. The amount of wild-type PBP 5 expressed from a single chromosomal copy of the dacA gene was regarded to be the 1× expression level, and amounts of overexpression of the wild-type and truncated PBP 5 forms are expressed as multiples of this baseline figure. PBP 5 (1× expression), ⧫; PBP 5 (12× expression), ■; PBP 5 (65× expression), ▾; carboxy-truncated PBP 5 (4× expression), ○; carboxy-truncated PBP 5 (7× expression), ▵.
DISCUSSION
Despite 25 years of work, little is known about the biologically relevant roles of the dd-carboxypeptidase PBPs in E. coli. Previously, we noted that deletion of PBP 5 altered the antibiotic resistance and morphology of several PBP mutants (22), representing the first phenotype associated with the loss of one of the dd-carboxypeptidases in an otherwise wild-type background. These observations enabled us to perform a direct test of the idea that several of these proteins have similar or identical functions, an idea that we call the equivalent substitution hypothesis. To date, little or no evidence contradicts this assertion of interchangeability (with the possible exception of two reports describing the effects of dd-carboxypeptidases on non-PBP mutants [4, 5]). Our findings indicate that PBP 5 has a unique role among the dd-carboxypeptidases in the development of normal bacterial shape and that no other PBP can completely substitute for PBP 5 in this regard.
PBP mutant combinations and abnormal morphology.
Although all strains with morphological abnormalities lacked PBP 5, the extent of deviation in diameter, contour, branching, and other defects varied greatly among mutants from which PBP 5 was deleted. The most dramatic aberrations appeared in cells lacking at least two additional PBPs and were correlated with two prominent combinations of mutations: deletion of PBP 5 and one of the endopeptidases (PBP 4 or 7), or deletion of PBP 5 and one of the class C β-lactamases (AmpC or AmpH). The most significant effects were in cells lacking PBP 5 and either PBP 4 or PBP 7 in combination with a deletion of either PBP 6 (in strains exhibiting the greatest alterations) or DacD, suggesting that active PBP 6 or DacD might moderate the morphological effects of losing PBP 5. Since these latter two enzymes are those most closely related to PBP 5, the results may be consistent with a weak form of the substitution hypothesis in which PBP 6 or DacD might perform some of the functions of PBP 5 with lower efficiency.
An important observation was that the mutational patterns associated with altered morphology were strongly correlated with the classical groupings of PBPs based on sequence similarity and biochemical activity; that is, the dd-carboxypeptidases, endopeptidases, and class C β-lactamases. This is a credible indication that similar enzymes may perform similar functions within the cell. However, it is clear that no one enzyme can carry out all of the physiological functions of the other members in its group. For example, deletion of either PBP 4 or PBP 7 from a mutant lacking PBPs 5 and 6 produces remarkable morphological effects, even though one or the other of the endopeptidases remains active. The same can be said of PBP 5 mutants lacking one or the other of the class C β-lactamases (AmpC or AmpH). Thus, for each biochemical group, losing the activity of a single PBP creates a noticeable phenotype, indicating that the enzymes within each classification are not equivalent and do not simply duplicate one another's functions. This is a compelling argument against the strong form of the substitution hypothesis.
Possible model for PBP 5 action.
How might PBP 5 contribute to the construction of a normally shaped cell? Theoretically, PBP 5 could contribute a specific and necessary enzymatic activity or the protein could be a structural component of a larger morphological complex (although the two possibilities are not mutually exclusive). We approached this question by creating two PBP 5 derivatives: one enzymatically inactive, and the other truncated to remove its membrane anchor. Neither derivative complemented dacA mutants, suggesting that PBP 5 must be both enzymatically active and properly localized to create the normal morphological form of E. coli. Although the data do not address whether PBP 5 acts in concert with other proteins, they do diminish the possibility that PBP 5 acts solely as a structural protein.
This is the first demonstration that membrane localization of a LMW PBP has physiological significance, because removal of the membrane anchor prevented truncated PBP 5 from performing its normal morphological function. An alternate explanation is that removing the carboxyl terminus altered the enzymatic activity of PBP 5 so that losing the membrane anchor had only an indirect effect. However, two pieces of evidence argue against this interpretation. First, truncated PBP 5 retained the ability to bind radiolabeled penicillin, a property of the active site and an indication that the enzymatic activity was unchanged. Second, the shortened protein remained lethal when overexpressed, a characteristic that also requires a functional dd-carboxypeptidase. In fact, the lethality of anchorless PBP 5 was increased approximately 10-fold over its wild-type counterpart, suggesting not only that membrane localization of PBP 5 is necessary for proper morphological control but that such localization may regulate the numbers or types of substrates on which PBP 5 can act.
Given these results, there are at least two ways by which PBP 5 may function. First, the carboxy-terminal membrane anchor may play a relatively nonspecific role by restricting access of PBP 5 to its peptidoglycan substrate; second, the membrane anchor may play a more active role by interacting with proteins or specific regions of the inner membrane to confine PBP 5's activity to localized sites. Since there is no evidence for the second possibility, we will discuss only the first possibility at more length.
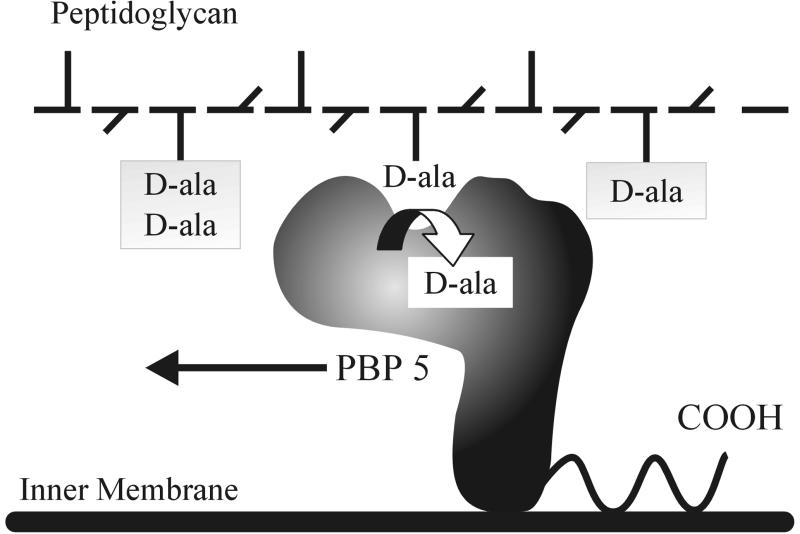
The recent determination of the crystal structure of a truncated form of PBP 5 (7) allows us to visualize a simple, speculative model by which the activity of PBP 5 might be constrained (Fig. 6). As reported by Davies et al. PBP 5 is composed of a globular domain, which contains the dd-carboxypeptidase active site on its outer face, and a domain of unknown function, which is composed almost entirely of β sheets (7) (represented schematically in Fig. 6). These two domains are connected so that they are oriented at right angles to one another, forming a head (the globular active site) and a stalk (the β-sheet domain) (7). Although absent from crystallized PBP 5, the extreme carboxy-terminal membrane-associated domain is predicted to be located at the base of the stalk, where the amphipathic helix could tether the protein to the outer face of the inner membrane (Fig. 6). If positioned as shown, PBP 5 might hydrolyze only those terminal d-alanine residues on peptide side chains that extend toward the inner membrane while being unable to hydrolyze residues on side chains in the plane of the cell wall or on the outer face of the peptidoglycan (Fig. 6). In this scenario, removing the membrane anchor may release PBP 5 from the membrane, allowing it to diffuse freely throughout the periplasm with increased access to all areas of the sacculus. This may explain the increased lethality of small amounts of the truncated protein because unregulated hydrolysis of normally protected side chains may prevent formation of essential structural cross-links.
FIG. 6.
Speculative structure and orientation of PBP 5 in the periplasm of E. coli. The schematic representation of PBP 5 (black inverted L-shaped form) is based on the crystal structure of PBP 5 as determined by Davies et al. (7). The dd-carboxypeptidase active site is depicted as an indentation in the predominately α-helical head portion of the molecule. The β-helical stalk is connected to the head to form a rough right angle, and the entire protein is tethered to the outer face of the inner membrane by a carboxy-terminal 18-amino-acid amphipathic helix (curved line). One peptidoglycan chain of N-acetylglucosamine-N-acetylmuramic acid subunits (horizontal dashes) is depicted. The peptide side chains attached to the N-acetylmuramic acid residues are illustrated by short lines extending above, below, or in the plane of the cell wall. As PBP 5 moves (arrow), the protein removes the terminal d-alanine residue (d-ala) from peptide side chains extending toward the inner membrane. Other side chains may be inaccessible to the normal activity of PBP 5.
This speculative model suggests a mechanism by which PBP 5 might maintain the smooth rod shape of the cell. In the absence of PBP 5, peptide side chains extending toward the inner membrane may remain available for inappropriate cross-linking with newly synthesized glycan chains, giving rise to misaligned sections of cell wall which manifest themselves as altered shapes of the peptidoglycan sacculus. If present, an endopeptidase (PBP 4 or 7) might remove these irregular cross-links, thus reversing or masking the morphological effects of losing PBP 5. Such a two-step pathway would be consistent with the fact that the most severely affected mutants lack PBP 5 and at least one of the endopeptidases. Of course, we do not know enough to explain why one endopeptidase cannot fulfill the role of the other, nor do we know enough about AmpC or AmpH to fit them into this scheme. Although the specifics of this putative morphological pathway must await further experimentation, the results do reinforce the argument that bacterial shape is not determined simply by general physical forces but that, instead, cell shape is monitored and manipulated by specific cellular proteins.
Overexpression of any dd-carboxypeptidase lyses E. coli.
As discussed above, excessive dd-carboxypeptidase activity may inhibit formation of a normal sacculus by interfering with the cross-linking of peptide side chains. Previous work supports this hypothesis for PBP 5 because overexpression of the protein causes E. coli to assume a rounded morphology, blocks cell division, and eventually leads to lysis after the cells grow to greatly increased diameters (17). On the other hand, overexpression of PBPs 4 and 6 was reported to be nonlethal (17, 18), and Baquero et al. reported no lysis after overexpression of DacD (3). However, with the exception of a single report calling the activity of PBP 6 into question (33), it has been consistently reported that PBPs 4, 5, and 6 and DacD are active dd-carboxypeptidases (1–3). Therefore, why should only PBP 5 induce cell lysis?
In contrast to earlier reports, we observed that overexpression of each of these other dd-carboxypeptidases was lethal and lytic. The difference between our results and those reported by other laboratories is most easily explained by the fact that in previous experiments PBP 4, PBP 6, and DacD were expressed late in logarithmic growth or near the beginning of stationary phase. However, we observed that these PBPs lysed cells only if the proteins were overexpressed in the early stages of logarithmic growth. This behavior is reminiscent of the lytic effects of β-lactams, to which cells also become more resistant as they grow slowly or as they approach stationary phase (31, 32). This phenotypic resistance phenomenon has never been explained on the molecular level, and it is unclear why the dd-carboxypeptidases would be less lethal during late log or stationary growth since peptidoglycan synthesis and turnover still occurs at these times (6).
Biological relevance of the dd-carboxypeptidases.
The dd-carboxypeptidases are not essential for bacterial viability (8), nor do they appear to completely substitute for one another. Therefore, ignoring the possibility that novel dd-carboxypeptidases remain undiscovered in E. coli, we entertain two scenarios which might explain the difficulty of ascribing functions to these proteins. First, a dd-carboxypeptidase activity may be essential in E. coli's normal ecological niche in the digestive tract of vertebrates but unnecessary in the zoo-like environment of laboratory culture. Thus, the phenotype could be subtle or undetectable except under special conditions. Second, the enzymes might contribute an incremental growth advantage to E. coli in the wild. In this case, the significance of defects caused by inactivation of one or more dd-carboxypeptidases might be overlooked. For example, there are small, but definite, differences in growth rates among our collection of multiple PBP mutants (unpublished results). Although a 1% difference in growth rate between two strains grown in flask culture would probably be inconsequential in the laboratory, the long-term selective advantage of such a difference in the wild can be significant.
ACKNOWLEDGMENTS
We thank Avery Paulson for help in evaluating the extent of morphological abnormalities among the mutants.
This work was supported by grant GM61019 from the National Institutes of Health. D. Nelson was supported by a North Dakota EPSCoR doctoral fellowship from the National Science Foundation.
REFERENCES
- 1.Amanuma H, Strominger J L. Purification and properties of penicillin-binding proteins 5 and 6 from Escherichia coli membranes. J Biol Chem. 1980;255:11173–11180. [PubMed] [Google Scholar]
- 2.Amanuma H, Strominger J L. Purification and properties of penicillin-binding proteins 5 and 6 from the dacA mutant strain of Escherichia coli (JE11191) J Biol Chem. 1984;259:1294–1298. [PubMed] [Google Scholar]
- 3.Baquero M-R, Bouzon M, Quintela J C, Ayala J A, Moreno F. dacD, an Escherichia coli gene encoding a novel penicillin-binding protein (PBP6b) with dd-carboxypeptidase activity. J Bacteriol. 1996;178:7106–7111. doi: 10.1128/jb.178.24.7106-7111.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Begg K J, Dewar S J, Donachie W D. A new Escherichia coli cell division gene, ftsK. J Bacteriol. 1995;177:6211–6222. doi: 10.1128/jb.177.21.6211-6222.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Begg K J, Takasuga A, Edwards D H, Dewar S J, Spratt B G, Adachi H, Ohta T, Matsuzawa H, Donachie W D. The balance between different peptidoglycan precursors determines whether Escherichia coli cells will elongate or divide. J Bacteriol. 1990;172:6697–6703. doi: 10.1128/jb.172.12.6697-6703.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blasco B, Pisabarro A G, de Pedro M A. Peptidoglycan biosynthesis in stationary-phase cells of Escherichia coli. J Bacteriol. 1988;170:5224–5228. doi: 10.1128/jb.170.11.5224-5228.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davies C, White S W, Nicholas R A. Crystal structure of a deacylation-defective mutant of penicillin-binding protein 5 at 2.3-Å resolution. J Biol Chem. 2001;276:616–623. doi: 10.1074/jbc.M004471200. [DOI] [PubMed] [Google Scholar]
- 8.Denome S A, Elf P K, Henderson T A, Nelson D E, Young K D. Escherichia coli mutants lacking all possible combinations of eight penicillin binding proteins: viability, characteristics, and implications for peptidoglycan synthesis. J Bacteriol. 1999;181:3981–3993. doi: 10.1128/jb.181.13.3981-3993.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghuysen J-M, Dive G. Biochemistry of the penicilloyl serine transferases. In: Ghuysen J-M, Hakenbeck R, editors. Bacterial cell wall. Amsterdam, The Netherlands: Elsevier Science B. V.; 1994. pp. 103–129. [Google Scholar]
- 10.Goffin C, Ghuysen J-M. Multimodular penicillin-binding proteins: an enigmatic family of orthologs and paralogs. Microbiol Mol Biol Rev. 1998;62:1079–1093. doi: 10.1128/mmbr.62.4.1079-1093.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guzman L-M, Belin D, Carson M J, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henderson T A, Templin M, Young K D. Identification and cloning of the gene encoding penicillin-binding protein 7 of Escherichia coli. J Bacteriol. 1995;177:2074–2079. doi: 10.1128/jb.177.8.2074-2079.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Henderson T A, Young K D, Denome S A, Elf P K. AmpC and AmpH, proteins related to the class C β-lactamases, bind penicillin and contribute to the normal morphology of Escherichia coli. J Bacteriol. 1997;179:6112–6121. doi: 10.1128/jb.179.19.6112-6121.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Höltje J-V. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol Mol Biol Rev. 1998;62:181–203. doi: 10.1128/mmbr.62.1.181-203.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jackson M E, Pratt J M. An 18 amino acid amphiphilic helix forms the membrane-anchoring domain of the Escherichia coli penicillin-binding protein 5. Mol Microbiol. 1987;1:23–28. doi: 10.1111/j.1365-2958.1987.tb00522.x. [DOI] [PubMed] [Google Scholar]
- 16.Joris B, Ghuysen J-M, Dive G, Renard A, Dideberg O, Charlier P, Frère J-M, Kelly J A, Boyington J C, Moews P C, Knox J R. The active-site-serine penicillin-recognizing enzymes as members of the Streptomyces R61 dd-peptidase family. Biochem J. 1988;250:313–324. doi: 10.1042/bj2500313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Korat B, Mottl H, Keck W. Penicillin-binding protein 4 of Escherichia coli: molecular cloning of the dacB gene, controlled overexpression, and alterations in murein composition. Mol Microbiol. 1991;5:675–684. doi: 10.1111/j.1365-2958.1991.tb00739.x. [DOI] [PubMed] [Google Scholar]
- 18.Markiewicz Z, Broome-Smith J K, Schwarz U, Spratt B G. Spherical E. coli due to elevated levels of d-alanine carboxypeptidase. Nature. 1982;297:702–704. doi: 10.1038/297702a0. [DOI] [PubMed] [Google Scholar]
- 19.Matsuhashi M. Utilization of lipid-precursors and the formation of peptidoglycan in the process of cell growth and division: membrane enzymes involved in the final steps of peptidoglycan synthesis and the mechanism of their regulation. In: Ghuysen J-M, Hakenbeck R, editors. Bacterial cell wall. Amsterdam, The Netherlands: Elsevier Science B. V.; 1994. pp. 55–71. [Google Scholar]
- 20.Miller J H. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 21.Mottl H, Nieland P, de Kort G, Wierenga J J, Keck W. Deletion of an additional domain located between SXXK and SXN active-site fingerprints in penicillin-binding protein 4 from Escherichia coli. J Bacteriol. 1992;174:3261–3269. doi: 10.1128/jb.174.10.3261-3269.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nelson D E, Young K D. Penicillin binding protein 5 affects cell diameter, contour, and morphology of Escherichia coli. J Bacteriol. 2000;182:1714–1721. doi: 10.1128/jb.182.6.1714-1721.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pratt J M, Jackson M E, Holland I B. The C terminus of penicillin-binding protein 5 is essential for localisation to the E. coli inner membrane. EMBO J. 1986;5:2399–2405. doi: 10.1002/j.1460-2075.1986.tb04510.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Romeis T, Höltje J-V. Penicillin-binding protein 7/8 of Escherichia coli is a dd-endopeptidase. Eur J Biochem. 1994;224:597–604. doi: 10.1111/j.1432-1033.1994.00597.x. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 26.Schiffer G, Höltje J-V. Cloning and characterization of PBP 1C, a third member of the multimodular class A penicillin-binding proteins of Escherichia coli. J Biol Chem. 1999;274:32031–32039. doi: 10.1074/jbc.274.45.32031. [DOI] [PubMed] [Google Scholar]
- 27.Spratt B G. Distinct penicillin binding proteins involved in the division, elongation, and shape of Escherichia coli K12. Proc Natl Acad Sci USA. 1975;72:2999–3003. doi: 10.1073/pnas.72.8.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stoker N G, Broome-Smith J K, Edelman A, Spratt B G. Organization and subcloning of the dacA-rodA-pbpA cluster of cell shape genes in Escherichia coli. J Bacteriol. 1983;155:847–853. doi: 10.1128/jb.155.2.847-853.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tamaki S, Matsuzawa H, Matsuhashi M. Cluster of mrdA and mrdB genes responsible for the rod shape and mecillinam sensitivity of Escherichia coli. J Bacteriol. 1980;141:52–57. doi: 10.1128/jb.141.1.52-57.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tamura T, Suzuki H, Nishimura Y, Mizoguchi J, Hirota Y. On the process of cellular division in Escherichia coli: isolation and characterization of penicillin-binding proteins 1a, 1b, and 3. Proc Natl Acad Sci USA. 1980;77:4499–4503. doi: 10.1073/pnas.77.8.4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tuomanen E. Phenotypic tolerance: the search for β-lactam antibiotics that kill nongrowing bacteria. Rev Infect Dis. 1986;8:S279–S291. doi: 10.1093/clinids/8.supplement_3.s279. [DOI] [PubMed] [Google Scholar]
- 32.Tuomanen E, Cozens R, Tosch W, Zak O, Tomasz A. The rate of killing of Escherichia coli by β-lactam antibiotics is strictly proportional to the rate of bacterial growth. J Gen Microbiol. 1986;132:1297–1304. doi: 10.1099/00221287-132-5-1297. [DOI] [PubMed] [Google Scholar]
- 33.van der Linden M P, de Haan L, Hoyer M A, Keck W. Possible role of Escherichia coli penicillin-binding protein 6 in stabilization of stationary-phase peptidoglycan. J Bacteriol. 1992;174:7572–7578. doi: 10.1128/jb.174.23.7572-7578.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van der Linden M P G, de Haan L, Dideberg O, Keck W. Site-directed mutagenesis of proposed active-site residues of penicillin-binding protein 5 from Escherichia coli. Biochem J. 1994;303:357–362. doi: 10.1042/bj3030357. [DOI] [PMC free article] [PubMed] [Google Scholar]