Abstract
Miniature inverted-repeat transposable elements (MITEs) are widely distributed in the plant genome and can be methylated. However, whether DNA methylation of MITEs is associated with induced allelic expression and drought tolerance is unclear. Here, we identified the drought-inducible MdRFNR1 (root-type ferredoxin-NADP+ oxidoreductase) gene in apple (Malus domestica). MdRFNR1 plays a positive role in drought tolerance by regulating the redox system, including increasing NADP+ accumulation and catalase and peroxidase activities and decreasing NADPH levels. Sequence analysis identified a MITE insertion (MITE-MdRF1) in the promoter of MdRFNR1-1 but not the MdRFNR1-2 allele. MdRFNR1-1 but not MdRFNR1-2 expression was significantly induced by drought stress, which was positively associated with the MITE-MdRF1 insertion and its DNA methylation. The methylated MITE-MdRF1 is recognized by the transcriptional anti-silencing factors MdSUVH1 and MdSUVH3, which recruit the DNAJ domain-containing proteins MdDNAJ1, MdDNAJ2, and MdDNAJ5, thereby activating MdRFNR1-1 expression under drought stress. Finally, we showed that MdSUVH1 and MdDNAJ1 are positive regulators of drought tolerance. These findings illustrate the molecular roles of methylated MITE-MdRF1 (which is recognized by the MdSUVH–MdDNAJ complex) in induced MdRFNR1-1 expression as well as the drought response of apple and shed light on the molecular mechanisms of natural variation in perennial trees.
Allelic expression of a ferredoxin-NADP+ oxidoreductase gene in apple is promoted by a complex that recognizes methylation in a MITE insertion in its promoter in response to drought stress.
In a Nutshell.
Background: Global climate change and rapid population growth have been increasing the shortage of water resources, enhancing the effects of natural disasters such as drought. Drought stress seriously affects the growth and fruit yield of apples. The redox regulatory pathway is important for plant responses to drought. We previously showed that the expression of the root-type ferredoxin-NADP+ oxidoreductase 1 gene (RFNR1) is induced by polyethylene glycol in apple, but whether MdRFNR1 is involved in the drought tolerance of this crop and the molecular mechanism behind its induction by drought are unclear.
Question: How does drought stress affect the expression of MdRFNR1 in apple? Does MdRFNR1 affect the drought tolerance of apple?
Findings: We found that MdRFNR1 increases the drought tolerance of apple by regulating the redox system, including facilitating NADP+ accumulation and catalase and peroxidase activities and decreasing NADPH levels. Sequence analysis revealed that a MITE transposon element, MITE-MdRF1, is inserted in the promoter of an MdRFNR1 allele (MdRFNR1-1). MITE-MdRF1 plays an essential role in simulated drought-induced MdRFNR1-1 expression and this induction is associated with methylated MITE-MdRF1. Further study revealed that a protein complex comprising the transcriptional anti-silencing factor MdSUVH and the DNAJ domain-containing protein DNAJ promotes the expression of the MdRFNR1-1 allele in response to drought via methylated MITE-MdRF1. Thus, our data reveal the molecular mechanism of the MdSUVH–MdDNAJ–MITE-MdRF1–MdRFNR1-1 regulatory module in promoting drought tolerance in apple.
Next steps: Our study revealed a mechanism by which methylated MITE-MdRF1 promotes the expression of MdRFNR1-1. Whether other factors are involved in this process is unclear. For instance, which transcription factors recognize the cis-element in MITE-MdRF1? We will continue to explore the potential mechanism behind this process in the future.
Introduction
Transposable elements (TEs) are mobile units of DNA fragments that can replicate autonomously in the genome and play important roles in gene regulation and chromosome architecture (Slotkin and Martienssen, 2007; Sigman and Slotkin, 2016). TEs are widely distributed in the genomes of eukaryotes and can be divided into two classes: class I (retrotransposons) TEs and class II TEs (DNA transposons), based on the presence or absence of an RNA transposition intermediate. Class II TEs can also be divided into autonomous TEs and non-autonomous TEs based on whether or not they encode transposases (autonomous TEs) (Slotkin and Martienssen, 2007). Most TEs are inactive to maintain genome stability (Lisch, 2012), whereas some play important roles in plant evolution and environmental adaption. For example, a Copia-like retrotransposon enhances the expression of the MYB transcription factor gene Ruby to produce more anthocyanin in Sicilian blood orange (Citrus × sinensis “Blood orange”) (Butelli et al., 2012). The Harbinger-like element, which is located 57 kb upstream of the CCT transcription factor gene ZmCCT9, represses ZmCCT9 expression to promote flowering in maize (Zea mays) under long days and contributes to the adaptation of maize to higher latitudes (Huang et al., 2017).
TEs can be localized in different sites of the genome, such as within a gene, near a gene, in a pericentromere/TE island, or at the centromere core (Sigman and Slotkin, 2016). TEs near genes tend to be small non-autonomous DNA transposons, particularly Mutator, hAT, Helitron family TEs, and miniature TE derivatives, which usually influence gene regulation (Sigman and Slotkin, 2016). Miniature inverted-repeat TEs (MITEs; ∼50–500 bp) are non-autonomous DNA TEs with terminal inverted repeats (TIRs) and a high copy number in plant genomes (Wessler et al., 1995). Several studies have demonstrated that MITEs are related to the biotic or abiotic stress tolerance in plants. A MITE insertion in the intron of the transcription factor gene WRKY45-1 generates a small interfering RNA (siRNA; TE-siR815) that is responsible for the negative role of WRKY45-1 in suppressing the expression of siR815 Target 1 (ST1) (Zhang et al., 2016). A MITE inserted into the ZmNAC111 promoter is significantly associated with natural variation in maize drought tolerance by repressing the expression of this transcription factor gene via RNA-directed DNA methylation (RdDM) and H3K9 dimethylation (Mao et al., 2015).
MITEs are often methylated through the RdDM pathway (Wei et al., 2014; Mao et al., 2015), which results in the methylation of cytosine in three different sequence contexts: CG, CHG, and CHH (H = A, T, or C) (Matzke and Mosher, 2014). RdDM is an important de novo DNA methylation pathway mediated by small RNAs in plants (Zhang and Zhu, 2011; Matzke and Mosher, 2014). The canonical RdDM pathway includes RNA polymerase IV (Pol IV)-dependent siRNA biogenesis and RNA polymerase V (Pol V)-mediated de novo methylation (Matzke and Mosher, 2014). Targeted sequences are transcribed into single-stranded RNA (ssRNA) by Pol IV (Herr et al., 2005; Kanno et al., 2005; Onodera et al., 2005) and then converted into double-stranded RNA (dsRNA) by RNA-DEPENDENT RNA POLYMERASE2 (RDR2) (Haag et al., 2012), which is further processed into 24-nucleotide (nt) siRNA by Dicer-like 3 (DCL3) (Blevins et al., 2015; Zhai et al., 2015) and loaded into ARGONAUTE 4 (AGO4) (Li et al., 2006; Qi et al., 2006; Ye et al., 2012). The siRNA/AGO4 complex then recruits DOMAIN REARRANGED METHYLTRANSFERASE 2 (DRM2) to target loci for de novo DNA methylation (Cao and Jacobsen, 2002; Wierzbicki et al., 2008; Zhong et al., 2014).
DNA methylation usually represses gene expression, especially DNA methylation in gene promoters (Mao et al., 2015). However, a few studies have demonstrated the positive role of DNA methylation in gene expression. A MITE insertion in the promoter of the sunflower (Helianthus annuus) transcription factor gene HaWRKY6 promoter triggers DNA methylation and affects chromatin topology, thereby improving HaWRKY6 expression in cotyledons (Gagliardi et al., 2019). DNA methylation in the REPRESSOR OF SILENCING1 (ROS1) promoter activates the expression of this gene in Arabidopsis thaliana (Lei et al., 2015). DWARF14 (D14) is positively regulated by RdDM at a nearby MITE in rice (Oryza sativa) (Xu et al., 2020).
In Arabidopsis, two transcriptional anti-silencing factors, SUVH1 and SUVH3, are SU(VAR)3-9 homologs that function as methyl readers that bind to methylated DNA. SUVH1 and SUVH3 recruit the DNAJ domain-containing homologs DNAJ1 and DNAJ2 to form a complex to enhance proximal gene expression, thereby repressing the effects of TEs (Li et al., 2016; Harris et al., 2018; Xiao et al., 2019; Zhao et al., 2019). In rice (O. sativa), OsSUVH7, the homolog of AtSUVH1 and AtSUVH3, recognizes the methylated MITE in the promoter of rice HIGH AFFINITY K+ TRANSPORTER1;5 (OsHKT1;5) and then forms a complex with rice BCL-2-ASSOCIATED ATHANOGENE4 (OsBAG4) and R2R3-type MYB transcription factor OsMYB106, which facilitates the binding of OsMYB106 to the cis-element in the OsHKT1;5 promoter and activates its expression, thereby playing a positive role in salinity stress (Wang et al., 2020). However, whether enhanced gene expression triggered by a MITE insertion plays a role in plant drought stress responses is largely unknown.
The redox regulatory network has six functional elements: input elements; transmitters; target proteins; buffer proteins; sensors; and final electron acceptors (Dietz, 2013). The first elements, such as NADPH, ferredoxin (Fd), and glutathione (GSH), transfer electrons to the redox regulatory network via redox transmitters. The last elements, including reactive oxygen species (ROS), reactive nitrogen species (RNS), reactive sulfur species (RSS), and reactive carbonyl species (RCS), can abstract electrons from the appropriate elements and reach a nonreactive oxidation level (Dietz, 2013). ROS are produced in plants at all times, and therefore, the production and removal of ROS must be balanced. However, conditions in the external environment can easily disrupt the ROS equilibrium, such as drought stress (Apel and Hirt, 2004). Drought and other environmental stresses can limit CO2 fixation and reduce the regeneration of NADP+ by the Calvin cycle. Consequently, superoxide radicals and singlet oxygen can be generated in the chloroplast since the photosynthetic electron transport chain is over-reduced (Shao et al., 2007).
Fd-NADP+ oxidoreductase (FNR) plays an important role in redox metabolism in plastids (Hanke and Mulo, 2013). There are two types of FNRs in plants: leaf-type FNR (LFNR), which is localized to chloroplasts, and root-type FNR (RFNR), which is localized in non-photosynthetic plastids (Benz et al., 2010). LFNR catalyzes the electron transfer from reduced Fd (Fdred) to reduce NADP+ to NADPH, whereas RFNR plays the opposite role. In non-photosynthetic plastids, RFNR catalyzes the NADPH-dependent reduction of Fd to generate reducing power for various biosynthetic processes, such as assimilatory pathways of nitrogen and sulfur and the desaturation of fatty acids (Onda et al., 2000; Benz et al., 2010; Mulo, 2011). Fdred is involved in the removal of ROS by preventing the accumulation of extra hydrogen peroxide (H2O2) in the chloroplast. A number of proteins participate in the reaction of H2O2 to H2O in chloroplasts, such as peroxidase (POD), which uses ascorbate as one of the electron donors (Mittler et al., 2004). In addition, Fdred is used to regenerate ascorbate (Asada, 1999). Although LFNR proteins have been extensively studied, little is known about the functions of RFNR proteins. Arabidopsis AtRFNR is essential for reducing and detoxifying nitrite absorbed by roots (Hachiya et al., 2016) and for plant development and survival under ozone and low temperature stress (Grabsztunowicz et al., 2021). Whether plant RFNRs, especially their natural variations, are involved in drought stress responses is largely unclear.
Water deficits (mainly caused by drought stress) limit apple (Malus domestica) growth and production (Geng et al., 2018; Wang et al., 2019). Due to climate change, water resources will become increasingly limited (Gupta et al., 2020). Therefore, it is imperative to understand the mechanisms underlying how apple trees deal with drought stress. TEs account for approximately 59.5% of the apple genome (Daccord et al., 2017). However, their regulatory roles in the drought response in apple remain unclear. In this study, we found that a natural variation of MdRFNR1, the MdRFNR1-1 allele, which contains a MITE (MITE-MdRF1) insertion in its promoter, is induced by drought stress in apple, whereas the MdRFNR1-2 allele is not. The promoted expression of MdRFNR1-1 under drought was correlated with its DNA methylation level at MITE-MdRF1. Further studies revealed that methylated MITE-MdRF1 in the MdRFNR1-1 promoter could be recognized by the MdSUVH–MdDNAJ complex. Our findings indicate that MdRFNR1, MdSUVH1, and MdDNAJ1 play positive roles in regulating drought tolerance in apple by maintaining the function of the redox system.
Results
Drought-induced MdRFNR1 has in vitro oxidoreductase activity
We previously determined that the homolog of MDP0000292058 was induced in the roots of the wild apple Malus sieversii by polyethylene glycol (PEG) treatment, as revealed by RNA-seq analysis (Supplemental Figure S1A; Geng et al., 2019). This gene is homologous to RFNR in Arabidopsis and we therefore named it RFNR1. To examine the expression pattern of RFNR1 under drought stress, we performed qRT-PCR analysis using drought-treated M. domestica. MdRFNR1 was induced by drought stress in M. domestica leaves (Supplemental Figure S1B). In addition, Malus RFNR1 was highly expressed in roots, followed by hypocotyls, shoots, and leaves (Supplemental Figure S1C). Phylogenetic analysis showed that RFNR was conserved in all plant species from the Chlorophyta to Eudicots (Supplemental Figure S2B and Supplemental Table S1), with two conserved domains in its N- and C-termini (Supplemental Figure S2A).
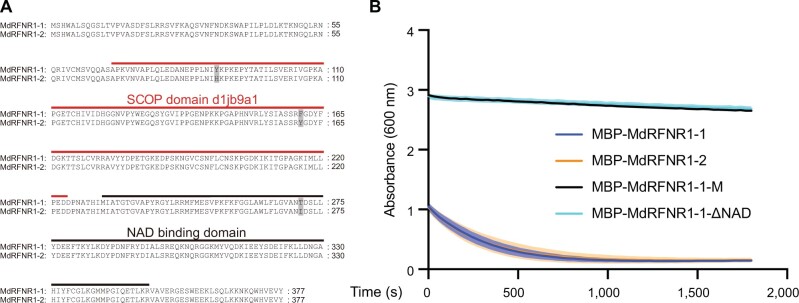
To investigate the function of MdRFNR1, we cloned MdRFNR1 from GL-3 (the progeny of Royal Gala apple, which was used as the background for apple transformation). There were two alleles encoding MdRFNR1 with three amino acid differences. We named these proteins MdRFNR1-1 and MdRFNR1-2 (Figure 1A). We fused these two MdRFNR1 proteins with MBP and purified the two recombinant MdRFNR1-MBP proteins from Escherichia coli. We also purified the recombinant proteins of the catalytically dead version of MdRFNR1, which contained two amino acid mutations (MBP-MdRFNR1-1-M, Arg299Gln, and Lys308Gln; Supplemental Figure S2A) (Kimata-Ariga et al., 2019), and a version of MdRFNR1-1 without the NAD domain (MBP-MdRFNR1-1-△NAD) (Supplemental Figure S3, A and B). Diaphorase activity analysis with 2,6-dichlorophenol indophenol (DCPIP) as an electron acceptor demonstrated that both MdRFNR1-1 and MdRFNR1-2 recombinant proteins could catalyze the oxidation of NADPH to NADP+ and transfer the electron to DCPIP, but MBP-MdRFNR1-1-M and MBP-MdRFNR1-1-△NAD could not (Figure 1B and Supplemental Table S2), suggesting that MdRFNR1-1 and MdRFNR1-2 play the same roles in catalyzing the conversion of NADPH to NADP+.
Figure 1.
NADPH-dependent enzyme activities of the proteins encoded by the two MdRFNR1 alleles. A, Amino acid sequence alignment of MdRFNR1-1 and MdRFNR1-2. B, NADPH-dependent enzyme activities of the MdRFNR1 proteins with DCPIP. The absorbance curve was monitored at 600 nm by a microplate reader at a concentration of 25 mM NADPH. Lines indicate means and shadowed areas indicate sd (n = 9, from three technical and three biological replicates, purified fusion proteins from one batch as a biological replicate).
MdRFNR1 confers in vivo oxidoreductase activity and drought tolerance
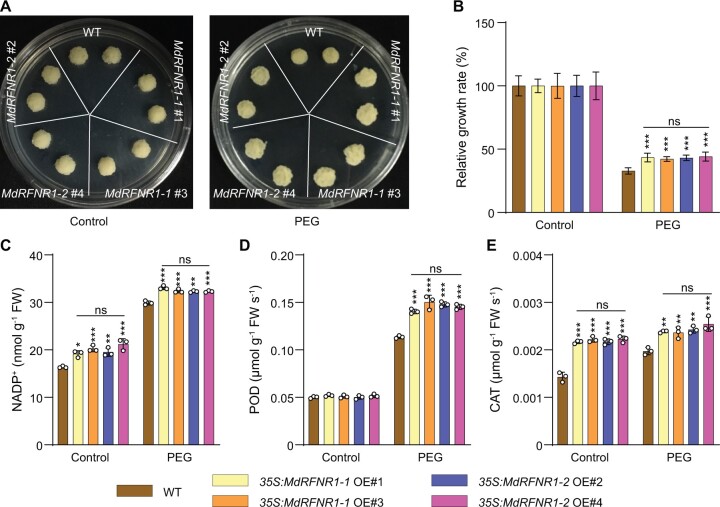
Since MdRFNR1 is responsible for the in vitro catalysis of NADPH to NADP+ and is responsive to drought stress, we next asked if MdRFNR1 has in vivo oxidoreductase activity and contributes to the drought tolerance of apple trees. We first produced transgenic apple calli with increased expression of MdRFNR1-1 and MdRFNR1-2 under the control of the 35S promoter (Supplemental Figure S4A). We then treated wild-type and transgenic calli with PEG to simulate drought stress. Under control or PEG treatment for 20 days, both MdRFNR1-1 and MdRFNR1-2 overexpression (OE) calli contained more NADP+ than the wild type (Figure 2C), indicating the oxidoreductase activities of both MdRFNR1 proteins in vivo. In addition, NADP+ levels in MdRFNR1-1 OE calli were comparable to that in MdRFNR1-2 OE under control and PEG treatment.
Figure 2.
Two MdRFNR1 variants play the same positive role in plant responses to simulated drought stress. A, Morphology of 35S:MdRFNR1-1 OE, 35S:MdRFNR1-2 OE, and wild-type calli in response to PEG treatment. Calli were cultured on MS medium (left) or MS medium supplemented with PEG (right) for 20 days. B, Relative growth rates of WT, 35S:MdRFNR1-1 OE, and 35S:MdRFNR1-2 OE transgenic calli under control and PEG treatment. Error bars indicate sd, n = 8. C–E, NADP+ contents (C) and POD (D) and CAT (E) activities in response to PEG treatment. WT, wild type. Error bars indicate sd, n = 3. Asterisks indicate significant differences between the transgenic lines and the WT in each group (control or PEG treatment). One-way ANOVA (Tukey’s test) was performed and statistically significant differences are indicated by *P ≤ 0.05, **P ≤ 0.01, or ***P ≤ 0.001. ns, no significant difference.
After PEG treatment, the fresh weights of all calli decreased; however, the MdRFNR1-1 and MdRFNR1-2 OE lines had greater fresh weights than the wild type. Moreover, there was no significant difference in the fresh weights of the two OE transgenic lines after PEG treatment (Figure 2, A and B). Considering that both MdRFNR1 proteins showed oxidoreductase activity, we analyzed the activities of two antioxidant enzymes, POD and catalase (CAT), under PEG treatment in both transgenic callus lines. As shown in Figure 2, D and E, the enzyme activities of MdRFNR1-1 and MdRFNR1-2 OE calli were higher than those of the wild type. Similar to fresh weight, MdRFNR1-1 OE calli did not show a significant difference in POD or CAT activity compared with MdRFNR1-2 OE calli under PEG treatment (Figure 2, D and E). Taken together, our data indicate that both the MdRFNR1-1 and MdRFNR1-2 alleles play critical roles in redox regulation and drought stress tolerance in apple. More importantly, both proteins have similar effects on oxidoreductase activity and drought tolerance in apple.
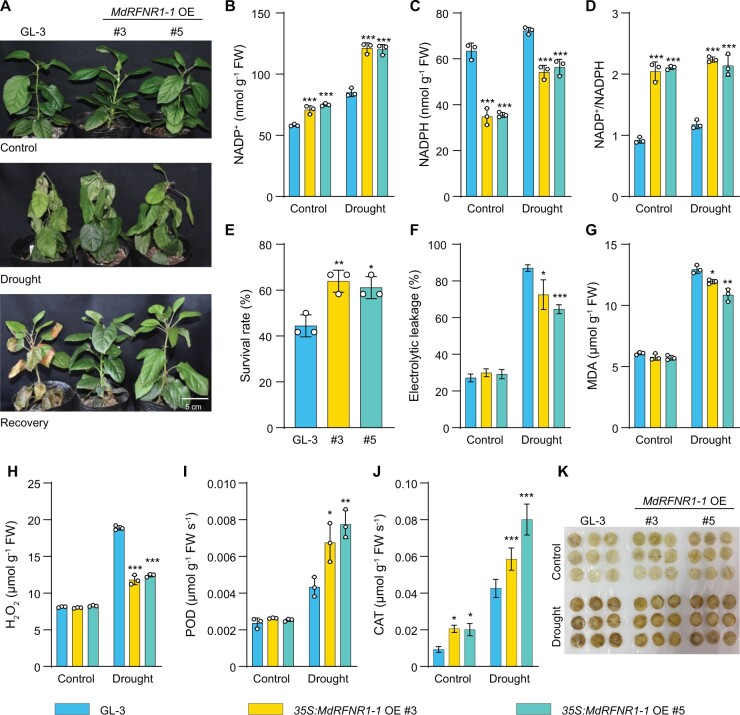
To further confirm the in vivo oxidoreductase activity and the positive role of MdRFNR1 in drought tolerance in apple, we generated transgenic plants with increased expression of MdRFNR1-1 (Supplemental Figure S4B). We treated the transgenic MdRFNR1-1 OE plants with a 15-day drought treatment, with the well-watered plants as a control (Figure 3A). Under control conditions, the MdRFNR1-1 OE plants contained more NADP+ and less NADPH than the non-transgenic (GL-3) plants, thereby showing an increased NADP+/NADPH ratio (Figure 3, B–D). These results are consistent with the in vitro catalytic activity of MdRFNR1-1. Similar results were obtained under drought conditions (Figure 3, B–D), suggesting that MdRFNR1-1 has in vivo oxidoreductase activity under control and drought conditions. After drought stress, the MdRFNR1-1 OE plants had a higher survival rate and photosynthetic rate than GL-3 plants (Figure 3E and Supplemental Figure S5A). In addition, the MdRFNR1-1 OE plants had a lower ion leakage rate and lower malondialdehyde (MDA) contents than GL-3 plants (Figure 3, F and G), suggesting that MdRFNR1-1 OE plants had better membrane integrity under drought stress. Moreover, MdRFNR1-1 OE plants accumulated less H2O2 and had higher POD and CAT activities under drought stress than the wild type (Figure 3, H–K).
Figure 3.
35S:MdRFNR1-1 OE transgenic plants are more tolerant to drought than the wild type. A, Morphology of 35S:MdRFNR1-1 OE and non-transgenic GL-3 plants under control and drought conditions. Water was withheld from 3-month-old plants for 15 days, followed by recovery for 7 days. B–D, Contents of NADP+ (B), NADPH (C), and NADP+/NADPH ratio (D) of GL-3 and 35S:MdRFNR1-1 OE transgenic plants under control and drought treatment. E, The survival rates of plants shown in (A). F–K, Leaf ion leakage (F), MDA contents (G), H2O2 contents (H), POD (I) and CAT (J) activities, and DAB staining (K) of GL-3 and 35S:MdRFNR1-1 OE transgenic plants under control and drought treatment. Error bars indicate sd, n = 3 in (B, C, D, G, H, and I), n = 36 in (E, 36 plants were used and divided into three biological replications), and n = 8 in (F and J). Asterisks indicate significant differences between the transgenic lines and GL-3 plants in each group (control and drought treatment). One-way ANOVA (Tukey’s test) was performed and statistically significant differences are indicated by *P ≤ 0.05, **P ≤ 0.01, or ***P ≤ 0.001.
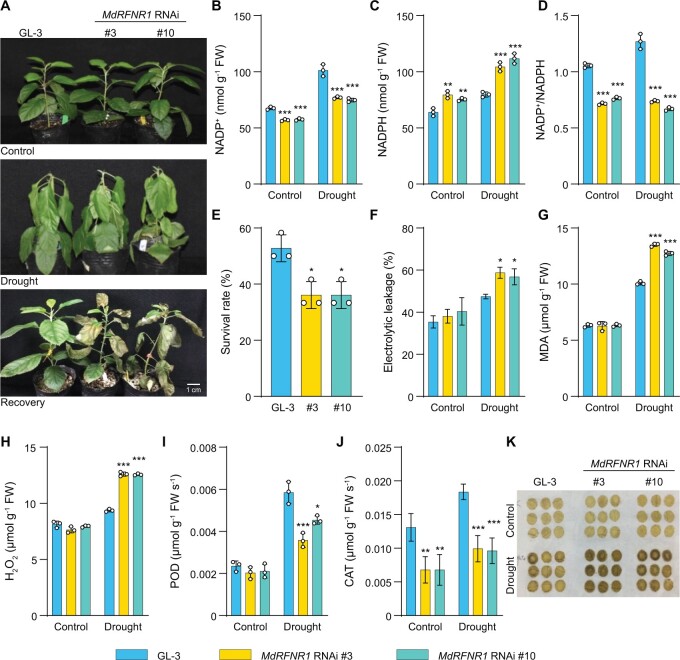
We also generated transgenic plants with repressed expression of MdRFNR1 (Supplemental Figure S4C). In response to short-term drought treatment, NADP+ accumulated in both GL-3 and MdRFNR1 RNAi plants; however, NADP+ levels were significantly lower in MdRFNR1 RNAi than GL-3 plants (Figure 4, A and B). NADPH contents had a similar pattern in MdRFNR1 RNAi and GL-3 plants under control and drought conditions (Figure 4C). In addition, the NADP+/NADPH ratio was lower in MdRFNR1 RNAi than in GL-3 plants under control and drought conditions (Figure 4D). These results further support the in vivo oxidoreductase activity of MdRFNR1 under control and drought conditions. After drought stress, the MdRFNR1 RNAi lines had a lower survival rate and photosynthetic rate than GL-3 plants (Figure 4E and Supplemental Figure S5B). Ion leakage and MDA analysis showed that MdRFNR1 knockdown resulted in greater cell membrane damage than GL-3 after drought stress (Figure 4, F and G). In addition, the H2O2 contents were higher in MdRFNR1 RNAi lines than in GL-3 plants under drought stress (Figure 4, H and K). Consistently, POD and CAT activities were lower in MdRFNR1 RNAi lines than in GL-3 plants after drought treatment (Figure 4, I and J). All of these results suggest that MdRFNR1-1 and MdRFNR1-2 have in vivo oxidoreductase activity and play positive roles in drought tolerance.
Figure 4.
MdRFNR1 RNAi plants are sensitive to drought treatment. A, Morphology of MdRFNR1 RNAi and GL-3 plants under control and drought conditions. Water was withheld from 3-month-old plants for 10 days, followed by recovery for 7 days. B–D, Contents of NADP+ (B), NADPH (C), and the NADP+/NADPH ratio (D) of GL-3 and MdRFNR1 RNAi transgenic plants under control and drought treatment. E, The survival rates of plants shown in (A). F–K, Leaf ion leakage (F), MDA contents (G), H2O2 contents (H), POD (I) and CAT (J) activities, and DAB staining (K) of GL-3 and MdRFNR RNAi transgenic plants under control and drought treatment. Error bars indicate sd, n = 3 in (B, C, D, G, H, and I), n = 36 in (E, 36 plants were used and divided into three biological replications), and n = 8 in (F and J). Asterisks indicate significant differences between the transgenic lines and GL-3 plants in each group (control and drought treatment). One-way ANOVA (Tukey’s test) was performed and statistically significant differences are indicated by *P ≤ 0.05, **P ≤ 0.01, or ***P ≤ 0.001.
A MITE insertion in the promoter of MdRFNR1-1 is essential for its induced expression and its positive role under drought stress
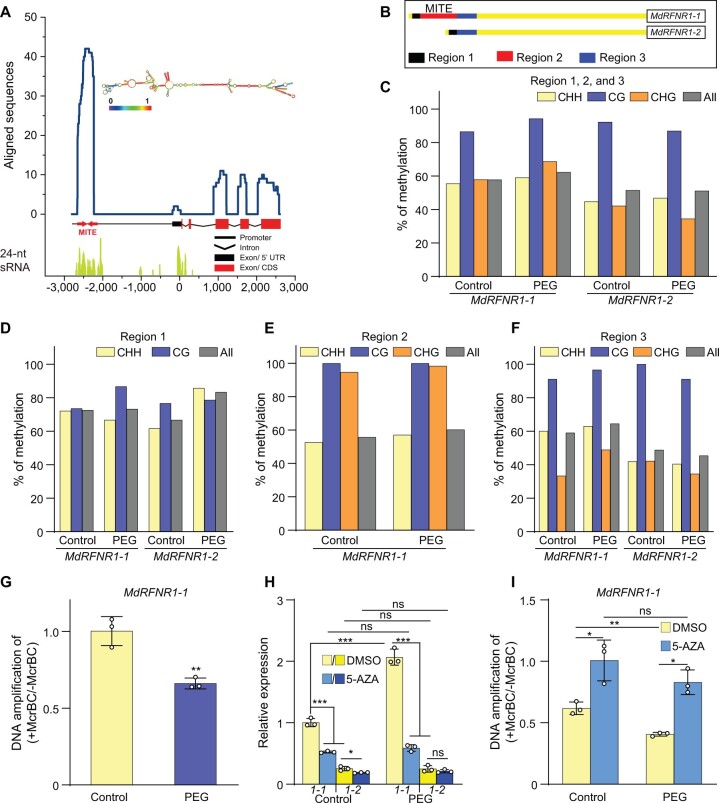
Since drought stress enhanced the expression of MdRFNR1, we cloned the promoters of MdRFNR1-1 and MdRFNR1-2 from GL-3. After sequencing and alignment, we identified a 430-bp MITE insertion in the promoter of MdRFNR1-1, which is located 2,234 bp upstream of the translation start codon (Supplemental Figure S6 and Figure 8A). We named this insertion MITE-MdRF1. We performed BLAST analysis against the plant MITE database (http://pmite.hzau.edu.cn) (Chen et al., 2014) using the 430-bp sequence as a query and found that it was most similar to DTH_Mad4, a member of the PIF/Harbinger superfamily. To investigate the presence of MITE-MdRF1 in Malus, we tested 371 Malus accessions, including 275 wild accessions and 96 domesticated cultivars. MITE-MdRF1 was detected in 30 accessions, including 17 wild accessions and 13 cultivars (Supplemental Figure S7 and Supplemental Table S3), pointing to the general presence of MITE-MdRF1 in Malus. Surprisingly, we did not observe homozygous MITE-MdRF1 in any of the accessions tested (Supplemental Figure S7).
Figure 8.
DNA methylation of the MITE-MdRF1 insertion in the MdRFNR1-1 promoter is positively associated with MdRFNR1-1 expression in response to simulated drought. A, Enrichment of stem–loop structure RNA and 24-nt sRNAs in the MITE-MdFR1 region of the MdRFNR1-1 promoter. The line chart represents the alignment of the MdRFNR1-1 locus (−2,798 to 2,584 bp) against an apple ESTs database. The EST reads with the highest similarity to MITE-MdRF1 were used for structural analysis, which was predicted using the ViennaRNA Web servers (http://rna.tbi.univie.ac.at/). For alignment of 24-nt sRNAs, reads of 24-nt small-RNAs were mapped to the MdRFNR1-1 locus. B, Schematic representation of locus-specific BS-seq analysis in (C–F). Region 1, upstream of MITE-MdRF1 in the MdRFNR1-1 promoter or the corresponding fragment of the MdRFNR1-2 promoter; Region 2, MITE-MdRF1; Region 3, downstream of MITE-MdRF1 in the MdRFNR1-1 promoter or the corresponding fragment of the MdRFNR1-2 promoter. C–F, The methylation percentage of Regions 1, 2, and 3 (C); Region 1 (D); Region 2 (E); and Region 3 (F) of the MdRFNR1 promoter in GL-3 leaves in response to simulated drought stress. G, McrBC–qPCR showing the DNA methylation level of MITE-MdRF1 in the MdRFNR1-1 promoter in GL-3 leaves under control and PEG treatment. Leaves were collected from 4-month-old hydroponically cultured GL-3 plants that were treated with 20% PEG8000 for 0 or 6 h, and locus-specific BS-seq (C–F) or McrBC-qPCR (G) was then performed. H and I, Allelic expression of MdRFNR1-1 and MdRFNR1-2 (H) and DNA methylation level of MITE-MdRF1 in the MdRFNR1-1 promoter (I) in leaves of GL-3 plants treated with DMSO or 5-AZA in response to simulated drought. 1-1 and 1-2 in (H) represent MdRFNR1-1 and MdRFNR1-2, respectively. Subcultured GL-3 plants were transferred to MS medium supplemented with 7 μg/mL 5-AZA or DMSO for an additional 3 weeks, treated with 20% PEG8000 (w/v) for 0 or 6 h, and the leaves were harvested. Error bars indicate sd, n = 3. Statistical analyses were performed by Student’s t test and statistically significant differences are indicated by *P ≤ 0.05, **P ≤ 0.01, or ***P ≤ 0.001. ns, no significant difference.
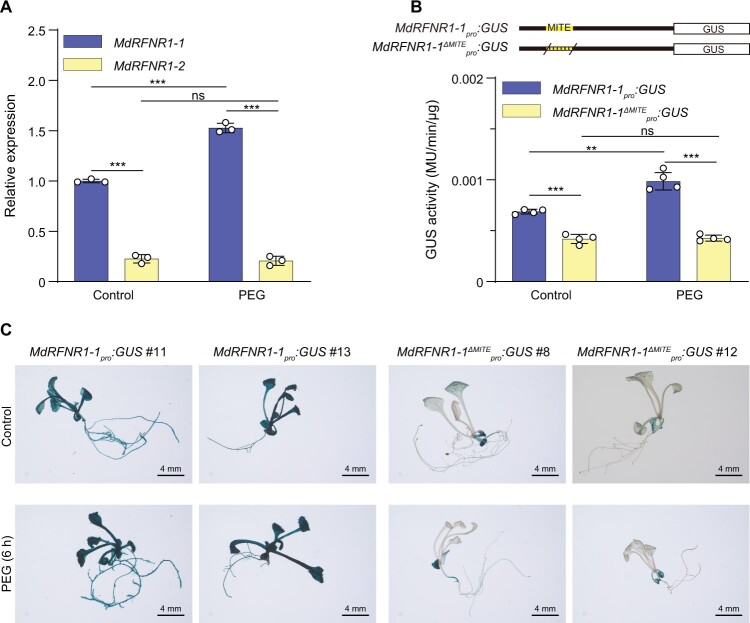
To investigate the impacts of MITE-MdRF1 on MdRFNR1 expression, we performed allele-specific qPCR of MdRFNR1 in GL-3. Using an allele-specific forward primer, we found that MdRFNR1-1 (with MITE-MdRF1) was expressed at a higher level in roots than MdRFNR1-2 (without MITE-MdRF1) (Figure 5A). We also noticed that MdRFNR1-1 was induced by PEG treatment whereas MdRFNR1-2 was not (Figure 5A). These results suggest that MITE-MdRF1 may induce MdRFNR1 expression in response to drought stress.
Figure 5.
A MITE insertion in the MdRFNR1-1 promoter is associated with its allelic induction by simulated drought stress. A, Allelic expression of MdRFNR1 in GL-3 under control and PEG treatment. Error bars indicate sd, n = 3. B, GUS activity of transgenic calli carrying MdRFNR1-1pro:GUS or MdRFNR1-:GUS under control and PEG treatment. Error bars indicate sd, n = 4. C, GUS staining of Arabidopsis plants carrying MdRFNR1-1pro:GUS or MdRFNR1-:GUS in response to PEG treatment. Bars = 4 mm. Statistical analyses were performed by the Student’s t test. Statistically significant differences are indicated by **P ≤ 0.01 or ***P ≤ 0.001, ns, no significant difference.
To further confirm the induction of MdRFNR1 expression by MITE-MdRF1, we transformed apple calli and Arabidopsis with MdRFNR1-1pro:GUS and MdRFNR1-:GUS. The GUS activity of MdRFNR1-1pro:GUS transgenic calli increased after PEG treatment. However, MdRFNR1-:GUS transgenic calli did not display induced GUS activity in response to PEG treatment, suggesting that MITE-MdRF1 is essential for the induction of MdRFNR1 by drought stress (Figure 5B). GUS staining of transgenic Arabidopsis also showed a similar pattern (Figure 5C and Supplemental Figure S8). These data further verify the role of MITE-MdRF1 in promoting MdRFNR1 expression under drought stress.
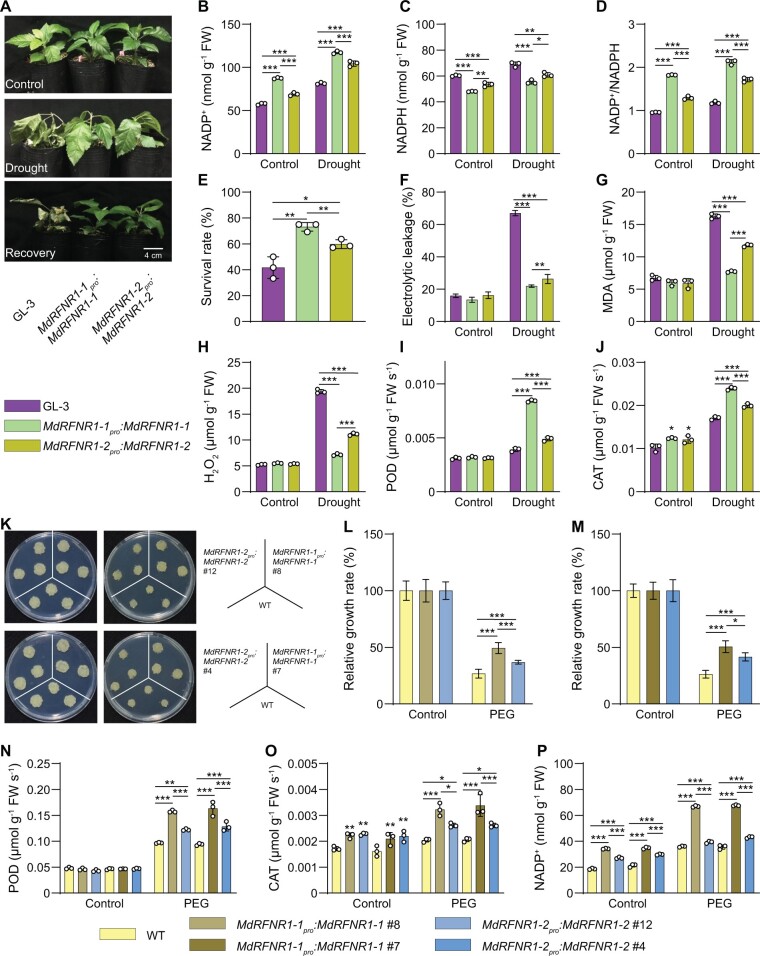
To investigate whether MITE-MdRF1-induced MdRFNR1 expression is associated with drought tolerance, we transformed apple plants and calli with MdRFNR1-1pro:MdRFNR1-1 and MdRFNR1-2pro:MdRFNR1-2 (Supplemental Figure S9). Transgenic plants or calli carrying MdRFNR1-1pro:MdRFNR1-1 had higher MdRFNR1 expression than those carrying MdRFNR1-2pro:MdRFNR1-2 (Supplemental Figure S9). After 15 days of drought stress, both transgenic plants had more NADP+ and less NADPH contents, resulting in a higher NADP+/NADPH ratio in response to drought (Figure 6, A–D). In addition, both types of transgenic plants performed better than wild type GL-3 plants, as indicated by higher survival rates, higher POD and CAT activities, lower electrolyte leakage, and lower MDA and H2O2 contents under drought stress (Figure 6, E–J). Compared with transgenic plants carrying MdRFNR1-2pro:MdRFNR1-2, transgenic plants carrying MdRFNR1-1pro:MdRFNR1-1 had a higher survival rate, higher POD and CAT activities, lower electrolyte leakage rates, and lower MDA and H2O2 contents under drought stress (Figure 6, E–J). Moreover, MdRFNR1-1pro:MdRFNR1-1 transgenic plants contained more NADP+ and less NADPH, thereby having a higher NADP+/NADPH ratio in response to drought (Figure 6, B–D).
Figure 6.
The MdRFNR1-1 allele plays a more important role than MdRFNR1-2 in drought stress tolerance. A, Morphology of GL-3 and transgenic plants carrying MdRFNR1-1pro:MdRFNR1-1 or MdRFNR1-2pro:MdRFNR1-2 under control and drought conditions. Water was withheld from 1.5-month-old plants for 15 days, followed by recovery for 7 days. B–D, Contents of NADP+ (B), NADPH (C), and NADP+/NADPH ratio (D) of GL-3 and transgenic plants carrying MdRFNR1-1pro:MdRFNR1-1 or MdRFNR1-2pro:MdRFNR1-2 under control and drought treatment. E, The survival rates of plants shown in (A). F–J, Leaf ion leakage (F), MDA (G) and H2O2 (H) contents, POD (I) and CAT (J) activities of GL-3, and transgenic plants carrying MdRFNR1-1pro:MdRFNR1-1 or MdRFNR1-2pro:MdRFNR1-2 under control and drought treatment. K, Morphology of WT and transgenic calli after 20 days on MS medium (left) or MS medium supplemented with PEG (right). L–M, Relative growth rates of WT and transgenic calli carrying MdRFNR1-1pro:MdRFNR1-1 or MdRFNR1-2pro:MdRFNR1-2. N–P, POD (N) and CAT (O) activities and NADP+ contents (P) of WT and transgenic calli carrying MdRFNR1-1pro:MdRFNR1-1 or MdRFNR1-2pro:MdRFNR1-2 under control and PEG treatment. Error bars indicate sd, n = 3 in (B, C, D, G, H, I, J, N, O, and P), n = 36 in (E, 36 plants were used and divided into three biological replications), n = 8 in (F), and n = 9 in (L and M). Asterisks indicate significant differences between the transgenic lines and GL-3 plants in each group (control and drought treatment). One-way ANOVA (Tukey’s test) was performed and statistically significant differences are indicated by *P ≤ 0.05, **P ≤ 0.01, or ***P ≤ 0.001.
Transgenic calli showed a similar pattern. After PEG treatment for 20 days, the fresh weights of both types of transgenic calli were greater than that of the wild type. However, the fresh weights of transgenic calli carrying MdRFNR1-1pro:MdRFNR1-1 were greater than that of MdRFNR1-2pro:MdRFNR1-2 calli under PEG treatment (Figure 6, K–M). Furthermore, MdRFNR1-1pro:MdRFNR1-1 calli had higher POD and CAT activities under PEG treatment than MdRFNR1-2pro:MdRFNR1-2 calli (Figure 6, N and O). We also observed that MdRFNR1-1pro:MdRFNR1-1 transgenic calli contained more NADP+ than MdRFNR1-2pro:MdRFNR1-2 calli (Figure 6P). These results further support the notion that MdRFNR1-1pro:MdRFNR1-1 improves drought tolerance and this improvement is associated with the induced expression of MdRFNR1-1 due to the presence of MITE-MdRF1 in its promoter (Figure 6).
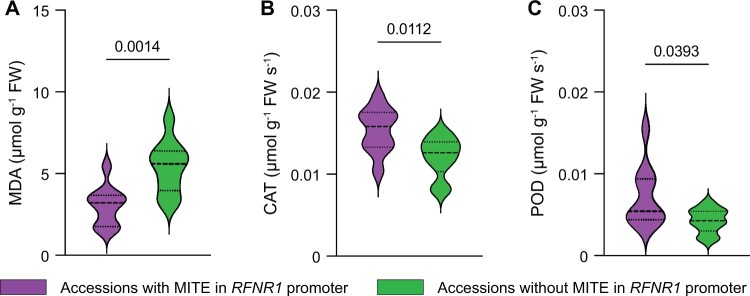
To further confirm the association of the MITE-MdRF1 insertion of RFNR1 with drought tolerance in an ecological context, we selected 20 Malus accessions, including 10 accessions with MITE-MdRF1 and 10 accessions without MITE-MdRF1 in the promoter of MdRFNR1. Following dehydration, we measured MDA contents as well as CAT and POD activities. As shown in Figure 7, accessions with the MITE-MdRF1 insertion in the RFNR1 promoter contained less MDA, indicating less membrane damage in response to drought stress. In addition, accessions with MITE-MdRF1 in the RFNR1 promoter had higher POD and CAT activities, suggesting a higher ability to scavenge ROS under drought stress. On the contrary, accessions without the MITE-MdRF1 insertion in the RFNR1 promoter had higher MDA contents and lower POD and CAT activities, indicating a lower ability to tolerate drought stress (Figure 7).
Figure 7.
Association of MDA contents and POD and CAT activities with the MITE-MdRF1 insertion in the RFNR1 promoter in Malus accessions under dehydration conditions. Twenty Malus accessions were subjected to dehydration treatment. A, MDA contents of Malus accessions. B, CAT activity of Malus accessions. C, POD activity of Malus accessions. Statistical analyses were performed by the Student’s t test and P values between two columns are indicated.
Induced expression of MdRFNR1-1 in response to drought stress is associated with DNA methylation of MITE-MdRF1
MITEs can be methylated by the RdDM pathway (Wei et al., 2014; Mao et al., 2015). Blastn against the Malus EST database revealed that many sequences could be mapped to MITE-MdRF1 in the MdRFNR1-1 promoter. In addition, this MITE-derived RNA could form a stem–loop structure (Figure 8A). We mapped the reads of several small (s)RNA-seq data sets to the genomic sequence of MdRFNR1-1 (promoter, exon, and intron), finding that numerous 24-nt sRNAs could be mapped to the MITE-MdRF1 sequence (Figure 8A). MITE-derived sRNAs can result in DNA methylation through the RdDM pathway (Mao et al., 2015).
To explore the methylation status of MITE-MdRF1, its upstream region and downstream region in the MdRFNR1 promotor, we divided the regions flanking MITE-MdRF1 into three regions: Region 1, Region 2, and Region 3. Region 1 is the upstream sequence of MITE-MdRF1 in the MdRFNR1-1 promoter or the corresponding fragment in the MdRFNR1-2 promoter; Region 2 is MITE-MdRF1; and Region 3 is the downstream sequence of MITE-MdRF1 in the MdRFNR1-1 promoter or the corresponding fragment in the MdRFNR1-2 promoter (Figure 8B and Supplemental Figure S10A). Using locus-specific bisulfite sequencing (BS-seq), we found that the average DNA methylation level of the MdRFNR1-1 promoter containing MITE-MdRF1 and the flanking sequence was higher than that of the MdRFNR1-2 promoter containing the corresponding sequence in both GL-3 leaves and roots under control and drought-stress conditions (Figure 8C and Supplemental Figure S10B). In addition, the DNA methylation level of the MdRFNR1-1 promoter increased in response to drought stress, while that of the MdRFNR1-2 promoter did not, in both leaves and roots (Figure 8C and Supplemental Figure S10B).
We compared the methylation percentage of Region 1, Region 2, and Region 3 of MdRFNR1-1 and MdRFNR1-2 in GL-3 leaves and roots in response to PEG treatment. Following PEG treatment, the methylation level of MITE-MdRF1, and its upstream and downstream regions in MdRFNR1-1, increased in both leaves and roots, whereas the methylation level of the corresponding regions of MdRFNR1-2 increased or decreased in leaves and roots (Figure 8, D–F and Supplemental Figures S10, C–E and S11). We also performed McrBC-qPCR analysis of the MdRFNR1-1 promoter containing the MITE-MdRF1 region in GL-3 leaves and roots under control and PEG treatment; McrBC is an endonuclease that cleaves DNA containing methylcytosine. Consistent with the results of locus-specific BS-seq, McrBC-qPCR showed that the methylation level of the MITE-MdRF1 region in the MdRFNR1-1 promoter increased after PEG treatment (Figure 8G and Supplemental Figure S10F), which further verified the increased overall methylation percentage of MdRFNR1-1 in response to drought conditions. These data suggest that MITE-MdRF1 is positively associated with DNA methylation under drought stress.
The DNA methylation inhibitor 5-Aza-2′-deoxycytodine (5-AZA) reduces the methylation level of DNA (Figure 8I). To further confirm the association of MITE-MdRF1 methylation and MdRFNR1 expression in response to drought stress, we treated GL-3 plants with 5-AZA under control or drought conditions. Following 5-AZA treatment, the expression levels of these two genes decreased under control or PEG treatment, but the expression of MdRFNR1-1 decreased the most, whereas only a slight reduction of MdRFNR1-2 expression was observed (Figure 8H). In response to drought, the induced expression of MdRFNR1-1 by PEG treatment was inhibited by 5-AZA, whereas there was little change in MdRFNR1-2 expression (Figure 8H). Consistently, McrBC-qPCR showed that the methylation level of the MITE region, which is present in the promoter of MdRFNR1-1, increased after PEG treatment (Figure 8I). However, 5-AZA treatment abolished this induction (Figure 8I). These results further support the notion that MdRFNR1-1 expression is positively associated with MITE-MdRF1-mediated DNA methylation in response to drought stress.
The MdSUVH–MdDNAJ complex recognizes methylated MITE-MdRF1 in the MdRFNR1-1 promoter and facilitates its expression under drought stress
Arabidopsis SUVH1 and SUVH3, two transcriptional anti-silencing factors and methyl readers, bind to methylated DNA and recruit DNAJ1 and DNAJ2 (Li et al., 2016; Harris et al., 2018). However, DNAJ1 and DNAJ2 do not directly bind methylated DNA but can act as transcriptional activators (Harris et al., 2018). We identified four homologs of SUVH1 and SUVH3 in the apple genome: MdSUVH1, MdSUVH1-like, MdSUVH3, and MdSUVH3-like. MdSUVH and MdSUVH-like appear to be duplicated genes based on phylogenetic analysis (Velasco et al., 2010; Supplemental Figure S12, A and C and Supplemental Table S4). The two proteins most closely related to SUVH1 and SUVH3 (based on Blastp analysis) were named MdSUVH1 and MdSUVH3, respectively, whereas the other two proteins were named MdSUVH1-like and MdSUVH3-like. We also identified five homologs of DNAJ1 and DNAJ2 in the apple genome: MdDNAJ1, MdDNAJ2, MdDNAJ3, MdDNAJ4, and MdDNAJ5 (Supplemental Figure S12, B and D and Supplemental Table S5). Yeast-two hybrid (Y2H) and split-luciferase (split-LUC) analysis revealed that MdSUVH1 and MdSUVH3 interacted with MdDNAJ1, MdDNAJ2, and MdDNAJ5 (Supplemental Figure S13, A and B). Thus, we selected MdDNAJ1, MdDNAJ2, MdDNAJ5, MdSUVH1, and MdSUVH3 for further analysis.
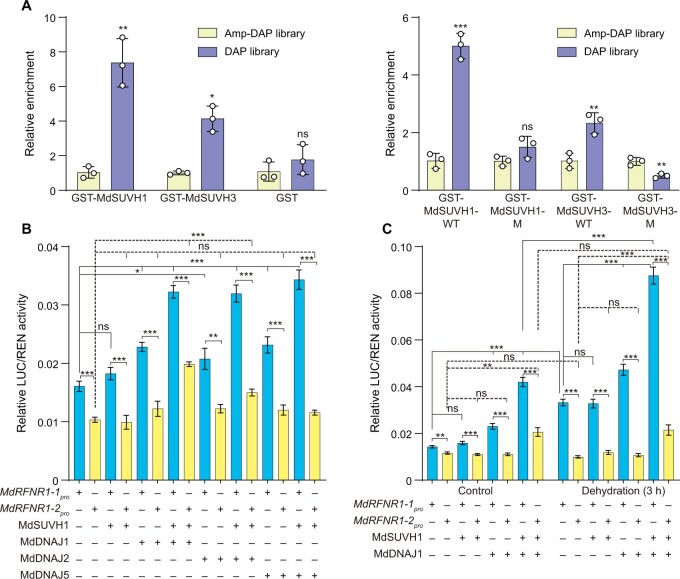
To determine if apple SUVH1 and SUVH3 could recognize methylated DNA, we performed DNA affinity purification (DAP)-qPCR analysis. Indeed, MdSUVH1 and MdSUVH3 bound to methylated MITE-MdRF1 in the MdRFNR1-1 promoter, with MdSUVH1 having a stronger binding ability (Figure 9A). The key amino acids determining the binding of SUVH1 and SUVH3 to the methylated DNA are Tyr290 and Tyr317, respectively (Harris et al., 2018). When these two amino acids of MdSUVH1 (Tyr290) and MdSUVH3 (Tyr317) were mutated to alanine (Ala), the binding of these two proteins to methylated DNA was abolished (Figure 9A), further suggesting that MdSUVH1 and MdSUVH3 bind to methylated MITE-MdRF1 in the MdRFNR1-1 promoter.
Figure 9.
The MdSUVH–MdDNAJ complex binds to methylated MITE-MdRF1 and activates MdRFNR1-1 expression in response to drought stress. A, Binding of MdSUVH1 and MdSUVH3 to methylated MITE-MdRF1 in the MdRFNR1-1 promoter, as revealed by DAP-qPCR. DAP, methylated DNA library. Amp-DAP, amplified DNA library, which contained non-methylated DNA. WT, wild type; M, mutant version of MdSUVH1 or MdSUVH3, in which Tyr290 of MdSUVH1 was mutated to Ala290 and Tyr317 of MdSUVH3 was mutated to Ala317. Error bars indicate sd, n = 3. B and C, Relative luciferase activity from the dual luciferase reporter assays in N. benthamiana leaves. Quantification was performed by normalizing firefly luciferase activity to that of Renilla luciferase, 35S:REN was used as an internal control. Error bars indicate se, n = 8. Statistical analyses were performed by the Student’s t test and statistically significant differences are indicated by *P ≤ 0.05, **P ≤ 0.01, or ***P ≤ 0.001. ns, no significant difference.
To further investigate the influence of the MdSUVH–MdDNAJ complex on the expression of MdRFNR1, we performed a dual-luciferase assay (DUAL-LUC) analysis. Under control conditions, individual MdSUVH1 did not significantly improve the expression of MdRFNR1-1 or MdRFNR1-2. However, MdDNAJ1, MdDNAJ2, or MdDNAJ5 alone significantly improved the expression of MdRFNR1-1 but not MdRFNR1-2. When both MdSUVH1 and MdDNAJ1 or MdDNAJ2 or MdDNAJ5 were present, the expression of MdRFNR1-1 was markedly induced (Figure 9B). We also noticed a significant induction of MdRFNR1-2 by MdSUVH1 and MdDNAJ1 or MdDNAJ2 (Figure 9B). After dehydration treatment, MdRFNR1-1 expression was markedly induced, which is consistent with our observation in Figures 5A and 8H. Similar to the pattern shown in Figure 9B, MdSUVH1 alone did not induce the expression of MdRFNR1-1 under dehydration conditions, whereas MdDNAJ1 did (Figure 9C). The presence of both MdSUVH1 and MdDNAJ1 markedly induced MdRFNR1-1 expression (Figure 9C). However, the expression of MdRFNR1-2 was not induced by MdSUVH1 or MdDNAJ1 alone but slightly increased in the presence of both MdSUVH1 and MdDNAJ1 (Figure 9C). The above data suggest that the MdSUVH1–MdDNAJ complex recognizes methylated MITE-MdRF1 and promotes MdRFNR1-1 expression under control and dehydration conditions.
To further confirm the effect of the MdSUVHs–MdDNAJs complex in activating MdRFNR1-1 expression, we generated NbSUVH1- and NbSUVH3-silenced Nicotiana benthamiana plants using a virus-induced gene silencing (VIGS) approach (Supplemental Figure S14, A and B). Considering the high sequence similarity between NbSUVH and MdSUVH (Supplemental Figure S12, A and C and Supplemental Table S4), we co-infiltrated wild-type and NbSUVH1- and NbSUVH3-silenced N. benthamiana plants with 35S:MdDNAJs and MdRFNR1-1pro:LUC and performed DUAL-LUC analysis. In the presence of NbSUVH1 and NbSUVH3, the expression of MdRFNR1-1 was activated by MdDNAJs. However, the activation of MdRFNR1-1 expression by MdDNAJs was abolished in the absence of NbSUVH1 and NbSUVH3 (Supplemental Figure S14C). These results further indicate the importance of the MdSUVHs–MdDNAJs complex in the activated expression of MdRFNR1-1.
MdSUVH and MdDNAJ are positive regulators of drought stress tolerance
Since MdSUVH–MdDNAJ recognizes methylated MITE-MdRF1 in the MdRFNR1-1 promoter and facilitates its expression in response to drought stress, we next asked whether MdSUVH and MdDNAJ proteins function in drought stress tolerance in apple. We first evaluated the expression of MdSUVH1, MdSUVH3, MdDNAJ1, MdDNAJ2, and MdDNAJ5 in response to drought stress. All these genes were induced by drought, except that the expression of MdDNAJ5 was too low to be detected (Supplemental Figure S15, A–H). We therefore selected MdDNAJ1 and MdSUVH1 as representatives of these genes.
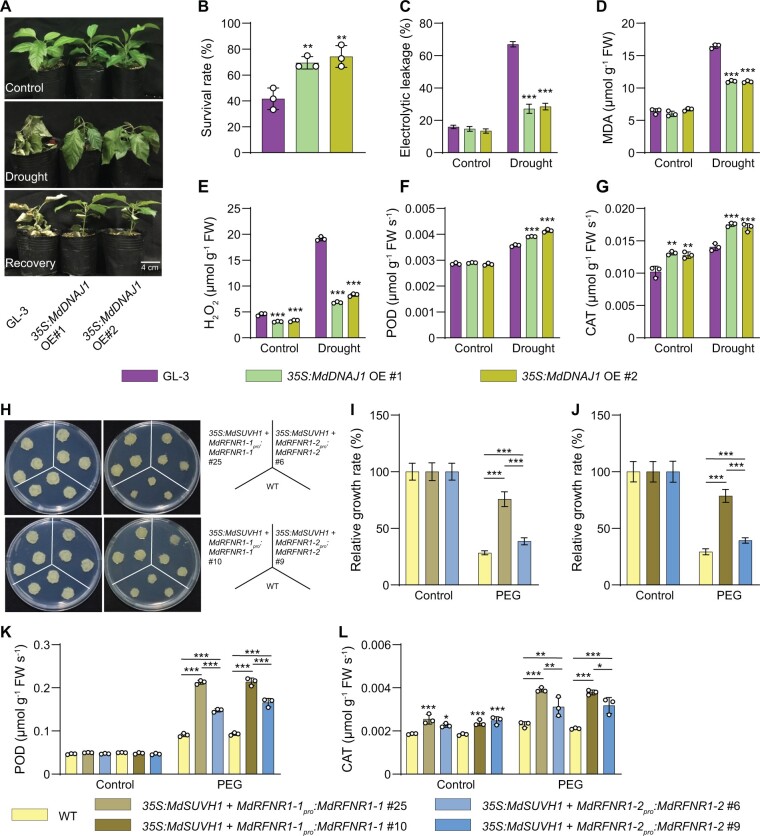
We stably transformed apple plants with 35S:MdDNAJ1 (Supplemental Figure S16A). After 15 days of drought treatment, ∼70% of MdDNAJ1 OE plants survived, whereas only ∼43% of GL-3 plants were still alive (Figure 10, A and B). In addition, under drought stress, the MdDNAJ1 OE plants had lower electrolyte leakage rates, lower MDA and H2O2 contents, and higher POD and CAT activities compared with GL-3 plants (Figure 10, C–G). We also found that MdRFNR1-1 expression was higher in MdDNAJ1 OE plants than in GL-3, but MdRFNR1-2 expression was not (Supplemental Figure S17, A and B). These results indicate that MdDNAJ1 OE plants are more tolerant to drought stress than GL-3 plants, suggesting the MdDNAJ1 plays a positive role in the drought stress response. These data also imply that the drought tolerance of MdDNAJ1 could be attributed to increased MdRFNR1-1 expression and ROS detoxification.
Figure 10.
MdDNAJ1 and MdSUVH1 play positive roles in drought stress tolerance. A, Morphology of GL-3 and transgenic 35S:MdDNAJ1 OE plants under control and drought conditions. Water was withheld from 1.5-month-old plants for 15 days, followed by recovery for 7 days. B, The survival rates of plants shown in (A). C–G, Leaf ion leakage (C), MDA (D) and H2O2 (E) contents, and POD (F) and CAT (G) activities of GL-3 and 35S:MdDNAJ1 OE transgenic plants under control and drought treatment. H, Morphology of WT and transgenic calli, which were co-transformed with 35S:MdSUVH1 and MdRFNR1-1pro:MdRFNR1-1 or MdRFNR1-2pro:MdRFNR1-2 and incubated for 20 days on MS medium (left) or MS medium supplemented with PEG (right). I and J, Relative growth rates of WT and transgenic calli in response to PEG treatment. K and L, POD and CAT activities in WT and transgenic calli in response to PEG treatment. Error bars indicate sd, n = 3 in (D, E, F, G, K, and L), n = 36 in (B, 36 plants were used and divided into three biological replications), n = 8 in (C), and n = 9 in (I and J). Asterisks indicate significant differences between the transgenic lines and GL-3 plants in each group (control and drought treatment). One-way ANOVA (Tukey’s test) was performed and statistically significant differences are indicated by *P ≤ 0.05, **P ≤ 0.01, or ***P ≤ 0.001.
To explore the biological function of MdSUVH1, we stably transformed apple calli with MdSUVH1. Since MdSUVH1 binds to methylated MITE-MdRF1, which is not present in the promoter of MdRFNR1 in apple calli (Supplemental Figure S7A), we co-transformed apple calli with 35S:MdSUVH1 and MdRFNR1-1pro:MdRFNR1-1 (Supplemental Figure S16B). As a control, we co-transformed apple calli with 35S:MdSUVH1 with MdRFNR1-2pro:MdRFNR1-2 (Supplemental Figure S16B). MdRFNR1 expression was higher in MdRFNR1-1pro:MdRFNR1-1/35S:MdSUVH1 than in MdRFNR1-2pro:MdRFNR1-2/35S:MdSUVH1 transgenic calli (Supplemental Figure S17C). After PEG treatment, the fresh weights of transgenic calli carrying 35S:MdSUVH1 and MdRFNR1-1pro:MdRFNR1-1 was greater than that of 35S:MdSUVH1 and MdRFNR1-2pro:MdRFNR1-2 transgenic calli under PEG treatment, although both transgenic calli had greater fresh weights than the wild type (Figure 10, H–J). POD and CAT activities were also higher in both transgenic calli than in the wild type after PEG treatment, and transgenic calli carrying 35S:MdSUVH1 and MdRFNR1-1pro:MdRFNR1-1 had even higher activities than transgenic calli carrying 35S:MdSUVH1 and MdRFNR1-2pro:MdRFNR1-2 (Figure 10, K and L). These results suggest that MdSUVH1 improves drought tolerance and that this improvement is associated with the induced expression of MdRFNR1-1 and ROS removal.
Discussion
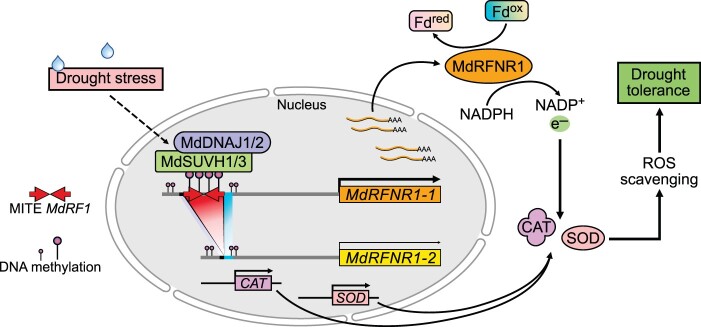
To deal with the redox imbalance caused by drought stress, plants have developed various mechanisms to detoxify ROS, including enzymatic and nonenzymatic mechanisms (Apel and Hirt, 2004). In this study, we characterized the role of a root-type FNR 1 (RFNR1) in redox regulation and drought tolerance. There are two alleles of MdRFNR1 in apple, MdRFNR1-1 and MdRFNR1-2. We found that the MdRFNR1-1 allele, but not MdRFNR1-2, is induced by drought stress. Further study revealed a MITE-MdRF1 insertion in the MdRFNR1-1 promoter, which can be methylated under drought stress. The methylated MITE is thereby recognized by MdSUVH1 and MdSUVH3, which recruit MdDNAJ1, MdDNAJ2, and MdDNAJ5 to activate MdRFNR1-1 expression in response to drought stress (Figure 11).
Figure 11.
A proposed working model of the role of the MdSUVH–MdDNAJ complex–MdRFNR1 module in the drought stress response in apple. Drought stress induces the expression of MdSUVH1, MdSUVH3, MdDNAJ1, and MdDNAJ2. The encoded proteins form a DNA methylation reader complex that binds to methylated MITE-MdRF1 in the MdRFNR1-1 promoter, thereby improving the expression of MdRFNR1-1. MdRFNR1-1 is responsible for redox balance by catalyzing the conversion of NADPH to NADP+ and promoting POD and CAT activities, leading to enhanced ROS removal, thereby playing a positive role in drought tolerance.
FNR proteins in plants are divided into two types, LFNR and RFNR, based on their functions and expression patterns. LFNR is not found in Arabidopsis root tissue, but RFNR proteins are found in both roots and shoots (Hanke et al., 2005; Grabsztunowicz et al., 2021). Similarly, we found that Malus RFNR1 was highly expressed in roots as well as in leaves (Supplemental Figure S1C). In leaves, Arabidopsis RFNR1 is expressed in leaf veins while RFNR2 is expressed in leaf tips, as revealed by observing GUS expression in RFNRpro:GUS transgenic plants. Notably, no gene expression signals of either RFNR were detected in green tissues (Grabsztunowicz et al., 2021). Strong MdRFNR1-1 signals were detected in every type of leaf tissue, including leaf veins, leaf tips, and green tissues. However, MdRFNR1-2 was only detected in cotyledons and the tips of mature leaves (Supplemental Figure S8), similar to RFNR2. These results suggest that the MITE-MdRF1 insertion in the MdRFNR1-1 promoter is critical for its tissue-specific expression in leaves.
Phylogenetic analysis and sequence alignment showed that RFNR proteins are highly conserved in all plants, even algae (Supplemental Figure S2), suggesting that RFNR might be an ancient gene with fundamental roles in plants. The high sequence similarity of RFNR among various species also suggests that RFNR might share similar functions in other species. RFNR catalyzes the conversion of NADPH, which is derived from the oxidative pentose phosphate pathway (oxPPP), to reduce oxidized Fd, resulting in NADP+ and reduced Fd. The latter participates in various plant processes, including ROS removal (Onda et al., 2000; Benz et al., 2010; Mulo, 2011; Kozuleva et al., 2016). In the current study, enzyme activity analysis demonstrated that MdRFNR1 has the ability to catalyze the conversion of NADPH to NADP+ in vivo and in vitro (Figures 1B, 2C, 3, B–D, 4, B–D, and 6, B–D), which is consistent with the enzyme activity of RFNR in maize (Onda et al., 2000).
FNR and RFNR play important roles in stress tolerance (Kozuleva et al., 2016; Grabsztunowicz et al., 2021), as their catalytic substrates Fd and NADPH feed electrons to the redox regulatory network (Dietz, 2013). After short-term drought treatment, the survival rates of transgenic MdRFNR1 RNAi lines were lower than that of GL-3, while the survival rates of the MdRFNR1-1 OE lines were higher (Figures 3E and 4E), suggesting that MdRFNR1 plays a positive role in drought tolerance. In plants, both enzymatic and nonenzymatic pathways can be used for ROS scavenging (Apel and Hirt, 2004). CAT and POD belong to enzymatic pathways and play very important roles in drought tolerance (Chakrabarty et al., 2016). Based on previous (Onda et al., 2000) and current results (Figures 1B, 2C, 3, B–D, 4, B–D, and 6, B–D), we hypothesized that MdRFNR1 catalyzes the reduction of Fd and that Fdred is involved in the removal of ROS by regenerating ascorbate for the electron donor of POD (Asada, 1999; Mittler et al., 2004). Indeed, POD and CAT activities were higher in MdRFNR1-1 OE lines and lower in MdRFNR1 RNAi lines under drought treatment (Figures 3 and 4, I and J), suggesting that MdRFNR1 positively regulates POD and CAT activities under drought stress. The substrate of POD and CAT, H2O2, accumulated to higher levels in MdRFNR1 RNAi lines (Figure 4H) and lower levels in MdRFNR1-1 OE lines compared with GL-3 under drought conditions (Figure 3H). Drought stress results in the accumulation of small hydrocarbons due to the peroxidation of membrane lipids, such as MDA (Gharibi et al., 2016), and an increase in ion leakage (Sun et al., 2018). In this study, we found the MDA levels and electrolyte leakage rates were higher in MdRFNR1 RNAi lines and lower in MdRFNR1-1 OE lines than in GL-3 under drought conditions (Figures 3 and 4, F and G), demonstrating that MdRFNR1 contributes to drought tolerance by alleviating cell membrane damage. Taken together, these data indicate that MdRFNR1 positively regulates drought tolerance, at least in part, by maintaining membrane integrity and by attenuating ROS production in response to drought.
The MITE in a promoter can result in altered gene expression and stress tolerance (Naito et al., 2009; Mao et al., 2015; Wang et al., 2020). Miniature Ping (mPing), a member of the PIF/Harbinger MITE superfamily in rice, which belongs to the same superfamily as MITE-MdRF1, was the first active MITE identified in any organism (Jiang et al., 2003; Kikuchi et al., 2003; Nakazaki et al., 2003). Sequences within mPing provide new binding sites for transcription factors or other regulatory proteins that act as enhancers for the cold stress response (Naito et al., 2009). In rice, a MITE insertion in the OsHKT1;5 promoter enhances its expression, which plays a positive role in plant tolerance to salinity stress (Wang et al., 2020). In maize, a MITE insertion in the ZmNAC111 promoter represses ZmNAC111 expression, which negatively affects drought tolerance (Mao et al., 2015).
In the current study, we identified two alleles of MdRFNR1 in GL-3, MdRFNR1-1, and MdRFNR1-2. The encoded proteins of the two alleles differ by three amino acids (Figure 1A); however, they had the same enzyme activity for catalyzing the NADPH-dependent reduction of Fd in vitro (Figure 1B) and share the same function in drought tolerance (Figure 2). The MdRFNR1-1 promoter harbors a MITE insertion, but the MdRFNR1-2 promoter does not. Allele-specific qPCR showed that drought improved the expression of MdRFNR1-1 but not MdRFNR1-2 and that MdRFNR1-1 was expressed at higher levels than MdRFNR1-2 even under normal conditions (Figure 5A), suggesting that MdRFNR1-1 has stronger promoter activity than MdRFNR1-2. GUS activity and GUS staining assays using transgenic calli and Arabidopsis plants carrying MdRFNR1-1pro:GUS or MdRFNR1-:GUS showed that MITE-MdRF1 plays an essential role in drought-induced expression of MdRFNR1-1 (Figure 5, B and C), indicating that MITE-MdRF1 functions as an enhancer for MdRFNR1-1 expression. Our results also showed that the MdRFNR1-1 allele containing the MITE-MdRF1 insertion in its promoter plays a larger role in drought tolerance by maintaining redox balance and membrane integrity under drought conditions (Figure 6). In addition, using natural populations, we found that Malus accessions with or without MITE-MdRF1 display distinct antioxidant enzyme activities under drought stress. The accessions containing the MITE-MdRF1 insertion in the RFNR1-1 promoter showed higher POD and CAT activities and better membrane integrity under drought than those without MITE-MdRF1 (Figure 7), indicating that natural variation of MITE-MdRF1 is associated with drought tolerance in natural populations.
TE insertions in the promoters of genes can usually be methylated by the RdDM pathway (Law and Jacobsen, 2010; Matzke and Mosher, 2014; Mao et al., 2015; Gagliardi et al., 2019; Xu et al., 2020). In maize, a MITE insertion in the ZmNAC111 promoter is methylated via the RdDM pathway (Mao et al., 2015). In rice, defects in the RdDM pathway affect the DNA methylation of TEs, including the MITE in the promoters of OsMIR156d and OsMIR156j and the MITE downstream of D14, resulting in a high-tillering phenotype (Xu et al., 2020). A MITE insertion in the promoter of sunflower HaWRKY6 can be transcribed into non-coding RNA and processed into 24-nt siRNAs, triggering DNA methylation at its locus through the RdDM pathway (Gagliardi et al., 2019). In the current study, structural analysis of MITE-MdRF1 in the MdRFNR1-1 promoter and an EST alignment assay suggested that non-coding RNAs derived from MITE-MdRF1 could generate sRNAs. Further analysis revealed that 24-nt sRNAs were abundant in MITE-MdRF1 and its flanking region (Figure 8A). In addition, locus-specific BS-seq showed that the DNA sequence of MITE-MdRF1 could be methylated (Figure 8C and Supplemental Figure S10B), especially the CHH and CHG contents, the hallmarks of RdDM (Sigman and Slotkin, 2016), indicating that MITE-MdRF1 in the MdRFNR1-1 promoter could be methylated by the RdDM pathway.
DNA methylation usually represses gene expression (Law and Jacobsen, 2010; Matzke and Mosher, 2014; Sigman and Slotkin, 2016; Ichino et al., 2021). The methyl readers MBD5 and MBD6 are recruited to chromatin by recognizing CG methylation. They then recruit the transcriptional repressor SILENZIO, which represses gene expression downstream of DNA methylation (Ichino et al., 2021). In maize, a methylated MITE insertion in the promoter of ZmNAC111 represses the expression of this gene (Mao et al., 2015). In rice, a methylated MITE in the OsMIR156d and OsMIR156j promoters represses the expression of miR156d and miR156j (Xu et al., 2020). However, several studies indicated that DNA methylation also facilitates the expression of nearby genes. RNA-seq on Osnrpd1a/b (mutants of NRPD1a and NRPD1b, two orthologs of the largest subunit of RNA Pol IV) (Xu et al., 2020) and OsDCL3a RNAi plants (Wei et al., 2014) suggested that RdDM plays a positive role in regulating gene expression. In Arabidopsis, DNA methylation in the ROS1 promoter can activate its expression (Lei et al., 2015). In the current study, we uncovered an association between increased MITE-MdRF1 methylation in the MdRFNR1-1 promoter and its expression in response to drought (Figure 8), providing further evidence for the activation of gene expression by DNA methylation.
Previous studies have revealed the molecular mechanisms of enhanced gene expression by DNA methylation (Harris et al., 2018). SUVH1 and SUVH3 act as methyl readers that bind to methylated DNA and recruit DNAJ1 and DNAJ2 to form an enhancer complex, thereby counteracting the repressive effects of TE insertions (Harris et al., 2018). In rice, OsSUVH7 recognizes the methylated MITE and forms a complex with OsBAG4 and OsMYB106, which facilitates the binding of OsMYB106 to the cis-element in the OsHKT1;5 promoter and activates the expression of this gene (Wang et al., 2020). Here, we found that the homologous proteins of SUVH1 and SUVH3 in apple bound to methylated but not non-methylated MITE-MdRF1 (Figure 9A).
In apple, three homologous proteins of DNAJ1 and DNAJ2, MdDNAJ1, MdDNAJ2, and MdDNAJ5, interact with MdSUVH1 and MdSUVH3 in vitro and in vivo (Supplemental Figure S13), forming a functional complex. Complexes comprising MdSUVH1 and MdDNAJ1, MdDNAJ2, or MdDNAJ5 could improve the promoter activity of MdRFNR1-1. In the absence of a methyl DNA reader, MdDNAJ1, MdDNAJ2, or MdDNAJ5 alone also activated the promoter activity of MdRFNR1-1 in N. benthamiana. We speculate that N. benthamiana SUVH1 and SUVH3 might contribute to the methyl DNA reader function due to the high sequence similarity of SUVH1 and SUVH3 between apple and N. benthamiana (Supplemental Figure S12A and Supplemental Table S4). However, DNAJ1 and DNAJ2 are not conserved between N. benthamiana and apple (Supplemental Figure S12B and Supplemental Table S5); therefore, MdSUVH1 alone failed to activate the MdRFNR1-1 promoter in N. benthamiana. In addition, MdRFNR1-2 expression was induced in N. benthamiana epidermal cells when MdSUVH1 and MdDNAJ1 or MdDNAJ2 were present (Figure 9), indicating that other proteins in N. benthamiana might be recruited to contribute to this induced expression of MdRFNR1-2.
DNAJ1 but not DNAJ2 and DNAJ3 have transcriptional activation activity in Saccharomyces cerevisiae (Harris et al., 2018; Zhao et al., 2019); however, DNAJ2 and DNAJ3 can activate gene expression in planta (Zhao et al., 2019). In the current study, we found that MdDNAJ1, MdDNAJ2, MdDNAJ3, and MdDNAJ5 lacked transcriptional activation activity in yeast (Supplemental Figure S13A). However, MdDNAJ1, MdDNAJ2, and MdDNAJ5 induced MdRFNR1-1 expression in N. benthamiana in response to drought (Figure 9, B and C). In addition, the expression of MdRFNR1-1 was elevated in MdDNAJ1 overexpressing apple plants (Supplemental Figure S17A). Considering that more members of the SUVH–DNAJ complex exist and can induce gene expression (Xiao et al., 2019; Zhao et al., 2019), it is possible that other members of the MdSUVH–MdDNAJ complex are responsible for the transcriptional activation of MdRFNR1-1 in apple in response to drought. It is also possible that other factors, including transcriptional activators, might be involved in the activated expression of MdRFNR1-1 in response to drought.
In conclusion, we identified the redox regulation enzyme MdRFNR1, which positively regulates drought tolerance in apple. Sequence analysis identified its two natural alleles, MdRFNR1-1 and MdRFNR1-2, with the former being responsive to drought and containing a MITE-MdRF1 insertion in its promoter. Furthermore, we revealed that MITE-MdRF1 is essential for the induced expression of MdRFNR1-1 in response to drought. We also showed that the MdRFNR1-1 promoter could be methylated and recognized by the MdSUVH–MdDNAJ complex, which activated the expression of MdRFNR1-1 under drought. In addition, MdRFNR1, MdSUVH1, and MdDNAJ1 are positive regulators of drought tolerance. Our study provides information about the role of the natural variation of MdRFNR1 in the response of apple to drought stress, which will be useful for breeding drought-tolerant plants by genetic engineering in the future.
Materials and methods
Plant material and growth conditions
Tissue-cultured GL-3 (the progeny of Royal Gala apple, which was used as the background for apple transformation) and transgenic plants were subcultured every 4 weeks on Murashige & Skoog (MS) medium (4.43 g/L MS salts, 30 g/L sucrose, 0.2 mg/L 6-Benzylaminopurine [6-BA], 0.2 mg/L 3-Indoleacetic acid [IAA], and 7.5 g/L agar, pH 5.8) under long-day conditions (14 h light [cool white, ∼100 μmol m−2 s−1, T5 LED batten]: 10 h dark) at 25°C (Xie et al., 2018). Tissue-cultured plants were rooted, transplanted into soil, and grown in a growth chamber at Northwest A&F University, Yangling, China (34°20′N, 108°24′E) (16-h light:8-h dark, 25°C, ∼55% relative humidity).
Apple calli (M. domestica cv. Orin) were grown on MS medium (4.43 g/L MS salts, 1.5 mg/L 2,4-dichlorophenoxyacetic acid [2,4-D], 0.4 mg/L 6-BA, 30 g/L sucrose, and 8 g/L agar, pH 5.8) in darkness at 25°C as previously described (Niu et al., 2019).
Vector construction and genetic transformation
The leaves of GL-3 were used for gene cloning of MdRFNR1-1 and MdRFNR1-2. The full-length sequence of MdRFNR1-1 or MdRFNR1-2 was introduced into the binary vector pK2GW7 (TAIR accession number: 6531113855) through Gateway technology to generate the OE vector. A 321-bp fragment of MdRFNR1 was introduced into the pK7GWIWG2D vector (TAIR accession number: 6531113856) to generate the RNAi plasmid. All plasmids were transformed into Agrobacterium tumefaciens strain EHA105, followed by transformation into GL-3 (Dai et al., 2013) or wild-type calli (Niu et al., 2019). Briefly, to generate transgenic apple plants, A. tumefaciens carrying the plasmid was transformed into GL-3 plants, which were used as the genetic background for genetic transformation. Transgenic plants were screened on MS medium (4.43 g/L MS salts, 30 g/L sucrose, 0.2 mg/L 6-BA, 0.2 mg/L IAA, and 7.5 g/L agar, pH 5.8) supplemented with 50 mg/L kanamycin and 250 mg/L cefotaxime. After propagation, transgenic plants were verified by qRT-PCR analysis. To generate transgenic calli, wild-type “Orin” apple calli were used as the background for Agrobacterium-mediated transformation. Transgenic calli were screened on MS medium (4.43 g/L MS salts, 1.5 mg/L 2,4-D, 0.4 mg/L 6-BA, 30 g/L sucrose, and 8 g/L agar) containing 50 mg/L kanamycin and 250 mg/L cefotaxime.
To construct the MdRFNR1-1pro:MdRFNR1-1 or MdRFNR1-2pro:MdRFNR1-2 plasmid, the genomic sequence of MdRFNR1-1 or MdRFNR1-2 (including the promoter, intron, and exon sequence) was cloned from GL-3 leaves and introduced into a modified pCAMBIA1300 vector (with the kanamycin resistance gene). The constructed MdRFNR1-1pro:MdRFNR1-1 (modified pCAMBIA1300 vector, kanamycin resistance) or MdRFNR1-2pro:MdRFNR1-2 (modified pCAMBIA1300 vector, kanamycin resistance) was transformed into A. tumefaciens strain EHA105, followed by GL-3 plants or calli as previously described (Niu et al., 2019).
To generate MdDNAJ1 or MdSUVH1 overexpressing apple plants or calli, the coding sequence (CDS) of MdDNAJ1 or MdSUVH1 was cloned and introduced into the pK2GW7 or pCAMBIA1300 vector. The resulting 35S:MdDNAJ1 (pK2GW7 vector) was transformed into A. tumefaciens strain EHA105, then transformed into GL-3 plants by an Agrobacterium-mediated transformation. The 35S:MdSUVH1 (pCAMBIA1300 vector, hygromycin resistance) plasmid was transformed into A. tumefaciens strain EHA105, then co-transformed with MdRFNR1-1pro:MdRFNR1-1 (modified pCAMBIA1300 vector, kanamycin resistance) or MdRFNR1-2pro:MdRFNR1-2 (modified pCAMBIA1300 vector, kanamycin resistance) into apple calli as previously described (Niu et al., 2019).
To generate NbSUVH1- and NbSUVH3-silenced N. benthamiana plants, the VIGS assay was performed as previously described (Zhang and Liu, 2014). Briefly, the 300 bp segments of NbSUVH1 and NbSUVH3, which were analyzed on the VIGS tool website (https://vigs.solgenomics.net/), were separately introduced into the pTRV2 vector (TAIR accession number: 6530604964). The pTRV1 plasmid (TAIR accession number: 5019327237), the empty vector pTRV2, the pTRV2-NbSUVH1 construct, and the pTRV2-NbSUVH3 construct were individually transformed into A. tumefaciens strain GV3101. Equal volumes of A. tumefaciens cultures at OD600 = 0.5 harboring pTRV1, pTRV2-NbSUVH1, and pTRV2-NbSUVH3 (or pTRV2, as a negative control) were mixed and infiltrated into 3-week-old N. benthamiana plants. After 20 days, the expression levels of NbSUVH1 and NbSUVH3 were analyzed by qRT-PCR. Nicotiana benthamiana Actin served as the reference gene for normalization (Wang et al., 2013) and the silenced plants were used for further study. The primers used are listed in Supplemental Data Set S1 and primer efficiency is shown in Supplemental Figure S18.
Drought treatment
PEG treatment of hydroponically cultured plants was performed as previously described (Geng et al., 2018, 2019). In brief, 3-month-old soil-grown plants were transferred to Hoagland solution for an additional month, followed by Hoagland solution containing 20% (w/v) PEG8000 (Sigma, P2139, USA) for 6 h. As a control, plants were grown in standard Hoagland solution. Leaves and roots were washed quickly and collected. Experiments were performed at least twice and 12 plants were used for each experiment, with 4 plants as one replicate.
For PEG treatment of apple calli, wild-type or transgenic calli were grown on MS medium for 20 days and transferred to MS agar medium supplemented with 1.2 g/L 2-(N-morpholino) ethane sulfonic acid (MES) pretreated with PEG8000 (−0.7 MPa) or standard MS agar medium supplemented with MES (Verslues et al., 2006; Chen et al., 2021). All calli had the same weight at the beginning of the treatment. The calli were grown in the dark at 25°C for 20 days for further measurement. Experiments were performed at least twice and eight–nine plates were used for each experiment.
For PEG treatment of tissue-cultured plants, 1-month-old plants grown on MS medium were soaked in 40 mL of MS medium (without agar) supplemented with 20% (w/v) PEG8000 for 0 or 6 h. Experiments were performed at least twice and 12 plants were used for each experiment, with pooled leaves from 4 plants as one biological replicate.
Short-term drought treatment was carried out as previously described (Li et al., 2020). Briefly, water was withheld from the plants for 10 or 15 days, the plants were recovered for 7 days, and the survival rates were calculated. After drought treatment, leaves were collected for measurement of H2O2, POD activity, CAT activity, MDA, NADP+ and NADPH contents, and ion leakage. The photosynthetic rates were measured with an LI-6400XT portable photosynthesis system (LI-COR, USA) from 8 a.m. to 10 a.m. in the growth chamber (room temperature: 25°C ± 2°C, humidity: ∼55%) after 10 or 7 days of drought. The photosynthesis photon flux density was 300 μmol m−2 s−1 and the airflow rates were a constant 500 μmol s−1. Experiments were performed at least three times.
Sequence alignment and phylogenetic analysis
For RFNR1 protein identification, the full-length amino acid sequences of RFNR1 from different species were obtained from the National Center for Biotechnology Information (NCBI, https://blast.ncbi.nlm.nih.gov/Blast.cgi) by Blastp and the sequences with the highest identity were used for further analysis. To identify homologs of SUVH1, SUVH3, DNAJ1, and DNAJ2 from apple, Blastp was performed on the NCBI website and the proteins that had a high similarity with SUVH1, SUVH3, DNAJ1, and DNAJ2 were used for further analysis. Sequence alignments were performed using the MUSCLE program (http://www.ebi.ac.uk/Tools/msa/muscle) and the domain was predicted using the SMART program (http://smart.embl-heidelberg.de/). Using complete protein sequences, the phylogenetic tree was constructed by rooting at the midpoint using the neighbor‐joining method in the MEGA5.2.2 program (Tamura et al., 2011). The reliability of the tree was estimated by bootstrapping using 2,000 replications. The evolutionary distances were computed using the Poisson correction method. Alignments used for phylogenetic analysis are provided in Supplemental Files S1–S6.
For genomic analysis of MdRFNR1-1, the gene structure was drawn on the website GSDS v2.0 (http://gsds.gao-lab.org/). The alignment of the MdRFNR1-1 locus (−2,798 to 2,584 bp) against an apple ESTs database was performed on NCBI website by Blastn. The EST reads with the highest similarity to MITE-MdRF1 were used for structural analysis, which was predicted using the ViennaRNA Web servers (http://rna.tbi.univie.ac.at/). For 24-nt sRNA alignment, sRNA sequence data from GL-3 plants under normal conditions were used for analysis. First, the lengths of reads were determined and 24-nt reads were separated into new fasta format files. The 24-nt sRNAs were then mapped to the MdRFNR1-1 locus using bowtie with the parameters “-f -v 1 -m 20 -S -a –best –strata –no-unal” (Langmead et al., 2009). Finally, visual verification was performed using IGV software.
qRT-PCR and allele-specific qRT-PCR analysis
The CTAB method was used to extract RNA from Malus leaves (Chang et al., 1993) and DNA was removed by digestion with RNase-free Dnase I (Thermo Scientific, EN0521, USA). Four leaves collected from three trees were pooled as one biological replicate. Three biological replicates with three technical repeats were used.
To extract RNA from roots, an RNAprep Pure Plant PlusKit (TIANGEN, DP441, China) was used, following the manufacturer’s instructions. For qRT-PCR analysis, ∼1 μg of total RNA was reverse transcribed with oligo-dT to first-strand cDNA using a RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific, K1622, USA). ChamQ Universal SYBR qPCR Master Mix (Vazyme, C601, China) was used to perform qRT-PCR on the CFX96 real-time system (Bio-Rad, USA). Malate dehydrogenase (MdMDH) was used as the reference gene and relative expression was calculated by the 2−ΔΔCt method (Livak and Schmittgen, 2001). All the roots of three hydroponically cultured apple trees were pooled as one biological replicate. Three biological replicates with three technical repeats were used.
A specific forward primer based on the nucleotide difference at the 5′-UTRs of MdRFNR1-1 and MdRFNR1-2 and a common reverse primer were designed and used for qRT-PCR analysis to detect the differential expression of the two alleles.
All primers used are listed in Supplemental Data Set S1 and primer efficiency is shown in Supplemental Figure S18.
Tissue-specific expression of RFNR1 in Malus fusca
Fragments Per Kilobase Million of RFNR1 in different tissues of M. fusca were retrieved from the apple eFP browser: http://bar.utoronto.ca/∼asher/efp_apple/cgi-bin/efpWeb.cgi.
MdRFNR1 enzyme assay
The CDS of MdRFNR1-1 or MdRFNR1-2 was cloned into the pMAL-c5X plasmid (NEB, N8108S, USA). The resulting MBP-MdRFNR1-1 plasmid was used as a template to generate a catalytically dead version of MBP-MdRFNR1-1 by site-directed mutagenesis (Arg299Gln and Lys308Gln) (Kimata-Ariga et al., 2019), resulting in MBP-MdRFNR1-1-M. The CDS of MdRFNR1-1 without the NAD domain was also cloned into pMAL-c5X. The resulting plasmids (MBP-MdRFNR1-1, MBP-MdRFNR1-1-M, MBP-MdRFNR1-1-△NAD, and MBP-MdRFNR1-2) were individually transformed into E. coli strain Rosetta2 (DE3). Fusion protein production was induced at 28°C at 150 rpm for 8 h and the fusion protein was purified with amylose resin (NEB, E8021S, USA) following the manufacturer’s instructions. Fusion protein purification was performed three times.
The purified MBP-fused proteins were used for the enzyme assay. Diaphorase activity was measured using DCPIP, an electron acceptor (Onda et al., 2000). The reaction buffer contained 50 mM Tris–HCl (pH = 7.5), 1 mM MgCl2, 550 μM DCPIP, 3 mM Glucose-6-phosphate (Glc-6-P), 50 μg Glc-6-P dehydrogenase, 100 nM purified proteins, and a series concentration of NADPH (from 3.125 to 200 μM). The absorbance curve was monitored at 600 nm in a microplate reader (TECAN, M200PRO). Purified fusion proteins from three experiments were used for the enzyme activity assay. For each enzymatic reaction, two or three technical repeats were used. This experiment was repeated three times.
Determination of H2O2 contents, POD activity, CAT activity, MDA content, NADP+ and NADPH contents, and ion leakage
POD and CAT activities and MDA contents were measured as previously described (Wang et al., 2012; Xie et al., 2018; Chen et al., 2021). For POD and CAT activities analysis, ∼0.1 g of leaf or callus tissue was extracted with 1 mL of extraction buffer (100 mM phosphate buffer [pH = 7.0], 1 mM ethylenediaminetetraacetic acid [EDTA], 0.1% Triton-X-100, and 1% polyvinyl pyrrolidone [PVP]) at 4°C. The POD reaction solution contained 50 mM phosphate buffer (pH = 7.0), 100 mM H2O2, 100 mM guaiacol, and a small volume of enzyme extract. Changes in absorbance of the POD reaction solution were monitored at 470 nm in a microplate reader. The CAT reaction solution contained 50 mM phosphate buffer (pH = 7.8), 50 mM H2O2, and a small volume of enzyme extract. Changes in absorbance of the CAT reaction solution were monitored at 240 nm in a microplate reader. POD activity is defined as the amount of guaiacol oxidized by H2O2 to 3,3′-dimethoxy-4,4′-biphenylquinone per fresh weight (g) per second. CAT activity is defined as the amount of H2O2 decomposed by the enzyme to water and oxygen per fresh weight (g) per second. For MDA contents measurement, the extracts for the POD or CAT assay were mixed with 0.5% 2-thiobarbituric acid (TBA) and 4.5% trichloroacetic acid (TCA) and boiled for 20 min. The mixture was centrifuged and the absorbance of the supernatant was measured at 450, 532, and 600 nm in a microplate reader.
DAB staining was performed as previously described (Xie et al., 2018). H2O2 quantification was performed using a Hydrogen Peroxide Assay Kit (Comin, H2O2-1-Y, China). NADP+ and NADPH contents were determined using a Coenzyme II NADP(H) Content Assay Kit (Comin, NADP-1-Y, China) according to the manufacturer’s manual.
The ion leakage assay was performed as previously described (Chen et al., 2021). Experiments were performed at least three times.
DNA methylation assay, 5-AZA treatment, and McrBC-qPCR
For locus-specific BS-seq, genomic DNA was extracted from GL-3 leaves using the CTAB method (Chang et al., 1993) with the RNA removed by RNase A treatment (Thermo Scientific, EN0531, USA). DNA from GL-3 roots was extracted using a Super Plant Genomic DNA Kit (TIANGEN, DP360, China). Approximately 300 ng DNA was treated with bisulfite using an EZ DNA Methylation-Gold Kit (Zymo Research, D5005, USA) and amplified by PCR using ZymoTaq PreMix (Zymo Research, E2003, USA). The amplified products were recovered from an agarose gel using a GeneJET Extraction and DNA Cleanup Micro Kit (Thermo Scientific, K0832, USA) and cloned into the pClone007 vector using a Versatile Simple Vector Kit (TSINGKE, 007VS, China). At least 14 clones were sequenced per genotype. The final results were analyzed using Kismeth online software (Gruntman et al., 2008). All primers used are listed in Supplemental Data Set S1.
5-AZA (Sigma, A3656, USA) treatment was performed as previously described with minor modifications (Li et al., 2016). Subcultured GL-3 plants were transferred to MS medium supplemented with 7 μg/mL 5-AZA or DMSO for an additional 3 weeks under long-day conditions (14-h light: 10-h dark) at 25°C and then treated with 20% PEG8000 (w/v) for 0 or 6 h.
For McrBC-qPCR, genomic DNA from GL-3 plants, which were treated as described above, was extracted using a Super Plant Genomic DNA Kit (TIANGEN, DP360, China) as previously described (Mao et al., 2015; Ghoshal et al., 2021) with minor modifications. Briefly, ∼200 ng of DNA was digested for 16 h at 37°C with McrBC (Takara, 1234A, Japan), followed by 20 min at 65°C to inactivate the enzyme. A parallel reaction, in which equal amounts of DNA were placed in buffer without the enzyme, was used as a negative control. The digested DNA was diluted four times, followed by qPCR. The McrBC digestion at the MITE-MdRF1 region was normalized to the reference gene MdMDH and then to the undigested control. Experiments were performed three times. All the leaves collected from four tissue-cultured plants were pooled as one biological replicate. Three biological replicates were used for each experiment.
Promoter:GUS analysis
The promoter (2,759 bp) of MdRFNR1-1 was amplified from GL-3 and cloned into the binary vector pMDC164 (TAIR accession number: 1009003759) through Gateway technology, resulting in the MdRFNR1-1pro:GUS plasmid. The MdRFNR1-1 promoter without the MITE was amplified by PCR using the MdRFNR1-1pro:GUS plasmid as the template, and the products were introduced into the binary vector pMDC164, resulting in MdRFNR1-:GUS. Both plasmids were transformed into Arabidopsis (ecotype Columbia-0) or calli by A. tumefaciens-mediated transformation. Transgenic calli or Arabidopsis roots were soaked in 20% PEG8000 (w/v) solution for 6 h. GUS staining was performed as previously described (Guan et al., 2013). GUS activity was measured using a GUS Gene Quantitative Detection Kit (Coolaber, SL7161, China) according to the manufacturer’s manual. Both experiments were performed three times, with an individual callus as one biological replicate, and at least four biological replicates for each experiment were used.
DAP-qPCR analysis
DAP-qPCR was carried out as previously described (Bartlett et al., 2017), with minor modifications. To generate the methylated DAP library and the non-methylated amp-DAP library, genomic DNA was extracted from GL-3 leaves by the CTAB method (Chang et al., 1993), using RNase A (Thermo Scientific, EN0531, USA) to remove the residual RNA. The genomic DNA was diluted to 10 ng/μL in nuclease-free water and sonicated into 200–400 bp DNA fragments. Approximately 500 μL of sonicated DNA was transferred to a new tube, followed by the addition of the adapter to the DNA ends. Sonicated DNA amplified with Q5 High-Fidelity DNA Polymerase (NEB, M0491S, USA) was used to construct the non-methylated amp-DAP library, whereas the remaining sonicated DNA was used to construct the methylated DAP library.
To express GST-fused MdSUVH1 and MdSUVH3, the TNT SP6 High-Yield Wheat Germ System (Promega, L3261, USA) was used. Briefly, the full-length sequences of MdSUVH1 and MdSUVH3 were cloned into the pET42a(+) vector (Novagen, USA) and Q5 High-Fidelity DNA Polymerase was used to amplify GST-MdSUVH1 and GST-MdSUVH3 with the forward primer (5′-GACTCATATTTAGGTGACACTATAGAACAGACCACCATGGGTATGTCCCCTATACTAGG-3′) and the reverse primer (5′-TTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTAGTGGTGGTGGTGGTGGT-3′). Amplified DNA was recovered from the agarose gel using a GeneJET Extraction and DNA Cleanup Micro Kit (Thermo Scientific, K0832, USA), followed by agarose gel electrophoresis to verify the correct product. Approximately 1 μg of the PCR product (no more than 8 μL) was added to 30 μL TNT SP6 High-Yield Wheat Germ Master Mix, with nuclease-free water was added to the final volume of 50 μL. GST-MdSUVH1 (wild type or mutant), GST-MdSUVH3 (wild type or mutant), and GST protein were then expressed at 25°C for 2 h.
For DAP-qPCR, 30 μL MagneGST Glutathione Particles (Promega, V8611, USA) was used to bind 15 μL GST-fusion protein at 4°C for 1 h and washed three times with MagneGST Binding/Wash Buffer. After incubation with 100 ng of the DAP library or amp-DAP library at 4°C for 1 h, unbound DNA was removed by washing four times with MagneGST Binding/Wash Buffer. Finally, 100 μL of elution buffer was used to resuspend the complex, followed by boiling for 10 min, and the supernatant containing recovered DNA was transferred to a new tube and used for qPCR analysis. ChamQ Universal SYBR qPCR Master Mix (Vazyme, C601, China) was used to perform qPCR analysis on a CFX96 real-time system (Bio-Rad, USA) and the relative enrichment was calculated by the 2−ΔΔCt method (Livak and Schmittgen, 2001). A fragment located in the MdRFNR1 exon and intron was used as the reference gene. DNA enrichment by GST protein was used as the negative control. All primers used are listed in Supplemental Data Set S1 and primer efficiency is shown in Supplemental Figure S18. Experiments were performed three times. In each experiment, the protein was expressed three times and the same library was used for affinity purification by proteins expressed from different batches.
Dual-luciferase assay
The DUAL-LUC was performed as previously described (Hellens et al., 2005; Xie et al., 2018). The CDS of MdSUVH1, MdDNAJ1, MdDNAJ2, and MdDNAJ5 was individually cloned into the pGreen62-SK vector (Hellens et al., 2005). The promoter fragments of MdRFNR1-1 and MdRFNR1-2 were separately introduced into the pGreen0800-LUC vector (Hellens et al., 2005). The resulting constructs were individually transformed into A. tumefaciens strain GV3101 and the different combinations were injected into 4-week-old N. benthamiana leaves. Leaf samples were collected after 3 days of co-infiltration, and firefly luciferase and Renilla luciferase activity was detected with a Dual Luciferase Reporter Gene Assay Kit (Yeasen, 11402ES60, China). For the dehydration analysis, leaves of infiltrated N. benthamiana plants were detached and air dried for 3 h. Experiments were performed three times. Leaf discs taken from six leaves were pooled as a biological replicate, at least eight biological replicates for each experiment were used.
Y2H assay
The Y2H assay was performed according to the manual of the Matchmaker Gold Yeast Two-Hybrid System (Clontech, USA). Briefly, the CDS of MdSUVH1 or MdSUVH3 was amplified by PCR and cloned into the pGADT7 (AD, Clontech, 630442, USA) vector and the CDSs of MdDNAJ1, MdDNAJ2, MdDNAJ3, and MdDNAJ5 were individually cloned into the pGBKT7 (BD, Clontech, 630443, USA) vector. The different AD and BD combinations were co-transformed into yeast (S. cerevisiae) strain Y2H Gold. The transformed yeast cells were grown on SD medium, which lacked leucine and tryptophan, or leucine, tryptophan, histidine, and adenine. Experiments were performed three times.
Split-LUC assay
The split-LUC assay was performed as previously described (Chen et al., 2021). The CDS of MdSUVH1 was cloned into the pCAMBIA1300-cLuc vector (Addgene accession number: 174051) and the CDSs of MdDNAJ1, MdDNAJ2, MdDNAJ3, and MdDNAJ5 were individually cloned into the pCAMBIA1300-nLuc vector (Addgene accession number: 174050). The resulting constructs were then transformed into A. tumefaciens strain C58C1 and the different cLuc and nLuc combinations were co-injected with 35S:p19 into N. benthamiana leaves. After 3 days of co-infiltration, the luciferase signal was captured as previously described (Ishitani et al., 1997) with a CCD camera (Lumazone Pylon 2048B, Princeton, NJ, USA). Experiments were performed three times. The injected leaves within one plant were pooled as a biological replicate and six biological replicates for each experiment were used.
Statistical analysis
Statistical significance was determined by Student’s t test or one-way ANOVA (Tukey’s test) analysis using Prism 5.0 software (GraphPad Software, USA). Differences were considered significant if P < 0.05, 0.01, or 0.001. The statistical results are shown in Supplemental Data Set S2.
Accession numbers
Sequence data used in this article can be found in GenBank under the following accession numbers: OL763817 (MdRFNR-1-1pro:MdRFNR1-1), OL763818 (MdRFNR-1-2pro:MdRFNR1-2), OL763829 (MdSUVH1), OL763830 (MdSUVH3), OL763825 (MdDNAJ1), OL763826 (MdDNAJ2), OL763827 (MdDNAJ3), and OL763828 (MdDNAJ5).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1 . RFNR1 expression patterns under PEG and short-term drought treatment and in different tissues.
Supplemental Figure S2. Sequence alignment and phylogenetic analysis of RFNR in different species.
Supplemental Figure S3. MdRFNR1 expression and purification.
Supplemental Figure S4. Expression of MdRFNR1 in 35S:MdRFNR1-1 and 35S:MdRFNR1-2 transgenic calli, 35S:MdRFNR1-1 OE and MdRFNR1 RNAi transgenic plants.
Supplemental Figure S5. Photosynthetic rates of GL-3 and transgenic plants under control and drought treatment.
Supplemental Figure S6. The TE (MITE-MdRF1) insertion in the MdRFNR1-1 promoter.
Supplemental Figure S7. Detection of the MITE-MdRF1 insertion in the RFNR1 promoter in different Malus accessions.
Supplemental Figure S8 . GUS staining of Arabidopsis plants carrying MdRFNR1-1pro:GUS or MdRFNR1-:GUS under control conditions.
Supplemental Figure S9. Expression of MdRFNR1 in MdRFNR1-1pro:MdRFNR1-1 and MdRFNR1-2pro:MdRFNR1-2 transgenic plants and calli.
Supplemental Figure S10. DNA methylation of the MITE-MdRF1 insertion in the MdRFNR1-1 promoter is positively associated with MdRFNR1-1 expression in GL-3 roots in response to PEG.
Supplemental Figure S11. Methylation of the two MdRFNR1 alleles in leaves and roots of GL-3 under control conditions and PEG treatment.
Supplemental Figure S12. Sequence alignment and phylogenetic analysis of SUVH1, SUVH3, DNAJ1, and DNAJ2 in different species.
Supplemental Figure S13. Interaction of MdSUVH1 and MdSUVH3 with MdDNAJ1, MdDNAJ2, and MdDNAJ5 in Y2H and split-LUC assays.
Supplemental Figure S14. MdRFNR1-1 expression in NbSUVH1- and NbSUVH3-silenced N. benthamiana plants.
Supplemental Figure S15. MdDNAJ1, MdDNAJ2, MdSUVH1, and MdSUVH3 are induced by drought treatment.
Supplemental Figure S16. Expression of MdDNAJ1 or MdSUVH1 in 35S:MdDNAJ1 OE transgenic apple plants and transgenic calli carrying 35S:MdSUVH1 + MdRFNR1-1pro:MdRFNR1-1 or 35S:MdSUVH1 + MdRFNR1-2pro:MdRFNR1-2.
Supplemental Figure S17. MdRFNR1-1 and MdRFNR1-2 expression in 35S:MdDNAJ1 OE plants and MdRFNR1 expression in transgenic calli carrying 35S:MdSUVH1 and MdRFNR1-1pro:MdRFNR1-1 or MdRFNR1-2pro: MdRFNR1-2.
Supplemental Figure S18. Efficiency (E) of primers used for qRT-PCR.
Supplemental Table S1. Sequence similarity and identity of RFNR1 in different species.
Supplemental Table S2. NADPH-dependent enzyme activity of MdRFNR1 with DCPIP.
Supplemental Table S3. Names of the Malus accessions containing MITE-MdRF1 in the RFNR1 promoter.
Supplemental Table S4. Sequence similarity and identity of SUVH1 and SUVH3 in different species.
Supplemental Table S5. Sequence similarity and identity of DNAJ1 and DNAJ2 in different species.
Supplemental Data Set S1. List of primers used in this study.
Supplemental Data Set S2. Summary of statistical analyses.
Supplemental File S1. FNR sequence alignment.
Supplemental File S2. FNR phylogenetic tree.
Supplemental File S3. SUVH sequence alignment.
Supplemental File S4. SUVH phylogenetic tree.
Supplemental File S5. DNAJ sequence alignment.
Supplemental File S6. DNAJ phylogenetic tree.
Supplementary Material
Acknowledgments
We thank Dr. Chunxiang You from Shandong Agricultural University for providing wild-type “Orin” calli, Dr. Zhihong Zhang from Shenyang Agricultural University for providing tissue-cultured GL-3 plants, and the High Performance Computing (HPC) platform of Northwest A&F University (NWAFU) for providing computing resources. We also thank Dr. Xiangqiang Zhan and Ms. Fang Ma for providing the modified pCAMBIA1300 vector.
Funding
This work was supported by the National Natural Science Foundation of China (32172530), the Central Funds Guiding the Local Science and Technology Development of Shenzhen (2021SZVUP117), the Key S&T Special Projects of Shaanxi Province, China (2020zdzx03-01-02), and the China Postdoctoral Science Foundation (2021M702692).
Conflict of interest statement. None declared.
Contributor Information
Chundong Niu, State Key Laboratory of Crop Stress Biology for Arid Areas/Shaanxi Key Laboratory of Apple, College of Horticulture, Northwest A&F University, Yangling 712100, China; College of Life Science, Northwest A&F University, Yangling 712100, China.
Lijuan Jiang, State Key Laboratory of Crop Stress Biology for Arid Areas/Shaanxi Key Laboratory of Apple, College of Horticulture, Northwest A&F University, Yangling 712100, China.
Fuguo Cao, State Key Laboratory of Crop Stress Biology for Arid Areas/Shaanxi Key Laboratory of Apple, College of Horticulture, Northwest A&F University, Yangling 712100, China.
Chen Liu, State Key Laboratory of Crop Stress Biology for Arid Areas/Shaanxi Key Laboratory of Apple, College of Horticulture, Northwest A&F University, Yangling 712100, China.
Junxing Guo, State Key Laboratory of Crop Stress Biology for Arid Areas/Shaanxi Key Laboratory of Apple, College of Horticulture, Northwest A&F University, Yangling 712100, China.
Zitong Zhang, State Key Laboratory of Crop Stress Biology for Arid Areas/Shaanxi Key Laboratory of Apple, College of Horticulture, Northwest A&F University, Yangling 712100, China.
Qianyu Yue, State Key Laboratory of Crop Stress Biology for Arid Areas/Shaanxi Key Laboratory of Apple, College of Horticulture, Northwest A&F University, Yangling 712100, China.
Nan Hou, State Key Laboratory of Crop Stress Biology for Arid Areas/Shaanxi Key Laboratory of Apple, College of Horticulture, Northwest A&F University, Yangling 712100, China.
Zeyuan Liu, State Key Laboratory of Crop Stress Biology for Arid Areas/Shaanxi Key Laboratory of Apple, College of Horticulture, Northwest A&F University, Yangling 712100, China.
Xuewei Li, State Key Laboratory of Crop Stress Biology for Arid Areas/Shaanxi Key Laboratory of Apple, College of Horticulture, Northwest A&F University, Yangling 712100, China; College of Life Science, Northwest A&F University, Yangling 712100, China.
Muhammad Mobeen Tahir, State Key Laboratory of Crop Stress Biology for Arid Areas/Shaanxi Key Laboratory of Apple, College of Horticulture, Northwest A&F University, Yangling 712100, China.
Jieqiang He, State Key Laboratory of Crop Stress Biology for Arid Areas/Shaanxi Key Laboratory of Apple, College of Horticulture, Northwest A&F University, Yangling 712100, China.
Zhongxing Li, State Key Laboratory of Crop Stress Biology for Arid Areas/Shaanxi Key Laboratory of Apple, College of Horticulture, Northwest A&F University, Yangling 712100, China.
Chao Li, State Key Laboratory of Crop Stress Biology for Arid Areas/Shaanxi Key Laboratory of Apple, College of Horticulture, Northwest A&F University, Yangling 712100, China.
Fengwang Ma, State Key Laboratory of Crop Stress Biology for Arid Areas/Shaanxi Key Laboratory of Apple, College of Horticulture, Northwest A&F University, Yangling 712100, China.
Qingmei Guan, State Key Laboratory of Crop Stress Biology for Arid Areas/Shaanxi Key Laboratory of Apple, College of Horticulture, Northwest A&F University, Yangling 712100, China.
Q.G., C.N., and L.J. designed the experiments. C.N., L.J., F.C., C.Liu., J.G., Z.Z., Q.Y., N.H., X.L., ZY.L., J.H., and ZX.L. performed the experiments. Q.G., C.N., L.J., F.C., C.Li., and F.M. analyzed the data. M.M.T. provided helpful suggestions for improving the article. C.N. and Q.G. wrote the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) is: Qingmei Guan (qguan@nwafu.edu.cn).
References
- Apel K, Hirt H (2004) Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol 55: 373–399 [DOI] [PubMed] [Google Scholar]
- Asada K (1999) The water–water cycle in chloroplasts: scavenging of active oxygens and dissipation of excess photons. Annu Rev Plant Physiol Plant Mol Biol 50: 601–639 [DOI] [PubMed] [Google Scholar]
- Bartlett A, O’Malley RC, Huang SC, Galli M, Nery JR, Gallavotti A, Ecker JR, Bartlett A, O’Malley RC, Huang S-SC, et al. (2017) Mapping genome-wide transcription-factor binding sites using DAP-seq. Nat Protoc 12: 1659–1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benz JP, Lintala M, Soll J, Mulo P, Bölter B (2010) A new concept for ferredoxin-NADP(H) oxidoreductase binding to plant thylakoids. Trends Plant Sci 15: 608–613 [DOI] [PubMed] [Google Scholar]
- Blevins T, Podicheti R, Mishra V, Marasco M, Wang J, Rusch D, Tang H, Pikaard CS (2015) Identification of Pol IV and RDR2-dependent precursors of 24 nt siRNAs guiding de novo DNA methylation in Arabidopsis. eLife 4: e09591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butelli E, Licciardello C, Zhang Y, Liu J, Mackay S, Bailey P, Reforgiato-Recupero G, Martin C (2012) Retrotransposons control fruit-specific, cold-dependent accumulation of anthocyanins in blood oranges. Plant Cell 24: 1242–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, Jacobsen SE (2002) Role of the Arabidopsis DRM methyltransferases in de novo DNA methylation and gene silencing. Curr Biol 12: 1138–1144 [DOI] [PubMed] [Google Scholar]
- Chakrabarty A, Aditya M, Dey N, Banik N, Bhattacharjee S (2016) Antioxidant signaling and redox regulation in drought- and salinity-stressed plants. InHossain MA, Wani SH, Bhattacharjee S, Burritt DJ, Tran L-SP, eds, Drought Stress Tolerance in Plants. Vol 1: Physiology and Biochemistry, Springer International Publishing, Cham, pp 465–498 [Google Scholar]
- Chang S, Puryear J, Cairney J (1993) A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Rep 11: 113–116 [Google Scholar]
- Chen J, Hu Q, Zhang Y, Lu C, Kuang H (2014) P-MITE: a database for plant miniature inverted-repeat transposable elements. Nucleic Acids Res 42: D1176–D1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P, Zhi F, Li X, Shen W, Yan M, He J, Bao C, Fan T, Zhou S, Ma F, et al. (2021) Zinc-finger protein MdBBX7/MdCOL9, a target of MdMIEL1 E3 ligase, confers drought tolerance in apple. Plant Physiol 188: 540–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daccord N, Celton J-M, Linsmith G, Becker C, Choisne N, Schijlen E, Geest Hvd, Bianco L, Micheletti D, Velasco R, et al. (2017) High-quality de novo assembly of the apple genome and methylome dynamics of early fruit development. Nat Genet 49: 1099–1106 [DOI] [PubMed] [Google Scholar]
- Dai H, Li W, Han G, Yang Y, Ma Y, Li H, Zhang Z (2013) Development of a seedling clone with high regeneration capacity and susceptibility to Agrobacterium in apple. Sci Horticult 164: 202–208 [Google Scholar]
- Dietz K-J (2013) Redox regulation of transcription factors in plant stress acclimation and development. Antioxid Redox Signal 21: 1356–1372 [DOI] [PubMed] [Google Scholar]
- Gagliardi D, Cambiagno DA, Arce AL, Tomassi AH, Giacomelli JI, Ariel FD, Manavella PA (2019) Dynamic regulation of chromatin topology and transcription by inverted repeat-derived small RNAs in sunflower. Proc Natl Acad Sci USA 116: 17578–17583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng D, Chen P, Shen X, Zhang Y, Li X, Jiang L, Xie Y, Niu C, Zhang J, Huang X, et al. (2018) MdMYB88 and MdMYB124 enhance drought tolerance by modulating root vessels and cell walls in apple. Plant Physiol 178: 1296–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng D, Lu L, Yan M, Shen X, Jiang L, Li H, Wang L, Yan Y, Xu J, Li C, et al. (2019) Physiological and transcriptomic analyses of roots from Malus sieversii under drought stress. J Integr Agric 18: 1280–1294 [Google Scholar]
- Gharibi S, Tabatabaei BES, Saeidi G, Goli SAH (2016) Effect of drought stress on total phenolic, lipid peroxidation, and antioxidant activity of Achillea species. Appl Biochem Biotechnol 178: 796–809 [DOI] [PubMed] [Google Scholar]
- Ghoshal B, Picard CL, Vong B, Feng S, Jacobsen SE (2021) CRISPR-based targeting of DNA methylation in Arabidopsis thaliana by a bacterial CG-specific DNA methyltransferase. Proc Natl Acad Sci USA 118: e2125016118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabsztunowicz M, Rantala M, Ivanauskaite A, Blomster T, Koskela MM, Vuorinen K, Tyystjärvi E, Burow M, Overmyer K, Mähönen AP, et al. (2021) Root-type ferredoxin-NADP+ oxidoreductase isoforms in Arabidopsis thaliana: expression patterns, location and stress responses Plant , Cell Environ 44: 548–558 [DOI] [PubMed] [Google Scholar]
- Gruntman E, Qi Y, Slotkin RK, Roeder T, Martienssen RA, Sachidanandam R (2008) Kismeth: analyzer of plant methylation states through bisulfite sequencing. BMC Bioinformatics 9: 371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Q, Lu X, Zeng H, Zhang Y, Zhu J (2013) Heat stress induction of miR398 triggers a regulatory loop that is critical for thermotolerance in Arabidopsis. Plant J 74: 840–851 [DOI] [PubMed] [Google Scholar]
- Gupta A, Rico-Medina A, Caño-Delgado AI (2020) The physiology of plant responses to drought. Science 368: 266–269 [DOI] [PubMed] [Google Scholar]
- Haag JR, Ream TS, Marasco M, Nicora CD, Norbeck AD, Pasa-Tolic L, Pikaard CS (2012) In vitro transcription activities of Pol IV, Pol V, and RDR2 reveal coupling of Pol IV and RDR2 for dsRNA synthesis in plant RNA silencing. Mol Cell 48: 811–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachiya T, Ueda N, Kitagawa M, Hanke G, Suzuki A, Hase T, Sakakibara H (2016) Arabidopsis root-type ferredoxin:NADP(H) oxidoreductase 2 is involved in detoxification of nitrite in roots. Plant Cell Physiol 57: 2440–2450 [DOI] [PubMed] [Google Scholar]
- Hanke G, Mulo P (2013) Plant type ferredoxins and ferredoxin-dependent metabolism. Plant Cell Environ 36: 1071–1084 [DOI] [PubMed] [Google Scholar]
- Hanke GT, Okutani S, Satomi Y, Takao T, Suzuki A, Hase T (2005) Multiple iso-proteins of FNR in Arabidopsis: evidence for different contributions to chloroplast function and nitrogen assimilation. Plant Cell Environ 28: 1146–1157 [Google Scholar]
- Harris CJ, Scheibe M, Wongpalee SP, Liu W, Cornett EM, Vaughan RM, Li X, Chen W, Xue Y, Zhong Z, et al. (2018) A DNA methylation reader complex that enhances gene transcription. Science 362: 1182–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens RP, Allan AC, Friel EN, Bolitho K, Grafton K, Templeton MD, Karunairetnam S, Gleave AP, Laing WA (2005) Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herr AJ, Jensen MB, Dalmay T, Baulcombe DC (2005) RNA polymerase IV directs silencing of endogenous DNA. Science 308: 118–120 [DOI] [PubMed] [Google Scholar]
- Huang C, Sun H, Xu D, Chen Q, Liang Y, Wang X, Xu G, Tian J, Wang C, Li D, et al. (2017) ZmCCT9 enhances maize adaptation to higher latitudes. Proc Natl Acad Sci USA 115: E334–E341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichino L, Boone BA, Strauskulage L, Harris CJ, Kaur G, Gladstone MA, Tan M, Feng S, Jami-Alahmadi Y, Duttke SH, et al. (2021) MBD5 and MBD6 couple DNA methylation to gene silencing through the J-domain protein SILENZIO. Science 372: 1434–1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishitani M, Xiong L, Stevenson B, Zhu J (1997) Genetic analysis of osmotic and cold stress signal transduction in Arabidopsis: interactions and convergence of abscisic acid-dependent and abscisic acid-independent pathways. Plant Cell 9: 1935–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang N, Bao Z, Zhang X, Hirochika H, Eddy SR, McCouch SR, Wessler SR (2003) An active DNA transposon family in rice. Nature 421: 163–167 [DOI] [PubMed] [Google Scholar]
- Kanno T, Huettel B, Mette MF, Aufsatz W, Jaligot E, Daxinger L, Kreil DP, Matzke M, Matzke AJ (2005) Atypical RNA polymerase subunits required for RNA-directed DNA methylation. Nat Genet 37: 761–765 [DOI] [PubMed] [Google Scholar]
- Kikuchi K, Terauchi K, Wada M, Hirano H-Y (2003) The plant MITE mPing is mobilized in anther culture. Nature 421: 167–170 [DOI] [PubMed] [Google Scholar]
- Kimata-Ariga Y, Chikuma Y, Saitoh T, Miyata M, Yanagihara Y, Yamane K, Hase T (2019) NADP(H) allosterically regulates the interaction between ferredoxin and ferredoxin-NADP+ reductase. FEBS Open Bio 9: 2126–2136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozuleva M, Goss T, Twachtmann M, Rudi K, Trapka J, Selinski J, Ivanov B, Garapati P, Steinhoff H-J, Hase T, et al. (2016) Ferredoxin:NADP(H) oxidoreductase abundance and location influences redox poise and stress tolerance. Plant Physiol 172: 1480–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law JA, Jacobsen SE (2010) Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet 11: 204–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei M, Zhang H, Julian R, Tang K, Xie S, Zhu J (2015) Regulatory link between DNA methylation and active demethylation in Arabidopsis. Proc Natl Acad Sci USA 112: 3553–3557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CF, Pontes O, El-Shami M, Henderson IR, Bernatavichute YV, Chan SW-L, Lagrange T, Pikaard CS, Jacobsen SE (2006) An ARGONAUTE4-containing nuclear processing center colocalized with Cajal bodies in Arabidopsis thaliana. Cell 126: 93–106 [DOI] [PubMed] [Google Scholar]
- Li S, Liu L, Li S, Gao L, Zhao Y, Kim YJ, Chen X (2016) SUVH1, a Su(var)3-9 family member, promotes the expression of genes targeted by DNA methylation. Nucleic Acids Res 44: 608–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Chen P, Xie Y, Yan Y, Wang L, Dang H, Zhang J, Xu L, Ma F, Guan Q (2020) Apple SERRATE negatively mediates drought resistance by regulating MdMYB88 and MdMYB124 and microRNA biogenesis. Horticult Res 7: 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisch D (2012) Regulation of transposable elements in maize. Curr Opin Plant Biol 15: 511–516 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Mao H, Wang H, Liu S, Li Z, Yang X, Yan J, Li J, Tran L-SP, Qin F (2015) A transposable element in a NAC gene is associated with drought tolerance in maize seedlings. Nat Commun 6: 8326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke MA, Mosher RA (2014) RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nat Rev Genet 15: 394–408 [DOI] [PubMed] [Google Scholar]
- Mittler R, Vanderauwera S, Gollery M, Van Breusegem F (2004) Reactive oxygen gene network of plants. Trends Plant Sci 9: 490–498 [DOI] [PubMed] [Google Scholar]
- Mulo P (2011) Chloroplast-targeted ferredoxin-NADP+ oxidoreductase (FNR): structure, function and location. Biochim Biophys Acta 1807: 927–934 [DOI] [PubMed] [Google Scholar]
- Naito K, Zhang F, Tsukiyama T, Saito H, Hancock CN, Richardson AO, Okumoto Y, Tanisaka T, Wessler SR (2009) Unexpected consequences of a sudden and massive transposon amplification on rice gene expression. Nature 461: 1130–1134 [DOI] [PubMed] [Google Scholar]
- Nakazaki T, Okumoto Y, Horibata A, Yamahira S, Teraishi M, Nishida H, Inoue H, Tanisaka T (2003) Mobilization of a transposon in the rice genome. Nature 421: 170–172 [DOI] [PubMed] [Google Scholar]
- Niu C, Li H, Jiang L, Yan M, Li C, Geng D, Xie Y, Yan Y, Shen X, Chen P, et al. (2019) Genome-wide identification of drought-responsive microRNAs in two sets of Malus from interspecific hybrid progenies. Horticult Res 6: 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onda Y, Matsumura T, Kimata-Ariga Y, Sakakibara H, Sugiyama T, Hase T (2000) Differential interaction of maize root ferredoxin:NADP+ oxidoreductase with photosynthetic and non-photosynthetic ferredoxin isoproteins. Plant Physiol 123: 1037–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onodera Y, Haag JR, Ream T, Nunes PC, Pontes O, Pikaard CS (2005) Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell 120: 613–622 [DOI] [PubMed] [Google Scholar]
- Qi Y, He X, Wang X-J, Kohany O, Jurka J, Hannon GJ (2006) Distinct catalytic and non-catalytic roles of ARGONAUTE4 in RNA-directed DNA methylation. Nature 443: 1008–1012 [DOI] [PubMed] [Google Scholar]
- Shao H, Chu L, Lu Z, Kang C (2007) Primary antioxidant free radical scavenging and redox signaling pathways in higher plant cells. Int J Biol Sci 4: 8–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigman MJ, Slotkin RK (2016) The first rule of plant transposable element silencing: location, location, location. Plant Cell 28: 304–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin RK, Martienssen R (2007) Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet 8: 272–285 [DOI] [PubMed] [Google Scholar]
- Sun X, Wang P, Jia X, Huo L, Che R, Ma F (2018) Improvement of drought tolerance by overexpressing MdATG18a is mediated by modified antioxidant system and activated autophagy in transgenic apple. Plant Biotechnol J 16: 545–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco R, Zharkikh A, Affourtit J, Dhingra A, Cestaro A, Kalyanaraman A, Fontana P, Bhatnagar SK, Troggio M, Pruss D, et al. (2010) The genome of the domesticated apple (Malus × domestica Borkh.). Nat Genet 42: 833–839 [DOI] [PubMed] [Google Scholar]
- Verslues PE, Agarwal M, Katiyar‐Agarwal S, Zhu J, Zhu JK (2006) Methods and concepts in quantifying resistance to drought, salt and freezing, abiotic stresses that affect plant water status. Plant J 45: 523–539 [DOI] [PubMed] [Google Scholar]
- Wang J, Nan N, Li N, Liu Y, Wang T, Hwang I, Liu B, Xu Z (2020) A DNA methylation reader–chaperone regulator–transcription factor complex activates OsHKT1;5 expression during salinity stress. Plant Cell 32: 3535–3558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Liang D, Li C, Hao Y, Ma F, Shu H (2012) Influence of drought stress on the cellular ultrastructure and antioxidant system in leaves of drought-tolerant and drought-sensitive apple rootstocks. Plant Physiol Biochem 51: 81–89 [DOI] [PubMed] [Google Scholar]
- Wang Y, Liu L, Wang Y, Tao H, Fan J, Zhao Z, Guo Y (2019) Effects of soil water stress on fruit yield, quality and their relationship with sugar metabolism in ‘Gala’ apple. Sci Horticult 258: 108753 [Google Scholar]
- Wang Y, Yu B, Zhao J, Guo J, Li Y, Han S, Huang L, Du Y, Hong Y, Tang D, et al. (2013) Autophagy contributes to leaf starch degradation Plant Cell 25: 1383–1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L, Gu L, Song X, Cui X, Lu Z, Zhou M, Wang L, Hu F, Zhai J, Meyers BC, et al. (2014) Dicer-like 3 produces transposable element-associated 24-nt siRNAs that control agricultural traits in rice. Proc Natl Acad Sci USA 111: 3877–3882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessler SR, Bureau TE, White SE (1995) LTR-retrotransposons and MITEs: important players in the evolution of plant genomes. Curr Opin Genet Dev 5: 814–821 [DOI] [PubMed] [Google Scholar]
- Wierzbicki AT, Haag JR, Pikaard CS (2008) Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell 135: 635–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao X, Zhang J, Li T, Fu X, Satheesh V, Niu Q, Lang Z, Zhu J-K, Lei M (2019) A group of SUVH methyl-DNA binding proteins regulate expression of the DNA demethylase ROS1 in Arabidopsis. J Integr Plant Biol 61: 110–119 [DOI] [PubMed] [Google Scholar]
- Xie Y, Chen P, Yan Y, Bao C, Li X, Wang L, Shen X, Li H, Liu X, Niu C, et al. (2018) An atypical R2R3 MYB transcription factor increases cold hardiness by CBF-dependent and CBF-independent pathways in apple. New Phytol 218: 201–218 [DOI] [PubMed] [Google Scholar]
- Xu L, Yuan K, Yuan M, Meng X, Chen M, Wu J, Li J, Qi Y (2020) Regulation of rice tillering by RNA-directed DNA methylation at miniature inverted-repeat transposable elements. Mol Plant 13: 851–863 [DOI] [PubMed] [Google Scholar]
- Ye R, Wang W, Iki T, Liu C, Wu Y, Ishikawa M, Zhou X, Qi Y (2012) Cytoplasmic assembly and selective nuclear import of Arabidopsis Argonaute4/siRNA complexes. Mol Cell 46: 859–870 [DOI] [PubMed] [Google Scholar]
- Zhai J, Bischof S, Wang H, Feng S, Lee T-F, Teng C, Chen X, Park SY, Liu L, Gallego-Bartolome J (2015) A one precursor one siRNA model for Pol IV-dependent siRNA biogenesis. Cell 163: 445–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Zhu J-K (2011) RNA-directed DNA methylation. Curr Opin Plant Biol 14: 142–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Liu Y (2014) VIGS assays. Bio-Protocol 4 [Google Scholar]
- Zhang H, Tao Z, Hong H, Chen Z, Wu C, Li X, Xiao J, Wang S (2016) Transposon-derived small RNA is responsible for modified function of WRKY45 locus. Nat Plants 2: 16016. [DOI] [PubMed] [Google Scholar]
- Zhao Q-Q, Lin R-N, Li L, Chen S, He X-J (2019) A methylated-DNA-binding complex required for plant development mediates transcriptional activation of promoter methylated genes. J Integr Plant Biol 61: 120–139 [DOI] [PubMed] [Google Scholar]
- Zhong X, Du J, Hale CJ, Gallego-Bartolome J, Feng S, Vashisht AA, Chory J, Wohlschlegel JA, Patel DJ, Jacobsen SE (2014) Molecular mechanism of action of plant DRM de novo DNA methyltransferases. Cell 157: 1050–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.