Abstract
DNA methylation is now recognized as a regulator of multiple bacterial cellular processes. CcrM is a DNA adenine methyltransferase found in the alpha subdivision of the proteobacteria. Like the Dam enzyme, which is found primarily in Escherichia coli and other gamma proteobacteria, it does not appear to be part of a DNA restriction-modification system. The CcrM homolog of Agrobacterium tumefaciens was found to be essential for viability. Overexpression of CcrM is associated with significant abnormalities of cell morphology and DNA ploidy. Mapping of the transcriptional start site revealed a conserved binding motif for the global response regulator CtrA at the −35 position; this motif was footprinted by purified Caulobacter crescentus CtrA protein in its phosphorylated state. We have succeeded in isolating synchronized populations of Agrobacterium cells and analyzing their progression through the cell cycle. We demonstrate that DNA replication and cell division can be followed in an orderly manner and that flagellin expression is cyclic, consistent with our observation that motility varies during the cell cycle. Using these synchronized populations, we show that CcrM methylation of the chromosome is restricted to the late S phase of the cell cycle. Thus, within the alpha subdivision, there is a conserved cell cycle dependence and regulatory mechanism controlling ccrM expression.
DNA methylation, long known to exist in both eukaryotes and prokaryotes, is a widespread regulator of bacterial cellular processes. In eukaryotes, it plays important roles in transcriptional regulation and genomic imprinting and is essential for proper development in the mouse (2, 16). Among prokaryotes, it is primarily found in the context of restriction-modification systems, where it allows an organism to distinguish foreign DNA from its own. However, regulatory functions mediated by methylation have been extensively characterized for the Dam (for “DNA adenine methyltransferase”) methyltransferase of Escherichia coli. Dam, termed an ‘orphan’ methyltransferase because it has no known cognate restriction enzyme, is involved in methyl-directed mismatch DNA repair as well as ensuring that multiple origins of chromosomal replication within the cell fire synchronously (23).
Dam has also been shown to influence the transcription of a growing number of genes important in the pathogenesis of bacterial diseases. The primary example of methylation-influenced transcription is that of the pyelonephritis-associated pilus, or pap, operon in uropathogenic E. coli (3, 27, 42). Methylation has now been found to also regulate phase variation of two other surface proteins: the plasmid-encoded fimbriae (Pef) of the enteric bacterium Salmonella enterica serovar Typhimurium, which mediate adhesion to mouse intestinal epithelium, and the nonfimbrial E. coli outer membrane protein Ag43 (13, 25). Furthermore, Dam has been shown to influence virulence of S. enterica serovar Typhimurium, where a dam-negative strain has severely attenuated infectivity in a mouse model and confers protection against challenge with a wild-type strain (11, 14). In a cell culture model, dam-negative S. enterica serovar Typhimurium also exhibits defects in the invasion of epithelial cells, secretion of type III effector proteins, and cytotoxicity to M cells (11).
CcrM (for “cell cycle-regulated methyltransferase”) is the best-characterized orphan methyltransferase aside from Dam. It is an essential DNA methyltransferase of the dimorphic aquatic bacterium Caulobacter crescentus (40). Both CcrM and Dam catalyze the transfer of a methyl group from S-adenosylmethionine to the N-6 position of adenine in a specific target sequence. However, they belong to separate methyltransferase groups, since their catalytic and S-adenosylmethionine-binding domains are organized in a different linear order (21). Their target sequences also differ: GATC for Dam and GANTC for CcrM. CcrM is widely distributed in the alpha subdivision of proteobacteria, which includes C. crescentus, Sinorhizobium meliloti, Brucella abortus, and Agrobacterium tumefaciens (40). In contrast, Dam is found primarily in the enteric bacteria and other gamma proteobacteria (32).
In Caulobacter, CcrM is highly regulated: its presence is restricted to a narrow window of the cell cycle by the dual mechanisms of transcriptional control and proteolysis (41, 46, 48). The Caulobacter CtrA response regulator, which controls multiple cellular events such as initiation of flagellar biogenesis and repression of chromosomal replication initiation, also induces transcription of ccrM just prior to cell division (30). Rapid proteolysis of CcrM occurs via a Lon protease-mediated pathway (46). Thus, replicating Caulobacter DNA is maintained in the hemimethylated state until the completion of replication, and its transition to full methylation occurs as a consequence of a burst of CcrM synthesis late in the cell cycle. Without this tight control, normal coordination between DNA replication and cell division fails to occur. If ccrM is constitutively transcribed throughout the cell cycle or if Lon is absent, then active CcrM protein is present throughout the cell cycle and the chromosomal DNA is fully methylated at all times. When this occurs, cells have a filamentous morphology and an increase in their DNA content (46, 48).
DNA methyltransferases are reasonable antimicrobial drug targets. This has been highlighted by the discovery that dam-negative salmonellae are avirulent (15). Unlike Dam, CcrM has been found to be essential for viability in multiple bacteria, thus raising the possibility that inhibitors of methylation may even be bactericidal in some cases. Recent work has now implicated CcrM-mediated DNA methylation as a modulator of intracellular survival of the animal pathogen B. abortus (34). Characterizing the role of this methyltransferase in bacteria with varied ecological niches and growth cycles will allow a better understanding of its physiological importance and its potential as a drug target.
We describe here the cloning and characterization of the CcrM homolog from the plant pathogen A. tumefaciens. A. tumefaciens is the causative agent of crown gall disease in plants, and most work on this organism has focused on the mechanisms by which it induces tumor formation. Its recently sequenced Ti plasmid carries most of the genes required for tumorigenesis, which involves the conjugal transfer of an oncogenic piece of DNA (T-DNA) into the nuclei of plant cells (47). Because of this ability to export DNA, A. tumefaciens has become instrumental in the genetic engineering of plants. Its chromosomal complement is less well studied but includes genes important for virulence. These include a locus mediating attachment to plants and a two-component signal transduction system required for tumorigenesis (7, 24).
We show here that the A. tumefaciens ccrM homolog is a chromosomally encoded gene essential for viability. Its overexpression results in a significant increase in the level of genomic DNA methylation, which correlates with aberrations of cell division and DNA content. We have mapped the transcriptional start site and found that the promoter has extensive homology to the ccrM promoters of both C. crescentus and B. abortus. Significantly, it contains a conserved binding motif for the global response regulator CtrA, which has been found in several alpha proteobacteria (1). Purified C. crescentus CtrA protein footprints this binding site in a phosphorylation-dependent manner. Finally, we show that A. tumefaciens cells can be studied as synchronous populations. Using these synchronized cells, we show that the methylation state of the A. tumefaciens chromosome varies as a function of the cell cycle. These last observations suggest that within the alpha subdivision of proteobacteria, there exists a conserved cell cycle dependence and regulatory mechanism controlling the expression of this essential DNA methyltransferase.
MATERIALS AND METHODS
Bacterial growth conditions and media.
Strains and plasmids used are listed in Table 1. E. coli was grown at 37°C in Luria-Bertani (LB) medium containing 10 g of NaCl/liter. A. tumefaciens was grown at 28°C in YEB (0.5% beef extract, 0.1% yeast extract, 0.5% peptone, 0.5% sucrose, 2 mM MgSO4), LB containing 5 g of NaCl/liter, or AB minimal medium (6) supplemented with 0.5% glucose under normal conditions or with 5% sucrose when used for sacB counterselection (see below). For solid media, 15 g of agar was added per liter. Antibiotics were used at the following concentrations: nalidixic acid, 20 μg/ml; kanamycin, 25 μg/ml for A. tumefaciens and 50 μg/ml for E. coli; spectinomycin, 200 μg/ml for A. tumefaciens and 50 μg/ml for E. coli; tetracycline, 12 μg/ml; and rifampin, 100 μg/ml. Plasmids were introduced into A. tumefaciens either by electroporation or by mating with E. coli S17-1 as a donor strain (6).
TABLE 1.
Strains and plasmids
| Strain or plasmid | Relevant characteristic(s)a | Reference or source |
|---|---|---|
| E. coli | ||
| DH5αF′ | Cloning strain | Gibco-BRL |
| S17-1 | Conjugal transfer of plasmids to A. tumefaciens | 36 |
| A. tumefaciens | ||
| A136 | Strain C58 cured of pTi plasmid | S. Long |
| A348 | A136 containing pTiA6NC | P. Zambryski |
| NT1REB | Flagellum-free derivative of NT1RE (C58 derivative) | 8 |
| LS3303 | A348 containing pLK308 | This study |
| LS3304 | A348 containing pLK309 | This study |
| LS3307 | A348 with pLK311 integrated 5′ of spectinomycin cassette | This study |
| LS3308 | A348 with pLK311 integrated 3′ of spectinomycin cassette | This study |
| LS3314 | A348 containing pRK290(20R) | This study |
| LS3315 | A348 containing pJS71 | This study |
| Plasmids | ||
| pBluescript II SK(+) | Ampr cloning vector | Stratagene |
| pCR2.1-TOPO | Ampr Kanr cloning vector for PCR products | Invitrogen |
| pCRII | Ampr Kanr cloning vector for PCR products | Invitrogen |
| pRK290(20R) | pRK290 with pIC20R polylinker, Tetr | M. R. K. Alley |
| pJS71 | Spectr pBBR1MCS derivative | 38 |
| pHP45 | Source of Spectr Ω cassette | 29 |
| Agro3 | Cosmid containing A. tumefaciens ccrM homolog | This study |
| pCS263 | pCRII containing PCR of partial A. tumefaciens ccrM homolog | C. Stephens |
| pNPTS138 | pLitmus 38-derived vector with added nptI, sacB, and RK2 oriT, Kanr | 38 |
| pLK300 | pBluescript II SK(+) containing 2.2-kb BbsI fragment of Agro3 with A. tumefaciens ccrM homolog | This study |
| pLK307 | pBluescript II SK(+) containing 1.5-kb ApaLI-ScaI fragment of pLK300 with ccrM and its promoter | This study |
| pLK308 | pRK290(20R) + XbaI-XhoI fragment from pLK307 containing complete A. tumefaciens ccrM gene and its promoter | This study |
| pLK309 | pJS71 + XbaI-XhoI fragment from pLK307 containing complete A. tumefaciens ccrM gene and its promoter | This study |
| pLK310 | pLK300 derivative with replacement of part of ccrM by spectinomycin resistance cassette | This study |
| pLK311 | pNPTS138 with EcoRI-SpeI fragment containing disrupted A. tumefaciens ccrM homolog from pLK310 | This study |
Amp, ampicillin; Kan, kanamycin; Spect, spectinomycin; Tet, tetracycline.
Cloning the full-length A. tumefaciens ccrM gene.
Degenerate primers were designed based on regions of homology common to the C. crescentus and Haemophilus influenzae methyltransferases (45). Primer 1 (5′-AT[C/T]TT[C/T]GC[CGT]GA[C/T]CC[C/G/T]TA-3′) and primer 2 (5′-TC[A/G]TT[G/C]A[A/G][A/G]ATCCA[A/G]AA-3′) were used in PCR with genomic DNA from A. tumefaciens. The PCR fragment was cloned into the pCRII vector (Invitrogen) as pCS263, and the insert was used to screen an A. tumefaciens A136 genomic library (7). Two clones were isolated and characterized. A 2.2-kb BbsI fragment containing the entire A. tumefaciens ccrM homolog was isolated from the cosmid Agro3 and cloned into the SmaI site of pBluescript II SK(+) (Stratagene) to generate pLK300. The gene was sequenced using either a Thermosequenase kit (Amersham) or automated sequencing (Applied Biosystems 373A; Bio101, La Jolla, Calif.). Sequence analysis and alignment were performed with the Wisconsin Package, version 10.1 (Genetics Computer Group, Madison, Wis.). Alignment was performed with ClustalW, followed by shading of the alignment with Boxshade.
Construction of the A. tumefaciens ccrM null mutant.
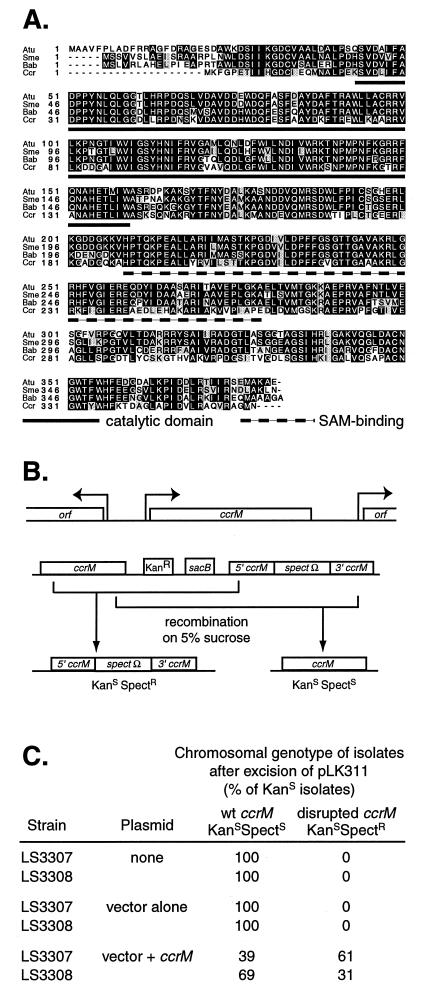
To determine whether ccrM is essential in A. tumefaciens, we replaced the nucleotide sequence encoding the catalytic domain of the methyltransferase with a spectinomycin resistance cassette (Fig. 1B). The spectinomycin resistance cassette from pHP45 (29) was cut out with SmaI and ligated into a blunt-ended PflMI-StuI digest of pLK300 to give pLK310. An EcoRI-SpeI fragment of pLK310 with the gene and cassette was then cloned into a suicide vector (pNPTS138) bearing a kanamycin resistance marker as well as the sacB gene of Bacillus subtilis, yielding pLK311. The levansucrase encoded by sacB produces a toxic metabolite when cells are grown on sucrose, allowing eventual counterselection against the presence of the plasmid. Only kanamycin-sensitive colonies were scored, because some sucrose-resistant colonies had inactivation of the sacB gene and therefore had not truly excised pLK311. For complementation, the intact gene and its upstream region were cloned into the low-copy-number vector pRK290(20R) to give pLK308. Tetracycline was used to maintain the control and complementing plasmids.
FIG. 1.
The A. tumefaciens ccrM homolog is essential for viability. (A) Homology diagram of the A. tumefaciens (Atu) CcrM homolog in relation to the other homologs from S. meliloti (Sme), B. abortus (Bab), and C. crescentus (Ccr). Black boxes denote identical amino acids and shaded boxes denote similar amino acids. The catalytic and S-adenosylmethionine-binding domains are marked. (B) (Top) Diagram of the ccrM locus. (Bottom) Diagram of the locus following integration of the plasmid pLK311, yielding tandem copies of the gene, one of which is disrupted by a spectinomycin resistance cassette. The resulting strain is resistant to both kanamycin and spectinomycin. Growth on 5% sucrose with excision of the vector sequences leads to two possible outcomes. If excision occurs on the opposite side of the Ω cassette from the original integration event, a disrupted copy of the gene will remain on the chromosome and the resulting strain will be spectinomycin resistant but kanamycin sensitive. If excision occurs on the same side as the original integration event, then the strain will be wild type and sensitive to both spectinomycin and kanamycin. (C) Only the presence of ccrM in trans allows viable colonies when the chromosomal copy of ccrM is disrupted. Strain LS3307 carries the pLK311 vector recombined on the 5′ side of the spectinomycin resistance cassette, and LS3308 carries pLK311 recombined on the 3′ side of the cassette. Results are shown for both strains with no plasmid, vector alone [pRK290(20R)], or vector plus ccrM (pLK308). A minimum of 100 isolates were scored for each experiment.
Microscopy and flow cytometry.
For analysis of mixed cultures, A. tumefaciens cells were grown to exponential phase (A600 ≈ 0.5) and then treated with 1/100 volume of fix solution (12.5% formaldehyde, 150 mM sodium phosphate [pH 7.5]) for 15 min at room temperature. For synchronized cell populations, 1/5 volume of fix solution was used to ensure immediate fixation. Cells were then washed twice in growth medium and stored at 4°C. For flow cytometry, cold ethanol was added to a final concentration of 70%. Without light formaldehyde fixation, the cells tended to lyse when ethanol was added. Samples were then visually examined under the light microscope to ensure preservation of the morphology as seen in the starting culture. For flow cytometry, cells were gently recentrifuged and resuspended in TMS buffer (10 mM Tris-HCl [pH 7.2], 1.5 mM MgCl2, 150 mM NaCl) containing 10 μg of chromomycin A3/ml (44). The DNA content of 10,000 cells from each sample was analyzed on a Becton Dickinson FACStar instrument with excitation at 458 nm and measurement of fluorescence at 495 nm. The data were collected and analyzed using FlowJo software (Tree Star Inc., San Carlos, Calif.).
For microscopy, the fixed, washed cells were placed on a 1% agarose pad and the coverslips sealed with melted Valap (1:1:1; vaseline-lanolin-paraffin wax). Samples were photographed using a Nikon E800 microscope with a 100× DIC (differential interference contrast) objective and either a Hamamatsu C2400 camera controlled by Scion Image software (Scion Corp., Frederick, Md.) or a MicroMAX cooled charge-coupled device camera (Princeton Instruments, Trenton, N.J.) controlled by METAMORPH 3.6A (Universal Imaging, Media, Pa.).
Determination of the methylation state of the chromosome in A. tumefaciens and assessment of the Ti plasmid and chromosomal DNA content.
The DNA methylation state of an overlapping HincII-HinfI site located in the att locus was assessed in the A. tumefaciens strains A348, LS3314, LS3315, LS3303, and LS3304. Genomic DNA was isolated (PureGene; Gentra Systems) at similar times in the exponential growth phase of the strains, and equal amounts were digested for each lane of a gel. Southern blots of the HincII-digested genomic DNA were probed with a 500-bp PCR product generated by using the primers Agro att forward (5′-GTAGAGGTTGAGAAGTGTAGG-3′) and Agro att reverse (5′-CGTGATGAGAGCGATACCG-3′). This DNA fragment was randomly labeled with [α-32P]dCTP using the T7 Quick-Prime kit (Amersham Pharmacia). All probes were verified by restriction digests and/or sequencing of the cloned PCR products. Blots were exposed and quantitated using a Molecular Dynamics PhosphorImager.
Primer extension.
The ccrM transcription start site was determined by primer extension analysis. Experiments were carried out with total RNA (20 to 40 μg) obtained from log-phase cultures of A136 and A348 using a standard hot phenol preparation method. The primer AgroMT17 (5′-GTCACTCTCGCCCGCACG-3′) was 5′ end labeled with [γ-32P]ATP using T4 polynucleotide kinase (Boehringer) and separated from free nucleotide using the QIAquick nucleotide removal kit (Qiagen). Samples were annealed at 50° for 20 min following denaturation. Synthesis of cDNA was performed using SuperscriptII (Gibco-BRL) at 42°C for 1 h. The reactions were stopped, and the products were desalted on Spin-X columns (Costar) using Sephadex G25 resin (Amersham Pharmacia) and subjected to polyacrylamide gel electrophoresis in parallel with a sequencing ladder generated using the same primer and pLK300 as a template (Thermosequenase kit; Amersham Pharmacia).
Footprinting.
DNase I protection experiments were performed as previously described with the purified Caulobacter CtrA response regulator (38). CtrA was phosphorylated using a maltose-binding protein–EnvZ fusion protein, also previously described (33). The template DNA was a 550-bp PCR product made using the primers AgMTFP1 (5′-CCGCACTTCAGGTTCCC-3′) and AgMTFP2 (5′-CGTTGAGGATCCAGAAATCG-3′). The PCR product was 5′ end labeled with [γ-32P]ATP using T4 polynucleotide kinase, and the 3′ end of the labeled template was removed by digestion at a naturally occurring BamHI site. A sequencing ladder generated with the AgMTFP1 primer was used to identify the protected region.
Synchronization of A. tumefaciens cultures.
Cultures of A. tumefaciens were synchronized using a modification of the method described by Evinger and Agabian (10). Strain A348 was grown in LB at 28°C to an A600 of approximately 0.35. This culture was mixed with the colloidal silica preparation Ludox (420840; Aldrich) which had been brought to neutral pH by the addition of 1 N HCl immediately before use. The final ratio was 1.8 volumes of culture to each volume of neutralized Ludox. It should be noted that this ratio was empirically determined and may vary somewhat between batches of Ludox, that outside an appropriate range of ratios the cells all either stay at the top or go to the bottom, that the viscosity of Ludox changes over time after it has been neutralized, and that using a denser culture resulted in more contamination of the small cells with larger cells. The mixture was centrifuged in 15-ml Corex tubes (Corning) in a JA-20 rotor at 9,000 rpm (i.e., 9,800 × g) for 5 min at 4°C. Small, motile cells were found at the very bottom of the gradient, just above the clear liquid at the bottom, while most of the larger cells floated on top. Cells of intermediate size could be found in between. The small, motile cells from the bottom of the tube were collected, pooled, visually examined under a light microscope, and quickly washed two or three times in cold 1× AB without glucose, with pelleting at 9,000 rpm for 2 min each time. After washing, the pellet was resuspended in LB at 28°C, and the culture was incubated at 28°C. Samples were withdrawn at the indicated times, examined by light microscopy, and immediately either fixed as described above or frozen as pellets for further processing. During a cell cycle experiment, equal volumes of cells were withdrawn at each time point. For Western blotting, equal volumes of the samples were loaded on each lane of a protein gel.
Western blotting of flagellins.
To examine flagellar protein levels by Western blotting, cell pellets from either mixed log-phase cultures or synchronous cultures were resuspended in 1× sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer and run on an SDS–12% PAGE protein gel (35). These were transferred onto Immobilon P membranes (Millipore) using standard Western blotting techniques. Blots were probed with primary rabbit antiserum (raised to pooled Caulobacter flagellins) at a dilution of 1:5,000 (39), followed by secondary antibody (donkey anti-rabbit immunoglobulin G–horseradish peroxidase conjugate; Jackson Laboratories) at a dilution of 1:10,000. Bound antibody was visualized by chemiluminescence. Autoradiograms were scanned using an Expression 800 scanner and Epson TWAIN Pro software (Epson America, Inc.).
Nucleotide sequence accession number.
The sequence of the A. tumefaciens ccrM homolog has been submitted to GenBank under the accession number AF327563.
RESULTS
The A. tumefaciens ccrM gene is essential for viability.
The full-length A. tumefaciens ccrM homolog was isolated from a genomic DNA library and sequenced as described in Materials and Methods. The encoded protein has 62% identity and 70% similarity to Caulobacter CcrM. While some of this homology is due to the conserved S-adenosylmethionine-binding and catalytic domains found in DNA methyltransferases, the C-terminal third of the protein is unique to the CcrM homologs. Compared to Caulobacter CcrM, the S. meliloti and B. abortus homologs have an N-terminal extension of 15 amino acids (45), while A. tumefaciens CcrM has an N-terminal extension of 20 amino acids (Fig. 1A). There are no open reading frames within 200 bp on either side of A. tumefaciens ccrM; a closely-linked open reading frame encoding a cognate restriction enzyme would be expected for a restriction-modification system (43). Thus, it is likely that, as in Caulobacter, ccrM is an orphan methyltransferase.
To determine whether ccrM is essential in A. tumefaciens, we replaced the sequence encoding the catalytic domain of the methyltransferase with a spectinomycin resistance cassette as described in Materials and Methods (Fig. 1B). This construct, on a vector which does not replicate in A. tumefaciens (pLK311), was transformed into strain A348. Integrants were isolated by selecting for the acquisition of kanamycin and spectinomycin resistance and then screened by Southern blotting. Strains LS3307 and LS3308 carry pLK311 integrated by recombination on the 5′ and 3′ sides of the spectinomycin resistance cassette, respectively. The low-copy-number vector pRK290(20R) alone or one containing ccrM was mated into LS3307 and LS3308, with subsequent selection on spectinomycin, kanamycin, and tetracycline. The resulting parent strains were grown without selection for either spectinomycin or kanamycin and then plated on minimal media containing 5% sucrose to select for a second recombination leading to excision of the sacB gene and vector. Isolates were then analyzed for spectinomycin and kanamycin resistance.
When no plasmid or the pRK290(20R) vector alone was present, recombination yielded only isolates sensitive to both kanamycin and spectinomycin, which therefore bore wild-type ccrM on the chromosome. However, if pLK308 was present, providing wild-type ccrM in trans, isolates bearing the disrupted copy of ccrM on the chromosome (spectinomycin resistant but kanamycin sensitive) could easily be obtained. Thus, ccrM is essential for viability in A. tumefaciens.
Overexpression of ccrM results in excess methylation of chromosomal DNA.
CcrM overexpression has been associated with cell division defects and aberrant DNA replication in other alpha proteobacteria (34, 45, 48). To evaluate the effects of ccrM overexpression in Agrobacterium, the gene and its upstream region were cloned into the low-copy-number vector pRK290(20R) (two to five copies per cell) and the high-copy-number vector pJS71 (20 to 30 copies per cell) (38). The CcrM homolog could not be detected, using anti-Caulobacter CcrM antibodies, in A. tumefaciens with a vector control or a low-copy-number plasmid bearing ccrM. However, in the strain with a high-copy-number plasmid bearing ccrM, a band of the appropriate size was detected, suggesting that a larger amount of CcrM protein was present (data not shown). To assess how this affected the methylation of genomic DNA, we used Southern blots to assay the methylation state of overlapping HincII-CcrM recognition sequences in the chromosomal att locus (Fig. 2A). The methylation state of the DNA determines its sensitivity to digestion at the overlapping HincII site; DNA which is fully methylated by CcrM cannot be cleaved by HincII. Passage of a replication fork through this fully methylated site yields two hemimethylated DNA products, only one of which can be cleaved by HincII.
FIG. 2.
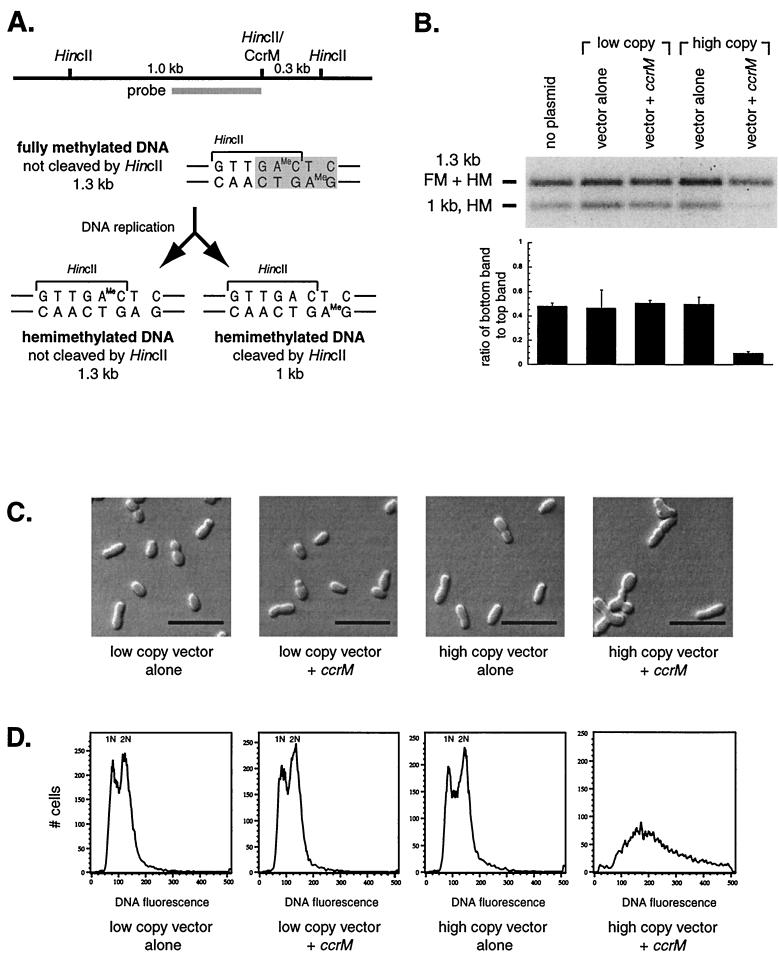
Overexpression of the ccrM homolog leads to an increase in the methylation of chromosomal DNA. (A) Schematic of the DNA methylation assay. CcrM methylates adenine in the sequence GANTC (shaded). This CcrM site overlaps the HincII restriction enzyme recognition site GTTGAC in the att locus. If the DNA is fully methylated by CcrM, HincII will not be able to cleave it. However, once this DNA is replicated, each replication product will be hemimethylated, as shown below. The top strand is methylated in one product (left); the bottom strand is methylated in the other product (right). Only one of these two replication products can be cleaved by HincII, as shown. Southern blotting with the probe shown will yield a 1.3-kb band or a 1-kb band, depending on the methylation state of the DNA. (B) Methylation state of the A. tumefaciens A348 DNA in strains carrying different plasmids, using the differential HincII cleavage assay described for panel A. Genomic DNA from mixed cultures in exponential-phase growth was digested with HincII. The low-copy-number vector is pRK290(20R); the low-copy-number vector with ccrM is pLK308 [pRK290(20R) with ccrM]; the high-copy-number vector is pJS71; the high-copy-number vector with ccrM is pLK309 (pJS71 with ccrM). The upper band contains fully methylated (FM) DNA and the hemimethylated (HM) DNA diagrammed on the left in panel A, while the lower band contains the hemimethylated DNA diagrammed on the right. Shown directly below is the ratio between bands for each lane of the gel. (C) Light microscopy of A. tumefaciens A348 carrying different plasmids, which are detailed for panel B above. Bar, 5 μm. (D) Flow cytometry of the strains in panel C. The y axis represents the number of cells with a given fluorescence intensity. The x axis shows the fluorescence intensity, which corresponds to DNA content.
For Fig. 2B, genomic DNA was digested with HincII. The top band contains fully methylated DNA and one hemimethylated replicative product; the lower band contains the other hemimethylated replicative product. In the wild-type strain, strains containing vector alone, and the strain carrying ccrM on a low-copy-number vector, there were similar ratios of counts in the lower band to counts in the upper band. However, when ccrM was present on a high-copy-number vector (pLK309), there was an 80 to 90% reduction in the amount of hemimethylated DNA, suggesting that following chromosome replication, the new DNA was rapidly brought to the fully methylated state. Southern blot assays of an overlapping ClaI-CcrM site in the chromosomal flagellar locus (9) confirmed this observation (data not shown). Therefore, when ccrM is present on a high-copy-number plasmid, GANTC sites are fully methylated much more often than under normal conditions.
Fully methylated chromosomal DNA is associated with morphological and flow-cytometric abnormalities.
Microscopy (Fig. 2C) and flow cytometry analysis (Fig. 2D) of the strains bearing the plasmids referred to in Fig. 2B revealed that those carrying vector alone or ccrM on a low-copy-number plasmid had a normal cell size with unilateral outgrowth of progeny cells, and flow cytometry showed two discrete peaks of DNA content. These peaks correspond to one and two genome equivalents, respectively, as confirmed by flow cytometry of separated cell populations (described below; also, see Fig. 4). One genome equivalent is likely to represent the circular chromosome, linear chromosome, cryptic plasmid, and Ti plasmid of A. tumefaciens A348 (12). The strain in which ccrM is present on a high-copy-number plasmid has grossly abnormal morphology, with branching, elongated cells (Fig. 2C). The corresponding flow cytometry demonstrates a significantly increased DNA content (Fig. 2D). In an effort to determine whether there was selective alteration of chromosomal versus Ti plasmid replication, Southern blots of total DNA in this strain were probed for both the Ti plasmid and the chromosome. However, the ratio between the two was the same as observed in the wild-type, low-copy-number ccrM, and plasmid controls (data not shown).
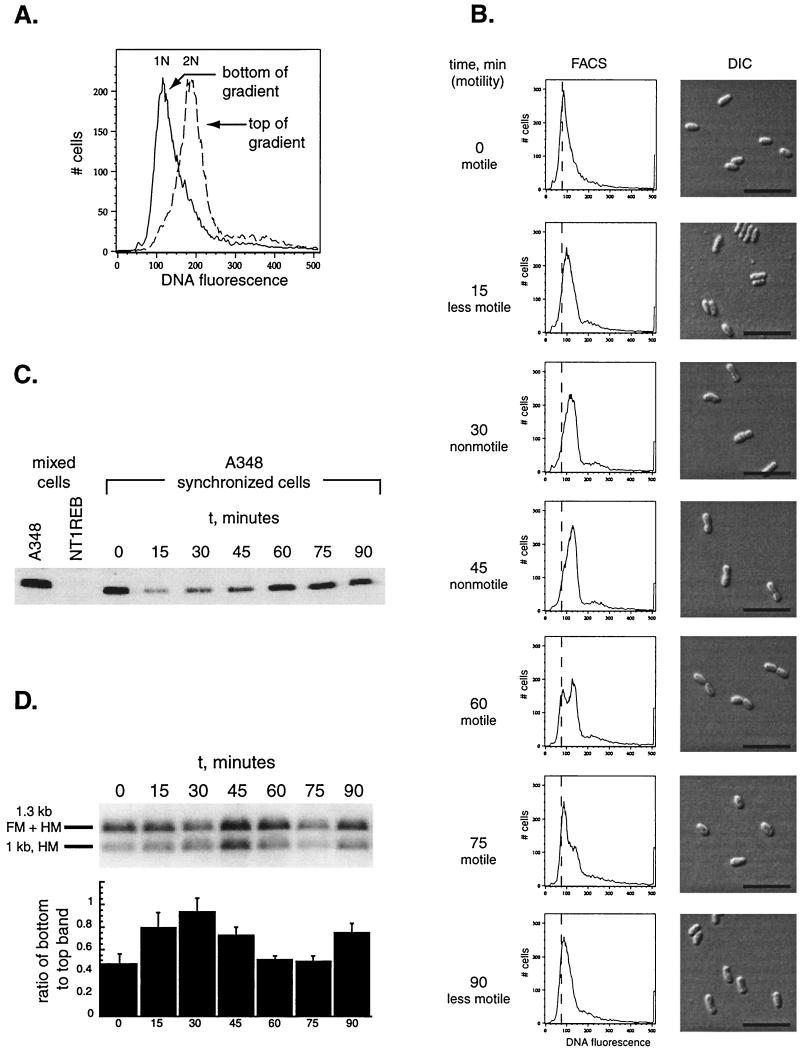
FIG. 4.
A. tumefaciens A348 can be synchronized, and the methylation state of the chromosome varies as a function of the cell cycle. (A) Cells were separated using density centrifugation, as described in Materials and Methods. Flow cytometry of the cells isolated from the bottom and the top of the gradient is shown. (B) The cells from the bottom of the gradient were isolated and allowed to grow as described in Materials and Methods. The changes in motility are indicated; DNA content, as assessed by flow cytometry, and cell morphology over time are shown. The assessment of motility reflects the behavior of the majority of the cell population, as observed by light microscopy, at the time the sample was taken. A vertical line has been arbitrarily placed at 75 fluorescence units on the x axis to aid in following the relative positions of the peaks over the course of the experiment. Bar, 5 μm. (C) Western blot of A. tumefaciens flagellin proteins using polyclonal antiserum to Caulobacter flagellins. The specific 32- to 33-kDa band is shown. Mixed cell populations from both wild-type (A348) and flagellin-negative (NT1REB) cells are shown in the first and second lanes as positive and negative controls. The remaining lanes track the flagellins over the course of the cell cycle in A348. (D) Genomic DNA was prepared from cells at the time points shown in panel B, and the methylation state of the att locus was assayed as described in Materials and Methods and for Fig. 2. A ratio of one between the bottom band and the top band corresponds to 100% hemimethylation of the DNA.
The A. tumefaciens ccrM homolog has a conserved promoter structure and CtrA binding site.
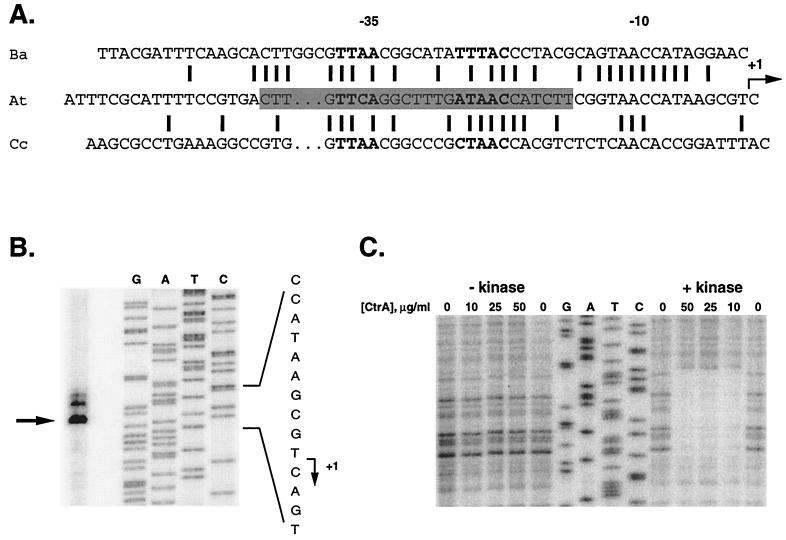
The start site of the ccrM transcript was mapped using primer extension and is shown in Fig. 3A and B. A strong signal was found 33 to 34 bases upstream from the putative translational start site, which is preceded by a good consensus ribosome binding site. The same result was obtained with a different primer as well as with RNA from A. tumefaciens A136, which is isogenic to A348 but lacks the Ti plasmid (data not shown). Three different primers located upstream from the start site did not give a signal. Sequence comparison revealed extensive homology to the promoters of the B. abortus and C. crescentus ccrM homologs. At the −35 position relative to the transcriptional start site, there was a conserved binding motif for the CtrA response regulator (Fig. 3A); this binding motif has been extensively characterized in Caulobacter (28).
FIG. 3.
The A. tumefaciens ccrM promoter has a highly conserved sequence and is bound by the CtrA response regulator. (A) Sequence of the ccrM promoter region and its alignment with the ccrM promoters of C. crescentus and B. abortus. The conserved CtrA binding site (consensus, TTAANTTAAC) is in bold type. The region protected from DNase I by phosphorylated CtrA is shaded. (B) Start site of ccrM transcription as determined by primer extension. A sequence ladder generated with the same primer was used to determine the start site. (C) DNase I protection of the ccrM promoter by CtrA. A sequence ladder generated by using the primer AgMTFP1 was used to identify the protected region. Protection was seen only if CtrA was phosphorylated by the histidine kinase EnvZ.
Although ctrA has not been cloned from Agrobacterium, a homolog of Caulobacter ctrA has been detected on Southern blots. CtrA is highly conserved in the other alpha proteobacteria from which it has been isolated (1, 19). A DNase I protection assay, described in Materials and Methods, was used to determine if the Caulobacter CtrA protein was able to bind to the conserved motif in the promoter region. Purified CtrA was phosphorylated in vitro using the E. coli histidine kinase EnvZ; this was verified in a parallel experiment using [γ-32P]ATP. CtrA was able to protect a 26-bp region from −17 to −42 relative to the transcriptional start site; the presence of the protection pattern required phosphorylation of CtrA (Fig. 3C).
Methylation of the A. tumefaciens chromosome varies as a function of the cell cycle.
A density centrifugation technique initially developed to synchronize cultures of Caulobacter (10) was used to separate small, motile cells from larger, mostly nonmotile cells of A. tumefaciens. As shown in Fig. 4A, fluorescence-activated cell sorter (FACS) analysis of these two populations showed that the small, motile cells from the bottom of the tube had a DNA content representing the 1N DNA fluorescence peak observed in a mixed cell population (Fig. 4A). The cells at the top had a DNA content superimposable on the 2N peak. We took the small, motile cells from the bottom of the tube and monitored their growth (Fig. 4B). These cells became nonmotile and began to elongate; they became motile again after they had doubled in size, and they divided at approximately 75 min following their initial resuspension in media. This was consistent with the growth rate of a mixed culture which did not undergo the separation process. As determined by FACS analysis, the DNA content gradually shifted from the 1N peak at the initial time point towards the 2N peak by 45 min. Reappearance of the 1N peak was associated with cell division and the reappearance of small motile cells in the culture, which was first observed at 60 min and was maximal at 75 min. At 90 min, the small cells had again begun to elongate and the DNA content to subtly increase. Thus, we successfully synchronized Agrobacterium cultures, allowing studies of the cell cycle.
We examined flagellin expression during the cell cycle to determine if it correlated with the variation in motility. Antibody to Caulobacter flagellins detected a protein(s) consistent with the size of A. tumefaciens flagellins (32 to 33 kDa); this band was absent in the NT1REB strain, from which the flaA, flaB, and flaC genes are deleted (8) (Fig. 4C). Western blots of samples taken from synchronized cultures showed that the expression of A. tumefaciens flagellar proteins is cell cycle dependent and maximal at times when the cells are motile.
The methylation state of the DNA at the att locus was assessed by the restriction enzyme digestion assay as described in Fig. 2. DNA was prepared from samples taken from synchronized cultures. Figure 4D shows that the degree of DNA methylation at this locus varied over the course of the cell cycle: as DNA replication proceeded, the att locus became completely hemimethylated by 30 min. A ratio of one between the bottom and top band on the blot indicates 100% hemimethylation of the DNA. After the completion of replication at 45 min, the DNA became newly methylated and was maximally so at 60 and 75 min, when cell division occurred. As the cells began to replicate their DNA at 90 min, the amount of hemimethylated DNA increased again. Thus, the activity of the CcrM methyltransferase is restricted to late in S phase of the cell cycle of A. tumefaciens, prior to cell division.
DISCUSSION
We have characterized the ccrM homolog from A. tumefaciens and have shown that it is essential for viability. This requirement is unlikely to be due to restriction of DNA: the gene has no linked open reading frames as would be expected of a restriction-modification system (43). In Caulobacter, furthermore, ccrM has no adjacent open reading frames; promiscuously replicating plasmid DNA can be isolated which is unmethylated at HinfI sites, suggesting that there is in fact no restriction endonuclease with the same sequence specificity as CcrM (48). Like its homologs in C. crescentus, S. meliloti, and B. abortus, therefore, A. tumefaciens ccrM appears to be an essential orphan methyltransferase.
In addition to the requirement for CcrM methylation in all bacteria where it has been isolated thus far, its overexpression causes significant abnormalities of morphology and DNA content (34, 45). In Agrobacterium, branched, elongated cells with multiple chromosomes were observed when ccrM was present on a high-copy-number plasmid. In this strain, DNA was maintained in a fully methylated state. Thus, inappropriate DNA methylation by CcrM interferes with DNA replication and cell division, or the coordination between the two. In E. coli, temporally synchronized initiation at several origins of replication within each rapidly growing cell is dependent on the level of Dam methyltransferase and becomes uncoordinated when the level of Dam is either too high or too low (4). It has been established that the hemimethylated origin in E. coli is incompetent for replication initiation and that its physical sequestration and delayed methylation due to the SeqA system are important in ensuring synchronous timing of initiations (5). While the recently completed Caulobacter genome (26) does not contain a SeqA homolog, CcrM methylation sites are found in the Caulobacter origin of replication at a frequency higher than expected by random chance, and studies to address whether they are important in the initiation of DNA replication are ongoing. As discussed below, there may be some parallels between Agrobacterium and Caulobacter replication, although the Agrobacterium chromosomal origin of replication has not yet been characterized.
No difference from wild-type levels of methylation or any obvious morphological abnormality was seen when ccrM was present on a low-copy-number plasmid. This is probably due to dual regulation of CcrM levels, as in Caulobacter, where a more robust transcriptional increase is needed to overcome proteolysis. However, it is possible that low-level, yet temporally inappropriate, DNA methylation can affect other cellular processes which are more difficult to assay in A. tumefaciens. In support of this hypothesis is the observation that ccrM on a low-copy-number plasmid can, without causing morphologic abnormalities, cause attenuation of intracellular bacterial proliferation in a macrophage model of B. abortus infection (34). Thus, as in the enteric bacteria, methylation may regulate diverse cellular functions among members of the alpha subdivision.
It is possible that CcrM DNA methylation in Agrobacterium also affects transcription of a number of genes. In Caulobacter, CcrM exerts negative control on its own promoter, repressing transcription in the swarmer cell (40). The abnormal branching morphology we observed in A. tumefaciens or in S. meliloti (45) overexpressing CcrM is similar to that associated with blockage of the cell cycle in S. meliloti by treatment with either DNA-damaging agents like nalidixic acid and mitomycin C or inhibition of septation by cephalexin (20). Thus, in addition to any effects it may have on chromosomal replication, inappropriate DNA methylation in Agrobacterium may also up- or down-regulate genes involved in normal cell cycle progression.
We have mapped the transcriptional start site of Agrobacterium ccrM and found that its upstream region is highly homologous to the promoters of the Caulobacter and Brucella ccrM genes. A notable feature of this region is a conserved binding motif for the global response regulator CtrA. While the CtrA homolog has not yet been cloned from Agrobacterium, it has been found in several other alpha proteobacteria and is highly conserved, with ≥80% identity between Caulobacter CtrA and its Sinorhizobium and Brucella homologs (1). CtrA has been shown to be essential for viability in both Caulobacter and S. meliloti (1, 30). It positively regulates ccrM transcription in Caulobacter, and it footprints the ccrM promoter in both Caulobacter and Brucella (33, 34). We have demonstrated that purified Caulobacter CtrA footprints the region of the A. tumefaciens ccrM promoter containing the conserved CtrA binding motif, which overlaps the −35 position relative to the transcriptional start site. This interaction, as for known CtrA targets in Caulobacter, occurs in a phosphorylation-dependent manner (17, 31, 33, 38). Thus, it is likely that in A. tumefaciens, CtrA also regulates ccrM transcription. In conjunction with what is known about Caulobacter and Brucella, this suggests conservation among the alpha bacteria of a regulatory mechanism governing ccrM expression. Site-directed mutation of methylation sites in the 5′ untranslated region of Caulobacter ccrM has been found to prolong its expression during the cell cycle, suggesting possible autoregulation (41). In contrast to the Caulobacter promoter, the promoters of the A. tumefaciens and B. abortus ccrM homologs do not have methylation sites. This may reflect a need for more stringent temporal modulation of CcrM expression in Caulobacter, perhaps due to its asymmetric cell division.
In this work, we demonstrate for the first time that it is feasible to observe the growth of synchronized cultures of A. tumefaciens, thus establishing it as another model organism for studying the bacterial cell cycle. We monitored DNA replication, cell division, DNA methylation state, motility, and flagellar protein expression over the course of the cell cycle. Cells doubled in length as they doubled their DNA content; this was followed by cell division and a concomitant reduction in ploidy. Because of innate drug resistance, we could not treat the cultures with antibiotics which would block initiation and allow us to count the number of origins per cell. However, we consistently observed only 1N and 2N peaks on flow cytometry of wild-type cells, rendering it likely that DNA replication in Agrobacterium occurs just once per cell cycle. DNA replication in Caulobacter has been shown to occur once and only once per cell cycle (22), in contrast to the multiple rounds which can be seen in E. coli (37). Our findings suggest that this precise control of DNA replication may be common to other alpha proteobacteria as well. We also observed that motility varied during the Agrobacterium cell cycle and correlated well with flagellin expression. Flagellation may facilitate virulence in Agrobacterium (8) and is downregulated in a strain-dependent manner under conditions inducing T-pilus biogenesis (18). In light of our work, motility also appears to be influenced by innate cell cycle cues, and studying temporal aspects of its regulation is now possible.
Finally, we demonstrate that methylation of the A. tumefaciens chromosome is constrained to a specific period of the cell cycle, as is the case in Caulobacter. In E. coli, it has been established that, with the exception of the origin of replication, the daughter chromosomes are swiftly methylated by Dam after the replication fork passes. Due to the rapidity of this process, hemimethylated DNA is difficult to detect at the sites outside the origin, and complete hemimethylation of the DNA is never attained (5). In contrast, it is clear that in Caulobacter, the restriction of CcrM to a narrow window of the cell cycle leads to prolonged hemimethylation of the chromosome during the cell cycle. Although we are currently unable to assess cell cycle dependence of A. tumefaciens ccrM transcription and CcrM protein levels, our DNA methylation assay evaluates CcrM activity directly. Our inability to knock out the gene makes it unlikely that there is functional redundancy of this methylating activity. We demonstrate here that the att locus becomes completely hemimethylated 15 min before the time at which all cells in the population have replicated their DNA, presumably due to passage of the replication fork. Methylation only occurs coincident with or immediately following completion of DNA replication, indicating that the presence and/or activity of A. tumefaciens CcrM is similarly temporally restricted.
In Caulobacter, the swarmer cells isolated at the beginning of a synchrony are unable to initiate replication. Hemimethylated DNA is detected only later in the cell cycle, as mature swarmer cells develop into replication-competent stalked cells. However, it is unclear whether A. tumefaciens is ever in an initiation-incompetent form. Immediate reinitiation could account for our observation that the A. tumefaciens DNA was not fully methylated at the beginning of the synchrony. It is also possible that we are unable to obtain starting cell populations as synchronized as in C. crescentus.
It has been established that CcrM is widespread in the alpha subdivision of bacteria (45). Here, we present evidence that temporally constrained DNA methylation activity may be a conserved means of regulating fundamental aspects of the cell cycle common to all these bacteria. Thus, CcrM appears to be an orphan methyltransferase which, like Dam, provides important regulatory information. The alpha proteobacteria include the plant and animal pathogens Agrobacterium, Brucella, and Bartonella; more distant CcrM homologs which are part of restriction-modification systems have been found in H. influenzae and Helicobacter pylori. Thus, inhibition of CcrM function may prove lethal to such bacteria, either through disruption of an essential regulatory function or by allowing digestion of unmethylated DNA to occur. Just as Dam has become a therapeutic target in bacteria, CcrM may be another attractive candidate target for the development of methyltransferase inhibitors as novel antibiotics.
ACKNOWLEDGMENTS
This work was supported by grant K08-AI-01510 from the National Institute for Allergy and Infectious Diseases, National Institutes of Health (L.S.K.), NIH grant GM51426 (L.S.), and Defense Advanced Research Projects Agency (DARPA) grant MDA972-97-1-0008 (L.S.).
We are grateful to Sharon Long and Patricia Zambryski for their gifts of strains, to Eugene Nester for the A. tumefaciens A136 genomic library, and to Craig Stephens for the plasmid used to screen the library. We also thank the members of the Shapiro lab, particularly Kathleen Ryan, Ken Keiler, and Jeff Skerker, for their critical reading of the manuscript and helpful discussions.
REFERENCES
- 1.Barnett M J, Hung D Y, Reisenauer A, Shapiro L, Long S R. A homolog of the CtrA cell cycle regulator is present and essential in Sinorhizobium meliloti. J Bacteriol. 2000;183:3204–3210. doi: 10.1128/JB.183.10.3204-3210.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bird A P, Wolffe A P. Methylation-induced repression—belts, braces, and chromatin. Cell. 1999;99:451–454. doi: 10.1016/s0092-8674(00)81532-9. [DOI] [PubMed] [Google Scholar]
- 3.Blyn L B, Braaten B A, Low D A. Regulation of pap pilin phase variation by a mechanism involving differential Dam methylation states. EMBO J. 1990;9:4045–4054. doi: 10.1002/j.1460-2075.1990.tb07626.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boye E, Lobner-Olesen A. The role of dam methyltransferase in the control of DNA replication in E. coli. Cell. 1990;62:981–989. doi: 10.1016/0092-8674(90)90272-g. [DOI] [PubMed] [Google Scholar]
- 5.Campbell J L, Kleckner N. E. coli oriC and the dnaA gene promoter are sequestered from dam methyltransferase following the passage of the chromosomal replication fork. Cell. 1990;62:967–979. doi: 10.1016/0092-8674(90)90271-f. [DOI] [PubMed] [Google Scholar]
- 6.Cangelosi G A, Best E A, Martinetti G, Nester E W. Genetic analysis of Agrobacterium. Methods Enzymol. 1991;204:384–397. doi: 10.1016/0076-6879(91)04020-o. [DOI] [PubMed] [Google Scholar]
- 7.Charles T C, Nester E W. A chromosomally encoded two-component sensory transduction system is required for virulence of Agrobacterium tumefaciens. J Bacteriol. 1993;175:6614–6625. doi: 10.1128/jb.175.20.6614-6625.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chesnokova O, Coutinho J B, Khan I H, Mikhail M S, Kado C I. Characterization of flagella genes of Agrobacterium tumefaciens, and the effect of a bald strain on virulence. Mol Microbiol. 1997;23:579–590. doi: 10.1046/j.1365-2958.1997.d01-1875.x. [DOI] [PubMed] [Google Scholar]
- 9.Deakin W J, Parker V E, Wright E L, Ashcroft K J, Loake G J, Shaw C H. Agrobacterium tumefaciens possesses a fourth flagellin gene located in a large gene cluster concerned with flagellar structure, assembly and motility. Microbiology. 1999;145:1397–1407. doi: 10.1099/13500872-145-6-1397. [DOI] [PubMed] [Google Scholar]
- 10.Evinger M, Agabian N. Envelope-associated nucleoid from Caulobacter crescentus stalked and swarmer cells. J Bacteriol. 1977;132:294–301. doi: 10.1128/jb.132.1.294-301.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garcia-Del Portillo F, Pucciarelli M G, Casadesus J. DNA adenine methylase mutants of Salmonella typhimurium show defects in protein secretion, cell invasion, and M cell cytotoxicity. Proc Natl Acad Sci USA. 1999;96:11578–11583. doi: 10.1073/pnas.96.20.11578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goodner B W, Markelz B P, Flanagan M C, Crowell C B, Jr, Racette J L, Schilling B A, Halfon L M, Mellors J S, Grabowski G. Combined genetic and physical map of the complex genome of Agrobacterium tumefaciens. J Bacteriol. 1999;181:5160–5166. doi: 10.1128/jb.181.17.5160-5166.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haagmans W, van der Woude M. Phase variation of Ag43 in Escherichia coli: Dam-dependent methylation abrogates OxyR binding and OxyR-mediated repression of transcription. Mol Microbiol. 2000;35:877–887. doi: 10.1046/j.1365-2958.2000.01762.x. [DOI] [PubMed] [Google Scholar]
- 14.Heithoff D M, Sinsheimer R L, Low D A, Mahan M J. An essential role for DNA adenine methylation in bacterial virulence. Science. 1999;284:967–970. doi: 10.1126/science.284.5416.967. [DOI] [PubMed] [Google Scholar]
- 15.Heithoff D M, Sinsheimer R L, Low D A, Mahan M J. In vivo gene expression and the adaptive response: from pathogenesis to vaccines and antimicrobials. Philos Trans R Soc Lond B. 2000;355:633–642. doi: 10.1098/rstb.2000.0604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jaenisch R. DNA methylation and imprinting: why bother? Trends Genet. 1997;13:323–329. doi: 10.1016/s0168-9525(97)01180-3. [DOI] [PubMed] [Google Scholar]
- 17.Kelly A J, Sackett M J, Din N, Quardokus E, Brun Y V. Cell cycle-dependent transcriptional and proteolytic regulation of FtsZ in Caulobacter. Genes Dev. 1998;12:880–893. doi: 10.1101/gad.12.6.880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lai E M, Chesnokova O, Banta L M, Kado C I. Genetic and environmental factors affecting T-pilin export and T-pilus biogenesis in relation to flagellation of Agrobacterium tumefaciens. J Bacteriol. 2000;182:3705–3716. doi: 10.1128/jb.182.13.3705-3716.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lang A S, Beatty J T. Genetic analysis of a bacterial genetic exchange element: the gene transfer agent of Rhodobacter capsulatus. Proc Natl Acad Sci USA. 2000;97:859–864. doi: 10.1073/pnas.97.2.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Latch J N, Margolin W. Generation of buds, swellings, and branches instead of filaments after blocking the cell cycle of Rhizobium meliloti. J Bacteriol. 1997;179:2373–2381. doi: 10.1128/jb.179.7.2373-2381.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Malone T, Blumenthal R M, Cheng X. Structure-guided analysis reveals nine sequence motifs conserved among DNA amino-methyltransferases, and suggests a catalytic mechanism for these enzymes. J Mol Biol. 1995;253:618–632. doi: 10.1006/jmbi.1995.0577. [DOI] [PubMed] [Google Scholar]
- 22.Marczynski G T. Chromosome methylation and measurement of faithful, once and only once per cell cycle chromosome replication in Caulobacter crescentus. J Bacteriol. 1999;181:1984–1993. doi: 10.1128/jb.181.7.1984-1993.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marinus M G. Methylation of DNA. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella. 2nd ed. Washington, D.C.: American Society for Microbiology; 1996. pp. 782–791. [Google Scholar]
- 24.Matthysse A G, Yarnall H A, Young N. Requirement for genes with homology to ABC transport systems for attachment and virulence of Agrobacterium tumefaciens. J Bacteriol. 1996;178:5302–5308. doi: 10.1128/jb.178.17.5302-5308.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nicholson B, Low D. DNA methylation-dependent regulation of pef expression in Salmonella typhimurium. Mol Microbiol. 2000;35:728–742. doi: 10.1046/j.1365-2958.2000.01743.x. [DOI] [PubMed] [Google Scholar]
- 26.Nierman W, et al. Complete genome sequence of the Caulobacter crescentus strain CB15. Proc Natl Acad Sci USA. 2001;98:4136–4141. doi: 10.1073/pnas.061029298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nou X, Braaten B, Kaltenbach L, Low D A. Differential binding of Lrp to two sets of pap DNA binding sites mediated by Pap I regulates Pap phase variation in Escherichia coli. EMBO J. 1995;14:5785–5797. doi: 10.1002/j.1460-2075.1995.tb00267.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ouimet M C, Marczynski G T. Analysis of a cell-cycle promoter bound by a response regulator. J Mol Biol. 2000;302:761–775. doi: 10.1006/jmbi.2000.4500. [DOI] [PubMed] [Google Scholar]
- 29.Prentki P, Krisch H M. In vitro insertional mutagenesis with a selectable DNA fragment. Gene. 1984;29:303–313. doi: 10.1016/0378-1119(84)90059-3. [DOI] [PubMed] [Google Scholar]
- 30.Quon K C, Marczynski G T, Shapiro L. Cell cycle control by an essential bacterial two-component signal transduction protein. Cell. 1996;84:83–93. doi: 10.1016/s0092-8674(00)80995-2. [DOI] [PubMed] [Google Scholar]
- 31.Quon K C, Yang B, Domian I J, Shapiro L, Marczynski G T. Negative control of bacterial DNA replication by a cell cycle regulatory protein that binds at the chromosome origin. Proc Natl Acad Sci USA. 1998;95:120–125. doi: 10.1073/pnas.95.1.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reisenauer A, Kahng L S, McCollum S, Shapiro L. Bacterial DNA methylation: a cell cycle regulator? J Bacteriol. 1999;181:5135–5139. doi: 10.1128/jb.181.17.5135-5139.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reisenauer A, Quon K, Shapiro L. The CtrA response regulator mediates temporal control of gene expression during the Caulobacter cell cycle. J Bacteriol. 1999;181:2430–2439. doi: 10.1128/jb.181.8.2430-2439.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robertson G T, Reisenauer A, Wright R, Jensen R B, Jensen A, Shapiro L, Roop R M., II The Brucella abortus CcrM DNA methyltransferase is essential for viability, and its overexpression attenuates intracellular replication in murine macrophages. J Bacteriol. 2000;182:3482–3489. doi: 10.1128/jb.182.12.3482-3489.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 36.Simon R, Priefer U, Puhler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Bio/Technology. 1983;1:784–790. [Google Scholar]
- 37.Skarstad K, Boye E, Steen H B. Timing of initiation of chromosome replication in individual Escherichia coli cells. EMBO J. 1986;5:1711–1717. doi: 10.1002/j.1460-2075.1986.tb04415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Skerker J M, Shapiro L. Identification and cell cycle control of a novel pilus system in Caulobacter crescentus. EMBO J. 2000;19:3223–3234. doi: 10.1093/emboj/19.13.3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stephens C, Mohr C, Boyd C, Maddock J, Gober J, Shapiro L. Identification of the fliI and fliJ components of the Caulobacter flagellar type III protein secretion system. J Bacteriol. 1997;179:5355–5365. doi: 10.1128/jb.179.17.5355-5365.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stephens C, Reisenauer A, Wright R, Shapiro L. A cell cycle-regulated bacterial DNA methyltransferase is essential for viability. Proc Natl Acad Sci USA. 1996;93:1210–1214. doi: 10.1073/pnas.93.3.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stephens C M, Zweiger G, Shapiro L. Coordinate cell cycle control of a Caulobacter DNA methyltransferase and the flagellar genetic hierarchy. J Bacteriol. 1995;177:1662–1669. doi: 10.1128/jb.177.7.1662-1669.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weyand N J, Low D A. Regulation of Pap phase variation. Lrp is sufficient for the establishment of the phase off pap DNA methylation pattern and repression of pap transcription in vitro. J Biol Chem. 2000;275:3192–3200. doi: 10.1074/jbc.275.5.3192. [DOI] [PubMed] [Google Scholar]
- 43.Wilson G G. Organization of restriction-modification systems. Nucleic Acids Res. 1991;19:2539–2566. doi: 10.1093/nar/19.10.2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Winzeler E, Shapiro L. Use of flow cytometry to identify a Caulobacter 4.5S RNA temperature-sensitive mutant defective in the cell cycle. J Mol Biol. 1995;251:346–365. doi: 10.1006/jmbi.1995.0439. [DOI] [PubMed] [Google Scholar]
- 45.Wright R, Stephens C, Shapiro L. The CcrM DNA methyltransferase is widespread in the alpha subdivision of proteobacteria, and its essential functions are conserved in Rhizobium meliloti and Caulobacter crescentus. J Bacteriol. 1997;179:5869–5877. doi: 10.1128/jb.179.18.5869-5877.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wright R, Stephens C, Zweiger G, Shapiro L, Alley M R. Caulobacter Lon protease has a critical role in cell-cycle control of DNA methylation. Genes Dev. 1996;10:1532–1542. doi: 10.1101/gad.10.12.1532. [DOI] [PubMed] [Google Scholar]
- 47.Zhu J, Oger P M, Schrammeijer B, Hooykaas P J, Farrand S K, Winans S C. The bases of crown gall tumorigenesis. J Bacteriol. 2000;182:3885–3895. doi: 10.1128/jb.182.14.3885-3895.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zweiger G, Marczynski G, Shapiro L. A Caulobacter DNA methyltransferase that functions only in the predivisional cell. J Mol Biol. 1994;235:472–485. doi: 10.1006/jmbi.1994.1007. [DOI] [PubMed] [Google Scholar]