SUMMARY
Disruption of antagonism between SWI/SNF chromatin remodelers and Polycomb repressor complexes drives formation of numerous cancer types. Recently, an inhibitor of Polycomb protein EZH2 was approved for treatment of sarcomas mutant in SWI/SNF subunit SMARCB1, but resistance occurs. Here we performed CRISPR screens in SMARCB1-mutant rhabdoid tumor cells to identify genetic contributors to SWI/SNF-Polycomb antagonism and potential resistance mechanisms. We found that loss of the H3K36 methyltransferase NSD1 caused resistance to EZH2 inhibition. We show that NSD1 antagonizes Polycomb via cooperation with SWI/SNF and identify co-occurrence of NSD1 inactivation in SWI/SNF-defective cancers, indicating in vivo relevance. We demonstrate that H3K36me2 itself has an essential role in activation of Polycomb target genes as inhibition of the H3K36me2 demethylase KDM2A restores efficacy of EZH2 inhibition in SWI/SNF-deficient cells lacking NSD1. Together our data expand the mechanistic understanding of SWI/SNF and Polycomb interplay and identify NSD1 as key for coordinating this transcriptional control.
INTRODUCTION
Regulation of the chromatin landscape plays key roles in the execution of differentiation programs and the control of cellular progression through development. The importance of these roles is highlighted by the finding that mutations in the genes encoding chromatin regulatory proteins are frequently the basis of both cancer and developmental disorders (Allis and Jenuwein, 2016). EZH2 is an enzymatic subunit of Polycomb Repressive Complex 2 (PRC2), which catalyzes methylation of Lys27 of histone 3 (H3K27me1/me2/me3) to promote transcriptional silencing to modulate differentiation (Deevy and Bracken, 2019; Kloet et al., 2016; Oliviero et al., 2016; Yu et al., 2019). Hyperactivity of EZH2 has been implicated in the genesis of numerous types of cancer (Kim and Roberts, 2016). Recurrent hot spot mutations at Tyr646 (Y646) result in gain-of-function amplification of the enzymatic activity of EZH2 in non-Hodgkin’s lymphoma (NHL), diffuse large B-cell lymphoma (DLBCL), follicular lymphoma (FL) and cutaneous melanoma (Majer et al., 2012; McCabe et al., 2012a; Richart and Margueron, 2020; Sneeringer et al., 2010). Hyperactivity of EZH2 also results from loss-of-function mutations in genes that encode subunits of SWI/SNF chromatin remodeling complexes as these complexes antagonize Polycomb function (Kim and Roberts, 2016).
Rhabdoid tumor (RT) is a highly aggressive cancer of early childhood that has yielded great insight into both SWI/SNF function and antagonism between SWI/SNF and Polycomb complexes. This genomically simple malignancy was the first cancer found to be driven by mutation of a SWI/SNF encoding gene, SMARCB1 (also known as SNF5, INI1, and BAF47), which is inactivated in nearly all RT (Lee et al., 2012; Versteege et al., 1998). Subsequently, SWI/SNF mutations were found to be remarkably prevalent in cancer, present in over 20% of malignancies. SWI/SNF complexes remodel chromatin structure via ATP hydrolysis to generate nucleosome-free regions that in part facilitate transcription factor binding (Mittal and Roberts, 2020; Wilson and Roberts, 2011). SMARCB1 facilitates recruitment of SWI/SNF complexes to enhancers. This activity enables transcriptional activation by displacing nucleosomes, evicting EZH2, thereby reducing H3K27me3 (Kadoch et al., 2017; Wilson et al., 2010), and engaging p300, causing acetylation of H3K27 (Alver et al., 2017). SWI/SNF mutations drive cancer in part due to impaired opposition to Polycomb leading to silencing of transcription programs required for differentiation (Alver et al., 2017; Kadoch et al., 2017; Nakayama et al., 2017; Wang et al., 2017; Wilson et al., 2010). The demonstration that genetic deletion of EZH2 preferentially killed SWI/SNF mutant cancers contributed to the adoption of use of EZH2 inhibitors for therapy of SWI/SNF mutant cancers (Knutson et al., 2013).
Recently, the small molecule EZH2 inhibitor Tazemetostat gained FDA approval for the treatment of sarcomas that carry inactivating mutations in the SWI/SNF subunit SMARCB1 (Rothbart and Baylin, 2020), and soon thereafter for lymphomas that harbor activating mutations in EZH2 (Knutson et al., 2012; McCabe et al., 2012b). Numerous additional trials remain ongoing, but as with any monotherapy, primary and secondary resistance to EZH2 inhibitors is quite common as the large majority of treated cancers progress. To expand understanding of the mechanisms of resistance to EZH2 inhibition and of SWI/SNF and Polycomb antagonism we performed a near genome-wide CRISPR screen in RT cells treated with EZH2 inhibitor and identified NSD1 as required for sensitivity to EZH2 inhibition. Mechanistically we establish that H3K36me2 written by NSD1 is essential for gene activation caused by EZH2 inhibition as inhibition of the H3K36me2 demethylase KDM2A re-establishes efficacy of EZH2 inhibition even in the absence of NSD1.
RESULTS
NSD1 loss confers resistance to EZH2 inhibition
To identify genes that when inactivated confer resistance to EZH2 inhibition, we performed a near genome-wide CRISPR-Cas9 drug resistance screen using two SMARCB1-deficient RT cell lines, G401 and G402, in the presence of the EZH2 inhibitor GSK126 (Figures 1A and S1A). We infected cells with a pool of over 70,000 small guide RNAs (sgRNAs) targeting approximately 18,000 genes (four sgRNAs per gene). Cells were treated with GSK126 and after three weeks of selection, sgRNA-barcoded sequences were amplified from the surviving population. Enriched and depleted sgRNAs were quantified relative to pretreatment abundance. Aside from TP53 and RB1, whose deletion confer growth advantages in many contexts, NSD1 scored as a top enriched target in both G401 and G402 cells (Figures 1B and S1B). Since NSD1 is a H3K36me2 methyltransferase, we evaluated whether other enzymes capable of methylating H3K36 (NSD2, NSD3, ASH1L) caused resistance to EZH2 inhibition in our screen. Although sgRNAs for all were equally represented within the screen library (Figure S1C), only NSD1 scored as an enrichment after EZH2 inhibition (Figure S1D and S1E). To validate these findings, we generated pools of G401 RT cells knocked out for NSD1, NSD2 or NSD3. We again found that only NSD1 knockout cells displayed resistance to EZH2 inhibition (Figure S1F-S1H).
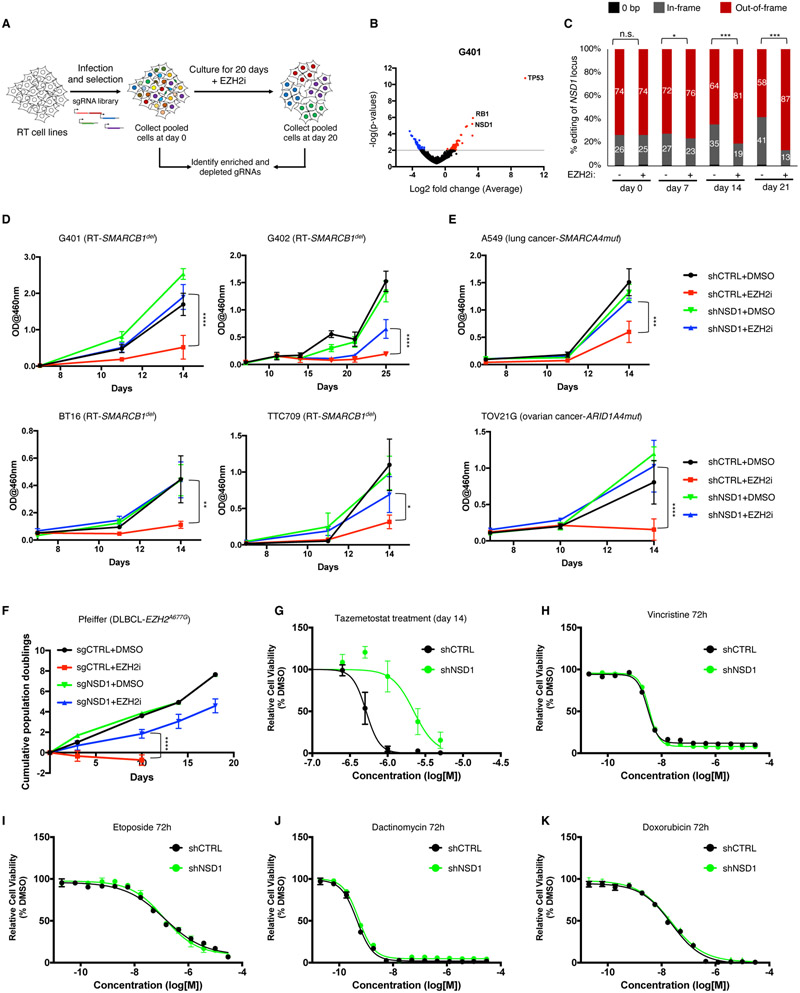
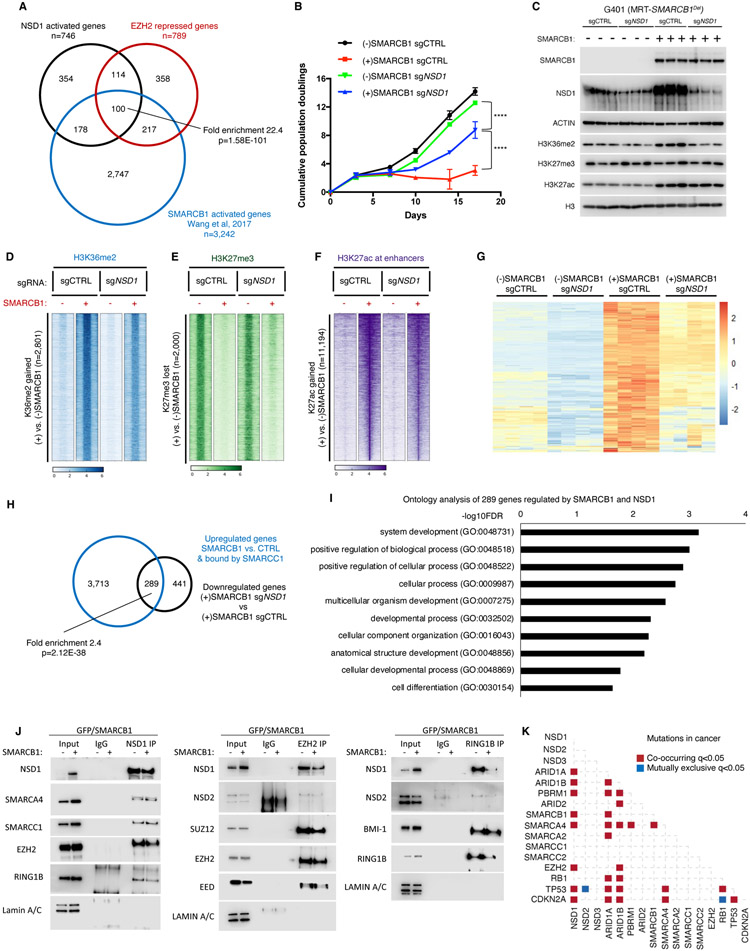
Figure 1. NSD1 loss confers resistance to EZH2 inhibition.
(A) Schematic overview of the CRISPR screen.
(B) Differential enrichment of gRNAs in the CRISPR-Cas9 screen of EZH2i treated G401 cells. The x-axis represents the log fold change of targets based on averaged sgRNA abundance at day 20 vs. day 0 in the presence of GSK126. Positive (red) values represent genes that when deleted provide resistance to GSK126 whereas negative (blue) values represent gene deletions that sensitize cells to GSK126 treatment. The screen was performed with two RT cell lines (G401 and G402) with n=3 biological replicates.
(C) CRISPR-based competitive fitness. NSD1 was targeted with sgRNA in G401 cells and genomic DNA sequenced after 0, 7, 14- and 21-days treatment with GSK126. All indels were binned into in-frame, out-of-frame, or 0-bp. Student’s t-test,*=<0.01, ***=<0.0001, n=3 biological replicates.
(D and E) Proliferation curves of RT (D) and non-RT SWI/SNF mutant (E) cell lines after NSD1 knockdown and treatment with EZH2 inhibitor GSK126. Proliferation was assessed by a modified MTT assay (MTS).
(F) Proliferation of the Pfeiffer lymphoma cell line after NSD1 KO and treatment with either DMSO or GSK126. Proliferation was assessed by cell counting.
(G) Sensitivity of G401 NSD1 KO cell lines to EZH2 inhibitor Tazemetostat.
(H-K) Proliferation of G401 control and NSD1-depleted cells after treatment with standard of care therapeutics vincristine (H), etoposide (I), dactinomycin (J) and doxorubicin (K). Proliferation was assessed by Cell Titer Glo assay at 72 hours of treatment.
For all viability assays, two-way Anova with Turkey’s test for multiple hypothesis correction was used. Error bars represent means ±s.d. (n=2-3 biological replicates, 3 technical replicates). *p < 0.05; **p < 0.01; ***p < 0.001.
To further confirm these results, we developed a CRISPR-based competitive fitness assay to determine whether EZH2 inhibition exerted selective pressure for loss-of-function mutations in NSD1. We performed CRISPR-Cas9 sgRNA targeting of NSD1 in G401 RT cells to induce insertions and deletions. Treatment with GSK126 enriched for out-of-frame/loss-of-function mutations in NSD1, confirming that NSD1 inactivation confers resistance to EZH2 inhibition (Figure 1C). We next performed shRNA mediated knockdown of NSD1 in four SMARCB1-deficient RT cell lines, which in all cases conferred resistance to EZH2 inhibition (Figure 1D), a finding additionally confirmed by CRISPR-mediated inactivation of NSD1 in G401 cells (Figure S1H).
While SMARCB1 was the first SWI/SNF subunit whose loss was shown to create dependence upon EZH2 (Wilson et al., 2010), loss of other SWI/SNF subunits also results in EZH2 dependence (Kim et al., 2015). We therefore asked whether inactivation of NSD1 could confer resistance to GSK126 in a SMARCA4-mutant lung carcinoma line and an ARID1A-mutant ovarian carcinoma (Wiegand et al., 2010), both of which depend upon EZH2 (Kim et al., 2015). NSD1 knockdown abrogated the anti-growth effects of EZH2 inhibition in these cell lines as well (Figures 1E and S1I).
EZH2 inhibitors were originally developed in part to treat lymphomas that carry gain-of-function mutations in EZH2 (Knutson et al., 2014; Knutson et al., 2012; McCabe et al., 2012b). We found that NSD1 loss conferred resistance to EZH2 inhibition in Pfeiffer diffuse large B-cell lymphoma cells (McCabe et al., 2012b) as well (Figure 1F and S1I).
To determine whether the resistance conferred by NSD1 loss extended to the recently FDA approved EZH2 inhibitor Tazemetostat, we treated G401 RT cells and found that inactivation of NSD1 similarly provided resistance to Tazemetostat (Figure 1G).
Finally, we evaluated the effect of NSD1 knockdown upon response to chemotherapeutics that represent standard of care in RT including vincristine, etoposide, dactinomycin and doxorubicin. Unlike the resistance to EZH2 inhibitors, NSD1 knockdown did not alter the response to any of these agents in G401 RT cells (Figures 1H-1K). These results show that loss of NSD1 provides resistance to EZH2 inhibition but does not provide general drug resistance in SWI/SNF mutant cells.
NSD1 depletion leads to H3K36me2 loss and concomitant expansion of H3K27me3 domains
To evaluate the mechanism by which NSD1 loss confers resistance to EZH2 inhibition, we first assessed the effect of NSD1 knockdown upon chromatin modifications in RT cells. NSD1 writes H3K36me2 in active chromatin (Bennett et al., 2017; Weinberg et al., 2019),. H3K36me2 has been shown to inhibit H3K27me3 deposition by EZH2 and in ES cells to facilitate demarcation of PRC2-mediated H3K27me3 domains (Streubel et al., 2018).
Given that NSD1 is only one of several enzymes capable of methylating H3K36 we evaluated whether loss of NSD1 affected total levels of H3K36me2. Western blot analysis of both G401 and BT16 SMARCB1-deficient RT cell lines revealed that NSD1 knockdown partially reduced H3K36me2 levels and elevated H3K27me3 (Figure 2A), consistent with NSD1 opposing EZH2 in RT cells. We note that differences in NSD1 knockdown efficiency and compensation by other H3K36me2 writers (such as NSD2 or NSD3, see below) likely contribute variability to the effect on histone modifications detected by immunoblot. To evaluate the effect of NSD1 knockdown using an orthogonal technique we performed ChIP-seq analyses in G401 RT cells and again found that NSD1 loss reduced H3K36me2 and increased H3K27me3 (Figure 2B).
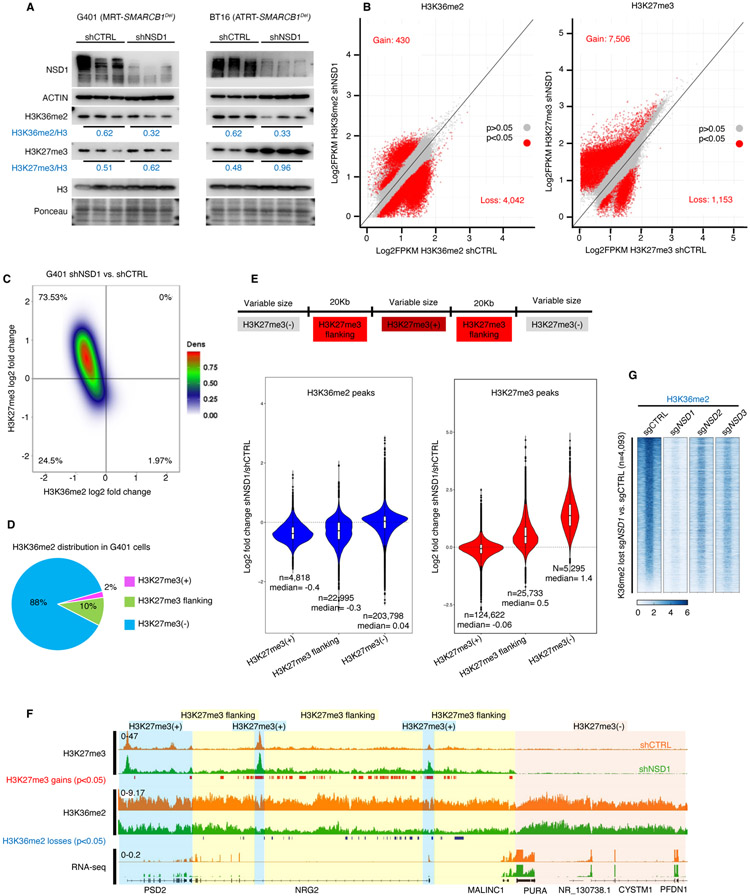
Figure 2. NSD1 depletion leads to H3K36me2 loss and concomitant expansion of H3K27me3 domains.
(A) Immunoblot analysis of G401 and BT16 RT cells upon NSD1 knockdown, with quantification of histone marks normalized to total H3.
(B) Scatter plot analysis of sites differentially enriched for H3K36me2 and H3K27me3 in ChIP-seq analyses of G401 cells subject to NSD1 knockdown. Significance computed using empirical Bayesian statistical tests and linear fitting, see methods for details.
(C) Density plot analysis using 10Kb window segmentation to reveal the relationship between sites of change in H3K27me3 and H3K36me2 upon NSD1 knockdown in G401 RT cells. The genome was segmented into 10Kb bins, and the log2 fold changes were calculated. The plot represents log2 fold change of H3K36me2 and H3K27me3 signal in 10Kb windows.
(D) H3K36me2 distribution in H3K27me3-defined genomic regions. The genome was classified into 3 groups based on H3K27me3 levels in shCTRL cells (see text for details). H3K36me2 signal across four conditions (shCTRL G401, shNSD1 G401 +/− EZH2 inhibitor, see methods) was defined and assigned to H3K27me3-based domains.
(E) Violin plots show differentially enriched H3K36me2 and H3K27me3 peaks (log2 fold change) in shNSD1 vs. shCTRL G401 cells, in each H3K27me3-based domain.
(F) Genome browser tracks for H3K27me3 and H3K36me2 in G401 RT cells treated with shCTRL or shNSD1. Significant H3K27me3 gains (red) and H3K36me2 losses (blue) are depicted. Blue shaded domain=H3K27me3(+), yellow=H3K27me3(+) flanking and pink=H3K27me3(−) domains
(G) Region-scaled heatmap visualization of H3K36me2 normalized ChIP-seq coverage, rank-ordered based on losses in sgNSD1 vs. control G401 cells, including averaged normalized coverage for sgNSD2 and sgNSD3 cells (n=2).
To evaluate the spatial relationships between these marks that often occupy broad swaths rather than tight peaks, we first segmented the genome into 10 Kb bins and plotted changes for the binned H3K36me2 and H3K27me3 ChIP-seq data upon NSD1 knockdown. For bins displaying both marks, there was a tight correlation between loss of H3K36me2 and gain of H3K27me3 upon NSD1 knockdown (Figure 2C). To further examine this relationship, we separated H3K36me2 and H3K27me3 peaks into three groups based on their distance to H3K27me3 peaks in G401 cells: regions encompassing H3K27me3 peaks (H3K27me3(+)), regions immediately flanking H3K27me3 peaks (within 20Kb upstream or downstream; termed H3K27me3 flanking), and regions that lack H3K27me3 and do not flank positive regions (H3K27me3(−)) (Figures 2D and 2E). NSD1 knockdown reduced H3K36me2 both within H3K27me3 peak regions (loss at 87% of the 4,818 peaks) and the H3K27me3 flanking regions (loss at 76% of the 22,995 peaks) but had minimal effect upon H3K36me2 levels in the 200,000+ regions that lacked H3K27me3 (Figures 2E and 2F; Table S1), suggesting a tight association between the activity of NSD1 and EZH2. With respect to H3K27me3, NSD1 loss did not significantly enhance previously defined peaks (124,622 peaks), but rather caused a gain of H3K27me3 at sites flanking H3K27me3(+) regions (25,733 peaks), as well as de-novo gain of 5,295 peaks at regions previously devoid of H3K27me3 (Figures 2E and 2F; Table S1).
To test whether NSD1 alone writes H3K36me2 at the sites most affected by NSD1 loss, we assessed H3K36me2 in cells depleted for NSD2 and NSD3. While H3K36me2 levels were strongly dependent upon NSD1, there was a more modest effect of either NSD2 or NSD3 depletion (Figure 2G), consistent with our finding that NSD2 or NSD3 cannot functionally compensate for loss of NSD1 (Figure S1H).
Thus, on loss of NSD1 there is a tight correlation between H3K36me2 loss and H3K27me3 gain occurring largely at sites that flank H3K27me3 peaks, suggesting that loss of NSD1 results in the spreading of H3K27me3.
H3K27me3 erasure by EZH2 inhibition is largely independent of NSD1
Next, we focused on the mechanism by which NSD1 loss confers resistance to EZH2 inhibition. Treatment with EZH2i leads to H3K27me3 erasure (Figure 3A), so we asked if this was impaired by NSD1 loss. Western blotting analysis revealed that in Pfeiffer cells, which contain a hyper-active mutant of EZH2, NSD1 was able to modestly blunt the EZH2 inhibitor’s erasure of H3K27me3, highlighting the interplay between EZH2 and NSD1 (Figure 3A). However, in SWI-SNF mutant cell lines, knockdown of NSD1 had minimal effect upon the erasure of H3K27me3 by EZH2 inhibition (Figure 3A). Similar results were obtained over a time course with a titration of EZH2 inhibitor (Figures S2A and S2B). Of note, the effects of NSD1 depletion on the global levels of H3K36me2 detected by immunoblot were in some cases modest, potentially due to our inability to fully knockdown NSD1, and because NSD1 controls H3K36me2 at only a small fraction of the genome (i.e., those regions adjacent to Polycomb domains).
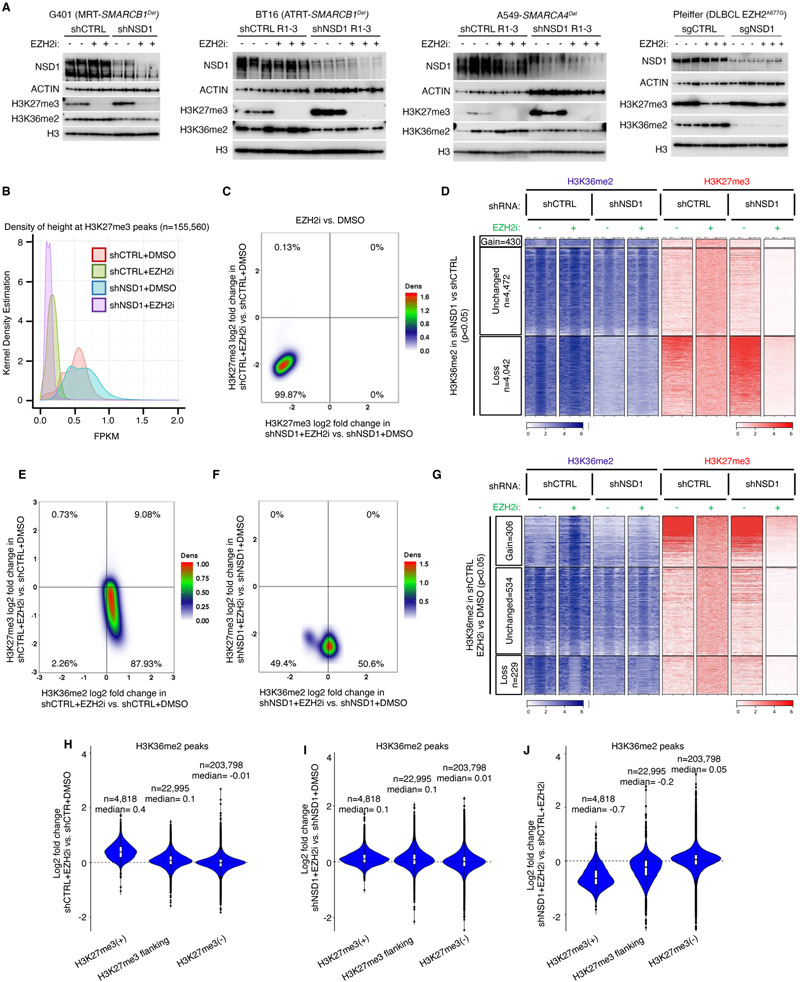
Figure 3. H3K27me3 erasure by EZH2 inhibition is independent of NSD1.
(A) Immunoblot analysis of either control or NSD1 depleted G401 (RT), BT16 (RT), A549 (SMARCA4Del) and Pfeiffer (EZH2 hypermorph) cells upon EZH2 inhibition.
(B) Kernel density plot of H3K27me3 density (fragment length per kilobase/million read (FPKM)) for control and NSD1 depleted cells EZH2 inhibition.
(C) Density plot showing the relationship between the genome-wide changes in H3K27me3 in control vs NSD1 depleted cells upon EZH2 inhibition, using 10kb genome segmentation analysis. Each dot is a 10-Kb window with x/y-axis as log2 fold change.
(D) Region scaled heatmap of H3K36me2, and H3K27me3 ChIP-seq data, of H3K36me2 peaks +/− 3Kb and grouped by changes in K36me2 upon NSD1 knockdown. Significance computed using empirical Bayesian statistical tests and linear fitting, see methods for details.
(E and F) Density plots of relationship between genome-wide changes in H3K27me3 and H3K36me2 using genome segmentation analysis in control (E) and NSD1 depleted (F) cells upon EZH2 inhibition.
(G) Region scaled heatmap visualization of H3K36me2 and H3K27me3 ChIP-seq data, of the H3K36me2 peak regions +/− 3Kb and grouped by changes upon EZH2 inhibition in shCTRL cells. Significance computed using empirical Bayesian statistical tests and linear fitting, see methods for details.
(H-J) Violin plots show differentially enriched H3K36me2 peaks (log2 fold change) in either G401 control (H), NSD1 depleted (I) or both (J), upon EZH2 inhibition, in each H3K27me3-based domain.
For all EZH2 inhibition experiments, cells were treated with either DMSO or 1uM GSK126 for 3 days.
To further probe this relationship, we used two approaches: quantitative mass spectrometry (MS) and ChIP-seq. MS analyses confirmed that NSD1 loss did not influence the EZH2 inhibition-mediated reduction in H3K27me3 in G401 RT cells (Figures S3A-S3D; Table S2), whereas in Pfeiffer cells, NSD1 loss again blunted H3K27me3 loss (Figures S3E-S3H; Table S2). For SMARCB1-deficient G401 cells, the changes in H3K27me3 and H3K36me2 upon EZH2i and NSD1 loss occurred predominantly on the H3.1 and H3.2 histone variants (Figure S3C and S3D). Upon EZH2 inhibition, H3.1/H3.2 K36me2 levels rose in both control and NSD1 depleted cells (Figure S3C), but less so in cells lacking NSD1. This suggests that the upregulation of NSD2 that occurs upon NSD1 knockdown (Figure S1F) is unable to fully compensate for the loss of NSD1.
ChIP-seq analyses showed GSK126 drastically reduced H3K27me3 levels (Figure 3B) and that this was unaffected by NSD1 loss (Figures 3C and S4A, S4B). Furthermore, we found no evidence of retention or de novo accumulation of pockets of H3K27me3 in GSK126 treated NSD1 knockdown cells (Figures 3D and S4A, S4B). Thus, while knockdown of NSD1 confers resistance to EZH2 inhibition in RT cells, surprisingly the mechanism appears independent of the direct activity of EZH2 upon H3K27me3.
Since NSD1 mediates dimethylation of H3K36, we next asked whether alteration of H3K36 methylation may underlie the mechanism of resistance. We first asked whether EZH2 inhibition alters H3K36me2 levels. Using ChIP-seq we found a modest gain of H3K36me2 upon EZH2 inhibition (Figures 3E and S4C), which was dependent upon NSD1 (Figure 3F). We next investigated the localization of the changes. We found that EZH2i induced a gain of H3K36me2 specifically at H3K27me3(+) regions (Figures 3G and 3H), and that this gain was dependent upon NSD1 (Figures 3G, 3I and 3J).
Our results thus far show that while NSD1 loss does not affect the removal of H3K27me3 following EZH2 inhibition in RT cells, its function is necessary to prevent spreading of H3K27me3 positive regions, and to deposit H3K36me2 in Polycomb regions upon H3K27me3 removal.
H3K27ac gain upon EZH2 inhibition shows partial dependence on NSD1
We and others have previously demonstrated that loss of SMARCB1, as occurs in RT, results in a reduction in Polycomb opposition accompanied by a gain of H3K27me3 and decreased H3K27ac at enhancers (Alver et al., 2017; Mathur et al., 2017; Nakayama et al., 2017; Wang et al., 2017). As methylation and acetylation of H3K27 are mutually exclusive, we asked whether inhibition of EZH2 in SMARCB1-deficient RT cells could restore H3K27ac levels. Upon EZH2 inhibition, H3K27ac was strongly gained at a modest number of sites (n=253) which were mostly at enhancers (Figures S5A-S5C). A subset of enhancers that gained H3K27ac significantly overlapped with genes upregulated upon SMARCB1 restoration in G401 cells, suggesting that EZH2 inhibition can partially ameliorate the effects of SMARCB1 loss upon acetylation of H3K27 in RT (Figure S5D). Next, we asked whether the EZH2 inhibitor-induced changes in H3K27ac were dependent on NSD1 and found that loss of NSD1 reduced H3K27ac gain at most enhancers (Figures S5B, S5C and S5E). Overall, we show that EZH2 inhibition has only a modest effect on H3K27ac gain and that this partially depends on NSD1.
Transcriptional de-repression following EZH2 inhibition depends on NSD1
Our results suggest that NSD1 loss confers resistance to EZH2 inhibitors largely via perturbation of H3K36me2 regulation. Since H3K36me2 is associated with transcriptional activation, we asked whether NSD1 is essential for the transcriptional activation that occurs following EZH2 inhibition. We performed RNA sequencing (RNA-seq) of G401 control and NSD1 depleted cells treated with or without GSK126. Consistent with EZH2’s known role as a transcriptional repressor, the predominant effect of EZH2 inhibition was transcriptional induction (Figure 4A, Table S3). Ontology analysis revealed activation of pathways associated with development and differentiation (Figure 4B; Table S4). Knockdown of NSD1 markedly blunted this gene activation (Figure 4A), indicating that NSD1 is required for the expression of lineage specific differentiation pathways following EZH2 inhibition. To evaluate whether this was a direct effect we performed ChIP for EZH2, which revealed highly significant binding of EZH2 adjacent to the affected genes (Figure 4C), supporting direct roles for both EZH2 and NSD1 in modulation of these genes. Of note, EZH2 inhibition had minimal effect upon PRC2 binding, as measured by ChIP-seq for SUZ12, suggesting that the changes in transcription arose from altered enzymatic activity rather than altered PRC2 binding (Figure 4D).
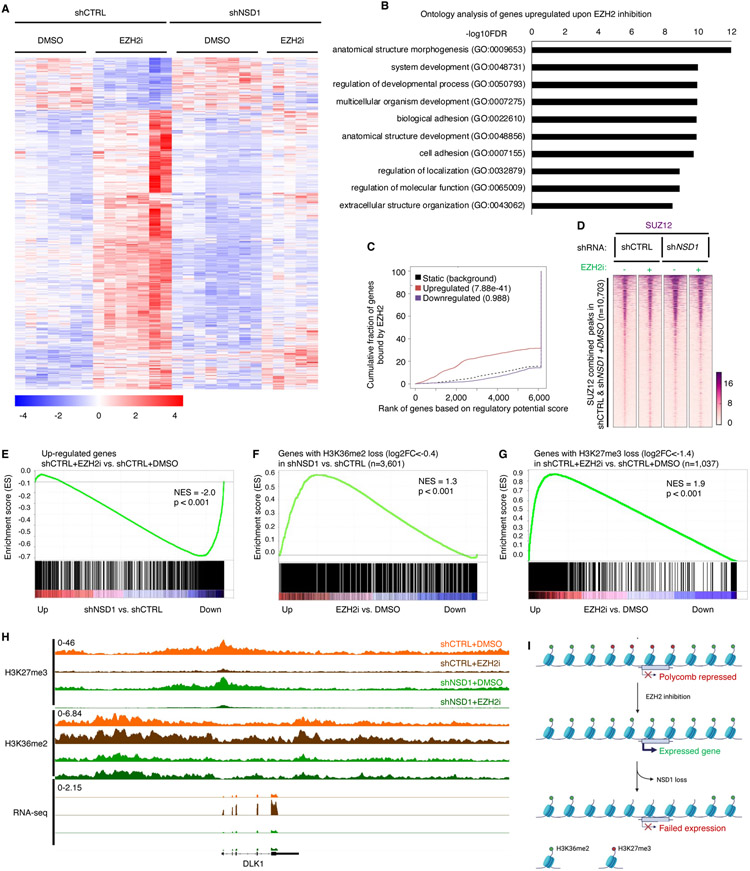
Figure 4. Transcriptional de-repression following EZH2 inhibition depends on NSD1.
(A) Heatmap of 1,041 differentially expressed genes (789 Up, 252 Down, log2 fold change>0 or<0, adjusted p value <0.05, significance computed using empirical Bayesian statistical tests and linear fitting) in control or NSD1 depleted G401 cells upon EZH2 inhibition. Each column represents a biological replicate and rows are hierarchically clustered based on gene expression z-scores (see methods for details).
(B) Gene ontology analysis of the 789 genes upregulated upon EZH2 inhibition in control G401 cells in (A). Only the top 10 out of the 363 total pathways with FDR<0.05 are shown.
(C) Binding and expression target analysis (BETA) of genes upregulated upon EZH2 inhibition and bound by EZH2 in untreated control cells. Each gene is assigned a regulatory potential score based on its expression and binding by EZH2. The red and blue lines represent genes up and down regulated upon EZH2 inhibition, respectively. The dashed line represents genes unchanged or static upon EZH2 inhibition and represent the background. Genes are aggregated based on the “regulatory potential score” and ranked from high to low. P-values are calculated using the Kolmogorov-Smirnov test.
(D) Peak centered heatmap of SUZ12 normalized ChIP-Seq coverage, rank-ordered based on peak signal in untreated control and NSD1 depleted cells (n=1 replicate).
(E) Preranked GSEA using a gene set defined as genes up-regulated (adjusted p-value < 0.05 and log2FC > 0) upon EZH2 inhibition, compared to log2FC ranked genes differentially expressed after NSD1 depletion.
(F and G) Preranked GSEA using a gene set defined as genes losing (log2FC < −0.4) H3K36me2 upon NSD1 depletion (F) or EZH2 inhibition (G) compared to log2FC ranked genes differentially expressed after EZH2 inhibition.
(H) Genome browser tracks depicting H3K27me3, H3K36me2 and RNA-seq reads upon EZH2 inhibition in both control and NSD1 depleted cells.
(I) Schematic representation of the H3K36me2 role in transcriptional activation of Polycomb repressed genes upon EZH2 inhibition.
For all EZH2 inhibition experiments, cells were treated with either DMSO or 1uM GSK126 for 3 days. For GSEA analysis, statistics were calculated by the GSEA tool ( methods for details). NES=normalized enrichment score.
To further investigate the transcriptional relationship between NSD1 knock down and EZH2 inhibition, we performed Gene Set Enrichment Analysis (GSEA). This analysis revealed a significant correlation between genes downregulated upon NSD1 loss and those upregulated upon EZH2 inhibition (Figure 4E and Table S5). Further, we found a significant association between genes overlapping sites that lose H3K36me2 following NSD1 knockdown and genes whose transcription is activated following EZH2 inhibition (Figure 4F). Additionally, genes activated following EZH2 inhibition were significantly enriched for genes overlapping regions that lost H3K27me3, further supporting that they are direct EZH2 targets (Figure 4G). Collectively, this data demonstrates that NSD1 is required for the activation of genes that occurs following erasure of H3K27me3 by EZH2 inhibition. A representative example is the gene DLK1. Loss of NSD1 blocked transcriptional activation of DLK1 that was otherwise caused by EZH2 inhibition, and reduced levels of H3K36me2 without affecting the loss of H3K27me3 caused by EZH2 inhibition (Figure 4H). Collectively, our results thus far suggest that NSD1-mediated H3K36me2 is critical for induction of Polycomb-repressed genes following inhibition of EZH2 (Figure 4I).
NSD1 cooperates with SWI/SNF to activate transcription and induce differentiation
Given our finding that NSD1 opposes EZH2 function in RT and our previous demonstration that SWI/SNF opposes EZH2 function in RT, we asked whether NSD1 cooperates with SWI/SNF in gene activation. We compared the 746 genes downregulated on loss of NSD1 and 789 genes upregulated by EZH2 inhibition to 3,242 genes that we had previously identified as directly activated by SMARCB1 (Wang et al., 2017) and bound by SWI/SNF (Nakayama et al., 2017; Wang et al., 2017) (Figure 5A). Despite overlap in this analysis being limited to the subset of genes that are moderately expressed such that both down-regulation (upon NSD1 knockdown) and up-regulation (upon SMARCB1 re-expression or EZH2 inhibition) can be detected, we found a highly significant relationship (p=1.58E-101) (Figures 5A and S6A). We additionally identified significant pairwise overlaps between genes activated by SMARCB1 and repressed by EZH2 (p=1.73E-64), genes activated by SMARCB1 and NSD1 (p=1.05E-48) and genes activated by NSD1 and repressed by EZH2 (p=1.19E-131), suggesting strong cooperativity between SMARCB1 and NSD1 in opposing Polycomb function (Figure S6A).
Figure 5. NSD1 cooperates with SWI/SNF to activate transcription and induce differentiation.
(A) Analysis of intersection between genes downregulated in shNSD1 vs. shCTRL, upregulated upon SMARCB1 addback + bound by SMARCC1, and upregulated upon EZH2 inhibition by Venn diagram and enrichment statistics (p-value computed using standard Fisher’s exact Test for pairwise overlaps and “Exact Test of Multi-set Intersections” for multi-list overlaps).
(B) Proliferation assays for G401 control and SMARCB1 cells with and without NSD1.
(C) Immunoblot analysis of G401 isogenic cell lines ectopically expressing GFP or SMARCB1.
(D-F) Region-scaled heatmap visualizations of normalized ChIP-Seq coverage, rank-ordered based on changes in SMARCB1-induced G401 cells vs control cells depicting sites of H3K36me2 gain (D), H3K27me3 loss (E) and H3K27ac gain at enhancers (F). n=2 biological replicates.
(G) Heatmap of the 289 genes that are both upregulated by SMARCB1 addition and downregulated by NSD1 loss. n=4 biological replicates per condition.
(H) Venn diagram of the 4,002 genes upregulated by SMARCB1 and bound by SMARCC1 and the 730 genes downregulated by loss of NSD1. There are 289 genes with significant overlap (Fisher’s exact Test) among the 2 datasets (p=2.12E-38).
(I) Significantly enriched biological processes based on the 289 genes that require NSD1 for upregulation by SMARCB1.
(J) Co-immunoprecipitation analysis of NSD1, EZH2 and RING1B in G401 cells upon SMARCB1 addback and schematic representation of the results. Representative immunoblots from n=2 biological replicates.
(K) Analysis of mutation co-occurrence and mutual exclusivity in cancer patients using cBioportal (see methods for details). Fisher’s Exact Test was used.
To directly test whether NSD1 cooperates with SWI/SNF, we generated NSD1 knockout G401 cells with inducible expression of SMARCB1. We and others have shown that restoration of SMARCB1 expression inhibits the growth of RT cell lines (Erkek et al., 2019; Kia et al., 2008; Nakayama et al., 2017; Wang et al., 2017; Weissmiller et al., 2019; Wilson et al., 2010). We thus tested whether this function of SMARCB1 was dependent upon NSD1. Loss of NSD1 indeed ameliorated the growth arrest otherwise caused by SMARCB1 restoration but did not affect proliferation of G401 RT cells that lack SMARCB1 (Figure 5B). Further supporting a dependent relationship, expression of SMARCB1 elevated levels of the NSD1 protein (Figure 5C). Together these data demonstrate that NSD1 is required for the growth arrest caused by SMARCB1 re-expression.
We next sought to elucidate the mechanism underlying rescue of the growth arrest otherwise induced by SMARCB1 expression. Upon SMARCB1 re-expression in RT cells, H3K36me2 was gained at 2,801 sites. Loss of NSD1 substantially blunted, but did not fully eliminate, this gain (Figure 5D). Thus, NSD1 contributes to the gain in H3K36me2 upon SMARCB1 reintroduction. We next examined sites at which SMARCB1 re-expression resulted in loss of H3K27me3 and found that NSD1 loss modestly blunted this effect (Figure 5E). In contrast, the loss of NSD1 had relatively minimal effect upon the gain of H3K27ac at enhancers, driven by SMARCB1 (Figure 5F). We evaluated the converse relationship by identifying regions with the greatest reduction of H3K36me2 upon NSD1 loss and found that re-expression of SMARCB1 modestly rescued the reduction of H3K36me2 levels (Figure S6B).
Loss of antagonism between SWI/SNF and PRC2 has been shown to be a central mechanism by which SMARCB1 inactivation drives RT formation (Nakayama et al., 2017; Wilson et al., 2010). In mESCs NSD1 loss leads to increased PRC2 binding (Streubel et al., 2018) and SWI/SNF antagonizes both PRC2 and PRC1 mediated silencing(Kadoch et al., 2017). Therefore, we evaluated how SWI/SNF activation and NSD1 loss affected targeting of PRC1 and PRC2 subunits RING1B and SUZ12 respectively. We focused on NSD1-dependent H3K36me2 sites and analyzed PRC1/2 occupancy by SUZ12 ChIP-seq and RING1B CUT&RUN (Figure S6C and S6D). Upon NSD1 knockdown the intensity of SUZ12 binding increased at the most strongly bound sites (Figure 4D). Re-expression of SMARCB1 resulted in substantial reduction in SUZ12 binding, which was partially ameliorated by knockout of NSD1 (Figure S6C). In contrast, RING1B binding at NSD1-dependent H3K36me2 sites was not affected by SMARCB1 reintroduction (Figure S6D).
We next investigated the effects of NSD1 loss upon SMARCB1-induced transcriptional activation. Reintroduction of SMARCB1 in G401 cells upregulated 4,002 genes that were direct SWI/SNF targets (bound by SMARCC1, Table S6)(Nakayama et al., 2017). Separately, knockdown of NSD1 resulted in the downregulation of 730 genes (Table S7). Of the 730, 289 (40%) were also directly activated by SMARCB1 (p=2.12E-38) (Figures 5G and 5H; Table S8). Ontology analysis of the 4,002 genes upregulated by SMARCB1 expression, of the 730 genes downregulated by NSD1 loss, and of the 289 overlapping genes in each case revealed significant enrichment of pathways associated with cell differentiation and development (Figures 5I and S6E, S6F, Table S9). This suggests that loss of NSD1 impairs the SMARCB1-dependent activation of genes that facilitate differentiation.
To better understand how loss of NSD1 blocks expression of differentiation-associated genes that are activated by SWI/SNF, we investigated PRC1 and PRC2 binding and histone modification changes at their gene bodies and flanking regions (Figures S6G-S6J). NSD1 depletion reduced H3K36me2 across these genomic regions and caused gain of H3K27me3 particularly at the promoters of these genes. SMARCB1 reintroduction had a smaller effect but increased H3K36me2 particularly at promoters in an NSD1-dependent manner and caused loss of H3K27me3 from promoters, also in an NSD1-dependent manner (Figures S6G and S6H).
Consistent with the effects on H3K27me3, NSD1 loss promoted SUZ12 (PRC2) association particularly at promoters, and SMARCB1 addback reduced SUZ12 signal (Figure S6I). RING1B (PRC1) binding was also reduced by SMARCB1 expression, an effect that was partially dependent upon NSD1. Paradoxically, NSD1 loss alone resulted in a reduction in RING1B binding, perhaps due to competition between PRC2 and PRC1 (Figure S6J). Thus, SMARCB1-mediated antagonism of binding of both PRC1 and PRC2 appears partially dependent upon NSD1. Collectively, despite their distinct biochemical activities, our findings demonstrate cooperative function between SMARCB1 and NSD1 in regulation of transcription and Polycomb opposition.
NSD1 physically interacts with SWI/SNF and PRC1/2 complexes
Finally, we sought to investigate physical interactions among SWI/SNF, Polycomb/EZH2 and NSD1. SWI/SNF has been shown to primarily interact with PRC1 to facilitate its eviction from chromatin and further promote eviction of PRC2 (Kadoch et al., 2017; Stanton et al., 2017). Separately, in mouse ESCs, NSD1 has been shown to associate with EZH2 (Streubel et al., 2018). To ask if Polycomb opposition by SWI/SNF involved interactions between NSD1, SWI/SNF or PRC1/2 members in RT cells, we immunoprecipitated NSD1. We found that NSD1 co-immunoprecipitated with SWI/SNF core subunits SMARCC1 and SMARCA4 as well as EZH2 (Figures 5J and S7A, S7B), demonstrating physical interactions between these chromatin regulators. We also identified a novel interaction between NSD1 and PRC1 (RING1B) (Figure 5J). On restoration of SWI/SNF by reintroduction of SMARCB1, NSD1 levels rose and NSD1 maintained association with SWI/SNF but displayed reduced association with PRC1/2 (Figure 5J). Interactions with PRC1 and PRC2 components were specific for NSD1 and did not extend to NSD2 (Figure 5J). Consequently, the loss of SMARCB1 as a driving event in the genesis of RT results in enhanced physical association of NSD1 with both PRC1 (RING1B) and PRC2 (EZH2), providing a mechanism by which NSD1 loss may directly have effects upon EZH2 inhibition and response.
NSD1 inactivation co-occurs with SWI/SNF mutations in cancer
We next sought to search for in vivo relevance of our findings in primary human cancers. Our data thus far demonstrate that NSD1 physically interacts with both SWI/SNF and Polycomb and that its function is necessary for SWI/SNF to activate transcription of a subset of Polycomb-silenced lineage specification genes. Because inactivating mutations in genes that encode SWI/SNF subunits drive cancer in part by reducing antagonism to EZH2, we sought to evaluate whether mutation of NSD1 is selected for in the formation of SWI/SNF-mutant human cancers to further reduce antagonism to EZH2. Because re-biopsy of progressive solid tumors is rarely performed, we are unable to test whether NSD1 mutations are selected for in tumors that develop secondary resistance to EZH2i. However, given the occurrence of primary resistance to EZH2 in many SMARCB1-mutant cancers (Kim et al., 2015), we hypothesized that mutation of NSD1 would be enriched in SWI/SNF mutant cancers as a mechanism of further reducing Polycomb antagonism. To test this, we first queried all available cBioportal databases (Cerami et al., 2012; Gao et al., 2013) spanning over 60,000 samples and many types of cancer (Figure 5K). We looked for putative pathogenic mutations in all NSD paralogs, SWI/SNF subunits frequently mutated in cancer (Mittal and Roberts, 2020) and as controls tumor suppressors TP53, CDKN2A and RB1 (see methods for details). Our analysis revealed that NSD1 mutations significantly co-exist with mutations in SWI/SNF encoding genes (Figure 5K Table S10). Seeking to validate this finding in a second data set, we analyzed clinical sequencing data generated by MSK (see methods for details) and again focused on NSD1 frameshift mutations. Our analysis confirmed our initial findings and showed that NSD1 inactivating mutations significantly co-exist with ARID1A, ARID1B, ARID2 and SMARCA4 but not with mutations in tumor suppressors TP53, CDKN2A and RB1 (Figures S8A-S8D and Tables S11-14).
SMARCB1 is mutated less frequently in cancer than ARID1A, ARID1B, ARID2 and SMARCA4 so we next directly evaluated studies that analyzed SMARCB1-mutant cancers (Chun et al., 2016). We found that NSD1 is among genes silenced via DNA hypermethylation in SMARCB1-mutant brain tumors compared to normal brain and that this correlated with reduced mRNA levels (Johann et al., 2016). Similarly, we found that NSD1 is hypermethylated in non-CNS RT patients compared to control human embryonic stem cells (hESCs) and its mRNA levels are higher in hESCs compared to MRT tumors (Chun et al., 2016).
To further investigate physiologic relevance of NSD1 mutation directly in patient samples we analyzed genomic data from 50 patients with SMARCB1-mutant RT treated at St. Jude Children’s Research Hospital (Upadhyaya et al., 2020) to determine whether mutation of NSD1 was present in any of these cancers. While none of the 50 samples were from patients who had been treated with EZH2i, we identified a patient whose tumor contained an inactivating mutation in NSD1. This was the sole pathogenic mutation identified in addition to SMARCB1 loss (Upadhyaya et al., 2020). Given the significant association of inactivating NSD1 mutations with SWI/SNF mutations, and the remarkable simplicity of rhabdoid tumor genomes (Lee et al., 2012), this case provides further evidence of physiologic relevance of NSD1 mutations in primary human cancers. Furthermore, NSD1 mutations have been found in SMARCB1-mutant epithelioid malignant peripheral nerve sheath tumors (Richer et al., 2017) and renal medullary carcinomas (Jia et al., 2019), reinforcing the co-occurrence of defects in NSD1 and SWI/SNF function in cancer.
KDM2A perturbation restores sensitivity of NSD1 depleted cells to EZH2 inhibition
We have shown that NSD1 loss confers resistance to EZH2 inhibition by reducing H3K36me2 at EZH2-target genes (Figure 3J and 4F). We thus asked whether raising levels of H3K36me2 could restore sensitivity to EZH2i in NSD1-depleted cells. KDM2A is a demethylase that erases H3K36me2 (Blackledge et al., 2010; He et al., 2008). We tested whether the novel KDM inhibitor TC-E5002, which inhibits KDM2A could restore sensitivity to EZH2i in growth assays. In RT cells, TC-E5002 did not affect cell proliferation or the growth-inhibitory effects of EZH2 inhibition (Figure 6A-6C). In contrast, in NSD1 knockdown RT cells the addition of TC-E5002 restored sensitivity to EZH2 inhibition, (Figure 6A-C). The same combination of inhibitors lacked cooperativity in control HFF-1 (human foreskin fibroblasts) and HEK-293T cell lines (Figure 6A and 6B), suggesting that this combination does not have a non-specific toxicity but rather an on-target effect on NSD1-depleted cells. Further, TC-E5002 effects appeared to be specific to inhibition of KDM2A, since cells knocked down for KDM2A were similarly sensitized to EZH2 inhibition (Figure 6C). Immunoblot analysis of G401 cells treated with EZH2i, KDM2Ai or a combination revealed increased levels of H3K36me2 in NSD1 depleted cells, indicating that the combined inhibition of EZH2 and KDM2A at least partially restores H3K36me2 levels (Figure 6D). Additionally, NSD2 or NSD3 depletion causes some loss of H3K36me2 from sites where H3K36me2 is written by NSD1 (Figure 2G). This strongly suggests that H3K36me2 can be written to some degree by NSD2 and NSD3 at sites where NSD1 is the prominent H3K36me2 methyltransferase, and suggests that on loss of NSD1, the residual H3K36me2 provides substrate for KDM2A activity.
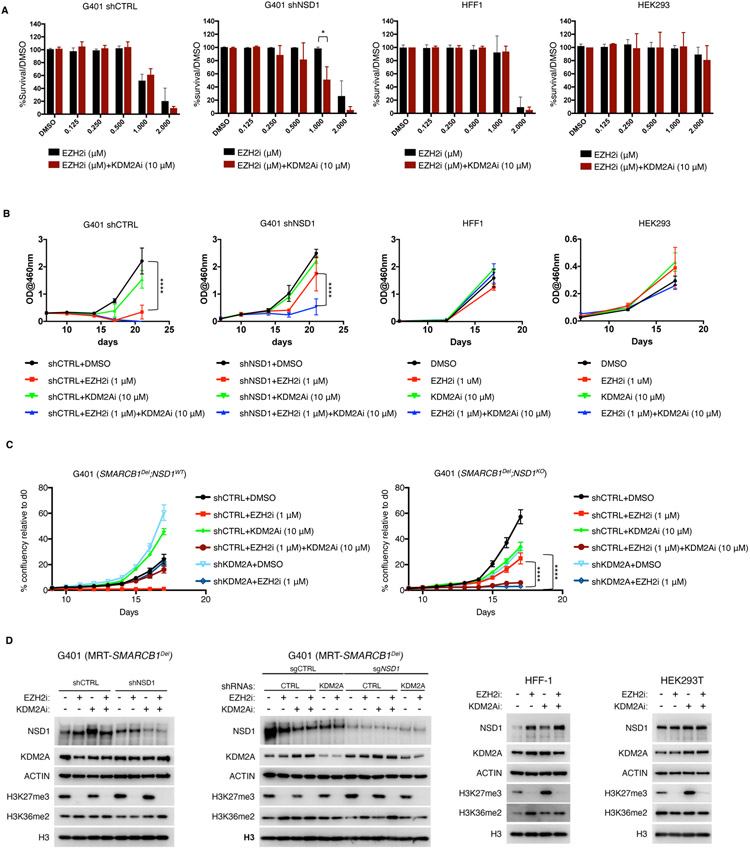
Figure 6. KDM2A inhibition restores sensitivity of NSD1-depleted cells to EZH2 inhibition.
(A) Clonogenic assay results from EZH2 and KDM2A inhibition of G401 control and NSD1 depleted cells treated for 14 days. Human fibroblasts (HFF-1) and HEK293T cells served as controls. Each experiment was performed with n=2 biological replicates and n=3 technical replicates and scanned by densitometry.
(B) TT-based proliferation curves of G401 control or NSD1 depleted cells treated with either DMSO, EZH2i (GSK126) 1uM, KDM2Ai (TC-E5002) 10uM or a combination (n=3 biological replicates).
(C) Incucyte-based proliferation curves of G401 control and NSD1 depleted cells expressing either shCTRL or shKDM2A and treated with DMSO, EZH2i (GSK126) 1uM, KDM2Ai (TC-E5002) 10uM or a combination. Two-way Anova with Turkey’s test for multiple hypothesis correction. Error bars represent means ±s.d. (n=3 biological replicates). *p < 0.05; **p < 0.01; ***p < 0.001.
(D) Immunoblot analysis of G401, HFF-1 and HEK293T cells treated with a combination of 1uM GSK126 and 10uM TC-E5002 or with KDM2A shRNA, after 7 days of treatment. Representative blots are shown from n=2 biological replicates.
Collectively these findings demonstrate the essential role of K36 methylation in the efficacious response to EZH2 inhibition and raise the possibility that therapeutic inhibition of KDM2A can enhance the efficacy of, and overcome resistance to, EZH2 inhibition.
DISCUSSION
Polycomb repressor complexes are central to the transcriptional control of developmental fate specification (Margueron and Reinberg, 2011). Key roles of Polycomb genes in human disease have emerged, including identification of unchecked EZH2 activity in numerous types of cancer (Bracken et al., 2019). The central role of EZH2 in driving these phenotypes has been established in animal models and pre-clinical studies and subsequently reinforced by the recent approval of an EZH2 inhibitor for cancers with hyper-active EZH2, either due to mutation of the SWI/SNF subunit SMARCB1 or direct gain-of-function mutations in EZH2 (Krug et al., 2021).
Despite the key roles for Polycomb complexes and EZH2 in development and disease, the mechanisms by which alterations in Polycomb disrupt transcriptional control to yield disease are poorly understood. Seeking to gain mechanistic insight into chromatin regulation of transcription, to better understand disease mechanisms, and to gain insights into potential mechanisms of resistance for EZH2 inhibitors, we performed a near genome-wide CRISPR screen in RT cells to identify genes that when inactivated confer resistance to EZH2 inhibition.
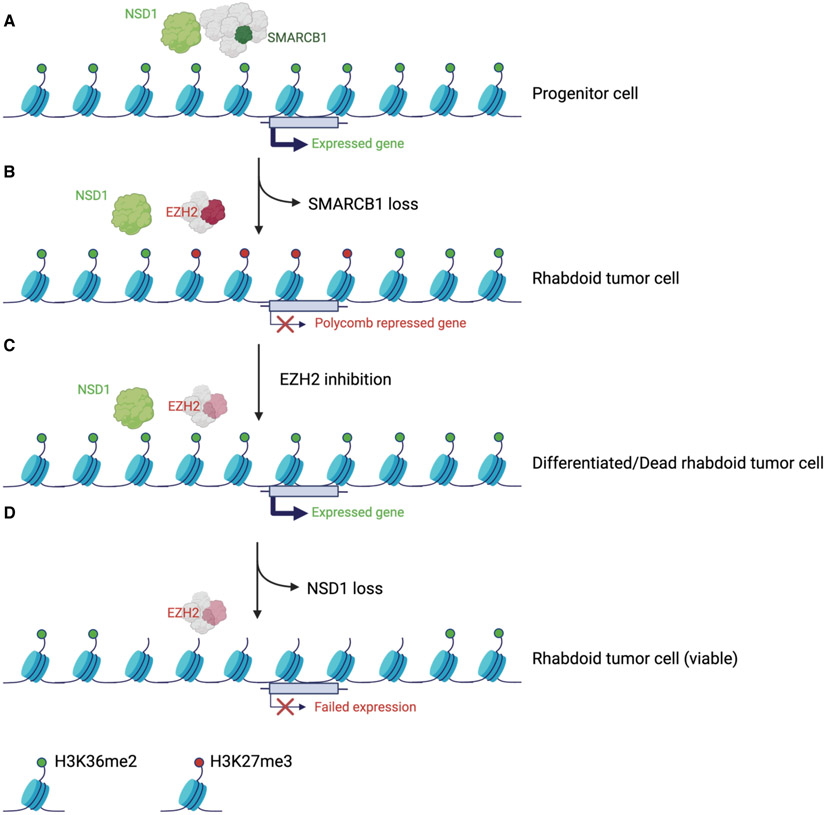
Our CRISPR screen leveraging SMARCB1-mutant RT cell lines revealed that loss of NSD1 conferred resistance to EZH2 inhibition. The mechanism of resistance conferred by NSD1 loss was specific to EZH2 inhibition and extended broadly to several other disease models.
Given that we and others have previously shown that SWI/SNF antagonizes Polycomb activity, which formed the basis of the recent FDA approval of an EZH2 inhibitor for SMARCB1-mutant cancers, we investigated how SWI/SNF complexes functionally relate to these other chromatin regulators during gene activation. We found that functional restoration of SWI/SNF in G401 cells via re-expression of SMARCB1 induced robust changes in H3K36me2, H3K27me3 and H3K27ac, and that gains in H3K36me2 were partially ameliorated by the loss of NSD1. Similarly, the reduction of H3K36me2 that resulted from NSD1 loss was only partially rescued by SMARCB1 re-expression. Thus, NSD1 is required for some, but not all, of the increase in H3K36me2 and H3K27ac resulting from restoration of SMARCB1-containing SWI/SNF complexes. With respect to H3K27me3, we found that NSD1 is needed for H3K27me3 removal driven by SMARCB1 as NSD1 loss resulted in H3K27me3 retention at the promoter and body of genes whose upregulation by SWI/SNF was dependent on NSD1.
Our data suggests that transcriptional activation following EZH2 inhibition depends upon H3K36me2 levels at Polycomb adjacent regions. There are several possible explanations for the role of transcriptional activation by H3K36me2 and/or NSD1. In this study we demonstrate that NSD1 binds SWI/SNF, which may provide a direct link between NSD1 and transcriptional activation. Additionally, NSD1 has been shown to facilitate binding of transcription factors, for example GATA-1, with the loss of NSD1 resulting in reduced GATA-1 binding and reduced expression of target genes (Leonards et al., 2020). Moreover, NSD1 has been shown to contribute to the transition of RNA Pol II from an initiation state to a fully elongation- competent state at its target genes (Lucio-Eterovic et al., 2010).
Given that H3K27ac and H3K27me3 are mutually exclusive, we queried the extent to which H3K27ac is required for the activation of EZH2 target genes. Loss of NSD1 has been associated with both loss (Farhangdoost et al., 2021; Rajagopalan et al., 2021) and gain (Fang et al., 2021) of H3K27ac at enhancers, suggesting a more complex and perhaps indirect relationship. In RT cells we found that EZH2 inhibition resulted in the gain of H3K27ac at only a small subset of genes, an effect that was modestly blunted in the absence of NSD1. Consequently, while there is some interplay of NSD1 with H3K27ac levels in the RT model, the effects are modest in magnitude and restricted to a relatively small subset of genes.
We find that NSD1, but not NSD2 or NSD3, interacts with PRC1/2 to restrict Polycomb-mediated repression, an interaction that is disrupted by SWI/SNF activity, potentially via SWI/SNF mediated eviction of PRC1/2. Moreover, we provide evidence that NSD1 writes H3K36me2 at specific genomic regions and its loss is minimally compensated by NSD2 or NSD3. During development/differentiation, SWI/SNF-driven removal of Polycomb leads to NSD1 and SWI/SNF-coupled activation of lineage specification gene expression.
In addition to causing Sotos syndrome (Tatton-Brown et al., 2011), NSD1 mutations are frequently found in cancer and are associated with a cancer predisposition syndrome (Jia et al., 2019). Our identification of NSD1 inactivating mutations as significantly co-occurring with SWI/SNF mutations further supports functional cooperativity between these genes in opposition to Polycomb mediated silencing. Therefore, it is possible that in patients treated with EZH2 inhibitors, resistance may occur via genetic or epigenetic silencing of NSD1, preventing the re-expression of repressed genes upon treatment with an EZH2 inhibitor. Further, NSD1 mutation status may be useful in predicting response to EZH2 inhibition with the presence of an NSD1 mutation predicting a poor response. Thus, our data indicate that NSD1 mutation is likely to be relevant both as a predictor of non-response as well as a mechanism of resistance.
Previously, NSD1-mediated H3K36me2 has been shown to localize at active chromatin, to flank Polycomb regions and block non-specific placement of H3K27me3 in ES cells (Streubel et al., 2018; Weinberg et al., 2019). Such blockage of Polycomb function by H3K36me2 formed the basis of the model for NSD1-EZH2 antagonism. However, we now show that even when EZH2 activity is completely blocked and H3K27me3 lost that NSD1 remains essential for activation of Polycomb silenced targets. Consequently, loss of H3K27me3 is insufficient to activate many EZH2 targets, which instead require active writing of H3K36me2 by NSD1. We provide evidence for the importance of H3K36me2, by showing that inhibition of KDM2A, one of the demethylases responsible for removing H3K36me2, restores the effects of EZH2 inhibition in NSD1 knockout cells.
Collectively, our data demonstrate that SWI/SNF, NSD1 and EZH2 are part of an integrated chromatin regulatory network whereby their distinct biochemical activities serve coordinated roles in transcriptional regulation.
Limitations of the study
Biopsy samples are not available from pediatric RT patients who have been treated with EZH2 inhibitors, and whose tumors have subsequently progressed, so we are presently unable to screen for the occurrence of therapy-related mutations in NSD1. Moreover, we cannot perform NSD1 ChIP so we cannot assess the localization of NSD1 in relation to H3K36me2 and H3K27me3.
STAR Methods
Resource availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Charles W.M. Roberts (Charles.Roberts@STJUDE.ORG)
Materials availability
Plasmids and cell lines generated in this study will be available upon request.
Data and code availability
The next-generation-sequencing data generated by this study have been deposited to GEO database under accession number GEO: GSE178490
Publicly available ChIP and RNA-Seq were obtained from GEO accession: GSE90631 and GSE71506 and analyzed with consistent methods.
Original western blot images have been deposited at Mendeley. The DOI is listed in the key resources table.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-NSD1 | Antibodies, Inc. | Cat#75-280; RRID: AB_11001827 |
| Mouse monoclonal anti-NSD2 29D1 | Abcam | Cat#ab75359; RRID: AB_1310816 |
| Rabbit monoclonal anti-NSD3 D4N9N | Cell Signaling | Cat# 92056, RRID:AB_2800178 |
| Rabbit monoclonal anti-BRG1 | Cell Signaling | Cat#49360; RRID: AB_2728743 |
| Rabbit monoclonal anti-BAF155 | Cell Signaling | Cat# 11956; RRID: AB_2797776 |
| Rabbit monoclonal anti-SMARCB1 | Cell Signaling | Cat#91735; RRID: AB_2800172 |
| Mouse monoclonal anti-EZH2 | Active Motif | Cat# 39875; RRID: AB_2561022 |
| Rabbit monoclonal anti-EZH2 | Cell Signaling | Cat#5246; RRID: AB_10694683 |
| Rabbit monoclonal anti-SUZ12 | Cell Signaling | Cat#3737; RRID: AB_2196850 |
| Mouse monoclonal anti-EED | Millipore Sigma | Cat#05-1320; RRID: AB_1586999 |
| Rabbit polyclonal anti-EED | Millipore Sigma | Cat#09-774; RRID: AB_1587000 |
| Rabbit monoclonal anti-BMI1 | Cell Signaling | Cat#6964; RRID: AB_10828713 |
| Rabbit monoclonal anti-RING1B | Cell Signaling | Cat#5694; RRID: AB_10705604 |
| Mouse monoclonal anti-b-Actin | Sigma Aldrich | Cat#A5441; RRID: AB_476744 |
| Rabbit polyclonal anti-HSP90 | Cell Signaling | Cat# 4874, RRID:AB_2121214 |
| Rabbit monoclonal anti-KDM2A | Abcam | Cat#ab191387 |
| Mouse monoclonal anti-Lamin A/C | Cell Signaling | Cat# 4777, RRID:AB_10545756 |
| Rabbit monoclonal anti-Histone H3 | Cell Signaling | Cat#4620; RRID: AB_1904005 |
| Rabbit polyclonal H3K27ac | Abcam | Cat#ab4729; RRID: AB_2118291 |
| Rabbit monoclonal H3K27me3 C36B11 | Cell Signaling | Cat#9733; RRID: AB_2616029 |
| Rabbit polyclonal H3K36me2 | Abcam | Cat#ab9049; RRID: AB_1280939 |
| Rabbit monoclonal Rabbit Control IgG | Cell Signaling | Cat#3900; RRID: AB_1550038 |
| Normal mouse IgG | Santa Cruz | Cat#sc-2025; RRID: AB_737182 |
| Drosophila spiked in antibody | Active Motif | Cat# 61686; RRID: AB_27370 |
| Monoclonal mouse anti-Rabbit IgG, light chain specific | Jackson | Cat#211-032-171; RRID: AB_2339149 |
| Goat anti-mouse IgG, light chain specific | Jackson | Cat#115-035-174; RRID: AB_2338512 |
| Chemicals, peptides, and recombinant proteins | ||
| GSK126 | ChemieTek | Cat#CT-GSK126 |
| TC-E5002 | Tocris | Cat#5089 |
| Vincristine (Tocris #1257 | Tocris | Cat#1257 |
| Doxorubicin (CarboSynth #AC15377 | CarboSynth | Cat#AC15377 |
| D-actinomycin (ApexBio #A4448 | ApexBio | Cat#A4448 |
| Etoposide (AG Chemicals #E2004 | AG Chemicals | Cat#E2004 |
| Dimethylsulfoxide (DMSO) | ATCC | Cat#4-X |
| Critical commercial assays | ||
| DirectZol Mini Prep Plus kit | Zymo Research | Cat#R2070 |
| Quant-iT RiboGreen assay | ThermoFisher | Cat#R11490 |
| Epicypher Cutana Cut&Run Kit v2.0 | Epicypher | Cat# 14-1048 |
| Covaris True- ChIP kit | Covaris | Cat#520154 |
| Drosophila chromatin spike in antibody | Active Motif | Cat#61686 |
| Dynabeads™ Protein G for Immunoprecipitation | ThermoFisher | Cat#10004D |
| RNase A | ThermoFisher | Cat#EN0531 |
| Ptoteinase K | ThermoFisher | Cat#4333793 |
| Nuclease | ThermoFisher | Cat#88702 |
| Cell titer aqueous reagent | Promega | Cat#G3582 |
| CellTiter-Glo | Promega | Cat#G9241 |
| Halt™ Protease and Phosphatase Inhibitor Cocktail (100X) | ThermoFisher | Cat# 78446 |
| Deposited data | ||
| RNAseq data | This study | GSE90631 |
| ChIP-seq data | This study | GSE90631 |
| Cut&Run data | This study | GSE90631 |
| Uncropped immunoblot data | This study | Mendeley data doi: 10.17632/fvzm88tm3v.2 |
| Experimental models: Cell lines | ||
| G401 | ATCC | CVCL_0270 |
| G402 | ATCC | CVCL_1221 |
| A549 | ATCC | CVCL_0023 |
| TOV21G | ATCC | CVCL_3613 |
| HFF-1 | ATCC | CVCL_3285 |
| HEK293T | ATCC | CVCL_0063 |
| Pfeiffer | ATCC | CVCL_3326 |
| BT16 | Laboratory of Dr. C. David James | CVCL_M156 |
| TTC709 | Laboratory of Dr. B.E. Weissman. | CVCL_8007 |
| Oligonucleotides | ||
| sgRNA for NSD1 knockout: SP185.g12 5- UCUCCAACUUCAUACUUCAG-3 |
This study | NA |
| Forward primer for Next Generation Sequence (NGS) of the NSD1 gene and assessment of indels: 5-CATACATTGCTTTTTCAGAAGGCT-3 |
This study | NA |
| Reverse primer for Next Generation Sequence (NGS) of the NSD1 gene and assessment of indels: 5- ACACCAATTCATTCACAAAATGTTC-3 |
This study | NA |
| sgRNA for NSD2 knockout: g7 5-CCAAAGUGUCGGGUUACCCU-3 |
This study | NA |
| Forward primer for Next Generation Sequence (NGS) of the NSD2 gene and assessment of indels: 5- AGCCTGGTGAATCCTTGGTCTTCTGG-3 |
This study | NA |
| Reverse primer for Next Generation Sequence (NGS) of the NSD2 gene and assessment of indels: 5-GCTGCAAAAATGAACCATAGCAGGGC-3 |
This study | NA |
| sgRNA for NSD3 knockout: g16 5-AAACUGGACAUGAUAUUCUC-3 |
This study | NA |
| Forward primer for Next Generation Sequence (NGS) of the NSD3 gene and assessment of indels: 5-ACAGGTGCCCGAGAATATCA-3 |
This study | NA |
| Reverse primer for Next Generation Sequence (NGS) of the NSD3 gene and assessment of indels: 5- ACGATTAAAACAGCTGAAAGGGG-3 |
This study | NA |
| Recombinant DNA | ||
| Plasmid: pLKO_005-shNSD1-1 | Sigma | TRCN0000238371 |
| Plasmid: pLKO_005-shNSD1-2 | Sigma | TRCN0000238372 |
| Plasmid: pLKO.1-shKDM2A-1 | Sigma | TRCN0000022000 |
| Plasmid: pLKO_005-shKDM2A-2 | Sigma | TRCN0000359598 |
| Plasmid: pLV[Exp]-Bsd-CMV>Tet3G | VectorBuilder | 180621-1181ctz |
| Plasmid: pLV[Exp]-Puro-TRE3G>EGFP | VectorBuilder | 180621-1204vsc |
| Plasmid: pLV[Exp]-Puro-TRE3G>V5/hSMARCB1 | VectorBuilder | 180622-1078xvz |
| Plasmid: pVSV-G | Addgene | Addgene #138479 |
| Plasmid: psPAX2 | Addgene | Addgene #12260 |
| Software and algorithms | ||
| Prism 7.0d | GraphPad | www.graphpad.com |
| Image J 2.1.0/1.53c | NIH | https://imagej.nih.gov/ij |
| Integrative Genomics Viewer (IGV_2.6.2) | The Broad Institute | https://software.broadinstitute.org/software/igv/ |
Experimental Model and Subject Details
Cell lines
G401 (male), G402 (female), A549 (male), TOV21G (female), HFF-1 (male), HEK293T (female) and Pfeiffer (male) cell lines were obtained from ATCC. BT16 (male) cells were obtained from C. David James, and TTC709 (unknown) cells were obtained from B.E. Weissman. The cells were grown in their recommended media supplemented with 10% fetal bovine serum and 1% Glutamax (Gibco) at 37°C and 5% CO2. All cell lines tested negative for mycoplasma contamination.
Method Details
Drug treatment and cell proliferation assay
For longer-term EZH2 and KDM2 inhibition assays, 100,000 cells were seeded in 10-cm dishes and were treated with DMSO (ATCC), GSK126 (ChemieTek, CT-GSK126), TC-E5002 (Tocris, Cat. No. 5089), or a combination of these agents. After 1 week of growth, the cells were trypsinized and quantified with a Countess automated cell counter (Thermo Fisher), and 200 treated cells were reseeded in each well of 96 well plates. Cell growth was monitored either every 3–4 days by adding 20 μL of the cell titer aqueous reagent (Promega, Catalog#G3582) to each well and measuring absorbance at 520 nm after 4 h, or by measuring cell confluency every 12 hours and calculating % confluency. Media with DMSO or drugs was changed every 3 days, and cells were maintained in culture for 2–3 weeks.
For short-term treatments with standard chemotherapeutics, 10 mM stocks in 100% DMSO of vincristine (Tocris #1257), doxorubicin (CarboSynth #AC15377), D-actinomycin (ApexBio #A4448), or etoposide (AG Chemicals #E2004) were dispensed in triplicate into 384-well flat-bottom tissue culture-treated solid white plates (Perkin Elmer, #6007688) using a Labcyte Echo 555 acoustic Liquid Dispenser (Beckman Coulter Life Sciences, USA). Fourteen-point dose-response curves were generated using 1:3 dilution intervals for each drug with a range of 30 μM to 0.02 nM. The final DMSO concentration of all wells was 0.313%, and an additional 24 wells of DMSO alone were used for normalization. Into the compound-containing plates, the shControl and shNSD1 cell lines were added at density of 2000 cells/well in volumes of 40 μL using an Integra multichannel pipette and settled for 20 seconds at 100x g in an Eppendorf 5810 centrifuge. Plates were then incubated at 37°C in 5% CO2 for 72 hours in a Thermo Scientific Stericult incubator. Following incubation, the plates were placed at room temperature for 20 minutes prior to viability assessment. 25 μL of CellTiter-Glo (Promega Catalog# G9241) was then added to each well to measure viability using a Multidrop Combi and the plates then incubated for an additional 25 minutes at room temperature. Luminescence was then detected with an EnVision 2102 Multilabel Plate Reader (PerkinElmer Life Sciences, USA). All raw luminescent values were normalized to the mean of 24 wells of 0.313% DMSO, the triplicate values plotted, and regression performed in GraphPad Prism to generate EC50s.
CRISPR-Cas9 screen and target identification
G401 and G402 RT cells stably expressing Cas9 (each cell line in triplicate) were infected with the Avana library of 74,687 barcoded sgRNAs including guides targeting 18,454 genes (with an average of 4 guides per gene) and 55 intergenic control sites, and 1003 non-targeting guides. Cells were infected at a low MOI (0.3) to ensure the majority of the cells was infected with only one guide and selected with puromycin for 3 days. After selection (d0) each pool of cells was split into 2 aliquots: one was harvested (d0) and the 2nd was further cultured in the presence of 5uM GSK126 for 20 days, prior to harvesting (d20). During the culture of cells in GSK126, at least 20 million cells in each replicate were passaged to ensure adequate library representation. gDNA was isolated from cells harvested at d0 and d20, and barcoded sgRNAs were PCR amplified and sequenced. The average Log fold change (LFC) for each guide RNA was calculated in d20 versus d0 samples using the hypergeometric analysis tool (https://portals.broadinstitute.org/gpp) and a threshold of p<0.01 (−log_p value >2.0) was applied to identify significant changes. Furthermore, the 2 target lists (G401, G402) were manually annotated to identify sgRNAs that scored in both cell lines and to exclude enrichments identified in RT cell lines by the Achilles project (https://depmap.org/). Candidate targets were defined as having 4 perturbations, a log.2. pvalue >=2, an average log2FC > 0 for enrichment and average log2FC < 0 for depletion. Enriched and depleted candidates were defined for each G401 and G402 and a Venn diagram was used to evaluate the overlap. The supertest function (SuperExactTest v1.0.7) was used to evaluate if the overlap was significant setting n=20,687 (for all protein coding genes).
CRISPR-Cas9 pools and growth fitness assay
G401 NSD1 knockdown pools were generated by using CRISPR-Cas9 technology. Briefly, 300,000 G401 cells were transiently transfected with precomplexed ribonuclear proteins consisting of 150 pmol of chemically modified sgRNA (5′-UCUCCAACUUCAUACUUCAG-3′, Synthego) and 50 pmol of Cas9 protein (St. Jude Protein Production Core) via nucleofection (Lonza, 4D-Nucleofector™ X-unit) with solution P3 and program EH-100 in a small (20-μl) cuvette according to the manufacturer protocol. A portion of the cells was collected at days 7, 14, and 21 post nucleofection, harvested for genomic DNA, and sequenced via targeted deep sequencing with gene-specific primers with partial Illumina adapter overhangs (NSD1.F, 5′-catacattgctttttcagaaggct-3′ and NSD1.R, 5′-acaccaattcattcacaaaatgttc-3′; overhangs not shown). Next-generation sequencing analysis of the edited cell pools was performed with CRIS.py (Connelly and Pruett-Miller, 2019). Protein knockdown was confirmed by immunoblot analysis. In the growth fitness assays, all indels were binned into in-frame, out-of-frame, or 0-bp indels.
Plasmids and lentivirus production for cell culture.
shRNA targeting human NSD1 (#1: 5′-CCGAGACGTCTCAGGTTAATC-3′, #2: 5′-TCCAGTGAGAACTCGTTAATA-3), KDM2A (#1: 5′-GCTTGAGAGATCCTCTGATTT-3′, #2: 5′-GCTTACTCCACCGGCTGATAA-3′, #3: 5′- TTCGCTATCCATTCTACTATG-3′) and control shRNAs (shCTRL:5’-CCTAAGGTTAAGTCGCCCTCG-3’) were cloned into the pLKO.1 or pLKO.5-puro vectors (Sigma). Inducible expression vectors for N-terminal V5-tagged SMARCB1 or GFP as control used TET3G (pLV[Exp]-Bsd-CMV>Tet3G (VB180621-1181ctz)) regulatory protein and TRE3G-driven expression vectors (pLV[Exp]-Puro-TRE3G>V5/hSMARCB1 (vectorbuilder 180622-1078xvz) and pLV[Exp]-Puro-TRE3G>EGFP (vectorbuilder 180621-1204vsc)).To generate lentiviral particles, 293-RTV (ATCC) cells were co-transfected with an envelope plasmid (pVSVG), packaging vector (psPAX2), and shRNA, TET3G or SMARCB1 expression vectors with PEI Pro (Polysciences Inc.). The medium containing the lentiviral particles was harvested 48 and 72 h after transfection and centrifuged at 20,000 r.p.m. for 2h at 4 °C. Virus-containing pellets were resuspended in PBS and the viral titer was determined. 48 h after transduction, transduced cells were selected in the presence of 1 μg/mL puromycin. pVSV-G was a gift from Akitsu Hotta (Gee et al., 2020)(Addgene plasmid # 138479 ; http://n2t.net/addgene:138479 ; RRID:Addgene_138479). psPAX2 was a gift from Didier Trono (Addgene plasmid # 12260 ; http://n2t.net/addgene:12260 ; RRID:Addgene_12260).
Generation of SMARCB1 inducible G401 cells
60K G401 cells were infected for 24hrs with TRE3G virus at an MOI of 5. Cells were washed with PBS, maintained for 2 days in absence of selection, followed by 7 days of Blasticidine treatment (2 μg/ml). G401-TET3G cells were then infected for 24hrs at an MOI of 5 with either Dox-inducible TRE3G driven GFP or V5-SMARCB1 viruses. After 24hrs, cells were washed with PBS, maintained in absence of selection for 48 hours followed by 7 days of Blasticidine (2 μg/ml) and Puromycin (1μg/ml) selection.
Immunoblotting.
Fractionated or whole-cell lysates were dissolved in 1x LDS sample buffer containing 2.5% β-mercaptoethanol and resolved by SDS–PAGE on 4%–12% Bis-Tris-polyacrylamide gels with 1x MOPS buffer. Proteins were transferred to a nitrocellulose membrane in 1x Tris-Glycine buffer overnight at 30V at 4°C. The transferred proteins were blocked in 5% nonfat milk in TBS containing 0.05% Tween-20, probed with primary antibodies overnight at 4°C, washed, and detected with horseradish peroxidase-conjugated anti-rabbit or anti-mouse secondary antibodies (GE Healthcare). Primary and secondary antibodies are described in Key Resource Table. Immunoblot bands for histone modifications and total H3 were quantified using ImageJ and the average ratio of each mark to total H3 is displayed.
Histone acid extraction, histone derivatization, and analysis of posttranslational modifications by nano-liquid chromatography/mass spectrometry
Acid extraction of histones was performed as described by Bhanu NV et al. (Bhanu et al., 2020). In brief, adherent cells were washed once in 1x ice-cold PBS, scraped, and pelleted. Cell pellets were lysed in nuclear isolation buffer (15 mM Tris-HCL, pH 7.5; 60 mM KCl; 15 mM NaCl; 5 mM MgCl2; 1 mM CaCl2; 250 mM sucrose; and 1× protease and phosphatase inhibitor cocktail (Thermo Fisher) containing 0.3% NP-40 on ice for 5 min. Nuclei were pelleted and resuspended in 0.2 M H2SO4, followed by 1.5 h rotation at 4°C. After centrifugation, supernatants were collected, and proteins were precipitated in 33% TCA for 2 h on ice, washed with acetone, air-dried, and resuspended in deionized water. Protein concentrations were measured via BCA assay (Pierce/Thermo).
Approximately 20 μg of acid-extracted histones were chemically derivatized to propionylate the unmodified lysine residues. The derivatized histones were digested in 1 μg trypsin/20 μg histones at 37°C overnight. The resulting peptides were desalted and eluted in 70% acetonitrile/0.1% formic acid. The peptide samples were injected directly for MS without liquid chromatography at a 1 μg/μL concentration with a TriVersa NanoMate (Advion) (Bhanu et al., 2020). The samples were programmed for analysis and acquired by contact closure in the Orbitrap Fusion Tribrid (Thermo Scientific). The samples were injected with voltage of 1.7 kV and gas pressure of 0.5 psi in the Nanomate. All scans were acquired in the Orbitrap at 240,000 resolution for full MS and at 120,000 resolution for MS/MS. MS2 scans were performed in HCD for accurate identification of approximately 44 isobaric peptides. The automatic gain control target for the tSIM-MSX scans was 106 ions (Bhanu et al., 2020).
Histone peptide quantification:
The in-house software EpiProfileLite was customized for direct infusion MS and is available on GitHub at https://github.com/zfyuan/EpiProfileLite (includes the user guide). The raw files were searched for intensities of histone peptides in MS scans and were used to calculate the relative ratios of all forms and PTMs for each histone peptide. Targeted preset MS/MS scans were analyzed to identify unique fragment ions in the MS/MS scans and were extracted to discriminate isobaric peptide intensities from the MS scans. In all, we quantified 29 peptide sequences with 45 PTMs (methylations, acetylations, and phosphorylations) for a total of 151 histone marks plus 16 unmodified histone peptides.
Chromatin fractionation.
Chromatin fractionation was performed using approximately 2 x 107 cells per fractionation. Cells were washed once using room temperature PBS, then scraped and collected in 1mL cold PBS with protease and phosphatase inhibitors. Cells were then resuspended in 1 mL Buffer A (20mM HEPES pH 7.4, 10mM KCl, 0.2mM EDTA, 1x protease and phosphatase inhibitor cocktail) and incubated on ice for 10 minutes. Then cells were incubated with 0.5% NP-40 for one minute and centrifuged. Supernatant was removed, and nuclei were resuspended in cell lysis buffer (20mM Phospho-Buffer pH7.5, 150mM NaCl, 1x protease and phosphatase inhibitor cocktail) and incubated for 20 minutes at 4°C with rotation. Nuclei were centrifuged and supernatant was kept as the nuclear soluble fraction. The pellet was suspended in 250ul 2xLDS+5% b-mercaptoethanol, heated for 10 minutes at 95°C, then passed through a 26g needle to homogenize the sample, and finally heated again for 10 minutes at 95°C. The nuclear soluble fraction was quantified using a BCA assay.
Total cell lysis
Approximately 2 X 106 cells were collected, washed once in PBS and resuspended in 500ul 2xLDS+5% B-mercaptoethanol. Samples were heated at 95°C for 10 minutes, passed through a 26G needle, then heated again for 10 minutes at 95°C.
Immunoprecipitation
Cellular fractionation was performed using approximately 2 x 107 cells per fractionation. Cells were washed once using room temperature PBS, then scraped and collected in 1mL cold PBS with protease and phosphatase inhibitors. Cells were resuspended in 1 mL Buffer A (20mM HEPES pH 7.4, 10mM KCl, 0.2mM EDTA, 1x protease and phosphatase inhibitor cocktail) and incubated on ice for 10 minutes, incubated with 5% NP-40 for one minute and centrifuged. Supernatant was removed, and nuclei were resuspended in cell lysis buffer (20mM Phospho-Buffer pH7.5, 150mM NaCl, 1x protease and phosphatase inhibitor cocktail). Approximately 1% nuclease (Thermo Fisher 88702) was added to each sample and incubated for 20 minutes in the cold room with rotation. Samples were incubated for 20 minutes at room temperature, pipetting every 5 minutes. Finally, they were centrifuged for 10 minutes at 4°C and the supernatant concentration was quantified using BCA. Approximately 250ug of protein was used for each IP, with 10% taken for input. Primary antibody was pre-bound to 50 μL Protein G Dynabeads (Thermo Fisher 10004D) for 10 minutes at room temperature. Lysates were combined with the antibody-bound beads and incubated for one hour at 4°C. Then, the beads were washed three times with Cell Lysis Buffer and resuspended in 100ul 2xLDS+5% B-mercaptoethanol. They were eluted at 95°C for 10 minutes, and then transferred to a new tube.
Chromatin immunoprecipitation.
Cross-linking ChIP in RT and Pfeiffer cell lines was performed using approximately 2 x 107 cells per immunoprecipitation and the Covaris True-ChIP kit per manufacturer’s instructions. Before fixation, the medium was aspirated, and cells were washed once with room temperature PBS. Cells were cross-linked directly on the plate using 1% paraformaldehyde for 10 min at room temperature with gentle shaking and glycine added to quench (final concentration 125 mM, incubated for 5 min at room temperature). Cells were washed once with cold PBS, scraped off the plates and pelleted. Nuclei were then prepared for sonication according to the Covaris True- ChIP kit manufacturer’s instructions. Chromatin extracts were sonicated for 8 min using a Covaris E220 focused ultrasonicator at a peak power of 140, a duty factor of 5 and 200 cycles per burst. For ChIP with reference exogenous genome (ChIP-Rx) for histone PTMs, after centrifugation soluble chromatin was spiked with Drosophila chromatin (Active Motif). The lysates were incubated with primary antibodies bound to 50 μL protein G Dynabeads (Invitrogen) and incubated 4h at 4 °C with 5% kept as input DNA. Magnetic beads were sequentially washed with low-salt buffer (150 mM NaCl, 0.1% SDS, 1% Triton X-100, 1 mM EDTA and 50 mM Tris-HCl), high-salt buffer (500 mM NaCl, 0.1% SDS, 1% Triton X-100, 1 mM EDTA and 50 mM Tris-HCl), LiCl buffer (150 mM LiCl, 0.5% sodium deoxycholate, 0.1% SDS, 1% Nonidet P-40, 1 mM EDTA and 50 mM Tris-HCl) and TE buffer (1 mM EDTA and 10 mM Tris-HCl). Beads were resuspended in elution buffer (1% SDS, 1mM NaHCO3) and incubated for 30 min at 65 °C, with shaking. The eluate was then reverse-cross-linked overnight at 65 °C with 5M NaCl. Next, the eluate was treated with RNaseA for 30 minutes at 37 °C, no shaking, and with Proteinase K (Invitrogen) for 2 h at 65 °C, shaking. The DNA was recovered using a Zymo ChIP DNA elution kit.
Cut & Run
Cut & Run was performed using the Epicypher Cutana Cut&Run Kit v2.0. Spike-in controls and buffers were prepared according to the manufacturer’s protocol. Streptavidin (SA) beads were resuspended in pre-wash buffer (20 mM HEPES pH 7.5, 150 mM NaCl) and aliquoted to incubate for 30 minutes at room temperature on a nutator with Cutana Spike-in Control dNucs (unmodified, H3K4me1, H3K4me2, H3K4me3). Cutana H3K4 MetStat Spike-in controls were washed with pre-wash buffer, combined, and resuspended in Antibody Buffer (20 mM HEPES pH 7.5, 0.5 mM Spermidine, 0.08% digitonin (Cell Signaling Technology), 2 mM EDTA). Concanavalin A (ConA) beads were resuspended and washed with Bead Activation Buffer (20 mM HEPES pH 7.9, 10 mM KCl, 1 mM CaCl2, 1 mM MnCl2). For each sample including positive and negative controls, 500,000 cells were harvested, washed with Wash Buffer (20 mM HEPES pH 7.5, 150 mM NaCl, 0.5 mM Spermidine), and combined with activated beads. Cells were bound to beads for 10 minutes at room temperature. Cells were resuspended in Antibody Buffer (20 mM HEPES pH 7.5, 150 mM NaCl, 0.5 mM Spermidine, 0.08% Digitonin, 2 mM EDTA). Cutana H3K4 MetStat Spike-in controls were added to H3K4me3 (positive control) and IgG (negative control) samples. Samples were incubated with 0.5 μg antibody overnight on the nutator at 4°C.
Cells were washed with Cell Permeabilization Buffer (20 mM HEPES pH 7.5, 150 mM NaCl, 0.5 mM Spermidine, 0.08% Digitonin), before incubating with pAG-MNase for 10 minutes at room temperature. Cells were again washed with Cell Permeabilization Buffer and incubated with 2 mM Calcium Chloride on the nutator for 2 hours at 4°C. Stop Buffer (340 mM NaCl, 20 mM EDTA, 4 mM EGTA, 50 μg/mL RNase A, 50 μg/mL Glycogen) and E. coli spike-in DNA was added and incubated for 10 minutes at 37°C. Supernatant was transferred and DNA was extracted using Phenol Chloroform. To the supernatant, SDS was added to a final concentration of 0.1% and proteinase K was added to incubate for 10 minutes at 65 °C. One volume of 4°C Phenol:Chloroform:isoamyl-alcohol was added, and supernatant was centrifuged at maximum speed for 5 minutes in phase-lock tubes. The aqueous layer was precipitated by adding 1/10 volume 3M sodium acetate, glycogen and 2 volumes 100% ethanol and by incubating for 2 hours at −80°C. Samples were centrifuged at maximum speed for 45 minutes at 4°C. DNA pellet was washed with 70% ethanol. Pellet was then resuspended in TE Buffer.
RNA sequencing
RNA was extracted with Trizol and the DirectZol Mini Prep Plus kit (Zymo Research, R2070). RNA was quantified by using the Quant-iT RiboGreen assay (Life Technologies) and quality checked with a 2100 Bioanalyzer RNA 6000 Nano Assay (Agilent) or the LabChip RNA Pico Sensitivity Assay (PerkinElmer) before library generation. The libraries were prepared from total RNA with the TruSeq Stranded mRNA Library Prep kit according to the manufacturer instructions (Illumina). The libraries were analyzed for insert size distribution with a 2100 BioAnalyzer High Sensitivity kit (Agilent Technologies) or Caliper LabChip GX DNA High Sensitivity Reagent kit (PerkinElmer). Libraries were quantified by using the Quant-iT PicoGreen dsDNA Assay (Life Technologies) or low-pass sequencing with a MiSeq Nano kit (Illumina). One hundred cycle paired-end sequencing was performed with a NovaSeq 6000 instrument (Illumina). The RNA-seq reads were mapped to the human GRCh37-lite reference genome with STAR (Dobin et al., 2013). Gene level counts were quantified by RSEM (v1.3.1) with GENCODE version 19 Ensembl 74 annotation. The read counts were further normalized with the trimmed mean of the M values (Robinson and Oshlack, 2010) method from the R package EdgeR. Differential gene expression analysis was performed with the lmFit and eBayes functions of the R package limma (v3.42.2 voom function) including a variable to model the preparation batch effect (Law et al., 2014). The genes were considered differentially expressed if their adjusted P values were lower than 0.05 and the FC was higher than 0. Heat maps were generated using z-score, row scaled, normalized expression for DEG. Transcription factor target gene enrichment analysis was performed on defined gene lists (up or down regulated genes) using Enrichr (v2.1) and the ChEA 2016 database. Gene ontology enrichment analysis on up or down regulated gene lists was performed using the GO Enrichment Analysis, by Panther (http://geneontology.org/). GSEAPreranked (v3.0) was used to evaluate the relationship between transcriptional changes in response to loss of NSD1 and upon EZH2 inhibition. To achieve this, custom gene sets (in the gmt format) were generated for up and down regulated genes in response to loss of NSD1. To generate the rnk file, representing expression changes upon EZH2 inhibition, all genes were ranked based on log2FoldChange.
ChIP-seq data analysis
Single-end reads of 50 bp were mapped to the human genome hg19 (GRCh37-lite) and the Drosophila melanogaster genome dm6 (Ensembl r97 BDGP6.22) hybrid by BWA (version 0.7.12-r1039, default parameter) (Li and Durbin). The duplicated reads were then marked by biobambam2 (version 2.0.87) (Tischler and Leonard, 2014). After the duplicated reads were marked, bam files were then split by either the human bam file or the Drosophila melanogaster spike-in bam file. For the spike-in bam files, we counted the nonduplicated reads with samtools. For the human bam files only, the nonduplicated reads were maintained by samtools (version 1.2, parameter -q 1 -F 1024) (Li et al.). We then followed the ENCODE guideline (Landt et al.) for quality control, as we have previously described (Yang et al.). After confirming that the quality of the reads were adequate, we extended the reads to fragment size (detected by SPP v1.1) (Kharchenko et al., 2008) and generated bigwig tracks for visualization. Expecting genome-wide equivalent changes in H3K27me3, we normalized the bigwig tracks to 100 kb uniquely mapped spike-in reads (i.e., we doubled the track heights for the human genes if the samples had 50 kb of uniquely mapped spike-in reads). For the other samples, we normalized them to 15 × 106 nonduplicated reads and further divided them by scaling factors estimated with ChIPseqSpikeInFree (Jin et al., 2019). For example, ChIPseqSpikeInFree estimated a scaling factor of 2 for 45 × 106 nonduplicated reads; therefore, the height for bigwig would be divided by 1.5 (45/1.5/2).
We called the peaks with MACS2 (version 2.1.1.20160309, parameters --nomodel --extsize fragment size) and called peaks with SICER (version 1.1; default parameters: false discovery rate [FDR] cutoff = 0.00001) for H3K27me3 and H3K36me2. To assure replicability, we first finalized the reproducible peaks for each group as only a retained peak if it was called with a stringent cutoff (FDR correct P < 0.05 in MACS2) in one sample and at least called with lower cutoff (FDR correct P < 0.5 in MACS2) in the other sample. The peaks were further merged between the groups, and we counted the reads extended to the fragment size from each of the samples with bedtools (version 2.24.0) (Quinlan and Hall, 2010). For H3K27me3 and H3K36me2, we retained the best resolution and broad domains by further merging the reproducible peaks called by MACS2 and SICER so that the SICER peaks overlapping the MACS2 peaks were excluded and the remaining SICER peaks were merged into the MACS2 peaks to create the reference peak set. Correlation plots confirmed that the libraries were reproducible between biological replicates. We then counted the extended reads to estimate the fragment size by SPP for each sample at these reference peaks. For H3K27me3, the spike-in read counts were provided as a scaling factor for normalization before the tests; otherwise TMM normalizations were used. After normalization, empirical Bayesian statistical tests were performed, along with linear fitting with the voom package (R 3.23, edgeR 3.12.1, limma 3.26.9) (Law et al.) to find the differential modified binding regions. We used a P value of 0.05 and FC >2 as cutoffs. Differentially modified regions were annotated with all genes living within 2-50kb Gencode annotation database (version 24lift37) (Harrow et al., 2012). Heatmaps and metaplots were generated with deeptools using either the scale-region of reference point depending on mark (version 2.5.7) (Ramírez et al., 2016) with the averaged, normalized coverage and custom region sets. Region sets include differentially modified regions upon loss of NSD1 and differentially expressed genes.
For the window-based analyses, we generated 10-kb window bed files with bedtools and extracted mean signals from bigwig files by bwtool (Pohl and Beato, 2014). We added the pseudo-value 0.01 to avoid a 0 value for calculating the log2 fold change. We plot the dots density calculated by smoothScatter function from R (geom_raster function from ggplot2) such that each dot is 10-kb window with x/y-axis as log2 fold change. For gene-based annotation, we used the Gencode annotation database (version 24lift37) (Harrow et al., 2012).
For density plots, FPKM values for each reference peak have been extracted from differential analysis, we further separated the peaks by their annotation groups and plot their kernel density for control and shNSD1.
CUT & RUN data analysis
Paired-end reads of 75 bp were mapped to the human genome hg19 (GRCh37-lite) and the Escherichia coli genome r46 (Ensembl r46 ASM584v2) hybrid by BWA (version 0.7.12-r1039, default parameter) (Li and Durbin, 2009). The duplicated reads were then marked by biobambam2 (version 2.0.87) (Tischler and Leonard, 2014). After the duplicated reads were marked, bam files were then split by either the human bam file or the Escherichia coli spike-in bam file. For the spike-in bam files, we counted the nonduplicated reads with samtools. For the human bam files only, the uniquely, properly paired reads were extracted by samtools, (version 1.2, parameter -q 1 -F 1804) (Li et al., 2009) sorted based on read name, (biobambam2) and bedtools was used to convert to BEDPE format. Fragment sizes <2000bp were extracted and the center 80bp of each fragment were used to generate normalized (to 10 million fragments) bigwig tracks for visualization. Peaks were called using MACS2 (-f BEDPE -g hs –keep-dup -q 0.05). The union of all peaks identified in G401 GFP sgCTRL and sgNSD1 were taken using bedtools merge. Normalized coverage was plotted across union peak centers and illustrated in a heat map using deeptools.
Data integration analysis
Averaged bigwigs were generated for ChIP or RNA-Seq by (1) combining replicates UCSC-bigWigMerge, (2) sorting combined bedgraphs (LC_COLLATE=C sort -k1,1 -k2,2n), (3) averaging the 4th bedgraph column based on number of input replicates using awk, and (4) converting to bigwig format using UCSC bedGraphToBigWig and hg19 chromosome sizes. Venn Diagrams were generated in R (v3.6.1) using VennDiagram (v1.6.20). Statistical significance and fold enrichment of overlap were computed using SuperExactTest (v1.0.7) setting n = 20,687 for the number of total protein coding genes. A custom gene set was created based on genes overlapping 10kb windows of H3K36me2 loss (log2FC < −0.52, the first quartile value) (shNSD1 vs. shCTRL). Pre-ranked GSEA was used to evaluate the relationship of these genes and expression changes in response to EZH2 inhibition (DEG EZH2i vs. DMSO). SMARCB1 target genes were defined as (1) significantly up regulated upon SMARCB1 add back and (2) contained a SMARCC1 (GSE90631) binding peak (bedtools intersect). To identify differentially expressed genes that live nearby differentially modified regions, candidate gene coordinates (biomaRt v2.45.7) were extended (bedtools v2.29.2 slop -b 20000) and intersected (bedtools intersect) with previously defined differentially modified regions of H3K36me2. Prior to evaluating if distributions (log2FC or FPKM) significantly differ, normality was evaluated using the Shapiro-Wilk test. When distributions significantly differed from the normal distribution, the Wilcoxon rank sum test was applied. IGV (Robinson et al., 2011) (v2.6.2) was used to visualize RNA-Seq and ChIP-Seq average normalized coverage at candidate loci.
Analysis of co-mutation and exclusivity in focused cohort
For our initial analysis we used cBioportal and queried almost all existing datasets consisting of 63,676 samples from 60,905 patients. We looked for mutations in NSD paralogs NSD1, NSD2, and NSD3, SWI/SNF subunits frequently mutated in cancer (Mittal and Roberts, 2020) and known tumor suppressors TP53, CDKN2A and RB1 as “negative” controls. To increase the strength of our analysis we excluded alterations (mutations, structural variants and copy number) of unknown significance. To account for the heterogeneous nature of the large number of different studies hosted by cBioportal, as well as the challenges in determining pathogenicity of somatic variants, we next performed a focused analysis by using the clinical sequencing data generated by MSK (Zehir et al., 2017), which included 10945 samples (last accessed July 15, 2021) profiled by the MSK-IMPACT platform and are expected to have minimal analytical bias as well as large enough cohort size. 9418 variants were downloaded from cBioportal (July 15, 2021; Tables S11 and S12). We first analyzed putative pathogenicity of these variants by using PeCanPIE/MedalCeremony (Edmonson et al., 2019), and compared the pathogenicity classification with the “Driver” status provided by cBioportal. We found that gene ARID2 has substantial under calling of pathogenicity by cBioportal (15 variants called as driver mutations by CBioportal versus 214 additional variants called as pathogenic/likely pathogenic by PeCanPIE/MedalCeremony). Together, we rescued 348 pathogenic and likely pathogenic SNV/Indel variants and also identified 9 potential false positives (homopolymer Indels) (Table S13). Upon removing duplicates (a gene may have more than 1 mutation in a given sample (ARID1A in P-0000069-T01-IM3)), these data (Table S13) were next used to detect co-mutations and mutual exclusivity (Table S14) by using a similar procedure as cBioportal. Briefly, for each gene pair A and B, we counted samples with mutation status of 1) Both mutated, 2) A mutated but not B, 3) B mutated but not A, and 4) Neither mutated. These four numbers were then subject to two-sided Fisher’s Exact Test (R version 3.4.2). The P values were then corrected for multiple testing by using package “qvalue” (R version 3.4.2). This focused analysis further supports the findings from our initial analysis.
Supplementary Material
Supplemental Figure S1. NSD1 scores as a top target in a CRISPR Cas9 GSK126 resistance screen in RT cell lines.
(A) Schematic overview of the antagonistic relationship between SWI/SNF and EZH2, the action of EZH2 inhibitors and the expected targets from the CRISPR Cas9 GSK126 resistance screen performed in SMARCB1-deficient rhabdoid cell lines.
(B) Venn diagram depicting the number of perturbed genes associated with enriched sgRNAs in G401 and G402 cells.
(C) sgRNA sequencing reads for all genes encoding H3K36me2 writers/erasers in G401 cells at the beginning (d0) and end of the screen (d20).
(D and E) Log2 fold changes of all sgRNAs targeting genes encoding H3K36me2 writers/erasers in the G401 (D) and G402 (E) CRISPR screens.
(F) Immunoblot analysis of the G401 NSD1, NSD2 and NSD3 knockout pools.
(G) Next generation sequencing analysis of the NSD1, NSD2 and NSD3 loci to assess knockout efficiency. Genes were targeted with sgRNA in G401 cells and genomic DNA sequenced after selection and expansion of the targeted pools (see methods for details). All indels were binned into in-frame, out-of-frame, or 0-bp.
(H) Cell proliferation assay of G401 NSD1/2/3 and control knockout pools
(I) NSD1 knockdown/knockout levels of cell lines used in Figures 1C-F.
Supplemental Figure S2. NSD1 loss does not affect H3K27me3 removal by EZH2 inhibition
(A and B) Immunoblot analysis of control and NSD1 depleted G401 cells treated with various concentration of Tazemetostat and analyzed at days 3 (A) and 7 (B). Viability is calculated for each time point using cell count.
For viability analysis, p values were calculated with Fisher’s test. ***: p<0.001, ****: p<0.0001.
Supplemental Figure S3. Mass spec analysis of histone modification changes upon NSD1 knockdown and EZH2 inhibition.
(A-B) Volcano plots of all histone modifications detected in G401 control (A) or NSD1 depleted (B) cells treated with DMSO or 1uM GSK126 for 3 days. The histone modifications that were significantly up or downregulated are marked in the plots with red and green color respectively.
(C-D) Relative abundance of H3K27me2, H3K27me3 and H3K36me2 in G401 H3.1/2 (C) and H3.3 (D) histones after treatment with DMSO or 1uM GSK126 for 3 days
(E-F) Volcano plot of all histone modifications detected in Pfeiffer control (E) or NSD1 depleted (F) cells treated with DMSO or 1uM GSK126 for 3 days. The histone modifications that were significantly up or downregulated are marked in the plots with red and green color respectively.
(G-H) relative abundance of H3K27me2, H3K27me3 and H3K36me2 in Pfeiffer H3.1/2 (G) and H3.3 (H) histones after treatment with DMSO or 1uM GSK126 for 3 days.
For volcano plots, values are considered significant if they have a p<0.05 (Student’s t-test) and a log2FC>1. For bar graphs, p values are calculated using Fisher’s test. *:p<0.05, **:p<0.01, ***:p<0.001.
Supplemental Figure S4. Differential enrichment analysis of H3K27me3 and H3K36me2 upon NSD1 knockdown and EZH2 inhibition.
(A-B) Scatter plots of regions differentially enriched for H3K27me3 as obtained by ChIP-seq, in control (A) or NSD1 depleted (B) G401 cells treated with 1uM GSK126 for 3 days.
(C) Scatter plot of regions differentially enriched for H3K36me2 as obtained by ChIP-seq, in control G401 cells treated with 1uM GSK126 for 3 days.
Significance computed using empirical Bayesian statistical tests and linear fitting.
Supplemental Figure S5. H3K27ac gain upon EZH2 inhibition shows partial dependence on NSD1.
(A) Scatter plot of the differentially enriched regions for H3K27ac upon NSD1 knockdown in G401 cells, obtained by ChIP-seq. Significance computed using empirical Bayesian statistical tests and linear fitting.
(B-C) Heatmaps of enhancers (B) and promoters (C) showing differential H3K27ac enrichment between control and NSD1 depleted cells ranked by gained, shared, and lost H3K27ac signal in control cells treated with 1uM GSK126 for 3 days.
(D) Venn diagram of genes gaining H3K27ac upon EZH2 inhibition, overlapping with genes activated by SMARCB1 in G401 cells. Significance computed using standard Fisher’s exact Test.
(E) Box plot of H3K27ac log2FC for the same sites that gain H3K27ac in shCTRL and shNSD1 cells with EZH2 inhibition for the same genomic regions. p value was calculated using Wilcoxon test.
Supplemental Figure S6. NSD1 cooperates with SWI/SNF to activate transcription.
(A) Intersection of gene sets displaying p-values (computed using standard Fisher’s exact Test for pairwise overlaps and “Exact Test of Multi-set Intersections” for multi-list overlaps) and enrichment statistics (see text for details) for Venn diagram in Figure 5A.
(B) Region-scaled heatmap visualization of H3K36me2 normalized ChIP-seq coverage, rank-ordered based on changes in control vs. NSD1 depleted G401 cells without SMARCB1. Significance computed using empirical Bayesian statistical tests and linear fitting.
(C) Peak centered heatmap visualization of normalized SUZ12 ChIP-Seq coverage, rank-ordered based on changes in H3K36me2 sites lost in sgNSD1 vs sgCTRL, as shown in (B).
(D) Peak centered heatmap visualization of normalized RING1B CUT&RUN coverage, rank-ordered based on changes in H3K36me2 sites lost in sgNSD1 vs sgCTRL, as shown in (B).
(E) Significantly enriched biological processes based on the 977 genes upregulated by SMARCB1 and bound by SMARCC1 (FDR < 0.05 and log2FC > 1).
(F) Significantly enriched biological processes based on the 730 genes downregulated by loss of NSD1.
(G-J) Metaplots of normalized, average coverage for H3K36me2 (G), H3K27me3 (H), SUZ12 (I) and RING1B (J) at +/****−20kb of the 289 genes activated by NSD1 and SMARCB1.
(K) Genome browser tracks depicting H3K27me3, H3K36me2, H3K27ac and RNA-seq reads upon SMARCB1 addback in control and NSD1 KO G401 RT cells.
Supplemental Figure S7. NSD1 physically interacts with SWI/SNF and PRC1/2 complex members.
(A and B) Co-immunoprecipitation analysis of SWI/SNF core subunits SMARCC1 (A) and SMARCA4 (B) in G401 cells upon SMARCB1 addback.
(C and D) Co-immunoprecipitation analysis of PRC1/2 subunits EZH2-PRC2 (C) and RING1B-PRC1 (D) in G401 cells treated with DMSO or 1uM GSK126 for 3 days.
(E) Co-immunoprecipitation analysis of SWI/SNF ATPase SMARCA4 in G401 cells treated with DMSO or 1uM GSK126 for 3 days.
(F) Schematic representation of the co-IP results.
Supplemental Figure S8. NSD1 and SWI/SNF mutations co-exist in cancer patients.
(A) Analysis of co-mutation and exclusivity in focused cohort of cancer patients (see methods for details). Fisher’s Exact Test (R version 3.4.2) was used for mutational analysis and p values were then corrected for multiple testing by using package “qvalue” (R version 3.4.2).
(B) Pathogenic mutations in NSD1 gene used for the cBioportal analysis.
(C and D) Venn diagram of cancer samples with NSD1 and ARID1A (C) or SMARCA4 (D) co-occurring mutations in focused analysis. Fisher’s Exact Test (R version 3.4.2) was used for mutational analysis and p values were then corrected for multiple testing by using package “qvalue” (R version 3.4.2).
Supplemental Table S1. H3K36me2 and H3K27me3 Log2FC in shNSD1 vs. shNSD1
Supplemental Table S2. Histone post translational modifications in G401 and Pfeiffer cells with EZH2 inhibition and NSD1 knockdown
Supplemental Table S3. Differentially expressed genes in shCTRL EZH2i vs. DMSO
Supplemental Table S4. Gene Ontology analysis of genes upregulated in shCTRL + EZH2i vs. DMSO
Supplemental Table S5. Differentially expressed genes in shNSD1 vs. shCTRL G401
Supplemental Table S6. Differentially expressed genes in G401 SMARCB1_sgCTRL vs. GFP_sgCTRL
Supplemental Table S7. Differentially expressed genes in G401 SMARCB1_sgNSD1 vs. SMARCB1_sgCTRL
Supplemental Table S8. 289 genes downregulated in G401 SMARCB1_sgNSD1 vs. SMARCB1_sgCTRL
Supplemental Table S9. Gene Ontology analysis of the 289 genes activated by SWI/SNF and NSD1
Supplemental Table S10. Co-occurring and mutual exclusive mutations in cBioportal analysis
Supplemental Table S11. Raw mutation data (columns A-P) downloaded from CBioportal (July 15, 2021) for study Nat Med 2017 (PMID 28481359)
Supplemental Table S12. Mutated gene list at sample level. Data derived from Table S13 by consolidating multiple datasets
Supplemental Table S13. Analysis of pathogenicity of mutations analyzed in Tables S13, S14 for study Nat Med 2017 (PMID 28481359)
Supplemental Table S14. Co-occurring and mutual exclusive mutations in focused analysis
Figure 7. Proposed model of NSD1 action.
(A) In a proliferating progenitor cell, the activation of differentiation related genes results in cessation of proliferation.
(B) The loss of SMARCB1 in RT cells results in the replacement of SWI/SNF complexes with Polycomb silencing complexes at differentiation driving genes, thus preventing differentiation.
(C) Treatment of RT cells with EZH2 inhibitors leads to removal of H3K27me3 modifications and results in NSD1 writing H3K36me2 activating modifications in its place, activating differentiation associated genes and causing cell arrest/death.
(D) The loss of NSD1 impairs the gain of H3K26me2 and blocks the cell death otherwise caused by EZH2 inhibition.
Figure created with BioRender.com
ACKNOWLEDGEMENTS
We thank all members of the Roberts lab for insightful discussions. This work was supported by the National Cancer Institute (NCI) R01 CA113794 and R01 CA172152 to C.W.M.R, R01 CA196539 to B.A.G., CURE AT/RT Now to C.W.M.R, Garrett B. Smith Foundation to C.W.M.R, St. Jude Children's Research Hospital Collaborative Research Consortium on Chromatin Regulation in Pediatric Cancer to B.A.G. and C.W.M.R. SR-J is funded by the St. Jude Graduate School of Biomedical Sciences and NCI Ruth L. Kirschstein NRSA for Individual Predoctoral Fellows (F31 CA261150). We thank Protein Production Core for Cas9 and Hartwell Center Core for library preparation and sequencing. We also thank St Jude Center for Applied Bioinformatics (CAB) for help with alignment of RNA-seq and ChIP-seq reads. St Jude cores are supported via Cancer Center support grant (NCI CCSG 2 P30 CA021765) and ALSAC of St. Jude Children’s Research Hospital.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Allis CD, and Jenuwein T (2016). The molecular hallmarks of epigenetic control. Nature Reviews Genetics 17, 487–500. [DOI] [PubMed] [Google Scholar]
- Alver BH, Kim KH, Lu P, Wang X, Manchester HE, Wang W, Haswell JR, Park PJ, and Roberts CW (2017). The SWI/SNF chromatin remodelling complex is required for maintenance of lineage specific enhancers. Nat Commun 8, 14648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett RL, Swaroop A, Troche C, and Licht JD (2017). The Role of Nuclear Receptor–Binding SET Domain Family Histone Lysine Methyltransferases in Cancer. Cold Spring Harbor Perspectives in Medicine 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhanu NV, Sidoli S, and Garcia BA (2020). A Workflow for Ultra-rapid Analysis of Histone Post-translational Modifications with Direct-injection Mass Spectrometry. Bio Protoc 10, e3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackledge NP, Zhou JC, Tolstorukov MY, Farcas AM, Park PJ, and Klose RJ (2010). CpG Islands Recruit a Histone H3 Lysine 36 Demethylase. Molecular Cell 38, 179–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken AP, Brien GL, and Verrijzer CP (2019). Dangerous liaisons: interplay between SWI/SNF, NuRD, and Polycomb in chromatin regulation and cancer. Genes Dev 33, 936–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. (2012). The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2, 401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun HE, Lim EL, Heravi-Moussavi A, Saberi S, Mungall KL, Bilenky M, Carles A, Tse K, Shlafman I, Zhu K, et al. (2016). Genome-Wide Profiles of Extra-cranial Malignant Rhabdoid Tumors Reveal Heterogeneity and Dysregulated Developmental Pathways. Cancer Cell 29, 394–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly JP, and Pruett-Miller SM (2019). CRIS.py: A Versatile and High-throughput Analysis Program for CRISPR-based Genome Editing. Sci Rep 9, 4194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deevy O, and Bracken AP (2019). PRC2 functions in development and congenital disorders. Development 146, dev181354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmonson MN, Patel AN, Hedges DJ, Wang Z, Rampersaud E, Kesserwan CA, Zhou X, Liu Y, Newman S, Rusch MC, et al. (2019). Pediatric Cancer Variant Pathogenicity Information Exchange (PeCanPIE): a cloud-based platform for curating and classifying germline variants. Genome Res 29, 1555–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erkek S, Johann PD, Finetti MA, Drosos Y, Chou HC, Zapatka M, Sturm D, Jones DTW, Korshunov A, Rhyzova M, et al. (2019). Comprehensive Analysis of Chromatin States in Atypical Teratoid/Rhabdoid Tumor Identifies Diverging Roles for SWI/SNF and Polycomb in Gene Regulation. Cancer Cell 35, 95–110 e118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y, Tang Y, Zhang Y, Pan Y, Jia J, Sun Z, Zeng W, Chen J, Yuan Y, and Fang D (2021). The H3K36me2 methyltransferase NSD1 modulates H3K27ac at active enhancers to safeguard gene expression. Nucleic Acids Research. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farhangdoost N, Horth C, Hu B, Bareke E, Chen X, Li Y, Coradin M, Garcia BA, Lu C, and Majewski J (2021). Chromatin dysregulation associated with NSD1 mutation in head and neck squamous cell carcinoma. Cell Reports 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. (2013). Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 6, pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gee P, Lung MSY, Okuzaki Y, Sasakawa N, Iguchi T, Makita Y, Hozumi H, Miura Y, Yang LF, Iwasaki M, et al. (2020). Extracellular nanovesicles for packaging of CRISPR-Cas9 protein and sgRNA to induce therapeutic exon skipping. Nat Commun 11, 1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, et al. (2012). GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 22, 1760–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Kallin EM, Tsukada Y.-i., and Zhang Y (2008). The H3K36 demethylase Jhdm1b/Kdm2b regulates cell proliferation and senescence through p15Ink4b. Nature Structural & Molecular Biology 15, 1169–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia L, Carlo MI, Khan H, Nanjangud GJ, Rana S, Cimera R, Zhang Y, Hakimi AA, Verma AK, Al-Ahmadie HA, et al. (2019). Distinctive mechanisms underlie the loss of SMARCB1 protein expression in renal medullary carcinoma: morphologic and molecular analysis of 20 cases. Mod Pathol 32, 1329–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Kasper LH, Larson JD, Wu G, Baker SJ, Zhang J, and Fan Y (2019). ChIPseqSpikeInFree: a ChIP-seq normalization approach to reveal global changes in histone modifications without spike-in. Bioinformatics 36, 1270–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johann PD, Erkek S, Zapatka M, Kerl K, Buchhalter I, Hovestadt V, Jones DTW, Sturm D, Hermann C, Segura Wang M, et al. (2016). Atypical Teratoid/Rhabdoid Tumors Are Comprised of Three Epigenetic Subgroups with Distinct Enhancer Landscapes. Cancer Cell 29, 379–393. [DOI] [PubMed] [Google Scholar]
- Kadoch C, Williams RT, Calarco JP, Miller EL, Weber CM, Braun SM, Pulice JL, Chory EJ, and Crabtree GR (2017). Dynamics of BAF-Polycomb complex opposition on heterochromatin in normal and oncogenic states. Nat Genet 49, 213–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharchenko PV, Tolstorukov MY, and Park PJ (2008). Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat Biotech 26, 1351–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kia SK, Gorski MM, Giannakopoulos S, and Verrijzer CP (2008). SWI/SNF Mediates Polycomb Eviction and Epigenetic Reprogramming of the <em>INK4b-ARF-INK4a</em> Locus. Molecular and Cellular Biology 28, 3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KH, Kim W, Howard TP, Vazquez F, Tsherniak A, Wu JN, Wang W, Haswell JR, Walensky LD, Hahn WC, et al. (2015). SWI/SNF-mutant cancers depend on catalytic and non-catalytic activity of EZH2. Nat Med 21, 1491–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KH, and Roberts CW (2016). Targeting EZH2 in cancer. Nat Med 22, 128–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloet SL, Makowski MM, Baymaz HI, van Voorthuijsen L, Karemaker ID, Santanach A, Jansen PWTC, Di Croce L, and Vermeulen M (2016). The dynamic interactome and genomic targets of Polycomb complexes during stem-cell differentiation. Nature Structural & Molecular Biology 23, 682–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutson SK, Kawano S, Minoshima Y, Warholic NM, Huang KC, Xiao Y, Kadowaki T, Uesugi M, Kuznetsov G, Kumar N, et al. (2014). Selective inhibition of EZH2 by EPZ-6438 leads to potent antitumor activity in EZH2-mutant non-Hodgkin lymphoma. Mol Cancer Ther 13, 842–854. [DOI] [PubMed] [Google Scholar]
- Knutson SK, Warholic NM, Wigle TJ, Klaus CR, Allain CJ, Raimondi A, Porter Scott M, Chesworth R, Moyer MP, Copeland RA, et al. (2013). Durable tumor regression in genetically altered malignant rhabdoid tumors by inhibition of methyltransferase EZH2. Proceedings of the National Academy of Sciences 110, 7922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutson SK, Wigle TJ, Warholic NM, Sneeringer CJ, Allain CJ, Klaus CR, Sacks JD, Raimondi A, Majer CR, Song J, et al. (2012). A selective inhibitor of EZH2 blocks H3K27 methylation and kills mutant lymphoma cells. Nat Chem Biol 8, 890–896. [DOI] [PubMed] [Google Scholar]
- Krug B, Harutyunyan AS, Deshmukh S, and Jabado N (2021). Polycomb repressive complex 2 in the driver's seat of childhood and young adult brain tumours. Trends Cell Biol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landt SG, Marinov GK, Kundaje A, Kheradpour P, Pauli F, Batzoglou S, Bernstein BE, Bickel P, Brown JB, Cayting P, et al. (2012). ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 22, 1813–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law CW, Chen Y, Shi W, and Smyth GK (2014). voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biology 15, R29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RS, Stewart C, Carter SL, Ambrogio L, Cibulskis K, Sougnez C, Lawrence MS, Auclair D, Mora J, Golub TR, et al. (2012). A remarkably simple genome underlies highly malignant pediatric rhabdoid cancers. J Clin Invest 122, 2983–2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonards K, Almosailleakh M, Tauchmann S, Bagger FO, Thirant C, Juge S, Bock T, Méreau H, Bezerra MF, Tzankov A, et al. (2020). Nuclear interacting SET domain protein 1 inactivation impairs GATA1-regulated erythroid differentiation and causes erythroleukemia. Nature Communications 11, 2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, and Durbin R (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics (Oxford, England) 25, 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, and Durbin R (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucio-Eterovic AK, Singh MM, Gardner JE, Veerappan CS, Rice JC, and Carpenter PB (2010). Role for the nuclear receptor-binding SET domain protein 1 (NSD1) methyltransferase in coordinating lysine 36 methylation at histone 3 with RNA polymerase II function. Proceedings of the National Academy of Sciences, 201002653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majer CR, Jin L, Scott MP, Knutson SK, Kuntz KW, Keilhack H, Smith JJ, Moyer MP, Richon VM, Copeland RA, et al. (2012). A687V EZH2 is a gain-of-function mutation found in lymphoma patients. FEBS Letters 586, 3448–3451. [DOI] [PubMed] [Google Scholar]
- Margueron R, and Reinberg D (2011). The Polycomb complex PRC2 and its mark in life. Nature 469, 343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur R, Alver BH, San Roman AK, Wilson BG, Wang X, Agoston AT, Park PJ, Shivdasani RA, and Roberts CW (2017). ARID1A loss impairs enhancer-mediated gene regulation and drives colon cancer in mice. Nat Genet 49, 296–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCabe MT, Graves AP, Ganji G, Diaz E, Halsey WS, Jiang Y, Smitheman KN, Ott HM, Pappalardi MB, Allen KE, et al. (2012a). Mutation of A677 in histone methyltransferase EZH2 in human B-cell lymphoma promotes hypertrimethylation of histone H3 on lysine 27 (H3K27). Proceedings of the National Academy of Sciences 109, 2989–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCabe MT, Ott HM, Ganji G, Korenchuk S, Thompson C, Van Aller GS, Liu Y, Graves AP, Della Pietra A 3rd, Diaz E, et al. (2012b). EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature 492, 108–112. [DOI] [PubMed] [Google Scholar]
- Mittal P, and Roberts CWM (2020). The SWI/SNF complex in cancer - biology, biomarkers and therapy. Nat Rev Clin Oncol 17, 435–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama RT, Pulice JL, Valencia AM, McBride MJ, McKenzie ZM, Gillespie MA, Ku WL, Teng M, Cui K, Williams RT, et al. (2017). SMARCB1 is required for widespread BAF complex-mediated activation of enhancers and bivalent promoters. Nat Genet 49, 1613–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliviero G, Brien GL, Waston A, Streubel G, Jerman E, Andrews D, Doyle B, Munawar N, Wynne K, Crean J, et al. (2016). Dynamic Protein Interactions of the Polycomb Repressive Complex 2 during Differentiation of Pluripotent Cells. Molecular & Cellular Proteomics 15, 3450–3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohl A, and Beato M (2014). bwtool: a tool for bigWig files. Bioinformatics 30, 1618–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, and Hall IM (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics (Oxford, England) 26, 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan KN, Chen X, Weinberg DN, Chen H, Majewski J, Allis CD, and Lu C (2021). Depletion of H3K36me2 recapitulates epigenomic and phenotypic changes induced by the H3.3K36M oncohistone mutation. Proceedings of the National Academy of Sciences 118, e2021795118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F, Ryan DP, Grüning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dündar F, and Manke T (2016). deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Research 44, W160–W165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richart L, and Margueron R (2020). Drugging histone methyltransferases in cancer. Curr Opin Chem Biol 56, 51–62. [DOI] [PubMed] [Google Scholar]
- Richer W, Masliah-Planchon J, Clement N, Jimenez I, Maillot L, Gentien D, Albaud B, Chemlali W, Galant C, Larousserie F, et al. (2017). Embryonic signature distinguishes pediatric and adult rhabdoid tumors from other SMARCB1-deficient cancers. Oncotarget 8, 34245–34257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, and Mesirov JP (2011). Integrative genomics viewer. Nat Biotech 29, 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, and Oshlack A (2010). A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 11, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothbart SB, and Baylin SB (2020). Epigenetic Therapy for Epithelioid Sarcoma. Cell 181, 211. [DOI] [PubMed] [Google Scholar]
- Sneeringer CJ, Scott MP, Kuntz KW, Knutson SK, Pollock RM, Richon VM, and Copeland RA (2010). Coordinated activities of wild-type plus mutant EZH2 drive tumor-associated hypertrimethylation of lysine 27 on histone H3 (H3K27) in human B-cell lymphomas. Proceedings of the National Academy of Sciences 107, 20980–20985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanton BZ, Hodges C, Calarco JP, Braun SM, Ku WL, Kadoch C, Zhao K, and Crabtree GR (2017). Smarca4 ATPase mutations disrupt direct eviction of PRC1 from chromatin. Nat Genet 49, 282–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streubel G, Watson A, Jammula SG, Scelfo A, Fitzpatrick DJ, Oliviero G, McCole R, Conway E, Glancy E, Negri GL, et al. (2018). The H3K36me2 Methyltransferase Nsd1 Demarcates PRC2-Mediated H3K27me2 and H3K27me3 Domains in Embryonic Stem Cells. Molecular Cell 70, 371–379.e375. [DOI] [PubMed] [Google Scholar]
- Tatton-Brown K, Hanks S, Ruark E, Zachariou A, Duarte Sdel V, Ramsay E, Snape K, Murray A, Perdeaux ER, Seal S, et al. (2011). Germline mutations in the oncogene EZH2 cause Weaver syndrome and increased human height. Oncotarget 2, 1127–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischler G, and Leonard S (2014). biobambam: tools for read pair collation based algorithms on BAM files. Source Code for Biology and Medicine 9, 13. [Google Scholar]
- Upadhyaya S, Campagne O, Robinson GW, Onar-Thomas A, Orr B, Billups CA, Tatevossian RG, Broniscer A, Kilburn LB, Baxter PA, et al. (2020). Phase II study of alisertib as a single agent in recurrent or progressive atypical teratoid rhabdoid tumors. Journal of Clinical Oncology 38, 10542–10542. [Google Scholar]
- Versteege I, Sevenet N, Lange J, Rousseau-Merck MF, Ambros P, Handgretinger R, Aurias A, and Delattre O (1998). Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature 394, 203–206. [DOI] [PubMed] [Google Scholar]
- Wang X, Lee RS, Alver BH, Haswell JR, Wang S, Mieczkowski J, Drier Y, Gillespie SM, Archer TC, Wu JN, et al. (2017). SMARCB1-mediated SWI/SNF complex function is essential for enhancer regulation. Nat Genet 49, 289–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg DN, Papillon-Cavanagh S, Chen H, Yue Y, Chen X, Rajagopalan KN, Horth C, McGuire JT, Xu X, Nikbakht H, et al. (2019). The histone mark H3K36me2 recruits DNMT3A and shapes the intergenic DNA methylation landscape. Nature 573, 281–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissmiller AM, Wang J, Lorey SL, Howard GC, Martinez E, Liu Q, and Tansey WP (2019). Inhibition of MYC by the SMARCB1 tumor suppressor. Nature Communications 10, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegand KC, Shah SP, Al-Agha OM, Zhao Y, Tse K, Zeng T, Senz J, McConechy MK, Anglesio MS, Kalloger SE, et al. (2010). ARID1A Mutations in Endometriosis-Associated Ovarian Carcinomas. New England Journal of Medicine 363, 1532–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson BG, and Roberts CW (2011). SWI/SNF nucleosome remodellers and cancer. Nat Rev Cancer 11, 481–492. [DOI] [PubMed] [Google Scholar]
- Wilson BG, Wang X, Shen X, McKenna ES, Lemieux ME, Cho Y-J, Koellhoffer EC, Pomeroy SL, Orkin SH, and Roberts CWM (2010). Epigenetic Antagonism between Polycomb and SWI/SNF Complexes during Oncogenic Transformation. Cancer Cell 18, 316–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Xu B, Mulvey B, Evans M, Jordan S, Wang Y-D, Pagala V, Peng J, Fan Y, Patel A, et al. (2019). Differentiation of human pluripotent stem cells into neurons or cortical organoids requires transcriptional co-regulation by UTX and 53BP1 cofi. Nat Neurosci 22, 362–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu JR, Lee CH, Oksuz O, Stafford JM, and Reinberg D (2019). PRC2 is high maintenance. Genes Dev 33, 903–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehir A, Benayed R, Shah RH, Syed A, Middha S, Kim HR, Srinivasan P, Gao J, Chakravarty D, Devlin SM, et al. (2017). Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med 23, 703–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure S1. NSD1 scores as a top target in a CRISPR Cas9 GSK126 resistance screen in RT cell lines.
(A) Schematic overview of the antagonistic relationship between SWI/SNF and EZH2, the action of EZH2 inhibitors and the expected targets from the CRISPR Cas9 GSK126 resistance screen performed in SMARCB1-deficient rhabdoid cell lines.
(B) Venn diagram depicting the number of perturbed genes associated with enriched sgRNAs in G401 and G402 cells.
(C) sgRNA sequencing reads for all genes encoding H3K36me2 writers/erasers in G401 cells at the beginning (d0) and end of the screen (d20).
(D and E) Log2 fold changes of all sgRNAs targeting genes encoding H3K36me2 writers/erasers in the G401 (D) and G402 (E) CRISPR screens.
(F) Immunoblot analysis of the G401 NSD1, NSD2 and NSD3 knockout pools.
(G) Next generation sequencing analysis of the NSD1, NSD2 and NSD3 loci to assess knockout efficiency. Genes were targeted with sgRNA in G401 cells and genomic DNA sequenced after selection and expansion of the targeted pools (see methods for details). All indels were binned into in-frame, out-of-frame, or 0-bp.
(H) Cell proliferation assay of G401 NSD1/2/3 and control knockout pools
(I) NSD1 knockdown/knockout levels of cell lines used in Figures 1C-F.
Supplemental Figure S2. NSD1 loss does not affect H3K27me3 removal by EZH2 inhibition
(A and B) Immunoblot analysis of control and NSD1 depleted G401 cells treated with various concentration of Tazemetostat and analyzed at days 3 (A) and 7 (B). Viability is calculated for each time point using cell count.
For viability analysis, p values were calculated with Fisher’s test. ***: p<0.001, ****: p<0.0001.
Supplemental Figure S3. Mass spec analysis of histone modification changes upon NSD1 knockdown and EZH2 inhibition.
(A-B) Volcano plots of all histone modifications detected in G401 control (A) or NSD1 depleted (B) cells treated with DMSO or 1uM GSK126 for 3 days. The histone modifications that were significantly up or downregulated are marked in the plots with red and green color respectively.
(C-D) Relative abundance of H3K27me2, H3K27me3 and H3K36me2 in G401 H3.1/2 (C) and H3.3 (D) histones after treatment with DMSO or 1uM GSK126 for 3 days
(E-F) Volcano plot of all histone modifications detected in Pfeiffer control (E) or NSD1 depleted (F) cells treated with DMSO or 1uM GSK126 for 3 days. The histone modifications that were significantly up or downregulated are marked in the plots with red and green color respectively.
(G-H) relative abundance of H3K27me2, H3K27me3 and H3K36me2 in Pfeiffer H3.1/2 (G) and H3.3 (H) histones after treatment with DMSO or 1uM GSK126 for 3 days.
For volcano plots, values are considered significant if they have a p<0.05 (Student’s t-test) and a log2FC>1. For bar graphs, p values are calculated using Fisher’s test. *:p<0.05, **:p<0.01, ***:p<0.001.
Supplemental Figure S4. Differential enrichment analysis of H3K27me3 and H3K36me2 upon NSD1 knockdown and EZH2 inhibition.
(A-B) Scatter plots of regions differentially enriched for H3K27me3 as obtained by ChIP-seq, in control (A) or NSD1 depleted (B) G401 cells treated with 1uM GSK126 for 3 days.
(C) Scatter plot of regions differentially enriched for H3K36me2 as obtained by ChIP-seq, in control G401 cells treated with 1uM GSK126 for 3 days.
Significance computed using empirical Bayesian statistical tests and linear fitting.
Supplemental Figure S5. H3K27ac gain upon EZH2 inhibition shows partial dependence on NSD1.
(A) Scatter plot of the differentially enriched regions for H3K27ac upon NSD1 knockdown in G401 cells, obtained by ChIP-seq. Significance computed using empirical Bayesian statistical tests and linear fitting.
(B-C) Heatmaps of enhancers (B) and promoters (C) showing differential H3K27ac enrichment between control and NSD1 depleted cells ranked by gained, shared, and lost H3K27ac signal in control cells treated with 1uM GSK126 for 3 days.
(D) Venn diagram of genes gaining H3K27ac upon EZH2 inhibition, overlapping with genes activated by SMARCB1 in G401 cells. Significance computed using standard Fisher’s exact Test.
(E) Box plot of H3K27ac log2FC for the same sites that gain H3K27ac in shCTRL and shNSD1 cells with EZH2 inhibition for the same genomic regions. p value was calculated using Wilcoxon test.
Supplemental Figure S6. NSD1 cooperates with SWI/SNF to activate transcription.
(A) Intersection of gene sets displaying p-values (computed using standard Fisher’s exact Test for pairwise overlaps and “Exact Test of Multi-set Intersections” for multi-list overlaps) and enrichment statistics (see text for details) for Venn diagram in Figure 5A.
(B) Region-scaled heatmap visualization of H3K36me2 normalized ChIP-seq coverage, rank-ordered based on changes in control vs. NSD1 depleted G401 cells without SMARCB1. Significance computed using empirical Bayesian statistical tests and linear fitting.
(C) Peak centered heatmap visualization of normalized SUZ12 ChIP-Seq coverage, rank-ordered based on changes in H3K36me2 sites lost in sgNSD1 vs sgCTRL, as shown in (B).
(D) Peak centered heatmap visualization of normalized RING1B CUT&RUN coverage, rank-ordered based on changes in H3K36me2 sites lost in sgNSD1 vs sgCTRL, as shown in (B).
(E) Significantly enriched biological processes based on the 977 genes upregulated by SMARCB1 and bound by SMARCC1 (FDR < 0.05 and log2FC > 1).
(F) Significantly enriched biological processes based on the 730 genes downregulated by loss of NSD1.
(G-J) Metaplots of normalized, average coverage for H3K36me2 (G), H3K27me3 (H), SUZ12 (I) and RING1B (J) at +/****−20kb of the 289 genes activated by NSD1 and SMARCB1.
(K) Genome browser tracks depicting H3K27me3, H3K36me2, H3K27ac and RNA-seq reads upon SMARCB1 addback in control and NSD1 KO G401 RT cells.
Supplemental Figure S7. NSD1 physically interacts with SWI/SNF and PRC1/2 complex members.
(A and B) Co-immunoprecipitation analysis of SWI/SNF core subunits SMARCC1 (A) and SMARCA4 (B) in G401 cells upon SMARCB1 addback.
(C and D) Co-immunoprecipitation analysis of PRC1/2 subunits EZH2-PRC2 (C) and RING1B-PRC1 (D) in G401 cells treated with DMSO or 1uM GSK126 for 3 days.
(E) Co-immunoprecipitation analysis of SWI/SNF ATPase SMARCA4 in G401 cells treated with DMSO or 1uM GSK126 for 3 days.
(F) Schematic representation of the co-IP results.
Supplemental Figure S8. NSD1 and SWI/SNF mutations co-exist in cancer patients.
(A) Analysis of co-mutation and exclusivity in focused cohort of cancer patients (see methods for details). Fisher’s Exact Test (R version 3.4.2) was used for mutational analysis and p values were then corrected for multiple testing by using package “qvalue” (R version 3.4.2).
(B) Pathogenic mutations in NSD1 gene used for the cBioportal analysis.
(C and D) Venn diagram of cancer samples with NSD1 and ARID1A (C) or SMARCA4 (D) co-occurring mutations in focused analysis. Fisher’s Exact Test (R version 3.4.2) was used for mutational analysis and p values were then corrected for multiple testing by using package “qvalue” (R version 3.4.2).
Supplemental Table S1. H3K36me2 and H3K27me3 Log2FC in shNSD1 vs. shNSD1
Supplemental Table S2. Histone post translational modifications in G401 and Pfeiffer cells with EZH2 inhibition and NSD1 knockdown
Supplemental Table S3. Differentially expressed genes in shCTRL EZH2i vs. DMSO
Supplemental Table S4. Gene Ontology analysis of genes upregulated in shCTRL + EZH2i vs. DMSO
Supplemental Table S5. Differentially expressed genes in shNSD1 vs. shCTRL G401
Supplemental Table S6. Differentially expressed genes in G401 SMARCB1_sgCTRL vs. GFP_sgCTRL
Supplemental Table S7. Differentially expressed genes in G401 SMARCB1_sgNSD1 vs. SMARCB1_sgCTRL
Supplemental Table S8. 289 genes downregulated in G401 SMARCB1_sgNSD1 vs. SMARCB1_sgCTRL
Supplemental Table S9. Gene Ontology analysis of the 289 genes activated by SWI/SNF and NSD1
Supplemental Table S10. Co-occurring and mutual exclusive mutations in cBioportal analysis
Supplemental Table S11. Raw mutation data (columns A-P) downloaded from CBioportal (July 15, 2021) for study Nat Med 2017 (PMID 28481359)
Supplemental Table S12. Mutated gene list at sample level. Data derived from Table S13 by consolidating multiple datasets
Supplemental Table S13. Analysis of pathogenicity of mutations analyzed in Tables S13, S14 for study Nat Med 2017 (PMID 28481359)
Supplemental Table S14. Co-occurring and mutual exclusive mutations in focused analysis
Data Availability Statement
The next-generation-sequencing data generated by this study have been deposited to GEO database under accession number GEO: GSE178490
Publicly available ChIP and RNA-Seq were obtained from GEO accession: GSE90631 and GSE71506 and analyzed with consistent methods.
Original western blot images have been deposited at Mendeley. The DOI is listed in the key resources table.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-NSD1 | Antibodies, Inc. | Cat#75-280; RRID: AB_11001827 |
| Mouse monoclonal anti-NSD2 29D1 | Abcam | Cat#ab75359; RRID: AB_1310816 |
| Rabbit monoclonal anti-NSD3 D4N9N | Cell Signaling | Cat# 92056, RRID:AB_2800178 |
| Rabbit monoclonal anti-BRG1 | Cell Signaling | Cat#49360; RRID: AB_2728743 |
| Rabbit monoclonal anti-BAF155 | Cell Signaling | Cat# 11956; RRID: AB_2797776 |
| Rabbit monoclonal anti-SMARCB1 | Cell Signaling | Cat#91735; RRID: AB_2800172 |
| Mouse monoclonal anti-EZH2 | Active Motif | Cat# 39875; RRID: AB_2561022 |
| Rabbit monoclonal anti-EZH2 | Cell Signaling | Cat#5246; RRID: AB_10694683 |
| Rabbit monoclonal anti-SUZ12 | Cell Signaling | Cat#3737; RRID: AB_2196850 |
| Mouse monoclonal anti-EED | Millipore Sigma | Cat#05-1320; RRID: AB_1586999 |
| Rabbit polyclonal anti-EED | Millipore Sigma | Cat#09-774; RRID: AB_1587000 |
| Rabbit monoclonal anti-BMI1 | Cell Signaling | Cat#6964; RRID: AB_10828713 |
| Rabbit monoclonal anti-RING1B | Cell Signaling | Cat#5694; RRID: AB_10705604 |
| Mouse monoclonal anti-b-Actin | Sigma Aldrich | Cat#A5441; RRID: AB_476744 |
| Rabbit polyclonal anti-HSP90 | Cell Signaling | Cat# 4874, RRID:AB_2121214 |
| Rabbit monoclonal anti-KDM2A | Abcam | Cat#ab191387 |
| Mouse monoclonal anti-Lamin A/C | Cell Signaling | Cat# 4777, RRID:AB_10545756 |
| Rabbit monoclonal anti-Histone H3 | Cell Signaling | Cat#4620; RRID: AB_1904005 |
| Rabbit polyclonal H3K27ac | Abcam | Cat#ab4729; RRID: AB_2118291 |
| Rabbit monoclonal H3K27me3 C36B11 | Cell Signaling | Cat#9733; RRID: AB_2616029 |
| Rabbit polyclonal H3K36me2 | Abcam | Cat#ab9049; RRID: AB_1280939 |
| Rabbit monoclonal Rabbit Control IgG | Cell Signaling | Cat#3900; RRID: AB_1550038 |
| Normal mouse IgG | Santa Cruz | Cat#sc-2025; RRID: AB_737182 |
| Drosophila spiked in antibody | Active Motif | Cat# 61686; RRID: AB_27370 |
| Monoclonal mouse anti-Rabbit IgG, light chain specific | Jackson | Cat#211-032-171; RRID: AB_2339149 |
| Goat anti-mouse IgG, light chain specific | Jackson | Cat#115-035-174; RRID: AB_2338512 |
| Chemicals, peptides, and recombinant proteins | ||
| GSK126 | ChemieTek | Cat#CT-GSK126 |
| TC-E5002 | Tocris | Cat#5089 |
| Vincristine (Tocris #1257 | Tocris | Cat#1257 |
| Doxorubicin (CarboSynth #AC15377 | CarboSynth | Cat#AC15377 |
| D-actinomycin (ApexBio #A4448 | ApexBio | Cat#A4448 |
| Etoposide (AG Chemicals #E2004 | AG Chemicals | Cat#E2004 |
| Dimethylsulfoxide (DMSO) | ATCC | Cat#4-X |
| Critical commercial assays | ||
| DirectZol Mini Prep Plus kit | Zymo Research | Cat#R2070 |
| Quant-iT RiboGreen assay | ThermoFisher | Cat#R11490 |
| Epicypher Cutana Cut&Run Kit v2.0 | Epicypher | Cat# 14-1048 |
| Covaris True- ChIP kit | Covaris | Cat#520154 |
| Drosophila chromatin spike in antibody | Active Motif | Cat#61686 |
| Dynabeads™ Protein G for Immunoprecipitation | ThermoFisher | Cat#10004D |
| RNase A | ThermoFisher | Cat#EN0531 |
| Ptoteinase K | ThermoFisher | Cat#4333793 |
| Nuclease | ThermoFisher | Cat#88702 |
| Cell titer aqueous reagent | Promega | Cat#G3582 |
| CellTiter-Glo | Promega | Cat#G9241 |
| Halt™ Protease and Phosphatase Inhibitor Cocktail (100X) | ThermoFisher | Cat# 78446 |
| Deposited data | ||
| RNAseq data | This study | GSE90631 |
| ChIP-seq data | This study | GSE90631 |
| Cut&Run data | This study | GSE90631 |
| Uncropped immunoblot data | This study | Mendeley data doi: 10.17632/fvzm88tm3v.2 |
| Experimental models: Cell lines | ||
| G401 | ATCC | CVCL_0270 |
| G402 | ATCC | CVCL_1221 |
| A549 | ATCC | CVCL_0023 |
| TOV21G | ATCC | CVCL_3613 |
| HFF-1 | ATCC | CVCL_3285 |
| HEK293T | ATCC | CVCL_0063 |
| Pfeiffer | ATCC | CVCL_3326 |
| BT16 | Laboratory of Dr. C. David James | CVCL_M156 |
| TTC709 | Laboratory of Dr. B.E. Weissman. | CVCL_8007 |
| Oligonucleotides | ||
| sgRNA for NSD1 knockout: SP185.g12 5- UCUCCAACUUCAUACUUCAG-3 |
This study | NA |
| Forward primer for Next Generation Sequence (NGS) of the NSD1 gene and assessment of indels: 5-CATACATTGCTTTTTCAGAAGGCT-3 |
This study | NA |
| Reverse primer for Next Generation Sequence (NGS) of the NSD1 gene and assessment of indels: 5- ACACCAATTCATTCACAAAATGTTC-3 |
This study | NA |
| sgRNA for NSD2 knockout: g7 5-CCAAAGUGUCGGGUUACCCU-3 |
This study | NA |
| Forward primer for Next Generation Sequence (NGS) of the NSD2 gene and assessment of indels: 5- AGCCTGGTGAATCCTTGGTCTTCTGG-3 |
This study | NA |
| Reverse primer for Next Generation Sequence (NGS) of the NSD2 gene and assessment of indels: 5-GCTGCAAAAATGAACCATAGCAGGGC-3 |
This study | NA |
| sgRNA for NSD3 knockout: g16 5-AAACUGGACAUGAUAUUCUC-3 |
This study | NA |
| Forward primer for Next Generation Sequence (NGS) of the NSD3 gene and assessment of indels: 5-ACAGGTGCCCGAGAATATCA-3 |
This study | NA |
| Reverse primer for Next Generation Sequence (NGS) of the NSD3 gene and assessment of indels: 5- ACGATTAAAACAGCTGAAAGGGG-3 |
This study | NA |
| Recombinant DNA | ||
| Plasmid: pLKO_005-shNSD1-1 | Sigma | TRCN0000238371 |
| Plasmid: pLKO_005-shNSD1-2 | Sigma | TRCN0000238372 |
| Plasmid: pLKO.1-shKDM2A-1 | Sigma | TRCN0000022000 |
| Plasmid: pLKO_005-shKDM2A-2 | Sigma | TRCN0000359598 |
| Plasmid: pLV[Exp]-Bsd-CMV>Tet3G | VectorBuilder | 180621-1181ctz |
| Plasmid: pLV[Exp]-Puro-TRE3G>EGFP | VectorBuilder | 180621-1204vsc |
| Plasmid: pLV[Exp]-Puro-TRE3G>V5/hSMARCB1 | VectorBuilder | 180622-1078xvz |
| Plasmid: pVSV-G | Addgene | Addgene #138479 |
| Plasmid: psPAX2 | Addgene | Addgene #12260 |
| Software and algorithms | ||
| Prism 7.0d | GraphPad | www.graphpad.com |
| Image J 2.1.0/1.53c | NIH | https://imagej.nih.gov/ij |
| Integrative Genomics Viewer (IGV_2.6.2) | The Broad Institute | https://software.broadinstitute.org/software/igv/ |