Abstract
NSD1, NSD2, and NSD3 constitute the nuclear receptor-binding SET Domain (NSD) family of histone 3 lysine 36 (H3K36) methyltransferases. These structurally similar enzymes mono- and di-methylate H3K36, which contribute to the maintenance of chromatin integrity and regulate the expression of genes that control cell division, apoptosis, DNA repair, and epithelial–mesenchymal transition (EMT). Aberrant expression or mutation of members of the NSD family is associated with developmental defects and the occurrence of some types of cancer. In this review, we discuss the effect of alterations in NSDs on cancer patient’s prognosis and response to treatment. We summarize the current understanding of the biological functions of NSD proteins, focusing on their activities and the role in the formation and progression in solid tumors biology, as well as how it depends on tumor etiologies. This review also discusses ongoing efforts to develop NSD inhibitors as a promising new class of cancer therapeutic agents.
Keywords: NSD1/KMT3B, NSD2/MMSET/WHSC1, NSD3/WHSC1L1, H3K36 methyltransferases, Histone methylation, Cancer, Solid tumors
Introduction
Tumorigenesis is accompanied by extensive changes in gene expression, which depend on the reorganization of chromatin to allow differential access to transcription factors. Chromatin is organized by octameric nucleosomes, containing two copies each of four types of histones (H2A, H2B, H3, and H4), which wrap DNA [1]. Chromatin can exist in “closed” and “open” configurations, with the open configuration supporting transcription. The transition between open and closed chromatin is regulated by reversible post-translational modifications (PTMs) on some of the histones, with H3 most frequently targeted. PTMs occur on both the flexible, protruding N-terminal histone tails and on the main globular domains, and include acetylation, methylation, phosphorylation, ubiquitylation, and ADP ribosylation [2]. These various chemical PTM "marks" on histones are associated with the availability of DNA to trans-acting factors [3].
Histone methylation typically occurs on positively charged lysine (K) or (less commonly) arginine (R) amino acids, influencing the interaction of the histone with DNA or associated proteins. The effect of histone methylation on transcription can be positive or negative, depending on several factors: the degree of histone methylation, the specific lysine site within the histone, and the location of the specific target nucleosome within transcriptionally active or silent regions of the genome [4]. Lysine 36 of histone 3 is subject to mono-, di-, and trimethylation (H3K36me1, H3K36me2, and H3K36me3). H3K36me1 is considered an intermediate state, without a functional role in transcription [4], and is broadly distributed in the genome [5]. H3K36me2 and H3K36me3 localize to specific genomic regions associated with active transcription [1]. H3K36me2 marks are concentrated in intergenic and regulatory regions, where these modified forms of H3K36 constitute 20–45% of total H3 histone [6–8]. In contrast, H3K36me3 marks are more concentrated in gene bodies (protein-coding sequences), where they constitute only 5% of total H3 histone [7, 9]. Appropriate control of methylation of histone H3K36 is critical for chromatin integrity and regulation of gene expression.
The different distribution of H3K36me2 and H3K36me3 throughout the genome is regulated by the action of enzymes that add or remove methyl groups [8]. These enzymes are known as lysine methyltransferases (KMTs) and lysine demethylases (KDMs). There are several distinct groups of KMTs encoded in the human genome, which catalyze the transfer of various numbers of methyl groups to H3K36. KMTs that perform mono- and dimethylation of H3K36 include the NSD proteins (NSD1-3), ASH1L, SMYD2, SETMAR, and SETD3 [10]. Among these, enzymatic activity has been most rigorously analyzed for NSD1, NSD2, NSD3, and ASH1L [3]. After dimethylation, trimethylated H3K36 is generated by only one enzyme, SETD2 [3]. H3K36 histone demethylases, which oppose the action of these KMTs, belong to the Jumonji C (JmjC) domain-containing family of histone demethylases (JHDMs) [11]. Two specific JHDMs demethylate H3K36: JHDM1 (also known as KDM2A-B) is H3K36me1- and H3K36me2-specific and JHDM3 (JMJD2/KDM4A-D) demethylates H3K36me2 and H3K36me3 [12], as well as some other methylated residues (e.g., H3K9Me3 [13]).
NSD1 (also known as KMT3B), NSD2 (WHSC1/MMSET), and NSD3 (WHSC1L1) together comprise the family of nuclear receptor-binding SET Domain (NSD) protein KMTs [14, 15]. The intact function of both NSD1 and NSD2 is critical for early mammalian development. Mice heterozygous for a defective allele of Nsd1 are viable and fertile, whereas homozygosity for inactive Nsd1 alleles causes lethality at embryonal day E10.5, due to profound mesodermal defects [16]. Male mice with germline-specific Nsd1 deficiency show severe defects in spermatogenesis, although females with Nsd1 deficiency are fertile [17]. In humans, NSD1 haplodeficiency is the cause of Sotos syndrome, a severe developmental defect characterized by excessive growth before and after birth, advanced bone age, dolichocephalic head with a typical facial feature, and neurological disorders with brain anomalies that result in intellectual disability [18]. In addition, some cases of Beckwith-Wiedemann Syndrome, which phenotypically presents with macroglossia, abdominal wall defects, and embryonic tumors, may also be associated with deletion or point mutations of NSD1[19].
Homozygous loss of Nsd2 in mice causes developmental defects occurring at later embryonal stages, resulting in neonates with smaller body sizes and problems breathing, that die within 10 days of birth [20]. Mice heterozygous for Nsd2 loss are viable and fertile. However, these mice have growth retardation and significant midline defects, which are phenotypically expressed as abnormal craniofacial development, skeletal anomalies, and congenital heart defects [20]. Mice selectively lacking Nsd2 in both alleles in hematopoietic stem cells develop immunodeficiency due to defects in B cell lineage specification [21], based on experiments in which bone marrow was transplanted from Nsd2−/−embryos into wild-type mice [21]. In humans, a heterozygous chromosomal deletion encompassing NSD2/WHSC1 causes Wolf-Hirschhorn syndrome, characterized by prenatal and postnatal growth deficiency, a characteristic facial appearance, intellectual disability, and seizures [22]. To date, there are no reports of phenotypes in humans or mice associated with mutation or loss of NSD3. However, several studies have shown that NSD3 is required for early neural crest formation in chick embryos models [23–25], suggesting it may also play a developmental role.
In addition to these developmental defects associated with loss of NSD protein function, somatic mutations affecting the NSD proteins have been recognized to promote tumor formation. The first indications of the involvement of NSDs in human cancer were reported in 2001, with the identification of an oncogenic gene fusion between NUP98 and NSD1 [26, 27] in hematologic malignancies and the discovery that NSD3 was amplified and dysregulated in breast cancer [28]. There are now many studies investigating dysregulation of NSD KMTs in many forms of cancer and characterizing the mechanistic consequences of NSD loss on H3K36 methylation and downstream signaling events. In this review, we focus on the role of NSD family proteins in solid tumors.
General structure and function of NSD KMTs
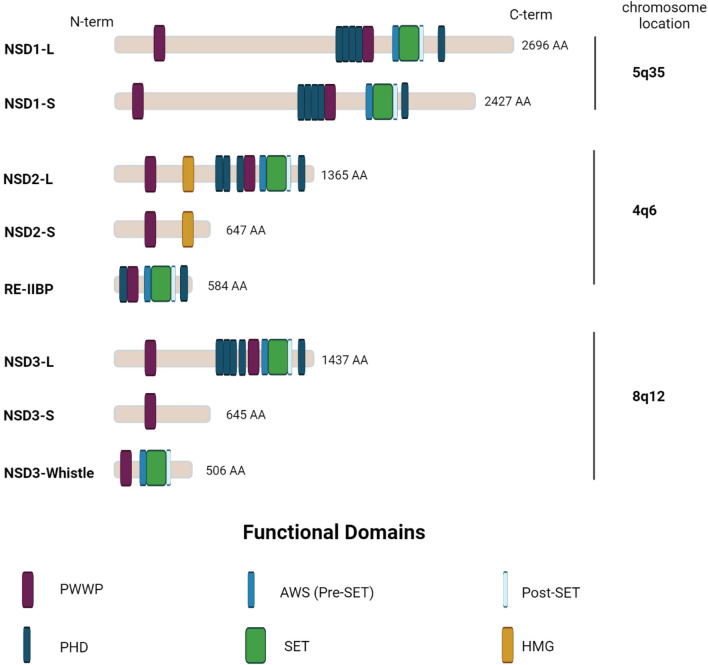
The human genes encoding NSD1, NSD2 and NSD3 proteins are located on chromosome 5q35 [26], 4p16 [22], and 8p12 [28], respectively. These three proteins are highly homologous, with 70–75% identity in their amino acid sequences, and each contains an enzymatic SET domain, which catalyzes the transfer of the methyl group from S-adenosylmethionine (SAM) to the substrate protein [29]. NSD family proteins exist as both long (–L) and short (–S) isoforms. In addition to the SET domain, the long isoforms contain a high-mobility-group (HMG) box, two PWWP (proline-tryptophan-tryptophan-proline) motifs, and five plant homeodomain (PHD) zinc finger domains, each of which contributes to the interactions with chromatin and selected target proteins (Fig. 1). In the case of NSD1, the predominant form is short; however, the short isoform retains all key domains [30]. The short isoforms of NSD2 and NSD3 proteins lack some of these domains [15, 31–33]; most importantly, the NSD2-S and NSD3-S proteins lack the catalytic SET domain, abolishing their primary methylation function [33]. NSD2 has an additional isoform, RE-IIBP (interleukin-5 response element II-binding protein) [34], which retains the catalytic domain and is an active KMT; however, this isoform has an altered substrate preference for H3K79, instead of H3K36 [34, 35]. NSD3 has a third “Whistle” isoform (WHSC1-like 1 isoform 9 with methyltransferase activity to lysine) [36], which is a short alternatively spliced version with altered substrate preference, targeting H3K4 and H3K27 [36]. Overall, the relative expression of long versus short isoforms of NSD enzymes has not been systematically studied in normal tissue or tumors.
Fig. 1.
The schematic structure of NSD proteins. Abbreviations of domain names: PWWP proline–tryptophan–tryptophan–proline, PHD plant homeo domain, SET suppressor of variegation, enhancer of zeste, and trithorax, HMG high-mobility group
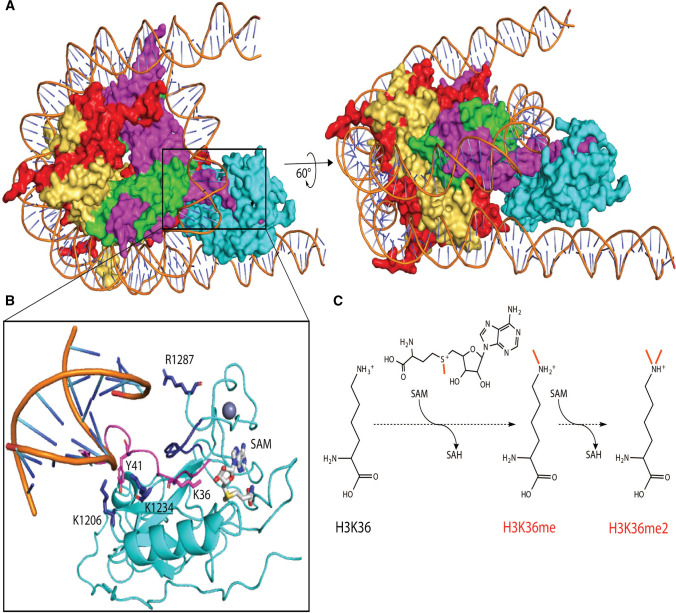
NSD proteins transition between inactive and active states in response to nucleosome binding (Fig. 2A). The inactive catalytic state of NSD proteins is maintained by the loop region, which connects the SET and post-SET domains [32, 37] and thus blocks access to the catalytic domain. The enzymes convert to a catalytically active state as a result of a conformation shift caused by the binding of the protein to nucleosomal DNA, which serves as an allosteric effector [38]. The inactive NSD protein uses its PHD zinc finger domains as modules for the recognition of histone-lysine labels, initially positioning the SET catalytic domain proximal to lysine substrates on histones [38]. This promotes partial DNA unwinding, allowing the SET domain to bind to the unfolded DNA; this, in turn, causes a conformational shift in the autoinhibitory loop, converting the NSD protein into an active catalytic state [38, 39]. As an additional result of this DNA unfolding, a groove between DNA and the histone octamer forms, where the histone tail, carrying the lysine substrate, enters in the catalytic channel. Salt bridges between the SET and post-SET domains with the phosphate backbone of the DNA groove form an additional stabilizing bond between NSD proteins and DNA. In turn, interactions between the histone H3 tail and the SET domain are mediated by hydrogen bonds and π-stacking interactions [39] (Fig. 2B). Once bound, NSDs can transfer up to two methyl groups from S-adenosylmethionine (SAM), to its substrate, the K36 ε-amino group [40] (Fig. 2C).
Fig. 2.
H3K36 methylation by NSD family proteins. A Nucleosome-bound NSD3 (cyan) in a catalytically active state (PDB ID 7CRR). Included in this complex are H2A (red), H2B (yellow), H4 (green), and H3 (magenta). B Allosteric regulation of nucleosome binding leads to a 2.4 Å movement of the SET domain loop (dark blue) which allows access of the H3 tail (magenta) and proximity of K36 to SAM (gray). Salt bridge contacts between Lys1206 and Lys1234 of NSD3, further stabilized by π-stacking with Tyr41 of H3, enforce tight binding in this complex. Arg1287 on the post-SET loop of NSD3 makes additional phosphate backbone contacts to the anti-parallel DNA strand. C The cofactor SAM serves as a methyl donor for single or double H3K36 methylation events. H3K36me2, in turn, regulates gene expression and DNA damage repair, and interplays with other histone modifications
In general, histone modifications exert their effects in two main ways. The first includes modifications that directly affect the overall chromatin structure, such as the acetylation that neutralizes the positive charge of histone, weakening electrostatic interactions with DNA and resulting in relaxed chromatin that becomes more accessible for transcription [41]. In contrast to acetylation, the NSD-mediated addition of methyl groups to H3K36 does not significantly change histone charge and does not directly affect chromatin structure; however, methylation creates binding sites for various downstream effector proteins [41]. In turn, H3K36 methylation acts on gene expression in several ways.
First, in contexts where this methylation promotes gene expression, this PTM on H3 opposes the ability of the Polycomb repressor complex (PRC2) to methylate H3K27 to produce H3K27me3, a repressive mark at the gene promoters that contributes to gene silencing [9, 42]. The interaction of NSD proteins and the PRC2 complex is bidirectional, as H3K36me2 and H3K27me3 levels antagonize each other in the cell [43]; in the absence of NSD1, PRC2 deposits H3K27me3 in the intergenic regions from which H3K36me2 is depleted [44, 45], reducing gene expression.
Second, NSD1 binding to nucleosomes in the vicinity of gene promoters helps the recruitment of RNA polymerase II (RNAP II) onto gene promoters [30], and contributes to the transition of RNAP II from initiation to elongation. Separately, methylation of H3K36 reduces interrupted initiation of transcription in the gene body and contributes to transcriptional elongation. During transcriptional elongation, there is a need to keep the chromatin structure open and accessible for RNAP II, and histones are maintained in an acetylated state. As the RNAP II moves along the DNA, it is important that the chromatin left behind the RNAP II is reorganized as nucleosomes and a repressive chromatin configuration to repress aberrant transcription. This is accomplished through the mechanism of histone deacetylation [46]. For this, SET2 interacts with S2-phosphorylated RNAP II and methylates H3K36 in the transcribed region. In turn, Esa1-associated factor 3 (Eaf3) and Rco1 [subunits of the histone deacetylase Reduced Potassium Dependency 3 (Rpd3)], directly bind to the methylated histone tail [47, 48], and RPD3 deacetylates local histones after the passage of RNAP II to slow elongation and suppress cryptic initiation [49].
Third, the mechanism of how H3K36me2 functions to repress gene expression of some genes has not been explored in detail. However, it has been demonstrated that NSD1 recruits the H3K27ac deacetylase HDAC1 to chromatin at active enhancers in mouse embryonic stem cells (mESCs). Conversely, NSD1 knockout in these cells decreases H3K36me2 and simultaneously increases H3K27ac at active enhancers of genes related to mesoderm differentiation genes, which causes their activation [50].
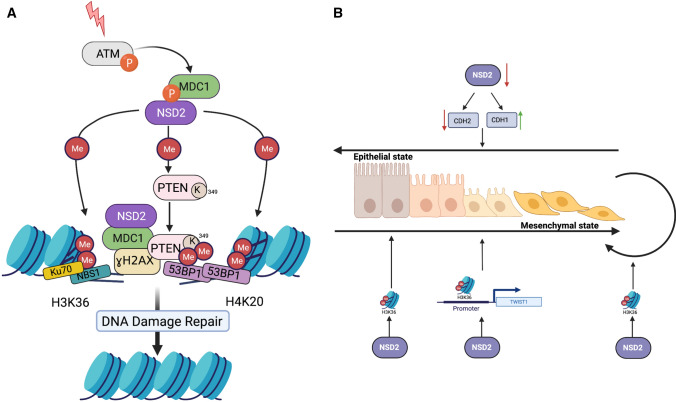
Fourth, besides its function in controlling gene expression, H3K36me2 plays a role in maintaining genomic stability by participating in DNA repair [51] (Fig. 3A). This mark rapidly accumulates at DNA double-strand breaks (DSB) after ionizing radiation (IR) [51]. Chromatin immunoprecipitates bearing the H3K36me2 mark contain Nijmegen breakage syndrome 1 (NBS1) protein and Ku70, suggesting this modification signals recruitment of these early DNA repair components at DSBs [51]. The effectiveness of non-homologous end-joining (NHEJ) repair has been shown to depend on the levels of H3K36me2 mediated by NSD2 [52]. Some data suggest that NSD proteins also recognize and methylate other histone substrates (H3K4, H3K9, H3K27, H3K79, and H4K20) in addition to H3K36, and methylates non-histone substrates in some settings [38], with relevance to processes including DNA repair. For example, NSD2-dependent methylation of H4K20 promotes DNA damage response (DDR) by recruiting P53-binding protein 1 (53BP1) [53]. In parallel, NSD2 methylates the protein phosphatase and tensin homolog (PTEN), allowing it to interact with 53BP1; this recruits PTEN to DNA damage sites, where it contributes to DNA repair through dephosphorylating histone H2A variant H2AX [54]. Phosphorylation of the H2AX variant on Ser 139 (γH2AX) is one of the most rapid events in response to DNA double-strand breaks and promotes the recruitment of many proteins that compose the DNA repair machine [55]. H2AX dephosphorylation at the late stages of DDR is necessary to remove unresolved γH2AX and complete repair; it has been shown that disruption of dephosphorylation of γH2AX also reduces the DNA repair efficiency [56, 57]. Another study has reported NSD2 involvement in a γH2AX-MDC1 repair pathway [58]. Consistent with these roles, depletion of NSD2 reduces DDR efficiency [54].
Fig. 3.
Role of NSD2 in DNA damage response and epithelial–mesenchymal transition. A NSD2 participates in DNA damage response and DNA repair. H3K36me2 mediated by NSD2 accumulates at DNA double-strands breaks (DSBs) and recruits early DNA repair components NBS1 and Ku70 at DSBs. Also, NSD2 is involved in γH2AX-MDC1 pathway: MDC1 interacts with phosphorylated Ser 102 of NSD2, in turn NSD2 methylates H4K20 and recruits 53BP1 at the sites of DNA damage. NSD2 also dimethylates PTEN protein, then dimethylated PTEN is recognized by the 53BP1 and is recruited to DNA damage sites. B H3K36me2 plays role in promoting EMT and to maintain cells in mesenchymal state. In the cells in epithelial state, NSD2 mediates H3K36me2 and promotes EMT to change the cells from epithelial state to mesenchymal state. NSD2 mediates H3K36me2 at the promoter of TWIST1 gene that maintains its expression, which supports mesenchymal state of cells. Similarly, inhibition of NSD2 downregulates N-cadherin and upregulates E-cadherin promoting epithelial states of cells. ATM Ataxia-Telangiectasia Mutated kinase, MDC1 mediator of DNA damage Checkpoint protein 1, PTEN phosphatase and tensin homolog, NBS1 Nijmegen breakage syndrome 1, 53BP1 P53-binding protein 1, CDH1 Cadherin-1 or E-Cadherin, CDH2 Cadherin-2 or N-Cadherin, TWIST1 Twist family bHLH transcription factor 1
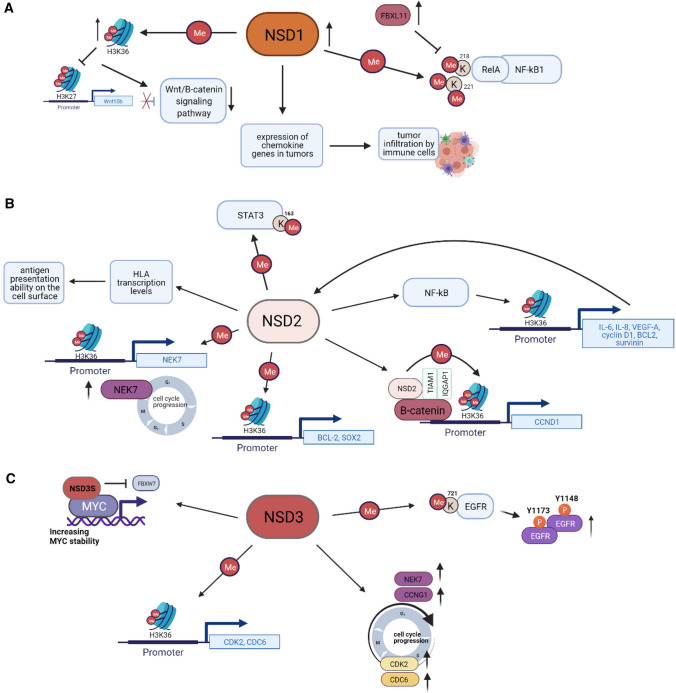
Finally, in addition to their role in protein methylation, NSD proteins have been reported to have some non-catalytic functions based on their scaffolding function. For example, NSD1 binds to the androgen receptor (AR) and functions as a co-activator to mediate AR-dependent transcription [59]. NSD2 binds directly to β-catenin and two functionally interacting proteins, IQ motif-containing GTPase-activating protein 1 (IQGAP1) and TIAM Rac1-associated GEF 1 (TIAM1) [60]. The short NSD3-S isoform (which is catalytically inactive) binds the MYC oncoprotein and may influence its function [61] (Fig. 4C). Collectively, these varied roles may contribute to the activities of NSD functions in solid tumors, as discussed below.
Fig. 4.
The scheme of signaling cancer pathways involving the NSD family proteins. A NSD1 methylates lysines 218 and 221 of RelA, resulting in NF-kB activation, while FBXL11 demethylates lysines 218 and 221 of RelA, which results in an inactivation of NF-kB in colon cancer cells. NSD1 also activates Wnt/β-catenin-signaling pathway, regulating expression of Wnt10b in HCC. Moreover, NSD1 is involved in regulation of chemokine genes expression and thus influences T-cell infiltration in HNSCC tumors. B NSD2 directly activates NF-kB and additionally increases expression of NF-kB target genes by dimethylation of H3K36 on their promoters in CRPC. Also, by dimethylating H3K36 on gene specific promoters, NSD2 increases expression of CCND1, BCL-2 in CRPC, NEK-7 in HNSCC, SOX2 in osteosarcoma; NSD2 methylates STAT3 at K163, thus promoting the activation of STAT3 pathway in colorectal adenocarcinoma; besides, NSD2 plays the role in the antigen presentation ability on the cell surface in prostate cancer. C NSD3 methylates EGFR at K721, which subsequently leads to increasing of phospho-Y1148 and phospho-Y1173 EGFR levels in HNSCC. NSD3 regulates expression of cell cycle-related genes as CDK2, CDC6 in HNSCC, NEK7, and CCNG1 in bladder cancer lung cancer. NSD3 also stabilizes and supports transcriptional activity of MYC oncoprotein. Wnt10b Wingless Type MMTV Integration Site Family, Member 10B, NF-kB Nuclear Factor-kappa B, FBXL11 F-box and leucine-rich repeat protein 11, STAT3 Signal transducer and activator of transcription 3, HLA human leukocyte antigen, NEK7 NIMA-related Kinase-7, BCL-2 B-Cell Lymphoma 2, SOX2 SRY-box transcription factor 2, TIAM1 TIAM Rac1 Associated GEF 1, IQGAP1 IQ Motif-containing GTPase-activating protein 1, CCND1 Cyclin D1, IL-6,8 Interleukin-6,-8, VEGF-A Vascular endothelial growth factor A, FBXW7 F-box and WD repeat domain-containing 7, CDK2 Cyclin-Dependent Kinase 2, CDC6 Cell Division Cycle 6, CCNG1 Cyclin G1, EGFR Epidermal Growth Factor Receptor
Dysregulation and mutational inactivation of NSD family genes in solid tumors: connection to prognosis
Dysregulation of NSD family proteins through somatic alteration has been reported in various cancers arising in a number of different tissues and occurs in between 3.3 and 21.36% of tumors (depending on the tumor type) (Table 1). Based on existing studies, somatic alterations in these proteins are associated with both positive and negative effects on prognosis.
Table 1.
Frequency and types of NSD genes dysregulation in different cancer types
| Gene | Cancer type | Frequency of gene alterations | Type of gene alterations | Data source | ||||
|---|---|---|---|---|---|---|---|---|
| Mutations | Amplifications | Deletion | Structural variant | Multiple alterations | TCGA PanCancer Atlas studies, 10,967 samples | |||
| NSD1 | Uterine corpus endometrial carcinoma | 19.1% | 17.6% | 1.6% | 529 cases | |||
| Head and neck squamous cell carcinoma | 12.6% | 11.5% | – | 1.2% | – | – | 523 cases | |
| Skin cutaneous melanoma | 10.6% | 9% | 0.7% | 0.2% | 0.7% | 444 cases | ||
| Uterine carcinosarcoma | 10.5% | 5.3% | 1.8% | – | 1.8% | 1.8% | 57 cases | |
| Kidney renal clear cell carcinoma | 8.6% | 1.2% | 7.4% | – | – | – | 511 cases | |
| Adrenocortical carcinoma | 7.7% | 2.2% | 4.4% | – | – | 1.1% | 91 cases | |
| Lung squamous cell carcinoma | 7.2% | 5.3% | 0.2% | 1.2% | 0.2% | 0.2% | 487 cases | |
| Lung adenocarcinoma | 6.7% | 4% | 1.6% | 1% | – | – | 566 cases | |
| Stomach adenocarcinoma | 6.6% | 5.5% | 0.2% | 0.7% | – | 0.2% | 440 cases | |
| Bladder urothelial carcinoma | 6.6% | 5.4% | 0.2% | 0.7% | – | 0.2% | 411 cases | |
| NSD2 | Uterine carcinosarcoma | 10.5% | – | 8.8% | – | – | 1.8% | 57 cases |
| Uterine corpus endometrial carcinoma | 10.2% | 7.4% | 2.7% | 0.2% | – | – | 529 cases | |
| Skin cutaneous melanoma | 7.4% | – | 0.7% | 0.5% | – | – | 444 cases | |
| Bladder urothelial carcinoma | 7% | 3.7% | 2.9% | 0.5% | – | – | 411 cases | |
| Stomach adenocarcinoma | 6.8% | 5% | 0.5% | 0.9$ | 0.2% | 0.2% | 440 cases | |
| Ovarian serous cystadenocarcinoma | 5.5% | 0.2% | 4.5% | 0.5% | 0.2% | 0.2% | 584 cases | |
| Esophageal adenocarcinoma | 3.9% | 0.6% | 1.7% | 1.7% | – | – | 182 cases | |
| Colorectal adenocarcinoma | 3.5% | 3% | 0.3% | 0.2% | – | – | 594 cases | |
| Cervical squamous carcinoma | 3.4% | 1.7% | 0.3% | 1.4% | – | – | 297 cases | |
| Adrenocortical carcinoma | 3.3% | 1.1% | 2.2% | – | – | – | 91 cases | |
| NSD3 | Lung squamous cell carcinoma | 21.4% | 2.7% | 16.2% | 0.6% | 0.4% | 1.4% | 487 cases |
| Bladder urothelial carcinoma | 13.6% | 2.7% | 8.5% | 2% | – | 0.5% | 411 cases | |
| Breast invasive carcinoma | 13.5% | 0.9% | 9.9% | 0.9% | 0.6% | 1.2% | 1084 cases | |
| Uterine corpus endometrial carcinoma | 12.7% | 9.5% | 2.3% | 0.8% | – | 0.2% | 529 cases | |
| Uterine carcinosarcoma | 10.5% | – | 7% | 3.5% | – | – | 57 cases | |
| Colorectal adenocarcinoma | 9% | 3.4% | 4.4% | 1.2% | – | 0.2% | 594 cases | |
| Head and neck squamous cell carcinoma | 9% | 0.6% | 6.3% | 1.5% | – | 0.6% | 523 cases | |
| Esophageal adenocarcinoma | 8.2% | 0.6% | 6% | 1.1% | 0.6% | – | 182 cases | |
| Prostate adenocarcinoma | 7.5% | 0.4% | 2.8% | 4% | – | 0.2% | 494 cases | |
| Ovarian serous cystadenocarcinoma | 6% | 0.7% | 2.9% | 1.9% | 0.3% | 0.2% | 584 cases | |
As some examples, in head and neck squamous cell cancer (HNSCC), nonsense and missense inactivating mutations of NSD1 and NSD2 mutations have been reported to affect prognosis for patients, but there are some disparate findings [62–64]. For example, loss-of-function mutations of NSD1 and NSD2 have been reportedly associated with a considerable survival benefit in laryngeal human papillomavirus (HPV)-negative HNSCC patients [63], with 2-year recurrence-free survival (RFS) of 77.5% for patients with mutated NSD1 compared to 44.7% for patients who do not have NSD1 mutations, and overall survival (OS) probability of 77.5% vs. 53%, respectively [63]. In contrast, in a study of 92 HNSCC patients, NSD2 protein expression has been reported to be significantly upregulated in many HPV-positive and HPV-negative HNSCC tumors versus in dysplastic tissue and normal epithelium, with higher expression correlated with poor outcomes in each case; however, there was no statistically significant difference in NSD2 staining between the HPV-positive and HPV-negative patient groups [65]. In contrast, another study found that low expression of NSD1 in HPV-positive HNSCC correlated with reduced survival for patients [66], while another found that HPV-positive HNSCC patients with NSD1-inactivating mutations were reported to have worse outcomes than HPV-positive patients without mutated NSD1 [62]. However, the set of HPV-positive patients was limited to 65 patients [62], and more studies are needed to unambiguously state the role of NSD1 mutation in HPV-positive HNSCC.
In pancreatic ductal adenocarcinoma (PDAC), an immunohistochemical (IHC) study detected significantly higher protein expression of NSD1 in metastatic tumors compared to benign ducts, primary PDAC, or all other lesions combined. In this case, higher expression of NSD1 was associated with stage III/IV disease compared to stage I/IIA disease, but no significant difference in OS or progression-free survival (PFS) was reported [67]. NSD1 mRNA expression was elevated in hepatocellular carcinoma (HCC) versus adjacent normal tissue and correlated with poor clinical outcomes for patients [68]. Elevated NSD1 gene expression has also been found in other tumor types—for instance, discriminating nonmalignant from prostate tumor tissues [69]—but the effect on prognosis has not been established.
High expression of NSD2 mRNA and protein in tumor tissue compared to normal tissue has been associated with poor clinical outcomes for patients with colorectal cancer [70, 71], cisplatin-resistant osteosarcoma [72], prostate cancer [73, 74], non-small cell lung cancer (NSCLC) [75, 76], stomach, colon, and anal canal carcinomas [77], breast cancer [78], neuroblastoma [79], and clear cell renal cell carcinoma (ccRCC) [80], NSD2 is amplified in pancreatic and lung tumors [81]. In sum, NSD2 mRNA is overexpressed in 15 different cancer types compared to their normal counterparts based on the analyses from publicly available datasets [82]. Whether this elevated expression is regulated at the transcriptional or post-transcriptional level is not well understood.
In contrast, in some tumors, NSD1 expression is sometimes silenced epigenetically by DNA methylation of the promoter region. This has been reported in neuroblastoma and glioma cells [83], and in ccRCC [84]. Hypermethylation of NSD1 promoter in neuroblastoma tumors correlates with higher patient mortality [83]. NSD1 silencing by promoter DNA hypermethylation in ccRCC is more frequent in metastatic RCC compared to localized cancer, based on analysis of specimens in TCGA [84]. Furthermore, in an additional independent cohort, 100% of grade IV ccRCC exhibited NSD1 DNA methylation compared to none in grade I–III tumors, although the number of stage IV cases was small in this study [84]. Methylation of the NSD1 gene and loss of protein expression is associated with worse OS for ccRCC patients, as well as elevated ccRCC metastasis [85].
The NSD3 gene is amplified and overexpressed in breast cancer [28, 86–88], and squamous cell lung cancer [89]. Elevated expression of NSD3 protein has been observed in several cancer types: in HNSCC, elevated expression correlated with poor differentiation grade [88]; in colorectal cancer, with poor prognosis [90]; and in breast cancer, with recurrence, distant metastasis, and poor survival [91, 92]. In bladder cancer, although overexpression has been noted, no correlations with any clinical and pathologic parameters have been described [93]. Of particular interest, a fusion of NSD3 to the NUT gene occurs in NUT midline carcinoma (NMC) [94]. NMC, defined by rearrangement of the NUT midline carcinoma family member 1 (NUTM1 or NUT) gene, is an extremely rare, poorly differentiated subtype of squamous cell carcinoma, which is highly aggressive and correlates with a low survival rate [95]. NUT carcinoma can originate in almost any body site [96]. Usually, the NUT gene is fused with Bromodomain Containing 4 (BRD4) in 75–78% of cases [97, 98], with BRD3 from 4.2 up to 15%, and NSD3 in 6–8.2% in different studies [97, 98]. The NSD3–NUT fusion protein acts as an oncogenic driver that blocks differentiation and stimulates cell growth [94]; however, the tumor-promoting mechanism is not associated with the catalytic ability of NSD3, as the NSD3 portion of the NSD3–NUT fusion lacks the SET domain and depleting wild-type part of NSD3 that is not included in the NSD3–NUT does not lead to changes in differentiation. The authors suggest that the function of NSD3–NUT as an oncogenic driver is related to its interaction with BRD4 [94], but additional studies are warranted. Thus, genetic dysregulation of the NSD family proteins occurs among many types of solid tumors. However, the correlation between dysregulations of the NSD proteins and prognosis for cancer patients depends not only on the specific member of this protein family but also on the type of tumor. This aspect of tumor specificity, which will also mention below when discussing the specific role of proteins in proliferation, growth, and cell signaling, is not yet fully investigated for each protein and may become more understood as knowledge accumulates.
NSD regulation of tumor growth, epithelial–mesenchymal transition (EMT) and metastasis
In tumor types where NSD expression correlates with tumor stage, functional evidence is needed to definitively ascertain whether NSD expression changes reflect driver or passenger events with respect to stage and prognosis. However, extensive evidence supports the idea that NSD proteins actively function in directly promoting tumor growth, tumor progression, and metastasis.
There are reports that NSD1 knockout leads to inhibition of cell proliferation in HCC cells [68], indicating an active role in modulating cell growth in certain tumor types. Knockdown of NSD2 suppresses cancer cell growth in castration-resistant prostate cancer (CRPC) cells [73], bladder cancer and lung cancer cells [60], SCCHN cell lines [65], colorectal cell models [71], and breast cancer cell lines [99]. Overexpression of NSD2 in multiple myeloma contributes to clonogenicity and cell proliferation [100, 101]. NSD2 interacts with the DNA binding domain of AR, mediates AR-mediated transcriptional regulation and promotes prostate carcinogenesis [101]. NSD3 knockdown inhibits proliferation in breast cancer [86], bladder and lung cancer [93], and squamous HNCC cell lines [88]. Conversely, cells with overexpression of NSD3 had a higher colony formation ability in soft agar and formed abnormal acini [86]. NSD3 depletion in PDAC [102] and in small-cell lung cancer (SCLC) with 8p11-12 amplification also decreased cell viability [103]. The NSD3–NUT fusion oncoprotein is required for proliferation, and the expression of this fusion protein blocks the differentiation in NMC cancer cells [94].
Some of the genes and proteins regulated by NSD proteins offer hypotheses for direct mechanisms of effects on tumor growth. For example, the binding of NSD3 to the oncoprotein MYC [61] and the MYC-associated protein BRD4 [104] enhances these proteins' stability and transcriptional activity, driving the expression of genes necessary for transition through the G1/S phases of the cell cycle. Similarly, signaling through the epidermal growth factor receptor (EGFR) is an essential stimulus to proliferation in many tumor types. NSD3 methylation of EGFR on K721 is associated with increased proliferation. It increases phosphorylation of EGFR on Y1148 and Y1173 in HNSCC cells, triggering a signaling activity that activates the pro-proliferative MEK/ERK and PI3K/AKT signaling cascades [105] (Fig. 4C). In ccRCC cells, NSD2 promotes proliferation through AKT/ERT1/2 signaling as silencing of NSD2 resulted in a decrease in cell proliferation (in vitro and in vivo mouse xenografts) and phosphorylation levels of AKT and ERK1/2, and NSD2 knockdown leads to induction of apoptosis by regulating B-Cell Lymphoma 2 (BCL2) and BCL2 Associated X (BAX) expression [80]. NSD2 regulation of apoptosis was also noted in colorectal cancer (CRC), based on a mechanism of enhancing BCL2 expression by mediating H3K36me2 of the BCL2 promoter [70]. Similarly, NSD2 depletion in osteosarcoma cells causes a higher level of apoptosis due to a mechanism proposed to involve decreased H3K36me2 at the BCL2 and SRY-box transcription factor 2 (SOX2) loci [72]. In addition, it has been demonstrated that in colorectal adenocarcinoma cells, NSD2 methylates Signal Transducer and Activator of Transcription 3 (STAT3) at lysine 163 (K163), thus promoting the activation of the STAT3 pathway and enhancing tumor angiogenesis in vitro and in vivo [106] (Fig. 4B).
Other studies show that NSD proteins directly control the mRNA expression of genes regulating cell cycle progression in various tumor types. However, the current literature is discordant regarding the stage of cell cycle arrest induced by loss of NSDs, which may reflect differences in tumor types, different functions of NSDs, or other factors. As some examples, in HNSCC, NSD2 was found to directly regulate the expression of NIMA-related kinase-7 (NEK7) at the mRNA and protein levels, and NSD2 knockdown in an HNSCC cell line caused cell cycle arrest at G0/G1 stage and induced apoptosis [65] (Fig. 4B). Another group found that NSD3 knockdown leads to G2/M-cell cycle arrest in bladder and lung cancer cell lines, based on a mechanism proposed to involve cell cycle-related genes Cyclin G1 (CCNG1) and NEK7 [93]. Depletion of NSD3 contributes to G2/M arrest and apoptosis in osteosarcoma cells [107], whereas NSD3 knockdown in HNSCC cells contributes to inhibition of cell cycle progression in G0/G1 phase through transcriptional regulation of (Cell Division Cycle 6) CDC6 and Cyclin-Dependent Kinase 2 (CDK2) [88] (Fig. 4C). No changes in the cell cycle were observed due to NSD2 depletion in CRC [70] and osteosarcoma [72], but at the same time, the apoptotic level was increased [70, 72].
NSDs regulation of progression and metastasis reflects its activity in regulating genes that contribute to EMT and invasion. In prostate cancer patients, NSD2 and H3K36me2 were found to be upregulated in metastatic tumors compared to the primary tumors, and a study also identified NSD2 as a conserved driver of lethal and metastatic prostate cancer progression [108]. Gene Set Enrichment analyses following CRISPR/Cas9 NSD2 knockout in PDAC indicated that EMT genes were enriched in the most significantly affected downregulated gene set and significant dysregulation of multiple genes involved in the process of EMT was found [109]. Overexpression of NSD2 promoted migration, invasion, and EMT in prostate cancer cell lines, whereas NSD2 knockdown had opposing effects [74]. Suggesting one mechanism for these activities, chromatin immunoprecipitation studies using prostate cancer cell lines showed that NSD2 directly binds to the promoter of the pro-EMT gene Twist family bHLH transcription factor 1 (TWIST1); TWIST1, in turn, induces the mesenchymal cell-associated N-cadherin (CDH2) [74]. Similarly, in human ccRCC cell lines, inhibition of NSD2 downregulated expression of the mesenchymal markers CDH2 and vimentin (VIM) and upregulated the epithelial marker E-cadherin (CDH1) [110]. Nsd2-deficient murine tumors exhibited an enhanced epithelial phenotype associated with the loss of H3K36me2, a histone mark linked to the expression of genes that induce EMT [109] (Fig. 3B).
NSD2 knockdown in KRAS-mutated lung cancer cells contributed to the downregulation of a group of H3K36me2-marked genes, including the invasion-promoting matrix metalloproteases MMP1 and MMP16 [75]. Although there are fewer studies of NSD3, one study showed that overexpression of NSD3 in epithelial-like breast cancer cells promotes EMT, invasion and metastases, whereas NSD3 knockdown in mesenchymal-like breast cancer cells results in reversal of EMT, impairing migration and invasion [91].
In summary, the above facts may indicate a cancer promoting role of the NSD proteins in several solid tumor types by acting on signaling pathways leading to the activation of cell proliferation, the cell cycle, and regulation of apoptotic pathways. Also, NSD proteins mediate a shift towards a mesenchymal cell phenotype, which defines proteins as participants in tumor progression through EMT.
Additional cancer-relevant signaling pathways regulated by NSD proteins
NF-κB
A role for NSD1 has been described in the activation of transcription factor Nuclear Factor-kappa B (NF-κB), which is composed of a heterodimer including a p50/p52 subunit encoded by NFKB1/NFKB2 associated with a p65 subunit encoded by a REL family member. A study in colon cancer cells connected NSD1 expression with methylation of K218 and K221 on p65, while elevated expression of the KDM F-box and leucine-rich repeat protein 11 (FBXL11) was associated with loss of methylation of the K218 and K221 sites. This study also showed that activation of NF-κΒ by cytokines Interleukin 1 Beta (IL-1β) and Tumor Necrosis Factor-Alpha (TNF-α) was dependent on NSD1 expression, with the authors suggesting reduced NSD1 expression may lead to a decrease in NF-κB activation in colon cancer [111]. However, a subsequent study did not observe direct methylation of RelA at K218 and K221 by NSD1 [112], raising the possibility that the initially reported result [111] described indirect signaling effects (Fig. 4A).
NSD2 also has been found to modulate the NF-κB pathway, serving as a direct co-activator of NF-κB transcriptional targets in CRPC through modification of histones [73]. Knockdown of NSD2 protein decreased H3K36me2 level at the promoter regions of the downstream target genes of NF-κB, including interleukin-6 (IL-6), IL-8, vascular endothelial growth factor A (VEGFA), Cyclin D1 (CCND1), BCL-2, and survivin, reducing their expression [73]. In addition to increasing histone methylation, NSD2 promotes the recruitment of NF-κB and p300 complexes for histone acetylation at the promoters of NF-κB target genes, which also leads to an increase in their expression and provides additional activation of NF-κB pathway. Interestingly, knockdown of RelA, a subunit of NF-κB, also decreased the expression of NSD2. These data suggest that NSD2 regulates the activation of NF-κB, and NF-κB may, in turn, enhance transcription of NSD2, thus forming a positive feedback loop [73] (Fig. 4B).
Wnt/β-catenin (CTNN1)
A role for NSD1 has been identified in the activation of the Wnt/β-catenin-signaling pathway. In one study, CRISPR/Cas9-mediated NSD1 knockout in HCC cells leads to decreased total dimethylation level of H3K36 and a decrease in Wingless Type MMTV Integration Site Family, Member 10B (Wnt10b) protein expression together with an increase in H3K27me3 in the promoter region of the WNT10B gene (Fig. 4A). Although a direct mechanism of NSD1 action was not defined in this study, the fact that the mark H3K27me3 is conferred by PRC2 is suggestive of direct action, given the known antagonism of NSD1 and PRC2. This study also showed that NSD1 knockdown reduced tumor cell proliferation in vitro and in vivo [68].
NSD2 has been found to be involved in the regulation of the Wnt/β-catenin-signaling pathway in lung and bladder cancers; however, the proposed mechanism that governs this regulation is different from that reported for NSD1. In this work, NSD2 was found to bind directly to β-catenin in the nucleus and to β-catenin-interacting proteins (IQGAP1 and TIAM1) that enhance β-catenin/Wnt-signaling. The NSD2-β-catenin complex induced H3K36me2 at the CCND1 promoter, increasing its transcription in bladder and adenocarcinoma cell lines [60]. Another study found that knockdown of NSD2 decreased levels of the β-catenin and cyclin D1 proteins in breast cancer cell lines, correlating this to the decrease in proliferation and regulation of G1–S cell cycle transition by NSD2 silencing [99] (Fig. 4B).
Like other NSD family members, NSD3 is thought to enhance WNT signaling. In 8p11-12 amplified breast cancer, where NSD3 is overexpressed, Iroquois Homeobox 3 (IRX3) and Transducin Beta Like 1 X-Linked (TBL1X), positive upstream regulators of the WNT pathway, are upregulated. Knockdown of NSD3 with shRNA inhibits cell proliferation in 8p11-12 amplified breast cancer cell lines and decreases expression of IRX3 and TBL1X [86]. In addition, NSD3 overexpression promoted tumorigenesis in 8p11–12 amplified lung squamous cell carcinoma in xenograft mice models [89], and deletion of NSD3 in LUSC mice models extended survival of mice, and reduced cell proliferation and increased apoptosis level in tumor tissues [89].
Tumor microenvironment and immunomodulation
The activity of the immune system and the ability of immune cells to infiltrate tumors are appreciated as major contributors to prognosis and response to immunotherapy in cancer patients [113]. The formation of the immunosuppressive tumor microenvironment suppresses cellular immunity and thus promotes the progression and invasion of this tumor. Investigations of NSD1 in HNSCC have suggested a potential contribution of this gene to the formation of the tumor microenvironment, finding a positive correlation between NSD1 RNA expression in HNSCC cells and T-cell infiltration into tumors [114]. This study also showed that the knockdown of NSD1 downregulated chemokine genes that enhance immune cell recruitment [114], implying a direct role in immune responses (Fig. 4A). The same study found the subtype of tumors defined by the presence of inactivating NSD1 mutations has an “immune cold” microenvironment that is characterized by the low level of tumor-associated leukocytes (including CD8+/CD4+ T cells, and M1 macrophages) and low expression of genes that are targets for immunotherapy (such as PD-1, PD-L1 and PD-L2); it was proposed that NSD1 loss may promote tumors to be more resistant to immunotherapy approaches in contrast to the “immune hot” phenotype that has higher infiltrating PD-1+ CD8+ T cells [114]. There are also other studies that describe similar trend. It was shown that HNSCC cells with the loss of NSD1 have reduced expression of genes responsible for the inflammatory response [44]. In another study, tumors are divided according to the degree of immune cell infiltration: TCIP-H and TCIP-L (high and low CD8+ T-cell inflamed phenotypes, respectively), considering that among other gene alterations the NSD1 gene was frequently mutated in TCIP-L tumors [115]. However, the association between infiltration phenotype of the tumors and survival was weak [115]. Since the relationship between the loss of NSD1 function and the formation of the tumor microenvironment has not been fully explored, we can only conclude that the presence of NSD1 mutation may be a factor to consider when choosing a patient therapy strategy, including immunotherapy. The biology and the value of these finding is currently not fully understood, and more research will be needed to better understand it, using patients treated with immune checkpoints, to see if NSD1 mutation status could be a biomarker predicting immune responses.
A positive correlation has been identified between the level of NSD2 and the presence of an immunosuppressive tumor microenvironment in prostate cancer [116]. One study predicting infiltration levels of immune cells based on analysis of the TCGA database found that patients with higher level NSD2 expression have a more immunosuppressive profile of tumor environment with the high level of Th2 and Treg cells and lower levels of Th1 cells, natural killer T (NKT) cells, and M1 macrophages. Increased expression of NSD2 also correlated with low levels of human leukocyte antigen (HLA) class I and class II transcripts, which would reduce the ability of a tumor to present tumor antigens to the immune system [116]. This study also showed that HLA transcription levels increased after the knockdown of NSD2/WHSC1 in a human prostate cell line, and also increased at the protein level after pharmacological inhibition of NSD2 [114]. These results suggest that inhibition of NSD2 increases the antigen presentation ability on the cell surface of prostate cancer cells (Fig. 4B).
Relationship between NSD proteins and the global DNA methylation landscape in cancer; implications for DNA damaging therapies
Although NSD proteins' primary function is to directly modify histones, they also can impact the methylation of DNA. KMTs, including SET domain-containing proteins, influence de novo DNA methylation during embryonic development by recruiting DNA methyltransferases (DNMTs) [117]. Mechanistically, areas of H3K36 methylation provide a recruitment signal for SETD2, which binds and directs DNMTs to methylate CpGs in target regions [118]. Additional mechanisms of cross-talk between the histone methylation machinery and DNA methylation, and roles for this cross-talk in regulating genome-wide epigenome landscapes, have been described [119, 120]. The direct interaction between the KMTs and DNMTs is reflected by their co-localization at specific promoter regions in cancer cells [121] to regulate CpG methylation. CpG methylation not only provides a distinct mechanism by which methyltransferases influence gene expression [122, 123], but also has other consequences, including reducing susceptibility to mutation [124]. NSD1 regulates DNA methylation in cancer and other diseases; for example, in Sotos syndrome, associated with inherited loss of NSD1 function, extensive areas of DNA hypomethylation are observed [114, 124–127]. In HNSCC, NSD1 mutation is significantly associated with the global DNA hypomethylation in gene promoters and reduction of H3K36me2 levels [63, 126]. Targeted disruption of NSD1 in HNSCC models, either associated with mutation or induced using CRISPR, led to global CpG hypomethylation and loss of H3K36me2 demonstrating that NSD1 promotes CpG methylation and regulates DNA methylome landscapes in HNSCC [44, 64]. Similarly, NSD1 loss promoted global DNA hypomethylation in lung squamous carcinoma (LUSC) [114]. It is interesting to see that the loss of H3K36me2 (an active mark) through the loss of NSD1 results in global DNA hypomethylation. This shows that there may be an additional layer of regulation involved that is evident by previous reports of different states of methylation of H3K36 marks [128, 129]. Furthermore, H3K36 methylation has also been implicated in transcriptional repression and other various regulatory processes such as alternative splicing, dosage compensation and DNA repair and recombination [14, 128].
DNMTs are recruited to the sites of double-stranded DNA breaks in the DNA repair pathways [130, 131]. As DNA hypomethylation is associated with greater sensitivity to some classes of DNA damaging therapies, the function of NSDs in enhancing DNA methylation may play a role in providing protection to chemotherapy and radiation therapy [130, 131]. Peri et al. first suggested that one reason for the association of NSD1 and NSD2 mutation with better prognosis in HNSCC may be the greater responsiveness of mutated tumors to radiation and chemotherapy [63]. It was also directly demonstrated that loss of NSD1 expression level increased sensitivity to cisplatin, carboplatin [62, 64], and radiation [64] in HNSCC [62]. Furthermore, it was also demonstrated that upregulated levels of NSD2 protein, but not NSD1 and NSD3, correlated with resistance to cisplatin treatment and preventing cisplatin-induced apoptosis in esophageal squamous cell carcinoma (ESCC) [132]. Knockdown of NSD2 with shRNA resulted in sensitivity to cisplatin in in vitro and in vivo models of ESCC [132] and in vitro in osteosarcoma cells [72]. Additionally, depletion of NSD2 increased sensitivity to commonly used chemotherapeutic drugs oxaliplatin and 5-fluorouracil (5-FU) in human colorectal cancer cell lines [70]. It has been suggested that NSD2 expression may be a useful predictive biomarker of sensitivity to different chemotherapy drugs such as docetaxel, 5-FU and oxaliplatin in gastric cancer [133].
NSD proteins as potential therapeutic targets
Numerous studies in the last 2 decades have implied that the NSDs and other KMTs are promising therapeutic targets for the treatment of many types of cancer [134]. However, the development of catalytic inhibitors of these proteins has been challenging, as the catalytic SET domain in the NSD family of proteins exists in an auto-inhibited conformation that lacks pockets for the binding of small molecules [135].
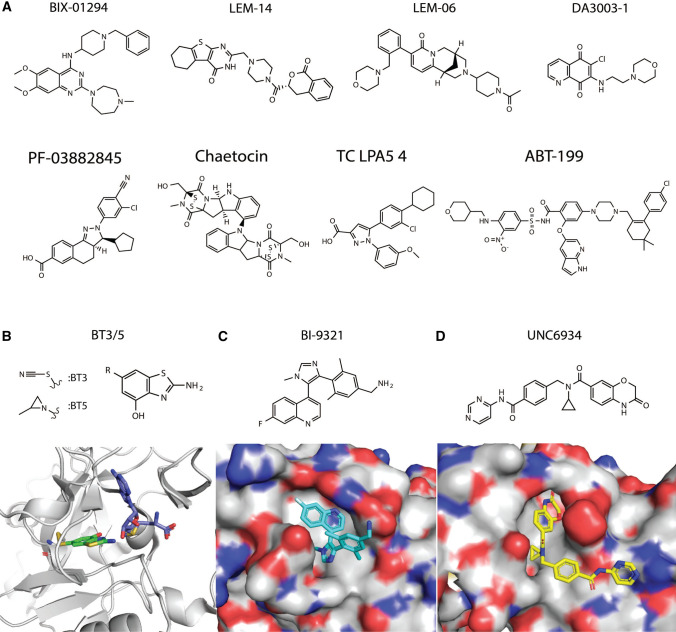
Despite these challenges, several compounds have been reported to inhibit the NSD family of KMTs (Fig. 5A). The first described inhibitor, BIX-01294 [136], was highly active against the histone KMT G9a, but also bound and inhibited NSD1, NSD2 and NSD3 (with the highest inhibition of NSD2) [137]. Two compounds, LEM-06 and LEM-14, have been reported as NSD2-specific inhibitors on the basis of enzyme assays using purified NSD proteins, but their extremely weak potency (IC50 890 µM for LEM-06 and 132 µM for LEM-14) stands as a barrier to testing activity in cells [138, 139]. Another study identified five compounds that inhibit NSD2 (DA3003-1, PF03882845, chaetocin, TC, LPA5, and ABT-199) using a HotSpot methyltransferase assay; however, these lacked specificity to NSD2 [140], as they broadly inhibited many other methyltransferases.
Fig. 5.
Chemical diversity of compounds reported to show inhibition of NSD family proteins. A Numerous diverse chemical scaffolds have been reported to show broad spectrum activity against NSD family members. B Covalent inhibitors BT3 and BT5; a crystal structure of BT3 (green) was solved in complex with NSD1 (gray) and SAM (purple) (PDB ID 6KQQ). C Inhibitor BI-9321 (cyan) was solved in complex with NSD3-PWWP1 (PDB ID 6G2O). D Inhibitor UNC6934 (yellow) was solved in complex with NSD2-PWWP1 (PDB ID 6XCG)
The NSD2 inhibitor PTD2 is a norleucine-containing peptide derived from histone H4 sequences [141]; this peptide exhibited micromolar affinity towards NSD2/WHSC1 in radiometric and SPR (surface plasmon resonance)-binding assays but has not advanced into further testing. Small molecule BT5 (Fig. 5B) was reported recently as an NSD1 inhibitor that decreases H3K36me2 level in cells, and exhibits high inhibitory activity specifically in cells driven by NUP98-NSD1 gene fusion [135]. Despite good selectivity in binding to the SET domain of the NUP98-NSD1 gene fusion, BT5 has a narrow application in the clinic, as it is only possible to use it against NUP98-NSD1 driven leukemia.
In contrast to inhibitors that bind the catalytic SET domain, another class of NSD inhibitors targets NSD family members by binding to the PWWP domain, which mediates interaction with nucleosomal DNA adjacent to H3K36 [142, 143]. For example, BI-9321 (Fig. 5C) inhibits the methyl-lysine binding site of the PWWP1 domain of NSD3 with sub-micromolar in vitro activity and cellular target engagement at 1 µM, as determined by FRAP, cellular fractionation and nanoBRET [144]. As a result, BI-9321 downregulates MYC mRNA and reduces the proliferation of acute myeloid leukemia (AML) cell lines. Another compound, UNC6934 (Fig. 5D), binds the NSD2 PWWP1 domain, as shown by SPR, thermal shift assay and cell fractionation assays, but does not affect cell viability or expression of select target genes [145].
In terms of the clinical utility of effective NSD-targeted therapy, one clear and immediate application would be to serve as a sensitizer to DNA damaging therapies, including platins and radiation. Given its additional roles in signaling, inhibition of NSDs may be useful in other combination settings. For instance, it has been reported that NSD2 depletion increases sensitivity to EGFR inhibitor gefitinib in triple-negative breast cancer (TNBC) [78]; the combination of an NSD inhibitor with EGFR inhibitors would be of interest to investigate. As another example, genetic or chemical depletion of BRD4 leads to differentiation and inhibition of proliferation of cells with NSD3-NUT-expression [94], and NSD3-depleted LUSC patient-derived xenografts were more sensitive to treatment with the BET (bromodomain and extra terminal protein family) inhibitor AZD5153 [89]. Hence, NSD/BRD4 inhibitor combinations would also be of interest to assess. Thus, there are currently two main strategies for the development of NSD inhibitors: targeting the catalytic domain of the NSD proteins and targeting domains that mediates the interaction of NSD proteins and nucleosomal DNA. Both strategies are not a universal solution to this challenge today and require further development, as inhibitors of epigenetic regulators remain an attractive target for improving therapeutic options.
Discussion and concluding remarks
In summary, dysregulation of the NSD family proteins occurs in multiple solid tumors, driven by changes that include amplification, loss-of-function mutation, and gene fusion. NSD proteins actively modulate tumor formation and progression in many tumor types, although there is evidence for differing functions depending on the tumor type. Either dysregulations of NSD1/2/3 expression or mutations of these genes correlate with PFS, RFS, and OS, again dependent on tumor type. Differences in prognosis associated with altered NSDs may, in part, reflect the therapies most commonly used for different tumor types; for instance, the fact that loss-of-function mutations in NSD1 and NSD2 correlate with dramatically improved survival in laryngeal cancer patients may reflect the common treatment of these patients with cisplatin and radiation therapy [63]. Hence, although the expression of NSD proteins may be useful as predictive biomarkers of response to therapy, it will be important to establish this for specific tumor types, in the context of discrete treatment regimens [77, 132, 133]. There is clearly a need for additional extensive clinical studies to explore the role of NSD proteins as potential biomarkers in the clinic.
Despite a similar protein structure and a common H3K36 methylation function, it is not clear whether all three NSD proteins utilize identical or similar signaling effectors. Genetic evidence in any biological system on whether the NSDs have a redundant function and whether they can compensate for each other is lacking, and this merits further investigations. At present, mechanisms that have been identified as pertinent to individual members of the family include activation of NF-κB and upregulation of NF-κB transcriptional targets [73, 111], regulation of the Wnt/B-catenin pathway [60, 68], direct regulation of NEK7 and others cell cycle-related genes [65, 93], and other mechanisms summarized above. To some extent, this likely reflects the currently incomplete degree of investigation of all three members of the family in parallel studies of signaling activity. Similarly, although the role of NSD proteins as players in the immune response and their participation in the formation of the immune microenvironment of tumors has been reported [114, 116], knowledge of this function remains limited. This information will be essential to guide ongoing efforts to develop targeted inhibitors of NSD proteins, given recent developments in this area [135, 145] suggest effective compounds may be within reach. Overall, much work remains to be done.
Acknowledgements
Author contributions
IT, RP and IB conducted the literature review and wrote the first draft. IT, RP and SM prepared the figures and tables. JK, JY, EI, EG, and YB conducted reviewing proofreading and content processing.
Funding
NIH R01 CA218802 grant, and the NCI NIH Core Grant P30 CA060553 to the Robert H Lurie Comprehensive Cancer Center at Northwestern University, Translational Bridge Award from Northwestern University (to Y.B.). This study was also partially supported by 1P50 DE030707-01 and USAMRAA W81XWH2110487 (to E.G.), NIH/NCI R01CA227918 (to J.Y.) Department of Defense grants W81XWH-17-1-0405 and W81XWH-17-1-0578 (to J.Y.), R01DE027809 to E. I., and Prostate Cancer Foundation 2017CHAL2008 (to J.Y.); J.K., and E.G. were in part supported by the NCI Core Grant P30 CA006927 (to Fox Chase Cancer Center).
Data availability
Not applicable.
Declarations
Conflict of interest
The author declares he has no conflict of interest.
Ethical approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol. 2005;6(11):838–849. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- 2.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403(6765):41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 3.Husmann D, Gozani O. Histone lysine methyltransferases in biology and disease. Nat Struct Mol Biol. 2019;26(10):880–889. doi: 10.1038/s41594-019-0298-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zaghi M, Broccoli V, Sessa A. H3K36 methylation in neural development and associated diseases. Front Genet. 2019;10:1291. doi: 10.3389/fgene.2019.01291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim A, Kiefer CM, Dean A. Distinctive signatures of histone methylation in transcribed coding and noncoding human beta-globin sequences. Mol Cell Biol. 2007;27(4):1271–1279. doi: 10.1128/MCB.01684-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bannister AJ, et al. Spatial distribution of di- and tri-methyl lysine 36 of histone H3 at active genes. J Biol Chem. 2005;280(18):17732–17736. doi: 10.1074/jbc.M500796200. [DOI] [PubMed] [Google Scholar]
- 7.Fang D, et al. The histone H3.3K36M mutation reprograms the epigenome of chondroblastomas. Science. 2016;352(6291):1344–1348. doi: 10.1126/science.aae0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li J, Ahn JH, Wang GG. Understanding histone H3 lysine 36 methylation and its deregulation in disease. Cell Mol Life Sci. 2019;76(15):2899–2916. doi: 10.1007/s00018-019-03144-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Popovic R, et al. Histone methyltransferase MMSET/NSD2 alters EZH2 binding and reprograms the myeloma epigenome through global and focal changes in H3K36 and H3K27 methylation. PLoS Genet. 2014;10(9):e1004566. doi: 10.1371/journal.pgen.1004566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rogawski DS, Grembecka J, Cierpicki T. H3K36 methyltransferases as cancer drug targets: rationale and perspectives for inhibitor development. Future Med Chem. 2016;8(13):1589–1607. doi: 10.4155/fmc-2016-0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsukada Y-I, et al. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439(7078):811–816. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- 12.Hyun K, et al. Writing, erasing and reading histone lysine methylations. Exp Mol Med. 2017;49(4):e324–e324. doi: 10.1038/emm.2017.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Whetstine JR, et al. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell. 2006;125(3):467–481. doi: 10.1016/j.cell.2006.03.028. [DOI] [PubMed] [Google Scholar]
- 14.Wagner EJ, Carpenter PB. Understanding the language of Lys36 methylation at histone H3. Nat Rev Mol Cell Biol. 2012;13(2):115–126. doi: 10.1038/nrm3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bennett RL, et al. The role of nuclear receptor-binding SET domain family histone lysine methyltransferases in cancer. Cold Spring Harb Perspect Med. 2017;7(6):a026708. doi: 10.1101/cshperspect.a026708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rayasam GV. NSD1 is essential for early post-implantation development and has a catalytically active SET domain. EMBO J. 2003;22(12):3153–3163. doi: 10.1093/emboj/cdg288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shirane K, et al. NSD1-deposited H3K36me2 directs de novo methylation in the mouse male germline and counteracts Polycomb-associated silencing. Nat Genet. 2020;52(10):1088–1098. doi: 10.1038/s41588-020-0689-z. [DOI] [PubMed] [Google Scholar]
- 18.Kurotaki N, et al. Haploinsufficiency of NSD1 causes Sotos syndrome. Nat Genet. 2002;30(4):365–366. doi: 10.1038/ng863. [DOI] [PubMed] [Google Scholar]
- 19.Baujat G, et al. Paradoxical NSD1 mutations in Beckwith–Wiedemann syndrome and 11p15 anomalies in Sotos syndrome. Am J Hum Genet. 2004;74(4):715–720. doi: 10.1086/383093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nimura K, et al. A histone H3 lysine 36 trimethyltransferase links Nkx2-5 to Wolf–Hirschhorn syndrome. Nature. 2009;460(7252):287–291. doi: 10.1038/nature08086. [DOI] [PubMed] [Google Scholar]
- 21.Campos-Sanchez E, et al. Wolf–Hirschhorn syndrome candidate 1 is necessary for correct hematopoietic and B cell development. Cell Rep. 2017;19(8):1586–1601. doi: 10.1016/j.celrep.2017.04.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stec I, et al. WHSC1, a 90 kb SET domain-containing gene, expressed in early development and homologous to a Drosophila dysmorphy gene maps in the Wolf–Hirschhorn syndrome critical region and is fused to IgH in t(4;14) multiple myeloma. Hum Mol Genet. 1998;7(7):1071–1082. doi: 10.1093/hmg/7.7.1071. [DOI] [PubMed] [Google Scholar]
- 23.Adams MS, Gammill LS, Bronner-Fraser M. Discovery of transcription factors and other candidate regulators of neural crest development. Dev Dyn. 2008;237(4):1021–1033. doi: 10.1002/dvdy.21513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jacques-Fricke BT, Gammill LS. Neural crest specification and migration independently require NSD3-related lysine methyltransferase activity. Mol Biol Cell. 2014;25(25):4174–4186. doi: 10.1091/mbc.e13-12-0744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jacques-Fricke BT, et al. Profiling NSD3-dependent neural crest gene expression reveals known and novel candidate regulatory factors. Dev Biol. 2021;475:118–130. doi: 10.1016/j.ydbio.2021.02.015. [DOI] [PubMed] [Google Scholar]
- 26.Jaju RJ, et al. A novel gene, NSD1, is fused to NUP98 in the t(5;11)(q35;p15.5) in de novo childhood acute myeloid leukemia. Blood. 2001;98(4):1264–1267. doi: 10.1182/blood.V98.4.1264. [DOI] [PubMed] [Google Scholar]
- 27.Panarello C, Rosanda C, Morerio C. Cryptic translocation t(5;11)(q35;p15.5) with involvement of the NSD1 and NUP98 genes without 5q deletion in childhood acute myeloid leukemia. Genes Chromosomes Cancer. 2002;35(3):277–281. doi: 10.1002/gcc.10119. [DOI] [PubMed] [Google Scholar]
- 28.Angrand PO, et al. NSD3, a new SET domain-containing gene, maps to 8p12 and is amplified in human breast cancer cell lines. Genomics. 2001;74(1):79–88. doi: 10.1006/geno.2001.6524. [DOI] [PubMed] [Google Scholar]
- 29.Saloura V, et al. The role of protein methyltransferases as potential novel therapeutic targets in squamous cell carcinoma of the head and neck. Oral Oncol. 2018;81:100–108. doi: 10.1016/j.oraloncology.2018.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lucio-Eterovic AK, et al. Role for the nuclear receptor-binding SET domain protein 1 (NSD1) methyltransferase in coordinating lysine 36 methylation at histone 3 with RNA polymerase II function. Proc Natl Acad Sci. 2010;107(39):16952–16957. doi: 10.1073/pnas.1002653107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He C, et al. The methyltransferase NSD3 has chromatin-binding motifs, PHD5-C5HCH, that are distinct from other NSD (nuclear receptor SET domain) family members in their histone H3 recognition. J Biol Chem. 2013;288(7):4692–4703. doi: 10.1074/jbc.M112.426148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qiao Q, et al. The structure of NSD1 reveals an autoregulatory mechanism underlying histone H3K36 methylation. J Biol Chem. 2011;286(10):8361–8368. doi: 10.1074/jbc.M110.204115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vougiouklakis T, et al. The NSD family of protein methyltransferases in human cancer. Epigenomics. 2015;7(5):863–874. doi: 10.2217/epi.15.32. [DOI] [PubMed] [Google Scholar]
- 34.Garlisi CG, et al. A unique mRNA initiated within a middle intron of WHSC1/MMSET encodes a DNA binding protein that suppresses human IL-5 transcription. Am J Respir Cell Mol Biol. 2001;24(1):90–98. doi: 10.1165/ajrcmb.24.1.4224. [DOI] [PubMed] [Google Scholar]
- 35.Woo Park J, et al. RE-IIBP methylates H3K79 and induces MEIS1-mediated apoptosis via H2BK120 ubiquitination by RNF20. Sci Rep. 2015;5:12485. doi: 10.1038/srep12485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim SM, et al. Characterization of a novel WHSC1-associated SET domain protein with H3K4 and H3K27 methyltransferase activity. Biochem Biophys Res Commun. 2006;345(1):318–323. doi: 10.1016/j.bbrc.2006.04.095. [DOI] [PubMed] [Google Scholar]
- 37.Graham SE, Tweedy SE, Carlson HA. Dynamic behavior of the post-SET loop region of NSD1: implications for histone binding and drug development. Protein Sci. 2016;25(5):1021–1029. doi: 10.1002/pro.2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morishita M, Mevius D, di Luccio E. In vitro histone lysine methylation by NSD1, NSD2/MMSET/WHSC1 and NSD3/WHSC1L. BMC Struct Biol. 2014;14:25. doi: 10.1186/s12900-014-0025-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li W, et al. Molecular basis of nucleosomal H3K36 methylation by NSD methyltransferases. Nature. 2020;590:498–503. doi: 10.1038/s41586-020-03069-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Han D, et al. Lysine methylation of transcription factors in cancer. Cell Death Dis. 2019;10(4):290. doi: 10.1038/s41419-019-1524-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21(3):381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmitges FW, et al. Histone methylation by PRC2 is inhibited by active chromatin marks. Mol Cell. 2011;42(3):330–341. doi: 10.1016/j.molcel.2011.03.025. [DOI] [PubMed] [Google Scholar]
- 43.Yuan W, et al. H3K36 methylation antagonizes PRC2-mediated H3K27 methylation. J Biol Chem. 2011;286(10):7983–7989. doi: 10.1074/jbc.M110.194027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Farhangdoost N, et al. Chromatin dysregulation associated with NSD1 mutation in head and neck squamous cell carcinoma. Cell Rep. 2021;34(8):108769. doi: 10.1016/j.celrep.2021.108769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Streubel G, et al. The H3K36me2 methyltransferase Nsd1 demarcates PRC2-mediated H3K27me2 and H3K27me3 domains in embryonic stem cells. Mol Cell. 2018;70(2):371–379.e5. doi: 10.1016/j.molcel.2018.02.027. [DOI] [PubMed] [Google Scholar]
- 46.Woo H, et al. Modulation of gene expression dynamics by co-transcriptional histone methylations. Exp Mol Med. 2017;49(4):e326–e326. doi: 10.1038/emm.2017.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carrozza MJ, et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123(4):581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 48.Keogh MC, et al. Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell. 2005;123(4):593–605. doi: 10.1016/j.cell.2005.10.025. [DOI] [PubMed] [Google Scholar]
- 49.Li B, et al. Infrequently transcribed long genes depend on the Set2/Rpd3S pathway for accurate transcription. Genes Dev. 2007;21(11):1422–1430. doi: 10.1101/gad.1539307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fang Y, et al. The H3K36me2 methyltransferase NSD1 modulates H3K27ac at active enhancers to safeguard gene expression. Nucleic Acids Res. 2021;49(11):6281–6295. doi: 10.1093/nar/gkab473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fnu S, et al. Methylation of histone H3 lysine 36 enhances DNA repair by nonhomologous end-joining. Proc Natl Acad Sci U S A. 2011;108(2):540–545. doi: 10.1073/pnas.1013571108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.De Krijger I, et al. H3K36 dimethylation by MMSET promotes classical non-homologous end-joining at unprotected telomeres. Oncogene. 2020;39(25):4814–4827. doi: 10.1038/s41388-020-1334-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hajdu I, et al. Wolf–Hirschhorn syndrome candidate 1 is involved in the cellular response to DNA damage. Proc Natl Acad Sci U S A. 2011;108(32):13130–13134. doi: 10.1073/pnas.1110081108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang J, et al. PTEN methylation by NSD2 controls cellular sensitivity to DNA damage. Cancer Discov. 2019;9(9):1306–1323. doi: 10.1158/2159-8290.CD-18-0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mah LJ, El-Osta A, Karagiannis TC. γH2AX: a sensitive molecular marker of DNA damage and repair. Leukemia. 2010;24(4):679–686. doi: 10.1038/leu.2010.6. [DOI] [PubMed] [Google Scholar]
- 56.Chowdhury D, et al. A PP4-phosphatase complex dephosphorylates gamma-H2AX generated during DNA replication. Mol Cell. 2008;31(1):33–46. doi: 10.1016/j.molcel.2008.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee DH, et al. A PP4 phosphatase complex dephosphorylates RPA2 to facilitate DNA repair via homologous recombination. Nat Struct Mol Biol. 2010;17(3):365–372. doi: 10.1038/nsmb.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pei H, et al. MMSET regulates histone H4K20 methylation and 53BP1 accumulation at DNA damage sites. Nature. 2011;470(7332):124–128. doi: 10.1038/nature09658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang X, et al. Identification and characterization of a novel androgen receptor coregulator ARA267-alpha in prostate cancer cells. J Biol Chem. 2001;276(44):40417–40423. doi: 10.1074/jbc.M104765200. [DOI] [PubMed] [Google Scholar]
- 60.Toyokawa G, et al. Histone lysine methyltransferase Wolf–Hirschhorn syndrome candidate 1 is involved in human carcinogenesis through regulation of the Wnt pathway. Neoplasia. 2011;13(10):887–898. doi: 10.1593/neo.11048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gonzalez-Pecchi V, et al. NSD3S stabilizes MYC through hindering its interaction with FBXW7. J Mol Cell Biol. 2020;12(6):438–447. doi: 10.1093/jmcb/mjz098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pan C, et al. NSD1 mutations by HPV status in head and neck cancer: differences in survival and response to DNA-damaging agents. Cancers Head Neck. 2019;4(1):1–13. doi: 10.1186/s41199-019-0042-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Peri S, et al. NSD1- and NSD2-damaging mutations define a subset of laryngeal tumors with favorable prognosis. Nat Commun. 2017;8(1):1–10. doi: 10.1038/s41467-017-01877-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bui N, et al. Disruption of NSD1 in head and neck cancer promotes favorable chemotherapeutic responses linked to hypomethylation. Mol Cancer Ther. 2018;17(7):1585–1594. doi: 10.1158/1535-7163.MCT-17-0937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Saloura V, et al. WHSC1 Promotes oncogenesis through regulation of NIMA-related kinase-7 in squamous cell carcinoma of the head and neck. Mol Cancer Res. 2015;13(2):293–304. doi: 10.1158/1541-7786.MCR-14-0292-T. [DOI] [PubMed] [Google Scholar]
- 66.Gameiro SF, et al. Low expression of NSD1, NSD2, and NSD3 define a subset of human papillomavirus-positive oral squamous carcinomas with unfavorable prognosis. Infect Agents Cancer. 2021;16(1):13. doi: 10.1186/s13027-021-00347-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ettel M, et al. Expression and prognostic value of NSD1 and SETD2 in pancreatic ductal adenocarcinoma and its precursor lesions. Pathology. 2019;51(4):392–398. doi: 10.1016/j.pathol.2019.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang S, et al. CRISPR/Cas9-mediated knockout of NSD1 suppresses the hepatocellular carcinoma development via the NSD1/H3/Wnt10b signaling pathway. J Exp Clin Cancer Res. 2019;38(1):1–14. doi: 10.1186/s13046-019-1462-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bianco-Miotto T, et al. Global levels of specific histone modifications and an epigenetic gene signature predict prostate cancer progression and development. Cancer Epidemiol Biomark Prev. 2010;19(10):2611–2622. doi: 10.1158/1055-9965.EPI-10-0555. [DOI] [PubMed] [Google Scholar]
- 70.Wang Y, et al. Histone methyltransferase WHSC1 inhibits colorectal cancer cell apoptosis via targeting anti-apoptotic BCL2. Cell Death Discov. 2021;7(1):1–9. doi: 10.1038/s41420-021-00416-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhao L-H, et al. Identification of histone methyltransferase NSD2 as an important oncogenic gene in colorectal cancer. Cell Death Dis. 2021;12(11):974. doi: 10.1038/s41419-021-04267-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.He C, et al. Histone methyltransferase NSD2 regulates apoptosis and chemosensitivity in osteosarcoma. Cell Death Dis. 2019;10(2):1–13. doi: 10.1038/s41419-019-1347-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yang P, et al. Histone methyltransferase NSD2/MMSET mediates constitutive NF-B signaling for cancer cell proliferation, survival, and tumor growth via a feed-forward loop. Mol Cell Biol. 2012;32(15):3121–3131. doi: 10.1128/MCB.00204-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ezponda T, et al. The histone methyltransferase MMSET/WHSC1 activates TWIST1 to promote an epithelial–mesenchymal transition and invasive properties of prostate cancer. Oncogene. 2013;32(23):2882–2890. doi: 10.1038/onc.2012.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.García-Carpizo V, et al. NSD2 contributes to oncogenic RAS-driven transcription in lung cancer cells through long-range epigenetic activation. Sci Rep. 2016;6(1):32952. doi: 10.1038/srep32952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sengupta D, et al. NSD2 dimethylation at H3K36 promotes lung adenocarcinoma pathogenesis. Mol Cell. 2021;81(21):4481–4492.e9. doi: 10.1016/j.molcel.2021.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hudlebusch HR, et al. The histone methyltransferase and putative oncoprotein MMSET is overexpressed in a large variety of human tumors. Clin Cancer Res. 2011;17(9):2919–2933. doi: 10.1158/1078-0432.CCR-10-1302. [DOI] [PubMed] [Google Scholar]
- 78.Wang JJ, et al. Histone methyltransferase NSD2 mediates the survival and invasion of triple-negative breast cancer cells via stimulating ADAM9-EGFR-AKT signaling. Acta Pharmacol Sin. 2019;40(8):1067–1075. doi: 10.1038/s41401-018-0199-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hudlebusch HR, et al. MMSET is highly expressed and associated with aggressiveness in neuroblastoma. Cancer Res. 2011;71(12):4226–4235. doi: 10.1158/0008-5472.CAN-10-3810. [DOI] [PubMed] [Google Scholar]
- 80.Han X, et al. NSD2 promotes renal cancer progression through stimulating Akt/Erk signaling. Cancer Manag Res. 2020;12:375–383. doi: 10.2147/CMAR.S222673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tonon G, et al. High-resolution genomic profiles of human lung cancer. Proc Natl Acad Sci U S A. 2005;102(27):9625–9630. doi: 10.1073/pnas.0504126102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kassambara A, Klein B, Moreaux J. MMSET is overexpressed in cancers: link with tumor aggressiveness. Biochem Biophys Res Commun. 2009;379(4):840–845. doi: 10.1016/j.bbrc.2008.12.093. [DOI] [PubMed] [Google Scholar]
- 83.Berdasco M, et al. Epigenetic inactivation of the Sotos overgrowth syndrome gene histone methyltransferase NSD1 in human neuroblastoma and glioma. Proc Natl Acad Sci U S A. 2009;106(51):21830–21835. doi: 10.1073/pnas.0906831106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Su X, et al. NSD1 inactivation and SETD2 mutation drive a convergence toward loss of function of H3K36 writers in clear cell renal cell carcinomas. Cancer Res. 2017;77(18):4835–4845. doi: 10.1158/0008-5472.CAN-17-0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Su X, et al. NSD1 inactivation and SETD2 mutation drive a convergence toward loss of function of H3K36 writers in clear cell renal cell carcinomas. Can Res. 2017;77(18):4835–4845. doi: 10.1158/0008-5472.CAN-17-0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yang ZQ, et al. Transforming properties of 8p11-12 amplified genes in human breast cancer. Cancer Res. 2010;70(21):8487–8497. doi: 10.1158/0008-5472.CAN-10-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Irish JC, et al. Amplification of WHSC1L1 regulates expression and estrogen-independent activation of ERα in SUM-44 breast cancer cells and is associated with ERα over-expression in breast cancer. Mol Oncol. 2016;10(6):850–865. doi: 10.1016/j.molonc.2016.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Saloura V, et al. WHSC1L1 drives cell cycle progression through transcriptional regulation of CDC6 and CDK2 in squamous cell carcinoma of the head and neck. Oncotarget. 2016;7(27):42527–42538. doi: 10.18632/oncotarget.9897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yuan G, et al. Elevated NSD3 histone methylation activity drives squamous cell lung cancer. Nature. 2021;590(7846):504–508. doi: 10.1038/s41586-020-03170-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yi L, et al. Downregulation of NSD3 (WHSC1L1) inhibits cell proliferation and migration via ERK1/2 deactivation and decreasing CAPG expression in colorectal cancer cells. Onco Targets Ther. 2019;12:3933–3943. doi: 10.2147/OTT.S191732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jeong GY, et al. NSD3-induced methylation of H3K36 activates NOTCH signaling to drive breast tumor initiation and metastatic progression. Cancer Res. 2021;81(1):77–90. doi: 10.1158/0008-5472.CAN-20-0360. [DOI] [PubMed] [Google Scholar]
- 92.D'Afonseca V, et al. Computational analyses on genetic alterations in the NSD genes family and the implications for colorectal cancer development. Ecancermedicalscience. 2020;14:1001. doi: 10.3332/ecancer.2020.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kang D, et al. The histone methyltransferase Wolf–Hirschhorn syndrome candidate 1-like 1 (WHSC1L1) is involved in human carcinogenesis. Genes Chromosomes Cancer. 2013;52(2):126–139. doi: 10.1002/gcc.22012. [DOI] [PubMed] [Google Scholar]
- 94.French CA, et al. NSD3-NUT fusion oncoprotein in NUT midline carcinoma: implications for a novel oncogenic mechanism. Cancer Discov. 2014;4(8):928–941. doi: 10.1158/2159-8290.CD-14-0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bauer DE, et al. Clinicopathologic features and long-term outcomes of NUT midline carcinoma. Clin Cancer Res. 2012;18(20):5773–5779. doi: 10.1158/1078-0432.CCR-12-1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Agaimy A, et al. Misleading germ cell phenotype in pulmonary NUT carcinoma harboring the ZNF532-NUTM1 fusion. Am J Surg Pathol. 2021;46:281–288. doi: 10.1097/PAS.0000000000001774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Stevens TM, et al. NUTM1-rearranged neoplasia: a multi-institution experience yields novel fusion partners and expands the histologic spectrum. Mod Pathol. 2019;32(6):764–773. doi: 10.1038/s41379-019-0206-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chau NG, et al. An anatomical site and genetic-based prognostic model for patients with nuclear protein in testis (NUT) midline carcinoma: analysis of 124 patients. JNCI Cancer Spectr. 2020;4(2):pkz094. doi: 10.1093/jncics/pkz094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhao X, et al. Downregulation of MMSET impairs breast cancer proliferation and metastasis through inhibiting Wnt/β-catenin signaling. Onco Targets Ther. 2019;12:1965–1977. doi: 10.2147/OTT.S196430. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 100.Huang Z, et al. NSD2 is recruited through its PHD domain to oncogenic gene loci to drive multiple myeloma. Cancer Res. 2013;73(20):6277–6288. doi: 10.1158/0008-5472.CAN-13-1000. [DOI] [PubMed] [Google Scholar]
- 101.Kang HB, et al. The histone methyltransferase, NSD2, enhances androgen receptor-mediated transcription. FEBS Lett. 2009;583(12):1880–1886. doi: 10.1016/j.febslet.2009.05.038. [DOI] [PubMed] [Google Scholar]
- 102.Sun Y, et al. Elevated expression of nuclear receptor-binding SET domain 3 promotes pancreatic cancer cell growth. Cell Death Dis. 2021;12(10):913. doi: 10.1038/s41419-021-04205-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mahmood SF, et al. PPAPDC1B and WHSC1L1 are common drivers of the 8p11-12 amplicon, not only in breast tumors but also in pancreatic adenocarcinomas and lung tumors. Am J Pathol. 2013;183(5):1634–1644. doi: 10.1016/j.ajpath.2013.07.028. [DOI] [PubMed] [Google Scholar]
- 104.Rahman S, et al. The Brd4 extraterminal domain confers transcription activation independent of pTEFb by recruiting multiple proteins, including NSD3. Mol Cell Biol. 2011;31(13):2641–2652. doi: 10.1128/MCB.01341-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Saloura V, et al. WHSC1L1-mediated EGFR mono-methylation enhances the cytoplasmic and nuclear oncogenic activity of EGFR in head and neck cancer. Sci Rep. 2017;7:40664. doi: 10.1038/srep40664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Song D, et al. NSD2 promotes tumor angiogenesis through methylating and activating STAT3 protein. Oncogene. 2021;40(16):2952–2967. doi: 10.1038/s41388-021-01747-z. [DOI] [PubMed] [Google Scholar]
- 107.Liu Z, et al. Silencing of histone methyltransferase NSD3 reduces cell viability in osteosarcoma with induction of apoptosis. Oncol Rep. 2017;38(5):2796–2802. doi: 10.3892/or.2017.5936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Aytes A, et al. NSD2 is a conserved driver of metastatic prostate cancer progression. Nat Commun. 2018;9(1):5201. doi: 10.1038/s41467-018-07511-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yuan S, et al. Global regulation of the histone mark H3K36me2 underlies epithelial plasticity and metastatic progression. Cancer Discov. 2020;10(6):854–871. doi: 10.1158/2159-8290.CD-19-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Han X, et al. Knockdown of NSD2 suppresses renal cell carcinoma metastasis by inhibiting epithelial–mesenchymal transition. Int J Med Sci. 2019;16(10):1404–1411. doi: 10.7150/ijms.36128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lu T, et al. Regulation of NF-B by NSD1/FBXL11-dependent reversible lysine methylation of p65. Proc Natl Acad Sci. 2010;107(1):46–51. doi: 10.1073/pnas.0912493107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kudithipudi S, et al. Substrate specificity analysis and novel substrates of the protein lysine methyltransferase NSD1. Chem Biol. 2014;21(2):226–237. doi: 10.1016/j.chembiol.2013.10.016. [DOI] [PubMed] [Google Scholar]
- 113.Murciano-Goroff YR, Warner AB, Wolchok JD. The future of cancer immunotherapy: microenvironment-targeting combinations. Cell Res. 2020;30(6):507–519. doi: 10.1038/s41422-020-0337-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Brennan K, et al. NSD1 inactivation defines an immune cold, DNA hypomethylated subtype in squamous cell carcinoma. Sci Rep. 2017;7(1):1–12. doi: 10.1038/s41598-017-17298-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Saloura V, et al. Immune profiles in primary squamous cell carcinoma of the head and neck. Oral Oncol. 2019;96:77–88. doi: 10.1016/j.oraloncology.2019.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Want MY, et al. WHSC1/NSD2 regulates immune infiltration in prostate cancer. J Immunother Cancer. 2021;9(2):e001374. doi: 10.1136/jitc-2020-001374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet. 2009;10(5):295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- 118.Baubec T, et al. Genomic profiling of DNA methyltransferases reveals a role for DNMT3B in genic methylation. Nature. 2015;520(7546):243–247. doi: 10.1038/nature14176. [DOI] [PubMed] [Google Scholar]
- 119.Pangeni RP, et al. G9a regulates tumorigenicity and stemness through genome-wide DNA methylation reprogramming in non-small cell lung cancer. Clin Epigenet. 2020;12(1):88. doi: 10.1186/s13148-020-00879-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhang K, et al. Targeting histone methyltransferase G9a inhibits growth and Wnt signaling pathway by epigenetically regulating HP1alpha and APC2 gene expression in non-small cell lung cancer. Mol Cancer. 2018;17(1):153. doi: 10.1186/s12943-018-0896-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Li H, et al. The histone methyltransferase SETDB1 and the DNA methyltransferase DNMT3A interact directly and localize to promoters silenced in cancer cells. J Biol Chem. 2006;281(28):19489–19500. doi: 10.1074/jbc.M513249200. [DOI] [PubMed] [Google Scholar]
- 122.Castillo-Aguilera O, et al. DNA methylation targeting: the DNMT/HMT crosstalk challenge. Biomolecules. 2017;7(1):3. doi: 10.3390/biom7010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Neri F, et al. Intragenic DNA methylation prevents spurious transcription initiation. Nature. 2017;543(7643):72–77. doi: 10.1038/nature21373. [DOI] [PubMed] [Google Scholar]
- 124.Lee ST, Wiemels JL. Genome-wide CpG island methylation and intergenic demethylation propensities vary among different tumor sites. Nucleic Acids Res. 2016;44(3):1105–1117. doi: 10.1093/nar/gkv1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Choufani S, et al. NSD1 mutations generate a genome-wide DNA methylation signature. Nat Commun. 2015;6:10207. doi: 10.1038/ncomms10207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Cancer Genome Atlas Network Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517(7536):576–582. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Brennan K, et al. Identification of an atypical etiological head and neck squamous carcinoma subtype featuring the CpG island methylator phenotype. EBioMedicine. 2017;17:223–236. doi: 10.1016/j.ebiom.2017.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Suzuki S, Murakami Y, Takahata S. H3K36 methylation state and associated silencing mechanisms. Transcription. 2017;8(1):26–31. doi: 10.1080/21541264.2016.1246076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.DiFiore JV, et al. Unique and shared roles for histone H3K36 methylation states in transcription regulation functions. Cell Rep. 2020;31(10):107751. doi: 10.1016/j.celrep.2020.107751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.O'Hagan HM, Mohammad HP, Baylin SB. Double strand breaks can initiate gene silencing and SIRT1-dependent onset of DNA methylation in an exogenous promoter CpG island. PLoS Genet. 2008;4(8):e1000155. doi: 10.1371/journal.pgen.1000155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Mortusewicz O, et al. Recruitment of DNA methyltransferase I to DNA repair sites. Proc Natl Acad Sci U S A. 2005;102(25):8905–8909. doi: 10.1073/pnas.0501034102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Xue W, et al. Long non-coding RNAs MACC1-AS1 and FOXD2-AS1 mediate NSD2-induced cisplatin resistance in esophageal squamous cell carcinoma. Mol Ther Nucleic Acids. 2021;23:592–602. doi: 10.1016/j.omtn.2020.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wei J, et al. Differential effect of MMSET mRNA levels on survival to first-line FOLFOX and second-line docetaxel in gastric cancer. Br J Cancer. 2014;110(11):2662–2668. doi: 10.1038/bjc.2014.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150(1):12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 135.Huang H, et al. Covalent inhibition of NSD1 histone methyltransferase. Nat Chem Biol. 2020;16(12):1403–1410. doi: 10.1038/s41589-020-0626-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kubicek S, et al. Reversal of H3K9me2 by a small-molecule inhibitor for the G9a histone methyltransferase. Mol Cell. 2007;25(3):473–481. doi: 10.1016/j.molcel.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 137.Morishita M, et al. BIX-01294 inhibits oncoproteins NSD1, NSD2 and NSD3. Med Chem Res. 2017;26(9):2038–2047. doi: 10.1007/s00044-017-1909-7. [DOI] [Google Scholar]
- 138.di Luccio E. Inhibition of nuclear receptor binding SET domain 2/multiple myeloma SET domain by LEM-06 implication for epigenetic cancer therapies. J Cancer Prev. 2015;20(2):113–120. doi: 10.15430/JCP.2015.20.2.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Shen Y, et al. Identification of LEM-14 inhibitor of the oncoprotein NSD2. Biochem Biophys Res Commun. 2019;508(1):102–108. doi: 10.1016/j.bbrc.2018.11.037. [DOI] [PubMed] [Google Scholar]
- 140.Coussens NP, et al. High-throughput screening with nucleosome substrate identifies small-molecule inhibitors of the human histone lysine methyltransferase NSD2. J Biol Chem. 2018;293(35):13750–13765. doi: 10.1074/jbc.RA118.004274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Morrison MJ, et al. Identification of a peptide inhibitor for the histone methyltransferase WHSC1. PLoS One. 2018;13(5):e0197082. doi: 10.1371/journal.pone.0197082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Qin S, Min J. Structure and function of the nucleosome-binding PWWP domain. Trends Biochem Sci. 2014;39(11):536–547. doi: 10.1016/j.tibs.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 143.Vermeulen M, et al. Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell. 2010;142(6):967–980. doi: 10.1016/j.cell.2010.08.020. [DOI] [PubMed] [Google Scholar]
- 144.Böttcher J, et al. Fragment-based discovery of a chemical probe for the PWWP1 domain of NSD3. Nat Chem Biol. 2019;15(8):822–829. doi: 10.1038/s41589-019-0310-x. [DOI] [PubMed] [Google Scholar]
- 145.Dilworth D, et al. A chemical probe targeting the PWWP domain alters NSD2 nucleolar localization. Nat Chem Biol. 2022;18(1):56–63. doi: 10.1038/s41589-021-00898-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.