Abstract
In Saccharomyces cerevisiae, the addition of glucose to cells growing on galactose induces internalization of the galactose transporter Gal2p and its subsequent proteolysis in the vacuole. Here we report that the essential step in Gal2p down-regulation is its ubiquitination through the Ubc1p-Ubc4p-Ubc5p triad of ubiquitin-conjugating enzymes and Npi1/Rsp5p ubiquitin-protein ligase. Moreover, Gal2p appears to be stabilized in mutant cells defective in the ubiquitin-hydrolase Npi2p/Doa4p, and the mutant phenotype can be reversed by overexpression of ubiquitin. An analysis of the fate of Gal2p in cells overexpressing wild-type ubiquitin as well as its variants incompetent to form polyubiquitin chains showed that monoubiquitination of Gal2p is sufficient to signal internalization of the protein into the endocytic pathway.
The covalent linkage of ubiquitin to a variety of eukaryotic proteins targets them for degradation. This process involves sequential transfer of the ubiquitin moiety to specific lysine residues of substrate proteins via an E1-E2-E3 enzyme (ubiquitin-activating enzyme, ubiquitin-conjugating enzyme, and ubiquitin-protein ligase) thioester cascade and culminates in formation of an isopeptide bond between the C-terminal glycine of ubiquitin and the ε-amino group of lysine(s) on the substrate protein. Specificity in substrate recognition resides largely at the level of E3′s, and an additional degree of combinatorial specificity probably results from specific E2-E3 interactions. Attachment of ubiquitin to substrate proteins has distinct mechanistic roles in two completely different intracellular proteolytic pathways. Ubiquitination of short-lived cytosolic and nuclear proteins, as well as proteins that undergo endoplasmic reticulum-associated degradation, marks them for hydrolysis by the multicatalytic, multisubunit protease, the 26S proteasome (15, 18, 20, 37, 38). For most of the 26S proteasome substrates, the attachment of a polyubiquitin chain in which at least four ubiquitin molecules must be linked by amide bonds between Lys48 of ubiquitin molecules and the C-terminal carboxyl group of the next following ubiquitin facilitates their binding to the proteasome (15, 20, 37, 54). Ubiquitination of many plasma membrane receptors and nutrient transporters, however, signals their internalization into the endocytic pathway and subsequent proteolysis in the vacuole (5, 16, 43). For all short-lived plasma membrane proteins examined to date, a single ubiquitin moiety or a very short ubiquitin chain appears to suffice to trigger their endocytotic uptake. These plasma membrane proteins are often internalized constitutively. Modulation of the rate of their internalization, induced by a change in nutrient availability, usually from less desirable nitrogen or carbon sources or starvation conditions to preferred sources or rich medium, by binding of their own substrates, or by cell stress, plays the primary role in controlling nutrient uptake (21). Because the ubiquitin itself is a long-lived protein in the yeast Saccharomyces cerevisiae, it must be removed from the substrate conjugates before or during substrate degradation. It appears that both major intracellular degradation pathways (e.g., proteasomal and vacuolar) share the requirement for protein deubiquitination by the deubiquitinating enzyme Npi2/Doa4 (1, 36, 51).
The GAL2-encoded galactose transporter Gal2p belongs to the family of conditionally short-lived plasma membrane proteins, which are inducible by their own substrates (34, 39, 52). Its activity undergoes tight regulation according to the carbon source in the culture medium, so that its activity is maximal in cells growing on galactose as the sole carbon source. Upon addition of glucose or any other easily fermentable carbon source, however, GAL2 transcription is repressed by multiple mechanisms, and presynthesized Gal2p transporter is inactivated by a process referred to as glucose or catabolite inactivation (6, 7, 22, 32). The overall effect of these glucose-regulated processes is thought to speed up the cell transition from utilization of galactose to the fermentation of the preferred sugar glucose. Previously, we (22) and Chiang et al. (6) have shown that during glucose-induced inactivation, a relatively rapid and irreversible loss of both Gal2p transport activity and Gal2p amount occurs due to its internalization by endocytosis from the plasma membrane. Once internalized, the protein is targeted to the vacuole, where it is degraded by vacuolar proteases with no assistance of the 26S proteasome. Moreover, our previous finding of Gal2p-ubiquitin conjugates under the conditions resulting in Gal2p proteolysis suggested the possible role of ubiquitin in this process (22).
Here we show that ubiquitin actually plays a primary role in the Gal2p proteolysis. Our results indicate that the ubiquitin-conjugating enzymes Ubc1p, Ubc4p, and Ubc5p, as well as the ubiquitin-protein ligase Npi1/Rsp5p, are required for Gal2p degradation. Consistent with this view, we find that loss of the free intracellular pool of ubiquitin due to a gene mutation of NPI2/DOA4 severely impairs glucose-induced Gal2p proteolysis and that this defect can be suppressed by the overexpression of ubiquitin. We also find that overexpression of mutant ubiquitins carrying Lys-to-Arg mutations that prevent the formation of various kinds of ubiquitin chains in the npi2/doa4 mutant restores Gal2p proteolysis to nearly the wild-type level. Taken together, the data suggest that monoubiquitination of Gal2p through the enzymes Ubc1p, Ubc4p, Ubc5p, and Npi1/Rsp5p of the ubiquitination machinery is sufficient to signal Gal2p for effective internalization by endocytosis and subsequent proteolysis in the vacuole.
MATERIALS AND METHODS
Yeast strains and growth conditions.
The S. cerevisiae strains used were 23344c (MATα ura3) and its isogenic derivatives 27088a (MATα ura3 npi1) and 270716 (MATα ura3 trp1 npi2). The ubc mutant strains used in this study are congenic to wild-type strain YWO1 (MATα lys2–801 leu2–3,2–112 ura3–52 his3–200 trp1–1) (am) (ubc1, ubc4, ubc5, and ubc4,5) or W303–1B (MATα ade2–1 leu2–3,112 his3–11,15 trp1–1 ura3-1) (ubc6,7 and ubc8), YWO9 (ubc1::URA3), YWO13 (ubc4::HIS3), YWO17 (ubc5::LEU2), YWO23 (ubc4::HIS3 ubc5::LEU2), CPQ (ubc6::LEU2, ubc7::LEU2), and YTS2 (ubc8::kanMX). The yeast cell cultures were grown in either rich medium (1% yeast extract, 2% Bacto peptone) or minimal SD medium (0.67% Difco nitrogen base without amino acids), supplemented with 2% glucose or 2% galactose plus 0.02% glucose and auxotrophic requirements. Solid media were supplemented with 2% Bacto agar. Where indicated, 100 μM CuSO4 was added to induce expression of genes under the CUP1 promoter. The cells were grown aerobically at 30°C on a rotary shaker, and their growth was monitored according to the optical density at 600 nm (OD600). For Western analyses and measurements of transport activity, the yeast strains were grown to an OD600 of 0.5 to 1.0. To induce inactivation, cells were harvested by centrifugation (2,500 rpm, 4 min [Jovan BR4]), washed, and resuspended in 0.17% yeast nitrogen base without ammonium and amino acids plus 2% glucose to an OD600 of 3. The samples were taken at the times indicated below over a 4- to 6-h period, and for each sample, galactose transport activity was determined and total cell extracts were prepared for Western analysis.
Plasmids and DNA manipulations.
The plasmid YEp96 is a 2μm S. cerevisiae-based shuttle vector that contains a synthetic ubiquitin gene under the control of the copper-inducible CUP1 promoter (9). Plasmids encoding mutant ubiquitin variants, in which Lys29 (UbK29R) (9), Lys48 (UbK48R) (19), Lys63 (UbK63R) (9), and all seven lysines (Lys6, -11, -27, -29, -33, -48, and -63; Ub-noLys) (53) have been replaced by arginine, are also derivatives of YEp96. Overexpression of CUP1-containing DNA was induced with 100 μM CuSO4 for at least 3 h. pS25 is a 2μm S. cerevisiae-Escherichia coli vector that carries the GAL2 gene under the control of its own promoter (23). Yeast transformation was performed by the lithium acetate procedure (23) or by electroporation. E. coli DH5α was used for propagation and isolation of plasmids as described previously (3).
Western blotting analysis.
For cell lysis, 1 ml of the cell suspension (OD600 of 3) was incubated for 10 min with 150 μl of freshly prepared 1.85 M NaOH and 7.5% β-mercaptoethanol. Proteins were precipitated for 10 min on ice by addition of 150 μl of 50% trichloroacetic acid, and the precipitates were collected by centrifugation for 10 min at 13,000 × g. The pellet was quickly rinsed with 1 M Tris base and resuspended in 120 to 150 μl of 50 mM Tris-HCl buffer (pH 6.8) containing 8 M urea, 5% sodium dodecyl sulfate (SDS), 0.1 mM EDTA, and 1.5% dithiothreitol. No addition of a protease inhibitor was required. The samples were incubated for 15 min at 37°C and centrifuged for 20 min at 13,000 × g. Equal amounts of protein in supernatants were loaded onto each lane of a standard 10% acrylamide gel, subjected to SDS-polyacrylamide gel electrophoresis, and electroblotted on polyvinylidene difluoride (PVDF) filters (Amersham). The filters were blocked with 5% nonfat milk in Tris-buffered saline (TBS-T buffer; 50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 0.1% Tween 20) by shaking for 2 h at room temperature (or overnight at 4°C) and were further treated either with a polyclonal anti-Gal2p antibody (22) or with antiserum directed against the first 20 N-terminal residues of Gal2p conjugated to ovalbumin (a gift of A. Kruckeberg, E. C. Slater Insituut, Amsterdam, The Netherlands) (diluted 1:500 and 1:750 in TBS-T containing 1% nonfat milk, respectively) for 1 h. After being washed five times in TBS-T, the PVDF sheets were incubated with the secondary goat anti-rabbit antibody (Medac, Hamburg, Germany) coupled to peroxidase, which was diluted 1:5,000 in TBS-T buffer for 1 h. The membranes were washed several times with TBS-T, and the peroxidase activity was detected with the ECL enhanced chemiluminescence system (Amersham). The relative intensities of the Gal2p bands at each time point were quantified by scanning densitometry on the ECL-Hyperfilms. Quantification was performed in the range in which the signal intensity was found to be proportional to the protein concentration.
Transport assays.
Gal2p transport activity was examined by using brief incubations of the yeast cell suspensions with 3 mM tritiated galactose at 30°C essentially as described previously (22).
Lucifer yellow carbohydrazide assays.
Endocytosis of lucifer yellow was performed essentially as described by Dulic et al. (8). Briefly, yeast cells were grown in minimal SD medium with 2% galactose and appropriate supplements to an OD600 of 0.8 to 1.0, harvested by centrifugation, concentrated to an OD600 of about 20 in fresh growth medium, and incubated for 1 h in the dark in the presence of 6 to 8 mg of lucifer yellow carbohydrazide (Sigma) per ml. Then the cells were harvested by low-speed centrifugation, washed three times in 1 ml of ice-cold buffer (50 mM sodium phosphate, 20 mM sodium azide, 20 mM sodium fluoride [pH 7.0]), and finally resuspended in the same buffer. Luciferase yellow was visualized by fluorescence microscopy with fluorescein isothiocyanate optics.
RESULTS
The enzymes Ubc1p, Ubc4p, Ubc5p, and Npi1p/Rsp5p of the ubiquitin system are responsible for efficient internalization and degradation of the Gal2p transporter.
Our previous finding of Gal2p ubiquitination in response to glucose added to cells growing on galactose, followed by its endocytosis and vacuolar proteolysis (22), pointed to the possibility of activity of the components of the ubiquitin system in these processes. If this is true, then mutations that inactivate the relevant components of the ubiquitination machinery should inhibit Gal2p endocytosis and degradation. Therefore, we measured Gal2p turnover by using the standard inactivation assay with yeast strains having deletions in various UBC genes as well as in the mutated NPI1/RSP5 gene.
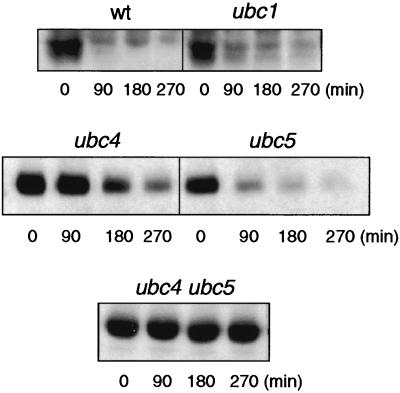
In S. cerevisiae, 11 E2 ubiquitin-conjugating enzymes (Ubcs) and two Ubc-like proteins have been identified, and at least for some of them, specific physiological functions have been uncovered (20). The UBC1, UBC4, and UBC5 genes encode a functionally overlapping group of ubiquitin-conjugating enzymes that together are required for multiple cell functions, including ubiquitination and/or endocytosis of several yeast plasma membrane proteins (13, 17, 25, 33, 41). Our immunoblot analysis for monitoring the fate of Gal2p indicated a half-life of about 1 h for the Gal2p transporter in wild-type cells (22) (Fig. 1A) and showed that its degradation in response to glucose is somewhat inhibited in ubc1 and partially inhibited in ubc4 and ubc5 single mutants. Degradation is strongly impaired in ubc4 ubc5 double mutant cells. Analysis of the data shows that deletions of the UBC1, UBC4, and UBC5 genes lead to 1.5- to 2-fold, 4-fold, and 2- to 3-fold increases in the half-life of Gal2p, respectively, while the half-life of Gal2p was increased up to 10-fold compared to that of the wild type in the ubc4 ubc5 double mutant. When measurement of Gal2p-mediated transport activity was used as an indirect assay of protein internalization, similar results were obtained under the same inactivation conditions (data not shown). In similar experiments, we also examined Gal2p internalization and proteolysis in other ubc deletion mutant strains. Consistent with their specific roles in endoplasmic reticulum-associated protein degradation in the case of Ubc6p and/or Ubc7p (38, 47), and Ubc8p, which is specifically involved in proteolysis of fructose-1,6-bisphosphatase (44), neither Gal2p internalization nor degradation was affected in mutant cells lacking the corresponding UBC genes (data not shown). The data presented above are thus in accordance with the view that the Ubc1p, Ubc4p, and Ubc5p enzymes play an overlapping role in ubiquitination and subsequent endocytosis of Gal2p. However, the ubc4 ubc5 double mutant grows much more slowly than its isogenic wild-type strain. Therefore, an alternative explanation for disturbed Gal2p degradation could be that growth defects observed in the ubc4 ubc5 double mutant are, in fact, a consequence of a general retardation of endocytosis. To test this possibility, we examined bulk endocytosis in all of the ubc mutants described above by using the fluid-phase (bulk) endocytosis marker lucifer yellow carbohydrazide. No defects in accumulation of this dye in the ubc mutants were observed (results not shown).
FIG. 1.
Ubiquitination of Gal2p transporter in response to glucose requires the Ubc1p, Ubc4p, and Ubc5p ubiquitin-conjugating enzymes. Wild-type (wt) and ubc1, ubc4, ubc5, and ubc4/5 mutant cells were grown to the exponential phase in SD galactose medium. The cells were harvested and resuspended in inactivation medium as described in Materials and Methods. At the indicated times, total cell extracts were prepared and the proteins were subjected to Western analysis with Gal2p-specific antibodies. The experiments were repeated at least twice with similar results.
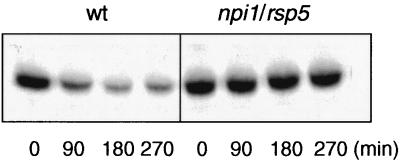
We also tested the role of Npi1p/Rsp5p, one of the known or putative E3′s that belong to the Hect domain family of ubiquitin-protein ligases (20, 43, 55) in Gal2p degradation. Because npi1/rsp5 null mutations are lethal for cells, we used cells carrying a promoter mutation that expresses less than 10% of the wild-type protein level of Npi1/Rsp5p (49). Both internalization and proteolysis of Gal2p (Fig. 2) are substantially reduced in the mutant cells. Our results are thus consistent with previous data that had revealed a requirement of Npi1p/Rsp5p for ubiquitination and/or endocytosis of all yeast plasma membrane proteins with which this aspect has been studied (4, 12–14, 26, 29, 33).
FIG. 2.
Effect of reduced expression of Npi1/Rsp5 ubiquitin-protein ligase on glucose-induced down-regulation of Gal2p. Analysis of Gal2p in wild-type (wt) and in npi1/rsp5 mutant cells was done as described in the legend to Fig. 1.
A defective Npi2/Doa4 deubiquitinating enzyme abolishes degradation of the Gal2p transporter.
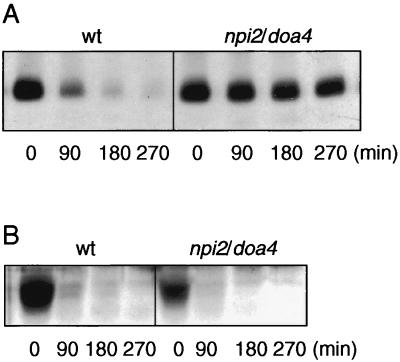
To further confirm the involvement of ubiquitin in glucose-induced proteolysis of the Gal2p transporter, we examined its turnover in an npi2/doa4 mutant, which is also defective in ubiquitination. NPI2/DOA4 (34) encodes one of 17 known deubiquitination enzymes (Dubs) that are in principle able to generate free ubiquitin from ubiquitin-protein conjugates (20, 57). The absence of functional Npi2p/Doa4p in npi2/doa4 mutant cells makes them defective in proteolysis of several plasma membrane proteins marked with ubiquitin (11, 29, 31, 33, 50, 53), probably as a result of their defect in replenishing ubiquitin into the free intracellular ubiquitin pool. To assess whether Npi2p/Doa4p is also involved in Gal2p proteolysis, we compared both the steady-state levels of Gal2p and its galactose transport activity in wild-type and npi2/doa4 cells. As shown in Fig. 3A, the Gal2p degradation is strongly impaired in the mutant cells compared to that of the wild-type strain. Similarly, we found that inactivation of Gal2p-mediated galactose transport in response to glucose is reduced to a comparable extent in the mutant (data not shown).
FIG. 3.
Stabilization of galactose transporter Gal2p in npi2/doa4 cells is reversed by overexpression of wild-type ubiquitin. Wild-type (wt) and npi2/doa4 cells, either untransformed (Fig. 3A) or transformed with plasmid YEp96 (B), encoding wild-type ubiquitin, were grown as described in the legend to Fig. 1. Cells were collected 4 h later after addition of CuSO4 (0.1 mM) and resuspended in inactivation medium, and protein extracts were prepared at the times indicated. Gal2p was detected by Western immunoblotting.
A number of intracellular defects observed in the npi2/doa4 mutant cells, including strongly impaired proteolysis of some plasma membrane proteins, can be overcome at least partially by restoration of normal intracellular ubiquitin levels (11, 30, 50, 53). Based on this knowledge, we overexpressed ubiquitin in wild-type and npi2/doa4 cells transformed with a multicopy plasmid bearing the ubiquitin gene under the control of the copper-inducible CUP1 promoter. Figure 3B shows that overexpression of ubiquitin does not affect Gal2p turnover in wild-type cells, while when overexpressed in npi2/doa4 mutant cells, the defect in Gal2p turnover is nearly fully restored.
Monoubiquitination of the Gal2p transporter suffices for initiating its internalization.
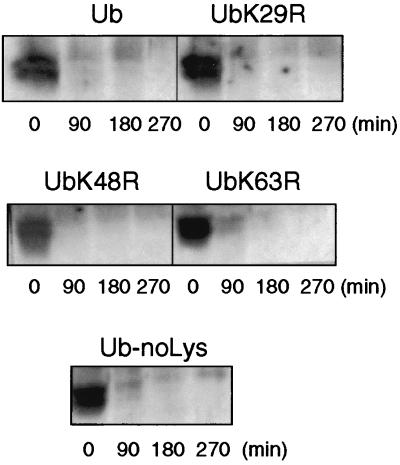
Our previous analysis of the Gal2p ubiquitination pattern had revealed a complex set of Gal2p-ubiquitin conjugates formed transiently in the cells growing on galactose in response to glucose addition (22). However, whether this modification corresponds to the formation of polyubiquitin chains on Gal2p or an attachment of single ubiquitin moieties on its multiple lysine residues remained unresolved. To distinguish between these alternatives, both steady-state levels and internalization of the Gal2p were examined in npi2/doa4 mutant cells overexpressing wild-type ubiquitin or mutant ubiquitins (bearing a single Lys-to-Arg mutation in Lys29, -48, and -63, respectively, which prevents the formation of polyubiquitin chains in vivo in yeast) (2, 10, 24, 48). Overexpression of all ubiquitin molecules carrying Lys-to-Arg mutations restored proteolysis in npi2/doa4 mutant cells (Fig. 4) as well as Gal2p internalization (data not shown) to an extent identical to that observed upon overexpression of wild-type ubiquitin (Fig. 3). These results suggest that polyubiquitin chains formed via Lys29, Lys48, and Lys63 are not required for Gal2p internalization and degradation. However, ubiquitin carries four other lysine residues in its sequence, namely Lys6, -11, -27, and -33. To test if Gal2p internalization and degradation require polyubiquitin chain formation via a lysine(s) other than Lys29, -48, or -63, we analyzed the fate of Gal2p in a yeast strain carrying a ubiquitin lacking all its lysine residues (Ub-noLys). Figure 4 shows that this is not the case and that Gal2p monoubiquitination is sufficient for degradation.
FIG. 4.
Mutant ubiquitins restore normal glucose-induced Gal2p turnover in npi2/doa4 cells. npi2/doa4 cells transformed with plasmids encoding wild-type ubiquitin (Ub), Ub29R, Ub48R, Ub63R, and Ub-no-Lys were grown in SD galactose medium as described in the legend to Fig. 1 and then induced for 3 h with CuSO4 (0.1 mM). The cells were collected and resuspended in inactivation medium, and protein extracts were prepared at the times indicated. Gal2p was detected by Western immunoblotting.
DISCUSSION
The results described in this report support our previous view (22) that ubiquitination of the galactose transporter Gal2p is a prerequisite for rapid posttranslational regulation of its activity by glucose-triggered proteolysis. This is based mainly on the following observations. First, mutations that inactivate (ubc mutations) or at least strongly reduce activity (npi1/rsp5 mutation) of some components of the ubiquitin conjugation pathway substantially slow down Gal2p degradation. Thus, we have shown that reduced levels of the Npi1p/Rsp5p E3 ubiquitin-protein ligase correlate well with the dramatic decrease in the rate of glucose-induced Gal2p endocytosis and proteolysis (Fig. 2). Npi1/Rsp5p appears to be necessary for the degradation of all yeast plasma membrane transporters for which it has been studied: e.g., the sugar transporters Mal11p, Mal61p, and Hxt6p/7p; the amino acid transporters Tat2p and Gap1p and Candida albicans Can1p when expressed in S. cerevisiae; the uracil transporter Fur4p; and the zinc transporter Zrt1p (4, 13, 14, 26, 29, 31, 33). Therefore, Npi1p/Rsp5p may be the key player in this posttranslational modification directing plasma membrane proteins for degradation. We also demonstrated the need for a functionally redundant family of E2′s (ubiquitin-conjugating enzymes), Ubc4p and Ubc5p (and probably also Ubc1p), for Gal2p ubiquitination (Fig. 1) and endocytosis. In addition, our results show that an ubc4 ubc5 double mutant is even more defective for the Gal2p internalization and proteolysis than the single mutants. This finding is consistent with the known overlapping functions of Ubc4p and Ubc5p. However, the functions of the three E2′s Ubc1p, Ubc4p, and Ubc5p are limited to date to only a subset of known or supposed Npi1p/Rsp5p targets, including Ste2p and Ste3p, the receptors for α- and a-factor, a-factor transporter Ste6p, and Zrt1p and Mal61p (13, 17, 33, 42). Moreover, for Mal61p, the E2′s described above were implicated in an as yet unknown posttranslational role in expression of the maltose transporter rather than its proteolysis (33). The rate of glucose-triggered proteolysis is strongly impaired in an npi2/doa4-mutated strain (Fig. 3). NPI2/DOA4 encodes a ubiquitin-protein hydrolase that appears to function in recycling of ubiquitin from ubiquitinated substrates targeted to the proteasome (35, 36, 51) and, rather surprisingly, from substrates targeted to the vacuole (1, 51). According to the current model, Npi2/Doa4p functions late in the ubiquitin-proteasome pathway, probably by releasing ubiquitin from ubiquitin-substrate conjugates associated with the 26S proteasome (36). Removal of ubiquitin from ubiquitinated plasma membrane proteins that are en route to the vacuole occurs at the level of the late endosome or prevacuolar compartment (1). Consistent with this view is the restoration of Gal2p degradation in npi2/doa4 mutant cells, in which ubiquitin was overexpressed (Fig. 3B).
Analysis of the mode of ubiquitination of α- and a-factor receptors (Ste2p and Ste3p) (17, 42, 53) and some nutrient transporters (Fur4p, Gap1p, Zrt1p, Mal11p, and Mal61p) (11, 13, 30, 50) revealed a scenario, according to which one to three of their target lysines accept one to three ubiquitin moieties, depending on the protein studied and/or the experimental conditions used. In addition, for Gap1p and Fur4p, it has been shown that they both bear two target lysine residues capable of accepting up to two or three ubiquitin residues linked via Lys63 (11, 50). To test if the short di- and triubiquitin chains or a single ubiquitin molecule might be sufficient to direct the internalization of plasma membrane proteins, truncated Ste2p and Ste3p lacking all ubiquitination sites were fused in frame with a ubiquitin monomer (42, 46, 53). Analysis of their fate strongly suggested that monoubiquitin provides a signal sufficient to initiate their internalization into the endocytic pathway. No required contribution from the sequence of the protein is sufficient to trigger this process.
Although the galactose transporter Gal2p displays a set of a high-molecular-mass ubiquitin conjugates formed in response to glucose, it obviously escapes recognition and subsequent proteolysis by the 26S proteasome (22). To find an explanation for this apparent paradox, we tested the two alternatives formation of polyubiquitin chains on Gal2p, in which ubiquitin moieties are not linked through Lys48, and/or modification of Gal2p by the addition of single ubiquitins on multiple lysine residues of the protein. To decide between these possibilities, we examined the fate of Gal2p in npi2/doa4 mutant cells overexpressing either wild-type or mutant ubiquitins incompetent to form ubiquitin chains via K29, K48, K63, and all other lysines, respectively. Overexpression of any of the ubiquitin mutant forms described above restored proteolysis of the Gal2p transporter (Fig. 4) as well as its internalization (results not shown) in npi2/doa4 mutant cells to the same extent as overexpression of wild-type ubiquitin. Taken together with our previous data (22), these results suggest that monoubiquitination, e.g., binding of a single ubiquitin moiety to multiple lysine residues in Gal2p, is sufficient to signal its efficient internalization and proteolysis. There is now increasing evidence that in response to specific physiological signals, ubiquitination may also regulate sorting of at least some yeast plasma membrane proteins from the endosome and/or trans-Golgi network to the vacuole without passing through the plasma membrane (4, 27, 40). Experiments to investigate whether this sorting mechanism is also operating in case of the Gal2 transporter are under way.
ACKNOWLEDGMENTS
We thank B. Andre, R. Haguenauer-Tsapis, M. Hochstrasser, and A. Kruckeberg for kindly providing strains, plasmids, and anti-Gal2p antibodies.
This work was supported by grant 204/98/0475 of the Grant Agency of the Czech Republic; grant IAA 5011005 of the Grant Agency of the Academy of Sciences of the Czech Republic; the Deutsche Forschungsgemeinschaft, Bonn, SFB 495; and the Fonds der Chemischen Industrie, Frankfurt.
REFERENCES
- 1.Amerik A Y, Nowak J, Swaminathan S, Hochstrasser M. The Doa4 deubiquitinating enzyme is functionally linked to the vacuolar protein-sorting and endocytic pathways. Mol Biol Cell. 2000;11:3365–3380. doi: 10.1091/mbc.11.10.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arnason T, Ellison M J. Stress resistance in Saccharomyces cerevisiae is strongly correlated with assembly of a novel type of multiubiquitin chain. Mol Cell Biol. 1994;14:7876–7883. doi: 10.1128/mcb.14.12.7876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley and Sons; 1995. [Google Scholar]
- 4.Beck T A, Schmidt A, Hall M N. Starvation induces vacuolar targeting and degradation of the tryptophan permease in yeast. J Cell Biol. 1999;146:1227–1237. doi: 10.1083/jcb.146.6.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonifacino J S, Weissman A M. Ubiquitin and the control of protein fate in the secretory and endocytic pathways. Annu Rev Cell Dev Biol. 1998;14:19–57. doi: 10.1146/annurev.cellbio.14.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chiang H-L, Schekman R, Hamamoto S. Selective uptake of cytosolic, peroxisomal, and plasma membrane proteins into the yeast lysosome for degradation. J Biol Chem. 1996;271:9934–9941. doi: 10.1074/jbc.271.17.9934. [DOI] [PubMed] [Google Scholar]
- 7.DeJuan C, Lagunas R. Inactivation of the galactose transport system. FEBS Lett. 1986;207:258–261. doi: 10.1016/0014-5793(86)81500-9. [DOI] [PubMed] [Google Scholar]
- 8.Dulic V, Egerton M, Elguindi I, Rath S, Singer B, Riezman H. Yeast endocytosis assays. Methods Enzymol. 1991;194:697–710. doi: 10.1016/0076-6879(91)94051-d. [DOI] [PubMed] [Google Scholar]
- 9.Ecker D J, Khan M I, Marsh J, Butt T R, Crooke S T. Chemical synthesis and expression of a cassette adapted ubiquitin gene. J Biol Chem. 1987;262:3524–3527. [PubMed] [Google Scholar]
- 10.Finley D, Sadis S, Monia B P, Boucher P, Ecker D J, Crooke S T, Chau V. Inhibition of proteolysis and cell cycle progression in a multiubiquitination-deficient yeast mutant. Mol Cell Biol. 1994;14:5501–5509. doi: 10.1128/mcb.14.8.5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galan J-M, Haguenauer-Tsapis R. Ubiquitin Lys63 is involved in ubiquitination of a yeast plasma membrane protein. EMBO J. 1997;16:5847–5884. doi: 10.1093/emboj/16.19.5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galan J-M, Moreau V, Andre B, Volland C, Haguenauer-Tsapis R. Ubiquitination mediated by the Npi1/Rsp5 ubiquitin-protein ligase is required for endocytosis of the uracil permease. J Biol Chem. 1996;271:10946–10952. doi: 10.1074/jbc.271.18.10946. [DOI] [PubMed] [Google Scholar]
- 13.Gitan R S, Eide D J. Zinc-regulated ubiquitin conjugation signals endocytosis of the yeast ZRT1 zinc transporter. Biochem J. 2000;346:329–336. [PMC free article] [PubMed] [Google Scholar]
- 14.Hein C, Springael J-Y, Volland C, Haguenauer-Tsapis R, Andre B. NPI1, an essential yeast gene involved in induced degradation of Gap1 and Fur4 permeases, encodes the Rsp5 ubiquitin-protein ligase. Mol Microbiol. 1995;18:77–87. doi: 10.1111/j.1365-2958.1995.mmi_18010077.x. [DOI] [PubMed] [Google Scholar]
- 15.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 16.Hicke L. Gettin' down with ubiquitin: turning off cell-surface receptors, transporters and channels. Trends Cell Biol. 1999;9:107–112. doi: 10.1016/s0962-8924(98)01491-3. [DOI] [PubMed] [Google Scholar]
- 17.Hicke L, Riezman H. Ubiquitination of a yeast plasma membrane receptor signals its ligand-stimulated endocytosis. Cell. 1996;84:277–287. doi: 10.1016/s0092-8674(00)80982-4. [DOI] [PubMed] [Google Scholar]
- 18.Hilt W, Wolf D H. Proteasomes: destruction as a program. Trends Biochem Sci. 1996;21:96–102. [PubMed] [Google Scholar]
- 19.Hochstrasser M, Ellison M J, Chau V, Varshavsky A. The short-lived MATα2 transcriptional regulator is ubiquitinated in vivo. Proc Natl Acad Sci USA. 1991;88:4606–4610. doi: 10.1073/pnas.88.11.4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hochstrasser M. Ubiquitin-dependent protein degradation. Annu Rev Genet. 1996;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- 21.Horak J. Yeast nutrient transporters. Biochim Biophys Acta. 1997;1331:41–79. doi: 10.1016/s0304-4157(96)00015-9. [DOI] [PubMed] [Google Scholar]
- 22.Horak J, Wolf D H. Catabolite inactivation of the galactose transporter in the yeast Saccharomyces cerevisiae: ubiquitination, endocytosis, and degradation in the vacuole. J Bacteriol. 1997;179:1541–1549. doi: 10.1128/jb.179.5.1541-1549.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ito H, Fukuda Y, Muruta K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson E S, Ma P C M, Ota I M, Varshavsky A. A proteolytic pathway that recognizes ubiquitin as a degradation signal. J Biol Chem. 1995;270:17442–17456. doi: 10.1074/jbc.270.29.17442. [DOI] [PubMed] [Google Scholar]
- 25.Kölling R, Hollenberg C P. The ABC-transporter Ste6 accumulates in the plasma membrane in a ubiquitinated form in endocytosis mutants. EMBO J. 1994;13:3261–3271. doi: 10.1002/j.1460-2075.1994.tb06627.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krampe S, Stamm O, Hollenberg C P, Boles E. Catabolite inactivation of the high-affinity transporters Hxt6 and Hxt7 of Saccharomyces cerevisiae occurs in the vacuole after internalization by endocytosis. FEBS Lett. 1998;441:343–347. doi: 10.1016/s0014-5793(98)01583-x. [DOI] [PubMed] [Google Scholar]
- 27.Lemmon S K, Traub L M. Sorting in the endosomal system in yeast and animal cells. Curr Opin Cell Biol. 2000;12:457–466. doi: 10.1016/s0955-0674(00)00117-4. [DOI] [PubMed] [Google Scholar]
- 28.Loayza D, Michaelis S. Role of the ubiquitin-proteasome system in the vacuolar degradation of Ste6, the a-factor transporter in Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:779–789. doi: 10.1128/mcb.18.2.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lucero P, Lagunas R. Catabolite inactivation of the yeast maltose transporter requires ubiquitin-ligase npi1/rsp5 and ubiquitin-hydrolase npi2/doa4. FEMS Lett. 1997;147:273–277. doi: 10.1111/j.1574-6968.1997.tb10253.x. [DOI] [PubMed] [Google Scholar]
- 30.Lucero P, Penalver E, Vela L, Lagunas R. Monoubiquitination is sufficient to signal internalization of the maltose transporter in Saccharomyces cerevisiae. J Bacteriol. 2000;182:241–243. doi: 10.1128/jb.182.1.241-243.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matejckova-Forejtova A, Kinclova O, Sychrova H. Degradation of Candida albicans Can1 permease expressed in Saccharomyces cerevisiae. FEMS Microbiol Lett. 1999;176:257–262. doi: 10.1111/j.1574-6968.1999.tb13670.x. [DOI] [PubMed] [Google Scholar]
- 32.Matern H, Holzer H. Catabolite inactivation of the galactose uptake system. J Biol Chem. 1977;252:6399–6402. [PubMed] [Google Scholar]
- 33.Medintz I, Jiang H, Michels C A. The role of ubiquitin conjugation in glucose-induced proteolysis of Saccharomyces maltose permease. J Biol Chem. 1998;273:34454–34462. doi: 10.1074/jbc.273.51.34454. [DOI] [PubMed] [Google Scholar]
- 34.Nehlin J O, Carlberg M, Ronne H. Yeast galactose permease is related to yeast and mammalian glucose transporters. Gene. 1989;85:313–319. doi: 10.1016/0378-1119(89)90423-x. [DOI] [PubMed] [Google Scholar]
- 35.Papa F R, Hochstrasser M. The yeast DOA4 gene encodes a deubiquitinating enzyme related to a product of the human tre-2 oncogene. Nature. 1993;366:313–319. doi: 10.1038/366313a0. [DOI] [PubMed] [Google Scholar]
- 36.Papa F R, Amerik A Y, Hochstrasser M. Interactions of the Doa4 deubiquitinating enzyme with the yeast 26S proteasome. Mol Biol Cell. 1999;10:741–756. doi: 10.1091/mbc.10.3.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pickart C M. Targeting of substrates to the 26S proteasome. FASEB J. 1997;11:1055–1066. doi: 10.1096/fasebj.11.13.9367341. [DOI] [PubMed] [Google Scholar]
- 38.Plemper R K, Wolf D H. Retrograde protein translocation: ERADication of secretory proteins in health and disease. Trends Biochem Sci. 1999;24:266–270. doi: 10.1016/s0968-0004(99)01420-6. [DOI] [PubMed] [Google Scholar]
- 39.Ramos J, Szkutnicka K, Cirillo V P. Characteristics of galactose transport in Saccharomyces cerevisiae cells and reconstituted lipid vesicles. J Bacteriol. 1989;171:3539–3544. doi: 10.1128/jb.171.6.3539-3544.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roberg K J, Rowley N, Kaiser C A. Physiological regulation of membrane protein sorting late in the secretory pathway of Saccharomyces cerevisiae. J Cell Biol. 1997;137:1469–1482. doi: 10.1083/jcb.137.7.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roth A F, Davis N G. Ubiquitination of the yeast a-factor receptor. J Cell Biol. 1996;143:661–674. doi: 10.1083/jcb.134.3.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roth A F, Davis N G. Ubiquitination of the PEST-like endocytosis signal of the yeast a-factor receptor. J Biol Chem. 2000;275:8143–8153. doi: 10.1074/jbc.275.11.8143. [DOI] [PubMed] [Google Scholar]
- 43.Rotin D, Staub O, Haguenauer-Tsapis R. Ubiquitination and endocytosis plasma membrane proteins: role of Nedd4/Rsp5 family of ubiquitin-protein ligase. J Membr Biol. 2000;176:1–17. doi: 10.1007/s00232001079. [DOI] [PubMed] [Google Scholar]
- 44.Schüle T, Rose M, Entian K-D, Thumm M, Wolf D H. Ubc8p functions in catabolite degradation of fructose-1,6-bisphosphatase. EMBO J. 2000;19:2161–2171. doi: 10.1093/emboj/19.10.2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seufert W, Jentsch S. Ubiquitin conjugating enzymes UBC4 and UBC5 mediate selective degradation of short-lived and abnormal proteins. EMBO J. 1990;9:543–550. doi: 10.1002/j.1460-2075.1990.tb08141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shih S C, Sloper-Mould K E, Hicke L. Monoubiquitin carries a novel internalization signal that is appended to activated receptors. EMBO J. 2000;19:187–198. doi: 10.1093/emboj/19.2.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sommer T, Wolf D H. Endoplasmic reticulum degradation: reverse protein flow of no return. FASEB J. 1997;11:1227–1233. doi: 10.1096/fasebj.11.14.9409541. [DOI] [PubMed] [Google Scholar]
- 48.Spence J, Sadis S, Haas A L, Finley D. A ubiquitin mutant with specific defects in DNA repair and multiubiquitination. Mol Cell Biol. 1995;15:1265–1273. doi: 10.1128/mcb.15.3.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Springael J-Y, Andre B. Nitrogen-regulated ubiquitination of the Gap1p permease of Saccharomyces cerevisiae. Mol Biol Cell. 1998;9:1253–1263. doi: 10.1091/mbc.9.6.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Springael J-Y, Galan J-M, Haguenauer-Tsapis R, Andre B. NH4+-induced down-regulation of the Saccharomyces cerevisiae Gap1 permease involves its ubiquitination with lysine-63-linked chains. J Cell Sci. 1999;112:1375–1383. doi: 10.1242/jcs.112.9.1375. [DOI] [PubMed] [Google Scholar]
- 51.Swaminathan S, Amerik A Y, Hochstrasser M. The Doa4 deubiquitinating enzyme is required for ubiquitin homeostasis. Mol Biol Cell. 1999;10:2583–2594. doi: 10.1091/mbc.10.8.2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Szkutnicka K, Tschopp J F, Andrews L, Cirillo V P. Sequence and structure of the yeast galactose transporter. J Bacteriol. 1989;171:4486–4493. doi: 10.1128/jb.171.8.4486-4493.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Terrel J, Shih S, Dunn R, Hicke L. A function for monoubiquitination in the internalization of a G protein-coupled receptor. Mol Cell. 1998;1:193–202. doi: 10.1016/s1097-2765(00)80020-9. [DOI] [PubMed] [Google Scholar]
- 54.Thrower J S, Hofman L, Rechsteiner M, Pickart C M. Recognition of the polyubiquitin proteolytic signal. EMBO J. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang G, Yang J, Huibregtse J M. Functional domains of the Rsp5 ubiquitin-protein kinase. Mol Cell Biol. 1999;19:342–358. doi: 10.1128/mcb.19.1.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wiederkehr A, Avaro S, Prescianotto-Baschong C, Haguenauer-Tsapis R, Riezman H. The F-box protein Rcy1p is involved in endocytic membrane traffic and recycling out of an early endosome in Saccharomyces cerevisiae. J Cell Biol. 2000;149:397–410. doi: 10.1083/jcb.149.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wilkinson K D. Regulation of ubiquitin-dependent processes by deubiquitinating enzymes. FASEB J. 1997;11:1245–1256. doi: 10.1096/fasebj.11.14.9409543. [DOI] [PubMed] [Google Scholar]