Abstract
Two well-characterized enzymes in Salmonella enterica serovar Typhimurium and Escherichia coli are able to hydrolyze N-terminal aspartyl (Asp) dipeptides: peptidase B, a broad-specificity aminopeptidase, and peptidase E, an Asp-specific dipeptidase. A serovar Typhimurium strain lacking both of these enzymes, however, can still utilize most N-terminal Asp dipeptides as sources of amino acids, and extracts of such a strain contain additional enzymatic activities able to hydrolyze Asp dipeptides. Here we report two such activities from extracts of pepB pepE mutant strains of serovar Typhimurium identified by their ability to hydrolyze Asp-Leu. Although each of these activities hydrolyzes Asp-Leu at a measurable rate, the preferred substrates for both are N-terminal isoAsp peptides. One of the activities is a previously characterized isoAsp dipeptidase from E. coli, the product of the iadA gene. The other is the product of the serovar Typhimurium homolog of E. coli ybiK, a gene of previously unknown function. This gene product is a member of the N-terminal nucleophile structural family of amidohydrolases. Like most other members of this family, the mature enzyme is generated from a precursor protein by proteolytic cleavage and the active enzyme is a heterotetramer. Based on its ability to hydrolyze an N-terminal isoAsp tripeptide as well as isoAsp dipeptides, the enzyme appears to be an isoAsp aminopeptidase, and we propose that the gene encoding it be designated iaaA (isoAsp aminopeptidase). A strain lacking both IadA and IaaA in addition to peptidase B and peptidase E has been constructed. This strain utilizes Asp-Leu as a leucine source, and extracts of this strain contain at least one additional, as-yet-uncharacterized, peptidase able to cleave Asp dipeptides.
The intracellular hydrolysis of peptides in Salmonella enterica serovar Typhimurium is carried out by at least 12 different enzymes. Some of these enzymes hydrolyze a wide range of peptides, while others are very specific for particular amino acids. Of the well-characterized peptidases in serovar Typhimurium, only three hydrolyze aspartyl (Asp)-X (where X is any amino acid) dipeptides: peptidase B, a broad-specificity aminopeptidase, hydrolyzes both Asp and non-Asp peptides (14); peptidase Q (15), an X-Pro-specific dipeptidase, hydrolyzes Asp-Pro (R. A. Larsen, unpublished results) but not other N-terminal Asp peptides; and peptidase E, an Asp-specific dipeptidase, hydrolyzes almost all Asp-X dipeptides (4, 5).
A strain of serovar Typhimurium (TN1300) lacking four broad-specificity peptidases, peptidases N, A, B, and D, as well as peptidase E and peptidase Q is still able to use Asp-Leu as a leucine source (4). In fact, such a mutant could grow on all of the Asp-X peptides that were tested except Asp-Pro. These results indicate that additional Asp peptide hydrolases are present in serovar Typhimurium. Previous work showed that crude cell extracts of serovar Typhimurium peptidase mutants contain at least two additional Asp-X-hydrolyzing activities (4). Mutants lacking these activities have not been isolated, however, and it is not clear if either or both of them are responsible for the ability of pepB pepE mutants to grow on Asp dipeptides.
This paper reports the characterization of two additional serovar Typhimurium peptidases that are capable of hydrolyzing N-terminal Asp peptides. One of these enzymes is the product of the serovar Typhimurium homolog of the Escherichia coli iadA gene (7). IadA is a dipeptidase that hydrolyzes N-terminal isoAsp dipeptides. Our data suggest that it also has low but detectable activity toward some but not all Asp-X peptides. The other enzyme is an isoAsp-hydrolyzing peptidase encoded by the serovar Typhimurium homolog of the E. coli ybiK gene which can also hydrolyze N-terminal Asp dipeptides at a low but detectable rate. Mutants lacking PepE, PepB, IadA, and YbiK have been constructed and shown to contain an additional Asp-X-specific peptidase.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The Salmonella enterica serovar Typhimurium strains used in this work are derivatives of strain LT2 and are listed in Table 1. E. coli DH5α was routinely used for DNA cloning experiments. E. coli strain BL21(DE3) was used for overexpression of plasmid-encoded IaaA. Strains were typically grown at 37°C in LB medium (Lennox L broth; Gibco BRL). E medium (24) containing 0.4% glucose was used as a minimal medium and was supplemented as indicated below with amino acids or peptides at 0.3 mM. When required, either ampicillin or chloramphenicol was added at 50 or 20 μg/ml, respectively.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| Strains | ||
| Serovar Typhimurium | ||
| TN1246 | leuBCD485 pepN90 pepA16 pepB11 supQ302 Δ(proAB pepD) pepP1 pepQ1 | Laboratory collection |
| TN1300 | leuBCD485 pepN90 pepA16 pepB11 supQ302 Δ(proAB pepD) pepP1 pepQ1 pepE1 | Laboratory collection |
| TN1379 | leuBCD485 | Laboratory collection |
| TN2373 | ara-9 pol-2 | Laboratory collection |
| TN2540 | metE551 metA22 ilv-452 trpB2 hisC527(Am) galE496 xyl-404 rpsL120(StrA) flaA66 hsdL6 hsdSA29(r−m+) | Laboratory collection |
| TN5131 | leuBCD485 pepN90 pepA16 pepB11 pepD3 pepP1 pepQ1 pepE8::MudJ | Laboratory collection |
| TN5347 | leuBCD485 pepN90 pepA16 pepB11 pepD3 pepP1 pepQ1 pepE8::MudJ/pCM429 | This study |
| TN5378 | ara-9 pol-2 iadA1::Knr | This study |
| TN5379 | leuBCD485 iadA1::Knr | This study |
| TN5391 | leuBCD485 pepB11 pepE1 zce850::Tn10 iadA1::Knr | |
| TN5426 | leuBCD485 pepN90 pepA16 pepB11 pepD3 pepP1 pepQ1 pepT1 iadA1::Knr pepE8::MudJ zja::Tn10 | This study |
| TN5507 | metE551 metA22 ilv-452 trpB2 hisC527(Am) galE496 xyl-404 rpsL120(StrA) flaA66 hsdL6 hsdSA29(r−m+)/pBR328 | |
| TN5538 | leuBCD485 iaaA1::Cmr | This study |
| TN5542 | leuBCD485 pepB11 pepE1 zce850::Tn10 iadA1::KnriaaA1::Cmr | This study |
| TN5549 | metE551 metA22 ilv-452 trpB2 hisC527(Am) galE496 xyl-404 rpsL120(StrA) flaA66 hsdL6 hsdSA29(r−m+)/pCM499 | |
| TN5617 | leuBCD485 iadA1::KnriaaA1::Cmr | This study |
| TN5784 | leuBCD485 orf2::Cmr | This study |
| E. coli | ||
| DH5α | F φ80dlacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17(rK− mk+) phoA supE44 λ− thi-1 gyrA96 relA1 | Gibco BRL |
| BL21(DE3) | F−omp T hsdSB(rB− mB−) gal dcm (DE3) | Novagen |
| TN5628 | BL21(DE3)/pCM532 | This study |
| Plasmids | ||
| pBR322 | Plasmid cloning vector; Tcr Apr | New England Biolabs |
| pBR328 | Plasmid cloning vector (a derivative of pBR322); Tcr Apr Cmr | New England Biolabs |
| pBluescript II KS(+) | Plasmid cloning vector with a T7 promoter; Apr | Stratagene |
| pET11b | Plasmid cloning vector with a T7 promoter; Apr | Novagen |
| pGEM-T | PCR cloning vector; Apr | Promega |
| pKRB9 | pBR322-based plasmid with a Cmr cassette; Apr Cmr | G. Philips |
| pSE380 | pBR322-based cloning vector with a superpolylinker following a trc promoter; Apr | 3 |
| pUC4K | pBR322-based vector carrying a Knr cassette | Pharmacia |
| pCM411 | iadA gene amplified by PCR from TN1379 and cloned into pGEM-T | This study |
| pCM429 | iadA excised from pCM411 using NcoI and NotI and cloned into pSE380 | This study |
| pCM435 | Knr cassette excised from pUC4K using BamHI and inserted into the BamHI site within iadA on pCM411 | This study |
| pCM499 | 8-kb serovar Typhimurium genomic fragment including iaaA carried on pBR328 | This study |
| pCM500 | 8-kb serovar Typhimurium genomic fragment including iaaA carried on pBR328 | This study |
| pCM529 | 2.3-kb serovar Typhimurium genomic fragment excised from pCM499 using HindIII and BamHI inserted into the HindIII and BamHI sites of pBluescript II KS(+) | This study |
| pCM531 | Cmr cassette from pKRB9 excised with BamHI, blunted with Klenow, and cloned into the SmaI site within iaaA on pCM529 | This study |
| pCM532 | iaaA amplified from TN1379 using primers ybiK1-3 and ybiK2-s, digested with NdeI and BamHI, and cloned into the NdeI and BamHI sites of pET11b | This study |
DNA techniques.
DNA manipulations were performed using standard techniques (13). Plasmids were purified using a QiaPrep Spin plasmid kit, and DNA fragments were prepared for cloning by agarose gel extraction using a Qiaquick gel extraction kit (Qiagen). Restriction enzymes were from Gibco BRL or New England Biolabs. PCR was performed using either Taq polymerase (Qiagen) or Pfu polymerase (Stratagene) and deoxynucleoside triphosphates from Qiagen. T4 DNA ligase was from Gibco BRL, and shrimp alkaline phosphatase was from Amersham. Oligonucleotide synthesis and DNA sequencing (on a Perkin Elmer ABI 377A automated DNA sequencer) were carried out by the W. M. Keck Center for Comparative and Functional Genomics at the University of Illinois. Sequencing of pCM499 was carried out using primers BamHI CW and BamHI CCW (New England Biolabs), which anneal near the BamHI sites of pBR322-derived plasmids.
Plasmids and chromosomal mutations were transferred between serovar Typhimurium strains using the generalized transducing phage P22HT 12/4 int-3 (21).
Cloning of peptidase genes.
Plasmids containing 8- to 12-kb fragments of serovar Typhimurium DNA from strain TN1246 cloned into pBR328 were introduced by transduction into TN5426 (leuBCD485 pepN90 pepA16 pepB11 pepD3 pepP1 pepQ1 pepT1 iadA1::Knr pepE8::MudJ zja::Tn10), selecting for growth on LB ampicillin medium. Transductants were replica plated on to minimal medium containing either leucine or isoAsp-Leu as the leucine source in order to identify colonies able to grow on isoAsp-Leu. Of approximately 1,000 colonies screened, 15 appeared to grow on isoAsp-Leu. Crude extracts of seven of these strains were tested for isoAsp-Leu-hydrolyzing activity after nondenaturing polyacrylamide gel electrophoresis (PAGE). Two of these strains overexpressed IadA, and three overexpressed IaaA. No overexpressed activity was observed for the remaining two clones and they were not further characterized. Plasmids (pCM499, pCM500, and pCM501) isolated from each of the strains overexpressing IaaA were used to transform TN5426, and the ability of the transformants to use isoAsp-Leu was confirmed. Each of these three clones was mapped using restriction endonucleases, but only pCM499 was used for sequencing.
Construction of an iadA overexpression plasmid.
Serovar Typhimurium iadA was amplified by PCR using primers designed for the E. coli iadA sequence, iadA-1 (5′-ATGATTGATTATACCGCAGCCGGTTTTAC-3′) and iadA-2 (5′-TTAAGCCGTTTCAAACGTTCCTTTCACGCAGGCTT-3′). The 1.1-kb product was cloned into pSE380 under the control of an inducible promoter (Ptrc), resulting in pCM429. To overexpress IadA, strain TN5347 (TN5131/pCM429) was grown in E minimal medium containing 0.4% glucose, 1% Casamino Acids, and ampicillin to mid-exponential phase (optical density at 600 nm = 0.8); 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) was added, and growth was continued overnight.
Construction of an iaaA overexpression plasmid.
The iaaA open reading frame was amplified using primers ybiK1-3 (5′-GGAACTCCATATGAATAAAGCAGTGATTG-3′) and ybiK2-s (5′-ATCGGATCCTATCCAGTTCATCGCTGTG-3′), both derived from the S. enterica serovar Typhi sequence using serovar Typhimurium genomic DNA as the template. The 1-kb product was cloned into the NdeI and BamHI sites of vector pET11b (Novagen), creating pCM532, in which iaaA replaced the phage T7 gene 10 exactly at the translational start site. This construct was transformed into E. coli strain BL21(DE3) containing an inducible T7 RNA polymerase gene (TN5628).
Construction of an iadA disruption.
The iadA gene was amplified by PCR as described above using Taq polymerase and cloned directly into pGEM-T (Promega), producing pCM411. A Knr cassette, prepared by BamHI excision from pUC4K (Pharmacia), was inserted into a unique BamHI site within the iadA gene. The resulting construct, pCM435, was then transformed into TN2540, from which it was transduced into TN2373 (polA), selecting for resistance to kanamycin. An Aps transductant, which had undergone a double-crossover event resulting in a single disrupted copy of iadA in the chromosome, was isolated (TN5378). Amplification of iadA from TN5378 using primers designed for the serovar Typhimurium sequence (which had become available during the course of these experiments), iadA-3 (5′-GCATCTGCGATGTTCTACTCGCGAAT-3′) and iadA-4 (5′-TACACCTGCTCAATGCGTAAATCT-3′), resulted in a single 2.3-kb product, which was the expected size of iadA plus the kanamycin resistance gene. Use of the same primers to amplify DNA from strain TN2373 resulted in a 1-kb fragment, which is the predicted size of the wild-type iadA gene.
Construction of an iaaA disruption.
A 2.3-kb HindIII/BamHI fragment from pCM499, including the iaaA gene, was cloned into the HindIII and Bam HI sites of pBluescript II KS(+) (Stratagene). A Cmr gene, which was excised from pKRB9 (G. Philips) using BamHI and had the ends blunted using the Klenow fragment, was then cloned into a unique SmaI site of the iaaA gene. Using the resulting plasmid, pCM531, a disruption of the chromosomal iaaA gene was constructed (TN5538) as described above for iadA. Amplification of iaaA using primers ybiK1-3 and ybiK2-s (described above) led to a 1-kb product from the wild-type strain (TN1379) and a single 2-kb product from the disrupted mutant (TN5538), demonstrating that in the mutant, the disrupted copy had replaced the wild-type gene.
Gel electrophoresis.
Sodium dodecyl sulfate-PAGE (SDS-PAGE) was performed as previously described (20). Molecular weight markers were Mark12 wide-range protein standards (Novex). Nondenaturing PAGE and staining for peptidase activity were carried out essentially as previously described (16). The stain solution contained 2 mg of amino acid oxidase, 4 mg of peroxidase, and 1 mg of o-dianisidine in 10 ml of 0.1 M Tris-Cl, pH 8; the solution was mixed with an equal volume of 3.5% agar and was poured directly onto the gel.
Peptidase assays.
A semiquantitative l-amino acid oxidase-based assay was used for determining peptidase activity in column fractions and for preliminary characterization of peptidase activities. Assays were carried out in the wells of plastic depression plates as described previously (4). The stain solution was prepared as described above for the staining of PAGE gels.
Hydrolysis of chromogenic substrates was tested as follows. A sample of purified IaaA diluted in 0.1 M Tris-Cl, pH 8.0, was incubated with either 1 mM isoAsp-p-nitroanilide (isoAsp-pNA; Bachem) or 1 mM isoAsp–7-amido-4-methylcoumarin (isoAsp-AMC; Bachem) for 30 min at 37°C. Hydrolysis of the substrates was detected as either a yellow color from the released p-nitroaniline or a purple color from the released AMC visible under UV light.
Assays for determining the specificity and kinetics of IaaA using high-performance liquid chromatography (HPLC) to monitor product formation were carried out essentially as previously described (10). Peptides were obtained from Sigma, Bachem, or Research Plus. isoAsp-Leu-Ala and Ala-isoAsp-Leu-Ala were synthesized by the Protein Science Facility at the University of Illinois Biotechnology Center.
Purification of IaaA.
Strain TN5628 [BL21(DE3)/pCM532] was grown in LB ampicillin medium overnight at 37°C. In preliminary experiments 1 mM IPTG was added at the mid-exponential phase of growth (optical density at 600 nm = 0.8); however, this step was omitted from the final purification protocol. Cells were disrupted by sonication in 0.1 M Tris-Cl, pH 8.0, and the resulting lysate was centrifuged for 30 min at 30,000 × g. The soluble fraction was then subjected to anion-exchange chromatography using a Q Sepharose HiLoad 26/10 column (Pharmacia) and was eluted with a linear gradient of 0 to 1.0 M NaCl in 20 mM Tris-Cl, pH 8.0. isoAsp-Leu-hydrolyzing activity, as determined by the amino acid oxidase assay described above, was eluted at approximately 0.28 M NaCl. Fractions with this activity were pooled and concentrated using a Centriprep 10 concentrator (Amicon) and subjected to gel filtration on a HiPrep Sephacryl HR S-100 (320-ml) column (Pharmacia) in a solution containing 20 mM Tris-Cl, pH 8.0, and 150 mM NaCl. The IaaA activity, which was eluted at an estimated molecular mass of 60 kDa, was concentrated and desalted again using a Centriprep 10 concentrator. Finally, this activity was chromatographed on a DEAE Sepharose fast-flow 10/10 column (Pharmacia) using a linear gradient of 0 to 1.0 M NaCl. IaaA activity was eluted at a concentration of approximately 0.22 M NaCl. The purified enzyme was analyzed by SDS-PAGE and was visually estimated to be greater than 90% pure.
Amino acid sequencing and mass spectrometry.
The N-terminal amino acid sequence of each subunit of IaaA was determined using a Perkin-Elmer Applied Biosystems Procise 494 HT protein sequencer. For each polypeptide, an unambiguous sequence of the first 10 residues was obtained. Mass spectrometric analysis of IaaA was performed on a PerSeptive Biosystems Voyager-DE matrix-assisted laser desorption ionization–time of flight spectrometer. Both procedures were carried out at the Protein Science Facility at the University of Illinois Biotechnology Center.
RESULTS
Asp-Leu-hydrolyzing activities in serovar Typhimurium peptidase mutants.
When a crude extract of a multiply peptidase-deficient (pepNABDPQE) strain (TN5131) of serovar Typhimurium was subjected to nondenaturing PAGE and the gel was stained to detect Asp-Leu hydrolysis, two distinct bands of activity (Rfs, 0.25 and 0.8) were observed (data not shown). These two activities were not resolved by anion-exchange chromatography (Q Sepharose) alone, but they were separated by subsequent gel filtration chromatography (Superdex 200 HR 16/60). The apparent native molecular masses of each were 200 kDa (Rf, 0.25) and 60 kDa (Rf, 0.8). Partially purified fractions from gel filtration chromatography were assayed in the presence and absence of the metal chelator EDTA. The activity of the 200-kDa enzyme was completely inhibited by EDTA, suggesting that it is a metallopeptidase. The 60-kDa enzyme was not inhibited by EDTA and was presumed to belong to a mechanistic class different from that of the 200-kDa enzyme.
The substrate specificity for each of the two activities separated by gel filtration was tested qualitatively using the l-amino acid oxidase assay. Both enzymes hydrolyzed isoAsp peptides (isoAsp-Leu and isoAsp-Ala) more rapidly than Asp-Leu and Asp-His, and neither enzyme had significant activity towards Asp-X, where X is Tyr, Val, Lys, Gly, Trp, or Pro. The observed preference for isoAsp-Leu was confirmed by staining nondenaturing PAGE gels to detect either Asp-Leu or isoAsp-Leu hydrolysis. Color developed more rapidly with isoAsp-Leu as a substrate than with Asp-Leu at the positions where each of the two enzymes migrated (data not shown).
The 200-kDa enzyme with an Rf of 0.25 is IadA.
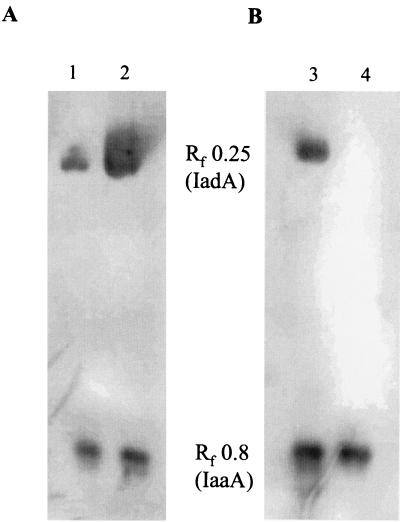
The properties of the enzyme with an Rf of 0.25 led us to suspect that it might be the serovar Typhimurium homolog of E. coli IadA, an isoAsp dipeptidase encoded by iadA and described by Gary and Clarke (7). To test this hypothesis, a clone carrying the serovar Typhimurium iadA gene (pCM429) was constructed. When a crude extract of a strain (TN5347) induced for IadA expression was subjected to nondenaturing PAGE and the gel was stained for isoAsp-Leu-hydrolyzing activity, the intensity of the band corresponding to the Rf of 0.25 was markedly increased (Fig. 1A), suggesting that the observed activity was IadA. Further confirmation of this assignment was obtained by constructing a strain containing an iadA::Knr chromosomal disruption (TN5379) and subjecting an extract of this strain to nondenaturing PAGE. The isoAsp-Leu-hydrolyzing activity that migrated at an Rf of 0.25 was absent in the iadA mutant strain (Fig. 1B). Although IadA has been reported to be specific for isoAsp peptides (9), our amino acid oxidase assays suggested that it hydrolyzed Asp-Leu and Asp-His. By HPLC analysis (using the overexpression strain described below) we determined that the rate of Asp-Leu hydrolysis is approximately 1% of the rate of isoAsp-Leu hydrolysis.
FIG. 1.
Nondenaturing PAGE of strains either overexpressing or lacking IadA. Soluble cell extracts of each strain were loaded as follows: lane 1, TN5131 (leuBCD485 pepN90 pepA16 pepB11 pepD3 pepP1 pepQ1 pepE8::MudJ); lane 2, TN5347 (leuBCD485 pepN90 pepA16 pepB11 pepD3 pepP1 pepQ1 pepE8::MudJ/pCM429); lane 3, TN1379 (leuBCD485); and lane 4, TN5379 (leuBCD485 iad1::Knr). Hydrolysis of isoAsp-Leu was detected using the amino acid oxidase assay.
Overexpression of IadA allows growth on isoAsp-Leu.
Because both IadA and the 60-kDa enzyme with an Rf of 0.8 hydrolyzed isoAsp-Leu, we expected that this peptide could be utilized by a leucine auxotroph as a source of leucine. Surprisingly, neither a wild-type strain, TN1379 (leuBCD485), nor a peptidase-deficient strain, TN5131 (leuBCD485 pepN90 pepA16 pepB11 pepD3 pepP1 pepQ1 pepE8::MudJ), grew on isoAsp-Leu as a leucine source (no visible growth 48 h after streaking on minimal medium with isoAsp-Leu). This suggests that either the rate of peptide uptake or the level of isoAsp-Leu-hydrolyzing activity was too low to support growth. When a strain carrying the IadA overexpression plasmid (TN5347) was streaked onto minimal medium containing isoAsp-Leu as the leucine source, however, small colonies were clearly visible after overnight incubation. This result suggests that peptide hydrolysis is rate limiting for utilization of isoAsp-Leu.
Identification of the gene encoding the 60-kDa isoAsp peptidase with an Rf of 0.8.
In order to facilitate further characterization of the enzyme with an Rf of 0.8, it was necessary to clone the gene encoding this enzyme. Because overexpression of IadA allowed growth on isoAsp-Leu as a leucine source, it was reasonable to assume that overexpression of the enzyme with an Rf of 0.8 might also allow growth on this peptide. If so, then it should be possible to clone the gene specifying this activity by screening a genomic library for plasmids that allow growth of a leucine auxotroph on isoAsp-Leu. Libraries of pBR328 carrying chromosomal DNA from TN1246 were screened as described in Materials and Methods. One of the three isolates that overexpressed the enzyme with an Rf of 0.8 was characterized further. The specific activity of isoAsp-Leu hydrolysis of an extract of this strain (TN5549, containing pCM499) was found to be 3.8 nmol of Asp min−1 μg−1, which is 20-fold higher than that of TN5507, a strain containing pBR328 alone (0.182 nmol of Asp min−1 μg−1).
Restriction mapping of pCM499, pCM500, and pCM501 indicated that they carried overlapping genomic fragments. One of these clones, pCM499, which carried an approximately 8-kb insert, was sequenced using primers that anneal near the BamHI site of pBR328. A comparison of each sequence with the GenBank database showed that the cloned region corresponded to a segment of the E. coli chromosome located at 869.5 kb (18 min) (Fig. 2). This segment of the E. coli chromosome is also 8 kb, suggesting that there are no major insertions or deletions in the corresponding region of serovar Typhimurium. When the sequence from serovar Typhi, a more closely related organism, became available, it was clear that the number and order of genes in this region are identical to those of E. coli.
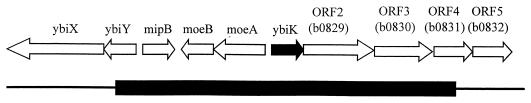
FIG. 2.
18-minute region of the E. coli genome. The portion present on plasmid pCM499, which carries iaaA, is indicated by the black bar.
The genomic DNA carried on pCM499 includes three genes with known functions (mipB, moeA, and moeB) and three genes of unknown function (ybiK, b0829, and b0830). A Blast search of the GenBank database using the amino acid sequence corresponding to ybiK revealed significant similarity between the product of this open reading frame and a group of enzymes including asparaginases from plants and glycosylasparaginases from both eukaryotic and prokaryotic sources. All of these enzymes hydrolyze an Asp side chain amide bond, a reaction similar to isoAsp peptide hydrolysis. The remaining genes carried by pCM499 showed similarity to sequences encoding proteins with distinct functions other than peptide bond hydrolysis. These results strongly suggest that the isoAsp peptidase activity is encoded by the serovar Typhimurium homolog of ybiK, hereafter referred to as iaaA for isoAsp aminopeptidase.
The asparaginases and glycosylasparaginases to which IaaA is related are members of the Ntn (N-terminal nucleophile) family of hydrolases or transferases. This family is characterized by an N-terminal amino acid residue, either threonine, serine, or cysteine, that functions as both the nucleophile and the proton donor for catalysis (2). For most of these enzymes the catalytic N-terminal residue is exposed only after an autoproteolytic processing step. Both the asparaginases and the glycosylasparaginases autoprocess in this manner, exposing a threonine as the catalytic residue. An amino acid sequence alignment of IaaA and a group of related sequences (Lupinus luteus l-asparaginase [GenBank accession number Q9ZSD6], Arabidopsis thaliana l-asparaginase [P50287], Drosophila melanogaster unknown open reading frame product [AAF48471], Flavobacterium meningosepticum glycosylasparaginase [JN0910], and Homo sapiens glycosylasparaginase [P20933]) revealed conservation of the proposed catalytic threonine residue (Thr 179 in IaaA) as well as other residues known to be important for catalysis and autoproteolysis.
A potential sigma 70 promoter was observed upstream of the iaaA open reading frame with near-consensus sequences (five out of six) for both the −10 and the −35 elements. iaaA is likely to be the first in a five-gene operon, which includes four genes with similarity to genes encoding ATP-binding cassette-type transporters. The open reading frame immediately downstream from iaaA has a predicted translational start site at GTG located 10 bp downstream from the stop codon of iaaA and has a putative ribosome binding site 8 bp upstream (overlapping the stop codon of iaaA). In order to confirm that the products of these downstream genes are not required for the isoAsp peptidase activity, an insertion mutation in orf2 (orf2::Cmr) was constructed. This disruption was predicted to have a polar effect on the remaining three downstream genes. A strain containing the orf2::Cmr (TN5784) had wild-type levels of IaaA activity as observed by nondenaturing PAGE analysis (data not shown), suggesting that none of the downstream genes are required for peptidase activity.
Cloning of iaaA and expression of its gene product.
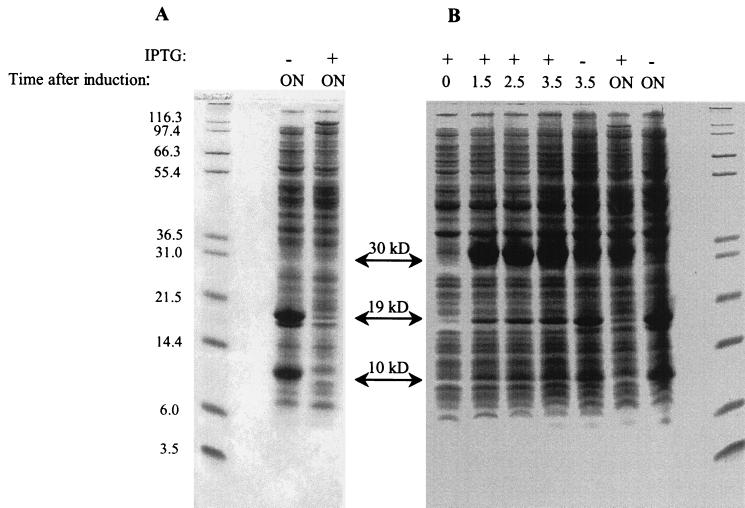
To test the hypothesis that iaaA encodes the peptidase with an Rf of 0.8 observed by native gel analysis, a clone with iaaA under the control of a phage T7 promoter was constructed (pCM532). The levels of IaaA protein expression (determined by SDS-PAGE) in a strain carrying this plasmid (TN5628) and grown in the presence or absence of an inducer (IPTG) were determined (Fig. 3A). Suprisingly, two overexpressed proteins, migrating at approximately the positions predicted for the two subunits of processed IaaA, were observed in extracts of the uninduced cells but not in extracts of induced cells. Correspondingly, the uninduced extract contained 70-fold-greater isoAsp-Leu-hydrolyzing activity: 168 nmol of Asp released min−1 mg−1, compared to 2.5 nmol of Asp min−1 mg−1 for the induced extract. In an attempt to explain these observations, we monitored the time course of protein expression after IPTG induction using whole cells rather than soluble cell extracts (Fig. 3B). In cultures induced with IPTG, high levels of an approximately 30-kDa protein were visible at the first time point (1.5 h) after induction. This protein is presumed to be the unprocessed iaaA gene product, which has a calculated molecular mass of 32.6 kDa. The same protein was visible at all subsequent time points. Proteins of approximately 19 kDa and 10 kDa appeared to increase in intensity after induction but were no longer present in cultures that were incubated overnight. The 30-kDa protein was not observed in soluble cellular extracts of induced cultures (Fig. 3A), suggesting that unprocessed IaaA forms inclusion bodies that are removed by the centrifugation step during extract preparation but are solubilized when whole-cell extracts are boiled in SDS. We hypothesized that low-level expression of the T7 polymerase gene in the absence of an inducer (22) leads to a level of IaaA expression that allows normal maturation of the enzyme. It appears that the high level of IaaA production that follows the induction of the T7 polymerase leads to aggregation rather than proper dimerization and processing.
FIG. 3.
SDS-PAGE analysis of IaaA overexpression. SDS-PAGE of soluble protein extracts (A) or whole cells (B) from TN5628 [BL21(DE3)pCM532] was carried out using cultures grown with or without IPTG, as indicated. The time after IPTG addition is indicated in hours. The positions at which unprocessed (30-kDa) and processed (19-kDa and 10-kDa) IaaA migrate are indicated with arrows. Sizes (in kilodaltons) of molecular mass markers are shown to the left of the gel. ON, overnight.
Purification and characterization of IaaA.
Based on the results described above, we concluded that overnight growth of TN5628 in the absence of IPTG provides significant overexpression of active IaaA. Extracts of TN5628 grown under these conditions were used to purify IaaA as described in Materials and Methods. SDS-PAGE analysis of samples from each step of the purification is shown in Fig. 4.
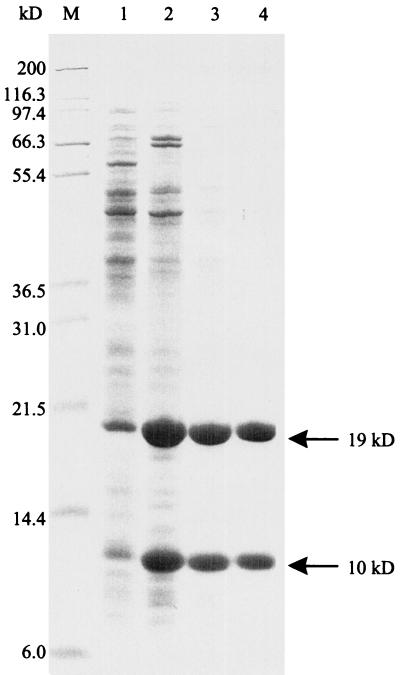
FIG. 4.
Purification of IaaA. IaaA was overexpressed in strain TN5628 and purified as described in Materials and Methods. Samples from each step in the purification were subjected to SDS-PAGE and loaded as follows: lane 1, soluble cell extract; lane 2, pooled Q Sepharose fractions; lane 3, pooled Superdex fractions:, and lane 4, pooled DEAE fractions. Molecular mass markers were loaded in lane M, and the corresponding sizes are shown to the left of the gel. The positions of each of the two bands corresponding to subunits of IaaA are indicated and are labeled with the approximate molecular mass as calculated from the migration on this gel relative to the mass standards.
Based on the site of posttranslational processing in related Ntn family enzymes, we predicted that the active form of IaaA would be generated by a cleavage between Gly178 and Thr179 to produce polypeptides of 19 and 10 kDa. Two proteins of the predicted size were observed by SDS-PAGE analysis (Fig. 4), and the N-terminal amino acid sequence of each was determined. The sequence of the 19-kDa band (MNKAVIAIHG) was identical to that predicted by the serovar Typhi sequence for the start of the N-terminal subunit of IaaA. The sequence of the 10-kDa polypeptide (TVGAVARDKF) was identical to the sequence beginning at Thr179. The calculated molecular mass of the larger protein, 18.7 kDa, is very similar to that determined by SDS-PAGE (19 kDa). The smaller protein, however, has a calculated molecular mass of 13.7 kDa, whereas SDS-PAGE suggests a mass of 10 kDa. To obtain a more accurate estimate of the molecular mass of each subunit, mass spectrometry was performed on purified IaaA. The results indicate that the two most abundant polypeptides, 13,719.9 and 18,787.4 Da, are very close in size to those predicted from the serovar Typhi sequence (13,711.7 and 18,782.1 Da), differing by only 8.2 and 5.3 Da from the predicted masses of the smaller and larger subunits, respectively. The differences in molecular mass are likely due to sequence differences between serovar Typhi, from which the masses were calculated, and serovar Typhimurium, from which the protein was isolated. From both the amino acid sequencing and the mass spectrometry data we concluded that the purified protein was encoded by iaaA, was processed into two polypeptides at the expected site, and did not require further modification or processing to be catalytically active.
Substrate specificity of IaaA.
Purified IaaA was used to determine the relative rates of hydrolysis of various peptide substrates. The substrates tested and the rate of hydrolysis relative to that of isoAsp-Leu are listed in Table 2. IaaA hydrolyzed asparagine poorly and did not hydrolyze N-acetylglucosaminylasparagine, indicating that its primary role is neither as an asparaginase nor as a glycosylasparaginase. Of the isoAsp substrates tested, isoAsp-Leu was hydrolyzed most rapidly and isoAsp-Phe-methyl ester (ME) was hydrolyzed least rapidly. The tripeptide isoAsp-Leu-Ala was a good substrate, indicating that IaaA is not strictly a dipeptidase. An internal isoAsp bond, Ala-isoAsp-Leu-Ala, however, was not hydrolyzed. Two chromogenic substrates, isoAsp-pNA and isoAsp-AMC, were both hydrolyzed (data not shown). The apparent Km of IaaA for the best substrate, isoAsp-Leu, was determined to be 0.3 mM. These results suggest that IaaA is an isoAsp aminopeptidase.
TABLE 2.
Substrate specificity of IaaA
| Peptide | Relative activitya |
|---|---|
| isoAsp-Leu | 100 |
| isoAsp-Ala | 83 |
| isoAsp-His | 69 |
| isoAsp-Phe | 24 |
| isoAsp-Lys | 39 |
| isoAsp-Phe-ME | 0.3 |
| isoAsp-Gly | 5 |
| isoAsp-Leu-Ala | 35 |
| Ala–isoAsp-Leu-Ala | <0.001 |
| Asp-Leu | 0.2 |
| Asp-Ala | 0.01 |
| Asp-Gly-Gly | <0.001 |
| Glu-Leu | <0.001 |
| γGlu-Leu | <0.001 |
| Asn-Leu | <0.001 |
| Asn | 1.6 |
| Asn-GlcNac | <0.001 |
Peptide hydrolysis was determined by HPLC analysis of reaction products as described in Materials and Methods and is expressed as a percentage of the rate of isoAsp-Leu hydrolysis (51.5 nmol of Asp released/min/μg).
Construction and properties of an iaaA disruption.
The iaaA gene was disrupted as described in Materials and Methods. When soluble cell extracts of a strain (TN5538) carrying the iaaA::Cmr mutation and a wild-type strain (TN2373) were compared by nondenaturing PAGE and staining for isoAsp-Leu-hydrolyzing activity, the band corresponding to IaaA was observed to be absent in the mutant (Fig. 5). This result confirms that the isoAsp peptidase activity is encoded by iaaA.
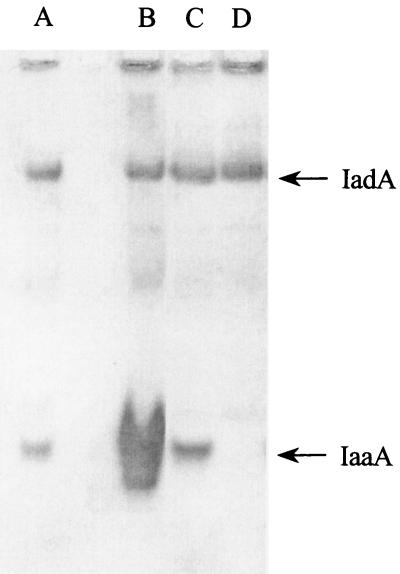
FIG. 5.
Nondenaturing PAGE of strains either overexpressing or lacking IaaA. Soluble cell extracts (10 μg each) were subjected to nondenaturing PAGE, after which the gel was stained for isoAsp-Leu hydrolysis using the amino acid oxidase assay. Strains tested were TN2373, wild type for peptidases (lane A); TN2540/pCM529, carrying an IaaA overexpression plasmid (lane B); TN2540/pCM531, carrying a plasmid with iaaA1::Cmr (lane C); and TN5379, containing a chromosomal iaaA disruption, iaaA::Cmr (lane D).
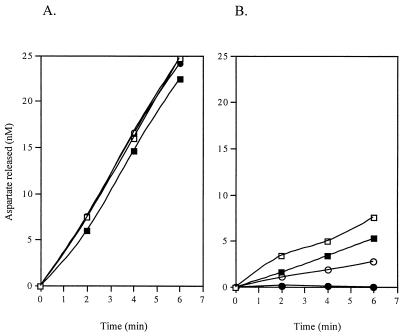
To test the hypothesis that IadA and IaaA are the only isoAsp-Leu-hydrolyzing enzymes in serovar Typhimurium, cell extracts of strains lacking one or both of these activities were measured for Asp peptide-hydrolyzing activity. The following strains were tested: TN1379 (wild type), TN5379 (iadA), TN5538 (iaaA), and TN5617 (iadA iaaA). The presence or absence of either or both of the proteins IadA and IaaA had no effect on α-Asp-Leu hydrolysis, suggesting that neither enzyme contributes significantly to Asp peptide hydrolysis (Fig. 6A). A mutation in either peptidase, however, led to a significant decrease in the rate of isoAsp-Leu hydrolysis from that of the wild-type strain, and a double mutant had no detectable activity (Fig. 6B). These results suggest that there are no additional isoAsp peptide-hydrolyzing enzymes in serovar Typhimurium that are detectable under the assay conditions used in this experiment.
FIG. 6.
Effects of iadA and iaaA mutations on Asp-Leu (A) and isoAsp-Leu (B) hydrolysis in cell extracts. Soluble cell extracts (100 μg) of strains containing no mutations (TN1379 [□]), mutations in either iadA or iaaA (TN5379 [■] and TN5538 [○], respectively), or mutations in both iadA and iaaA (TN5617 [●]) were assayed by HPLC for the rate of Asp-Leu and isoAsp-Leu hydrolysis.
As described earlier, serovar Typhimurium does not grow on isoAsp-Leu as a leucine source but will grow on Asp-Leu. Because IaaA has a low level of Asp-Leu-hydrolyzing activity in vitro, any contribution to growth on Asp-Leu was of interest. Strains with mutations in iaaA alone or in combination with mutations in genes encoding other Asp peptidases were compared to a wild-type strain. Each of four strains (TN1379, TN5538, TN5391, and TN5542) was grown in E minimal medium containing glucose and either leucine or Asp-Leu as the sole source of leucine. Growth rates and final cell densities of the mutant strains did not appear to vary significantly from that of the wild type (data not shown).
Identification of an additional Asp-X peptidase.
Although TN5542 (pepB pepE iadA iaaA) lacks all known enzymes that hydrolyze Asp-Leu, this strain still utilizes this peptide as a source of Leu. In addition, this strain can utilize other Asp-X peptides as nitrogen sources. These results suggest strongly that another Asp-X peptidase is present in serovar Typhimurium. Experiments undertaken during the course of characterizing IaaA demonstrated the existence of such an enzyme. When a crude cell extract of TN5426 was chromatographed on Q Sepharose and the fractions were assayed for Asp-Leu hydrolysis under the standard assay conditions, only one peak of activity corresponding to IaaA was observed. In an effort to test the effects of various divalent cations on IaaA activity, we assayed the column fractions in the presence of the cations and found that when Mn2+ (1 mM) was included in the reaction mixture an additional peak of activity distinct from IaaA was observed. This activity was not observed in the presence of other divalent ions (Mg2+, Co2+, Ca2+, or Zn2+ as the chloride salt). To provide a preliminary characterization of the substrate specificity of the activity we tested its ability to hydrolyze other peptides. Peptides with residues other than Asp at the N terminus (Glu-Leu, Leu-Leu, Leu-Pro, Asn-Leu, and Lys-Leu) were not hydrolyzed. In addition, no activity toward Asp-pNA, Met-Asp-Phe, or isoAsp-Leu was observed. Based on these limited data the activity appears to be specific for N-terminal Asp peptides and may be another Asp-specific peptidase. Based on gel filtration chromatography, the native molecular mass was estimated to be 70 kDa. When partially purified fractions containing the activity were stained for Asp-Leu hydrolysis after nondenaturing PAGE, only a very faint band could be observed, suggesting that the enzyme is inactivated by this procedure and explaining why it was not observed previously.
DISCUSSION
The goal of this work was to identify all of the Asp peptide-hydrolyzing activities in S. enterica serovar Typhimurium. We have shown that there are at least three enzymes in addition to peptidases B and E that can hydrolyze N-terminal Asp peptides. Two of these enzymes are more active toward isoAsp (β-Asp) peptides than toward peptides with the normal α-Asp linkage. One of these enzymes is the serovar Typhimurium homolog of E. coli IadA (isoAsp dipeptidase) (7). This enzyme was originally described by Haley (9), who reported that it did not hydrolyze α-Asp peptides. Because of these earlier observations, we considered the possibility that the apparent hydrolysis of Asp-Leu observed after nondenaturing gel electrophoresis might have resulted from contamination of the substrate with the isoAsp peptide. HPLC analysis of the lot of Asp-Leu used in these experiments showed that it contained about 3% isoAsp-Leu. It is possible, therefore, that some of the signal observed is generated by hydrolysis of the isoAsp peptide. The ability of IadA (and IaaA) to hydrolyze Asp-Leu was unambiguously established, however, by using HPLC to monitor the rate of substrate disappearance. Because Asp-Leu and isoAsp-Leu were easily separated by HPLC, we could monitor the rate of loss of each substrate independently. The data concerning the relative rates of hydrolysis of the two peptides by both IadA and IaaA were determined in this way, and these data establish clearly that both enzymes are able to hydrolyze Asp-Leu. The assay method used by Haley was much less sensitive than the HPLC assay used here, and it is not unreasonable to assume that the relatively slow hydrolysis of α-Asp peptides by this enzyme might not have been observed under the conditions of these earlier experiments.
The gene encoding the second isoAsp peptidase, iaaA, has been identified for the first time in this work as a homolog of ybiK in E. coli. Although IaaA and IadA share the ability to hydrolyze an N-terminal isoAsp linkage, the two enzymes show no amino acid sequence similarity and clearly belong to different structural and mechanistic families. Based on similarity to certain plant asparaginases, the annotators of the E. coli genome suggested that the ybiK gene product is an asparaginase (1). Although the amino acid sequence similarity between the ybiK gene product and these plant enzymes is significant (44 to 45% identity), our results indicate that IaaA has only a relatively weak l-asparaginase activity. The ybiK gene product also has significant amino acid sequence similarity to glycosylasparaginases from both Flavobacterium and eukaryotes, but it has no activity towards the substrate most rapidly hydrolyzed by glycosylasparaginases, N-acetylglucosaminyl-l-asparagine (Asn-GlcNAc). The human glycosylasparaginase has been shown to hydrolyze isoAsp peptides in addition to Asn-GlcNAc (18), suggesting that the primary difference between this enzyme and IaaA is the more restrictive substrate specificity of IaaA.
Both the lupin asparaginases and the glycosylasparaginases are members of the Ntn hydrolase enzyme family (2). Ntn hydrolases are generated from enzymatically inactive precursors by a proteolytic processing step that generates a new N-terminal serine, threonine, or cysteine residue. This residue functions as the catalytic nucleophile in the hydrolytic reaction. Crystal structures have been determined for several members of this family: Thermoplasma 20S proteasome (12), glutamine phosphoribosylpyrophosphate amidotransferase (17), human glycosylasparaginase (19), Flavobacterium glycosylasparaginase (8), and penicillin acylase (6). These structures reveal a common fold characteristic of the family.
Posttranslational processing of many Ntn hydrolases is thought to occur by autoproteolysis of a homodimeric precursor to generate an α2β2 heterotetramer as the active species. Our data show clearly that IaaA is a heterotetramer. The active form of IaaA has a native molecular mass of 60 kDa, twice the molecular mass predicted by the open reading frame (32 kDa). SDS-PAGE analysis showed that this species contains two polypeptides, and N-terminal amino acid sequencing indicates that they have been generated by proteolysis at a position equivalent to the processing site in similar enzymes (Gly178-Thr179). Although specific evidence for autoprocessing of IaaA has not been presented, it is likely that it shares this property with other Ntn hydrolases.
The physiological functions of IadA and IaaA are not clear. Both enzymes appear to preferentially hydrolyze isoAsp peptides, although they differ in that IadA is restricted to dipeptides whereas IaaA hydrolyzes tripeptides. It is generally assumed that isoAsp peptidases function in the hydrolysis of peptides generated by the intracellular degradation of isoAsp-containing proteins. Such proteins arise from the isomerization of Asp or Asn peptide bonds. These reactions are thought to occur most often in flexible regions of proteins, and because they introduce an additional CH2 group into the peptide backbone, they frequently affect the secondary structure and the functioning of the protein. A protein damaged in this manner is either repaired by l-isoAsp methyltransferase (encoded by pcm in E. coli) or degraded. Gary and Clarke (7) have shown that E. coli iadA mutations have no discernible effect on growth or on stationary-phase survival. Although the phenotype of an iaaA iadA double mutant has not been thoroughly characterized, our observations suggest that this strain grows normally.
The location of iaaA as the first gene of an operon that also encodes a putative ATP-binding cassette transporter suggests that peptide catabolism might be an additional function for IadA and IaaA. We speculate that the transporter may function to transport isoAsp peptides, allowing them to be used as sources of amino acids. The third open reading frame in the proposed operon encodes a periplasmic binding protein that on the basis of its sequence belongs to the peptide and nickel binding transporters (family 5 according to Tam and Saier [23] and family 2 according to Linton and Higgins [11]). With the exception of a nickel transporter, all of the characterized members of this family have been shown to bind and import peptides. Although wild-type serovar Typhimurium does not grow on isoAsp-Leu as a leucine source in glucose minimal medium, mutants that do grow can easily be isolated and at least one class of these mutants overproduced IadA (data not shown). It seems possible, therefore, that one or the other of these genes might be regulated to allow isoAsp peptide use under as-yet-unknown environmental conditions.
Serovar Typhimurium contains a seemingly large number of Asp and isoAsp peptidases. In the present study we have shown that the role of IaaA, like that of IadA, is to hydrolyze isoAsp peptides. Peptidase B, peptidase E, and a previously unreported peptidase all hydrolyze α-Asp peptides. The third, as-yet-unidentified peptidase has the unusual characteristic of requiring manganese for activity. It will be interesting to define the particular specificity of this enzyme and to determine whether or not it is the only remaining Asp peptidase in serovar Typhimurium.
ACKNOWLEDGMENTS
This work was supported by a grant (AI10333) from the National Institute of Allergy and Infectious Diseases. R.A.L. was supported in part by a training grant (5 T32 GMO7283) from the National Institute of General Medical Sciences.
REFERENCES
- 1.Blattner F R, Plunkett III G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coliK-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 2.Brannigan J A, Dodson G, Duggleby H J, Moody P C, Smith J L, Tomchick D R, Murzin A G. A protein catalytic framework with an N-terminal nucleophile is capable of self-activation. Nature. 1995;378:416–419. doi: 10.1038/378416a0. [DOI] [PubMed] [Google Scholar]
- 3.Brosius J. Superpolylinkers in cloning and expression vectors. DNA. 1989;8:759–777. doi: 10.1089/dna.1989.8.759. [DOI] [PubMed] [Google Scholar]
- 4.Carter T H, Miller C G. Aspartate-specific peptidases in Salmonella typhimurium: mutants deficient in peptidase E. J Bacteriol. 1984;159:453–459. doi: 10.1128/jb.159.2.453-459.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Conlin C A, Håkensson K, Liljas A, Miller C G. Cloning and nucleotide sequence of the cyclic AMP receptor protein-regulated Salmonella typhimurium pepEgene and crystallization of its product, an α-aspartyl dipeptidase. J Bacteriol. 1994;176:166–172. doi: 10.1128/jb.176.1.166-172.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duggleby H J, Tolley S P, Hill C P, Dodson E J, Dodson G, Moody P C. Penicillin acylase has a single-amino-acid catalytic centre. Nature. 1995;373:264–268. doi: 10.1038/373264a0. [DOI] [PubMed] [Google Scholar]
- 7.Gary J D, Clarke S. Purification and characterization of an isoaspartyl dipeptidase from Escherichia coli. J Biol Chem. 1995;270:4076–4087. doi: 10.1074/jbc.270.8.4076. [DOI] [PubMed] [Google Scholar]
- 8.Guo H C, Xu Q, Buckley D, Guan C. Crystal structures of Flavobacteriumglycosylasparaginase. An N-terminal nucleophile hydrolase activated by intramolecular proteolysis. J Biol Chem. 1998;273:20205–20212. doi: 10.1074/jbc.273.32.20205. [DOI] [PubMed] [Google Scholar]
- 9.Haley E E. Purification and properties of a beta-aspartyl peptidase from Escherichia coli. J Biol Chem. 1968;243:5748–5752. [PubMed] [Google Scholar]
- 10.Lassy R A L, Miller C G. Peptidase E, a peptidase specific for N-terminal aspartic dipeptides, is a serine hydrolase. J Bacteriol. 2000;182:2536–2543. doi: 10.1128/jb.182.9.2536-2543.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Linton K J, Higgins C F. The Escherichia coliATP-binding cassette (ABC) proteins. Mol Microbiol. 1998;28:5–13. doi: 10.1046/j.1365-2958.1998.00764.x. [DOI] [PubMed] [Google Scholar]
- 12.Lowe J, Stock D, Jap B, Zwickl P, Baumeister W, Huber R. Crystal structure of the 20S proteasome from the archaeon T. acidophilumat 3.4 A resolution. Science. 1995;268:533–539. doi: 10.1126/science.7725097. [DOI] [PubMed] [Google Scholar]
- 13.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 14.Mathew Z, Knox T M, Miller C G. Salmonella entericaserovar Typhimurium peptidase B is a leucyl aminopeptidase with specificity for acidic amino acids. J Bacteriol. 2000;182:3383–3393. doi: 10.1128/jb.182.12.3383-3393.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McHugh G L, Miller C G. Isolation and characterization of proline peptidase mutants of Salmonella typhimurium. J Bacteriol. 1974;120:364–371. doi: 10.1128/jb.120.1.364-371.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller C G, Mackinnon K. Peptidase mutants of Salmonella typhimurium. J Bacteriol. 1974;120:355–363. doi: 10.1128/jb.120.1.355-363.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muchmore C R, Krahn J M, Kim J H, Zalkin H, Smith J L. Crystal structure of glutamine phosphoribosylpyrophosphate amidotransferase from Escherichia coli. Protein Sci. 1998;7:39–51. doi: 10.1002/pro.5560070104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Noronkoski T, Stoineva I B, Ivanov I P, Petkov D D, Mononen I. Glycosylasparaginase-catalyzed synthesis and hydrolysis of beta-aspartyl peptides. J Biol Chem. 1998;273:26295–26297. doi: 10.1074/jbc.273.41.26295. [DOI] [PubMed] [Google Scholar]
- 19.Oinonen C, Tikkanen R, Rouvinen J, Peltonen L. Three-dimensional structure of human lysosomal aspartylglucosaminidase. Nat Struct Biol. 1995;2:1102–1108. doi: 10.1038/nsb1295-1102. [DOI] [PubMed] [Google Scholar]
- 20.Schagger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 21.Schmieger H. Phage P22 mutants with increased or decreased transduction abilities. Mol Gen Genet. 1972;119:75–88. doi: 10.1007/BF00270447. [DOI] [PubMed] [Google Scholar]
- 22.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 23.Tam R, Saier M H., Jr Structural, functional, and evolutionary relationships among extracellular solute-binding receptors of bacteria. Microbiol Rev. 1993;57:320–346. doi: 10.1128/mr.57.2.320-346.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vogel H J, Bonner D M. Acetylornithinase of Escherichia coli: partial purification and some properties. J Biol Chem. 1956;218:97–106. [PubMed] [Google Scholar]