Abstract
Cenchrus ciliaris is an apomictic, allotetraploid pasture grass widely distributed in the tropical and subtropical regions of Africa and Asia. In this study, we aimed to investigate the genomic organization and characterize some of the repetitive DNA sequences in this species. Due to the apomictic propagation, various aneuploid genotypes are found, and here, we analyzed a 2n = 4x + 3 = 39 accession. The physical mapping of Ty1-copia and Ty3-gypsy retroelements through fluorescence in situ hybridization with a global assessment of 5-methylcytosine DNA methylation through immunostaining revealed the genome-wide distribution pattern of retroelements and their association with DNA methylation. Approximately one-third of Ty1-copia sites overlapped or spanned centromeric DAPI-positive heterochromatin, while the centromeric regions and arms of some chromosomes were labeled with Ty3-gypsy. Most of the retroelement sites overlapped with 5-methylcytosine signals, except for some Ty3-gypsy on the arms of chromosomes, which did not overlap with anti-5-mC signals. Universal retrotransposon probes did not distinguish genomes of C. ciliaris showing signals in pericentromeric regions of all 39 chromosomes, unlike highly abundant repetitive DNA motifs found in survey genome sequences of C. ciliaris using graph-based clustering. The probes developed from RepeatExplorer clusters gave strong in situ hybridization signals, mostly in pericentromeric regions of about half of the chromosomes, and we suggested that they differentiate the two ancestral genomes in the allotetraploid C. ciliaris, likely having different repeat sequence variants amplified before the genomes came together in the tetraploid.
Keywords: repetitive DNA, methylation, retrotransposons, chromosomes, polyploidy, aneuploidy, breeding
Introduction
Cenchrus ciliaris L. (buffelgrass, buffel-grass, or African foxtail grass; syn. Pennisetum ciliare) is an apomictic, allotetraploid perennial C4 grass native to tropical and subtropical arid regions of Africa and Western Asia. Predominantly distributed in warmer tropical regions, C. ciliaris grows in a range of harsh conditions (Kharrat-Souissi et al., 2012, 2013, 2014; Marshall et al., 2012). Cenchrus species are the predominant pasture grasses in India due to their high fodder value (Meena and Nagar, 2019). They are of great ecological importance due to persistence in native arid ecosystems, associated with tolerance to heavy grazing and drought, with deep root systems, and they are also used for erosion control (Miller et al., 2010). C. ciliaris is recognized as an invasive species in Australia (where it is a “declared plant”), South America, and the USA (particularly in Arizona and the Sonoran Desert) and has become a threat to natural ecosystems (Jackson, 2005; Miller et al., 2010) both by displacement of native plants and by intensifying wildfires. It is important to study the genetic characteristics and genomic organization of C. ciliaris to understand the association between biological, agricultural, and ecological characteristics.
Cenchrus ciliaris, with a base chromosome number of x = 9, includes tetra-, penta-, hexa-, and septa-ploid cytotypes (Visser et al., 1998; Kharrat-Souissi et al., 2012, 2013; Dhaliwal et al., 2018), and aneuploid accessions with 2n = 32, 40, 43, 44, and 48 have also been found (Burson et al., 2012; Carloni-Jarrys et al., 2018). Most accessions are apomictic, although sexual plants are also found occasionally in nature (Fisher et al., 1954; Marshall et al., 2012; Yadav et al., 2012), with a DNA-based SCAR marker reported by Dwivedi et al. (2007). Apomictic seed production preserves superior genotypes, including heterozygotes and polyploids (Spillane et al., 2004; Bhat et al., 2005; Abdi et al., 2016), and can discover new polyploid lineages. The characteristic is well-represented in polyploid grasses, including members of Paniceae grass genera such as Panicum, Pennisetum, Cenchrus, and Paspalum (Worthington et al., 2016; Higgins et al., 2021; Tomaszewska et al., 2021b). In Urochloa (Brachiaria), there is a robust molecular marker discriminating sexual and apomictic plants. The genes involved in C. ciliaris have been examined and compared with other species (Roche et al., 1999; Akiyama et al., 2005; Yadav et al., 2012). Similar to the widely grown Urochloa (Brachiaria) tropical forage grasses (Tomaszewska et al., 2021a,b), apomicts at all ploidies produce seeds, as there is neither meiosis nor necessity for restitution of diploid pairing behavior (Sepsi et al., 2017). Although there is relatively weak evidence that polyploid plants as a group outperform diploids either in wild or agricultural settings (particularly where selection is strong enough to remove mildly deleterious mutations), there are many prospects to exploit polyploidy systematically using additional gene alleles and increased genetic variability, which enhance adaptability (Brits et al., 2003; Bhat et al., 2005; Kharrat-Souissi et al., 2014).
Cenchrus ciliaris has a genome size of approximately 3,000 Mbp in the tetraploid species, 50% larger than the 1,971 Mbp reported for the genome of C. purpureus (Elephant or Napier grass; 2n = 4x = 28) and the more distantly related but smaller 1,580 Mbp C. americanus (2n = 2x = 14; syn. Pennisetum americanum) genome (Yan et al., 2021). A major proportion of plant genomes consist of repeated sequences such as ribosomal DNAs, tandem repeats, satellites, and transposable elements, which play an important role in the shaping of genome organization (rDNA in Cenchrus: Kharrat-Souissi et al., 2012; other species: Heslop-Harrison and Schwarzacher, 2011; Macas et al., 2011; Biscotti et al., 2015). Yan et al. (2021) reported the composition of the C. purpureus genome as comprising 66.32% repetitive sequences, similar to the proportion of repeats in C. americanus (77.2%). Ty3-gypsy and Ty1-copia are two superfamilies of LTR-retroelements widely distributed at the centromeric region of chromosomes in many members of Poaceae and terminal heterochromatic chromosomal regions in Allium cepa, respectively (Kumar et al., 1997).
Members of the major groups of repetitive elements, tandemly repeated satellites, and transposable elements (Class I retrotransposons and Class II DNA transposons) differ in their abundance and genomic distribution in the genome. Tandemly repeated motifs may be located in subterminal, intercalary, or centromeric regions of chromosomes, or, apart from the 5S and 45S rDNA arrays, not be abundant. In contrast, all plants have relatively abundant retrotransposons, often with a dispersed distribution (Heslop-Harrison et al., 1997), and often dispersed throughout the euchromatin but absent from specialized regions such as heterochromatin, centromeres, telomeres, and nuclear organization regions (NOR). Retrotransposon families may also be restricted to characteristic genomic locations. In Arabidopsis thaliana, many Ty3-gypsy retroelements are present in the pericentromeric heterochromatin region (Brandes et al., 1997b). Centromeric retroelements in maize (CRM) are a lineage of Ty3-gypsy retroelements (Jin et al., 2004), and the diploid oat Avena longiglumis (2n = 2x = 14; 3,850 Gb genome size) has specific LTR retrotransposons located at the centromeres (Liu et al., 2019, 2022); in the rest of the cases, they are present with other retroelement families dispersed along all chromosomes. Some of these repetitive elements may have a role in centromere functionality, although they may also accumulate in regions of the genome where there are few genes.
The activity of retroelements is modulated by epigenetic mechanisms through DNA methylation (Slotkin and Martienssen, 2007; Le et al., 2015), wherein the methyl group is covalently added to the fifth carbon of cytosine residues to form 5-methylcytosine (5-mC) (Moore et al., 2013). The expression profile of retroelements can be correlated with the differential methylation pattern (Castilho et al., 1999). DNA methylation is mainly distributed in heterochromatic regions in the genome that are rich in repetitive sequences consisting of retroelements (Vining et al., 2012). Comparison of methylomes between the tissues or of different stages can be studied to determine genes regulated through methylation and the pathways involved in response to stress and developmental conditions (Li et al., 2020). Whole-genome methylation analysis can reveal the distribution pattern of methylation in the genome and the associated genes or regions. In A. thaliana, genome-wide methylation analysis found that one-third of genes are methylated within their transcribed regions, while in rice, approximately 16% of genes are enriched with methylation (Zilberman et al., 2007; He et al., 2010; Vining et al., 2012). Genome methylation analysis in Populus trichocarpa also indicated that Ty3-gypsy retroelements, which are abundant in centromeric, pericentromeric, and heterochromatic regions in plants, are enriched with DNA methylation (Douglas and Di Fazio, 2010; Vining et al., 2012). Therefore, the methylation status of retroelements in the genome can be assessed by combining the physical mapping of retroelements through fluorescent in situ hybridization (FISH) with an assessment of 5-methylcytosine through immunostaining, which enables us to understand the genome-wide distribution pattern of retroelements and their association with the methylation.
In this study, we aimed to characterize the nature of repetitive DNA in Cenchrus ciliaris, investigate the genomic organization, including tandemly repeated satellite DNA and LTR retroelements, and examine genome-wide methylation patterns in C. ciliaris, aiming to characterize features related to its contribution to the genome, evolution, and possible variation in the genomes.
Materials and methods
Fixation of plant material
Seeds of Cenchrus ciliaris (CcA7-5) were germinated in Petri dishes at 25°C for 3 days, and root tips were collected. Seedling root tips were pretreated in alpha-bromonaphthalene for 2 h to accumulate metaphases and then fixed in ethanol:glacial acetic acid (3:1) solution and stored at 4°C (Schwarzacher and Heslop-Harrison, 2000).
Preparation of chromosome spreads
Root tips were washed two times in enzyme buffer (10 mM citric acid/sodium citrate) for 15 min and then digested with enzyme solution (20 U/ml cellulase, 10 U/ml Onozuka RS cellulase, and 20 U/ml pectinase in enzyme buffer) for 45 min at 37°C. After digestion, root tips were again washed in enzyme buffer. The meristematic tissues were dissected and squashed in 60% acetic acid under the coverslip by using light thumb pressure. Slides were frozen on dry ice for 5–10 min and then coverslips were removed with a razor blade. Slides were air dried and used for FISH.
Isolation of genomic DNA and designing of probes for repetitive sequences
Genomic DNA was isolated from the apomictic genotype C. ciliaris (CcA7-5) using the Qiagen DNeasy Plant mini kit. Primers were designed from conserved regions of the reverse transcriptase domain of LTR retroelements (Ty1-copia and Ty3-gypsy) and 45S ribosomal DNA genes using the BLAST search to identify the conserved region (Sayers et al., 2019; Table 1). Whole-genome sequencing data from a Chinese variety of C. ciliaris (deposited in SRA Sequence Read Archive under accession SRR8666664, Illumina HiSeq 2500; Nevill et al., 2020) were downloaded and used to discover the most abundant repetitive DNA sequences. Similarity-based clustering, repeat identification, and classification were performed using RepeatExplorer2 (Novak et al., 2013). In detail, raw paired-end reads were preprocessed, including quality-filtering (quality score ≥10 over 95% of bases and no Ns allowed), trimming, excluding overlapping read pairs, and interlacing. A subset of reads was randomly sampled from a dataset as input for RepeatExplorer to identify repetitive sequences and tandem repeats (following Novak et al., 2013, including the use of the REXdb reference database of transposable element domains, Neumann et al., 2019). For selected repetitive elements, primers were designed (Table 1) using Primer3 (Rozen and Skaletsky, 1999). Conserved regions were amplified from genomic leaf DNA in a standard polymerase chain reaction (PCR) using specific primers synthesized commercially (Sigma). DNA probes for Ty3-gypsy, Ty1-copia, CL105, CL58, and 18S rDNA were obtained by PCR amplification from genomic DNA, and labeled with biotin-16-dUTP or digoxigenin-11-dUTP with the BioPrime CGH array labeling kit (Invitrogen), and then purified using BioPrime Purification Module (Invitrogen). Fluorochrome-labeled synthetic oligonucleotide probes for the clusters (CL), CenchCL58_869, CenchStCL1ConsS, CenchEnCL1ConsRC, and CenchCL22Copia were used as probes for FISH (Table 1).
Table 1.
List of PCR primers and commercial oligonucleotide probes used in the study.
| Primer name | Primer sequence | Oligonucleotide length [bp] | Product length [bp] | Probe name | References |
|---|---|---|---|---|---|
| CopiaQMDVKT _F CopiaYVDDML_R |
CARATGGARGTNAARAC ARCATRTCRTCNACRTA |
17 | 290 | universal Ty1-copia | Flavell et al., 1992a,b; Hirochika and Hirochika, 1993 |
| GypsyRT1RMCVDYR_F | MRNATGTGYGTNGAYTAYMG | 20 | 410 | universal Ty3-gypsy | Friesen et al., 2001 |
| GypsyRT4YAKLSKC_R | RCAYTTNSWNARYTTNGCR | 19 | |||
| 18S rDNA_F | CGAACTGTGAAACTGCGAATGGC | 23 | 900 | 18S rDNA | Chang et al., 2010 |
| 18S rDNA_R | TAGGAGCGACGGGCGGTGTG | 20 | |||
| CC_C105F76 | CGAGTAGCCCAAATCCAGGA | 20 | 590 | Gy105 | This paper |
| CC_C105R665 | TTGCTGAACCAACTCCCTGT | 20 | |||
| CC_C105F76 | CGAGTAGCCCAAATCCAGGA | 20 | 919 | Gy105 | This paper |
| CC_C105R994 | CAAGGAAGGGGAAACAACGG | 20 | |||
| CenchCL58_361F | GGCTGGGGGCGATAAGTAG | 19 | 350 | CenchCL58 | This paper |
| CenchCL58_710R | CAAGAAGGCGCAGATTGTGG | 20 | |||
| CenchCL58_869 | [Cy3]CTGGAGGTTGTAATTCCAAGGCCACGCATCTCAGAAGGGAGTCG | 44 | – | CenchCL58_869 | This paper |
| CenchStCL1ConsS | [Cy3]CCATTTTCACAACTTCGGTACCCTGATGTAGTGCATTCATGCAC | 44 | – | CenchStCL1ConsS | This paper |
| CenchEnCL1ConsRC | [FAM]GCCTACCCCATTAGATCCCAAACGATGTTTGAGAGTGTTTAGGAG | 45 | – | CenchEnCL1ConsRC | This paper |
| CenchCL22Copia | [FAM]GGGGTTAGAAATAGGACCCAACAAGAAAAACAATATGATGTAACCGCGAGG | 51 | – | CenchCL22Copia | This paper |
Fluorescence in situ hybridization
Slides were refixed in ethanol:glacial acetic acid (3:1) for 30 min, dehydrated in 100% ethanol two times for 5 min, and then air dried. Slides were treated with RNase (100 μg/ml) in 2X SSC for 1 h at 37°C in a humid chamber and then washed two times in 2X SSC for 5 min at room temperature. Then, the slides were incubated in 0.01 M HCl for 2 min, treated with pepsin (5 μg/ml) in 0.01 M HCl for 10 min at 37°C in a humid chamber, rinsed in distilled water for 1 min, and washed in 2X SSC for 5 min. Slides were refixed in 4% formaldehyde at room temperature for 10 min, washed in 2X SSC for 2 min, and then in 2X SSC for 10 min. Slides were then dehydrated in an ethanol series (70, 85, and 100% ethanol, 2 min each).
The hybridization mixture consisted of probes (2 ng/μl each; Table 1), 50% formamide (v/v), 2X SSC, 10% dextran sulfate (v/v), 200 ng/μl salmon sperm DNA, 0.125 mM EDTA, and 0.125% SDS; a total volume of 40 μl was denatured at 83°C for 10 min and then cooled down on the ice for 10 min. The hybridization mixture was then added to the slides and placed in a hybridization oven at 73°C for 6 min to denature the probe and chromosomes and then cooled to 37°C and left at that temperature overnight for hybridization.
Posthybridization washes were performed to remove the unincorporated and weakly hybridized probes. Slides were washed two times in 2X SSC for 2 min at 42°C and then washed in 0.1X SSC at 42°C for 2 min and again for 10 min at the same temperature before a final wash in 2X SSC for 5 min at room temperature. To detect probes, slides were incubated in a detection buffer (4X SSC containing 0.2% Tween 20) for 5 min and blocked by incubation with 5% BSA (bovine serum albumin) in a detection buffer for 10 min at 37°C in a humid chamber. Hybridization sites were detected by incubation with streptavidin conjugated to Alexa 594 streptavidin antibody or FITC (fluorescein isothiocyanate) anti-digoxigenin antibody in the detection buffer with 5% BSA for 1 h at 37°C in a humid chamber. Slides were washed in detection buffer three times at 40°C for 10 min each. Chromosomes were then counterstained with DAPI (4',6-diamidino-2-phenylindole) in antifade solution (AF1, Citifluor) and photographed.
Methylation analysis using anti-5-methylcytosine through immunostaining
Immunostaining was performed to visualize how whole-genome cytosine methylation is distributed in different chromosomal regions of C. ciliaris genome. Coverslips were removed from slides previously denatured, probed with the DNA, hybridized, and analyzed. After washing in detection buffer, the preparations were blocked by 1% BSA (w/v) in 1X PBS buffer/0.5% Tween 20 for 30 min at room temperature and incubated with primary anti-5-methylcytosine antibody (anti-5-mC; 1:500 in 1X PBS buffer/0.5% Tween 20) for 1 h at 37°C in a humid chamber. Slides were then washed in 1X PBS buffer/0.5% Tween 20 two times for 5 min each at room temperature and incubated with Alexa 488-conjugated goat anti-mouse secondary antibody (1:500) in 1X PBS buffer/0.5% Tween 20. Slides were then washed two times in 1X PBS buffer/0.5% Tween 20 for 5 min at room temperature and stained with DAPI in antifade solution (AF1, Citifluor) and metaphases previously analyzed with FISH were rephotographed after immunostaining.
Photography and imaging
Slides were examined with a Nikon Eclipse 80i epifluorescence microscope equipped with a DS-QiMc monochromatic camera and NIS-Elements version 2.34 software (Nikon). Images were analyzed using the Adobe Photoshop software.
Results
Chromosomes of Cenchrus ciliaris
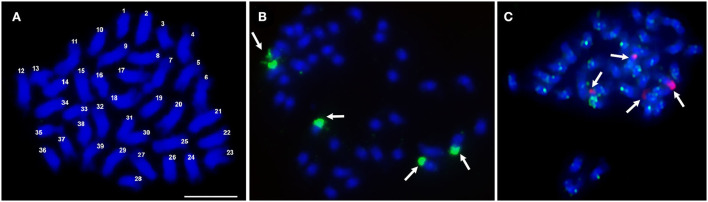
Most plants of the apomictic tetraploid C. ciliaris were 2n = 4x = 36, but one hyperploid 2n = 4x = 39 plant was found (Figure 1A); all chromosomes were submetacentric and of similar length. The 45S rDNA probe was located at four subterminal regions (Figures 1B,C arrows), also visible as the satellite at the nucleolar organizing region (NORs by DAPI staining). Similar results were also observed by Kharrat-Souissi et al. (2012).
Figure 1.
Number of chromosomes and rDNA sites in Cenchrus ciliaris. (A) Metaphase spread of C. ciliaris after DAPI staining (blue) showing a chromosome count of 2n = 4x + 3 = 39. (B) Fluorescent in situ hybridization with 45S rDNA probe fluorescing green (arrows). (C) 45S rDNA probe (red) showing four major hybridization sites and Gy105 probe (green) hybridizing to most parts of the chromosomes. Scale bar = 10 μm in (A).
The degenerate PCR product used as a “universal” Ty1-copia (Table 1) probe labeled broad regions of chromosomes, with sites often overlapping or spanning centromeric DAPI-positive heterochromatin (Figure 2Ai). Along with the centromeric regions, the arms of most chromosomes were labeled with Ty3-gypsy, but most distal ends lacked signal (Figure 2Bi). Probe Gy105 (Table 1), amplified from a sequence including a Cenchrus ciliaris Ty3-gypsy retrotransposon (Genbank JN559989, extended from Genbank ED545466 identified in a BAC by Conner et al., 2008) (Figure 1C, green signal), labeled regions near centromeres on most but not all chromosomes.
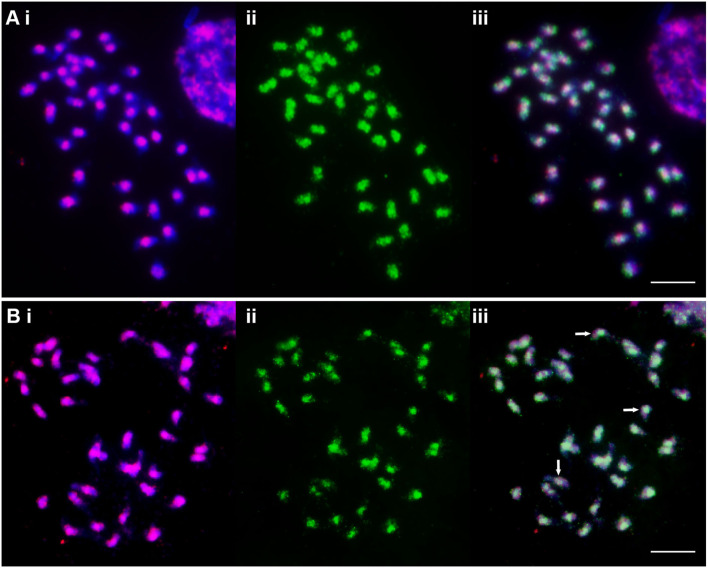
Figure 2.
Metaphase chromosomes of C. ciliaris (2n = 4x + 3 = 39) after FISH (red fluorescing, (i) and immunostaining with anti-5-methylcytosine (green fluorescing, (ii) and as an overlap image (iii). (A) The “universal” Ty1-copia probe shows concentrated signal around the centromeres of all chromosomes, while 5-mC signal is not only seen overlapping them but also further along the chromosome arms. 5-mC strength is variable between the chromosomes with most showing a small gap at the centromeres. (B) The “universal” Ty3-gypsy probe shows a dispersed signal over the chromosome arms, but is absent from the most distal parts. 5-mC signal covers most Ty3-gypsy signals but is weak or absent in some cases (arrows). Scale bar = 10 μm.
Immunostaining with anti-5-methylcytosine antibody showed genome-wide methylation patterns in apomictic C. ciliaris along chromosomes, with variability observed in the patterns. About one-third of chromosome arms were more weakly labeled, particularly at the ends of the arms; most centromeric regions showed small gaps, and a few chromosomes were strongly labeled (Figures 2Aii, Bii).
Chromosomal locations of the anti-5-methylcytosine antibody were compared with retroelement distribution by sequential labeling of the same metaphase preparations. The anti-5-mC signals overlapped the centromeric Ty1-copia probe signals of most of the chromosomes (Figure 2Aiii). Immunostaining of chromosomal spread previously probed with Ty3-gypsy retrotransposons showed methylation signals of anti-5-mC overlapped with Ty3-gypsy retroelements on most of the pericentromeric and centromeric regions of chromosomes. However, some Ty3-gypsy signals on the arms of chromosomes did not overlap with anti-5-mC signals (Figure 2Biii; examples show arrows). Along chromosome arms, the anti-5-mC signal was generally more widespread than retroelement.
Chromosomal organization of highly abundant repetitive DNA motifs
Graph-based clustering (RepeatExplorer) of the survey sequence of Cenchrus ciliaris (SRR8666664; Nevill et al., 2020) identified highly abundant repetitive DNA motifs in the genome (Supplementary Figure 1A). A total of 332,421 raw reads showed that approximately 50% of reads clustered into 13,278 superclusters and 165,079 singlets (Supplementary Figure 1A; full analysis including summary tables are available at http://dx.doi.org/10.25392/leicester.data.19798966). For in situ hybridization, three clusters with a circular graphical representation (CL22, CL58, and CL105) and one cluster (CL1, with a star-like graph pattern characteristic of tandem repeats) were used (Supplementary Figures 1B,C; Supplementary Table 1). Those with rDNA or organellar DNA similarities were excluded from further analysis. The extracted clusters were checked by BLAST for repeat identification (Sayers et al., 2019).
CL1 (Supplementary Table 1; Supplementary Figure 1B) accounted for 2.5% of the C. ciliaris genome and is a putative low-confidence satellite of 140 bp monomer length (Supplementary Figure 1C), part of which is homologous to a similarly sized satellite reported in Setaria viridis (Thielen et al., 2020). From two different parts of this sequence, two oligonucleotide probes (44 and 45 bp long) were designed, namely, CenchStCL1ConsS and CenchEnCL1ConsRC (Table 1). CL58 (Supplementary Table 1; Supplementary Figures 1B,C) is identified as a satellite with a repeat unit of 1,326 bp that represents a much smaller proportion, 0.061% of the genome, and PCR primers and a 44 bp probe were designed for this sequence (Table 1). CL22 (Supplementary Table 1; Supplementary Figures 1B,C) is identified as a 4,807 bp Ty1-copia retrotransposon, and the oligonucleotide probe CenchCL22Copia was designed to the RNaseH domain (RC) from this sequence (Table 1). The probes were localized on metaphase chromosomes of C. ciliaris using fluorescence in situ hybridization.
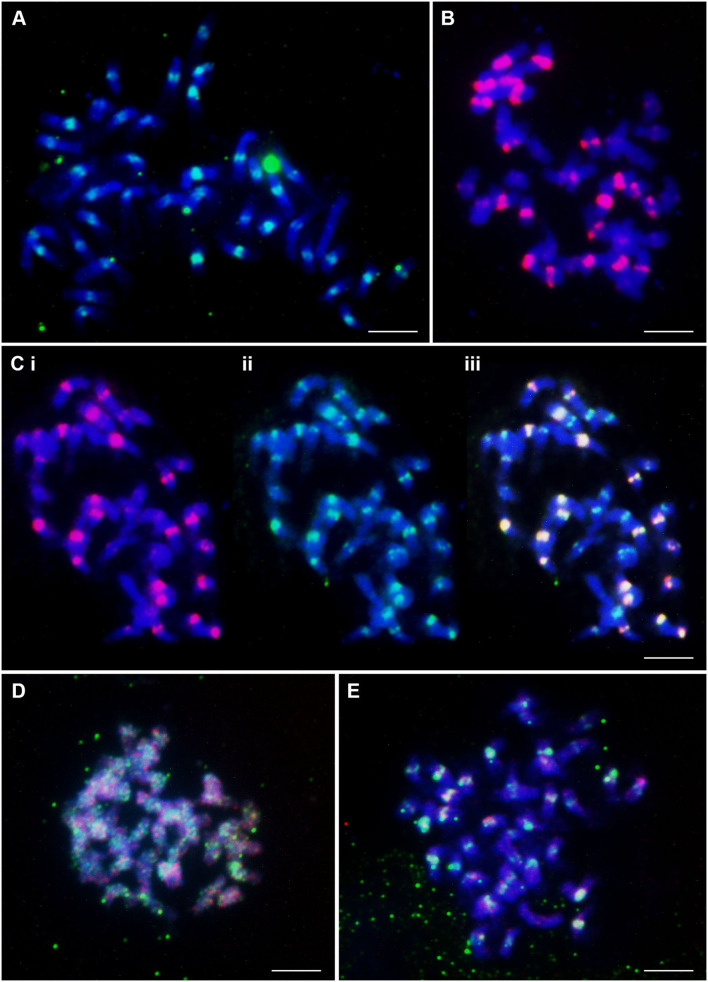
The CenchEnCL1ConsRC probe gave strong signals mostly in the pericentromeric regions of approximately 20 chromosomes from each metaphase, and the remaining chromosomes showed very weak signals (Figure 3A). In pericentromeric regions of chromosomes, 20 very strong signals of CenchStCL1ConsS probe were also detected, and some minor signals, mostly intercalary, were visible (Figure 3B). CenchStCL1ConsS colocalized with CenchCL22Copia signals (Figure 3C). Both probes are good chromosome markers, distinguishing about half of the chromosomes. The amplified CenchCL58 probe and commercially synthesized CenchCL58_869 showed dispersed signals along all chromosomes from metaphase (Figure 3D). Stronger signals of the CenchCL58_869 probe colocalized with CenchEnCL1ConsRC (Figure 3E), with 19 weaker and 20 stronger centromeric sites.
Figure 3.
Distribution of highly abundant repetitive DNA motifs on chromosomes of C. ciliaris (2n = 4x + 3 = 39). (A) Tandem repeat CenchEnCL1ConsRC probe labeled with FAM (green) showed strong signals in pericentromeric regions of about half of the chromosomes. The remaining chromosomes showed weak signals. (B) Twenty very strong and four weaker red signals of Tandem Repeat CenchStCL1ConsS probe labeled commercially with Cy3 (red). (C) Three pictures of the same metaphase showing colocalization of CL1 and CL22 signals: (i) CenchStCL1ConsS (labeled with Cy3; red) showing strong signal on about half of the chromosomes, (ii) CenchCL22Copia (labeled with FAM; green) with strong signals on about half the chromosomes also showing strong CL1 signal, and weaker CL22 signal on the remaining chromosomes, (iii) merged. (D) Metaphase chromosomes showing dispersed signals of amplified CenchCL58 probe (detected with FITC; green) and synthetic labeled CenchCL58_869 probe (Cy3, red). (E) Dispersed signals of CenchCL58_869 probe (Cy3, red) along chromosomes. Some of the stronger signals in the image colocalize with CenchEnCL1ConsRC (FAM; green). Scale bar = 10 μm.
Discussion
Aneuploidy and polyploidy in Cenchrus ciliaris—consequences for forage grasses
Cenchrus ciliaris is an allotetraploid with chromosome number 2n = 4x = 36 (Akiyama et al., 2011) and a primary base number of x = 9. In this study, we reported a new chromosomal count of C. ciliaris with 2n = 4x + 3 = 39. Aneuploidy in C. ciliaris has been widely reported (Visser et al., 1998), with Burson et al. (2012) detecting 99 aneuploids from 568 accessions, with different chromosome numbers from 2n = 18 to 2n = 56 (Vij and Chaudhary, 1981). Hyperploidy or monosomic addition lines are well-known in tetraploids and hexaploids in many Poaceae members, including C. ciliaris (e.g., 2n = 6x + 1 = 55). Meiotic irregularities have been widely reported in C. ciliaris, presumably because of the close relationships of the ancestral genomes in the polyploids (Visser et al., 1998; Burson et al., 2012). Where seed production is continued through apomixis, the predominant commercial reproduction pathway in C. ciliaris, there is low selective pressure for euploids. Polyploidization has significant conservational and environmental consequences where one species can become invasive due to the adaptation of a species in a new locality and can also change reproductive biology (Thompson and Pellmyr, 1992; John et al., 2004). Thus, characterization of the genome constitution of apomictic C. ciliaris is important both genetically and environmentally.
Aneuploid and polyploid cytotypes are widely distributed within tropical forage grasses, including species of the genera Cenchrus, Urochloa, Panicum, Pennisetum, and Paspalum (Carloni-Jarrys et al., 2018; Worthington et al., 2019; Tomaszewska et al., 2021a,b), often arising from spontaneous chromosome doubling following nondisjunction after meiotic or mitotic divisions (Gallo et al., 2007; Burson et al., 2012; Worthington et al., 2019), and fertilization with unreduced (2n) gametes. Where seed is produced through apomixis, many ploidies and aneuploid lines can be maintained and remain unchanged (also maintaining any heterozygosis or heterosis in the maternal genotype; Lippman and Zamir, 2007), enabling propagation of heterozygous genotypes, which is valuable for grass breeding (Ozias-Akins and van Dijk, 2007). Unlike many tropical forage grasses, including the segmental allopolyploids (Figures 1, 2); Jessup et al., 2003; Worthington et al., 2019; Tomaszewska et al., 2021b), it has proved difficult to develop agronomically acceptable genotypes of cereal grain-crops (including Triticum, Hordeum, and Avena genera). Identification of loci that first control heterotic phenotypes and second parthenogenesis or apomixis in tropical forage grasses is important for forage grass improvement, and further work may lead to developing suitable systems to exploit apomixis in grain crops, regarded as a significant challenge for wheat and rice (Dresselhaus et al., 2001; Conner and Ozias-Akins, 2017; Rathore et al., 2020).
Chromosomal localization of highly abundant repetitive DNA motifs
Various approaches have been used to identify and measure the abundance and genome organization of repetitive DNA in the plant genome. Assemblies from short reads will never provide the amount present because of the collapse of similar repeats during assembly (e.g., Yan et al., 2021). Graph-based clustering of unassembled raw reads, using RepeatExplorer (Novak et al., 2013), is proving to be a valuable reference-free approach to characterize all repeat families in a genome and identify those with genome specificity. Analysis of the frequency of k-mers—oligonucleotide sequences k bases long, where k is typically 16–64 bp—has also proved to be a robust method to identify repetitive motifs (Liu et al., 2019; Tomaszewska et al., 2021b).
In this study, we used both specific probes developed using graph-based sequence clustering compared with “universal” conserved regions of Ty1-copia and Ty3-gypsy retroelements. Similar to Urochloa tropical forage grasses, where genomic DNA and retroelements used as probes showed no substantial differences between the genomes (Santos et al., 2015; Tomaszewska et al., 2021b), parallel universal retroelement repeats used in this study were relatively equally abundant over the whole Cenchrus ciliaris genome (see Supplementary Figure 1). The Gy105 probe (see Figure 1D) labeled some chromosomes as more weaker. In contrast, CL1 (a putative satellite) and CL22 (an LTR-copia element) probes showed clear differential labeling of about half of the chromosomes (see Figure 3), and we suggest differences between the two ancestral genomes in the allopolyploid C. ciliaris having different retroelement (and/or satellite) sequence variants amplified before the genomes, which came together in the tetraploid. The “universal” retroelement probes amplified abundant sequences from both genomes and did not show any genome differentiation (see Figure 2).
Methylation pattern and major repeat distribution
The genome distribution and organization of retroelements are important for understanding retroelement dynamics, movement or amplification, chromosomal structure, and evolutionary processes. In this study, centromeric DAPI-staining bands (corresponding to constitutive heterochromatin; Siljak-Yakovlev et al., 2002) collocated with Ty1-copia probes, suggesting accumulation of these retroelements at heterochromatic regions. Both Ty1-copia and Ty3-gypsy tended to cluster at specific regions on most of the chromosomes (along with some intercalary and dispersed sites) contrasting with other species (Brandes et al., 1997a) while Ty1-copia was more uniformly distributed on the euchromatin regions in the genome. Whether the nonuniform distribution of retroelements is due to insertion-site preference in the genome (Belyayev et al., 2001), perhaps of particular element subfamilies, or purging from genome regions, will require further study.
A combination of in situ hybridization with immunostaining using anti-5-mC allows correlation of the distribution of DNA methylation and sequence location across the genome (Sepsi et al., 2014). 5-mC has been extensively studied for its role in the regulation of gene expression, genome imprinting, and suppression of transposable elements (Zhang et al., 2010). In this study, anti-5-methylcytosine immunofluorescence overlapped with most of the Ty1-copia elements and Ty3-gypsy, showing their location predominantly in the methylated genomic regions associated with lower transcriptional activity. With the majority of 5-mC signals associated with retrotransposon-rich regions, there was no further differentiation of groups of chromosomes, and, in particular, no indication of differences between ancestral genomes in the polyploid. In contrast, differential methylation of parental genomes has been observed in intraspecific A. thaliana hybrids and implicated in heterosis as a key epigenetic factor (Lauss et al., 2018). However, in rice tetraploid hybrids, Li et al. (2019) reported overall methylome stability with regional variation of cytosine methylation states, in part associated with gene expression changes. Methylation repatterning, often stimulated by hybridization and/or polyploidization, can potentially lead to increased genetic variation through facilitating somatic recombination, which could be adaptive in apomictic lineages (Verhoeven et al., 2010). Changes in methylation patterns were observed both in the newly formed and in the established apomicts of Taraxacum, suggesting that considerable methylation variation can increase over short evolutionary time scales.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: Previously published sequence data is available under accession SRR8666664 (Nevill et al., 2020). Results of the RepeatExplorer2 analysis of these sequence reads are archived on Figshare at http://dx.doi.org/10.25392/leicester.data.19798966.
Author contributions
PR, VB, JH-H, and PT: funding acquisition. PR, TS, and PT: investigation and methodology. VB and JH-H: supervision. All authors: conceptualization, writing original draft review and editing, and have read and agreed to the published version of the manuscript.
Funding
PR was supported under the Newton Bhabha Ph.D. placement program fellowship coordinated by the Department of Biotechnology, India, and the British Council, United Kingdom (BTIIN/UKJDBT-BC/2017-18). This study was supported under the RCUK-CIAT Newton-Caldas Initiative Exploiting biodiversity in Brachiaria and Panicum tropical forage grasses using genetics to improve livelihoods and sustainability, with funding from UK's Official Development Assistance Newton Fund, awarded by the UK Biotechnology and Biological Sciences Research Council (BB/R022828/1). PT has received support from the European Union's Horizon 2020 research and innovation program under the Marie Sklodowska-Curie grant agreement no. 844564 and no. 101006417 for the analysis of polyploid chromosomal evolution.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.952968/full#supplementary-material
Consensus sequences used to design primers and probes for fluorescence in situ hybridization. (A) RepeatExplorer graph showing number of raw sequencing reads forming abundant clusters; about 50% of the sequences form clusters, with <20 motifs representing one-third of the genome. (B) Graphical 2D projection of the structure of three graph-based clusters found in Cenchrus ciliaris genome using RepeatExplorer. Each node represents one of the sequence reads. The placement of the nodes reflects sequence similarity and overlaps. CL1 and CL58 are tandem repeats, while CL22 is an LTR-copia element. (C) Sequence LOGO representations showing lengths of repetitive motifs and conservation of sequences. CL1 is 140 bp tandem repeat; CL58 consensus is 1,326 bp and CL22 4,807 bp.
List of the most abundant clusters extracted from the Chinese variety of Cenchrus ciliaris SRR8666664 using RepeatExplorer.
References
- Abdi S., Dwivedi A., Bhat V. (2016). Harnessing apomixis for heterosis breeding in crop improvement, in Molecular Breeding for Sustainable Crop Improvement, eds Rajpal V. R., Rao S. R., Raina S. N. (Springer: Cham; ), 79–99. [Google Scholar]
- Akiyama Y., Goel S., Conner J. A., Hanna W. W., Yamada-Akiyama H., Ozias-Akins P. (2011). Evolution of the apomixis transmitting chromosome in Pennisetum. BMC Evol. Biol. 11, 289. 10.1186/1471-2148-11-289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama Y., Hanna W. W., Ozias-Akins P. (2005). High-resolution physical mapping reveals that the apospory-specific genomic region (ASGR) in Cenchrus ciliaris is located on a heterochromatic and hemizygous region of a single chromosome. Theoret. Appl. Genet. 111, 1042–1051. 10.1007/s00122-005-0020-5 [DOI] [PubMed] [Google Scholar]
- Belyayev A., Raskina O., Nevo E. (2001). Chromosomal distribution of reverse transcriptase-containing retroelements in two Triticeae species. Chromosome Res. 9, 129–136. 10.1023/A:1009231019833 [DOI] [PubMed] [Google Scholar]
- Bhat V., Dwivedi K. K., Khurana J. P., Sopory S. K. (2005). Apomixis: an enigma with potential applications. Curr. Sci. 10, 1879–1893. [Google Scholar]
- Biscotti M. A., Olmo E., Heslop-Harrison J. S. (2015). Repetitive DNA in eukaryotic genomes. Chromosome Res. 23, 415–420. 10.1007/s10577-015-9499-z [DOI] [PubMed] [Google Scholar]
- Brandes A., Heslop-Harrison J. S., Kamm A., Kubis S., Doudrick R. L., et al. (1997a). Comparative analysis of the chromosomal markers and genomic organization of Ty1-copia-like retrotransposons in pteridophytes, gymnosperms and angiosperms. Plant Mol. Biol. 33, 11–21. 10.1023/A:1005797222148 [DOI] [PubMed] [Google Scholar]
- Brandes A., Thompson H., Dean C., Heslop-Harrison J. S. (1997b). Multiple repetitive DNA sequences in the paracentromeric regions of Arabidopsis thaliana L. Chromosome Res. 5, 238–246. 10.1023/A:1018415502795 [DOI] [PubMed] [Google Scholar]
- Brits G., Trytsman M., Smith A., Ndlovu S., Mogale E., Molifi R. (2003). Screenings of subtropical grass species for drought tolerance, in Proceedings of the VII International Rangelands Congress (Durban, South Africa: ), 1348–1350. [Google Scholar]
- Burson B. L., Actkinson J. M., Hussey M. A., Jessup R. W. (2012). Ploidy determination of buffel grass accessions in the USDA national plant germplasm system collection by flow cytometry. S. Afr. J. Bot. 79, 91–95. 10.1016/j.sajb.2011.12.003 [DOI] [Google Scholar]
- Carloni-Jarrys E. J., Acosta-Bragato M. C., Grunberg-Fraga K. (2018). Nuclear DNA content and ploidy level in apomictic buffelgrass genotypes. Revista Fitotecnia Mexicana 41, 23–29. 10.35196/rfm.2018.1.23-29 [DOI] [Google Scholar]
- Castilho A., Neves N., Rufini-Castiglione M., Viegas W., Heslop-Harrison J. S. (1999). 5-Methylcytosine distribution and genome organization in Triticale before and after treatment with 5-azacytidine. J. Cell Sci. 112, 4397–4404. 10.1242/jcs.112.23.4397 [DOI] [PubMed] [Google Scholar]
- Chang K. D., Fang S. A., Chang F. C., Chung M. C. (2010). Chromosomal conservation and sequence diversity of ribosomal RNA genes of two distant Oryza species. Genomics 96, 181–190. 10.1016/j.ygeno.2010.05.005 [DOI] [PubMed] [Google Scholar]
- Conner J. A., Goel S., Gunawan G., Cordonnier-Pratt M.-M., Johnson V. E., Liang C., et al. (2008). Sequence analysis of bacterial artificial chromosome clones from the apospory-specific genomic region of Pennisetum and Cenchrus. Plant Physiol. 147, 1396–1411. 10.1104/pp.108.119081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conner J. A., Ozias-Akins P. (2017). Apomixis: engineering the ability to harness hybrid vigor in crop plants. Methods Mol. Biol. 1669, 17–34. 10.1007/978-1-4939-7286-9_2 [DOI] [PubMed] [Google Scholar]
- Dhaliwal A., Dhaliwal R. S., Kaur N., Gupta R. C. (2018). Cytomorphological study in genus Cenchrus L.: an important medicinal plant from north India (Family: Poaceae). Cytologia. 83, 45–52. 10.1508/cytologia.83.45 [DOI] [Google Scholar]
- Douglas C., Di Fazio S. (2010). Genetics and genomics of Populus, The Populus Genome and Comparative Genomics, eds Jansson S., Bhalerao R., Groover A. (Berlin: Springer-Verlag; ), 67–90. [Google Scholar]
- Dresselhaus T., Carman J. G., Savidan Y. (2001). Genetic engineering of apomixis in sexual crops: a critical assessment of the apomixis technology, in Flowering of Apomixis: From Mechanisms to Genetic Engineering, eds Savidan Y., Carman J. G., Dresselhaus T. (Mexico: CIMMYT, IRD, European Commission DG VI; ), 229–243. [Google Scholar]
- Dwivedi K. K., Bhat S. R., Bhat V., Bhat B. V., Gupta M. G. (2007). Identification of a SCAR marker linked to apomixis in buffelgrass (Cenchrus ciliaris L.). Plant Sci. 172, 788–795. 10.1016/j.plantsci.2006.12.006 [DOI] [Google Scholar]
- Fisher W. D., Bashaw E. C., Holt E. C. (1954). Evidence for apomixis in Pennisetum ciliare and Cenchrus setigerus. Agron. J. 46, 401–404. 10.2134/agronj1954.00021962004600090002x [DOI] [Google Scholar]
- Flavell A. J., Dunbar E., Anderson R., Pearce S. R., Hartley R., Kumar A. (1992b). Ty1-copia group retrotransposons are ubiquitous and heterogeneous in higher plants. Nucleic Acids Res. 20, 3639–3644. 10.1093/nar/20.14.3639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell A. J., Smith D. B., Kumar A. (1992a). Extreme heterogeneity of Ty1-copia group retrotransposons in plants. Mol. Gene Genet. 231, 233–242. 10.1007/BF00279796 [DOI] [PubMed] [Google Scholar]
- Friesen N., Brandes A., Heslop-Harrison J. S. (2001). Diversity, origin and distribution of retrotransposons (gypsy and copia) in conifers. Mol. Biol. Evol. 18, 1176–1188. 10.1093/oxfordjournals.molbev.a003905 [DOI] [PubMed] [Google Scholar]
- Gallo P. H., Micheletti P. L., Boldrini K. R., Risso-Pascotto C., Pagliarini M. S., do Valle C. B. (2007). 2n gamete formation in the genus Brachiaria (Poaceae: Paniceae). Euphytica 154, 255–260. 10.1007/s10681-006-9294-1 [DOI] [Google Scholar]
- He G., Zhu X., Elling A. A., Chen L., Wang X., Guo L., et al. (2010). Global epigenetic and transcriptional trends among two rice subspecies and their reciprocal hybrids. Plant Cell 22, 17–33. 10.1105/tpc.109.072041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heslop-Harrison J. S., Brandes A., Taketa S., Schmidt T., Vershinin A. V., Alkhimova E. G., et al. (1997). The chromosomal distributions of Ty1-copia group retrotransposable elements in higher plants and their implications for genome evolution. Genetica 100, 197–204. 10.1023/A:1018337831039 [DOI] [PubMed] [Google Scholar]
- Heslop-Harrison J. S., Schwarzacher T. (2011). Organisation of the plant genome in chromosomes. Plant J. 66, 18–33. 10.1111/j.1365-313X.2011.04544.x [DOI] [PubMed] [Google Scholar]
- Higgins J., Tomaszewska P., Pellny T. K., Castiblanco V., Arango J., Tohme J., et al. (2021). The five Urochloa spp. used in development of tropical forage cultivars originate from defined subpopulations with differentiated gene pools. BioRXiv. 10.1101/2021.07.21.453213 [DOI] [Google Scholar]
- Hirochika H., Hirochika R. (1993). Tyl-copia group retrotransposons as ubiquitous components of plant genomes. Jpn. J. Genet. 68, 35–46. 10.1266/ggs.68.35 [DOI] [PubMed] [Google Scholar]
- Jackson J. (2005). Is there a relationship between herbaceous species richness and buffel grass (Cenchrus ciliaris)? Austral. Ecol. 30, 505–517. 10.1111/j.1442-9993.2005.01465.x [DOI] [Google Scholar]
- Jessup R. W., Burson B. L., Burow G., Wang Y.-W., Chang C., Li Z., et al. (2003). Segmental allotetraploidy and allelic interactions in buffelgrass (Pennisetum ciliare (L.) Link syn. Cenchrus ciliaris L.) as revealed by genome mapping. Genome 46, 304–313. 10.1139/g03-005 [DOI] [PubMed] [Google Scholar]
- Jin W., Melo J. R., Nagaki K., Talbert P. B., Henikoff S., Dawe R. K., et al. (2004). Maize centromeres: organization and functional adaptation in the genetic background of oat. Plant Cell 16, 571–581. 10.1105/tpc.018937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- John N. T., Nuismer S. L., Merg K. (2004). Plant polyploidy and the evolutionary ecology of plant/animal interactions. Biol. J. Linn. Soc. 82, 511–519. 10.1111/j.1095-8312.2004.00338.x [DOI] [Google Scholar]
- Kharrat-Souissi A., Siljak-Yakovlev S., Brown S., Chaieb M. (2013). Cytogeography of Cenchrus ciliaris (Poaceae) in Tunisia. Folia Geobot. 48, 95–113. 10.1007/s12224-012-9137-x [DOI] [Google Scholar]
- Kharrat-Souissi A., Siljak-Yakovlev S., Brown S. C., Baumel A., Torre F., Chaieb M. (2014). The polyploid nature of Cenchrus ciliaris L. (Poaceae) has been overlooked: new insights for the conservation and invasion biology of this species – a review. Rangeland J. 36, 11–23. 10.1071/RJ13043 [DOI] [Google Scholar]
- Kharrat-Souissi A., Siljak-Yakovlev S., Pustahija F., Chaieb M. (2012). Physical mapping of 5S and 18S-5.8S-26S RNA gene families in polyploid series of Cenchrus ciliaris Linnaeus, 1771 (Poaceae). Comp. Cytogenet. 6, 273–286. 10.3897/compcytogen.v6i3.3380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A., Pearce S. R., McLean K., Harrison G., Heslop-Harrison J. S., Waugh R., et al. (1997). The Ty1-copia group of retrotransposons in plants: genomic organization, evolution, and use as molecular markers. Genetica 100, 205–217. 10.1023/A:1018393931948 [DOI] [PubMed] [Google Scholar]
- Lauss K., Wardenaar R., Oka R., van Hulten M. H., Guryev V., Keurentjes J. J., et al. (2018). Parental DNA methylation states are associated with heterosis in epigenetic hybrids. Plant Physiol. 176, 1627–1645. 10.1104/pp.17.01054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le T. N., Miyazaki Y., Takuno S., Saze H. (2015). Epigenetic regulation of intragenic transposable elements impacts gene transcription in Arabidopsis thaliana. Nucleic Acid Res. 43, 3911–3921. 10.1093/nar/gkv258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N., Xu C., Zhang A., Lv R., Meng X., Lin X., et al. (2019). DNA methylation repatterning accompanying hybridization, whole genome doubling and homoeolog exchange in nascent segmental rice allotetraploids. New Phytol. 223, 979–992. 10.1111/nph.15820 [DOI] [PubMed] [Google Scholar]
- Li R., Hu F., Li B., Zhang Y., Chen M., Fan T., et al. (2020). Whole genome bisulfite sequencing methylome analysis of mulberry (Morus alba) reveals epigenome modifications in response to drought stress. Sci. Rep. 10, 8013. 10.1038/s41598-020-64975-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippman Z. B., Zamir D. (2007). Heterosis: revisiting the magic. TRENDS Genet. 23, 60–66. 10.1016/j.tig.2006.12.006 [DOI] [PubMed] [Google Scholar]
- Liu Q., Li X., Zhou X., Li M., Zhang F., Schwarzacher T., et al. (2019). The repetitive DNA landscape in Avena (Poaceae): chromosome and genome evolution defined by major repeat classes in whole-genome sequence reads. BMC Plant Biol. 19, 226. 10.1186/s12870-019-1769-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Yuan H., Li M., Wang Z., Cui D., Ye Y., et al. (2022). Chromosome-scale genome assembly of the diploid oat Avena longiglumis reveals the landscape of repetitive sequences, genes and chromosome evolution in grasses. BioRXiv. 10.1101/2022.02.09.479819 [DOI] [Google Scholar]
- Macas J., Kejnovský E., Neumann P., Novák P., Koblížková A, Vyskot B. (2011). Next generation sequencing-based analysis of repetitive DNA in the model dioceous plant Silene latifolia. PLoS ONE 6, e27335. 10.1371/annotation/4ccaacb2-92d7-445a-87da-313cedf18feb [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall V. M., Lewis M. M., Ostendorf B. (2012). Buffel grass (Cenchrus ciliaris) as an invader and threat to biodiversity in arid environments: a review. J. Arid Environ. 78, 1–12. 10.1016/j.jaridenv.2011.11.005 [DOI] [Google Scholar]
- Meena S., Nagar R. P. (2019). Seasonal effects on fodder yield and quality in Cenchrus species under semi-arid climate. Indian J. Agric. Sci. 89, 1287–1292. [Google Scholar]
- Miller G., Friedel M., Adam P., Chewings V. (2010). Ecological impacts of Buffel grass (Cenchrus ciliaris L.) invasion in central Australia: does field evidence support a fire-invasion feedback? Rangeland J. 32, 353–365. 10.1071/RJ09076 [DOI] [Google Scholar]
- Moore L. D., Le T., Fan G. (2013). DNA methylation and its basic function. Neuropsychopharmacology 38, 23–38. 10.1038/npp.2012.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann P., Novak P., Hoštáková N., Macas J. (2019). Systematic survey of plant LTR-retrotransposons elucidates phylogenetic relationships of their polyprotein domains and provides a reference for element classification. Mobile DNA. 10, 1. 10.1186/s13100-018-0144-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevill P. G., Zhong X., Tonti-Filippini J., Byrne M., Hislop M., Thiele K., et al. (2020). Large scale genome skimming from herbarium material for accurate plant identification and phylogenomics. Plant Methods 16, 1–8. 10.1186/s13007-019-0534-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak P., Neumann P., Pech J., Steinhais J., Macas J. (2013). RepeatExplorer: a galaxy-based web server for genome-wide characterization of eukaryotic repetitive elements from next generation sequence reads. Bioinformatics 29, 792–793. 10.1093/bioinformatics/btt054 [DOI] [PubMed] [Google Scholar]
- Ozias-Akins P., van Dijk P. J. (2007). Mendelian genetics of apomixis in plants. Annu. Rev. Genet. 41, 509–537. 10.1146/annurev.genet.40.110405.090511 [DOI] [PubMed] [Google Scholar]
- Rathore P., Raina S. N., Kumar S., Bhat V. (2020). Retro-element Gypsy-163 is differentially methylated in reproductive tissues of apomictic and sexual plants of Cenchrus ciliaris. Front. Genet. 11, 795. 10.3389/fgene.2020.00795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche D., Cong P., Chen Z., Hanna W. W., Gustine D. L., Sherwood R. T., et al. (1999). An apospory-specific genomic region is conserved between Buffelgrass (Cenchrus ciliaris L.) and Pennisetum squamulatum Fresen. Plant J. 19, 203–208. 10.1046/j.1365-313X.1999.00514.x [DOI] [PubMed] [Google Scholar]
- Rozen S., Skaletsky H. (1999). Primer3 on the WWW for general users and for biologist programmers. Bioinform. Methods Protoc. 132, 365–386. 10.1385/1-59259-192-2:365 [DOI] [PubMed] [Google Scholar]
- Santos F. C., Guyot R., do Valle C. B., Chiari L., Techio V. H., Heslop-Harrison P., et al. (2015). Chromosomal distribution and evolution of abundant retrotransposons in plants: gypsy elements in diploid and polyploid Brachiaria forage grasses. Chromosome Res. 23, 571–582. 10.1007/s10577-015-9492-6 [DOI] [PubMed] [Google Scholar]
- Sayers E. W., Agarwala R., Bolton E. E., Brister R. J., Canese K., Clark K., et al. (2019). Database resources of the national center for biotechnology information. Nucleic Acids Res. 47, D23–D28. 10.1093/nar/gky1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzacher T., Heslop-Harrison J. S. (2000). Practical in situ Hybridization. Oxford: Bio, 203+xii. [Google Scholar]
- Sepsi A., Higgins J. D., Heslop-Harrison J. S., Schwarzacher T. (2017). CENH3 morphogenesis reveals dynamic centromere associations during synaptonemal complex formation and the progression through male meiosis in hexaploid exaploidy wheat. Plant J. 89, 235–249. 10.1111/tpj.13379 [DOI] [PubMed] [Google Scholar]
- Sepsi A., Shiura H., Okamoto A., Sasaki H., Abe K. (2014). Whole-mount MeFISH: a novel technique for simultaneous visualization of specific DNA methylation and protein/RNA expression. PLoS ONE 9, e95750. 10.1371/journal.pone.0095750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siljak-Yakovlev S., Cerbah M., Coulaud J., Stoian V., Brown S. C., Jelenic S., et al. (2002). Nuclear DNA content, base composition, heterochromatin and rDNA in Picea omorika and Picea abies. Theor. Appl. Genet. 104, 505–512. 10.1007/s001220100755 [DOI] [PubMed] [Google Scholar]
- Slotkin R. K., Martienssen R. (2007). Transposable elements and the epigenetic regulation of the genome. Nat. Rev. Genet. 8, 272–285. 10.1038/nrg2072 [DOI] [PubMed] [Google Scholar]
- Spillane C., Curtis M. D., Grossniklaus U. (2004). Apomixis technology development—virgin births in farmers' fields? Nat. Biotechnol. 22, 687–691. 10.1038/nbt976 [DOI] [PubMed] [Google Scholar]
- Thielen P. M., Pendleton A. L., Player R. A., Bowden K. V., Lawton T. J., Wisecaver J. H. (2020). Reference genome for the highly transformable Setaria viridis ME034V. G3 Genes Genom. Genet. 10, 3467–3478. 10.1534/g3.120.401345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. N., Pellmyr O. (1992). Mutualism with pollinating seed parasites amid co-pollinators: constraints on specialization. Ecology 73, 1780–1791. 10.2307/1940029 [DOI] [Google Scholar]
- Tomaszewska P., Pellny T. K., Hernández L. M., Mitchell R. A. C., Castiblanco V., de Vega J. J., et al. (2021a). Flow cytometry-based determination of ploidy from dried leaf specimens in genomically complex collections of the tropical forage grass Urochloa s. l. Genes 12, 957. 10.3390/genes12070957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomaszewska P., Vorontsova M. S., Renvoize S. A., Ficinski S. Z., Tohme J., Schwarzacher T., et al. (2021b). Complex polyploid and hybrid species in an apomictic and sexual tropical forage grass group: genomic composition and evolution in Urochloa (Brachiaria) species. Ann. Botany. (in press) 10.1093/aob/mcab147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhoeven K. J. F., Dijk J. D., Biere A. (2010). Changes in genomic methylation patterns during the formation of triploid asexual dandelion lineages. Mol. Ecol. 19, 315–324. 10.1111/j.1365-294X.2009.04460.x [DOI] [PubMed] [Google Scholar]
- Vij S. P., Chaudhary J. D. (1981). Cylological investigation in three species of Cenchrus L. (Gramineae). Cytologia 46, 661–669. 10.1508/cytologia.46.661 [DOI] [Google Scholar]
- Vining K. J., Pomraning K. R., Wilhelm L. J., Priest H. P., Pellegrini M., Mockler T. C., et al. (2012). Dynamic DNA cytosine methylation in the Populus trichocarpa genome: tissue-level variation and relationship to gene expression. BMC Genom. 13, 27. 10.1186/1471-2164-13-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser N. C., Spies J. J., Venter H. J. T. (1998). Aneuploidy in Cenchrus cilliaris (Poaceae, Panicoideae, Paniceae): truth or fiction? S. Afc. J. Bot. 64, 337–345. 10.1016/S0254-6299(15)30921-2 [DOI] [Google Scholar]
- Worthington M., Ebina M., Yamanaka N., Heffelfinger C., Quintero C., Zapata Y. P., et al. (2019). Translocation of a parthenogenesis gene candidate to an alternate carrier chromosome in apomictic Brachiaria humidicola. BMC Genom. 20, 41. 10.1186/s12864-018-5392-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worthington M., Heffelfinger C., Bernal D., Quintero C., Zapata P., Perez J. G., et al. (2016). A parthenogenesis gene candidate and evidence for segmental allopolyploidy in apomictic Brachiaria decumbens. Genetics 203, 1117–1132. 10.1534/genetics.116.190314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav B., Anuj C., Kumar S., Gupta M. G., Bhat V. (2012). Genetic linkage maps of the chromosomal regions associated with apomictic and sexual modes of reproduction in Cenchrus ciliaris. Mol. Breed. 30, 239–250. 10.1007/s11032-011-9614-6 [DOI] [Google Scholar]
- Yan Q., Wu F., Xu P., Sun Z., Li J., Gao L., et al. (2021). The elephant grass (Cenchrus purpureus) genome provides insights into anthocyanidin accumulation and fast growth. Mol. Ecol. Resour. 21, 526. 10.1111/1755-0998.13271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Kimatu J. N., Xu K., Liu B. (2010). DNA cytosine methylation in plant development. Genet. Genom. 37, 1–12. 10.1016/S1673-8527(09)60020-5 [DOI] [PubMed] [Google Scholar]
- Zilberman D., Gehring M., Tran R. K., Ballinger T., Henikoff S. (2007). Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat. Genet. 39, 61–69. 10.1038/ng1929 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Consensus sequences used to design primers and probes for fluorescence in situ hybridization. (A) RepeatExplorer graph showing number of raw sequencing reads forming abundant clusters; about 50% of the sequences form clusters, with <20 motifs representing one-third of the genome. (B) Graphical 2D projection of the structure of three graph-based clusters found in Cenchrus ciliaris genome using RepeatExplorer. Each node represents one of the sequence reads. The placement of the nodes reflects sequence similarity and overlaps. CL1 and CL58 are tandem repeats, while CL22 is an LTR-copia element. (C) Sequence LOGO representations showing lengths of repetitive motifs and conservation of sequences. CL1 is 140 bp tandem repeat; CL58 consensus is 1,326 bp and CL22 4,807 bp.
List of the most abundant clusters extracted from the Chinese variety of Cenchrus ciliaris SRR8666664 using RepeatExplorer.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: Previously published sequence data is available under accession SRR8666664 (Nevill et al., 2020). Results of the RepeatExplorer2 analysis of these sequence reads are archived on Figshare at http://dx.doi.org/10.25392/leicester.data.19798966.