Abstract
The bacterium Photorhabdus luminescens is a symbiont of the entomopathogenic nematode Heterorhabditis bacteriophora. The nematode requires the bacterium for infection of insect larvae and as a substrate for growth and reproduction. The nematodes do not grow and reproduce in insect hosts or on artificial media in the absence of viable P. luminescens cells. In an effort to identify bacterial factors that are required for nematode growth and reproduction, transposon-induced mutants of P. luminescens were screened for the loss of the ability to support growth and reproduction of H. bacteriophora nematodes. One mutant, NGR209, consistently failed to support nematode growth and reproduction. This mutant was also defective in the production of siderophore and antibiotic activities. The transposon was inserted into an open reading frame homologous to Escherichia coli EntD, a 4′-phosphopantetheinyl (Ppant) transferase, which is required for the biosynthesis of the catechol siderophore enterobactin. Ppant transferases catalyze the transfer of the Ppant moiety from coenzyme A to a holo-acyl, -aryl, or -peptidyl carrier protein(s) required for the biosynthesis of fatty acids, polyketides, or nonribosomal peptides. Possible roles of a Ppant transferase in the ability of P. luminescens to support nematode growth and reproduction are discussed.
Photorhabdus luminescens (Enterobacteriaceae) bacteria are symbiotic with entomopathogenic rhabditid nematodes of the family Heterorhabditidae, with which they cooperate in infecting a wide variety of insect larvae (38, 45; for reviews, see references 25 and 26). The nematode requires P. luminescens for insect pathogenicity (34), while the bacteria depend on the nematodes for transmission between insect prey. The infective juvenile (IJ)-stage nematodes specifically retain symbiotic P. luminescens cells in their gut mucosa, and transmission of the bacteria is a requisite for insect pathogenicity (31, 32, 34). The nematodes require P. luminescens cells as a substrate for growth and reproduction (2, 21, 22, 30). It was suggested previously that symbiotic P. luminescens cells provide favorable nutritional conditions for Heterorhabditis bacteriophora nematodes to grow and reproduce (45).
During prolonged laboratory culture, P. luminescens strains show a tendency to undergo an apparent phase variation phenomenon (8, 9, 36). The native form of the bacteria, termed primary phase, is isolated from the IJ stage of the nematode. The secondary-phase variants appear at high frequency during prolonged culturing, while more rare is the generation of primary-phase cells from secondary phase (6). The secondary-phase cells differ from the primary-phase cells in colony morphology, cell size, and dye uptake characteristics (6, 7, 9, 52). Also, typical primary-phase characteristics such as bioluminescence, pigment synthesis, phospholipase and siderophore activities, and production of intracellular crystalline inclusion proteins are depressed or absent in secondary-phase cells. The mechanism and role of phase variation in P. luminescens are unknown. Particularly significant for the subject of this investigation is the inability of secondary-phase variant cells to support nematode growth and reproduction (22, 30).
Because H. bacteriophora nematodes have a strict requirement for P. luminescens for growth and reproduction, it seems likely that P. luminescens provides some nutrients and/or other factors to the nematode. Dead cells or culture supernatants of P. luminescens cells do not provide the nutrients and/or factors required for nematode growth and reproduction (34; T. A. Ciche, personal observation), suggesting that actively metabolizing P. luminescens cells are required. To better understand the contribution of P. luminescens to nematode growth and reproduction, we developed a genetic screen to identify P. luminescens genes necessary for nematode growth and reproduction.
Genetic studies of P. luminescens have been limited, probably because of a low frequency of transformation (7) and the current inability to introduce recombinant DNA into P. luminescens by conjugation (52). Success has been achieved in using allelic exchange to construct disruptions in the genes encoding intracellular inclusion proteins CipA and CipB (7) and insecticidal toxin genes tca, tcb, tcc, and tcd (10) of P. luminescens.
Here we describe the construction of a mini-Tn5-based transposon and a delivery vector for efficient mutagenesis and gene characterization in P. luminescens. We used this system to identify genes that are required for P. luminescens to support growth and reproduction of its nematode host, H. bacteriophora. We identified a transposon mutant that has lost the ability to support nematode growth and reproduction, and we describe the analyses of the disrupted gene region.
MATERIALS AND METHODS
Microbiological methods.
Sources of strains and plasmids are listed in Table 1. Dye reagents, Tween types, and antibiotics were purchased from Sigma Chemical Corp. (St. Louis, Mo.), and bacteriological growth media were purchased from Difco (Detroit, Mich.). Cells of P. luminescens were grown in 2% Proteose Peptone 3 (PP3), with 1.5% agar added when required, at 28°C in the dark. Kanamycin (15 μg/ml), streptomycin (25 μg/ml), spectinomycin (25 μg/ml), and sucrose (7.5% [wt/vol]) were added when required. Escherichia coli strains were grown in Luria-Bertani (LB) broth or on LB agar (1.5% agar) at 37°C with ampicillin (100 μg/ml), kanamycin (50 μg/ml), streptomycin (25 μg/ml), spectinomycin (25 μg/ml), 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) (40 μg/ml), chloramphenicol (35 μg/ml), and sucrose (5% [wt/vol]) added when required.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Nematode strains | ||
| H. bacteriophora | Strain NC1, symbiotic with NC1/1 bacteria | 38 |
| H. megidis | Strain Meg | H. Kaya (46) |
| Bacterial strains | ||
| P. luminescens | ||
| NC1/1 | Primary phase, isolated from H. bacteriophora (identical to ATCC 29304, strain NC-19) | 38 |
| NC1/2 | Secondary phase, isolated from NC1/1 cells | This study |
| Meg/1 | Primary phase, isolated from H. megidis | 46 |
| NP394 | NC1/1 with pUB394 | This study |
| NGR209 | NC1/1 ngrA::mini-Tn5 | This study |
| NGR209A | NC1/1 with allelic-exchange vector p209A | This study |
| NGR209C | NC1/1 ngrA::mini-Tn5 via allelic exchange | This study |
| NPngrA::ΩKm | NC1/1 ngrA::ΩKm | This study |
| E. coli | ||
| DH5α | Cloning strain | Gibco BRL |
| EC393 | DH5α with pUB394 | This study |
| EC209R | DH5α with p209, Kmr | This study |
| Plasmid vectors | ||
| pGEM-3Z | Cloning vector | Promega |
| pGEM-7Zf(+) | Cloning vector | Promega |
| pBC SK(+) | Cloning vector | Stratagene |
| pUT-mTn5 | Mini-Tn5; transposase | 19, 35 |
| pSU19 | P15A replicon, Cmr | 5, 43 |
| pSU39 | P15A replicon, Kmr | 5, 43 |
| pSB101 | pBC SK(+) with 2.6-kb PstI fragment containing sacR/sacB | 7 |
| pHP45Ω | Source of interposon containing streptomycin resistance | 24 |
| pHP45Ω-Km | Source of interposon containing kanamycin resistance | 24 |
| pG325 | pGEM-7 containing Spr Smr and sacB/sacR | This study |
| pUB394 | Mini-Tn5 delivery vector, Kmr Smr Spr sucroses | This study |
| p209 | Retrieved plasmid with mini-Tn5 and flanking P. luminescens DNA | This study |
| p209A | p209 with Spr Smr and sacB/sacR PstI fragment into single NsiI site of p209 | This study |
| pngrA::ΩKm | pBCSK containing ngrA, Smr Spr, and sacB/sacR PstI fragment | This study |
| pngrA | pBCSK containing ngrA | This study |
Secondary-phase (NC1/2) cells of P. luminescens were obtained and distinguished from the primary-phase (NC1/1) cells as described previously (7). Siderophore activity was determined using chrome azurol S (CAS) agar (49), and lipase activity was determined on spirit blue agar containing 0.5% (vol/vol) Tween 20, Tween 40, Tween 60, Tween 80, or Tween 85 (50). Antibiotic activity was determined by placing a 5-mm-diameter plug, taken 5 mm away from confluent growth of a 96-h culture of P. luminescens on PP3 agar, onto a plate of antibiotic medium 3 (Difco) that had been inoculated with Micrococcus luteus cells.
Nematode propagation.
IJ nematodes were propagated by infecting greater wax moth larvae, Galleria mellonella (Ja-Da Bait Co., Antigo, Wis.), or by adding approximately 20 IJ nematodes to lawns of P. luminescens cells on lipid agar (LA) (53) (2.5% nutrient broth, 1.5% agar, 1% corn oil), nematode growth medium (12), or liquid culture medium (LCM) (22) as described previously (7).
The IJ nematodes were collected by flooding the LA plates or infected larvae at the time of IJ release and separated from adult nematodes by the water trap method of Wouts (53). Alternatively, IJ nematodes were grown on P. luminescens cells that were seeded onto LA or nematode growth medium contained on one side of a divided petri dish (Fisher Scientific, Pittsburgh, Pa.). After 14 days, newly formed IJ nematodes migrated into the sterile saline contained on the other side. IJ nematodes were surface sterilized as described previously (41).
Axenic nematodes.
Axenic nematodes were obtained using a modification of the procedure of Han and Ehlers (33). H. bacteriophora nematodes were propagated on a P. luminescens strain, Meg/1, that was isolated from Heterorhabditis megidis nematodes. The H. bacteriophora nematodes grow and reproduce normally on Meg/1 bacteria, but the resulting surface-sterilized IJ nematodes do not retain Meg/1 bacteria and are therefore axenic (33).
Retention of bacteria by nematodes.
The numbers of P. luminescens cells in the intestine of IJ nematodes were determined. For some experiments, 50 to 100 surface-sterilized nematodes were disrupted using an 0.1-ml microtissue grinder (Kontes, Vineland, N.J.). The homogenate was then serially diluted and plated on PP3 agar. Alternatively, a 10-μl sample of a water suspension containing 10 to 50 IJ nematodes was placed in the depression of a sterile hanging drop slide and dried in a laminar flow hood for 5 to 10 min, and then each nematode was disrupted with a sterile scalpel while being examined under a 40× dissecting microscope. The disrupted nematodes were suspended in 0.1 ml of PP3 broth, and the scalpel blade was rinsed in this suspension. The material was then transferred to a tube containing 0.9 ml of PP3 broth. The slide depression and scalpel were rinsed three times before plating serial dilutions of the tube onto PP3 agar. Colonies were counted following incubation at 28°C for 3 days.
DNA manipulations.
Plasmid purification from P. luminescens and E. coli was carried out using Wizard Mini and Midi preps (Promega Corp., Madison, Wis.). Restriction enzymes and T4 ligase were used according to the manufacturer's instructions (Promega Corp.). When required, DNA fragments were extracted from agarose gels using the Qiagen Gel extraction kit (Qiagen Inc., Valencia, Calif.). The bacterial DNA was purified using a modified cetyltrimethylammonium bromide (CTAB) method (14). Southern hybridization was performed under high-stringency conditions using the Genius kit (Boehringer Mannheim Corp., Indianapolis, Ind.). Transformation of E. coli and P. luminescens was done by electroporation using a Bio-Rad gene pulser according to the conditions suggested for E. coli by the supplier (Bio-Rad Laboratories, Hercules, Calif.).
Construction of pUB394.
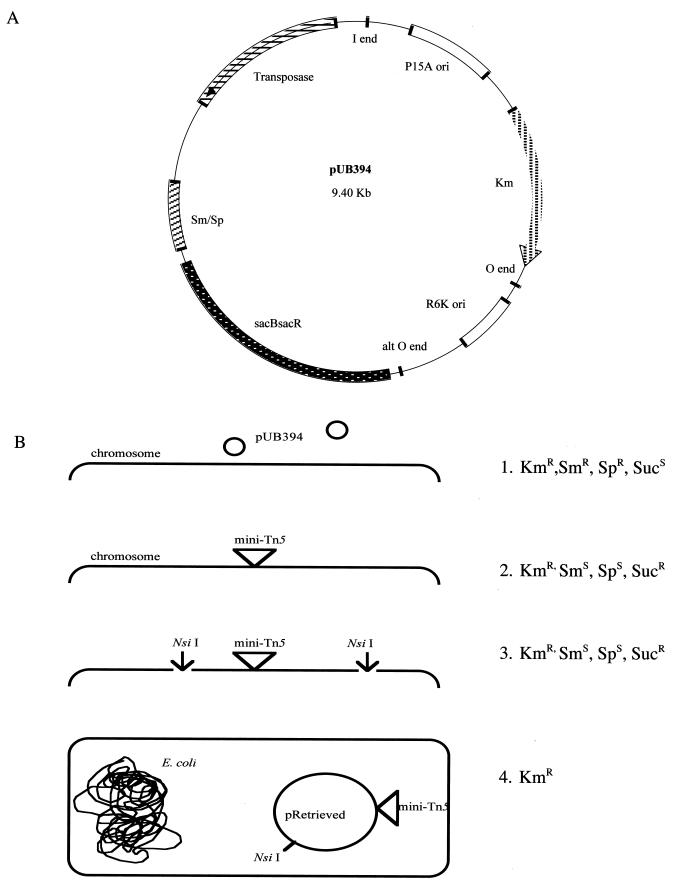
The structure of the transposon delivery vector, pUB394, is shown in Fig. 1A. A pSU39 plasmid (5, 43) was inserted between the I and O ends that define the termini of the mini-Tn5 by removing the chloramphenicol resistance gene from the mini-Tn5 of pUT-mTnCm (24) by digesting it with SmaI and replacing it with a HincII and SmaI fragment of pSU39. The resulting plasmid is named pUS39. The correct orientation of the insert was determined. Because P. luminescens organisms are resistant to ampicillin and conjugation techniques have not been established with these bacteria, the ampicillin resistance and Mob RP4 genes were removed by BamHI and SfiI digestion of pUS39 followed by self-ligation, resulting in pUSF39. pGTn, pGHP, and pGHS were constructed to isolate the transposase, streptomycin and spectinomycin resistance, and levansucrase genes, respectively. Plasmid pGTn was constructed by removing a 1.5-kb SalI fragment containing the transposase gene from pUT-mTn5Cm (19, 35) and inserting it into pGEM-7Zf(+) (Promega). The orientation of the insert was verified. The transposase-encoding gene was inserted into the XbaI site located outside the I-end inverted repeat of the mini-Tn5 of pUB39 as a XbaI fragment from plasmid pGTn. The resulting plasmid is named pUGS394. The orientation of the insert was verified. pUGS394 was digested with BamHI to remove the ampicillin resistance and Mob RP4 genes and self-ligated. The resulting plasmid is named pUS394. The streptomycin and spectinomycin resistance gene was removed as a 2.0-kb HindIII fragment from pHP45Ω (24) and inserted into pGEM-7Zf(+). The resulting plasmid is named pGHP. Plasmid pGHS was constructed by inserting a 2.6-kb XbaI fragment from pBS101 (7) containing sacB/sacR (27, 47) into XbaI-digested pGHP. The orientation of the insert was verified. The streptomycin and spectinomycin resistance and the sacB/sacR sucrose sensitivity genes were removed as a 4.6-kb BamHI fragment from pGHS and inserted into a BamHI site of pUS394, yielding pUB394. A second, alternative O end was located 1.7 kb 5′ to the O end of the mini-Tn5. Between the two O ends are the R6K ori and DNA encoding the N-terminal (amino acids 1 to 202) region of the Tn5 transposase-encoding genes. No difference was observed in the stability of these and normal O-end mini-Tn5 insertions.
FIG. 1.
(A) The structure of pUB394. Genes conferring resistance to kanamycin (Km) and streptomycin and spectinomycin (Sm/Sp) and the sacB/sacR gene conferring sensitivity to sucrose are shown. The mini-Tn5 is located between the I and O ends. (B) Use of pUB394 for transposon mutagenesis of P. luminescens. (Line 1) Transformants containing pUB394 were selected for by resistance to streptomycin. (Line 2) Transposon-induced mutants containing the mini-Tn5 mutants that have lost pUB394 were selected for by resistance to kanamycin and sucrose. (Line 3) The mini-Tn5 was retrieved from the mutants by NsiI digestion of mutant DNA, self-ligation, and transformation into E. coli. (Line 4) Transformants containing the mini-Tn5 were selected for by their resistance to kanamycin. The DNA sequence flanking the insertion was obtained using M13 forward and reverse priming sites present in the mini-Tn5. Causality of the mini-Tn5 insertion was determined by inserting a PstI causality cassette (containing the sacB/sacR sucrose sensitivity genes and a gene conferring resistance to streptomycin and spectinomycin) into the NsiI site present in the retrieved plasmids. Allelic exchange was then performed with this plasmid and the wild-type allele as described previously (7).
Transposon mutagenesis.
The use of pUB394 for transposon mutagenesis and retrieval of DNA containing mini-Tn5 insertions is shown in Fig. 1B. Low transformation efficiency and the inability to conjugate or transduce DNA in P. luminescens make the standard transposon mutagenesis techniques utilizing suicide plasmids inefficient. The sacB gene, conferring sucrose sensitivity, allows selection against cells containing pUB394 and, when used with selection for the transposon (resistance to kanamycin), allows cells containing insertions, but not pUB394, to be selected. Cells of P. luminescens NC1/1 were transformed with the transposon delivery vector pUB394 to create strain NP394. As a consequence of pUB394 replication in NP394, mini-Tn5 insertions may accumulate. NP394 containing a mini-Tn5 insertion and pUB394 will cause, at high frequency (10−3), kanamycin- and sucrose-resistant cells by loss of pUB394. To select mini-Tn5 mutants that have lost pUB394, cells of NP394 (checked for the absence of a mini-Tn5 insertion prior to mutant generation) were grown overnight in PP3 containing kanamycin, and 10−1 and 10−2 dilutions were plated on PP3 agar containing kanamycin and sucrose to select for cells containing the mini-Tn5 but not pUB394. The mutants were transferred to PP3 agar containing kanamycin to verify the resistance to kanamycin, PP3 agar containing streptomycin and spectinomycin to verify the absence of pUB394, M9 minimal medium to determine auxotrophy, and eosin-methylene blue to determine the phase state of the mutants. Mutant cells unable to grow on M9 medium were assumed to be auxotrophs, and those not accumulating dye on eosin-methylene blue were assumed to be secondary-phase cells. Secondary-phase cells do not support nematode growth and reproduction and were not characterized, because they are likely to be spontaneous phase variants of NP394 and not transposon-induced secondary-phase cells.
Screening for the ability of mutants to support growth and reproduction of H. bacteriophora nematodes
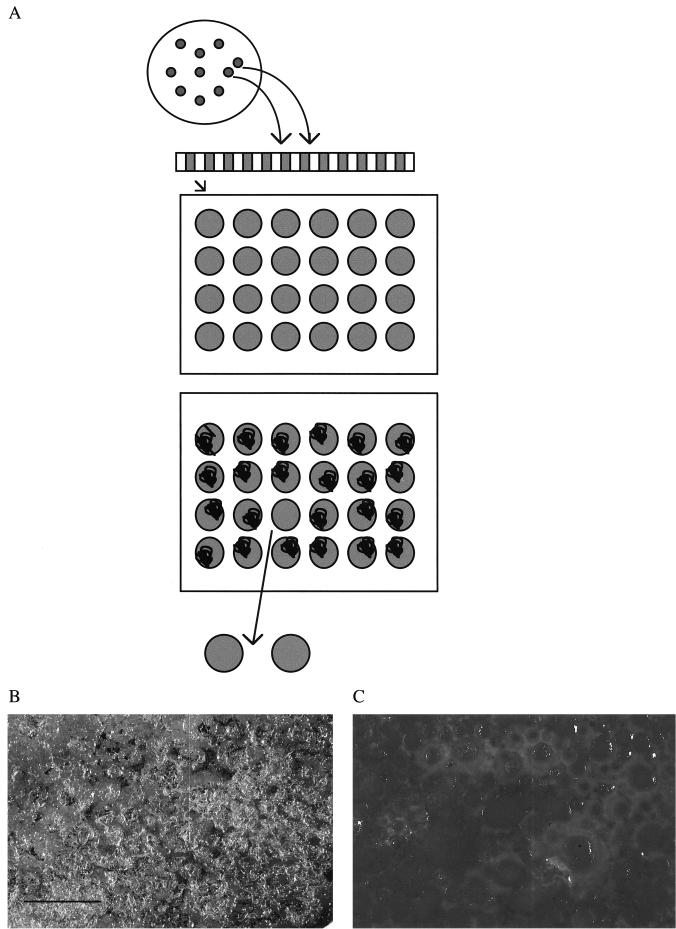
The screen for the ability of bacteria to support nematode growth and reproduction is shown in Fig. 2A. Individual colonies of transposon-induced mutants of P. luminescens were inoculated into 0.25 ml of PP3 with 10 μg of kanamycin/ml and were incubated statically overnight at 28°C. Samples of 0.05 ml were added to individual wells of 24-well tissue culture plates (Falcon 1143; Becton Dickinson Labware, Lincoln Park, N.J.) with 1.5 ml of LA-containing kanamycin. Following incubation overnight at 28°C, an average of 12 axenic IJ nematodes was added to each well. Bacteria able to support nematode growth and reproduction were detected by the appearance of a white mass of nematodes 21 days later. NP394 and secondary-phase (NC1/2) P. luminescens containing pUB394 were included in the assay as positive and negative controls, respectively. Putative nematode growth and reproduction mutants were verified by repeating the nematode growth and reproduction experiments twice, each with 12 replicates. Mutants were named NGR for nematode growth and reproduction mutants.
FIG. 2.
Illustration of the in vitro screen for the ability of mutants to support nematode growth and reproduction. (A) Transposon-induced mutants were grown overnight in liquid culture, and a sample was placed onto LA in titer dishes and incubated overnight. An average of 12 IJ nematodes was added to the wells. Following incubation for 20 days, the wells were then observed for the presence of nematode growth. Mutant cells in wells not showing nematode growth were further characterized. (B and C) Photographs of the surface of LA wells showing growth of nematodes on NC1/1 cells (B) and no growth on NGR209 cells (C). Bar, 1 cm.
Retrieval of DNA flanking the mini-Tn5 of NGR209.
DNA from mutant NGR209 was purified, restriction enzyme digested with NsiI (the mini-Tn5 contains no NsiI site), intramolecularly and ethanol precipitated, and transformed by electroporation into E. coli DH5α (Fig. 1B). Transformants containing the mini-Tn5 were selected by being resistant to kanamycin. The retrieved plasmid, p209, was purified and restriction enzyme digested with NsiI and SfiI to verify that the plasmid contained a single NsiI restriction fragment and the mini-Tn5 (determined by the presence of a 2.9-kb SfiI restriction fragment), respectively.
Causality determination of the mini-Tn5 insertions.
A 4.6-kb PstI fragment from pG325 (Table 1) containing the sacB/sacR genes and a gene conferring resistance to streptomycin and spectinomycin was ligated into the NsiI site of the retrieved mini-Tn5 plasmids to create the allelic-exchange plasmid p209C. The plasmid was transformed into wild-type NC1/1 cells. Allelic exchange was selected for by growing NC1/1 cells containing the allelic-exchange plasmid overnight in PP3 containing kanamycin and then plating the cells on PP3 containing kanamycin and sucrose. The mutants were tested for sensitivity to streptomycin and spectinomycin to verify the loss of the allelic-exchange plasmid. The phenotype of the allelic-exchange mutant, NGR209A, was compared to that of the original mini-Tn5 mutant, NGR209.
Sequence analysis of p209.
The sequence of DNA flanking the transposon insertion of p209 was obtained by using M13 forward and reverse primers located 60 or 40 bp from the inverted repeat termini of the transposon and by primer walking. If the sequence obtained from the M13 forward primer revealed the alternative O-end insertion, the DNA sequence flanking the alternative O end was obtained by using the oligonucleotide primer (5′ TAAGCGCCTTCCTGCATGGCTT 3′). Dye terminator cycle sequencing using ABI terminator mix was performed using the conditions suggested by the supplier (Perkin-Elmer Corp., Foster City, Calif.), and then the reaction products were analyzed on an ABI 377 automated sequencer (Perkin-Elmer Corp.) at the University of Wisconsin Biotechnology Center. Comparison of the DNA sequence to database sequences was done using BLAST programs using nonredundant databases (4).
Complementation of NGR209 with pNgrA.
The disrupted allele of NGR209 was designated ngrA. Intact ngrA was obtained by PCR amplification using the oligonucleotide primers Edf (5′ ATTAAGTATAGACTGTAGGATA 3′) and Edr (5′ TGATCAGGGACGGTATCAGCT 3′) and Pfu polymerase (Stratagene Cloning Systems, La Jolla, Calif.). The Edf primer was designed to include the intergenic region between ngrA and the phfB gene that might contain a promoter element (see Fig. 4). An 0.8-kb band was extracted from an agarose gel and blunt end ligated into pBC SK− (Stratagene) that had been treated with HincII and shrimp alkaline phosphatase (United States Biochemical Corp., Cleveland, Ohio). Clones containing intact ngrA were obtained by screening transformants on LB medium containing chloramphenicol and X-Gal. Plasmid preparations were made on white colonies, resulting in a clone containing an 0.8-kb insert with a sequence identical to ngrA of p209 and in the same orientation as the lacZ gene to allow the lac promoter to be utilized for transcription of ngrA. The resulting plasmid, pNgrA, was transformed into NGR209, and the phenotype of the resulting clone was determined.
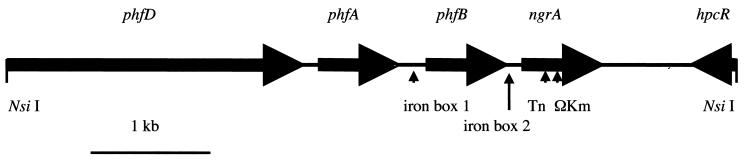
FIG. 4.
Physical structure of DNA flanking the mini-Tn5 insertion of mutant NGR209. Shown are the insertion site of the mini-Tn5 transposon, the location of two potential iron boxes, the site of an ΩKm insertion, and NsiI sites at the termini of the retrieved DNA.
Phenotypic characterization.
Analyses of phase-dependent characteristics and the ability of the cells to support nematode growth and reproduction or to be retained by IJ nematodes were performed as described above. The pathogenicity of bacteria for insects was determined as described previously (11). Positive insect pathogenicity was defined 72 h postinjection as 50% mortality of insect larvae resulting from a dose of less than 30 cells. To test for oral insecticidal activity (11), growth liquors from 72-h PP3 cultures were filter sterilized and concentrated 15 times using a Microcon 40 (Amicon, Inc., Beverly, Mass.) microconcentrator. An 0.05-ml sample of retentate was added to a 1.0-g portion of gypsy moth diet (ICN Pharmaceuticals Inc., Costa Mesa, Calif.), and a single first- or second-instar larva of Manduca sexta was then added. Larvae were observed for weight gain and/or death following 72 h of incubation. Experiments were performed twice with 12 replicates each.
Nucleotide sequence accession number.
The GenBank accession number for DNA sequence flanking the mini-Tn5 insertion of mutant NGR209 is AF288077.
RESULTS
Transposon mutagenesis.
The frequency of transposition and loss of the pUB394 delivery vector, defined as the ratio of cells resistant to kanamycin and sucrose to total cells present, was 1.7 × 10−7. A large number of strains with putative mini-Tn5 insertions were obtained. Southern analysis of 20 randomly picked mutants showed that more than 90% of the putative transposon mutants contained single insertions on different locations of DNA (data not shown). Less than 2% of the strains with putative insertions were resistant to streptomycin and spectinomycin. This suggests that resistance to sucrose in these rare strains occurred by a mechanism other than loss of the delivery vector pUB394. Approximately 1 to 3% of the mutants were auxotrophic. This frequency would be expected if the mini-Tn5 was inserted randomly into the genome of P. luminescens, assuming the genome to be approximately the same size as that of E. coli.
Mutant screening.
Of 2,800 transposon-induced mutants screened, a mutant (NGR209) was obtained that consistently failed to support nematode growth and reproduction. A white mass of nematodes is evident when nematodes are grown on lawns of NC1/1 (Fig. 2B), while no adult nematodes are seen on lawns of NGR209 (Fig. 2C).
Cloning and analyses of DNA flanking the mini-Tn5 insertion of mutant NGR209.
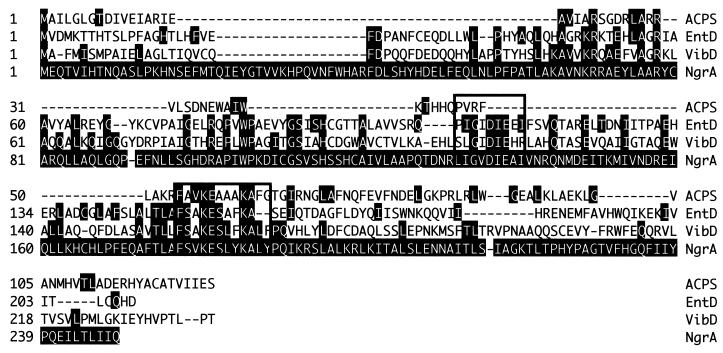
A 10.6-kb plasmid containing the mini-Tn5 was retrieved from NGR209. The ngrA gene, disrupted by the mini-Tn5 in mutant NGR209, appears to encode a 4′-phosphopantetheinyl (Ppant) transferase enzyme. The deduced protein product shows a significant degree of similarity to the two Ppant transferase motifs (boxed residues) characteristic of these proteins (39) (Fig. 3). NgrA is most similar to the Ppant transferases EntD from Salmonella enterica serovar Typhimurium and E. coli and VibD from Vibrio cholerae, which are required for the biosynthesis of the catechol siderophores enterobactin (16, 39) and vibriobactin (54), respectively (residues identical to NgrA are shaded). Ppant transferases transfer the Ppant moiety from coenzyme A to acyl carrier proteins (ACP), aryl carrier proteins, and peptidyl carrier proteins (39). The Ppant-modified carrier proteins are required for the biosynthesis of lipids, lipoproteins, polyketide, and nonribosomal peptides, many with siderophore, antibiotic, or pharmacological activities (37, 42).
FIG. 3.
Alignment of NgrA with Ppant transferase proteins. Shaded residues indicate identity to NgrA. Boxed are the two Ppant transferase motifs (39). Proteins were aligned using Clustal W (DNAStar, Madison, Wis.): ACPS (accession no. sp P24224); EntD, enterobactin synthetase component D (accession no. P19925), E. coli; VibD, phosphopantetheinyl transferase (accession no. AAD48884), V. cholerae.
The sequence of the entire 5.9 kb of p209 flanking the mini-Tn5 was analyzed. The physical map of this DNA (Fig. 4) shows that three open reading frames similar to those for fimbrial proteins, phfB, phfA, and phfD, are located 5′ and on the same strand as ngrA. The phfB gene is located 54 bp from ngrA and is 1,014 bp in length, with residues 215 to 337 of the predicted protein product being 37% identical and 53% similar to the type I fimbrial subunit (FimA/PapA family) (spP12903) (29). The next gene 5′ of ngrA, phfA, is 966 bp in length with residues 115 to 320 of the predicted protein product being 28% identical and 42% similar to the fimbrial adhesin MrkD from Klebsiella pneumoniae (spP21648) (3). The phfD gene is at least 2,397 bp in length (the start codon was not retrieved). Its protein product is 38% identical and 58% similar to the S-fimbrial usher SfaF (prf1713397E) (48). Two possible iron boxes (ferric iron uptake [Fur] regulator-binding sites) (13, 18) were identified. Iron box 1 is located between phfA and phfB and contains 15 bases identical, with a 3-bp insertion, to the palindromic 19-bp consensus iron box (13, 17, 23). Iron box 2 is located between phfB and ngrA and contains 10 bases identical to the 19-bp consensus iron box.
Phenotypic characterization of mutant NGR209.
The phenotype of NGR209 was compared to those of the primary (NC1/1)- and secondary (NC1/2)-phase cells and to NGR209 reconstituted with pNgrA (complementation of NGR209 with intact ngrA) (Table 2). NGR209 is identical to NC1/1 in all properties except in not supporting nematode growth and reproduction or producing siderophore and antibiotic activities. Complementation of NGR209 with pNgrA restored these properties. The restoration of these properties was not due to gene replacement of the mini-Tn5-disrupted ngrA with intact ngrA, because NGR209 cured of pNgrA reverted to the NGR209 phenotype.
TABLE 2.
Phenotypic characterization of NC1/1, NC1/2, NGR209, and NGR209 reconstituted with pNgrA
| Phenotype assayed | Reaction of
strain:
|
|||
|---|---|---|---|---|
| NC1/1 (primary) | NC1/2 (secondary) | NGR209 | NGR209 + pNgrA | |
| Dye absorption | ||||
| Eosin Y-methylene blue | + | − | + | + |
| Neutral red | + | − | + | + |
| Bromthymol blue | + | − | + | + |
| Bioluminescence | + | − | + | + |
| Extracellular products | ||||
| Lipase activity | + | − | + | + |
| Hemolytic activity | + | − | + | + |
| Protease activity | + | − | + | + |
| Antibiotic activity | + | − | − | + |
| Siderophore activity | + | − | − | + |
| Colony morphology | Convex, mucoid | Flat, nonmucoid | Convex, mucoid | Convex, mucoid |
| Pigmentation | + | − | + | + |
| CipA and CipB production | + | − | + | + |
| Insect pathogenicity | ||||
| Injected cells | + | + | + | + |
| Oral cell-free activity | + | NDb | + | + |
| Support of nematode growth and reproductiona | + | − | − | + |
Nematode growth and reproduction on LA and LCM and in G. mellonella hosts.
ND, not determined.
The causality of the transposon for the phenotype of NGR209 was demonstrated by performing allelic exchange with the mini-Tn5-disrupted ngrA gene into the wild-type ngrA gene of P. luminescens cells. The resulting mutant, NGR209C, had all the phenotypic characteristics of NGR209 (data not shown). An omega cassette disruption of ngrA, NPngrA::ΩKm, also had a phenotype identical to that of NGR209 (data not shown).
The results of the analyses of growth, reproduction, and mortality of nematodes when grown on cells of NC1/1 and NGR209 and retention of NC1/1 and NGR209 by nematodes are shown in Table 3. Nematode development from the IJ to the J4 stage was reduced significantly when nematodes were grown on LA medium seeded with cells of NGR209 compared to the equivalent nematodes propagated on NC1/1 cells. The inability of nematodes to reproduce on NGR209 cells was shown by the observation that no IJ nematodes were present after 20 days of growth on the NGR209 cells. In contrast, large numbers of nematodes were produced by nematodes propagated on NC1/1 cells. The NGR209 cells are not toxic to the nematodes, as indicated by a similar percent mortality of IJ nematodes after incubation for 3 days with NGR209 or NC1/1 cells. Also, the numbers of IJ nematodes were essentially the same following 10 to 14 days of growth on 1:1 mixtures of NGR209 and NC1/1 or NGR209 and Meg/1 cells, again indicating that NGR209 cells are not inhibitory for nematode growth and reproduction.
TABLE 3.
Ability of NC1/1 and NGR209 to associate with H. bacteriophora
| Characteristic | Value for
straing:
|
|
|---|---|---|
| NC1/1 | NGR209 | |
| Nematode developmenta | 5.1 (2.5) | 1 (1.1) |
| Nematode yieldsb | 2,730 (1,750) | 0 (0) |
| % Nematode mortalityc | 13.7 (12.0) | 7.7 (7.0) |
| Retention by IJ nematodesd | ||
| Grown on NC1/1 | 106.6 (102) | —e |
| Grown on NGR209 and Meg/1 | —f | 78 (35) |
| Grown on NGR209 and NC1/1 | 83.5 (70) | 0 |
Average numbers of J4 or adult nematodes observed on LA 4 days following the addition of an average of 12 IJ nematodes (n = 12).
Average numbers of IJ nematodes observed on LA at day 20 (same cultures as for nematode development; n = 12).
Percent nonviable IJ nematodes per milliliter of LCM 5 days after addition of 150 IJ nematodes per ml, determined by observing no movement 1 min after the addition of 1.0% commercial bleach (n = 3).
IJ nematodes were propagated on LA seeded with bacteria on one half of divided petri dishes as described in Materials and Methods. Results show the average number of bacteria per disrupted IJ nematode (n = 3).
NGR209 was not present in the experiment.
NC1/1 was not present in the experiment.
Values in parentheses are standard deviations.
The addition of large numbers (250) of IJ nematodes did not overcome the inability of NGR209 cells to support nematode growth and reproduction. The addition of 110 IJ nematodes to NGR209 cells caused the development of IJ to J4 nematodes to increase from 1 to 5.7 (standard deviation, 2.1; n = 3). Although this is approximately equal to the number of J4 nematodes seen on NC/1 cells (Table 3), the J4 nematodes growing on NGR209 cells did not develop to hermaphrodites. This suggests that the decreased development from IJ to J4 observed on NGR209 cells was not the only cause for the inability of NGR209 to support nematode growth and reproduction. Furthermore, a mixture of developmental stages (J1 to adult) added to NGR209 cells did not grow and reproduce. Some nematode development and reproduction were initially seen but quickly ceased. This again indicates that the defect of NGR209 for nematode growth and reproduction does not involve only the development of IJ nematodes to reproductive hermaphrodites; other stages of nematodes were also unable to develop and reproduce.
Because of the close proximity of fimbrial genes to ngrA, it is conceivable that the defect in NGR209 for growth and reproduction might also affect colonization by NGR209 of the IJ nematode intestine. Therefore, the ability of NGR209 to be retained by the IJ nematodes was determined. To do this, the H. bacteriophora nematodes were first propagated on Meg/1 bacteria. This resulted in IJ nematodes that were nearly axenic (approximately one Meg/1 cell per 200 IJ nematodes). The nematodes were incubated for 10 to 14 days on LA inoculated with NC1/1 cells, a 1:1 (vol/vol) mixture of NGR209 and Meg/1 cells, and a 1:1 mixture of NGR209 and NC1/1 cells. The IJ nematodes cultured on NC1/1 cells retained an average of 106 NC1/1 cells (Table 3). The IJ nematodes retained an average of 78 NGR209 cells and no Meg/1 cells when cultured on a mixture of NGR209 and Meg/1. This is essentially the same as the amount of NC1/1 cells retained by IJ nematodes. However, IJ nematodes retained an average of 83 NC1/1 and no NGR209 cells when propagated on a mixture of NC1/1 and NGR209. At day 20, the proportions of bacteria on the LA were about the same as those added initially. Thus, competition of the bacterial strains on the LA prior to IJ retention is not the cause for the differential retention in the IJ nematodes. It is evident that the mini-Tn5 insertion in ngrA causes NGR209 not to be retained in the presence of NC1/1 cells but to be retained in the absence of NC1/1 and the presence of Meg/1 cells.
DISCUSSION
The entomopathogenic nematode H. bacteriophora will grow and reproduce only when feeding on living cells of its symbiotic bacterium, P. luminescens. Spontaneous phase variants of the bacterium that have lost expression of multiple characteristics will not support nematode growth. A screen of 2,800 transposon mutants of P. luminescens yielded only one mutant, NGR209, which lost the ability to support nematode growth and reproduction while retaining most primary-phase characteristics. The transposon is inserted into a gene, ngrA, which database analyses show to be most similar to the entD gene that encodes the enzyme Ppant transferase. The enzyme transfers the Ppant moiety from coenzyme A to EntB and EntF, which are required for the biosynthesis of the siderophore enterobactin (16, 28, 39).
The nematode growth and reproduction mutation also caused loss of detectable antibiotic and siderophore production. It is unlikely that loss of these properties is involved in the nematode growth phenotype, because adding growth liquor from a P. luminescens culture, which contained both activities, to the nematode growth medium did not overcome the growth defect. In addition, we isolated a transposon mutant of P. luminescens producing no detectable siderophore activity, and this mutant supports nematode growth and reproduction (15).
The ngrA gene is more likely to be involved in biosynthesis of a hormone or signal regulator of nematode development than in that of a nutritional factor. The gene is probably not involved in the biosynthesis of fatty acids or lipids because the Ppant transferase of E. coli that activates ACP required for fatty acid biosynthesis is essential (40, 51). Other possible functions of NgrA are the biosynthesis of polyketide or nonribosomally synthesized peptide molecules (39) that are good candidates for hormonal or signal molecules. One possible candidate is the quorum-sensing homoserine lactone molecules that require ACP and ACP synthase (ACPS) for biosynthesis (44). It is unlikely that NgrA is involved in homoserine lactone biosynthesis because NgrA is not equivalent to ACPS based on amino acid similarity.
The NgrA product might be very unstable or active at a critical threshold level, since adding growth liquor of exponential- or stationary-phase cultures of P. luminescens to LA does not restore nematode growth. It is also possible that the putative signal molecule is produced by the bacteria only when they are grown in the presence of the nematode.
The ngrA mutant cells retain most of the characteristics of the parent and have clearly not been converted to the secondary-phase variant. The parent and ngrA mutant cells produce two crystalline inclusion proteins, CipA and CipB (7). The secondary-phase cells do not produce them. Inactivation of either cipA or cipB by omega cassettes resulted in cells exhibiting secondary-phase characteristics (7). These mutants did not support nematode growth and reproduction. It thus seems clear that the ngrA mutant is specifically related to nematode growth and is not involved in the secondary-phase variation phenomenon.
The genes located near ngrA, having putative functions involving fimbrial biogenesis and adhesion and the iron boxes (Fig. 4), might be relevant to the nematode-bacterium symbiosis. Fur is a global regulator in E. coli and regulates some virulence genes (17). Fimbriae are often responsible for specific binding of bacterial cells to eukaryotic cells (20), which can signal changes in gene expression in both bacteria and host cells (1). Knowing the nucleotide sequence of these genes will allow us to specifically disrupt the genes to determine their possible roles in the symbiotic association. Our isolation of the mini-Tn5 insertion in the ngrA gene provides a starting point for genetic and physiological analysis of this symbiotic relationship.
ACKNOWLEDGMENTS
This research was partially supported by the S. C. Johnson Wax Distinguished Scientist Fellowship awarded to T.A.C. and funds from DowAgroSciences and the College of Agriculture and Life Sciences at the University of Wisconsin—Madison.
REFERENCES
- 1.Abraham S, Jonsson A B, Normark S. Fimbriae-mediated host-pathogen cross-talk. Curr Opin Microbiol. 1998;1:75–81. doi: 10.1016/s1369-5274(98)80145-8. [DOI] [PubMed] [Google Scholar]
- 2.Akhurst R J, Mourant R G, Baud L, Boemare N E. Phenotypic and DNA relatedness study between nematode-symbiotic and clinical strains of the genus Photorhabdus (Enterobacteriaceae). Int J Syst Bacteriol. 1996;43:249–255. doi: 10.1099/00207713-46-4-1034. [DOI] [PubMed] [Google Scholar]
- 3.Allen B L, Gerlach G F, Clegg S. Nucleotide sequence and functions of mrk determinants necessary for expression of type 3 fimbriae in Klebsiella pneumoniae. J Bacteriol. 1991;173:916–920. doi: 10.1128/jb.173.2.916-920.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 5.Bartolome B, Jubete Y, Martinez E, de la Cruz F. Construction and properties of a family of pACYC184-derived cloning vectors compatible with pBR322 and its derivatives. Gene. 1991;102:75–78. doi: 10.1016/0378-1119(91)90541-i. [DOI] [PubMed] [Google Scholar]
- 6.Bintrim S B. A study of the crystalline inclusion proteins of Photorhabdus luminescens. Ph.D. thesis. University of Wisconsin, Madison; 1994. [Google Scholar]
- 7.Bintrim S B, Ensign J C. Insertional inactivation of genes encoding the crystalline inclusion proteins of Photorhabdus luminescensresults in mutants with pleiotropic phenotypes. J Bacteriol. 1998;180:1261–1269. doi: 10.1128/jb.180.5.1261-1269.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bleakley B, Nealson K H. Characterization of primary and secondary forms of Xenorhabdus luminescensstrain Hm. FEMS Microbiol Ecol. 1988;53:241–250. [Google Scholar]
- 9.Boemare N E, Akhurst R J. Biochemical and physiological characterization of colony form variants in Xenorhabdus spp. (Enterobacteriaceae). J Gen Microbiol. 1988;134:1835–1845. doi: 10.1099/00221287-134-7-1835. [DOI] [PubMed] [Google Scholar]
- 10.Bowen D, Rocheleau T A, Blackburn M, Andreev O, Golubeva E, Bhartia R, ffrench-Constant R H. Insecticidal toxins from the bacterium Photorhabdus luminescens. Science. 1998;280:2129–2132. doi: 10.1126/science.280.5372.2129. [DOI] [PubMed] [Google Scholar]
- 11.Bowen D J, Ensign J C. Purification and characterization of a high-molecular-weight insecticidal protein complex produced by the entomopathogenic bacterium Photorhabdus luminescens. Appl Environ Microbiol. 1998;64:3029–3055. doi: 10.1128/aem.64.8.3029-3035.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Calderwood S B, Mekalanos J J. Confirmation of the Fur operator site by insertion of a synthetic oligonucleotide into an operator fusion plasmid. J Bacteriol. 1988;170:1015–1017. doi: 10.1128/jb.170.2.1015-1017.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chan J W Y F, Goodwin P H. Extraction of genomic DNA from extracellular polysaccharide-synthesizing Gram-negative bacteria. BioTechniques. 1994;18:419–422. [PubMed] [Google Scholar]
- 15.Ciche T A. Symbiotic interactions between the bacterium Photorhabdus luminescens and the entomopathogenic nematode Heterorhabditis bacteriophora. Ph.D. thesis. University of Wisconsin, Madison; 2000. [Google Scholar]
- 16.Coderre P E, Earhart C F. The entD gene of the Escherichia coliK12 enterobactin gene cluster. J Gen Microbiol. 1989;135:3043–3055. doi: 10.1099/00221287-135-11-3043. [DOI] [PubMed] [Google Scholar]
- 17.Crosa J H. Signal transduction and transcriptional and posttranscriptional control of iron-regulated genes in bacteria. Microbiol Mol Biol Rev. 1997;61:319–336. doi: 10.1128/mmbr.61.3.319-336.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Lorenzo V, Wee S, Herrero M, Neilands J B. Operator sequences of the aerobactin operon of plasmid ColV-K30 binding the ferric uptake regulator (fur) repressor. J Bacteriol. 1987;169:2624–2630. doi: 10.1128/jb.169.6.2624-2630.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Lorenzo V, Herrero M, Jakubzik U, Timmis K H. Mini-Tn5transposon derivatives for the insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J Bacteriol. 1990;172:6568–6572. doi: 10.1128/jb.172.11.6568-6572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Edwards R A, Puente J L. Fimbrial expression in enteric bacteria: a critical step in intestinal pathogenesis. Trends Microbiol. 1998;6:282–287. doi: 10.1016/s0966-842x(98)01288-8. [DOI] [PubMed] [Google Scholar]
- 21.Ehlers R D, Lunau S, Krasomil-Osterfeld K, Osterfeld J H. Liquid culture of the entomopathogenic nematode-bacterium-complex Heterorhabditis megidis/Photorhabdus luminescens. BioControl. 1998;43:77–86. [Google Scholar]
- 22.Ehlers R D, Stoessel S, Whyss U. The influence of phase variants of Xenorhabdus spp. and Escherichia coli (Enterobacteriaceae) on the propagation of entomopathogenic nematodes of the genera Steinernema and Heterorhabditis. Rev Nematol. 1990;13:417–424. [Google Scholar]
- 23.Escolar L, Pèrez-Martìn J, de Lorenzo V. Binding of Fur (ferric uptake regulator) repressor of Escherichia colito arrays of GATAAT sequence. J Mol Biol. 1998;283:537–547. doi: 10.1006/jmbi.1998.2119. [DOI] [PubMed] [Google Scholar]
- 24.Fellay R, Frey J, Krisch H. Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vitroinsertional mutagenesis of Gram-negative bacteria. Gene. 1987;52:147–154. doi: 10.1016/0378-1119(87)90041-2. [DOI] [PubMed] [Google Scholar]
- 25.Forst S, Nealson K H. Molecular biology of the symbiotic-pathogenic bacteria Xenorhabdus spp. and Photorhabdusspp. Microbiol Rev. 1996;60:21–43. doi: 10.1128/mr.60.1.21-43.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Forst S, Dowds B, Boemare N, Stackebrandt E. Xenorhabdus spp. and Photorhabdusspp.: bugs that kill bugs. Annu Rev Microbiol. 1997;51:47–72. doi: 10.1146/annurev.micro.51.1.47. [DOI] [PubMed] [Google Scholar]
- 27.Gay R, Le Coq D, Steinmetz M, Berkelman T, Kado C I. Positive selection procedure for entrapment of insertion sequence elements in gram-negative bacteria. J Bacteriol. 1985;164:918–921. doi: 10.1128/jb.164.2.918-921.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gehring A M, Bradley K A, Walsh C T. Enterobactin biosynthesis in Escherichia coli: isochorismate lyase (EntB) is a bifunctional enzyme that is phosphopantetheinylated by EntD and then acylated by EntE using ATP and 2,3-dihydroxybenzoate. Biochemistry. 1997;36:8495–8503. doi: 10.1021/bi970453p. [DOI] [PubMed] [Google Scholar]
- 29.Gerlach G F, Clegg S, Allen B L. Identification and characterization of the genes encoding type 3 and type 1 fimbrial adhesin of Klebsiella pneumoniae. J Bacteriol. 1989;171:1262–1270. doi: 10.1128/jb.171.3.1262-1270.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gerritsen L J M, Smits P H. Variation in pathogenicity of recombinations of Heterorhabditis and Xenorhabdus luminescensstrains. Fundam Appl Nematol. 1993;16:367–373. [Google Scholar]
- 31.Han R, Wouts W M, Li L. Development of Heterorhabditis spp. strains as characteristics of possible Xenorhabdus luminescenssubspecies. Rev Nematol. 1990;13:411–415. [Google Scholar]
- 32.Han R, Wouts W M, Li L. Development and virulence of Heterorhabditis spp. strains associated with different Xenorhabdus luminescensisolates. J Invertebr Pathol. 1991;58:27–32. [Google Scholar]
- 33.Han R C, Ehlers R-U. Cultivation of axenic Heterorhabditis spp. dauer juveniles and their response to non-specific Photorhabdus luminescensfood signals. Nematologica. 1998;44:425–435. [Google Scholar]
- 34.Han R C, Ehlers R-U. Pathogenicity, development, and reproduction of Heterorhabditis and Steinernema carpocapsae under axenic in vivoconditions. J Invertebr Pathol. 2000;75:55–58. doi: 10.1006/jipa.1999.4900. [DOI] [PubMed] [Google Scholar]
- 35.Herrero M, de Lorenzo V, Timmis K H. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J Bacteriol. 1990;172:6557–6567. doi: 10.1128/jb.172.11.6557-6567.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hurlbert R E, Xu J, Small C L. Colonial and cellular polymorphism in Xenorhabdus luminescens. Appl Environ Microbiol. 1989;55:1136–1143. doi: 10.1128/aem.55.5.1136-1143.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Keating T A, Walsh C T. Initiation, elongation, and termination strategies in polyketide and polypeptide antibiotics. Curr Opin Chem Biol. 1999;3:598–606. doi: 10.1016/s1367-5931(99)00015-0. [DOI] [PubMed] [Google Scholar]
- 38.Khan A, Brooks W M. A chromogenic bioluminescent bacterium associated with the entomophilic nematode Chromonema heliothidis. J Invertebr Pathol. 1976;29:253–261. [Google Scholar]
- 39.Lambalot R H, Gehring A M, Flugel R S, Zuber P, LaCelle M, Marahiel M A, Reid R, Khosla C, Walsh C T. A new enzyme superfamily—the phosphopantetheinyl transferases. Chem Biol. 1996;3:923–936. doi: 10.1016/s1074-5521(96)90181-7. [DOI] [PubMed] [Google Scholar]
- 40.Lambalot R H, Walsh C T. Cloning, overproduction, and characterization of the Escherichia coliholo-acyl carrier protein synthase. J Biol Chem. 1995;270:24658–24661. doi: 10.1074/jbc.270.42.24658. [DOI] [PubMed] [Google Scholar]
- 41.Lunau S, Stoessal S, Schmidt-Peisker A J, Ehlers R-U. Establishment of monoxenic inocula for scaling up in vitro cultures of the entomopathogenic nematodes Steinernema spp. and Heterorhabditisspp. Nematologica. 1993;39:385–399. [Google Scholar]
- 42.Marahiel M A, Stachelhaus T, Mootz H D. Modular peptide synthetases involved in nonribosomal peptide synthesis. Chem Rev. 1997;97:2651–2674. doi: 10.1021/cr960029e. [DOI] [PubMed] [Google Scholar]
- 43.Martinez E, Bartomole B, de la Cruz F. pACYC184-derived cloning vectors containing the multiple cloning site and lacZα reporter gene of pUC8/9 and pUC18/19 plasmids. Gene. 1988;68:159–162. doi: 10.1016/0378-1119(88)90608-7. [DOI] [PubMed] [Google Scholar]
- 44.More M I, Finger L, David L, Stryker J L, Fuqua C, Eberhard A, Winans S C. Enzymatic synthesis of a quorum-sensing autoinducer through use of defined substrates. Science. 1998;272:1655–1658. doi: 10.1126/science.272.5268.1655. [DOI] [PubMed] [Google Scholar]
- 45.Poinar G O, Jr, Thomas G M, Hess R. Characteristics of the specific bacterium associated with Heterorhabditis bacteriophora(Heterorhabditidae: Rhabditida) Nematologica. 1977;23:97–102. [Google Scholar]
- 46.Poinar G O, Jackson T, Klein M. Heterorhabditis megidis sp. n. (Heterorhabditae: Rhabditida), parasitic in the japanese beetle, Popillia japonica(Scarabaeidae: Coleoptera), in Ohio. Proc Helminthol Soc Wash. 1987;54:53–59. [Google Scholar]
- 47.Reid J L, Collmer A. An nptI-sacB-sacRcartridge for constructing directed, unmarked mutations in Gram-negative bacteria by marker exchange-eviction mutagenesis. Gene. 1987;57:239–246. doi: 10.1016/0378-1119(87)90127-2. [DOI] [PubMed] [Google Scholar]
- 48.Schmoll T, Morschhaeuser J, Ott J M, Ludwig B, Van Die I, Hacker J. Complete genetic organization and functional aspects of the Escherichia coli S fimbrial adhesin determinant. Nucleotide sequence of genes sfaB, C, D, E, F. Microb Pathol. 1990;9:331–343. doi: 10.1016/0882-4010(90)90067-z. [DOI] [PubMed] [Google Scholar]
- 49.Schwyn B, Neilands J B. Universal chemical assay for the detection and determination of siderophores. Anal Biochem. 1987;160:47–56. doi: 10.1016/0003-2697(87)90612-9. [DOI] [PubMed] [Google Scholar]
- 50.Sierra G. A simple method for the detection of lipolytic activity of microorganisms and some observations on the influence of the contact between cells and fatty acid substrates. J Microbiol Serol. 1957;23:15–22. doi: 10.1007/BF02545855. [DOI] [PubMed] [Google Scholar]
- 51.Takiff H E, Baker T, Copeland T, Chen S M, Court D L. Locating essential Escherichia coli genes by using mini-Tn10 transposons: the pdxJoperon. J Bacteriol. 1992;174:1544–1553. doi: 10.1128/jb.174.5.1544-1553.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang H, Dowds B C A. Phase variation in Xenorhabdus luminescens: cloning and sequencing of the lipase gene and analysis of its expression in the primary and secondary phases of the bacterium. J Bacteriol. 1993;175:1665–1673. doi: 10.1128/jb.175.6.1665-1673.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wouts W M. Mass production of the entomogenous nematode Heterorhabditis bacteriophoraon artificial media. J Nematol. 1981;13:467–469. [PMC free article] [PubMed] [Google Scholar]
- 54.Wyckoff E E, Stoebner J A, Reed K E, Payne S M. Cloning of a Vibrio choleraegene cluster: identification of genes required for early steps in siderophore biosynthesis. J Bacteriol. 1997;179:7055–7062. doi: 10.1128/jb.179.22.7055-7062.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]