Abstract
Pediatric acute myeloid leukemia (AML) remains a fatal disease for at least 30% of patients, stressing the need for improved therapies and better risk stratification. As proteins are the unifying feature of (epi)genetic and environmental alterations, and are often targeted by novel chemotherapeutic agents, we studied the proteomic landscape of pediatric AML. Protein expression and activation levels were measured in 500 bulk leukemic patients’ samples and 30 control CD34+ cell samples, using reverse phase protein arrays with 296 strictly validated antibodies. The multistep MetaGalaxy analysis methodology was applied and identified nine protein expression signatures (PrSIG), based on strong recurrent protein expression patterns. PrSIG were associated with cytogenetics and mutational state, and with favorable or unfavorable prognosis. Analysis based on treatment (i.e., ADE vs. ADE plus bortezomib) identified three PrSIG that did better with ADE plus bortezomib than with ADE alone. When PrSIG were studied in the context of cytogenetic risk groups, PrSIG were independently prognostic after multivariate analysis, suggesting a potential value for proteomics in combination with current classification systems. Proteins with universally increased (n=7) or decreased (n=17) expression were observed across PrSIG. Certain proteins significantly differentially expressed from normal could be identified, forming a hypothetical platform for personalized medicine.
Introduction
Pediatric acute myeloid leukemia (AML) is a heterogeneous disease resulting from clonal expansion of myeloid precursors that have lost the ability to differentiate normally.1 Despite improvements in outcome, the 5-year overall survival of affected patients approximates 70% and serious long-term complications are common among survivors.2 With the exception of acute promyelocytic leukemia, leukemia with FLT3-internal tandem duplication (ITD) mutations, and mixed phenotype acute leukemia, pediatric AML has been treated as a homogeneous disease, as therapy does not differ based on the underlying mutations.
Many genetic “drivers” have been implicated in pediatric AML disease pathology and risk stratification. However, only a minority of these drivers have been exploited by targeted therapeutic interventions.3 Current risk-stratification considers genetic abnormalities (e.g., inv (16), monosomy 7), but otherwise relies on early response to therapy (minimal residual disease status). However, risk-stratification is imperfect and outcome within risk groups is heterogeneous. Since many drivers may prove to be directly “undruggable”, targeting downstream proteins may be efficacious. This requires knowledge of the proteomic landscape that emerges from the combined “net” consequences of genetic and epigenetic events. However, little is known about the proteomic landscape in pediatric AML. Improved understanding of this might enhance pretreatment risk stratification and guide the selection of therapies against targetable molecular lesions, especially agents targeting protein function.
Genomic mutations influence cellular physiology via altered protein abundance or activity, but several factors diminish the correlation between genetic alterations and protein effects, including the general lack of correlation between cellular messenger RNA (mRNA) abundance and protein expression,4,5 and the inability to assess posttranslational modifications of proteins with genomic techniques. In other tumor types, protein quantification commonly influences diagnosis, classification and therapy (e.g., estrogen and progestin receptors, programmed cell death ligand 1).6,7 Despite these advantages, proteomics has not been used to guide AML therapy.
We previously performed a pilot study of the proteomic landscape examining 194 proteins in 95 de novo pediatric AML patients using an approach that recognized protein expression patterns within protein functional groups (PFG).8 After determining the characterization of each patient’s PFG, we built higher order structures based on strong correlations between PFG patterns, recognizing eight protein expression signatures (PrSIG) that were prognostic. Here, we used this same approach to prospectively examine 500 pediatric AML patients treated on a Children’s Oncology Group (COG) randomized phase III clinical trial (AAML1031). The hypothesis tested in this trial was that the addition of the proteasome inhibitor bortezomib could improve therapy based on Ara-C (cytarabine), daunorubicin, and etoposide (ADE). This trial closed early when it was determined that adding bortezomib to ADE (ADEB) did not improve either event-free or overall survival at 3 years across the entire group of patents. We aimed to: (i) validate the ability to classify pediatric AML patients based on proteomics in a larger cohort, with significantly more protein targets; (ii) determine whether protein classification could enhance risk stratification; (iii) identify patients who could benefit from ADEB; and (iv) identify additional targets for potential combination therapy.
Methods
Patients’ samples
Peripheral blood samples were collected from 500 pediatric patients with de novo AML participating in the COG AAML1031 (#NCT01371981) phase III clinical trial (n=483)9 or in older clinical trials (n=17), and 30 control CD34+ bone marrow samples from healthy donors (20 children and 10 adults) between July 2011 and February 2017.9 Samples with <80% blasts were enriched for leukemic cells by CD3/CD19 depletion. Samples were collected before the start of chemotherapy (n=500), and 10 hours (h) (n=463) and 24 h (n=466) after the start of induction chemotherapy. At both 10 h and 24 h, one dose of each chemotherapeutic agent had been administered.10 Written informed consent was obtained in accordance with the Declaration of Helsinki and local institutional review boards.
Outcome data were restricted to 410 of the 483 patients enrolled on the AAML1031 trial. Outcome was not determined for 69 patients treated with ADE after trial closure by the Data and Safety Monitoring Board, and for four patients who did not meet eligibility criteria. Two-hundred patients received standard ADE induction therapy, including 36 who also received sorafenib (ADES), while 210 patients received ADEB. Induction therapy produced complete remission by the end of induction II in 348 (85%) patients, 31 (8%) were either refractory or died early. Relapse occurred in 156 (45%) patients, and 286 (70%) were still alive with a median follow-up of 4.4 years (range, 0.3-7.5 years). Outcome data were calculated as published previously.9 Mutation data were available for CEBPA, NPM1, KIT (exons 8 and 17) and FLT3-ITD.11
Reverse phase protein microarray methodology
The methodology and validation of the reverse phase protein microarray (RPPA) technique, including antibody validation, are described elsewhere.12-14 Briefly, slides were probed with 301 validated antibodies (Online Supplementary Table S1). Stained slides were analyzed using Microvigene® software version 3.0 (VigeneTech, Inc., Carlisle, MA, USA) to produce quantified data. Samples were printed in five serial dilutions and SuperCurve algorithms were used to generate a single protein concentration value in log2 format.15 Loading control16 and topographical normalization17 were performed to account for protein concentration and background staining variations. Replicate-based normalization18 was used to align samples from two different slides. Five antibodies were excluded for different reasons yielding a final set of 296 antibodies used for analysis.9 Median expression levels of the normal bone marrow CD34+ samples were subtracted from the expression in the patients’ samples to equalize the median of the control samples to zero.
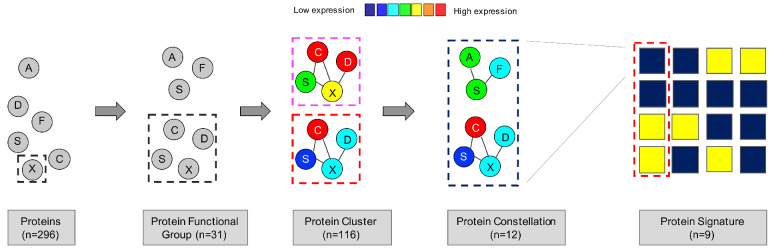
Figure 1.
Computational MetaGalaxy work flow. Multistep analysis that starts with relative expression of 296 protein targets. Proteins were allocated into 31 protein functional groups (PFG) based on their known functionality or strong correlation from the dataset. The progeny clustering algorithm identified an optimal number of protein clusters (PrCL) in each PFG. Block clustering was applied to a binary matrix indicating PrCL-membership for all patients, and identified the existence of protein constellations (PrCON) (i.e., strongly correlated PrCL from various PFG). Patients who expressed similar combinations of PrCON were defined as having a given protein expression signature (PrSIG).
Computational analysis
The MetaGalaxy computational analysis was done as published previously (Figure 1).8,21,22 It takes a multistep approach that starts with separating proteins into 31 PFG based on prior knowledge from the literature or strong correlations within this dataset (Online Supplementary Table S1). K-means23 coupled with the progeny clustering algorithm24 was applied to identify an optimal number of protein clusters (PrCL), i.e., patient subgroups with a similar correlated protein expression profile within each PFG. Collective PrCL-memberships for each patient were combined into a binary matrix. Block-clustering25 was used to search for correlations between PrCL (protein expression constellation [PrCON]), and to cluster patients with similar PrCON-membership into a PrSIG. Statistical analyses were performed in R version 1.3.959 2009-2020 (RStudio, Inc., Boston, MA, USA) or SAS version 9.4 (SAS Institute, Inc., Cary, NC, USA).
The remaining parts of the methods are described in the Online Supplementary Material. Online Supplementary Table S2 summarizes the Cox analyses for outcome reported in Figures 3D, 5, and Online Supplementary Figures S3, S7 and S8.
Results
Correlation of protein expression identifies functional protein paterns within a protein functional group
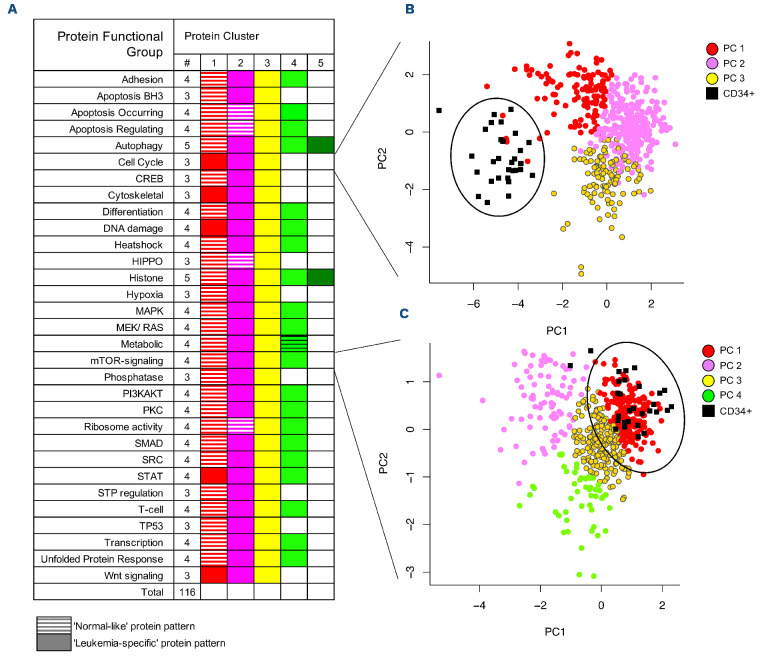
The 296 proteins that were analyzed in this study were allocated into 31 PFG (autophagy, cell cycle, metabolism, etc.).8,20,21 Clustering analysis identified an optimal number of patterns (i.e., PrCL) of similar correlated protein expression (negatively or positively) between patients within each PFG. PrCL numbers ranged from three to five per PFG (Figure 2A) and the total number of PrCL was 116.
Principal component analysis was applied to graphically compare patients’ PrCL expression patterns to those of non-malignant CD34+ cells. Although the overall proteomic profiles of the pediatric AML patients were distinct from those of normal CD34+ cells (Online Supplementary Figure S1), we found overlapping “normal-like” expression patterns for 31 (27%) of the PrCL (Figure 2A). In four PFG, more than one cluster was defined as “normal-like”, and in five PFG no “normal-like” pattern was found. PrCL without dominant co-localization to CD34+ samples on the principal component plot were defined as “leukemia-specific”.
To visually map how proteins interact with other PFG core-members and RPPA dataset proteins, networks were generated for each PrCL. Proteins were connected if they were known to interact with other proteins based on the STRING database or correlation in our dataset. The median protein expression was calculated for each PrCL and overlaid onto the networks to visualize relative expression. Networks can be viewed online at: http://leukemiaproteinatlas.org/pediatric-aml/.
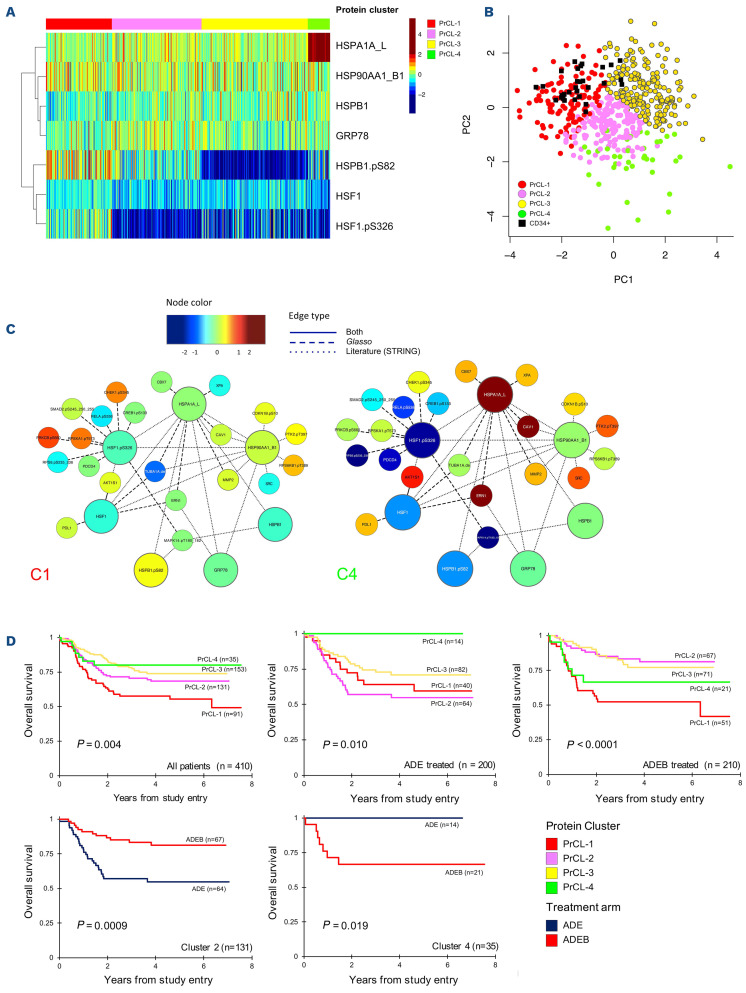
As an example, the PFG “Heat shock” comprises five antibodies recognizing total proteins and two recognizing phosphorylated proteins. We discerned four “Heat shock” PrCL (Figure 3A). Expression levels in PrCL-1 were identified as most “normal-like” (Figure 3B). Protein networks were generated for the seven heat shock protein members. For PrCL-2 to PrCL-4, expression of HSPA1A_L and HSBP1-pSer82 changed between the four PrCL, as did AKT1S1 (connected to HSF1 and HSF1-pSer326) and CAV1 (connected to HSP90AA1_B1 and HSPA1_L1), showing that associated nodes correlate with core-protein PFG-members. Simplified versions of the networks are shown for PrCL-1 and PrCL-4 (Figure 3C). It is important to note that heatmaps presented in other analyses have typically been median normalized to 0 with the variance set from -1 to +1, so that all variables are shown as ranging from the minimum to the maximum of the scale (color range). In contrast, our expression levels are shown relative to normal, and therefore may only use a portion of the scale (color) range.
Figure 2.
Protein functional group classification and similarity to that of normal CD34+ cells. (A) The progeny clustering algorithm was applied to the 31 protein functional groups (PFG) and identified an optimal number of protein clusters (PrCL). PrCL were compared with those of normal CD34+ cells using principal component analysis, (PCA) and classified as either “normallike” or “leukemia-like”. “Normal-like” patterns are represented by hatched boxes, “leukemia-specific” patterns by full boxes. (B, C) PCA with each PrCL being assigned to a color within the PFG to illustrate its similarity to, or difference from, normal CD34+ cells. Two examples of PCA mapping include (B) cell cycle and (C) mTOR-signaling Normal CD34+ samples are represented by small black squares and large black circles. There was no co-localization with CD34+ cells for cell cycle.
Protein clusters correlate with clinical outcome
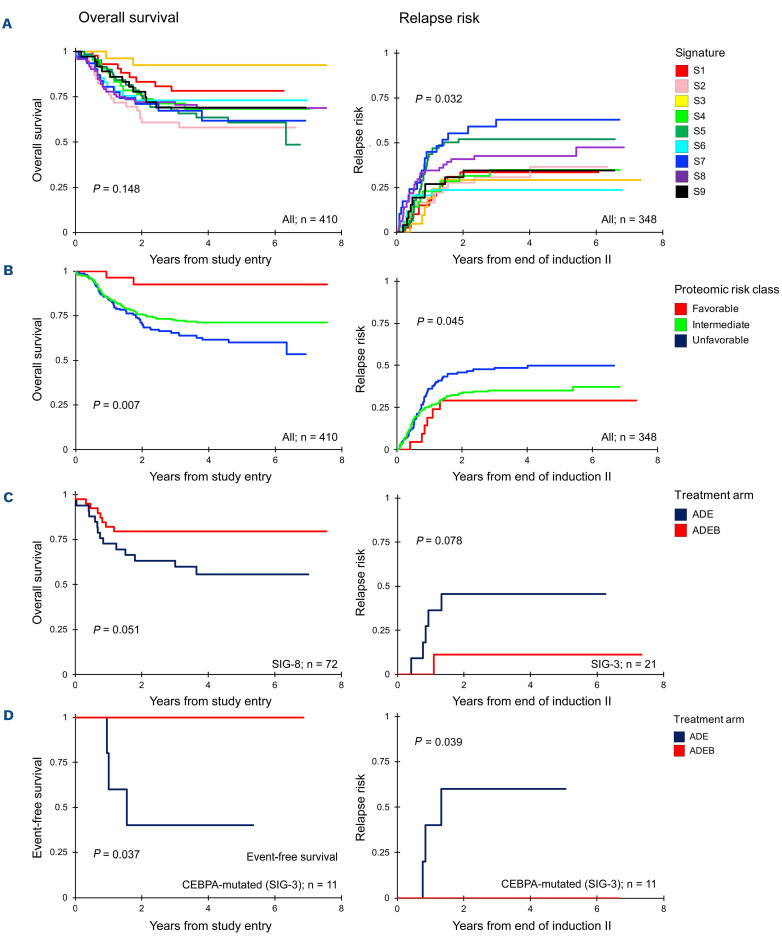
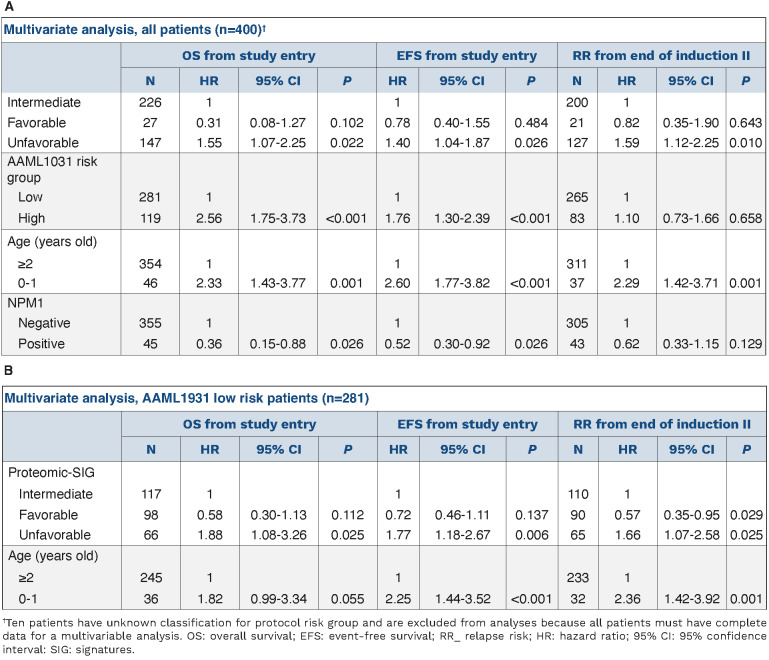
To evaluate the effect of protein expression alone on prognosis, PrCL were correlated to outcome. Seven (23%) PFG were found to be significantly associated with outcome (Online Supplementary Figure S2). For instance, heat shock PrCL were prognostic, in the whole group of patients, for overall survival (P=0.004), event-free survival (P=0.0009), and relapse risk (P=0.0016), as well as in patients treated with either ADE or ADEB (ADE: overall survival, P=0.0035; event-free survival, P=0.0097; relapse risk, P=0.0207; ADEB: overall survival, P=0.0002; event-free survival, P<0.0001; relapse risk, P=0.0009) (Figure 3D). When we compared outcome after ADE to that after ADEB, patients with PrCL-2 significantly benefited from ADEB (n=131, 5-year overall survival, 54% vs. 81%, P=0.00087), whereas patients with PrCL-4 did worse (n=35, 5-year overall survival, 100% vs. 67%, P=0.019). Bortezomib had no effect on patients with PrCL-1 (n=91) (P=0.190), which was an unfavorable prognostic indicator after both ADE and ADEB; this cluster was characterized by high-HSF1-pSer326 and HSB1-pSer82. Event-free survival and relapse risk curves are shown in Online Supplementary Figure S3. Online Supplementary Table S3 shows the distribution across the PrCL and the different treatment arms.
Figure 3.
Analysis of the heat shock protein functional group. (A) Optimal number of four protein clusters (PrCL) was identified as shown by the heatmap (annotation bar: PrCL-1 [red], PrCL-2 [pink], PrCL-3 [yellow] and PrCL-4 [light green]). (B) Principal component analysis shows relative expression of the four clusters in relation to normal CD34+ cells (black squares). (C) Simplified version of the protein networks for PrCL-1 and PrCL-4. Networks illustrate the median expression of protein functional group (PFG) core-protein members (large nodes) in relation to associated proteins (small nodes). Interactions between nodes are based on the literature (…), reverse phase protein microarray data (- - -) or both (—). Associated nodes with most differences between PrCL-1 and PrCL-4 are selected. As an example, AKT1S1 and CAV1 both have relatively normal expression in PrCL-1, while their expression is relatively high in PrCL-4. (D) Outcome data stratified by PrCL. Outcomes for all patients (upper left panel), patients treated with ADE (upper middle panel) and those treated with ADEB (upper right panel). Lower panels show outcome data in cluster 2 (left) and cluster 4 (right) for patients treated with ADE (dark blue) or ADEB (red).
We previously published that low HSF1-pSer326 was associated with a better outcome after ADEB.10 In the current analysis, this effect was true for PrCL-2 and PrCL-3 (characterized by low HSF1-pSer326), but was absent for PrCL-4, which also had concomitant increased expression of HSPA1A_L, emphasizing that a simultaneous integrated analysis of multiple proteins, rather than a single protein, could identify more detailed protein expression patterns and better characterize subpopulations that could benefit from the addition of novel agents.
Recurrences in protein patterns classify patients into nine protein expression signatures
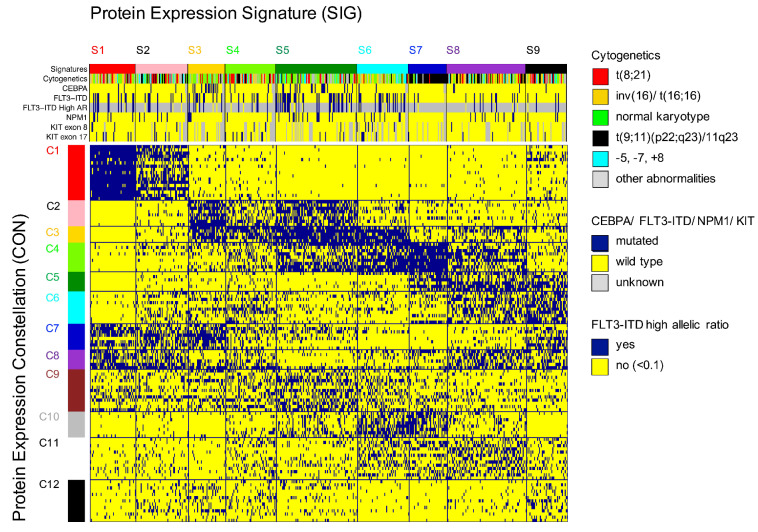
To obtain a more systemic understanding of the 116 identified PrCL, pattern recognition of the relations between PrCL from various PFG was done using co-clustering. PrCL-memberships for the 31 PFG were assigned to all 500 pediatric AML patients and compiled in a binary matrix (MetaGalaxy) (Figure 4). Optimization calculations8,22 identified 12 patterns of recurrent (i.e., correlated) PrCL, defined as PrCON. From this, nine PrSIG were defined as clusters of patients who expressed similar combinations of PrCON. Robustness of the PrCON and PrSIG was tested on a training set (n=355) and test set (n=145) and showed high reproducibility (P<0.0001) (Online Supplementary Figure S4). Sets were created by using random sampling.26 None of the PrCON was predominantly associated with the “normal-like” clusters (P=0.200). The PrCL in each PrCON are listed in Online Supplementary Table S4.
Protein expression partially correlates with cytogenetics and mutational state, but not with gene expression
Correlation between mRNA expression from RNA-sequencing and RPPA protein abundance was determined for 205 total-proteins in 390 samples, with a mean correlation of 0.17. Thirty-four (17%) proteins were negatively correlated, while 83% were positively correlated (Online Supplementary Figure S5).
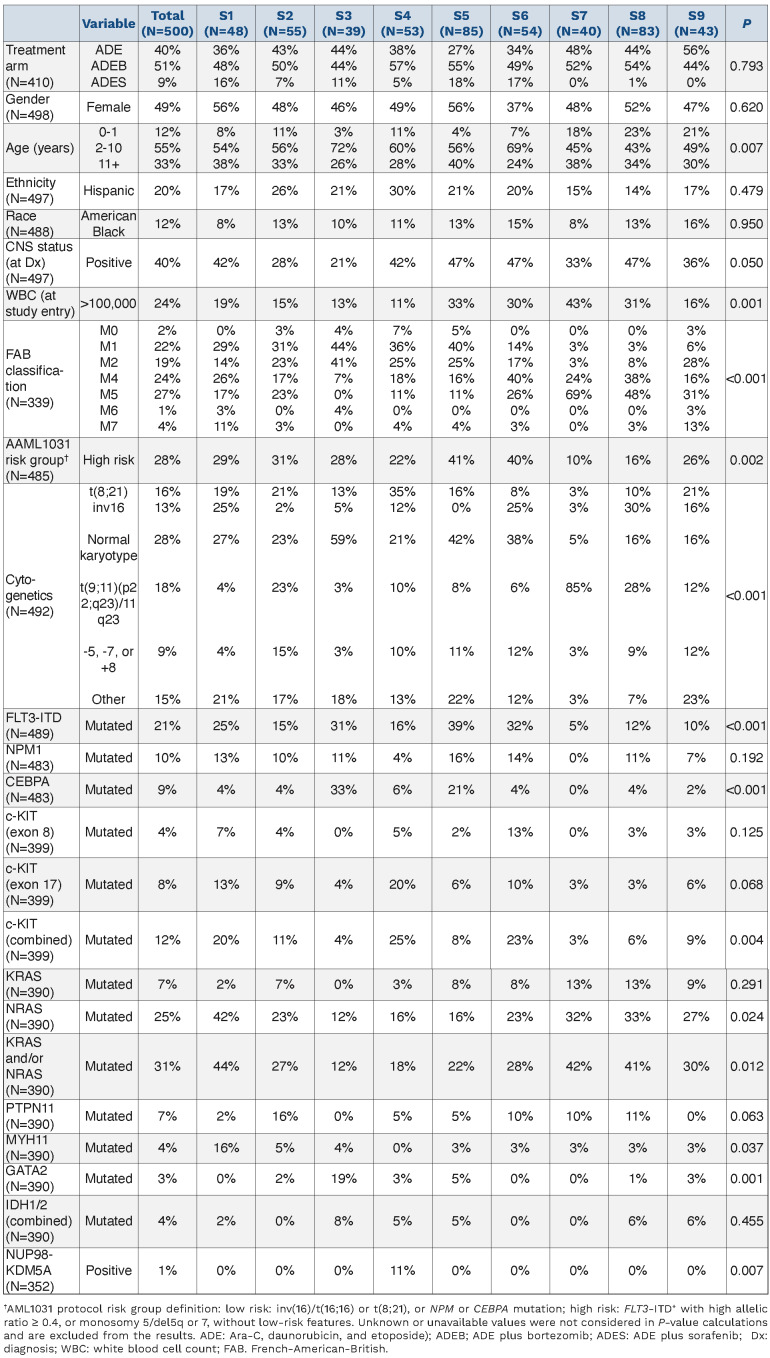
We found associations between PrSIG and cytogenetics and mutation states. Data were available for CEBPA, NPMI, FLT3-ITD, KIT (exons 8 and 17), KRAS, NRAS, GATA2, PTPN11, MYH11 and IDH1/2. Mutations present in ≤10 of the patients were not analyzed (Table 1). Translocation t(8;21) was more frequently detected in PrSIG-4 (35% vs. 6% overall) (P=0.001). Inversion (16) was associated with PrSIG-1, -6 and -8 (25%, 25%, 30% vs. 14%, overall), but scarcely seen in PrSIG-2, -3, -5 and -7 (2%, 5%, 0% and 3%) (P<0.001). Normal karyotype (diploid) was enriched in PrSIG-3, -5 and -6 (59%, 42%, 38% vs. 28% overall) that shared PrCON-3. Those three also had the highest frequencies of CEBPA mutation (PrSIG-3) and FLT3-ITD (PrSIG-3, -5 and -6). While the MLL-rearrangement (11q23) was not exclusive to the PrSIG-7 protein expression pattern, 85% of patients with this signature harbored the MLL-rearrangement (vs. 18% overall). KIT mutations were mostly in patients with PrSIG-4 and -6 (P=0.004), and NRAS and MYH11 in those with PrSIG-1 (P=0.024, P=0.037, respectively). Although only 3% (n=12) of the patients had mutated GATA2, 19% of those with PrSIG-3 had this mutation. Patients with the fusion gene NUP98-KDM5A (n=4) were all present in PrSIG-4 (P=0.007). NPM1, KRAS, PTPN11 and IDH were not associated with specific PrSIG.
Correlation with patients’ characteristics and clinical variables
Patients aged ≤1 year at the time of diagnosis were most frequently clustered in PrSIG-7 to -9, which are associated with PrCON-5. Low white blood cell count (≤100,000 cells/μL) strongly correlated with PrCON-7, with 81-89% of the patients in PrSIG-1 to -4 and -9 having a low white blood cell count versus 76% overall (P=0.001). Gender, ethnicity, race and central nervous system status at the time of diagnosis were not associated with any PrSIG or PrCON (Online Supplementary Figure S6).
Figure 4.
MetaGalaxy analysis identified the existence of 12 protein constellations and nine protein expression signatures. Block clustering was applied to a binary matrix constructed from 116 protein clusters (PrCL) from 31 protein functional groups (PFG). Each column indicates one patient (n=500) and his or her PrCL-membership. This identified the existence of 12 protein constellations (PrCON; horizontally); i.e., PrCL that strongly correlated with each other. Patients with a similar pattern of PrCON were defined as having a given a protein expression signature (PrSIG; vertically). The annotation bar shows nine PrSIG, cytogenetics (t(8;21) [red], inv(16) [yellow], normal karyotype [green]), MLL (11q23) rearrangement [black], -5, -7, +8 abnormalities [light blue], and other [gray]), and mutational status for CEBPA, FLT3-ITD, FLT3-ITD high allelic ratio (≥0.1), NPM1 and KIT (exons 8 and 17) (wildtype [blue], mutated [yellow]). Those with unknown mutational status are represented in gray.
Protein expression signatures provide prognostic information
PrSIG were associated with response to therapy (Figure 5A) with greater spread in 5-year overall survival in ADEB-treated cases compared to ADE-treated cases (Online Supplementary Figure S7). Similar to what was previously done with cytogenetics prognostication, we identified PrSIG associated with favorable (PrSIG-3), intermediate (PrSIG-1, -4, -6, -8, -9) and unfavorable (PrSIG-2, -5, -7) prognosis (overall survival, P=0.007; event-free survival, P=0.046; relapse risk, P=0.045) (Figure 5B). PrSIG-risk groups did not correlate with AAML1031 risk group stratification or conventional risk group stratification (Online Supplementary Table S5). Unfavorable PrSIG remained an independent prognostic factor using multivariate Cox regression analysis in overall and event-free survival and relapse risk (Table 2A). PrSIG with poorest prognosis in ADE-treated patients were PrSIG-5 to -8, all characterized by PrCON-4-membership. Addition of bortezomib was beneficial for 5-year overall survival in PrSIG-6 (62% vs. 84%, n=41, P=0.07, hazard ratio [HR]=0.32) and PrSIG-8 (56% to 79%, n=72, P=0.06, HR=0.43) (Figure 5C), and a similar trend was observed in PrSIG-7. As PrSIG-6 to -8 were most strongly associated with PrCON-11, we compared ADE versus ADEB in the PrCON-11 PrSIG. Overall survival at 5 years increased from 58% to 78% (P=0.011, HR=0.46). Across the nine PrSIG, relapse risk showed a significant dispersion, ranging from 24% to 63% at 5-year complete remission (P=0.03). In PrSIG-3, relapse risk decreased from 45% to 11% with ADEB (n=21, P=0.09, HR=0.18), while PrSIG-6 patients did worse with ADEB (36% vs. 12%, n=34, P=0.10, HR=3.54) (Figure 5C). While analysis of CEBPA-mutated patients as a group did ot show benefit from bortezomib,12 none of the patients in PrSIG-3 relapsed or died after ADEB versus a 60% event and relapse-rate (n=3/5, P=0.039, P=0.037, respectively) with ADE (Figure 5D). Survival analysis stratified by PrSIG-risk restricted to patients with a normal karyotype again identified low- versus high-risk AML patients (P=0.044) (Online Supplementary Figure S8). Of note, while heat shock proteins were strongly associated with outcome in the above analysis, they were not among the main drivers of PrCON and PrSIG clustering.
Table 1.
Patients’ characteristics stratified by protein expression signatures.
Proteomics augment cytogenetic risk stratification
We analyzed AAML1031 low-risk patients (defined by inv(16)/t(16;16), t(8;21), NPM1 or CEBPA mutations) separately to determine whether proteomics were informative for outcome. We found stratification for event-free survival and relapse risk by PrSIG with favorable prognosis in PrSIG-1, -3, -6 and -9 and unfavorable prognosis in PrSIG-5 and -7 (overall survival, P=0.071; event-free survival, P=0.027; relapse risk, P=0.014) (Online Supplementary Figure S9). Cox proportional hazards regression models identified unfavorable proteomic signatures as a significant independent prognostic factor in multivariate analysis (Table 2B). Within the AAML1031 high-risk patients (i.e., those with FLT3-ITD+ high allelic ratio, monosomy 5 or 7 or del(5q), or minimal residual disease >0.1% at end of induction 1) PrSIG were also significantly prognostic. While prognosis of PrSIG-1, -3 and -6 was consistent between AAML1031 risk groups, prognosis of PrSIG-9 was favorable among the low-risk, but highly unfavorable among the high-risk patients.
Proteins significantly different from normal can be identified for each protein expression signature and protein constellation
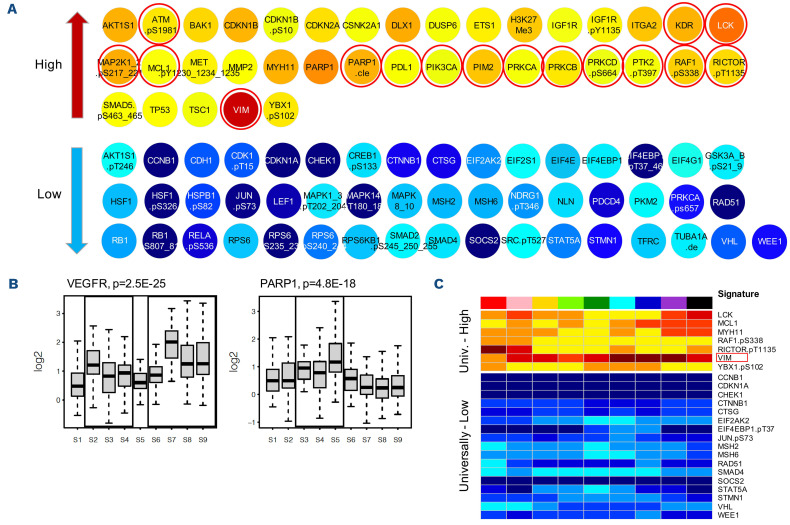
The majority of targeted drugs in development act on protein function. Recognition of proteins with an abnormal expression could identify targets for therapy across AML subgroups. We identified proteins significantly different from normal for each PrSIG/PrCON (Figure 6) (available online at: http://leukemiaproteinatlas.org/pediatric-aml/). As an example, two proteins, VEGFR and PARP1, are shown in particular, as they may also function as potential therapeutic targets for inhibitory drugs. Twenty-four proteins were identified as having universally higher (n=7) or lower (n=17) expression across all PrSIG with vimentin (VIM) most strongly expressed.
Proteins involved in cell cycle regulation and DNA damage change following exposure to chemotherapy
To assess whether cells would adapt their PrSIG differentially following chemotherapy, unsupervised hierarchical clustering of pre-treatment and post-treatment samples was performed. None of the expression patterns was specific to a given time-point or treatment arm. Comparison of individual protein expression levels identified 87 (29%) proteins that had changed by 10 h after treatment and 173 (58%) by 24 h after treatment. Sixty-seven (77%) proteins were changed at both time-points, and were predominantly involved in the TP53 pathway (TP53, MDM4), DNA damage response (ATM, Chek2) and cell cycle regulation (Wee1, CCND3, RB1-pSer) (Online Supplementary Table S6).
Figure 5.
Kaplan-Meier curves for overall survival, event-free survival and relapse risk. Left upper three panels: overall survival curves (log-rank), right panels: relapse risk (Gray statistics). (A) Overall survival and relapse risk stratified by the nine protein expression signatures (PrSIG). (B) Proteomic risk groups defined as “favorable”, “intermediate”, “unfavorable”. (C) Comparison of outcomes in patients treated with ADE or. ADEB in PrSIG-8 (left) and PrSIG-3 (right). (D) Event-free survival and relapse risk for CEBPA-mutated patients in PrSIG-3 treated with ADE (blue) or ADEB (red).
Table 2.
Multivariable analysis for overall survival, event-free survival and relapse risk, including proteomic-based signatures.
When survival analysis was performed for each individual protein (stratified by median, thirds or quartiles), a similar number of proteins was prognostic within each group regardless of whether the pre-treatment or the post-treatment expression was assessed. Approximately 20% of the proteins that were significantly prognostic before treatment, remained prognostic after treatment. Seven proteins were significantly prognostic at all three time-points when considering all patients together; BCL2A1, CCND3, CD74, EIF2S1, GSK3A_B, HSPB1.pSer82, and MKNK1. Nine other proteins, mostly involved in protein translation or signal transduction, were prognostic at both 10 h and 24 h after treatment, but not before treatment. These proteins were: ATF3, EIF2S1.pS51, EIF4EBP1, EIF4G1, HSF1, MET.pY1230_1234_1235, PTEN, RPS6KB1, and YAP1.
Discussion
In this study, to our knowledge the largest proteomic study in pediatric AML, we validated our central hypothesis that the genetic heterogeneity of pediatric AML coalesces into a finite number of recurrent protein expression patterns. Unique to this study is the use of a multistep protein analysis, which moves beyond individual protein expression and activation, to a combined analysis in functionally related protein groups, and then into a system spanning structure based on strongly correlated PrCL. We believe that this is a more complete approach, utilizing known functional interactions, and is superior to conventional unsupervised clustering which weights all proteins equally and ignores known relationships. Furthermore, this study uses proteomics for the first time in samples collected from a phase III randomized clinical trial, and identified patients who responded well to a specific therapy.
Figure 6.
Protein expression significantly different from normal. (A) Protein expression deviated significantly from normal in protein expression signature (PrSIG)-3 (P<0.05, and log2 change ≥0.50 log2). Proteins indicated by red circles are potentially targetable. (B) Relative VEGFR (KDR) (left, potential target for PrSIG-2 to 4, and PrSIG-6 to -9) and PARP1 (right, target for patients in PrSIG-3 to -5) protein expression levels across the nine PrSIG. (C) Twenty-four proteins with universally higher (n=7) or lower (n=17) expression (P<0.05, and log2 change ≥0.50) compared to that in normal CD34+ cells. VIM (indicated by the red box) was most strongly upregulated across the nine PrSIG.
Traditional risk stratification in pediatric AML considers selected cytogenetics and molecular features, and early response to induction chemotherapy, but predicts outcome for only 40% of patients.32 When prognostically similar PrSIG were grouped as “favorable”, “intermediate”, or “unfavorable”, we demonstrated increased prognostic significance when added to traditional risk stratifying factors in multivariate analysis. PrSIG were more strongly predictive when combined with AAML1031 risk groups, demonstrating that adding proteomics to genetic risk-stratification can direct therapy leading to improved outcome. Proteins were also individually significantly prognostic, including several that had previously been published as being so in adult AML.
We identified three PrSIG (PrSIG-3, -6, and -8; 34% of all patients) that benefited from proteasome inhibitor therapy, a finding not appreciated by analysis of the entire cohort. This finding suggests that proteomic analysis is able to predict a-priori those who would benefit from a specific therapy. PrSIG-6 and -8 both contained PrCON-11 and were characterized by upregulation of autophagy proteins (ATG3, ATG7, BECN1).33 We hypothesize that these autophagy effectors are required for bortezomib-induced autophagy, given that the ubiquitin-proteasome system has active crosstalk with autophagy, and bortezomib stimulates this compensatory autophagy mechanism resulting in cell death.34 PrCON-11 was associated with a prevalence of M4/M5 (monocytic) patients and least M0/M1/M2 cases (P<0.001) as well as a high frequency of high white blood cell counts (P<0.001). In PrSIG-7, which shares PrCON-11, a similar but less strong beneficial effect of bortezomib was observed. Unlike PrSIG-6 and -8, PrSIG-7 had higher phosphorylation levels of heat shock binding protein 1 (HSPB1-pSer82), a strongly unfavorable prognostic factor in our dataset, but without a benefit from proteasome inhibitor therapy. This protein could possibly work by preventing the toxic build-up of misfolded proteins due to bortezomib.35 PrSIG-7 had a protein expression profile suggesting high cell cycle turnover (high CCND3, CDKN1B-pThr198, and RB1-phospho), high white cell count, and a high frequency of MLL-rearrangements. This highlights the need to apply a holistic system approach to be able to predict response to drugs.
The importance of studying protein expression and activation is stressed by the low correlation (r=0.17) between protein and mRNA expression and the inability of mRNA data to replicate protein-determined PrSIG. The lack of correlation was expected, as the presence of mRNA does not imply that translation is occurring (non-coding RNA could impede it); nor does it dictate the rate of translation or protein longevity after translation. Moreover, environmental effects from mesenchymal stromal cells, including chemokine and cytokine production, affect how emerging leukemia cells develop and behave. Nonetheless, PrSIG were partially correlated with cytogenetics and mutational state. “Driver” mutations would be expected to have a defining effect on biology/protein expression even though the combination of other events might further alter these signals. For instance, the majority of PrSIG-7 (85%) had MLL-rearrangements and, similar to the recognition of Ph+-like acute lymphoblastic leukemia, this may point toward the existence of MLL-like cases based on protein expression.
This study demonstrates that the “hallmarks of cancer”36,37 are achieved via different patterns of protein utilization within the defined PrCON. As an example of the five apoptosis PFG used in this study, PrCON-6 demonstrated high BH3 pro-apoptotic member activation (BAX, BBC3, and BCL2L11) and simultaneously high expression of the antiapoptotic BH3-member MCL1, a protein associated with resistance to chemotherapy.38 PrCON-11 demonstrated a different pathway with high expression of autophagy proteins, but no associations with the other apoptotic PFG. PrCON-9 had modest upregulation of autophagy proteins and evidence of increased spontaneous apoptosis proteins (high PARP1-cleavage, apoptosis-occurring PrCL-2, and PrCL-3).
Targeted therapies offer the promise of improved outcome, often with less toxicity, compared to current chemotherapy, but an effective means of matching the appropriate patient to the correct agent hampers implementation. To select drugs rationally for specific subgroups of patients, we identified proteins that were expressed significantly differently between cases of AML and normal subjects, raising the hypothesis that those could be targeted by inhibitory/replacement or (re)activation agents, potentially even combined with targeting genetic molecular events (e.g., midostaurin [FLT3] enasidenib [IDH2], or ivosidenib [IDH1]). Online Supplementary Table S7 lists matched proteins and drugs used in the clinical setting. If validated, rapid real-time classification, based on measuring expression of a restricted number of proteins with the highest discriminative value between PrSIG (a “classifier set”), could enable PrSIG determination and facilitate initial therapy selection as well as classification.
We identified 24 universally altered proteins, identifying novel potential targets for all patients. The most highly overexpressed protein was VIM, a protein involved in epithelial-to-mesenchymal transition. The role for VIM in AML is uncertain, but there is evidence that epithelial-to-mesenchymal transition occurs in hematologic malignancies.39,40 Fluvastatin targets VIM via caspase-3-mediated proteolysis,41,42 and prior trials in AML demonstrated that the addition of pravastatin to idarubicin and high-dose cytarabine counteracted the chemoresistance associated with the defensive adaptation of increasing cellular cholesterol.43,44 MCL1, previously found to be upregulated in six of the eight PrSIG in another study,8 was also universally highly expressed, most prominently in PrSIG-7 to -9, which had the highest frequency of infants, a historically chemoresistant population.8,45 MCL1 is also strongly associated with chemoresistance to venetoclax. Although clinical trials have evaluated the benefit of (combinational) treatment with venetoclax in adults and relapsed pediatric AML, no studies evaluating the effect of venetoclax in de novo pediatric AML have yet been published. Our finding predicts for lower efficacy in pediatric de novo AML, and suggests that venetoclax could be combined with MCL1 inhibitors.
A final feature of this RPPA study was the measurement of therapy effects on protein expression over time. We expected to find specific treatment-induced changes in protein expression, and different adaptation of proteins across the PrSIG; however, changes were limited to DNA damage, cell cycle regulation, protein translation and histone modification pathways. This likely reflects the presence of many pre-apoptotic cells trying to repair DNA damage. Single-cell proteomics might better profile post-treatment AML heterogeneity and predict which changes are associated with resistance or relapse by identifying “survivor cells”, which cannot be identified in studies of bulk cells.46 Proteomics may also enable identification and analysis of stem cell proteomics, which differ from bulk cells.47-49 Moreover, unbiased approaches such as mass spectrometry, which allow evaluation of global proteomics, might also be of use.50
In summary, we confirmed the existence of recurrent protein patterns in pediatric AML which enabled separation of AML patients into recurrent PrSIG that were prognostic, particularly when combined with known pediatric AML risk factors. We identified PrSIG that benefited from ADEB, and postulate that recognition of abnormal proteins can aid in risk stratification and therapy selection in pediatric, and perhaps adult AML.
Supplementary Material
Funding Statement
Funding: TMH, RBG, ASG, RA, EAK, and RAA were funded by the NIH COG grants U10 CA98543, U10 CA98413, U10 CA180886, U24 CA196173 and U10 CA180899. TMH was funded by the NCI R01-CA164024, a grant from Hope on Wheels, Hyundaii Foundation, the St. Baldrick’s Foundation, and a grant from Takeda Pharmaceuticals. FWH was funded by the Junior Scientific Masterclaas and the Van der Meer-Boerema Stichting
References
- 1.Estey E, Dohner H. Acute myeloid leukaemia. Lancet. 2006;368(9550):1894-1907. [DOI] [PubMed] [Google Scholar]
- 2.Elgarten CW, Aplenc R. Pediatric acute myeloid leukemia: updates on biology, risk stratification, and therapy. Curr Opin Pediatr. 2020;32(1):57-66. [DOI] [PubMed] [Google Scholar]
- 3.Eryılmaz E, Canpolat C. Novel agents for the treatment of childhood leukemia: an update. Onco Targets Therapy. 2017;10:3299-3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maier T, Güell M, Serrano L. Correlation of mRNA and protein in complex biological samples. FEBS Lett. 2009;583(24):3966-73. [DOI] [PubMed] [Google Scholar]
- 5.Gygi SP, Rochon Y, Franza BR, Aebersold R. Correlation between protein and mRNA abundance in yeast. Mol Cell Biol. 1999;19(3):1720-1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu X, Gu Z, Chen Y, et al. Application of PD-1 blockade in cancer immunotherapy. Comput Struct Biotechnol J. 2019;17:661-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brunnström H, Johansson A, Westbom-Fremer S, et al. PD-L1 immunohistochemistry in clinical diagnostics of lung cancer: inter-pathologist variability is higher than assay variability. Mod Pathol. 2017;30(10):1411-1421. [DOI] [PubMed] [Google Scholar]
- 8.Hoff FW, Hu CW, Qiu Y, et al. Recognition of recurrent protein expression patterns in pediatric acute myeloid leukemia identified new therapeutic targets. Mol Cancer Res. 2018;16(8):1275-1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoff FW, van Dijk AD, Qiu Y, et al. Heat shock factor 1 (HSF1-pSer326) predicts response to bortezomib-containing chemotherapy in pediatric AML: a COG report. Blood. 2021;137(8):1050-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horton TM, Hoff FW, van Dijk AD, et al. The effects of sample handling on proteomics assessed by reverse phase protein arrays (RPPA): functional proteomic profiling in leukemia. J Proteomics. 2021;233:104046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aplenc R, Meshinchi S, Sung L, et al. Bortezomib with standard chemotherapy for children with acute myeloid leukemia does not improve treatment outcomes: a report from the Children's Oncology Group. Haematologica. 2020;105(7):1879-1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kornblau SM, Singh N, Qiu Y, Chen W, Zhang N, Coombes KR. Highly phosphorylated FOXO3A is an adverse prognostic factor in acute myeloid leukemia. Clin Cancer Res. 2010;16(6):1865-1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kornblau SM, Tibes R, Qiu YH, et al. Functional proteomic profiling of AML predicts response and survival. Blood. 2009;113(1):154-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tibes R, Qiu Y, Lu Y, et al. Reverse phase protein array: validation of a novel proteomic technology and utility for analysis of primary leukemia specimens and hematopoietic stem cells. Mol Cancer Ther. 2006;5(10):2512-2521. [DOI] [PubMed] [Google Scholar]
- 15.Hu J, He X, Baggerly KA, Coombes KR, Hennessy BT, Mills GB. Non-parametric quantification of protein lysate arrays. Bioinformatics. 2007;23(15):1986-1994. [DOI] [PubMed] [Google Scholar]
- 16.Neeley ES, Kornblau SM, Coombes KR, Baggerly KA. Variable slope normalization of reverse phase protein arrays. Bioinformatics. 2009;25(11):1384-1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neeley ES, Baggerly KA, Kornblau SM. Surface adjustment of reverse phase protein arrays using positive control spots. Cancer Inform. 2012;11:77-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Akbani R, Ng PK, Werner HM, et al. A pan-cancer proteomic perspective on The Cancer Genome Atlas. Nat Commun. 2014;5:3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith JL, Ries RE, Hylkema T, et al. Comprehensive transcriptome profiling of cryptic CBFA2T3–GLIS2 fusion–positive AML defines novel therapeutic options: a COG and TARGET pediatric AML study. Clin Cancer Res. 2020;26(3):726-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bolouri H, Farrar JE, Triche J, et al. The molecular landscape of pediatric acute myeloid leukemia reveals recurrent structural alterations and age-specific mutational interactions. Nat Med. 2018;24(1):103-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu CW, Qiu Y, Ligeralde A, et al. A quantitative analysis of heterogeneities and hallmarks in acute myelogenous leukaemia. Nat Biomed Eng. 2019;3(11):889-901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoff FW, Hu CW, Qiu Y, et al. Recurrent patterns of protein expression signatures in pediatric acute lymphoblastic leukemia: recognition and therapeutic guidance. Mol Cancer Res. 2018;16(8):1263-1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hartigan JA, Wong MA. Algorithm AS 136: a K-means clustering algorithm. J Royal Stat Soc Series C. 1979;28(1):100-108. [Google Scholar]
- 24.Hu CW, Kornblau SM, Slater JH, Qutub AA. Progeny clustering: a method to identify biological phenotypes. Sci Rep. 2015;5:12894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Govaert G, Nadif M. Clustering with block mixture models. Pattern Recognit. 2003;36(2):463-473. [Google Scholar]
- 26.Kuhn M. Building predictive models in R using the caret package. J Statistic Software. 2008;28(5). [Google Scholar]
- 27.Szklarczyk D, Franceschini A, Wyder S, et al. STRING v10: proteinprotein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao T, Liu H, Roeder K, Lafferty J, Wasserman L. The huge package for high-dimensional undirected graph estimation in R. J Mach Learn Res. 2013;13(1):1059-1062. [PMC free article] [PubMed] [Google Scholar]
- 29.Liu H, Roeder K, Wasserman L. Stability approach to regularization selection (StARS) for high dimensional graphical models. Adv Neural Inf Process Syst. 2010;24(2):1432-1440. [PMC free article] [PubMed] [Google Scholar]
- 30.Lopes CT, Franz M, Kazi F, Donaldson SL, Morris Q, Bader GD. Cytoscape web: an interactive web-based network browser. Bioinformatics. 2010;26(18):2347-2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498-2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cooper TM, Ries RE, Alonzo TA, et al. Revised risk stratification criteria for children with newly diagnosed acute myeloid leukemia: a report from the Children's Oncology Group. Blood. 2017;130 (Suppl 1):407. [Google Scholar]
- 33.Li X, Zhou Y, Li Y, et al. Autophagy: a novel mechanism of chemoresistance in cancers. Biomed Pharmacother. 2019;119:109415. [DOI] [PubMed] [Google Scholar]
- 34.Zhuang L, Ma Y, Wang Q, et al. Atg3 overexpression enhances bortezomib-induced cell death in SKM-1 cell. PloS One. 2016;11(7):e0158761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ciocca DR, Calderwood SK. Heat shock proteins in cancer: diagnostic, prognostic, predictive, and treatment implications. Cell Stress Chaperones. 2005;10(2):86-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100(1):57-70. [DOI] [PubMed] [Google Scholar]
- 37.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646-674. [DOI] [PubMed] [Google Scholar]
- 38.Michels J, Obrist F, Vitale I, et al. MCL-1 dependency of cisplatin-resistant cancer cells. Biochem Pharmacol. 2014;92(1):55-61. [DOI] [PubMed] [Google Scholar]
- 39.Stavropoulou V, Kaspar S, Brault L, et al. MLL-AF9 expression in hematopoietic stem cells drives a highly invasive AML expressing EMT-related genes linked to poor outcome. Cancer Cell. 2016;30(1):43-58. [DOI] [PubMed] [Google Scholar]
- 40.Chen S, Liao T, Yang M. Emerging roles of epithelial-mesenchymal transition in hematological malignancies. J Biomed Sci. 2018;25(1):37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Strouhalova K, Přechová M, Gandalovičová A, Brábek J, Gregor M, Rosel D. Vimentin intermediate filaments as potential target for cancer treatment. Cancers. 2020;12(1):184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kanugula AK, Dhople VM, Völker U, Ummanni R, Kotamraju S. Fluvastatin mediated breast cancer cell death: a proteomic approach to identify differentially regulated proteins in MDA-MB-231 cells. PLoS One. 2014;9(12):e108890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kornblau SM, Banker DE, Stirewalt D, et al. Blockade of adaptive defensive changes in cholesterol uptake and synthesis in AML by the addition of pravastatin to idarubicin + high dose Ara-C: a phase 1 study. Blood. 2007;109(7):2999-3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sassano A, Katsoulidis E, Antico G, et al. Suppressive effects of statins on acute promyelocytic leukemia cells. Cancer Res. 2007;67(9):4524-4532. [DOI] [PubMed] [Google Scholar]
- 45.Bose P, Gandhi V, Konopleva M. Pathways and mechanisms of venetoclax resistance. Leuk Lymphoma. 2017;58(9):2026-2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kornblau SM, Qiu YH, Bekele BN, et al. Studying the right cell in acute myelogenous leukemia: dynamic changes of apoptosis and signal transduction pathway protein expression in chemotherapy resistant ex-vivo selected "survivor cells". Cell Cycle. 2006;5(23):2769-77. [DOI] [PubMed] [Google Scholar]
- 47.Kornblau SM, Qutub A, Yao H, et al. Proteomic profiling identifies distinct protein patterns in acute myelogenous leukemia CD34+CD38- stem-like cells. PLoS One. 2013;8(10):e78453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Majeti R, Becker MW, Tian Q, et al. Dysregulated gene expression networks in human acute myelogenous leukemia stem cells. Proc Natl Acad Sci U S A. 2009;106(9):3396-3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou J, Chng W. Identification and targeting leukemia stem cells: the path to the cure for acute myeloid leukemia. World J Stem Cells. 2014;6(4):473-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mertins P, Tang LC, Krug K, et al. Reproducible workflow for multiplexed deep-scale proteome and phosphoproteome analysis of tumor tissues by liquid chromatography–mass spectrometry. Nat Protoc. 2018;13(7):1632-1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.