Background.
Characterization of anti-HLA versus anti-severe acute respiratory syndrome coronavirus 2 (anti–SARS-CoV-2) immune globulin isotypes in organ transplant recipients after coronavirus disease 2019 (COVID-19) infection has not been reported. We aimed to determine changes in anti-HLA antibodies in renal transplant patients with COVID-19 and compare the immunoglobulin and epitope-binding pattern versus anti–SARS-CoV-2 antibodies.
Methods.
This is a cross-sectional study of 46 kidney transplant recipients including 21 with longitudinal sampling. Using a semi-quantitative multiplex assay, we determined immunoglobulin (Ig) M, IgA, IgG, and IgG1-2-3-4 antibodies against Class I and Class II HLA, and 5 SARS-CoV-2 epitopes including the nucleocapsid protein and multiple regions of the spike protein.
Results.
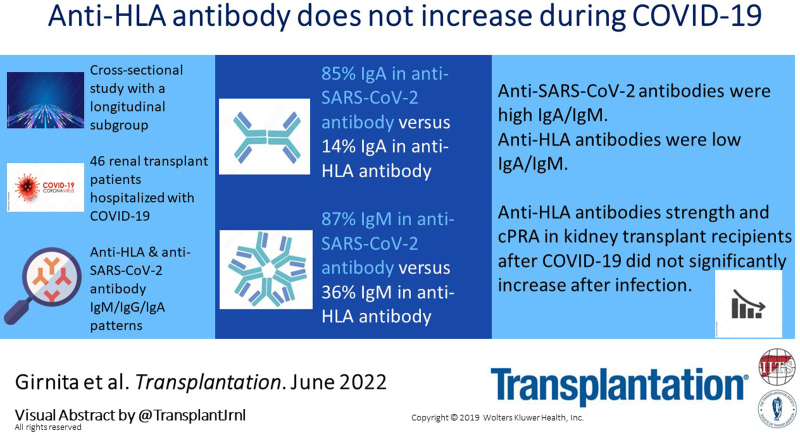
Fourteen of 46 (30%) patients had donor-specific anti-HLA antibodies (donor-specific antibody [DSA]), 12 (26%) had non-DSA anti-HLA antibodies and 45 (98%) had anti–SARS-CoV-2 antibodies. Most DSAs targeted HLA-DQ (71%), with a dominant IgG isotype and IgG1 subtype prevalence (93%), and/or IgG3 (64%), followed by IgG2 (36%). Comparatively, there was a higher prevalence of IgA (85% versus 14%, P = 0.0001) and IgM (87%, versus 36%, P = 0.001) in the anti–SARS-CoV-2 antibody profile, when compared to DSAs, respectively. Anti–SARS-CoV-2 antibody profile was characterized by increased prevalence of IgM and IgA, when compared to DSAs. The median calculated panel reactive antibody before COVID-19 diagnosis (24%) tended to decrease after COVID-19 diagnosis (10%) but it was not statistically significant (P = 0.1).
Conclusions.
Anti-HLA antibody strength and calculated panel reactive antibody in kidney transplant recipients after COVID-19 do not significantly increase after infection. Although the IgG isotype was the dominant form in both HLA and SARS-CoV-2 antigens, the alloimmune response had a low IgA pattern, whereas anti–SARS-CoV-2 antibodies were high IgA/IgM.
INTRODUCTION
The development of donor-specific anti-HLA alloantibody (DSA) has been recognized as a major risk factor for allograft rejection and/or graft loss after renal transplantation.1-3 Immunoglobulin G (IgG) is the dominant isotype involved in early or late antibody-mediated rejection, and the 4 IgG1/G2/G3/G4 subtypes have been associated with various allograft outcomes.4-7 Additionally, viral infection has been also associated with allograft rejection.8-11 The novel coronavirus disease 2019 (COVID-19) produced by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has significantly affected the transplant community, whose mortality is higher than in the general population, potentially as a result of immunosuppression and comorbidities in this population.12-14 Intriguingly, reduction of immunosuppression during COVID-19 has not resulted in a major increase in the risk of allograft rejection. Whether infection and concomitant reduction of immunosuppression increase the risk of DSA development has only been addressed in a very limited cohort of 7 transplant recipients.15
The inter-relations between infection and allo-recognition depend on the possibility that various acute viral infections can trigger allograft rejection through the cross-reactive potential of virus-specific T cells targeting allogeneic HLA molecules.16 Using single-HLA molecule expressing target cells, Amir et al have shown 45% and 80% of influenza, varicella zoster, cytomegalovirus, or Epstein-Barr virus–specific memory T-cell lines exhibit cross-reactivity with HLA Class I and/or Class II molecules.16 This mechanism can also explain generation of allo-reactive T cells in non-sensitized individuals.17 Furthermore, other studies have shown that a single virus can substantially enlarge the allo-HLA memory T-cell repertoire.18 Infection and inflammation can increase the strength and diversity of anti-HLA antibody responses in previously sensitized individuals.
Because both anti-HLA and anti–SARS-CoV-2 antibodies target protein antigens, the aims of the present study were (1) to determine if the allo-antibody response is modified by the COVID-19 infection in a cohort of renal transplanted patients with COVID-19 diagnosis and (2) to characterize and compare the immunoglobulin isotype and subtype profiles and the epitope binding patterns of anti-HLA and anti–SARS-CoV-2 antibodies.
MATERIALS AND METHODS
Study Population
Our study included consecutive consenting adult kidney transplant recipients followed-up from April 2020 to February 2021 during hospitalization or at follow-up clinic visits at Mount Sinai and Montefiore Medical Center in New York with an ongoing or prior SARS-CoV-2 infection diagnosed through real-time polymerase chain reaction (RT-PCR) of nasopharyngeal swab samples as previously described.19 We recorded epidemiological, clinical, and laboratory data in an ad hoc database.
The study received appropriate approval from the ethics and scientific committees of the participating centers (institutional review board titles/numbers: STUDY-20-01922, Mount Sinai Medical Center; IRB-2020-11662 Montefiore Medical Center; IRB-56413 Stanford University). All patients provided informed consent.
Specimens
Blood was collected in sterile tubes, allowed to clot, and then centrifuged to separate the serum. Samples were aliquoted and stored at −20° C until analyses.
Anti-HLA and Anti–SARS-CoV-2 Antibody
All 46 patients did not exhibit pre-formed DSAs, and all had negative T- and B-cell flow crossmatches before transplantation. Twelve patients developed DSAs before the diagnosis of COVID-19, whereas 2 patients developed DSAs after the diagnosis of COVID-19.
For all renal transplant recipients, anti–SARS-CoV-2 [trimeric spike protein (spike), spike S1 (S1), spike receptor binding domain (RBD), spike 2 (S2), and nucleocapsid (NC) epitopes] IgG, IgA, IgM, and IgG1/G2/G3/G4 antibodies were identified as previously described (LABScreen COVID Plus, One Lambda, Canoga Park, CA), and then analyzed on a Luminex FLEXMAP 3D instrument (Luminex Corp, Austin, TX).19,20 The time interval between the diagnosis of COVID-19 and testing for anti–SARS-CoV-2 antibody was 29.64 ± 21.28 d in the DSA group (N = 14), versus 36.25 ± 65.47 in patients without DSA (N = 32, P = 0.72). Anti-HLA (-A, -B, -C, -DRB1, -DRB3/4/5, -DQA1, -DQB1, -DPA1, and -DPB1) IgG antibodies were also tested by multiplex single-antigen bead array (LABScreen Single Antigen; One Lambda, Inc, Canoga Park, CA). Anti-HLA antibodies with strength value greater or equal to 1000 mean fluorescence intensity units were considered for analysis. During the entire study, we used the same controls and cutoff values for the alloantibody detection and identification by single-antigen bead assay. We obtained by chart review the donor and recipient HLA molecular (reverse sequence-specific oligonulceotide) typing data for all mentioned loci. Calculated panel reactive antibody (cPRA) and antibody strength of prior donor-specific antibodies were also obtained from the medical record on a subset of 76 samples in 21 subjects. All 46 kidney recipients have been tested for circulating IgG, IgM, and IgA donor-specific anti-HLA antibodies after SARS-CoV-2 infection diagnosis, and those detected DSA positive were further tested for IgG1/G2/G3/G4 as previously described.6
IgG Isotypes and Subtypes
The anti-HLA and anti–SARS-CoV-2 IgA, IgM, and IgG1/G2/G3/G4 antibody detection was obtained with R-Phycoerythrin AffiniPure Goat Anti-Human Serum IgA, α-chain specific (Cat. # 109-115-011; Jackson ImmunoResearch, West Grove, PA), PE-Conjugated Anti-Human IgM (One Lambda, West Grove, PA: Cat. #: IGM-PEC1), Mouse anti-Human IgG1 (Cat. # MH1013; Invitrogen), Mouse anti-Human IgG2 (Cat. # 05-3500; Invitrogen), Mouse anti-Human IgG3 (Cat. # 05-3600; Invitrogen), and Mouse anti-Human IgG4 (Cat. # A-10651; Invitrogen), respectively.6 We established the cutoff (mean + 3 SD) based on 27 COVID-19 negative sera. These 27 sera were obtained from non-transplanted, non-transfused males, and were determined by Luminex single-antigen bead assay to be negative for anti-HLA antibody. To avoid cross-reactivity, our subtype antibody identification was performed on a Luminex single-antigen bead array, with multiple concomitant positivity criteria (mean fluorescence intensity [MFI], normalized background ratio, and epitope/cross-reactivity pattern), with a <2.5% cross-reactivity between subclasses. All 46 patients were tested for IgA, IgM, and IgG/G1/G2/G3/G4 antibody toward anti–SARS-CoV-2.
Statistical Analysis
For descriptive statistics, categorical variables were reported as percentages and continuous variables as median and interquartile ranges or mean and SD. For univariate analysis, we used 2-tailed chi-square/Fisher’s exact testing ± Yates correction for categorical variables, and one-way analysis of variance for continuous variables. All statistical analyses were performed using STATISTICA statistical software (TIBCO), and an alpha level less 5% was considered statistically significant.
RESULTS
The study cohort included 46 kidney transplant recipients with PCR-confirmed diagnoses of SARS-CoV-2 infection (Table 1). Among all patients, 59% were male, 52% were Hispanic, and median age was 56 (IQR 42-65). Of the 46 patients, 20 did not have any anti-HLA antibodies, 14 had DSAs, and 12 had non-DSA anti-HLA antibodies. Immunosuppression at time of sample draw mainly consisted of calcineurin inhibitors and steroids. Most patients (80%) were taking mycophenolate mofetil (MMF) before diagnosis, but anti-metabolite was held, or dose decreased in 89% of the patients at time of first antibody level draw. Patient characteristics were generally similar across the various antibody groups with notable exceptions of pre-diagnosis peak cPRA level and mean pre-diagnosis MMF dose. Those with anti-HLA antibodies but no DSA had a higher dose of pre-diagnosis MMF than those with either (1) no HLA antibodies or (2) DSA. Those with DSA had similar pre-diagnosis MMF dose compared to those with no anti-HLA antibodies. There were no significant differences between the groups in terms of rejection episodes and/or changes in immunosuppressive medications (Tables 1S and 2S, SDC, http://links.lww.com/TP/C503).
TABLE 1.
Demographics of the study population
| All patients (n = 46) | DSA (n = 14) | HLA antibodies, no DSA (n = 12) | No HLA antibodies (n = 20) | P Value for differences between subgroups | |
|---|---|---|---|---|---|
| Age (IQR) | 56 (42–65) | 59.5 (44–64) | 43 (39–53) | 61.5 (45–67.5) | 0.1a |
| Sex (% male) | 59 | 50 | 75 | 55 | 0.4b |
| Race/ethnicity (%) | 0.8b | ||||
| Non-Hispanic White | 9 | 7 | 0 | 15 | |
| Non-Hispanic Black | 33 | 36 | 42 | 25 | |
| Hispanic | 52 | 50 | 50 | 55 | |
| Other | 7 | 7 | 8 | 5 | |
| Years since transplant (IQR) | 3.5 (1.0–8.5) | 5.0 (1.0–9.3) | 3.7 (0.9–6.1) | 3.0 (1.3–8.5) | 0.9b |
| Percent deceased donor transplant | 68 | 64 | 75 | 67 | 0.8b |
| Percent with history of rejection | 20 | 7 | 17 | 30 | 0.2b |
| Mean peak PRA (SD) | 26 (35) | 47 (31) | 40 (42) | 4 (10) | 0.04c |
| Mean MMF dose pre-COVID-19 in Mg (SD) | 1021 (730) | 892 (738) | 1416 (764) | 875 (646) | 0.1b |
| MMF dose decreased before first COVID-19 antibody sample drawn (%) | 89 | 80 | 91 | 94 | 0.5 b |
| Steroid administration before first COVID-19 antibody sample drawn (%) | 91 | 100 | 92 | 85 | 0.3b |
| Mean A, B, DR mismatches (SD) | 4.4 (1.6) | 5 (0.9) | 4.6 (1.7) | 4 (1.9) | 0.5b |
| Mean DQ mismatches (SD) | 1.1 (0.8) | 1.3 (0.6) | 0.8 (0.9) | 1.3 (0.8) | 0.3b |
aP >0.05 for (1) DSA compared with HLA antibodies, no DSA; (2) DSA compared with No HLA antibodies; and (3) HLA antibodies, no DSA compared with no HLA antibodies.
bP = 0.6 for DSA compared with HLA antibodies, no DSA; P = 0.01 for DSA compared with no HLA antibodies, no DSA; and P = 0.05 for HLA antibodies, no DSA compared with no HLA antibodies.
cP = 0.03 for DSA compared with HLA antibodies, no DSA; P = 0.6 for DSA compared with no HLA antibodies; and P = 0.03 for HLA antibodies, no DSA compared with no HLA antibodies.
COVID-19, coronavirus disease 2019; DSA, donor-specific antibodies; IQR, interquartile range; MMF, mycophenolate mofetil; PRA, panel reactive antibody.
Antibody Specificity
Anti-HLA antibodies were predominantly directed against HLA Class II (20/26, 77%). Fourteen subjects had DSAs. The dominant specificity of DSAs targeted HLA-DQ molecules, 71% in DSA versus 8% in non-DSA, P = 0.001. Anti-HLA antibodies in patients with non-DSA exhibited a more equilibrated distribution toward HLA-Class I and anti-HLA DR specificities: 42% anti-HLA Class I, 50% anti-HLA-DR, and 8% anti-HLA-DQ (Table 2).
TABLE 2.
Antibody specificity
| Antibody specificity | Patients with DSA (n = 14) | Patients with non-DSA (n = 11) | P Value |
|---|---|---|---|
| Anti-HLA Class I | 1 (7%) | 5 (45%) | 0.06 |
| Anti-HLA DR | 3 (21%) | 6 (55%) | 0.13 |
| Anti-HLA DQ | 10 (71%) | 1 (9%) | 0.001 |
| Anti-S | 9 (64%) | 11 (100%) | 0.16 |
| Anti-S1 | 7 (50%) | 7 (64%) | 1 |
| Anti-RBD | 7 (50%) | 11 (100%) | 0.03 |
| Anti-S2 | 6 (43%) | 9 (82%) | 0.13 |
| Anti-NC | 13 (93%) | 6 (55%) | 0.02 |
DSA, donor-specific anti-HLA antibody; S, trimeric spike protein; RBD, receptor binding domain; NC, nucleocapsid.
The most prevalent immunoglobulin isotype was IgG, both in anti-HLA and in anti–SARS-CoV-2 antibodies (Table 3). DSA immunoglobulin isotype/subtype frequently belonged to IgG1 (13/14, 93%) and/or IgG3 (9/14, 64%), followed by IgG2 (5/14, 36%), IgM (5/14, 36%), and IgA (2/14, 14%) heavy chain types. Comparatively, there was a higher prevalence of IgA [39/46 (85%) versus 2/14 (14%), P = 0.0001] and IgM [(40/46 (87%), versus 5/14 (36%) P = 0.001] immunoglobulin heavy chain isotype in the anti–SARS-CoV-2 versus DSAs. A similar higher prevalence of IgA [39/46 (85%) versus 8/32 (25%), P = 0.00001] and IgM [40/46 (87%) versus 7/32 (22%), P = 0.00001] was found when we compared immunoglobulin isotype in anti–SARS-CoV-2 antibodies with anti-HLA antibodies in patients without DSA.
TABLE 3.
Immunoglobulin isotype and subtypes
| Total number of cases (n = 46) | Patients DSA (n = 14) | Patients with anti–SARS-CoV-2 antibody (n = 45) | P Value |
|---|---|---|---|
| IgM | 5 (36%) | 40 (89%) | 0.001 |
| IgA | 2 (14%) | 39 (87%) | 0.0001 |
| IgG | 14 (100%) | 45 (100%) | 1 |
| IgG1 | 13 (93%) | 41 (91%) | 0.85 |
| IgG2 | 5 (36%) | 10 (22%) | 0.2 |
| IgG3 | 9 (64%) | 31 (69%) | 0.83 |
| IgG4 | 1 (7%) | 0 | 0.49 |
DSA, donor-specific anti-HLA antibody; Ig, immunoglobulin; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Anti-HLA Antibody cPRA Did Not Increase During COVID-19
We next assessed whether antibody strength or diversity changed in the setting of natural SARS-CoV-2 infection. cPRA was available for a subgroup of 21 patients at 4 time-points: before transplantation, 1–3 mo before COVID-19–positive RT-PCR, early (average 42 ± 23 d) after COVID-19 diagnosis, and 1-y after COVID-19 diagnosis. Within this group, no patients had pre-formed DSA before transplantation, and all had negative pretransplant T- and B-cell crossmatches. Of the 21 patients with longitudinal follow-up, 6 developed de novo DSAs before COVID-19 diagnosis (4 anti-HLA DQ, 1 anti-HLA-DR51, and 1 toward HLA-DR53), 2 patients developed DSA after COVID-19 diagnosis (actually after the allograft was removed and the immunosuppression was stopped, see below), 7 patients had non-DSA anti-HLA antibody, and 8 patients had no detectable anti-HLA antibodies at all time-points.
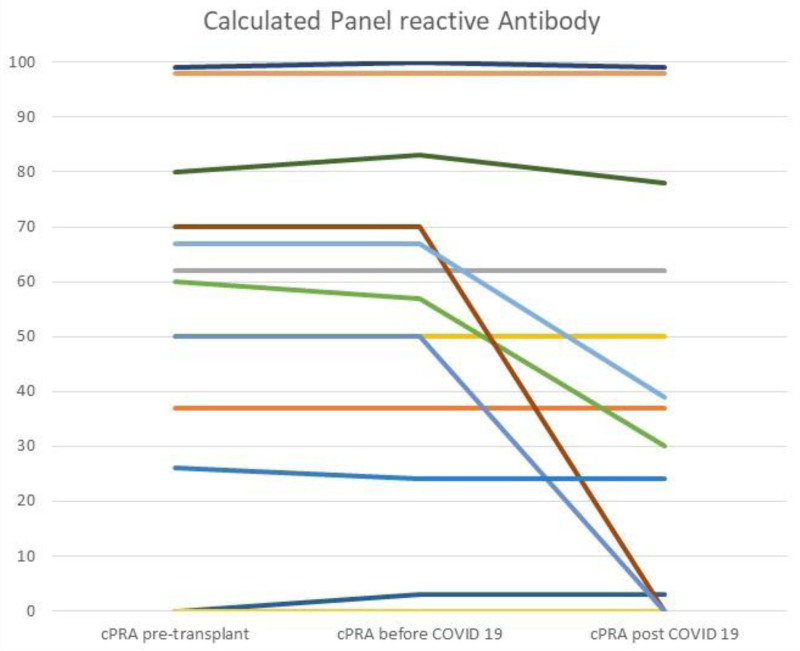
Changes in cPRA were used to assess COVID-19–related changes in overall sensitization. The median cPRA before COVID-19 diagnosis (24%) tended to decrease after COVID-19 diagnosis (10%, P = 0.1). As shown in Figure 1, out of 21 patients with longitudinal data available, cPRA did not change after COVID-19 in 14 cases (67%). Unexpectedly, in 5 patients (24%) we found a decrease in cPRA, from 65.4 ± 12.6% before COVID-19, to 29.4 ± 33.6% after COVID-19 (P < 0.05, Figure 1). Furthermore. we found a non-statistical trend of higher peak clinical COVID-19 severity score in patients with decreased cPRA (3.8 ± 1.3) when compared to patients with no decrease in cPRA (3.1 ± 1.1, P = 0.2, Figure S2, SDC, http://links.lww.com/TP/C503).19 In contrast, the follow-up cPRA increased in 2 cases (10%), both after allograft nephrectomy and immunosuppression discontinuation. No patient developed de novo DSA after COVID-19 diagnosis, except for those 2 cases with graft failure during the post-COVID 19 follow-up. Both patients lost their allograft after COVID 19 diagnosis and underwent transplant nephrectomy. Patients had their immunosuppression discontinued, and DSAs (anti-HLA-DQ4) developed thereafter. The DSA strength for the first case changed from negative (100 MFI) before COVID-19 to 11 587 MFI after nephrectomy, whereas the DSA strength for the second case jumped from negative (128 MFI) before COVID-19 to 18 564 MFI after nephrectomy. The corresponding cPRA changed from 0% pre-COVID-19 to 19% post-nephrectomy for the first case, and for the latter case cPRA increased from 0% pre-COVID-19 to 99% after nephrectomy.
FIGURE 1.
Calculated panel reactive antibody (%) before and after COVID-19 diagnosis. COVID-19, coronavirus disease 2019; cPRA, calculated panel reactive antibody; RT-PCR, real-time polymerase chain reaction.
Donor-specific Antibody Strength Did Not Increase During COVID-19
Six DSA subjects with longitudinal MFI data had detectable antibodies at one or more time points assessed. The 2 subjects who developed de novo DSA after COVID-19 diagnosis had DSA detected only after transplant nephrectomy and immunosuppression discontinuation. Out of the 4 remaining cases, 3 patients had a decrease in DSA strength after COVID-19 diagnosis (Figure S3, SDC, http://links.lww.com/TP/C503): anti-HLA-DQ6 from 4029 MFI to 105 MFI, anti-HLA-DR51 from 3214 MFI to 2102 MFI, and anti-HLA DR53 from 2347 MFI to 58 MFI, respectively. One patient had a slight increase in DSA strength, anti-HLA DQ7 from 2000 MFI before COVID-19 to 5094 MFI after COVID-19. Notably, immunosuppression levels decreased in all 4 DSA cases‚ and, at the same time, those patients developed anti–SARS-CoV-2 antibodies. Unlike HLA-specific antibody dynamics, the anti–SARS-CoV-2 antibody strength significantly increased in late (>14 d) versus early (<14 d) post-COVID-19 diagnosis periods for the entire cohort.19
Anti–SARS-CoV-2 Antibodies
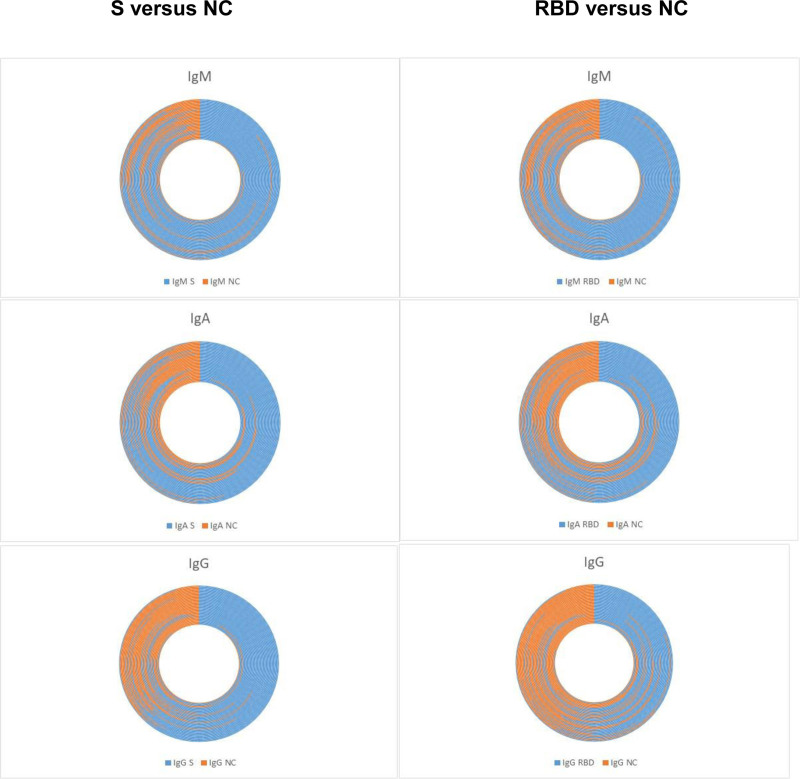
For the entire group, anti–SARS-CoV-2 antibody strength averaged 28 326 ± 16 031 (range 250–62 866) MFI for anti-S protein (full trimeric); 14 487 ± 12 361 (range 177–47 451) MFI for anti-S1; 17 194 ± 12 679 (range 141–40 496) MFI for anti-RBD; 17 027 ± 14 612 (range 76–75 328) MFI for anti-S2; 14 467 ± 15 549 (range 212–91 971) MFI for anti-NC proteins. Anti–SARS-CoV-2 antibodies exhibited a stronger MFI reactivity toward full trimeric spike protein (S) and RBD when compared with NC proteins (Figure 2).
FIGURE 2.
Anti–SARS-CoV-2 IgM, IgA, or IgG antibody specificity for the trimeric spike protein (S) and/or receptor binding domain (RBD) compared with the nucleocapsid (NC) protein. Sectors are represented based on mean fluorescence intensity values. Ig, immunoglobulin.
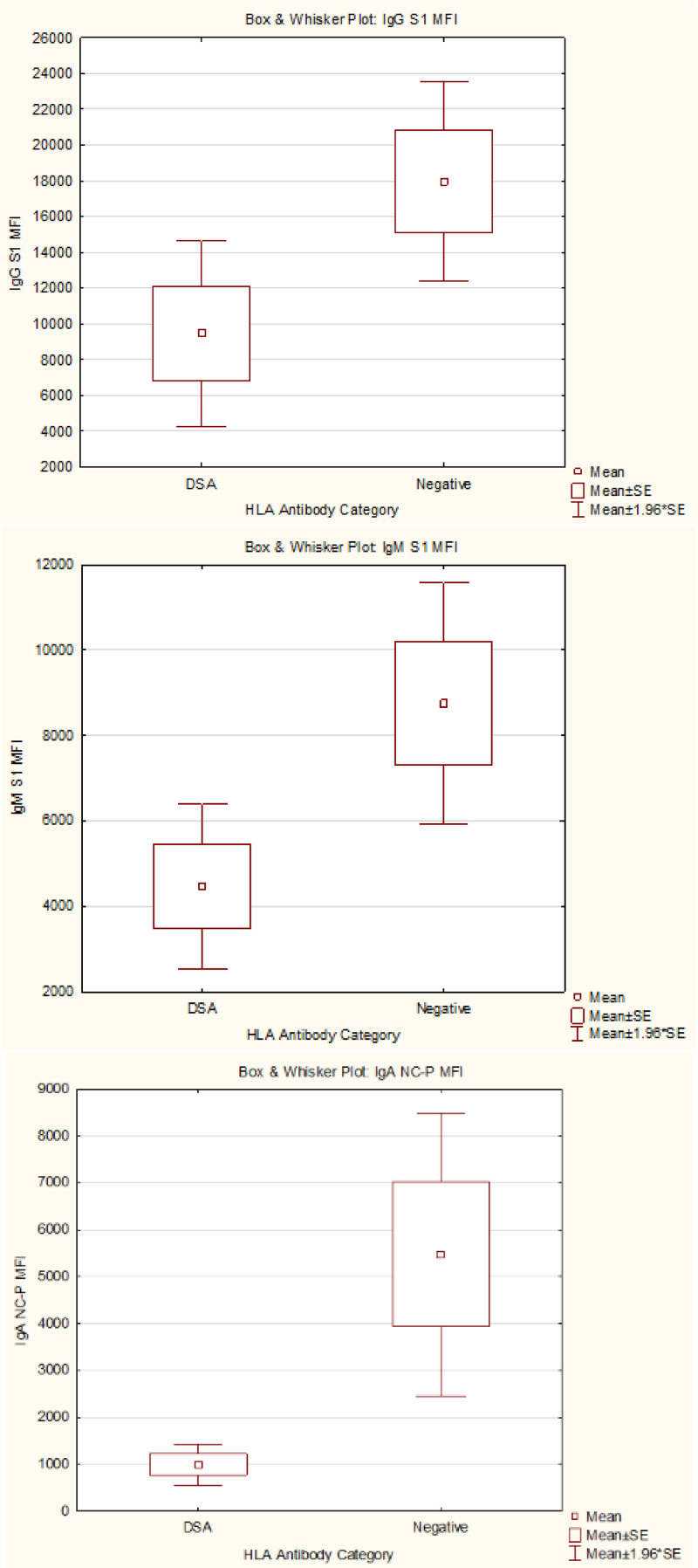
The 14 patients with DSA had significantly lower anti–SARS-CoV-2 antibody strength when compared to 32 patients without DSA (Figure 3). The anti-spike S1 IgG was 9453 ± 9945 MFI in the DSA group versus 17 975 ± 12 792 MFI in the no DSA group, P = 0.04; anti–SARS-CoV-2 spike S1 IgM strength was 4464 ± 3693 MFI in the DSA group versus 8751 ± 6468 MFI in non-DSA, P = 0.03, and anti–SARS-CoV-2 NC IgA was 998 ± 835 MFI in DSA versus 5476 ± 6895 MFI in non-DSA, P = 0.02. Furthermore, anti–SARS-CoV-2 antibody strength in patients with DSA was lower than in the non-DSA group when comparing other binding specificities (RBD, S2, full-spike protein) as well (Figure S1, SDC, http://links.lww.com/TP/C503). Because anti–SARS-CoV-2 antibody strength was higher in late (>14 d) versus early samples, we sought if patients with DSA had earlier sample draw after positive PCR, but the difference was not significant (29.64 ± 21.28 d in the DSA group versus 36.25 ± 65.47 d in the non-DSA group, P = 0.72). Additionally, the immunosuppression levels were not different between the groups (Table 1).
FIGURE 3.
Anti–SARS-CoV-2 antibody strength (MFI) in patients with DSA vs patients without anti-HLA antibody. DSA, anti-HLA donor-specific antibody; Ig, immunoglobulin; MFI, mean fluorescence intensity; NCP, nucleocapsid protein; S, trimeric spike protein; SARSCoV-2, severe acute respiratory syndrome coronavirus 2.
DISCUSSION
We found no increase in HLA-specific humoral sensitization in renal transplant recipients with COVID-19, expressed either as cPRA, or as antibody strength by MFI. In fact, we noticed a decrease in cPRA and/or DSA MFI in a subgroup of patients. To our knowledge, this is the first description of decreased levels of alloantibody after an infection. This antibody strength pattern of HLA-specific antibodies is different from the antibody strength pattern of anti–SARS-CoV-2 antibody, which shows a significant, albeit delayed increment in late versus early post-COVID diagnosis samples.19 The fact that alloantibodies did not increase is remarkable, because over 80% of the patients underwent either significant reduction or withdrawal of MMF after COVID-19 diagnosis. Furthermore, we did not observe a significant IgG3 antibody strength representation in the DSA group. IgG3 subclass is independently associated with significant future graft dysfunction even in patients with similar histology and/or graft function at the time of assessment.6 The trend for higher clinical severity scores in patients with a decrease in cPRA might reflect COVID-19–associated suppression of adaptive T-cell response.21
The lack of an increase in DSA response to COVID-19 is consistent with several other studies. In a recent single-center report on renal transplant patients with COVID-19, DSAs did not change in 94% of cases, despite immunosuppression modulation (holding off the anti-metabolite) was encountered in 68.6% of cases. Furthermore, it has been shown that the COVID-19 and/or SARS-CoV-2 mRNA vaccines do not induce de novo anti-HLA antibody and/or increase existing anti-HLA antibody and do not change the virtual crossmatch eligibility in previously sensitized or non-sensitized solid-organ transplant candidates.22-26
In our study, DSA predominantly targeted HLA Class II, mainly HLA-DQ molecules. Although the number of polymorphic amino acid residues is not higher on HLA-DQ molecules when compared to HLA-Class I or HLA-DR, this DSA specificity is consistent with previous publications, showing a higher prevalence of donor-specific anti-HLA class II, especially anti-HLA DQ alloantibody in solid-organ transplantation.27,28 The complexity of epitopes on HLA-DQ epitopes is greater than HLA-Class I epitopes, due to polymorphism on both alpha and beta chains, and cis- or trans-heterodimer configurations.28-31 Compared to anti-HLA antibodies, the COVID-19–induced anti–SARS-CoV-2 antibodies have a larger epitope variability. The higher prevalence of anti-spike and anti-RBD epitopes might be associated with the neutralization potential of anti–SARS-CoV-2 antibodies and it has been reported in solid-organ transplant recipients.32
Our results indicate that anti–SARS-CoV-2 antibodies have a higher IgA isotype prevalence than anti-HLA DSAs. The dominant epitopes in DSAs belong to HLA-DQ, whereas the strongest reactivity in anti–SARS-CoV-2 antibody is detected against the spike-S and -RBD SARS-CoV-2 antigens.
To our knowledge, this is the first study to parallel the anti-HLA and anti–SARS-CoV-2 antibody immunoglobulin isotype and subtype profiles in renal transplant patients with a COVID-19 diagnosis.
Limitations
This study has limited statistical power due to a relatively small size and the absence of pre- and post-COVID 19 serial samples for most cases. However, the trends observed in the subgroup with longitudinal serum collection parallel the findings obtained in the cross-sectional data. Furthermore, both anti–SARS-CoV-2 and anti-HLA IgM/A/G1/G2/G3/G4 antibody tests were performed in single large batches to avoid inter-run variability. Finally, our results may not be relevant to non-renal allografts under different immunosuppression protocols.
CONCLUSIONS
There was no consistent increase in either cPRA or in HLA-antibody strength (MFI) after PCR-confirmed COVID-19 diagnosis in renal transplant recipients; in a subset of subjects, we found a decline in cPRA and DSA strength at the time of symptomatic COVID-19 infection. The lack of increase in alloantibody response is quite remarkable because over 80% of the patients underwent either significant reduction or withdrawal of MMF after COVID-19 diagnosis.
Anti-HLA donor-specific antibodies exhibited a predominant IgG1 specificity toward HLA-DQ and a low IgM/IgA isotype profile, whereas anti–SARS-CoV-2 antibody displayed a predominant spike protein/RBD IgG specificity and a higher IgA/IgM prevalence. Patients with DSA had reduced anti–SARS-CoV-2 antibody strength when compared to patients without DSA.
ACKNOWLEDGMENTS
The authors acknowledge the contributions of the Histocompatibility, Immunogenetics and Disease Profiling Laboratory, Stanford University, and the Histocompatibility Laboratory of Montefiore Medical Center.
Supplementary Material
Footnotes
P.C., J.S.M., and E.A. are co-senior authors.
J.S.M. has received honoraria from One Lambda, Inc./ThermoFisher, serves on the Qihan Biotech SAB, and has a family member who is employed by and has an equity interest in Genentech/Roche. E.A. has received research grant support from the National Institutes of Health (NIH), CareDx, Immucor and Advisory Board at CareDx, Immucor, Exosome, and Takeda. M.C.M. and P.C. have received research grant support from NIH National Institute of Allergy and Infectious Diseases (NIAID), ancillary mechanistic grant associated with Grant 3U01AI063594-17S1. P.A. has received research grant support from NIH NIAID, R25 AI147369. The other authors declare no conflicts of interest.
This work was funded in part by a grant from One Lambda/Thermo Fisher (to J.S.M.). The funders had no input into the study design or decision to publish these findings.
A.L.G. contributed new reagents or analytic tools, participated in research design, the performance of the research, data analysis, and the writing of the article. L.W., A.I.C., H.M.G., Y.A., M.F.-V., M.C.M., and E.S.W. participated in the performance of the research and in article review. P.A. participated in data analysis and in article review. P.C. participated in research design, in data analysis and in the writing of the article. J.S.M. participated in research design, in data analysis and in the writing of the article. E.A. participated in research design, in data analysis and in the writing of the article.
Supplemental Visual Abstract; http://links.lww.com/TP/C580.
Supplemental digital content (SDC) is available for this article. Direct URL citations appear in the printed text, and links to the digital files are provided in the HTML text of this article on the journal’s Web site (www.transplantjournal.com).
REFERENCES
- 1.Terasaki PI, Ozawa M. Predicting kidney graft failure by HLA antibodies: a prospective trial. Am J Transplant. 2004;4:438–443. [DOI] [PubMed] [Google Scholar]
- 2.Lee PC, Terasaki PI, Takemoto SK, et al. All chronic rejection failures of kidney transplants were preceded by the development of HLA antibodies. Transplantation. 2002;74:1192–1194. [DOI] [PubMed] [Google Scholar]
- 3.Zeevi A, Girnita A, Duquesnoy R. HLA antibody analysis: sensitivity, specificity, and clinical significance in solid organ transplantation. Immunol Res. 2006;36:255–264. [DOI] [PubMed] [Google Scholar]
- 4.Arnold ML, Ntokou IS, Doxiadis II, et al. Donor-specific HLA antibodies: evaluating the risk for graft loss in renal transplant recipients with isotype switch from complement fixing IgG1/IgG3 to noncomplement fixing IgG2/IgG4 anti-HLA alloantibodies. Transpl Int. 2014;27:253–261. [DOI] [PubMed] [Google Scholar]
- 5.Böhmig GA, Eskandary F, Doberer K, et al. The therapeutic challenge of late antibody-mediated kidney allograft rejection. Transpl Int. 2019;32:775–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hamdani G, Goebel JW, Brailey P, et al. IGG3 anti-HLA donor-specific antibodies and graft function in pediatric kidney transplant recipients. Pediatr Transplant. 2018;22:e13219. [DOI] [PubMed] [Google Scholar]
- 7.Valenzuela NM, Schaub S. The biology of IgG subclasses and their clinical relevance to transplantation. Transplantation. 2018;102:S7–S13. [DOI] [PubMed] [Google Scholar]
- 8.Batal I, Girnita A, Zeevi A, et al. Clinical significance of the distribution of C4d deposits in different anatomic compartments of the allograft kidney. Mod Pathol. 2008;21:1490–1498. [DOI] [PubMed] [Google Scholar]
- 9.Batal I, Zainah H, Stockhausen S, et al. The significance of renal C4d staining in patients with BK viruria, viremia, and nephropathy. Mod Pathol. 2009;22:1468–1476. [DOI] [PubMed] [Google Scholar]
- 10.Brehm MA, Daniels KA, Priyadharshini B, et al. Allografts stimulate cross-reactive virus-specific memory CD8 T cells with private specificity. Am J Transplant. 2010;10:1738–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gupta G, Shapiro R, Girnita A, et al. Neutrophilic tubulitis as a marker for urinary tract infection in renal allograft biopsies with C4d deposition. Transplantation. 2009;87:1013–1018. [DOI] [PubMed] [Google Scholar]
- 12.Cravedi P, Mothi SS, Azzi Y, et al. COVID-19 and kidney transplantation: Results from the TANGO International Transplant Consortium. Am J Transplant. 2020;20:3140–3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maggiore U, Riella LV, Azzi J, et al. Mortality in solid organ transplant recipients with COVID-19: More than meets the eye. Am J Transplant. 2021;22:1496–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heldman MR, Kates OS, Safa K, et al. ; UW COVID-19 SOT Study Team. Changing trends in mortality among solid organ transplant recipients hospitalized for COVID-19 during the course of the pandemic. Am J Transplant. 2022;22:279–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gandolfini I, Zanelli P, Palmisano A, et al. Anti-HLA and anti-SARS-CoV-2 antibodies in kidney transplant recipients with COVID-19. Transpl Int. 2021;34:596–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amir AL, D’Orsogna LJ, Roelen DL, et al. Allo-HLA reactivity of virus-specific memory T cells is common. Blood. 2010;115:3146–3157. [DOI] [PubMed] [Google Scholar]
- 17.D’Orsogna LJ, Roelen DL, Doxiadis II, et al. Alloreactivity from human viral specific memory T-cells. Transpl Immunol. 2010;23:149–155. [DOI] [PubMed] [Google Scholar]
- 18.van den Heuvel H, van der Meer-Prins EMW, van Miert PPMC, et al. Infection with a virus generates a polyclonal immune response with broad alloreactive potential. Hum Immunol. 2019;80:97–102. [DOI] [PubMed] [Google Scholar]
- 19.Cravedi P, Ahearn P, Wang L, et al. Delayed kinetics of IgG, but not IgA, antispike antibodies in transplant recipients following SARS-CoV-2 infection. J Am Soc Nephrol. 2021;32:3221–3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bray RA, Lee JH, Brescia P, et al. Development and validation of a multiplex, bead-based assay to detect antibodies directed against SARS-CoV-2 proteins. Transplantation. 2021;105:79–89. [DOI] [PubMed] [Google Scholar]
- 21.Sun Z, Zhang Z, Banu K, et al. Blood transcriptomes of SARS-CoV-2 infected kidney transplant recipients demonstrate immune insufficiency. medRxiv. 2022. Preprint. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roll GR, Lunow-Luke T, Braun HJ, et al. COVID-19 does not impact HLA antibody profile in a series of waitlisted renal transplant candidates. Hum Immunol. 2021;82:568–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Russo G, Lai Q, Poli L, et al. SARS-COV-2 vaccination with BNT162B2 in renal transplant patients: Risk factors for impaired response and immunological implications. Clin Transplant. 2022;36:e14495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herrera S, Colmenero J, Pascal M, et al. Cellular and humoral immune response after mRNA-1273 SARS-CoV-2 vaccine in liver and heart transplant recipients. Am J Transplant. 2021;21:3971–3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumar D, Kimball P, Gupta G. COVID-19 vaccine does not alter panel reactive antibody or flow cytometric cross match in kidney transplant candidates. Transpl Immunol. 2021;69:101469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Masset C, Benotmane I, Dantal J, et al. A fourth SARS-CoV-2 mRNA vaccine in strictly seronegative kidney transplant recipients. Kidney Int. 2022;101:825–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walsh RC, Brailey P, Girnita A, et al. Early and late acute antibody-mediated rejection differ immunologically and in response to proteasome inhibition. Transplantation. 2011;91:1218–1226. [DOI] [PubMed] [Google Scholar]
- 28.Duquesnoy RJ, Awadalla Y, Lomago J, et al. Retransplant candidates have donor-specific antibodies that react with structurally defined HLA-DR, DQ, DP epitopes. Transpl Immunol. 2008;18:352–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kramer CSM, Roelen DL, Heidt S, et al. Defining the immunogenicity and antigenicity of HLA epitopes is crucial for optimal epitope matching in clinical renal transplantation. HLA. 2017;90:5–16. [DOI] [PubMed] [Google Scholar]
- 30.Tambur AR, Leventhal JR, Friedewald JJ, et al. The complexity of human leukocyte antigen (HLA)-DQ antibodies and its effect on virtual crossmatching. Transplantation. 2010;90:1117–1124. [DOI] [PubMed] [Google Scholar]
- 31.Habig DF, Gaspari JL, Lokhandwala PM, et al. Donor-specific antibody to trans-encoded donor HLA-DQ heterodimer. Hum Immunol. 2015;76:587–590. [DOI] [PubMed] [Google Scholar]
- 32.Karaba AH, Zhu X, Liang T, et al. A third dose of SARS-CoV-2 vaccine increases neutralizing antibodies against variants of concern in solid organ transplant recipients. medRxiv. 2021;22:1253–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.