Abstract
Erwinia chrysanthemi is a phytopathogenic enterobacterium causing soft rot disease in a wide range of plants. Osmoregulated periplasmic glucans (OPGs) are intrinsic components of the gram-negative bacterial envelope. We cloned the opgGH operon of E. chrysanthemi, encoding proteins involved in the glucose backbone synthesis of OPGs, by complementation of the homologous locus mdoGH of Escherichia coli. OpgG and OpgH show a high level of similarity with MdoG and MdoH, respectively, and mutations in the opgG or opgH gene abolish OPG synthesis. The opg mutants exhibit a pleiotropic phenotype, including overproduction of exopolysaccharides, reduced motility, bile salt hypersensitivity, reduced protease, cellulase, and pectate lyase production, and complete loss of virulence. Coinoculation experiments support the conclusion that OPGs present in the periplasmic space of the bacteria are necessary for growth in the plant host.
Osmoregulated periplasmic glucans (OPGs) are a family of oligosaccharides found in the periplasmic space of gram-negative bacteria. Their two common features are the presence of glucose as the sole constituent sugar and their increased level in media of low osmolarity (5).
Members of the family Enterobacteriaceae and related bacteria synthesize a family of linear and branched OPGs that are variously substituted. The linear backbone is constituted by glucose units joined by β,1-2 linkages, and the branches are made of one glucose unit linked to the main chain by a β,1-6 linkage. In Escherichia coli, the backbone, containing 7 to 13 glucose units, is substituted with phosphoglycerol, phosphoethanolamine, and succinyl residues (19). In Erwinia chrysanthemi, the backbone contains 5 to 12 glucose units substituted with succinyl and acetyl residues (9), and in Pseudomonas syringae, the backbone, consisting of 6 to 13 glucose units, is not substituted (38). In E. coli, the OPG backbone is synthesized by the products of the mdoGH operon located in the vicinity of pyrC, a gene involved in the biosynthesis of uracil (6). In this bacterium, the defect in OPG synthesis does not confer an easily selectable phenotype in laboratory conditions. Thus, the mdoGH locus was cloned using the linked selectable genetic marker pyrC (23).
Many factors are involved in the virulence of pathogenic bacteria, and OPGs appear to be among them. In P. syringae pv. syringae, the causal agent of brown spot disease of the common bean (Phaseolus vulgaris), the hrpM mutant, obtained after transposon mutagenesis, was isolated because it failed to incite disease on the host plant and to cause the hypersensitive reaction on a non-host plant such as tobacco (29). The hrpM mutant does not synthesize OPGs, and the hrpM locus complements the OPG synthesis defect of the mdoH200::Tn10 mutant of E. coli. The amino acid sequences of HrpM and MdoH are 75.5% identical and 87.5% similar (25). More recently, a transposon insertion in a gene similar to hrpM/mdoH was isolated because it severely reduces the virulence in Pseudomonas aeruginosa PA14, an opportunistic pathogen for human, mice, and the worm Caenorhabditis elegans (26).
The pectinolytic bacterium E. chrysanthemi, a member of the Enterobacteriaceae family, causes soft rot disease in a wide range of plant species. The virulence of this bacterium is strongly associated with the synthesis and secretion of a set of enzymes that degrade plant cell wall components, such as pectinases, cellulases, and proteases. The action of these enzymes, particularly the pectinases, leads to the extension of the disease, causes maceration of the plant tissues, and supplies the bacteria with carbon sources derived from the degraded polymers. The combination of pectin lyase, polygalacturonase, pectate lyase, pectin acetyl esterase, and pectin methyl esterase activities is needed for the degradation of pectin, reflecting the structural complexity of this polymer (2, 36). Among these proteins, endo pectate lyases play the most important role in the maceration process.
The variability of the plant cell wall is reflected by the great number of pectinase genes and correlated with the great number of stimuli affecting the differential expression of the pel genes (15). These genes seem to be, at least in part, the reason for the broad host range of this bacterium (3). E. chrysanthemi 3937 synthesizes five major endo pectate lyases, encoded by pelA to pelE (39), and a set of minor ones. Transcription of the pel genes is induced by pectin catabolic products, the stationary growth phase, host plant metabolites (7), and iron depletion (28). Transcription of these genes is repressed by nitrogen starvation, high temperature, and high osmolarity and is under catabolic repression (16). Several proteases and the cellulase EGZ, encoded by the celZ gene, are secreted by E. chrysanthemi 3937, but they seem to play a minor role in pathogenesis. The cellulase and most of the pectinases are secreted in the medium by the specific Out secretion system encoded by the 15 out genes (34). Synthesis and secretion of pectinases are tightly associated. Thus, expression of the out genes is regulated in the same way and by the same factors as the pel genes (11). Because of iron depletion found in plant fluids, two iron-scavenging systems are needed for full virulence (27).
In this paper, we describe the molecular analysis of the opgGH operon involved in OPG backbone synthesis in E. chrysanthemi. opgG and opgH mutants were obtained by reverse genetic manipulation and analyzed for the synthesis and secretion of plant cell wall-degrading enzymes and for virulence.
MATERIALS AND METHODS
Bacterial strains and plasmids.
E. chrysanthemi and E. coli K-12 strains are listed in Table 1. Plasmids used for subcloning experiments were pYZ4 (kanamycin resistance) and pUC18 (ampicillin resistance) (25).
TABLE 1.
Bacterial strains.
| Strain | Relevant genotype | Sourcea or reference |
|---|---|---|
| E. chrysanthemi | ||
| 3937 | Wild type | 20 |
| A350 (= L2) | lacZ2 | 16 |
| A655 | lacZ2 arg-10 met-2 outC::lacZ-amp | 18 |
| A1383 | ura-2 arg-1 pro-546 rpsL | UMG collection |
| A1703 | lacZ2 pelE::lacZ-cml | UMG collection |
| A1706 | lacZ2 pelC::lacZ-cml | UMG collection |
| A2575 | lacZ2 epsG::lacZ-tet | 10 |
| A3144 | 3937, outS::kan | UMG collection |
| RH7006 | 3937, msrA::cml | |
| NFB3500 | 3937, opgG500::uidA-kan | This study |
| NFB3501 | 3937, opgH501::uidA-kan | This study |
| NFB3502 | A1703, opgG500::uidA-kan | This study |
| NFB3503 | A1706, opgG500::uidA-kan | This study |
| NFB3504 | A655, opgG500::uidA-kan | This study |
| NFB3511 | A2575, opgG500::uidA-kan | This study |
| E. coli | ||
| NFB223 | F−pyrC46 argE3 Δ(lac)U169 thi araD139 mdoH200::Tn10 flbB5301 ptsF25 relA1 rpsL150 rbsR deoC1 rpoB | 20 |
| NFB216 | pyrC46 Δ(lac-pro) thi ara mdoH200::Tn10 rpsL (φ80ΔlacZΔM15) | 23 |
| NFB702 | pyrC46 Δ(lac-pro) thi ara mdoG202::neo rpsL (φ80ΔlacZΔM15) | 20 |
UMG, Unité de Microbiologie Génétique.
Media and growth conditions.
Bacteria were grown at 30°C (E. chrysanthemi) or 37°C (E. coli) in Luria-Bertani broth (LB) or in minimal medium M63 supplemented with the required metabolites and a carbon source added at a concentration of 2 g/liter (29). Polygalacturonate was added at a concentration of 4 g/liter. Potato extract was added at a concentrations of 4 or 8% (vol/vol). When low-osmolarity medium was required, LB without NaCl or low-osmolarity medium (LOS) was used (23). Solid media were obtained by adding agar at 15 g/liter.
Potato extract was obtained as follows. Potato tubers were crushed in a domestic mixer, and the resulting suspension was passed through a French pressure cell at 20,000 lb/in2 (1.4 × 107 Pa) and filter sterilized (0.22-μm; Millipore).
Motility tests were made on LB plates containing agar at 0.4 g/liter. The MIC of bile salts (no. 3; Difco Laboratories) was determined on LB plates. The solid media used to test the pectinase and cellulase activities have been described previously (16, 28), and protease activity was detected by adding 1% milk to LB solid medium.
Antibiotics were used in media at the following concentrations: streptomycin, 100 μg/ml; ampicillin and kanamycin, 50 or 10 μg/ml; and tetracycline, 25 and 5 μg/ml for E. coli and E. chrysanthemi, respectively. X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) was used in media at a concentration of 20 μg/ml.
Transduction and transformation.
Transduction of E. chrysanthemi with phage φEC2 was carried out according to Resibois et al. (32).
Transformation of E. coli and E. chrysanthemi cells was carried out by the rubidium chloride technique (29) and by electroporation (35), respectively.
Transfer of RP4 derivative plasmids by mating.
Plasmid pULB110, a kanamycin-sensitive RP4::mini-Mu derivative (41), was used to generate R-prime derivatives containing bacterial DNA. Matings between recipient and donor strains carrying plasmids were performed by spreading 0.2 ml of overnight cultures of the strains on 63 minimal medium plates and incubating for 5 h at 30°C. Bacteria were resuspended in 1 ml of 63 minimal medium and spread on the appropriate selective media.
Recombinant DNA techniques.
Standard procedures were performed for genomic and plasmid DNA extraction (35). Restriction enzymes (Eurogentec) and T4 DNA ligase (Gibco-BRL) were used according to the manufacturer's recommendations. DNA sequences were determined with the Sequenase version 2.0 kit (U.S. Biochemical Corporation). For DNA hybridization, 10 μg of DNA was digested with the appropriate restriction enzyme(s) and treated as previously described (35). DNA fragments were transferred and fixed onto a Biotrans nylon membrane as recommended by the manufacturer (ICN). The digoxigenin DNA-labeling and detection kit was used for labeling of probes, hybridization, and detection of hybrids according to the manufacturer's recommendations (Boehringer Mannheim).
The DNA sequences and deduced amino acid sequences were analyzed by using computer programs made available from Infobiogen (http://www.infobiogen.fr/).
Construction of opgG::uidA-kan and opgH::uidA-kan insertions and marker exchange recombination.
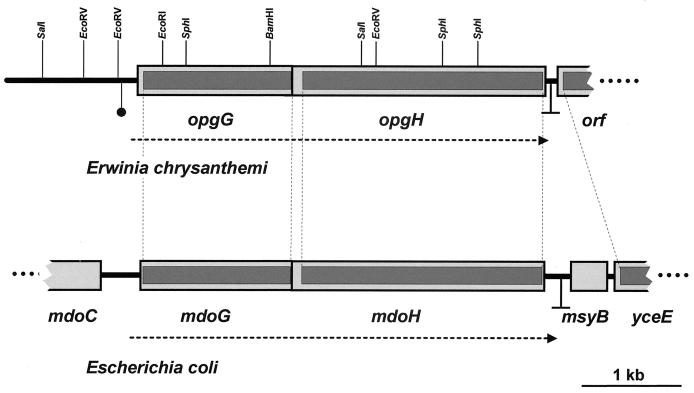
The uidA-kan cassette was liberated by SphI digestion of plasmid pUIDK11 (1). In order to inactivate the opgG or opgH gene, this SphI DNA fragment was then introduced into pNFW32, containing the opgGH operon, partially digested by SphI, either in the SphI site located in opgG or in the first of the two SphI sites located in opgH (Fig. 1). The resulting plasmids were introduced into E. chrysanthemi cells by electroporation. The insertions were then integrated into the E. chrysanthemi chromosome by marker exchange recombination after successive cultures in low-phosphate medium in the presence of kanamycin (33).
FIG. 1.
Genetic structure of homologous regions of the chromosomes of E. chrysanthemi and E. coli in the vicinity of the opgGH and mdoGH operons. Boxes represent open reading frames. Dark boxes represent the regions where the nucleotide sequences are 75% identical (on average) between both organisms. A restriction map is given at the top of the opgGH region of E. chrysanthemi. Transcriptional terminators of the opgGH and mdoGH operons are indicated by an inverted T. The horizontal dotted arrows indicate the location and direction of transcription of the genes. The solid circle upstream from opgG indicates the location of 13-bp direct repeats (see text).
OPG analysis.
To measure OPG synthesis in E. chrysanthemi, cells were grown overnight in LB without NaCl (50 ml). Bacteria were collected by centrifugation at 4°C for 15 min at 8,000 × g. Cell pellets were resuspended in 1 ml of distilled water and extracted with 5% trichloroacetic acid (TCA). The TCA extracts were neutralized with ammonium hydroxide 10%, desalted on a Sephadex G15 column (Pharmacia), and concentrated by rotary evaporation. The resulting material (2 ml) was then fractionated by gel filtration on a Bio-Gel P-4 (Bio-Rad). The column (55 by 1.6 cm) was equilibrated with 0.5% acetic acid. The column was eluted in the same buffer at a flow rate of 15 ml/h, and fractions of 2.5 ml were collected. Sugar content in each fraction was determined colorimetrically by the anthrone reagent procedure (37).
To measure OPG synthesis in E. coli, cultures (5 ml) were made in LOS containing 0.45 mM [2-3H]-glycerol (296 MBq/mmol; New England Nuclear), and labeled OPGs were extracted by the charcoal adsorption procedure; 0.2 ml from the 2 ml of pyridine extract obtained by this procedure was used for counting (23). When necessary, 1.5 ml of pyridine extract was chromatographed on a Sephadex G25 medium column (Pharmacia). The column (2 by 45 cm) was equilibrated with 0.15 M ammonium acetate in 7% (vol/vol) aqueous propanol. The column was eluted with the same buffer at a rate of 17 ml/h. Fractions (1.5 ml) were collected, and radioactivity was counted.
Determination of enzyme activities.
Enzymatic assays were performed on toluene-treated cells. Pectate lyase activity was determined by monitoring spectrophotometrically the formation of unsaturated products from polygalacturonate at 230 nm. The assay mixture consisted of 0.1 M Tris-HCl (pH 8.5), 0.1 mM CaCl2, and polygalacturonate at 0.5 mg/ml. One unit of activity was defined as the amount of enzyme required to produce 1 μM unsaturated product per min (30). β-Galactosidase activity was determined in Z buffer (29).
Pathogenicity test.
Potato tubers were inoculated as previously described (24). Sterile pipette tips containing a bacterial suspension of 107 cells in 5 μl were inserted into the tuber parenchyma. After 24 to 72 h of incubation in a dew chamber, tubers were sliced vertically through the inoculation point, and the weight of decayed tissue was taken as the characteristic of disease severity. Moreover, in coinoculation tests containing both Kanr (Opg−) and Kans (Opg+) bacteria, all the macerated tissues (ranging from 0.3 to 2.0 g) were collected, weighed, and diluted to a concentration of 1 g of macerated tissue in 2 ml of sterile physiological distilled water. The numbers of bacteria originating from each of the two strains were determined by spreading appropriate dilutions of the cell suspensions onto LB plates with or without kanamycin. Chicory leaves were slightly wounded prior to inoculation (106 bacteria). After 24 h of incubation in a dew chamber, the length of rotted tissue was measured to estimate disease severity. These tests were repeated in at least three independent experiments.
Nucleotide sequence accession number.
Sequence data will appear in the EMBL/GenBank/DDBJ nucleotide sequence data libraries under accession number AJ294718.
RESULTS
Complementation of the mdoH200::Tn10 mutation of E. coli with an RP4 derivative of E. chrysanthemi.
Since plasmids harboring the P. syringae hrpM gene complemented the E. coli mdoH200::Tn10 mutation, we postulated that the genes involved in OPG backbone synthesis in E. chrysanthemi may also complement such mutations. The absence of a selectable phenotype in mdoGH mutants of E. coli prevented direct selection for Opg+ complementation. We decided to use the selectable marker pyrC, located 13 kb downstream from the mdoGH locus in E. coli. Despite some important genomic rearrangements, the genetic organization of several genomic regions is quite similar in E. coli and E. chrysanthemi (17).
Plasmid pULB110, a kanamycin-sensitive RP4::mini-Mu derivative (41), was used to generate R-prime derivatives containing an insert of bacterial DNA from E. chrysanthemi complementing the pyrC mutation of E. coli strain NFB223 (pyrC mdoH200::Tn10). Ura+ (Pyr+) transconjugants were isolated on M63 medium plates supplemented with streptomycin. The plasmid contents of 12 Pyr+ transconjugants were analyzed. Four plasmids with a DNA insert of 15 to 44 kb were tested for the Opg phenotype that they conferred. One of them, pR24, complemented the mdoH mutation and conferred an Opg+ phenotype. Moreover, the amount of OPGs produced by the mdoH200::Tn10 strain harboring pR24 was 1.5 times higher than in the wild-type E. coli strain.
Cloning of opgGH locus of E. chrysanthemi.
Plasmid pR24 was digested with EcoRI, and the resulting DNA fragments were ligated into the EcoRI site of pYZ4. Plasmid pNFW3, containing a 9.5-kb DNA insert, was Opg+ (Fig. 1) and thus complemented the mdoH200::Tn10 mutation of NFB216, but only at the wild-type level. The sequences of both ends of the insert were established. The region adjacent to the lac promoter present in the vector was highly similar to the beginning of the open reading frame of the E. coli mdoG gene (starting at amino acid number 72). Then, a truncated promoterless version of the E. chrysanthemi locus has been cloned, and complementation of the mdoH200::Tn10 mutation was most probably the result of transcription from the lac promoter. A similar observation was made during the cloning of the mdoGH operon (23).
In order to restore a complete version of the operon, a 3.3-kb SalI DNA fragment of pR24 overlapping the pNFW3 insert was detected by Southern blot using the 1.1-kb EcoRI-BamHI fragment of the pNFW3 insert as a probe (Fig. 1) and cloned into pUC18. The resulting plasmid, pNFW27, introduced into NFB702 complemented the mdoG202::neo mutation. The reconstitution of the whole operon was done as follows. pNFW3 was digested with BamHI and partially digested with EcoRV to obtain a 4.1-kb BamHI-EcoRV DNA fragment (Fig. 1). This DNA fragment was ligated into pNFW27 digested with BamHI and SmaI (using a site originating from the vector polylinker) to give pNFW32 containing a 6.4-kb DNA insert. Complementation of mdoG202::neo and mdoH200::Tn10 was observed when pNFW32 was introduced into strains NFB702 and NFB216, respectively. Moreover, strains harboring pNFW32 contained an OPG level twice that of the wild type. The same increasing effect was observed when E. coli mdoGH was cloned into a multicopy plasmid (23). These results clearly indicated that the pNFW32 insert contains the entire E. chrysanthemi locus necessary for complementation of the mdoGH operon of E. coli.
Analysis of nucleotide sequence of opgGH genes of E. chrysanthemi.
The 5.0 kb starting from the first EcoRV (Fig. 1) and encompassing the opgGH locus were sequenced in both strands. The analysis of this sequence confirmed the genetic analysis, and two open reading frames were detected. The first open reading frame, opgG, encoding a 522-amino-acid polypeptide, begins with a putative start codon (GTG) and ends at a TAA codon. The GTG start codon represents 11.5% (versus 86.5% for ATG) of the start codons found in the 88 sequences of E. chrysanthemi available in the EMBL sequence database (release 62, March 2000). The second open reading frame, opgH, encoding an 850-amino-acid polypeptide, begins with a putative start codon (ATG) and ends at a TGA codon. Moreover, the end of the opgG sequence overlaps the start of opgH (ATGAATAA). An identical sequence was observed in E. coli, leading to the same overlapping between mdoG and mdoH (25). The putative ribosome-binding site is located 8 nucleotides upstream from the GTG start codon of opgG. The −10 and −35 regions of the putative promoter are separated by 18 nucleotides, and the GTG start codon is located 32 nucleotides downstream from the putative transcriptional start site. In E. coli, the same promoter sequence was found except for one nucleotide in the −10 region. Thus, the two genes most probably form an operon functionally homologous to the mdoGH operon.
Comparison of the nucleotide sequences of opgGH (E. chrysanthemi) and mdoGH (E. coli) shows strong sequence alignments beginning just before the promoter and ending just after the stop codon of the second gene. Two 13-bp directly repeated sequences (TTAAGCGCGTCAATTAAGCGCGTCAA) are found upstream from the putative promoter of opgGH, and the nucleotide sequence found upstream from these direct repeats does not show similarity to any sequence present in the nucleotide sequence data libraries. Nineteen nucleotides downstream from the stop codon of opgH are found two inverted repeats of 11 nucleotides (AAGCAACGGCCAGTCCGTTGCTT), followed by a stretch of 5 T's characteristic of an intrinsic transcription terminator. However, the terminator sequences are divergent (Fig. 1). The opgH gene is followed by an open reading frame transcribed in the opposite orientation which shows similarity to the E. coli yceE gene of unknown function.
Alignment between the MdoG and OpgG proteins shows 75.5% identity (386 of 511) and 87.5% similarity (447 of 511) over the whole sequence, and alignment between MdoH and OpgH shows 74.7% identity (625 of 829) and 85.8% similarity (711 of 829). Similar results were previously observed between MdoH and HrpM of P. syringae (25). Comparison between OpgH and HrpM shows 68.1% (524 of 769) identity and 78.5% (623 of 769) similarity.
Construction of opgG and opgH mutants and localization of opgGH locus on chromosome of E. chrysanthemi.
Both gene disruptions were obtained by insertion of a uidA-kan cassette into pNFW32 followed by transfer into the chromosome (see Materials and Methods) to give NFB3500 (opgG500::uidA-kan) and NFB3501 (opgH501::uidA-kan). Recombination of these constructions with the homologous locus into the chromosome was confirmed by Southern blot hybridization using the 3.3-kb SalI DNA fragment of pNFW27 (data not shown).
OPG synthesis was measured in both mutant strains. OPGs were extracted from overnight cultures grown in LB without NaCl, and the extracts were loaded onto a Bio-gel P-4 column (see Materials and Methods). The peak containing OPGs, normally observed in extracts of the wild-type strain 3937 (9), was absent in both extracts, indicating that these mutants are affected in OPG backbone synthesis (data not shown). Chromosomal mobilization mediated by plasmid pULB110 was used for conjugation with various polyauxotrophic recipients to localize the opgGH locus on the E. chrysanthemi chromosome. Using the opgG500::uidA-kan fusion, the Kanr marker cotransferred at 4% with ade-377, 10% with ogl, and 96% with ura-2. Thus, the opgGH locus is located very near the ura-2 marker on the E. chrysanthemi chromosome (17). In addition, the auxotrophy of the ura-2 mutant is complemented by the four pyrC+ R-prime plasmids isolated above, strongly suggesting that ura-2 is a mutation affecting the pyrC gene of E. chrysanthemi. Moreover, in a generalized transduction by phage φEC2, the Kanr resistance of the opgG500::uidA mutant was cotransduced at 80% with ura-2.
Pleiotropic phenotypes of opg mutants of E. chrysanthemi.
Colonies of the opg mutants have a mucoid aspect on minimal medium agar plates, a typical sign of increased exopolysaccharide (EPS) biosynthesis. We analyzed the effect of the opgG mutation on the transcription of the EPS biosynthetic operon of E. chrysanthemi using the epsG::lacZ fusion present in the eps-29 mutant (A2575). The β-galactosidase activity was increased about threefold in the opgG500::uidA-kan derivative (data not shown) grown with either glycerol or glucose as the sole carbon source. Induction of transcription of the EPS genes was previously observed in opg mutants of E. coli (12) and Agrobacterium tumefaciens (8).
On semisolid agar plates, swarm diameters of opgG and opgH mutants and of wild-type strains were 0.50 ± 0.25 cm and 2.25 ± 0.25 cm, respectively (data not shown). Thus, even if we cannot exclude a defect in flagellum functioning, we consider that flagellum apparatus synthesis or assembly is most probably affected in opg mutants of E. chrysanthemi, since this was previously observed in opg mutants of E. coli (14) and Agrobacterium tumefaciens (8).
Since the opg mutations seemed to have a pleiotropic effect on several envelope components, we decide to test the bile salt sensitivity of the mutants. The MIC was 2.5 ± 0.3 g/liter for both opg mutants, while it was 18.3 ± 0.3 g/liter for the wild-type strain. Thus, the opg mutants were sevenfold more sensitive to bile salts than the wild-type strain, confirming that the envelope structure was severely affected in these mutants.
Exoenzyme synthesis and secretion in opgG mutant of E. chrysanthemi.
Pectate lyase activity was first estimated on plates containing polygalacturonate and a second carbon source. No halo derived from polygalacturonate degradation was observed when the opg mutants were grown in the presence of glycerol, glucose, or sucrose. A reduced halo, compared with that of the wild-type strain, was observed when succinate or galacturonate was added or in the absence of a second carbon source. Thus, pectate lyase activity appeared to be reduced at various levels in the opg mutants, depending on the carbon source used by the bacteria (data not shown).
Assay of the global pectate lyase activity demonstrated that production of these enzymes decreased by twofold in the presence of the opgG mutation in either the presence or absence of an inducer (Table 2). A similar decrease was observed when pelC::lacZ or pelE::lacZ transcriptional fusions were used to analyze the expression of these genes (Table 2).
TABLE 2.
Expression of pel and out genes in the presence of an opgG mutation in E. chrysanthemia
| Enzyme | Fusion | Inducer | Sp act (nmol of product liberated/min/mg (dry wt) ± SD
|
|
|---|---|---|---|---|
| opgG+ | opgG | |||
| Pectate lyase | − | 0.05 ± 0.02 | 0.02 ± 0.01 | |
| + | 19.7 ± 3.2 | 8.4 ± 2.1 | ||
| β-Galactosidase | pelE-lacZ | − | 7 ± 3 | 5 ± 2 |
| + | 132 ± 18 | 55 ± 9 | ||
| pelC-lacZ | − | 5 ± 2 | 2 ± 2 | |
| + | 863 ± 82 | 432 ± 25 | ||
| outC-lacZ | − | 87 ± 5 | 58 ± 6 | |
| + | 340 ± 20 | 156 ± 32 | ||
Cultures were grown in minimal medium to late log phase in the presence of either glycerol (−) or the inducing compounds polygalacturonate and galacturonate (+). The results reported are averages of at least three independent experiments ± standard deviation. Pectate lyase specific activity reflects the expression of the five major pel genes (pelA to pelE). β-Galactosidase specific activity reflects the expression of the corresponding lacZ fusion.
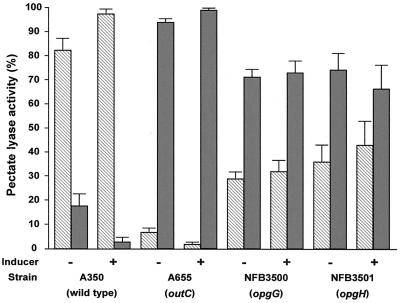
We also tested the effect of the opgG mutation on the efficiency of the Out secretion machinery (Fig. 2). While more than 80% of the pectate lyase activity was found in the supernatant of the parental strain, only 30 to 50% of the pectate lyase activity was observed in the supernatant of the opgG and opgH mutants. Comparison with a nonsecretory outC mutant indicated that secretion is only partially affected by the opg mutations. When the transcription of the out operon was analyzed using an outC::lacZ transcriptional fusion (Table 2), a 30 to 50% decrease was observed in the presence of the opgG mutation.
FIG. 2.
Pectate lyase activity of outC, opgG, and opgH mutants compared to the wild-type strain. Cultures were grown in minimal medium to late log phase in the presence of either glycerol (−) or the inducing compounds polygalacturonate and galacturonate (+). Cells were harvested, and pectate lyase activities in the pellet (gray bars) and the culture supernatant (hatched bars) were determined (see Materials and Methods). The results reported are averages of three independent experiments. Pectate lyase activity reflects the expression of the five major pel genes (pelA to pelE).
Cellulase and protease activities were only estimated on plates. In the cellulase plate test, haloes derived from carboxymethylcellulose degradation by opg and out mutants were similar, showing severely reduced sizes compared to that of the wild-type strain (data not shown). In the protease plate test, no halo derived from milk protein degradation could be detected around the opg mutants (data not shown). This suggested that cellulase and protease secretions, like pectinase secretion, were affected in the opg mutants.
Pathogenicity of opg mutants of E. chrysanthemi.
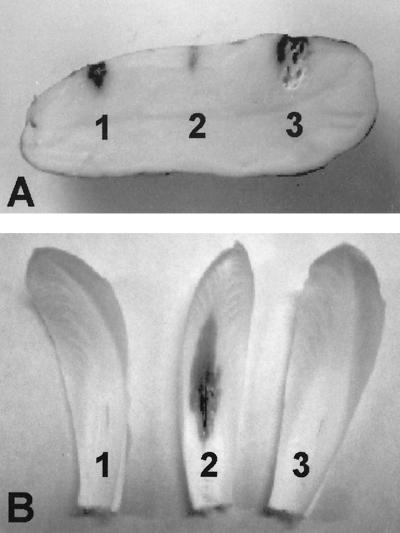
Inoculation with the opgG mutant incited no tissue maceration on potato tubers after 24 h, while an outS mutant produced reduced symptoms (Fig. 3A). Identical results were obtained when the opgG and opgH mutants were used to inoculate chicory leaves (Fig. 3B). Moreover, after a 10-day incubation, plants inoculated with opg mutants did not exhibit any symptoms. The opg mutants were thus completely nonvirulent.
FIG. 3.
Pathogenicity of outS, opgG, and opgH mutant strains on potato tubers and chicory leaves. (A) Bacteria (107) of the outS strain A3144 (spot 1), the opgG strain NFB3500 (spot 2), and the wild-type strain 3937 (spot 3) were inoculated into holes on potato tubers. Disease symptoms were observed after 72 h. (B) Bacteria (106) of the opgG strain NFB3500 (spot 1), the wild-type strain 3937 (spot 2), and the opgH strain NFB3501 (spot 3) were inoculated into scarified chicory leaves. Disease symptoms were observed after 48 h.
Among the various phenotypic characters altered in the opg mutants, two are necessary for virulence: secretion of cell wall-degrading enzymes (2) and motility, since a nonmotile msrA mutant strain exhibits reduced symptoms on chicory leaves (13). Coinoculation experiments were done in order to determine if the defect in virulence of the opg mutants could be complemented by wild-type bacteria present in the close environment. Potato tubers were inoculated with a mix of wild-type and mutant strains in various relative concentrations. Three different mutants were tested: opgG, outS, and msrA. The three different initial percentages of mutant bacteria were 10, 90, and 99%. After 24 to 72 h of incubation, depending on the wild-type strain concentration, wild-type and mutant cells present in the macerated tissue were counted (see Materials and Methods).
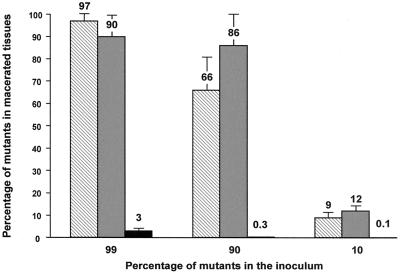
The percentages of outS and msrA mutant cells were similar in the inoculum and in the macerated tissue (Fig. 4). This indicates that growth occurred at a similar rate for the outS, msrA, and wild-type bacteria. Thus, intercellular complementation occurred even when the inoculum contained only 1% wild-type cells. The percentage of opgG mutant cells was reduced by a factor of 22 to 330 depending on the ratio of mutant cells in the inoculum (Fig. 4). Nevertheless, the viability of the opg cells was unaffected, because the number of bacteria was similar in the inoculum and in the macerated tissue. For example, when the inoculum contained 90% opg cells, the total macerated tissue contained an average of 1.2 × 107 ± 0.3 × 107 Opg− bacteria, while the inoculum contained 9 × 106 Opg− bacteria. Since the population would have increased only by a factor of 1.3 (in the range of error), one can consider that no growth of the mutant strain occurred in the plant tissues. During the same time, the wild-type population increased by a factor of 4,580. Thus, intercellular complementation did not occur between the opgG mutant and wild-type strains, indicating that the loss of virulence of opgG mutants did not result simply from motility or exoenzyme deficiencies. After plant infection, the opg mutant stopped its growth but remained viable.
FIG. 4.
Coinoculation of potato tubers with wild-type strain 3937 and outS strain A3144 (hatched bars), msrA strain RH7006 (gray bars), or opgG strain NFB3500 (black bars). Mixed bacteria (107) were inoculated into holes in potato tubers. After 72 h, macerated tissue was collected, weighed, and diluted in physiological distilled water. The percent mutant bacteria was determined by the ratio of the number of mutant bacteria (Kanr) to the number of total bacteria per gram of tissue. The results reported are averages of three independent experiments performed on at least two potato tubers.
In order to determine whether the wild-type strain could inhibit the growth of the opgG mutant, we performed coinoculation tests in M63 minimal liquid medium. In these conditions, both strains grew equally, and the mutant/wild-type ratio remained constant throughout the experiment, indicating that the wild-type strain did not inhibit growth of NFB3500 (data not shown).
In order to determine whether some potato tuber molecules could inhibit the growth of NFB3500, M63 minimal liquid medium was supplemented with potato tuber extracts (see Materials and Methods). The growth rate and the stationary-phase plateau were identical when various amounts of potato tuber extracts were present, indicating that potato tuber does not contain factors that inhibit the growth of the opg mutant (data not shown).
DISCUSSION
This paper describes the molecular organization of the opgGH operon from E. chrysanthemi, the phenotypes observed when these genes are disrupted, and their implication in its pathogenicity.
As in E. coli, the two genes necessary for OPG backbone synthesis in E. chrysanthemi are organized in an operon, opgGH, in the vicinity of the pyrC gene. In both organisms, yceE, a gene of unknown function, is found downstream from mdoH or opgH. In E. coli, this gene is transcribed with msyB, which is only known to encode a suppressor of a secY thermosensitive mutation. Nevertheless, in E. chrysanthemi, msyB is not present at this locus (Fig. 1). The DNA sequences of the two organisms are conserved over the entire sequences of the opgGH and mdoGH operons, including the promoter region. In E. coli, the mdoC gene, involved in OPG succinylation, is located just upstream from the mdoGH operon (21). Despite the fact that the OPGs of E. chrysanthemi are succinylated (9), no gene homologous to mdoC was found upstream of the opgGH operon. The 13-bp direct repeats found upstream from opgGH could be the limit of a chromosomal rearrangement mediated by a transposable element. Moreover, the gene encoding the OPG succinylation activity could differ strongly from mdoC, since several attempts to obtain PCR amplification of that gene failed (data not shown).
The opg mutants exhibit a pleiotropic phenotype. Colonies of opg mutants are mucoid, indicating increased biosynthesis of EPS. This correlates with a threefold increase in the expression of the epsG::lacZ fusion in an opgG mutant background. This increase could be the result of a direct action of OPGs on eps gene transcription via a regulatory protein(s). Actually, in E. coli, the cps genes, involved in colanic acid capsular polysaccharide biosynthesis, are regulated by a two-component sensor-regulator system, RcsB/RcsC. In an mdoH mutant, cps gene transcription is increased but remains regulated in an RcsB/RcsC-dependent fashion. This suggests that RcsC, the sensor of the two-component system, could sense the level of OPG in the periplasm in a still unknown way (12). In Erwinia amylovora and Erwinia stewartii, the RcsB and RcsC proteins are involved in the regulation of EPS biosynthesis and show strong similarities with the RcsB and RscC proteins of E. coli, indicating the existence of a family of related capsule activator proteins (4). Thus, in E. chrysanthemi, OPGs could also participate in EPS biosynthesis regulation by acting via a similar two-component regulatory system. Alternatively, this increase could be one of the responses resulting from a disorganization of the cell envelope induced by the lack of OPGs. Disorganization of the cell envelope is suggested by the other phenotypes of the opg mutants, particularly bile salt hypersensivity and defective chemotaxis. Lack of OPGs could result in abnormal assembly of several envelope components due to an unknown structural function of the glucans, or OPG concentration could be detected and used to regulate the synthesis of several envelope components, especially those which are subjected to osmotic regulation, like the flagellum apparatus (14) and colanic acid synthesis (12) in E. coli.
Among the various effects observed in opg mutants, the most important seems to be the reduction in pectate lyase production and secretion, because they are known to play a crucial role in virulence. opg mutations result in a twofold decrease in pectate lyase production associated with a similar decrease in pel gene transcription. OPGs could modulate pel gene expression via the two-component sensor-regulator system PecM/PecS (15) in a way which remains to be elucidated, but the existence of new regulatory factors cannot be excluded. The reduction in Pel protein synthesis is associated with a reduction in their secretion through the cell envelope. This defect in the Out secretory apparatus could be explained in part by a decrease in its synthesis and in part by an alteration in its assembly and/or functioning resulting from the cell envelope disorganization.
The opg mutants are completely nonvirulent when inoculated into potato tubers or chicory leaves. This phenotype has already been observed in planta with the hrpM mutant of P. syringae pv. syringae (28) and for the hrpM mutant of P. aeruginosa PA14 (26). The major consequence of pel and out mutations is the severely reduced virulence of strains harboring these mutations. However, the complete loss of virulence of the opg mutants cannot be explained only by a defect in pectate lyase secretion, since the outS mutant (severely impaired in pectate lyase secretion) induces reduced symptoms, while the opg mutant, which secretes more pectate lyases than the outS mutant, induces no symptoms.
The outS (altered secretion) and msrA (altered motility and peptide methionine sulfoxide reductase) mutant cells could be sustained by coinoculation with no more than 1% of the wild-type strain in potato tubers. These mutant strains grew poorly in planta and produced few macerated tissues by themselves, but they are able to use the macerated tissues produced by the wild-type strain for their growth. This clearly suggests that only the plant-bacterium interaction is disrupted in these mutants. On the other hand, the opg mutant cells cannot grow when coinoculated in potato tubers with as much as 90% wild-type cells, while growth remains unaffected when the coinoculation occurs in laboratory conditions. However, the opg mutants which are unable to use macerated tissues for their growth can survive in these conditions. These results indicate that OPGs are necessary not only for plant colonization, but also for bacterial multiplication in macerated tissues. When E. chrysanthemi is grown in a liquid medium, a large fraction of the OPGs produced can be recovered in the medium, at least under certain circumstances (9). We have no indication whether this is true when bacteria grow in planta. However, our experiments clearly show that if OPGs are released in the plant tissues during co-inoculation with wild-type cells, these external OPGs cannot complement the growth defect of the opg mutants. Moreover, OPG-defective mutants of A. tumefaciens remain nonvirulent when inoculated in planta with purified OPGs (8). This strongly suggests that OPGs need to be inside the periplasmic space and not in the external environment during the infection process.
ACKNOWLEDGMENTS
We are grateful to F. Barras and to our colleagues at Unité de Microbiologie Génétique for providing us with bacterial strains.
This research was supported by grants from the Centre National de la Recherche Scientifique and the Ministère de l'Education Nationale, de la Recherche et de la Technologie.
REFERENCES
- 1.Bardonnet N, Blanco C. ′uidA-antibiotic-resistance cassettes for insertion mutagenesis, gene fusions and genetic constructions. FEMS Microbiol Lett. 1992;93:243–248. doi: 10.1016/0378-1097(92)90469-5. [DOI] [PubMed] [Google Scholar]
- 2.Barras F, Van Gijsegem F, Chatterjee A K. Extracellular enzymes and pathogenesis of soft-rot Erwinia. Annu Rev Phytopathol. 1994;32:201–234. [Google Scholar]
- 3.Beaulieu C, Boccara M, Van Gijsegem F. Pathogenic behavior of pectinase-defective Erwinia chrysanthemi mutants on different plants. Mol Plant-Microbe Interact. 1993;6:197–202. [Google Scholar]
- 4.Bereswill S, Geider K. Characterization of the rcsB gene from Erwinia amylovora and its influence on exopolysaccharide synthesis and virulence of the fire blight pathogen. J Bacteriol. 1997;179:1354–1361. doi: 10.1128/jb.179.4.1354-1361.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bohin J-P. Osmoregulated periplasmic glucans in proteobacteria—a minireview. FEMS Microbiol Lett. 2000;186:11–19. doi: 10.1111/j.1574-6968.2000.tb09075.x. [DOI] [PubMed] [Google Scholar]
- 6.Bohin J-P, Kennedy E P. Mapping of a locus (mdoA) that affects the biosynthesis of membrane-derived oligosaccharides in Escherichia coli. J Bacteriol. 1984;157:956–957. doi: 10.1128/jb.157.3.956-957.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bourson C, Favey S, Reverchon S, Robert-Baudouy J. Regulation of the expression of a pelA::uidA fusion in Erwinia chrysanthemi and demonstration of the synergistic action of plant extract with polygalacturonate on pectate lyase synthesis. J Gen Microbiol. 1993;172:6950–6958. doi: 10.1099/00221287-139-1-1. [DOI] [PubMed] [Google Scholar]
- 8.Cangelosi G A, Martinetti G, Nester E W. Osmosensitivity phenotypes of Agrobacterium tumefaciens mutants that lack periplasmic β-1,2-glucans. J Bacteriol. 1990;172:2172–2174. doi: 10.1128/jb.172.4.2172-2174.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cogez V, Talaga P, Lemoine J, Bohin J-P. Osmoregulated periplasmic glucans of Erwinia chrysanthemi. J Bacteriol. 2001;183:3127–3133. doi: 10.1128/JB.183.10.3127-3133.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Condemine G, Castillo A, Passeri F, Enard C. The PecT repressor coregulates synthesis of exopolysaccharides and virulence factors in Erwinia chrysanthemi. Mol Plant-Microbe Interact. 1999;12:45–52. doi: 10.1094/MPMI.1999.12.1.45. [DOI] [PubMed] [Google Scholar]
- 11.Condemine G, Robert-Baudouy J. Synthesis and secretion of Erwinia chrysanthemi virulence factors are coregulated. Mol Plant-Microbe Interact. 1995;8:632–636. [Google Scholar]
- 12.Ebel W, Vaughn G J, Peters III H K, Trempy J E. Inactivation of mdoH leads to increased expression of colanic acid capsular polysaccharide in Escherichia coli. J Bacteriol. 1997;179:6858–6861. doi: 10.1128/jb.179.21.6858-6861.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.El Hassouni M, Chambost J-P, Expert D, Van Gijsegem F, Barras F. The minimal gene set member msrA encoding peptide methionine sulfoxide reductase is a virulence determinant of the plant pathogen Erwinia chrysanthemi. Proc Natl Acad Sci USA. 1999;96:887–892. doi: 10.1073/pnas.96.3.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fiedler W, Rotering H. Properties of Escherichia coli mutants lacking membrane-derived oligosaccharides. J Biol Chem. 1988;263:14684–14689. [PubMed] [Google Scholar]
- 15.Hugouvieux-Cotte-Pattat N, Condemine G, Nasser W, Reverchon S. Regulation of pectinolysis in Erwinia chrysanthemi. Annu Rev Microbiol. 1996;50:213–257. doi: 10.1146/annurev.micro.50.1.213. [DOI] [PubMed] [Google Scholar]
- 16.Hugouvieux-Cotte-Pattat N, Dominguez H, Robert-Baudouy J. Environmental conditions affect transcription of the pectinases genes of Erwinia chrysanthemi 3937. J Bacteriol. 1992;174:7807–7818. doi: 10.1128/jb.174.23.7807-7818.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hugouvieux-Cotte-Pattat N, Reverchon S, Robert-Baudouy J. Expanded linkage map of Erwinia chrysanthemi strain 3937. Mol Microbiol. 1989;3:573–581. doi: 10.1111/j.1365-2958.1989.tb00204.x. [DOI] [PubMed] [Google Scholar]
- 18.Ji J, Hugouvieux-Cotte-Pattat N, Robert-Baudouy J. Molecular cloning of the outJ gene involved in pectate lyase secretion by Erwinia chrysanthemi. Mol Microbiol. 1989;3:285–293. doi: 10.1111/j.1365-2958.1989.tb00173.x. [DOI] [PubMed] [Google Scholar]
- 19.Kennedy E P. Membrane-derived oligosaccharides (periplasmic beta-d-glucans) of Escherichia coli. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Maganasik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: American Society for Microbiology; 1996. pp. 1064–1074. [Google Scholar]
- 20.Kotoujanski A, Lemattre M, Boistard P. Utilization of a thermosensitive episome bearing transposon Tn10 to isolate Hfr donor strains of Erwinia carotovora subsp. chrysanthemi. J Bacteriol. 1982;150:122–131. doi: 10.1128/jb.150.1.122-131.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lacroix J-M, Lanfroy E, Cogez V, Lequette Y, Bohin A, Bohin J-P. The mdoC gene of Escherichia coli encodes a membrane protein that is required for succinylation of osmoregulated periplasmic glucans. J Bacteriol. 1999;181:3626–3631. doi: 10.1128/jb.181.12.3626-3631.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lacroix J-M, Loubens I, Tempete M, Menichi B, Bohin J-P. The mdoA locus of Escherichia coli consist of an operon under osmotic control. Mol Microbiol. 1991;5:1745–175. doi: 10.1111/j.1365-2958.1991.tb01924.x. [DOI] [PubMed] [Google Scholar]
- 23.Lacroix J-M, Tempête M, Menichi B, Bohin J-P. Molecular cloning and expression of a locus (mdoA) implicated in the biosynthesis of the membrane-derived oligosaccharides in Escherichia coli. Mol Microbiol. 1989;3:1173–1182. doi: 10.1111/j.1365-2958.1989.tb00267.x. [DOI] [PubMed] [Google Scholar]
- 24.Lojkowska E, Masclaux C, Boccara M, Robert-Baudouy J, Hugouvieux-Cotte-Pattat N. Characterization of the pelL gene encoding a novel pectate lyase of Erwinia chrysanthemi 3937. Mol Microbiol. 1995;16:1183–1195. doi: 10.1111/j.1365-2958.1995.tb02341.x. [DOI] [PubMed] [Google Scholar]
- 25.Loubens I, Debarbieux L, Bohin A, Lacroix J-M, Bohin J-P. Homology between a genetic locus (mdoA) involved in the osmoregulated biosynthesis of periplasmic glucans in Escherichia coli and a genetic locus (hrpM) controlling the pathogenicity of Pseudomonas syringae. Mol Microbiol. 1993;10:329–340. doi: 10.1111/j.1365-2958.1993.tb01959.x. [DOI] [PubMed] [Google Scholar]
- 26.Mahajan-Miklos S, Tan M-W, Rahme L G, Ausubel F M. Molecular mechanisms of bacterial virulence elucidated using a Pseudomonas aeruginosa-Caenorhabditis elegans pathogenesis model. Cell. 1999;96:47–56. doi: 10.1016/s0092-8674(00)80958-7. [DOI] [PubMed] [Google Scholar]
- 27.Masclaux C, Expert D. Signaling potential iron plant microbe interaction: the pathogenic switch of iron transport in Erwinia chrysanthemi. Plant J. 1995;7:121–128. [Google Scholar]
- 28.Masclaux C, Hugouvieux-Cotte-Pattat N, Expert D. Iron is a triggering factor for differential expression of Erwinia chrysanthemi strain 3937 pectate lyase in pathogenesis of African violets. Mol Plant-Microbe Interact. 1996;9:198–205. [Google Scholar]
- 29.Miller J H. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 30.Moran F, Nasuno S, Starr M P. Extracellular and intracellular polygalacturonic acid transeliminase of Erwinia carotovora. Arch Biochem Biophys. 1968;123:293–306. doi: 10.1016/0003-9861(68)90138-0. [DOI] [PubMed] [Google Scholar]
- 31.Mukhopadhyay P, Williams J, Mills D. Molecular analysis of a pathogenicity locus in Pseudomonas syringae pv. syringae. J Bacteriol. 1988;170:5479–5488. doi: 10.1128/jb.170.12.5479-5488.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Resibois A, Colet M, Faelen M, Schoonejans M, Toussaint A. Phi-EC2, a new generalized transducing phage of Erwinia chrysanthemi. Virology. 1984;137:102–112. doi: 10.1016/0042-6822(84)90013-8. [DOI] [PubMed] [Google Scholar]
- 33.Roeder D L, Collmer A. Marker-exchange mutagenesis of a pectate lyase isoenzyme gene in Erwinia chrysanthemi. J Bacteriol. 1985;164:51–56. doi: 10.1128/jb.164.1.51-56.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salmond G P C. Secretion of extracellular virulence factors by plant pathogenic bacteria. Annu Rev Phytopathol. 1994;32:181–200. [Google Scholar]
- 35.Sambrook J E, Fritsch F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 36.Shevchik V E, Hugouvieux-Cotte-Pattat N. Identification of a bacterial pectin acetyl esterase in Erwinia chrysanthemi. Mol Microbiol. 1997;24:1285–1301. doi: 10.1046/j.1365-2958.1997.4331800.x. [DOI] [PubMed] [Google Scholar]
- 37.Spiro R G. Analysis of sugars found in glycoproteins. Methods Enzymol. 1966;8:3–27. [Google Scholar]
- 38.Talaga P, Fournet B, Bohin J-P. Periplasmic glucans of Pseudomonas syringae pv. syringae. J Bacteriol. 1994;176:6538–6544. doi: 10.1128/jb.176.21.6538-6544.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tardy F, Nasser W, Robert-Baudouy J, Hugouvieux-Cotte-Pattat N. Comparative analysis of the five major Erwinia chrysanthemi pectate lyases: enzyme characteristics and potential inhibitors. J Bacteriol. 1997;179:2503–2511. doi: 10.1128/jb.179.8.2503-2511.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Teather R M, Wood P J. Use of Congo red polysaccharide interactions in enumeration and characterization of cellulolytic bacteria from bovine rumen. Appl Environ Microbiol. 1982;43:777–780. doi: 10.1128/aem.43.4.777-780.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van Gijsegem F, Toussaint A, Schoonejans E. In vivo cloning of pectate lyase and cellulase genes of Erwinia chrysanthemi. EMBO J. 1985;4:787–792. doi: 10.1002/j.1460-2075.1985.tb03698.x. [DOI] [PMC free article] [PubMed] [Google Scholar]