Abstract
FliM is part of the flagellar switch complex. Interaction of this protein with phospho-CheY (CheY-P) through its N terminus constitutes the main information relay point between the chemotactic system and the flagellum. In this work, we evaluated the role of the N terminus of FliM in the swimming behavior of Rhodobacter sphaeroides. Strains expressing the FliM protein with substitutions in residues previously reported in Escherichia coli as being important for interaction with CheY showed an increased stop frequency compared with wild-type cells. In accordance, we observed that R. sphaeroides cells expressing FliM lacking either the first 13 or 20 amino acids from the N terminus showed a stopped phenotype. We show evidence that FliMΔ13 and FliMΔ20 are stable proteins and that cells expressing them allow flagellin export at levels indistinguishable from those detected for the wild-type strain. These results suggest that the N-terminal region of FliM is required to promote swimming in this bacterium. The role of CheY in controlling flagellar rotation in this organism is discussed.
The flagellar switch complex is composed of multiple subunits of three different proteins, FliG, FliN, and FliM (8, 9, 15–17, 21, 44, 45). These proteins interact with each other and are required for flagellar assembly and normal swimming (13, 23–25, 36, 38, 43). FliG is thought to be directly involved in torque generation (20). FliM receives the output of the chemotactic system through its binding to the phosphorylated form of the response regulator CheY (CheY-P) (7, 25, 37, 38, 41). In the peritrichous bacteria Escherichia coli and Salmonella enterica serovar Typhimurium, binding of CheY-P to FliM induces a change in the direction of flagellar rotation from counterclockwise (CCW) to clockwise (CW) (5, 27, 42). When most of the flagella of a cell rotate in the CCW direction, they coalesce in a bundle that pushes the cell body in a linear trajectory called a run. When flagella switch from CCW to CW rotation, the bundle loses stability, and the uncoordinated motion of the filaments produces a tumble that reorients the cell. The direction of the next run is randomly determined (40). The frequency of tumble determines the overall swimming direction of the cell (for a review, see reference 22).
It has been shown that the main CheY-binding domain of FliM corresponds to the first 16 amino acids of the N terminus. This domain is sufficient to bind CheY-P in vitro (7). Strains expressing FliM lacking this N-terminal region show a smooth-swimming phenotype (25, 38). In addition, specific single-amino-acid substitutions in this region have been reported to weaken FliM-CheY-P interaction in vitro; accordingly, cells expressing any of these FliM mutations also show smooth swimming (7, 33). These results support the idea that the N terminus of FliM is the main CheY-P-binding target that controls flagellar switching.
The flagellum of the monoflagellated bacterium Rhodobacter sphaeroides rotates only in the CW direction, alternating with brief stop periods in which the cell reorients. During the stop periods, the filament loses its typical helicoidal morphology and retracts into a coiled form (2). It has been suggested that the slow motion of the filament in this conformation helps to reorient the cell (3). Chemotaxis towards some compounds has been reported. An increase in attractant concentration results in a decrease in the stopping frequency, and vice versa (12). Besides this response, R. sphaeroides also shows chemokinetic behavior, which consists of an increase of the rate of flagellar rotation in response to a positive stimulus (26).
Several copies of most of the chemotactic genes have been found in R. sphaeroides, including five copies of cheY and two each of cheA and cheR, whereas the cheZ gene has not been identified. It has been suggested that this reiteration and the absence of the CheY-specific phosphatase CheZ may contribute to the complexity of the chemotactic system in this bacterium (for a review, see reference 1). It has been shown that some of these CheY proteins alter the chemotactic response when they are expressed in Escherichia coli (29). However, the role of these proteins in R. sphaeroides has only just begun to be characterized. Recently, it was proposed that CheY4 and CheY5 are the motor-binding response regulators, whereas CheY3 would be a phosphate sink (30).
Since in other bacteria the main control of the chemotactic response relies on the binding of CheY-P to FliM (6, 22), we decided to investigate the effect of altering the putative CheY-binding domain on the FliM protein of R. sphaeroides (FliMRs). In our first approach, three different amino acid substitutions were made that are known to be relevant for CheY-P interaction with FliM in E. coli and S. enterica. Analysis of the free-swimming behavior of strains carrying these mutations showed an increase in the stopping frequency compared with that observed for wild-type cells. In agreement with this result, strains expressing FliM lacking the N-terminal region showed a stopped phenotype. These results allow us to propose that, in contrast to the situation found in E. coli and S. enterica, in R. sphaeroides the N-terminal region of FliM is involved in promoting flagellar rotation, probably depending on CheY binding.
MATERIALS AND METHODS
Plasmids and strains.
The bacterial strains and plasmids used in this work are described in Table 1.
TABLE 1.
Bacterial strains and plasmids used in this work
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| E. coli | ||
| JM103 | hsdR4 Δ(lac-pro) F′ traD36 proAB lacIqZΔM15 | 4 |
| S17-1 | recA endA thi hsdR RP4-2-Tc::Mu::Tn7 Tpr Smr | 31 |
| R. sphaeroides | ||
| WS8 | Wild type; spontaneous Nalr | 34 |
| SP4 | WS8 fliM::aadA, Spcr Nalr | This work |
| SP5 | WS8 fliMΔ1 | This work |
| Plasmids | ||
| pJQ200mpl8 | Suicide vector used for gene replacement in gram-negative bacteria | 28 |
| pRK415 | pRK404 derivative used for expression in R. sphaeroides | 14 |
| pRS100(wt) | pTZ19R carrying fliM+ as a 1.3-kb SacII fragment | This work |
| pRS75 | 4.4-kb SalI fragment expanding from fliK to fliP cloned in pTZ19R9(mut1) | 10 |
| pFliM-wt | pRK415 carrying wild-type fliM under control of vector promoters | This work |
| pFliM-wt(His) | pRK415 carrying wild-type fliM encoding His tag at the C terminus of FliM | This work |
| pFliM8LI, pFliM9SY, and pFliM12EG | pRK415 carrying fliM encoding the indicated FliM mutant protein | This work |
Media and growth conditions.
R. sphaeroides cell cultures were grown in liquid or solid Sistrom's medium (32) at 30°C. Aerobic growth conditions were achieved in the dark with strong shaking (300 rpm). Photoheterotrophic cultures were grown under constant illumination in completely filled screw-cap tubes. Motility plates were prepared using 0.3% Bacto-Agar. Strains of E. coli were grown in Luria-Bertani (LB) medium (4). When needed, antibiotics were added at the following concentrations: spectinomycin 10 μg/ml; gentamicin, 30 μg/ml; and tetracycline, 1 μg/ml. For E. coli, the antibiotics used were ampicillin (100 μg/ml), tetracycline (15 μg/ml), and spectinomycin (100 μg/ml).
Isolation of strains carrying fliMΔ1 or fliM::aadA.
Two chromosomal fliM mutations were isolated for use as recipients of plasmids carrying the different fliM alleles studied in this work. To isolate the fliM::aadA strain, a 4.6-kb SalI fragment carrying a DNA fragment from fliK up to fliP was cloned into a pTZ19R derivative in which the EcoRI site was previously mutated (pTZ19Rmut1). This plasmid was named pRS75 (10). An internal portion of the omega-Spcr cassette, obtained by PCR, containing no known transcriptional termination signals, was then inserted into the unique EcoRI site of pRS75, which is located in the middle of fliM. The 6.6-kb SalI fragment carrying fliM::aadA was then subcloned into pJQ200 (28). The resulting plasmid was introduced by transformation into E. coli S-17 and subsequently transferred to R. sphaeroides by conjugation. Since pJQ200 cannot replicate in R. sphaeroides, the double recombination event was selected on LB plates in the presence of spectinomycin and 5% sucrose. The fliMΔ1 allele was constructed by removing a 431-bp NcoI-EcoRI fragment from plasmid pRS75. The resultant 4.2-kb SalI fragment carrying fliMΔ1 was subcloned into pJQ200. To obtain the chromosomal replacement with this allele, the cells were grown on LB plates in the presence of 5% sucrose. Individual single colonies were subsequently tested for motility. For both strains, the presence of the correct replacement was confirmed by Southern blotting.
Deletion of 5′ end of fliM.
The fliMΔ13 and fliMΔ20 alleles were constructed by PCR using plasmid pRS100(wt) as the substrate for amplification. This plasmid carries the complete fliM gene in a 1.3-kb chromosomal SacII fragment cloned into pTZ19Rmut1 within the SmaI site. In this construction, the KpnI site from the polylinker is located near the 3′ end of fliM. The forward oligonucleotides were designed to prime immediately downstream of the last amino acid to be deleted. The sequence corresponding to amino acids 13 and 20 was modified in the corresponding oligonucleotide to encode methionine; this residue represents the fliM start codon in the final constructions. In both cases, this methionine codon was designed as part of an NcoI recognition site. The reverse oligonucleotide was complementary to the sequence located at the junction between fliM and fliN, carrying the fliM stop codon. A KpnI recognition site was included at the 5′ end. Each PCR product was cloned into pRS100(wt), replacing the wild-type NcoI-KpnI fragment. These plasmids were subsequently sequenced to corroborate the presence of the correct allele and also to discard any possible errors that might have occurred during the amplification reaction.
Site-directed mutagenesis of fliM.
Site-directed mutagenesis was carried out on uracil-containing single-stranded DNA from pRS100(wt), using Kunkel's method (19). The presence of the desired mutation in the resultant plasmid was confirmed by sequencing. The DNA fragment carrying the fliM gene was then transferred to pRK415 and introduced into R. sphaeroides by conjugation.
Motility assays.
A 5-μl sample of a stationary-phase culture was placed on the surface of swarm plates and incubated aerobically in the dark. Swarming ability was recorded as the ability of bacteria to move away from the inoculation point after 36 to 72 h. Free-swimming motility was evaluated in an aliquot from an aerobic or photosynthetic culture at the mid-logarithmic growth phase. Swimming behavior was recorded, and segments of the video were analyzed by computerized image analysis as described below.
Behavioral assays.
The samples were observed with a Nikon E-600 dark-field microscope, mounted with a charge-coupled device (CCD) video camera (Ikegami ICD-42A). Swimming cells were recorded for several minutes in an S-VHS video tape recorder. For each sample, segments of 2 s at 30 frames per second were analyzed. These segments were digitized and converted to a digital movie file (avi format) using a Pinnacle Miro (DC30 Pro) video capture card and the Adobe Premier 5.1 software. Further analysis of the swimming behavior was done automatically using the ImagePro-Plus analyzer (version 4.0, running in a Pentium III personal computer under Windows 98) in conjunction with a specific bacterium-tracking macro file designed for this work. The 60 images of the 2-s movie .avi file were saved as single images to be processed individually. The images were converted to 8-bit monochrome, followed by background flattening and high gauss filter application. After this global process, object labeling by automatic white object thresholding was done once in the first image. Objects with a mean surface value of a bacterial cell body ± 3 standard deviations were considered artifacts and eliminated from the labeled list. The bacterium-tracking process begins here for each labeled object among the full sequence of 60 images. The trajectory of each bacterium was automatically followed in the sequence of processed images and marked with points (x and y coordinates) corresponding to the center of mass of the bacterium in each new position. For further processing, these coordinates were stored in an Excel workbook. Only bacteria not leaving the field of view during the complete length of the movie were considered. Overlapping trajectories as well as those produced by bacteria leaving the field were automatically discarded. The stored x and y coordinates of the trajectories were then used to calculate the mean stop frequency, the mean stop time, the mean velocity, and the run length distribution. The results obtained with this program were verified using a subsample of data for which the above parameters were calculated manually using a semiautomatic program (F. Caviedes, technical report MME0005, Centro de Instrumentos, Universidad Nacional Autónoma de México).
Immunoblotting.
Cellular levels of the FliM protein were examined on immunoblots. For this, a His tag was introduced by PCR at the 3′ end of the open reading frame of wild-type fliM and fliMΔ13 alleles. The PCR products were cloned into plasmid pTZ19R to confirm the correct sequence. Subsequently, fliM-wt(His) and fliMΔ13(His) alleles were transferred to pRK415. R. sphaeroides cells carrying these plasmids were grown to the mid-logarithmic phase, harvested, and resuspended in protein sample buffer. Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose membranes as described elsewhere (4). The membranes were then probed with an anti-His polyclonal antibody (Pierce). Detection was carried out by enhanced chemiluminescence using an ECL immunoblotting detection kit (Amersham International).
RESULTS
Isolation of R. sphaeroides strains carrying the chromosomal alleles fliM::aadA and fliMΔ1.
To study the effect of different fliM alleles on the swimming behavior of R. sphaeroides, we isolated two different mutant strains. The strain carrying the chromosomal allele fliM::aadA was isolated by a double recombination event as described in Materials and Methods. This strain was named SP4 and showed a Fla− phenotype. Full motility was recovered when a plasmid expressing only fliM+ (pFliM-wt) was introduced into these cells. However, when different mutant plasmid-borne fliM alleles were introduced into this strain, recombinant cells showing the wild-type phenotype always appeared early in the culture (data not shown). To avoid this problem, we isolated a strain carrying the chromosomal mutation fliMΔ1, which we named SP5. This mutation is a deletion, expanding from the ATG initiation codon of the fliM open reading frame to codon 143. As expected, strain SP5 (fliMΔ1) showed a Fla− phenotype, and the introduction of pFliM-wt restored flagellation and motility. The analysis of the fliM alleles studied in this work was carried out using this strain.
Isolation and motility behavior of strains expressing mutant proteins FliM8LI, FliM9SY, and FliM12EG.
A detailed study of Salmonella mutants isolated as cheZ suppressors allowed the identification of several residues on FliM involved in promoting CCW rotation (33). In a later study, it was shown that FliM6LI, FliM7SY, and FliM10EG mutant proteins bind CheY-P to a lesser extent than the wild-type protein. These residues are conserved in the amino acid sequence of FliMRs (10). Thus, to obtain some insight into the possible role of the N terminus of FliMRs as the possible target of the chemotactic system, we investigated whether the same amino acid substitutions could affect the free-swimming behavior of R. sphaeroides cells.
Site-directed mutagenesis of the fliMRs gene was carried out to obtain fliM alleles encoding FliM8LI, FliM9SY, and FliM12EG. DNA fragments carrying the wild-type and mutant alleles were cloned into pRK415 under control of the plasmid promoters and transferred into strain SP5 by conjugation.
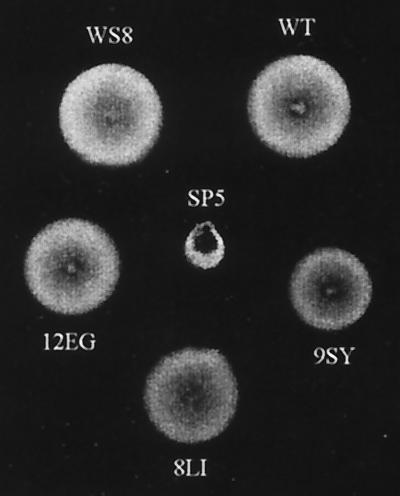
Cells expressing FliM-wt, FliM8LI, and FliM12EG were able to swarm, and the size of the ring was very similar to that observed for the wild-type strain WS8. The mean swarm diameters after 72 h of incubation were 1.65, 1.70, 1.60, and 1.60 cm, respectively (Fig. 1). However, the strain expressing FliM9SY formed a denser ring, with a reduction in diameter of approximately 20% compared with those formed by the WS8 and SP5/pFliM-wt strains. By observation under the microscope, we detected that only cells expressing FliM9SY had a clearly recognizable phenotype. Surprisingly, instead of the smooth-swimming phenotype that this mutation provokes in Salmonella cells, a clear increase in the stop frequency and also in the length of the stopping period was observed.
FIG. 1.
Swarming of SP5 cells expressing different FliM proteins with single amino acid substitutions. WS8 cells were included as a positive control. SP5 cells expressing FliM-wt, FliM12EG, FliM9SY, or FliM8LI are labeled WT, 12EG, 9SY, and 8IL, respectively. An SP5 strain without a plasmid was also included (SP5).
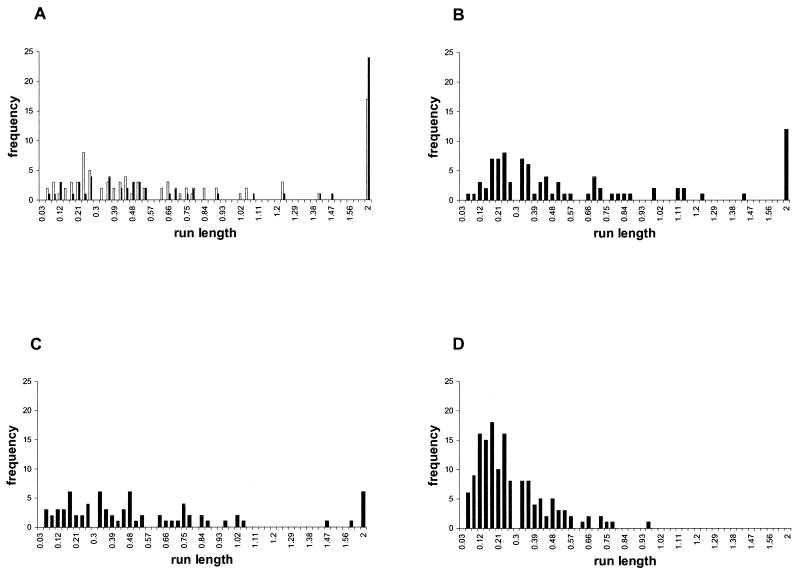
To analyze in detail the free-swimming behavior of these strains, data from at least 30 cells were collected for each culture, and three independent cultures of each strain were used. As shown in Table 2, strains WS8, SP5/pFliM-wt, and SP5/pFliM8LI showed similar values for both stop frequency and stop length. Although we observed a slight increase in the average stop frequency for SP5/pFliM8LI compared with SP5/pFliM-wt and WS8 cells, this did not seem to be significant. In contrast, strains SP5/pFliM12EG and SP5/pFliM9SY showed a significant increase in average stopping frequency, this trait being more pronounced for SP5/pFliM9SY cells. Moreover, this strain also showed a clear increase in the length of the stop periods. The effect of these changes on the swimming behavior is reflected in the run length distributions shown in Fig. 2. It can be observed that cells expressing FliM9SY have a high frequency of short running periods. To a lesser extent, the same can be observed for cells expressing FliM12EG. However, cells expressing FliM8LI were similar to the wild-type cells. In addition, a reduction in the number of cells swimming for the complete time analyzed was also noticed for all these mutants (Fig. 2). The mean velocity observed was similar among these strains, suggesting that the amino acid substitutions tested in this experiment had no effect on the rotation speed of the flagellum (Table 2). Assuming that these mutations could affect the binding of CheY to FliM in a similar way as in E. coli and S. enterica, these results would suggest that, in R. sphaeroides, the binding of CheY to FliM promotes swimming, and the absence of this interaction promotes a stop. Alternatively, the increased stopping frequency could also be explained if these mutations, instead of reducing binding between FliM and CheY, would favor it.
TABLE 2.
Swimming behavior of strains expressing different FliM mutants
| Strain | Mean stop frequencya (stops s−1) ± SD | Mean stop timeb (s) ± SD | Mean velocityc (μm s−1) ± SD |
|---|---|---|---|
| WS8 | 0.7 ± 0.12 | 0.31 ± 0.22 | 28.9 ± 8.6 |
| SP5/pFliM-wt | 0.6 ± 0.11 | 0.38 ± 0.26 | 32.1 ± 9.1 |
| SP5/pFliM8LI | 0.8 ± 0.10 | 0.35 ± 0.27 | 26.1 ± 8.9 |
| SP5/pFliM12EG | 0.9 ± 0.05 | 0.41 ± 0.25 | 33 ± 11.4 |
| SP5/FliM9SY | 1.3 ± 0.03 | 0.45 ± 0.36 | 23 ± 6.4 |
Mean stop frequency calculated for 100 cells. Data for approximately 30 cells from three independent cultures were used. Free-swimming cells were analyzed, as described in Materials and Methods, for 2 s. The samples were taken from aerobically grown cells and diluted on fresh culture medium.
Mean time per stop. Data were collected and analyzed as described for stop frequency.
Mean velocity during running events.
FIG. 2.
Distribution of run lengths from cells expressing different FliM proteins. Free-swimming cells diluted in growth medium were analyzed for 2 s. (A) WS8 (open bars) and SP5/pFliM-wt cells (solid bars). (B) SP5 cells expressing FliM8LI (C) SP5/pFliM12EG. (D) SP5/pFliM9SY. Run length was determined using a computerized swimming behavior analyzer and is expressed in seconds.
Isolation and characterization of fliMΔ13 and fliMΔ20 alleles.
If, in R. sphaeroides, the N-terminal region of FliM acts as the target for the CheY proteins to promote a run, it would be expected that deletion of this region would deviate the flagellar rotational bias to the stop mode. However, if the contrary is true, a smooth-swimming phenotype should be observed. To study these possibilities, we decided to generate deletions of the first 13 and 20 amino acids to completely disrupt the N terminus of FliMRs.
Deletion mutants were constructed by PCR as described in Materials and Methods. The alleles fliMΔ13 and fliMΔ20 were cloned into pRK415 under control of the plasmid promoters and introduced into strain SP5 by conjugation.
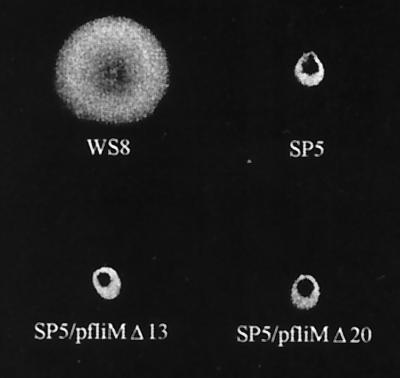
SP5 cells (fliMΔ1) expressing either FliMΔ13 or FliMΔ20 from photoheterotrophic or aerobic cultures were observed under the microscope. When the culture was sufficiently diluted to allow an appropriate view of bacteria in the field, all the cells were nonmotile. However, if a large number of cells were placed on the slide (for instance, an aliquot from an undiluted mid-log-phase culture), a few swimming cells could be observed. We estimate that motile cells do not represent more than 0.0002% of the entire population, and this ratio does not change even after several subcultures or purification steps. These cells consistently showed a running time several seconds longer than that of the wild-type cells. Nonetheless, when individual cells were followed, stop periods could be detected. These usually lasted for several seconds, although some shorter events were also noticed. As expected from these observations, when SP5/pFliMΔ13 and SP5/pFliMΔ20 were tested in a swarm plate, both strains were unable to form a swarm ring, even after several days of incubation (Fig. 3). These results confirm that this fraction of swimming bacteria does not behave like wild-type cells and suggest that these cells do not represent a different population that may have emerged by a recombination event. Therefore, we consider that the few swimming cells of the SP5/pFliMΔ13 and SP5/pFliMΔ20 cultures might represent spontaneous motor-switching events. This phenomenon has been observed in E. coli cells in the absence of CheY, but only at low temperatures (39). In contrast, we detected this event at room temperature. This may be explained on the basis of structural differences between these two motors, which could favor lower activation energies between the two possible states of the R. sphaeroides motor. In addition, an intracellular metabolite such as fumarate could also contribute to this effect, as has been observed for E. coli (7).
FIG. 3.
Swarm plate showing phenotype of SP5 cells expressing FliMΔ13 or FliMΔ20 protein. WS8 and SP5 cells were included as controls. The plate was incubated for 72 h aerobically in the dark.
Since most of the cells expressing FliM lacking the N terminus were stopped, we propose that this region is essential to promote swimming. It seems plausible that switching between the two possible motor states (i.e., stop and CW) can be mediated by this domain, perhaps through CheY binding.
Analysis of amounts of FliM and flagellin expressed from wild-type, fliMΔ13, and fliMΔ20 cells.
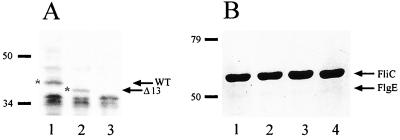
It has been reported for E. coli that a low level of expression of FliM produces nonmotile cells because the C-ring is unable to assemble and a stable export apparatus is probably not formed (18, 35). We know from complementation of strain SP5 with pFliM-wt that the expression level of the fliM gene from the vector promoters is sufficient to restore full motility in these cells (Table 2). However, this result does not rule out the possibility that deletion of the N terminus may alter FliM stability or its ability to form a functional C-ring and therefore an efficient export apparatus. To address the first question, we performed a Western blot experiment to detect the amount of FliMΔ13 protein versus FliM-wt present in SP5 cells. To do this, His-tagged versions of the fliM+ and fliMΔ13 alleles were constructed and cloned into pRK415. These plasmids were introduced into strain SP5, and a cell extract was analyzed with anti-His antibodies. As shown in Fig. 4A, extracts from SP5/pFliM-wt(His) and SP5/pFliMΔ13(His) showed a band of approximately 42.3 and 40.2 kDa, respectively; no specific signal was apparent for the SP5 strain alone. Since similar amounts of FliM-wt and FliMΔ13 were detected and no visible degradation was present, we concluded that FliMΔ13 must be approximately as stable as FliM-wt. It should be noted that the phenotype of the SP5 strain carrying either of these His-tagged versions of FliM is the same as that previously described for untagged FliM proteins.
FIG. 4.
(A) Western blot of SP5 cells expressing FliM-wt(His) and FliMΔ13(His) proteins. Total extracts were subjected to SDS-PAGE, and proteins were transferred to nitrocellulose. The membrane was then blotted with anti-His antibodies. The arrows and asterisks indicate the bands corresponding to FliM-wt (WT) and FliMΔ13 (Δ13). Lane 1, SP5/pFliM-wt; lane 2, SP5/pFliM-Δ13; lane 3, SP5. The positions of molecular mass markers are indicated (in kilodaltons). (B) SDS-PAGE of sheared filaments. Lane 1, WS8 cells; lane 2, SP5/pFliM-wt; lane 3, SP5/pFliMΔ13; lane 4, SP5/pFliMΔ20 cells. Arrows show the band corresponding to flagellin (FliC) and the faint band corresponding to the hook protein (FlgE). Positions of molecular mass markers are indicated.
To obtain indirect evidence that FliMΔ13 and FliMΔ20 are capable of forming a functional C-ring and thereby a functional export apparatus, flagellin was purified by shearing from cultures of SP5 cells expressing FliMΔ13, FliMΔ20, or FliM-wt; a culture of WS8 cells was also included as a control. As shown in Fig. 4B, the flagellin and hook protein yields from all these strains were very similar. This result suggests that FliMΔ13 and FliMΔ20 are both able to form a functional export apparatus.
DISCUSSION
At least in terms of the components described so far, the flagellum of R. sphaeroides is similar to that of E. coli and S. enterica. In contrast, the chemotactic system shows particular characteristics, such as reiteration of several genes, which add complexity to this system. In this work, we present evidence suggesting that the N-terminal region of FliM is essential to promote swimming in R. sphaeroides. This idea is supported by the results from strains expressing either FliMΔ13 or FliMΔ20 protein. These strains consisted mainly of stopped cells and a few swimming cells. We showed evidence that the stopped phenotype is not due to low levels of FliMΔ13 in the cytoplasm. In addition, we observed that the amount of flagellin obtained from cultures of cells expressing FliMΔ13 or FliMΔ20 was similar to that obtained from cultures of WS8 or SP5/pFliM-wt cells. These results allow us to conclude that all the nonswimming cells expressing FliMΔ13 and FliMΔ20 had flagella but were unable to rotate them, indicating a role of the N terminus of FliM in promoting CW rotation.
From studies carried out with FliM from E. coli (FliMEc), it is known that deletion of the first 10 or 38 residues from the N terminus produces flagellated but nonchemotactic cells. This phenotype is due to loss of the main CheY-binding domain on FliM; consequently, flagella rotate exclusively in the CCW direction, producing smooth-swimming cells (25, 37). On the basis of the good similarity observed between FliMRs and FliMEc (10), we expected that deletion of the N-terminal region of FliMRs would not effect the stability of the protein or its ability to assemble the flagellum. Our results confirmed this; however, the stopped phenotype was unexpected. To explain why these deletions affected flagellar rotation, we hypothesized that the N terminus of FliMRs might be the main target for CheY binding, which would be required to promote CW rotation and therefore swimming. In the simpler model, this motor would have two functional states, i.e., stopped and CW rotation, with the stopped state being favored in the absence of CheY binding.
In this work, we also studied the swimming behavior of cells expressing FliM proteins carrying a single-amino-acid substitution in the N-terminal region. As mentioned in the previous section, FliM8LI, FliM9SY, and FliM12EG were selected because each of the corresponding substitutions in E. coli binds CheY to a lesser extent than does FliM-wt. Therefore, when expressed in a fliM mutant strain, they promote smooth swimming (7, 33). In contrast, when R. sphaeroides cells expressed FliM9SY or FliM12EG, an increase in the stop frequency was observed. These changes are less likely to affect the overall secondary structure of FliMRs, therefore, the increase in the stop frequency should be related to the decreased ability of these FliM mutants to bind CheY. Surprisingly, the increased stop frequency for each of these mutants correlated with the relative ability reported for the FliMEc mutants to bind CheY in vitro. Specifically, FliM6LI, FliM10EG, and FliM7SY bound 7-, 11-, and 35-fold less CheY than wild-type FliMEc (7). From these results, we believe that some determinants involved in CheY binding by this domain might be conserved between these organisms. In fact, it can be observed that most of the CheY residues that in S. enterica have been proposed to interact with FliM are conserved in the deduced sequence of the CheY proteins from R. sphaeroides (29).
In summary, from the results obtained with the different FliM mutants isolated in this work, we propose that the N-terminal region of FliMRs is required to promote CW rotation and consequently swimming, perhaps through CheY binding. In terms of the motor, this situation is similar to that found in E. coli and S. enterica, in which binding of CheY-P to FliM induces CW rotation. However, since the helical sense of the flagellar filament in R. sphaeroides is opposite to that of the above-mentioned enteric bacteria, the consequence of CW rotation on the swimming behavior would also be opposite, i.e., running for R. sphaeroides and tumbling for E. coli and S. enterica. Under this view, the default state of these motors (stopped versus CCW) shows a more pronounced difference, which remains to be investigated.
On the other hand, binding of CheY-P to the motor in order to produce swimming has been observed in Bacillus subtilis. In this case, CheA is activated in response to an attractant, so that the final result is net movement of bacteria towards a positive stimulus (6, 11). Whether this is the case for R. sphaeroides is still unknown.
A recent study with R. sphaeroides strains carrying different combinations of deleted cheY genes showed that a certain combination (i.e., cheY1, cheY2, and cheY3) produced a stopped phenotype. However, this phenotype reverted upon the deletion of another cheY gene (cheY4) (30). It should be noted that, given that the different CheY proteins could compete for the interaction with CheA(s) and FliM, it is not possible at this point to establish a direct correlation between our results and those reported by Shah et al. (30).
ACKNOWLEDGMENTS
We thank Blanca Itzel Taboada Ramírez for her contribution to the development of command macro files for automatic bacteria tracking and analysis. We also thank Francisco Caviedes for development of the semiautomatic tracking program and Francisco Dela Mora for technical support.
This work was funded partially by DGAPA grant IN221598 to G.D. and L.C.
REFERENCES
- 1.Armitage J P. Bacterial tactic responses. Adv Microb Physiol. 1999;41:229–289. doi: 10.1016/s0065-2911(08)60168-x. [DOI] [PubMed] [Google Scholar]
- 2.Armitage J P, Macnab R M. Unidirectional, intermittent rotation of the flagellum of Rhodobacter sphaeroides. J Bacteriol. 1987;169:514–518. doi: 10.1128/jb.169.2.514-518.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Armitage J P, Pitta T P, Vigeant M A, Packer H L, Ford R M. Transformations in flagellar structure of Rhodobacter sphaeroides and possible relationship to changes in swimming speed. J Bacteriol. 1999;181:4825–4833. doi: 10.1128/jb.181.16.4825-4833.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley and Sons; 1987. [Google Scholar]
- 5.Barak R, Eisenbach M. Correlation between phosphorylation of the chemotaxis protein CheY and its activity at the flagellar motor. Biochemistry. 1992;31:1821–1826. doi: 10.1021/bi00121a034. [DOI] [PubMed] [Google Scholar]
- 6.Bischoff D S, Ordal G W. Bacillus subtilis chemotaxis: a deviation from the Escherichia coli paradigm. Mol Microbiol. 1992;6:23–28. doi: 10.1111/j.1365-2958.1992.tb00833.x. [DOI] [PubMed] [Google Scholar]
- 7.Bren A, Eisenbach M. The N terminus of the flagellar switch protein, FliM, is the binding domain for the chemotactic response regulator, CheY. J Mol Biol. 1998;278:507–514. doi: 10.1006/jmbi.1998.1730. [DOI] [PubMed] [Google Scholar]
- 8.Francis N R, Irikura V M, Yamaguchi S, DeRosier D J, Macnab R M. Localization of the Salmonella typhimurium flagellar switch protein FliG to the cytoplasmic M-ring face of the basal body. Proc Natl Acad Sci USA. 1992;89:6304–6308. doi: 10.1073/pnas.89.14.6304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Francis N R, Sosinsky G E, Thomas D, DeRosier D J. Isolation, characterization and structure of bacterial flagellar motors containing the switch complex. J Mol Biol. 1994;235:1261–1270. doi: 10.1006/jmbi.1994.1079. [DOI] [PubMed] [Google Scholar]
- 10.Garcia N, Campos A, Osorio A, Poggio S, González-Pedrajo B, Camarena L, Dreyfus G. The flagellar switch genes fliM and fliN of Rhodobacter sphaeroides are contained in a large flagellar gene cluster. J Bacteriol. 1998;180:3978–3982. doi: 10.1128/jb.180.15.3978-3982.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garrity L F, Ordal G W. Activation of the CheA kinase by asparagine in Bacillus subtilis chemotaxis. Microbiology. 1997;143:2945–2951. doi: 10.1099/00221287-143-9-2945. [DOI] [PubMed] [Google Scholar]
- 12.Hamblin P A, Maguire B A, Grishanin R N, Armitage J P. Evidence for two chemosensory pathways in Rhodobacter sphaeroides. Mol Microbiol. 1997;26:1083–1096. doi: 10.1046/j.1365-2958.1997.6502022.x. [DOI] [PubMed] [Google Scholar]
- 13.Irikura V M, Kihara M, Yamaguchi S, Sockett H, Macnab R M. Salmonella typhimurium fliG and fliN mutations causing defects in assembly, rotation, and switching of the flagellar motor. J Bacteriol. 1993;175:802–810. doi: 10.1128/jb.175.3.802-810.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keen N T, Tamaki S, Kobayashi D, Trollinger D. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene. 1988;70:191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
- 15.Khan I H, Reese T S, Khan S. The cytoplasmic component of the bacterial flagellar motor. Proc Natl Acad Sci USA. 1992;89:5956–5960. doi: 10.1073/pnas.89.13.5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khan S, Khan I H, Reese T S. New structural features of the flagellar base in Salmonella typhimurium revealed by rapid-freeze electron microscopy. J Bacteriol. 1991;173:2888–2896. doi: 10.1128/jb.173.9.2888-2896.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khan S, Zhao R, Reese T S. Architectural features of the Salmonella typhimurium flagellar motor switch revealed by disrupted C-rings. J Struct Biol. 1998;122:311–319. doi: 10.1006/jsbi.1998.3999. [DOI] [PubMed] [Google Scholar]
- 18.Kubori T, Yamaguchi S, Aizawa S. Assembly of the switch complex onto the MS ring complex of Salmonella typhimurium does not require any other flagellar proteins. J Bacteriol. 1997;179:813–817. doi: 10.1128/jb.179.3.813-817.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kunkel T A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lloyd S A, Tang H, Wang X, Billings S, Blair D F. Torque generation in the flagellar motor of Escherichia coli: evidence of a direct role for FliG but not for FliM or FliN. J Bacteriol. 1996;178:223–231. doi: 10.1128/jb.178.1.223-231.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lux R, Kar N, Khan S. Overproduced Salmonella typhimurium flagellar motor switch complexes. J Mol Biol. 2000;298:577–583. doi: 10.1006/jmbi.2000.3703. [DOI] [PubMed] [Google Scholar]
- 22.Macnab R M. Flagella and motility. In: Neidhart F C, Curtiss III R I, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: American Society for Microbiology; 1996. pp. 123–145. [Google Scholar]
- 23.Marykwas D L, Berg H C. A mutational analysis of the interaction between FliG and FliM, two components of the flagellar motor of Escherichia coli. J Bacteriol. 1996;178:1289–1294. doi: 10.1128/jb.178.5.1289-1294.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marykwas D L, Schmidt S A, Berg H C. Interacting components of the flagellar motor of Escherichia coli revealed by the two-hybrid system in yeast. J Mol Biol. 1996;256:564–576. doi: 10.1006/jmbi.1996.0109. [DOI] [PubMed] [Google Scholar]
- 25.Mathews M A, Tang H L, Blair D F. Domain analysis of the FliM protein of Escherichia coli. J Bacteriol. 1998;180:5580–5590. doi: 10.1128/jb.180.21.5580-5590.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Packer H L, Armitage J P. The chemokinetic and chemotactic behavior of Rhodobacter sphaeroides: two independent responses. J Bacteriol. 1994;176:206–212. doi: 10.1128/jb.176.1.206-212.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parkinson J S. Complementation analysis and deletion mapping of Escherichia coli mutants defective in chemotaxis. J Bacteriol. 1978;135:45–53. doi: 10.1128/jb.135.1.45-53.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Quandt J, Hynes M F. Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene. 1993;127:15–21. doi: 10.1016/0378-1119(93)90611-6. [DOI] [PubMed] [Google Scholar]
- 29.Shah D S, Porter S L, Harris D C, Wadhams G H, Hamblin P A, Armitage J P. Identification of a fourth cheY gene in Rhodobacter sphaeroides and interspecies interaction within the bacterial chemotaxis signal transduction pathway. Mol Microbiol. 2000;35:101–112. doi: 10.1046/j.1365-2958.2000.01680.x. [DOI] [PubMed] [Google Scholar]
- 30.Shah D S, Porter S L, Martin A C, Hamblin P A, Armitage J P. Fine tuning bacterial chemotaxis: analysis of Rhodobacter sphaeroides behaviour under aerobic and anaerobic conditions by mutation of the major chemotaxis operons and cheY genes. EMBO J. 2000;19:4601–4613. doi: 10.1093/emboj/19.17.4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology. 1983;1:37–45. [Google Scholar]
- 32.Sistrom W R. The kinetics of the synthesis of photopigments in Rhodopseudomonas sphaeroides. J Gen Microbiol. 1962;28:607–616. doi: 10.1099/00221287-28-4-607. [DOI] [PubMed] [Google Scholar]
- 33.Sockett H, Yamaguchi S, Kihara M, Irikura V M, Macnab R M. Molecular analysis of the flagellar switch protein FliM of Salmonella typhimurium. J Bacteriol. 1992;174:793–806. doi: 10.1128/jb.174.3.793-806.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sockett R E, Foster J C A, Armitage J P. Molecular biology of the Rhodobacter sphaeroides flagellum. FEMS Symp. 1990;53:473–479. [Google Scholar]
- 35.Tang H, Blair D F. Regulated underexpression of the FliM protein of Escherichia coli and evidence for a location in the flagellar motor distinct from the MotA/MotB torque generators. J Bacteriol. 1995;177:3485–3495. doi: 10.1128/jb.177.12.3485-3495.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tang H, Braun T F, Blair D F. Motility protein complexes in the bacterial flagellar motor. J Mol Biol. 1996;261:209–221. doi: 10.1006/jmbi.1996.0453. [DOI] [PubMed] [Google Scholar]
- 37.Toker A S, Kihara M, Macnab R M. Deletion analysis of the FliM flagellar switch protein of Salmonella typhimurium. J Bacteriol. 1996;178:7069–7079. doi: 10.1128/jb.178.24.7069-7079.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Toker A S, Macnab R M. Distinct regions of bacterial flagellar switch protein FliM interact with FliG, FliN and CheY. J Mol Biol. 1997;273:623–634. doi: 10.1006/jmbi.1997.1335. [DOI] [PubMed] [Google Scholar]
- 39.Turner L, Caplan S R, Berg H C. Temperature-induced switching of the bacterial flagellar motor. Biophys J. 1996;71:2227–2233. doi: 10.1016/S0006-3495(96)79425-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Turner L, Ryu W S, Berg H C. Real-time imaging of fluorescent flagellar filaments. J Bacteriol. 2000;182:2793–2801. doi: 10.1128/jb.182.10.2793-2801.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Welch M, Oosawa K, Aizawa S, Eisenbach M. Phosphorylation-dependent binding of a signal molecule to the flagellar switch of bacteria. Proc Natl Acad Sci USA. 1993;90:8787–8791. doi: 10.1073/pnas.90.19.8787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wolfe A J, Conley M P, Kramer T J, Berg H C. Reconstitution of signaling in bacterial chemotaxis. J Bacteriol. 1987;169:1878–1885. doi: 10.1128/jb.169.5.1878-1885.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamaguchi S, Aizawa S, Kihara M, Isomura M, Jones C J, Macnab R M. Genetic evidence for a switching and energy-transducing complex in the flagellar motor of Salmonella typhimurium. J Bacteriol. 1986;168:1172–1179. doi: 10.1128/jb.168.3.1172-1179.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao R, Amsler C D, Matsumura P, Khan S. FliG and FliM distribution in the Salmonella typhimurium cell and flagellar basal bodies. J Bacteriol. 1996;178:258–265. doi: 10.1128/jb.178.1.258-265.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao R, Pathak N, Jaffe H, Reese T S, Khan S. FliN is a major structural protein of the C-ring in the Salmonella typhimurium flagellar basal body. J Mol Biol. 1996;261:195–208. doi: 10.1006/jmbi.1996.0452. [DOI] [PubMed] [Google Scholar]