Abstract
Potent immunosuppressive drugs have been introduced into clinical care for solid organ transplant recipients. It is now time to guide these drugs on an individual level to optimize their efficacy. An ideal tool simultaneously detects overimmunosuppression and underimmunosuppression, is highly standardized, and is straightforward to implement into routine. Randomized controlled interventional trials are crucial to demonstrate clinical value. To date, proposed assays have mainly focused on the prediction of rejection and were based on the assessment of few immune compartments. Recently, novel tools have been introduced based on a more integrated approach to characterize the immune function and cover a broader spectrum of the immune system. In this respect, the quantification of the plasma load of a highly prevalent and apathogenic virus that might reflect the immune function of its host has been proposed: the torque teno virus (TTV). Although TTV control is driven by T cells, other major immune compartments might contribute to the hosts’ response. A standardized in-house polymerase chain reaction and a conformité européenne-certified commercially available polymerase chain reaction are available for TTV quantification. TTV load is associated with rejection and infection in solid organ transplant recipients, and cutoff values for risk stratification of such events have been proposed for lung and kidney transplantation. Test performance of TTV load does not allow for the diagnosis of rejection and infection but is able to define at-risk patients. Hitherto TTV load has not been used in interventional settings, but two interventional randomized controlled trials are currently testing the safety and efficacy of TTV-guided immunosuppression.
CLINICAL PROBLEM AND INTRODUCTION
Allograft transplantation (TX) is the preferred treatment for patients with end-stage solid organ disease. Immunosuppressive drugs are crucial for reducing the risk of organ rejection. Despite this desired effect, the compromised immune system of the recipient increases the risk for infectious and oncologic disease. Moreover, current immunosuppression regimens are unable to control chronic subclinical allorecognition of the graft, which leads to graft damage and loss.1,2 Thus, the optimal management of immunosuppressive drug dosing requires a delicate balance between inadequate and excessive immunosuppression. At present, there is no diagnostic test or algorithm for the optimal guidance of immunosuppressive drugs.3,4 Monitoring in routine post-TX care largely relies on the quantification of calcineurin inhibitor through levels in the peripheral blood, which correlate more closely with the risk of drug-related toxicity than with the effectiveness of immunosuppression.5 Thus, there is an urgent need for tools to personalize immunosuppression to reduce the risk of infectious and oncologic disease as well as graft rejection.
A broad range of assays for immunologic monitoring has been proposed in recent decades.6-8 However, only one noninvasive test system has been broadly accepted for routine post-TX care and implemented in diagnostic guidelines: donor-specific antibodies (DSAs).9,10 Notably, DSAs are associated with antibody-mediated rejection (AMR) only and are not applicable for the detection of T cell-mediated rejection (TCMR), which is the dominant rejection type in the early phase after TX.11 Moreover, DSAs are not useful to assess susceptibility to infections or other consequences of excessive immunosuppression. Indeed, most assays analyzing blood and urine for immunologic monitoring currently under investigation—including cell-free donor-derived DNA,12,13 chemokines, gene expression- and proteomics-based assays, and Enzyme-linked-immuno-Spot6,8,14—focus on the prediction of graft rejection only. However, some biomarkers—including immunoglobulins, leukocyte subsets, components of the complement system, and viral nucleic acids—have been proposed for the prediction of infections in solid organ transplantation (SOT).7 Notably, the value of any immunologic monitoring tool—including DSA—has not been sufficiently proven in an interventional randomized controlled clinical setting.4
The ideal marker for the guidance of immunosuppressive drugs would simultaneously predict the consequences of both overimmunosuppression and underimmunosuppression. To address this need, two commercially available assays have been proposed: a test of leucocyte function known as the QuantiFERON Monitor (Qiagen, Germany; now T-SPOT.PRT, Oxford Immunotec, United Kingdom),15 and the assessment of lymphocyte function using the ImmuKnow (ImmuKnow; Cylex, Germany; now Eurofins Viracor, United States).16 The latter has been tested in an interventional randomized controlled setting, with test results suggesting a reduction of infectious events in liver TX (LTX) patients treated by ImmuKnow-guided immunosuppression. However, the trial design warrants careful interpretation of these preliminary data. Recently, a more functional and holistic assessment of the immune system has proposed the assessment of virus-specific T cells (Tvis) as a marker of immunosuppression. In a phase II trial of Tvis-guided immunosuppression in pediatric recipients of a kidney allograft, no major safety signals were noted.17 However, the complex laboratory technique required for Tvis analysis might complicate a broad implementation in routine post-TX care.
Monitoring torque teno virus (TTV) in the peripheral blood is a promising new strategy to characterize the immune function. TTV can be detected in up to 90% of healthy individuals and has not been linked to any human disease.18 The prevalence of TTV in immuno-compromised patients after TX is up to 100% and the virus is unaffected by conventional antiviral drug therapies used in the post-TX setting.18 TTV copy number is directly associated with the amount and type of immunosuppressive drugs administered to TX recipients and additional major factors determining the immune function of its host (eg, age and sex); thus, it is indirectly associated with graft rejection and infectious disease.18 Two randomized controlled interventional trials are currently investigating the value of TTV-guided immunosuppression19-21 in kidney (KTX) and lung transplant (LuTX) recipients.
With novel and innovative tools to characterize the immune status potentially entering post-TX routine care in the next years, a review covering these assays is both timely and essential. Due to the differences in applied test systems and study designs as well as the paucity of high-quality prospective trials, it is not reasonable to perform a quantitative systemic review. We aimed to cover novel approaches for comprehensive immunologic monitoring by means of a narrative review with a clear focus on TTV.
NOVEL CONCEPTS
An ideal marker for the guidance of immunosuppressive drugs is able to simultaneously predict the consequences of both overimmunosuppression and underimmunosuppression, allows for comparability across assessment centers, and is supported by interventional studies based on biomarker-guided drug dosing.
In this respect, a commercially available assay measuring the concentration of ATP from CD4+ cells after stimulation (ImmuKnow) has been proposed. Three meta-analyses, including noninterventional studies, have been published: one demonstrating a benefit for immunosuppression guided by ImmuKnow in SOT22 and two with a negative result.23,24 Consequently, the ImmuKnow assay was tested in a single-center randomized controlled trial including 202 LTX recipients.16 In the intervention group, the tacrolimus dose was reduced if the ImmuKnow values were low and vice versa. Infection events after the first 2 wk post-TX were lower in the interventional group, and there was no difference in acute rejection. The trial design complicates a reliable analysis concerning safety and efficacy: (1) multiple endpoints were used, (2) no sufficientdefinitions of infection and rejection were provided, (3) no assessor blinding was performed, (4) uncertainty about the impact of “clinical experience” on ImmuKnow-guided tacrolimus dosing exists, and (5) a post hoc analysis (exclusion of the first 2 weeks post-TX) was performed.
Recently, a broader assessment of the immune function—a commercially available test of interferon gamma in whole blood after T cell and Toll-like receptor stimulation, known as the QuantiFERON Monitor (Qiagen)—was tested in a single-center cohort study.15 A total of 151 KTX, LTX, LuTx, and small bowel TX recipients were enrolled. Notably, no statement on blinding was available and no primary outcome was predefined. Patients with an infection had lower interferon gamma values detected in the samples drawn before the event compared with those without infection. However, potential confounders and effect modifiers were not considered. No differences were observed concerning rejections. Notably, such finding might have been overlooked due to the different levels of immunosuppression required to prevent rejection in the 4 types of SOT included. Despite these encouraging preliminary results, no further prospective study or interventional trial testing the value of QuantiFERON Monitor-guided drug dosing are currently registered.
A more comprehensive assessment of the immune system might be feasible by assessing the ability of the immune system to control opportunistic viruses. A single-center cohort study analyzed 383 KTX recipients.25 Epstein-Barr virus (EBV) viremia was more frequent among patients experiencing at least 1 opportunistic infection (including zoster, Pneumocystis jirovicii, aspergillosis, legionellosis, tuberculosis, and nocardiosis). No primary outcome was predefined, multiple testing was performed, and no sufficient effect size adjustment was performed. Moreover, the association between EBV viremia and infections might have been disguised due to the per-protocol decrease in immunosuppression in the case of EBV polymerase chain reaction (PCR) >4 log10 copies/milliliter (c/mL) in two consecutive measurements. Notably, EBV-based monitoring might be complicated due to insufficient post-TX prevalence, reactivity towards the antiviral drugs routinely used in the post-TX setting, and the association with disease requiring EBV infection treatment (eg, lymphoma).
Expanding upon the idea of a functional readout for the characterization of the immune system, a German 4-centered, open-label, randomized controlled trial involving 64 pediatric KTX recipients tested the steering of immunosuppressive therapy by levels of virus-specific CD4+ cells (Tvis) directed towards adenovirus, cytomegalovirus (CMV), and herpes simplex virus.17 In the active group, the dosage of immunosuppressive drugs was decreased if Tvis were low and vice versa. Tvis were analyzed in 4 steps over a total of 6 h, including (1) stimulation, (2) fixation, (3) immunostaining, and (4) flow-cytometry. No difference in the estimated glomerular filtration rate (cystatin C based on Schwartz and Filler) in the intention-to-treat population 24 mo post-TX was observed (adjusted mean difference: 1.7; 95% confidence interval [CI], −10.2 to 13.6). In the intervention group, dose reductions of cyclosporine and everolimus following Tvis levels were performed in 28 of 31 patients, while dose increases were performed for 2 patients. In the intervention group, lower drug trough levels and doses of everolimus and cyclosporine were noted. Additionally, more patients were free of glucocorticoid treatment. In the intervention group, 11 acute rejection episodes (including borderline changes suspicious for acute TCMR) were documented in comparison with 19 rejections in the control group. Moreover, fewer patients with EBV viremia were detected in the interventional group. The overall numbers of adverse events in the intervention and control groups were comparable. Considering the limitations of an open-label protocol with a surrogate endpoint in combination with a short-term follow-up, this trial provides evidence for the potential of Tvis measurements to detect patients in which a reduction in immunosuppressive therapy might be feasible without an increased risk of rejection. These unique and promising data justify a phase III randomized controlled trial to determine the efficacy of Tvis-guided immunosuppression. Notably, the complexity of Tvis monitoring might pose an obstacle for further large-scale efficacy trials and introduction into clinical routine.
TTV—AN INTRODUCTION
Recently, a highly prevalent and nonpathogenic virus has been introduced for immunologic monitoring in SOT recipients. TTV is a small nonenveloped DNA virus that was discovered in Japan in 1997.26 TTV contains a single-stranded circular DNA of negative polarity that is approximately 3800 bases in length with at least 4 overlapping open reading frames (ORFs).27 This small genome exhibits a strikingly high genomic diversity.28,29 All TTV sequences known to date can be phylogenetically grouped into 21 distinct TTV species under the genus Alphatorquevirus of the family of Anelloviridae. Notably, a taxonomic update of the Anellovirus classification was recently published.30
TTV is highly prevalent in a wide variety of mammalian species.31 In the human population, TTV-DNA is persistently detectable in the peripheral blood of up to 95% of healthy persons throughout their lifetime.32-36 TTV, together with Beta- and Gammatorqueviruses, are considered the most abundant eukaryotic viruses in the human virome.28,29,37 TTV was identified in blood and other samples taken from various body sites of healthy persons, as well as in environmental samples, which highlights its omnipresence.38 The main TTV transmission routes in humans are thought to be fecal-oral and airway-mediated. Notably, initial infections seem to occur very early in life.37 It is believed that TTV mainly replicates in T cells; however, as a polytrophic virus, its DNA was detectable in all leukocyte subsets and many other cell types.39-42 TTV is a very actively replicating virus, and it has been estimated that >10 log10 c/mL of virions are generated per day in a healthy human body, with >90% of these being cleared by the immune system. Consequently, in cases with detectable TTV, viral load varies over time but remains at approximately 2 log10 to 8 log10 c/mL.40,43-45 Multiple TTV strains can accumulate in a human host, which may be acquired simultaneously or serially over time, thereby leading to a mixture of TTV strains copersisting.46-48
In healthy immunocompetent individuals, plasma TTV levels are maintained at a well-balanced steady state that is controlled by host immunity.49 However, only limited data exist on the specific immune response toward TTV as well as on the evasion mechanisms of TTV. Various immune compartments have been suggested to contribute to virus control: TTV-specific IgM and IgG antibodies directed against ORF1- and ORF2-encoded proteins can be found in TTV-positive individuals.50-52 The high TTV loads detected in SOT recipients with interleukine-2 signal blocking immunosuppression suggest T cells being crucial for the virus control.18 Evidence of the contribution of natural killer cells and antigen-presenting cells has also been provided.53 Disease-modifying drugs (eg, anti-CD20) have been shown to increase TTV load in patients with rheumatoid arthritis, thereby suggesting that B cells contribute to TTV control.54 Toll-like receptor TTV antigen-recognition55 and interferon-α has been suggested to limit TTV replication.56-58 Viral immune evasion is partly based on the ability of ORF2 to interfere with the host’s inflammatory response via the suppression of NF-κB translocation.59 TTV particles circulating in exosomes being less exposed to neutralizing antibodies might serve as another immune evasion mechanism.60
Due to the lack of a cell culture or well-established serological assay, the diagnosis of TTV infection is based on molecular methods. The measurement of TTV-DNA via quantitative PCR represents a sensitive and rapid method that has been applied to testing various samples, including serum and whole blood. Of note, TTV load in whole blood is >1 log level higher compared with plasma because of the high viral loads in leukocytes.61 Currently, an in-house PCR developed by Maggi et al62 and a commercially available PCR—both optimized and standardized for plasma—are mostly in use.45 Despite high sequence diversity among TTV species, there is a short conserved ≈150-base sequence stretch at the 5′ nontranslated region of the TTV genome. This region is suited for the detection and quantification of most—if not all—TTV species known to date.63 However, caution should be exercised when plasma TTV loads quantified in different laboratories and by different PCR assays, respectively, are compared.45 TTV quantified by the in-house PCR produces results, which might differ up to 2 log10 levels between laboratories. In this respect, it is important to note that these differences are constant and thus linear across the whole range of TTV load; therefore, results from different labs are comparable. A standardization process has been performed via the External Quality Assessment pilot study by Quality Control for Molecular Diagnostics, with accuracy in TTV quantification being observed in all participating laboratories.64 Besides the in-house PCR, a commercially available PCR (Real-time detection and quantification kit, TTV R-GENE; bioMérieux SA, France) was introduced and conformité européenne-certified for routine clinical application in 2021.45 In the same year, an EU-funded project (TTVguideTX) was initiated to implement, harmonize, and quality control this assay (in addition to the necessary process already performed during the conformité européenne certification) in 13 TX centers across Europe in preparation for a randomized controlled trial.19,65 First results from the TTVguideTX project show low intercenter variability (data under review).
TTV is considered a nonpathogenic virus, and no substantial association with clinical symptoms has been detected to date.66,67 The virus is unaffected by conventional antiviral drug therapies68 and highly resistant to inactivation procedures.18 Most importantly, the TTV load in peripheral blood might mirror the immune status of its host. A group at Stanford was the first to demonstrate an association between TTV load and allograft rejection in SOT.68 Subsequently, other groups verified this finding and additionally provided evidence for the association between TTV load and infection.69 TTV load was shown to directly associate with the amount and type of immunosuppressive drugs and thus with allograft rejection and infectious disease.24,68 TTV is also associated with other major determinants of immune function, including age and sex of the host.18 In addition to infectious events in SOT, peaks of TTV replication have been observed to occur during solid cancer growth, and associations have been described between TTV load and the use of chemotherapy.70,71 In patients infected with HIV, TTV load was predictive for the course of immune recovery.72 Recently, an association has been shown between TTV load and response to severe acute respiratory syndrome coronavirus type 2 vaccination in KTX recipients.73 According to these findings, the central hypothesis of immune monitoring using TTV in SOT has formed: if the immune system is strong, the TTV load is low; this indicates a risk for organ rejection. If the immune system is weak, the TTV load is high; this indicates a risk for infection.
TTV IN KIDNEY TRANSPLANTATION
The majority of the reports on the value of TTV for immunologic monitoring in SOT have been focused on KTX. Reproducible evidence has been provided for an association between TTV load and rejection and infection, respectively (Table 1). Notably, all existing studies were observational and from single centers, and only half of them followed a prospective design. Moreover, some of their results must be interpreted with caution due to potential biases in study design, including selection bias, small numbers of events, post hoc analyses, multiple testing, and insufficient effect size adjustments (Tables 1 and 2). However, sufficient evidence exists for a linear, robust, and independent association between TTV load and all types of clinically overt and subclinical rejection, including TCMR and AMR,21,74-79 as well as infectious events,21,74,75,79-85 including all common posttransplant pathogens in adult KTX recipients (opportunistic infections, CMV, BK polyomavirus [BKV], and bacterial infections), respectively. For easier interpretation of the studies presented next, we included the section Application of TTV Cutoff Values in Clinical Routine of Kidney Transplant Care and Table 3. Therein, we converted all relevant TTV cutoff values to correspond a commercially available PCR. With only two studies available in pediatric KTX,85,86 this review will focus on adult cohorts.
TABLE 1.
Studies that evaluated the association between TTV load and allograft rejection in kidney transplant recipients
| Study designa | TX period | Included patients | Endpoint; timing | Total BX; BX proven rejection | PCR | Main association | Limitationsb |
|---|---|---|---|---|---|---|---|
| Cohort74 | 2014–2016 | 221 | Clinically overt rejection; <3 mo post-TX | 10c | C | TTV at TX–rejection | Secondary endpoint; BX not mandatory; multiple testing; missing information on model design/some major determinants of TTV not included |
| Cohort75 | 2016–2018 | 37 | Rejection (iBX); months 4–12 post-TX | 39; 11 | IH | TTV 2 wk before BX–TCMR, AMR, mixed | Limited number of events |
| Cohort76 | 2016–2018 | 82 | Rejection (pBX); month 12 post-TX | 82; 19 | IH | TTV at BX–TCMR, AMR | High loss to follow-up |
| Cross-sectional77 | 1973–2014 | 715 | Rejection (pBX); 6 y post-TX | 86; 46 | IH | TTV at BX–AMR | Cross-sectional design; possible selection bias due to missing BX in DSA-positive subjects |
| Case-control78 | 2012–2017 | 113 | Rejection (iBX); months 4–12 post-TX | 113; 33 | IH | TTV 1 mo before BX–TCMR, AMR | Case-control design |
| Case-control79 | 2012–2014 | 63 | Rejection (iBX); <2 y post-TX | 12d | C | TTV pre-TX–TCMR, AMR, mixed | Possible selection bias; nonrejection not BX proven; multiple testing; no effect size adjustment |
| Case-control21 | 2003–2013 | 389 | Clinically overt rejection; <12 mo post-TX | 80; 54e | IH | TTV kinetic–time to rejection | Secondary endpoint; BX not mandatory; possible misclassification of rejection: nonrejection in BX categorized as rejection |
The studies are listed according to the design and date of their online publication.
All studies followed a noninterventional and single-center design.
The total number of biopsies was not stated; 11 events were scored as rejection.
The total number of biopsies was not stated; 14 biopsies were available for the posttransplant month 1 evaluation.
Eighty-eight events were scored as rejection.
AMR, antibody-mediated rejection; BX, biopsy; C, commercial; DSA, donor-specific antibody; iBX, indication biopsy; IH, in-house; pBX, protocol biopsy; PCR, polymerase chain reaction; TCMR, T cell–mediated rejection; TTV, torque teno virus; TX transplantation.
TABLE 2.
Studies that evaluated the association between TTV load and infection in kidney transplant recipients
| Study designa | TX period | Included patients | Endpoint; timing | Patients with event; infectious events | PCR | Main association | Limitationsb |
|---|---|---|---|---|---|---|---|
| Cohort80 | 2016 | 71 | Infection leading to medical measure; months 4–12 post-TX | 22; 41 | IH | TTV 1 mo before event–infection | Interim analysis; secondary endpoint |
| Cohort74 | 2014–2016 | 221 | Infection leading to medical measure/opportunistic infection + malignancy; <12 mo post-TX | 51; 65 | C | TTV 1 mo post-TX–subsequent event | Two main endpoints; multiple testing; missing information on model design/some major determinants of TTV not included |
| Cohort81 | 2015–2016 | 116 | BKV viremia; <12 mo post-TX | 24; 24 | C | NA | Multiple testing; no effect size adjustment |
| Cohort75 | 2016–2018 | 274 | Infection leading to medical measure; months 4–12 post-TX | 127; 193 | IH | TTV 1 mo before event–infection | Secondary endpoint |
| Case-control82,c | 2011–2016 | 145 | CMV viremia; <4 mo post-TX | 35; 35 | IH | TTV days 0 to 10 post-TX–CMV | Possible selection bias; main analyses include LTX; multiple testing; no effect size adjustment |
| Case-control79 | 2012–2014 | 66 | BKV viremia; <2 y post-TX | 50; 50 | C | TTV–BKV month 6 post-TX | Possible selection bias; multiple testing; no effect size adjustment |
| Case-control83 | 2014–2016 | 215 | BKV viremia; <12 mo post-TX | 47; 47 | C | TTV 1 mo post-TX–subsequent BKV | No data on subject selection; multiple testing; missing information on model design/some major determinants of TTV not included |
| Case-control21 | 2003–2013 | 389 | BKV and CMV viremia; <12 mo post-TX | 182; 105/77d | IH | TTV kinetic–time to infection | CMV secondary endpoint |
The studies are listed according to their design and date of their online publication.
All studies followed a noninterventional and single-center design.
Kidney and liver transplant recipients.
One hundred five BKV and 77 CMV.
BKV, BK polyomavirus; C, commercial; CMV, cytomegalovirus; IH, in-house; LTX, liver transplantation; NA, not available; PCR, polymerase chain reaction; TTV, torque teno virus; TX, transplantation.
TABLE 3.
Proposed plasma TTV load cutoff values determined in kidney transplant recipients for the risk prediction of allograft rejection and infection, respectively
| Citation | Event; timing | Predictor | TTV cutoffa | AUC | Sensitivity | Specificity | PPV | NPV |
|---|---|---|---|---|---|---|---|---|
| Rejection | ||||||||
| 77 | AMR (pBX); 6 y post-TX | TTV at BX | <3.6 log10 | NA | NA | NA | NA | NA |
| 79 | Rejection; <2 y post-TX (iBX) | TTV pre-TX/TTV 1 mo post-TX | <3.4 log10 | NA | NA | NA | 0.63 | 0.92 |
| <4.2 log10 | 0.48 | 0.92 | ||||||
| 75 | Rejection (iBX); months 4–12 post-TX | TTV 2 wk before BX | <4.6 log10 | 0.73 | 0.36 | 0.89 | 0.56 | 0.77 |
| 76 | Rejection (pBX); month 12 post-TX | TTV at BX | <4.6 log10 | NA | 0.63 | 0.51 | 0.27 | 0.82 |
| Infection | ||||||||
| 82 | CMV viremia; <4 mo post-TX | TTV days 0 to 10 post-TX | >3.8 log10 | 0.72 | 0.83 | 0.56 | NA | NA |
| 74 | Infection/opportunistic infection or malignancy; <12 mo post-TX | TTV 1 mo post-TX | >3.2 log10 | 0.62 | 0.90 | 0.31 | 0.54 | 0.77 |
| >4.6 log10 | 0.70 | 0.76 | 0.66 | 0.41 | 0.90 | |||
| 83 | BKV viremia; <12 mo post-TX | TTV 1 mo post-TX | >5.0 log10 | 0.75 | 0.77 | 0.75 | 0.31 | 0.96 |
| 84 | Death due to infectious cause | TTV 5 y post-TX | >3.4 log10 | NA | 0.55 | 0.67 | NA | NA |
| 75 | Infection; months 4–12 post-TX | TTV 1 mo before infection | >6.6 log10 | 0.62 | 0.41 | 0.76 | 0.36 | 0.80 |
To facilitate comparison of the proposed TTV cutoffs, values have been converted to values that correspond to the commercial PCR.
AMR, antibody-mediated rejection; AUC, area under the curve; BKV, BK polyomavirus; BX, biopsy; CMV, cytomegalovirus; iBX, indication biopsy; NA, not available; NPV, negative predictive value; pBX, protocol biopsy; PPV, positive predictive value; TTV, torque teno virus; TX, transplantation.
TTV and Kidney Transplant Rejection
An association between TTV load and rejection in KTX recipients was first described in a cross-sectional study by the Vienna Group.77 A total of 1165 KTX recipients were subjected to screening for AMR and TTV, and 86 DSA-positive patients were subjected to a protocol biopsy. A total of 46 were diagnosed with AMR (median AMR diagnosis: 6 y posttransplant).87 TTV load at the time of biopsy was lower in AMR-positive recipients when compared with patients who had no AMR (6.6 × 104 c/mL, interquartile range [IQR], 3.0 × 103–7.2 × 105 versus 2.6 × 105 c/mL, IQR, 2.2 × 104–2.1 × 106). Statistical tests, including multivariate analysis, revealed a robust and independent linear association between TTV load and AMR (risk ratio, 0.94, 95% CI, 0.90-0.99).
Subsequently, the Vienna Group provided evidence for an association between acute rejection detected upon indication biopsies and TTV load in a case-control study,78 screening 1010 consecutive renal allograft recipients. Inclusion criteria were an indication biopsy performed between months 4 and 12 post-TX and adequately stored blood samples for retrospective TTV quantification taken between month 4 post-TX and the date of the transplant biopsy. The median time between TTV quantification and biopsies was 43 d. Patients with rejection (n = 33; 14 AMR and 19 TCMR) had lower levels of TTV, with a median of 3.1 × 107 c/mL (IQR, 4.9 × 105–2.3 × 108 c/mL) compared with patients without rejection (n = 80; 2.3 × 108 c/mL, IQR, 1.4 × 107–3.6 × 109 c/mL). The risk for rejection decreased by 11% per log level increase in TTV load (risk ratio, 0.89, 95% CI, 0.82-0.96) in a multivariate model, including all potential confounders selected based on background knowledge.
More evidence on the association between TTV and rejection was provided by the Strasbourg Group in a case-control study.79 A total of 14 patients experienced rejection: 6 TCMR, 2 AMR, and 6 mixed rejections. Patients without biopsy were categorized as rejection-negative. The TTV loads pretransplant and at month 1 posttransplant were lower in patients who subsequently developed graft rejection than in graft rejection-free patients. TTV loads of <3.4 log10 c/mL (negative predictive value [NPV] = 92%, positive predictive value [PPV] = 63%) and <4.2 log10 c/mL (NPV = 92% and PPV = 48%), respectively, were suggested as cutoff values for risk stratification.
Recently, a case-control study by the Leiden Group linked TTV load to rejection21 screening 519 recipients. A total of 88 recipients were categorized as rejection positive within 1 y after TX: 80 had undergone a biopsy, and 54 had clear histological evidence for rejection. The patients classified as rejection-negative were not subjected to biopsy. A predictive linear mixed-effects model showed a decreased risk of rejection with increasing TTV load (hazard ratio [HR], 0.74 per logTTV c/mL, 95% CI, 0.71-0.76).
Evidence of an association between acute rejection and TTV load was first provided in a prospective setting by the Madrid Group.74 The study was designed to test the association between TTV and infections, and organ rejection was the secondary endpoint. A cohort of 221 patients was analyzed, and 10 showed biopsy-proven rejection in the first 3 mo posttransplant. A biopsy for the diagnosis of rejection was not mandatory but relied on clinical course. The patients classified as “no rejection” were not subjected to biopsy. Multivariate analysis revealed an association between baseline TTV and rejection (HR per logTTV 0.69, 95% CI, 0.49-0.97).
The value of TTV in clinically overt rejection was also evaluated by the Vienna Group in an observational cohort study including 386 consecutive adult kidney graft recipients.75 All TTV measurements taken after TTV load stabilization at the end of post-TX month 3 with an available subsequent for cause biopsy (ie, the primary endpoint) were analyzed. Samples for TTV quantification were taken at a median of 154 d after TX and preceded subsequent biopsies, with a median of 14 d. Of the 39 biopsies, 11 showed signs of allograft rejection: 5 TCMR, 2 AMR, and 4 mixed. Patients with allograft rejection had lower levels of TTV compared with patients without rejection (3.5 × 106 c/mL, IQR, 1.7 × 105–1.3 × 108 c/mL versus 2.5 × 108 c/mL, IQR, 5.8 × 106–9.3 × 108 c/mL) in subsequent biopsies. The odds for rejection decreased by 22% with every log level increase of TTV (odds ratio, 0.78, 95% CI, 0.62-0.97). An area under the curve (AUC) of 0.73 (IQR, 0.54–0.92) was calculated to classify rejection by TTV level. A TTV load cutoff of 1.5 × 106 c/mL corresponded to a specificity of 89%, sensitivity of 36%, NPV of 77%, and a PPV of 50%. Multivariate testing found no confounding variables.
Recently, the association of TTV and subclinical rejection was observed in a cohort study from the Vienna Group with a 1 y protocol biopsy.76 The primary outcome (ie, allograft rejection) was diagnosed in 19 of the 82 available cases (15 TCMR and 4 AMR). Patients with rejection had lower TTV loads when compared with patients without rejection (2 × 105 c/mL, IQR, 3 × 103–2 × 106 versus 7 × 105 c/mL, IQR, 1 × 105–2 × 107). A multivariate analysis demonstrated an independent inverse association between TTV and rejection (risk ratio, 0.92, 95% CI, 0.86-0.98). The study also demonstrated that an increase in chronic graft damage between the month 3 and month 12 protocol biopsies was associated with the number of days with a TTV load <106 c/mL within the same period of time (coefficient: 0.07, 95% CI, 0.01-0.14).
TTV and Infection in Kidney Transplant Recipients
In the interim analysis80 of the full data set described in detail previously,75 the Vienna Group was the first to provide evidence for the association between TTV and infection in a prospective setting. Infection was the secondary endpoint of the study and defined as any bacterial, fungal, or viral infection requiring antimicrobial or antiviral treatment, reduction of immunosuppressive drugs, hospitalization, or prolongation of hospital stay. All patients who continued to be followed after month 3 posttransplant and had a TTV infection were included in the analysis. For these patients, TTV was quantified at 785 time points. TTV measurements were followed by an infectious event in 193 of the observed periods in 127 patients, whereas no infectious event was documented 592 times. TTV was quantified over a median of 27 d before the onset of infection. The likelihood of infection increased by 11% with every log level increase in TTV (odds ratio, 1.11, 95% CI, 1.06-1.15). A comparable effect size was described for infections that did not require hospitalization. The largest effect size was calculated for BKV infections, followed by CMV disease and infections restricted to opportunistic pathogens. A smaller effect size was found for infections with extracellular bacteria. An AUC of 0.62 (IQR, 0.58–0.67) was calculated when classifying infections by TTV level. A TTV level >5.8 × 109 c/mL corresponded to a specificity of 90%, sensitivity of 18%, NPV of 77%, and a PPV of 37% to detect infection.
In the same year, the Pisa Group provided further evidence on the association between CMV viremia and TTV in a case-control study including 145 patients.82 CMV infection was defined as the presence of viral DNA above the detection threshold (800 viral genomes/mL of whole blood) within the first 4 mo posttransplant. TTV load between days 0 and 10 after TX was higher in the CMV-positive patients (n = 35) when compared with CMV-negative patients (n = 110). The optimal cutoff value for TTV load to detect CMV reactivation in the first 10 d was determined as 3.5 log10 c/mL (sensitivity: 83% and specificity: 56%) Similar differences were observed when subjects were stratified by the type of transplanted organ.
Additional evidence of an association between infection and TTV load was provided by a trial of the Madrid Group discussed in detail previously.74 The authors defined two primary outcomes: infection (need for hospitalization and intravenous antimicrobial therapy) and immunosuppression-related adverse events (iRAE; occurrence of opportunistic infection: intracellular bacteria, herpesviruses [eg, CMV disease], BKV [PVAN and presumptive PVAN], invasive yeasts/molds, parasites, and posttransplant de novo malignancy). A total of 51 patients had 65 episodes of iRAE. Upon analyzing TTV at discrete time points, associations between subsequent study outcomes were detected at months 1, 3, and 6 (only iRAE) but not on day 0, day 7, and month 12 posttransplant. The AUCs for diagnosing infection and iRAE via TTV load at month 1 were 0.62 (95% CI, 0.52-0.73) and 0.70 (95% CI, 0.59-0.82). After applying cutoff values of 3.2 log10 c/mL and 4.5 log10 c/mL for infection and iRAE, respectively, a sensitivity of 90% and specificity of 31% were calculated to detect infection, while a specificity of 76% and sensitivity of 66% were calculated to detect iRAE beyond month 1 posttransplant. The association between TTV and infection/iRAE remained significant after multivariate adjustment. The authors also analyzed TTV AUCs and doubling times and described associations with the endpoints. However, the complexity of calculation-based cutoffs using longitudinal data might limit the implementation in clinical practice.
Possible limitations of TTV quantified in the early phase of post-TX were reported in a cohort study from the Amiens Group.81 Overall, 24 of the 116 patients (21%) had positive plasma BKV during the first year after TX. TTV load at months 1, 2, and 3 post-TX were not associated with BKV viremia. Limited evidence on the association between TTV and BKV was provided from a study by the Strasbourg group, as reported in detail here previously.79 A total of 50 patients with detectable levels of BKV and 16 without were selected. Compared with BKV-negative recipients, TTV loads were only higher at month 6 post-TX in patients with BKV replication. More evidence for an association between TTV and BKV was provided in a case-control study including 215 patients by the Madrid Group.83 TTV load at month 1 posttransplant was higher among patients developing BKV viremia (n = 47) thereafter when compared with those who remained free from BKV viremia. For TTV loads >5.0 log10 c/mL at posttransplant month 1, a sensitivity of 77%, specificity of 75%, positive predictive value of 31%, and negative predictive value of 96% to predict subsequent BKV viremia were calculated. A study by the Leiden Group described in detail here analyzed the association between TTV and BKV (primary endpoint) and CMV viremia (secondary endpoint), respectively.21 Of the 389 recipients, 27% (n = 105) developed BKV viremia, and 20% (n = 77) developed CMV viremia within 1 y after TX. A predictive linear mixed-effects model showed an increased risk of viremia with increasing TTV (HR, 1.03 per logTTV c/mL and 95% CI, 1.03-1.04 for BKV; HR, 1.01 and 95% CI, 1.01-1.01 for CMV).
Application of TTV Cutoff Values in Clinical Routine of Kidney Transplant Care
Several TTV cutoff values for the detection of rejection and infection have been proposed (Table 3). In this respect, it is important to note that the limited diagnostic test performance of TTV load does not allow for the prediction of subsequent rejection and infection; instead, it defines at-risk patients. Notably, the cutoff values have not been evaluated in an interventional setting.
Using an in-house PCR, a TTV load <6 log10 c/mL quantified after stabilization in month 3 post-TX was suggested as a risk factor for subsequent rejection in the first year after TX75,76,78 by the Vienna Group. This value corresponds to a TTV load of 4.6 log10 c/mL detected by the commercial PCR (the Vienna in-house PCR quantifies TTV at 1.4 log10 c/mL higher than the commercial PCR; data under review). Specificity for prediction of rejection increased for TTV loads <5 log10 c/mL (corresponding to 3.6 log10 c/mL for the commercial PCR), whereas sensitivity decreased. For long-term, stable transplant recipients, the risk of rejection might be acceptable for a TTV load as low as 5 log10 c/mL (corresponding to 3.6 log10 c/mL for the commercial PCR).77 Additionally, cutoff values for the risk prediction of rejection within 2 y post-TX by pre-TX (TTV load <3.4 log10 c/mL) and month 1 post-TX TTV assessment (TTV load <4.2 log10 c/mL), respectively, have been proposed by applying the commercial PCR.79 In this respect, it is interesting to note that TTV is quantified in the plasma of healthy individuals without immunosuppression with a median of 2.3 log10 c/mL by the commercial PCR.61 Due to the great variety of noninvasive assays for the diagnosis of graft rejection tested in a multitude of trials with a large range of designs published in current literature, our review is not able to provide a comparison of the test performance between the proposed TTV cutoff values and these assays. However high-quality and up-to date reviews and comments are covering such assay systems in detail.6,8,12-14
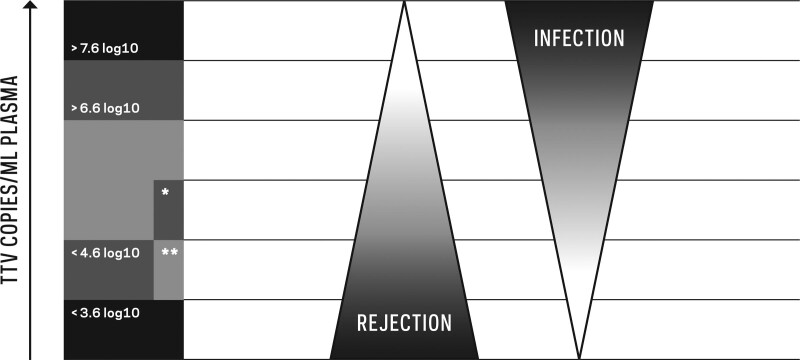
Using an in-house PCR, the Vienna Group suggested a TTV load >8 log10 c/mL, quantified after stabilization in month 3 post-TX, as a risk factor for subsequent infection in the first year after TX. This cutoff changes to 6.6 log10 c/mL if converted to values that correspond to the commercial PCR.75,80 Specificity for the prediction of infection increased for TTV loads >9 log10 c/mL (>7.6 log10 c/mL for the commercial PCR), whereas sensitivity decreased. Using an in-house PCR, a TTV load >3.5 log10 c/mL in the first 10 d posttransplant was associated with a risk for CMV viremia in the first 4 mo post-TX,82 which changes to a value of 3.8 log10 c/mL if converted to values corresponding to the commercial PCR (the in-house PCR performed in Pisa quantifies TTV at 0.3 log10 c/mL below the commercial PCR45). TTV loads >3.2 log10 c/mL and >4.6 log10 c/mL quantified by the commercial PCR at month 1 post-TX were associated with a risk of infection and immunosuppression-related adverse events such as opportunistic infections in the first year post-TX, respectively.74 A TTV load >5.0 log10 c/mL quantified by the commercial PCR at post-TX month 1 was proposed as a risk factor for the development of BKV viremia in the first year post-TX.83 Taken together, a TTV load between 4.6 log10 c/mL and 6.6 log10 c/mL—corresponding to values obtained by the commercial PCR—detected after TTV stabilization in month 3 post-TX might be an optimal range to reduce rejection and infection in the first year (Figure 1).
FIGURE 1.
The hosts’ plasma torque teno virus (TTV) load in relation to the risk of allograft rejection and infection in kidney (KTX) and lung transplant (LuTX) recipients. A high TTV load indicates a risk of infection, and a low TTV load indicates a risk of rejection. Proposed cutoff values for risk stratification have been converted to values that correspond to the commercial polymerase chain reaction (PCR) to facilitate comparison of the published data. The risk for infection increases above 6.6 log10 c/mL for both KTX and LuTX recipients. For KTX patients, the risk for rejection increases at TTV loads below 4.6 log10 c/mL. The field including an asterisk represents TTV loads below 5.6 log10 c/mL, which already indicate a risk in the LuTX setting due to the higher level of immunosuppression needed to prevent rejection compared with KTX. In KTX recipients transplanted >1 y ago, a TTV load above 3.6 log10 c/mL might indicate sufficient immunosuppression to prevent rejection (double asterisk).
A randomized controlled single-blinded interventional trial—TTVguideIT—involving 260 KTX recipients from 13 centers in 6 countries across Europe will test the value of TTV-guided immunosuppression based on these suggestions.19,65 The trial is sponsored by the Medical University Vienna and financed by the European Union. Immunological low-risk and stable adult KTX recipients will be randomized at month 4 post-TX to receive either TTV-guided tacrolimus dosing or conventionally dosed immunosuppression in the first year post-TX. The primary composite outcome at month 12 post-TX includes infection and graft rejection assessed by personnel blinded to the randomization sequence. The trial will begin in 2022, and results are expected in 2026.
TTV IN LUNG TRANSPLANTATION
Convincing evidence for associations between TTV load and rejection and infection, respectively, has been provided in the LuTX setting 3 y earlier than for the KTX setting.63,69,88-90 However, less data are available, and the same caveats concerning study design apply (Table 4).
TABLE 4.
Studies of lung transplant recipients’ evaluations of association between TTV load detected by in-house PCR and allograft rejection and infection, respectively
| Study designa | TX period | Included patients | Endpoint; timing | Number of events | Main association | Limitationsb |
|---|---|---|---|---|---|---|
| Rejection | ||||||
| Cohort89 | 2013–2015 | 143 | CLAD/AR (iBX); month 6–y 5 post-TX | 22 CLAD | TTV 3 mo before event–rejection | Three endpoints; some major determinants of TTV not included in the effect size adjustment |
| 11 AR | ||||||
| Case-control88 | 2003–2013 | 47 | CLAD; month 4–y 3 post-TX | 20 | TTV at event–rejection | Possible selection bias; rejection not BX proven; no effect size adjustment |
| Case-control90 | 2006–2015 | 34 | AR (pBX); months 4–12 post-TX | 13 | TTV before event–rejection | Possible selection bias; 2 endpoints; 3 suspected rejection episodes in the control group; no effect size adjustment |
| Infection | ||||||
| Cohort69 | 2008 | 31c | Infectious events; month 4–y 2 post-TX | 13 | TTV before event–infection | Insufficient definition of outcome; no effect size adjustment |
| Cohort89 | 2013–2015 | 143 | Infections requiring hospitalization; month 6–y 5 post-TX | 28 | TTV 3 mo before event–infection | Three endpoints; some major determinants of TTV not included in the effect size adjustment |
| Case-control90 | 2006–2015 | 34 | Infection leading to medical measure; months 4–12 post-TX | 19 | TTV months 4 to 12 post-TX–infection | Possible selection bias; 2 endpoints; no effect size adjustment |
The studies are listed according to their design and date of their online publication.
All studies followed a noninterventional and single-center design.
A total of 24 patients analyzed.
AR, acute rejection; BX, biopsy; CLAD, chronic lung allograft dysfunction; iBX, indication biopsy; pBX, protocol biopsy; PCR, polymerase chain reaction; TTV, torque teno virus; TX, transplantation.
The Vienna Group was the first to describe the association between TTV and infections in a cohort study including 31 patients.69 TTV loads were analyzed during the steady state of TTV kinetics after month 3 post-TX in patients before the first episode of microbial infection (n = 13). TTV loads were higher when compared with the TTV load detected in patients who did not experience clinical complications (n = 11). A cutoff of 9.3 log10 c/mL was predictive for the development of infection, with a sensitivity of 54% and specificity of 91%. The same groups also exhibited an association between TTV load and chronic lung allograft dysfunction (CLAD) for the first time.88 A case-control study included 20 patients developing CLAD (forced expiratory pressure in 1 s ≤80%) within 3 y post-TX and 27 matched controls. Recipients with CLAD showed lower TTV loads, and a cutoff of 7.0 log10 c/mL detected CLAD with a sensitivity of 65% and specificity of 82%.
The first prospective cohort study on CLAD was also published by the Vienna Group.89 During 3 y post-TX, 28 of 143 patients developed an infection requiring hospitalization. Overall, 22 patients with CLAD and 11 with acute rejection were registered. The maximum TTV load during the 3 mo before an event was associated with infection. The risk increased within a rate of 5.05 with every log increase in TTV load (HR; 95% CI, 2.94-8.67). Moreover, the minimum TTV load during the 3 mo before an event was associated with CLAD and acute rejection. The risk for CLAD and acute rejection decreased within a rate of 0.71 and 0.48, respectively, with every log increase in TTV load (HR; 95% CI, 0.54-0.93 and 0.26-0.88). TTV levels between 7 log10 c/mL and 9.5 log10 c/mL were described cutoff values to avoid high risk of rejection or infection.
In the same year, the Freiburg Group presented a case-control study including 34 patients with 13 patients experiencing biopsy-proven rejection between 4 and 12 mo after LuTX90 and 21 matched patients without rejection. TTV load before the event was lower in patients with rejection compared with patients without rejection. Additionally, TTV load decreased before rejection. The sensitivity of 1 log decrease in TTV load for a subsequent rejection episode within 1 mo was 74%, with a specificity of 99%. Within this cohort, 19 patients had an infection leading to medical measures (antibiotic treatment, hospitalization, or change in immunosuppression). The TTV load during months 3 and 12 post-TX was higher in the group of patients with infectious complications when compared with those of other recipients.
Based on the cutoff values determined in their noninterventional studies, the Vienna Group—together with the Hannover LuTX center—initiated a two-center, open-label, randomized, controlled, and investigator-driven trial including 144 LuTX recipients to investigate the safety and preliminary efficacy of immunosuppression guided by TTV monitoring as an add-on to conventional therapeutic drug monitoring (VIGILung).20 The study is sponsored by the University of Marburg and financed by the Deutsche Forschungsgemeinschaft. Adult de novo LuTX recipients with tacrolimus-based immunosuppression and stable graft function are randomized 1:1 to receive either (1) tacrolimus guided by TTV monitoring in addition to drug trough level (active group) or (2) tacrolimus according to conventional therapeutic drug monitoring (control group) after month 3 post-TX. In the active group, the tacrolimus target range will be adjusted according to TTV load following predefined steps. If TTV is above the predefined optimal range, tacrolimus will be reduced by 1 step; if it is below the optimal range, tacrolimus will be increased by 1 step. Outcomes will be assessed 12 mo after randomization with the change in estimated glomerular filtration rate as the primary endpoint. Main secondary endpoints will include allograft function, allograft rejection, and infections. Trial results are expected in 2024.
TTV IN LIVER TRANSPLANTATION
Compared with KTX and LuTX, the association between TTV and immunologic events in LTX recipients is not supported by the same amount and quality of data, while the same caveats concerning study design also apply (Table 5). However, preliminary data suggest an association between TTV load and infectious events and organ rejection.82,91-93
TABLE 5.
Studies in liver transplant recipients’ evaluations of association between TTV load detected by in-house PCR and allograft rejection and infection, respectively
| Study designa | TX period | Included patients | Endpoint; timing | Events | Main association | Limitationsb |
|---|---|---|---|---|---|---|
| Rejection | ||||||
| Cohort91 | NA | 39 | BX proven rejection; <12 mo post-TX | 13c | TTV pre-TX–rejection | Possible selection bias; insufficient endpoint definition; multiple testing; possible model overfitting |
| Cohort92 | 2014–2017 | 63 | BX proven rejection; <12 mo post-TX | 19c | TTV pre-BX–rejection | Missing data on BX without rejection; 2 endpoints; no effect size adjustment |
| Infection | ||||||
| Cohort92 | 2014–2017 | 63 | CMV viremia/ disease; <12 mo post-TX | 26d | TTV at event–CMV viremia/ disease | Two endpoints; no effect size adjustment |
| Cross-sectional93 | 1982–2016 | 136 | BKV events; 10 y post-TX | 23 | TTV at event–urinary BKV | Possible selection bias; missing data on sampling; multiple testing; no effect size adjustment |
| Case-control82,e | 2011–2016 | 90 | CMV viremia; <4 mo post-TX | 64 | TTV days 0 to 10 post-TX–CMV viremia | Possible selection bias; main analysis includes KTX; multiple testing; no effect size adjustment |
The studies are listed according to the date of their design and online publication.
All studies followed a noninterventional and single-center design.
The numbers of total biopsies were not stated.
Five of the cases were diagnosed with CMV disease.
Kidney and liver transplant recipients.
BKV, BK polyomavirus; BX, biopsy; CMV, cytomegalovirus; KTX, kidney transplantation; NA, not available; PCR, polymerase chain reaction; TTV, Torque Teno virus; TX, transplantation.
TTV and Liver Transplant Rejection
The Swiss Transplant Cohort Study was the first to describe an association between TTV and biopsy-proven rejection in LTX recipients.91 A total of 39 recipients were dichotomized according to TTV positivity at TX. The cumulative incidence of rejection in recipients with detectable TTV at TX was lower (21%, 95% CI, 8-37) than in patients with undetectable TTV (70%, detection limit: 25 c/mL; 95% CI, 28-90) in the first year after TX. A prospective study from Spain including 63 LTX recipients provides more data on the association between TTV and acute biopsy-proven rejection.92 A total of 20 rejection episodes were diagnosed in 19 patients: 12 upon indication biopsy and 8 upon protocol biopsy. No differences in the TTV load of plasma obtained closest to the rejection event were observed between episodes of rejection and nonrejection. However, in the subgroup of biopsies performed upon clinical indication, TTV was lower in patients with rejection compared with patients without rejection. A cutoff value of 5.6 × 104 c/mL yielded sensitivity, specificity, NPV, and PPV values of 100%, 77%, 100%, and 38%, respectively.
TTV and Infection in Liver Transplant Recipients
A German group was the first to describe a correlation between urinary BKV and serum TTV load in 136 LTX patients in a cross-sectional study.93 A positive correlation was described between urinary BKV DNA and serum TTV load. However, no association was found between BKV viremia and TTV. The Pisa Group provided evidence of an association between CMV viremia and TTV in a case-control study including LTX and NTX patients (as previously described in detail in the context of KTX).82 A total of 90 subjects with LTX were included, and 64 patients (65%) showed CMV reactivation. TTV loads between days 0 and 10 after TX were higher in the CMV-positive patients when compared with the CMV-negative patients.
Within the data set described here in the section on TTV and rejection in LTX patients, the Spanish group also reported an association between TTV load and CMV.92 During CMV disease (n = 5) and CMV infection (n = 26), TTV load was higher when compared with the remaining time points (1.6 × 108 versus 7.1 × 105 c/mL and 3.9 × 106 versus 6.2 × 105 c/mL). However, based on linear regression, no associations were observed between TTV load and CMV DNA during episodes of infection.
Taken together, existing evidence of an association between TTV and infectious events remains limited. Larger prospective studies with longitudinal monitoring are encouraged to determine whether findings concerning TTV and infectious events in other SOTs are also applicable to LTX. Moreover, such future studies could also define clinically useful cutoffs.
SUMMARY AND OUTLOOK
Novel concepts of immunologic monitoring in SOT have emerged to guide—and thus optimize—immunosuppressive drugs. The apathogenic and highly prevalent TTV represents a promising candidate in this regard. The viral copy number in the peripheral blood of its host has been shown to associate with organ rejection and infectious disease. Although TTV cutoff values for the guidance of immunosuppression have been proposed in the adult KTX and LuTX settings, more data are needed for LTX and heart TX. PCR-based assays or TTV quantification offer a simple and standardized implementation. Diagnostic test performance of TTV load does not allow for the prediction of subsequent rejection and infection; instead, it defines at-risk patients. Hitherto TTV load has not been tested in an interventional setting. Currently, two multinational, investigator-driven, randomized controlled interventional trials are testing the safety and efficacy of TTV-guided immunosuppression in KTX and LuTX recipients.
ACKNOWLEDGMENTS
The authors would like to thank Frederik Haupenthal for lecturing.
Supplementary Material
Footnotes
Funded by European Union Horizon2020 research and innovation programme, grant agreement number 896932.
The authors declare no conflicts of interest.
The data that support the findings of this study are available from the corresponding author upon reasonable request.
G.B. wrote the manuscript. P.J. wrote the section Torque Teno Virus in Lung Transplantation. I.G. and E.P.-S. wrote the section Torque Teno Virus—an Introduction.
Supplemental Visual Abstract; http://links.lww.com/TP/C418.
Supplemental digital content (SDC) is available for this article. Direct URL citations appear in the printed text, and links to the digital files are provided in the HTML text of this article on the journal’s Web site (www.transplantjournal.com).
REFERENCES
- 1.Chand S, Atkinson D, Collins C, et al. The spectrum of renal allograft failure. PLoS One. 2016;11:e0162278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bos S, Vos R, Van Raemdonck DE, et al. Survival in adult lung transplantation: where are we in 2020? Curr Opin Organ Transplant. 2020;25:268–273. [DOI] [PubMed] [Google Scholar]
- 3.Tian D, Huang H, Wen HY. Noninvasive methods for detection of chronic lung allograft dysfunction in lung transplantation. Transplant Rev (Orlando). 2020;34:100547. [DOI] [PubMed] [Google Scholar]
- 4.Naesens M, Anglicheau D. Precision transplant medicine: biomarkers to the rescue. J Am Soc Nephrol. 2018;29:24–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andrews LM, Li Y, De Winter BCM, et al. Pharmacokinetic considerations related to therapeutic drug monitoring of tacrolimus in kidney transplant patients. Expert Opin Drug Metab Toxicol. 2017;13:1225–1236. [DOI] [PubMed] [Google Scholar]
- 6.Anglicheau D, Naesens M, Essig M, et al. Establishing biomarkers in transplant medicine: a critical review of current approaches. Transplantation. 2016;100:2024–2038. [DOI] [PubMed] [Google Scholar]
- 7.Dendle C, Mulley WR, Holdsworth S. Can immune biomarkers predict infections in solid organ transplant recipients? A review of current evidence. Transplant Rev (Orlando). 2019;33:87–98. [DOI] [PubMed] [Google Scholar]
- 8.Tissot A, Danger R, Claustre J, et al. Early identification of chronic lung allograft dysfunction: the need of biomarkers. Front Immunol. 2019;10:1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haas M, Loupy A, Lefaucheur C, et al. The Banff 2017 Kidney Meeting Report: revised diagnostic criteria for chronic active T cell-mediated rejection, antibody-mediated rejection, and prospects for integrative endpoints for next-generation clinical trials. Am J Transplant. 2018;18:293–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roux A, Levine DJ, Zeevi A, et al. Banff Lung Report: current knowledge and future research perspectives for diagnosis and treatment of pulmonary antibody-mediated rejection (AMR). Am J Transplant. 2019;19:21–31. [DOI] [PubMed] [Google Scholar]
- 11.Konvalinka A, Tinckam K. Utility of HLA antibody testing in kidney transplantation. J Am Soc Nephrol. 2015;26:1489–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oellerich M, Sherwood K, Keown P, et al. Liquid biopsies: donor-derived cell-free DNA for the detection of kidney allograft injury. Nat Rev Nephrol. 2021;17:591–603. [DOI] [PubMed] [Google Scholar]
- 13.Tsuji N, Agbor-Enoh S. Cell-free DNA beyond a biomarker for rejection: biological trigger of tissue injury and potential therapeutics. J Heart Lung Transplant. 2021;40:405–413. [DOI] [PubMed] [Google Scholar]
- 14.Nissaisorakarn V, Lee JR, Lubetzky M, et al. Urine biomarkers informative of human kidney allograft rejection and tolerance. Hum Immunol. 2018;79:343–355. [DOI] [PubMed] [Google Scholar]
- 15.Mian M, Natori Y, Ferreira V, et al. Evaluation of a novel global immunity assay to predict infection in organ transplant recipients. Clin Infect Dis. 2018;66:1392–1397. [DOI] [PubMed] [Google Scholar]
- 16.Ravaioli M, Neri F, Lazzarotto T, et al. Immunosuppression modifications based on an immune response assay: results of a randomized, controlled trial. Transplantation. 2015;99:1625–1632. [DOI] [PubMed] [Google Scholar]
- 17.Ahlenstiel-Grunow T, Liu X, Schild R, et al. Steering transplant immunosuppression by measuring virus-specific T cell levels: the randomized, controlled IVIST Trial. J Am Soc Nephrol. 2021;32:502–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Focosi D, Antonelli G, Pistello M, et al. Torquetenovirus: the human virome from bench to bedside. Clin Microbiol Infect. 2016;22:589–593. [DOI] [PubMed] [Google Scholar]
- 19.CORDIS. Personalisation of immunosuppression by monitoring viral load post kidney transplantation - a randomised controlled phase II trial. Available at https://cordis.europa.eu/project/id/896932. Accessed March 19, 2022.
- 20.Gottlieb J, Reuss A, Mayer K, et al. Viral load-guided immunosuppression after lung transplantation (VIGILung)-study protocol for a randomized controlled trial. Trials. 2021;22:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Rijn AL, Wunderink HF, Sidorov IA, et al. Torque teno virus loads after kidney transplantation predict allograft rejection but not viral infection. J Clin Virol. 2021;140:104871. [DOI] [PubMed] [Google Scholar]
- 22.Kowalski RJ, Post DR, Mannon RB, et al. Assessing relative risks of infection and rejection: a meta-analysis using an immune function assay. Transplantation. 2006;82:663–668. [DOI] [PubMed] [Google Scholar]
- 23.Rodrigo E, López-Hoyos M, Corral M, et al. ImmuKnow as a diagnostic tool for predicting infection and acute rejection in adult liver transplant recipients: a systematic review and meta-analysis. Liver Transpl. 2012;18:1245–1253. [DOI] [PubMed] [Google Scholar]
- 24.Ling X, Xiong J, Liang W, et al. Can immune cell function assay identify patients at risk of infection or rejection? A meta-analysis. Transplantation. 2012;93:737–743. [DOI] [PubMed] [Google Scholar]
- 25.Bamoulid J, Courivaud C, Coaquette A, et al. Subclinical Epstein-Barr virus viremia among adult renal transplant recipients: incidence and consequences. Am J Transplant. 2013;13:656–662. [DOI] [PubMed] [Google Scholar]
- 26.Nishizawa T, Okamoto H, Konishi K, et al. A novel DNA virus (TTV) associated with elevated transaminase levels in posttransfusion hepatitis of unknown etiology. Biochem Biophys Res Commun. 1997;241:92–97. [DOI] [PubMed] [Google Scholar]
- 27.Peng YH, Nishizawa T, Takahashi M, et al. Analysis of the entire genomes of thirteen TT virus variants classifiable into the fourth and fifth genetic groups, isolated from viremic infants. Arch Virol. 2002;147:21–41. [DOI] [PubMed] [Google Scholar]
- 28.Moustafa A, Xie C, Kirkness E, et al. The blood DNA virome in 8,000 humans. Plos Pathog. 2017;13:e1006292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tisza MJ, Pastrana DV, Welch NL, et al. Discovery of several thousand highly diverse circular DNA viruses. Elife. 2020;9:e51971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Varsani A, Opriessnig T, Celer V, et al. Taxonomic update for mammalian anelloviruses (family Anelloviridae). Arch Virol. 2021;166:2943–2953. [DOI] [PubMed] [Google Scholar]
- 31.Okamoto H, Takahashi M, Nishizawa T, et al. Genomic characterization of TT viruses (TTVs) in pigs, cats and dogs and their relatedness with species-specific TTVs in primates and tupaias. J Gen Virol. 2002;83:1291–1297. [DOI] [PubMed] [Google Scholar]
- 32.Spandole S, Cimponeriu D, Berca LM, et al. Human anelloviruses: an update of molecular, epidemiological and clinical aspects. Arch Virol. 2015;160:893–908. [DOI] [PubMed] [Google Scholar]
- 33.Maggi F, Bendinelli M. Human anelloviruses and the central nervous system. Rev Med Virol. 2010;20:392–407. [DOI] [PubMed] [Google Scholar]
- 34.Hsiao KL, Wang LY, Lin CL, et al. New phylogenetic groups of torque teno virus identified in Eastern Taiwan indigenes. PLoS One. 2016;11:e0149901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vasilyev EV, Trofimov DY, Tonevitsky AG, et al. Torque teno virus (TTV) distribution in healthy Russian population. Virol J. 2009;6:134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brajao de Oliveira K. Torque teno virus: a ubiquitous virus. Rev Bras Hematol Hemoter. 2015;37:357–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tyschik EA, Rasskazova AS, Degtyareva AV, et al. Torque teno virus dynamics during the first year of life. Virol J. 2018;15:96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaczorowska J, van der Hoek L. Human anelloviruses: diverse, omnipresent and commensal members of the virome. FEMS Microbiol Rev. 2020;44:305–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maggi F, Focosi D, Albani M, et al. Role of hematopoietic cells in the maintenance of chronic human torquetenovirus plasma viremia. J Virol. 2010;84:6891–6893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maggi F, Fornai C, Zaccaro L, et al. TT virus (TTV) loads associated with different peripheral blood cell types and evidence for TTV replication in activated mononuclear cells. J Med Virol. 2001;64:190–194. [DOI] [PubMed] [Google Scholar]
- 41.Takahashi M, Asabe S, Gotanda Y, et al. TT virus is distributed in various leukocyte subpopulations at distinct levels, with the highest viral load in granulocytes. Biochem Biophys Res Commun. 2002;290:242–248. [DOI] [PubMed] [Google Scholar]
- 42.Kosulin K, Kernbichler S, Pichler H, et al. Post-transplant replication of torque teno virus in granulocytes. Front Microbiol. 2018;9:2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maggi F, Bendinelli M. Immunobiology of the Torque teno viruses and other anelloviruses. Curr Top Microbiol Immunol. 2009;331:65–90. [DOI] [PubMed] [Google Scholar]
- 44.Kulifaj D, Durgueil-Lariviere B, Meynier F, et al. Development of a standardized real time PCR for torque teno viruses (TTV) viral load detection and quantification: a new tool for immune monitoring. J Clin Virol. 2018;105:118–127. [DOI] [PubMed] [Google Scholar]
- 45.Macera L, Spezia PG, Medici C, et al. Comparative evaluation of molecular methods for the quantitative measure of torquetenovirus viremia, the new surrogate marker of immune competence. J Med Virol. 2022;94:491–498. [DOI] [PubMed] [Google Scholar]
- 46.Abbas AA, Diamond JM, Chehoud C, et al. The perioperative lung transplant virome: torque teno viruses are elevated in donor lungs and show divergent dynamics in primary graft dysfunction. Am J Transplant. 2017;17:1313–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Segura-Wang M, Görzer I, Jaksch P, et al. Temporal dynamics of the lung and plasma viromes in lung transplant recipients. PLoS One. 2018;13:e0200428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arze CA, Springer S, Dudas G, et al. Global genome analysis reveals a vast and dynamic anellovirus landscape within the human virome. Cell Host Microbe. 2021;29:1305–1315.e6. [DOI] [PubMed] [Google Scholar]
- 49.Freer G, Maggi F, Pifferi M, et al. The virome and its major component, anellovirus, a convoluted system molding human immune defenses and possibly affecting the development of asthma and respiratory diseases in childhood. Front Microbiol. 2018;9:686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tsuda F, Takahashi M, Nishizawa T, et al. IgM-class antibodies to TT virus (TTV) in patients with acute TTV infection. Hepatol Res. 2001;19:1–11. [DOI] [PubMed] [Google Scholar]
- 51.Kakkola L, Bondén H, Hedman L, et al. Expression of all six human torque teno virus (TTV) proteins in bacteria and in insect cells, and analysis of their IgG responses. Virology. 2008;382:182–189. [DOI] [PubMed] [Google Scholar]
- 52.Chen T, Väisänen E, Mattila PS, et al. Antigenic diversity and seroprevalences of Torque teno viruses in children and adults by ORF2-based immunoassays. J Gen Virol. 2013;94:409–417. [DOI] [PubMed] [Google Scholar]
- 53.Giacconi R, Maggi F, Macera L, et al. Torquetenovirus (TTV) load is associated with mortality in Italian elderly subjects. Exp Gerontol. 2018;112:103–111. [DOI] [PubMed] [Google Scholar]
- 54.Studenic P, Bond G, Kerschbaumer A, et al. Torque teno virus quantification for monitoring of immunomodulation with biological compounds in the treatment of rheumatoid arthritis. Rheumatology (Oxford). 2022;61:2815–2825. [DOI] [PubMed] [Google Scholar]
- 55.Rocchi J, Ricci V, Albani M, et al. Torquetenovirus DNA drives proinflammatory cytokines production and secretion by immune cells via toll-like receptor 9. Virology. 2009;394:235–242. [DOI] [PubMed] [Google Scholar]
- 56.Lai YC, Hu RT, Yang SS, et al. Coinfection of TT virus and response to interferon therapy in patients with chronic hepatitis B or C. World J Gastroenterol. 2002;8:567–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Moen EM, Sagedal S, Bjøro K, et al. Effect of immune modulation on TT virus (TTV) and TTV-like-mini-virus (TLMV) viremia. J Med Virol. 2003;70:177–182. [DOI] [PubMed] [Google Scholar]
- 58.Maggi F, Pistello M, Vatteroni M, et al. Dynamics of persistent TT virus infection, as determined in patients treated with alpha interferon for concomitant hepatitis C virus infection. J Virol. 2001;75:11999–12004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zheng H, Ye L, Fang X, et al. Torque teno virus (SANBAN isolate) ORF2 protein suppresses NF-kappaB pathways via interaction with IkappaB kinases. J Virol. 2007;81:11917–11924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Martelli F, Macera L, Spezia PG, et al. Torquetenovirus detection in exosomes enriched vesicles circulating in human plasma samples. Virol J. 2018;15:145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Focosi D, Spezia PG, Macera L, et al. Assessment of prevalence and load of torquetenovirus viraemia in a large cohort of healthy blood donors. Clin Microbiol Infect. 2020;26:1406–1410. [DOI] [PubMed] [Google Scholar]
- 62.Maggi F, Pifferi M, Fornai C, et al. TT virus in the nasal secretions of children with acute respiratory diseases: relations to viremia and disease severity. J Virol. 2003;77:2418–2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Görzer I, Jaksch P, Kundi M, et al. Pre-transplant plasma torque teno virus load and increase dynamics after lung transplantation. PLoS One. 2015;10:e0122975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McCulloch E, Montgomery D, Maggi F, et al. External quality assessment (EQA) pilot study for molecular diagnostics of torque teno virus (TTV). 22nd Annual Meeting of the European Society for Clinical Virology. Copenhagen, DK, 2019. [Google Scholar]
- 65.TTV Guide TX. Our story. Available at https://www.ttv-guide.eu. Accessed March 19, 2022.
- 66.Reshetnyak VI, Maev IV, Burmistrov AI, et al. Torque teno virus in liver diseases: on the way towards unity of view. World J Gastroenterol. 2020;26:1691–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lolomadze EA, Rebrikov DV. Constant companion: clinical and developmental aspects of torque teno virus infections. Arch Virol. 2020;165:2749–2757. [DOI] [PubMed] [Google Scholar]
- 68.De Vlaminck I, Khush KK, Strehl C, et al. Temporal response of the human virome to immunosuppression and antiviral therapy. Cell. 2013;155:1178–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Görzer I, Haloschan M, Jaksch P, et al. Plasma DNA levels of torque teno virus and immunosuppression after lung transplantation. J Heart Lung Transplant. 2014;33:320–323. [DOI] [PubMed] [Google Scholar]
- 70.Zhong S, Yeo W, Tang MW, et al. Gross elevation of TT virus genome load in the peripheral blood mononuclear cells of cancer patients. Ann N Y Acad Sci. 2001;945:84–92. [DOI] [PubMed] [Google Scholar]
- 71.Sawata T, Bando M, Nakayama M, et al. Clinical significance of changes in torque teno virus DNA titer after chemotherapy in patients with primary lung cancer. Respir Investig. 2018;56:173–178. [DOI] [PubMed] [Google Scholar]
- 72.Schmidt L, Jensen BO, Walker A, et al. Torque teno virus plasma level as novel biomarker of retained immunocompetence in HIV-infected patients. Infection. 2021;49:501–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Reindl-Schwaighofer R, Heinzel A, Mayrdorfer M, et al. Comparison of SARS-CoV-2 antibody response 4 weeks after homologous vs heterologous third vaccine dose in kidney transplant recipients: a randomized clinical trial. JAMA Intern Med. 2022;182:165–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fernández-Ruiz M, Albert E, Giménez E, et al. Monitoring of alphatorquevirus DNA levels for the prediction of immunosuppression-related complications after kidney transplantation. Am J Transplant. 2019;19:1139–1149. [DOI] [PubMed] [Google Scholar]
- 75.Doberer K, Schiemann M, Strassl R, et al. Torque teno virus for risk stratification of graft rejection and infection in kidney transplant recipients-a prospective observational trial. Am J Transplant. 2020;20:2081–2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Doberer K, Haupenthal F, Nackenhorst M, et al. Torque teno virus load is associated with subclinical alloreactivity in kidney transplant recipients: a prospective observational trial. Transplantation. 2021;105:2112–2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schiemann M, Puchhammer-Stöckl E, Eskandary F, et al. Torque teno virus load-inverse association with antibody-mediated rejection after kidney transplantation. Transplantation. 2017;101:360–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Strassl R, Doberer K, Rasoul-Rockenschaub S, et al. Torque Teno Virus for risk stratification of acute biopsy-proven alloreactivity in kidney transplant recipients. J Infect Dis. 2019;219:1934–1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Solis M, Velay A, Gantner P, et al. Torquetenovirus viremia for early prediction of graft rejection after kidney transplantation. J Infect. 2019;79:56–60. [DOI] [PubMed] [Google Scholar]
- 80.Strassl R, Schiemann M, Doberer K, et al. Quantification of torque teno virus viremia as a prospective biomarker for infectious disease in kidney allograft recipients. J Infect Dis. 2018;218:1191–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Handala L, Descamps V, Morel V, et al. No correlation between torque teno virus viral load and BK virus replication after kidney transplantation. J Clin Virol. 2019;116:4–6. [DOI] [PubMed] [Google Scholar]
- 82.Maggi F, Focosi D, Statzu M, et al. Early post-transplant torquetenovirus viremia predicts cytomegalovirus reactivations in solid organ transplant recipients. Sci Rep. 2018;8:15490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fernández-Ruiz M, Albert E, Giménez E, et al. Early kinetics of torque teno virus DNA load and BK polyomavirus viremia after kidney transplantation. Transpl Infect Dis. 2020;22:e13240. [DOI] [PubMed] [Google Scholar]
- 84.Gore EJ, Gomes-Neto AW, Wang L, et al. Torquetenovirus serum load and long-term outcomes in renal transplant recipients. J Clin Med. 2020;9:E440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Uhl P, Heilos A, Bond G, et al. Torque teno viral load reflects immunosuppression in paediatric kidney-transplanted patients-a pilot study. Pediatr Nephrol. 2021;36:153–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Eldar-Yedidia Y, Ben-Shalom E, Hillel M, et al. Association of post-transplantation anellovirus viral load with kidney transplant rejection in children. Pediatr Nephrol. 2022;37:1905–1914. [DOI] [PubMed] [Google Scholar]
- 87.Eskandary F, Regele H, Baumann L, et al. A randomized trial of bortezomib in late antibody-mediated kidney transplant rejection. J Am Soc Nephrol. 2018;29:591–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Görzer I, Jaksch P, Strassl R, et al. Association between plasma torque teno virus level and chronic lung allograft dysfunction after lung transplantation. J Heart Lung Transplant. 2017;36:366–368. [DOI] [PubMed] [Google Scholar]
- 89.Jaksch P, Kundi M, Görzer I, et al. Torque teno virus as a novel biomarker targeting the efficacy of immunosuppression after lung transplantation. J Infect Dis. 2018;218:1922–1928. [DOI] [PubMed] [Google Scholar]
- 90.Frye BC, Bierbaum S, Falcone V, et al. Kinetics of torque teno virus-DNA plasma load predict rejection in lung transplant recipients. Transplantation. 2019;103:815–822. [DOI] [PubMed] [Google Scholar]
- 91.Simonetta F, Pradier A, Masouridi-Levrat S, et al. ; Swiss Transplant Cohort Study (STCS). Torque teno virus load and acute rejection after orthotopic liver transplantation. Transplantation. 2017;101:e219–e221. [DOI] [PubMed] [Google Scholar]
- 92.Ruiz P, Martínez-Picola M, Santana M, et al. Torque teno virus is associated with the state of immune suppression early after liver transplantation. Liver Transpl. 2019;25:302–310. [DOI] [PubMed] [Google Scholar]
- 93.Herrmann A, Sandmann L, Adams O, et al. Role of BK polyomavirus (BKV) and torque teno virus (TTV) in liver transplant recipients with renal impairment. J Med Microbiol. 2018;67:1496–1508. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.