Abstract
Bacterial two-component regulatory systems control the expression of target genes through regulated changes in protein phosphorylation. Signal reception alters the ability of a membrane-bound histidine kinase (HK) protein to transfer phosphate from ATP to a highly conserved histidine residue. The transfer of phosphate from the histidine to an aspartate residue on the cognate response regulator (RR) changes the ability of the latter protein to bind to target DNA sequences and to alter gene transcription. UhpB is the HK protein which controls production of the sugar phosphate transporter UhpT. Elevated expression of full-length UhpB or of a soluble hybrid protein, GST-Bc, which is glutathione S-transferase (GST) fused to the cytoplasmic C-terminal portion of UhpB, results in complete blockage of uhpT expression in a uhp+ strain. This dominant-negative interference could result from the ability of GST-Bc to bind and sequester the RR UhpA and to accelerate its dephosphorylation. The portion of GST-Bc responsible for the interference phenotype was localized using truncation, linker insertion, and point mutations to the region between residues 293 and 366 flanking His-313, the putative site of autophosphorylation. Point mutations which allow GST-Bc to activate uhpT expression or which relieve the interference phenotype were obtained at numerous sites throughout this region. This region of UhpB is related to the phosphoryl transfer domain of EnvZ, which forms half of an interdimer four-helix bundle and is responsible for dimerization of its cytoplasmic domain. The expression of GST fusion proteins carrying the corresponding portions of EnvZ strongly interfered with the activation of porin gene expression by OmpR. The GST-Bc protein accelerated dephosphorylation of P-UhpA. Reverse transfer of phosphate from P-UhpA to GST-Bc was observed in the presence of the metal chelator EDTA and depended on the presence of His-313. Phosphate transfer from P-UhpA to the liberated phosphoryl transfer domain also occurred. Taken together, these results indicate that the phosphoryl transfer-dimerization domain of UhpB participates in the specific binding of UhpA, in the control of autokinase activity, and in the dephosphorylation of P-UhpA.
In Escherichia coli, a simple genetic circuit controls production of the organophosphate-Pi antiporter UhpT, which is responsible for the uptake of many phosphorylated sugars. Induction of uhpT gene expression in response to the presence of extracellular glucose-6-phosphate (Glc6P) is mediated by an unusual two-component regulatory system comprised of three proteins, the histidine kinase (HK) UhpB, the response regulator (RR) UhpA, and the membrane receptor UhpC (20). Unlike most HK proteins, the N-terminal portion of UhpB is hydrophobic and contains 6 to 10 transmembrane segments (18). The transmembrane portion of UhpB (residues 1 to 272) is thought to operate in complex with UhpC, so that binding of periplasmic Glc6P to UhpC results in stimulation of the autophosphorylation activity of UhpB (17). The cytoplasmic C-terminal portion of UhpB (residues 273 to 500) contains the conserved sequence motifs characteristic of the catalytic portion of HK proteins. Phosphate transfer from UhpB to UhpA stimulates the ability of UhpA to bind at the uhpT promoter and to activate its transcription.
Information on the domain organization, structure, regulation, and phylogenetic relationships of HK and RR proteins is emerging (for recent reviews, see references 10 and 41). HK proteins typically contain two distinct parts. The N-terminal signaling region is variable in sequence and in the number of transmembrane segments. Some HK proteins are cytoplasmic, many possess two transmembrane segments which divide the intracellular and extracellular portions of the protein, and some have more complex transmembrane topology. The well-studied membrane-inserted HK proteins function as homodimers in which the two transmembrane segments from each monomer combine to form a four-helix signaling structure (reviewed in reference 7). The relative orientations of these transmembrane helices are affected by the binding of specific effectors to the periplasmic domain and transmit a conformational signal to the cytoplasmic HK portion. The cytoplasmic C-terminal portion contains several key elements and conserved homology motifs. The linker region is predicted to form a coiled coil, follows the last transmembrane segment, and participates in signal transduction (reviewed in reference 48). The linker region is typically followed by a domain which forms two amphipathic α-helices which can pair with each other and with the corresponding region from the other monomer to form a four-helix bundle which is responsible for dimerization of the cytoplasmic portion of the HK (44). Except in CheA and related proteins, the dimerization domain contains the highly conserved site of phosphorylation, typically a histidine, and the flanking conserved H box sequences. This segment is called the phosphoryl transfer domain. The other major functional domain of HK proteins is the nucleotide-binding kinase domain, whose structures in EnvZ (43) and CheA (2) resemble the ATP-binding domains of DNA gyrase and chaperone Hsp90 but not other protein kinases (6). Examples of cross talk between noncognate HKs and RRs have been documented (1, 8, 14, 32), but under physiological conditions there appears to be remarkable specificity in allowing communication only between cognate pairs of HK and RR proteins. Identification of the determinants which specify this recognition should further our understanding of the action of two-component regulatory systems.
The function of all three uhp regulatory genes is required for uhpT expression in E. coli, indicating that UhpC must allow activation of UhpB (18). In agreement, we have found that expression of uhpB at a higher gene copy number than that of uhpC prevents the induction of uhpT (49), whereas co-overexpression of uhpB and uhpC allows normal inducibility by Glc6P. In addition, expression of the cytoplasmic C-terminal portion of UhpB (residues 273 to 500) fused to glutathione S-transferase (GST-Bc) conferred a strong dominant-negative effect and completely blocked the expression of uhpT from the wild-type chromosomal uhpABCT locus. The liberated HK domain carried in GST-Bc was found to prevent both the binding of UhpA at the uhpT promoter and transcription activation in vitro. This interference phenotype cannot be solely a consequence of the ability of GST-Bc to accelerate dephosphorylation of P-UhpA. The inhibitory effect of GST-Bc was seen even when UhpA could drive uhpT transcription in its unphosphorylated state, as occurs with the phosphorylation-independent H170Y variant of UhpA carrying the replacement of His-170 with Tyr or upon overexpression of UhpA. In the latter case, GST-Bc even inhibited the activity of overexpressed UhpA D54N, which is unable to be phosphorylated (4, 49). We interpreted from these results that UhpB can sequester UhpA in an inactive state.
Several variant forms of UhpB which were selected for their ability to allow uhpT expression in the absence of UhpC function had a reduced inhibitory effect in vivo when expressed as GST-Bc fusions (49). Although GST-Bc does not exhibit autokinase activity, a variant carrying the replacement of Arg-324 with Cys (R324C), which confers UhpC independence in the context of full-length UhpB, exhibited both the ability to phosphorylate itself and to transfer that phosphate to UhpA. The GST-Bc R324C variant had not lost the ability to sequester UhpA, which was revealed by the restoration of the dominant-negative behavior when its autokinase activity was eliminated by mutations affecting conserved residues in the catalytic N or G boxes. The GST-Bc R324C variant also appeared to possess the ability to dephosphorylate P-UhpA, since the phosphate transferred to UhpA by the action of GST-Bc was much more labile than expected from the half-life of P-UhpA formed in the absence of UhpB by phosphate transfer from acetyl phosphate. The phosphatase activity of GST-Bc was examined here. The region of GST-Bc required for the interference or squelching phenotype was localized by analysis of linker substitution, truncation, and point mutations to the phosphoryl transfer domain. A similar specific squelching phenotype was seen when the corresponding region of the HK EnvZ was expressed. This finding suggests that the interference phenotype is a general manifestation of the interaction of cognate HK-RR protein pairs.
MATERIALS AND METHODS
Strains and plasmids.
The E. coli strain RK1309 (uhp+) is derived from MC4100 [Δ(argF-lac)U169 araD139 rpsL 150 relA1 flbB5301 deoC1 ptsF25 rbsR22] but is recA and carries the λRZ5 uhpT-lacZ transcriptional fusion as a lysogen at the attλ site. Strain RK1307 is RK1309 Δuhp(B60-C437). All plasmids used in this study were derived from pGEX3x-TEV and pJSW-B141 (49), which encodes a hybrid protein in which residues 273 to 500 of UhpB are fused to the C terminus of GST.
Purification of GST and His-tag fusions.
Procedures for the expression and purification of UhpA and UhpB proteins fused to His6 and GST tags were described previously (49).
β-Galactosidase assays.
Regulation of the uhpT promoter was determined from the level of β-galactosidase expressed from a uhpT-lacZ fusion carried as a single lysogen of phage λRZ5 (28). β-Galactosidase assays were performed as described previously (18). Overnight cultures were diluted 1:100 in minimal medium A supplemented with 1% glycerol (vol/vol), 0.5% Casamino Acids, and 1.5 mM MgSO4. When indicated, IPTG (isopropyl β-d-thiogalactopyranoside) was added at the time of subculture. Cells in exponential growth were induced with 0.25 mM Glc6P in 96-well microtiter plates containing 200 μl of culture per well. After 40 min of induction at 37°C, cells were lysed with CHCl3-sodium dodecyl sulfate (SDS). The permeabilized cells were mixed with Z buffer (29) and 2 mM o-nitrophenyl-β-d-galactopyranoside, and the time course of hydrolysis was measured at 415 nm over 5 min at 37°C in a microplate reader (Molecular Devices). The rate of the enzyme reaction was normalized for cell density. Reported values are the averages of at least three experiments, with a variation of <10%.
Construction of linker insertions, truncations, and chimeras.
The isolation of uhpB linker insertions carrying a 12-bp oligonucleotide which introduced a PstI recognition site and resulted in the insertion of a tetrapeptide after UhpB residues 288, 345, 387, 411, 473, and 489 was previously described (18). The numbering of all amino acid coordinates corresponds to the wild-type protein. These insertions were transferred into the coding region for GST-Bc. The PstI site in the bla gene of plasmid pGEX3x-TEV was first removed by restriction fragment exchange of the PstI-less bla gene of plasmid pQE32 (Qiagen). As described previously for the cloning of GST-Bc (49), linker insertions in the uhpB coding region were cloned by PCR using appropriate primers and VENT polymerase (New England Biolabs) from plasmid templates which contain the PstI insertions in the uhpABCT operon (17). The sequences of all PCR primers are available upon request. Cloning of the PCR amplimers yielded plasmids encoding variants of GST-Bc (UhpB residues 273 to 500) carrying each insertion. The presence of the expected changes was confirmed by PstI digestion and by DNA sequencing of the coding region. Sequence determination was provided by the University of Virginia Biomolecular Research Facility.
C-terminal truncations of GST-Bc removed sequences distal to the site of the PstI linker and were constructed by digesting the plasmids carrying GST-Bc linker insertion mutants with PstI and EcoRI, removing the overhangs by the addition of VENT polymerase and nucleoside triphosphates at 70°C for 20 min, and recircularizing the plasmid by ligation. The C-terminal truncations removed the normal uhpB stop codon, but the pGEX3x vector contains stop codons in all three reading frames directly after the EcoRI site, resulting in the addition of a few nonspecific residues to the C terminus of the truncated UhpB sequence. N-terminal truncations were created by PCR with VENT polymerase using primers which added a 5′ BamHI site to allow the fusion of GST to UhpB at residue 282, 288, or 293.
To facilitate the construction of hybrid proteins combining segments of UhpB and EnvZ, an EagI site was introduced into homologous sequences of both genes. The nucleotide substitutions which introduced the EagI site after the coding region for residue 366 of GST-Bc required no change in amino acid sequence and created plasmid pJSW142. The coding region for residues 296 to 450 of EnvZ was amplified from the chromosome of strain MC4100 by PCR with VENT DNA polymerase using primers 1 (5′-CCGGGCAGGAGCGGCCGATGGAAAT-3′ introducing an EagI site) and 2 (5′-GATTTGAAGCTGGAGAATTCCTATCCAGTATCTT-3′ introducing an EcoRI site) (new restriction sites are underlined). The EcoRI/EagI-digested amplimer was cloned into similarly digested pJSW142 to create plasmid pJSW143, which encodes the hybrid protein designated GST-BZc. Similarly, the coding region for residues 181 to 295 of EnvZ was amplified with primers 3 (5′-GGGGCGTGGCTGTTTATTCGGATCCAGAACCGAC-3′ introducing a BamHI site) and 4 (5′-CCGCCATTTCCATCGGCCGCTCCTGCCCGGT-3′ introducing an EagI site), and the EagI/BamHI-digested amplimer was cloned into similarly digested pJSW142 to create plasmid pJSW144, which encodes the hybrid protein designated GST-ZBc. The introduced EagI site in envZ resulted in a change of methionine-294 to arginine. Plasmid pJSW145, which encodes GST-Zc, was made by appropriate restriction fragment exchange. The correct nucleotide sequences of all plasmid inserts were confirmed.
C-terminal truncations in which GST-Bc ends at residue 366 or GST-Zc ends at residue 296 were created by digestion with EagI and EcoRI, as described above for the construction of the other C-terminal truncations.
PCR mutagenesis.
Localized random mutagenesis of the portion of the uhpB sequence encoding residues 293 to 366 was performed by PCR with Taq polymerase (Life Technologies) using modifications of a mutagenic reaction (3) as described by Zhou and Blair (50). In brief, this method takes advantage of the increased misincorporation exhibited by Taq polymerase in the presence of Mn2+ and suboptimal ratios of nucleoside triphosphate substrates. Each amplified PCR fragment was digested with BamHI and EagI and ligated into plasmid pJSW142 digested with the same enzymes.
Western blot analysis.
Cell samples were adjusted to the same optical density at 415 nm (OD415) and mixed with an equal volume of 2× sample buffer, boiled for 5 min, and resolved by SDS-polyacrylamide gel electrophoresis (SDS-PAGE). Proteins were transferred to nitrocellulose filters and blocked in Tris-buffered saline with 5% milk solids and 0.05% Tween 20 overnight at 4°C. For detection of GST fusion proteins the membranes were developed with anti-GST monoclonal antibody 9D9 at 1:25,000 (gift of Amy Ma and J. T. Parsons, University of Virginia) and horseradish peroxidase-labeled rabbit anti-mouse immunoglobulin G antisera (Sigma Immunochemicals) at 1:5,000. Peroxidase activity was localized using the ECL bioluminescence kit (Kirkegaard and Perry) and exposed to film (Kodak X-OMAT).
Assay of porin expression.
Overnight cultures of cells expressing GST fusion constructs were diluted 1:100 into minimal medium A supplemented with 1% glycerol (vol/vol), 0.5% Casamino Acids, and 1.5 mM MgSO4. Sucrose was added to a final concentration of 20% (vol/vol) for high-osmolarity conditions. After growth to mid-exponential phase, cell volumes adjusted to the same OD415 were harvested by centrifugation and disrupted by sonication in 50 mM HEPES, pH 8.0. Following low-speed centrifugation to remove unbroken cells, Triton X-100 was added to a final concentration of 0.5% to solubilize the cytoplasmic membrane components. The insoluble outer membrane fraction was pelleted by ultracentrifugation at 110,000 × g for 60 min. The outer membrane proteins were resolved by SDS-PAGE with 4 M urea in 11% polyacrylamide gels (24) and stained with CodeBlue (Pierce).
Protein phosphatase assay.
Acetyl [32P]phosphate was prepared by the chemical method as described previously (26). The concentration of acetyl phosphate was determined as described by Skarstedt and Silverstein (40) in comparison to a standard sample (Sigma).
His6-UhpA with an extension containing six His residues at the N terminus of UhpA (49) was phosphorylated with 20 to 25 mM acetyl [32P]phosphate by incubation at 37°C for 2 to 3 h in assay buffer (50 mM HEPES [pH 8.0], 5 mM MgCl2, 1 mM dithiothreitol). Ni2+-conjugated agarose beads (100 μl of 50% slurry) were added and incubated for 10 min at 25°C. Beads were washed three times with 100 ml of assay buffer until the eluted radioactivity decreased to background levels. His6-UhpA was eluted with 320 μl of assay buffer containing 250 mM imidazole. When indicated, EDTA or unlabeled ATP was added to the eluted protein at a final concentration of 15 or 1 mM, respectively. The phosphatase assay was initiated by the addition of GST-Bc fusion proteins at 10% of the initial molar concentration of His6-UhpA and incubation of the mixture at 25°C. Samples were removed at intervals and mixed with 2 μl of 6× sample buffer and 1 μl of 150 mM EDTA and placed on ice. Proteins were resolved by SDS-PAGE, and radioactive proteins on dried gels were detected by a PhosphorImager (Molecular Dynamics).
RESULTS
Localization of the UhpA-docking region on UhpB.
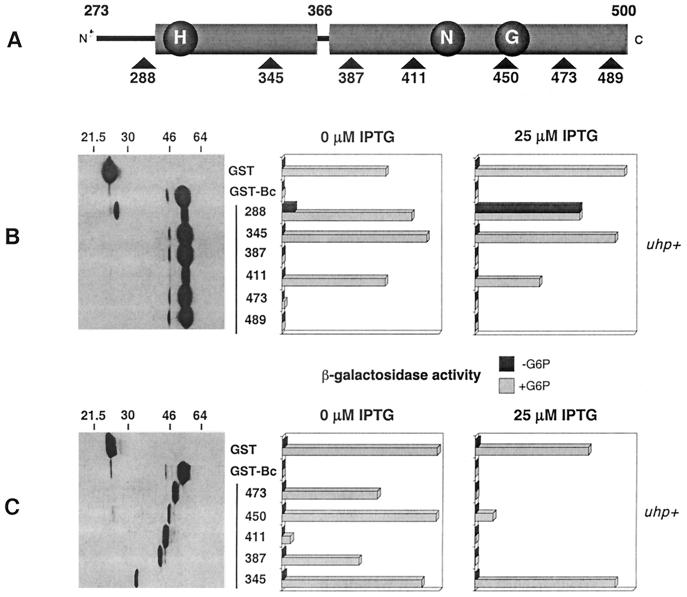
To help localize the portion of GST-Bc (containing UhpB residues 273 to 500) which is necessary for the inhibition of UhpA action, we took advantage of several Pstl linker insertions in this region (18). When present in full-length uhpB, the five mutations which introduce tetrapeptide insertions after residues 288 (designated UhpB-288::Ω4), 345, 411, 473, and 489 resulted in an uninducible Uhp− phenotype, whereas insertion after residue 387 resulted in normal inducibility (17). Each of these insertions was transferred into the coding sequence for GST-Bc (Fig. 1A) and examined for its effect on uhpT-lacZ expression in the uhp+ strain RK1309 (Fig. 1B). Western blot analysis using antibody to GST showed that most of the variant proteins were produced at comparable levels, with the exceptions that a substantial proportion of the GST-Bc-288::Ω4 protein was cleaved near the fusion junction and the level of GST-Bc-411::Ω4 was reduced. Three regulatory patterns were observed. When expressed in the absence of IPTG induction, the GST-Bc derivatives with insertions at residues 387, 473, and 489 exhibited the same strong dominant-negative interference as did GST-Bc, suggesting that these insertions did not affect the region involved in the docking of UhpA. In contrast, GST-Bc variants carrying insertions at residues 288, 345, and 411 showed no interference effect. The GST-Bc-288::Ω4 variant even conferred a low level of expression in the absence of Glc6P.
FIG. 1.
Effect of tetrapeptide insertions and C-terminal truncations on the interference phenotype exerted by GST-Bc. (A) Schematic representation of the UhpB transmitter region indicating the location of the tetrapeptide insertions and truncation endpoints relative to conserved motifs in HK proteins (H, N, and G boxes). (B) Basal expression levels of GST-Bc tetrapeptide insertion variants detected by Western immunoblot with detection of the GST moiety by monoclonal antibody 9D9 and their effect on uhpT-lacZ expression in RK1309 (uhp+) cells in the absence or presence of IPTG (25 μM). (C) The data shown are as in panel B but are for the GST-Bc C-terminal truncations. Cells were grown in the absence or presence of Glc6P (G6P).
Expression of the fusion proteins was increased by the addition of 25 μM IPTG to derepress the tac promoter driving their transcription. The uhpT-lacZ activity was then measured (Fig. 1B). Under these conditions, the GST-Bc-288::Ω4 variant conferred high basal expression comparable to the Glc6P-induced level. This variant was able to restore constitutive Uhp expression in strain RK1307 (ΔuhpBC) (data not shown), which suggests that this insertion in the putative linker region might activate the autokinase activity of GST-Bc. A similar constitutive phenotype had been observed for the E295G/E302K double substitution in the same vicinity (49). The insertion at residue 345 lacked the interference phenotype even when amplified, and the insertion at residue 411, which confers some lability, showed a modest increase in inhibition of uhpT-lacZ expression following induction with IPTG. The same patterns of dominant-negative behavior were observed when the GST-Bc::Ω4 variants were expressed in a strain carrying the phosphorylation-independent and constitutively active uhpA H170Y allele (data not shown). These results showed that the insertion at position 288 altered regulation and that at position 345 abrogated the interference with UhpA function.
Interference by truncated UhpB regions.
C-terminal truncations of GST-Bc were made that deleted UhpB sequences beyond the site of each linker insertion (Fig. 1C). Western immunoblot analysis which detected the GST moiety showed that all truncated GST-Bc variants produced polypeptides which were of the expected size but were present in reduced amounts relative to GST-Bc. In comparison to the full-length derivatives with linker insertions, most of the C-terminal truncations showed a reduced ability to inhibit uhpT-lacZ expression in the presence of Glc6P but without IPTG, except GST-Bc(273-411), which showed strong inhibition. Upon induction with 25 μM IPTG, all truncations except the shortest one, GST-Bc(273-345), showed strong inhibition of uhpT expression. As was found with the linker insertion derivatives, amplified expression of all truncated derivatives except GST-Bc(273-345) also strongly interfered with transcription driven by the phosphorylation-independent UhpA H170Y variant (data not shown). Thus, the portion of UhpB from residues 273 to 387, containing the phosphoryl transfer domain, is sufficient for the interference phenotype.
The UhpB region from residues 273 to 305 is predicted to form a coiled-coil motif (38) and shows some similarity to linker regions of other HK proteins. Some GST-Bc truncation derivatives in which the UhpB sequences started at residue 282, 288, or 293 were made (Table 1). Each of the N-terminal truncation variants blocked uhpT expression as completely as did the intact GST-Bc fusion, indicating that residues 273 to 293 were not required for the interference phenotype. Interestingly, when these N-terminal truncations were combined with the R324C substitution, which results in constitutive, UhpC-independent activation of autokinase activity, the resulting uhpT-lacZ expression was increased more than 10-fold relative to the GST-Bc R324C protein. The elevated activity of all three truncation derivatives was independent of Glc6P induction or the presence of the chromosome-encoded UhpB or UhpC protein. Thus, truncations into the linker region, which may extend from residues 273 to 305, resulted in enhanced autokinase or phosphotransfer activity when the autokinase activity was triggered by the R324C substitution, but the truncations did not allow autokinase activity by themselves. Other changes in this linker region could activate UhpB function by themselves, namely the insertion at residue 288, the double substitution at residues 295 and 302, and the L293P substitution described below.
TABLE 1.
Effects of UhpB N-terminal truncations on uhpT-lacZ expression
| Fusion | UhpB truncation | β-Galactosidase activity (U) in strain:
|
|||
|---|---|---|---|---|---|
| RK1309 (uhp+)
|
RK1307 (ΔuhpBC)
|
||||
| −Glc6P | +Glc6P | −Glc6P | +Glc6P | ||
| GST | 3 | 850 | 4 | 3 | |
| GST-Bc | 273–500 | 1 | 2 | ||
| 282–500 | 1 | 1 | |||
| 288–500 | 1 | 1 | |||
| 293–500 | 3 | 4 | |||
| GST-Bc R324C | 273–500 | 260 | 310 | 390 | 300 |
| 282–500 | >3,700 | >3,700 | 4,520 | 2,930 | |
| 288–500 | >3,700 | >3,700 | 5,180 | 3,240 | |
| 293–500 | >3,700 | >3,700 | >3,700 | 3,990 | |
Hybrids of UhpB and EnvZ.
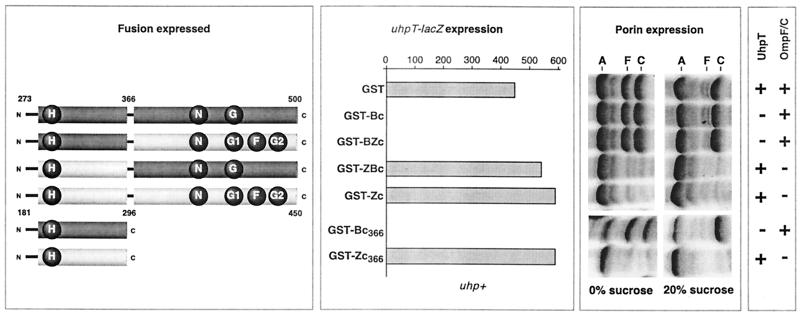
Several chimeric proteins which joined portions of UhpB and EnvZ were made to test whether replacement of the nucleotide-binding domain of UhpB with that from EnvZ would allow Uhp expression and to test for consequences of overexpression of the homologous domains from another two-component regulated system. The fusion junction was chosen on the basis of the domain boundaries seen in the structure of the EnvZ transmitter (34, 43). A proline residue in the linker region between the dimerization domain and the nucleotide-binding domain of EnvZ was also found to occur in a region of sequence similarity in UhpB. The coding regions around both proline residues were altered by the introduction of an EagI restriction site at comparable portions of both genes, thereby facilitating construction of the desired hybrid proteins by restriction fragment exchange. The hybrids carried the four possible combinations of the phosphoryl transfer domains of UhpB (residues 273 to 366) and EnvZ (residues 181 to 296) with the C-terminal nucleotide-binding HK domains of UhpB (residues 367 to 500) and EnvZ (residues 297 to 450) (Fig. 2, first panel). These regions were coupled to GST as in GST-Bc.
FIG. 2.
Expression of chimeric proteins containing segments of the UhpB and EnvZ transmitter domains and their effect on UhpT and OmpF-OmpC expression. The left panel shows a schematic portraying the domain organization of the GST fusion chimeras between UhpB (dark region) and EnvZ (light region). The second panel shows the effect of fusion protein expression on uhpT-lacZ expression in RK1309 (uhp+) cells measured by β-galactosidase activity in the presence of Glc6P. The third panel displays the effect of chimera fusion expression on outer membrane porin expression levels (A, OmpA; F, OmpF; C, OmpC) in RK1309 cells under moderate (0% sucrose) and high (20% sucrose) osmolarity conditions. The results of these experiments are summarized in the fourth panel and show whether expression of the reporter protein occurred (+) or was inhibited (−).
Western immunoblot analysis showed that all four fusions were expressed at levels comparable to that of GST-Bc (data not shown). Both GST-Bc (UhpB sequence from 273 to 500) and GST-BZc (UhpB sequence from 273 to 366 and EnvZ sequence from 297 to 450) completely blocked Glc6P-induced uhpT-lacZ expression in strain RK1309 (uhp+) (Fig. 2, second panel). In contrast, GST-ZBc (EnvZ sequence from 181 to 296 and UhpB sequence from 367 to 500) and GST-Zc (EnvZ sequence from 181 to 450) had no apparent effect on uhpT expression. This result confirmed that the region required for the inhibitory effect of GST-Bc lies between UhpB residues 273 and 366. In addition, the EnvZ ATP-binding domain, which is active for phosphate transfer (5, 51), showed no phosphate transfer to the H box of UhpB, as deduced from the lack of uhpT-lacZ expression in the presence of the GST-BZc variant.
Cells expressing these hybrid proteins were assayed for their effect on the levels of the porins OmpF and OmpC, whose transcription depends on the phosphorylation of OmpR by EnvZ. Changes in medium osmolarity alter the kinase and phosphatase activities of EnvZ, resulting in the expression of OmpF at a low osmolarity but of OmpC at a high osmolarity (35). Here, cells were grown in minimal medium A in the absence or presence of 20% sucrose as an osmolyte. In this minimal medium, roughly equal amounts of OmpF and OmpC were expressed by cells grown in the absence of sucrose, and OmpC was the major species after growth with sucrose. Strikingly, the presence of either GST-ZBc or GST-Zc almost completely abolished expression of both OmpF and OmpC under both growth conditions (Fig. 2, third panel). In contrast, GST-Bc and GST-BZc had no apparent effect on porin levels or on their osmoregulation. As expected from their decreased porin expression, cells expressing GST-ZBc and GST-Zc grew slowly in liquid media and formed small colonies on Luria-Bertani agar plates. The lack of cross-inhibition of Uhp signaling by GST-Zc or GST-ZBc and of porin regulation by GST-Bc or GST-BZc indicates that the specific recognition of the cognate RR is determined by the phosphoryl transfer domain.
The GST-BZc hybrid protein was modified to carry the R324C substitution, which activates the autokinase activity of GST-Bc, to test whether this substitution would allow the EnvZ nucleotide-binding domain to phosphorylate the UhpB-derived H box. This GST-BZc R324C variant showed no uhpT-lacZ expression in RK1307 (ΔuhpBC) and exerted strong interference in RK1309 (uhp+), just like the GST-BZc hybrid did (data not shown). This absence of phosphate transfer between noncognate phosphoryl transfer and nucleotide-binding domains in the same molecule points to a high specificity for their interaction as well as for interaction with the RR protein.
Activity of the liberated H box domains.
Derivatives of GST-Bc and GST-Zc with truncations of the nucleotide-binding domain terminating at residues 366 and 296, respectively, were examined. They were assayed for their effect on uhpT-lacZ expression and porin synthesis (Fig. 2). The truncated form of GST-Bc expressing UhpB residues 273 to 366, called GST-Bc366, exhibited a complete dominant-negative effect on Uhp expression even without IPTG induction. This interference effect was stronger than that exerted by the other C-terminal truncations shown in Fig. 1, perhaps owing to the greater stability of this domain without the appended portions of the kinase domain. As was seen with the chimeric GST-Bc and GST-BZc proteins, the GST-Bc366 variant had no effect on porin expression. An even shorter GST-Bc variant containing residues 293 to 366 of UhpB was also able to block Uhp signaling completely (data not shown).
The corresponding domain liberated from EnvZ, called GST-Zc296, had the same strong interference on OmpR signaling for porin expression as did the GST-Zc protein. It had no effect on uhpT-lacZ expression. These results show that the expression of the liberated phosphoryl transfer domain is sufficient for the interference behavior of the full-length chimeric proteins.
Variants of GST-Bc with altered signaling.
Genetic analysis of the functional role of amino acid residues in the phosphoryl transfer domain used two selection procedures for the isolation of mutants with altered regulatory behavior. The coding region for UhpB residues 293 to 366 was mutagenized by PCR and resected into the unmutagenized plasmid to restore the sequence of GST-Bc (residues 293 to 500). In the first selection, variants of GST-Bc which activate uhpT expression were selected as Uhp+ transformants able to grow on fructose-6-phosphate as a carbon source following introduction of the mutagenized plasmid into the ΔuhpBC strain RK1307. Plasmids isolated from these colonies were transformed into naïve RK1307 cells to confirm that the mutant phenotype was associated with the plasmid. Sequencing of the coding region for the phosphoryl transfer domain from the variant plasmids showed that 12 of 24 variants carried the previously described R324C substitution (49). The remaining 12 variants carried 10 unique single and 1 double substitution between residues 293 and 358. The nature of the amino acid changes in these gain-of-function mutations, which may also stimulate autokinase activity like the R324C substitution does, are presented in Table 2. All of the single amino acid substitutions affected residues in the two predicted α-helical regions or the joining loop of the phosphoryl transfer domain, except for the L293P mutation in the proposed linker region (see below).
TABLE 2.
Changes in the H box region of GST-Bc which activated uhpT-lacZ expressiona
| Mutant | No. of isolates | Presence of UhpT expression |
|---|---|---|
| GST | −b | |
| GST-Bc (293–500) | − | |
| L293P | 1 | + |
| I316F | 1 | + |
| T319I | 1 | + |
| I323M | 1 | + |
| R324C | 12 | + |
| I329F/A338T | 1 | + |
| V340M | 1 | + |
| S351T | 2 | + |
| L352Q | 1 | + |
| Y355H | 1 | + |
| D356A | 1 | + |
| D356G | 1 | + |
Growth of strain RK1307 (ΔuhpBC) on minimal A salts agar with 0.2% fructose-6-phosphate as the carbon source.
−, no uhpT-lacZ expression; +, uhpT-lacZ expression was present.
The second genetic selection procedure involved the isolation of GST-Bc variants which had lost the interference phenotype. The pool of mutagenized plasmids described above was introduced into the uhp+ strain RK1309. Ampicillin-resistant variants able to grow on fructose-6-phosphate as a carbon source were selected. This loss of interference is the same null phenotype which would be given by the empty vector or one encoding an inactive GST-Bc protein. As expected, this selection yielded many more colonies than did the selection for gain of autokinase function. Hence, 50 colonies were randomly chosen for analysis by Western immunoblot, which showed that only 14 of the 50 isolates produced full-length GST-Bc at levels comparable to that of the transformant carrying the unmutagenized plasmid. Plasmids obtained from these 14 isolates were transformed into naïve RK1309 for phenotypic analysis. The growth properties and levels of uhpT-lacZ expression indicated that all 14 variants had lost their dominant-negative character, even when their expression was increased by the addition of 25 μM IPTG (Table 3).
TABLE 3.
Changes in the H box region which relieved the dominant-negative effect of GST-Bc
| GST-Bc variant | No. of isolates | Presence of uhpT-lacZ expression in strain RK1309 (uhp+)
|
Uhp phenotypeb | ||
|---|---|---|---|---|---|
| −Glc6P | +Glc6P | +Glc6P and IPTGa | |||
| GST | −c | + | + | I | |
| GST-Bc (293–500) | − | − | − | DN | |
| L293P/D356A | 1 | + | + | + | C |
| I316T | 1 | − | + | + | I |
| I316N/S351L | 1 | − | + | + | I |
| T321S/T325P | 1 | − | + | + | I |
| R324C | 3 | + | + | + | C |
| A327E/V340G | 1 | − | + | + | I |
| V330D | 1 | − | + | + | I |
| G344R | 1 | − | + | + | I |
| L352P | 2 | − | + | + | I |
| Y355C | 1 | + | + | + | C |
| V358E | 1 | − | + | + | I |
25 μM IPTG.
I, inducible; DN, dominant negative; C, constitutive.
−, no uhpT-lacZ expression; +, uhpT-lacZ expression was present.
As expected, some of the noninterfering mutants were able to confer constitutive, Glc6P-independent activation of Uhp expression. These included one isolate carrying the double substitution L293P plus D356A, one isolate carrying the Y355C substitution, and three isolates carrying the R324C substitution. Note that substitutions in the same positions, namely L293P, R324C, Y355H, and D356A, had been identified in the previous selection for constitutive uhpT expression. The other nine noninterfering mutants were defective in both activation and interference and resulted from five unique single substitutions and three double substitutions (Table 3). These positions are scattered among the same positions as the up-regulated mutations. Some substitutions for the same amino acid had different phenotypes, namely I316F and L352Q conferred constitutive activation, whereas I316T and L352P resulted in loss of interference without activation of expression. Thus, numerous positions in the putative helical regions of the phosphoryl transfer domain appear to affect both the regulation of autokinase activity and the ability to interact with UhpA.
Dephosphorylation of P-UhpA by GST-Bc.
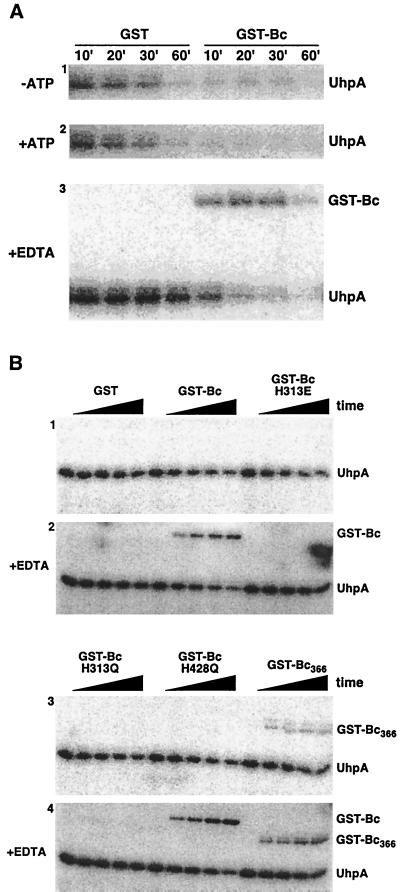
Many HK proteins can accelerate the dephosphorylation of their cognate RR protein. The cophosphatase activity of GST-Bc was expected to exist based on the finding that the turnover of P-UhpA was much more rapid when it was formed by phosphate transfer from GST-Bc R324C than when it was produced in the absence of GST-Bc by phosphate transfer from acetyl phosphate (4, 49). To demonstrate directly this cophosphatase activity, GST-Bc was added to 32P-labeled His6-UhpA prepared by incubation with acetyl [32P]phosphate and then purified by elution from an Ni2+-chelate matrix. The addition of the wild-type form of GST-Bc greatly increased the rate of dephosphorylation of P-UhpA compared to the rate in the presence of GST (Fig. 3A). Dephosphorylation of P-UhpA appeared to be increased somewhat in the presence of 1 mM ATP. The phosphatase activities of the NarX and NarQ proteins are independent of ATP (36), whereas those of NtrB (22) and EnvZ (15) depend on ATP as a cofactor.
FIG. 3.
Dephosphorylation of P-UhpA by GST-Bc. (A) UhpA (15 μM) was phosphorylated by incubation with 20 mM acetyl [32P]phosphate as described in Materials and Methods. P-UhpA was isolated by adsorption to Ni2+-conjugated agarose beads, washed, and eluted to remove excess acetyl [32P]phosphate. P-UhpA dephosphorylation was measured following further incubation in the presence of 1.5 μM GST or GST-Bc in the absence of ATP (first panel), in the presence of 1 mM ATP (second panel), and in the presence of 15 mM EDTA (third panel). Samples were removed at the indicated times and separated by SDS-PAGE. The location of phosphorylated proteins was determined by PhosphorImager analysis. (B) Reverse phosphotransfer was detected by incubation of P-UhpA with GST-Bc variants carrying substitutions in the H box (H313E and H313Q) or the catalytic domain (H428Q) (49) or with the nucleotide-binding domain removed (GST-Bc366). Following incubation with P-UhpA, portions were removed at 0, 5, 10, 20, and 30 min and analyzed as described for panel A. Hydrolysis of P-UhpA was determined in the absence of 15 mM EDTA (panels 1 and 3) and in its presence (panels 2 and 4).
Reverse phosphotransfer from UhpA to GST-Bc.
Transfer of phosphate from P-UhpA to GST-Bc was observed to occur under certain conditions. The removal of Mg2+ typically prevents phosphorylation of the receiver domain of RR proteins, and chelation of Mg2+ once phosphotransfer has occurred can stabilize the phosphorylated form of the protein (25). When the phosphatase assay with GST-Bc was carried out in the presence of Mg ions, all of the label released from P-UhpA was of low molecular weight and was presumably Pi. When the phosphatase assay was carried out in the presence of the metal chelator EDTA, the half-life of P-UhpA in the sample incubated with GST was greatly extended, from 10 to 20 min to around 60 min (Fig. 3A). In these experiments, the amount of label on UhpA was fairly low and extended exposure on the PhosphorImager screen was required for detection. The resultant reduction in the signal-to-background ratio prevented convincing quantification of the rates of phosphate release, although the general differences in lifetimes are clear. The rate of dephosphorylation of P-UhpA by GST-Bc was substantially slowed in the presence of EDTA but still showed a half-life of <10 min. Strikingly, label was seen to be transferred from P-UhpA to GST-Bc when EDTA was present. This labeling of GST-Bc was transient and the label was released from both proteins by 60 min.
The effect of several mutations affecting conserved residues in GST-Bc on the dephosphorylation of P-UhpA and on reverse phosphate transfer to GST-Bc was studied (Fig. 3B) using shorter time intervals than in the previous experiment. In this experiment, GST-Bc accelerated dephosphorylation of P-UhpA, but at a much lower rate than was seen for Fig. 3A. As expected, phosphate transfer from P-UhpA to GST-Bc was seen only when EDTA was present. Two variant forms of GST-Bc in which the presumed site of phosphorylation, His-313, was changed to Glu or Gln (H313E or H313Q) did not show formation of phosphorylated GST-Bc. These two variants seemed to retain some ability to accelerate dephosphorylation of P-UhpA. In contrast, the GST-Bc variant with the H428Q substitution in the N box, which is also completely defective for autokinase activity (49), showed dephosphorylation of P-UhpA comparable to that of GST-Bc. Phosphate transfer to the H428Q variant of GST-Bc occurred in the presence of EDTA.
The GST-Bc fusion carrying UhpB residues 273 to 366 (GST-Bc366) and lacking the HK domain appeared to be able to dephosphorylate P-UhpA, but at a lower rate than the other GST-Bc proteins. This truncated protein was able to accept phosphate from P-UhpA in the presence of EDTA and displayed a low level of phosphate transfer in the absence of EDTA. Taken together, these results show that GST-Bc can accelerate dephosphorylation of P-UhpA and that reverse phosphotransfer to GST-Bc can be trapped in the presence of EDTA or the absence of the HK nucleotide-binding domain. Reverse phosphotransfer requires the presence of His-313, but the data do not allow us to conclude whether His-313 is the site of phosphorylation or whether reverse transfer is an obligate step in P-UhpA dephosphorylation.
DISCUSSION
UhpA-docking domain.
The expression of UhpB or its cytoplasmic C-terminal segment as a GST fusion protein results in specific blockage of the Uhp signaling process by inactivation of the transcription activator UhpA. Dephosphorylation of P-UhpA is shown to be catalyzed by GST-Bc and presumably also occurs with the full-length protein, but the squelching phenotype occurs even under conditions where phosphorylation of UhpA is not involved. The most likely explanation for the squelching phenotype involves the binding or sequestration of UhpA rather than dephosphorylation of P-UhpA or the formation of inactive mixed multimers of GST-Bc with the wild-type UhpB. Localization of the inhibitory portion of UhpB, whose liberated expression prevents UhpA action, might identify the UhpA-docking surface on UhpB by a method analogous to the domain liberation approach of Morrison and Parkinson (30).
Probable recognition surfaces on several RR proteins for their cognate HK proteins have been identified (e.g., see references 11, 42, and 45), but there is less detailed knowledge about the recognition determinants on HK proteins for the cognate RR proteins. The CheY-docking domain on the HK CheA was identified as the P2 domain by the domain liberation technique (30) and was subsequently cocrystallized with CheY (27, 47). Most other HK proteins lack a P2 domain, as shown in the structure of the EnvZ protein (43, 44). Our studies here analyzing the regulatory consequences of expressing various segments of the C-terminal half of UhpB localized the region responsible for the interference phenotype to between residues 293 and 366. The insertion at residue 288 affected the interference phenotype by activating UhpB function, hence affecting regulation rather than UhpA docking. Several other substitutions in the proposed linker region upstream of the phosphoryl transfer domain also changed UhpB regulation from its default state of kinase-off. The phosphoryl transfer domain of UhpB contains the conserved His-313 residue, which is required for all known phosphotransfer processes, and the flanking residues in the H box. The corresponding region of EnvZ conferred the same phenotype by strongly inhibiting the action of its cognate RR, OmpR. This interference by docking and sequestration of the RR may be a general feature of HK proteins but may not be detected in wild-type cells owing to the considerable excess of OmpR molecules relative to EnvZ molecules (35).
The lack of activation of porin expression by GST-Zc was unexpected because some soluble forms of EnvZ exhibit high and unregulated autokinase activity in vitro (5, 33, 39). These forms of EnvZ retain substantial amounts of the input domain (16). To our knowledge, the activity of the cytoplasmic portion of EnvZ (residues 181 to 450) as used here has not been previously studied. The lack of activation could result from the presence of the appended GST moiety or of the M294R mutation formed by the EagI site substitution. It remains to be shown whether the dominant-negative effect of GST-Zc or GST-ZBc on porin expression results from sequestration of OmpR (34), as proposed for the effect by GST-Bc on UhpA, or from its action as P-OmpR phosphatase (15, 51). Nonetheless, the portion of UhpB which appears to be involved in the binding of UhpA overlaps the homologous region of a VanS-derived peptide which binds VanR (46), and portions of NtrB and EnvZ which are necessary for phosphatase activity on NtrC and OmpR, respectively (19, 23).
The interference by the phosphoryl transfer domains present in GST-Bc or GST-Zc with UhpA or OmpR action was highly specific: expression of the UhpB domain had no effect on OmpR function, or conversely, expression of the EnvZ domain had no effect on UhpA function. Thus, the interference phenotype is not a general disruption of two-component signaling but must reflect a specific interaction between the HK and its cognate RR. Whether the interference reflects a process that occurs in wild-type cells expressing full-length proteins at normal levels needs further study.
Phosphate transfer.
The ability of GST-Bc to accelerate dephosphorylation of P-UhpA was demonstrated using P-UhpA prepared by incubation with acetyl phosphate. This activity is seen in both GST-Bc and its constitutively active R324C variant (49). The activation of autokinase activity in the R324C variants thus appears not to result from the loss of its cophosphatase activity. Reverse transfer of phosphate from P-UhpA to GST-Bc was found to occur in the presence of EDTA and to be dependent on His-313. There are many possible explanations for this behavior. Metal chelation strongly retards the rate of spontaneous or GST-Bc-catalyzed dephosphorylation of UhpA as well as the rate of phosphate transfer to UhpA (4, 49). It is possible that Mg chelation by EDTA changes the reaction mechanism so that reverse transfer can occur only under these conditions. This seems unlikely, because reverse phosphotransfer has been observed for other HK-RR pairs in the absence of EDTA. Chelation may trap phosphate transferred to GST-Bc by preventing its transfer back to UhpA. Perhaps P-GST-Bc is subject to Mg-dependent hydrolysis by its nucleotide-binding portion. Unlike the situation with EnvZ and NtrB (14, 22, 31), dephosphorylation of P-UhpA by GST-Bc did not require the presence of ATP, although ATP may stimulate it somewhat (but see reference 51). Interestingly, the truncated species GST-Bc366, which lacks the nucleotide-binding domain, exhibited a low level of reverse phosphotransfer from P-UhpA in the absence of EDTA. Our conclusion at this time is that the phosphoryl transfer domain of UhpB can recognize and bind UhpA and can accept phosphate from P-UhpA.
A role for reverse phosphotransfer in RR dephosphorylation has been proposed for EnvZ, PhoR, and ArcA (5, 9, 37). Some HK proteins lose phosphatase activity when the histidine at the site of phosphorylation is changed to certain residues but not to others (13, 21, 39, 51). This variable dependence on the identity of the substitute for the histidine residue questions the role of the His in the phosphatase reaction and suggests that reverse phosphotransfer is not the only route by which HK proteins can accelerate dephosphorylation of their cognate RR protein. Some substitutions for the histidine might disrupt the surrounding active site and alter the protein conformation, which is important for phosphatase action or RR binding. In the case of GST-Bc, replacement of His-313 with Glu or Gln prevented both autokinase activity and reverse phosphotransfer from P-UhpA but did not prevent the interference phenotype, which probably requires only the binding of UhpA. It remains to be determined how these substitutions affect UhpB activity in the context of the full-length protein.
Model for dimerization domain.
In many HK proteins, the H box is located downstream of the linker region and is part of the dimerization domain which, in the cases of EnvZ and Spo0B, was shown to form part of a four-helix bundle. The linker region plays an important role in coupling transmembrane signal reception to regulation of autokinase and/or phosphatase activity (48). This region is predicted to form a coiled-coil structure based on its heptad repeat character. Interestingly, EnvZ does not require the integrity of this coiled-coil region for autophosphorylation, phosphotransfer, or phosphatase activity in vitro, suggesting that the points of contact with OmpR do not lie in this region (33). For UhpB, various structure prediction models suggest that the likely linker region of residues 273 to 305 following the last transmembrane segment can form a coiled-coil structure. GST-Bc variants with truncations of this region with fusion junctions at UhpB residues 282, 288, and 293 did not confer activation of GST-Bc kinase activity and still showed the strong interference phenotype, as did the GST-Bc(273–500) species, indicating that the putative linker region is not involved in contact with UhpA or in keeping the kinase activity in a silent state. However, these truncations in combination with the constitutively active R324C substitution showed about 10 times more activity than the R324C variant with the intact linker region. This finding, together with the constitutive phenotypes of the 288-Ω4, E295G/E302K, and L293P variants, supports the importance of the linker region in regulating the phosphate transfer process.
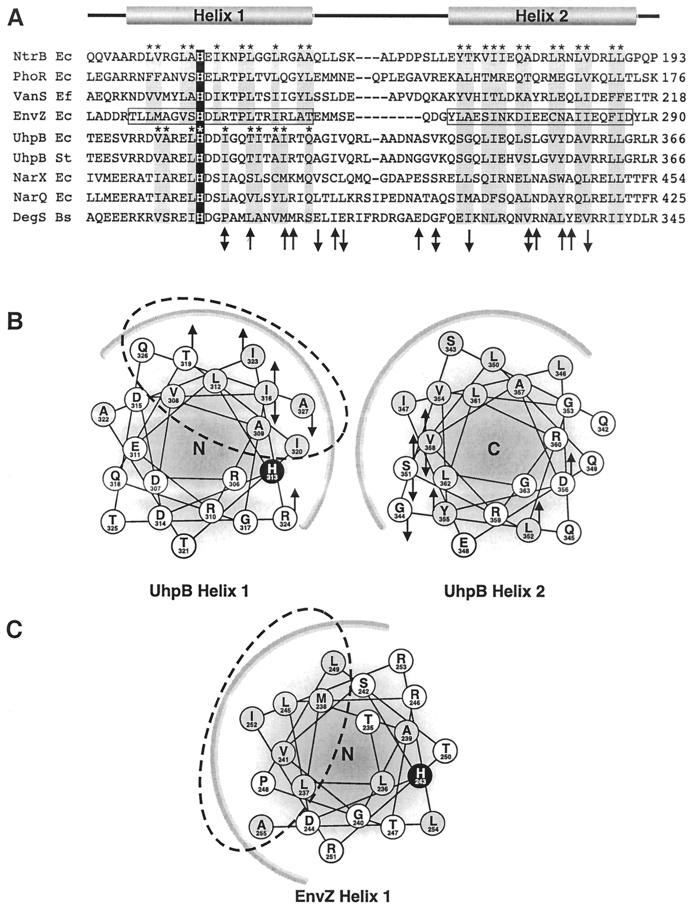
Although the greatest sequence similarity in this region of HK proteins is in the H box, there is substantial relatedness throughout the phosphoryl transfer and dimerization domain, as shown in Fig. 4 (12). Several secondary structure prediction programs suggested that the regions of UhpB from residues 305 to 327 and 342 to 363 could form two α-helices. The ends of these putative helices were not convincingly assigned, and the position of the loop separating the homologous helices in EnvZ was not correctly identified by these programs. Nevertheless, display of the residues forming the predicted four-helix bundle in UhpB as helical wheels revealed some interesting features (Fig. 4). The helix containing residues 305 to 327 (helix 1) displays helical faces of three distinct properties, one hydrophobic, one basic, and one acidic. The second helix, containing residues 342 to 363 (helix 2), also shows strong amphipathic character, with a hydrophobic face and a polar face but without the prominent charge segregation seen in helix 1. It is likely that the interfaces between helices in the four-helix bundles are through their hydrophobic surfaces, as in EnvZ.
FIG. 4.
Sequence alignment and secondary structure prediction of the UhpB four-helix bundle domain. (A) Alignment of the four-helix bundle regions of representative transmitters. Helix 1 and helix 2 correspond to the helices defined in the structure of this region of EnvZ (44). Based on the EnvZ structure, the residues in helix 1 and helix 2 that contribute to the interhelical and dimer packing interface are denoted with an asterisk and shaded gray. The phosphorylated histidine is shaded black. (B) Helical wheel diagram of proposed interhelical interface of the UhpB monomer. Helix 1 is viewed from its N terminus (N) and helix 2 is viewed from its C terminus (C). Hydrophobic residues are shaded gray and His-313 is shaded black. The heavy arc along both helical wheel diagrams corresponds to the putative interhelical hydrophobic packing interface with the other UhpB monomer. Substitutions in the UhpB H box domain from Tables 2 and 3 that result in gain of function (upward arrow) or loss of function (downward arrow) or different substitutions that result in both phenotypes (two-headed arrow) are shown on the helical wheels and on the primary sequence in panel A. (C) Helical wheel diagram of EnvZ helix 1. The dashed oval corresponds to the residues involved in the interhelical and dimer interface of EnvZ helix 1 as determined by nuclear magnetic resonance. The relative positions of the active site His-243 in EnvZ and His-313 in UhpB helix 1 can be compared with the location of the hydrophobic interfaces denoted by the dashed oval.
Strikingly, His-313 in UhpB is exposed at the boundary between the hydrophobic face and the basic face of helix 1. Similar exposure of this highly conserved histidine at the boundary between the hydrophobic and polar faces was also predicted for all other HK proteins of the same homology family as UhpB, which we examined. On the other hand, in EnvZ and related HK proteins, this histidine is exposed in the middle of the polar face. The suggested structure of the four-helix bundle shows that most of the mutations which affected GST-Bc function fall on or near the hydrophobic face of each helix. In EnvZ several amino acid substitutions which affected autokinase regulation were located in the X region (12) at the C-terminal end of helix 2. It is likely that disruption of helical packing might increase or decrease access of the histidine to the nucleotide-binding domain, thereby altering its activity as an autokinase. We suggest that the His in UhpB is normally not accessible to the nucleotide-binding domain, thereby accounting for the lack of autokinase activity in the wild-type protein. The UhpA-binding residues of UhpB have not been identified yet. However, the UhpA-binding surface is likely to include the poorly conserved residues in the loop and adjoining helices in the four-helix bundle. It is intriguing that a few gain-of-activity mutations affected residues on the polar faces of both helices. Future studies will address the effect of these changes on UhpA binding and test whether the access of the histidine to phosphorylation can be affected by the binding of inducer and the presence of UhpC.
ACKNOWLEDGMENTS
We appreciate helpful discussions with Qing Chen, Igor Olekhnovich, Bob Nakamoto, and Phil Matsumura. We thank Amy Ma and J. T. Parsons for the monoclonal antibody to GST.
This work was supported by research grant GM38681 from the National Institute of General Medical Sciences. J.S.W. was a predoctoral trainee supported in part by training grant CA09091 from the National Cancer Institute.
REFERENCES
- 1.Amemura M, Makino K, Shinagawa H, Nakata A. Cross talk to the phosphate regulation of Escherichia coli by PhoM protein: PhoM is a histidine protein kinase and catalyzes phosphorylation of PhoB and PhoM-open reading frame 2. J Bacteriol. 1990;172:6300–6307. doi: 10.1128/jb.172.11.6300-6307.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bilwes A M, Alex L A, Crane B R, Simon M I. Structure of CheA, a signal-transducing histidine kinase. Cell. 1999;96:131–141. doi: 10.1016/s0092-8674(00)80966-6. [DOI] [PubMed] [Google Scholar]
- 3.Caldwell R C, Joyce G F. Randomization of genes by PCR mutagenesis. PCR Methods Appl. 1992;2:28–33. doi: 10.1101/gr.2.1.28. [DOI] [PubMed] [Google Scholar]
- 4.Dahl J L, Wei B Y, Kadner R J. Protein phosphorylation affects binding of the Escherichia coli transcription activator UhpA to the uhpT promoter. J Biol Chem. 1997;272:1910–1919. doi: 10.1074/jbc.272.3.1910. [DOI] [PubMed] [Google Scholar]
- 5.Dutta R, Inouye M. Reverse phosphotransfer from OmpR to EnvZ in a kinase−/phosphatase+ mutant of EnvZ (EnvZ · N347D), a bifunctional signal transducer of Escherichia coli. J Biol Chem. 1996;271:1424–1429. doi: 10.1074/jbc.271.3.1424. [DOI] [PubMed] [Google Scholar]
- 6.Dutta R, Qin L, Inouye M. Histidine kinases: diversity of domain organization. Mol Microbiol. 1999;34:633–640. doi: 10.1046/j.1365-2958.1999.01646.x. [DOI] [PubMed] [Google Scholar]
- 7.Falke J J, Bass R B, Butler S L, Chervitz S A, Danielson M A. The two-component signaling pathway of bacterial chemotaxis: a molecular view of signal transduction by receptors, kinases, and adaptation enzymes. Annu Rev Cell Dev Biol. 1997;13:457–512. doi: 10.1146/annurev.cellbio.13.1.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fisher S L, Jiang W, Wanner B L, Walsh C T. Cross-talk between the histidine protein kinase VanS and the response regulator PhoB. J Biol Chem. 1995;270:23142–23149. doi: 10.1074/jbc.270.39.23143. [DOI] [PubMed] [Google Scholar]
- 9.Georgellis D, Kwon O, De Wulf P, Lin E C C. Signal decay through a reverse phosphorelay in the Arc two-component signal transduction system. J Biol Chem. 1998;273:32864–32869. doi: 10.1074/jbc.273.49.32864. [DOI] [PubMed] [Google Scholar]
- 10.Grebe T W, Stock J B. The histidine protein kinase superfamily. Adv Microb Phys. 1999;41:139–227. doi: 10.1016/s0065-2911(08)60167-8. [DOI] [PubMed] [Google Scholar]
- 11.Haldimann A, Prahalad M K, Fisher S L, Kim S-K, Walsh C T, Wanner B L. Altered recognition mutants of the response regulator PhoB: a new genetic strategy for studying protein-protein interactions. Proc Natl Acad Sci USA. 1996;93:14361–14366. doi: 10.1073/pnas.93.25.14361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hsing W, Russo F D, Bernd K K, Silhavy T J. Mutations that alter the kinase and phosphatase activities of the two-component sensor EnvZ. J Bacteriol. 1998;180:4538–4546. doi: 10.1128/jb.180.17.4538-4546.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hsing W, Silhavy T J. Function of conserved histidine-243 in phosphatase activity of EnvZ, the sensor for porin osmoregulation in Escherichia coli. J Bacteriol. 1997;179:3729–3735. doi: 10.1128/jb.179.11.3729-3735.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Igo M M, Ninfa A J, Silhavy T J. A bacterial environmental sensor that functions as a protein kinase and stimulates transcriptional activation. Genes Dev. 1989;3:598–605. doi: 10.1101/gad.3.5.598. [DOI] [PubMed] [Google Scholar]
- 15.Igo M M, Ninfa A J, Stock J B, Silhavy T J. Phosphorylation and dephosphorylation of a bacterial transcriptional activator by a transmembrane receptor. Genes Dev. 1989;3:1725–1734. doi: 10.1101/gad.3.11.1725. [DOI] [PubMed] [Google Scholar]
- 16.Igo M M, Silhavy T J. EnvZ, a transmembrane environmental sensor of Escherichia coli K-12, is phosphorylated in vitro. J Bacteriol. 1988;170:5971–5973. doi: 10.1128/jb.170.12.5971-5973.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Island M D, Kadner R J. Interplay between the membrane-associated UhpB and UhpC regulatory proteins. J Bacteriol. 1993;175:5028–5034. doi: 10.1128/jb.175.16.5028-5034.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Island M D, Wei B-Y, Kadner R J. Structure and function of the uhp genes for the sugar phosphate transport system in Escherichia coli and Salmonella typhimurium. J Bacteriol. 1992;174:2754–2762. doi: 10.1128/jb.174.9.2754-2762.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang P, Atkinson M R, Srisawat C, Sun Q, Ninfa A J. Functional dissection of the dimerization and enzymatic activities of Escherichia coli nitrogen regulator II and their regulation by the PII protein. Biochemistry. 2000;39:13433–13449. doi: 10.1021/bi000794u. [DOI] [PubMed] [Google Scholar]
- 20.Kadner R J. Expression of the Uhp sugar-phosphate transport system of Escherichia coli. In: Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington, D.C.: ASM Press; 1995. pp. 263–274. [Google Scholar]
- 21.Kamberov E S, Atkinson M R, Chandran P, Ninfa A J. Effect of mutations in Escherichia coli ginL (ntrB), encoding nitrogen regulator II (NRII or NtrB), on the phosphatase activity involved in bacterial nitrogen regulation. J Biol Chem. 1994;269:28294–28299. [PubMed] [Google Scholar]
- 22.Keener J, Kustu S. Protein kinase and phosphoprotein phosphatase activities of nitrogen regulatory proteins NtrB and NtrC of enteric bacteria: roles of conserved amino terminal domain of NtrC. Proc Natl Acad Sci USA. 1988;85:4976–4980. doi: 10.1073/pnas.85.14.4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kramer G, Weiss V. Functional dissection of the transmitter module of the histidine kinase NtrB in Escherichia coli. Proc Natl Acad Sci USA. 1999;96:604–609. doi: 10.1073/pnas.96.2.604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lugtenberg B, Meijers J, Peters R, van der Hoek P, van Alphen L. Electrophoretic resolution of the “major outer membrane protein” of Escherichia coli K12 into four bands. FEBS Lett. 1975;58:254–258. doi: 10.1016/0014-5793(75)80272-9. [DOI] [PubMed] [Google Scholar]
- 25.Lukat G S, Stock A M, Stock J B. Divalent metal ion binding to the CheY protein and its significance to the phosphotransfer in bacterial chemotaxis. Biochemistry. 1990;29:5436–5442. doi: 10.1021/bi00475a004. [DOI] [PubMed] [Google Scholar]
- 26.McCleary W R, Stock J B. Acetyl phosphate and the activation of two-component response regulators. J Biol Chem. 1994;269:31567–31572. [PubMed] [Google Scholar]
- 27.McEvoy M M, Hausrath A C, Randolph G B, Remington S J, Dahlquist F W. Two binding modes reveal flexibility in kinase/response regulator interactions in the bacterial chemotaxis pathway. Proc Natl Acad Sci USA. 1998;95:7333–7338. doi: 10.1073/pnas.95.13.7333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Merkel T J, Nelson D M, Brauer C L, Kadner R J. Promoter elements required for positive control of transcription of the Escherichia coli uhpT gene. J Bacteriol. 1992;174:2763–2770. doi: 10.1128/jb.174.9.2763-2770.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller J H. A short course in bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 30.Morrison T B, Parkinson J S. Liberation of an interaction domain from the phosphotransfer region of CheA, a signaling kinase of Escherichia coli. Proc Natl Acad Sci USA. 1994;91:5485–5489. doi: 10.1073/pnas.91.12.5485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ninfa A J, Magasanik B. Covalent modification of the glnG product, NRI, by the glnL product, NRII, regulates the transcription of the glnALG operon in Escherichia coli. Proc Natl Acad Sci USA. 1986;83:5909–5913. doi: 10.1073/pnas.83.16.5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ninfa A J, Ninfa E G, Lupas A N, Stock A, Magasanik B, Stock J. Crosstalk between bacterial chemotaxis signal transduction proteins and regulators of transcription of the Ntr regulon: evidence that nitrogen assimilation and chemotaxis are controlled by a common phosphotransfer mechanism. Proc Natl Acad Sci USA. 1988;85:5492–5496. doi: 10.1073/pnas.85.15.5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park H, Inouye M. Mutational analysis of the linker region of EnvZ, an osmosensor in Escherichia coli. J Bacteriol. 1997;179:4382–4390. doi: 10.1128/jb.179.13.4382-4390.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park H, Saha S K, Inouye M. Two-domain reconstitution of a functional protein histidine kinase. Proc Natl Acad Sci USA. 1998;95:6728–6732. doi: 10.1073/pnas.95.12.6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pratt L A, Silhavy T J. Porin regulon of Escherichia coli. In: Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington, D.C.: ASM Press; 1995. pp. 105–127. [Google Scholar]
- 36.Schröder I, Wolin C D, Cavicchioli R, Gunsalus R P. Phosphorylation and dephosphorylation of the NarQ, NarX, and NarL proteins of the nitrate-dependent two-component regulatory system of Escherichia coli. J Bacteriol. 1994;176:4985–4992. doi: 10.1128/jb.176.16.4985-4992.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shi L, Liu W, Hulett F M. Decay of activated Bacillus subtilis Pho response regulator, PhoP-P, involves the PhoR-P intermediate. Biochemistry. 1999;38:10119–10125. doi: 10.1021/bi990658t. [DOI] [PubMed] [Google Scholar]
- 38.Singh M, Berger B, Kim P S, Berger J M, Cochran A G. Computational learning reveals coiled coil-like motifs in histidine kinase linker domains. Proc Natl Acad Sci USA. 1998;95:2738–2743. doi: 10.1073/pnas.95.6.2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Skarphol K, Waukau J, Forst S A. Role of His-243 in the phosphatase activity of EnvZ in Escherichia coli. J Bacteriol. 1997;179:1413–1416. doi: 10.1128/jb.179.4.1413-1416.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Skarstedt M T, Silverstein E. Escherichia coli acetate kinase mechanism studied by net initial rate, equilibrium, and independent isotopic exchange kinetics. J Biol Chem. 1976;251:6775–6783. [PubMed] [Google Scholar]
- 41.Stock A M, Robinson V L, Goudreau P N. Two-component signal transduction. Annu Rev Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 42.Swanson R V, Lowry D F, Matsumura P, McEvoy M M, Simon M I, Dahlquist F W. Localized pertubations in CheY structure monitored by NMR identify a CheA binding interface. Nat Struct Biol. 1995;2:906–910. doi: 10.1038/nsb1095-906. [DOI] [PubMed] [Google Scholar]
- 43.Tanaka T, Saha S K, Tomomori C, Ishima R, Liu D, Tong K I, Park H, Dutta R, Qin L, Swindells M B, Yamazaki T, Ono A M, Kainosho M, Inouye M, Ikura M. NMR structure of the histidine kinase domain of the E. coli osmosensor EnvZ. Nature. 1998;396:88–92. doi: 10.1038/23968. [DOI] [PubMed] [Google Scholar]
- 44.Tomomori C, Tanaka T, Dutta R, Park H, Saha S K, Zhu Y, Ishima R, Liu D, Tong K I, Kurokawa H, Qian H, Inouye M, Ikura M. Solution structure of the homodimeric core domain of Escherichia coli histidine kinase EnvZ. Nat Struct Biol. 1999;6:729–734. doi: 10.1038/11495. [DOI] [PubMed] [Google Scholar]
- 45.Tzeng Y-L, Hoch J A. Molecular recognition in signal transduction: the interaction surfaces of the SpoOF response regulator with its cognate phosphorelay proteins revealed by alanine scanning mutagenesis. J Mol Biol. 1997;272:200–212. doi: 10.1006/jmbi.1997.1226. [DOI] [PubMed] [Google Scholar]
- 46.Ulijasz A T, Kay B K, Weisblum B. Peptide analogues of the VanS catalytic center inhibit VanR binding to its cognate promoter. Biochemistry. 2000;39:11417–11424. doi: 10.1021/bi0012888. [DOI] [PubMed] [Google Scholar]
- 47.Welch M, Chinardet N, Mourey L, Birck C, Samama J-P. Structure of the CheY-binding domain of histidine kinase CheA in complex with CheY. Nat Struct Biol. 1997;5:25–29. doi: 10.1038/nsb0198-25. [DOI] [PubMed] [Google Scholar]
- 48.Williams S B, Stewart V. Functional similarities among two-component sensors and methyl-accepting chemotaxis proteins suggest a role for linker region amphipathic helices in transmembrane signal transduction. Mol Microbiol. 1999;33:1093–1102. doi: 10.1046/j.1365-2958.1999.01562.x. [DOI] [PubMed] [Google Scholar]
- 49.Wright J S, Olekhnovich I N, Touchie G A, Kadner R J. The histidine kinase domain of UhpB inhibits UhpA action at the Escherichia coli uhpT promoter. J Bacteriol. 2000;182:6279–6286. doi: 10.1128/jb.182.22.6279-6286.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou J, Blair D F. Residues of the cytoplasmic domain of MotA essential for torque generation in the bacterial flagellar motor. J Mol Biol. 1997;273:428–439. doi: 10.1006/jmbi.1997.1316. [DOI] [PubMed] [Google Scholar]
- 51.Zhu Y, Qin L, Yoshida T, Inouye M. Phosphatase activity of histidine kinase EnvZ without kinase catalytic domain. Proc Natl Acad Sci USA. 2000;97:7808–7813. doi: 10.1073/pnas.97.14.7808. [DOI] [PMC free article] [PubMed] [Google Scholar]