Summary
Cilia or eukaryotic flagella are microtubule-based organelles found across the eukaryotic tree of life. Their very high aspect ratio and crowded interior are unfavorable to diffusive transport of most components required for their assembly and maintenance. Instead, a system of intraflagellar transport (IFT) trains moves cargo rapidly up and down the cilium (Figure 1A).1, 2, 3 Anterograde IFT, from the cell body to the ciliary tip, is driven by kinesin-II motors, whereas retrograde IFT is powered by cytoplasmic dynein-1b motors.4 Both motors are associated with long chains of IFT protein complexes, known as IFT trains, and their cargoes.5, 6, 7, 8 The conversion from anterograde to retrograde motility at the ciliary tip involves (1) the dissociation of kinesin motors from trains,9 (2) a fundamental restructuring of the train from the anterograde to the retrograde architecture,8,10,11 (3) the unloading and reloading of cargo,2 and (4) the activation of the dynein motors.8,12 A prominent hypothesis is that there is dedicated calcium-dependent protein-based machinery at the ciliary tip to mediate these processes.4,13 However, the mechanisms of IFT turnaround have remained elusive. In this study, we use mechanical and chemical methods to block IFT at intermediate positions along the cilia of the green algae Chlamydomonas reinhardtii, in normal and calcium-depleted conditions. We show that IFT turnaround, kinesin dissociation, and dynein-1b activation can consistently be induced at arbitrary distances from the ciliary tip, with no stationary tip machinery being required. Instead, we demonstrate that the anterograde-to-retrograde conversion is a calcium-independent intrinsic ability of IFT.
Keywords: intraflagellar transport, total-internal reflection microscopy, TIRF microscopy, micromanipulator, cilia and flagella, ciliary tip
Graphical abstract
Highlights
-
•
Anterograde IFT trains can change direction without aid of a ciliary tip
-
•
IFT trains turn around normally in the absence of free calcium ions
-
•
Disengagement from the microtubule likely triggers anterograde-to-retrograde conversion
Intraflagellar transport (IFT) allows the assembly of cilia by moving components from the cell to the ciliary tip and back. Nievergelt et al. demonstrate that, contrary to what was previously hypothesized, no stationary tip machinery is required at the tip for IFT turnaround, which is instead a calcium-independent intrinsic ability of IFT trains.
Results and discussion
The hypothesis of the existence of dedicated machinery at the ciliary tip that is responsible for the conversion of trains from their anterograde to their retrograde structure and motility is supported by several lines of evidence. First, anterograde intraflagellar transport (IFT) trains travel consistently to the tip and seldom, if ever, reverse direction before reaching this destination,9,14, 15, 16, 17, 18 implying that the tip is uniquely specialized for this function. Second, the ciliary tip is characterized by large protein complexes that cap the microtubule ends14,19, 20, 21, 22 and that may contain machinery involved in converting anterograde to retrograde trains. Third, ciliogenesis associated kinase 1 (CILK1, also called ICK) and related kinases are found at the ciliary tip, although not exclusively, and have been implicated in regulating IFT turnaround and release of kinesins and cargo.23,24 However, whether the switch from anterograde to retrograde IFT requires dedicated machinery at the ciliary tip is not understood.
Here, we directly analyze the requirement for the ciliary tip in Chlamydomonas cells by blocking anterograde transport at intermediate positions along the cilium/flagellum together with imaging of IFT by total internal reflection microscopy (TIRFM).
We first set out to physically block and observe IFT in Chlamydomonas cilia. In order to apply a gentle localized force onto a specific spot on the cilia of gliding cells, we fabricated a silicone wedge by drop casting silicone resin onto the end of a glass capillary mounted to a glass slide. After curing the drop on a hot plate and trimming it, the wedge was detached from the glass slide (Figure 1B). Consequently, this wedge was mounted on a hydraulic micro-manipulator, which was installed on a TIRF microscope. During TIRFM imaging, the wedge was carefully lowered onto the cilium of a gliding cell (Figure 1C). The soft wedge, when carefully controlled by the micro-manipulator, allows fine control over the degree to which IFT trains are blocked. Insufficient pressure would result in IFT trains passing underneath the partial barrier created by the wedge. Excessive pressure, instead, would damage or even sever the cilium, resulting in a rapid tearing of the ciliary membrane and the spread of all visible IFT trains onto the nearby glass surface. When using just the right amount of pressure, IFT trains can no longer pass through the blockage (Figures 1D and 2A). As a consequence, we observed no fluorescence underneath the wedge and a striking accumulation of trains on both sides of it (Figures 1D and 2A [upper part of the kymogram]). After blocking the cilium with the wedge, the trains between wedge and tip continued normally until they got stuck at the tip side of the wedge, where they remained visible as a dot of constant brightness (Figure 2A [upper part of the kymogram]). The rest of the space between the wedge and the tip became progressively devoid of fluorescence signal, since no new anterograde trains could arrive to the tip from the cell body. This observation emphasizes (1) the completeness of the blockage and (2) that retrograde trains do not spontaneously convert back into anterograde trains.
Figure 1.
The intraflagellar transport (IFT) machinery of gliding Chlamydomonas can be observed and manipulated in total internal reflection microscopy (TIRFM)
(A) ① Anterograde trains, driven by the heterotrimeric motor kinesin-II, move towards the ciliary tip. ② At the ciliary tip, kinesin-II dissociates from the train, and the trains convert into retrograde form by an unknown mechanism. ③ Reassembled retrograde trains move away from the tip towards the cell body by the dynein-1b motor specific for IFT.
(B) A sharp but soft wedge is made by drop casting silicone onto a supporting capillary, followed by trimming off excess after curing.
(C) Illustration of mechanical blockage by lowering a silicone wedge onto the cilium of a Chlamydomonas reinhardtii cell by micro-manipulator on a TIRF microscope.
(D) Individual frames from a TIRFM movie show how IFT is locally and reversibly blocked inside a cilium by the force applied using a silicone wedge (dashed line, non-blocking in blue, blocking in red). See also Video S1.
Figure 2.
Kymograms visualizing the time-dependent response of the IFT machinery to external manipulation
(A) Compressing the cilium with a soft wedge creates a small band where IFT cannot pass (horizontal white lines in the kymogram, created by the accumulation of stopped fluorescent trains). After the blockage is applied, trains build up and shortly after start moving in retrograde direction. The approximate location of the ciliary tips is outlined for clarity in purple. See also Video S1.
(B) Detail view of the region between the cell body and the block (white square) in (A). About 2.5 s after IFT is locally blocked, retrograde trains can be seen moving towards the cell body at normal speed (white arrows).
(C) Kymogram of wedge blocking experiment in severely calcium-depleted cilia (see also Figure S1 and Videos S2 and S3). Transport and conversion show no discernable difference to (A) where calcium is available.
(D) In lithium-treated cells, the motors are chemically inhibited and pile up at variable distances from the ciliary tip. Gliding of the cell dislodges the stuck trains, which immediately move in retrograde direction at high speed (≈10 μm s−1) (insert). Anterograde trains are colored in green, while retrograde trains are colored in magenta.
Anterograde trains, in contrast, were still injected from the cell body into the cilium and traveled up to the barrier where they also formed a bright fluorescence accumulation spot. Notably, on the cell-body side of the barrier (proximal side), about 2.5 s post blocking, retrograde IFT trains could be observed as they departed from just below the barrier and moved toward the cell (Figure 2B). Soon after the onset of this phenomenon, the frequency of departing retrograde trains returned to a rate that we would consider normal at the ciliary tip. When the wedge was retracted, the motion of the artificially arrested trains on both sides of the wedge reproducibly returned to regular anterograde and retrograde motion without any noticeable residual effect (Video S1). These data clearly show that the conversion from anterograde to retrograde IFT can and does take place in the absence of the ciliary tip.
It has been proposed that calcium-dependent kinases are involved in the dissociation of kinesin-II from anterograde trains,25 leading to conversion to retrograde motility. Thus, we reasoned that the application of the wedge could induce a localized tension on the ciliary membrane and potentially open nearby calcium channels. If that were the case, the resulting calcium influx from the buffer would activate the calcium-dependent kinases and locally induce the anterograde-to-retrograde conversion. Therefore, to test whether free calcium is a cofactor in IFT turnaround at the barrier, we performed the wedge blocking experiment in calcium-free tris-acetate-phosphate (TAP) medium with ethylene glycol-bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid (EGTA) added as a strong calcium chelator. This medium is further referred to as TAP-Ca. We confirmed the depletion of calcium by using a Chlamydomonas strain with a fluorescent free-calcium reporter (GCaMP6), expressed as a fusion protein to the dynein regulatory complex protein 4 (DRC4). In TAP medium, these cells exhibited periodic flashes of free calcium in their cilia, which were also observed when the cilia were only lightly touched by the wedge (Figure S1; Video S2). In contrast, we observed that in TAP-Ca, cells exhibited a total lack of spontaneous or induced flashes (Video S3), indicating that in such an environment, the free calcium is below the level of detection. When repeating the wedge blocking experiment in TAP-Ca medium, anterograde IFT trains still converted to retrograde trains, and no noticeable differences to the observations made in regular TAP medium could be observed (Figure 2C). Therefore, we conclude that calcium is not required for IFT train turnaround and for activation of IFT dyneins. These results suggest that the mere stalling of the trains at the wedge is sufficient to induce anterograde-to-retrograde conversion.
It remains possible that application of the silicon wedge has some additional unidentified effects on the structure of the trains or the microtubules that might be involved in the conversion. To address this point, we treated Chlamydomonas cells with 13 mM LiCl, which inhibits IFT motors,26 thereby causing frequent pauses in the motion of trains (Figure 2D) without mechanical interference. This inhibition of motility resulted in the accumulation of anterograde IFT trains along the cilium, which were unable to dislodge on their own. However, we observed that the onset of the gliding motion of a cell was often associated with the restored motility of stuck trains. Interestingly, as can be seen in Figure 2D, stuck trains always reactivated in a retrograde direction, even if they were anterograde trains when stalling. Hence, treatment of Chlamydomonas with LiCl also can trigger anterograde-to-retrograde conversion and shows that, irrespective of the method, stopping and accumulation of trains induces conversion.
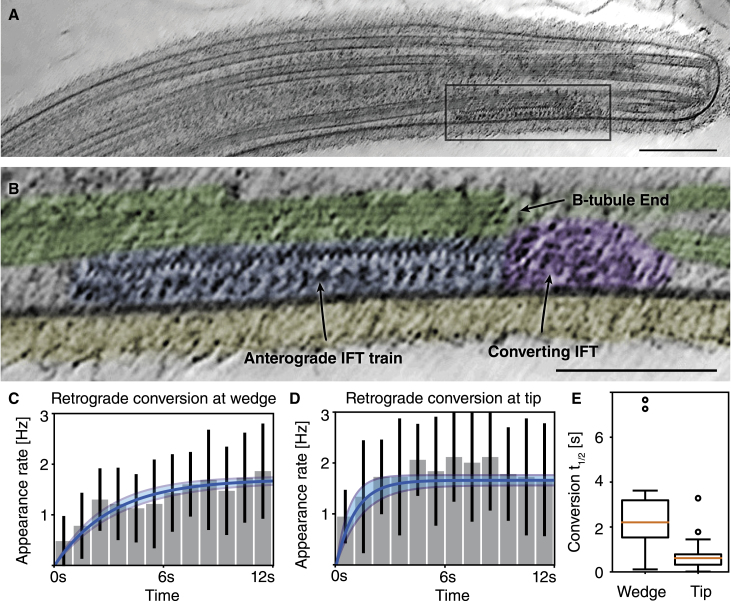
Having established that the ability to convert from the anterograde to retrograde form is intrinsic to IFT trains, we wanted to better understand what happens when the trains reach the ciliary tip. In order to visualize trains that are in the process of anterograde-to-retrograde conversion at the ciliary tip, we performed cryo-electron tomography on cilia of intact cells (Figure 3A). In Figure 3B, we show a denoised27 tomographic slice through a train located to the distal end of the axoneme (Video S4). Such trains show the expected repeating densities of anterograde trains8 as long as they are engaged with the B-tubule of the microtubule doublet they are walking on.10 Interestingly, this structure is rapidly lost after the train disengages from the microtubule, supporting the idea that this disengagement is part of the mechanism that initiates IFT turnaround. Likewise, in Caenorhabditis elegans, IFT turnaround occurs within the distal micron of cilia, apparently corresponding to the ends of individual microtubules that do not quite reach the tip.28
Figure 3.
Kinetics of anterograde to retrograde transport conversion of IFT trains
(A and B) Tomographic slice through ciliary tip showing an anterograde train in the process of disassembly (scale bar, 200 nm). Highlighted rectangular region is magnified in (B), showing loss of structural cohesion of the train localized at the end of the B-tubule (scale bar, 100 nm). The structured part of the train is colored in blue, the disordered part of the train in violet, the microtubule in green, and the ciliary coat in yellow. See also Video S4.
(C) Retrograde train appearance frequency at physical blockage. Transport recovers to normal steady-state level with a half-time of 2.6 ± 1.7 s (first-order response fit in blue, uncertainty bounds in light gray).
(D) Retrograde train appearance frequency at the ciliary tip after releasing the physical blockage. Steady-state retrograde transport recovers with a half-time of 0.77 ± 0.74 s (fit same as C; error bars in C and D represent SEM).
(E) Box plot of train recovery half-time at the wedge and at the ciliary tip after release.
Data at the wedge (C and E) from 895 trains in 23 experiments on 12 individual cells.
Data at the tip (D and E) from 580 individual trains in 18 experiments on 10 individual cells.
See also Figure S2.
We reasoned that if the microtubule end and "walking off the rail" at the tip are relevant for anterograde-to-retrograde conversion, this process should be in some way facilitated at the tip. Hence, we tested whether conversion at the ciliary tip and at the wedge exhibit different dynamics. To this end, we quantified the frequency of retrograde trains forming at (1) the proximal side of the wedge directly after onset of wedge blocking and (2) at the ciliary tip after first depleting the tip of all IFT material by wedge blocking and then releasing anterograde trains by removing the wedge.
Figures 3C and 3E show the rate at which retrograde trains depart from the wedge area after the wedge blocks transport. While very rapidly converting trains were occasionally observed, the conversion of most of the trains required several seconds. When this transition was modeled as a first-order system (fitted blue line with light grey uncertainty bounds), we found that IFT returned to its nominal conversion frequency with a recovery half-time of 2.6±1.7 s after blocking. This recovery time does not significantly depend on the position of the wedge on the cilium (Figure S2). Therefore, the turnaround is not dependent on the time the IFT train has been in the cilium, because in that case, we would expect trains to linger longer at blockages placed closer to the cell body.
In order to compare this to natural conversion at the tip, we repeated the same annotation and analysis for the departure times of retrograde trains at the IFT-depleted tip. Figures 3D and 3E show the analysis of 18 such experiments on 10 different cells. In contrast to conversion at the wedge, IFT returned to the steady state about three times faster, with a recovery half-time of 0.77±0.74 s, in line with previously reported values.9 Hence, while anterograde-to-retrograde conversion can happen in the absence of the ciliary tip, our findings also show that the rate of conversion is higher at the tip than at the wedge. The disengagement of the trains from the microtubule at the ciliary tip, where they literally walk off their rails, seems to speed up IFT turnaround.
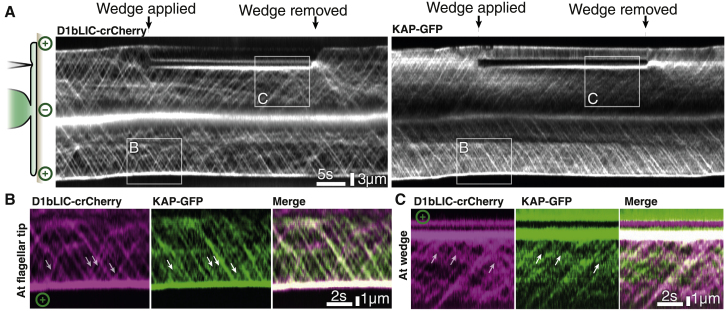
In order to further understand the mechanism of train rearrangement at the wedge with respect to the two IFT motors, we recorded TIRFM movies of the cilia of CC-5502, which is dual labeled for kinesin-II (by the subunit kinesin accessory protein [KAP-GFP]) and dynein (by the subunit dynein-1b light intermediate chain [D1bLIC-crCherry]).9 While dynein is visible on both anterograde and retrograde trains departing from the ciliary tip, those same retrograde trains usually lack co-localized signal of KAP-GFP (Figures 4A–4C). We only occasionally observed KAP-GFP signals moving with retrograde IFT trains, while the majority of trains show no discernable KAP-GFP signal, as the kinesin-II returns to the cell body by diffusion (Figure S3), as previously reported.9,16 Likewise, dynein, but not kinesin, was associated with retrograde trains that arose from the wedge indicating that, as at the tip, the kinesin detached from the train and dynein was activated to drive retrograde IFT (Figures 4B and 4C). As such, only dynein remains part of IFT trains after conversion. The release of kinesin motors from the train, together with the dissociation from the microtubule, could also contribute to the destabilization and fragmentation of the highly ordered polymeric structure of anterograde trains at the tip as well as at the wedge blockage. Phosphorylation of kinesin is known to affect cargo binding and activity of the motor.29 Kinases that phosphorylate and inhibit kinesin-II, such as CILK1 in mammals and CrCDPK1 in Chlamydomonas, have been reported to localize at the tip. However, CILK1 is not exclusive to the ciliary tip, and it moves along the cilium as if carried by IFT.30 Likewise, a related Chlamydomonas kinase, LF4, is not found preferentially at the tip even though it is also implicated in IFT turnaround.31 Thus, kinases that move together with IFT trains remain potential attractive players in IFT turnaround. On the contrary, it seems unlikely that CrCDPK1, a calcium-dependent kinase, is responsible for the IFT turnaround, as the process can take place in the absence of calcium also.
Figure 4.
Dynamics of the IFT motors kinesin-II and dynein during anterograde to retrograde conversion
(A) Denoised kymograms of dual-labeled cilia show distinct behavior between dynein (labeled with D1bLIC-mCherry) and kinesin-II (labeled with KAP-GFP), both in the undisturbed flagellum and when blocked by a wedge. Inserts illustrate regions highlighted in (B) and (C).
(B) Background-subtracted detail at the ciliary tip. Positions of some prominent retrograde trains are marked with white arrows, with the same positions shown in the KAP-GFP detail for reference.
(C) Background-subtracted detail at the wedge. White arrows denote the location of visible retrograde trains, the positions of which are duplicated in the KAP-GFP detail for reference.
See also Figure S3.
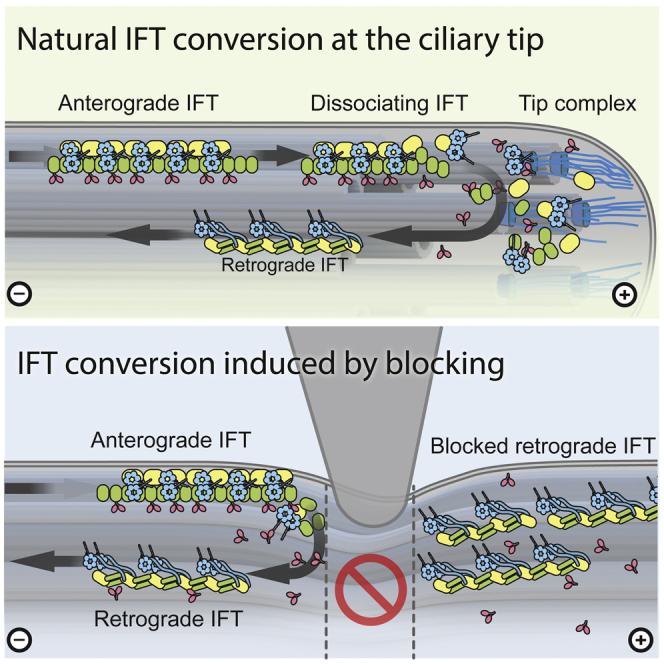
In this work, we demonstrate that anterograde IFT trains can reverse their direction in the absence of the ciliary tip. When mechanically blocked, anterograde trains spontaneously and independently of free calcium convert into retrograde trains. In complement, by observing chemically inhibited trains, we offer another way by which IFT turnaround can be induced at locations distant from the ciliary tip. We assume that the chemical inhibition of motors causes a different form of mechanical blockage—one that does not require an external force to be applied but that also results in turnaround of IFT trains.
It is a possibility that the IFT trains that pile up at the wedge carry the means to assemble a fresh tip complex structure, which then mediates anterograde-to-retrograde conversion. However, two observations in our data suggest that this is likely not the case: first, the fact that only a few piled up trains are necessary before the first retrograde trains appear would mean that the components of hypothetical tip machinery are abundant cargo of the trains. However, all ciliary components that are known as IFT cargos are only found in small amounts and on a small percentage of IFT trains and in full-length cilia.32, 33, 34 Second, when the wedge is released, we did not observe any passing trains spontaneously stopping or even converting at the position of the former blockage, indicating that no stationary tip complex structure was formed at the wedge blockage.
We hypothesize that anterograde trains at the wedge detach from the microtubule because of a buckling instability caused by the motors compressing the trains against the wedge from the side of the cell body. Anterograde trains might be inherently unstable and might require the physical interaction with the ciliary membrane on one side and the microtubule through the kinesin motor on the opposite side to maintain their structure. Being pushed out of the microtubule track at the wedge or running off the end of the microtubule at the tip could be sufficient to trigger the fragmentation of an anterograde train and the exposure to train-associated kinases and other enzymes that might be involved in the inhibition of the kinesin motors, uncoupling of other cargo from the IFT machinery, or activation of the dynein motors for retrograde transport.
In contrast to anterograde trains, retrograde trains were never seen to reverse directions at the wedge: they remained stationary until the wedge was removed and then continued toward the cell body. A trivial explanation for this in Chlamydomonas is that kinesin motors are not present on retrograde trains.9,16 Additional factors may also be involved because in C. elegans, kinesins are carried on retrograde trains, yet they only turn around at the ciliary base.35 The dramatic rearrangement of IFT trains that occurs during anterograde-to-retrograde conversion8,10 may further constrain the turnaround of retrograde trains. Retrograde trains may need to be completely disassembled as (or after) they leave the cilium before their subunits can repolymerize in the anterograde configuration.
Our data indicate that the conversion from anterograde to retrograde transport is an intrinsic property of IFT trains that can happen all along the axoneme without any dedicated stationary tip machinery required. Even though the ciliary tip is not necessary for conversion, this process is roughly three times faster at the tip than it is at a wedge blockage site. Based on this kinetic difference, we propose a conversion-after-derailment model, in which the switch from anterograde to retrograde IFT requires disengagement of trains from the microtubule doublet.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-IFT46 | Rosenbaum lab | 17600 |
| anti-GFP | Roche | 11814460001; RRID:AB_390913 |
| Deposited data | ||
| TIRF Microscopy Timeseries | This study | https://doi.org/10.5281/zenodo.6787231 |
| Chemicals, peptides, and recombinant proteins | ||
| PDMS | Dow Corning | Sylgard 184 |
| Lithium Chloride | Sigma-Aldrich | 203637 |
| Tris Base | Merck | 93362 |
| Ethylene glycol-bis(2-aminoethylether)-N,N,N′,N′-tetraacetic acid | Sigma | E4378 |
| NH4Cl | Sigma | A9434 |
| MgSO4 | Unknown | N/A |
| KCl | Sigma | 1.04936 |
| K2HPO4 | Sigma | P8281 |
| KH2PO4 | Sigma | P0662 |
| Hutner's Trace Elements | Chlamydomonas Resource Center | N/A |
| Experimental models: Organisms/strains | ||
| Chlamydomonas: ift46-1::NIT | Chlamydomonas Resource Center | CC-4375 |
| Chlamydomonas: pf2 | Chlamydomonas Resource Center | CC-1025 |
| Chlamydomonas: KAP-GFP D1bLIC-crCherry | Chlamydomonas Resource Center | CC5502 |
| Chlamydomonas: mat3-4 | Chlamydomonas Resource Center | CC3994 |
| Recombinant DNA | ||
| IFT46-mNeonGreen | This work | N/A |
| PF2-GFP | Porter lab | N/A |
| pSI103 | Chlamydomonas Resource Center | N/A |
| pE345 | Unknown | N/A |
| Software and algorithms | ||
| FRAP analysis script | This work | makeFrapPlots.py |
| Python 3 | Python Software Foundation | https://www.python.org |
| Geneious | Biomatters | https://www.geneious.com |
| Train analysis script | This work | makeDirectionalMasks.py |
| iQ3 imaging software | Andor | https://andor.oxinst.com/products/iq-live-cell-imaging-software/ |
| Fiji | Fiji | https://imagej.net/software/fiji/ |
| IMOD | Mastonarde group | https://bio3d.colorado.edu/imod/ |
| CryoCare | Jug Lab | https://github.com/juglab/cryoCARE_T2T |
| CSBDdeep Noise2Void | Fiji | https://csbdeep.bioimagecomputing.com/ |
| SerialEM | Mastonarde group | https://bio3d.colorado.edu/SerialEM/ |
| MotionCor2 | UCSF | https://emcore.ucsf.edu/ucsf-software |
| ICON | Sun Lab | http://www.ibp.cas.cn/feilab/research/md/202111/t20211115_6256123.html |
| Other | ||
| Glass Capillaries | Sutter instru-ments | BF100–50–7.5 |
| Glass slides | Marienfeld | 1000000 |
| Holey Carbon Grids | Quantifoil | Q350AR-35 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Gaia Pigino (gaia.pigino@fht.org).
Materials availability
-
•
Strains generated in this study have been deposited to the Chlamydomonas Resource Center as CC-5900 and CC-5901.
-
•
Plasmids generated in this study have been deposited to the Chlamydomonas Resource Center as pIFT46-mNeonGreen-PAR and pPF2-GcAMP6-PAR.
Experimental model and subject details
Chlamydomonas cell culture
IFT54-mNeon-IFT139-mCherry cells were a kind gift from Karl Lechtreck, University of Georgia. CC-5502 KAP-GFP D1bLIC-crCherry cells were purchased from the Chlamydomonas Resource Center. The cells were inoculated from TAP–agar plates into TAP medium and grown in aeration bottles connected to clean air under circadian conditions to early- to mid-log phase. Cells for wedge-experiments were harvested at 1000×g for 10 min and subsequently resuspended in TAP medium supplemented with 1 mM EGTA (ethylene glycol-bis(β-aminoethyl ether)-N,N,N’,N’-tetraacetic acid, E3889, Sigma Aldrich, USA). Cells for LiCl experiments were likewise washed in just TAP buffer.
Method details
Creation of IFT46-mNeonGreen
The EcoRI-EcoRV YFP containing fragment in pE345 (IFT46-YFP under control of PSAD promoter and IFT46 5’ UTR and unknown 3’UTR; contains IFT46 genomic DNA; paramomycin resistance) was replaced by an EcoRI-EcoRV fragment encoding the mNeonGreen ORF. The fragment was codon optimized for expression in Chlamydomonas. Codon optimization was done with GenSmart™ Codon optimization tool (GenScript) and synthesized by Integrated DNA Technology. The resulting plasmid was linearized with PsiI.
CC-4375 ift46-1::NIT1 mt+ cells were obtained from the Chlamydomonas Resource Center and grown in TAP to a dense colony. Cells were harvested at 800RCF and subsequently had their cell walls removed with autolysin. 400uL of concentrated cells were then mixed with ≈600 ng of linearized plasmid and vortexed with 0.5 mm Zirconia/Glass beads (11079105z, BioSpecProducts, Bartlesville, OK, United States) for 30 s before plating on TAP agar plates with 10 μg mL−1 Paromomycin for positive selection. Colonies were subsequently analyzed by western blotting for IFT46.
Creation of PF2-GcAMP6
The PF2-GFP plasmid obtained from Mary Porter as a gracious gift. The Chlamydomonas PF2 (DRC4) was amplified by PCR using the PF2-GFP plasmid as a template and cloned into pBR25 using XhoI and BamHI sites. GCaMP6 was amplified using PCR and cloned in-frame at the unique BamHI at the c-terminal of PF2.
The paralyzed flagella strain pf2 (CC-1025, Chlamydomonas Resource Center), was cotransformed with 1 μg of PF2::pGCaM6 and 1 μg pSI103, encoding an aphVIII paromomycin resistance cassette,36 using the glass bead method,37 but without the addition of polyethylene glycol. Following transformation, the cells were plated on TAP agar plates containing 10 μg/ml paromomycin and resulting colonies were screened for motility after transfer to TAP medium in microtiter wells. Cilia of swimming cells were screened by western blotting with anti-GFP for the PF2::pGCaMP6.
Wedge preparation
Glass capillaries (BF100–50–7.5, Sutter instruments, USA) were placed on microscope slides with double-sided adhesive tape and a drop of polydimethylsiloxane (PDMS, Sylgard 184, Dow Corning, USA), premixed at 10:1 monomer-crosslinker ratio, applied to the end of the capillary. The slice was subsequently placed on a hot plate at 120 °C to 140 °C to cure the PDMS for several minutes. Finally, the sides of the PDMS drop were trimmed off with a scalpel and the device carefully delaminated from the microscope slide right before use.
TAP-Ca medium preparation
Calcium-free TAP-Ca medium was prepared by dissolving 2.42 g of Tris-Base (93362, Merck KGaA, Darmstadt, Germany) to 850 mL deionized water. 25 mL of TAP-Ca salts (15 g L−1 NH4Cl, 4 g L−1 MgSO4, 2 g L−1 KCl) and 1 mL of Phosphate solution (288 g L−1 K2 HPO4 , 2 g L−1 KH2 PO4) and 1 mL of Hutner’s trace elements (Chlamydomonas resource center) were added sequentially. Finally, the pH was adjusted to 7 with 1 mL of glacial acetic acid and the total volume adjusted to 1L. The medium was then aliquoted in glass bottles and autoclaved. 1 mM EGTA was added immediately prior to TIRF imaging. TAP-Ca is nominally Ca2+ free (free Ca2+ <0.1 nM).
TIRF microscopy
All imaging was performed on an inverted TIRF microscope (IX71, Olympus, Japan with an Olympus UApochromat 150x/1.45 Oil-immersion TIRFM objective) equipped with an EMCCD camera (iXon Ultra 897 BV, Andor, Belfast, Northern Ireland). The microscope is equipped with a custom-built TIRF condenser with manual TIRF-angle adjustment. TIRF illumination is provided by a fiber coupled 491 nm diode-pumped solid state laser, operated at nominally 260 μW. The microscope is fitted with a water hydraulic micromanipulator (WR-6, Narishige, Japan), which was used to hold and position the wedge. All images were recorded at 81 ms frame time and 512x512 pixels at a resolution of 0.107 μm px−1. FRAP and FLIP data was acquired at 100 ms frame time and 1024x1024 pixels at a resolution of 0.086 μm px−1. In the case of cells treated with Lithium, 100 μL of 33 mM LiCl was added to 0.5 mL of cells directly on the glass slide on the TIRF microscope without mixing.
For photo-bleaching experiments, cells were imaged on a Ti2 Eclipse (Nikon, Japan) equipped with a 100x/1.49NA TIRF objective (MRD01991, Nikon, Japan) with a custom orbital TIRF consisting of a FRAP/TIRF combination system (iLas2, Gataca Systems, France) coupled to a multi-laser combiner (VS-LMS-MOT100, Visitron Systems, Germany). The system is equipped with multiple EMCCD cameras (iXon life, Oxford Instruments, UK) for simultaneous acquisition and is controlled by VisiView software (Visitron Systems, Germany). The wedge was manipulated by a combination of a coarse manipulator (MN-4, Narishige, Japan) and a fine manipulator (MMO-4, Narishige, Japan). FRAP pulses were applied for 40 ms at 8 mW of 488 nm laser light with manual point-and-shoot.
Cryo electron tomography
Whole C. reinhardtii cells (mat3-4, CC3994) were frozen by applying a light cell suspension directly onto the front side of glow-discharged holey carbon grids (Q350AR-35, Quantifoil Micro Tools, Germany). Cells were pretreated with 5 mM LiCl to increase the length of cilia. 10 nm gold beads were added in suspension as fiducial markers and the grids blotted from the back side before plunge freezing into liquid ethane.
Grids were imaged at 300 kV on an FEI Titan Halo equipped with a field-emission gun, an imaging energy filter operated at 20 eV and a K2 direct detection camera (both Gatan, USA). Cilia were located on full-grid low-magnification images and imaged bi-directionaly in 2° steps from −20° to 60° and −22° to −60° in SerialEM.38 Images were recorded at a nominal magnification of 30 000 in dose-fractioned super-resolution stacks of 10 images per tilt angle with a total of 3.2 e−A−2, resulting in a nominal pixel size of 2.35 A px−1.
The tilt-series data was motion-corrected and binned once with MotionCor2-1.2.6.39 Resulting tilt-stacks were aligned, CTF-corrected in Etomo in IMOD-4.10.32.40 To denoise the data, raw frame stacks were split into motion-corrected even/odd tilt frames. The previously generated alignment was applied to both even and odd stacks. These fine-aligned stacks were reconstructed into tomograms with ICON-GPU-v1.2.941 on cluster nodes with two NVidia M2090 Tesla GPUs. Finally, the two tomograms were used as input to cryoCARE27,42 in order to generate a denoised tomogram.
Quantification and statistical analysis
Analysis of TIRFM image data
Recorded TIRFM images were opened in Fiji,43 a de-speckle filter applied, and the background subtracted with a rolling-ball filter with a radius of 20 pixels. To correct for cells not sitting still, the stacks were maximum-projected in groups in order to identify all possible locations of the cilia, which were then traced individually using splines. Thus, selected splines were used to create kymograms from the filtered and background-subtracted movies using the multi-kymograph plugin (https://imagej.net/Multi_Kymograph), using a linewidth of 3 pixels. Resulting kymograms were aligned using StackReg (http://bigwww.epfl.ch/thevenaz/stackreg/) registration and finally maximum-projected to result in a kymogram covering the entire temporal duration of the recording. Finished kymograms were analyzed with a Python script using OpenCV-v4.1.244 to identify anterograde and retrograde trains by template matching with 8px × 8px templates of lines at angles of ±35° to ±55°, respectively. Resulting maps were curated for false positives and used to false color the original kymogram data.
Data used for Figures 4A–4C has been denoised with the CSBDeep module Noise2Void45 using a network trained with TIRF recordings of fluorescently labelled IFT trains and motors (40 epochs).
Measurements of train-recovery during and after wedge blocking are based on manual annotation of appearing retrograde trains. Individual experiments have been fitted in integral space to convert the stochastic appearance of trains into a cumulative number of total trains per time. Each experiment was fitted to a cumulative recovery function of the form
where A is an arbitrary scaling constant. Fitting was performed with a Pythom script using NumPy-1.22.2, SciPy-1.8.0 and plotted using Matplotlib-3.5.1.
Photo-bleaching experiments were analyzed with a custom Python script. In short, regions of kymograms after a photo-bleaching pulse were cropped out and a region of about 0.5 μm to 1.2 μm averaged together to be fitted in intensity along the time axis with the function
and recovery or loss half-times calculated as
Acknowledgments
We would like to thank the Light Microscopy Facility (LMF) as well as the Electron Microscopy Facility (EMP) at MPI-CBG for their support. We would also like to thank Aliona Bogdanova from the Protein Expression and Purification (PEPC) Facility for cloning of the IFT46-mNeonGreen plasmid. Additionally, we would like to thank Iain K. Patten for fruitful discussions and corrections to the manuscript and Oscar Gonzales for assistance with high-performance computing resources. A.P.N was supported by an EMBO long–term fellowship under ALTF number 891-2018 as well as by an HFSP cross-disciplinary fellowship with reference number LT000515/2019. I.Z. was supported by the IMPRS Student Research Internship program. L.S. was supported by the Dresden International Graduate School for Biomedicine and Bioengineering (DIGS-BB), granted by the German Research Foundation (DFG) in the context of the Excellence Initiative. Finally, we would like to acknowledge the DFG grant GACR-DFG Cooperation 2019 no. PI1218/3-1 and the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (grant agreement no. 819826) to G.P.
Author contributions
Conceptualization, A.P.N., L.S., and G.P.; methodology, A.P.N., D.D., L.S., and G.P.; investigation, A.P.N., I.Z., A.C., D.D., M.D., T.B., and L.S.; data curation, A.P.N.; visualization, A.P.N.; formal analysis, A.P.N.; writing – original draft, A.P.N. and G.P.; writing – review & editing, A.P.N., D.D., T.B., M.D., S.D., F.J., L.S., and G.P.; funding acquisition, A.P.N., S.D., F.J., and G.P.; resources, S.D., F.J., and G.P.; supervision, S.D., F.J., and G.P.
Declaration of interests
The authors declare no competing interests.
Published: August 3, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.cub.2022.07.033.
Contributor Information
Luděk Štěpánek, Email: ludek.stepanek@img.cas.cz.
Gaia Pigino, Email: gaia.pigino@fht.org.
Supplemental information
Data and code availability
-
•
TIRFM time series data used in this paper has been deposited to Zenodo with https://doi.org/10.5281/zenodo.6787231. All other original data for this work are available upon request from the lead contact.
-
•
Code generated in this study is available upon request from the lead contact.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Kozminski K.G., Johnson K.A., Forscher P., Rosenbaum J.L. A motility in the eukaryotic flagellum unrelated to flagellar beating. Proc. Natl. Acad. Sci. USA. 1993;90:5519–5523. doi: 10.1073/pnas.90.12.5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lechtreck K.F. IFT–Cargo Interactions and Protein Transport in Cilia. Trends Biochem. Sci. 2015;40:765–778. doi: 10.1016/j.tibs.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Picariello T., Brown J.M., Hou Y., Swank G., Cochran D.A., King O.D., Lechtreck K., Pazour G.J., Witman G.B. A global analysis of IFT-A function reveals specialization for transport of membrane-associated proteins into cilia. J. Cell Sci. 2019;132:jcs220749. doi: 10.1242/jcs.220749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rosenbaum J.L., Witman G.B. Intraflagellar transport. Nat. Rev. Mol. Cell Biol. 2002;3:813–825. doi: 10.1038/nrm952. [DOI] [PubMed] [Google Scholar]
- 5.Cole D.G., Diener D.R., Himelblau A.L., Beech P.L., Fuster J.C., Rosenbaum J.L. Chlamydomonas Kinesin-II-dependent Intraflagellar Transport (IFT): IFT Particles Contain Proteins Required for Ciliary Assembly in Caenorhabditis elegans Sensory Neurons. J. Cell Biol. 1998;141:993–1008. doi: 10.1083/jcb.141.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pigino G., Geimer S., Lanzavecchia S., Paccagnini E., Cantele F., Diener D.R., Rosenbaum J.L., Lupetti P. Electron-tomographic analysis of intraflagellar transport particle trains in situ. J. Cell Biol. 2009;187:135–148. doi: 10.1083/jcb.200905103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taschner M., Lorentzen E. The Intraflagellar Transport Machinery. Cold Spring Harb. Perspect. Biol. 2016;8:a028092. doi: 10.1101/cshperspect.a028092.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jordan M.A., Diener D.R., Stepanek L., Pigino G. The cryo-EM structure of intraflagellar transport trains reveals how dynein is inactivated to ensure unidirectional anterograde movement in cilia. Nat. Cell Biol. 2018;20:1250–1255. doi: 10.1038/s41556-018-0213-1. http://www.nature.com/articles/s41556-018-0213-1 [DOI] [PubMed] [Google Scholar]
- 9.Chien A., Shih S.M., Bower R., Tritschler D., Porter M.E., Yildiz A. Dynamics of the IFT machinery at the ciliary tip. Elife. 2017;6 doi: 10.7554/eLife.28606. e28606–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stepanek L., Pigino G. Microtubule doublets are double-track railways for intraflagellar transport trains. Science. 2016;352:721–724. doi: 10.1126/science.aaf4594. https://www.sciencemag.org/lookup/doi/10.1126/science.aaf4594 [DOI] [PubMed] [Google Scholar]
- 11.Wingfield J.L., Mekonnen B., Mengoni I., Liu P., Jordan M., Diener D., Pigino G., Lechtreck K. In vivo imaging shows continued association of several IFT-A, IFT-B and dynein complexes while IFT trains U-turn at the tip. J. Cell Sci. 2021;134:jcs259010. doi: 10.1242/jcs.259010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Webb S., Mukhopadhyay A.G., Roberts A.J. Intraflagellar transport trains and motors: Insights from structure. Semin. Cell Dev. Biol. 2020;107:82–90. doi: 10.1016/j.semcdb.2020.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Collingridge P., Brownlee C., Wheeler G.L. Compartmentalized calcium signaling in cilia regulates intraflagellar transport. Curr. Biol. 2013;23:2311–2318. doi: 10.1016/j.cub.2013.09.059. [DOI] [PubMed] [Google Scholar]
- 14.Dentler W. Intraflagellar transport (IFT) during assembly and disassembly of Chlamydomonas flagella. J. Cell Biol. 2005;170:649–659. doi: 10.1083/jcb.200412021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shih S.M., Engel B.D., Kocabas F., Bilyard T., Gennerich A., Marshall W.F., Yildiz A. Intraflagellar transport drives flagellar surface motility. Elife. 2013;2:e00744. doi: 10.7554/elife.00744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Engel B.D., Lechtreck K.F., Sakai T., Ikebe M., Witman G.B., Marshall W.F. First edit edition. Vol. 93. Elsevier; 2009. Total Internal Reflection Fluorescence (TIRF) Microscopy of Chlamydomonas Flagella; pp. 157–177. (Methods in cell biology). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Absalon S., Blisnick T., Kohl L., Toutirais G., Doré G., Julkowska D., Tavenet A., Bastin P. Intraflagellar Transport and Functional Analysis of Genes Required for Flagellum Formation in Trypanosomes. Mol. Biol. Cell. 2008;19:929–944. doi: 10.1091/mbc.e07-08-0749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Besschetnova T.Y., Roy B., Shah J.V. Methods in Cell Biology. Elsevier; 2009. Imaging Intraflagellar Transport in Mammalian Primary Cilia; pp. 331–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dentler W.L. Structures linking the tips of ciliary and flagellar microtubules to the membrane. J. Cell Sci. 1980;42:207–220. doi: 10.1242/jcs.42.1.207. [DOI] [PubMed] [Google Scholar]
- 20.Miller J.M., Wang W., Balczon R., Dentler W.L. Ciliary microtubule capping structures contain a mammalian kinetochore antigen. J. Cell Biol. 1990;110:703–714. doi: 10.1083/jcb.110.3.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fisch C., Dupuis-Williams P. Ultrastructure of cilia and flagella – back to the future. Biol. Cell. 2011;103:249–270. doi: 10.1042/BC20100139. https://onlinelibrary.wiley.com/doi/abs/10.1042/BC20100139 [DOI] [PubMed] [Google Scholar]
- 22.Satish Tammana T.V., Tammana D., Diener D.R., Rosenbaum J. Centrosomal protein CEP104 (Chlamydomonas FAP256) moves to the ciliary tip during cilia assembly. J. Cell Sci. 2013;126:5018–5029. doi: 10.1242/jcs.133439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chaya T., Omori Y., Kuwahara R., Furukawa T. ICK is essential for cell type-specific ciliogenesis and the regulation of ciliary transport. EMBO J. 2014;33:1227–1242. doi: 10.1002/embj.201488175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chaya T., Furukawa T. Post-translational modification enzymes as key regulators of ciliary protein trafficking. J. Biochem. 2021;169:633–642. doi: 10.1093/jb/mvab024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liang Y., Pang Y., Wu Q., Hu Z., Han X., Xu Y., Deng H., Pan J. FLA8/KIF3B phosphorylation regulates kinesin-II interaction with IFT-B to control IFT entry and turnaround. Dev. Cell. 2014;30:585–597. doi: 10.1016/j.devcel.2014.07.019. [DOI] [PubMed] [Google Scholar]
- 26.Wilson N.F., Lefebvre P.A. Regulation of flagellar assembly by glycogen synthase kinase 3 in Chlamydomonas reinhardtii. Eukaryot. Cell. 2004;3:1307–1319. doi: 10.1128/EC.3.5.1307-1319.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Buchholz T.O., Krull A., Shahidi R., Pigino G., Jékely G., Jug F. Content-aware image restoration for electron microscopy. Methods Cell Biol. 2019;152:277–289. doi: 10.1016/bs.mcb.2019.05.001. [DOI] [PubMed] [Google Scholar]
- 28.Mijalkovic J., van Krugten J., Oswald F., Acar S., Peterman E.J.G. Single-Molecule Turnarounds of Intraflagellar Transport at the C. elegans Ciliary Tip. Cell Rep. 2018;25:1701–1707.e2. doi: 10.1016/j.celrep.2018.10.050. [DOI] [PubMed] [Google Scholar]
- 29.Hirokawa N., Noda Y. Intracellular Transport and Kinesin Superfamily Proteins, KIFs: Structure, Function, and Dynamics. Physiol. Rev. 2008;88:1089–1118. doi: 10.1152/physrev.00023.2007. [DOI] [PubMed] [Google Scholar]
- 30.Broekhuis J.R., Verhey K.J., Jansen G. Regulation of Cilium Length and Intraflagellar Transport by the RCK-Kinases ICK and MOK in Renal Epithelial Cells. PLoS One. 2014;9:e108470. doi: 10.1371/journal.pone.0108470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao Q., Li S., Shao S., Wang Z., Pan J. FLS2 is a CDK-like kinase that directly binds IFT70 and is required for proper ciliary disassembly in Chlamydomonas. PLoS Genet. 2020;16:e1008561. doi: 10.1371/journal.pgen.1008561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wren K.N., Craft J.M., Tritschler D., Schauer A., Patel D.K., Smith E.F., Porter M.E., Kner P., Lechtreck K.F. A Differential Cargo-Loading Model of Ciliary Length Regulation by IFT. Curr. Biol. 2013;23:2463–2471. doi: 10.1016/j.cub.2013.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Craft J.M., Harris J.A., Hyman S., Kner P., Lechtreck K.F. Tubulin transport by IFT is upregulated during ciliary growth by a cilium-autonomous mechanism. J. Cell Biol. 2015;208:223–237. doi: 10.1083/jcb.201409036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hunter E.L., Lechtreck K., Fu G., Hwang J., Lin H., Gokhale A., Alford L.M., Lewis B., Yamamoto R., Kamiya R., et al. The IDA3 adapter, required for intraflagellar transport of I1 dynein, is regulated by ciliary length. Mol. Biol. Cell. 2018;29:886–896. doi: 10.1091/mbc.e17-12-0729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prevo B., Mangeol P., Oswald F., Scholey J.M., Peterman E.J.G. Functional differentiation of cooperating kinesin-2 motors orchestrates cargo import and transport in C. elegans cilia. Nat. Cell Biol. 2015;17:1536–1545. doi: 10.1038/ncb3263. [DOI] [PubMed] [Google Scholar]
- 36.Sizova I., Fuhrmann M., Hegemann P. A Streptomyces rimosus aphVIII gene coding for a new type phosphotransferase provides stable antibiotic resistance to Chlamydomonas reinhardtii. Gene. 2001;277:221–229. doi: 10.1016/S0378-1119(01)00616-3. [DOI] [PubMed] [Google Scholar]
- 37.Kindle K.L. High-frequency nuclear transformation of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA. 1990;87:1228–1232. doi: 10.1073/pnas.87.3.1228. https://www.pnas.org/content/87/3/1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mastronarde D.N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. https://www.sciencedirect.com/science/article/pii/S1047847705001528 [DOI] [PubMed] [Google Scholar]
- 39.Zheng S.Q., Palovcak E., Armache J.P., Verba K.A., Cheng Y., Agard D.A. MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. http://msg.ucsf.edu/em/software/index.html [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mastronarde D.N., Held S.R. Automated tilt series alignment and tomographic reconstruction in IMOD. 16. J. Struct. Biol. 2017;197:102–113. doi: 10.1016/j.jsb.2016.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen Y., Wang Z., Zhang J., Li L., Wan X., Sun F., Zhang F. Accelerating electron tomographyreconstruction algorithm ICON with GPU. Biophys. Rep. 2017;3:36–42. doi: 10.1007/s41048-017-0041-z. https://link.springer.com/article/10.1007/s41048-017-0041-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Buchholz T.O., Jordan M., Pigino G., Jug F. Vol. 2019. IEEE Computer Society; 2019. Cryo-CARE: Content-aware image restoration for cryo-transmission electron microscopy data; pp. 502–506. (Proceedings - International Symposium on Biomedical Imaging). [Google Scholar]
- 43.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Itseez Open Source Computer Vision Library. 2015. https://github.com/itseez/opencv
- 45.Weigert M., Schmidt U., Boothe T., Müller A., Dibrov A., Jain A., Wilhelm B., Schmidt D., Broaddus C., Culley S., et al. Content-aware image restoration: pushing the limits of fluorescence microscopy. Nat. Methods. 2018;15:1090–1097. doi: 10.1038/s41592-018-0216-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
TIRFM time series data used in this paper has been deposited to Zenodo with https://doi.org/10.5281/zenodo.6787231. All other original data for this work are available upon request from the lead contact.
-
•
Code generated in this study is available upon request from the lead contact.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.