Abstract
The cytoskeleton not only deals with numerous interaction and communication mechanisms at the cellular level but also has a crucial role in the viral infection cycle. Although numerous aspects of SARS-CoV-2 virus interaction at the cellular level have been widely studied, little has been reported about the structural and functional response of the cytoskeleton. This work aims to characterize, at the ultrastructural level, the modifications in the cytoskeleton of infected cells, namely, its participation in filopodia formation, the junction of these nanostructures forming bridges, the viral surfing, and the generation of tunnel effect nanotubes (TNT) as probable structures of intracellular viral dissemination. The three-dimensional reconstruction from the obtained micrographs allowed observing viral propagation events between cells in detail for the first time. More profound knowledge about these cell–cell interaction models in the viral spread mechanisms could lead to a better understanding of the clinical manifestations of COVID-19 disease and to find new therapeutic strategies.
Keywords: Cytoskeleton, Actin, Filopodia, Ultrastructure, TNT, SARS-CoV-2
Introduction
SARS-CoV-2 is an enveloped virus belonging to the Orthocoronavirinae subfamily of the order Nidovirales. This seventh member of human-infected beta-CoV was rapidly sequenced by Wu et al. (2020) through samples isolated from human airway epithelial cells. It contemplates a single-stranded RNA of ~ 29,900 base pairs with a methylated cap at the 5' end and a polyadenylated (poly-A) tail at the 3' end. The viral genome of SARS-CoV-2 encodes structural; namely, the S protein (commonly known as “Spike”), the membrane protein (M), the nucleocapsid proteins (N), and the envelope protein (E), and non-structural proteins (NSP1 to NSP16) encoded by genes located within the 5′-region of viral RNA genome and with different enzymatic activities (Huang et al. 2020; Klein et al. 2020; Wang et al. 2020; Raj 2021).
Several studies have already characterized cell alterations at the ultrastructural level during SARS-CoV-2 infection. The accumulation of cytoplasmic lipid droplets, the thickening of the rough endoplasmic reticulum (RER), and its associated proliferation of double-membrane vesicles (DMV), the mitochondrial alterations, and the formation of filopodia as membrane projections have been well described (Goldsmith et al. 2004; Qinfen et al. 2004; Tseng et al. 2005; Snijder et al. 2006; Knoops et al. 2008; Klein et al. 2020; Barreto-Vieira et al. 2022). Nevertheless, cytoskeleton alterations have been poorly assessed.
In cells, the cytoskeleton plays a crucial role in numerous viral processes, including cell entry, transport to cellular regions for viral replication and assembly, and release of new viruses (Foo and Chee 2015; Denes et al. 2018; Bedi and Ono 2019). In the early phases of the infection, various entry routes of coronavirus into the cell have already been described, such as clathrin-mediated endosomal (CME) entry or fusion between viral and cellular membranes (cell surface entry) (Jackson et al. 2022). The contribution of actin polymerization to membrane deformation (i.e., endocytic invagination) is well known. In the CME process, receptor recognition and clathrin aggregation typically begin on the cytosolic side of the membrane, establishing the layer that generates the initial curvature and recruiting cargo to be internalized (Chen et al. 1998; Carlsson and Bayly 2014). Below the membrane, the nucleation promoters activate the Arp2/3 complex, polymerizing new actin filaments attached to the existing filaments, creating a branched framework (Carlsson and Bayly 2014), whose geometric structure will depend on the biomechanical demands of the process (Akamatsu et al. 2020). Together with the type I myosin motor activity, this growth generates the force necessary for the membrane invagination and the tubule extension into the cytoplasm. Subsequently, the endocytic neck contracts, and the vesicle is generated by membrane fusion (Mooren et al. 2012; Carlsson and Bayly 2014). Upon entry into the cell, actin filaments and microtubules with dyneins and kinesins reorganize and allow endocytic vesicles to traffic with viral particles to replication sites (Granger et al. 2014). Although still unknown, it has been suggested that actin could also have a regulatory role in the replication and transcription of SARS-CoV-2. Nevertheless, it has been described in previous studies using parainfluenza virus models (Bishnu and Banerjee 1999). In other coronaviruses, reorganizing actin filaments close to the membrane to form rings could support the replication of the viral genome and the synthesis of viral proteins (Wen et al. 2020). In the late stages of infection, actin restructuring provides force for membrane bending during exocytosis (Ng et al. 2004; Wen et al. 2020).
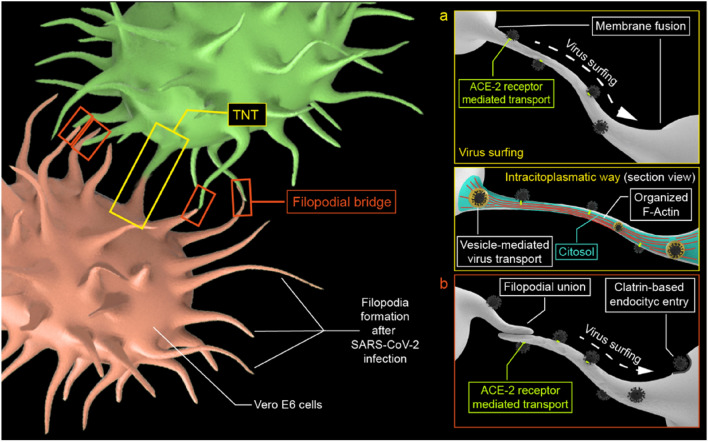
The cytoskeleton-dependent structures have a crucial activity in viral infection processes. However, its role in viral propagation has been poorly described, especially for the SARS-CoV-2 virus. At the cellular level, enveloped viruses can spread either through diffusion from the aqueous intercellular environment or by cell–cell interaction. Diffusion-based viral propagation requires high viral loads to be effective, and it is essential for human-to-human spread. Cell-to-cell propagation is more efficient than diffusion due to its speed and immune evasion (Mothes et al. 2010). Lehman et al. (2005) associated filopodia proliferation with an efficient infectious pathway, noting that rapid navigation of viruses into cells through filopodia, which is mediated by the underlying myosin II-dependent actin cytoskeleton. Three mechanisms of filopodia associated with viral infection have been described (Chang et al. 2016): viral surfing, filopodial retraction, and tunneling nanotube formation.
Firstly, viral surfing (Fig. 1a) consists of the orderly movement of viral particles toward the cell body through membrane projections before their internalization by fusion with the cell membrane or by receptor-mediated endocytosis (Lehmann et al. 2005). Secondly, retraction consists of the attraction of the virus to the cell through a mechanical impulse of retraction of the filopodium (Bornschlögl 2013). Thirdly, tunneling nanotubes (TNT) (Fig. 1b) correspond to intercellular ducts that connect cells for the intracytoplasmatic transfer of molecules, organelles, and small bodies (Jansens et al. 2020). TNTs were originally described by Rustom et al. as "open-ended channels mediating membrane continuity between connected cells" (Rustom et al. 2004). Given the difficulty in interpreting these structures, some essential criteria that define TNTs have been proposed: have a diameter between 50 and 700 nm, have a length up to hundreds of microns, be dynamic and hover on top of the substratum, contain actin, provide cargo transport, have membrane fusion, are open-ended (Zurzolo 2021). Moreover, these structures could contain the three elements of the cytoskeleton: actin filaments, intermediate filaments, and microtubules (Resnik et al. 2019). Its composition may vary depending on the diameter of the TNT (Önfelt et al. 2006).
Fig. 1.
Conceptual scheme of a viral surfing and the intracellular pathway in TNT. b Viral surfing along filopodial bridges
Ultrastructural analyses of morphogenesis in SARS-CoV-2-infected cells point out filopodia formation as plasma membrane projections (Ng et al. 2004; Caldas et al. 2020; Bideau et al. 2021; Barreto-Vieira et al. 2022). However, uncertainty remains about the involvement of the cellular cytoskeleton in SARS-CoV-2 infection. This article aims to characterize filopodia and TNT formation due to cytoskeleton restructuring in SARS-CoV-2-infected Vero E6 cells utilizing transmission electron microscopy (TEM). Additionally, the role of these subcellular structures in the cell–cell virus spread is discussed.
Materials and methods
Cell culture and SARS-CoV-2 infection
SARS-CoV-2 virus was isolated from a COVID-19 patient at the Hospital Clínico Universitario Lozano Blesa (Zaragoza, Spain). Viral stocks were prepared and quantified using the epithelial cell line, Vero E6, (kindly provided by Julia Vergara from Centro de Investigación en Sanidad Animal IRTA-CReSA, Barcelona, Spain) as previously described in Santiago et al. (2021) Vero E6 cells were cultured in Dulbecco’s modified Eagle’s medium, obtained from Sigma-Aldrich (Darmstadt, Germany) and supplemented with 10% fetal bovine serum (FBS) (Sigma), 2 mM Glutamax (Gibco), 100 U/ml penicillin (Sigma), 100 µg/ml streptomycin (Sigma), 0.25 µg/ml amphotericin B (Sigma), 1% non-essential amino acids (Gibco), and 25 mM HEPES (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid; Biowest), referred as complete medium and used for cell expansion. Vero E6 cells were kept at 37 ºC, in a 5% CO2 humidified incubator and for the antiviral assays, complete medium with 2% FBS was used. Control cells were cultured following the same procedure, excluding SARS-CoV-2 infection.
TEM and 3D reconstruction
For electron microscopy studies, cells were seeded at 100,000 cells/cm2 in a Lab-Tek chamber slides of four wells (Nalge Nunc International, Naperville, IL, USA) and infected with 500 TCID50/ml SARS-CoV-2 for 48 h. Then, the cells were washed three times for 2 min with phosphate buffer (PB) fixed in 2.5% glutaraldehyde in 0.1 M PB for 5 min at 37 ºC and then 2 h at 4 ºC. Afterwards, samples were washed in 0.1 M PB five times and stored at 4 ºC. The samples were postfixed in 2% OsO4 for 1 h at room temperature and stained in 2% uranyl acetate in the dark for 2 h at 4 °C. Then, the samples were rinsed in distilled water, dehydrated in ethanol, and infiltrated overnight in Durcupan resin (Sigma-Aldrich, St. Louis, MO, USA). Following polymerization, embedded cultures were detached from the wells and glued to Durcupan blocks. Finally, ultra-thin sections (0.08 µm) were cut with an Ultracut UC-6 (Leica Microsystems, Wetzlar, Germany), stained with lead citrate (Reynolds solution), and examined under a transmission electron microscope JEOL 1010 (Deben UK Ltd, Edmunds, United Kingdom). Pictures were taken using Radius software (Version 2.1) with a Gatan Bioscan (Gatan Inc, Pleasanton, USA).
Three-dimensional reconstruction from serial electron microscopy sections were performed using ultra-thin sections with 0.05-µm thickness. The subcellular structures were segmented manually using FIJI ImageJ software (Schindelin et al. 2012). Additionally, for the statistical analysis, the length and the diameter were measured using ImageJ software.
Results
SARS-CoV-2 viral particles identification
Virus particles in the extracellular aqueous medium (Fig. 2a), adhered to the cell membrane (Fig. 2b), and grouped inside vesicles have been observed (Fig. 2c), and frequently isolated individually (Fig. 2d). We found an average diameter of 63 ± 11 nm for n = 50 viral capsids, although the measurements are underestimated given the poor resolution of the membrane proteins.
Fig. 2.
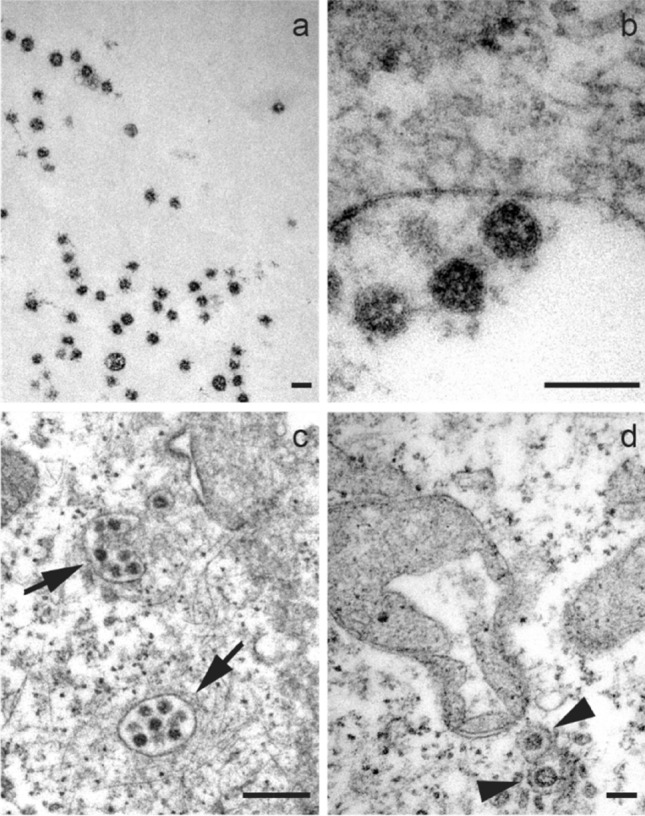
Micrographs show a free viral particles hoving in the extracellular medium. b Viruses adhered to the plasma membrane. c Clusters of multiple viral particles (arrows). d Individual viruses within vesicles in the cytosol (arrowhead). A mitochondrial alteration induced by SARS-CoV-2 infection is also shown in d. Scale bars represent 0.1 μm in a–d
Formation of filopodia and cytoskeleton rearrangement
We confirmed the formation of filopodia induced by SARS-CoV-2 infection. In uninfected Vero E6 cells (Fig. 3a), there are tiny membrane projections with no intercellular interaction. In contrast, infected cells showed more developed filopodia, recurrently joined forming between cells (Fig. 3b). In the cytosol of infected cells, DMV structures show a thick membrane which consists of two or more layers closely apposed (Fig. 3c, d).
Fig. 3.
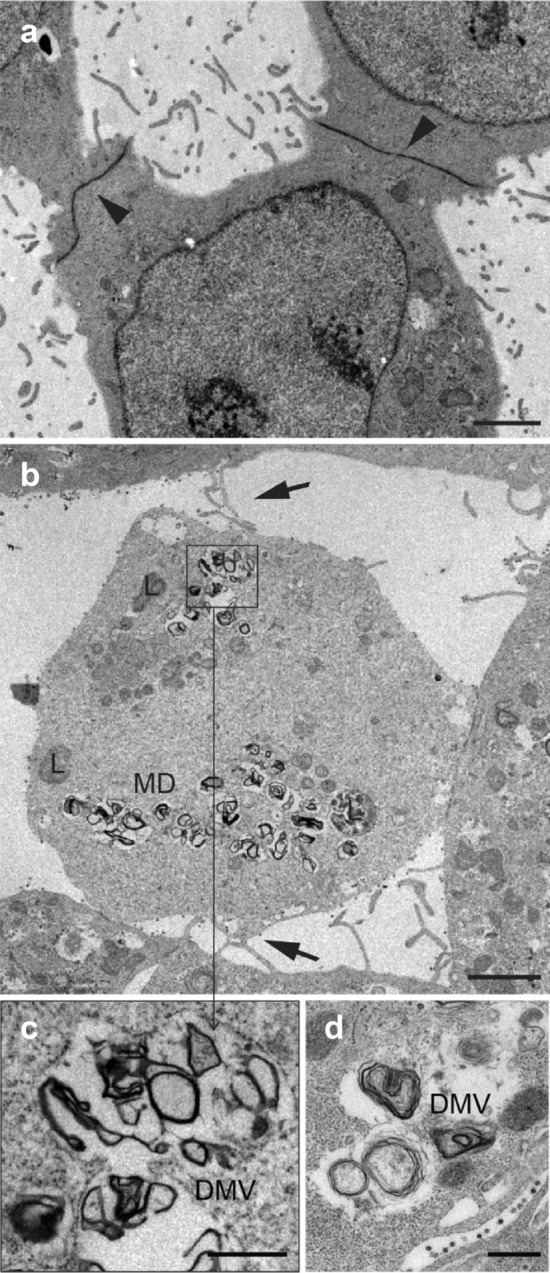
a Uninfected control cells establish extensive junctions between them, with a low number of filopodia and no filopodial bridges. b Filopodia formation induced by SARS-CoV-2 is observed in the infected cells, where the formation of filopodium bridges (arrows) is usually identified. c, d Detail of double membrane vesicles (DMVs). Scale bars represent 2 μm in a, b, and 0.5 μm in c, d
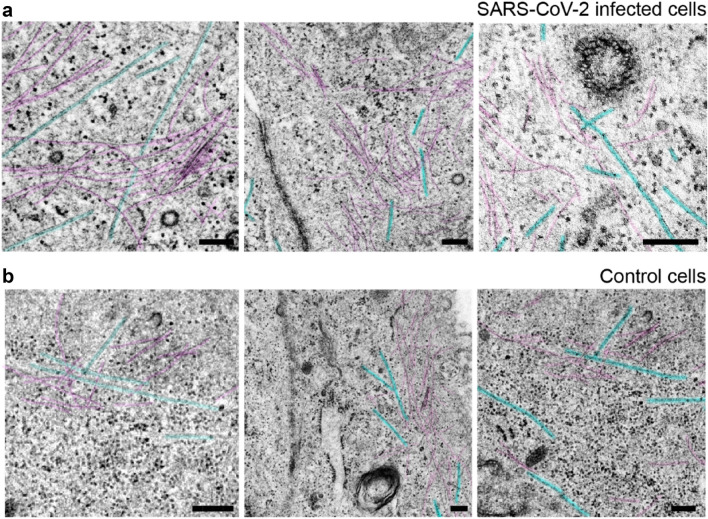
The infected cells exhibit a higher density of actin/intermediate filaments and microtubules (Fig. 4a) than control cells (Fig. 4b). We found an average length of 2.39 ± 1.53 μm (from 780 nm to 6.58 μm), and a diameter of 120 ± 47 nm (from 60 to 245 nm) for n = 30 filopodial bridges in infected cells. In control cells, there was a lack of filopodial bridges and entire filopodia. Therefore, it was only possible to determine the average diameter, which was 104 ± 22 nm (from 73 to 148 nm) for n = 30. The filopodia of control cells were observed to be slightly finer with a more homogeneous size polydispersity than in infected cells.
Fig. 4.
Comparative cytoskeleton micrographs from infected and control cells. a Microtubules (blue) and actin (or intermediate) filaments (purple) in the juxtamembranous region from three SARS-CoV-2-infected cells. b Cytoskeleton of three non-infected cells. It can be also appreciated nearby endocytic vesicles probably coated with clathrin and free ribosomal units. Scale bars represent 0.2 μm in a, b
Viral particles over filopodial bridges
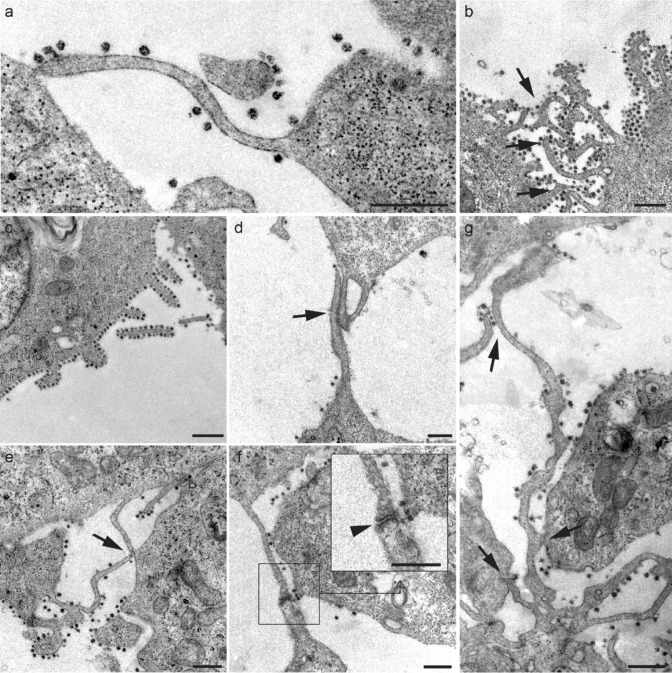
It was common to find viral particles along the filopodial membranes (Fig. 5a–g). A higher viral load was observed covering the filopodial surface in comparison with the extracellular medium (Fig. 5b, c).
Fig. 5.
a-g Micrographs of various filopodial structures found in SARS-CoV-2-infected Vero E6 cells. a Filopodial extension whose end adheres to the plasma membrane of a neighboring cell. b-c The number of viral particles attached to the filopodia was variable, although they were frequently observed in high density surrounding the filopodial surface. d Filopodial bridges were constituted through wide intermembrane junctions. e Junctions of filopodial bridges were often found in the distal extremes. f In many cases, bridge filopodial junctions showed electron-dense reinforcements (arrowhead; detail). g Commonly, larger filopodia contact more than one neighboring cell. Scale bars represent 0.5 μm in a-g
Even though filopodia came into direct contact with the plasma membrane of one (Fig. 5a) or several (Fig. 5g) cells, most frequent interaction appeared to be the formation of filopodial bridges (Fig. 5b, d–g). These bridges are often built with large overlapping surfaces of filopodia (Fig. 5d), and more regularly with a reduced contact through the distal extreme (Fig. 5e). It was common to find binding points between filopodia with high electron density, associated with protein aggregation (Fig. 5f), as discussed in later sections. The connection between more than two cells has also been observed (Fig. 5g).
Cell projections ultrastructural features
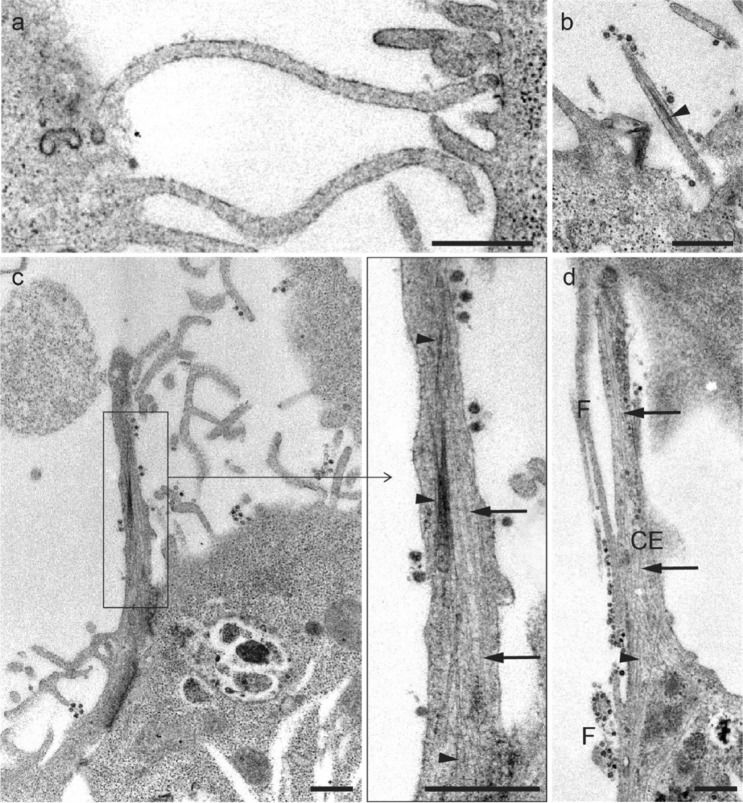
Filopodia present a characteristic cytoskeleton arrangement, being longitudinally traversed by actin filaments (Fig. 6a, b). Additionally, cells emit the thickest cytoplasmic extensions (typically > 0.25 μm) that characteristically also have microtubules arranged in parallel with the actin (or intermediate) filaments (Fig. 6c, d).
Fig. 6.
a, b Longitudinal section of filopodia, characterized by the absence of microtubules. c Longitudinal section of thick cytoplasmic extensions showing SARS-CoV-2 virus-like particles in the plasma membrane, and microtubules (arrows) and filaments (arrowheads) arranged parallelly. d Comparison between thick cytoplasmic extensions (CE) and filopodium (F) structures. Scale bars represent 0.5 μm a-d
Cell membranes fusion
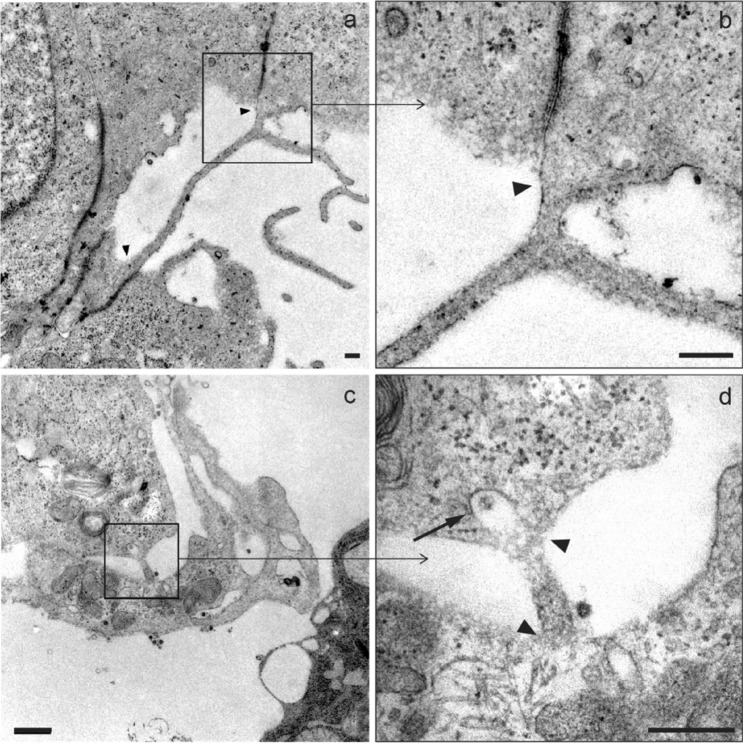
In addition to intercellular filopodial bridges, it was possible to identify TNT-like structures mainly characterized by a continuity of the plasma membrane of the connected cells (Fig. 7a, b). These types of structures were less frequently observed, and only once an event similar to a clathrin-mediated endocytic process inside a short nanotube structure was identified (Fig. 7c, d).
Fig. 7.
Micrographs of structures similar to TNT. a A nanotube > 2.5 μm in length and 70–90 nm in diameter promoting direct contact between the cytosols of two adjacent cells is observed. b Membrane fusion (arrowhead) detail of (a). c A smaller filopodium (< 0.5 μm in length and 90–100 nm in diameter), where a structure similar to a clathrin-coated vesicle (arrow) can be seen endocytosing a viral particle. d Detail of the fusion membranes from two neighboring cell bodies (arrowheads) from c. Scale bars represent 0.2 μm in a–d
Viral extrusion
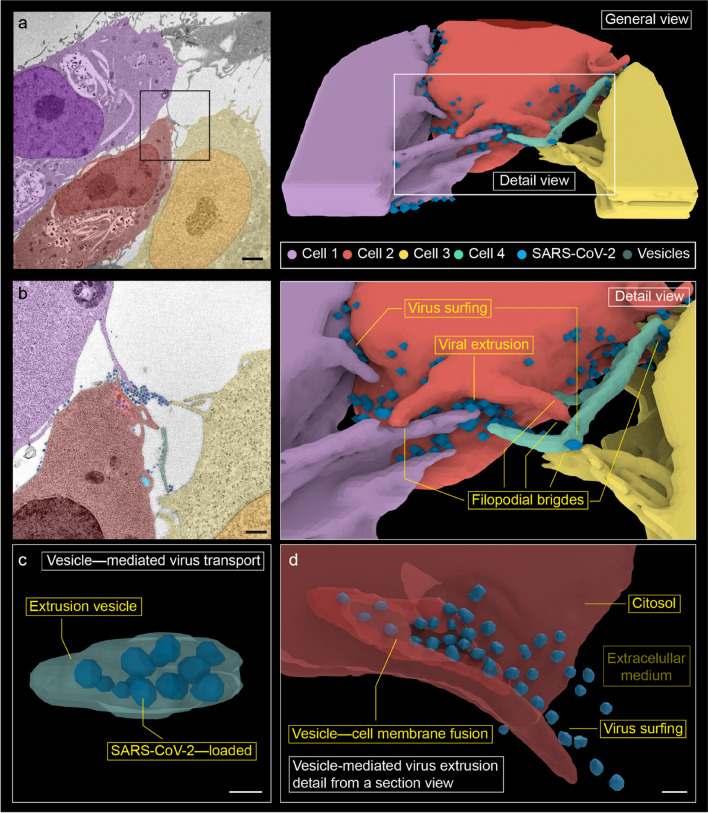
Extrusion of replicated viruses was identified in the three-dimensional reconstruction (Fig. 8a, b). In our observations, vesicles loaded with multiple viruses (Fig. 8c) fuse with the plasma membrane allowing an outwards virus release (Fig. 8d). At least three neighboring cells filopodia cluter around the extruded viruses. While the majority of these viruses are attached to filopodia, a few are located in the extracellular medium.
Fig. 8.
Three-dimensional reconstruction from 14 serial electron micrographs. a Overview of cell–cell interaction in viral propagation. b Detail of the membrane projections, where filopodial connections and viral surfing are observed. Numerous membrane projections from (at least) three different cells converge in a region of high viral load. We designate as 'viral extrusion' the region where the membrane of the extrusion vesicle fuses with the plasma membrane. c A vesicle with internalized viruses from cell 2 has been reconstructed. It is observed 12 viral particles that occupy practically the entire vesicle. It is probably an extrusion vesicle externalizing the virus. d A detail of viral extrusion has been reconstructed using eight micrographs. The micrographs have been selected to show how a vesicle fuses with the membrane to extrude viruses along a filopodium. This finding supports the hypothesis that filopodia are formed to promote the spread of the virus between cells. Scale bars represent 2 μm in a, 0.5 μm in b, and ~ 0.1 μm in c, d
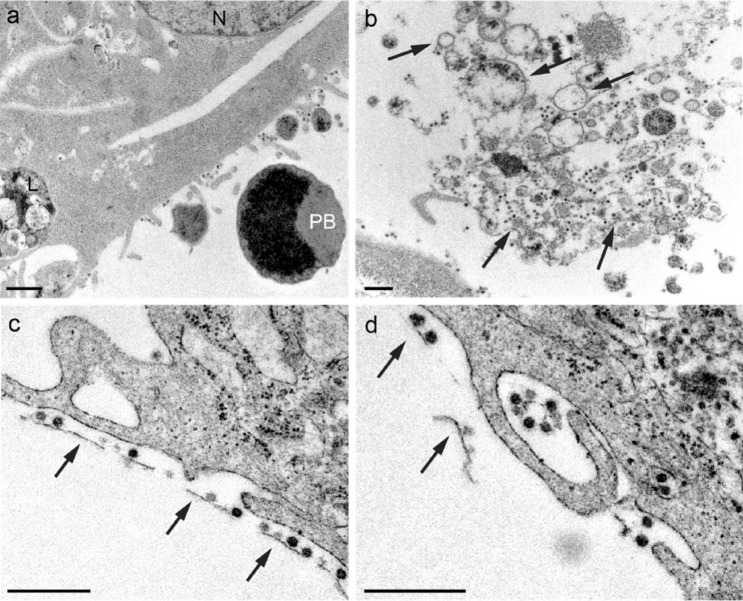
Cell membrane fragments in the extracellular space
Cell debris and pyroptotic bodies are visible in the extracellular space (Fig. 9a, b). Membrane fragments are typically surrounded by viruses and it was common to find them in regions close to the cell surface (Fig. 9c, d). So far, we mainly observed viruses attached to filopodial surfaces (Fig. 5), in the extracellular medium (Fig. 2a), adhered to plasma membranes (Fig. 2b), and inside vesicles (Fig. 2c, d). More rarely, the viral particles were arranged linearly along the membrane debris (Fig. 9b–d).
Fig. 9.
Micrographs showing a fragments of a pyroptotic body in the extracellular space (PB pyroptotic body, N nucleus, L lysosome). b, c Virions adhered to a membrane fragments (arrows). Subcellular fragments in the extracellular space have been observed as a consequence of SARS-CoV-2 infection. In membrane fragments, it has been possible to identify adherent virions. d Virus adhered to remains of membranes are occasionally encovered by the filopodia (possible macropinocytosis). Scale bars represent 0.5 μm in (a-d)
Discussion
Throughout the COVID-19 pandemic, numerous studies focused on the ultrastructural analysis of cell morphogenesis induced by SARS-CoV-2 infection. However, the overwhelming number of articles published in this regard, together with the scant knowledge of the biology of the newly discovered virus, has represented an obstacle when it comes to interpreting electron microscopy studies. As a result, numerous TEM-based reports have continuously confused SARS-CoV-2 viral particles with normal cellular structures. Akilesh et al. (2021) pointed to five cellular structures typically confused with enveloped viruses: clathrin-coated vesicles, secretory vesicles and granules, coatomer- and ER-coated vesicles, multivesicular bodies and exosomes, and microvilli. Expressly, clathrin-coated vesicles represent the structures most frequently misinterpreted as viral particles in TEM (Calomeni et al. 2020; Kniss 2020; Miller and Brealey 2020; Brealey and Miller 2021). Our microscopic observation identified clathrin-containing vesicles whose sizes ranged from 90 to 115 nm, similar to SARS-CoV-2 (60–140 nm). However, vesicles could be free in the cytoplasm and do not have electron density so they can be quickly differentiated from viral particles. In this work, we found viral particles arranged in three ways: isolated or grouped in the extracellular space, inside vesicles in the intracellular space, and bounded both to the membrane of the cell body and the filopodia projected as a consequence of infection with SARS-CoV-2.
Viral-infected cells rearranged their cytoskeleton. An increase in ribosomal units and polyribosomes was observed in the cytosol for the protein monomers synthesis that make up the cytoskeleton (G-actin and α and β tubulins). Filopodia are cell membrane projections rich in actin filaments arranged in parallel (Mattila and Lappalainen 2008). There are two alternative models that explain filopodia origins. The convergent elongation model postulates that filopodia arise from the filopodium tip complex (FTP) assembly in the plasma membrane or its vicinity (Faix and Rottner 2006), which contain actin polymerases, such as Ena proteins VASP, Dia2, and myosin X that accelerate filament growth (Cheng and Mullins 2020). An alternative model proposes that actin filaments are nucleated at the tips of filopodia by formins (Mattila and Lappalainen 2008).
Filopodia growth induced by SARS-CoV-2 infection in cells has been described in previous studies (Barreto-Vieira et al. 2022). These nanostructures were slightly finer and more homogeneous in the control cells (104 ± 22 nm) compared to the infected cells (120 ± 47 nm). Similar numbers of filopodia were found in both cultures (around 15–20 filopodia/section/cell). Nonetheless, only in infected cells did filopodia organize in bridges. These membrane projections have played a relevant role in probing the extracellular environment, directed migration, and cell–cell communication processes (Mattila and Lappalainen 2008). Additionally, intercellular communication pathway mediated by filopodia as an effective cell–cell viral propagation model. The role of filopodia was initially described in 2003 as a spread pathway for the human T-cell leukemia virus type 1 (HTLV-1) (Igakura et al. 2003), and was subsequently associated with other enveloped viruses (Lehmann et al. 2005; Sherer et al. 2007; Choudhary et al. 2013). Furthermore, Oh et al. (2010) found a higher viral load in filopodia than in other regions of the cell membrane. Accordingly, our observations suggest a more significant presence of viral particles adhered to the filopodial surface than in other membrane regions. Viral binding may be mediated by positively charged glycoprotein moieties that recognize negative domains of cellular receptors. Some authors point to a high expression of receptor-negative domains in filopodia, as the heparan sulfate receptors (HS), which could convert them into preferential binding regions (Oh et al. 2010; Tiwari et al. 2012).
Once the viruses bonded in the filopodial surface, they can slide or move laterally along these membrane projections; this phenomenon is known as surfing. Although viruses can enter the cell through other membrane regions, filopodium-dependent viral entry is more efficient. Physical intercellular connection is 2–3 orders of magnitude more efficient than diffusion spread due to the speed of transmission and the evasion of the immune system (Dimitrov et al. 1993; Carr et al. 1999; Miranda-Saksena et al. 2018). This mechanism could also promote the virus propagation between distant cells. In this work, we observed an average length of 2.39 ± 1.53 μm (from 780 nm to 6.58 μm) in filopodial bridges. However, cell culture requires proximity between cells; therefore, longer bridges with more connections could appear in vivo. SARS-CoV-2 virus could bind to ACE-2 receptors to navigate through the filopodial membrane all the way until their eventual entry to the cell (Kloc et al. 2022). Viral envelope proteins have high specificity and affinity to cellular receptors, which are associated with actin filaments. Hence, viral surfing is propelled by microfilament contraction through internal myosin motors (Medeiros et al. 2006) to regions of the membrane with high endocytic activity (Taylor et al. 2011). Furthermore, Seyran et al. (2021) reported that SARS-CoV-2 protein S interacts with the sialic acid layer of the epithelium, which may allow its transport across the epithelial surface until it encounters an ACE-2 receptor.
There is still no consensus on how to refer to the different types of membrane projections. We use the term TNT to refer to filopodial bridges with membrane fusion (Zurzolo 2021; Kloc et al. 2022). Additionally, some authors only mention the intracellular transport through TNT and they do not rule out a dual transfer by surfing (coupling to HS receptors, and entering via the endocytic pathway). In the first reference to a membrane nanotube, membrane fusion was not specified (Ramírez-Weber and Kornberg 1999). The term ‘TNT’ was coined to refer to an intracytoplasmic transport pathway (Önfelt et al. 2004; Rustom et al. 2004). However, as can be seen in some published micrographs (e.g., Figure 2g of (Rustom et al. 2004)), the section of filopodium obtained in the ultrathin section can lead to misinterpretations (e.g., Figure 1g1 from (Rustom et al. 2004)). Therefore, unless the physical separation between filopodia is evident (e.g., Fig. 5d, e of this article), confusion between filopodia bridges and tunneling nanotubes cannot be ruled out (e.g., Figures 5a, b, g of this article). When the ultra-thin section cut longitudinally the entire filopodium (e.g., Fig. 6), the cell structure can be interpreted with a lower uncertainty. To avoid these limitations, serial ultra-thin sections could represent an appropriate option. Despite TEM limitations for TNT identification, ultrastructural studies using scanning electron microscopy (SEM) entail more difficulty since membrane fusion cannot be clearly identified [as for e.g., in (Caldas et al. 2020), (Lu et al. 2019), or (Pontes et al. 2008)].
Given the high viral load attached to filopodia, we suggest that they could play a crucial role in the SARS-CoV-2 viral cycle. The filopodia formation induced by SARS-CoV-2 was promptly described during the beginning of the COVID-19 pandemic (2020) using Vero E6 cells (Bouhaddou et al. 2020). However, no further ultrastructure analyses were performed until two recent studies. In the first report, not yet peer-reviewed, Swain et al. (2022) again suggested that filopodia could contribute to the cell–cell transmission of Betacoronavirus. In the second study, Merolli et al. (2022) commented little on its presence in infected cells and described tunneling nanotubes (TNT) as a novel intracytoplasmic route of the virus, despite being previously reported (Kumar et al. 2017; Jansens et al. 2020; Tiwari et al. 2021). Our work found that surfing was the predominant route of viral propagation. Vesicle-mediated cytosolic transport of the virus has also been found less frequently. We have identified the cytosolic pathway in two ways: through TNT and filopodia. Firstly, we have only identified transport through TNT in Fig. 7c. According to this hypothesis, the virus could be internalized in vesicle and transported until being endocytosed by the host cell (Jansens et al. 2020). Note that the viral transport pathways mentioned are not mutually exclusive but could coexist and increase the yield of intercellular viral propagation.
The findings in Fig. 8 confirm that filopodia act as means of intercellular transport of the virus; herein it is possible to see extrusion of virus replicated from the host cell. Firstly, the large vesicle is loaded with numerous viral particles together with the absence of clathrin accumulation at the vesicular edge (Fig. 8c), makes endocytosis to be unlikely. Secondly, as depicted in Fig. 8a, the host cell (cell 2) is altered by viral infection (numerous replicating DMVs, lysosomes, and autophagic-like events). Given the above, it could be interpreted that viruses have already been replicated in this cell and try to escape to colonize nearby cells. Finally, those viruses are unlikely released from cell 1 and cell 3, since a significant interaction between them coexists in a specific area of cell 2. No nearby exocytic vesicles have been observed in these two neighboring cells. This clustering of filopodia in the high viral load region may suggest underlying molecular mechanisms (Jansens et al. 2020). Additionally, the viral particles are extruded in the direction of the filopodium (see detail in Fig. 8d), which reinforces the hypothesis of cellular transport mediated by filopodia.
Numerous viral particles bound to membranous debris were observed in the extracellular environment (Fig. 9b, c). Pyroptosis occurs in cells infected by SARS-CoV-2, although the mechanisms that trigger it are still unknown in detail (Bittner et al. 2022). One theory is that membranous debris could be associated with the rupture of these cells. The rearrangement of viruses along these structures could have implications for propagation dynamics, so future work should identify its implications in viral processes.
SARS-CoV-2 virus, like other respiratory viruses, targets the ciliated cells of the respiratory epithelium, facilitating dissemination to the lungs. Preferential budding of coronavirus towards membrane projections, as cilia, has also been suggested using in vivo samples. For example, Afzelius et al. (Afzelius 1994) observed virions inside and outside hair cells, but not in goblet cells or other human nasal mucosa cells. Ciliary loss has been widely reported both in SARS-CoV-2 and in other coronaviruses that preceded it (such as MERS-CoV or HCoV-229E) (Chilvers et al. 2001; Haverkamp et al. 2018; Schreiner et al. 2022), and it is associated with the internalization of the cilia during the virus entry to the cell. Consequently, deciliation could be related to the recurrent anosmia induced by SARS-CoV-2 infection; correlation between histological damage and olfactory disfunction is still debated, though (Reyna et al. 2022). This phenomenon of ciliary invagination is similar to the filopodial retraction, where cell filopodia internalize the virus through membrane traction mechanisms (Bornschlögl 2013; Chang et al. 2016).
Given the importance that filopodia and TNT acquires in the viral infection, it is essential to know in greater depth the interface between viruses and filopodia to understand the viral pathogenesis of SARS-CoV-2, as well as the cellular and molecular mechanisms that govern the course of the disease by COVID-19.
Conclusions
In this work, we have ultrastructurally characterized filopodia formation as a mechanism of cell interconnection in the late stages of SARS-CoV-2 infection (48 hpi) using Vero E6 cells in the primitive variant of the virus. SARS-CoV-2 infected cells shows a more developed cytoskeleton in contrast with control cells, which might be associated with viral cell–cell spread or defense mechanisms linked to virus extrusion. Communication between two or more neighboring cells was evident, finding filopodial bridges (filopodia joined but not fused) and tunneling nanotubes (filopodia with fused membranes).
Viral particles were commonly adhered to the filopodial surface, suggesting a preference of the virus towards these membrane regions.
Viruses surf along membrane projections. Surfing seems to predominate in the viral spread between cells. Evidence of vesicle-mediated intracellular transport was also found via TNT and via filopodia. Vesicular transport by filopodia probably include exocytosis in the host cell and endocytosis in the neighboring cell. Viral spread through TNT occurs intracellularly. Those cytosolic events have been found less frequently. However, it may occur more times since can be hidden in the ultra-thin section.
To the best of our knowledge, no prior studies have reported three-dimensional reconstructions of viral cell–cell propagation. Extrusion of numerous viral particles and the clustering of filopodia from neighboring cells around them was observed. We probably would have not found this event in a single ultrathin section. Therefore, serial sections have been especially useful in this work.
Given the importance of the cytoskeleton for the proper cellular function, it is crucial to characterize in greater depth its alterations to understand the SARS-CoV-2 pathogenesis.
Acknowledgements
The authors would like to acknowledge the use of Servicio General de Apoyo a la Investigación-SAI, Universidad de Zaragoza and to Mario Soriano from Centro de Investigación Príncipe Felipe, Valencia, Spain. This research was funded by the Instituto de Investigación Sanitaria Aragón: Campaña Investiga COVID-19.
Author contributions
MB and CJ: Investigation, Original manuscript text preparation, Methodology, Validation. CJ: Project administration, Manuscript review. IU, MA., and EM: Investigation, sample preparation (including cell culture), virus isolation, Manuscript review.
Declarations
Conflict of interest
The authors declare no conflicts of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Marta Baselga, Email: mbaselga@iisaragon.es.
Eduardo Moreo, Email: ejmoreo@gmail.com.
Iratxe Uranga-Murillo, Email: iratxe.um@gmail.com.
Maykel Arias, Email: maykelariascabrero@gmail.com.
Concepción Junquera, Email: cjunquer@unizar.es.
References
- Afzelius BA. Ultrastructure of human nasal epithelium during an episode of coronavirus infection. Virchows Arch. 1994;424:295–300. doi: 10.1007/BF00194614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akamatsu M, Vasan R, Serwas D, et al. Principles of self-organization and load adaptation by the actin cytoskeleton during clathrin-mediated endocytosis. Elife. 2020;9:1–40. doi: 10.7554/eLife.49840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akilesh S, Nicosia RF, Alpers CE, et al. Characterizing viral infection by electron microscopy: lessons from the coronavirus disease 2019 pandemic. Am J Pathol. 2021;191:222–227. doi: 10.1016/j.ajpath.2020.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreto-Vieira DF, da Silva MAN, de Almeida ALT, et al. SARS-CoV-2: ultrastructural characterization of morphogenesis in an in vitro system. Viruses. 2022 doi: 10.3390/v14020201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedi S, Ono A. Friend or foe: the role of the cytoskeleton in influenza a virus assembly. Viruses. 2019 doi: 10.3390/v11010046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bideau ML, Wurtz N, Baudoin JP, La Scola B. Innovative approach to fast electron microscopy using the example of a culture of virus-infected cells: an application to SARS-CoV-2. Microorganisms. 2021 doi: 10.3390/microorganisms9061194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishnu P, Banerjee AK. Involvement of actin microfilaments in the transcription/replication of human parainfluenza virus type 3: possible role of actin in other viruses. Microsc Res Tech. 1999;47:114–123. doi: 10.1002/(SICI)1097-0029(19991015)47:2<114::AID-JEMT4>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Bittner ZA, Schrader M, George SE, Amann R. Pyroptosis and its role in SARS-CoV-2 infection. Cells. 2022;11:1717. doi: 10.3390/cells11101717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornschlögl T. How filopodia pull: what we know about the mechanics and dynamics of filopodia. Cytoskeleton. 2013;70:590–603. doi: 10.1002/cm.21130. [DOI] [PubMed] [Google Scholar]
- Bouhaddou M, Memon D, Meyer B, et al. The global phosphorylation landscape of SARS-CoV-2 infection. Cell. 2020;182:685–712.e19. doi: 10.1016/j.cell.2020.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brealey JK, Miller SE. SARS-CoV-2 has not been detected directly by electron microscopy in the endothelium of chilblain lesions. Br J Dermatol. 2021;184:186. doi: 10.1111/bjd.19572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldas LA, Carneiro FA, Higa LM, et al. Ultrastructural analysis of SARS-CoV-2 interactions with the host cell via high-resolution scanning electron microscopy. Sci Rep. 2020;10:1–8. doi: 10.1038/s41598-020-73162-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calomeni E, Satoskar A, Ayoub I, et al. Multivesicular bodies mimicking SARS-CoV-2 in patients without COVID-19. Kidney Int. 2020;98:233–234. doi: 10.1016/j.kint.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsson AE, Bayly PV. Force generation by endocytic actin patches in budding yeast. Biophys J. 2014;106:1596–1606. doi: 10.1016/j.bpj.2014.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr JM, Hocking H, Li P, Burrell CJ. Rapid and efficient cell-to-cell transmission of human immunodeficiency virus infection from monocyte-derived macrophages to peripheral blood lymphocytes. Virology. 1999;265:319–329. doi: 10.1006/viro.1999.0047. [DOI] [PubMed] [Google Scholar]
- Chang K, Baginski J, Hassan SF, et al. Filopodia and viruses: an analysis of membrane processes in entry mechanisms. Front Microbiol. 2016;7:1–13. doi: 10.3389/fmicb.2016.00300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Fre S, Slepnev CI, et al. Epsin is an EH-domain-binding protein implicated in clathrin-mediated endocytosis. Nat Lett. 1998;394:793–797. doi: 10.1038/255243a0. [DOI] [PubMed] [Google Scholar]
- Cheng KW, Mullins RD. Initiation and disassembly of filopodia tip complexes containing VASP and lamellipodin. Mol Biol Cell. 2020;31:2021–2034. doi: 10.1091/mbc.E20-04-0270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chilvers MA, McKean M, Rutman A, et al. The effects of coronavirus on human nasal ciliated respiratory epithelium. Eur Respir J. 2001;18:965–970. doi: 10.1183/09031936.01.00093001. [DOI] [PubMed] [Google Scholar]
- Choudhary S, Burnham L, Thompson JM, et al. Role of filopodia in HSV-1 entry into zebrafish 3-O-sulfotransferase-3-expressing cells. Open Virol J. 2013;7:41–48. doi: 10.2174/1874357901307010041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denes CE, Miranda-Saksena M, Cunningham AL, Diefenbach RJ. Cytoskeletons in the closet—subversion in alphaherpesvirus infections. Viruses. 2018;10:1–25. doi: 10.3390/v10020079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrov DS, Willey RL, Sato H, et al. Quantitation of human immunodeficiency virus type 1 infection kinetics. J Virol. 1993;67:2182–2190. doi: 10.1128/jvi.67.4.2182-2190.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faix J, Rottner K. The making of filopodia. Curr Opin Cell Biol. 2006;18:18–25. doi: 10.1016/j.ceb.2005.11.002. [DOI] [PubMed] [Google Scholar]
- Foo KY, Chee HY. Interaction between flavivirus and cytoskeleton during virus replication. Biomed Res Int. 2015 doi: 10.1155/2015/427814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldsmith CS, Tatti KM, Ksiazek TG, et al. Ultrastructural characterization of SARS coronavirus. Emerg Infect Dis. 2004;10:320–326. doi: 10.3201/eid1002.030913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granger E, McNee G, Allan V, Woodman P. The role of the cytoskeleton and molecular motors in endosomal dynamics. Semin Cell Dev Biol. 2014;31:20–29. doi: 10.1016/j.semcdb.2014.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haverkamp AK, Lehmbecker A, Spitzbarth I, et al. Experimental infection of dromedaries with Middle East respiratory syndrome-coronavirus is accompanied by massive ciliary loss and depletion of the cell surface receptor dipeptidyl peptidase. Sci Rep. 2018;8:1–15. doi: 10.1038/s41598-018-28109-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Yang C, Xu XF, et al. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol Sin. 2020;41:1141–1149. doi: 10.1038/s41401-020-0485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igakura T, Stinchcombe JC, Goon PKC, et al. Spread of HTLV-I between lymphocytes by virus-induced polarization of the cytoskeleton. Science (80-) 2003;299:1713–1716. doi: 10.1029/2001PA-000633. [DOI] [PubMed] [Google Scholar]
- Jackson CB, Farzan M, Chen B, Choe H. Mechanisms of SARS-CoV-2 entry into cells. Nat Rev Mol Cell Biol. 2022;23:3–20. doi: 10.1038/s41580-021-00418-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansens RJJ, Tishchenko A, Favoreel HW. Bridging the gap: virus long-distance spread via tunneling nanotubes. J Virol. 2020;94:e02120–e2219. doi: 10.1128/JVI.02120-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein S, Cortese M, Winter SL, et al. SARS-CoV-2 structure and replication characterized by in situ cryo-electron tomography. Nat Commun. 2020;11:1–10. doi: 10.1038/s41467-020-19619-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloc M, Uosef A, Wosik J, et al. Virus interactions with the actin cytoskeleton—what we know and do not know about SARS-CoV-2. Arch Virol. 2022;167:737–749. doi: 10.1007/s00705-022-05366-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kniss DA. Alternative interpretation to the findings reported in visualization of severe acute respiratory syndrome coronavirus 2 invading the human placenta using electron microscopy. Am J Obstet Gynecol. 2020;223:785–786. doi: 10.1016/j.ajog.2020.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoops K, Kikkert M, Van Den Worm SHE, et al. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 2008;6:1957–1974. doi: 10.1371/journal.pbio.0060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Kim JH, Ranjan P, et al. Influenza virus exploits tunneling nanotubes for cell-to-cell spread. Sci Rep. 2017;7:1–14. doi: 10.1038/srep40360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann MJ, Sherer NM, Marks CB, et al. Actin- and myosin-driven movement of viruses along filopodia precedes their entry into cells. J Cell Biol. 2005;170:317–325. doi: 10.1083/jcb.200503059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu JJ, Yang WM, Li F, et al. Tunneling nanotubes mediated microRNA-155 intercellular transportation promotes bladder cancer cells’ invasive and proliferative capacity. Int J Nanomed. 2019;14:9731–9743. doi: 10.2147/IJN.S217277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattila PK, Lappalainen P. Filopodia: molecular architecture and cellular functions. Nat Rev Mol Cell Biol. 2008;9:446–454. doi: 10.1038/nrm2406. [DOI] [PubMed] [Google Scholar]
- Medeiros NA, Burnette DT, Forscher P. Myosin II functions in actin-bundle turnover in neuronal growth cones. Nat Cell Biol. 2006;8:215–226. doi: 10.1038/ncb1367. [DOI] [PubMed] [Google Scholar]
- Merolli A, Kasaei L, Ramasamy S, et al. An intra-cytoplasmic route for SARS-CoV-2 transmission unveiled by helium-ion microscopy. Sci Rep. 2022;12:1–11. doi: 10.1038/s41598-022-07867-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SE, Brealey JK. Visualization of putative coronavirus in kidney. Kidney Int. 2020;98:231–232. doi: 10.1016/j.kint.2020.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda-Saksena M, Denes CE, Diefenbach RJ, Cunningham AL. Infection and transport of herpes simplex virus type 1 in neurons: role of the cytoskeleton. Viruses. 2018;10:1–20. doi: 10.3390/v10020092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooren OL, Galletta BJ, Cooper JA. Roles for actin assembly in endocytosis. Annu Rev Biochem. 2012;81:661–686. doi: 10.1146/annurev-biochem-060910-094416. [DOI] [PubMed] [Google Scholar]
- Mothes W, Sherer NM, Jin J, Zhong P. Virus cell-to-cell transmission. J Virol. 2010;84:8360–8368. doi: 10.1128/jvi.00443-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng ML, Lee JWM, Leong MLN, et al. Topographic changes in SARS coronavirus-infected cells during late stages of infection. Emerg Infect Dis. 2004;10:1907–1914. doi: 10.3201/eid1011.040195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh MJ, Akhtar J, Desai P, Shukla D. A role for heparan sulfate in viral surfing. Biochem Biophys Res Commun. 2010;391:176–181. doi: 10.1016/j.bbrc.2009.11.027.A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Önfelt B, Nedvetzki S, Yanagi K, Davis DM. Cutting edge: membrane nanotubes connect immune cells. J Immunol. 2004;173:1511–1513. doi: 10.4049/jimmunol.173.3.1511. [DOI] [PubMed] [Google Scholar]
- Önfelt B, Nedvetzki S, Benninger RKP, et al. Structurally distinct membrane nanotubes between human macrophages support long-distance vesicular traffic or surfing of bacteria. J Immunol. 2006;177:8476–8483. doi: 10.4049/jimmunol.177.12.8476. [DOI] [PubMed] [Google Scholar]
- Pontes B, Viana NB, Campanati L, et al. Structure and elastic properties of tunneling nanotubes. Eur Biophys J. 2008;37:121–129. doi: 10.1007/s00249-007-0184-9. [DOI] [PubMed] [Google Scholar]
- Qinfen Z, Jinming C, Xiaojun H, et al. The life cycle of SARS coronavirus in Vero E6 cells. J Med Virol. 2004;73:332–337. doi: 10.1002/jmv.20095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj R. Analysis of non-structural proteins, NSPs of SARS-CoV-2 as targets for computational drug designing. Biochem Biophys Reports. 2021;25:100847. doi: 10.1016/j.bbrep.2020.100847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez-Weber FA, Kornberg TB. Cytonemes: cellular processes that project to the principal signaling center in drosophila imaginal discs. Cell. 1999;97:599–607. doi: 10.1016/S0092-8674(00)80771-0. [DOI] [PubMed] [Google Scholar]
- Resnik N, Erman A, Veranič P, Kreft ME. Triple labelling of actin filaments, intermediate filaments and microtubules for broad application in cell biology: uncovering the cytoskeletal composition in tunneling nanotubes. Histochem Cell Biol. 2019;152:311–317. doi: 10.1007/s00418-019-01806-3. [DOI] [PubMed] [Google Scholar]
- Reyna RA, Kishimoto-Urata M, Urata S, et al. Recovery of anosmia in hamsters infected with SARS-CoV-2 is correlated with repair of the olfactory epithelium. Sci Rep. 2022;12:1–8. doi: 10.1038/s41598-021-04622-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustom A, Saffrich R, Markovic I, et al. Nanotubular highways for intercellular organelle transport. Science (80-) 2004;303:1007–1010. doi: 10.1126/science.1093133. [DOI] [PubMed] [Google Scholar]
- Santiago L, Uranga-Murillo I, Arias M, et al. Determination of the concentration of igg against the spike receptor-binding domain that predicts the viral neutralizing activity of convalescent plasma and serum against SARS-CoV-2. Biology (Basel) 2021 doi: 10.3390/biology10030208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiner T, Allnoch L, Beythien G, et al. SARS-CoV-2 infection dysregulates cilia and basal cell homeostasis in the respiratory epithelium of hamsters. Int J Mol Sci. 2022;2:1–20. doi: 10.3390/ijms23095124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyran M, Takayama K, Uversky VN, et al. The structural basis of accelerated host cell entry by SARS-CoV-2. FEBS J. 2021;288:5010–5020. doi: 10.1111/febs.15651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherer N, Lehmann MJ, Jimenez-Soto LF, et al. Retroviruses can establish filopodial bridges for efficient cell-to-cell transmission. Nat Cell Biol. 2007;9:310–315. doi: 10.1038/ncb1544.Retroviruses. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder EJ, van der Meer Y, Zevenhoven-Dobbe J, et al. Ultrastructure and origin of membrane vesicles associated with the severe acute respiratory syndrome coronavirus replication complex. J Virol. 2006;80:5927–5940. doi: 10.1128/jvi.02501-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder EJ, Limpens RWAL, de Wilde AH, et al. A unifying structural and functional model of the coronavirus replication organelle: tracking down RNA synthesis. PLoS Biol. 2020;18:1–25. doi: 10.1371/journal.pbio.3000715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain J, Merida P, Rubio K, et al. Reorganization of F-actin nanostructures is required for the late phases of SARS-CoV-2 replication in pulmonary cells. bioRxiv. 2022 doi: 10.1101/2022.03.08.483451. [DOI] [Google Scholar]
- Taylor MP, Koyuncu OO, Enquist LW. Subversion of the actin cytoskeleton during viral infection. Nat Rev Microbiol. 2011;9:427–439. doi: 10.1038/nrmicro2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari V, Maus E, Sigar IM, et al. Role of heparan sulfate in sexually transmitted infections. Glycobiology. 2012;22:1402–1412. doi: 10.1093/glycob/cws106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari V, Koganti R, Russell G, et al. Role of tunneling nanotubes in viral infection, neurodegenerative disease, and cancer. Front Immunol. 2021;12:1–17. doi: 10.3389/fimmu.2021.680891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng C-TK, Tseng J, Perrone L, et al. Apical entry and release of severe acute respiratory syndrome-associated coronavirus in polarized calu-3 lung epithelial cells. J Virol. 2005;79:9470–9479. doi: 10.1128/jvi.79.15.9470-9479.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang MY, Zhao R, Gao LJ, et al. SARS-CoV-2: structure, biology, and structure-based therapeutics development. Front Cell Infect Microbiol. 2020;10:1–17. doi: 10.3389/fcimb.2020.587269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Z, Zhang Y, Lin Z, et al. Cytoskeleton - a crucial key in host cell for coronavirus infection. J Mol Cell Biol. 2020;12:968–979. doi: 10.1093/jmcb/mjaa042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F, Zhao S, Yu B, et al. Complete genome characterisation of a novel coronavirus associated with severe human respiratory disease in Wuhan. China Biorxiv. 2020 doi: 10.1101/2020.01.24.919183. [DOI] [Google Scholar]
- Zurzolo C. Tunneling nanotubes: reshaping connectivity. Curr Opin Cell Biol. 2021;71:139–147. doi: 10.1016/j.ceb.2021.03.003. [DOI] [PubMed] [Google Scholar]