Abstract
Protein P19 encoded by the conjugative resistance plasmid R1 has been identified as being one member of a large family of muramidases encoded by bacteriophages and by type III and type IV secretion systems. We carried out a mutational analysis to investigate the function of protein P19 and used in vivo complementation assays to test those of several P19 mutants. The results indicated that conserved residues present in the presumed catalytic center of P19 are absolutely essential for its function in conjugation of plasmid R1 and infection by the RNA phage R17. Overexpression of protein P19 in an early growth phase resulted in a massive lysis of Escherichia coli cells in liquid culture, as indicated by a rapid and distinct decrease in cell culture densities after induction. Change of the proposed catalytic glutamate at position 44 to glutamine completely abolished this effect. P19-induced cell lysis was directly shown by transmission and scanning electron microscopy. Typically, P19-overexpressing cells showed bulges protruding from the cell surfaces. Our interpretation is that these protrusions arose from a localized and spatially confined disruption of the bacterial cell wall. To our knowledge such an effect has not previously been documented for any member of the lytic transglycosylase family. From the data presented here, we conclude that protein P19 possesses the proposed localized peptidoglycan-hydrolyzing activity. This activity would be a prerequisite for efficient penetration of the cell envelope by the DNA translocation complex encoded by the conjugative plasmid.
Gram-negative bacteria possess a cell envelope that consists of two membranes and an intermembrane space, the periplasm. To cope with the high osmotic pressure generated by the uptake of nutrients, these bacteria maintain a cell wall that is located in the periplasm. The cell wall is composed of a unique biopolymer called murein or peptidoglycan which surrounds the whole bacterial cell as a single macromolecule (12). Peptidoglycan is unique to bacteria, and its disruption typically leads to lysis and cell death. Thus, it is essential for a bacterium to maintain the integrity of its cell wall. The balanced action of murein-synthesizing and -degrading enzymes allows growth and determines the specific shape of the bacterial cell. One of the glycan strand-degrading enzymes is the soluble lytic transglycosylase Slt70 (5). Slt70 is only one of several muramidases in Escherichia coli which are involved in maintaining the integrity and plasticity of the organism's cell wall. The protein has been crystallized (27), and a conserved fingerprint typical for lytic transglycosylases has been proposed (9). Sequence similarity searches revealed that this sequence fingerprint is present in an even larger family of proteins (3, 8, 13, 17, 21). Intriguingly, the newly identified members of the lytic transglycosylase family are encoded by either conjugative DNA transfer systems, virulence factor export systems, or bacteriophages. Thus, notwithstanding the large number and possible redundancy of these enzymes in gram-negative bacteria, macromolecular transport systems obviously encode their own specialized lytic transglycosylases. Although the contribution of a functional specialized lytic transglycosylase to pathogenicity could be established only in the case of VirB1 from the plant pathogen Agrobacterium tumefaciens (4, 18, 21), it is tempting to speculate that these specialized muramidases facilitate efficient transenvelope movement of DNA and/or protein substrates in other systems as well (8). Our working hypothesis is that these proteins support the correct assembly of the respective transport apparatus in the cell envelope by locally opening the peptidoglycan. Such an activity is thought to be necessary since the peptidoglycan meshwork is not wide enough to allow penetration of the cell wall by large protein complexes (8, 31). In support of this hypothesis is the recent finding that the peptidoglycan-hydrolyzing activity of the FlgJ protein is essential for flagellar rod formation in Salmonella enterica serovar Typhimurium (22). A suggested function for the lytic transglycosylases found in bacteriophages like T7 (20) or PRD1 (24) is to facilitate the entry step during the phage life cycle.
Our aim was to characterize the function of one member of this protein family, P19, in the conjugative DNA transfer of the F-like resistance plasmid R1. Gene 19 is highly conserved among F-like plasmids; its expression is posttranscriptionally down-regulated by RNase III (14). The immediate translation product of gene 19 mRNA of plasmid R1 is a precursor protein possessing a typical signal sequence. Pulse-chase experiments demonstrated the processing of P19 and a hybrid BlaP19 protein—in which the signal sequence of P19 was replaced by the signal sequence of TEM-1 β-lactamase—by the signal peptidase I of E. coli (2). Localization and gene fusion studies with phoA unambiguously demonstrated that P19 is transported through the cytoplasmic membrane into the periplasmic compartment (2). Earlier experiments from our laboratory revealed that P19 is important for efficient conjugal DNA transfer and RNA phage infection (3). We speculated that P19 is associated with the proposed DNA transport complex (32) and that it could be involved in its correct assembly by a temporally and spatially defined opening of the peptidoglycan. Here we show that P19 can indeed locally disrupt the cell envelope of E. coli upon overexpression. This observation is consistent with the results of mutational analyses and the proposed role of P19 in DNA transport.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The E. coli strains used in this work were BL21(DE3) (Novagen) for expression of P19 and P19 variants in the growth experiments, J5 (F− pro met λ+), and MC1061 [F− araD139 Δ(ara-leu)7697 galE15 galK16 Δ(lac)X74 rpsL (Strr) hsdR2(rK− mK+) mcrA mcrB1] (6). E. coli cells were grown in 2 ×TY medium (16 g of tryptone, 10 g of yeast extract, and 5 g of NaCl per liter) or in minimal medium M9 (6 g of Na2HPO4, 3 g of K2HPO4, 1 g of NH4Cl, and 0.5 g of NaCl per liter, 1 mM MgSO4, 0.4% [wt/vol] glucose, 100 μg of thiamine per ml). Antibiotics, when needed, were added at the following concentrations: ampicillin, 100 μg ml−1; tetracycline, 15 μg ml−1; and kanamycin, 40 μg ml−1.
Enzymes, chemicals, and oligonucleotides.
Restriction endonucleases and other enzymes used for standard cloning and DNA modification procedures were purchased from Roche Diagnostics. The thermostable DNA polymerases DynaZyme and Expand high-fidelity PCR System were purchased from Finnzymes and Roche Diagnostics, respectively. Oligonucleotides were purchased from the Vienna Biocenter or MWG Biotech.
DNA manipulations.
Recombinant DNA techniques were performed according to published methods (1, 25) or the manufacturers' protocols.
Recombinant plasmids and site-specific mutagenesis.
Oligonucleotide-directed site-specific mutagenesis of gene 19 was carried out by the method of Kunkel (15). Single-stranded DNA produced from plasmid pCK217 (14) was used with the mutagenic primers G63E (5′-GGGATATGGCAGCGAGCTGATGCAGGTAG-3′), and Q66E (5′-GCGGACTGATGGAGGTAGAT-3′ (mutagenic bases are in italics). The signal sequence of P19 was removed by oligonucleotide-directed deletion mutagenesis and replaced by the TEM-1 β-lactamase signal sequence as described previously (2). β-lactamase signal sequence was then deleted by using single-stranded DNA produced from plasmid pKSMBlaP19, which contains the HindIII/EcoRI fragment of pCKBlaP19 (2) cloned into the vector pKSM717 (19) and the oligonucleotide P19BlaΔ1 (5′-GCAAGATCAAAGCACATGGTAAAAACCC-3′). The resulting plasmid was designated pKSMP19ns. The C-terminal His6-tagged P19 protein was generated as follows. The oligonucleotides HISTAG1 (5′-CACCACCATCACCATCACTAATAAG-3′) and HISTAG2 (5′-AATTCTTATTAGTGATGGTGATGGTGGTG-3′) were annealed and cloned into the TthI/EcoRI-restricted plasmid pCK217 (14), creating the recombinant plasmid pCK217-6His. The in-frame fusion of gene 19 with the His6, tag was performed by inverse PCR using the oligonucleotides HISPCR3. (5′-CACCACCATCACCATCACTAA-3′) and HISPCR5 (5′-ATTATTCTGCACGCTGTTAATTTCC-3′). The resulting recombinant plasmid was designated pCK217His. The sequence of the mutagenic oligonucleotide that was used to reconstitute the NcoI restriction site by PCR in the recombinant plasmid pKSMP19ns was 5′-GGGTTTTTACCATGGGCTTTGATC-3′. The recognition sequence for the restriction endonuclease is underlined, and the mutagenic base is in italics. pKSMBlaP19His was constructed by cloning the AatII/EcoRI fragment from pCK217His into pKSMBlaP19. Reconstitution of the BglII restriction site in the recombinant plasmid pKSMBlaP19His was carried out by sequential PCR steps using the mutagenic primer bglprimerI (5′-GCTCACCCAGAAAAGATCTGCC-3′). Different point mutations were introduced in the fusion protein BlaP19 in the same manner. To create an E44Q mutation, primer 5′-CATGGAAACAATCCCGTTAC-3′ was used; a G61D mutation was introduced by using primer 5′-GGGATATGACAGCGGACTG-3′; use of the mutagenic oligonucleotide 5′-GATGCAGGTAAATTCCCAG-3′ resulted in a D68N mutation; an E83Q mutation was created by using primer 5′-GCCACAACATCTGACAAC-3′; the Y117F mutation was introduced by using the mutagenic oligonucleotide 5′-GTTGGTGCATTCAATGCAGG-3′. Amplification of gene 19 with the oligonucleotides 5′-ACGCGGATCCATGAAAAAATGGATGTTAGC-3′ and 5′-CATTGCGGCCGCTTATTACCGGTAAACCTCTG-3′ using pCK217 as a template generated a DNA fragment containing a C-terminally deleted gene 19 flanked by the restriction sites BamHI and NotI. After cloning of the digested fragment into pBluescript SK(+), the recombinant plasmid called pIRP19_R140 carried a mutated gene 19 encoding a truncated protein P19 (amino acids [aa] 1 to 140). Another C-terminal deletion of protein P19 (aa 1 to 126) was generated by the same method, using 5′-CATTGCGGCCGCTTATTAGCGTTCGGATTTCCTG-3′ as the second primer. The recombinant plasmid was named pIRP19_R126. All mutations and deletions generated by site-directed mutagenesis were verified by semiautomatic DNA sequencing using an ALF Express DNA sequencer. DNA sequence analysis and comparisons were performed using the Genetics Computer Group software package, version 10.
For growth experiments and demonstration of the lysis phenotype by electron microscopy, the following pET28a(+) (Novagen)-derived plasmids were used: pETBlaP19 and pETBlaP19His+ for expression of the BlaP19 and BlaP19His fusion proteins, respectively; and pETP19ns (no signal sequence), pETBlaP19_R140+ (fusion protein with P19 truncated after aa 140), and pETBlaP19mut1 (fusion protein with P19 truncated after aa 46).
Conjugation assays.
E. coli MC1061 donor strain cultures, harboring either the R1-16 (wild-type P19) or R1-16/mut1 (nonfunctional, truncated P191-46) plasmid, were grown in 2× TY medium supplemented with kanamycin. Overnight cultures of donor strains carrying both the large resistance plasmid and a second compatible test plasmid based on pKSM717/718 vectors (19) were grown in the presence of kanamycin and ampicillin; 10 to 40 μl of the donor culture was diluted in 0.9 ml of fresh medium prewarmed to 37°C and incubated for 30 min without shaking. Then 100 μl of recipient E. coli J5 cells was added directly from an overnight culture to the donor culture. Mating was allowed to proceed for 30 min at 37°C without shaking. DNA transfer was stopped by vigorous mixing for 1 min and rapid cooling on ice. Aliquots were diluted and plated on MacConkey lactose agar containing kanamycin. The conjugation frequency was determined by counting white donor and red transconjugant colonies and is expressed as the number of transconjugants per donor cell.
Infection studies with the RNA phage R17.
Male-specific R17 phages were propagated on the host strain, i.e., MC1061 with R1-16 or one of its derivatives, as described elsewhere (25). Essentially, the same plaque assay as described previously (3) was applied. The same test plasmids as used for the conjugation assays were used in the phage infection studies.
Growth curves.
E. coli BL21(DE3), harboring wild-type and mutated gene 19 expressed from pET28a(+) vectors, was grown in 50 ml of minimal medium M9 supplemented with kanamycin at 37°C. The T7 promoter that is present in the recombinant plasmids was used to drive expression of the various BlaP19 hybrid proteins by the addition of 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) (26). Aliquots of the culture were taken at the indicated time points, and the culture optical densities at 600 nm (OD600) were measured in a spectrophotometer.
Electron microscopy.
For investigations by transmission electron microscopy, E. coli cells were cultivated in M9 as described above. When the OD600 reached 0.25, 100-μl aliquots were mixed with top agar (M9 with 0.9% agar agar) supplemented with kanamycin and 1 mM IPTG to induce expression of the various BlaP19 constructs. The mixture was poured onto an agar plate and incubated overnight at 37°C. Agar blocks (1 mm in diameter) were cut with the tip of a Pasteur pipette, and the samples were fixed in 3% glutaraldehyde–60 mM phosphate buffer (pH 7.3) for 2 h at room temperature. The samples were then rinsed several times in 60 mM phosphate buffer (pH 7.3) and postfixed for 1 h with 1% osmium tetroxide buffered with 100 mM phosphate buffer (pH 7.3). Subsequently, the material was rinsed twice in 100 mM phosphate buffer (pH 7.3), dehydrated in a graded series of acetone including en bloc staining with 1% uranyl acetate in 70% acetone for 2 h, and embedded in agar 100 epoxy resin. Ultrathin (70-nm) sections were stained with lead citrate and viewed with a Philips CM10 electron microscope. For morphological studies by scanning electron microscopy, samples were taken 15 min after induction of protein expression and the cells were fixed overnight at room temperature with 2.5% glutardialdehyde in 75 mM sodium cacodylate–1 mM MgCl2 (pH 7.2). Thereafter, the cells were rinsed twice with 75 mM sodium cacodylate–1 mM MgCl2 (pH 7.2) and then dehydrated with a graded series of ethanol solutions. After a final dehydration step with 100% methanol, cells were bound to poly-l-lysine-coated coverslips, which were mounted with double-sided tape on aluminum stubs. Samples were critical point dried with liquid CO2 using a Baltec CPD 030. The samples were sputtered with gold by using an agar sputter coater. All scanning electron micrographs were taken with a Philips XL30 ESEM (20° tilt angle; 20-kV accelerating voltage).
RESULTS
Biological activities of P19 and P19 mutants.
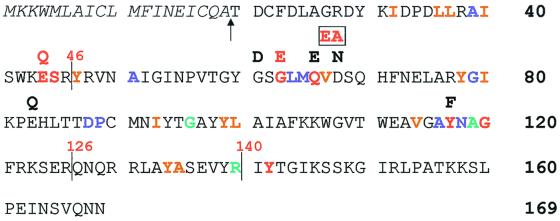
Mutational inactivation of gene 19 on the transfer-derepressed R1 derivative R1-16 by introduction of two nonpolar stop codons (R1-16/mut1) resulted in a 10-fold decrease in the conjugation frequency and the inability of bacteriophage R17 to efficiently infect E. coli cells harboring the mutated R1-16 plasmid. Both the conjugation-/and infection-attenuated phenotypes caused by the defective gene 19 could be complemented in trans by the presence of gene 19 alleles encoding the wild-type protein (3). Here we use these assays to investigate the effects of deletions and amino acid exchanges in protein P19. Based on sequence similarities between protein P19 and the 70-kDa E. coli soluble lytic transglycosylase Slt70 (8) and other members of the lytic transglycosylase family (17), gene 19 was subjected to mutational analysis and tested for the ability to complement the above-mentioned defects associated with R1-16/mut1. Mutations in P19 were introduced by site-specific mutagenesis leading to amino acid exchanges in highly conserved sequence elements (3, 13, 17, 21). Specific amino acid exchanges as well as deletions of P19 that were subjected to the analyses presented in this report are shown in Fig. 1. Mutations that were introduced and tested mainly affected conserved residues in P19. These mutations were expected to have negative effects on the function of P19. Other mutations were introduced in nonconserved residues to show that mutations per se or similar amino acid exchanges do not disturb the protein's function (compare E44Q and E83Q in Fig. 1). N- and C-terminally truncated versions were tested because it was theoretically possible that one of these truncations did not have an effect on the protein's function. By this approach, we wished to determine the minimal essential region of the protein.
FIG. 1.
Mutations in P19. The amino acid sequence of protein P19 encoded by the resistance plasmid R1 is shown. The numbering of amino acids refers to the wild-type sequence of protein P19 (SwissProt accession number P17738). The signal sequence is denoted by italic letters; the proposed signal sequence cleavage site is indicated by an arrow. As determined in reference 17, residues that are more than 85% conserved among the lytic transglycosylases are shown in red, those conserved between 65 and 85% are shown in blue, and those conserved between 50 and 65% are shown in green. Conserved hydrophobic residues are shown in brown. C-terminal deletions of the P19 protein are indicated by vertical lines together with the position of the last residue in the truncated protein. Amino acid changes in P19 are written above the corresponding residues in the P19 sequence. Loss-of-function mutations in P19 are indicated by red coloration.
Complementation of the conjugation-attenuated phenotype.
A BlaP19 fusion protein, consisting of 27 aa of TEM-1 β-lactamase and 154 C-terminal aa of protein P19, can complement the defect in a fashion indistinguishable from the wild-type protein (Table 1). This result demonstrated that the signal sequence of P19 can be functionally replaced by the β-lactamase signal sequence without disturbing the biological function of the protein. This fact and a faster processing of BlaP19 (2) were the reasons that further analyses were performed using this hybrid protein as a reference construct. P19ns, a P19 variant devoid of its own signal sequence, could not complement the conjugation deficiency. Both results are consistent with the finding that P19 is formed as a preprotein which contains a typical signal sequence directing its transport across the inner membrane into the periplasmic space (2). Two different C-terminally truncated versions of BlaP19, extending to either arginine at position 126 or arginine at position 140 (Fig. 1), were nonfunctional in the complementation studies. On the other hand, the addition of a His6 tag to the C terminus of BlaP19 did not impair the protein's activity (Table 1). The replacement of glutamate at position 44 by glutamine completely abolished the activity of P19, confirming the vital role of this invariantly conserved amino acid residue. In contrast, the replacement of glutamine at position 66 by glutamate had no negative effect. To test for a theoretically possible functional replacement of the putative catalytic glutamate at position 44 by the newly introduced glutamate at position 66, a double mutant (BlaP19E44Q;Q66EHis) was created. However, the glutamate at position 66 could not perform the function of the catalytic amino acid at position 44 (Table 1). In another mutant, a nonconserved glutamate (E83) was replaced by glutamine. This mutation turned out to be fully functional and did not influence the ability to complement the conjugation deficiency of R1-16/mut1. Furthermore, point mutations resulting in an amino acid change from glycine at position 61 to aspartate, aspartate at position 68 to asparagine, or tyrosine at position 117 to phenylalanine complemented the phenotype associated with the defective gene 19 in trans. In contrast, the amino acid glycine at position 63 or valine at position 67 together with aspartate at position 68 is essential for the biological activity of protein P19. Replacement of these amino acids by glutamate or by glutamate and alanine, respectively, resulted in a complete loss of the complementation ability.
TABLE 1.
Complementation of conjugation and phage infection defects
| Relevant feature of coresident plasmida | Relative conjugation frequencyb (mean ± SD) | Activity
|
|
|---|---|---|---|
| Conjugationc | Phage Infectiond | ||
| Vector control | 22.2 ± 5.1 | − | − |
| BlaP19 | 120.9 ± 34.4 | + | + |
| P19ns | 28.5 ± 13.8 | − | −/+ |
| BlaP19_R126+ | 9.5 ± 6.8 | − | − |
| BlaP19_R140+ | 12.4 ± 3.5 | − | − |
| BlaP19His+ | 83.8 ± 25.8 | + | + |
| BlaP19E44QHis+ | 2.8 ± 1.6 | − | − |
| BlaP19Q66EHis+ | 130.8 ± 59.8 | + | + |
| BlaP19E44Q;Q66EHis+ | 13.4 ± 5.5 | − | − |
| BlaP19E83QHis+ | 132.3 ± 56.5 | + | + |
| BlaP19G61DHis+ | 143.0 ± 66.3 | + | + |
| BlaP19G63EHis+ | 15.0 ± 3.9 | − | − |
| BlaP19V67E;D68AHis+ | 13.2 ± 8.1 | − | − |
| BlaP19D68NHis+ | 72.5 ± 18.1 | + | + |
| BlaP19Y117FHis+ | 145.5 ± 72.9 | + | + |
Relevant features of the pKSM-derived plasmids coresident in the R1-16/mut1 carrying donor strain are indicated.
Based on the conjugation frequency of the wild-type plasmid R1-16 (typically 5 × 10−1 transconjugants per donor cell), which was taken as 100%. Values represent combined results of at least three independent experiments.
+, full complementation to the level of wild-type plasmid R1-16; −, no complementation.
The male-specific RNA bacteriophage R17 was propagated on E. coli MC1061 cells harboring the defective R1-16/mut1 derivative. +, full complementation to the level of wild-type plasmid R1-16 (clear plaques); −, no complementation (turbid and fewer plaques).
Complementation of the RNA phage infection-attenuated phenotype.
To substantiate the data from the conjugation assays, we tested the same mutants of P19 for the ability to restore the phage infection defect associated with a defective P19 (3). The results of these tests (Table 1) are fully consistent with the conjugation data described above. The bacteriophage development was rescued when R1-16/mut1 was complemented in trans with gene 19 alleles encoding either BlaP19, BlaP19His, BlaP19G61DHis, BlaP19Q66EHis, BlaP19D68NHis, BlaP19E83QHis, or BlaP19Y117FHis. In these instances phage R17 formed clear plaques on lawns of the complemented strains. No complementation could be observed with gene 19 alleles encoding either of the C-terminally deleted fusion proteins BlaP19_R126 and BlaP19_R140. Furthermore, no complementation was obtained by the use of the BlaP19His fusion proteins carrying the E44Q or G63E mutation as well as the double mutant BlaP19E44Q;Q66E or BlaP19V67E;D68A. Similarly, the gene 19 allele encoding P19ns did not complement the infection defect, although the plaques were less turbid than in the cases described above.
P19 overexpression causes cell lysis.
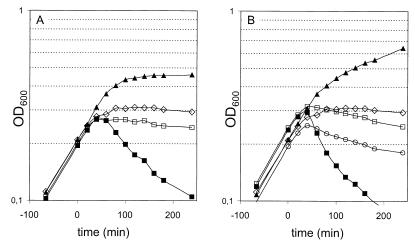
To study the effects of protein P19 overexpression on cell growth and cell morphology, we used a strong T7 polymerase/promoter-driven expression system. Growth curves were used to monitor the effects caused by overproduction of P19 and P19 derivatives (Fig. 2). Approximately 60 min after induction, a sharp decrease in the turbidity of the cell culture indicated lysis of E. coli cells expressing the BlaP19 protein (Fig. 2A). The addition of six His residues to the C-terminal end of this protein did not significantly alter its cell-lysing activity except for a slightly earlier decrease in the OD (compare pETBlaP19 and pETBlaP19His+ in Fig. 2A and B, respectively). No such detrimental effect on the cell growth in liquid culture could be observed in the case of the vector control [pET28a(+)] and the P19 deletion mutant expressed from pETP19ns, pETBlaP19_R140+, or pETBlaP19mut1. These results unambiguously showed that the lysing activity was due to the P19 part of the fusion protein and not to the β-lactamase signal sequence, which was necessary only to direct the transport of the P19ns into the periplasm. Again, the effects of mutations in the three conserved boxes on P19-induced cell lysis were investigated. As an example, the effects of two amino acid changes in P19 are shown in Fig. 2B. The E44Q mutation shifted the growth curve to the position of the vector control, showing that in this case the lysis activity of P19 was lost. In contrast, the Q66E change allowed growth between that of BlaP19His and the vector control, showing that this P19 variant still possessed lysis activity, albeit weaker than that of the wild type.
FIG. 2.
Effects of P19 expression on bacterial growth. Growth curves of E. coli BL21(DE3) harboring different pET28a(+) derivatives are shown. Expression of the P19 variants was induced by the addition of 1 mM IPTG at the time zero. A decline in the growth curve indicates cell lysis. (A) P19-mediated lysis requires a functional signal sequence and sequences beyond arginine at position 140. Growth curves of cells harboring the vector control plasmid pET28a(+) (open diamonds), pET19P19ns (filled triangles), pETBlaP19_R140 (open rectangles), or pET BlaP19 (filled rectangles) are shown. (B) P19-mediated lysis requires glutamine at position 44. Growth curves of cells harboring the vector control plasmid pET28a(+) (open diamonds), pETBlaP19mut1 (filled triangles), pETBlaP19E44QHis+ (open rectangles), pETBlaP19Q66EHis+ (open circles), or pET BlaP19His+ (filled rectangles) are shown.
Visualization of P19-mediated cell lysis by electron microscopy.
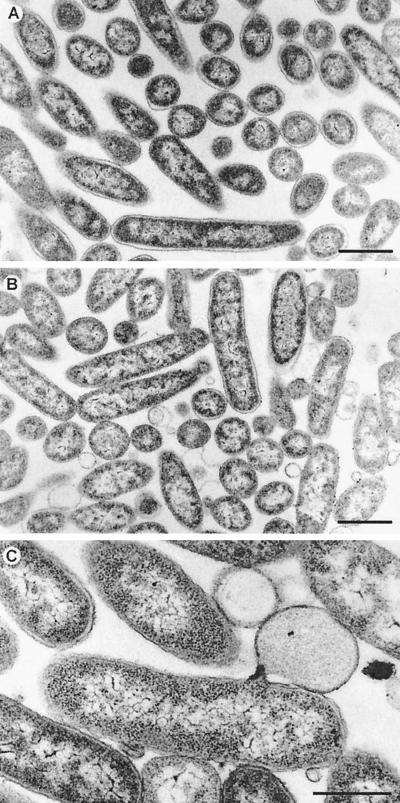
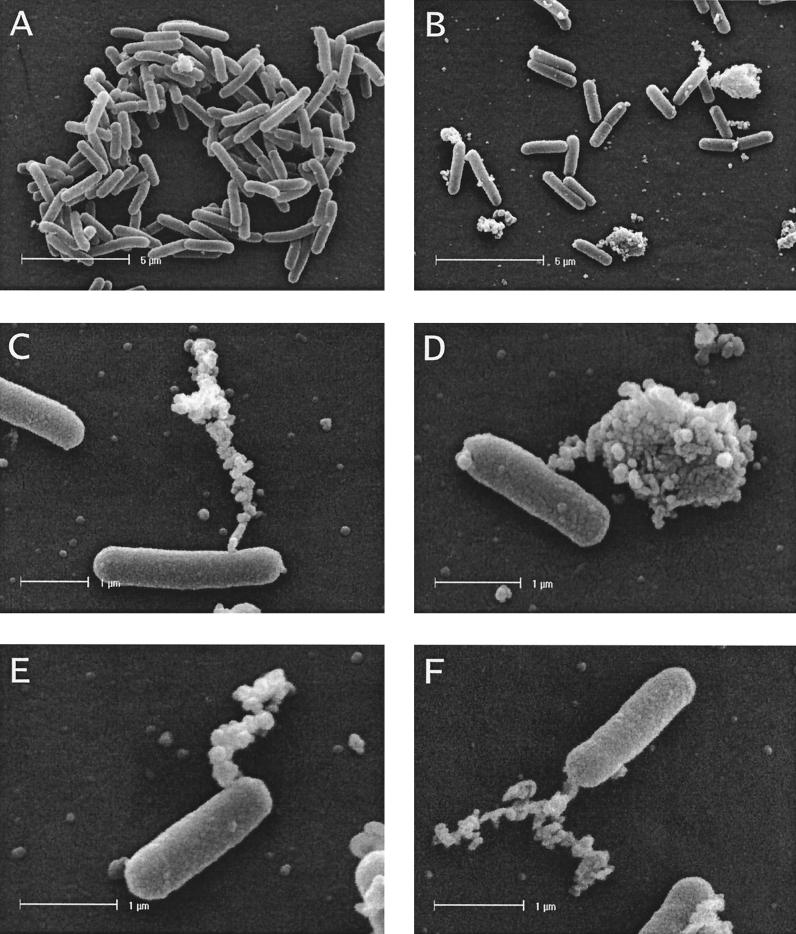
To investigate the effects of P19 expression on the morphology of E. coli cells, we prepared samples for electron microscopy. When aliquots of cell cultures used for the growth curves were subjected to electron microscopy, it was apparent that expression of P19 induced a specific lysis phenotype (Fig. 3 and 4). Transmission electron microscopy images showed a dramatic change in the morphology of E. coli cells overexpressing the BlaP19His fusion protein (Fig. 3B and C). Cells with protruding bulges in the cell envelope, membrane vesicles, and empty ghost cells were observed frequently. In most cases the vesicles were surrounded by a 6- to 7-nm-thick electron-dense material which we assumed to represent a single phospholipid bilayer. In some sections, clear connections between the cell envelope and the extruded bulges were visible. It is noteworthy that ribosomes that were clearly visible in the interior of the cells were not present in the interior of the bulges (Fig. 3C). In a control culture expressing the mut1 allele resulting in the truncated protein (aa 1 to 46), the typical morphology of E. coli cells was observed (Fig. 3A). Further evidence for P19's cell lysing activity was obtained by scanning electron microscopy of E. coli cells expressing the BlaP19His fusion protein (Fig. 4B to F). The images also showed that at this early stage of P19 expression, only 15 min after induction, the cells largely retain their integrity. The visible protrusions suggested a limited and spatially confined disruption of the cell wall. Roughly one-third of the cells were affected by a massive efflux of cellular material. The position of the extravasations was uniformly distributed over the cells. No preferred location at the cell poles or in the center of the cell could be observed. Besides the large primary sites of the protrusions, smaller secondary sites on the surface of the cells were visible. Additionally, many small vesicles were present on these preparations. The overexpression of the BlaP19/mut1His protein in a control culture did not produce any of the above-mentioned morphological changes (Fig. 4A).
FIG. 3.
Lysis phenotype of E. coli cells overexpressing the fusion protein BlaP19His visualized by transmission electron microscopy. E. coli BL21(DE3) cells harboring pETBlaP19His+ (B and C) or pETBlaP19/mut1 (A) as used for the growth curves shown in Fig. 2B were used for morphological studies by transmission electron microscopy. Typical morphological changes such as membrane vesicles are clearly visible in samples from cells expressing P19. Note the large protrusion in panel C which is still connected to the cell. Scale bars represent 1.0 μm (A and B) and 0.5 μm (C).
FIG. 4.
Lysis phenotype of E. coli cells overexpressing the fusion protein BlaP19His visualized by scanning electron microscopy. E. coli BL21(DE3) cells harboring pETBlaP19His+ (B to F) or pETBlaP19/mut1 (A) as used for the growth curves shown in Fig. 2B were used for morphological studies by scanning electron microscopy. Single large and several minor protrusions on the cell surfaces of the cells expressing P19 are visible. Also note the numerous small vesicles that are present in images B to F.
DISCUSSION
The results presented here show that protein P19 encoded by the conjugative resistance plasmid R1 has the ability, if overexpressed in high quantities, to disrupt the cell envelope of E. coli cells. Although such an effect is to be expected from a protein that has been categorized as belonging to a large family of lytic transglycosylases (3, 8, 13, 17, 21), it has not been shown by using electron microscopy for any member of this protein family so far. The observed lysis seems to be quite distinct from other lysis phenotypes, for instance, the large disruptions in the cell envelope when growing cells are treated with β-lactam antibiotics. In the case of P19, the small lesions indicate a localized opening in the peptidoglycan meshwork, leading to the observed efflux of intracellular material which is most likely driven by the osmotic pressure inside the cell. The appearance of vesicles surrounded by a membrane bilayer also suggests that it is not the membranes that are affected or disrupted by P19. The localized protrusions from the cells expressing P19 raise the question of whether P19 is targeted to a certain location in the cell envelope. Although the bulges on the cells are not confined to certain areas on the cell surface, data obtained in studies of the subcellular localization of P19 showed that mature P19 is a periplasmic protein and suggested that it might be attached to the proposed membrane-spanning DNA transport complex (2). To clarify whether P19 interacts with one of the transport proteins, detailed protein-protein interaction studies will be required. A potential interaction domain could be represented by the C-terminal amino acid residues of P19 beginning beyond the highly conserved tyrosine at position 142. This notion is supported by a high variation of the different lytic transglycosylase sequences in this region (17) and by the high sensitivity of P19 toward proteases which is, at least to some extent, specified by this part of the protein (unpublished observations).
The results of the mutational analyses together with the complementation assays are in good agreement with the proposed enzymatic lytic transglycosylase activity of protein P19. First, it was shown that a functional signal sequence is absolutely required for P19 activity, which means that the protein must be transported across the cytoplasmic membrane to fulfill its function as a cell wall-degrading enzyme. The P19 signal sequence can be functionally replaced by the β-lactamase signal sequence, which demonstrates that its only role is to direct the protein to the periplasm. A His6 extension at the C terminus does not significantly affect P19's biological function, although the conjugation frequency in the complementation tests was somewhat lower than that of the corresponding protein without the His6 sequence. One explanation for this slight reduction might be that the His6 sequence reduces the ability of the protein to interact with its postulated partner protein. On the other hand, the addition of the His6 sequence to the C terminus of P19 seemed to enhance the cell-lysing activity, as reflected by an earlier and more pronounced decrease in the growth curve. This enhanced activity may be due to a reduced protease sensitivity of this P19 variant. Both C-terminal deletion mutants that were tested completely lost their biological and also their cell lysing activities. Therefore, an essential element for the enzymatic activity of P19 is present beyond the arginine residue at position 140. This finding was surprising since all conserved sequence elements identified in earlier studies (3, 8, 13, 21) are present in this deletion mutant. From a recently published comparison of lytic transglycosylase sequences of bacterial, bacteriophage, and plasmid origins (17), however, it became clear that another amino acid residue, namely, tyrosine at position 142 in P19, is present in virtually all of the compared sequences. It is therefore very likely that tyrosine at position 142 is essential for the enzymatic function of the protein. Indeed, the proposed steps in the catalytic mechanism of Slt70 involve the participation of tyrosine at position 587 (28), which is equivalent to tyrosine at position 142 of P19. A newly constructed deletion variant of P19 extending to leucine at position 153 which includes this tyrosine but lacks 16 C-terminal aa turned out to be fully active in the complementation assays (unpublished observations).
Other mutants of P19 with specific single amino acid changes that were tested in the experiments behaved exactly as expected for an enzyme belonging to the lytic transglycosylase family. For instance, the E44Q mutation led to a completely inactive form of the protein with no detectable activity. Since glutamate at position 44 corresponds to the catalytic glutamate at position 587 in α-helix 2 of the C domain of Slt70 (30), we suggest that P19 indeed possesses the proposed catalytic cleavage activity of the glycosidic bonds between N-acetylmuramic acid and N-acetylglucosamine residues in peptidoglycan. The corresponding exchange in the VirB1 protein of A. tumefaciens also led to complete inactivation of this protein, which was found to be required for efficient tumor formation in plants (21). Similarly, an E587Q mutation in Slt70 (27) and also the mutation of Glu162 to glutamine in the catalytic core domain of Slt35 (29) resulted in completely inactive enzymes. Another residue that was found to be essential for P19's activity was glycine at position 63, which resides in a highly conserved sequence element (the GLMQ box) which is essential for active-site architecture in Slt70 (9). A surprising result was that the change of the highly conserved tyrosine at position 117 to phenylalanine did not influence P19's activity. We assume that this Y117F mutation does not create a rearrangement of the active-site cleft or alter the substrate binding specificity of P19.
Taken together, our results strongly support the hypothesis that P19 acts as a specialized lytic transglycosylase that facilitates a timely and spatially controlled opening of the peptidoglycan during the process of conjugation, a bacterial type IV secretion system (7, 16). The data presented in this paper are substantiated by recently performed zymogram analyses of purified MalE-P19 fusion protein. The results of these analyses unambiguously demonstrate P19's peptidoglycan-hydrolyzing activity (unpublished observations).
Furthermore, we believe that our data represent a very strong argument for the existence of such specialized lytic transglycosylases in macromolecular transport systems in general. Besides the already recognized related proteins present in type III (for instance, IpgF of Shigella flexneri) and type IV (for instance, VirB1 of A. tumefaciens) secretion systems, there are a number of novel members identified as belonging to the rapidly growing lytic transglycosylase family, such as the protein encoded by rorf3 found in the locus of enterocyte effacement from enteropathogenic E. coli (10), VirB1 encoded by the virB operon of the animal pathogen Brucella suis (23), Hpa2 from the hrp gene cluster of Xanthomonas oryzae (33), and YsaH from a chromosomally encoded type III secretion pathway in Yersinia enterocolitica (11). Thus, knowledge of how P19 encoded by the conjugative resistance plasmid R1 acts at the molecular level will probably lead to understanding the role of specialized lytic transglycosylases in other transenvelope transport systems and possibly provide the basis for the development of new anti-infective molecules.
ACKNOWLEDGMENTS
This work was supported by the Austrian Science Foundation (grant P11844-MED) and the EU-BIOTECH concerted action MECBAD.
We thank Friederike Turnowsky for critically reading the manuscript.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Smith J A, Seidman J G, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1987. [Google Scholar]
- 2.Bayer M, Bischof K, Noiges R, Koraimann G. Subcellular localization and processing of the lytic transglycosylase of the conjugative plasmid R1. FEBS Lett. 2000;466:389–393. doi: 10.1016/s0014-5793(00)01088-7. [DOI] [PubMed] [Google Scholar]
- 3.Bayer M, Eferl R, Zellnig G, Teferle K, Dijkstra A J, Koraimann G, Högenauer G. Gene 19 of plasmid R1 is required for both efficient conjugative DNA transfer and bacteriophage R17 infection. J Bacteriol. 1995;177:4279–4288. doi: 10.1128/jb.177.15.4279-4288.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berger B R, Christie P J. Genetic complementation analysis of the Agrobacterium tumefaciens virB operon: virB2 through virB11 are essential virulence genes. J Bacteriol. 1994;176:3646–3660. doi: 10.1128/jb.176.12.3646-3660.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Betzner A S, Keck W. Molecular cloning, overexpression and mapping of the slt gene encoding the soluble lytic transglycosylase of Escherichia coli. Mol Gen Genet. 1989;219:489–491. doi: 10.1007/BF00259625. [DOI] [PubMed] [Google Scholar]
- 6.Casadaban M J, Cohen S N. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980;138:179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- 7.Christie P J, Vogel J P. Bacterial type IV secretion: conjugation systems adapted to deliver effector molecules to host cells. Trends Microbiol. 2000;8:354–360. doi: 10.1016/s0966-842x(00)01792-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dijkstra A J, Keck W. Peptidoglycan as a barrier to transenvelope transport. J Bacteriol. 1996;178:5555–5562. doi: 10.1128/jb.178.19.5555-5562.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dijkstra B W, Thunnissen A M. ‘Holy’ proteins. II. The soluble lytic transglycosylase. Curr Opin Struct Biol. 1994;4:810–813. doi: 10.1016/0959-440x(94)90261-5. [DOI] [PubMed] [Google Scholar]
- 10.Elliott S J, Wainwright L A, McDaniel T K, Jarvis K G, Deng Y K, Lai L C, McNamara B P, Donnenberg M S, Kaper J B. The complete sequence of the locus of enterocyte effacement (LEE) from enteropathogenic Escherichia coli E2348/69. Mol Microbiol. 1998;28:1–4. doi: 10.1046/j.1365-2958.1998.00783.x. [DOI] [PubMed] [Google Scholar]
- 11.Haller J C, Carlson S, Pederson K J, Pierson D E. A chromosomally encoded type III secretion pathway in Yersinia enterocolitica is important in virulence. Mol Microbiol. 2000;36:1436–1446. doi: 10.1046/j.1365-2958.2000.01964.x. [DOI] [PubMed] [Google Scholar]
- 12.Höltje J V. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol Mol Biol Rev. 1998;62:181–203. doi: 10.1128/mmbr.62.1.181-203.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koonin E V, Rudd K E. A conserved domain in putative bacterial and bacteriophage transglycosylases. Trends Biochem Sci. 1994;19:106–107. doi: 10.1016/0968-0004(94)90201-1. [DOI] [PubMed] [Google Scholar]
- 14.Koraimann G, Schroller C, Graus H, Angerer D, Teferle K, Högenauer G. Expression of gene 19 of the conjugative plasmid R1 is controlled by RNase III. Mol Microbiol. 1993;9:717–727. doi: 10.1111/j.1365-2958.1993.tb01732.x. [DOI] [PubMed] [Google Scholar]
- 15.Kunkel T A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lai E M, Kado C I. The T-pilus of Agrobacterium tumefaciens. Trends Microbiol. 2000;8:361–369. doi: 10.1016/s0966-842x(00)01802-3. [DOI] [PubMed] [Google Scholar]
- 17.Lehnherr H, Hansen A M, Ilyina T. Penetration of the bacterial cell wall: a family of lytic transglycosylases in bacteriophages and conjugative plasmids. Mol Microbiol. 1998;30:454–457. doi: 10.1046/j.1365-2958.1998.01069.x. [DOI] [PubMed] [Google Scholar]
- 18.Llosa M, Zupan J, Baron C, Zambryski P. The N- and C-terminal portions of the Agrobacterium VirB1 protein independently enhance tumorigenesis. J Bacteriol. 2000;182:3437–3445. doi: 10.1128/jb.182.12.3437-3445.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maneewannakul S, Maneewannakul K, Ippen-Ihler K. The pKSM710 vector cassette provides tightly regulated lac and T7lac promoters and strategies for manipulating N-terminal protein sequences. Plasmid. 1994;31:300–307. doi: 10.1006/plas.1994.1032. [DOI] [PubMed] [Google Scholar]
- 20.Moak M, Molineux I J. Role of the Gp16 lytic transglycosylase motif in bacteriophage T7 virions at the initiation of infection. Mol Microbiol. 2000;37:345–355. doi: 10.1046/j.1365-2958.2000.01995.x. [DOI] [PubMed] [Google Scholar]
- 21.Mushegian A R, Fullner K J, Koonin E V, Nester E W. A family of lysozyme-like virulence factors in bacterial pathogens of plants and animals. Proc Natl Acad Sci USA. 1996;93:7321–7326. doi: 10.1073/pnas.93.14.7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nambu T, Minamino T, Macnab R M, Kutsukake K. Peptidoglycan-hydrolyzing activity of the FlgJ protein, essential for flagellar rod formation in Salmonella typhimurium. J Bacteriol. 1999;181:1555–1561. doi: 10.1128/jb.181.5.1555-1561.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O'Callaghan D, Cazevieille C, Allardet-Servent A, Boschiroli M L, Bourg G, Foulongne V, Frutos P, Kulakov Y, Ramuz M. A homologue of the Agrobacterium tumefaciens VirB and Bordetella pertussis Ptl type IV secretion systems is essential for intracellular survival of Brucella suis. Mol Microbiol. 1999;33:1210–1220. doi: 10.1046/j.1365-2958.1999.01569.x. [DOI] [PubMed] [Google Scholar]
- 24.Rydman P S, Bamford D H. Bacteriophage PRD1 DNA entry uses a viral membrane-associated transglycosylase activity. Mol Microbiol. 2000;37:356–363. doi: 10.1046/j.1365-2958.2000.01996.x. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 26.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 27.Thunnissen A M, Dijkstra A J, Kalk K H, Rozeboom H J, Engel H, Keck W, Dijkstra B W. Doughnut-shaped structure of a bacterial muramidase revealed by X-ray crystallography. Nature. 1994;367:750–753. doi: 10.1038/367750a0. [DOI] [PubMed] [Google Scholar]
- 28.Thunnissen A M, Rozeboom H J, Kalk K H, Dijkstra B W. Structure of the 70-kDa soluble lytic transglycosylase complexed with bulgecin A. Implications for the enzymatic mechanism. Biochemistry. 1995;34:12729–12737. doi: 10.1021/bi00039a032. [DOI] [PubMed] [Google Scholar]
- 29.van Asselt E J, Dijkstra A J, Kalk K H, Takacs B, Keck W, Dijkstra B W. Crystal structure of Escherichia coli lytic transglycosylase Slt35 reveals a lysozyme-like catalytic domain with an EF-hand. Struct Fold Des. 1999;7:1167–1180. doi: 10.1016/s0969-2126(00)80051-9. [DOI] [PubMed] [Google Scholar]
- 30.van Asselt E J, Thunnissen A M, Dijkstra B W. High resolution crystal structures of the Escherichia coli lytic transglycosylase Slt70 and its complex with a peptidoglycan fragment. J Mol Biol. 1999;291:877–898. doi: 10.1006/jmbi.1999.3013. [DOI] [PubMed] [Google Scholar]
- 31.Yao X, Jericho M, Pink D, Beveridge T. Thickness and elasticity of gram-negative murein sacculi measured by atomic force microscopy. J Bacteriol. 1999;181:6865–6875. doi: 10.1128/jb.181.22.6865-6875.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zechner E L, de la Cruz F, Eisenbrand R, Grahn A M, Koraimann G, Lanka E, Muth G, Pansegrau W, Thomas C M, Wilkins B M, Zatyka M. Conjugative DNA transfer processes. In: Thomas C M, editor. The horizontal gene pool: bacterial plasmids and gene spread. Amsterdam, The Netherlands: Harwood Academic Publishers; 1999. pp. 87–173. [Google Scholar]
- 33.Zhu W, MaGbanua M M, White F F. Identification of two novel hrp-associated genes in the hrp gene cluster of Xanthomonas oryzae pv. oryzae. J Bacteriol. 2000;182:1844–1853. doi: 10.1128/jb.182.7.1844-1853.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]