Abstract
Studies of citrate synthase (CitA) were carried out to investigate its role in morphological development and biosynthesis of antibiotics in Streptomyces coelicolor. Purification of CitA, the major vegetative enzyme activity, allowed characterization of its kinetic properties. The apparent Km values of CitA for acetyl coenzyme A (acetyl-CoA) (32 μM) and oxaloacetate (17 μM) were similar to those of citrate synthases from other gram-positive bacteria and eukaryotes. CitA was not strongly inhibited by various allosteric feedback inhibitors (NAD+, NADH, ATP, ADP, isocitrate, or α-ketoglutarate). The corresponding gene (citA) was cloned and sequenced, allowing construction of a citA mutant (BZ2). BZ2 was a glutamate auxotroph, indicating that citA encoded the major citrate synthase allowing flow of acetyl-CoA into the tricarboxylic acid (TCA) cycle. Interruption of aerobic TCA cycle-based metabolism resulted in acidification of the medium and defects in morphological differentiation and antibiotic biosynthesis. These developmental defects of the citA mutant were in part due to a glucose-dependent medium acidification that was also exhibited by some other bald mutants. Unlike other acidogenic bald strains, citA and bldJ mutants were able to produce aerial mycelia and pigments when the medium was buffered sufficiently to maintain neutrality. Extracellular complementation studies suggested that citA defines a new stage of the Streptomyces developmental cascade.
The unique developmental characteristics of members of the genus Streptomyces have generated considerable interest in their genetics and physiology. These gram-positive soil bacteria grow initially as substrate mycelium that differentiates both morphologically and metabolically. Unlike unicellular bacteria, where daughter cells separate, Streptomyces cells remain associated, thus forming long filaments. Such hyphae elongate by apical growth at the tips and at the same time branch into a complex network of interconnected substrate mycelium. Streptomyces exhibits a biphasic growth pattern correlated with a developmental switch (9, 42). Thus, an initial phase of growth is followed by a brief interruption; biomass accumulation then resumes, accompanied by the emergence of aerial hyphae on the colony surface and the onset of secondary metabolism. Many of the characterized Streptomyces coelicolor mutants defective for aerial hyphal erection (bald [bld]mutants) are also unable to make the pigmented antibiotics actinorhodin and undecylprodigiosin (4, 21).
Aerial hyphae formation can be triggered by extracellular factors, a phenomenon first observed in Streptomyces griseus. Purification, structural determination, and chemical synthesis defined Factor A as one of the first pheromone-like molecules described for bacteria (15). Using similar assays, physiological interactions and developmental signals between pairwise combinations of a selected series of S. coelicolor bld mutants have been described. When two such mutants were grown adjacent to each other, one was able to restore the ability of its neighbor to erect aerial hyphae (30, 31, 44, 45). The fact that this response was partner dependent suggested a model in which these complementation groups represented an ordered signaling cascade leading to aerial mycelium erection. In principle, the events underlying the extracellular complementation phenomenology may reflect either a positively or negatively acting compound(s). As in the case of Factor A, compounds excreted by the complementing strain may trigger the formation of aerial hyphae. Alternatively, recent data have suggested that the differentiating colonies might be able to detoxify growth-inhibitory conditions, such as acidogenesis, that arrest differentiation (42).
Acidification of the growth medium by organic acids generated from glucose has been well documented for various Streptomyces species cultivated in either liquid or solid medium (6, 11, 24, 35, 42). Accumulation of acids leads to a gradual drop in pH; eventually, acids are consumed, thus avoiding growth-inhibitory pH levels (<5 in S. coelicolor). On unbuffered glucose-based minimal medium, this cyclic AMP-dependent transition is coincident with and is essential for aerial mycelium formation and biosynthesis of the polyketide antibiotic actinorhodin (42). While an adenylate cyclase mutant (cya) was bald and pigmentless on classical glucose minimal medium, it was able to differentiate if the pH was neutralized by the inclusion of buffer (42).
Interestingly, many S. coelicolor bld mutants have been identified by their inability to form aerial hyphae on unbuffered glucose minimal media. The developmental block can be overcome by replacing glucose with permissive, nonacidogenic carbon sources such as mannitol (42). Organic acid oxidation is carried out via the tricarboxylic acid (TCA) and/or glyoxylate cycle, both of which are initiated by citrate synthase.
Bacillus subtilis requires a functional TCA cycle to initiate a developmental program leading to spore formation (8). Sporulation is dependent on a phosphorelay cascade that ultimately activates the master regulatory protein, Spo0A. Loss of various TCA enzymes blocks sporulation, each at different stages: citrate synthase (mutations in both citA and citZ) at stage III, aconitase (citB) at stage 0, and isocitrate dehydrogenase (citC) at stage I (5, 16–20). A priori, this could mean that the levels of the TCA cycle intermediates serve as nutritional checkpoints in the developmental cascade. Alternatively, the sporulation defect might merely reflect the inhibitory effects of an artificially imposed metabolic imbalance. A mutant that lacks the first three TCA cycle enzymes (citrate synthase, aconitase, and isocitrate dehydrogenase) has a severe sporulation defect (16). This developmental block might reflect a biological sporulation signal, since it can be suppressed by a spo0A mutant that does not require the phosphorelay for activation (16). These studies established the concept that TCA metabolism is intimately connected with the development of spores in B. subtilis (5, 16–20).
This suggested a potential role of citrate synthase in Streptomyces morphogenesis. In addition, one of its substrates, acetyl coenzyme A (acetyl-CoA), is a precursor for polyketide antibiotics. Thus, blocking metabolic flux at the level of citrate synthase might allow diversion of acetyl-CoA to increase the fermentative output of commercially important polyketide antibiotics.
In order to investigate the role of the TCA cycle in development and antibiotic biosynthesis, we identified the primary citrate synthase gene (citA) and constructed a citA null mutation. This mutant accumulated organic acids, indicating a reduced TCA cycle activity, and was conditionally defective both for aerial hypha formation and synthesis of the pigmented antibiotics undecylprodigiosin and actinorhodin.
MATERIALS AND METHODS
Bacterial strains, plasmids, media and cultivation.
S. coelicolor A3 (2) derivatives used were MT1110 (SCP1− SCP2−) and J1501 (hisA1 uraA1 strA1 pgl SCP1− SCP2−). The developmentally blocked S. coelicolor strains J1700 (bldA39), J701 (bldB15), J660 (bldC18), J774 (bldD18), C103 (bldG103), C109 (bldH109), NS17 (bldK1), and HU61 (bldJ) were kindly provided by R. Losick's laboratory. The brgA mutant was provided by K. Ochi.
Streptomyces lividans 1326 and Escherichia coli XL1-Blue (Stratagene) were used as hosts for cloning. When S. coelicolor was transformed, nonmethylated DNA was prepared from E. coli ET 12567 (provided by D. MacNeil and T. MacNeil) in order to circumvent the restriction barrier.
YEME (14), minimal medium (MM) (14), MS (mannitol soy flour, also known as SFM) (10), J medium (33), NE (28), and R2YE medium (14) have been described previously.
Standard conditions were used for extracellular complementation experiments (44). Bald strains were grown on R2YE plates for 7 days at 30°C.
S. coelicolor and E. coli were grown as described by Hopwood et al. (14) and Sambrook et al. (37).
For antibiotic selection in E. coli, Luria broth was supplemented with ampicillin (100 μg/ml), kanamycin (50 μg/ml), spectinomycin (10 μg/ml), or apramycin (50 μg/ml). For antibiotic selection in Streptomyces, solid R2YE medium was supplemented with: thiostrepton (50 μg/ml), apramycin (100 μg/ml), spectinomycin (500 μg/ml), or kanamycin (250 μg/ml). YEME liquid medium was supplemented with thiostrepton (5 μg/ml), apramycin (20 μg/ml), or spectinomycin (100 μg/ml). All antibiotics were purchased from Sigma.
DNA techniques.
General DNA techniques were performed as described by Sambrook et al. (37). Minipreps and recovery of DNA fragments from agarose gels were done using the Qiaprep or Qiaquick system from Qiagen.
Media composition analyses.
To remove liquid from agar medium, contents of a plate were placed in a 30-ml tube, frozen at −80°C for 1 h, thawed for 2 h at room temperature, and finally centrifuged at 20,000 rpm in a Sorvall SS34 rotor. Ammonium concentrations were determined using the Dr. Lange LCK303 cuvette test in combination with the Dr. Lange CADAS 30 Photometer (Dr. Lange AG, Hegnau, Switzerland). The NH4+ concentration was determined after a 30-min incubation at 30°C. Other metabolites (fumarate, succinate, and malate) were measured using enzyme-linked assays. Analyses were preformed in an automatic enzymatic analyzer (Beckman Synchron CX5CE clinical system; Beckman Instruments, Inc., Fullerton, Calif.). All tests were based on determination of quantitative changes in NADH content reflected by changes in absorption at 340 nm. Commercial test kits were used to determine concentrations of glucose (Synchron systems glucose reagent 442640; Beckman Instruments), acetate (TC acetic acid 148261; Roche Molecular Biochemicals, Basel, Switzerland) and citric acid (TC citric acid 139076; Roche). Pyruvate was assayed using standard protocols (3).
Purification of citrate synthase (CitA).
S. coelicolor J1501 grown in YEME (200 ml) for 48 h served as a seed culture to inoculate 9 liters of YEME. After 40 h of growth, mycelia were harvested by centrifugation to yield a pellet (30 g [wet weight]). The pellet was resuspended in 150 ml of MB buffer (20 mM Tris-HCl [pH 7.6], 1 mM MgCl2, 1 mM EDTA, 1 mM dithiothreitol, 10% [vol/vol] glycerol) containing 50 mM NaCl and lysed by sonication at 4°C. All subsequent chromatographic purification steps were performed at 4°C. The disrupted cells were diluted with 150 ml of MB buffer containing 150 mM NaCl, and the cell debris was removed by centrifugation in a GSA rotor (Sorvall) at 11,000 rpm. The supernatant was fractionated by fast protein liquid chromatography (FPLC) on a DEAE Sepharose FF column (Pharmacia) equilibrated in MB buffer containing 50 mM NaCl. After the column was washed extensively with loading buffer, bound proteins were eluted with a continuous gradient ranging from 50 to 500 mM NaCl. Citrate synthase activity was assayed spectrophotometrically (41). Active fractions were pooled, diluted to a NaCl concentration equal to 50 mM in MB buffer, and subjected to chromatography on Q Sepharose FF (Pharmacia) equilibrated in the same buffer. Citrate synthase activity was eluted with a linear NaCl gradient (50 mM to 1 M). The active fractions were pooled and dialyzed in MB buffer to give a final conductivity equal to 50 mM. The dialyzed sample was loaded on a MonoQ HR5/5 column (Pharmacia) and eluted with 80 ml of a linear gradient of 50 mM to 1 M NaCl. One-milliliter fractions were collected and assayed for citrate synthase activity. Finally, an affinity chromatography step was performed by applying the citrate synthase active fractions onto a Matrex gel red A (Amicon) column in MB buffer containing 50 mM NaCl. After the column was washed extensively with MB containing 50 mM NaCl, the retained citrate synthase activity was eluted in MB buffer supplemented with 0.2 mM oxaloacetate, 0.2 mM CoA, and 50 mM NaCl.
Protein analyses.
Protein concentrations were determined by the Bradford technique using a kit purchased from Bio-Rad using bovine serum albumin as the standard. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed as described by Laemmli (22). In order to determine the N-terminal sequence of the 43-kDa protein, 10 μg was run on SDS–12.5% PAGE and blotted onto a polyvinylidene difluoride Immobilon membrane (Millipore) using 10 mM CAPS (3-[cyclohexyl-amino]-1-propanesulfonic acid), pH 11.0, buffer containing 10% methanol.
Cloning of citA.
A degenerate oligonucleotide based on the N-terminal sequence (CitN-term, 5′-GT[G/C]GT[G/C]CT[G/C]CGITACGGIGA-3′; I = inosine) and a “guessmer” (Cit2, (5′-AC[G/C]CGGTGICCGAAICCCAT) based on a conserved region of citrate synthases were synthesized to PCR amplify a fragment of citA from MT1110 genomic DNA. Taq DNA polymerase (5 U; Boehringer) was used in a PCR mixture (consisting of 100 pmol of both primers, 1 μg of template DNA, 10% dimethyl sulfoxide, and 0.2 mM deoxynucleoside triphosphates in 1× PCR buffer supplemented with 0.15 mM MgCl2) to amplify a 900-bp fragment. This fragment was gel purified using the Quiex II system (Qiagen) and, following treatment with T4 DNA polymerase, was cloned into the EcoRV site of pBluescript SKI (Stratagene) to yield pVHP90.
Sequencing the ends of the insert in pVHP90 and subsequent database searches of the translated sequence showed that the cloned fragment was highly homologous to other citrate synthase genes. pVHP90 was digested with HindIII and EcoRI (flanking the EcoRV site in pBluescript SK) in order to release the insert. However, two fragments (420 and 510 bp) were generated (a HindIII site was present on the insert), both of which were isolated and 32P labeled using the random primer labeling technique (Boehringer). Southern blotting of BamHI-digested S. coelicolor MT1110 genomic DNA gave a 3.3-kb hybridization signal. Hence, BamHI-restricted genomic fragments were size fractionated, isolated, and ligated into BamHI-digested pK18. A citA-carrying clone was identified by colony hybridization and designated pVHP92.
Sequencing strategy.
The nucleotide sequence of a 2.1-kb region encompassing the entire citA open reading frame (ORF) was determined by subcloning and primer walking. Sequencing was performed on an ABI 373 sequencer (Applied Biosystems, Inc.) using dye terminator cycle sequencing ready reactions (Perkin Elmer) according to the manufacturer's instructions. Sequences were assembled and analyzed using the Lasergene package from DNA Star.
Disruption of citA.
A PCR approach was employed to create a gene replacement of citA. A 465-bp fragment, which contains 390 bp upstream of the N terminus and the first 75 bp of the citA reading frame, was amplified using oligonucleotides CitA1 (5′-TGTACACGGAATTCACCC-3′) and CitA2 (5′-GAAGCCCTTGGATCCGAC-3′) (EcoRI and BamHI sites are underlined) (5 U of Taq polymerase; Roche Biochemicals). Likewise, a 380-bp C-terminal fragment, containing the last 230 bp of the citA reading frame as well as 150 bp of the downstream region, was amplified by PCR. The two C-terminal oligomers used in this reaction were CitA3 (5′-CTCGCGGATCCTCTACC-3′) and CitA4 (5′-GGCGCTCTAGATCGTCGG-3′). The BamHI/EcoRI-restricted N-terminal fragment and BamHI/XbaI-cleaved C-terminal fragment were inserted via a three-way ligation into EcoRI/XbaI-cut pOK12. The resulting plasmid, pVHP96, was linearized with BamHI, and an apramycin resistance gene cassette was inserted. The resulting plasmid, pVHP97, carried a mutated citA allele, in which most of the citA-coding region had been replaced by an apramycin resistance gene. pVHPA22 was created by isolating the mutated citA allele from SpeI-digested pVHP97 and ligating it into the XbaI site of pGM9. pGM9 contains a Streptomyces origin of replication which is temperature sensitive and has been used efficiently in gene replacement experiments by selecting for homologous recombination between the genome and the plasmid at nonpermissive temperatures (29). MT1110 spores containing pVHPA22 were plated on NE medium containing 100 μg of apramycin/ml and incubated at 39°C for 5 days. A thiostrepton- and apramycin-resistant transformant was isolated and grown in J medium containing 1% glutamate for 48 h. The culture was diluted 1/50 and grown for another 36 h. In order to ensure segregation of individual clones, the mycelium was lysozyme digested to produce protoplasts that were regenerated on R2YE. From several hundred colonies having a wild-type phenotype (thiostrepton and apramycin resistant), two bald (thiostrepton-sensitive and apramycin-resistant) colonies were isolated. The chromosomal citA locus of both isolates was probed by PCR and Southern blot hybridization. PCR was performed on colonies and purified genomic DNA with an Expand long-template kit (Roche Biochemicals) and oligonucleotides CitA6 (5′-GTCCTCGTCGGGTCGCGCAA-3′) and CitA7 (5′-GCGGTACGGGACTCACCAGA-3′). Southern blot hybridization was performed on BamHI- and PstI-cleaved genomic DNA that had been separated by 0.8% agarose gel electrophoresis and transferred to Hybond N nylon membranes (Amersham). The 870-bp insert of pVHP96 was liberated by restriction with EcoRI and XbaI, gel purified, 32P labeled by random primer labeling (Roche Biochemicals), and used to probe Southern blots (37). After confirmation of the citA gene replacement, one of the two isolates was selected and designated BZ2.
The SCP2 plasmid derivative pIJ903 (23) (one to two copies per chromosome) served as a vector to complement the citA lesion. The 3.4-kb BamHI insert in pVHP92 was isolated and ligated into BamHI-cut pIJ903 to give pIJ903-cit.
pVHP190 is a derivative of pPM927 (40) containing citA on a 1.7-kb BamHI fragment obtained by Expand PCR (Roche Biochemicals) using the primers comp_cit1 (5′-GAGGGATCCTTCTGCTGTCCGGGTCCGTTC-3′) and comp_cit2 (5′-GAGGGATCCTGGATGATGCTCCAGCGGAA-3′) and pVHP91 as a template.
Enzyme assays and kinetic analysis.
Citrate synthase was assayed at 25°C using acetyl-CoA and oxaloacetate as substrates (41). This assay indirectly measures the rate of formation of CoA, which is liberated from acetyl-CoA, by splitting 5,5′-dithiobis-(2-nitro-benzoic acid). Changes in absorption were measured spectrophotometrically at A412 (extinction coefficient [E] = 136,000).
Citrate synthase was calculated as the rate of change in A412 per minute and expressed as specific activity (units or milliunits per milligram of protein).
The purified CitA described above was used in the reaction assays to calculate the apparent Km and Vmax values (13a). When limiting concentrations of either substrate were used in the assay, the other substrate was maintained at saturating levels.
Nucleotide sequence accession number.
The sequence determined in this study has been assigned accession number AF181118_1.
RESULTS
Purification of the major citrate synthase (CitA) of S. coelicolor.
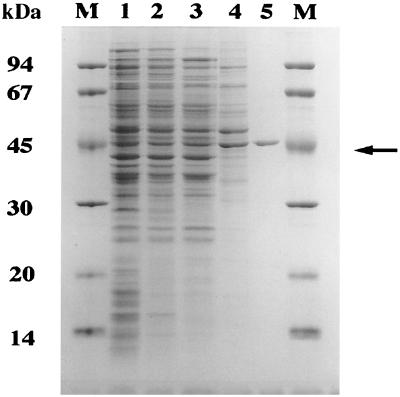
The major S. coelicolor citrate synthase activity was purified from crude cytoplasmic protein extracts (Fig. 1). Ion-exchange chromatography (DEAE, Q-Sepharose, MonoQ), followed by affinity chromatography (Matrex gel red A) (19, 26, 36, 39), enriched the enzyme ca. 70-fold (Table 1). A Matrex red A fraction containing the majority of enzyme activity was essentially pure citrate synthase as judged by SDS-PAGE (Fig. 1). The activity of the purified enzyme (21.0 U/mg) corresponded well with that of citrate synthases from Streptomyces hygroscopicus, Bacillus megaterium, and B. subtilis. Furthermore, CitA subunits migrated with an apparent molecular mass on SDS-PAGE (45 kDa) (Fig. 1) consistent with that described for other citrate synthases (19, 26, 36, 39). In order to isolate the citA gene, purified CitA was subjected to sequential Edman N-terminal degradation. The sequence of the first 10 residues was NH2-S-D-N-X-V-V-L-R-Y-G-D (X represents an unassigned amino acid). Identification of the primary citrate synthase activity later allowed us to focus on the major vegetative gene rather than other paralogous loci that have since been identified in the S. coelicolor genome database.
FIG. 1.
Purification of CitA from S. coelicolor crude extracts. Representative samples from each chromatography step (Table 1) were analyzed by SDS–12.5% PAGE. S. coelicolor crude extract (lane 1) was fractionated by DEAE-Sepharose (lane 2), Q-Sepharose (lane 3), and MonoQ anion-exchange chromatography (lane 4) and Matrex gel red A affinity chromatography to yield a pure preparation of CitA (lane 5; arrow). The molecular masses of the protein marker depicted in lane M are shown on the left.
TABLE 1.
Purification of citrate synthase from S. coelicolora
| FPLC column fraction | Vol (ml) | Total activity (U) | Total protein (mg) | Sp act (U/mg) | Yield (%) |
|---|---|---|---|---|---|
| Crude extract | 150 | 945 | 3,150 | 0.3 | 100 |
| DEAE-Sepharose | 100 | 294 | 490 | 0.6 | 31 |
| Q-Sepharose | 25 | 213 | 305 | 0.7 | 22.5 |
| MonoQ | 30 | 149 | 18.6 | 8 | 15.7 |
| Matrex gel red A | 15 | 134 | 6.4 | 21 | 14.2 |
The primary citrate synthase detected in protein extracts of S. coelicolor J1501 grown in YEME was purified by FPLC.
Kinetic properties of CitA.
Reaction kinetics of CitA-catalyzed conversion of acetyl-CoA and oxaloacetic acid to citrate were analyzed as described previously (13a). The apparent Km values for acetyl-CoA (32 μM) and oxaloacetate (17 μM) were similar to those citrate synthases from other gram-positive bacteria and eukaryotes but were quite distinct from the low affinities of citrate synthases from gram-negative organisms (i.e., GltA from E. coli and citrate synthase isoenzyme I from Pseudomonas aeruginosa (7, 19, 26, 36, 39).
CitA was tested for allosteric effects of various metabolic intermediates known to affect enzyme activity in other bacteria. While citrate synthases of gram-negative bacteria are strongly inhibited by NADH, citrate synthases isolated from gram-positive bacteria and eukaryotes are generally less affected (26, 27). CitA, like CitZ of B. subtilis and GltA of Corynebacterium glutamicum, was not inhibited by NADH concentrations up to 10 mM (7, 19, 20). Inhibition of S. coelicolor CitA by ATP was less pronounced (40% inhibition at 10 mM ATP) (19, 20) than that reported for B. subtilis CitZ (50% inhibition at 0.81 mM ATP). Likewise, citrate synthases of S. hygroscopicus and C. glutamicum are only weakly inhibited by ATP (7, 39). S. coelicolor CitA activity was also unaffected by NAD+ (5 mM), ADP (5 mM), isocitrate (10 mM), or EDTA (0.05 mM). Interestingly, and in contrast to the citrate synthase of S. hygroscopicus (39), S. coelicolor CitA activity was not inhibited by α-ketoglutarate (10 mM).
Cloning and sequencing of citA.
Two degenerate primers were used to PCR amplify a citA internal fragment from S. coelicolor MT1110 genomic DNA. Degenerate primers were designed from the CitA N-terminal amino acid sequence as well as an internal peptide sequence that is highly conserved among all known citrate synthases (MGFGHRV). The PCR fragment was used to probe Southern blots of restricted MT1110 genomic DNA. A strongly hybridizing 3.4-kb BamHI fragment of genomic DNA was subsequently cloned into pK18 (a pUC18 derivative). A combination of primer walking and subcloning was used to determine a 2.1-kb sequence.
The sequence included an ORF (1,290 nucleotides) predicting a protein of 429 amino acids (PID g6492340) and with a potential GTG translational initiation site (nucleotides 621 to 623) followed immediately by a region encoding an N-terminal sequence indistinguishable from that of CitA determined by Edman degradation. The predicted CitA protein had a molecular mass of 47 kDa and a pI of 5.8, in agreement with values determined by two-dimensional gel analysis of CitA (reference 43 and data not shown). Database searches showed that its highest similarity was to citrate synthases of Mycobacterium tuberculosis (CisY; 68% identity) and C. glutamicum (GltA; 63% identity) (7).
The citA gene encodes the primary citrate synthase supporting growth.
The TCA cycle intermediate α-ketoglutarate is an essential precursor for glutamate biosynthesis in bacteria growing on glucose minimal medium. Therefore, if the TCA cycle is blocked, strains become glutamate auxotrophs. Although CitA was the major citrate synthase activity in crude extracts, more than one citrate synthase activity might have been present. Two other citrate synthase-like genes (SC5B8.21c and SC5B8.22) distinct from citA have been identified as a part of the S. coelicolor genome sequencing project.
To decide whether CitA was the principal citrate synthase determining flux through the TCA cycle during growth, a citA-disrupted mutant was constructed in which most of the citA ORF was deleted and replaced with an apramycin resistance cassette (see Materials and Methods). This mutated allele was inserted at the chromosomal citA locus of S. coelicolor MT1110 using pGM9, a temperature-sensitive delivery system containing the thiostrepton resistance gene (29) (see Materials and Methods). A mutant colony (BZ2) was isolated, and its citA disruption was verified by Southern blotting and PCR (data not shown).
In order to determine whether BZ2 was a glutamate auxotroph, SMM+Asn plates containing glucose as the primary carbon source and asparagine as a nitrogen source were supplemented with 0, 5, 50, or 500 μg of glutamate/ml. While the isogenic parent (MT1110) was able to grow on all plates, BZ2 was strictly dependent on the presence of at least 50 μg of glutamate/ml in the medium. Similarly, glutamate was required for growth on aspartate (probably incorporated into the TCA cycle by transamination, which converts it to oxaloacetate).
These genetic experiments confirmed biochemical indications that citA encoded the major vegetative citrate synthase and that paralogous genes in the genome probably do not significantly contribute to the formation of citrate during the early phases of growth.
The citA mutation causes a bald phenotype.
In the endospore-forming gram-positive bacterium B. subtilis, loss of the major citrate synthase (citZ) impaired sporulation when the organism was grown in rich sporulation medium. This phenotype was strongly enhanced when the gene encoding the residual citrate synthase activity (citA) was disrupted (19, 20). Thus, a B. subtilis citA citZ double mutant was a glutamate auxotroph and arrested during sporulation (19, 20), even in the presence of sufficient glutamate to support growth. Inactivation of citA alone rendered the strain auxotrophic for glutamate (BZ2). However, even when high concentrations (500 μg/ml) of glutamate were supplied, BZ2 had developmental defects on R2YE.
While MT1110 had begun to produce red pigment (presumably undecylprodigiosin) and aerial hyphae after 40 h of growth, the citA mutant remained unpigmented and bald. MT1110 continued to differentiate over the next few days and produced blue pigment (presumably gamma-actinorhodin) and spores. In contrast, BZ2 showed none of these signatures of development on R2YE or glucose minimal medium (SMMS).
These BZ2 phenotypes were solely attributable to the citA mutation. Auxotrophy and developmental defects were eliminated when the citA gene was supplied in trans on pIJ903 (23) (pIJ903-cit) or when citA was integrated into the pSAM att site using pVHP190. This proved that the phenotypes observed were due to citA disruption and not to polar effects on downstream genes.
The bald phenotype of the citA mutant was glucose dependent. On media lacking glucose, BZ2 aerial hypha morphogenesis was comparable to that of MT1110. Media tested included R2YE, SMMS minimal media in which glucose was replaced with mannitol, and mannitol soya (MS). BZ2 was not able to differentiate on MS supplemented with glucose (1%). These experiments indicated that the bald phenotype of BZ2, like that of most other bald mutants, was mediated by glucose.
BZ2 defects were also suppressed by genetic manipulations of glucose kinase expression (data not shown) that block glucose utilization and alter catabolite control (2). Paradoxically, when the glucose kinase gene (glkA) is cloned on a high-copy-number plasmid (pIJ702 or pIJ2427), S. coelicolor cannot grow on glucose and catabolite repression is relieved (2). BZ2/pIJ2427 cultures produced aerial mycelium and pigment on R2YE. The fact that it retained its glutamate auxotrophy requirement indicated that the effect was not due to activation of a cryptic citrate synthase during growth. In a second series of experiments, five mutant derivatives of BZ2 that could produce aerial mycelium were isolated. These suppressors all retained their glutamate auxotrophy; however, four were unable to grow on glucose as the primary carbon source.
Effect of buffer on the development of BZ2 and other bald mutants.
The glucose-dependent bald phenotype of BZ2 and the frequently described accumulation of organic acids in cultures of Streptomyces growing on glucose-rich medium (6, 11, 24, 35, 42) suggested that acidogenesis might be responsible for the developmental arrest in BZ2 and perhaps other bald mutants. The pH of BZ2 cultures on two glucose-containing media, SMMS and R2YE, where it was bald, decreased and remained below 5 (Fig. 2). In contrast, the pH of MT1110-conditioned medium was rather constant (Fig. 2). When BZ2 was grown on SMMS or R2YE containing mannitol instead of glucose, development was not impaired and the medium pH remained neutral.
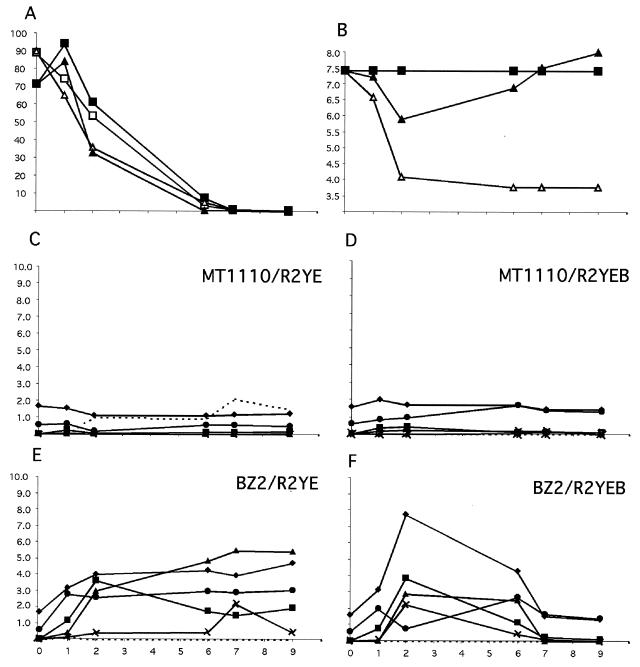
FIG. 2.
The developmental block of the citrate synthase mutant was accompanied by alterations in metabolism. MT1110 or BZ2 was grown on normal R2YE plates (25 mM TES buffer, pH 7.2) or the same medium supplemented with additional buffer (to 100 mM, R2YEB) for nine days (x axes) at 30°C. MT1110 produced aerial mycelium and pigmented antibiotics beginning at day 2 and spores beginning at day 4 on both types of plates. BZ2 produced neither aerial mycelium nor pigmented antibiotics on R2YE but was able to undergo a developmental program similar to that of MT1110 on R2YEB. Extracts of the agar medium were prepared as described in Materials and Methods in order to measure glucose concentration (millimolar) (A), pH (B), concentrations (millimolar) of organic acids (citrate, acetate, malate, pyruvate, succinate) (C to F), and concentrations (millimolar) of ammonium (C to F). (A and B) Squares, MT1110; triangles, BZ2; open symbols, R2YE; filled symbols, R2YEB. (C to F) Diamonds, acetate; squares, pyruvate; triangles, malate; exes, fumarate; circles, succinate; dotted line, NH4.
The consequences of the citA mutation on glucose metabolism and acidification of R2YE-glucose were analyzed by comparing BZ2 and MT1110 cultures grown on R2YE medium. BZ2 consumed glucose more rapidly than MT1110 and accumulated much higher concentrations of acetate, pyruvate, malate, succinate, and fumarate (Fig. 2).
If the low pH contributed as a direct cause rather than a consequence of the BZ2 developmental arrest, then buffer might suppress the bald phenotype. While the presence of buffer (25 mM TES, pH 7.2) in glucose minimal medium (SMMS) fully restored the developmental defects of the S. coelicolor cya mutant (BZ1), very high concentrations of buffer (100 or 125 mM TES) were required to maintain the medium pH of BZ2 near neutrality and suppress the bald phenotype (Fig. 3A). Interestingly, under these conditions, in which aerial mycelia are formed, BZ2 was able to reassimilate the organic acids it had previously excreted into the medium (Fig. 2F).
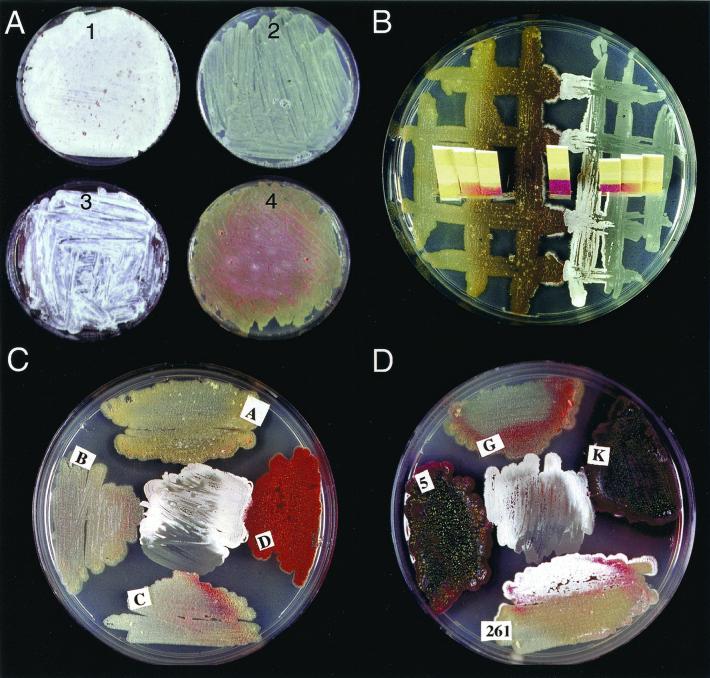
FIG. 3.
Rescue of developmentally blocked mutants by buffer or extracellular complementation. (A) pH control rescues morphological differentiation of the citA and bldJ (bld261) mutants. BZ2 (citA; plates 1 and 2) and HU61 (bldJ; plates 3 and 4) were grown at 30°C either on R2YE plates (plates 2 and 4) or R2YE supplemented with 100 mM TES (plates 1 and 3). (B) Aerial hypha formation of BZ2 (citA; right half of plate) was induced when the strain was grown next to the acidogenic bldA mutant. The pH of both mutant cultures, recorded on pH papers, dropped to less than 5.0 (yellow pH paper) and remained acidic at the edge of the plate, where differentiation was blocked. In closer proximity, interactions between the strains first allowed differentiation of the citA mutant associated with neutralization of the medium (pH 6.0, red pH paper). Several days later, the bldA mutant produced few aerial mycelia in response to differentiating citA mutant mycelium. (C) Developmentally blocked mutants citA (BZ2; center), bldA (A), bldD (D), bldC (C), and bldB (B). All mutants except bldC induced aerial mycelium formation in the citA mutant. (D) Developmentally blocked mutants citA (BZ2; center), bldG (G), bldK (K), bldJ (261), and bld5 (5, a previously undescribed S. coelicolor bald mutant isolated by K. Nguyen). All mutants except bldJ induced aerial mycelium formation in the citA mutant. In contrast, bldJ was induced to form aerial mycelium by the citA mutant.
Medium acidification caused by S. coelicolor bald mutants.
S. coelicolor wild-type strain MT1110 and strains with the bald mutation bldA, bldB, bldC, bldD, bldF, bldH, bldK, brgA, or bldJ were plated at a high cell density on complex (R2YE, a medium containing 25 mM TES, pH 7.2) or minimal (SMMS) glucose-based media. The pH of the cultures was monitored on a daily basis using pH paper. On this complex medium, MT1110 did not display the documented decrease in pH observed transiently during development on a minimal medium (SMMS) (42). Thus, significant pH changes reflecting a metabolic shift occurred on minimal medium but not on complex media routinely used to characterize bald mutants by extracellular complementation. However, lawns of bldA, bldB, bldG, bldH, and bldJ strains irreversibly acidified both R2YE and SMMS media from pH 7.2 to ≤5 shortly after growth became visible. In contrast, lawns of bldD, bldF, and bldK strains did not alter the pH of R2YE. Like the citA strain, all acidogenic mutants failed to produce pigments, while the mutants that maintained neutral pH were all pigmented.
Quantitative analysis of organic acids excreted by bldA, bldB, bldG, bldH, and bldJ strains indicated that the pH drop was accompanied by increased accumulation of organic acids. Liquid extracts were assayed for acetate, pyruvate, malate, succinate, fumarate, and citrate (Table 2). While three independent experiments have shown that wild-type strain MT1110 never accumulated more than 0.6 mM pyruvate during growth (1 to 9 days), all bald mutant culture supernatants contained elevated concentrations (>2.5 mM) after 2 days of culture. In addition, whereas MT1110 accumulated only 0.2 mM succinate, acidogenic bald mutants accumulated 1.8 to 2.6 mM.
TABLE 2.
Organic acid accumulation in developmental mutantsa
| Strain | Concn (mM) of:
|
|||||
|---|---|---|---|---|---|---|
| Pyruvate | Succinate | Fumarate | Malate | Citrate | Acetate | |
| None (R2YE alone) | 0.01 | 0.5 | <0.1 | <0.1 | 0.0 | 1.7 |
| MT1110 | 0.6 | 0.2 | 0.4 | <0.1 | <0.1 | 1.0 |
| bldA mutant | 2.6 | 1.8 | 0.2 | <0.1 | <0.1 | 1.7 |
| bldB mutant | 2.6 | 2.5 | 0.2 | <0.1 | <0.1 | 1.5 |
| bldG mutant | 2.6 | 2.6 | 0.2 | <0.1 | <0.1 | 0.9 |
| bldH mutant | 2.6 | 2.2 | 0.1 | <0.1 | <0.1 | 2.3 |
| bldJ mutant | 2.6 | 2.3 | 0.4 | <0.1 | <0.1 | 1.3 |
Extracts were prepared from 48-h cultures grown on R2YE and analyzed for organic acid concentrations as described in Materials and Methods.
These results suggested that bald mutant phenotypes might also be due to acid arrest of the developmental program. Therefore, bldA, bldB, bldC, bldH, bldD, bldF, bldJ, and bldK strains were tested for their ability to differentiate on strongly buffered medium. Only bldJ was rescued by pH control (Fig. 3A, plate 1).
Extracellular complementation characteristics of the citA mutant detected using the standard agar diffusion assay.
Extracellular complementation was first examined by growing BZ2 adjacent to various Streptomyces species under standard conditions (44). The BZ2 developmental defect was readily complemented extracellularly by wild-type S. coelicolor as well as other species, including S. lividans, Streptomyces parvulus, and Streptomyces griseolus.
In order to find out whether BZ2 could be assigned to one of the S. coelicolor extracellular complementation groups, cross-feeding experiments were carried out between BZ2 and other developmental mutants (Fig. 3C and D show results for strains carrying bldJ [=bld261], bldA, bldB, bldC, bldD, bldG, and bldK; bldH and bldF were also tested). These experiments showed that most bld mutants (except those with bldC and bldJ) were able to rescue the BZ2 morphological defect by extracellular complementation. Given that bldK complemented BZ2 (Fig. 3D) and, in turn, BZ2 complemented bldJ, together with the assumption that these interactions reflected positively acting factors, suggested that citA defines a new complementation group located between bldJ and bldK in the signaling cascade model.
A correlation between developmental changes and pH changes in the medium during cultivation of the citA mutant adjacent to other bald mutants suggested that a positively acting factor was involved. During the first three days of the complementation tests, BZ2 differentiation was blocked and the underlying medium was acidic (pH < 5). After seven days, the BZ2 culture became more neutral (pH increased from <5 to 6.5) and differentiated (Fig. 3B) in response to all adjacent bld mutants (except bldJ and bldC). In principle, this could result from consumption of organic acids by the adjacent bald mutant. However, the fact that the adjacent bald mutants were also acidic during the arrested period (Fig. 3B), suggested that the effect was at least partially a result of a novel BZ2 metabolism triggered by a compound provided by its partner.
DISCUSSION
Citrate synthase acts at the junction of glycolytic and TCA pathways and is thus ideally situated to maintain physiological balance, allowing morphological development and antibiotic biosynthesis. The citA mutant was unable to erect aerial mycelium or produce pigmented antibiotics (actinorhodin and undecylprodigiosin), and this mutation maps to a location different from all previously reported developmental mutations (encoded by cosmid StC57 [34] in the Sanger Center Streptomyces genome database). Glutamate starvation did not appear to cause the developmental arrest. While the growth defect of the citA mutant on minimal medium was suppressed by glutamate, it remained developmentally blocked. Similar observations have led to the attractive concept that TCA cycle activity serves as a developmental signal in B. subtilis (5, 16–20).
The discovery that developmentally blocked cya, bldA, bldB, bldG, bldH, and bldJ mutants were acidogenic on glucose-based media suggested that control of TCA metabolism might be part of the S. coelicolor developmental program. The fact that acidification was easily monitored and reversed by buffer allowed experiments to determine the causal relationships between metabolism and aerial mycelium defects.
Maintaining a constant pH did not suppress the developmental defects of most bald mutants (bldA, bldB, bldG, and bldH strains), suggesting that acidogenesis was the effect rather than the cause of their phenotype. Accumulation of organic acids by these mutants may have been a consequence of limited oxygen availability in older, densely packed substrate mycelium. Bacterial colonies can generate steep oxygen gradients such that even cells relatively near the air-surface interface are in a microaerobic environment (46); projection of hyphae into the air may increase oxygen availability and thereby allow renewed TCA metabolic activity that consumes organic acids. Aerial mycelium affords better surface exposure and may thereby increase oxygen availability. If this is the case, these bald mutants undergo a mixed acid metabolism as a function of their morphology. In addition, this predicts aerobic TCA-based metabolism in aerial mycelium, perhaps involving the other S. coelicolor citrate synthase paralogs. In contrast, buffer allowed citA and bldJ mutants to produce aerial mycelium and pigments (Fig. 3A), suggesting that in these cases, metabolic imbalance leading to acidosis was a primary cause of their developmental defects.
The acidosis and developmental defect of BZ2, similar to those of most other bald mutants, was dependent on glucose. Previous studies have shown that organic acid excretion reflects imbalance between glycolysis and TCA cycle activities (13) and can be induced by oxygen limitation (12). The citrate synthase mutant consumed glucose at slightly higher rates and was unable to oxidize glycolytic products via the TCA cycle. While the underlying biochemical pathways in Streptomyces are not defined, BZ2 accumulated numerous organic acids, including pyruvate, acetate, fumarate, succinate, and malate (Fig. 2). The BZ2 developmental defect was suppressed by removing glucose from the medium or genetic manipulations of glucose kinase that prevent the utilization of glucose.
Similarly, other metabolic perturbations, not necessarily related to the normal pathway of differentiation, may account for the blocks of other bald mutations. Pope et al. (32) have suggested that the primary defect in bald mutants is in the regulation of carbon metabolism. In both Aspergillus nidulans (1, 38) and Streptomyces (25), auxotrophic mutants can be developmentally impaired.
Extracellular complementation studies already indicate that such a plethora of nonspecific primary metabolic disorders are not overly represented among currently investigated bald mutants. These studies show that most bald mutations can be organized into an ordered developmental cascade, which would not be the case if they reflected a network of independent metabolic imbalances unrelated to the developmental program.
Using previously established criteria, the citA mutation fits into an early stage of this proposed extracellular complementation cascade (bldJ > bldK > bldA and bldH > bldG > bldC > bldD). citA and bldJ represent different extracellular complementation groups; citA would be placed between bldJ and bldK (Fig. 3D), assuming that the signal is positively acting. Alternatively, extracellular complementation resulting from inactivation of a negative signal produced by bldJ, for example, would imply a reversal of the order of the two genes. Therefore, we prefer not to definitively assign the position of citA in the cascade model until the nature of the extracellular “signal” and its activity as activator or inhibitor of development are established.
In cross complementation between BZ2 and acidogenic partners with mutations that are “downstream” in the cascade (bldA, bldB, bldG, bldH), the BZ2 sector increased its pH, often while its partner remained acidic. Thus, while acidosis was sufficient to account for the citA developmental defects, diffusible compounds, thought to reflect a S. coelicolor signaling cascade, apparently evoke changes in metabolism leading to medium neutralization coincident with aerial mycelium formation.
Our previous studies of S. coelicolor differentiation revealed that increases in medium pH were coincident with developmental changes on minimal media (42). Failure to carry out this shift in an adenylate cyclase mutant resulted in a low pH environment that inhibited both growth and development. Such dramatic changes in medium pH do not accompany development of wild-type S. coelicolor on R2YE. However, this homeostasis, determined by the cell's capacity to metabolize its glycolytic products, may be disrupted in citA and other bald mutants. In a neutral environment that allowed aerial mycelium formation of the citA mutant, organic acids accumulated and were eventually metabolized coincident with aerial mycelium formation (Fig. 2F). These observations indicate that maintenance of physiological pH within the environment of substrate mycelium reflects a metabolic regulatory system that is an integral part of development on this medium. It is possible that for some pairs of bald mutants, complementation assays may reflect activity of this program. However, the citA mutant responds to other bald mutants independently of pH neutralization in a pattern consistent with the proposed extracellular signaling model. This lends credibility to the extracellular complementation model and our tentative conclusion that this metabolic gene has been incorporated into the Streptomyces developmental program.
In conclusion, the accumulation of organic acids by citA and many bald mutants growing on glucose reflects imbalance between glycolytic and TCA flux. This may be avoided during normal development by converting fermentation products to less acidic compounds such as lipids or polyketides, decreasing glycolytic flux, or increasing TCA cycle flux. The fact that bald and citA mutants cannot do this may reflect a glucose-dependent metabolic perturbation(s) (32) and/or an important function of aerial mycelium for the uptake of oxygen.
ACKNOWLEDGMENTS
We are grateful to Paul Jenö for carrying out the N-terminal sequence analysis and especially to Kien Nguyen and Joanne Willey for helpful discussions and critical reading of the manuscript.
This work was supported by Swiss National Science Foundation grants to C. J. Thompson (3100-039669 and SPP Biotechnology 5002-046085) and W. Minas (SPP Biotechnology 5002-46086).
REFERENCES
- 1.Adams T H, Wieser J K, Yu J H. Asexual sporulation in Aspergillus nidulans. Microbiol Mol Biol Rev. 1998;62:35–54. doi: 10.1128/mmbr.62.1.35-54.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Angell S, Lewis C G, Buttner M J, Bibb M J. Glucose repression in Streptomyces coelicolor A3(2): a likely regulatory role for glucose kinase. Mol Gen Genet. 1994;244:135–143. doi: 10.1007/BF00283514. [DOI] [PubMed] [Google Scholar]
- 3.Bergmeyer J, Grassl M. Metabolites. 1. Carbohydrates. Weinheim, Germany: Verlag Chemie; 1984. [Google Scholar]
- 4.Chater K F. Taking a genetic scalpel to the Streptomyces colony. Microbiology. 1998;144:1465–1478. doi: 10.1099/00221287-144-6-1465. [DOI] [PubMed] [Google Scholar]
- 5.Craig J E, Ford M J, Blaydon D C, Sonenshein A L. A null mutation in the Bacillus subtilis aconitase gene causes a block in Spo0A-phosphate-dependent gene expression. J Bacteriol. 1997;179:7351–7359. doi: 10.1128/jb.179.23.7351-7359.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doskocil J, Sikyta B, Kasparova J, Doskocilova D, Zajicek J. Development of the culture of Streptomyces rimosus in submerged fermentations. J Gen Microbiol. 1958;18:302–314. doi: 10.1099/00221287-18-2-302. [DOI] [PubMed] [Google Scholar]
- 7.Eikmanns B J, Thum-Schmitz N, Eggeling L, Ludtke K U, Sahm H. Nucleotide sequence, expression and transcriptional analysis of the Corynebacterium glutamicum gltA gene encoding citrate synthase. Microbiology. 1994;140:1817–1828. doi: 10.1099/13500872-140-8-1817. [DOI] [PubMed] [Google Scholar]
- 8.Fortnagel P, Freese E. Analysis of sporulation mutants. II. Mutants blocked in the citric acid cycle. J Bacteriol. 1968;95:1431–1438. doi: 10.1128/jb.95.4.1431-1438.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Granozzi C, Billetta R, Passantino R, Sollazzo M, Puglia A M. A breakdown in macromolecular synthesis preceding differentiation in Streptomyces coelicolor A3(2) J Gen Microbiol. 1990;136:713–716. doi: 10.1099/00221287-136-4-713. [DOI] [PubMed] [Google Scholar]
- 10.Hobbs G, Frazer C M, Gardner D C J, Cullum J A, Oliver S G. Dispersed growth of Streptomyces in liquid culture. Appl Microbiol Biotechnol. 1989;31:272–277. [Google Scholar]
- 11.Hobbs G, Obanye A I, Petty J, Mason J C, Barratt E, Gardner D C, Flett F, Smith C P, Broda P, Oliver S G. An integrated approach to studying regulation of production of the antibiotic methylenomycin by Streptomyces coelicolor A3(2) J Bacteriol. 1992;174:1487–1494. doi: 10.1128/jb.174.5.1487-1494.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hockenhull D J D, Fantes K H, Herbert M, Whitehead B. Glucose utilization by Streptomyces griseus. J Gen Microbiol. 1954;10:353–370. doi: 10.1099/00221287-10-3-353. [DOI] [PubMed] [Google Scholar]
- 13.Hodgson D A. Primary metabolism and its control in Streptomyces: a most unusual group of bacteria. Adv Microb Physiol. 2000;42:47–238. doi: 10.1016/s0065-2911(00)42003-5. [DOI] [PubMed] [Google Scholar]
- 13a.Hofstee B H J. Non-inverted versus inverted plots in enzyme kinetics. Nature. 1959;184:1296–1298. doi: 10.1038/1841296b0. [DOI] [PubMed] [Google Scholar]
- 14.Hopwood D A, Bibb M J, Chater K F, Kieser T, Bruton C J, Kieser H M, Lydiate D J, Smith C P, Ward J M, Schrempf H. Genetic manipulation of Streptomyces. A laboratory manual. Norwich, United Kingdom: The John Innes Foundation; 1985. [Google Scholar]
- 15.Horinouchi S, Beppu T. A-factor as a microbial hormone that controls cellular differentiation and secondary metabolism in Streptomyces griseus. Mol Microbiol. 1994;12:859–864. doi: 10.1111/j.1365-2958.1994.tb01073.x. [DOI] [PubMed] [Google Scholar]
- 16.Ireton K, Jin S, Grossman A D, Sonenshein A L. Krebs cycle function is required for activation of the Spo0A transcription factor in Bacillus subtilis. Proc Natl Acad Sci USA. 1995;92:2845–2849. doi: 10.1073/pnas.92.7.2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jin S, De Jesus-Berrios M, Sonenshein A L. A Bacillus subtilis malate dehydrogenase gene. J Bacteriol. 1996;178:560–563. doi: 10.1128/jb.178.2.560-563.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jin S, Levin P A, Matsuno K, Grossman A D, Sonenshein A L. Deletion of the Bacillus subtilis isocitrate dehydrogenase gene causes a block at stage I of sporulation. J Bacteriol. 1997;179:4725–4732. doi: 10.1128/jb.179.15.4725-4732.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jin S, Sonenshein A L. Identification of two distinct Bacillus subtilis citrate synthase genes. J Bacteriol. 1994;176:4669–4679. doi: 10.1128/jb.176.15.4669-4679.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jin S, Sonenshein A L. Transcriptional regulation of Bacillus subtilis citrate synthase genes. J Bacteriol. 1994;176:4680–4690. doi: 10.1128/jb.176.15.4680-4690.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kelemen G H, Buttner M J. Initiation of aerial mycelium formation in Streptomyces. Curr Opin Microbiol. 1998;1:656–662. doi: 10.1016/s1369-5274(98)80111-2. [DOI] [PubMed] [Google Scholar]
- 22.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 23.Lydiate D J, Malpartida F, Hopwood D A. The Streptomyces plasmid SCP2*: its functional analysis and development into useful cloning vectors. Gene. 1985;35:223–235. doi: 10.1016/0378-1119(85)90001-0. [DOI] [PubMed] [Google Scholar]
- 24.Madden T, Ward J M, Ison A P. Organic acid excretion by Streptomyces lividans TK24 during growth on defined carbon and nitrogen sources. Microbiology. 1996;142:3181–3185. doi: 10.1099/13500872-142-11-3181. [DOI] [PubMed] [Google Scholar]
- 25.Meade H. Cloning of argG from Streptomyces: loss of gene in Arg mutants of S. cattleya. Bio/Technology. 1985;3:917–918. [Google Scholar]
- 26.Mitchell C G, Anderson S C, el-Mansi E M. Purification and characterization of citrate synthase isoenzymes from Pseudomonas aeruginosa. Biochem J. 1995;309:507–511. doi: 10.1042/bj3090507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morse D, Duckworth H W. A comparison of the citrate synthases of Escherichia coli and Acinetobacter anitratum. Can J Biochem. 1980;58:696–706. doi: 10.1139/o80-098. [DOI] [PubMed] [Google Scholar]
- 28.Murakami T, Anzai H, Imai S, Satoh A, Nagaoka K, Thompson C J. The bialaphos biosynthetic genes of Streptomyces hygroscopicus: molecular cloning of the gene cluster. Mol Gen Genet. 1986;205:42–50. [Google Scholar]
- 29.Muth G, Nussbaumer B, Wohlleben W, Pühler A. A vector system with temperature-sensitive replication for gene disruption and mutational cloning in streptomycetes. Mol Gen Genet. 1989;219:341–348. [Google Scholar]
- 30.Nodwell J R, Losick R. Purification of an extracellular signaling molecule involved in production of aerial mycelium by Streptomyces coelicolor. J Bacteriol. 1998;180:1334–1337. doi: 10.1128/jb.180.5.1334-1337.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nodwell J R, McGovern K, Losick R. An oligopeptide permease responsible for the import of an extracellular signal governing aerial mycelium formation in Streptomyces coelicolor. Mol Microbiol. 1996;22:881–893. doi: 10.1046/j.1365-2958.1996.01540.x. [DOI] [PubMed] [Google Scholar]
- 32.Pope K M, Green B D, Westpheling J. The bld mutants of Streptomyces coelicolor are defective in the regulation of carbon utilization, morphogenesis and cell-cell signalling. Mol Microbiol. 1996;19:747–756. doi: 10.1046/j.1365-2958.1996.414933.x. [DOI] [PubMed] [Google Scholar]
- 33.Puglia A M, Vohradsky J, Thompson C J. Developmental control of the heat-shock stress regulon in Streptomyces coelicolor. Mol Microbiol. 1995;17:737–746. doi: 10.1111/j.1365-2958.1995.mmi_17040737.x. [DOI] [PubMed] [Google Scholar]
- 34.Redenbach M, Kieser H M, Denapaite D, Eichner A, Cullum J, Kinashi H, Hopwood D A. A set of ordered cosmids and a detailed genetic and physical map for the 8 Mb Streptomyces coelicolor A3(2) chromosome. Mol Microbiol. 1996;21:77–96. doi: 10.1046/j.1365-2958.1996.6191336.x. [DOI] [PubMed] [Google Scholar]
- 35.Redshaw P A, McCann P A, Sankaran L, Pogell B M. Control of differentiation in streptomycetes: involvement of extrachromosomal deoxyribonucleic acid and glucose repression in aerial mycelia development. J Bacteriol. 1976;125:698–705. doi: 10.1128/jb.125.2.698-705.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robinson S M, Danson M J, Weitzman P D. Citrate synthase from a Gram-positive bacterium. Purification and characterization of the Bacillus megaterium enzyme. Biochem J. 1983;213:53–59. doi: 10.1042/bj2130053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 38.Serlupi-Crescenzi O, Kurtz M B, Champe S P. Developmental defects resulting from arginine auxotrophy in Aspergillus nidulans. J Gen Microbiol. 1983;129:3535–3544. doi: 10.1099/00221287-129-11-3535. [DOI] [PubMed] [Google Scholar]
- 39.Shimotohno K W, Imai S, Murakami T, Seto H. Purification and characterization of citrate synthase from Streptomyces hygroscopicus SF-1293 and comparison of its properties with those of 2-phosphinomethylmalic acid synthase. Agric Biol Chem. 1990;54:463–470. [PubMed] [Google Scholar]
- 40.Smokvina T, Mazodier P, Boccard F, Thompson C J, Guérineau M. Construction of a series of pSAM2-based integrative vectors for use in actinomycetes. Gene. 1990;94:53–59. doi: 10.1016/0378-1119(90)90467-6. [DOI] [PubMed] [Google Scholar]
- 41.Srere P A. Citrate synthase. Methods Enzymol. 1969;13:3–6. [Google Scholar]
- 42.Süsstrunk U, Pidoux J, Taubert S, Ullmann A, Thompson C J. Pleiotropic effects of cAMP on germination, antibiotic biosynthesis, and morphological development in Streptomyces coelicolor. Mol Microbiol. 1998;30:33–46. doi: 10.1046/j.1365-2958.1998.01033.x. [DOI] [PubMed] [Google Scholar]
- 43.Vohradsky J, Li X M, Thompson C J. Identification of procaryotic developmental stages by statistical analyses of two-dimensional gel patterns. Electrophoresis. 1997;18:1418–1428. doi: 10.1002/elps.1150180817. [DOI] [PubMed] [Google Scholar]
- 44.Willey J, Santamaria R, Guijarro J, Geistlich M, Losick R. Extracellular complementation of a developmental mutation implicates a small sporulation protein in aerial mycelium formation by S. coelicolor. Cell. 1991;65:641–650. doi: 10.1016/0092-8674(91)90096-h. [DOI] [PubMed] [Google Scholar]
- 45.Willey J, Schwedock J, Losick R. Multiple extracellular signals govern the production of a morphogenetic protein involved in aerial mycelium formation by Streptomyces coelicolor. Genes Dev. 1993;7:895–903. doi: 10.1101/gad.7.5.895. [DOI] [PubMed] [Google Scholar]
- 46.Wimpenny J W T. Responses of microorganisms to physical and chemical gradients. Philos Trans R Soc Lond. 1982;297:497–515. doi: 10.1098/rstb.1982.0057. [DOI] [PubMed] [Google Scholar]