Abstract
The studies of aconitase presented here, along with those of citrate synthase (P. H. Viollier, W. Minas, G. E. Dale, M. Folcher, and C. J. Thompson, J. Bacteriol. 183:3184–3192, 2001), were undertaken to investigate the role of the tricarboxylic acid (TCA) cycle in Streptomyces coelicolor development. A single aconitase activity (AcoA) was detected in protein extracts of cultures during column purification. The deduced amino acid sequence of the cloned acoA gene constituted the N-terminal sequence of semipurified AcoA and was homologous to bacterial A-type aconitases and bifunctional eukaryotic aconitases (iron regulatory proteins). The fact that an acoA disruption mutant (BZ4) did not grow on minimal glucose media in the absence of glutamate confirmed that this gene encoded the primary vegetative aconitase catalyzing flux through the TCA cycle. On glucose-based complete medium, BZ4 had defects in growth, antibiotic biosynthesis, and aerial hypha formation, partially due to medium acidification and accumulation of citrate. The inhibitory effects of acids and citrate on BZ4 were partly suppressed by buffer or by introducing a citrate synthase mutation. However, the fact that growth of an acoA citA mutant remained impaired, even on a nonacidogenic carbon source, suggested alternative functions of AcoA. Immunoblots revealed that AcoA was present primarily during substrate mycelial growth on solid medium. Transcription of acoA was limited to the early growth phase in liquid cultures from a start site mapped in vitro and in vivo.
Members of the genus Streptomyces are spore-forming gram-positive soil bacteria having a developmental program that coordinates changes in growth rate, metabolism, and morphology (reviewed in references 7 and 20). After germination, during the first growth phase (39), these filamentous organisms generate a dense multicellular network within the nutrient substrate (22, 36). Biomass accumulation then slows down transiently; resumption of growth is coordinated with formation of antibiotics and aerial hyphae (13, 39). Acidification of glucose-based minimal medium, which occurs during early growth, is followed by a cyclic AMP-dependent metabolic switch that neutralizes the medium (39) and thus allows differentiation and antibiotic biosynthesis.
Developmental mutants have been isolated that are unable to erect aerial mycelium (bald), a phenotype often accompanied by defects in antibiotic biosynthesis. Many bald genes are homologous to known regulatory elements; however, the developmental functions they control are largely unknown. Evidence that certain bald genes are parts of an organized developmental system comes from complementation tests involving physiological interactions or exchange of signals between pairwise combinations of bld mutants. When two mutants are grown adjacent to each other, one of the pair is able to restore the ability of its neighbor to erect aerial hyphae (28, 29, 44, 45). The fact that this response is partner dependent suggests a model in which complementation groups represent an ordered signaling cascade leading to aerial mycelium erection (28, 29, 44, 45).
Whereas many bald mutants are acidogenic on glucose-based media (39, 42), they are able to differentiate and remain neutral on mannitol-based media (39, 42). This may relate to their defects in catabolite repression (32) and accumulation of organic acids (39, 42). Therefore, carbon source and tricarboxylic acid (TCA) metabolism may play a central role in growth and development. In the accompanying article, we report that a mutant lacking the first enzyme of the TCA cycle, citrate synthase (CitA), is bald and does not produce antibiotics on glucose-containing media (42). citA could be placed between bldJ and bldK in the proposed signaling cascade.
The second enzyme of the TCA cycle, aconitase (EC 4.2.1.3), catalyzes the reversible isomerization of citrate to isocitrate (via cis-aconitate). In prokaryotes, aconitases have been most thoroughly studied in gram-negative bacteria, where they define two principal families (AcnA and AcnB) (14), sometimes present in the same organism. The AcnA- and AcnB-type aconitases in Escherichia coli are differentially regulated, suggesting separate physiological roles (9).
The recent discovery of novel biological roles and regulatory activities for aconitase suggested its possible alternative function in iron metabolism and mediating the bacterial oxidative stress response (1, 41). AcnA and AcnB in Escherichia coli, CitB (1), the only aconitase of Bacillus subtilis, and bifunctional eukaryotic aconitases (iron regulatory proteins) (33) contain an iron-sulfur center that senses low iron concentrations and oxidative stress. In response to these conditions, they bind to sequence motifs within mRNAs called iron regulatory elements (IRE) and thereby provide posttranscriptional control of gene expression. This activity may play a role in bacterial differentiation and virulence. In Legionella pneumophila and Xanthomonas campestris, AcnA-type aconitases maintain iron homeostasis important for pathogenicity (27, 46). The fact that an enzymatically inactive aconitase (CitB) able to bind mRNA promoted sporulation in B. subtilis indicated its alternative role in development (1).
citB transcription is activated preceding sporulation (10) in response to citrate accumulation. In addition to its activity in relieving catabolite repression of citB expression (12, 30), citrate is a strong chelator of divalent cations needed as cofactors for the Spo0A phosphorelay system that initiates spore formation. Since both citB mutations and starvation for one of these divalent cations (Mn2+) arrest initiation of development at a comparable stage, Craig and coworkers have proposed a unifying model in which citrate-mediated chelation of divalent metals impedes the activity of kinases within the Spo0A phosphorelay (8). This concept is supported by the observation that an aconitase (citB)-citrate synthase (citZ citA) double mutant could overcome the block in expressing Spo0A-dependent genes in the citB mutant (19). Recently, Schwartz et al. (37) demonstrated that Streptomyces viridochromogenes has multiple aconitase activities, at least one of which is needed for normal morphologic differentiation.
Here we describe diverse physiological roles for acoA, a Streptomyces coelicolor aconitase gene that encodes the primary vegetative aconitase. Our results show that inactivation of acoA results in growth and developmental defects related to medium acidification, citrate accumulation, and possibly an alternative function of aconitase.
MATERIALS AND METHODS
Bacterial strains and media.
E. coli XL1-Blue (Stratagene) was used as the host for cloning. When S. coelicolor was transformed, nonmethylated DNA was prepared from E. coli ET 12567 (provided by D. MacNeil and T. MacNeil) in order to circumvent a restriction barrier (24). E. coli was grown in Luria broth supplemented, as required, with antibiotics purchased from Sigma: ampicillin (100 μg/ml), kanamycin (50 μg/ml), spectinomycin (100 μg/ml), or apramycin (50 μg/ml).
S. coelicolor MT1110 is a prototrophic, plasmid-free (SCP1− SCP2−) derivative of S. coelicolor A3(2) (15, 16, 39). Construction of the citA mutant BZ2 is described in the accompanying paper (42). For S. coelicolor growth curves in liquid media, 10 ml of a seed culture previously grown for at least 48 h (stationary phase) in YEME (16) served to inoculate 500 ml of YEME in a 2-liter baffled flask. This medium was supplemented with thiostrepton (5 μg/ml), apramycin (20 μg/ml), or spectinomycin (100 μg/ml) as required. Solid complex media employed included R2YE, R5, MR5 (the same as R5 [16] except that glucose, sucrose, and proline were omitted). Minimal medium was based on MG (11) (maltose glutamate liquid medium containing 1.5% Difco agar) or SMMS (40) in which Casamino Acids were replaced by 0.5% asparagine. Test for auxotrophy were carried out on SMMS-based minimal medium (using 2% agarose [Sigma] instead of agar) with or without the addition of 50 μg of glutamate/ml. These solid media were supplemented as required with antibiotics purchased from Sigma: thiostrepton (50 μg/ml), apramycin (50 μg/ml), or spectinomycin (500 μg/ml).
Media composition analyses.
To remove liquid from agar medium, contents of a plate were placed in a 30-ml tube, frozen at −80°C for 1 h, thawed for 2 h at room temperature, and finally centrifuged at 20,000 rpm in an SS34 rotor. Ammonium concentrations were determined using the Dr. Lange LCK303 cuvette test in combination with the Dr. Lange CADAS 30 Photometer (Dr. Lange AG, Hegnau, Switzerland). The NH4+ concentration was determined after a 30-min incubation at 30°C. Other metabolites (acetate, citrate, and pyruvate) were measured using enzyme-linked assays. Analyses were preformed in an automatic enzymatic analyzer (Synchron CX5CE clinical system; Beckman Instruments, Inc., Fullerton, Calif.). All tests are based on quantitative changes in NADH content reflected by changes in absorption at 340 nm. Commercial test kits were used to determine concentration of glucose (Beckman Synchron systems glucose reagent 442640; Beckman Instruments), acetate (TC acetic acid 148261; Roche Molecular Biochemicals, Basel, Switzerland), and citric acid (TC citric acid 139076; Roche). Pyruvate was assayed using standard protocols (2).
Protein analyses.
Protein concentrations were determined by the Bradford technique (4) using a Bio-Rad kit and bovine serum albumin as a standard. Sodium dodecyl sulfate–10% polyacrylamide gel electrophoresis (SDS–10% PAGE) was performed as described by Laemmli (23). For immunoblots, crude cell lysates containing 20 μg of protein were separated by SDS-PAGE. Proteins were transferred to a polyvinylidene difluoride (PVDF) membrane (Millipore) at 400 mA in 5 liters of transfer buffer (15 mM Tris, 120 mM glycine, 10% methanol) for 90 min. The membranes were incubated in blocking buffer (TBS [20 mM Tris-HCl, pH 7.6; 130 mM NaCl] containing 5% nonfat dried milk and 0.1% Tween 20) for 2 h at room temperature. The antibody raised against E. coli AcnA (kindly provided by John Guest) was added at a 1/10,000 dilution to the blocking buffer and incubated with the membrane for 1 h. Unbound antibody was removed by four 5-min washes in TBS. The membrane was immersed into blocking buffer containing horseradish peroxidase conjugated to swine anti-rabbit immunoglobulin G (Gibco) at a dilution of 1/10,000, incubated for 1 h, and then given four washes (5 min each) in TBS. AcnA cross-reacting bands were visualized by chemiluminescence (ECL Plus; Amersham Pharmacia).
Catechol dioxygenase (XylE) activities (18) in crude extracts were measured at 37°C, calculated as the rate of change in optical density at 375 nm min−1, and expressed as specific activity (units or milliunits per milligram of protein).
Partial purification of AcoA.
Aconitase activity was assayed by measuring the conversion of isocitrate to cis-aconitate (43). All purification steps were performed at 4°C as rapidly as possible due to the lability of aconitase enzyme activity. S. coelicolor MT1110 mycelium (70 g [wet weight]) was lysed by sonication in 200 ml buffer A (20 mM Tris-HCl [pH 8.0], 0.1 mM dithiothreitol, 5% [vol/vol] glycerol) with 0.1 M NaCl supplemented with 1 mM citrate (to protect the enzyme) and complete protease inhibitors (Roche Molecular Biochemicals). Cell debris was removed by centrifugation at 15,000 rpm in a Sorvall GSA rotor. The supernatant was loaded onto a 50-ml Q-Sepharose Fast Flow column (XK26/20; Amersham Pharmacia) equilibrated in buffer A and eluted with a linear gradient from 0.1 to 1 M NaCl in buffer A. Active fractions were pooled, diluted in buffer A to give a conductivity equal to 0.1 M NaCl, and applied to an 8-ml Source Q column (XK16/20; Amersham Pharmacia) equilibrated in buffer A containing 0.1 M NaCl. The column was washed extensively, and adsorbed proteins were eluted using a linear gradient to 1 M NaCl in buffer A. Fractions that showed aconitase activity were collected and dialyzed against buffer P (50 mM NaH2PO4-Na2HPO4 [pH 7.5], 0.1 mM dithiothreitol, 5% [vol/vol] glycerol) containing 0.1 M NaCl and applied to an 8-ml Source S column (XK16/20; Amersham Pharmacia). Although this step contributed most to the purification, the enzyme activity was rapidly lost thereafter. After the aconitase activity was eluted from the cation exchanger with a linear gradient to 1 M NaCl in buffer P, the active fractions were concentrated by ultrafiltration (YM10; Amicon). The final purification step was achieved by gel filtration using a Superose 6 (Amersham Pharmacia) column equilibrated in buffer P containing 0.5 M NaCl, run at a flow rate of 0.5 ml/min. Fractions (1 ml) were assayed for aconitase activity and analyzed by SDS-PAGE.
N-terminal amino acid sequencing of AcoA.
In order to determine the N-terminal sequence of the 97-kDa protein, 10 μg was separated by SDS-PAGE, blotted onto a Millipore PVDF Immobilon membrane {using a buffer containing 10 mM CAPS [3-(cyclohexyl-amino)-1-propanesulfonic acid], pH 11, and 10% methanol}, and subjected to Edman degradation.
DNA techniques.
General DNA techniques were performed as described by Sambrook et al. (35). Minipreps and recovery of DNA fragments from agarose gels were done using Qiagen Qiaprep or Qiaquick systems.
Cloning and sequencing of acoA.
Taq DNA polymerase (5 U; Roche Molecular Biochemicals) and the degenerate primers Aco3 (5′-G[G/C][G/C][C/T]GAT[A/G]TGGTC[G/C]GT[G/C]GT-3′) and Aco6 (5′-CC[G/C]CT[G/C]GT[G/C]GT[G/C]GC[G/C]TACG-3′) were used in a PCR mixture (consisting of 100 pmol of both primers, 1 μg of template DNA, 10% dimethyl sulfoxide, and 0.2 mM deoxynucleoside triphosphates in 1× PCR buffer supplemented with 0.15 mM MgCl2) to amplify a 381-bp fragment from MT1110 genomic DNA. This fragment was gel purified, blunt ended with T4 DNA polymerase, and cloned into the EcoRV site of pBluescript SK (Stratagene) to yield pVHP80.
After sequence confirmation that the PCR fragment contained a part of the aconitase gene (acoA), pVHP80 was digested with HindIII and EcoRI (flanking the EcoRV site in pBluescript SK) to generate a 381-bp fragment that was gel purified and labeled with [α-32P]dATP using random primers (Roche Molecular Biochemicals). This probe identified a 7.2-kb band on Southern blots of BamHI-digested S. coelicolor MT1110 genomic DNA. BamHI genomic fragments were size fractionated on agarose gels to enrich for this fragment and ligated into BamHI-digested pK18. An acoA-carrying plasmid (pVHP81) was identified by colony hybridization. pVHP84 was constructed by isolating a 3.6-kb NdeI/BamHI fragment from pVHP81, treating the fragment with T4 DNA polymerase, and ligating it into EcoRV-cut pBluescript SK.
Sequencing was done on an ABI 373 sequencer (PE Applied Biosystems) using dye terminator cycle sequencing ready reactions (PE Applied Biosystems) according to the manufacturer's instructions. Standard universal and reverse primers were used to sequence the exonuclease III-deletion derivatives of pVHP84. Gaps were closed using acoA-specific primers. Sequences were assembled and analyzed with the aid of the Lasergene package (DNA Star).
Operon fusion constructs with the acoA upstream region.
The PacoA region (a 277-bp XmnI/NsiI fragment containing 269 bp upstream of acoA) was subcloned in order to make operon fusions to the promoterless xylE gene on the SCP2-derived shuttle vector pIJ2839 (constructed from pXE3 [18] by T. Clayton). This fragment was isolated from pVHP84, blunt ended with T4 DNA polymerase, and ligated into EcoRV-digested pBluescript SK to give pVHP85. The NsiI/XmnI fragment of pVHP85 containing PacoA was excised by HindIII (within the polylinker)/BamHI and cloned into similarly restricted pIJ2839, yielding pVHP171.
Plasmids used to disrupt S. coelicolor acoA.
pVHP184 was made by amplifying a 1.1-kb fragment from pVHP84 using primers Aco12 (5′-AAGGAATTCGGCGGCGACCCGAACAAGATCAA-3′) and Aco14 (5′-CTTCGAATTCAGGCCGTTCTCGACCGCCTTCTT-3′) and Taq DNA polymerase (5 U; Roche Molecular Biochemicals), digesting it with EcoRI (site is underlined), and cloning it into pKC1132 (containing the apramycin resistance gene). pVHP185 was constructed in a similar manner using Aco13 (5′-CCCGAATTCCCGGTCGACGACGAGACGCTGAA-3′) and Aco15 (5′-GAAGAATTCGCTGTAGGACTTGGAGAACA-3′) as primers to amplify a 1-kb acoA internal fragment from pVHP184. The PCR fragment amplified by Aco13 and Aco15 primers was also cloned into pSET151 (containing the thiostrepton resistance gene) to generate pVHP186.
Disruption of acoA and complementation.
MT1110 protoplasts were transformed with pVHP184 or pVHP185 extracted from the nonmethylating E. coli strain ET12567. After 18 h incubation at 30°C, regeneration plates were flooded with 1 ml of a solution containing 1 mg of apramycin/ml. Two apramycin-resistant transformants having the correct insertion of the plasmid gave a PCR product of about 2 kb (BZ3) or 3 kb (BZ4). BZ5, an acoA citA double mutant, was derived from BZ2 (citA) (see the accompanying paper [42]) by insertional mutagenesis using pVHP186. The constructions were also confirmed by PCR and Southern blotting.
The integrative plasmid pPM925 (38) was used as a vector to complement the acoA lesion. pPM925 mediates insertion of cloned DNA into Streptomyces chromosomes at its pSAM2 attachment site. The 3.8-kb insert of pVHP84 was released by digestion with KpnI and BamHI and ligated into pPM925, yielding pPM925-acoA. pPM925-acoA or pPM925 was introduced into BZ4 by conjugation (26), selecting for thiostrepton- and apramycin-resistant exconjugants.
Mapping transcriptional initiation sites.
In vitro transcription experiments were performed as described previously (6), and products were analyzed on 6% polyacrylamide–7 M urea gels.
RNA extracted from S. coelicolor MT1110 using hot phenol (25) was treated with RNase-free DNase I (Roche Molecular Biochemicals). One picomole of oligonucleotide that had been kinase labeled at the 5′ end was used to prime cDNA synthesis from 10 μg of total RNA isolated from cultures grown in YEME to early exponential phase (10 h). The extension reaction was carried out for 1 h at 42°C using Superscript II reverse transcriptase (Gibco BRL).
Purification of S. coelicolor RNA polymerase.
Total RNA polymerase (RNAP) was isolated from S. coelicolor MT1110 as described previously (5), with one modification: after the polymin P extractions and subsequent ammonium sulfate precipitation, heparin Sepharose chromatography was used instead of DNA cellulose chromatography. The sample was loaded onto a 25-ml heparin Sepharose Cl-6B (Amersham Pharmacia) column equilibrated in TGED buffer (0.1 M Tris-HCl [pH 7.9], 5% [vol/vol] glycerol, 0.1 mM EDTA, 0.1 mM dithiothreitol) containing 0.15 M NaCl. The column was washed extensively with TGED containing 0.2 M NaCl, and RNAP was step eluted with TGED containing 1 M NaCl. The eluate was dialyzed in TGED to a final salt concentration of 0.1 M NaCl and then applied to an 8-ml Source 15Q column (Amersham Pharmacia) equilibrated with the same buffer. After a washing step with TGED containing 0.2 M NaCl, RNAP was eluted with an 80-ml linear gradient of TGED buffer containing 0.7 M NaCl. One-milliliter fractions were collected and analyzed by SDS-PAGE. Fractions containing RNAP were identified by their α and ββ′ subunits stained with Coomassie brilliant blue. Fifty-microliter samples of each fraction containing RNAP were combined to yield the RNAP mixed holoenzymes that were used in the subsequent experiments.
Nucleotide sequence accession number.
The fragment of S. coelicolor genomic DNA containing acoA has been deposited in GenBank under accession number AF180948.
RESULTS
Isolation of AcoA, the primary S. coelicolor aconitase.
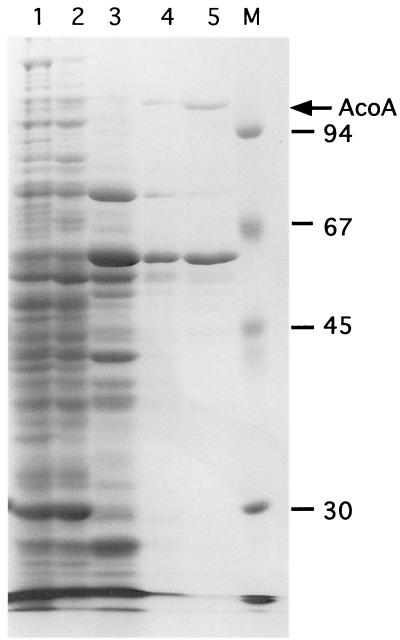
The major aconitase activity detected in S. coelicolor protein extracts (AcoA) was partially purified using an ordered series of ion-exchange (Q-Sepharose, Source Q, and Source S) and gel filtration (Superose 6) chromatography steps (see Materials and Methods). The most active fraction from the Superose 6 column contained only two major proteins (100 and 62 kDa) detected by SDS-PAGE (Fig. 1). All known prokaryotic aconitases are large monomeric enzymes with predicted molecular masses of approximately 100 kDa. The N-terminal sequence of the 100-kDa protein obtained by Edman degradation was NH2-X-A-N-X-F-D-A-R-X-T (X represents an unassigned amino acid).
FIG. 1.
Partial purification of AcoA from S. coelicolor crude extracts. S. coelicolor crude extract (lane 1) was fractionated by Q-Sepharose anion-exchange chromatography (lane 2), Source Q anion-exchange chromatography (lane 3), Source S cation-exchange chromatography (lane 4), and Superose 6 gel filtration chromatography (lane 5). Representative samples from each chromatographic step were resolved by SDS-PAGE and stained with Coomassie brilliant blue. The molecular masses of the protein marker present in lane M are indicated on the right (in kilodaltons). The arrow on the right designates the position of AcoA.
Only one aconitase activity peak was detected throughout purification, suggesting that it served as the primary enzyme in vivo. However, aconitases are characteristically unstable (21) (their [4Fe-4S] cluster is readily oxidized) and major losses of activity were observed during purification. Therefore, in vitro activity may not be a reliable measure of relative in vivo function, and these data did not rule out the possibility that another undetected aconitase played the primary metabolic role. Genetic analyses were used to address this question.
Isolation of the gene encoding AcoA.
To isolate the acoA gene, degenerate primers based on amino acids from two conserved regions of the AcnA family of aconitases (P-L-V-V-A-Y and T-D-H-I-S-P-A) allowed amplification of a 381-bp DNA fragment. This PCR fragment was used to probe Southern blots of BamHI-digested S. coelicolor genomic DNA. The hybridizing 7.2-kb fragment was cloned and a 3.2-kb region containing acoA was sequenced (all subsequent references to nucleotide positions are with respect to this sequence). The sequence (nucleotides 340 to 3053) encoded a protein (904 amino acids) highly homologous to aconitases of S. viridochromogenes (PID g6456849; 91% identity), Mycobacterium tuberculosis (PID g3261503; 65% identity) and Mycobacterium avium (PID g3121732; 65% identity), Corynebacterium glutamicum (PID g4587326; 65% identity), and X. campestris (PID g2661438; 55% identity). The protein's predicted molecular mass of 97 kDa and pI of 4.7 were in good agreement with the mass measured on SDS-PAGE gels (100 kDa), and its N-terminal sequence was indistinguishable from that of purified AcoA. The corresponding open reading frame (ORF) initiated at a GTG translational initiation site that was preceded by a purine-rich sequence (GAAGGAGA) that could serve as a ribosome-binding site. These data indicated that the ORF encoded AcoA.
Construction and morphology of S. coelicolor acoA mutants.
acoA mutants were constructed to investigate the possible role of aconitase in primary metabolism and growth, colonial development, or secondary metabolism. Repeated attempts to construct a stable mutant by a double-crossover event designed to replace acoA with an allele containing a resistance gene were unsuccessful. Disruption of the gene was achieved by insertion of plasmids containing internal fragments of acoA, via a single-crossover event. Two different ca. 1-kb fragments within the acoA ORF were amplified by PCR and cloned into plasmid pKC1132 (3) (generating pVHP184 and pVHP185; see Materials and Methods). Since pKC1132 is unable to replicate in Streptomyces spp., stable maintenance is most likely to occur by a single homologous recombination event between the cloned DNA and the acoA chromosomal locus. Apramycin-resistant colonies (BZ3 and BZ4, transformed with pVHP184 and pVHP185, respectively) were isolated. PCR and Southern hybridizations confirmed that integration events in BZ3 and BZ4 had disrupted the acoA gene (described in Materials and Methods). Immunoblots showed that the AcoA band was missing in BZ4 (see below).
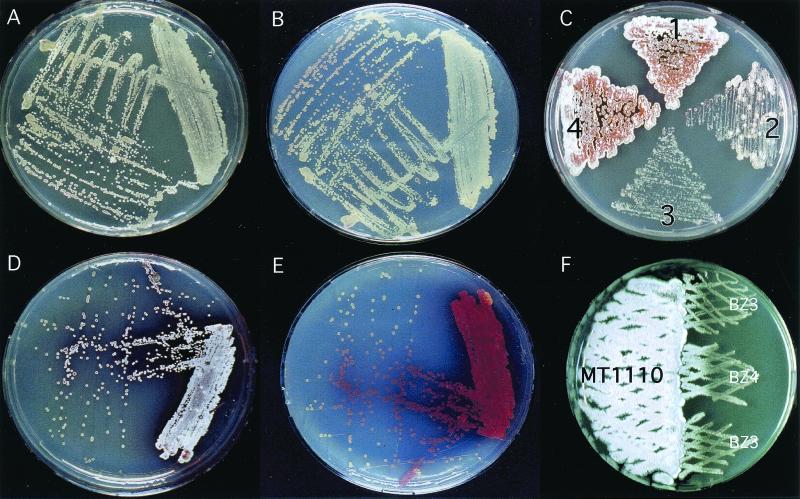
BZ3 and BZ4 had similar phenotypic defects on R2YE. Both formed small colonies and produced little pigment or aerial hyphae even after prolonged incubation (BZ4 is shown in Fig. 2A and B). Introduction of the integrative plasmid containing the acoA locus (pPM925-AcoA) restored BZ4 (Fig. 2C, sector 2) to its wild-type parental colonial phenotype (Fig. 2C, sector 4; BZ3 was not tested). This proved that the phenotypes observed were due to acoA disruption and not to polar effects on downstream genes.
FIG. 2.
Colonial morphology of the acoA mutant. The developmental processes of the acoA mutant, BZ4, was conditionally blocked on R2YE plates. BZ4 grew slowly and produced few white aerial hyphae (shown best in panel A, the top view) or pigmented antibiotics (shown best in panel B, the bottom view). When the medium was supplemented with 100 mM TES buffer, BZ4 colonies grew more rapidly but were still growth impaired compared to the wild type and were able to produce characteristic aerial mycelium (panel D, top view) and pigments (panel E, bottom view). Plate C illustrates complementation of the mutated acoA allele in BZ4: sector 1, MT1110; sector 2, BZ4; sector 3, BZ4/pPM925; sector 4, BZ4/pPM925-acoA. Note the appearance of several BZ4 revertants on plate C, sector 2. Developmental defects were also suppressed by growing either acoA mutant (BZ3 or BZ4) next to MT1110 (F). Since all plates had to be supplemented with 100 μg of apramycin/ml to maintain the mutant acoA allele, MT1110 was transformed with a plasmid conferring resistance (pKC1218).
The phenotypes of the acoA mutants were highly unstable. To maintain the mutant allele, they were grown on media containing apramycin. Without this selection, apramycin-sensitive strains appeared at high frequency, a phenomenon visualized as colonial sectors presumably reflecting a reversal of the integration event. Suppressor strains also appeared in the presence of apramycin, albeit at lower frequency (Fig. 2C, sector 2). Since these strains having wild-type morphology still had the plasmid inserted at the acoA locus (confirmed by PCR), they probably resulted from suppressive mutations that occurred elsewhere in the chromosome. Thus, the acoA mutants BZ3 and BZ4 could be reliably maintained only by frequent purification of small, bald, apramycin-resistant colonies on R2YE agar.
Effect of the acoA mutation on flux through the TCA cycle.
Metabolic flux through the TCA cycle not only serves as the optimal aerobic pathway for the generation of ATP and reducing compounds but also supplies ketoglutarate for the biosynthesis of glutamate. Therefore, the immediate physiological consequences of the lack of aconitase could include the arrest of the TCA cycle leading to accumulation of the AcoA substrate (citrate) and glutamate auxotrophy.
BZ4 could not grow on minimal medium in the absence of glutamate. This was the first proof that metabolic flow through the TCA cycle was severely disrupted and that the protein encoded by acoA was the primary vegetative aconitase.
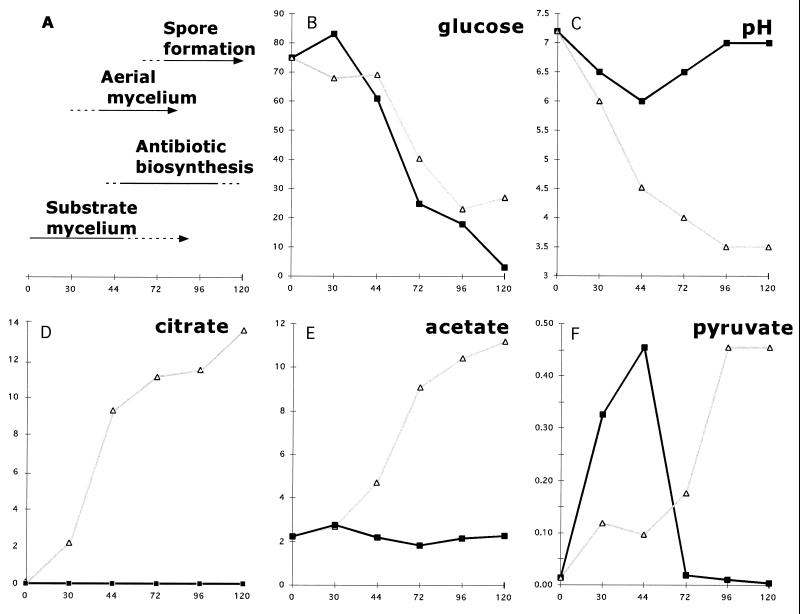
This was reconfirmed by analyses of the acids accumulated by the acoA mutant (BZ4) growing on glucose. MT1110 and BZ4 utilized glucose at about the same rate (Fig. 3B). While the wild type did not produce detectable amounts of citrate, BZ4 accumulated it continuously; levels had reached 14 mM and were increasing at 120 h (Fig. 3D). This provided direct physiological evidence that acoA encoded the aconitase with primary importance in oxidizing glucose via the TCA cycle. The accumulation of acetic acid and pyruvate in the medium (Fig. 3E and F) provided further evidence that TCA-based oxidative metabolism was blocked in the acoA mutant. Acetic acid is commonly produced from pyruvate by bacteria growing on glucose under oxygen-limited conditions when TCA cycle activity is minimal. During the course of development, wild-type MT1110 did not accumulate acetate (Fig. 3E).
FIG. 3.
The developmental block of BZ4 was accompanied by alterations in metabolism. MT1110/pKC1218 (closed squares) and BZ4 (open triangles) were grown on cellophane disks covering R2YE plates (supplemented with 100 μg of apramycin/ml to retain the mutant allele). MT1110/pKC1218 underwent typical developmental changes (A) (time in hours is indicated on the x axis), while BZ4 produced neither aerial mycelium nor pigmented antibiotics. An extract of the agar medium was prepared in order to measure pH and metabolite concentrations (glucose, citrate, acetate, and pyruvate were quantified [in millimolar units] as described in Materials and Methods).
Dissecting the underlying causes of the growth defect of an acoA mutant.
The above results suggested that the acoA mutant defects might relate to pH, citrate accumulation, or alternative functions of aconitase.
In wild-type MT1110 cultures growing on cellophane disks overlying R2YE (which contains 25 mM TES buffer), the medium pH gradually decreased during substrate mycelial growth from its initial pH of 7.2 to 6.0 at 44 h and then increased during later stages of development (Fig. 3C). White aerial hyphae appeared around 30 h, red pigment appeared at 44 h, and gray spores appeared at 72 h (Fig. 3A). This biphasic pH change associated with aerial hyphae and pigment formation on R2YE was observed only when the strain was growing on cellophane disks (i.e., it was not observed in the accompanying study, where cultures were grown directly on the agar surface [42]). The phenomenon was similar to that observed on a glucose minimal medium (39).
In contrast, BZ4 lawns irreversibly acidified the medium to pH 3.5 (Fig. 3C). Medium acidification to pH values of <5 inhibits S. coelicolor growth and differentiation (39). Only very high concentrations of buffer (100 mM TES, designated R2YE-HITES) allowed maintenance of neutral pH. Even under these conditions, the acoA mutant was growth retarded; however, after prolonged cultivation (10 to 14 days), colonial development was clearly accelerated on R2YE-HITES, especially within densely seeded areas of the plate (Fig. 2D and E). Single, well-isolated colonies remained bald, indicating that plating density was a factor contributing to the conditional developmental defects.
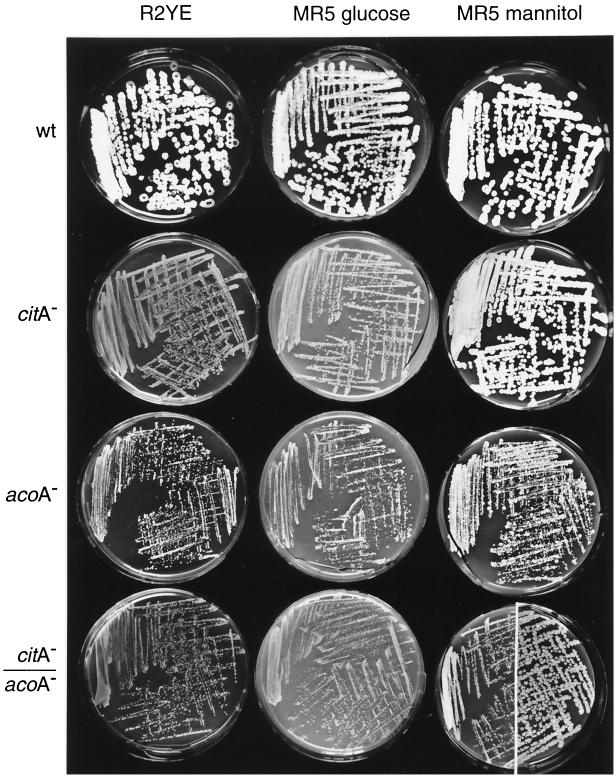
In order to determine whether the acoA mutant phenotypes were specifically related to aconitase rather than reflecting a defect due to impaired TCA metabolism or citrate accumulation, an acoA mutant (BZ4) was compared to a citrate synthase mutant (BZ2) (Fig. 4). The mutant colonies displayed similar retardation in growth and differentiation on media containing glucose. In addition, on nonacidogenic plates containing mannitol instead of glucose, the citA mutant grew and differentiated normally; the acoA mutant was defective in growth but able to differentiate after prolonged incubation. This indicated that, in addition to acidogenesis, the acoA mutant had other growth defects.
FIG. 4.
Colonial phenotypes of TCA cycle mutants. MT1110 (wild type [wt]), BZ2 (citA), BZ4 (acoA) and BZ5 (citA acoA) were streaked out on R2YE or MR5 containing 2% glucose (MR5 glucose) or mannitol (MR5 mannitol). Plates were incubated for 5 days at 30°C and photographed. Colonies had not undergone major changes in morphology upon reexamination 5 days later, with one significant exception. BZ5 colonies had continued to grow and produced gray spores between 5 and 10 days of incubation on MR5 mannitol (photographed and presented as a composite of the right side of the plate).
Pleiotropic effects of aconitase mutants have been attributed partially to chelation of divalent cations by citrate that may limit the activity of enzymes and regulatory components dependent on divalent cations (19, 41). Since these effects can be prevented by inactivation of the citrate synthase genes (19, 41), a citA acoA double mutant was constructed. citA, acoA, and citA acoA mutants were all growth inhibited on the acidogenic glucose-based medium (Fig. 4). Although the citA mutants grew like the wild type on mannitol, the acoA mutant remained severely growth inhibited. However, the acoA defect was partially suppressed in the citA acoA mutant; after long incubation, the double mutant grew into larger colonies than the acoA single mutant. Citrate was thus implicated as a second of at least three factors contributing to the growth defect of the acoA mutant.
In principle, chelator activity of citrate might be suppressed by supplementing the medium with appropriate concentrations of specific cations. However, the addition of various concentrations (1 to 100 mM) of CuSO4, MnSO4, or FeSO4 did not visibly restore growth or development of BZ4 colonies on R2YE medium. Similarly, application of these heavy metals in solution (1 to 100 mM) to the surface of R2YE seeded with BZ4 did not accelerate growth or development (data not shown). These negative results were difficult to interpret. Citrate has a variable affinity for a variety of essential divalent cations having diverse activities as cofactors. In addition, the potential toxicity of heavy metals at superoptimal concentrations may complicate efforts to suppress the BZ4 defects by supplementing the medium.
Finally, the fact that the double mutant remained growth impaired relative to both the wild type and citA strains on all media tested suggested that AcoA might have an activity independent of its role as an enzyme in the TCA cycle.
acoA expression during growth.
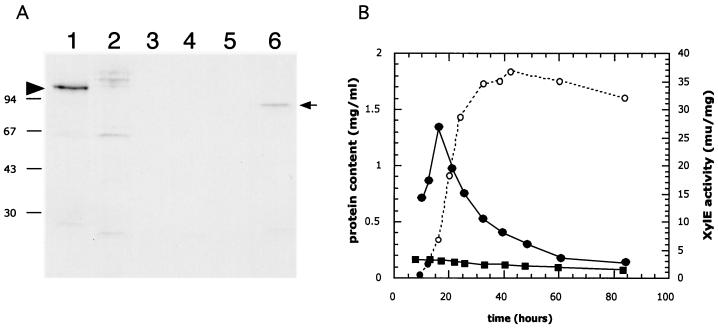
Developmentally regulated expression of the acoA gene was shown by immunoblot experiments (Fig. 5A) using a polyclonal antibody raised against E. coli AcnA to probe protein extracts of MT1110 cultures differentiating on solid R2YE covered by a cellophane membrane. A strongly cross-reacting species migrating with an apparent molecular mass of 97 kDa was detected before white aerial hyphae appeared (Fig. 5A, lane 1). This protein was no longer detected after aerial hypha emergence (Fig. 5A, lanes 2 through 5). While a 97-kDa cross-reactive band was not detected at any time during growth of BZ4, acoA mutant extracts (30 h) did contain a cross-reacting band (Fig. 5A, lane 6) that may be a truncation or degradation product of AcoA.
FIG. 5.
Developmental control of acoA. (A) Accumulation of AcoA in cultures grown on solid medium. AcoA was detected in extracts of surface-grown cultures by immunoblotting using antibodies against E. coli AcnA. MT1110/pKC1218 (lanes 1 through 5) and BZ4 (lane 6) were grown on R2YE (containing 100 μg of apramycin/ml) plates that had been covered with cellophane disks. Crude extracts prepared from samples harvested after 30 h (lane 1 and 6), 44 h (lane 2), 72 h (lane 3), 96 h (lane 4) and 120 h (lane 5) were resolved by SDS–10% PAGE and transferred to a PVDF nylon membrane. Immunodetection was performed as described in Materials and Methods. The left arrowhead indicates the position of the wild-type AcoA (MT1110/pKC1218), while the arrow on the right points to the presumed truncated or degraded AcoA detected in extracts of BZ4. The molecular masses of the protein marker are indicated on the left (in kilodaltons). (B) Activity of the acoA promoter in liquid cultures. S. coelicolor MT1110 strains carrying pIJ2839 (a XylE-based promoter probe vector) or a derivative (pVHP171) containing a 277-bp NsiI/XmnI fragment (Fig. 6A) extending from within the acoA ORF to 269 bp upstream were grown in YEME liquid medium. Crude extracts prepared from cultures throughout growth (open circles, protein content) were assayed for XylE specific activity (closed squares, pIJ2839; closed circles, pVHP171).
Mapping the acoA promoter in vivo and in vitro.
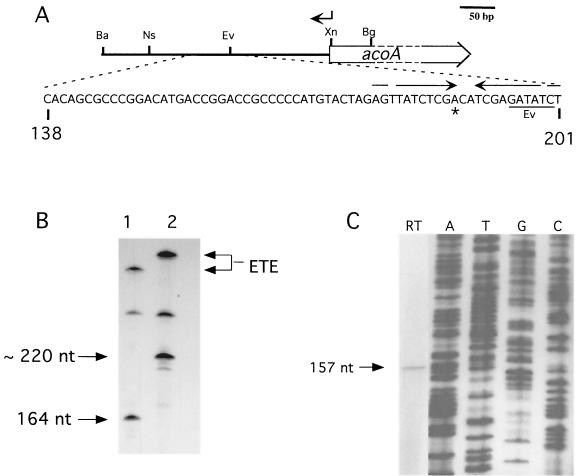
In order to locate the acoA promoter (PacoA), a 277-bp NsiI/XmnI fragment including 269 bp upstream of the acoA translational initiation site was cloned into the promoter probe vector pIJ2839 (pVHP171) (Fig. 6A). This vector allows estimation of promoter activity using a promoterless xylE reporter gene, whose enzyme product can be detected visually or measured enzymatically as a yellow compound (2-hydroxymuconic semialdehyde) (18).
FIG. 6.
Mapping the acoA transcriptional start site. (A) The acoA transcriptional start site locus. Nucleotide coordinates refer to the acoA locus sequence. The location of the oligonucleotide used in primer extension is shown by an arrow above the map. Restriction sites: Ba, BamHI; Bg, BglII; Ev, EcoRV; Ns, NsiI; Xn, XmnI. The transcriptional start site (asterisk) mapped in vitro (B) and in vivo (C) was located in a region containing an inverted repeat (opposing arrows). (B) In vitro transcription using two different acoA promoter fragments and S. coelicolor RNAP. Runoff transcription was carried out using either the 348-bp BamHI/XmnI fragment (lane 1) or the 398-bp BamHI/BglII fragment (lane 2). The lengths of the two template fragments differed at the acoA proximal end, thus fixing the orientation of the runoff transcript. A larger, undefined runoff signal was also obtained from a promoter oriented oppositely to PacoA. The sizes of the runoff transcripts (arrows on the left) were estimated by separating the samples on a 6% polyacrylamide–6 M urea gel alongside a 32P-labeled M13mp18 sequencing ladder generated with a standard universal primer (5′-GCCAGGGTTTTCCCAGTCACGACG-3′). The signals generated by “end-to-end” (ETE) transcription are indicated on the right. (C) Reverse transcription assay (lane RT) using RNA isolated from exponential-phase S. coelicolor grown in YEME. An oligonucleotide overlapping with the acoA ORF (coordinates 321 to 344; indicated above by arrow; RT-Aco1 [5′-GACACGACAGTCTCCTTCATTTAT-3′]) was used as the primer. The size of the cDNA transcript (arrow) was estimated by running the sample alongside a 32P-labeled M13mp18 sequencing ladder generated with a standard universal sequencing primer (5′-TGTAAAACGACGGCCAGT-3′; lanes A, T, G, and C).
When MT1110/pVHP171 was grown in YEME liquid cultures, XylE (promoter) specific activity increased during early exponential phase (Fig. 5B) and then steadily declined throughout later exponential and stationary phases (XylE may be a rather unstable enzyme [34]). By late stationary phase, the XylE activity reached a minimum value that was >10-fold lower (3 mU/mg) than the peak of activity during early exponential phase (27 mU/mg) (34). These experiments indicated that this fragment encoded a promoter activity (PacoA).
In order to map the promoter more precisely, primer extension was performed on total RNA isolated from early-exponential-phase cells (12 h) cultivated in YEME. The size of the extended cDNA (157 nucleotides) (Fig. 6C) suggested a transcriptional initiation site located 152 bp upstream of acoA (Fig. 6A).
In vitro transcription experiments (Fig. 6B) using a PacoA template confirmed this transcriptional start site. RNAP isolated from S. coelicolor MT1110 grown in YEME to late log phase synthesized an acoA-specific mRNA whose size mapped the PacoA transcriptional start site to the nucleotide found in primer extension experiments (Fig. 6C).
Extracellular suppression of the acoA defect by differentiating wild-type colonies.
When bald BZ4 lawns (high density) or colonies (low density) were grown close to the differentiating wild-type strain (Fig. 2F), they were able to slowly differentiate and produce small amounts of pigment after about a week. The appearance of large suppressor mutant colonies (Fig. 2C, sector 2) hindered the longer-term incubation required for “intercolonial complementation” studies of BZ4 with other bald mutants.
DISCUSSION
The TCA cycle is needed to support both growth and developmental in B. subtilis and S. coelicolor (8, 19, 42). Sonenshein and his collaborators have proposed that TCA cycle enzymes provide key developmental checkpoints in B. subtilis, with aconitase playing an important role. Our studies, as well as those of Schwartz et al. (37), showed that acoA mutants have pleiotropic phenotypes, including a defect in development.
Biochemical and genetic studies showed that AcoA was the primary aconitase supporting growth of S. coelicolor. Only one activity was observed during column purification of aconitase from liquid-grown mycelial extracts. Expression of acoA and accumulation of its product were largely limited to early vegetative growth. Inactivation of the acoA gene in BZ4 severely disrupted the TCA cycle activity, clearly indicated by the glutamate auxotrophy of BZ4. This was unlike the S. viridochromogenes acoA mutant, which was a prototroph. While the S. coelicolor mutant remained bald in the presence of glutamate, the S. viridochromogenes acoA mutant was able to produce malformed aerial mycelium (37). These differences probably reflected a second aconitase-like activity able to maintain flux through the TCA cycle in S. viridochromogenes. The S. coelicolor mutant, devoid of an alternative activity able to support vegetative growth, suffered more severe physiological and developmental defects.
The organic acids (citrate, pyruvate, and acetate) that accumulated to high levels on R2YE media conditioned by the S. coelicolor acoA mutant (Fig. 3) inhibited growth via at least two mechanisms. The most obvious inhibition was due to the decrease in pH to levels known to inhibit growth of S. coelicolor. As was the case with the citA mutant, this effect was partially suppressed by high concentrations of buffer on glucose-based media (Fig. 2). However, the fact that the acoA mutant remained growth retarded on these buffered plates as well as on mannitol-based media, in which the pH remained near neutrality, indicated other defects.
In the absence of AcoA, accumulation of its substrate, citrate, was partially responsible for the growth defect of BZ4. The acoA growth defects were partially relieved by the citA mutation on mannitol-based media, presumably by blocking citrate accumulation (Fig. 4). A mutation in the primary B. subtilis aconitase gene similarly resulted in high levels of citrate accumulation and was associated with developmental defects (8). The fact that these defects could be partly suppressed using a quantitative assay for sporulation suggested a model in which chelation of certain divalent cations by citrate was at least partially involved in the developmental phenotype.
A very interesting potential role for AcoA that we have not yet attempted to dissect is in regulating the oxidative stress response. This question has been addressed genetically in other organisms by preventing binding of aconitase to its IRE-like sequences. The ability of Acn to bind RNA and thus mediate a response to oxidative stress can be preferentially inactivated by a mutagenic change in B. subtilis (1). Alternatively, its IRE-like binding sites in upstream untranslated mRNA can be specifically inactivated. Such sites might be identified by searching the S. coelicolor genome (http://www.sanger.ac.uk/Projects/S coelicolor) for its sequence motif (1). As in E. coli, the acoA gene itself may contain an IRE-like sequence in its upstream untranslated region (41) which our data localize to a 152-bp sequence between the transcriptional and translational start sites.
Due to the diverse roles played by aconitase in bacteria, it is difficult to determine whether the defects of the aconitase null mutant reflect its direct role in Streptomyces development. Similar questions have been raised in studies of the role of TCA cycle enzymes in the B. subtilis sporulation program. Other criteria should be considered to help clarify this question.
Developmental changes correlated well with temporal patterns of acoA gene expression. On solid glucose-based R2YE medium, immunoblots demonstrated that AcoA accumulated in early substrate mycelium and then rapidly disappeared later during development as aerial mycelium was formed. Analyses of the underlying physiological or developmental effectors of these regulatory patterns will focus on its promoter region, mapped both in vivo and in vitro to a site 152 bp upstream of the acoA ORF. Future studies should clarify why the AcoA protein levels decreased in the colony during aerial mycelium formation while pyruvate was being consumed (Fig. 3). Catabolism of organic acids may involve TCA oxidation supported by low levels of AcoA or perhaps employ an as-yet-undetected aconitase activity expressed only during aerial mycelial growth (note: S. viridochromogenes has at least two aconitase activities [37]). Alternatively, they may be converted to less acidic products, such as lipids or polyketides (42).
Responsiveness to diffusible developmental signals provides the most readily applicable criterion for evaluating whether bald alleles such as acoA or citA represent an integral part of a programmed differentiation cascade. In Streptomyces, differentiation is coordinated by diffusion of well-characterized butyrolactone pheromones (17) and perhaps another positively acting developmental signaling molecule(s) (28, 29, 31, 39, 44, 45). Cultivation of the acoA mutants in proximity to the S. coelicolor wild type allowed them to differentiate but did not restore normal growth of the colony (Fig. 2F). Therefore, the positive effect of the wild type on differentiation of the acoA mutant could reflect such a signal. Alternatively, partial elimination of organic acids or other negatively acting factors by the wild type might allow BZ4 to form aerial mycelium. Studies of complementation between BZ4 and other acidogenic developmental mutants could in principle provide information to clarify this question; however, frequently arising suppressor strains (as have been observed in E. coli [41]) have so far hindered such analyses. Identification of these mutant alleles may provide further insights into the role of acoA in development.
Data presented here show that acoA is developmentally controlled to support TCA cycle activity during growth of substrate mycelium preceding differentiation. Like citrate synthase mutants (42), acoA mutants are TCA cycle arrested, resulting in acidogenesis, glutamate auxotrophy, and disruption of colonial development. Only some of the additional pleiotropic defects of the aconitase mutant could be attributed to citrate accumulation. It is therefore likely that AcoA plays auxiliary roles in growth, perhaps related to iron metabolism or oxidative stress adaptation as reported for E. coli (41) and B. subtilis (1). These studies, together with our accompanying paper (42), demonstrate the role of TCA cycle enzymes in maintaining metabolic balance, supporting not only growth but also morphologic development and secondary metabolism.
ACKNOWLEDGMENTS
We thank John Guest for aconitase antibody, X.-M. Li, J. Vohradsky, and J. Novotna for 2D gel analyses, P. Jenö for N-terminal sequencing, and W. Wohlleben, D. Schwartz, and A. L. Sonenshein for discussions.
This work was supported by Swiss National Science Foundation Grants to C. J. Thompson (3100-039669 and SPP Biotechnology 5002-046085) and W. Minas (SPP Biotechnology 5002-46086).
REFERENCES
- 1.Alen C, Sonenshein A L. Bacillus subtilis aconitase is an RNA-binding protein. Proc Natl Acad Sci USA. 1999;96:10412–10417. doi: 10.1073/pnas.96.18.10412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bergmeyer J, Grassl M. Metabolites. 1. Carbohydrates. Weinheim, Germany: Verlag Chemie; 1984. [Google Scholar]
- 3.Bierman M, Logan R, O'Brien K, Seno E T, Rao R N, Schoner B E. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene. 1992;116:43–49. doi: 10.1016/0378-1119(92)90627-2. [DOI] [PubMed] [Google Scholar]
- 4.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 5.Buttner J M, Brown N L. RNA polymerase-DNA interactions in Streptomyces: in vitro studies of a S. lividans plasmid promoter with S. coelicolor RNA polymerase. J Mol Biol. 1985;185:177–188. doi: 10.1016/0022-2836(85)90189-5. [DOI] [PubMed] [Google Scholar]
- 6.Buttner J M, Brown N L. Two promoters from Streptomyces plasmid pIJ101 and their expression in Escherichia coli. Gene. 1987;51:179–186. doi: 10.1016/0378-1119(87)90306-4. [DOI] [PubMed] [Google Scholar]
- 7.Chater K F. Taking a genetic scalpel to the Streptomyces colony. Microbiology. 1998;144:1465–1478. doi: 10.1099/00221287-144-6-1465. [DOI] [PubMed] [Google Scholar]
- 8.Craig J E, Ford M J, Blaydon D C, Sonenshein A L. A null mutation in the Bacillus subtilis aconitase gene causes a block in Spo0A-phosphate-dependent gene expression. J Bacteriol. 1997;179:7351–7359. doi: 10.1128/jb.179.23.7351-7359.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cunningham L, Gruer M J, Guest J R. Transcriptional regulation of the aconitase genes (acnA and acnB) of Escherichia coli. Microbiology. 1997;143:3795–3805. doi: 10.1099/00221287-143-12-3795. [DOI] [PubMed] [Google Scholar]
- 10.Dingman D W, Rosenkrantz M S, Sonenshein A L. Relationship between aconitase gene expression and sporulation in Bacillus subtilis. J Bacteriol. 1987;169:3068–3075. doi: 10.1128/jb.169.7.3068-3075.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doull J L, Vining L C. Culture conditions promoting dispersed growth and biphasic production of actinorhodin in shaken cultures of Streptomyces coelicolor A3(2) FEMS Microbiol Lett. 1989;65:265–268. doi: 10.1016/0378-1097(89)90228-0. [DOI] [PubMed] [Google Scholar]
- 12.Fouet A, Jin S F, Raffel G, Sonenshein A L. Multiple regulatory sites in the Bacillus subtilis citB promoter region. J Bacteriol. 1990;172:5408–5415. doi: 10.1128/jb.172.9.5408-5415.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Granozzi C, Billetta R, Passantino R, Sollazzo M, Puglia A M. A breakdown in macromolecular synthesis preceding differentiation in Streptomyces coelicolor A3(2) J Gen Microbiol. 1990;136:713–716. doi: 10.1099/00221287-136-4-713. [DOI] [PubMed] [Google Scholar]
- 14.Gruer J M, Guest J R. Two genetically-distinct and differentially-regulated aconitases (AcnA and AcnB) in Escherichia coli. Microbiology. 1994;140:2531–2541. doi: 10.1099/00221287-140-10-2531. [DOI] [PubMed] [Google Scholar]
- 15.Hindle Z, Smith C P. Substrate induction and catabolite repression of the Streptomyces coelicolor glycerol operon are mediated through the GylR protein. Mol Microbiol. 1994;12:737–745. doi: 10.1111/j.1365-2958.1994.tb01061.x. [DOI] [PubMed] [Google Scholar]
- 16.Hopwood D A, Bibb M J, Chater K F, Kieser T, Bruton C J, Kieser H M, Lydiate D J, Smith C P, Ward J M, Schrempf H. Genetic manipulation of Streptomyces. A laboratory manual. Norwich, United Kingdom: The John Innes Foundation; 1985. [Google Scholar]
- 17.Horinouchi S, Beppu T. A-factor as a microbial hormone that controls cellular differentiation and secondary metabolism in Streptomyces griseus. Mol Microbiol. 1994;12:859–864. doi: 10.1111/j.1365-2958.1994.tb01073.x. [DOI] [PubMed] [Google Scholar]
- 18.Ingram M M, Brawner M, Youngman P, Westpheling J. xylE functions as an efficient reporter gene in Streptomyces spp.: use for the study of galP1, a catabolite-controlled promoter. J Bacteriol. 1989;171:6617–6624. doi: 10.1128/jb.171.12.6617-6624.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ireton K, Jin S, Grossman A D, Sonenshein A L. Krebs cycle function is required for activation of the Spo0A transcription factor in Bacillus subtilis. Proc Natl Acad Sci USA. 1995;92:2845–2849. doi: 10.1073/pnas.92.7.2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kelemen G H, Buttner M J. Initiation of aerial mycelium formation in Streptomyces. Curr Opin Microbiol. 1998;1:656–662. doi: 10.1016/s1369-5274(98)80111-2. [DOI] [PubMed] [Google Scholar]
- 21.Kennedy C M, Emptage M H, Dreyer J L, Beinert H. The role of iron in the activation-inactivation of aconitase. J Biol Chem. 1983;258:11098–11105. [PubMed] [Google Scholar]
- 22.Kretschmer S. Dependence of the mycelial growth pattern on the individually regulated cell cycle in Streptomyces granaticolor. Z Allg Mikrobiol. 1982;22:335–347. doi: 10.1002/jobm.3630220507. [DOI] [PubMed] [Google Scholar]
- 23.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 24.MacNeil D J. Characterization of a unique methyl-specific restriction system in Streptomyces avermitilis. J Bacteriol. 1988;170:5607–5612. doi: 10.1128/jb.170.12.5607-5612.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Magni C, Marini P, de Mendoza D. Extraction of RNA from gram-positive bacteria. BioTechniques. 1995;19:880–884. [PubMed] [Google Scholar]
- 26.Mazodier P, Petter R, Thompson C. Intergeneric conjugation between Escherichia coli and Streptomyces species. J Bacteriol. 1989;171:3583–3585. doi: 10.1128/jb.171.6.3583-3585.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mengaud J M, Horwitz M A. The major iron-containing protein of Legionella pneumophila is an aconitase homologous with the human iron-responsive element-binding protein. J Bacteriol. 1993;175:5666–5676. doi: 10.1128/jb.175.17.5666-5676.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nodwell J R, Losick R. Purification of an extracellular signaling molecule involved in production of aerial mycelium by Streptomyces coelicolor. J Bacteriol. 1998;180:1334–1337. doi: 10.1128/jb.180.5.1334-1337.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nodwell J R, McGovern K, Losick R. An oligopeptide permease responsible for the import of an extracellular signal governing aerial mycelium formation in Streptomyces coelicolor. Mol Microbiol. 1996;22:881–893. doi: 10.1046/j.1365-2958.1996.01540.x. [DOI] [PubMed] [Google Scholar]
- 30.Ohne M. Regulation of aconitase synthesis in Bacillus subtilis: induction, feedback repression, and catabolite repression. J Bacteriol. 1974;117:1295–1305. doi: 10.1128/jb.117.3.1295-1305.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Onaka H, Nakagawa T, Horinouchi S. Involvement of two A-factor receptor homologues in Streptomyces coelicolor A3(2) in the regulation of secondary metabolism and morphogenesis. Mol Microbiol. 1998;28:743–753. doi: 10.1046/j.1365-2958.1998.00832.x. [DOI] [PubMed] [Google Scholar]
- 32.Pope K M, Green B D, Westpheling J. The bld mutants of Streptomyces coelicolor are defective in the regulation of carbon utilization, morphogenesis and cell-cell signalling. Mol Microbiol. 1996;19:747–756. doi: 10.1046/j.1365-2958.1996.414933.x. [DOI] [PubMed] [Google Scholar]
- 33.Rouault T, Klausner R. Regulation of iron metabolism in eukaryotes. Curr Top Cell Regul. 1997;35:1–19. doi: 10.1016/s0070-2137(97)80001-5. [DOI] [PubMed] [Google Scholar]
- 34.Salah-Bey K, Blanc V, Thompson C J. Stress-activated expression of a Streptomyces pristinaespiralis multidrug resistance gene (ptr) in various Streptomyces and Escherichia coli. Mol Microbiol. 1995;17:1001–1012. doi: 10.1111/j.1365-2958.1995.mmi_17051001.x. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 36.Schuhmann E, Bergter F. Microscopic studies of Streptomyces hygroscopicus growth kinetics. Z Allg Mikrobiol. 1976;16:201–205. doi: 10.1002/jobm.3630160305. [DOI] [PubMed] [Google Scholar]
- 37.Schwartz D, Kaspar S, Kienzlen G, Muschko K, Wohlleben W. Inactivation of the tricarboxylic acid cycle aconitase gene from Streptomyces viridochromogenes Tu494 impairs morphological and physiological differentiation. J Bacteriol. 1999;181:7131–7135. doi: 10.1128/jb.181.22.7131-7135.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smokvina T, Mazodier P, Boccard F, Thompson C J, Guérineau M. Construction of a series of pSAM2-based integrative vectors for use in actinomycetes. Gene. 1990;94:53–59. doi: 10.1016/0378-1119(90)90467-6. [DOI] [PubMed] [Google Scholar]
- 39.Süsstrunk U, Pidoux J, Taubert S, Ullmann A, Thompson C J. Pleiotropic effects of cAMP on germination, antibiotic biosynthesis, and morphological development in Streptomyces coelicolor. Mol Microbiol. 1998;30:33–46. doi: 10.1046/j.1365-2958.1998.01033.x. [DOI] [PubMed] [Google Scholar]
- 40.Takano E, Gramajo H C, Strauch E, Andres N, White J, Bibb M J. Transcriptional regulation of the redD transcriptional activator gene accounts for growth-phase dependent production of the antibiotic undecylprodigiosin in Streptomyces coelicolor A3(2) Mol Microbiol. 1992;6:2797–2804. doi: 10.1111/j.1365-2958.1992.tb01459.x. [DOI] [PubMed] [Google Scholar]
- 41.Tang Y, Guest J R. Direct evidence for mRNA binding and post-transcriptional regulation by Escherichia coli aconitases. Microbiology. 1999;145:3069–3079. doi: 10.1099/00221287-145-11-3069. [DOI] [PubMed] [Google Scholar]
- 42.Viollier P H, Minas W, Dale G E, Folcher M, Thompson C J. Role of acid metabolism in Streptomyces coelicolor morphological development and antibiotic biosynthesis. J Bacteriol. 2001;183:3184–3192. doi: 10.1128/JB.183.10.3184-3192.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vohradsky J, Li X M, Thompson C J. Identification of procaryotic developmental stages by statistical analyses of two-dimensional gel patterns. Electrophoresis. 1997;18:1418–1428. doi: 10.1002/elps.1150180817. [DOI] [PubMed] [Google Scholar]
- 44.Willey J, Santamaria R, Guijarro J, Geistlich M, Losick R. Extracellular complementation of a developmental mutation implicates a small sporulation protein in aerial mycelium formation by S. coelicolor. Cell. 1991;65:641–650. doi: 10.1016/0092-8674(91)90096-h. [DOI] [PubMed] [Google Scholar]
- 45.Willey J, Schwedock J, Losick R. Multiple extracellular signals govern the production of a morphogenetic protein involved in aerial mycelium formation by Streptomyces coelicolor. Genes Dev. 1993;7:895–903. doi: 10.1101/gad.7.5.895. [DOI] [PubMed] [Google Scholar]
- 46.Wilson T J, Bertrand N, Tang J L, Feng J X, Pan M Q, Barber C E, Dow J M, Daniels M J. The rpfA gene of Xanthomonas campestris pathovar campestris, which is involved in the regulation of pathogenicity factor production, encodes an aconitase. Mol Microbiol. 1998;28:961–970. doi: 10.1046/j.1365-2958.1998.00852.x. [DOI] [PubMed] [Google Scholar]