Abstract
During development of the symbiotic soil bacterium Sinorhizobium meliloti into nitrogen-fixing bacteroids, DNA replication and cell division cease and the cells undergo profound metabolic and morphological changes. Regulatory genes controlling the early stages of this process have not been identified. As a first step in the search for regulators of these events, we report the isolation and characterization of a ctrA gene from S. meliloti. We show that the S. meliloti CtrA belongs to the CtrA-like family of response regulators found in several α-proteobacteria. In Caulobacter crescentus, CtrA is essential and is a global regulator of multiple cell cycle functions. ctrA is also an essential gene in S. meliloti, and it is expressed similarly to the autoregulated C. crescentus ctrA in that both genes have complex promoter regions which bind phosphorylated CtrA.
The α-proteobacterium Sinorhizobium meliloti forms nitrogen-fixing nodules on the roots of certain legumes of the genera Medicago, Melilotus, and Trigonella. The bacterium and plant exchange chemical signals during initiation of this process (7, 24). Bacteria attach to root hairs and invade plant cells via an infection thread which encases the bacterial colony as the infection thread grows into the root; bacteria are released from the infection thread into the cytoplasm of plant cells (reviewed in references 4 and 15). After release, one or more rounds of bacterial cell division occur before the bacteria differentiate into nitrogen-fixing bacteroids. These bacteroids are four to seven times longer than vegetative bacteria and are often Y shaped. Bacteroids remain in the host cytoplasm, each one enclosed in a peribacteroid membrane, a structure formed from both bacterial and plant components (4). They continue to fix nitrogen until senescence of the plant cell. Nitrogen-fixing bacteroids are thought to represent a terminally differentiated state; however, it is possible that nondifferentiated bacteria are present in the plant cell and can multiply upon release (40, 44).
While much is known about the genetics and biochemistry of nitrogen fixation, little is known about bacterial release and differentiation. Specific regulatory circuits must exist to coordinate the cessation of DNA replication and cell division with the other processes of bacteroid differentiation (reviewed in reference 30). Some S. meliloti genes involved in cell division have been characterized, but neither the mechanisims of control of these genes nor their role in the differentiation process is known (25–27).
In contrast, control of the cell cycle in Caulobacter crescentus, a closely related member of the α-proteobacteria, is better understood. Several regulatory proteins that play essential roles in cell cycle progression have been identified in this organism (17). These include members of the two-component family of signal transduction proteins, the CtrA response regulator (32), the CckA histidine kinase that phosphorylates CtrA (18), and the DivK histidine kinase (43), as well as a DNA adenine methyltransferase, CcrM, that methylates newly replicated DNA at the end of S phase (39). CtrA is a global regulator that plays a pivotal role in orchestrating the cell cycle. It is involved in the control of a quarter of the cell cycle-regulated genes (23) including genes required for DNA replication (32, 33), DNA methylation (32, 34), cell division (20), and biogenesis of flagella and pili (32, 38). CtrA binds to the chromosomal origin of replication and inhibits DNA replication initiation; it must be cleared from the cell before it enters S phase (9, 33). Transcription of ctrA is activated after initiation of DNA replication and reaches threshold levels by an autoregulated positive feedback loop. ctrA gene is transcribed from two promoters: P1, a promoter that is active in early S phase and is negatively controlled by CtrA; and a stronger promoter, P2, which is active later in S phase and is positively controlled by CtrA (10). The level of intracellular CtrA is controlled not only by autoregulated transcription but also by specific degradation of this protein when the cells enter S phase and by cell cycle-controlled phosphorylation which activates the protein (9, 18).
Here we report the isolation and characterization of ctrA from S. meliloti. As a first step in dissecting the regulatory circuit controlling the cell cycle and bacteroid differentiation in S. meliloti, we show that ctrA is essential for viability of S. meliloti. The promoter region was identified and shown to contain multiple CtrA binding sites, suggesting that the S. meliloti gene may be autoregulated, as is the case in C. crescentus.
MATERIALS AND METHODS
Bacterial genetic techniques.
Strains and plasmids used in this study are listed in Table 1. Escherichia coli strains were grown in either Luria broth (LB) or 2XYT medium at 37°C. S. meliloti strains were grown on LB agar plates or in liquid TY medium at 30°C. Antibiotic concentrations were for S. meliloti, neomycin and spectinomycin, 200 μg/ml; gentamicin, 25 μg/ml; tetracycline, 10 μg/ml; streptomycin, 500 μg/ml; and trimethoprim, 600 μg/ml; for E. coli, gentamicin, 5 μg/ml; kanamycin, 25 μg/ml; tetracycline, 10 μg/ml; ampicillin, 50 or 100 μg/ml; chloramphenicol, 50 μg/ml; and spectinomycin, 40 μg/ml. Triparental conjugations were performed using MT616 as the helper plasmid strain (12). C. crescentus was grown in PYE broth as previously described (11). Kanamycin (20 μg/ml) was used for C. crescentus. Agrobacterium tumefaciens was grown in LB medium.
TABLE 1.
Strains, plasmids, and primers used
| Strain, plasmid or primer | Descriptiona | Reference or source |
|---|---|---|
| Strains | ||
| DH5α | endA1 hsdR17 supE44 thi-1 recA1 gyrA relA1 Δ(lacZYA-argF) U169 deoR | BRL Corp. |
| GV310(pMPg90) | Agrobacterium tumefaciens | 39 |
| LS2195 | NA1000 ctrA401(Ts) | 32 |
| MT616 | MM294 pRK600, Cmr | 12 |
| MB492X | Rm1021, single crossover of pMB492 into ctrA | This study |
| MB506 | Rm1021, ctrA mutant with pMB467 | This study |
| MB507 | Rm1021, ctrA mutant with pSAL290 | This study |
| Rm1021 | S. meliloti RCR2011 Smr | F. M. Ausubel |
| S2308 | Brucella abortus | R. M. Roop |
| Plasmids | ||
| pBBR1MCS | Cloning vector, pBBR origin | K. M. Peterson |
| pDYH218 | 1.6-kb XhoI-RsaI fragment containing S. meliloti ctrA in pMR10 | This study |
| pJQ200 | Cloning vector, Gmr, ColE1, oriT, sacB | 31 |
| pMB393 | Cloning vector, Spr Cmr version of pBBR1MCS | M. Barnett |
| pMB453 | 4.5-kb SalI fragment containing S. meliloti ctrA in pBS SK(−) | This study |
| pMB454 | Same as pMB453, opposite orientation | This study |
| pMB464 | 3-kb PstI fragment containing S. meliloti ctrA in pBS SK(−) | This study |
| pMB467 | 3-kb PstI fragment from pMB453 in pMB393 | This study |
| pMB492 | Mutated S. meliloti ctrA in pJQ200 | This study |
| pMR10 | Cloning vector, Kmr, RK2, oriT | C. Mohr, R. Roberts |
| pRK290 | Cloning vector, IncP | D. Helinski |
| pSAL290 | C. crescentus ctrA in pRK290 | 32 |
| Primers | ||
| PCR amplification of C. crescentus ctrA probe | 5′-ATGCGCGTACTGTTGATCGA | |
| 5′-TCAGGCGGCGTTAACCTGC | ||
| Primer extension | ||
| PE-1 | 5′-CGCGCTGTCGTCTTCGATCAGTAG | |
| PE-2 | 5′-GAGCTCGATGCTCTGAGCCGTCGCGC | |
| PE-3 | 5′-GGGAATGCCGTTAACTGACTCTTCCGAC | |
| DNase I protection | ||
| P1-for | 5′-GAAGGTGGGATAGGCGAAAT-3′ | |
| P1-rev | 5′-ATCTGAAATTGCGCTCCTCA-3′ | |
| P2-for | 5′-GCCGGTTCGCCCGGAATTCCCGTGA-3′ | |
| P2-rev | 5′-TCCCCTTTTCCGCCGCCCAAGG-3′ |
Apr, Cmr, Gmr, Kmr, Spr, Smr, Tcr, and Tpr denote resistance to ampicillin, chloramphenicol, gentamicin, kanamycin, spectinomycin, tetracycline, and trimethoprim, respectively. pBS, pBluescript.
Southern hybridization.
DNA from S. meliloti, C. crescentus, and A. tumefaciens was purified using the PureGene kit (Gentra systems), and Brucella abortus strain 2308 DNA was kindly provided by R. M. Roop, Louisiana State University. Genomic DNA was digested with various restriction enzymes and electrophoresed on a 1% agarose gel. The DNA was transferred to Hybond N+ (Amersham) as described previously (37). Hybridization was performed by the Church and Gilbert method (5); a random-primed EcoRI-SalI restriction fragment from pSALF1, containing C. crescentus ctrA, was the probe (32). The blot was hybridized for 4 h at 65°C and then washed once in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) for 1 h, twice in 1× SSC (20 min each time), and finally once in 0.1× SSC for 20 min.
Isolation of the S. meliloti ctrA clone.
An S. meliloti partial Sau3A genomic library, constructed in a λ FixII vector (1), was screened using Church and Gilbert hybridization (5). The probe was a random-primed 695-bp PCR product containing C. crescentus ctrA; the primers used to amplify this DNA are shown in Table 1. Following hybridization, filters were washed three times in 2X SSC–0.1% sodium dodecyl sulfate and then once in 0.5X SSC–0.1% sodium dodecyl sulfate. Positive plaques were purified and rescreened. DNA was isolated from putative ctrA-containing phage, mapped, subcloned, and sequenced. The University of Wisconsin Genetics Computer Group (8) and Sequencher (Gene Codes Corp.) software were used for sequence assembly and analysis.
Construction and complementation of S. meliltoti ctrA mutant.
To introduce an insertion mutation into the S. meliloti ctrA coding sequence we used pJQ200, which contains the levan sucrase (sacB) gene, which confers lethality when strains are grown on sucrose (31). This vector has been previously used to create null mutations in S. meliloti ccrM (42). A derivative of pJQ200 lacking polylinker sites from Ecl136 to SmaI was created. A 3-kb PstI-SalI fragment from pMB453 was cloned into this derivative to form pMB491. A blunted, BamHI-HindIII fragment containing a neomycin resistance (Nmr) cassette was ligated into pMB491 that had been digested with BamHI, thereby removing the 5′ half of ctrA, and blunted with Klenow to form pMB492. In pMB492, the Nmr cassette is oriented such that the neomycin promoter reads in the opposite direction from the ctrA promoter. 1.35 and 0.95 kb of S. meliloti DNA are present upstream and downstream, respectively, of the Nmr cassette. After conjugation into S. meliloti strain Rm1021, colonies were selected on LB agar plates containing gentamicin, neomycin, and streptomycin; these are the putative single recombinants containing both an intact and a disrupted copy of ctrA. Either a ctrA plasmid (pMB467 or pSAL290) or the vector only (pMB393 or pRK290) was introduced into this strain by conjugation. The resulting strains were selected on plates lacking gentamicin and containing 5% sucrose. Colonies viable on these plates were screened for gentamicin sensitivity (Gms) in the presence of sucrose (Gmr sucrose-resistant mutants likely contain mutations in the sacB gene or in host genes that suppress the sacB phenotype). Double recombinants, containing a single mutated ctrA, are expected to be Gms, Nmr, and sucrose resistant. The frequency of apparent double recombinants was calculated for a representative experiment by dividing the number of Gms Nmr sucrose-resistant colonies by the total number of Nmr sucrose-resistant CFU.
Complementation tests of C. crescentus ctrA mutants with S. meliloti ctrA.
To determine if the S. meliloti ctrA gene could replace the C. crescentus ctrA, the S. meliloti ctrA promoter and gene were ligated into the low-copy-number vector pMR10, generating pDYH218. Both pDYH218 and pMR10 were mated into strains NA1000 (wild-type C. crescentus) and LS2195 (NA1000 ctrA401, a temperature-sensitive allele of ctrA). Logarithmic-phase cultures were diluted serially, plated in duplicate on PYE agar containing kanamycin (20 μg/ml), and incubated at either 30 or 37°C. Viable cell number at the permissive (30°C) and restrictive (37°C) temperatures was measured as CFU per milliliter.
Primer extension analysis.
S. meliloti total RNA was purified from LB-grown cells, using Trizol reagent (BRL Corp.), as previously described (3). Primer extension reactions were performed as before (3), using an initial annealing temperature of 65°C for 1 h and then cooling to 40°C over 1.5 h. Extensions were performed at 49°C. The nucleotide sequences of PE-1, PE-2, and PE-3 used for primer extension are shown in Table 1. Extension products were analyzed on a urea-polyacrylamide sequencing gel as described elsewhere (13). Sequencing ladders using each of the three primers and pMB464 as template were used to determine the transcription start site.
DNase I protection experiments.
DNase I footprinting experiments were performed with purified C. crescentus CtrA that was phosphorylated using a maltose-binding protein (MBP)-EnvZ fusion protein as previously described (34). Native C. crescentus CtrA was overexpressed in E. coli using the pET expression system (Novagen), solubilized from inclusion bodies with 4 M guanidine-HCl, and purified by Q-Sepharose chromatography (K. Ryan and L. Shapiro, unpublished data). Template DNA for footprinting the P1 and P2 promoters was generated by PCR using the oligonucleotides in Table 1. Both reverse primers (P1-rev and P2-rev) were end labeled with [γ-32P]ATP and T4 DNA polynucleotide kinase prior to PCR amplification of the template.
RESULTS
CtrA is conserved among α-proteobacteria.
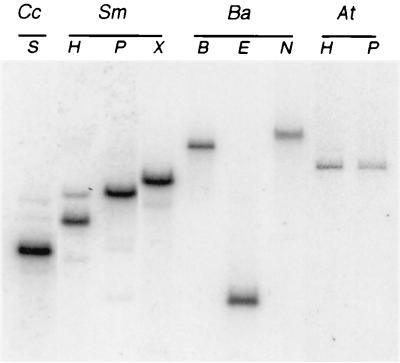
To determine the extent of conservation of the gene encoding the CtrA response regulator among the α-proteobacterial family, we generated a probe containing the entire C. crescentus ctrA coding region and part of the 5′ and 3′ flanking sequences (32). We hybridized this SalI-EcoRI DNA fragment to digested genomic DNA from S. meliloti, B. abortus, and A. tumefaciens (Fig. 1) using fairly stringent conditions. The C. crescentus ctrA probe hybridized to at least one band in genomic digests of all these organisms.
FIG. 1.
ctrA is present in several members of the α-proteobacteria. The Southern blot shows hybridization of the C. crescentus ctrA probe to chromosomal DNA from C. crescentus (Cc), S. meliloti (Sm), B. abortus (Ba), and A. tumifaciens (At). Restriction enzymes used are BamHI (B), EcoRI (E), HindIII (H), NotI (N), PstI (P), SalI (S), and XhoI (X).
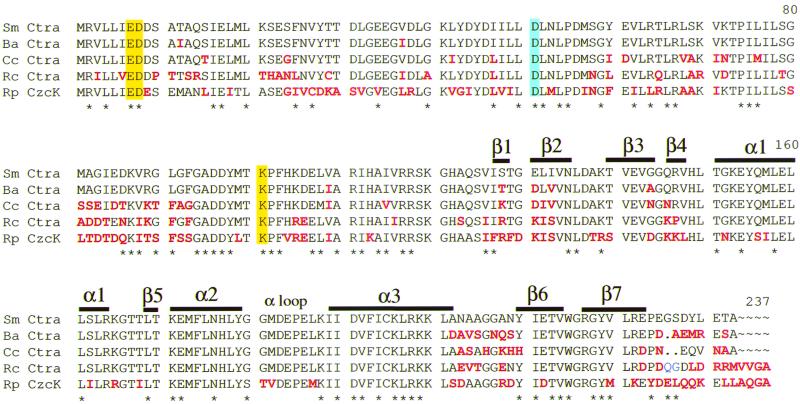
To isolate the putative ctrA homolog from S. meliloti, we screened a genomic library with a probe internal to the coding region of C. crescentus ctrA. Positive plaques were purified, and phage DNA was subcloned. Sequence analysis of this DNA revealed an open reading frame with an excellent pattern of S. meliloti codon usage and a high degree of similarity to C. crescentus ctrA (80% nucleotide sequence identity; 82% amino acid sequence identity) (Fig. 2). S. meliloti ctrA encodes a 26-kDa protein. Genes encoding CtrA-like proteins are also found in three other α-proteobacteria. Of these, S. meliloti CtrA is most similar to CtrA from B. abortus (86% amino acid identity) and less similar to the CtrA of Rhodobacter capsulatus and CzcR of Rickettsia prowazekii (77 and 59% identity, respectively.) Thus, CtrA homologs are present in at least six members of the α-proteobacteria, suggesting that CtrA is conserved in this group of bacteria.
FIG. 2.
Sequence alignment of S. meliloti CtrA with CtrA family proteins using Pileup (8). Residues that differ from S. meliloti CtrA are in red; the conserved aspartate phosphorylation site is boxed in blue; the acidic pocket conserved residues are boxed in yellow. Secondary structures (α helices and β strands) corresponding to those in the OmpR C-terminal domain are indicated (29). Wing1 is is positioned between the α1 helix and β4 (29). Wing2 connects β6 and β7 of the C-terminal hairpin (29). We aligned E. coli OmpR (accession no. P03025) with the CtrA family, ignoring the gap caused by differing lengths in the N terminus-C terminus linker region. Residues conserved in S. meliloti CtrA and OmpR are indicated by asterisks below the alignment. Conserved groupings were Val, Ile; Leu, Met; Asp, Glu; and Arg, Lys. Proteins included in this lineup are S. meliloti (Sm) CtrA (accession no. AF288464), B. abortus (Ba) CtrA (accession no. AAC69920), C. crescentus (Cc) CtrA (accession no. AAF1377), R. capsulatus (Rc) CtrA (accession no. AAF1377), and Rickettsia prowazekii (Rp) CzcR (accession no. CAA14542).
CtrA is a member of the OmpR family of winged helix-turn-helix response regulators (32). OmpR-type regulators contain an N-terminal regulatory domain and a C-terminal DNA binding domain. The five CtrA-like proteins shown in Fig. 2 form a distinct subgroup within the OmpR family, thus far unique to the α-proteobacteria. No other winged helix regulators show greater than 35% amino acid sequence identity to CtrA (data not shown). E. coli OmpR is only 31% identical to S. meliloti CtrA (Fig. 2). The activity of C. crescentus CtrA, like OmpR and other response regulators (41), is dependent on phosphorylation at a conserved aspartate residue (Fig. 2) (9). Additional conserved residues are hypothesized to lie within the acidic pocket active site, the phosphorylation site and acidic pocket residues are conserved in all five CtrA proteins (Fig. 2). The three dimensional structure of the C-terminal region of OmpR, including the DNA binding domain, has been determined by X-ray crystallography (21, 28). The OmpR amino acid sequence was used to predict the secondary structures in S. meliloti CtrA (Fig. 2). Profile network prediction of secondary structure (PHDsec [35, 36]), with the S. meliloti amino acid sequence as input, gave similar results (data not shown). OmpR contains three α helices and two antiparallel β strands. The α3 helix is hypothesized to recognize the major groove, and the two wings may be important for recognition of the minor groove (16). These are nearly identical in the CtrA subfamily (Fig. 2). The α loop interacts with the α subunit of RNA polymerase in other OmpR homologs and is well conserved in the CtrA subfamily, even though it is one of the least conserved regions in the OmpR family (29). The least conserved region in the CtrA subfamily is the C terminus of the protein. The amino acid sequence after the β7 strand shows little conservation.
S. meliloti ctrA encodes an essential function.
To determine if ctrA is essential for viability in S. meliloti, as is the case in C. crescentus (32), we disrupted the ctrA gene by deleting a 390-bp BamHI fragment by single-crossover, Campbell recombination. This deletion removes the first 112 amino acids of CtrA. Counterselection with the single-crossover strain MB492X was performed as described in Materials and Methods, with either S. meliloti ctrA present on a plasmid (pMB467) or the vector only (pMB393) (Table 2). Double recombinants (Nmr Gms sucrose resistant) were obtained only when S. meliloti ctrA was present on a complementing plasmid, indicating that ctrA is necessary for viability in S. meliloti.
TABLE 2.
Complementation of an S. meliloti ctrA mutant
| Origin of ctrA | Frequency
of Gms colonies
|
|
|---|---|---|
| MB492X + ctrA | MB492X + vector only | |
| S. meliloti | 0.35 (na = 145) | 0 (n = 162) |
| C. crescentus | 0.19 (n = 94) | 0 (n = 97) |
n is the total number of Nmr, a sucrose-resistant colonies screened.
Exchange of S. meliloti and C. crescentus ctrA.
To test if C. crescentus ctrA functions in S. meliloti, sucrose counterselection with MB492X was performed as described above, in the presence of either C. crescentus ctrA on a plasmid or the vector alone. Table 2 shows that Nmr Gms sucrose-resistant colonies were obtained only when ctrA was present on a plasmid (pSAL290); therefore, C. crescentus ctrA can substitute for S. meliloti ctrA for viability. To test if S. meliloti ctrA could complement a C. crescentus ctrA temperature-sensitive mutation, we mated a plasmid containing S. meliloti ctrA expressed from its own promoter (pDYH218) into the temperature-sensitive strain at the permissive temperature (Materials and Methods). This plasmid failed to complement the C. crescentus ctrA mutant at the restrictive temperature (data not shown). Since correct function of ctrA in C. crescentus requires coordinated expression of the gene, phosphorylation, and proteolysis of the gene product, the heterologous product may have failed at any one of these levels.
The CtrA response regulator binds the S. meliloti ctrA promoter.
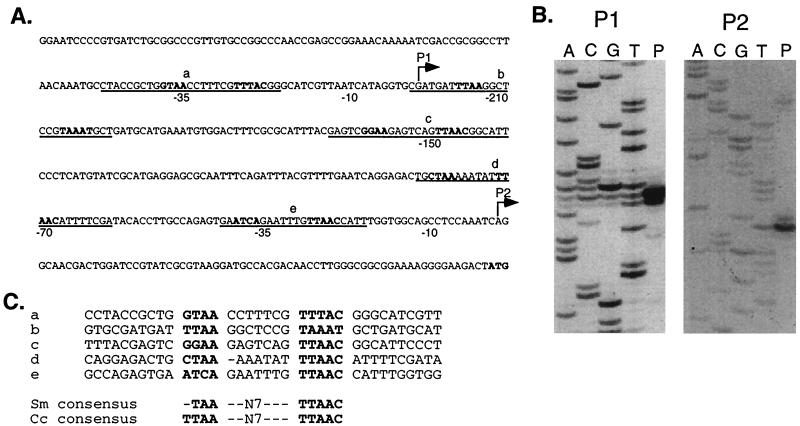
To define the S. meliloti ctrA promoter region, we determined the transcriptional start site of the ctrA gene by primer extension assays using RNA isolated from wild-type cells. We identified two transcripts initiating 69 and 291-bp upstream of the ctrA translational start site (Fig. 3B).
FIG. 3.
Identification of the ctrA transcription start sites. (A) Sequence upstream of ctrA. Locations of the P1 and P2 start sites are shown by bent arrows. The −10 and −35 regions of the two promoters are marked below the sequence. The sequence protected from DNase I digestion by CtrA∼P is underlined (see Fig. 4), and the CtrA recognition motifs are in bold. The final ATG is the presumed translational start codon. (B) Autoradiograms of primer extension products (P) and sequencing reactions (labeled A, C, G, and T) for the P1 and P2 start sites of the S. meliloti ctrA promoter. The P1 start site was mapped using primer PE-3 (left); the P2 promoter was mapped using primers PE-1 and PE-2. Results obtained with PE-2 are shown (right). (C) Alignment of the five S. meliloti CtrA binding sites in the S. meliloti ctrA promoter with the S. meliloti and C. crescentus CtrA consensus sequences (32).
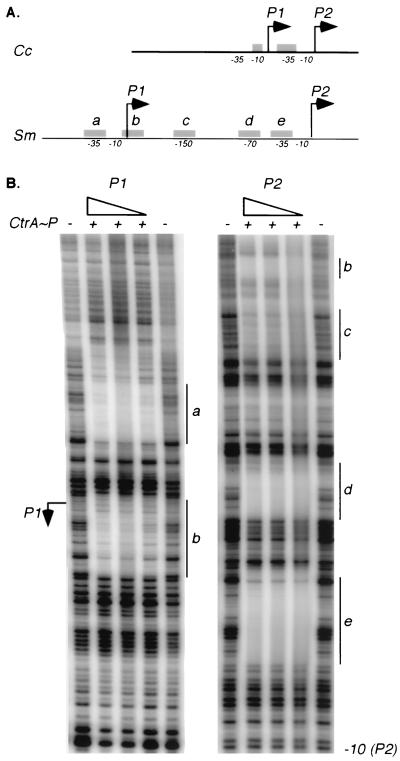
Analysis of the 5′ untranslated sequence upstream of ctrA revealed putative CtrA binding sites overlapping the −35 regions of both the ctrA P1 and P2 promoters (Fig. 3A). Because ctrA transcription is controlled by the CtrA response regulator in C. crescentus (10), we investigated the possibility that CtrA also binds to the S. meliloti ctrA promoter. We used DNase I footprinting analysis to assess the binding of phosphorylated CtrA (CtrA∼P) to the ctrA promoter (Fig. 4B). Separate DNA templates for P1 and P2 were generated by PCR and labeled at the 3′ end as described in Materials and Methods. We phosphorylated purified C. crescentus CtrA in vitro using the E. coli EnvZ histidine kinase and used it in the footprinting reactions (34). We chose to use C. crescentus CtrA, because this heterologous CtrA supports viability in S. meliloti (Table 2) and purified C. crescentus CtrA was available. As shown in Fig. 4B, CtrA∼P specifically protected five sites in the ctrA promoter. Not only were the −35 regions of the P1 and P2 promoters protected, but CtrA∼P also protected a region overlapping the +1 of the P1 promoter and the −70 and −150 regions of the P2 promoter. As shown in Fig. 3C, a consensus binding motif derived from the five CtrA binding sites in the S. meliloti ctrA promoter is almost identical to the C. crescentus consensus (32). These in vitro data are consistent with CtrA∼P regulation of ctrA transcription in S. meliloti.
FIG. 4.
P CtrA∼P binds to five sites in the ctrA promoter. (A) Diagram of the ctrA promoter regions in C. crescentus (top) and S. meliloti. Locations of the ctrA P1 and P2 transcription start sites are marked by arrows, and the regions protected from DNase I digestion by CtrA∼P are shown as gray boxes (labeled a to e to match the footprinted regions in panel B). (B) DNase I protection of the ctrA P1 and P2 promoters. CtrA purified from C. crescentus was phosphorylated with MBP-EnvZ and used in the footprinting reactions (0.5 to 1.7 μM). CtrA-P was omitted from the reactions in the lanes marked with a minus sign. The vertical lines labeled a to e mark the protected regions.
DISCUSSION
The CcrM methyltransferase is conserved in α-proteobacteria (42). Here we report the conservation of a second regulatory protein, the CtrA response regulator, in this group of bacteria. We isolated and characterized CtrA, a global regulator of cell cycle progression in C. crescentus (32), from S. meliloti. A ctrA homologue appears to be present in another member of the Rhizobiaceae, A. tumefaciens. Our data show that by sequence similarity CtrA belongs to a unique subfamily of the OmpR family of helix-turn-helix transcriptional regulators. Thus far, all members of this subfamily belong to the α-proteobacteria, which are characterized by specialized mechanisms to adapt to unique environments, including mechanisms for pathogenicity and symbiosis (2). Some, particularly the Rhizobiaceae, possess large, multipartite genomes (2, 19).
The primary sequence of S. meliloti CtrA is very similar to C. crescentus CtrA. In addition, antibodies to C. crescentus CtrA cross-react with S. meliloti CtrA (A. Reisenauer, unpublished data). The sequence conservation among CtrA-like proteins is especially strong for the C-terminal DNA binding domain, suggesting that the DNA target site recognized by these proteins is also conserved. In fact, the promoter region for S. meliloti ctrA has five CtrA binding motifs that are nearly identical to the C. crescentus consensus motif.
The S. meliloti and C. crescentus ctrA promoters are strikingly similar. In both species there are two transcriptional start sites, and each promoter (called P1 and P2) contains at least one CtrA binding motif. In addition, purified and phosphorylated C. crescentus CtrA binds the consensus CtrA motifs in vitro in each of the ctrA promoters, suggesting that ctrA transcription is autoregulated in both species. In fact, CtrA has been shown to repress transcription from P1 and activate transcription from P2 in vivo in C. crescentus (10). However, the spacing between P1 and P2 and the number of CtrA binding sites in the S. meliloti and C. crescentus ctrA promoters are distinct (Fig. 4A). P1 and P2 are separated by 222 bp in S. meliloti but only 57 bp in C. crescentus. The most distal promoter in S. meliloti (P1) is far upstream from the ATG translational start site; however, it is not unusual for S. meliloti genes to have long mRNA leader sequences (3, 13).
In the S. meliloti ctrA promoter, CtrA∼P protects five distinct sites from DNase I digestion; two overlap the −35 regions of P1 and P2, respectively, and there are three additional sites between P1 and P2. Moreover, the P1 transcriptional start site itself is protected from DNase I digestion by CtrA∼P. In contrast, CtrA protects only two regions of the C. crescentus ctrA promoter, one overlapping the −10 region of P1 and the second overlapping the −35 region of P2 (10). It is tempting to speculate that CtrA molecules bound to the different recognition motifs in each of these promoters act cooperatively to regulate ctrA transcription. The CtrA binding sites in the S. meliloti ctrA promoter are very similar to the C. crescentus CtrA consensus motif. Given the similarity of the C-terminal DNA binding domains and the fact that C. crescentus CtrA can substitute for S. meliloti CtrA in vivo, it is likely that CtrA recognizes these same binding sites in both species.
As in C. crescentus, S. meliloti ctrA is essential for viability. In contrast, ctrA is not essential in R. capsulatus where insertion mutations in ctrA were obtained (22). CtrA in R. capsulatus is responsible for expression of the gene transfer agent structural genes. The gene transfer agent is a small phage-like particle that can transfer genes between R. capsulatus cells. Possibly, ctrA is present in multiple copies in R. capsulatus or is not involved in critical cell cycle functions. It is not known if ctrA is an essential gene in B. abortus or R. prowazekii.
Based on sequence similarity, we thought it likely that C. crescentus ctrA could substitute for S. meliloti ctrA, as is the case for the conserved ccrM in these organisms (42). C. crescentus ctrA, expressed from its own promoter, complements a S. meliloti null mutant for viability. Thus, S. meliloti contains the cellular machinary necessary for transcription and phosphorylation of C. crescentus CtrA, at least to an extent that supports viability.
Little is known of cell division control in S. meliloti, but examination of symbiotic behavior suggests the likelihood of specific controls related to invasion and differentiation. We and others have suggested that bacterial cell division is coordinately controlled to match the expansion rate of the infection thread, in order to keep the infection thread filled but not overrun with bacteria (14, 15). After invasion is complete and bacteria are released into the plant cell cytoplasm, it appears that one or several rounds of bacterial cell division likely occur. Circuitry must exist to halt DNA replication and cell division after this step, as the bacteria differentiate. The essential functions of S. meliloti CtrA are not known, but it is an attractive candidate to study in regard to these inferred regulatory steps. In C. crescentus, CtrA is known to directly prevent DNA synthesis in the swarmer cell by binding to the origin of replication (33). It will be interesting to determine if perturbation of ctrA circuiting in free-living S. meliloti cells results in disruption of the invasion process.
ACKNOWLEDGMENTS
This work was funded by NIH grant GM-32506/5120MZ to L.S. and NIH grant GM-30692 to S.R.L. S.R.L. is an investigator of the Howard Hughes Medical Institute.
We thank J. J. Letesson for sharing B. abortus data prior to publication and for sharing strains and plasmids. We thank R. M. Roop for B. abortus DNA. We are grateful to K. Ryan for providing purified Caulobacter CtrA. R. Fisher helped with critical reading of the manuscript.
REFERENCES
- 1.Abola A P, Willits M G, Wang R C, Long S R. Reduction of adenosine-5′-phosphosulfate instead of 3′-phosphoadenosine-5′-phosphosulfate in cysteine biosynthesis by Rhizobium meliloti and other members of the family Rhizobiaceae. J Bacteriol. 1999;181:5280–5287. doi: 10.1128/jb.181.17.5280-5287.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K-H, editors. The prokaryotes. III. New York, N.Y: Springer-Verlag; 1992. [Google Scholar]
- 3.Barnett M J, Rushing B G, Fisher R F, Long S R. Transcription start sites for syrM and nodD3 flank an insertion sequence relic in Rhizobium meliloti. J Bacteriol. 1996;178:1782–1787. doi: 10.1128/jb.178.7.1782-1787.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brewin N J. Tissue and cell invasion by Rhizobium: the structure and development of infection threads and symbiosomes. In: Spaink H P, Kondorosi A, Hooykaas P J J, editors. The Rhizobiaceae. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1998. pp. 418–429. [Google Scholar]
- 5.Church G M, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cormack B P, Valdivia R H, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 7.Dénarié J, Debellé F, Rosenburg C. Signaling and host range variation in nodulation. Annu Rev Microbiol. 1992;46:497–531. doi: 10.1146/annurev.mi.46.100192.002433. [DOI] [PubMed] [Google Scholar]
- 8.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Domian I J, Quon K C, Shapiro L. Cell type-specific phosphorylation and proteolysis of a transcriptional regulator controls the G1-to-S transition in a bacterial cell cycle. Cell. 1997;90:415–424. doi: 10.1016/s0092-8674(00)80502-4. [DOI] [PubMed] [Google Scholar]
- 10.Domain I J, Reisenauer A, Shapiro L. Feedback control of a master bacterial cell-cycle regulator. Proc Natl Acad Sci USA. 1999;96:6648–6653. doi: 10.1073/pnas.96.12.6648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ely B. Genetics of Caulobacter crescentus. Methods Enzymol. 1991;204:372–384. doi: 10.1016/0076-6879(91)04019-k. [DOI] [PubMed] [Google Scholar]
- 12.Finan T M, Kunkel B, De Vos G F, Signer E R. Second symbiotic megaplasmid in Rhizobium meliloticarrying exopolysaccharide and thiamine synthesis genes. J Bacteriol. 1986;167:66–72. doi: 10.1128/jb.167.1.66-72.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fisher R F, Brierley H L, Mulligan J T, Long S R. Transcription of Rhizobium meliloti nodulation genes: identification of a nodD transcription initiation site in vitro and in vivo. J Biol Chem. 1987;262:6849–6855. [PubMed] [Google Scholar]
- 14.Gage D J, Bobo T, Long S R. Use of green fluorescent protein to visualize the early events of symbiosis between Rhizobium meliloti and alfalfa (Medicago sativa) J Bacteriol. 1996;178:7159–7166. doi: 10.1128/jb.178.24.7159-7166.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gage D J, Margolin W. Hanging by a thread: invasion of legume plants by rhizobia. Curr Opin Microbiol. 2000;3:613–617. doi: 10.1016/s1369-5274(00)00149-1. [DOI] [PubMed] [Google Scholar]
- 16.Harrison-McMonagle P, Denissova N, Martínez-Hackert E, Ebright R H, Stock A M. Orientation of OmpR monomers within an OmpR:DNA complex determined by DNA affinity cleaving. J Mol Biol. 1999;285:555–566. doi: 10.1006/jmbi.1998.2375. [DOI] [PubMed] [Google Scholar]
- 17.Hung D, McAdams H, Shapiro L. Regulation of the Caulobacter cell cycle. In: Brun Y V, Shimkets L J, editors. Prokaryotic development. Washington, D.C.: American Society for Microbiology; 2000. pp. 361–378. [Google Scholar]
- 18.Jacobs C, Domian I J, Maddock J R, Shapiro L. Cell cycle-dependent polar localization of an essential bacterial histidine kinase that controls DNA replication and cell division. Cell. 1999;97:111–120. doi: 10.1016/s0092-8674(00)80719-9. [DOI] [PubMed] [Google Scholar]
- 19.Jumas-Bilak E, Michaux-Charachon S, Bourg G, Ramuz M, Allardet-Servent A. Unconventional genomic organization in the alpha subgroup of the proteobacteria. J Bacteriol. 1998;180:2749–2755. doi: 10.1128/jb.180.10.2749-2755.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kelly A J, Sackett M J, Din N, Quardokus E, Brun Y V. Cell cycle-dependent transcriptional and proteolytic regulation of FtsZ in Caulobacter. Genes Dev. 1998;12:880–893. doi: 10.1101/gad.12.6.880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kondo H, Nakagawa A, Nishihira J, Nishimura Y, Mizuno T, Tanaka I. Escherichia coli positive regulator OmpR has a large loop structure at the putative RNA polymerase interaction site. Nat Struct Biol. 1997;4:28–31. doi: 10.1038/nsb0197-28. [DOI] [PubMed] [Google Scholar]
- 22.Lang A S, Beatty T J. Genetic analysis of a bacterial genetic exchange element: the gene transfer agent of Rhodobacter capsulatus. Proc Natl Acad Sci USA. 2000;97:859–864. doi: 10.1073/pnas.97.2.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laub M T, McAdams H H, Feldblyum T, Fraser C M, Shapiro L. Global analysis of the genetic network controlling a bacterial cell cycle. Science. 2000;290:2144–2148. doi: 10.1126/science.290.5499.2144. [DOI] [PubMed] [Google Scholar]
- 24.Long S R. Rhizobiumsymbiosis: nod factors in perspective. Plant Cell. 1996;8:1885–1898. doi: 10.1105/tpc.8.10.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Margolin W, Bramhill D, Long S R. The dnaA gene of Rhizobium melilotilies within an unusual gene arrangement. J Bacteriol. 1995;177:2892–2900. doi: 10.1128/jb.177.10.2892-2900.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Margolin W, Corbo J C, Long S R. Cloning and characterization of a Rhizobium meliloti homolog of the Escherichia coli cell division gene ftsZ. J Bacteriol. 1991;173:5822–5830. doi: 10.1128/jb.173.18.5822-5830.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Margolin W, Long S R. Rhizobium meliloti contains a novel second homolog of the cell division gene ftsZ. J Bacteriol. 1994;176:2033–2043. doi: 10.1128/jb.176.7.2033-2043.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martínez-Hackert E, Stock A M. The DNA-binding domain of OmpR: crystal structures of a winged helix transcription factor. Structure. 1997;5:109–124. doi: 10.1016/s0969-2126(97)00170-6. [DOI] [PubMed] [Google Scholar]
- 29.Martínez-Hackert E, Stock A M. Structural relationships in the OmpR family of winged-helix transcription factors. J Mol Biol. 1997;269:301–12. doi: 10.1006/jmbi.1997.1065. [DOI] [PubMed] [Google Scholar]
- 30.Oke V, Long S R. Bacteroid formation in the Rhizobium-legume symbiosis. Curr Opin Microbiol. 1999;2:641–646. doi: 10.1016/s1369-5274(99)00035-1. [DOI] [PubMed] [Google Scholar]
- 31.Quandt J, Hines M F. Versatile suicide vectors which allow direct selection for gene replacement in Gram-negative bacteria. Gene. 1993;127:15–21. doi: 10.1016/0378-1119(93)90611-6. [DOI] [PubMed] [Google Scholar]
- 32.Quon K C, Marczynski G T, Shapiro L. Cell cycle control by an essential bacterial two-component signal transduction protein. Cell. 1996;84:83–93. doi: 10.1016/s0092-8674(00)80995-2. [DOI] [PubMed] [Google Scholar]
- 33.Quon K C, Yang B, Domian I J, Shapiro L, Marczynski G T. Negative control of bacterial DNA replication by a cell cycle regulatory protein that binds at the chromosome origin. Proc Natl Acad Sci USA. 1998;95:120–125. doi: 10.1073/pnas.95.1.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reisenauer A, Quon K, Shapiro L. The CtrA response regulator mediates temporal control of gene expression during the Caulobactercell cycle. J Bacteriol. 1999;181:2430–2439. doi: 10.1128/jb.181.8.2430-2439.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rost B, Sander C. Combining evolutionary information and neural networks to predict protein secondary structure. Proteins. 1994;19:55–77. doi: 10.1002/prot.340190108. [DOI] [PubMed] [Google Scholar]
- 36.Rost B, Sander C. Prediction of protein structure at better than 70% accuracy. J Mol Biol. 1993;232:584–599. doi: 10.1006/jmbi.1993.1413. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook J, Fritsch E F, Maniatis T, editors. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 38.Skerker J M, Shapiro L. Identification and cell cycle control of a novel pilus system in Caulobacter crescentus. EMBO J. 2000;19:3223–3234. doi: 10.1093/emboj/19.13.3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stephens C, Reisenauer A, Wright R, Shapiro L. A cell cycle-regulated bacterial DNA methyltransferase is essential for viability. Proc Natl Acad Sci USA. 1996;93:1210–1214. doi: 10.1073/pnas.93.3.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sutton W D, Pankhurst C E, Craig A S. The Rhizobium bacteroid state. Int Rev Cyt Suppl. 1981;13:149–177. [Google Scholar]
- 41.Volz K. Structural conservation in the CheY superfamily. Biochemistry. 1993;32:11741–11753. doi: 10.1021/bi00095a001. [DOI] [PubMed] [Google Scholar]
- 42.Wright R, Stephens C, Shapiro L. The CcrM DNA methyltransferase is widespread in the alpha subdivision of proteobacteria, and its essential functions are conserved in Rhizobium meliloti and Caulobacter crescentus. J Bacteriol. 1997;179:5869–5877. doi: 10.1128/jb.179.18.5869-5877.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu J, Ohta N, Zhao J-L, Newton A. A novel bacterial tyrosine kinase essential for cell division and differentiation. Proc Natl Acad Sci USA. 1999;96:13068–13073. doi: 10.1073/pnas.96.23.13068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou J C, Tchan Y T, Vincent J M. Reproductive capacity of bacteroids in nodules of Trifolium repens L. and Glycine max(L.) Merr. Planta. 1985;163:473–482. doi: 10.1007/BF00392704. [DOI] [PubMed] [Google Scholar]