Significance
Mucins are densely O-glycosylated proteins found on the surfaces of many cells in the human body. In order to break down mucin glycodomains, cells must catabolize both the peptide backbone and the appended glycans. In mammals, these two processes were thought to occur largely independently. Here, we show that the human lysosomal enzyme cathepsin D can proteolyze fully glycosylated mucin domains into glycopeptides and that mucins accumulate in some tissues when cathepsin D activity is lost. These findings suggest a mammalian pathway for endogenous mucin glycodomain catabolism, with implications for major histocompatibility complex loading of glycopeptides and tumor extracellular matrix degradation, as well as potential therapies to reverse pathological mucin accumulation.
Keywords: O-glycosylation, mucin, catabolism, protease, cathepsin
Abstract
Mucins are functionally implicated in a range of human pathologies, including cystic fibrosis, influenza, bacterial endocarditis, gut dysbiosis, and cancer. These observations have motivated the study of mucin biosynthesis as well as the development of strategies for inhibition of mucin glycosylation. Mammalian pathways for mucin catabolism, however, have remained underexplored. The canonical view, derived from analysis of N-glycoproteins in human lysosomal storage disorders, is that glycan degradation and proteolysis occur sequentially. Here, we challenge this view by providing genetic and biochemical evidence supporting mammalian proteolysis of heavily O-glycosylated mucin domains without prior deglycosylation. Using activity screening coupled with mass spectrometry, we ascribed mucin-degrading activity in murine liver to the lysosomal protease cathepsin D. Glycoproteomics of substrates digested with purified human liver lysosomal cathepsin D provided direct evidence for proteolysis within densely O-glycosylated domains. Finally, knockout of cathepsin D in a murine model of the human lysosomal storage disorder neuronal ceroid lipofuscinosis 10 resulted in accumulation of mucins in liver-resident macrophages. Our findings imply that mucin-degrading activity is a component of endogenous pathways for glycoprotein catabolism in mammalian tissues.
Mammalian cells append glycans to the majority of their secreted and cell surface proteins (1). These extracellular glycoproteins are broadly categorized as Asn-linked N-glycoproteins and Ser/Thr-linked O-glycoproteins, both of which are typically elaborated into branching structures with many monosaccharide units strung together. Therefore, catabolism of a given extracellular protein typically involves hydrolase-mediated breakdown of both its peptide backbone and one or more complex glycans.
Much of our current understanding of glycoprotein catabolism arises from the study of human mutations that cause N-glycoprotein degradation pathways to go awry. Analysis of accumulation products in lysosomal storage disorders such as mannosidosis, aspartylglucosaminuria, sialidosis, Schindler (types I and II), galactosialidosis, and fucosidosis have provided a framework to understand N-glycoprotein catabolism (2). In brief, N-glycoproteins are extensively proteolyzed such that there are free alpha carboxyl and amino groups on the asparagine residue bearing the glycan. After fucose is removed by lysosomal α-L-fucosidase, N-glycanase aspartylglucosaminidase hydrolyzes the glycan-peptide bond. The free glycan can then be broken down from both reducing and nonreducing ends by a variety of hydrolases (2).
Lysosomal degradation of O-glycoproteins is understudied relative to that of N-glycoproteins, in part due to unique difficulties associated with structural analysis of O-glycopeptides (3), and is typically assumed to occur analogously to N-glycoprotein catabolism (2, 4). Mucins are a class of extracellular O-glycoproteins that have challenged this assumption. Mucins are characterized by repeating domains bearing a high frequency of N-acetylgalactosamine (GalNAc)-linked serine and threonine residues, such that the biomolecule as a whole can exceed 50% glycosylation by mass (5). The densely spaced glycans in mucin glycodomains endow them with unique properties, including a rigid, extended secondary structure and resistance to proteolysis (6). As such, mucin catabolism has been suggested to proceed in the reverse order of N-glycan catabolism, involving removal of glycans followed by proteolysis of the peptide backbone (2), or through shedding from cell surfaces into luminal spaces (7). Meanwhile, major histocompatibility complex (MHC) I and MHC II peptides bearing mucin-type O-glycans have been repeatedly observed (8–11), indicating that mucin domains can, under some circumstances, be proteolyzed with their glycans intact.
We and others have characterized proteases from the bacterial kingdom that cleave within densely O-glycosylated mucin domains without prior deglycosylation, termed mucinases (3, 12–14). The existence of bacterial mucinases indicates that access to the peptide backbone through densely spaced O-glycans is not impossible for a proteolytic enzyme. Given the biological ubiquity and clinical significance of mucins (15–20), we set out to systemically evaluate if mammals encode enzyme(s) with proteolytic activity toward glycosylated mucin domains. At the outset, we considered sequence and structure-based approaches to identify candidates from mammalian genome sequences. However, as bacterial mucinases share poor sequence homology (12, 21), we turned to a biochemical strategy.
Results
Our initial goal was to develop an assay for mucin-cleaving activity and then to use that assay for screening of mammalian tissue fractions. For the assay, we required 1) a mucin substrate with near native glycosylation and 2) a readout for mucin proteolysis. We purified the mucin MUC16 from the human ovarian cancer cell line OVCAR-3 for use as the substrate (SI Appendix, Fig. S1A) (22). MUC16, also known as CA-125, is used as a prognostic biomarker for ovarian cancer patients due to its high serum levels and the positive correlation between serum levels and disease burden (23). MUC16 derived from OVCAR-3 cells is approximately 3.5 million Daltons and 24–28% glycosylation by mass (24). The majority of these glycans are core 1 and core 2 O-GalNAc glycans, and many are elaborated with sialic acid and fucose modifications (24). For in vivo imaging and detection by in-gel fluorescence, we labeled purified MUC16 with a fluorophore via N-hydroxysuccinimide chemistry. The resulting material was high molecular weight (MW), sialylated, and stable to incubation overnight at 37 °C in buffers of pH 3 to 9 (SI Appendix, Fig. S1 B and C). Importantly, the MUC16 preparation was resistant to proteolysis by commonly employed proteases such as trypsin and chymotrypsin (6). By contrast, treatment of fluorophore-labeled MUC16 with two bacterial mucinases (3, 21), as well as proteinase K (25), resulted in digestion that was clearly distinguished using a gel shift assay (SI Appendix, Fig. S1D).
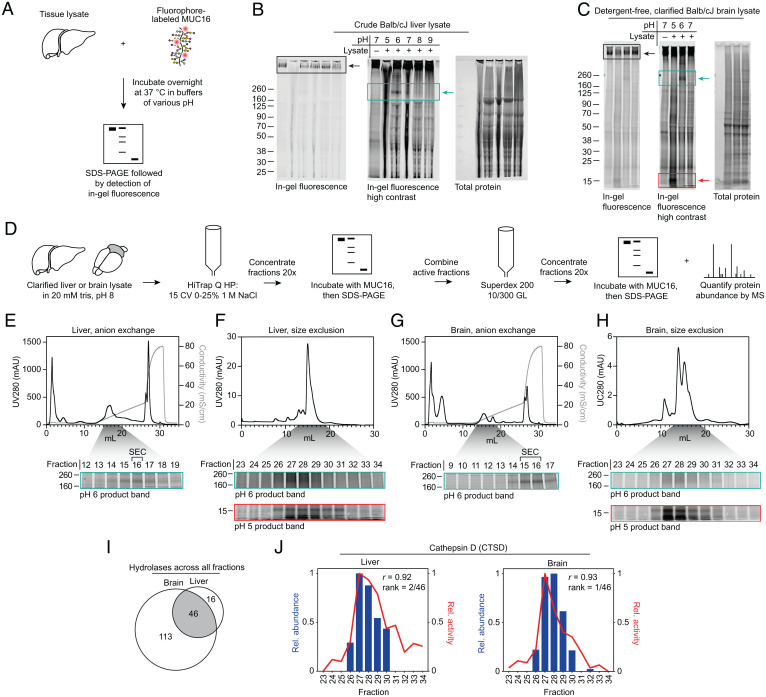
To screen for mucin-cleaving activity, we chose murine tissues for biochemical fractionation and testing. We injected our fluorophore-labeled MUC16 preparation retro-orbitally into nude mice and observed clearance over the course of 24 h by in vivo fluorescence imaging (SI Appendix, Fig. S2A). Imaging of dissected abdominal organs revealed accumulation in the liver, as previously observed (26), and excretion into the gastrointestinal tract (SI Appendix, Fig. S2B). Liver homogenate is a common starting point for biochemical fractionation experiments due to its high protein yield and multitude of enzymatic activities. Freshly dissected liver from Balb/cJ mice was Dounce homogenized in 20 mM Tris, pH 8, supplemented with 1% Nonidet P-40 (NP-40). The crude homogenate was diluted into buffers of various pHs supplemented with common biological ions (Materials and Methods), then incubated with fluorophore-labeled MUC16 overnight at 37 °C. The resulting material was solubilized in protein loading buffer, separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), and analyzed by in-gel fluorescence (Fig. 1A). Despite poor separation of the crude preparation by SDS-PAGE, we observed apparent loss of the high molecular weight substrate band at pH 5 (Fig. 1B, black arrow) and appearance of a band between 160 and 260 kDa at pH 6 (Fig. 1B, cyan arrow).
Fig. 1.
Mucin degradation correlates with cathepsin D abundance in fractionated mouse liver and brain. (A) Schematic of mucin proteolysis assay. (B) Crude liver lysate, generated by Dounce homogenization in 1% NP-40, 20 mM Tris, pH 8, without spin clarification, was incubated with MUC16-680 overnight at 37 °C in buffers with the indicated pH. Proteins were separated by SDS-PAGE, then visualized by in-gel fluorescence. Total protein was visualized with Coomassie stain. (C) Detergent-free, clarified brain lysate generated by Dounce homogenization in 20 mM Tris, pH 8, with 21,000 × g spin clarification was treated as in B. (D) Workflow for biochemical fractionation and fraction analysis, following the general strategy of Ogura et al. (27). (E) Top: UV280 and conductivity traces for liver lysate AEX. Bottom: AEX fractions 12 to 19 (gray shading) of 30 total were subjected to an activity assay at pH 6 (cyan box). (F) Top: UV280 trace for SEC on liver AEX fraction 16. Bottom: SEC fractions 23 to 34 (gray shading) of 48 total were subjected to activity assays at pH 6 (cyan box) and pH 5 (red box). (G) Brain lysate AEX, depicted as in E. (H) SEC of brain lysate AEX fractions, depicted as in F. (I) Hydrolases detected across all fractions, as defined by the GO molecular function annotation, grouped by tissue. (J) Pearson correlation of relative cathepsin D (CTSD) abundance and relative MUC16-degrading activity for each fraction. Abundance was measured using label-free quantification, and activity for liver and brain was measured via densitometry on the pH 5 product band in F, Bottom and H, Bottom, respectively. Black boxes highlight the substrate band; cyan boxes highlight appearance of a product band between 160 and 260 kDa; red boxes highlight appearance of a product band between 8 and 15 kDa. Rel., relative. CV, column volumes. mS/cm, milliSiemens per centimeter. mAU, milli-Absorbance Units.
Optimization of the lysis protocol revealed that the enzyme(s) responsible for both pH 5 and 6 activity were retained in the detergent-free, clarified tissue fraction (SI Appendix, Fig. S3A). To further explore the tissue distribution of the observed activities, we prepared detergent-free, clarified lysates from liver, kidney, spleen, lung, heart, and brain of Balb/cJ and C57BL/6J mice. In both mouse strains, the strongest activity was observed in liver and brain (SI Appendix, Fig. S3 B and C). In high-activity samples such as the brain, upon incubation at pH 5 we consistently observed appearance of an 8- to 15-kDa product band (Fig. 1C, red arrow), which later proved useful for quantification of MUC16-degradation activity.
Anti-MUC16 Western blot using clarified, detergent-free lysate confirmed that pH 5 and 6 product bands were derived from MUC16 (SI Appendix, Fig. S3D). The observed MUC16-degrading activity was dependent on lysate concentration (SI Appendix, Fig. S4A), as well as incubation time (SI Appendix, Fig. S4B). In time course experiments, the band between 160 and 260 kDa was present at pH 5 at shorter incubation times, then apparently degraded further at later time points, implying that the higher molecular weight pH 6 band could be an intermediate cleavage product (SI Appendix, Fig. S4B, cyan arrow).
To isolate the enzyme(s) responsible for MUC16 proteolysis, Balb/cJ mouse liver and brain were subjected to anion exchange chromatography (AEX) using an optimized elution method (Materials and Methods). AEX fractions with the highest MUC16-degrading activity were then separated by size exclusion chromatography (SEC). Fractions from SEC were subjected to another activity assay and mass spectrometry-based proteomics analysis, following the general procedure that was recently applied to discover the identity of the mammalian calcium-dependent N-acyltransferase (26) (Fig. 1D). Activities at pH 5 and 6 were read out by the product bands at 8 to 15 kDa (Fig. 1 E–H, red boxes) and at 160 to 260 kDa (Fig. 1 E–H, cyan boxes), respectively. Activity at pH 5 and 6 eluted at the same chromatographic retention times in both liver and brain lysate for both AEX and SEC, suggesting that a single enzyme could be responsible for the two product bands (for uncropped gels, lower contrast images, and total protein stain, see SI Appendix, Fig. S5).
Proteomics analysis of SEC fractions from both liver and brain identified hundreds of proteins across the fractions (Dataset S1). We found 46 hydrolases, as defined by the Gene Ontology (GO) molecular function annotation, to be common between liver and brain fractions (Fig. 1I). Relative abundance of the 46 proteins across fractions was determined through label-free quantification (Materials and Methods). Relative MUC16-proteolyzing activity across fractions was determined by densitometry on the clearly distinguishable pH 5 product band in Fig. 1 F and H (red boxes). The 46 hydrolases were ranked for both liver and brain via Pearson correlation of relative abundance and relative activity in each fraction (Dataset S2). Cathepsin D, encoded by the ctsd gene, was the highest ranked protein based on this analysis (Fig. 1J).
Cathepsin D is a soluble aspartic acid endopeptidase found across tissues of the body (28). It is synthesized in the endoplasmic reticulum as a 52-kDa precursor, then undergoes proteolytic maturation and modification with mannose-6-phosphate bearing N-glycans in the Golgi apparatus. Upon acidification in endosomes and lysosomes, the proprotein is proteolytically processed again to a 34-kDa heavy chain and a 14-kDa light chain that remain noncovalently associated and are present in lysosomes at up to 1 mM (29). Cathepsin D is reported to hydrolyze proteins with a pH optimum of 3.5 to 5, with preference for hydrophobic amino acids at the scissile bond (30). Loss-of-function mutations in the CTSD gene result in the human lysosomal storage disease neuronal ceroid lipofuscinosis 10 (CLN10), the most severe and early onset of the neuronal ceroid lipofuscinoses (31).
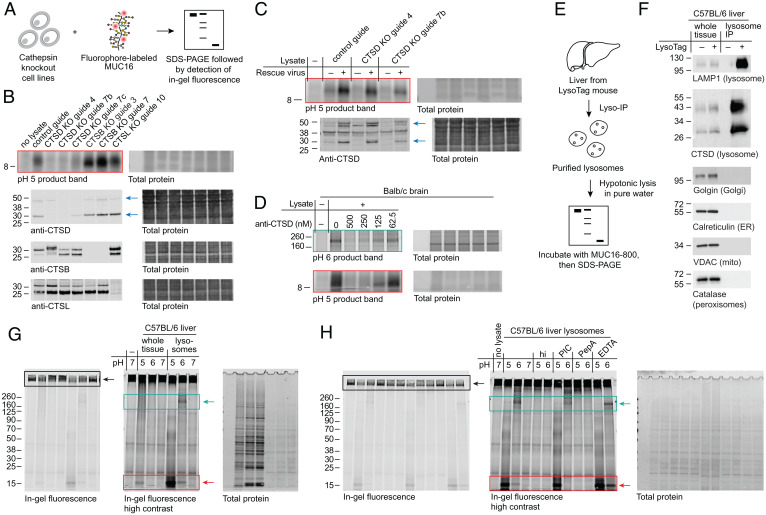
To validate cathepsin D as the enzyme responsible for the observed MUC16-degrading activity, we generated CRISPR-Cas9–mediated knockouts of CTSD and related cathepsins in the 8988T human pancreas adenocarcinoma cell line (Fig. 2A). Absence of cathepsin D protein resulted in complete loss of MUC16 degradation in 8988T lysate (Fig. 2B). A clone exhibiting a partial knockdown of CTSD (guide 7c) exhibited proportional decrease in activity against MUC16, and knockout of CTSB and CTSL had no apparent effect. Infection of knockout cell lines derived from two different guides with a cathepsin D-encoding lentiviral vector rescued loss of activity in lysate against MUC16 (Fig. 2C). Infection of the control guide cell line with the same vector resulted in an increase in cathepsin D expression alongside an increase in MUC16-degrading activity. Addition of polyclonal anti-cathepsin D antibodies to mouse brain lysate inhibited MUC16 proteolysis at pH 5 and 6 in a dose-dependent manner, further supporting the dependence of cleavage bands on cathepsin D activity (Fig. 2D). For uncropped gels corresponding to Fig. 2 B–D and F, see SI Appendix, Figs. S6 and S7.
Fig. 2.
Knockout and inhibition of cathepsin D in cellulo and in purified mouse liver lysosomes support degradation of MUC16 by lysosomal cathepsin D. (A) Schematic of mucin proteolysis assay with cathepsin knockout cell lines. (B) Activity assay at pH 5 (red box) and immunoblots of lysates derived from knockouts (KOs) of cathepsin D (CTSD), cathepsin B (CTSB), and cathepsin L (CTSL) in 8988T cells. See SI Appendix, Supplementary Methods for guide sequences. CTSD KO guide 4 appears to alter CTSD processing, possibly due to coregulation of these enzymes (32). Anti-cathepsin D antibodies are raised against the 34-kDa heavy chain of cathepsin D. Blue arrows indicate the ∼50-kDa cathepsin D proprotein and the ∼30-kDa heavy chain of the proteolytically processed form. (C) Activity assay at pH 5 (red box) and anti-CTSD immunoblot of 8988T control and CTSD knockout cells with and without rescue using a cathepsin D expression lentivirus (rescue virus). Blue arrows indicate the ∼50-kDa cathepsin D proprotein and the ∼30-kDa heavy chain of the proteolytically processed form. (D) Detergent-free, clarified mouse brain lysate was treated with various concentrations of a polyclonal goat antibody with specificity to mouse cathepsin D, then subjected to activity assays at pH 6 (cyan box) and pH 5 (red box). (E) Schematic of mucin proteolysis assay with purified mouse liver lysosomes. (F) Immunoblots of organelle markers comparing whole tissue lysate and Lyso-Tag IPs of livers from mice with and without the Lyso-Tag (33). (G) Lysosomes were purified from C57BL/6 mouse liver as described in Materials and Methods. Whole tissue lysate (input for lysosome enrichment) and purified lysosomes were subjected to activity assays at pH 5, 6, and 7. Densitometry of pH 5 product bands normalized to total protein indicates a 107-fold enrichment in lysosomal activity relative to whole tissue activity. (H) Lysosomes purified as in G were subjected to activity assays with heat inactivation at 65 °C for 10 min (hi), addition of EDTA-free protease inhibitor cocktail (PIC), addition of 10 µM PepA, or addition of 25 mM EDTA. Buffer pH was corrected after addition of each inhibitor. Black boxes highlight the substrate band; cyan boxes highlight appearance of a product band between 160 and 260 kDa; red boxes highlight appearance of a product band between 8 and 15 kDa. ER, endoplasmic reticulum. IP, immunoprecipitation. VDAC, voltage-dependent anion channel 1. Mito, mitochondria.
Given the observed acidic pH optimum for MUC16 degradation, we hypothesized that lysosomal cathepsin D was chiefly responsible for the observed activity in cell lysate. Using the Lyso-Tag method (33, 34), we purified lysosomes from C57BL/6 mouse liver and lysed them in pure water for activity assays or 1% Triton for immunoblotting (Materials and Methods and Fig. 2E). Immunoblots of markers for various organelles confirmed the purity of the lysosome preparations as well as enrichment of cathepsin D (Fig. 2F). Both pH 5 and 6 activity in lysosomes was increased relative to whole tissue homogenate from the same animal (Fig. 2G). Inhibition of cysteine and serine proteases with ethylenediaminetetraacetic acid (EDTA)–free protease inhibitor cocktail (PIC) and inhibition of metalloproteases with 25 mM EDTA had no effect on MUC16 proteolysis in lysosome extract, but heat inactivation of the extract or addition of 10 µM pepstatin A (PepA), a highly selective aspartic acid protease inhibitor, completely abrogated activity (Fig. 2H). These data provide genetic and biochemical evidence for degradation of MUC16 by the lysosomal form of cathepsin D.
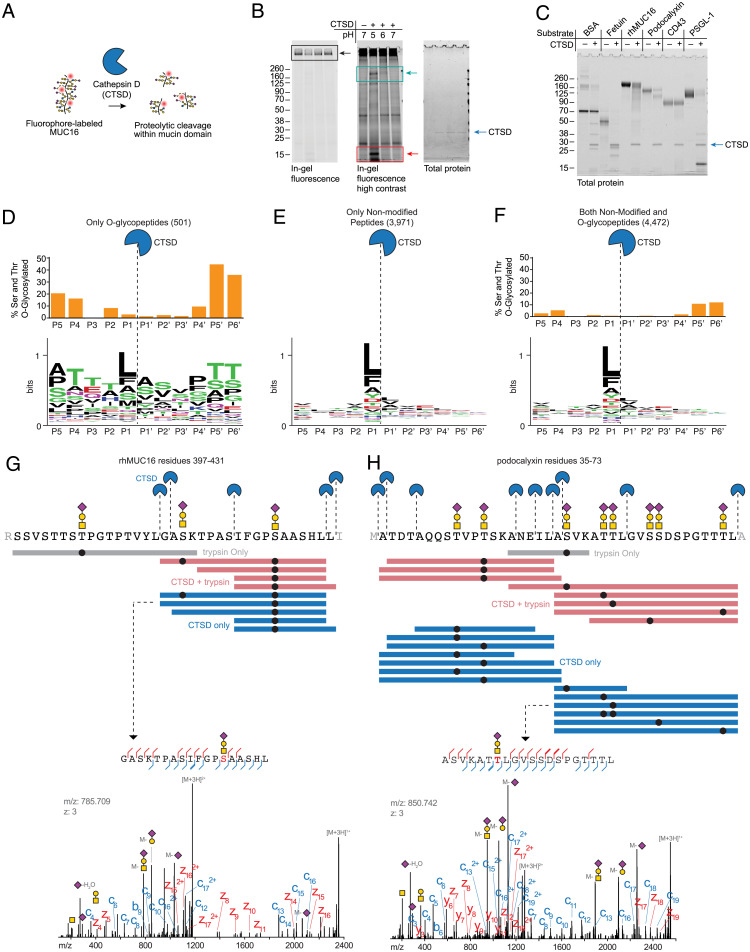
The lysosomal form of cathepsin D purified from human liver was obtained from a commercial source and incubated at 50 nM with MUC16 in buffers of various pHs (Fig. 3 A and B). The banding pattern of MUC16 degradation matched closely with what was observed in liver and brain (compare to Fig. 1C), implying that human lysosomal cathepsin D, in addition to murine lysosomal cathepsin D, directly digests the mucin substrate MUC16. Densitometry of the high molecular weight substrate band and 8- to 15-kDa product bands at pH 5 indicated ∼40% substrate loss over 24 h, which is consistent with the 20-h half-life of cellular mucins reported in primary murine cells (SI Appendix, Fig. S8) (36).
Fig. 3.
Purified human cathepsin D cleaves MUC16 and other mucins within their densely glycosylated mucin domains. (A) Schematic of mucin proteolysis assay with lysosomal cathepsin D purified from human liver. (B) A commercial preparation of cathepsin D purified from human liver (Molecular Innovations) was incubated at 50 nM with MUC16-800 at 37 °C overnight. Proteins were separated by SDS-PAGE and visualized by in-gel fluorescence followed by Coomassie (total protein). Black boxes highlight the substrate band; cyan boxes highlight appearance of a product band between 160 and 260 kDa; red boxes highlight appearance of a product band between 8 and 15 kDa. (C) Protein substrates were incubated with 500 nM human cathepsin D at pH 5 overnight at 37 °C. Proteins were separated by SDS-PAGE, then stained with Coomassie for total protein. (D–F) Cleavage motif of cathepsin D as determined by glycoproteomic analysis on the substrates shown in C (Dataset S3). WebLogo was used for logo plot generation (35). The percentage of O-glycosylated serine and threonine residues in peptide identifications resulting from CTSD-derived cleavage was determined by counting the number of modified residues at a given position relative to the total number of residues (SI Appendix, Supplementary Methods) and is shown as bar graphs above the logos for O-glycopeptides (D), nonmodified O-glycopeptides (E), and an aggregate of both (F). (G) Top: Visualization of cathepsin D cleavage sites in rhMUC16 residues 397 to 431 (see SI Appendix, Supplementary Methods for sequence). Purple diamond, sialic acid; yellow circle, galactose; yellow square, N-acetylgalactosamine; black circle, glycosylation site. Colored bars represent individual detected peptides from trypsin only, CTSD + trypsin, and CTSD only samples. Bottom: annotated spectrum for the indicated MUC16 peptide. (H) Top: visualization of cathepsin D cleavage sites in podocalyxin residues 35 to 73, represented as in E. Bottom: annotated spectrum for the indicated podocalyxin peptide.
We next asked whether human lysosomal cathepsin D cleaves MUC16 and other mucins within their densely O-glycosylated domains. Using methods previously employed for bacterial mucinases (3, 21), we determined the cleavage specificity of cathepsin D using a panel of mucin and nonmucin substrates. Bovine serum albumin (BSA; nonglycosylated), fetuin (sparsely O-glycosylated), and a panel of mucins, including recombinant human MUC16 (rhMUC16), podocalyxin, CD43, and P-selectin glycoprotein ligand 1 (PSGL-1), were digested with cathepsin D (Fig. 3C; see SI Appendix, Fig. S9 for additional substrates). The products were then analyzed using mass spectrometry (SI Appendix, Supplementary Methods and Dataset S3). We generated cleavage motifs using only O-glycopeptides (Fig. 3D), only nonmodified peptides (Fig. 3E), and both nonmodified and O-glycopeptides (Fig. 3F). Frequency of detected serine and threonine O-glycosylation is shown as bar graphs above each residue in the cleavage motifs. Confirming previous reports (37), these data supported broad and nonspecific cleavage propensity with a slight preference for hydrophobic residues at the P1 position. Importantly, glycosylation was permitted at the regions near the cleavage site, and analysis of glycopeptide spectra confirmed cleavage events within densely glycosylated mucin domains (Fig. 3 G and H). While cathepsin D is active at heavily O-glycosylated sequences, it should be noted that O-glycosylated serine and threonine residues were found at a reduced relative frequency near the scissile bond, indicating that glycans may not be preferred directly adjacent to the site of cleavage (Fig. 3H, Top). Indeed, enzymatically deglycosylated mucin substrates were cleaved more efficiently by purified human lysosomal cathepsin D (SI Appendix, Fig. S10). Taken together, these data suggest that in vitro, cathepsin D is not selective for mucins (see cleavage of BSA and fetuin, Fig. 3C) but is tolerant of dense O-glycosylation near the proteolytic site.
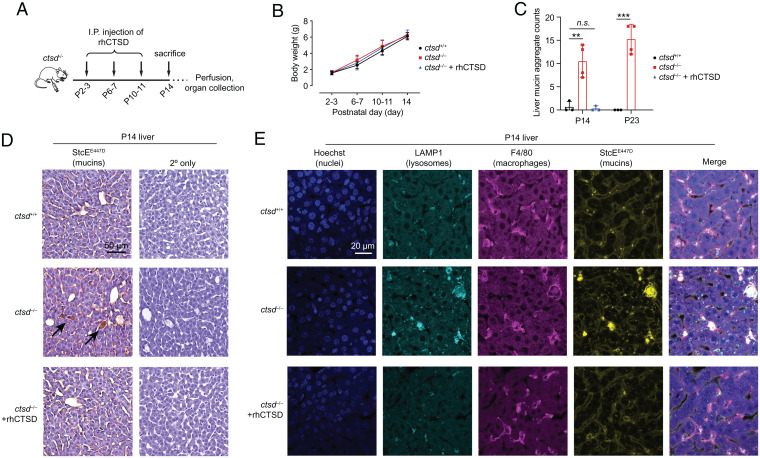
To explore whether cathepsin D is a critical enzyme in lysosomal mucin catabolism in mouse liver, we tested for mucin accumulation upon loss of cathepsin D. A mouse model of neuronal ceroid lipofuscinosis 10 (CLN10), caused by a deficiency of cathepsin D, recapitulates the human disease, which results in lethality soon after birth (37). Homozygous cathepsin D knockout animals develop normally during the first two weeks of life, stop thriving in the third week, and die at day 26 ± 1 (38). At postnatal day 14 (P14), cathepsin D knockout mice (ctsd−/−) have no gross morphological or behavioral changes relative to wild-type (ctsd+/+) littermates, but upon histological examination, they exhibit liver abnormalities stereotyped by the presence of large, apparently empty spaces in the tissues (38). In these animals, histological features resulting from cathepsin D knockout can be prevented by repeated administration of recombinant human procathepsin D (rhCTSD) (39) (Fig. 4A).
Fig. 4.
A mouse model of CLN10 caused by loss of cathepsin D activity exhibits mucin accumulation in tissues. (A) Schematic of dosing schedule for rescue of CLN10 via administration of rhCTSD. (B) Weights of wild-type (ctsd+/+), cathepsin D knockout (ctsd−/−), and rescued (ctsd−/− + rhCTSD) animals through P14. Error bars are SD. (C) Quantification of mucin aggregates in livers of P14 ctsd+/+, ctsd−/−, and ctsd−/− + rhCTSD animals and P23 ctsd+/+ and ctsd−/− animals. Replicates shown are from individual mice (see Materials and Methods and SI Appendix, Figs. S11–S13). Error bars are SD. **P < 0.01, ***P < 0.001, n.s., P > 0.05. (D) Representative images of fixed and paraffin-embedded liver from P14 ctsd+/+, ctsd−/−, and ctsd−/− + rhCTSD animals stained for mucins using Biotin-StcEE447D and counterstained with hematoxylin. Control sections from the same tissues omit Biotin-StcEE447D (2° only). For all images, see SI Appendix, Figs. S11–S13. (E) Fixed and paraffin-embedded liver slices from P14 ctsd+/+, ctsd−/−, and ctsd−/− + rhCTSD animals were dewaxed, hydrated, and subjected to antigen retrieval and permeabilization (Materials and Methods). Tissues were stained with the markers shown and imaged by confocal microscopy, taking 15 to 18 Z-stacks with 1-µm step size. Representative maximum intensity projections are shown with brightness and contrast normalized across images. For all images, see SI Appendix, Fig. S28. I.P., intraperitoneal.
Wild-type animals (n = 3), cathepsin D knockout animals (n = 4), and rhCTSD-rescued animals (n = 3) did not differ in weight through P14, consistent with prior observations (Fig. 4B) (39). Histological sections of liver, brain, spleen, lung, and intestine from all animals were prepared, and mucins were stained using a catalytically inactivated bacterial mucin-degrading enzyme (StcEE447D) (21). A separate set of wild-type (n = 3) and cathepsin D knockout (n = 4) animals were euthanized at P23, near the end of the expected lifetime for knockout animals, and their tissues were prepared in the same way. Accumulation of mucins was observed in CTSD knockout liver at P14 and P23 but was absent in livers of rhCTSD-rescued P14 animals (Fig. 4 C and D and SI Appendix, Figs. S11–S13). The same phenotype was observed in spleen and not observed in any of the other examined tissues (SI Appendix, Figs. S11 and S14–S21). Notably, conclusions are limited to the tissue regions that were explored, for which at the imaging resolution employed, only liver and spleen exhibited storage material plaques. Mucin accumulation was absent in tissues prepared from mice homozygous for loss of cln3 (cln3−/−), the causative mutation for a different neuronal ceroid lipofuscinosis, CLN3 (40) (SI Appendix, Figs. S22–S27), highlighting that mucin buildup is not a general phenotype of neuronal ceroid lipofuscinoses.
Next, we searched for cell type-specific mucin accumulation. We focused on liver cells rather than spleen cells, as cathepsin D knockout liver tissue exhibited robust mucin accumulation at P14 (SI Appendix, Fig. S12) prior to onset of disease symptoms, suggesting that the mucin buildup was a primary effect of cathepsin D loss rather than a secondary effect of tissue damage in this organ. Enlarged liver-resident macrophages, also called Kupffer cells, had been previously reported in cathepsin D knockout animals (39). Given that mucin accumulation in liver sections appeared to be concentrated inside enlarged cells (see Fig. 4D, unstained sections), we assayed tissue sections from P14 wild-type, knockout, and rescued animals for markers of macrophages (F4/80), lysosomes (LAMP-1), and mucins (StcEE447D) by immunofluorescence microscopy (Fig. 4E and SI Appendix, Fig. S28). Colocalization of StcEE447D-positive signal with LAMP-1–positive signal in F4/80-positive cells suggested that liver macrophages were the primary reservoir for the observed mucin accumulation. Thus, lysosomal cathepsin D is critical for mucin catabolism in a subset of liver-resident cells.
Discussion
In summary, we have identified a mode by which mammalian cells proteolyze mucin domains without prior deglycosylation. Further work is needed to determine the pathophysiological relevance, if any, of cathepsin D knockout-mediated mucin accumulation in the course of CLN10 progression and, by extension, in healthy development. Advances in tissue-level O-glycoproteomics (41) will be needed to determine the specific mucins that accumulate in ctsd-deficient animals, as mucin domains are found across too many proteins in the mammalian proteome to allow individual screening with antibodies (42).
In addition, our findings should spur further efforts to elucidate pathways responsible for O-glycoprotein catabolism in mammals. Indeed, our search for human mucinase activity was largely focused on the murine liver. Given that we did not observe mucin accumulation broadly across tissues in cathepsin D knockout animals (SI Appendix, Figs. S11–S21), we expect other proteases may be capable of digesting mucin glycodomains. For example, gel shifts of bulk mucin preparations by the secreted enzyme neutrophil elastase have been previously observed (43), and degradation of lubricin, a glycoprotein with a mucin-like domain, by cathepsin G has been recently reported (44). Second, we were initially motivated to search for mammalian mucinases in part due to repeated reports of MHC I and MHC II peptides bearing mucin-type O-glycans (8–11), which indicated to us that mucin glycodomains could, under some circumstances, be proteolyzed with their glycans intact. Further work, likely involving functional genomic screening, is needed to elucidate the mechanisms by which such MHC loading may occur.
Finally, cathepsin D’s mucin-degrading capability may play a role in the progression of mucin-overexpressing carcinomas. Cathepsin D is reported to be secreted in both preprocessed and fully processed forms by cancer cells and tumors, with its levels correlating with aggressiveness in breast and ovarian cancer (45). It is interesting to speculate that cathepsin D may be involved in the breakdown of extracellular mucins, especially in acidic tumor microenvironments. Additionally, the existence of a human enzyme that can cleave mucin glycodomains may aid therapeutic approaches for targeted degradation of mucins (46) in disorders such as mucinous carcinomas (47) and cystic fibrosis (15), in which mucin accumulation is functionally tied to disease severity and progression.
Materials and Methods
Tissue Homogenization.
All wild-type tissues were collected from Balb/cJ or C57BL/6J mice scheduled for euthanization under Stanford University Administrative Panel on Laboratory Animal Care (APLAC) protocol No. 31511. Liver, kidney, spleen, lung, heart, and brain were collected after euthanization by CO2 asphyxiation and either stored at −80 °C or processed fresh. Organs were weighed, minced with a razor, and then resuspended in 1 mL ice-cold 20 mM Tris, pH 8, per ∼200 mg tissue. The slurry was Dounce homogenized on ice using 60 strokes with pestle A (0.0030- to 0.0050-inch clearance), then passed through a 70-µm mesh. The resulting slurry was Dounce homogenized on ice using 60 strokes with pestle B (0.0005- to 0.0025-inch clearance) and then spun at 21,000 × g for 20 min at 4 °C. Clarified supernatant was passed through a Target2 PES 0.45-µm filter (Thermo Fisher Scientific, F2500-14). The resulting lysate was concentrated to a final volume of 1 mL per ∼1 g of starting tissue using Amicon Ultra 10,000 NMWL filters (Millipore Sigma, UFC801024), then used fresh for fractionation or activity assays or frozen at −80 °C.
Activity Assays.
MUC16 and MUC2 purification and labeling are described in SI Appendix. Recombinant MUC1-Fc chimera protein was purchased from R&D Systems (10332-MU). Buffers of various pH were made following the recipes of Li et al. (48). These were 100 mM NaOAc, pH 5; 100 mM NaOAc, pH 6; 100 mM N-2-hydroxyethylpiperazine-N′-2-ethane sulfonic acid (HEPES), pH 7; 100 mM Tris⋅HCl, pH 8; and 100 mM Tris⋅HCl, pH 9. In early experiments, these buffers were supplemented with common biological ions: 150 mM KCl, 2 mM MgCl2, 2 mM CaCl2, and 200 µM ZnCl2. These ions were omitted upon observation that the activity was not inhibited by 25 mM EDTA in ion-free buffer. Activity assays were performed in PCR tubes with 9 µL buffer, 2 µL ∼10 mg/mL tissue or cell lysate in 20 mM Tris, pH 8, and 1 µL MUC16-fluorophore in phosphate-buffered saline (PBS), pH 7. Reactions were incubated in a PCR block at 37 °C for the indicated time periods, then quenched by addition of 4 µL NuPAGE lithium dodecyl sulfate (LDS) Sample Buffer (4×) (Thermo Fisher Scientific) supplemented with 250 mM dithiothreitol (DTT). Samples were boiled for 5 min at 95 °C, then loaded into 18- or 26-well 4–12% Criterion XT Bis-Tris gels (Bio-Rad). Gels were run in 2-(N-morpholino)ethanesulfonic acid (MES) buffer for separation of low MW proteins and protein fragments or 3-(N-morpholino)propanesulfonic acid (MOPS) buffer for separation of high MW proteins and protein fragments. In-gel fluorescence was quantified using a LI-COR Odyssey Blot Imager. Total protein was stained using AcquaStain Coomassie stain (Bulldog Bio) and visualized either with a bright field gel imager or a LI-COR Odyssey Blot Imager. For antibody blocking assays, Balb/cJ brain lysate prepared as described above was supplemented with goat polyclonal anti–cathepsin D (Novus Biologicals, AF1029) to a final concentration of 500 nM, 250 nM, 125 nM, 62.5 nM, and 0 nM (control). Inhibitor experiments were performed with UltraPure 0.5 M EDTA, pH 8.0 (Thermo Fisher Scientific), PepA (Selleck Chemicals), and cOmplete EDTA-free Protease Inhibitor Mixture (Sigma-Aldrich). Heat inactivation of lysosome extract was performed in a PCR tube at 65 °C for 10 min. StcE, AM0627, and sialidase were expressed in-house as previously described (3, 21, 49) and used at 50 nM. Trypsin (Promega), chymotrypsin (Promega), AspN (New England Biolabs), GluC (Sigma-Aldrich), and proteinase K (Promega) were used at 50 nM. Enzymatic deglycosylation was performed using Deglycosylation Mix II (New England Biosciences) following manufacturer’s instructions for native substrate deglycosylation.
Western Blotting.
Lysates were subjected to SDS-PAGE and transferred to nitrocellulose membranes. Total protein was quantified using REVERT stain (Li-Cor Biosciences). Primary antibodies were used according to manufacturer recommendations: anti-MUC16 [X75] (Novus Biologicals, NB6001468), anti-human cathepsin D (Cell Signaling, #2284), anti-mouse cathepsin D (Novus Biologicals, AF1029), anti-cathepsin B [D1C7Y] (Cell Signaling, #31718), anti-cathepsin L [33/2] (Santa Cruz, sc-32320), anti-LAMP1 [1D4B] (Developmental Studies Hybridoma Bank), anti-VDAC1 (Cell Signaling, #4661), anti-GOLGIN-97 (Cell Signaling, #13192), anti-CALR (Cell Signaling #12238), and anti-CATLASE (Cell Signaling, #14097). Secondary antibodies were used according to manufacturer recommendations.
Tissue Fractionation.
Balb/cJ liver lysate (1 mL, 24 mg/mL) and Balb/cJ brain lysate (1 mL, 13 mg/mL) prepared as described above were subjected to AEX using 1 mL HiTrap Q HP columns (Cytiva Life Sciences) run on an ÄKTA pure FPLC. Bound proteins were eluted with a 15-column volume linear gradient from 0 to 25% 1 M NaCl in 20 mM Tris, pH 8. Fractions (0.5 mL) were concentrated to minimum volume (∼20 µL) using Amicon Ultra 0.5 mL 10,000 NMWL centrifugal filter unit (Millipore Sigma, UFC501096), then subjected to activity assays as described above. Fractions with the greatest activity were diluted to ∼200 µL with 20 mM Tris, pH 8, then subjected to SEC using a Superdex 200 10/300 GL (Cytiva Life Sciences) run on an ÄKTA Pure fast protein liquid chromatography (FPLC). Running buffer was 20 mM Tris, 150 mM NaCl, pH 8. Fractions (0.5 mL) were concentrated to minimum volume (∼20 µL) using Amicon Ultra 0.5-mL 10,000 NMWL centrifugal filter unit (Millipore Sigma, UFC501096), then subjected to activity assays as described above and mass spectrometry as described below. Mass spectrometry of tissue fractions is described in SI Appendix.
Pearson Correlation of Tissue Fractions.
Label-free quantitation of protein intensity values across fractions was performed using Perseus v 1.6.2.2 (50). Activity across fractions was assessed by densitometry on the product band of the activity gels shown in Fig. 1F (liver, pH 5) and Fig. 1H (brain, pH 5). Pearson’s two-sided product-moment correlation comparing relative protein intensity and relative activity across fractions was employed to give P-values for liver and brain.
Lyso-Tag Immunoprecipitation (IP) of Mouse Liver.
Lyso-Tag–expressing mice were generated via cross of Rosa26; lox-stop-lox-Tmem192-3xHA with CMV-Cre transgenic mice of the C57BL/6J background (The Jackson Laboratory, strain no. 035401) (33, 34). Tissues were collected from Lyso-Tag mice under Stanford APLAC protocol No. 33464. Following euthanasia, a small round piece (4-mm diameter) of mouse liver was obtained and gently homogenized in 1 mL ice-cold PBS supplemented with protease and phosphatase inhibitor mixtures (Roche) using 25 strokes. Homogenate was collected, and 25 μL was saved on ice as the input fraction while the remaining was spun down at 1,000 × g for 2 min at 4 °C. The supernatant was then incubated with anti-HA (Thermo Fisher Scientific) beads for 15 min on a gentle rotator shaker 4 °C and followed by three rounds of washing in the same cold buffer. The input fraction and lysosomal fraction were resuspended in ice-cold Triton lysis buffer (40 mM Hepes, pH 7.4, 1% Triton X-100, 10 mM β-glycerol phosphate, 10 mM pyrophosphate, 2.5 mM MgCl2, and protease and protease inhibitor mixtures) for Western blots or pure water without additives for activity assays. The input fraction was incubated for 10 min at 4 °C and centrifuged at maximum speed. The lysosomal fraction was incubated on the magnet and transferred into a new tube.
Treatment of CLN10 Knockout Mice with rhCTSD.
Animal handling and care were performed in agreement with the German animal welfare law according to the guidelines of the Christian-Albrechts-University of Kiel and the University Medical Center Hamburg-Eppendorf. Experiments involving animals were approved by the Ministry of Energy, Agriculture, the Environment and Rural Areas Schleswig-Holstein under the reference number V242–40536/2016 (81–6/16). Animal experiments were performed as previously described (39). Briefly, mice were genotyped at P0. Ctsd−/− and ctsd+/+ littermates were selected and dosed intraperitoneally with 50 mg/kg of recombinant human proCTSD between P2 and P3. Next, animals received another two intraperitoneal (I.P.) injections of 25 mg/kg at P6-7 and P10-11 and were euthanized at P14. At P14 to 23, mice were anesthetized by intraperitoneal injection of 10 mg/mL ketamine (Bremer Pharma GmbH, 26706) and 6 mg/mL Rompun (Bayer, KPOCCNU) in 0.9% (wt/vol) NaCl solution and transcardially perfused with 0.1 M phosphate buffer. The collected organs were postfixed in 4% paraformaldehyde for 72 h.
Immunohistochemistry.
Ctsd−/−, ctsd+/+, and procathepsin D rescued ctsd−/− animals were obtained as described above. Cln3−/− animals were purchased from The Jackson Laboratory (strain no. 029471, B6.129S6-Cln3tm1Nbm/J) under Stanford APLAC protocol No. 33464. In both cases, mouse tissues were fixed in 4% paraformaldehyde for 72 h prior to paraffin embedding and sectioning. Tissue slides were dried in an oven at 62 °C for 1 h. Slides were dewaxed in xylene (3 × 5 min) and then hydrated in a 100%, 95%, 75% ethanol gradient (2 × 3 min each). Antigen retrieval was performed in Antigen Unmasking Solution, Citric Acid Based (Vector Laboratories) for 15 min. Enzymatic treatment of tissue slides with 50 nM StcE was performed for 20 h at room temperature in PBS. For mucinase staining, endogenous peroxidases were quenched for 10 min at room temperature in Hydrogen Peroxide Blocking Reagent (Abcam). Slides were washed in PBS-T (0.05% Tween-20) and incubated in Carbo-Free Blocking Solution (Vector Laboratories) for 30 min at room temperature. Biotin-StcEE447D (10 μg/mL) was applied to slides in PBS at room temperature for 30 min. Following two washes with PBS-T, tissues were incubated with streptavidin–horseradish peroxidase (HRP) (Abcam) (1:10,000) in PBS for 30 min at room temperature. Slides were washed three times with PBS-T, and mucinase substrates were visualized using the DAB Substrate Kit (Abcam) according to manufacturer’s recommendations. Tissues were counterstained with hematoxylin (Abcam), dehydrated in a 75%, 95%, 100% ethanol gradient (2 × 3 min each), and cleared in xylene (3 × 5 min). Cover slides were mounted with Vectamount Permanent Mounting Medium (Vector Laboratories) prior to imaging using a Leica DM2000 microscope. Quantification of mucin aggregates and confocal microscopy are described in SI Appendix.
Supplementary Material
Acknowledgments
We thank Dr. Jonathan Z. Long (Stanford University) for foundational discussions. We thank Dr. Melissa Gray (Stanford University) for expression and purification of sialidases. We thank Dr. Natalia Vélez Alicea (Stanford University) for help with statistical analysis. This work was supported in part by National Cancer Institute Grant R01CA227942 (to C.R.B.). K.P. was supported by a US NSF Graduate Research Fellowship, a Stanford Graduate Fellowship, and the Stanford ChEM-H Chemistry/Biology Interface Predoctoral Training Program. D.J.S. was supported by a US NSF Graduate Research Fellowship and Stanford Graduate Fellowship. N.M.R. was funded through an NIH Predoctoral to Postdoctoral Transition Award (Grant K00 CA212454). P.S. was supported through the Deutsche Forschungsgemeinschaft (SFB877-A3 and Z3). N.N.L. and M.A.-R. were supported by BeatBatten (Netherlands) and Neuronal Ceroid Lipofuscinosis (NCL)-Stiftung (Germany) foundations. M.A-R. is a Terman Faculty Fellow at Stanford University.
Footnotes
Competing interest statement: M.A.-R. is a scientific advisory board member of Lycia Therapeutics. C.R.B. is a cofounder and scientific advisory board member of Lycia Therapeutics, Palleon Pharmaceuticals, Enable Bioscience, Redwood Biosciences (a subsidiary of Catalent), OliLux Bio, Grace Science LLC, and InterVenn Biosciences.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2117105119/-/DCSupplemental.
Data Availability
Mass spectrometry raw files have been deposited in ProteomeXchange (PXD027616) (51).
References
- 1.Steentoft C., et al. , Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. EMBO J. 32, 1478–1488 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Winchester B., Lysosomal metabolism of glycoproteins. Glycobiology 15, 1R–15R (2005) [DOI] [PubMed] [Google Scholar]
- 3.Malaker S. A., et al. , The mucin-selective protease StcE enables molecular and functional analysis of human cancer-associated mucins. Proc. Natl. Acad. Sci. U.S.A. 116, 7278–7287 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Varki A., et al., Eds., Essentials of Glycobiology (Cold Spring Harbor Laboratory Press, 2015). [PubMed] [Google Scholar]
- 5.Hattrup C. L., Gendler S. J., Structure and function of the cell surface (tethered) mucins. Annu. Rev. Physiol.70, 431–457 (2008). [DOI] [PubMed] [Google Scholar]
- 6.Madsen J. B.,Svensson B.,Abou Hachem M.,Lee S., Proteolytic degradation of bovine submaxillary mucin (BSM) and its impact on adsorption and lubrication at a hydrophobic surface. Langmuir 31, 8303–8309 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Kim Y. J., Borsig L., Han H.-L., Varki N. M., Varki A., Distinct selectin ligands on colon carcinoma mucins can mediate pathological interactions among platelets, leukocytes, and endothelium. Am. J. Pathol. 155, 461–472 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vlad A. M., et al. , Complex carbohydrates are not removed during processing of glycoproteins by dendritic cells: Processing of tumor antigen MUC1 glycopeptides for presentation to major histocompatibility complex class II-restricted T cells. J. Exp. Med. 196, 1435–1446 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Apostolopoulos V., et al. , A glycopeptide in complex with MHC class I uses the GalNAc residue as an anchor. Proc. Natl. Acad. Sci. U.S.A. 100, 15029–15034 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ninkovic T., et al. , Identification of O-glycosylated decapeptides within the MUC1 repeat domain as potential MHC class I (A2) binding epitopes. Mol. Immunol. 47, 131–140 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Malaker S. A., et al. , Identification of glycopeptides as posttranslationally modified neoantigens in leukemia. Cancer Immunol. Res.5, 376–384 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Noach I., et al. , Recognition of protein-linked glycans as a determinant of peptidase activity. Proc. Natl. Acad. Sci. U.S.A. 114, E679–E688 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haurat M. F., et al. , The glycoprotease CpaA secreted by medically relevant acinetobacter species targets multiple O-linked host glycoproteins. MBio 11, e02033-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang W., Ao M., Hu Y., Li Q. K., Zhang H., Mapping the O-glycoproteome using site-specific extraction of O-linked glycopeptides (EXoO). Mol. Syst. Biol. 14, e8486 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kreda S. M., Davis C. W., Rose M. C., CFTR, mucins, and mucus obstruction in cystic fibrosis. Cold Spring Harb. Perspect. Med. 2, a009589 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Delaveris C. S., Webster E. R., Banik S. M., Boxer S. G., Bertozzi C. R., Membrane-tethered mucin-like polypeptides sterically inhibit binding and slow fusion kinetics of influenza A virus. Proc. Natl. Acad. Sci. U.S.A. 117, 12643–12650 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bensing B. A., et al., Recognition of specific sialoglycan structures by oral streptococci impacts the severity of endocardial infection. PLoS Pathog. 15, e1007896 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hansson G. C., Mucins and the microbiome. Annu. Rev. Biochem. 89, 769–793 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pinho S. S., Reis C. A., Glycosylation in cancer: Mechanisms and clinical implications. Nat. Rev. Cancer 15, 540–555 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Sun X.-L., Chemical regulation of glycosylation processes. Trends Glycosci. Glycotechnol. 30, E179–E193 (2018). [Google Scholar]
- 21.Shon D. J., et al. , An enzymatic toolkit for selective proteolysis, detection, and visualization of mucin-domain glycoproteins. Proc. Natl. Acad. Sci. U. S. A. 117, 21299–21307 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wong N. K., et al. Characterization of the Oligosaccharides Associated with the Human Ovarian Tumor Marker CA125. J. Biol. Chem. 278, 28619–28634 (2003). [DOI] [PubMed] [Google Scholar]
- 23.Suh K. S., et al. , Ovarian cancer biomarkers for molecular biosensors and translational medicine. Expert Rev. Mol. Diagn. 10, 1069–1083 (2010). [DOI] [PubMed] [Google Scholar]
- 24.Goodell C. A. R., et al. , Characterization of the tumor marker muc16 (CA125) expressed by murine ovarian tumor cell lines and identification of a panel of cross-reactive monoclonal antibodies. J. Ovarian Res. 2, 8 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tytgat K. M., Opdam F. J., Einerhand A. W., Buller H. A., Dekker J., MUC2 is the prominent colonic mucin expressed in ulcerative colitis. Gut 38, 554–563 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wahrenbrock M. G., Varki A., Multiple Hepatic Receptors Cooperate to Eliminate Secretory Mucins Aberrantly Entering the Bloodstream: Are Circulating Cancer Mucins the “Tip of the Iceberg”? Cancer Res. 66, 2433–2441 (2006). [DOI] [PubMed] [Google Scholar]
- 27.Ogura Y., Parsons W. H., Kamat S. S., Cravatt B. F., A calcium-dependent acyltransferase that produces N-acyl phosphatidylethanolamines. Nat. Chem. Biol. 12, 669–671 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fusek M., Mares M., Vetvicka V., “Cathepsin D” in Handbook of Proteolytic Enzymes, N. D. Rawlings, G. Salvesen, Eds. (Academic Press, ed. 3, 2013), chap. 8, pp. 54–63. 10.1016/B978-0-12-382219-2.00008-9. [DOI] [Google Scholar]
- 29.Turk B., Turk D., Turk V., Lysosomal cysteine proteases: More than scavengers. Biochim. Biophys. Acta 1477, 98–111 (2000). [DOI] [PubMed] [Google Scholar]
- 30.T. Imoto, K. Okazaki, H. Koga, H. Yamada, Specificity of rat liver cathepsin D. J. Biochem. 101, 575–580 (1987). [DOI] [PubMed] [Google Scholar]
- 31.P. Benes, V. Vetvicka, M. Fusek, Cathepsin D--Many functions of one aspartic protease. Crit. Rev. Oncol. Hematol. 68, 12–28 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aghdassi A. A., et al. , Cathepsin D regulates cathepsin B activation and disease severity predominantly in inflammatory cells during experimental pancreatitis. J. Biol. Chem. 293, 1018–1029 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laqtom N. N., et al. , CLN3 is required for the clearance of gylcerophosphodiesters from lysosomes. Nature, in press. 10.1038/s41586-022-05221-y. [DOI] [PMC free article] [PubMed]
- 34.Abu-Remaileh M., et al. , Lysosomal metabolomics reveals V-AT Pase- and mTOR-dependent regulation of amino acid efflux from lysosomes. Science 358, 807–813 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pimental R. A., Julian J., Gendler S. J., Carson D. D., Synthesis and intracellular trafficking of Muc-1 and mucins by polarized mouse uterine epithelial cells. J. Biol. Chem. 271, 28128–28137 (1996). [DOI] [PubMed]
- 36.Crooks G. E., Hon G., Chandonia J. M., Brenner S. E., WebLogo: A sequence logo generator. Genome Res. 14, 1188–1190 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun H., et al. , Proteolytic characteristics of cathepsin D related to the recognition and cleavage of its target proteins. PLoS One 8, e65733 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saftig P., et al. , Mice deficient for the lysosomal proteinase cathepsin D exhibit progressive atrophy of the intestinal mucosa and profound destruction of lymphoid cells. EMBO J. 14, 3599–3608 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marques A. R. A., et al. , Enzyme replacement therapy with recombinant pro-CTSD (cathepsin D) corrects defective proteolysis and autophagy in neuronal ceroid lipofuscinosis. Autophagy 16, 811–825 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mitchison H. M., et al. , Targeted disruption of the Cln3 gene provides a mouse model for Batten disease. The Batten Mouse Model Consortium [corrected]. Neurobiol. Dis. 6, 321–334 (1999). [DOI] [PubMed] [Google Scholar]
- 41.Marx V., Tools to cut the sweet layer-cake that is glycoproteomics. Nat. Methods 18, 991–995 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Malaker S. A., et al. , Revealing the human mucinome. bioRxiv [Preprint] (2021). https://www.biorxiv.org/content/10.1101/2021.01.27.428510v1.
- 43.Van-Seuningen I., Aubert J. P., Davril M., Interaction between secretory leucocyte proteinase inhibitor and bronchial mucins or glycopeptides. Physiopathological implications for the protection of mucins against proteolysis by human leucocyte elastase. Biochem. J. 281, 761–766 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang S., et al. , Cathepsin g degrades both glycosylated and unglycosylated regions of lubricin, a synovial mucin. Sci. Rep. 10, 4215 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pranjol Z. I., Whatmore J. L., “Cathepsin D in the tumor microenvironment of breast and ovarian cancers” in Tumor Microenvironment: Molecular Players – Part A, Birbrair A., Ed. (Springer International Publishing, 2020), pp. 1–16. 10.1007/978-3-030-43093-1_1. [DOI] [PubMed] [Google Scholar]
- 46.Pedram K., et al. , Design of a mucin-selective protease for targeted degredation of cancer-associated mucins. bioRxiv [Preprint] (2022). https://www.biorxiv.org/content/10.1101/2022.05.20.492748v1 (Accessed 20 May 2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xie G.-D., Liu Y.-R., Jiang Y.-Z., Shao Z.-M., Epidemiology and survival outcomes of mucinous adenocarcinomas: A SEER population-based study. Sci. Rep. 8, 6117 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li L., et al. , Hydrolysis of 2‘3’-cGAMP by ENPP1 and design of nonhydrolyzable analogs. Nat. Chem. Biol. 10, 1043–1048 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gray M. A., et al. , Targeted glycan degradation potentiates the anticancer immune response in vivo. Nat. Chem. Biol. 16, 1376–1384 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tyanova S., et al. , The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 13, 731–740 (2016). [DOI] [PubMed] [Google Scholar]
- 51.Pedram K., et al. , Data from “Lysomal cathepsin D mediates endogenous mucin glycodomain catabolism in mammals. http://proteomecentral.proteomexchange.org/cgi/GetDataset?ID=PXD027616. ProteomeXchange database. Deposited 28 July 2021. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Mass spectrometry raw files have been deposited in ProteomeXchange (PXD027616) (51).