Significance
The multisubunit ATPase-dependent SWI/SNF complex plays an important role in chromatin remodeling. Large numbers of SWI/SNF subunit mutations have been identified in large variety of human cancers, suggesting that they function against tumorigenesis. Here we report long noncoding RNA TGFB2-AS1 correlates with prognosis in triple-negative breast cancer, the most aggressive cluster of all breast cancers. Especially, we show that TGFB2-AS1 interacts with SMARCA4, a core subunit of the SWI/SNF complex, and blocks the complex to approach its target promoters both in cis and in trans, thus inhibiting the expression of the target genes, TGFB2 and SOX2, eventually leading to the inhibition of breast cancer progression. These findings shed light on understanding regulation and roles of the SWI/SNF complex in carcinogenesis.
Keywords: TGFB2-AS1, cancer stem-like cell, SWI/SNF complex, TGFβ signaling, triple-negative breast cancer
Abstract
Triple-negative breast cancer (TNBC) is the most challenging breast cancer subtype for its high rates of relapse, great metastatic potential, and short overall survival. How cancer cells acquire metastatic potency through the conversion of noncancer stem-like cells into cancer cells with stem-cell properties is poorly understood. Here, we identified the long noncoding RNA (lncRNA) TGFB2-AS1 as an important regulator of the reversibility and plasticity of noncancer stem cell populations in TNBC. We revealed that TGFB2-AS1 impairs the breast cancer stem-like cell (BCSC) traits of TNBC cells in vitro and dramatically decreases tumorigenic frequency and lung metastasis in vivo. Mechanistically, TGFB2-AS1 interacts with SMARCA4, a core subunit of the SWI/SNF chromatin remodeling complex, and results in transcriptional repression of its target genes including TGFB2 and SOX2 in an in cis or in trans way, leading to inhibition of transforming growth factor β (TGFβ) signaling and BCSC characteristics. In line with this, TGFB2-AS1 overexpression in an orthotopic TNBC mouse model remarkably abrogates the enhancement of tumor growth and lung metastasis endowed by TGFβ2. Furthermore, combined prognosis analysis of TGFB2-AS1 and TGFβ2 in TNBC patients shows that high TGFB2-AS1 and low TGFβ2 levels are correlated with better outcome. These findings demonstrate a key role of TGFB2-AS1 in inhibiting disease progression of TNBC based on switching the cancer cell fate of TNBC and also shed light on the treatment of TNBC patients.
Metastasis is the main cause of death of cancer patients, accounting for more than 90% of tumor mortality (1). This event is a long and hard process requiring tumor cells to leave the primary tumor, migrate to a distant site, and colonize the new site to initiate their growth. Increasing lines of evidence suggest that metastasis is initiated by specialized tumor cells that present cancer stem cell (CSC) properties (2, 3). Single-cell lineage tracing and high-resolution sequencing of tumor samples provide the view that metastasis relies on epigenetic amplification of cell survival and self-renewal mechanisms (4, 5). Triple-negative breast cancer (TNBC) remains the most aggressive cluster of all breast cancers due to its rapid progression, high probabilities of early recurrence, distant metastasis, and resistance to standard treatment (6). However, the precise mechanisms of recurrence and metastasis remain unclear.
Long noncoding RNAs (lncRNAs) shape biological activity in various aspects and are involved in embryonic development, homeostasis maintenance, and promoting or inhibiting the development of many pathogeneses, including cancers (7). In the cytoplasm, lncRNAs act as sponges of microRNAs (8) and guide the translation repressor, RNA-binding proteins involving in mRNA decay, and active polysomes on the mRNAs to regulate gene expression after transcription (9–11). Signal transduction pathways can also be modulated by lncRNAs such as lnc-DC and NKILA, which mask the sites bound by posttranslational modification (PTM) enzymes or PTM sites (12, 13). In the nuclei, besides some “architectural RNAs” that function as the scaffold of nuclear bodies (14), a significant fraction of lncRNAs are associated with chromatin organization, transcription, and RNA processing (15). They can regulate transcription by influencing transcriptional factor activity (16), assembling Pol II machineries (17), and interacting with chromatin modulating proteins such as subunits of SWI/SNF or polycomb repressive complexes (PRC) (18–20).
Chromatin remodeling is one of the mechanisms essential in dynamic regulation of gene expression, which is performed by a number of different proteins/protein complexes, among which are multisubunit ATPase-dependent SWI/SNF (switch/sucrose nonfermentable) complexes. SWI/SNF complex recruits to DNA regions by transcription regulators and other proteins, upon which the complex moves the nucleosomes along the DNA and modifies its accessibility, allowing gene expression regulation through binding of the transcriptional regulators to exposed DNA (21). Therefore, SWI/SNF plays a critical role in coordinating chromatin architecture and is a fundamental epigenetic regulator of gene transcription gene expression (22, 23). The mammalian SWI/SNF complex exists in multiple forms characterized by different subunit compositions. These complexes usually include either SMARCA4 (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin A4)/BRG1 or SMARCA2/BRM and a set of 6 to 11 other proteins called Brg1/BRM-associated factors (BAFs) that are essential for binding to DNA or proteins. In humans, three of these subunits, SNF5 (INI1 for Integrase interactor 1 or SMARCB1), BAF170/SMARCC2, and BAF155/SMARCC1, are highly conserved “core” subunits because they are essential for the ATP-dependent chromatin remodeling activity of the SWI/SNF complexes together with SMARCA4 (22, 23). It is an essential event that chromatin status is shaped by the SWI/SNF complex during somatic cell reprogramming toward induced pluripotent stem cells by the traditional defined factors (Oct4, SOX2, myc, and klf4) (24, 25). Therefore, malignant transformation of tumor cells is always accompanied by acquisition of cancer stemness properties, in which the SWI/SNF complex plays an important role (18, 24, 26).
Both transforming growth factor β (TGFβ) and SOX2 play key roles in the turning point of cell fate (3, 24, 27–30). TGFβ induces epithelial mesenchymal transition (EMT), enhances the CSC potential in breast cancer cells (31), presents in invasive fronts and metastatic niches to support invasion and survival of tumor cells, and even induces the mesenchymal epithelial transition that facilitates distant metastatic colonization in breast cancer (31, 32). SOX2 is a key reprogramming factor (24), switching non-CSCs to CSCs to facilitate tumor initiatiation and self-renewal (33).
Herein, we report that lncRNA TGFB2-antisense RNA1 (TGFB2-AS1), which is down-regulated in the TNBC samples from the patients more prone to have distant metastasis, interacts with SMARCA4 and blocks the complex to approach its target promoters both in cis and in trans, thus inhibiting the expression of the target genes, TGFB2 and SOX2, eventually leading to the inhibition of breast cancer progression.
Results
TGFB2-AS1 Correlates with Prognosis in TNBC.
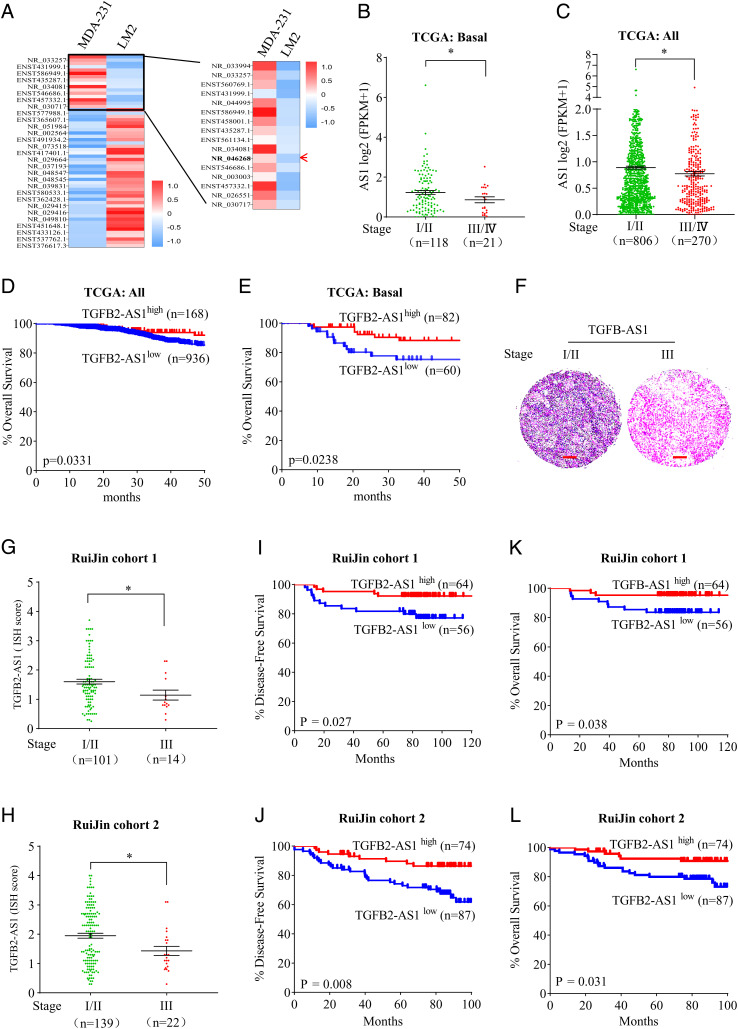
To identify lncRNAs that play a role in the progression of TNBC, RNA profiles were performed between human TNBC cell lines MDA-MB-231 cells (named MDA-231 cells for short) and MDA-MB-231-LM2 cells (named LM2 cells for short), the latter presenting significantly enhanced lung metastatic activity compared to the parental MDA-231 cells (34) (SI Appendix, Fig. S1). According to the primary data analysis, 5,913 lncRNAs were aberrantly expressed, including 2,855 down-regulated in the LM2 cells (SI Appendix, Table S1). According to the criteria as following, the ≥1 of RPKM (expected number of reads per kilobase of transcript sequence per million base pairs sequenced) of lncRNAs in at least one of two cell lines and twofold or greater of the expression level change in LM2 cells compared to MDA-231 cells, in total 16 lncRNAs were identified to be decreased (Fig. 1A and SI Appendix, Table S2). With the RNA-sequencing data from The Cancer Genome Atlas (TCGA) database, we investigated the correlation of expression levels of these down-regulated lncRNAs with clinical stages in all breast cancer tissues as well as in basal-like breast cancer, which lacks or shows low levels of ER, PR, and Her2 proteins. Among all 13 lncRNAs with available expression data in TCGA, expression of only TGFB2-AS1(NR_046268) but not the other 12 lncRNAs was negatively correlated to clinical stages in basal-like breast cancer tissues (Fig. 1B and SI Appendix, Fig. S2). On the other hand, the analysis from the GTEx database showed that TGFB2-AS1 was widely expressed in various normal tissues including normal breast and mammary tissue (SI Appendix, Fig. S3). Therefore, we focused on the potential roles of TGFB2-AS1 in breast cancers in the following work.
Fig. 1.
Identification of lncRNA TGFB-AS1, which is associated with good prognosis in TNBC patients. (A) Heat-map representation of lncRNAs that are differentially expressed in MDA-231 cells and LM2 cells. The red arrow points to TGFB2-AS1. (B and C) TGFB2-AS1 expression of stage I/II compared with stage III/IV in basal (B) and all subtypes of breast cancers (C) analyzed using the TCGA database. (D and E) Kaplan–Meier analyses of the correlation between TGFB2-AS1 levels and the overall survival in patients with all subtypes (D) and basal-like (E) breast cancer using the TCGA database (log rank test). (F) Representative images of ISH staining of TGFB2-AS1 in stage I/II and stage III in TNBC tissues from cohort 1. (Scale bar, 200 μm.) (G and H) TGFB2-AS1 ISH score quantitative data of stage I/II compared with stage III/IV in TNBC tissues from cohort 1 (G) and cohort 2 (H). (I–L) Kaplan–Meier analyses of the correlation between TGFB2-AS1 levels and the DFS (I and J) and OS (K and L) in two TNBC cohorts (log rank test). Error bars represent mean ± SEM, *P < 0.05. Statistical significance was assessed using two-tailed Student’s t test. Determination of the optimal cutoff value for predicting survival was performed using X-tile bioinformatics software version 3.6.1.
Although a negative correlation between TGFB2-AS1 expression and clinical stages could be seen in all types of breast cancer (Fig. 1C), it could not be seen in other subtypes of breast cancers (SI Appendix, Fig. S4). Similarly, the expression levels of TGFB2-AS1 were strongly negatively correlated to the prognosis in all breast cancers, especially in basal-like breast cancer patients, and patients with low levels of TGFB2-AS1 expression showed poorer prognosis (Fig. 1 D and E). To further validate the clinical relevance of TGFB2-AS1 expression, we detected the expression level of TGFB2-AS1 by in situ hybridization by LNA RNA probe in a TNBC tissue microarray including TNBC tumor cohort 1 with 120 samples and cohort 2 with 161 samples from Ruijin Hospital. The results showed that stage III tumors had less positive staining compared to stage I/II samples (Fig. 1 F–H). More importantly, Kaplan–Meier analysis demonstrated that lower TGFB2-AS1 expression was associated with worse disease-free survival (DFS) as well as overall survival (OS) in these two cohorts (Fig. 1 I–L). Taken together, these data implied that TGFB2-AS1 might play an inhibitory role in TNBC progression.
TGFB2-AS1 Suppresses TNBC Progression in an Orthotopical Mouse Model.
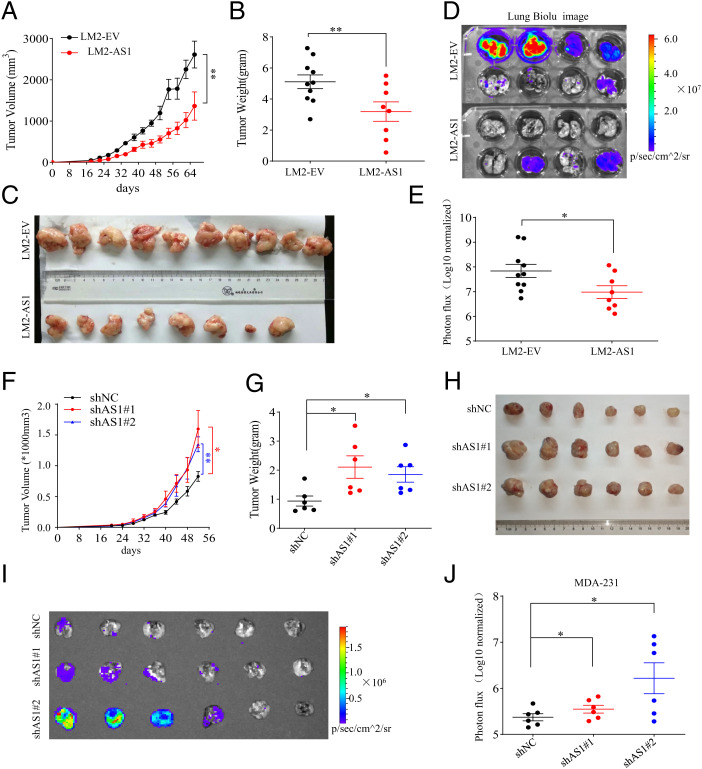
To evaluate the potential roles of TGFB2-AS1 in TNBC, we measured its expression level by real-time PCR and showed that TGFB2-AS1 was significantly reduced in the LM2 cells compared to MDA-231(SI Appendix, Fig. S5A). Then, we generated TGFB2-AS1 stably overexpressed LM2 cells (LM2-AS1) and control cells (LM2-EV) (SI Appendix, Fig. S5B) and injected them orthotopically into the mammary fat pad of female NOD-SCID mice. The primary tumor volumes and weights in LM2-AS1 cells were strikingly depressed compared to LM2-EV cells (Fig. 2 A–C). Furthermore, mice were given intraperitoneal injection of d-luciferin when alive and lungs were dissected quickly after death and monitored by bioluminescence imaging (BLI) with an IVIS system. The BLI quantitative data showed that the lung metastasis of LM2-AS1 cells was significantly reduced compared to LM2-EV cells (Fig. 2 D and E). The hematoxylin–eosin staining in the lung metastatic foci also demonstrated decreased lung metastasis by ectopic TGFB2-AS1 expression (SI Appendix, Fig. S5C). Reciprocally, MDA-231 cells with TGFB2-AS1 knockdown by two pairs of different TGFB2-AS1–targeting small interfering RNAs (siRNAs) (siAS1#1, siAS1#2) grew more quickly in female mammary fat pad of NOD-SCID mice and more actively disseminated to lung tissue (Fig. 2 F–J and SI Appendix, Fig. S5 D and E). Collectively, these data indicated that TGFB2-AS1 antagonizes TNBC progression in vivo.
Fig. 2.
TGFB2-AS1 suppresses breast cancer tumorigenesis and lung metastases in vivo. (A–E) LM2-AS1 cells and the LM2-EV cells were transplanted orthotopically into mammary fat pad of female NOD/SCID mice, and tumor growth (A), tumor weight (B), tumor morphology (C), BLI images of lung metastasis (D), and its quantitative data (E) at 68 d after orthotopic injection are shown. n = 8 or 10 per group. (F–J) MDA-231 cells with TGFB2-AS1 knockdown by shAS1#1 or shAS1#2 and their control cells (shNC) were transplanted orthotopically into mammary fat pad of female NOD/SCID mice, and tumor growth (F), tumor weight (G), tumor morphology (H), and BLI image of lung metastasis (I) and its quantitative data (J) at 52 d after orthotopic injection are shown (n = 6 per group). Data are presented as means ± SEM (n = 6 to 10). Statistical significance was assessed using two-tailed Student’s t test. *P < 0.05. **P < 0.01.
TGFB2-AS1 Attenuates CSC Self-Renewal Activity and Inhibits the Malignant Character of TNBC Cells.
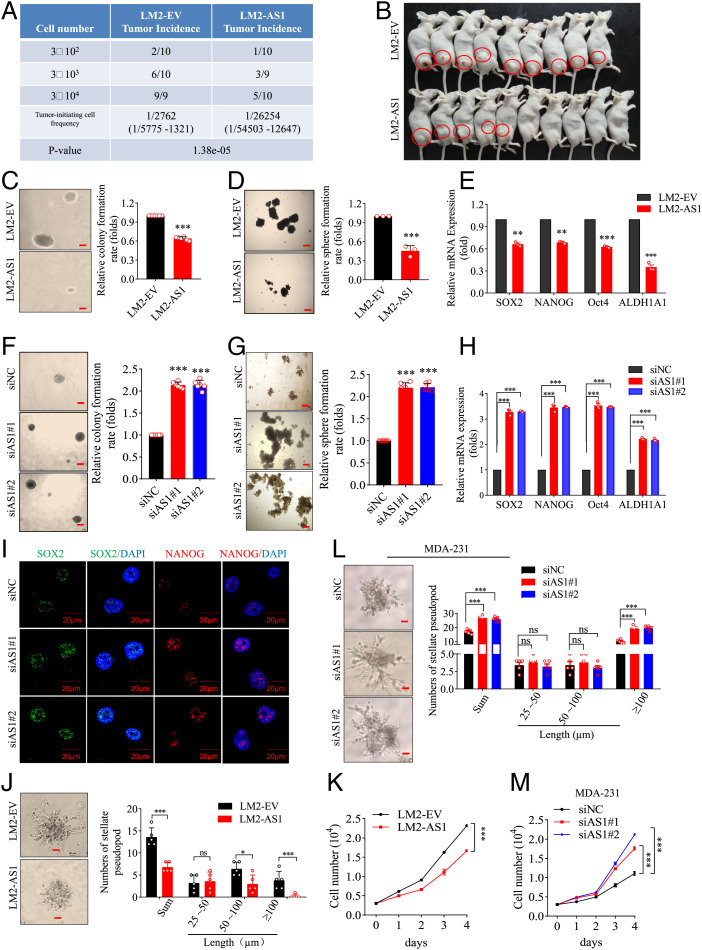
To investigate the effect of TGFB2-AS1 on cell fate transition in TNBC cells, LM2-EV and LM2-AS1 cells were diluted limitedly (3 × 104∼2) and inoculated subcutaneously into female BALB/c nude mice respectively to determine the tumor-initiating cell frequency. The results showed that TGFB2-AS1 cells presented dramatically decreased tumorigenic frequency with 1/5,775 to 1,321 and 1/54,503 to 12,647, respectively, for LM2-EV and LM2-AS1 cells, together with decreased tumor volumes and weights (Fig. 3 A and B and SI Appendix, Fig. S6 A and B). As is well known, CSC may contribute to tumor initiation, metastasis, and recurrence (35). Hence, we surveyed the CSC self-renewal activity between LM2-EV and LM2-AS1 cells and revealed that LM2-AS1 cells showed significantly weaker ability of soft agar colony formation (Fig. 3C), mammosphere formation (Fig. 3D), and plate colony formation (SI Appendix, Fig. S6C). In agreement, the messenger RNA (mRNA) and/or protein expression levels of CSC markers SOX2, NANOG, Oct4, and ALDH1A1 were decreased in the LM2-AS1 cells (Fig. 3E and SI Appendix, Fig. S6 D and E). On the other hand, MDA-231 cells with TGFB2-AS1 knockdown as aforementioned presented significantly enhanced ability of soft agar colony formation, mammosphere formation, and plate colony formation as well as increased expression levels of CSC markers SOX2, NANOG, Oct4, and ALDH1A1(Fig. 3 F–I and SI Appendix, Fig. S6 F and G). Collectively, TGFB2-AS1 attenuates CSC self-renewal activity of TNBC cells.
Fig. 3.
TGFB2-AS1 impairs breast cancer cell stemness. (A) LM2-AS1 and LM2-EV cells were diluted as indicated and subcutaneously implanted into BALB/c nude mice. Tumor-initiating cell frequency and P value for limiting dilution xenograft analysis was calculated on the website https://bioinf.wehi.edu.au/software/elda/. n = 9 or 10 for each group. (B) Representative image showing 3 × 104 group of limiting dilution xenograft analysis. (C and D) Soft agar colony formation assay (C) and mammosphere assay (D) between LM2-EV and LM2-AS1 cells with images (Left; scale bar, 200 μm) and values (Right). (E) The expressions of stemness-related genes were detected in LM2-AS1 cells using qPCR. (F–I) Representative images (Left; scale bar, 200 μm) and values (Right) of soft agar colony formation assay (F). Representative images (Left; scale bar, 200 μm) and values (Right) of mammosphere assay (G), the expression of stemness-related genes detected using qPCR (H), and the representative images of immunofluorescence staining for SOX2 and Nanog (I) in TGFB2-AS1–silenced MDA-231 cells and their control cells (MDA-231-siNC). (J) Representative images (Left; scale bar, 50 μm) and values (Right) of the stellate pseudopod of LM2-EV and LM2-AS1 cells cultured on Matrigel on day 6. (K) Cell proliferation curves in LM2-EV and LM2-AS1 cells. (L) The images (Left; scale bar, 50 μm) and values (Right) of stellate pseudopod in TGFB2-AS1 silenced MDA-231 and their control cells cultured on Matrigel on day 8. (M) Cell proliferation curves in TGFB2-AS1–silenced MDA-231 cells. Error bars represent mean ± SD (n = 3 to 6). Statistical significance was assessed using two-tailed Student’s t test. *P < 0.05. **P < 0.01. ***P < 0.001. ns, not significant.
We continued to survey other malignant characters of cancer cells. Toward this end, the LM2-EV and LM2-AS1 cells were cultured on Matrigel, which mimics a three-dimensional tumor microenvironment (36), and the results revealed that the numbers and invasion distance of pseudopods in LM2-AS1 cells were reduced than in LM2-EV cells (Fig. 3J). Also, the ability to pass through the transwell chambers with or without Matrigel were repressed in LM2-AS1 cells (SI Appendix, Fig. S6 H and I). Likewise, ectopic TGFB2-AS1 expression inhibited the growth of LM2 cells (Fig. 3K). Conversely, MDA-231 cells with TGFB2-AS1 knockdown showed increased numbers and invasion distance of pseudopods (Fig. 3L) and more easily passed through the chambers (SI Appendix, Fig. S6 J and K) and more increased proliferation (Fig. 3M). Additionally, we also detected two other TNBC cell lines, SUM159PT and BT-549, as well as one luminal cell line, BT474, and showed that the TGFB2-AS1 knockdown remarkably promoted colony formation, mammosphere formation, migration, and cell proliferation of both SUM159PT and BT-549 cells but not BT474 cells (SI Appendix, Fig. S7). Unexpectedly, TGFB2-AS1 knockdown also remarkably promoted colony formation, cell proliferation, and sphere formation of MCF-10A cells (SI Appendix, Fig. S8), an immortal, nontransformed cell line which has many features of basal progenitor cells and may represent a multipotent lineage (37). All these results suggested that TGFB2-AS1 impairs the malignant character of TNBC cells.
TGFB2-AS1 Interacts with Chromatin Remodeling Complex SWI/SNF.
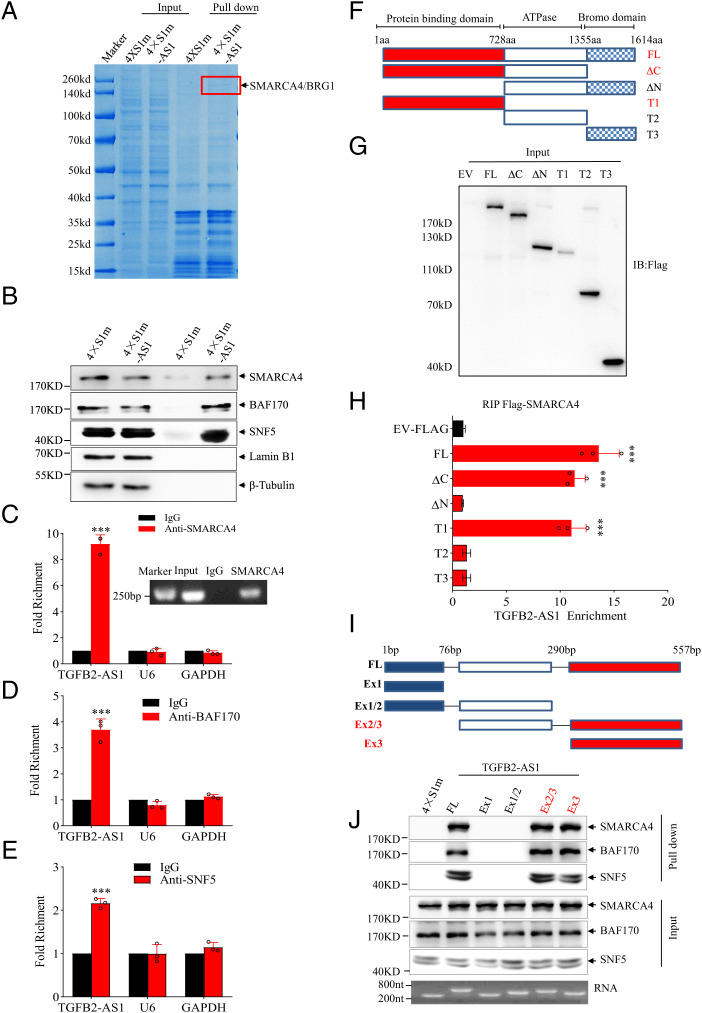
Considering that lncRNAs often exert their functions through RNA-interacting proteins (38), we employed RNA-pulldown assays between MDA-231 cells transfected with pCDNA3.1-TGFB2-AS1-4×S1m (4×S1m-AS1) and pCDNA3.1-4×S1m (4×S1m) (39) to explore possible TGFB-AS1–binding proteins. One overtly differential band around 200 kDa appeared by Coomassie bright blue staining, and mass spectrometry assay identified the band as SMARCA4 (Fig. 4A), a core subunit of chromatin remodeling complex SWI/SNF (7). Western blot analysis proved SMARCA4 as well as the other core subunits of SWI/SNF complex BAF170 and SNF5 in the TGFB2-AS1-pulled down complex (Fig. 4B). Consistently, the TGFB2-AS1–protein complex was further validated by anti-SMARCA4, BAF170, and SNF5 antibody-based RNA immunoprecipitation (RIP) followed by qPCR analysis (Fig. 4C–E). Subcellular fractionation showed that TGFB2-AS1 transcript distributes in both nucleus and cytoplasm of MDA-231 cells (SI Appendix, Fig. S9A) and fluorescence in situ hybridization (FISH) of TGFB2-AS1 combined with immunofluorescence of SMARCA4 indicated that TGFB2-AS1 and SMARCA4 could colocalize in the nucleus of MDA-231 cells (SI Appendix, Fig. S9B). Furthermore, to investigate if TGFB2-AS1 impacts the integrity of the SWI/SNF complex, we performed coimmunoprecipitation experiments of the three core subunits of the SWI/SNF complex in the TGFB2-AS1 knockdown MDA-231 cells. The result showed that TGFB2-AS1 knockdown had no obvious effect on the interaction between SMARCA4, BAF170, and SNF5, which indicated that TGFB2-AS1 may not impact the integrity of the SWI/SNF complex (SI Appendix, Fig. S9C). Collectively, these data suggested that TGFB2-AS1 interacts with the SWI/SNF chromatin remodeling complex.
Fig. 4.
TGFB2-AS1 interacts with SMARCA4. (A) RNA-pulldown assay to identify TGFB2-AS1 binding proteins in MDA-231 cells. The arrow and red bracket indicate the band representing the TGFB2-AS1 specific binding protein identified by mass spectrometry as SMARCA4. (B) Western blot identifying SMARCA4, BAF170, and SNF5 present in TGFB2-AS1 pull-down precipitates with Lamin B1 and β-tubulin as negative controls. (C–E) The interaction of TGFB2-AS1 with SMARCA4 (C, a semiquantitative RT-PCR was inserted), BAF170 (D), and SNF5 (E) was verified by an RIP assay together with U6 and GAPDH as negative controls. (F) Schematic representation of Flag-tagged full-length human SMARCA4 and its deletion mutants. (G) The anti-Flag Western blot images showing expressions of full-length or deleted SMARCA4 in MDA-231 cells. (H) RIP and subsequent RT-PCR assays in MDA-231 for TGFB2-AS1 enrichment of human full-length SMARCA4 and its deletion mutants. (I) Schematic representation of full-length and various truncated TGFB2-AS1. (J) Western blot showing SMARCA4, BAF170, and SNF5 pulled down by full-length and various truncated TGFB2-AS1. Error bars represent mean ± SD (n = 3). Statistical significance was assessed using two-tailed Student’s t test. ***P < 0.001.
TGFB2-AS1 Interacts with the N Terminus of SMARCA4 through its Exon 3.
To clearly decipher the binding region of SMARCA4 with TGFB2-AS1, MDA-231 cells were transfected respectively with full-length Flag-SMARCA4 or its five truncations fused to Flag tag, followed by the RIP assay. The results demonstrated that N-terminal fragment of SMARCA4 was essential for its interaction with TGFB2-AS1 (Fig. 4 F–H). We also mapped the TGFB2-AS1 functional motifs corresponding to SMARCA4 binding using a series of truncated TGFB2-AS1 fragments according to its secondary structure of TGFB2-AS1, which was predicted based on minimum free energy (Fig. 4 I and J and SI Appendix, Fig. S9D). In vitro RNA pull-down assay revealed that exon 3 of TGFB2-AS1 (TGFB2-AS1 290 to 557 nucleotides) was sufficient to interact with SMARCA4 protein (Fig. 4J). Collectively, TGFB2-AS1 interacts with the N-terminal region of SMARCA4 through its exon 3.
TGFB2-AS1 Antagonizes the Transcription-Regulating Activity of the SWI/SNF Complex.
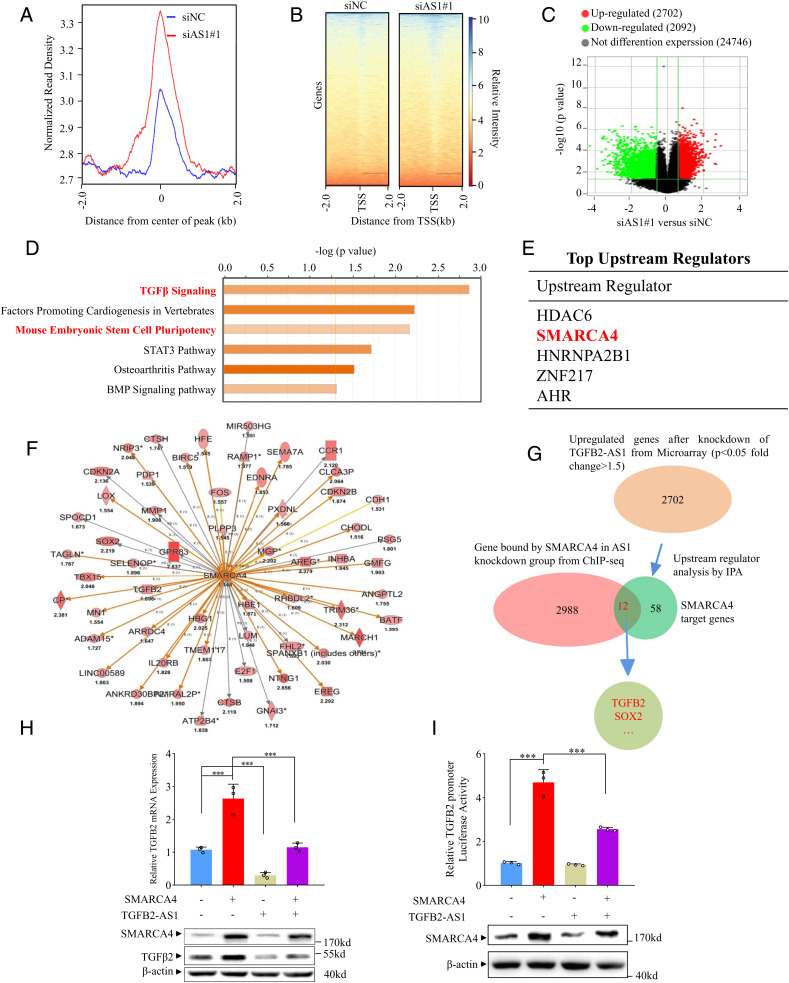
It is known that the SWI/SNF complex regulates gene transcription by binding to promoter loci to facilitate or hamper gene transcription (40). Hence, we performed chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-seq) for SMARCA4 in MDA-231-siAS1 and MDA-231-siNC cells. In contrast to the control group, TGFB2-AS1 silence significantly enhanced the activity of SMARCA4 binding to promoters (Fig. 5 A and B). In total 2,988 genes were occupied by SMARCA4 in TGFB2-AS1 knockdown cells according to ChIP-seq data. In order to identify the genes that are regulated together by TGFB2-AS1 and SMARCA4, we performed transcriptome microarray analysis in MDA-231-siAS1 and MDA-231-siNC cells (SI Appendix, Fig. S10 A) and found that 2,702 and 2,092 genes were respectively up-regulated and down-regulated in the MDA-231-siAS1 cells relative to MDA-231-siNC cells (fold change ≥1.5; P < 0.05) (Fig. 5C). Canonical signal pathway analysis revealed that the deregulated genes were enriched for TGFβ signaling, mouse embryonic stem cell pluripotency, and other signaling pathways (Fig. 5D). Furthermore, the top upstream regulators were predicted by Ingenuity Pathway Analysis (IPA) using the data of microarray assay. Of interest, SMARCA4 was listed as second-highest upstream regulator and regulated 58 target genes (Fig. 5 E and F). By overlapping 2,988 genes which were occupied by SMARCA4 in TGFB2-AS1 knockdown cells from ChIP-seq assay and the 58 SMARCA4 targeting genes, 12 genes emerged including TGFB2 and SOX2 (Fig. 5G).
Fig. 5.
TGFB2-AS1 inhibits TGFB2 expression via sequestering SMARCA4 away from TGFB2 promoter. (A) Global representation of SMARCA4 genomic binding over a 4-kb window centered on SMARAC4 ChIP-seq peak in MDA-231 with siAS1 or siNC transfected. (B) The heat map of SMARCA4 genomic binding around the transcription start site (TSS) in MDA-231 with siAS1 or siNC transfected. (C) Volcano plots of mRNA expression profiles in MDA-231cells with TGFB2-AS1 knockdown and control group. Red dots represent up-regulated genes and green dots represent down-regulated genes (fold change ≥2). (D) Canonical signal pathway analysis via IPA using genes which are altered expression in microarray profiles and occupied by SMARCA4 in ChIP-seq data simultaneously in MDA-231 cells with TGFB2-AS1 knockdown. (E) Upstream regulators analysis by IPA using mRNA microarray data from TGFB2-AS1 knockdown MDA-231 cells and control cells. (F) Graph showing 58 up-regulated SMARCA4 target genes from IPA; the color depth is correlated to the fold change which is represented under each gene. Orange lines with arrows indicate activation, yellow lines with arrows depict inconsistent effects, and gray lines with arrows depict no prediction. (G) Schematic view of the identification of SMARCA4 regulated genes in MDA-231 cells with siAS1 transfected. (H) Real-time PCR and Western blot analysis show TGFB2 mRNA and protein expression in MDA-231 cells transfected with SMARCA4 or/and TGFB2-AS1 overexpression plasmids. (I) Luciferase reporter assay of chromatinized TGFB2 promoter in MDA-231 cells transfected with SMARCA4 or/and TGFB2-AS1 overexpression plasmids. Error bars represent mean ± SD (n = 3). Statistical significance was assessed using two-tailed Student’s t test. ***P < 0.001.
To address whether TGFB2-AS1 antagonizes SMARCA4 activity in regulating TGFB2 transcription, we revealed that SMARCA4 overexpression up-regulated TGFB2 mRNA and protein expression. Of great interest, the SMARCA4-induced TGFB2 up-regulation could be significantly antigonized by TGFB2-AS1 overexpression in MDA-231 cells (Fig. 5H). To further testify the antagonizing effect of TGFB2-AS1 on SMARCA4 in facilitating TGFB2 transcription, we cloned the promoter of TGFB2 (−500 bp to +2,500 bp) into an episomal luciferase reporter, pREP4, which allows promoter chromatinization (41). As expected, the SMARCA4 activity was reduced by ectopic expressing TGFB2-AS1 (Fig. 5I).
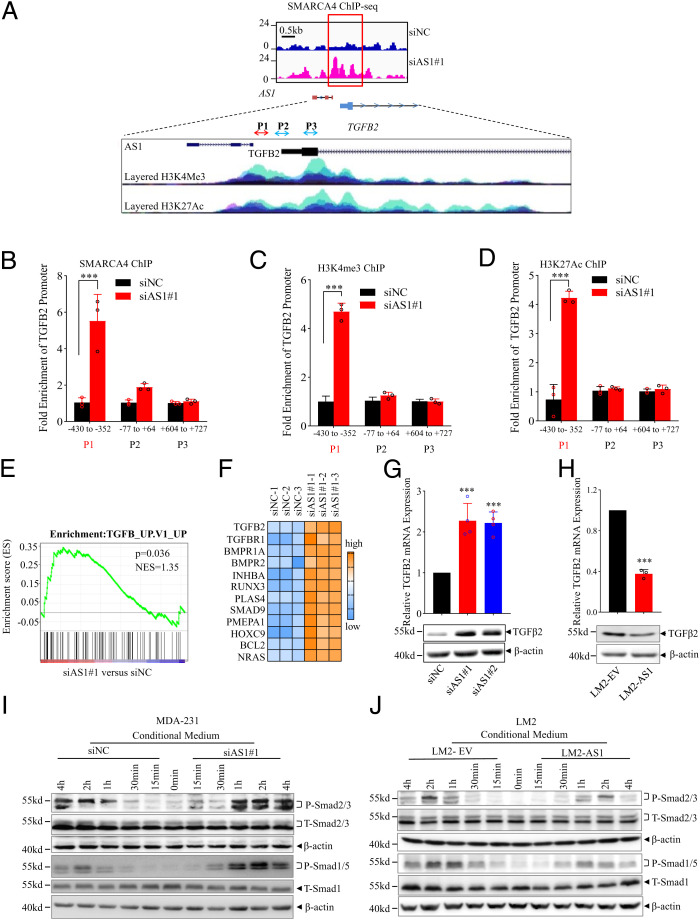
The ChIP-seq revealed that TGFB2-AS1 knockdown significantly increased SMARCA4 binding to TGFB2 promoter (Fig. 6A). The binding capacity of SMARCA4 to −430 to −352 bp segment of TGFB2 promoter was enriched in TGFB2-AS1 knockdown cells (Fig. 6B). Furthermore, H3K4Me3 and H3K27Ac were enriched at the gene promoter that denotes the activation of the TGFB2 gene (Fig. 6 C and D). Altogether, the above results demonstrated that TGFB2-AS1 antagonizes the transcription-regulating activity of the SWI/SNF complex.
Fig. 6.
TGFB2-AS1 suppresses TGFβ2 signaling in TNBC cells. (A) Schematic view of SMARCA4 occupancy around TGFB2-AS1 and TGFB2 genomic locus from ChIP-seq in MDA-231 cells with knockdown of TGFB2-AS1. Blue indicates siNC group and pink indicates siB2-AS1 group. H3K4Me3 and H3K27Ac histone mark were displayed on four cell lines from ENCODE, on the layered H3K4Me3 and the layered H3K27Ac tracks respectively. Each color represents one cell line. (B) ChIP-qPCR shows the binding efficiency of SMARCA4 to the TGFB2 promoter region using three primers in MDA-231 cells transfected siAS1. (C and D) ChIP-qPCR shows the H3K4me3 and H3K27Ac levels of the TGFB2 promoter region using three primers in MDA-231 cells transfected siAS1. Error bars represent mean ± SD (n = 3). (E) GSEA of microarray profiles from MDA-231 with TGFB2-AS1 knockdown matching with TGFβ activated gene set. (F) Heat-map representation of up-regulated genes relating TGFβ signaling pathway in TGFB2-AS1 knockdown group comparing control group from IPA. RT-PCR and Western blot assay show TGFB2 mRNA and protein expression in MDA-231 cells after transfected with two TGFB2-AS1 siRNAs (G) and LM2 cells overexpressed TGFB2-AS1 (H). Western blot analysis shows P-SMAD2/3 and P-SMAD1/5 protein levels after treated with CM collected from MDA-231 cells after TGFB2-AS1 knockdown (I) or LM2 cells TGFB2-AS1 overexpression (J), and SMAD1/5, SMAD2/3, and β-actin work as internal control. Error bars represent mean ± SD (n = 3). Statistical significance was assessed using two-tailed Student’s t test. ***P < 0.001.
TGFB2-AS1 Suppresses the TGFβ2 Signal Pathway in TNBC Cells.
To further investigate the whole landscape of the TGFB2-AS1–regulated genes and their effects on TNBC progression, we analyzed the two expression profiles of TGFB2-AS1 knockdown MDA-231 cells and TGFB2-AS1 overexpressed LM2 cells. Gene set enrichment analysis (GSEA) revealed that TGFB2-AS1 knockdown was closely associated with the activated TGFβ signal pathway (Fig. 6 E and F), while TGFB2-AS1 overexpression was related to the inhibited TGFβ signal pathway (SI Appendix, Fig. S10B). Of note, TGFβ2 rather other two TGFβ members, TGFβ1 and TGFβ3, was up-regulated in MDA-231-siAS1 cells and was down-regulated in LM2-AS1 cells (Fig. 6 G and H and SI Appendix, Fig. S10C). In addition, Western blot showed that the expression levels of pSmad2/3 and pSmad1/5 were significantly up-regulated in MDA-231 cells by the conditional medium (CM) from MDA-231-siAS1 cells compared to their control CM (Fig. 6I). On the contrary, the levels of pSmad2/3 and pSmad1/5 were reduced by the CM from LM2-AS1 cells compared to their control CM (Fig. 6J). Altogether, TGFB2-AS1 suppresses the TGFβ2 signal pathway in TNBC cells.
TGFB2-AS1 Reverses CSC Signaling in TNBC Cells.
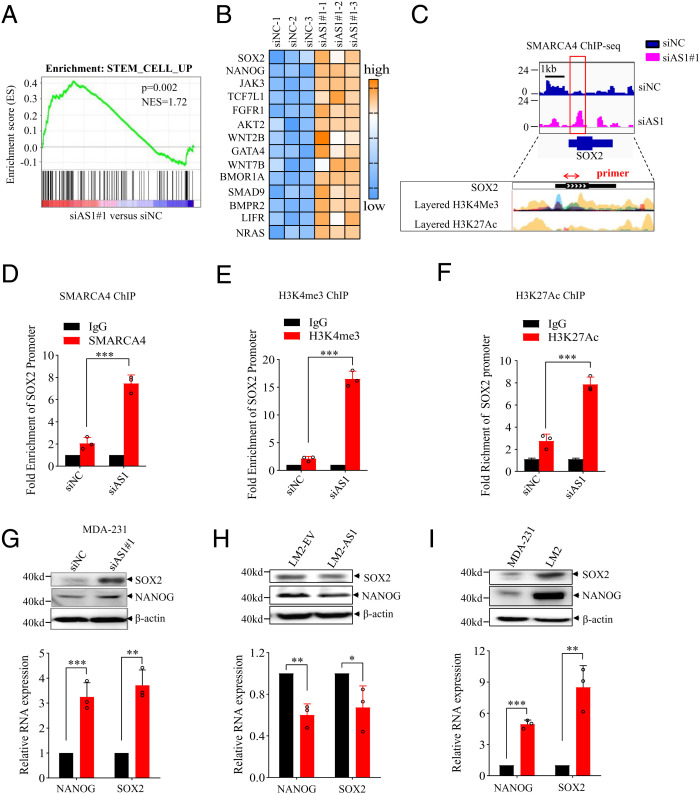
Furthermore, both IPA and GSEA indicated that TGFB2-AS1 knockdown was closely associated with stem cell traits (Figs. 5D and 7A), while the two iPS pioneer transcription factors SOX2 and NANOG were up-regulated by 2.2-fold and 3.89-fold, respectively, in the microarray profiles of MDA-231-siAS1/MDA-231-siNC cells (Fig. 7B). ChIP-seq data also revealed that SMARCA4 bound to the SOX2 gene promotor (Fig. 7C), which could be validated by ChIP-qPCR (Fig. 7D). H3K4Me3 and H3K27Ac enrichment at the SOX2 gene promotor in TGFB2-AS1 silenced cells which symbolized the active gene (Fig. 7 E and F). Moreover, the mRNA and protein of both SOX2 and NANOG were up-regulated in MDA-231-siAS1 cells and down-regulated in LM2-AS1 cells compared to their control cells (Fig. 7 G and H). As expected, SOX2 and NANOG were also up-regulated in the LM2 cells vs. their parent MDA-231 cells (Fig. 7I). Given that the SOX2 gene is located in chromosome 3 far from TGFB2-AS1, we suggest that TGFB2-AS1 restrained SOX2 expression in an in trans manner. Collectively, our results propose that TGFB2-AS1 inhibits both TGFβ2 and CSC signaling in TNBC cells.
Fig. 7.
TGFB2-AS1 reverses breast cancer cell stemness. (A) GSEA of microarray profiles from MDA-231 with TGFB2-AS1 knockdown matching with stem cell activated gene set. (B) Heat-map representation of up-regulated genes relating mammalian embryonic stem cell pluripotency from IPA pathway analysis in TGFB2-AS1 knockdown group and control group. The orange color scales represent up-regulated genes, and the blue color scales represent down-regulated genes. (C) Schematic view of SMARCA4 occupancy on locus of SOX2 from ChIP-seq in MDA-231 cells with TGFB2-AS1 knockdown. Blue indicates siNC group and pink indicates siB2-AS1 group. H3K4Me3 and H3K27Ac histone mark were displayed on seven cell lines from ENCODE, on the layered H3K4Me3 and the layered H3K27Ac tracks respectively. Each color represents one cell line. (D) ChIP-qPCR shows the binding efficiency of SMARCA4 to the SOX2 using primer in MDA-231 cells with TGFB2-AS1 knockdown. The red arrow indicates the target fragment. (E and F) ChIP-qPCR shows the H3K4me3 and H3K27Ac efficient occupancy on the SOX2 promoter region in MDA-231 cells with TGFB2-AS1 knockdown. The mRNA and protein expression of SOX2 and NANOG in MDA-231 cells after transfected with siAS1 in (G) LM2 cells overexpressed TGFB2-AS1 in (H) and in MDA-231 cells and LM2 cells (I). Error bars represent mean ± SD (n = 3). Statistical significance was assessed using 2-tailed Student’s t test. *P < 0.05. **P < 0.01. ***P < 0.001.
TGFB2-AS1 Impairs the Malignant Character of TNBC through TGFβ2 and SOX2.
To test whether the inhibitory effects of TGFB2-AS1 on TNBC are dependent on TGFB2 and/or SOX2, we abrogated the up-regulation of TGFB2 by siRNA targeting TGFB2 (siTGFB2) in MDA-231-siAS1 cells (SI Appendix, Fig. S11 A and B). Consequently, TGFB2 knockdown successfully abrogated the increased capability of migration, invasion, and colony formation induced by siAS1 (SI Appendix, Fig. S11 C–E). In the same way, when SOX2 expression was knocked down with high efficiency in MDA-231-siAS1 cells, the increased capability of invasion as well as the capability of colony formation induced by siAS1 were successfully abolished (SI Appendix, Fig. S12 A–C). Altogether, TGFB2-AS1 reverses the malignant character of TNBC by inhibiting both TGFβ2 signal pathway and CSC characteristics.
TGFB2-AS1 Reverses TGFβ2-Promoted TNBC Development.
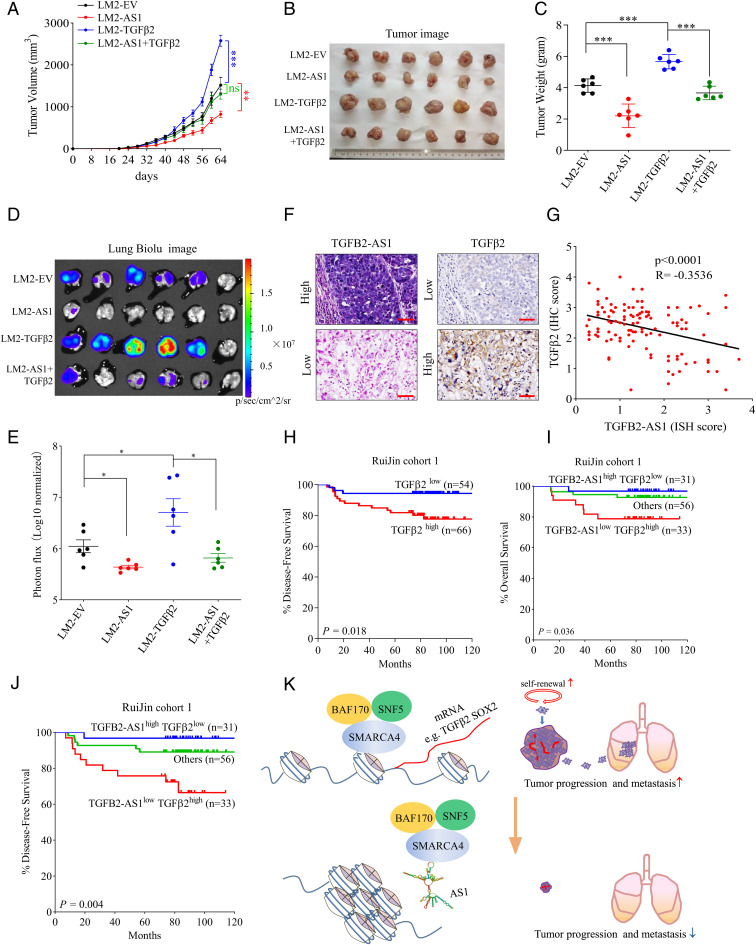
Finally, TGFβ2- or/and TGFB2-AS1–overexpressed LM2 cells (LM2-TGFβ2, LM2-AS1, and LM2-AS1+TGFβ2) were stably generated (SI Appendix, Fig. S13 A and B). Cells were injected orthotopically into the mammary fat pad of female NOD-SCID mice. TGFβ2 promoted orthotopic tumor growth (Fig. 8 A–C) and tumor cell diffusion to the lung tissues (Fig. 8 D and E). TGFB2-AS1 not only remarkably suppressed the tumor growth and lung metastasis of LM2 as the given results but also abrogated the enhancement of tumor growth (Fig. 8 A–C) and lung metastasis (Fig. 8 D and E) endowed by TGFβ2.
Fig. 8.
TGFB2-AS1 suppresses TNBC progression and metastasis via inhibiting TGFβ2. After tumor cells were injected into the mammary fat pad orthotopically, tumor growth curves were measured. (A) Tumors were harvested, photographed (B), and weighed (C) on day 64. IVIS images of lung metastasis (D) and luciferase quantitative data (E) on 64 d after orthotopic injection (n = 6 per group). Data are presented as means ± SEM (n = 6). Statistical significance was assessed using two-tailed Student’s t test. (F) Representative images (scale bar, 50 μm) of ISH of TGFB2-AS1 and immunohistochemical staining of TGFβ2 in Ruijin cohort 1. (G) Correlation between TGFB2-AS1 and TGFβ2 expression from RuiJin cohort 1 determined by Pearson correlation analysis. (H) Kaplan–Meier analysis of disease-free survival of TGFB2 from Ruijin cohort 1. (I and J) Kaplan–Meier analysis of the overall survival and disease-free survival of TGFB2-AS1high/TGFβ2low and TGFB2-AS1low/TGFβ2high from Ruijin cohort 1. (K) Schematic view of the regulatory mechanisms and roles of TGFB2-AS1 in suppressing TNBC progression. Error bars represent mean ± SEM. Statistical significance was assessed using two-tailed Student’s t test. *P < 0.05. **P < 0.01. ***P < 0.001. ns, not significant.
To investigate the clinical significance of TGFB2-AS1 inhibiting TGFβ2 signal, we performed in situ hybridization (ISH) analysis to detect TGFB2-AS1 expression and immunohistochemistry analysis to detect TGFβ2 expression in TNBC specimens. The results revealed that TGFB2-AS1 was negatively correlated with TGFβ2 (Fig. 8 F and G). As expected, patients with high TGFβ2 expression tended to have poorer prognosis, as demonstrated by log-rank tests of the Kaplan–Meier curves (Fig. 8H). Combined prognosis analysis of TGFB2-AS1 and TGFβ2 showed that patients with high TGFB2-AS1 and low TGFβ2 levels correlated with better OS and better DFS than the patients with low TGFB2-AS1 and high TGFβ2 levels (Fig. 8 I and J). In total, our results proposed that TGFB2-AS1 downregulating TGFβ2 plays a key role in inhibiting disease progression of TNBC.
Discussion
The malignant progression of tumors is the result of the accumulation of alterations of tumor-related genes, including activation of oncogenes or/and inactivation of tumor suppressor genes (42, 43). Herein, the transcriptomes from two breast cancer cell lines, LM2 and its parental cell MDA-231, were compared by deep sequencing and we found that TGFB2-AS1 was significantly down-regulated in LM2 cells which are strongly prone to lung metastasis (34). Consistently, lower TGFB2-AS1 expression levels are associated with advanced tumor stage and poor prognosis. Moreover, the potential of CSCs and lung metastasis was induced significantly when TGFB2-AS1 was knocked down by siRNA, while they were dramatically suppressed under exogenously TGFB2-AS1 overexpression in vitro and in vivo.
With the application of techniques such as next-generation sequencing, large numbers of heterozygous or homozygous SWI/SNF subunit mutations have been identified in a large variety of human cancers, suggesting that one or more of the multiple SWI/SNF functions protect against tumorigenesis, as reviewed in refs. 21 and 44. For example, loss-of-function mutations in genes encoding subunits of SWI/SNF complexes, such as the AT-rich interaction domain 1A (ARID1A), are prevalent in cancer, occurring in 20 to 25% of all human malignancies, and these mutations frequently drive oncogenic programs (45). SMARCA4 mutations, including class I SMARCA4 alterations (truncating mutations, fusions, and homozygous deletion) that cause loss of function and class II alterations (missense mutations) that have a dominant negative effect and/or loss of function have been identified in a variety of adult-onset epithelial and mesenchymal neoplasms (46). Therefore, drugs exploiting genetic and epigenetic mechanisms of SMARCA4 antagonism hold promise for future targeted therapies (47). Although these mutations in SWI/SNF subunits including SMARCA4 were present at low frequency (less than 10%) in TNBC (48), we identified that SMARCA4 interacted with TGFB2-AS1, which has been reported to associate with EED, a PRC2 adaptor (49). Besides SMARCA4, the other core subunits of the SWI/SNF complex including BAF170 and SNF5 were also shown to interact with TGFB2-AS1. However, whether these interactions are dependent on SMARCA4 remains to be further investigated. Especially, TGFB2-AS1 inhibits the transcription-regulating activity of SWI/SNF via interaction with SMARCA4 and results in transcriptional repression of its target genes including TGFB2 and SOX2. That should be an alternative mechanism for the chromatin remodeling besides the loss-of-function mutations of SWI/SNF subunits. Meanwhile, we have proved that TGFB2-AS1 directly interacted with the N-terminal region of SMARCA4 through its exon 3 and the N-terminal region contains at least three domains including the QLQ domain, HSA domain, and BRK domain. The HSA domain is reported to mediate intracomplex protein–protein interactions between SAMRCA4 and other subunits of the SWI/SNF complex and is required for SAMRCA4-dependent transcriptional activation (50, 51). Thus, there was a possibility that BAF170 and SNF5 interacted with TGFB2-AS1 through their interaction with SMARCA4, but further investigation needs to be conducted to decipher the detailed molecular mechanism (24, 25, 52).
This story suggested a more complicated regulating network TGFB2-AS1 might involve in the determination of TNBC cell fate. GSEA also revealed that TGFB2-AS1 was closely associated with EMT-related gene signature (SI Appendix, Fig. S14 A and B). In addition, TGFB2-AS1 down-regulation is correlated with WU_CELL_MIGRATION (migration) (53) and VANTVEER_BREAST_CANCER_POOR_PROGNOSIS as well (54) (SI Appendix, Fig. S14 C and D), and these gene sets are all closely related to tumor progression. Our results showed that mesenchymal markers like slug, vimentin, and fibronection were up-regulated and the epithelial marker β-catenin was down-regulated, as expected. However, E-cadherin, a recognized epithelial mark, was slightly up-regulated in MDA-231-siAS1 cells versus control cells (SI Appendix, Fig. S14E). This point is consistent with the need that cancer cells develop overt metastasis in distant organs (27, 32, 55–57) and are also strongly associated with success of cell reprogramming and self-renewal (24, 25).
The protein level of SMARCA4 in the MDA-231-siAS1 cells versus control cells was also investigated and the result indicated that knockdown TGFB2-AS1 had no influence on the protein level of SMARCA4 (SI Appendix, Fig. S14F). However, the protein level of SMARCA4 in LM2 cells was higher than that in MDA-231cells (SI Appendix, Fig. S14G), which suggested that the expressive regulation of SMARCA4 was complicated and further investigation needs to be conducted.
TGFβ signaling plays key roles in cancer progression through its effects on gene expression, release of immunosuppressive cytokines, and epithelial plasticity and thus enhances cancer cell invasion and dissemination, CSC properties, as well as therapeutic resistance (58). Drugs targeting TGFβ signaling have received tremendous attention for late-stage cancer treatment during the past decades. However, targeting TGFβ signaling for cancer therapy is challenging because of its double-edged sword effect in tumor initiation and progression. Different approaches toward selective inhibition of TGFβ signaling should be considered in order to enhance effectiveness and reduce its toxicity. Our results revealed that lncRNA TGFB2-AS1 remarkably abrogated the enhancement of tumor growth and lung metastasis endowed by TGFβ2 in an orthotopically injected TNBC mouse model, which could provide a new potential therapy strategy in TNBC treatment.
In summary, our work reveals a mechanism that TGFB2-AS1 switches cell fate depending on both TGFβ2 and CSC signaling through antagonizing the SWI/SNF complex, thereby suppressing TNBC metastasis (Fig. 8K). The importance of cooperative activity of TGFβ2 and CSC signal pathways emphasizes the regulatory mechanism of the lncRNA in cis and in trans in the same cellular event. The identification of TGFB2-AS1 determining cancer cell fate aids in advancing our understanding of the basic biology of cancer and also sheds light to the treatment of TNBC patients.
Materials and Methods
Clinical Samples.
Human breast cancer samples with informed consent were collected from the Comprehensive Breast Health Center, Shanghai Ruijin Hospital of Shanghai Jiao Tong University School of Medicine. Use of human tissues was approved by the research ethnics committee of Shanghai Jiao Tong University School of Medicine. The data for the TCGA and GTEx cohort of breast cancer was obtained from GEPIA (Gene Expression Profiling Interactive Analysis; http://gepia.cancer-pku.cn/) or downloaded from https://xena.ucsc.edu/public/.
ChIP-Seq.
ChIP-Seq was performed by Novogene. ChIP-Seq reads were mapped to the human reference genome (UCSC hg19) using Bowtie 2 version 2.3.5 with default parameters. Peaks were called with MACS2 version 2.1.2. Metagene profile plots and heat maps were generated from the Galaxy platform using deepTools2 version 3.3.0 (59).
Statistical Analyses.
The statistical analyses were performed using GraphPad Prism version 7 software. Significance was calculated using unpaired two-tailed Student’s t test. Data are represented as mean ± SEM or ± SD with at least three independent experiments. The survival curve was evaluated using the Kaplan–Meier method with a log-rank test. For all figures, statistical significance was represented as *P < 0.05, **P < 0.01, and ***P < 0.001.
Supplementary Material
Acknowledgments
We thank Proteomics Core of College of Basic Medical Sciences, Shanghai Jiao Tong University School of Medicine, for protein liquid chromatography–mass spectrometry analyses and Dr. Keji Zhao and Dr. Qingsong Tang (Systems Biology Center, National Heart, Lung, and Blood Institute, NIH) for providing the materials described in Materials and Methods. This study was funded by the National Natural Science Foundation of China (grants 81772831, 81721004, 81830091, and 91853206), National Key R&D Program of China (2020YFA0803403), Chinese Universities Scientific Fund, and Chinese Academy of Medical Sciences Innovation Fund for Medical Sciences (2019-I2M-5-051).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2117988119/-/DCSupplemental.
Data, Materials, and Software Availability
All study data are included in the article and/or supporting information.
References
- 1.Garrido-Castro A. C., Lin N. U., Polyak K., Insights into molecular classifications of triple-negative breast cancer: Improving patient selection for treatment. Cancer Discov. 9, 176–198 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Batlle E., Clevers H., Cancer stem cells revisited. Nat. Med. 23, 1124–1134 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Nassar D., Blanpain C., Cancer stem cells: Basic concepts and therapeutic implications. Annu. Rev. Pathol. 11, 47–76 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Vanharanta S., Massagué J., Origins of metastatic traits. Cancer Cell 24, 410–421 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Quinn J. J., et al. , Single-cell lineages reveal the rates, routes, and drivers of metastasis in cancer xenografts. Science 371, eabc1944 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu S. Y., Wang H., Shao Z. M., Jiang Y. Z., Triple-negative breast cancer: New treatment strategies in the era of precision medicine. Sci. China Life Sci. 64, 372–388 (2021). [DOI] [PubMed] [Google Scholar]
- 7.Saw P. E., Xu X., Chen J., Song E. W., Non-coding RNAs: The new central dogma of cancer biology. Sci. China Life Sci. 64, 22–50 (2021). [DOI] [PubMed] [Google Scholar]
- 8.Tay Y., Rinn J., Pandolfi P. P., The multilayered complexity of ceRNA crosstalk and competition. Nature 505, 344–352 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoon J. H., et al. , LincRNA-p21 suppresses target mRNA translation. Mol. Cell 47, 648–655 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carrieri C., et al. , Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature 491, 454–457 (2012). [DOI] [PubMed] [Google Scholar]
- 11.Yao R.-W., Wang Y., Chen L.-L., Cellular functions of long noncoding RNAs. Nat. Cell Biol. 21, 542–551 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Liu B., et al. , A cytoplasmic NF-κB interacting long noncoding RNA blocks IκB phosphorylation and suppresses breast cancer metastasis. Cancer Cell 27, 370–381 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Wang P., et al. , The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science 344, 310–313 (2014). [DOI] [PubMed] [Google Scholar]
- 14.Yamazaki T., et al. , Functional domains of NEAT1 architectural lncRNA induce paraspeckle assembly through phase separation. Mol. Cell 70, 1038–1053.e7 (2018). [DOI] [PubMed] [Google Scholar]
- 15.He P., et al. , Epithelial cells-enriched lncRNA SNHG8 regulates chromatin condensation by binding to Histone H1s. Cell Death Differ., 10.1038/s41418-022-00944-x (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li X. L., et al. , Long noncoding RNA PURPL suppresses basal p53 levels and promotes tumorigenicity in colorectal cancer. Cell Rep. 20, 2408–2423 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Espinoza C. A., Allen T. A., Hieb A. R., Kugel J. F., Goodrich J. A., B2 RNA binds directly to RNA polymerase II to repress transcript synthesis. Nat. Struct. Mol. Biol. 11, 822–829 (2004). [DOI] [PubMed] [Google Scholar]
- 18.Gupta R. A., et al. , Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464, 1071–1076 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Creamer K. M., Lawrence J. B., XIST RNA: A window into the broader role of RNA in nuclear chromosome architecture. Philos. Trans. R. Soc. Lond. B Biol. Sci. 372, 20160360 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jégu T., et al. , Xist RNA antagonizes the SWI/SNF chromatin remodeler BRG1 on the inactive X chromosome. Nat. Struct. Mol. Biol. 26, 96–109 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Savas S., Skardasi G., The SWI/SNF complex subunit genes: Their functions, variations, and links to risk and survival outcomes in human cancers. Crit. Rev. Oncol. Hematol. 123, 114–131 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Ribeiro-Silva C., Vermeulen W., Lans H., SWI/SNF: Complex complexes in genome stability and cancer. DNA Repair (Amst.) 77, 87–95 (2019). [DOI] [PubMed] [Google Scholar]
- 23.Wanior M., Krämer A., Knapp S., Joerger A. C., Exploiting vulnerabilities of SWI/SNF chromatin remodelling complexes for cancer therapy. Oncogene 40, 3637–3654 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Apostolou E., Hochedlinger K., Chromatin dynamics during cellular reprogramming. Nature 502, 462–471 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen K., et al. , Heterochromatin loosening by the Oct4 linker region facilitates Klf4 binding and iPSC reprogramming. EMBO J. 39, e99165 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mittal P., Roberts C. W. M., The SWI/SNF complex in cancer - biology, biomarkers and therapy. Nat. Rev. Clin. Oncol. 17, 435–448 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oskarsson T., Batlle E., Massagué J., Metastatic stem cells: Sources, niches, and vital pathways. Cell Stem Cell 14, 306–321 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hachim M. Y., Hachim I. Y., Dai M., Ali S., Lebrun J. J., Differential expression of TGFβ isoforms in breast cancer highlights different roles during breast cancer progression. Tumour Biol. 40, 1010428317748254 (2018). [DOI] [PubMed] [Google Scholar]
- 29.Abd El-Maqsoud N. M., Abd El-Rehim D. M., Clinicopathologic implications of EpCAM and Sox2 expression in breast cancer. Clin. Breast Cancer 14, e1–e9 (2014). [DOI] [PubMed] [Google Scholar]
- 30.Liu P., et al. , SOX2 promotes cell proliferation and metastasis in triple negative breast cancer. Front. Pharmacol. 9, 942 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Massagué J., TGFbeta in cancer. Cell 134, 215–230 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stankic M., et al. , TGF-β-Id1 signaling opposes Twist1 and promotes metastatic colonization via a mesenchymal-to-epithelial transition. Cell Rep. 5, 1228–1242 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boumahdi S., et al. , SOX2 controls tumour initiation and cancer stem-cell functions in squamous-cell carcinoma. Nature 511, 246–250 (2014). [DOI] [PubMed] [Google Scholar]
- 34.Minn A. J., et al. , Genes that mediate breast cancer metastasis to lung. Nature 436, 518–524 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shaw F. L., et al. , A detailed mammosphere assay protocol for the quantification of breast stem cell activity. J. Mammary Gland Biol. Neoplasia 17, 111–117 (2012). [DOI] [PubMed] [Google Scholar]
- 36.Saglam-Metiner P., Gulce-Iz S., Biray-Avci C., Bioengineering-inspired three-dimensional culture systems: Organoids to create tumor microenvironment. Gene 686, 203–212 (2019). [DOI] [PubMed] [Google Scholar]
- 37.Neve R. M., et al. , A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell 10, 515–527 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Anastasiadou E., Jacob L. S., Slack F. J., Non-coding RNA networks in cancer. Nat. Rev. Cancer 18, 5–18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leppek K., Stoecklin G., An optimized streptavidin-binding RNA aptamer for purification of ribonucleoprotein complexes identifies novel ARE-binding proteins. Nucleic Acids Res. 42, e13 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang B., Chambers K. J., Faller D. V., Wang S., Reprogramming of the SWI/SNF complex for co-activation or co-repression in prohibitin-mediated estrogen receptor regulation. Oncogene 26, 7153–7157 (2007). [DOI] [PubMed] [Google Scholar]
- 41.Liu R., et al. , Regulation of CSF1 promoter by the SWI/SNF-like BAF complex. Cell 106, 309–318 (2001). [DOI] [PubMed] [Google Scholar]
- 42.Bozic I., et al. , Accumulation of driver and passenger mutations during tumor progression. Proc. Natl. Acad. Sci. U.S.A. 107, 18545–18550 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ding L., et al. , Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature 464, 999–1005 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.St Pierre R., Kadoch C., Mammalian SWI/SNF complexes in cancer: Emerging therapeutic opportunities. Curr. Opin. Genet. Dev. 42, 56–67 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Helming K. C., Wang X., Roberts C. W. M., Vulnerabilities of mutant SWI/SNF complexes in cancer. Cancer Cell 26, 309–317 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mardinian K., Adashek J. J., Botta G. P., Kato S., Kurzrock R., SMARCA4: Implications of an altered chromatin-remodeling gene for cancer development and therapy. Mol. Cancer Ther. 20, 2341–2351 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chabanon R. M., Morel D., Postel-Vinay S., Exploiting epigenetic vulnerabilities in solid tumors: Novel therapeutic opportunities in the treatment of SWI/SNF-defective cancers. Semin. Cancer Biol. 61, 180–198 (2020). [DOI] [PubMed] [Google Scholar]
- 48.Shain A. H., Pollack J. R., The spectrum of SWI/SNF mutations, ubiquitous in human cancers. PLoS One 8, e55119 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Papoutsoglou P., et al. , The TGFB2-AS1 lncRNA Regulates TGF-β Signaling by Modulating Corepressor Activity. Cell Rep. 28, 3182–3198.e11 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Trotter K. W., Fan H. Y., Ivey M. L., Kingston R. E., Archer T. K., The HSA domain of BRG1 mediates critical interactions required for glucocorticoid receptor-dependent transcriptional activation in vivo. Mol. Cell. Biol. 28, 1413–1426 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu Q., et al. , The BRG1 ATPase of human SWI/SNF chromatin remodeling enzymes as a driver of cancer. Epigenomics 9, 919–931 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Valencia A. M., Kadoch C., Chromatin regulatory mechanisms and therapeutic opportunities in cancer. Nat. Cell Biol. 21, 152–161 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu Y., Siadaty M. S., Berens M. E., Hampton G. M., Theodorescu D., Overlapping gene expression profiles of cell migration and tumor invasion in human bladder cancer identify metallothionein 1E and nicotinamide N-methyltransferase as novel regulators of cell migration. Oncogene 27, 6679–6689 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van ’t Veer L. J., et al. , Gene expression profiling predicts clinical outcome of breast cancer. Nature 415, 530–536 (2002). [DOI] [PubMed] [Google Scholar]
- 55.Yousefi M., et al. , Organ-specific metastasis of breast cancer: Molecular and cellular mechanisms underlying lung metastasis. Cell Oncol. (Dordr.) 41, 123–140 (2018). [DOI] [PubMed] [Google Scholar]
- 56.Padmanaban V., et al. , E-cadherin is required for metastasis in multiple models of breast cancer. Nature 573, 439–444 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu Y., et al. , Long non-coding RNA NR2F1-AS1 induces breast cancer lung metastatic dormancy by regulating NR2F1 and ΔNp63. Nat. Commun. 12, 5232 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Derynck R., Turley S. J., Akhurst R. J., TGFβ biology in cancer progression and immunotherapy. Nat. Rev. Clin. Oncol. 18, 9–34 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Afgan E., et al. , The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res. 44 (W1), W3–W10 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.