Significance
Ribosome hibernation is a conserved mechanism used by both bacteria and eukaryotes to occlude translation and to extend organismal lifespan. Here, we describe a dual-function, hibernation factor hibernation-promoting factor (HPF) in pathogenic Staphylococcus aureus that dimerizes 70S ribosomes and represses YwlG-mediated glutamate dehydrogenase activity via a physical HPF–YwlG interaction. Conversely, YwlG binding to HPF reduces the formation of 100S ribosomes. Our data support the genetic and functional links between HPF and YwlG in the cold response and glucose tolerance. Both HPF and YwlG are validated virulence factors, and our data pave the way to further disentangle the interconnection between bifunctional HPF and YwlG and their multimer assembly.
Keywords: ribosome, glutamate dehydrogenase, Staphylococcus aureus, hibernation, moonlighting protein
Abstract
Bacterial hibernating 100S ribosomes (the 70S dimers) are excluded from translation and are protected from ribonucleolytic degradation, thereby promoting long-term viability and increased regrowth. No extraribosomal target of any hibernation factor has been reported. Here, we discovered a previously unrecognized binding partner (YwlG) of hibernation-promoting factor (HPF) in the human pathogen Staphylococcus aureus. YwlG is an uncharacterized virulence factor in S. aureus. We show that the HPF–YwlG interaction is direct, independent of ribosome binding, and functionally linked to cold adaptation and glucose metabolism. Consistent with the distant resemblance of YwlG to the hexameric structures of nicotinamide adenine dinucleotide (NAD)–specific glutamate dehydrogenases (GDHs), YwlG overexpression can compensate for a loss of cellular GDH activity. The reduced abundance of 100S complexes and the suppression of YwlG-dependent GDH activity provide evidence for a two-way sequestration between YwlG and HPF. These findings reveal an unexpected layer of regulation linking the biogenesis of 100S ribosomes to glutamate metabolism.
The dimerization of 70S ribosomes to form the translationally inactive hibernating 100S complex is a universal strategy employed by most, if not all, bacteria to maintain long-term viability and exit from quiescence (1, 2). Ribosome hibernation is also widespread among eukaryotes (see review (3)). While γ-proteobacteria (e.g., Escherichia coli and Pseudomonas aeruginosa) utilize both ribosome modulation factor (RMF) and hibernation-promoting factor (HPF) to induce 70S dimerization, the majority of bacteria use a single long form of HPF to promote the formation of the 100S complex. In the opportunistic human pathogen Staphylococcus aureus, the 190 amino acid–long HPF consists of a self-dimerizing carboxyl-terminal domain (CTD) and a translation-blocking N-terminal domain (NTD) connected by a 30-amino-acid (a.a.) unstructured linker (Fig. 1A). Cryoelectron microscopy structures of the S. aureus 100S ribosomes have shown that the NTD-HPF sterically occludes mRNA entry and transfer RNA binding to the A site and P site of the 30S decoding center, resulting in translational avoidance. The CTD-HPF at the interface of 30S-30S on one 70S directly interacts with another CTD-HPF that is tethered to the opposite copy of the 70S, resulting in “side-by-side” conjoining of the two 70S monomers (4, 5). In contrast, the E. coli 70S dimer adopts a “back-to-back” joining conformation (6, 7). Although S. aureus HPF forms a homodimer in vitro and binds both individual 30S subunits and 70S complexes (5), 30S-30S dimers have not been observed in vivo.
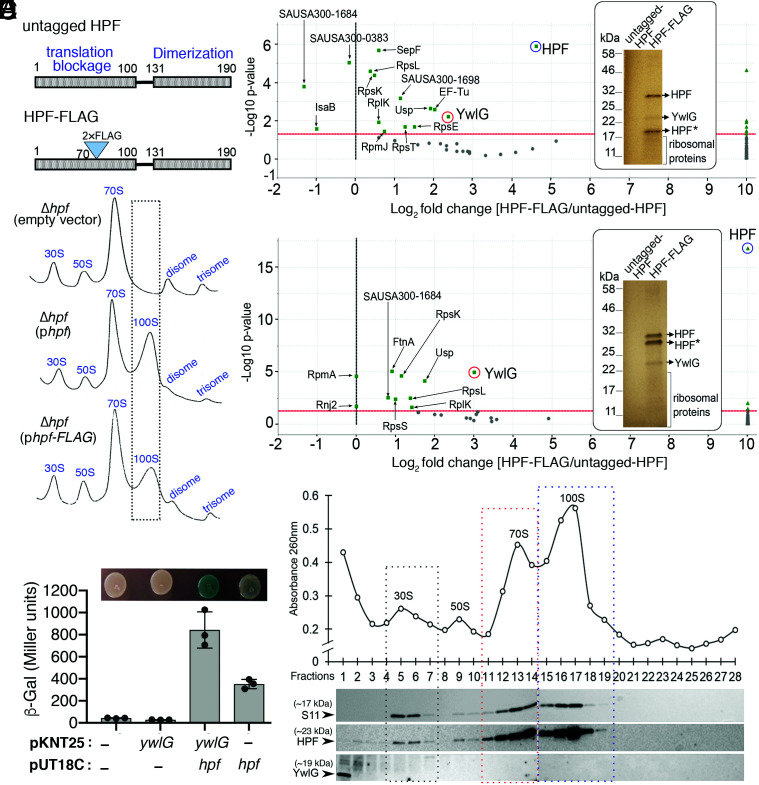
Fig. 1.
Identification of HPF interaction partners in S. aureus. (A) Schematic diagram showing the domain structures of S. aureus HPF and the position of the 2×FLAG in-frame fusion. The N-terminal domain (NTD) of HPF prevents translational initiation, whereas the carboxyl-terminal domain (CTD) promotes the dimerization of 70S ribosomes to form the 100S complex. (B) Ribosome sedimentation profiles of the Δhpf-null mutant bearing either empty vector, untagged HPF, or 2×FLAG-HPF. Five Abs260 units of RNA were loaded on a 5–30% sucrose gradient (x axes), and the levels of ribosomal complexes were monitored by the Abs260 (y axes). FLAG-tag insertion in HPF did not affect ribosome assembly and only modestly reduced the formation of 100S ribosomes. (C and D) Affinity copurification of HPF-binding proteins. TSB cultures of strains from A were harvested during the exponential (C) (4 h) or stationary phase (D) (18 h) at 37 °C. The total proteins were incubated with anti-FLAG M2 magnetic beads, and HPF and HPF-binding candidates were eluted as described in the Materials and Methods. Ten percent of the eluate was analyzed on an SDS-PAGE gel (Insets, images were pseudocolored), and protein bands that were unique in FLAG-tagged samples were identified by trypsin digestion followed by liquid chromatography (LC)-MS/MS. HPF* indicates degraded intermediates. The remaining eluate was directly subjected to gel-free quantitative MS, and differentially enriched proteins were visualized in Scaffold 4.11.0 and are presented in volcano plots (n = 2, P < 0.005, Fisher’s test with Benjamini–Hochberg parameters). (E) Validation of the HPF–YwlG interaction using a bacterial 2-hybrid (BACTH) system. T25-YwlG and T18-HPF chimeras are indicated on the x axis. High β-galactosidase activity is indicative of a direct YwlG–HPF interaction, consistent with chromogenic detection of LacZ accumulation on LB-X-Gal-isopropyl ß-D-1-thiogalactopyranoside solid agar after 24–30 h at 30 °C (Top). Each bar represents the average of three independent biological replicates (±SD). (F) YwlG does not associate with the mature ribosomes. Ribosome sedimentation profile of the wild-type JE2 strain grown in TSB to late exponential phase (optical density at 600 nm [OD600] = 1.6) at 37 °C. Five Abs260 units of RNA were ultracentrifuged through a 5–30% sucrose gradient (x axis), and the abundance of ribosomal complexes was monitored by Abs260 (y axis). Fractions were collected and probed with anti-HPF, anti-YwlG, and anti-S11 (30S subunit marker) on Western blots.
Despite the variation in hibernation factors and distinct dimeric architecture of the 100S complexes (5–11), the loss of HPF or RMF homologs leads to one or more phenotypes, including decreased long-term viability and regrowth (12–14), reduced antibiotic and stress tolerance (8, 15–17), and accelerated ribosome degradation and damage (12, 13, 18–23). Recent studies have shown that S. aureus HPF binding protects the 30S subunit and 70S ribosome from degradation by the 3′–5′ exoribonuclease (RNase) R and an unidentified RNase at three functionally important regions of 16S ribosomal RNA (rRNA) (24), whereas in E. coli lacking RMF, HPF, and YfiA, 16S rRNA is degraded by the concerted action of RNase R and the endonuclease YbeY at sites that are distinct from those found in S. aureus (25). The rapid ribosome turnover observed in an S. aureus Δhpf-null mutant is linked to ∼1,000-fold and ∼100-fold reductions in murine infections and viability in long-term cultures in vitro, respectively (13, 26). Intriguingly, S. aureus constitutively expresses hpf throughout the cell growth cycle (13, 27–29), overexpression of HPF does not significantly retard cell growth (13), and global translation is only modestly compromised in HPF-proficient cells (13). These observations have led to an emerging hypothesis that the primary role of HPF is to prevent ribosome degradation rather than translational inhibition (as previously envisioned) and that translational avoidance is a consequence of HPF-NTD occupancy at the 30S decoding center.
The reversal of ribosome hibernation via the disassembly of 100S complexes is thought to promote bacterial regrowth and long-term survival, during which the dissociated 30S, 50S, and 70S ribosomes are presumably recycled for translational restart. In S. aureus, the dissociation involves the major ribosome recycling factor/elongation factor G (EF-G) pair and an alternative guanosine triphosphatase (GTPase) HflX that split the 100S ribosomes in a GTP hydrolysis–dependent manner under nonstressed conditions and heat stress, respectively (26, 30, 31). This mechanism suggests that assembly and disassembly of 100S ribosomes are dynamic and partially explains the lack of translational derepression in a Δhpf-null mutant (13). S. aureus HPF is a highly abundant protein, so how the cells avoid rebinding of HPF to ribosomes following 100S complex disassembly is unclear. One possibility is that mRNAs may compete for the 30S subunits during translational initiation. This model has not been validated directly and cannot be applied to 70S ribosomes, which are the major targets of HPF. Rapid HPF turnover is unlikely, as HPF remains abundant even when no mature ribosomes are detected (13). Alternatively, the HPF–ribosome interaction may be interposed by a previously unrecognized factor that titrates HPF away from ribosomes. Motivated by this latter hypothesis, we identified an uncharacterized virulence factor, YwlG, that directly interacts with HPF and reduces the formation of 70S dimers in a methicillin-resistant S. aureus (MRSA). The HPF–YwlG interaction is independent of ribosome binding. Inactivation of ywlG suppresses the cold sensitivity and glucose susceptibility phenotypes of a Δhpf-null mutant. We solved the X-ray crystallography structure of YwlG to 1.75 Å and found that HPF-CTD is critical for HPF–YwlG interactions. Unexpectedly, we found that YwlG is able to functionally compensate for the loss of a NAD-specific glutamate dehydrogenase GudB. The surprising physical and functional association of HPF and YwlG unveils an additional layer of regulation controlling the cellular concentrations of 100S ribosomes and key metabolic pathway.
Results
HPF Directly Interacts with a Cytoplasmic YwlG Independent of Ribosome Association.
To test the hypothesis that HPF is sequestered by a hitherto unknown factor, we constructed a FLAG-tagged HPF as a bait to identify potential HPF binding partners using a whole-cell lysate affinity purification approach and compared the enrichment of copurified proteins with that of an untagged HPF control. HPF is extremely sensitive to mutational perturbation in the NTD–CTD linker region and cannot tolerate affinity tagging at its N and carboxyl termini (8, 13). We found that an insertion of a tandem FLAG tag at the position 70 of the protein resulted in less perturbation of 70S dimerization than various affinity-tagged fusions (Fig. 1A). The untagged HPF and 2×FLAG HPF were expressed on a medium-copy plasmid under the control of the native hpf promoter. We confirmed that 2×FLAG HPF is able to restore the formation of 100S ribosomes in a Δhpf knockout strain, albeit at a lower level than the untagged HPF (Fig. 1B). S. aureus Δhpf cells carrying the HPF-expressing plasmids were harvested at the exponential growth phase (Fig. 1C) and stationary phase (Fig. 1D). Our label-free quantitative mass spectrometry analyses revealed that, aside from the expected HPF bait, ribosomal proteins (RpxX, x indicates any letters), a universal stress protein (Usp homolog, SAUSA300_1652), and a translation factor (EF-Tu) were copurified. Additionally, a protein of unknown function (YwlG, locus tag SAUSA300_2068) was enriched by 5-fold to 16-fold during the logarithmic growth and stationary phases, respectively. YwlG enrichment was also detectable on a sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gel stained with EZ-Blue following mass spectrometric verification (Fig. 1 C and D, Insets). The results indicate that YwlG is a binding partner and potentially a sequestrator of HPF.
The HPF–YwlG association detected in the affinity pull-down assay could have been indirect. To rule out this possibility, we used the bacterial two-hybrid (BACTH) system to confirm a direct interaction between HPF and YwlG. The BACTH system is based on the interaction-mediated reconstruction of a cyclic adenosine monophosphate (cAMP) signaling cascade (32). YwlG and HPF are each genetically fused to either the T25 or T18 fragment of the adenylate cyclase (Cya) catalytic domain from Bordetella pertussis and coexpressed in an E. coli Δcya strain. Functional complementation between T25 and T18 via a direct interaction of heterologous gene products leads to the production of cAMP, in turn activating the expression of a β-galactosidase LacZ reporter. We found that high β-galactosidase activity was only observed when both T25-YwlG and T18-HPF were coexpressed, not when either was expressed by itself, although T18-HPF alone produced basal LacZ levels. The quantitative β-galactosidase activity results were consistent with the colorimetric visualization of LacZ production (Fig. 1E). The BACTH results confirm that the Cya catalytic activity is mediated by the physical interactions between HPF and YwlG.
S. aureus HPF predominantly binds to 70S and 100S ribosomes and to a lesser extent to the 30S subunits. It is possible that ribosomes act as mediators of HPF–YwlG binding. To exclude this possibility, we performed sucrose density gradient ultracentrifugation to fractionate ribosomes into subunits and complexes. Each fraction was then collected to probe for the presence of HPF and YwlG by Western blotting. The quality of fractionation was monitored by the distribution of the 30S ribosomal protein S11. We first confirmed that the newly raised polyclonal YwlG antibody is specific (SI Appendix, Fig. S1). As expected, HPF and S11 were cofractionated with 30S, 70S, and 100S ribosomes, but YwlG was only found in the ribosome-free, low-density fractions (Fig. 1F), indicating that YwlG is not associated with mature ribosomes and does not bind to HPF-bound ribosomes. Collectively, the data from the unbiased affinity copurification, targeted BACTH, and the ribosome fractionation confirm that the S. aureus HPF–YwlG interaction is direct and specific, occurs extraribosomally, and can be recapitulated heterologously in E. coli.
Distribution and Possible Roles of YwlG.
S. aureus YwlG is a 174-a.a.-long uncharacterized protein member of the UPF0340 family with a DUF436 domain of unknown function. A phylogenetic tree constructed for a selected set of 256 YwlG homologs from representative bacterial phyla revealed that YwlG homologs are almost exclusively found in the Firmicutes phylum, with a few exceptions found in the phyla of Deinococcota, Fibrobacteres, Bacteroidetes, and Spirochaeta (SI Appendix, Fig. S2). Incidentally, all bacterial species that carry YwlG homologs only harbor HPF homologs that have an extended carboxyl-terminal dimerization domain (Fig. 1A). Multiple sequence alignment of YwlG homologs revealed distinct regions of conservation (SI Appendix, Fig. S3). A previous chemical proteomic screen has suggested that S. aureus YwlG binds to pyridoxal phosphate (PLP) (33), a B6 vitamer that serves as the cofactor of many enzymes involved in a.a. transformation. However, YwlG–PLP binding has not been experimentally validated in vivo and the putative PLP binding residue (K58 in S. aureus) is not conserved among YwlG homologs (SI Appendix, Fig. S3). A ΔywlG knockout did not exhibit growth defects in laboratory rich medium, changes in cell morphology, or sensitivity to oxidative, osmotic, and heat stress. However, a ΔywlG mutant has been found to exhibit 50% compromised lung colonization in a model of acute murine pneumonia (34) and to exhibit reduced fitness during osteomyelitis (35), indicating that YwlG is critical for staphylococcal pathogenesis.
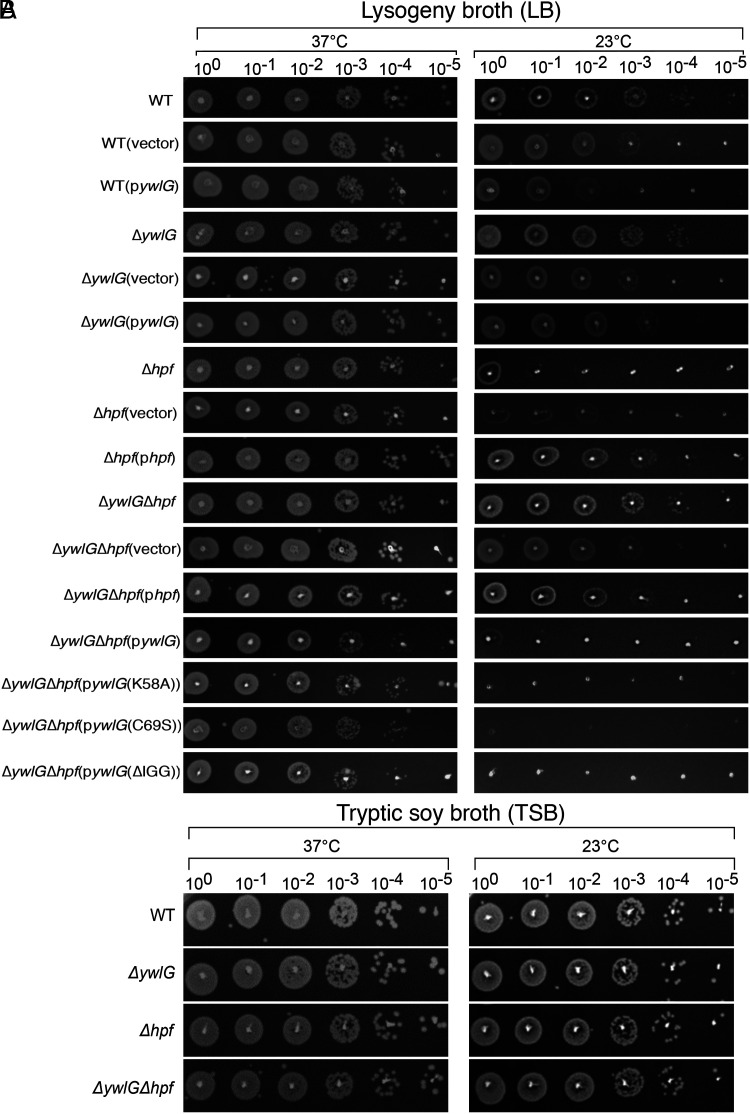
Inactivation of ywlG Suppresses the Cold Sensitivity of Δhpf Null Mutant in Lysogeny Broth (LB).
Cold sensitivity is a hallmark of defects in ribosome biogenesis and translation (36). It has been assumed that, at lower temperatures, rRNA folding is slowed, ribosome assembly is hindered, the enzymatic activity of translation factors is reduced, and mRNA secondary structure is increased. While a Δhpf-null mutant does not exhibit measurable growth defects under various laboratory conditions, the growth of a Δhpf-null mutant on LB agar at low temperature (23 °C) was reduced by four orders of magnitude, and the cold sensitivity could be fully rescued by expression of hpf in trans (Fig. 2A). These defects were not seen on a tryptic soy broth (TSB)-based agar (Fig. 2B). LB medium is known to undergo rapid depletion of carbon sources and to contain low concentrations of Mg2+ and Ca2+ (37, 38). The cold sensitivity of the Δhpf-null mutant was likely associated with defects in ribosome assembly due to Mg2+ deficiency and/or loss of cell-membrane strength as a result of insufficient Mg2+ and Ca2+. Importantly, introducing ΔywlG-null allele in the Δhpf-null mutant fully restored the growth of ΔhpfΔywlG to the wild-type (WT) levels. Genetic complementation with an hpf-expressing plasmid rendered cold tolerance of the ΔhpfΔywlG double mutant, whereas introducing a ywlG-expressing plasmid to the ΔhpfΔywlG double mutant restored cold sensitivity (Fig. 2A). Furthermore, overexpression of YwlG in the WT background sensitized cells at low temperature by ∼10-fold, presumably due to insufficiency of HPF (partially resembles a Δhpf) in excess of YwlG (Fig. 2A). Overexpression of YwlG in the WT also caused a growth delay (SI Appendix, Fig. S4A), implying that excess amount of YwlG is detrimental. Taken together, these results indicate that hpf mutation renders medium-specific cold sensitivity and that inactivation of ywlG suppresses the Δhpf phenotype, providing evidence that hpf and ywlG are genetically and functionally linked, in addition to engaging in direct physical interaction.
Fig. 2.
Inactivating ywlG suppresses the cold sensitivity of a Δhpf-null mutant in a medium-dependent manner. Complementation with plasmid-encoded ywlG and hpf confirmed the YwlG-mediated suppression and HPF-dependent cold response. Serial dilution spot assays were performed on (A) lysogeny broth (LB)-based or (B) tryptic soy broth (TSB)-based agar plates. Exponentially growing cells (OD600 = 0.6) in TSB were adjusted to OD600 = 0.2 after two washes in 1xPBS. After serial dilutions, 2 µL of each dilution was spotted on the agar plates. The results were recorded after 24 h and 48 h of incubation at 37 °C and 23 °C, respectively. The images are representative of three independent experiments.
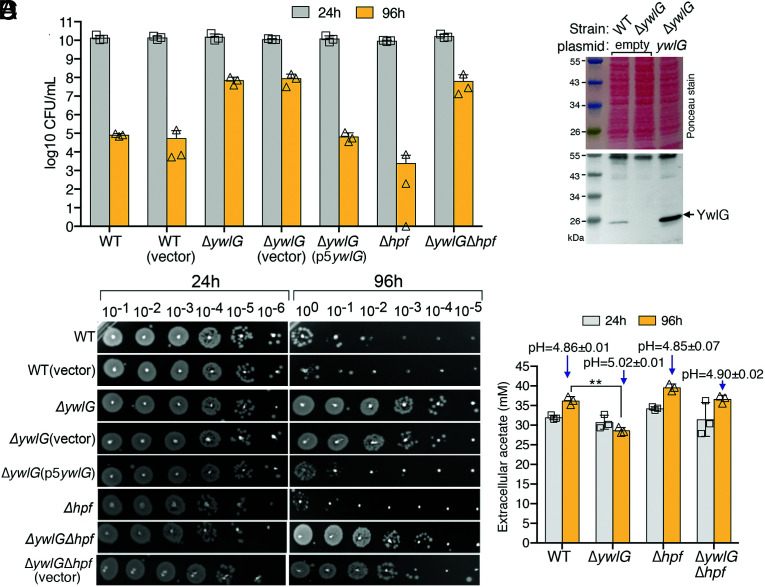
Inactivation of ywlG Promotes Stationary Phase Survival of Δhpf-Null Mutant under Conditions of Excess Glucose.
Aerobically grown S. aureus in TSB medium containing excess glucose (35–45 mM) enters stationary phase before the glucose is exhausted. However, bacteria continue to consume glucose and excrete acetate as a byproduct that, when not consumed as a secondary carbon source, potentiates cell death due to inflow of protonated acetate, which causes intracellular acidification and reactive oxygen species (ROS) production (39, 40). Motivated by these observations, we compared the survival of Δhpf and ΔywlG mutant strains in TSB-glucose medium by enumerating the colony forming units (CFUs) (CFUs/mL) (Fig. 3A) and by performing a dilution spot assay over a period of 4 d (Fig. 3B). Consistent with the findings of previous studies (39, 40), no significant differences in viable cell counts and growth rate were observed among all strains in the first 24 h of growth (Fig. 3 and SI Appendix, Fig. S4 B and C). However, the WT and a Δhpf-null mutant showed decreases of five and six orders of magnitude in cell viability after 96 h, respectively. In contrast, a ΔywlG-null mutant was more viable than the WT and the Δhpf-null strain by three and four orders of magnitude, respectively. Expressing ywlG from a plasmid converted the glucose tolerance of the ΔywlG-null mutant into cell death. Furthermore, glucose killing in the Δhpf-null mutant could be suppressed by combination with a ΔywlG-null mutant (Fig. 3 A–C). By an unknown mechanism, Bacillus subtilis YwlG is associated with 5-aminopentanoylation of the EF-P. EF-P functions to alleviate ribosome stalling at the polyproline sequence (41). That a Δefp mutant did not phenocopy the ΔywlG mutant (SI Appendix, Fig. S5) suggests that S. aureus YwlG and EF-P likely do not act in the same pathway during acetate-potentiated cell death. Finally, the degrees of cell death were only loosely correlated with acetate production and pH (Fig. 3D), suggesting that cytoplasmic acidification and pH were not the direct cause of cell death.
Fig. 3.
A ΔywlG-null mutant is resistant to glucose-dependent cell death in the stationary phase. (A) Excess glucose differentially impacts cell death in S. aureus mutant strains. A ΔywlG knockout promoted cell viability and suppressed cell death in a Δhpf-null mutant. Cell viability (CFU/mL, n = 3, mean ± SD) was monitored over a period of 4 d in TSB-45 mM glucose. (B) Dilution spot assays for cells collected in A. Each spot corresponds to 2 µL of serially diluted cells. Genetic complementation was achieved by expressing ywlG from a plasmid. The empty vector served as a control. (C) Western blot showing the expression levels of endogenous YwlG (WT lane) versus plasmid encoded ywlG in TSB-glucose medium at 24 h. (D) Acetate levels and pH of culture supernatant of TSB-glucose cultures at 24 h and 96 h (n = 3, mean ± SD). A moderate reduction in acetate and a slight increase in pH were observed in the ΔywlG backgrounds. **P < 0.01.
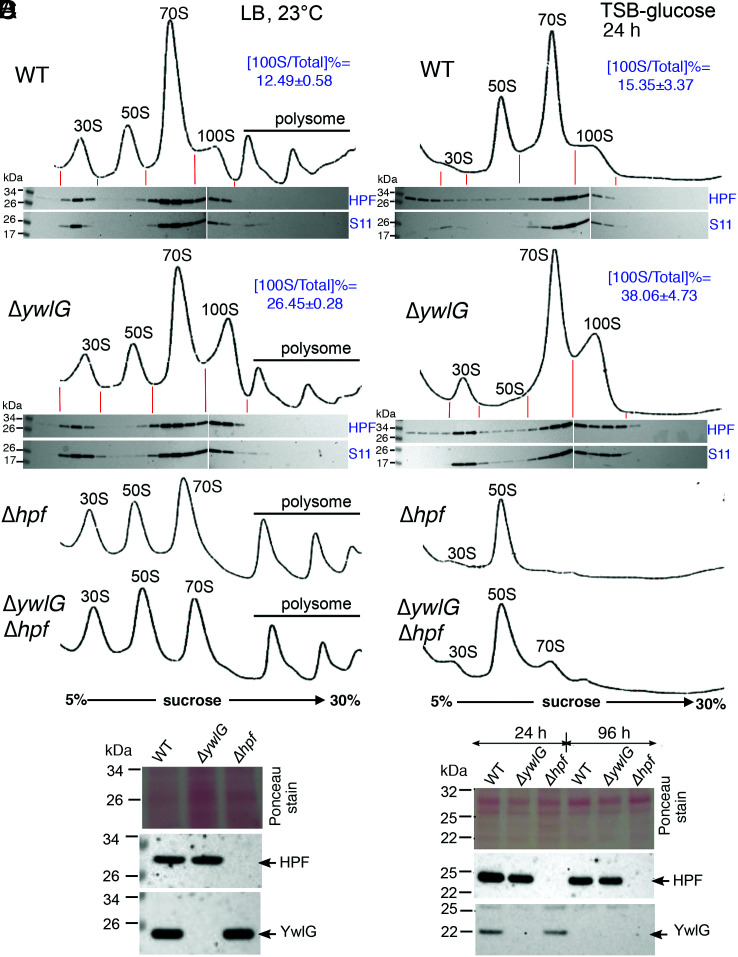
The ΔywlG Mutant Accumulates 100S Ribosomes.
To validate our initial hypothesis that YwlG sequesters HPF away from the ribosome binding, we examined the abundance of 100S ribosomes upon ywlG inactivation in LB (23 °C) and TSB-glucose cultures, two conditions that support the genetic and functional links between HPF and YwlG. Crude ribosomes were prepared by cryomilling to faithfully preserve ribosome complexes and translational states (13, 27). Next, we assessed the proportion of the 100S peak relative to the total mature ribosomes on a density gradient fractionation chromatogram. Under both conditions, the levels of 100S ribosomes in the ΔywlG increased by twofold relative to the WT strain (Fig. 4 A and C). We confirmed that the accumulation of 100S ribosomes was not due to an increase in HPF concentration (Fig. 4 B and D). Since YwlG likely binds to the free HPFs not associated with ribosome (Fig. 1F), Western blots show that significantly more free HPFs were found in the low-density fractions in WT than in the ΔywlG (Fig. 4C, Bottom). These results collectively indicate that HPF–YwlG binding prevents the formation of 100S ribosomes.
Fig. 4.
Inactivation of ywlG increases the formation of 100S ribosomes. (A and C) Ribosome sedimentation profiles of WT and its mutants. Cells were grown in LB at 23 °C until OD600 = 1.6 (A) or grown in TSB-glucose at 37 °C for 24 h (C). Crude ribosomes were extracted and subjected to 5–30% sucrose gradient fractionation (x axes), and ribosomal complexes were monitored by ultraviolet absorbance at 254 nm (y axes). Western blots show that significant amount of ribosome-free HPF was found in the WT than the ΔywlG mutant (C, Bottom). The ratios of 100S ribosome versus total mature ribosome content were quantitated according to the areas under the curve using ImageJ (n = 3, mean ± SD). The 100S ribosomes accumulate upon ywlG deletion by greater than twofold relative to the level in the WT. (B and D) Western blots showing that the levels of HPF were unaffected by ΔywlG knockout in the total lysates of LB cultures (A) and TSB-glucose cultures (D). Total proteins were resolved on a 4–12% Bis-Tris NuPAGE gel, and immunoblotting was performed with anti-YwlG (1/1,000) or anti-HPF (1/4,000).
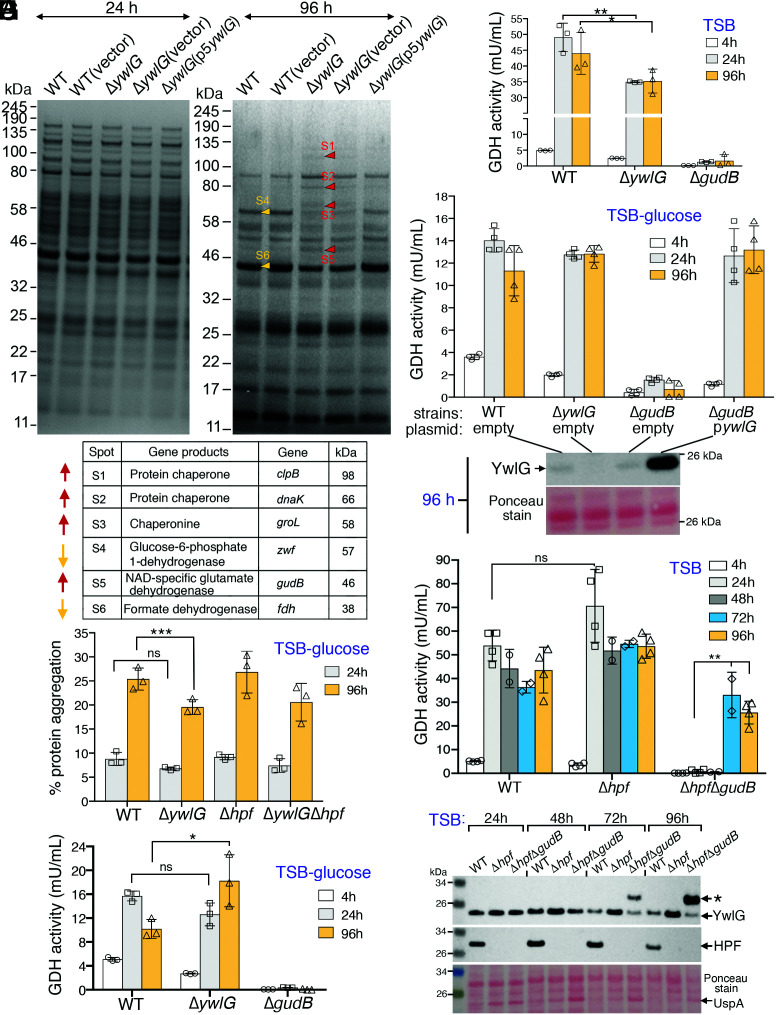
The ΔywlG Mutant Produces a Distinct Proteome under Excess Glucose Conditions.
Beyond serving as an HPF sequestrator and participating in cold- and glucose-response pathways, the actual biological activity of YwlG remains unknown. We sought to address this knowledge gap by comparing the protein profiles of WT, ΔywlG, Δhpf, and their derivatives grown in TSB-glucose medium, a condition that produces one of the strongest phenotypes (Fig. 3). Total soluble proteins extracted from 24 h and 96 h were resolved on an SDS-PAGE gel, and differentially produced proteins were identified by mass spectrometry. Despite the low resolution and sensitivity of this one dimensional gel–based approach, we found that at least six protein species were differentially produced in the ΔywlG and ΔhpfΔywlG strains at 96 h, whereas no differences were detectable at 24 h (Fig. 5A and SI Appendix, Fig. S6A), consistent with previous findings that all strains have similar viability at 24 h (Fig. 3A). The four upregulated proteins were the protein chaperones ClpB, DnaK, and GroL and the NAD-specific glutamate dehydrogenase GudB. Two metabolic enzymes, Zwf and Fdh, were downregulated (Fig. 5B). Importantly, the changes could be reversed by complementation with a plasmid encoding ywlG, confirming that these alterations were specific to the loss of ywlG. To rule out that the changes were technical artifacts, we validated the differential steady-state levels of one of the proteins (DnaK) by immunoblotting (SI Appendix, Fig. S6B).
Fig. 5.
Proteomic approach to elucidate the biological activity of YwlG. (A) Comparative protein profiles of the WT, the ΔywlG mutant, and the ΔywlG complementation strain. Total proteins were extracted from TSB-glucose cultures at 24 and 96 h, resolved on a 4–12% Bis-Tris NuPAGE gel, and stained with EZ-Blue. Differentially expressed proteins are marked with red arrows (upregulated) or orange arrows (downregulated). The protein identities were determined by LC/MS-MS following in-gel trypsin digestion. (B) Differentially expressed proteins identified in A. (C) Strains containing the ΔywlG mutant allele produce reduced amounts of protein aggregates. Total aggregates were measured with a PROTEOSTAT dye and the manufacturer’s supplied standards consisting of defined quantities of protein aggregates and nonaggregates. (D and E) Quantitation of glutamate dehydrogenase (GDH) activity in strains grown in TSB-glucose (D) and TSB (E). One unit of GDH is defined as the amount of enzyme that generates 1.0 µmol of NADH per minute at pH 7.6 at 37 °C. Excess glucose negatively impacted the production of GDH activity. (F) Overexpression of YwlG restored GDH activity in a ΔgudB-null mutant in TSB-glucose medium. Western blot showing the levels of YwlG protein at 96 h. (G) Combining the Δhpf allele and a ΔgudB null mutant alleviated the inhibition of HPF on YwlG-mediated GDH activity in plain TSB medium after prolonged culture. (H) Western blot showing expression levels of YwlG and HPF in strains harvested from G. An asterisk indicates potential modification of YwlG or a nonspecific band against anti-YwlG. A universal stress protein homolog (UspA, SAUSA300_1656), as identified by MS, is upregulated in the ΔhpfΔgudB mutant. ***P < 0.005, **P < 0.01, *P < 0.05, not significant (ns).
The bacterial chaperone ClpB solubilizes and disaggregates misfolded proteins in conjunction with DnaKJE, and GroELS aids in protein refolding (42). The enrichment of ClpB, DnaK, and GroL in the ΔywlG knockout mutant prompted us to test whether the loss of WT cell viability in TSB-glucose medium was due to accumulation of toxic protein aggregates. We used a thioflavin T–like analog, PROTEOSTAT, to quantitate the amount of protein aggregates. The analog is a molecular motor that spins in solution and produces no fluorescence. Intercalation of the analog into the amyloid fibril–like aggregates reduces rotation of the dye and generates strong fluorescence signals. Using this approach, we found that, indeed, strains bearing the ΔywlG allele had ∼7% lower total protein aggregate amounts than the WT and Δhpf strains (Fig. 5C). Although the effects were less pronounced than expected, we reasoned that an increase in the cell viability of the ΔywlG knockout mutant in TSB-glucose medium could have occurred due to the derepression of protein chaperones, leading to alleviation of toxic aggregation and improvement of cell survival. To test this possibility, we introduced a ΔclpB-null allele into a ΔywlG knockout mutant with the assumption that the double mutant would be less viable than the single ΔywlG mutant. However, the ΔclpBΔywlG double mutant was equally viable compared with the single ΔywlG mutant (SI Appendix, Fig. S5), suggesting that the accumulation of protein chaperones was likely a secondary byproduct or that inactivating one chaperone alone was insufficient to elicit cell death due to the redundancy of bacterial protein chaperones.
YwlG Compensates for Loss of the NAD-Specific Glutamate Dehydrogenase GudB.
An ≃2.5-fold increase in GudB protein levels was observed in the ΔywlG knockout mutant (Fig. 5A). GudB catalyzes the interconversion of glutamate to 2-oxoglutarate and ammonia while reducing NAD to NADH. Of note, the ΔgudB mutant was sensitive to glucose killing, similar to the WT and Δhpf mutant (SI Appendix, Fig. S5). Using the ΔgudB mutant as a negative control, we performed glutamate dehydrogenase (GDH) activity assays to confirm that higher GudB in the ΔywlG knockout mutant contributed to a modest increase in GDH activity in TSB-glucose medium at 96 h (Fig. 5D). In contrast, we found that GDH activity was moderately but significantly lower in the ΔywlG knockout mutant than in the WT in plain TSB medium throughout the 96-h time course (Fig. 5E). Overall, GDH activity was >2.5-fold lower in all strains under conditions of excess glucose (compare y axes of Fig. 5 D and E), possibly due to CcpA-mediated catabolic repression of GDH genes similar to that reported in B. subtilis (43, 44) and S. aureus (45).
Unlike B. subtilis, which carries two paralogous GDHs (RocG and GudB) (46), S. aureus possesses only GudB. Incidentally, S. aureus YwlG shares 11% sequence identity with the GudB homologs (SI Appendix, Fig. S7) and adopts a three-dimensional fold reminiscent of GDHs (Fig. 6). We hypothesized that YwlG and GudB may be functionally interchangeable, in part because we were unable to create a ΔywlGΔgudB double-knockout mutant despite numerous attempts. To test this hypothesis, we complemented a ΔgudB mutant with the plasmid-borne YwlG under control of ywlG native promoter. To our surprise, overexpressing YwlG in trans fully restored GDH activity to the WT levels in both TSB-glucose medium (Fig. 5F) and plain TSB (SI Appendix, Fig. S8A), while the mutant carrying the empty vector did not, implying that S. aureus ywlG may encode an alternative GDH.
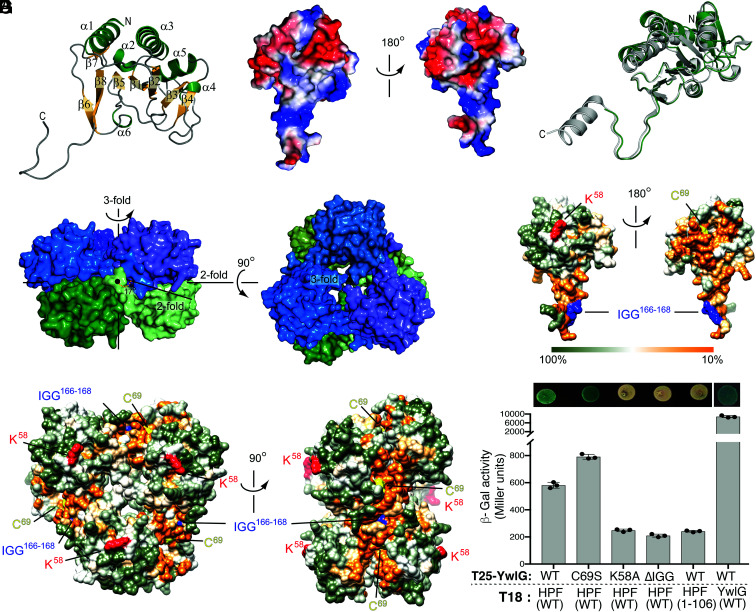
Fig. 6.
X-ray crystal structure of S. aureus YwlG and mutagenesis of YwlG and HPF. (A) Folding topology of YwlG shown on the monomer with secondary structural elements annotated. (B) Charges of amino acid side chains (red = negative; blue= positive) mapped onto the surface of a YwlG monomer. (C) Superposition of a YwlG monomer (green) with the homologous structure of the conserved hypothetical protein TT1679 from Thermus thermophilus (gray, PDB 1V8D). (D) Assembly of YwlG into a hexamer composed of two stacked trimeric rings. (E and F) Sequence conservation mapped onto the surface of the YwlG monomer (E) and hexamer (F), with K58, C69, and the 166IGG168 motif highlighted in red, yellow, and blue, respectively. (G) BACTH assay. HPF-CTD is important for HPF–YwlG interaction, whereas YwlG(K58A)-T25 and YwlG(ΔIGG)-T25 proteins are unstable (SI Appendix, Fig. S10), resulting in the lack of β-Gal activity. Interaction between YwlG-T25 and YwlG-18 independently validates the oligomerization status of YwlG in the crystals. The error bars indicate mean ± SD, n = 3.
If S. aureus YwlG processes GDH activity, the finding that the ΔgudB mutant completely lacked GDH activity suggests that YwlG activity might be repressed in the ΔgudB background, potentially due to the HPF–YwlG interaction. To test this possibility, we constructed a ΔhpfΔgudB double mutant and repeated the GDH assays. In partial agreement with our prediction, a significant increase in GDH activity was detected in the ΔhpfΔgudB mutant grown in prolonged TSB cultures (72–96 h) (Fig. 5G), but not in cells grown in TSB-glucose medium (SI Appendix, Fig. S8B). Immunoblotting showed an elevated YwlG protein level when hpf was deleted (Fig. 5H), suggesting that YwlG is destabilized by HPF in TSB. These results collectively suggest that YwlG is potentially a bona fide GDH that is only enzymatically active in the absence of HPF. Alternately, YwlG may serve as an activator of an otherwise silent GDH of unknown identity or is one of the components of an unidentified GDH complex (see Discussion).
Structural Determination of S. aureus YwlG and Mutagenesis Analyses.
We determined the X-ray crystallography structure of YwlG to 1.75-Å resolution and did not detect bound PLP or other ligands. The data and refinement statistics are summarized in SI Appendix, Table S1. The fold of YwlG is characterized by a central eight-stranded antiparallel β-sheet decorated with three large and three short α-helices with an unusual 12-residue-long carboxyl-terminal extension (Fig. 6 A–C). The oligomeric state was determined by size-exclusion chromatography (SEC), in which YwlG was eluted with a size corresponding to a complex equivalent to a hexamer (SI Appendix, Fig. S9A). Notably, the elution volume is largely impacted by the shape of the protein or protein complex, with globular-shaped complexes traveling faster. Consistent with the SEC analysis, the asymmetric unit of the crystal was six protomers that made up two stacked trimers (Fig. 6D). The carboxyl-terminal extension appeared to interlock the oligomer. Analysis of the multimeric state of the protein showed that the hexamer was a stable composition, with a solvent-accessible surface area of 39,710 Å2 and a surface area upon hexamer formation of 12,540 Å2 (estimated G: −56.6 kcal/mol). The S. aureus YwlG monomer was almost superimposable on a conserved hypothetical protein from Thermus thermophilus (Protein Data Bank [PDB] 1V8D) (Fig. 6C), with which it shared 40% sequence identity (SI Appendix, Fig. S3). YwlG also shares distant similarity to GDHs, e.g., PDB 1V9L (47) and PDB 3K8Z (48). Notably, YwlG is half the size of the GDHs (SI Appendix, Fig. S7) and is only superimposable with the CTD of GudB1 (SI Appendix, Fig. S9B).
Guided by the YwlG structure, we attempted to delineate regions that are important for HPF interaction. We substituted the nonconserved residue K58 with an alanine because S. aureus YwlG K58 has been previously predicted to bind PLP (33). We replaced the invariant residue C69 with a serine because C69 was oxidized in the crystals with unknown significance. We deleted the 166IGG168 sequence in the carboxyl-terminal extension because the 167Gly-Gly168 dipeptide is invariant in all YwlG homologs (Fig. 6 E and F and SI Appendix, Fig. S3) and may be involved in trimeric oligomerization and/or HPF interaction. YwlG homologs are absent from bacteria harboring a short version of HPF that lacks the equivalent S. aureus HPF-CTD dimerizing domain (Fig. 1A). We predicted that HPF-CTD is responsible for interacting with YwlG because these proteins may have coevolved over time. We introduced a premature stop codon at the E107 position of HPF, which resulted in truncation of the entire CTD. These mutations were introduced into the existing BACTH system. With the exception of the YwlG(C69S) substitution, which resulted in an increase in β-galactosidase (β-Gal) activity, all other mutations produced negligible low β-Gal activity (Fig. 6G). Immunoblotting showed that the instability of CyaA’-YwlG(K58A) and CyaA’-YwlG(ΔIGG) in E. coli contributes to the lack of β-Gal activity, although the same YwlG mutants in the native S. aureus are fully functional in reversing cold sensitivity of a Δhpf (Fig. 2A) and restoring GDH activity of a ΔgudB (SI Appendix, Fig. S8C). By contrast, the expression levels of CyaA’-HPF1–106 and CyaA’-YwlG(WT) were comparable (SI Appendix, Fig. S10); thus, the lack of β-Gal activity is due to the impaired HPF–YwlG interaction. YwlG–YwlG interaction was also validated in the BACTH system (Fig. 6G and SI Appendix, Fig. S10). While these experiments confirm the critical role of HPF-CTD in YwlG binding, more exhaustive separation-of-function mutagenesis will be needed to distinguish perturbation of protein oligomerization from defective partner binding.
Discussion
Bacterial hibernation factors are not known to interact with extraribosomal components. Here, we uncovered the genetic, physical, and functional links between the HPF and a previously uncharacterized YwlG protein in a MRSA. Two-way sequestration between HPF and YwlG is supported by (1) inactivation of ywlG suppressing cold sensitivity and glucose susceptibility of an Δhpf knockout, (2) deletion of hpf derepressing YwlG-dependent GDH activity in a ΔgudB knockout, (3) HPF–YwlG interaction reducing the levels of 100S ribosomes, and (4) HPF without the carboxyl-terminal dimerization domain failing to interact with YwlG. Several major protein chaperones and metabolic enzymes were differentially expressed in the ΔywlG mutant. Proteomic analysis, structural determination of YwlG, and functional complementation support a model in which YwlG is either a previously unrecognized GDH enzyme, a pseudo-GDH, or a stimulator of an unidentified GDH enzyme (SI Appendix, Fig. S8D). S. aureus YwlG has been found to be required for successful colonization in mouse models of pneumonia and osteomyelitis (34, 35). Our data reveal the moonlighting role of YwlG in regulating the abundance of 100S ribosomes (via HPF) and the important role of YwlG in glutamate metabolism (via GDH activity) (Fig. 7).
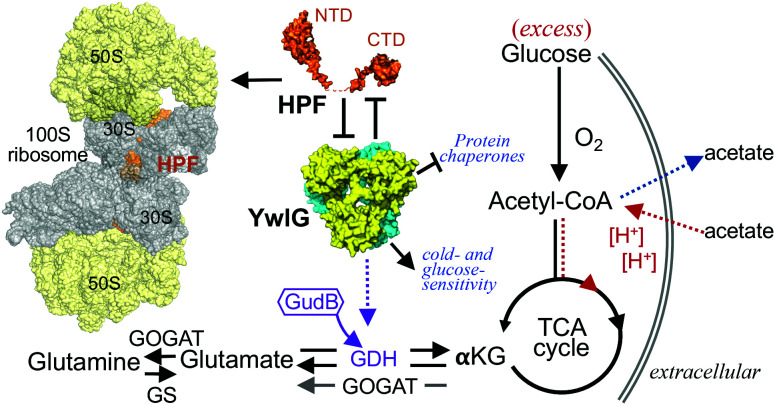
Fig. 7.
Interplay between HPF and YwlG. The carboxyl-terminal domain (CTD) of HPF promotes the dimerization of 70S monomers to form the translationally inactive 100S ribosomes. A fraction of HPF is sequestered by YwlG, diluting the levels of 100S complexes. Deletion of ywlG suppresses acetate-potentiated cell death and cold sensitivity in an hpf knockout, as well as derepresses the expression of protein chaperones. GDH resides at the crossroads of nitrogen- and carbon-assimilation pathways. YwlG structurally resembles distantly related NAD-specific GDHs and can functionally compensate for loss of the GudB GDH enzyme in S. aureus by an unresolved mechanism. A concentration threshold of YwlG is likely required for its GDH activity. GOGAT, glutamine oxoglutarate aminotransferase/glutamate synthase; GS, glutamate synthetase; αKG, α-ketoglutarate, Acetyl-CoA, acetyl-coenzyme A; TCA, Tricarboxylic acid cycle.
The precise orders of 70S ribosome dimerization and 100S complex disassembly are poorly understood. Consequently, how and when S. aureus YwlG outcompetes ribosomes for HPF remain to be determined. In B. subtilis, the resolution of 100S ribosomes is stimulated by active transcription that may involve competition between HPF and mRNA for ribosome binding (31). The identification of YwlG as the sequestrator of HPF provides an additional means to prevent the formation of translationally incompetent 70S dimers to meet the translational needs. Future studies focusing on the stoichiometry, stability, atomic structure, oligomeric state and kinetics of the HPF–YwlG interaction, as well as the timing of sequestration (pre-70S dimerization and/or post-100S dissociation) will offer a more holistic picture of the in vivo activity of the HPF–YwlG complex.
Very limited information is available about the biological activity of YwlG homologs aside from their possible roles in the modification of EF-P in B. subtilis (41) and PLP-dependent a.a. transformation (33). While these possibilities cannot be completely ruled out, a ΔywlG and a Δefp of S. aureus do not phenocopy each other, and PLP binding has not been validated in vivo or detected in vitro. The inability to detect PLP or other cofactor binding in the YwlG protein crystal could be due to instability of YwlG in its PLP-bound form or the missing component(s), e.g., HPF, in the heterologous E. coli from which recombinant YwlG proteins were isolated. Although E. coli harbors an HPF homolog, it lacks the hallmark self-dimerizing CTD uniquely found in YwlG-containing bacteria, implying a coevolution of YwlG and HPF-CTD.
The hexameric structure of S. aureus YwlG remotely resembles the structures of members of the catabolic, NAD-specific GDH family. Surprisingly, while GudB is the only known producer of GDH activity in S. aureus, overexpressing YwlG restored the GDH activity in a ΔgudB-null mutant, prompting our speculation that YwlG is a bona fide GDH. Further inspection of the protein domain structure revealed that YwlG did not conform to the conventional GDH domain assignment. First, YwlG is half of the typical size of a GDH and lacks the N-terminal catalytic domain and ligand binding sites (SI Appendix, Figs. S7 and S9B). Second, GDH activity was nearly undetectable upon gudB knockout, suggesting that YwlG alone was enzymatically inactive or that GDH activity was repressed by another factor in the ΔgudB mutant. One such repressor is HPF. However, inactivation of hpf led to only 50% derepression of YwlG-mediated GDH activity in prolonged TSB cultures, suggesting possible existence of an unknown repressor (SI Appendix, Fig. S8D). HPF and this repressor may sterically occlude GDH catalysis and substrate binding or prevent the formation of an active hexamer (46). Enzymatic repression of GDH via protein–protein interactions is not unprecedented. B. subtilis GudB is constitutively expressed, and its GDH activity is sequestered by the counter enzymes GltA–GltB under nutrient-specific conditions (49). A direct contribution of YwlG to GDH activity cannot be fully excluded. YwlG may serve as a pseudoenzyme to activate an unidentified GDH or may be a catalytically deficient GDH variant that requires a complementation fragment to fulfill GDH catalytic activity. The possible roles of YwlG in GDH activity are depicted in SI Appendix, Fig. S8D. Recombinant YwlG produced from E. coli by itself lacks dehydrogenase activity. Therefore, in vitro biochemical characterization of YwlG isolated from the native S. aureus is needed to discern all these possibilities.
The finding that the rescue of GDH activity in the ΔgudB-null mutant required YwlG overexpression suggests that the cellular concentrations of YwlG are critical in controlling GDH activity. Monomers and dimers of B. subtilis GudB are inactive. The oligomeric state of B. subtilis GudB is regulated by pH and glutamate levels (46). It is conceivable that overriding HPF repression and concentration-dependent hexameric assembly of YwlG drive GDH activity. B. subtilis ywlG is negatively regulated by the TnrA regulator in response to glutamine and ammonia availability (50). The regulation of S. aureus ywlG expression is unclear. An apparent TnrA binding motif is absent from the promoter region, and the YwlG protein, albeit not as abundant as HPF, was detectable under a wide range of laboratory conditions. YwlG appeared to be unstable in acidic environments in cells grown in TSB-glucose medium (Figs. 4D and 5F, barely detectable YwlG at 96 h), consistent with the notion that high pH promotes and/or stabilizes higher-order oligomers of GDH (46).
Cytoplasmic acidification causes S. aureus cell death under excess glucose conditions through generation of ROS (39, 40). Resistance to acetate-potentiated cell death in S. aureus carrying a ΔywlG mutant allele may appear counterintuitive (Fig. 3). Glutamate is the major nitrogen reservoir and one of the most abundant cytoplasmic anions in bacteria, with cellular concentrations of up to 150 mM (51). Glutamate has multiple roles; for example, it stabilizes interactions among biomolecules (protein–protein interactions and protein–nucleic acid interactions), acts as a counterion to potassium, buffers intracellular pH, and maintains osmolarity (49, 51, 52). Glutamate can also be converted into the antioxidants glutathione (S. aureus produces bacillithiols instead) and 2-oxoglutarate. Our results suggest that YwlG contributes to glutamate metabolism via GDH activity. Although GudB is the core GDH in S. aureus, it is possible that moderate accumulation of 2-oxoglutarate upon ΔywlG deletion provides some oxidative stress defense while not perturbing central GudB-dependent nitrogen and carbon metabolic circuits. An analogous anti-ROS survival strategy has been reported in a ΔclpP mutant that preserves antioxidant proteins otherwise degraded by the ClpXP protease (39).
Some eukaryotic and bacterial metabolic enzymes play dual roles as mRNA-binding proteins or ribosome-binding factors to modulate translation (53–55). In B. subtilis, the ribosome assembly checkpoint protein CpgA moonlights as a metabolite-proofreading enzyme (56). The discovery of bifunctional HPF and YwlG adds to a growing list of moonlighting proteins (57, 58) that regulate two of the most essential biochemical reactions (protein synthesis and glutamate metabolism) in all living cells.
Materials and Methods
Strains, Plasmids, Chemicals, and Growth Conditions.
Strains and plasmids were constructed as described in SI Appendix and are listed in SI Appendix, Table S2.
FLAG Affinity Pull-Down and Mass Spectrometric Analysis.
S. aureus carrying the pLI50 derivatives were grown in 200 mL TSB supplemented with 10 µg/mL chloramphenicol at 37 °C until optical density at 600 nm equals 1.4 (4 h) or ≥8.0 (18 h). Cells were disrupted using cryomilling method (13) on a Retsch MM400 miller in Buffer C (50 mM Tris-Cl [pH 7.5], 100 mM KCl, 1 mM phenylmethylsulfonyl fluoride). Approximately 1 mL of cell lysates was incubated with 50 µL anti-FLAG M2 magnetic beads (Sigma-Aldrich, M8823-5ML) at room temperature (∼22 °C) on a tube rotator for 1 h and additional 15 h at 4 °C. The magnetic beads were washed extensively with Buffer C (7 ×1 mL), and proteins were eluted with 100 µL glycine (pH 2.6). Samples were neutralized with 15 µL 1 M Tris base and analyzed on a 4–12% Bis-Tris NuPAGE gel (Invitrogen). Liquid chromatography–tandem mass spectrometry (MS) was performed on trypsin digested gel slices or gel-free whole eluate by the Northwestern University Proteomics Core to identify protein species. Scaffold (version Scaffold_4.11.0, Proteome Software Inc., Portland, OR) was used to validate MS/MS-based peptide and protein identifications.
Common molecular biology techniques.
BACTH system, ribosome sedimentation profiling, Western blots, TSB-glucose killing assays, quantitation of acetate and protein aggregates, and recombinant YwlG production are described in SI Appendix.
GDH Activity Assay.
One milliliter of bacterial cultures was collected, resuspended in sterile water, and lysed on a FastPrep-24 instrument using Lysing Matrix B beads (MP Biomedicals), and 10 µL cell lysates were immediately used in the reactions. GDH activity was measured by the amount of NADH production upon glutamate consumption using a colorimetric kit according to manufacturer’s procedures (Sigma-Aldrich No. MAK099). One unit of GDH is defined as the amount of enzyme that generates 1.0 µmol of NADH per minute at pH 7.6 at 37 °C.
YwlG Crystallization and Structural Determination.
Overexpression and purification of recombinant YwlG are described in SI Appendix. Prior to crystallization, YwlG was centrifuged (16,000 × g, 4 °C, 20 min) to remove aggregates and debris. Diffraction-quality crystals of S. aureus YwlG were obtained using the sitting-drop vapor diffusion in 0.2 M Na+K+ tartrate, 20% (wt/vol) PEG3350 and 0.18 M ammonium citrate, 20% (wt/vol) PEG3350 at 18 °C. For cryoprotection, crystals were soaked in mother liquor supplemented with 30% (wt/vol) ethylene glycol, flash cooled, and stored in liquid nitrogen. Data collection was carried out at the synchrotron beamline ID30B and ID30A1 at the European Synchrotron Radiation Facility, Grenoble, France. The data were processed with XDS (59), and the structure was solved by molecular replacement with PHASER (60), using the coordinates of the conserved hypothetical protein TT1679 from T. thermophilus (PDB 1V8D), which shares about 36% and 52% sequence identity and similarity, respectively, with YwlG from S. aureus, as a search model. Model building was done with COOT (61), and refinement of the coordinates was carried out with REFMAC5 (62, 63). The hexameric complex was analyzed using PdbePISA (64). Structural figures were prepared with PyMOL (65) and University of California, San Francisco Chimera (66).
Statistically Analysis.
Statistical significance was tested by unpaired two-tailed Student’s t test in GraphPad Prism 9.0. Only P < 0.05 is considered statistically significant.
Supplementary Material
Acknowledgments
Transposon mutants were obtained through Network on Antimicrobial Resistance in Staphylococcus aureus for distribution by BEI Resources, National Institute of Allergy and Infectious Diseases, NIH. Proteomics services were performed by Northwestern Proteomics Core Facility, supported by National Cancer Institute Cancer Center Support Grants P30CA060553, S10OD0235194, and P41GM108569. M.-N.F.Y. was supported by the NIH grants R01GM121359 and R01AI150986. S.S. was supported by the German Research Foundation DFG: GRK 2062 and SCHN 1273/10. S.A.S. received funding from the European Research Council (ERC) and the European Union’s Horizon 2020 research and innovation program (grant agreement no. 725085, CHEMMINE, ERC consolidator grant). M.P. acknowledges a PhD fellowship from the Studienstiftung des deutschen Volkes. M. F. is supported by Kekulé-Stipendium des Fonds der Chemischen Industrie.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2207257119/-/DCSupplemental.
Data, Materials, and Software Availability
Crystallographic data (atomic coordinates and structure factors) of YwlG have been deposited in the PDB with accession code 7Z06 (https://www.ebi.ac.uk/pdbe/entry/pdb/7Z06) (67).
[Protein structure] data have been deposited in [RCSB PDB] (7Z06; https://www.ebi.ac.uk/pdbe/entry/pdb/7Z06). All other data are included in the manuscript and/or SI Appendix.
References
- 1.Gohara D. W., Yap M. F., Survival of the drowsiest: The hibernating 100S ribosome in bacterial stress management. Curr. Genet. 64, 753–760 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prossliner T., Skovbo Winther K., Sørensen M. A., Gerdes K., Ribosome hibernation. Annu. Rev. Genet. 52, 321–348 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Smith P. R., Pandit S. C., Loerch S., Campbell Z. T., The space between notes: Emerging roles for translationally silent ribosomes. Trends Biochem. Sci. 47, 477–491 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khusainov I., et al. , Structure of the 70S ribosome from human pathogen Staphylococcus aureus. Nucleic Acids Res. 44, 10491–10504 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matzov D., et al. , The cryo-EM structure of hibernating 100S ribosome dimer from pathogenic Staphylococcus aureus. Nat. Commun. 8, 723 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beckert B., et al. , Structure of a hibernating 100S ribosome reveals an inactive conformation of the ribosomal protein S1. Nat. Microbiol. 3, 1115–1121 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Matzov D., Bashan A., Yap M. F., Yonath A., Stress response as implemented by hibernating ribosomes: A structural overview. FEBS J. 286, 3558–3565 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beckert B., et al. , Structure of the Bacillus subtilis hibernating 100S ribosome reveals the basis for 70S dimerization. EMBO J. 36, 2061–2072 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Franken L. E., et al. , A general mechanism of ribosome dimerization revealed by single-particle cryo-electron microscopy. Nat. Commun. 8, 722 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khusainov I., et al. , Structures and dynamics of hibernating ribosomes from Staphylococcus aureus mediated by intermolecular interactions of HPF. EMBO J. 36, 2073–2087 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flygaard R. K., Boegholm N., Yusupov M., Jenner L. B., Cryo-EM structure of the hibernating Thermus thermophilus 100S ribosome reveals a protein-mediated dimerization mechanism. Nat. Commun. 9, 4179 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Akanuma G., et al. , Ribosome dimerization is essential for the efficient regrowth of Bacillus subtilis. Microbiology (Reading) 162, 448–458 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Basu A., Yap M. N., Ribosome hibernation factor promotes Staphylococcal survival and differentially represses translation. Nucleic Acids Res. 44, 4881–4893 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Puri P., et al. , Lactococcus lactis YfiA is necessary and sufficient for ribosome dimerization. Mol. Microbiol. 91, 394–407 (2014). [DOI] [PubMed] [Google Scholar]
- 15.Kline B. C., McKay S. L., Tang W. W., Portnoy D. A., The Listeria monocytogenes hibernation-promoting factor is required for the formation of 100S ribosomes, optimal fitness, and pathogenesis. J. Bacteriol. 197, 581–591 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McKay S. L., Portnoy D. A., Ribosome hibernation facilitates tolerance of stationary-phase bacteria to aminoglycosides. Antimicrob. Agents Chemother. 59, 6992–6999 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Williamson K. S., et al. , Heterogeneity in Pseudomonas aeruginosa biofilms includes expression of ribosome hibernation factors in the antibiotic-tolerant subpopulation and hypoxia-induced stress response in the metabolically active population. J. Bacteriol. 194, 2062–2073 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Akiyama T., et al. , Resuscitation of Pseudomonas aeruginosa from dormancy requires hibernation promoting factor (PA4463) for ribosome preservation. Proc. Natl. Acad. Sci. U.S.A. 114, 3204–3209 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feaga H. A., Kopylov M., Kim J. K., Jovanovic M., Dworkin J., Ribosome dimerization protects the small subunit. J. Bacteriol. 202, e00009–e00020 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Y., et al. , Zinc depletion induces ribosome hibernation in mycobacteria. Proc. Natl. Acad. Sci. U.S.A. 115, 8191–8196 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shcherbakova K., Nakayama H., Shimamoto N., Role of 100S ribosomes in bacterial decay period. Genes Cells 20, 789–801 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Trauner A., Lougheed K. E., Bennett M. H., Hingley-Wilson S. M., Williams H. D., The dormancy regulator DosR controls ribosome stability in hypoxic mycobacteria. J. Biol. Chem. 287, 24053–24063 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Theng S., Williamson K. S., Franklin M. J., Role of hibernation promoting factor in ribosomal protein stability during Pseudomonas aeruginosa dormancy. Int. J. Mol. Sci. 21, E9494 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lipońska A., Yap M. F., Hibernation-promoting factor sequesters Staphylococcus aureus ribosomes to antagonize RNase R-mediated nucleolytic degradation. MBio 12, e0033421 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prossliner T., Gerdes K., Sørensen M. A., Winther K. S., Hibernation factors directly block ribonucleases from entering the ribosome in response to starvation. Nucleic Acids Res. 49, 2226–2239 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Basu A., Shields K. E., Eickhoff C. S., Hoft D. F., Yap M. N., Thermal and Nutritional Regulation of Ribosome Hibernation in Staphylococcus aureus. J. Bacteriol. 200, e00426–e00418 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davis A. R., Gohara D. W., Yap M. N., Sequence selectivity of macrolide-induced translational attenuation. Proc. Natl. Acad. Sci. U.S.A. 111, 15379–15384 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ueta M., et al. , Conservation of two distinct types of 100S ribosome in bacteria. Genes Cells 18, 554–574 (2013). [DOI] [PubMed] [Google Scholar]
- 29.Ueta M., Wada C., Wada A., Formation of 100S ribosomes in Staphylococcus aureus by the hibernation promoting factor homolog SaHPF. Genes Cells 15, 43–58 (2010). [DOI] [PubMed] [Google Scholar]
- 30.Basu A., Shields K. E., Yap M. F., The hibernating 100S complex is a target of ribosome-recycling factor and elongation factor G in Staphylococcus aureus. J. Biol. Chem. 295, 6053–6063 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feaga H. A., Dworkin J., Transcription regulates ribosome hibernation. Mol. Microbiol. 116, 663–673 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karimova G., Gauliard E., Davi M., Ouellette S. P., Ladant D., Protein-Protein Interaction: Bacterial Two-Hybrid. Methods Mol. Biol. 1615, 159–176 (2017). [DOI] [PubMed] [Google Scholar]
- 33.Hoegl A., et al. , Mining the cellular inventory of pyridoxal phosphate-dependent enzymes with functionalized cofactor mimics. Nat. Chem. 10, 1234–1245 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim G. L., et al. , Growth and stress tolerance comprise independent metabolic strategies critical for Staphylococcus aureus infection. MBio 12, e0081421 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilde A. D., et al. , Bacterial hypoxic responses revealed as critical determinants of the host-pathogen outcome by TnSeq analysis of Staphylococcus aureus invasive infection. PLoS Pathog. 11, e1005341 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y., Gross C. A., Cold shock response in bacteria. Annu. Rev. Genet. 55, 377–400 (2021). [DOI] [PubMed] [Google Scholar]
- 37.Papp-Wallace K. M., Maguire M. E., Magnesium transport and magnesium homeostasis. Ecosal Plus 3, (2008). [DOI] [PubMed] [Google Scholar]
- 38.Sezonov G., Joseleau-Petit D., D’Ari R., Escherichia coli physiology in Luria-Bertani broth. J. Bacteriol. 189, 8746–8749 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alqarzaee A. A., et al. , Staphylococcal ClpXP protease targets the cellular antioxidant system to eliminate fitness-compromised cells in stationary phase. Proc. Natl. Acad. Sci. U.S.A. 118, e2109671118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thomas V. C., et al. , A central role for carbon-overflow pathways in the modulation of bacterial cell death. PLoS Pathog. 10, e1004205 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Witzky A., et al. , EF-P posttranslational modification has variable impact on polyproline translation in Bacillus subtilis. MBio 9, e00306-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schramm F. D., Schroeder K., Jonas K., Protein aggregation in bacteria. FEMS Microbiol. Rev. 44, 54–72 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Belitsky B. R., Kim H. J., Sonenshein A. L., CcpA-dependent regulation of Bacillus subtilis glutamate dehydrogenase gene expression. J. Bacteriol. 186, 3392–3398 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Belitsky B. R., Sonenshein A. L., Role and regulation of Bacillus subtilis glutamate dehydrogenase genes. J. Bacteriol. 180, 6298–6305 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seidl K., et al. , Effect of a glucose impulse on the CcpA regulon in Staphylococcus aureus. BMC Microbiol. 9, 95 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Noda-Garcia L., Romero Romero M. L., Longo L. M., Kolodkin-Gal I., Tawfik D. S., Bacilli glutamate dehydrogenases diverged via coevolution of transcription and enzyme regulation. EMBO Rep. 18, 1139–1149 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bhuiya M. W., et al. , The first crystal structure of hyperthermostable NAD-dependent glutamate dehydrogenase from Pyrobaculum islandicum. J. Mol. Biol. 345, 325–337 (2005). [DOI] [PubMed] [Google Scholar]
- 48.Gunka K., et al. , Functional dissection of a trigger enzyme: Mutations of the bacillus subtilis glutamate dehydrogenase RocG that affect differentially its catalytic activity and regulatory properties. J. Mol. Biol. 400, 815–827 (2010). [DOI] [PubMed] [Google Scholar]
- 49.Jayaraman V., et al. , A counter-enzyme complex regulates glutamate metabolism in Bacillus subtilis. Nat. Chem. Biol. 18, 161–170 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yoshida K., et al. , Identification of additional TnrA-regulated genes of Bacillus subtilis associated with a TnrA box. Mol. Microbiol. 49, 157–165 (2003). [DOI] [PubMed] [Google Scholar]
- 51.Bennett B. D., et al. , Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat. Chem. Biol. 5, 593–599 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Young V. R., Ajami A. M., Glutamate: An amino acid of particular distinction. J. Nutr. 130 (4S Suppl), 892S–900S (2000). [DOI] [PubMed] [Google Scholar]
- 53.Simsek D., et al. , The mammalian ribo-interactome reveals ribosome functional diversity and heterogeneity. Cell 169, 1051–1065.e18 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Benjamin J. A., Massé E., The iron-sensing aconitase B binds its own mRNA to prevent sRNA-induced mRNA cleavage. Nucleic Acids Res. 42, 10023–10036 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hentze M. W., Castello A., Schwarzl T., Preiss T., A brave new world of RNA-binding proteins. Nat. Rev. Mol. Cell Biol. 19, 327–341 (2018). [DOI] [PubMed] [Google Scholar]
- 56.Sachla A. J., Helmann J. D., A bacterial checkpoint protein for ribosome assembly moonlights as an essential metabolite-proofreading enzyme. Nat. Commun. 10, 1526 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jeffery C. J., Enzymes, pseudoenzymes, and moonlighting proteins: Diversity of function in protein superfamilies. FEBS J. 287, 4141–4149 (2020). [DOI] [PubMed] [Google Scholar]
- 58.Singh N., Bhalla N., Moonlighting proteins. Annu. Rev. Genet. 54, 265–285 (2020). [DOI] [PubMed] [Google Scholar]
- 59.Kabsch W., XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McCoy A. J., et al. , Phaser crystallographic software. J. Appl. Cryst. 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Murshudov G. N., et al. , REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Winn M. D., et al. , Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Krissinel E., Henrick K., Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007). [DOI] [PubMed] [Google Scholar]
- 65.W. L. DeLano, The PyMOL Molecular Graphics System, Version 1.8 (Schrodinger, LLC, 2015). [Google Scholar]
- 66.Pettersen E. F., et al. , UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 67.S. Schneider, C. M. Scheidler, S. A. Sieber, Structure of YwlG (Q2FF14) from Staphylococcus aureus. Protein Data Bank. https://www.ebi.ac.uk/pdbe/entry/pdb/7Z06. Deposited 15 February 2022. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Crystallographic data (atomic coordinates and structure factors) of YwlG have been deposited in the PDB with accession code 7Z06 (https://www.ebi.ac.uk/pdbe/entry/pdb/7Z06) (67).
[Protein structure] data have been deposited in [RCSB PDB] (7Z06; https://www.ebi.ac.uk/pdbe/entry/pdb/7Z06). All other data are included in the manuscript and/or SI Appendix.