Significance
Shape variations of plant edible organs are the result of adaptive evolution and domestication. Fruit neck is an undesirable trait that affects fruit shape and commercial value in cucumber. Despite fruit neck length (FNL) varying greatly among cucumber germplasms, the molecular mechanisms controlling FNL remain unknown. Here, we demonstrate that HECATE1 (CsHEC1) positively regulates FNL by directly activating the auxin biosynthesis gene CsYUC4. CsOVATE, whose expression is negatively correlated with FNL, combats CsHEC1 to attenuate the CsHEC1-mediated CsYUC4 transcriptional activation. Our work not only paves a way to shorten FNL by manipulating the CsHEC1-CsOVATE module to decrease local auxin levels during cucumber breeding but also provides significant insights into fruit shape regulation in pepo fruits.
Keywords: cucumber, CsHEC1, CsOVATE, fruit neck, auxin biosynthesis
Abstract
Fruit neck is the proximal portion of the fruit with undesirable taste that has detrimental effects on fruit shape and commercial value in cucumber. Despite the dramatic variations in fruit neck length of cucumber germplasms, the genes and regulatory mechanisms underlying fruit neck elongation remain mysterious. In this study, we found that Cucumis sativus HECATE1 (CsHEC1) was highly expressed in fruit neck. Knockout of CsHEC1 resulted in shortened fruit neck and decreased auxin accumulation, whereas overexpression of CsHEC1 displayed the opposite effects, suggesting that CsHEC1 positively regulated fruit neck length by modulating local auxin level. Further analysis showed that CsHEC1 directly bound to the promoter of the auxin biosynthesis gene YUCCA4 (CsYUC4) and activated its expression. Enhanced expression of CsYUC4 resulted in elongated fruit neck and elevated auxin content. Moreover, knockout of CsOVATE resulted in longer fruit neck and higher auxin. Genetic and biochemical data showed that CsOVATE physically interacted with CsHEC1 to antagonize its function by attenuating the CsHEC1-mediated CsYUC4 transcriptional activation. In cucumber germplasms, the expression of CsHEC1 and CsYUC4 positively correlated with fruit neck length, while that of CsOVATE showed a negative correlation. Together, our results revealed a CsHEC1-CsOVATE regulatory module that confers fruit neck length variation via CsYUC4-mediated auxin biosynthesis in cucumber.
The fruit is the prominent edible organ in horticultural crops that is essential for seed development and sexual reproduction. Fruit shape directly affects appearance quality and market value, and thus explosion in fruit shape variation is one of the hallmarks during crop domestication (1–3). So far, most fruit shape studies have been performed in tomato bearing the berry fruit, such as SUN, an IQ67-domain protein involved in Ca2+ signal transduction, acting as a positive regulator for elongated fruit shape, whereas the CLAVATA3/EMBRYO SURROUNDING REGION–related family member FASCIATED (FAS) and the WUSCHEL homeodomain protein LOCULE NUMBER (LC) function coordinately in fruit locule number controlling the flat shape (4–8). Cucumber is an important vegetable crop bearing the pepo fruit that is harvested immature at 8 to 18 d after anthesis, and consumed fresh or processed into pickles (9, 10). Morphologically, cucumber fruit consists of the fruit neck at the proximal end, connecting with the peduncle and the tasty fruit at the distal end (Fig. 1A). Fruit neck, also known as stalk or gynophore, usually has no spines/tubercules on the surface and no placenta inside (11–13). In cucumber germplasms, the fruit neck length (FNL) varies from 1 to 12 cm (Fig. 1B), which accounts for up to 35% of the total fruit length (2, 12). Due to the undesirable taste and the reduced diameter compared with that of the fruit, resembling a constricted neck, the fruit neck has detrimental effects on fruit shape and commercial value in cucumber (14).
Fig. 1.
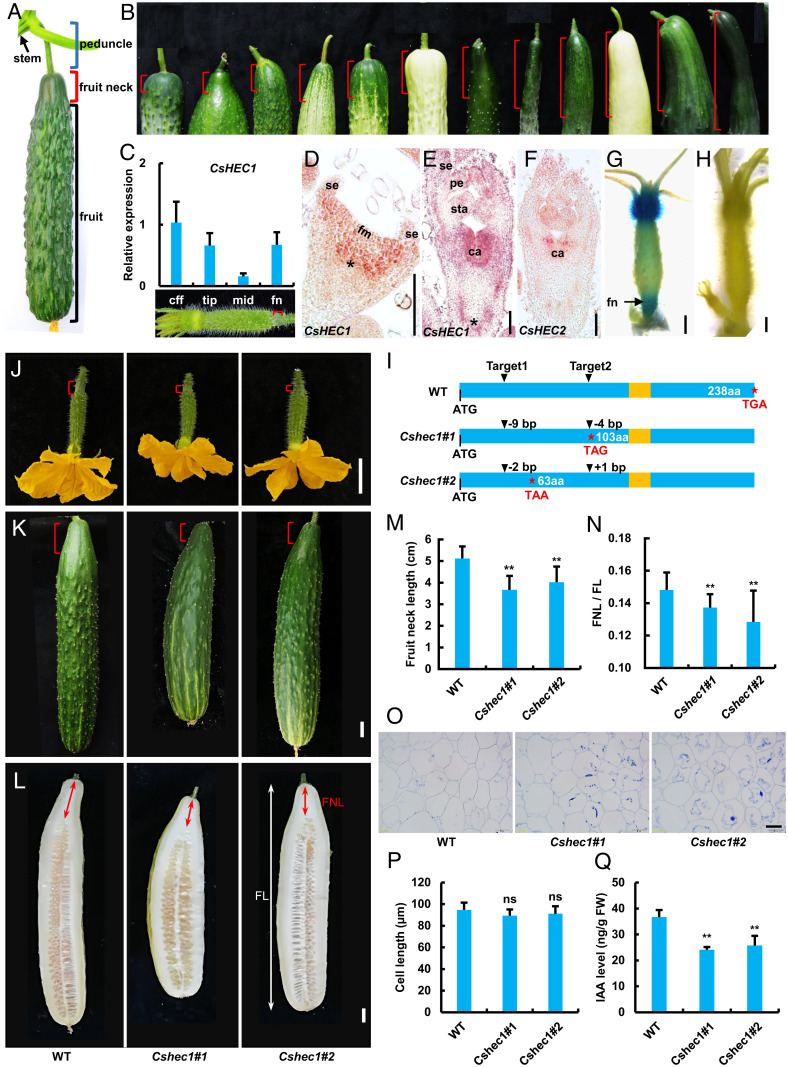
Expression analysis of CsHEC1 and phenotypic characterization of CsHEC1 knockout lines in cucumber. (A) Structure of cucumber fruit at the commercial harvest stage. Blue bracket, peduncle; red bracket, fruit neck; black bracket, fruit. (B) Variations in FNL (red brackets) in different cucumber germplasms. (C) The expression pattern of CsHEC1 in different parts of the ovary at 3 DBA. cff, corolla from the female flowers; fn, fruit neck; mid, middle. Values are means ± SD (n = 3). The red brackets indicate the fruit neck. (D–F) In situ hybridization analysis of CsHEC1 (D and E) or CsHEC2 (F) in cucumber. Longitudinal sections of developing flower primordium at stage 2 (D) and stage 6 (E and F). Asterisks indicate the developing fruit neck in cucumber. ca, carpel; fm, floral meristem; pe, petal; se, sepal; sta, stamen. (Scale bars, 100 µm.) (G and H) Tissue specificity of CsHEC1 expression was examined using the ProCsHEC1:GUS reporter system. GUS signals of CsHEC1 were highly enriched at the fruit neck and corolla of the female flower (G). Negative control showed no GUS signal (H). (Scale bars, 1 mm.) (I) Genotype identification of CsHEC1 knockout plants indicated the Cshec1#1 mutant with 9- and 4-bp deletions, and the Cshec1#2 mutant with a 2-bp deletion and 1-bp insertion. The stars represent the termination codon position and the yellow boxes indicate the bHLH domain. (J–L) Morphology of WT, Cshec1#1, and Cshec1#2 fruits at 0 DPP (J), 10 DPP (K), and 40 DPP (L). The red brackets indicate the fruit neck; the red and white double arrows represent the measured FNL and FL, respectively. (Scale bars, 2 cm.) (M and N) Quantification of FNL (M) and the ratio of FNL/FL (N) in WT and Cshec1 mutants at 40 DPP. Values are means ± SD (n = 10). (O and P) Representative cell morphology (O) and cell-length quantification (P) of longitudinal sections of the fruit necks in WT and Cshec1 fruits at 40 DPP. Values are means ± SD (n = 9). (Scale bar, 50 µm.) (Q) IAA content in the fruit necks of WT and Cshec1 mutants. FW, fresh weight. Values are means ± SD (n = 3). Significance analysis compared with WT was performed with the two-tailed Student’s t test (ns, no significant difference; **P < 0.01).
Previous studies showed that FNL variation is controlled by additive genetic rather than environmental factors in cucumber (15, 16). In 2008, a major-effect quantitative trait locus (QTL) accounting for 18.5% in FNL was identified as located on a 21.4-cM region on chromosome 1 (17). Subsequently, four QTLs on chromosomes 3, 6, and 7 were detected by QTL mapping using 160 recombinant inbred lines (18). The first gene cloned for fruit neck elongation is CsFnl7.1, encoding a protein of the late embryogenesis abundant family, that may regulate fruit neck development by modulating cell expansion in cucumber (19). However, the regulatory mechanism underlying FNL variation remains largely unknown in cucumber.
Fruit neck is an important part of the apical–basal patterning of the fruit developed from the gynoecium, in which the arrangement of stigma, style, ovary, and fruit neck along the distal end to the proximal end constitutes the apical–basal axis of the gynoecium (Fig. 1 A and C) (11, 20, 21). Mutations in HECATE (HEC) genes, members of the basic helix–loop–helix (bHLH) family, showed defects in transmitting tract and stigma formation leading to reduced fertility. Overexpression of HEC1 or HEC3 driven by the constitutive cauliflower mosaic virus 35S promoter occasionally produced gynoecia with defects in apical–basal polarity (22), resembling the loss of function in the auxin efflux carrier PIN-FORMED1 (PIN1) (23). Further study showed that HEC1 boosted auxin transport via directly stimulating the expression of PIN1 and PIN3 during gynoecium development in Arabidopsis (24).
OVATE family proteins (OFPs) were shown to regulate fruit shape and size in agricultural crops (25, 26). OVATE was the first gene identified in the OFPs as a repressor of fruit growth, and its loss-of-function mutation resulted in fruit transition from round- to pear-shaped with longitudinal elongation at the proximal end and obvious neck constriction in tomato (27, 28). SlOFP20, another member of the OFPs underlying the suppressor of ovate (sov1) locus, acted synergistically with OVATE resulting in a conspicuous pear-shaped tomato. OVATE and SlOFP20 interacted with several members of TONNEAU1 RECRUITMENT MOTIF (TRM) to regulate tomato fruit shape by fine-tuning cell-division patterns (26). In peach, transcriptional activation of PpOFP1 caused by the 1.7-Mb chromosomal inversion event was responsible for flat-shaped fruit. PpOFP1 was the homologous protein of both AtOFP1 and SlOFP20, which may function as a transcriptional repressor of cell division or elongation, interacting with the elongation activator PpTRM17 (26, 29, 30). In addition, QTL mapping showed that several homologous genes to OFP and TRM members were encompassed in the chromosome regions specifying the shape change from round to elongated fruit in melon, cucumber, and potato; despite this, functional validation and regulatory mechanisms are mysterious (26, 31, 32).
In this study, we found that cucumber CsHEC1 is highly expressed in the fruit neck region and plays a positive role in fruit neck elongation. Further analysis showed that CsHEC1 directly bound to the CsYUC4 promoter and enhanced its expression, resulting in elevated auxin accumulation and increased FNL. Moreover, our data indicated that CsOVATE functions as a negative regulator for fruit neck elongation through physical interaction with CsHEC1 to attenuate the CsHEC1-mediated CsYUC4 activation. A working model involved in the regulation of FNL variation by the CsHEC1-CsOVATE module through mediated auxin biosynthesis is proposed.
Results
Characterization of the CsHEC1 Transcription Factor in Cucumber.
In the dehiscent silique of Arabidopsis, HEC1/2/3 genes function redundantly during gynoecium development through controlling the formation of the transmitting tract and stigma (22, 24). In cucumber bearing fresh pepo fruit, our recent studies showed that CsHEC2 regulates fruit wart formation by stimulating cytokinin biosynthesis and irregular vasculature patterning (CsIVP); the CsHEC3 subfamily participates in both organ shape determination and downy mildew resistance via mediating vasculature development (33, 34). In this study, we characterized the function of CsHEC1 (Csa4G639900) during fruit development in cucumber. Phylogenetic analysis showed that Cucurbitaceae HEC1 and HEC2 belong to two different clades, and CsHEC1 is more closely related to HEC1 and HEC2 of Arabidopsis (SI Appendix, Fig. S1 and Table S1). Both CsHEC1 and CsHEC2 contain a conserved bHLH domain, but share only 41% amino acid identity across the full length of proteins (SI Appendix, Fig. S2) (35). Similar to the gene structure of HEC1 and HEC2 in Arabidopsis, the coding region of CsHEC1 is 717 bp with a single exon (SI Appendix, Fig. S3A) (22). Subcellular localization assay indicated that CsHEC1 was localized in the nucleus (SI Appendix, Fig. S3B). Dual-luciferase reporter (DLR) assay showed that CsHEC1 exhibited higher luciferase activity compared with the control vector, similar to that of VP16 positive control (SI Appendix, Fig. S3C) (36), implying that CsHEC1 may act as a transcriptional activator located in the nucleus.
CsHEC1 Is Highly Expressed in the Fruit Neck of Cucumber.
To explore the expression pattern of CsHEC1, three methods were performed in cucumber. qRT-PCR showed that CsHEC1 transcripts were prominently enriched in ovaries 3 d before anthesis (DBA) (SI Appendix, Fig. S4A). Particularly, CsHEC1 transcripts were highly accumulated in the corolla and fruit neck (Fig. 1C). RNA in situ hybridization analysis showed that CsHEC1 signal was observed throughout the shoot apical meristem and floral meristem (SI Appendix, Fig. S4B). In floral buds at stages 1 to 4, strong signals were detected beneath the rib zone where the fruit neck will initiate (Fig. 1D, asterisk and SI Appendix, Fig. S4 C–E) (37). Upon stages 6 to 8, CsHEC1 messenger RNAs were predominantly found in the developing carpels and fruit necks (Fig. 1E and SI Appendix, Fig. S4 F and G) and corolla (SI Appendix, Fig. S4 H and I). Importantly, CsHEC2, the paralogous gene of CsHEC1, was expressed in the carpel primordium, but not in the fruit neck (Fig. 1F and SI Appendix, Fig. S1). GUS (β-glucuronidase) reporter results further confirmed the high expression of CsHEC1 in the fruit neck, corolla, and developing ovules (Fig. 1 G and H and SI Appendix, Fig. S4 J and K). These results indicated a potential role of CsHEC1 in fruit neck development in cucumber.
CsHEC1 Promotes Fruit Neck Elongation through Mediating Auxin Accumulation.
To explore the function of CsHEC1 in cucumber, homozygous loss-of-function mutants were obtained using the CRISPR-Cas9 system in the inbred line XTMC, a North China–type cucumber with long fruits and medium FNL. Two mutant lines, Cshec1#1 (with 9- and 4-bp deletions) and Cshec1#2 (with a 2-bp deletion and 1-bp insertion) were identified for further characterizations (Fig. 1I and SI Appendix, Fig. S5A). No mutation was detected in the potential off-target sites of PCR sequencing products (SI Appendix, Table S2). Compared with wild-type (WT) plants, both mutants exhibited a shorter fruit neck from 0 d postpollination (DPP) to maturity in cucumber (Fig. 1 J–L). Quantification data analyses showed that the Cshec1 mutants had 21 to 28% decreases in FNL (Fig. 1M). Similarly, the fruit length (FL) was decreased, and the ratio of FNL to FL (FNL/FL) was significantly reduced in Cshec1 mutants (Fig. 1N and SI Appendix, Fig. S5B). Longitudinal sections of the fruit neck tissue from Cshec1 mutants and WT plants showed no significant difference in cell length (Fig. 1 O and P), suggesting that CsHEC1 may stimulate fruit neck elongation by mediating cell division. Previous studies indicated that auxin plays a unique role in regulating apical–basal fruit patterning in Arabidopsis and tomato (38–40). The auxin indole-3-acetic acid (IAA) level in fruit neck was greatly reduced in Cshec1 mutants (Fig. 1Q), implying that CsHEC1 may modulate local auxin accumulation during fruit neck development.
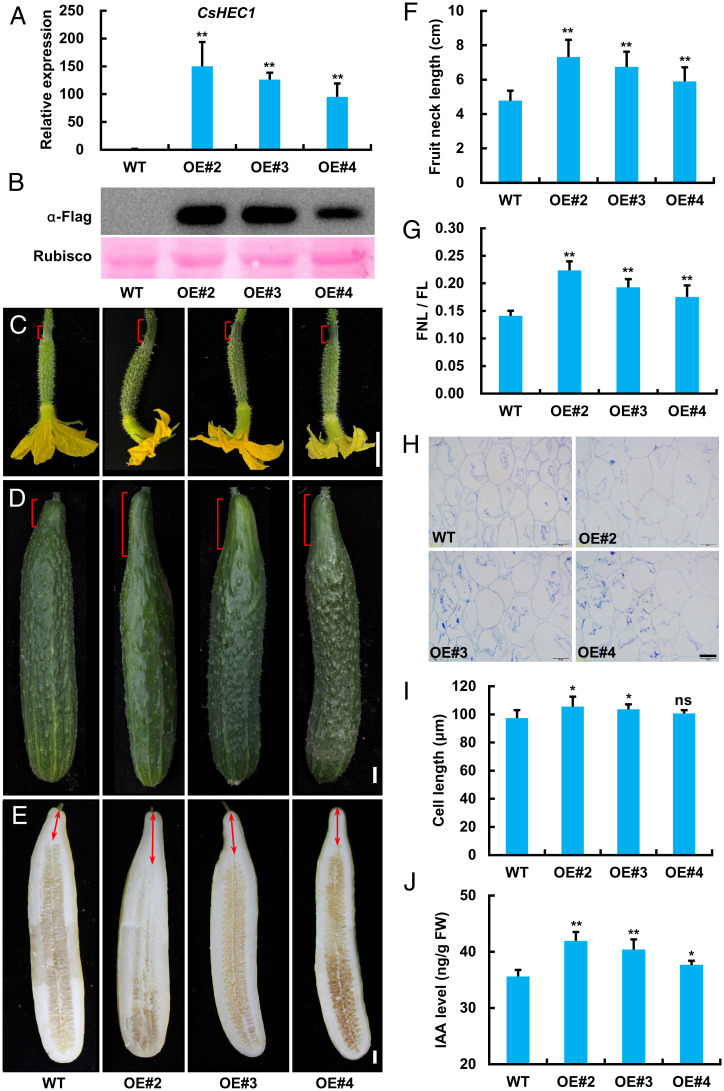
To further confirm CsHEC1 function in cucumber fruit, a CsHEC1 overexpression (OE) vector (Pro35S:CsHEC1-Flag) was constructed for genetic transformation. Five CsHEC1-OE transgenic lines were obtained and three representative lines (OE#2, OE#3, and OE#4) were selected for further analyses (Fig. 2). The qRT-PCR result indicated that the expression levels of CsHEC1 in the overexpression lines OE#2, OE#3, and OE#4 were 150-, 126-, and 95-fold higher than that of WT, respectively (Fig. 2A). Immunoblot assay demonstrated that the CsHEC1 protein had a high-level accumulation in all three OE transgenic lines (Fig. 2B). Compared with the WT, CsHEC1-OE fruits displayed remarkably elongated fruit neck from 0 DPP to maturity (Fig. 2 C–E). Quantification analyses showed that the CsHEC1-OE lines had 24 to 53% increases in FNL at maturity (Fig. 2F). No significant differences were detected in FL between WT and CsHEC1-OE lines (SI Appendix, Fig. S6A). Thus, the ratio of FNL/FL was significantly elevated in CsHEC1-OE lines (Fig. 2G). Longitudinal sections of the fruit neck at 40 DPP showed no consistent change in cell length (Fig. 2 H and I), suggesting that the main cause of fruit neck elongation may be increased cell numbers in CsHEC1-OE lines. Moreover, the IAA level in fruit neck was greatly elevated in CsHEC1-OE lines (Fig. 2J). To explore whether the effect of CsHEC1 on fruit neck elongation is ecotype-specific, overexpression of CsHEC1 in a South China–type cucumber, GFC, bearing short fruits was performed. Similarly, our data showed that both FNL and auxin level were increased upon overexpression of CsHEC1 in GFC (SI Appendix, Fig. S7), supporting that CsHEC1 positively regulated fruit neck elongation by promoting auxin accumulation in cucumber.
Fig. 2.
Overexpression of CsHEC1 results in elongated fruit neck in cucumber. (A) qRT-PCR analysis of CsHEC1 expression in CsHEC1-OE lines. Values are means ± SD (n = 3). (B) Immunoblot analysis of CsHEC1 protein levels in CsHEC1-OE lines using anti-Flag antibody. Rubisco large subunit stained by Ponceau S served as a loading control. (C–E) Fruit phenotype of CsHEC1-OE transgenic plants at 0 DPP (C), 10 DPP (D), and 40 DPP (E). The brackets indicate the fruit neck, and the double arrows represent the measured FNL. (Scale bars, 2 cm.) (F and G) Quantification of FNL (F) and ratio of FNL/FL (G) in WT and CsHEC1-OE lines at 40 DPP. Values are means ± SD (n = 6). (H and I) Representative cell morphology (H) and cell-length quantification (I) of longitudinal sections of the fruit necks in WT and CsHEC1-OE fruits at 40 DPP. Values are means ± SD (n = 9). (Scale bar, 50 µm.) (J) IAA content in the fruit neck of WT and CsHEC1-OE plants. Values are means ± SD (n = 3). Significance analysis compared with WT was performed with the two-tailed Student’s t test (ns, no significant difference; *P < 0.05, **P < 0.01).
CsHEC1 Directly Activates the Auxin Biosynthesis Gene CsYUC4 to Promote Fruit Neck Elongation.
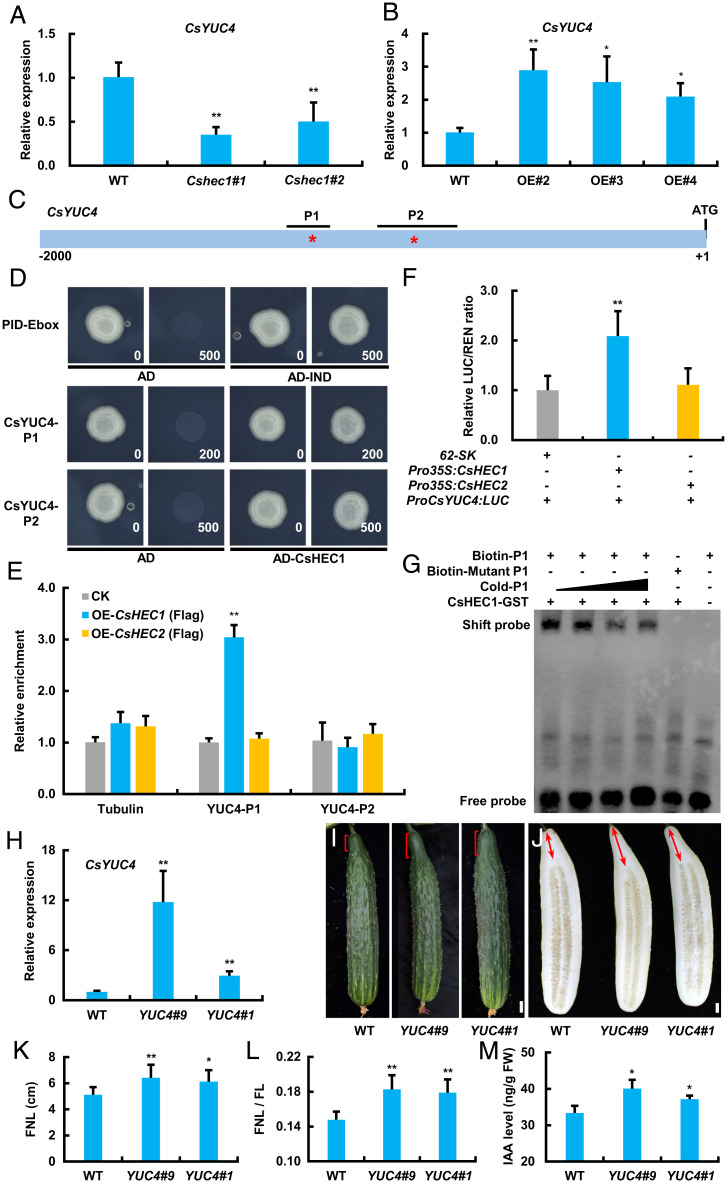
Members of the YUCCA (YUC) family mediate local auxin biosynthesis by catalyzing indole-3-pyruvic acid into IAA, the major naturally occurring auxin (41, 42). To explore whether CsHEC1 mediates auxin accumulation through auxin biosynthesis genes, the expression of 10 CsYUCs was examined in young stems and fruit necks in cucumber (43). Among them, the expression levels of CsYUC10a and CsYUC11 were undetectable. Interestingly, only CsYUC4 was highly expressed in the fruit neck compared with that in the stem (SI Appendix, Fig. S8). We further analyzed the expression level of CsYUC4 in CsHEC1 transgenic lines, and our data showed that transcripts of CsYUC4 were significantly decreased in Cshec1 mutants and greatly elevated in the CsHEC1-OE lines (Fig. 3 A and B and SI Appendix, Fig. S7H), suggesting that CsHEC1 may promote auxin accumulation through enhancing CsYUC4 expression.
Fig. 3.
CsHEC1 directly activates the expression of the auxin biosynthesis gene CsYUC4 to promote fruit neck elongation. (A and B) Gene expression analysis of CsYUC4 in Cshec1 knockout mutants (A) and CsHEC1-OE lines (B). Values are means ± SD (n = 3). *P < 0.05, **P < 0.01 (Student’s t test). (C) Schematic diagram of the putative E-box cis-elements in the CsYUC4 promoter. Asterisks represent the locations of putative E-box cis-elements. Black lines represent the fragments used for ChIP-PCR in E. (D) Yeast one-hybrid analysis of the interaction between the CsHEC1 protein and the variant E box from the CsYUC4 promoter. The interaction between IND-AD and the PID-E box was used as a positive control. (E) CsHEC1 binds to the cis-element P1 regions of the CsYUC4 promoter in vivo by ChIP-PCR analysis. Tubulin was used as a negative control. (F) Firefly luciferase (LUC) and renilla reiformis luciferase (REN) activity measurement was performed in N. benthamiana leaves by coexpression of Pro35S:CsHEC1 or Pro35S:CsHEC2 and ProCsYUC4:LUC. The empty vector pGreenII 62-SK was used as the control. Values are means ± SD (n = 3 in E; n = 5 in F). **P < 0.01 (Student’s t test). (G) Electrophoretic mobility-shift experiment showed that CsHEC1 binds to the P1 fragment of the CsYUC4 promoter. (H) Expression analysis of CsYUC4 in the ProCsHEC1:CsYUC4 transgenic lines. (I and J) Fruit morphology at 10 DPP (I) and 40 DPP (J) in WT and ProCsHEC1:CsYUC4 plants. The brackets indicate the fruit neck; the double arrows represent the measured FNL. (Scale bars, 2 cm.) (K and L) Quantification of FNL (K) and ratio of FNL/FL (L) in WT and ProCsHEC1:CsYUC4 lines at 40 DPP. (M) IAA content in the fruit neck of WT and ProCsHEC1:CsYUC4 plants. Values are means ± SD (n = 3 in H and M; n = 7 in K and L). Significance analysis compared with WT was performed with the two-tailed Student’s t test (*P < 0.05, **P < 0.01).
The bHLH transcription factors were shown to directly bind to the variant “E box” (5′-CANNTG-3′) in the gene regulatory regions (44, 45). Two putative E-box elements (P1, -1199, 5′-CATTTG-3′; P2, -889, 5′-CAAATG-3′) in the CsYUC4 promoter were identified (Fig. 3C). Yeast one-hybrid assay showed that CsHEC1 directly interacted with the P1 and P2 elements (Fig. 3D). Chromatin immunoprecipitation (ChIP)–PCR assays in transgenic cucumber plants showed that the P1 fragment was significantly enriched by CsHEC1-Flag, rather than by CsHEC2-Flag protein, after immunoprecipitation (Fig. 3E). A transactivation assay using the DLR system showed that the LUC/REN ratio was significantly increased upon coexpression of ProCsYUC4:LUC with Pro35S:CsHEC1 but not with the empty vector 62-SK or Pro35S:CsHEC2 (Fig. 3F). Furthermore, electrophoretic mobility-shift assay further supported the direct binding of CsHEC1 to CsYUC4 through the P1 element (Fig. 3G). These results suggest that CsHEC1 directly binds to the CsYUC4 promoter to activate its expression.
To verify CsHEC1 stimulates fruit neck elongation through CsYUC4-mediated auxin biosynthesis, the ProCsHEC1:CsYUC4 transgenic lines (YUC4#1 and YUC4#9) were generated. qRT-PCR analysis indicated the expression of CsYUC4 was significantly increased in YUC4#9 and YUC4#1 lines compared with WT plants (Fig. 3H). As expected, elevated expression of CsYUC4 resulted in a 17 to 25% increase in FNL and 12 to 24% elevation in the FNL/FL ratio (Fig. 3 I–L). The IAA level in fruit neck was also significantly increased in the ProCsHEC1:CsYUC4 lines (Fig. 3M). Further, two Csyuc4 mutants were generated using the CRISPR-Cas9 system, both of which resulted in significant reduction of FNL and decrease of fruit length, similar to the phenotype of Cshec1 mutants (SI Appendix, Fig. S9). Taken together, these results suggested that CsHEC1 directly activated CsYUC4 expression to enhance auxin accumulation and thus promoted fruit neck elongation in cucumber.
CsOVATE Negatively Regulates Fruit Neck Elongation in Cucumber.
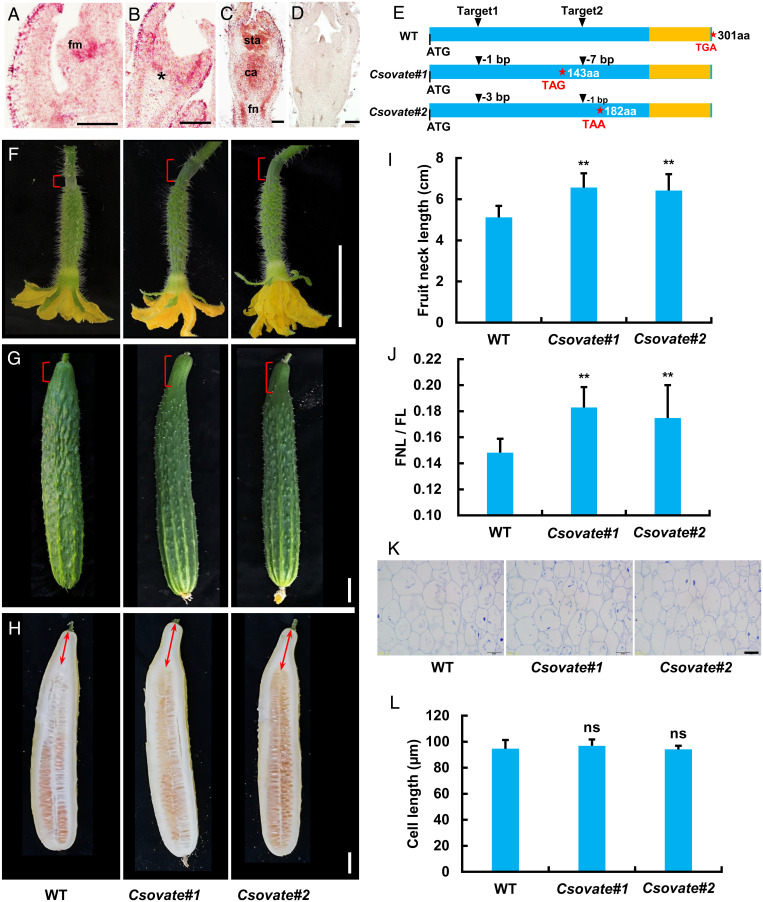
Overexpression of CsHEC1 resulted in fruits with elongated fruit necks (Fig. 2 and SI Appendix, Fig. S7), reminiscent of the phenotype of the tomato ovate mutant (1, 27). To investigate whether CsOVATE (31), a homologous protein of tomato OVATE, regulates fruit neck development in cucumber, we first analyzed the expression pattern using an in situ hybridization assay. Similar to CsHEC1, the results showed that the CsOVATE signal was predominantly accumulated in the floral meristem, fruit neck, and carpel primordia (Fig. 4 A–D). Knockout mutant lines were obtained using the CRISPR-Cas9 system, and two homozygous Csovate mutants (Csovate#1 with 1- and 7-bp deletions; Csovate#2 with 3- and 1-bp deletions) resulting in premature stop codons were identified for further characterization (Fig. 4E and SI Appendix, Fig. S10A and Table S3). Loss of function of CsOVATE led to elongated fruit neck from 0 DPP to maturity (Fig. 4 F–H), similar to that in CsHEC1-OE lines. Quantification analysis showed that Csovate mutants had a 25 to 28% increase in FNL (Fig. 4I), and a larger ratio of FNL/FL than that in WT (Fig. 4J and SI Appendix, Fig. S10B). Longitudinal sections of the fruit neck tissue from Csovate fruits and WT plants showed no significant difference in cell length (Fig. 4 K and L), suggesting that CsOVATE negatively regulated fruit neck elongation by decreasing cell numbers in cucumber.
Fig. 4.
CsOVATE negatively regulates fruit neck elongation in cucumber. (A–D) In situ hybridization analysis of CsOVATE expression in cucumber. Longitudinal sections showing floral meristem (A) and developing flower primordia at stage 2 (B) and stage 6 (C). The asterisk indicates the developing fruit neck in cucumber. The negative control hybridized with the sense CsOVATE probe (D). (Scale bars, 100 µm.) (E) CRISPR-Cas9–induced homozygous mutations in CsOVATE producing knockout alleles due to early appearance of stop codons. The stars represent the termination codon position and the yellow boxes indicate the OVATE domain. (F–H) Fruit morphology of WT and Csovate mutants at 0 DPP (F), 10 DPP (G), and 40 DPP (H). The brackets indicate the fruit neck and the double arrows represent the measured FNL. (Scale bars, 2 cm.) (I and J) Quantification of FNL (I) and ratio of FNL/FL (J) in WT and Csovate mutants at 40 DPP. Values are means ± SD (n = 9 to 14). (K and L) Representative cell morphology (K) and cell-length quantification (L) in the longitudinal sections of fruit necks in WT and Csovate fruits at 40 DPP. Values are means ± SD (n = 9). (Scale bar, 50 µm.) Significance analysis compared with WT was performed with the two-tailed Student’s t test (ns, no significant difference; **P < 0.01).
CsHEC1 Physically Interacts with CsOVATE at the Protein Level.
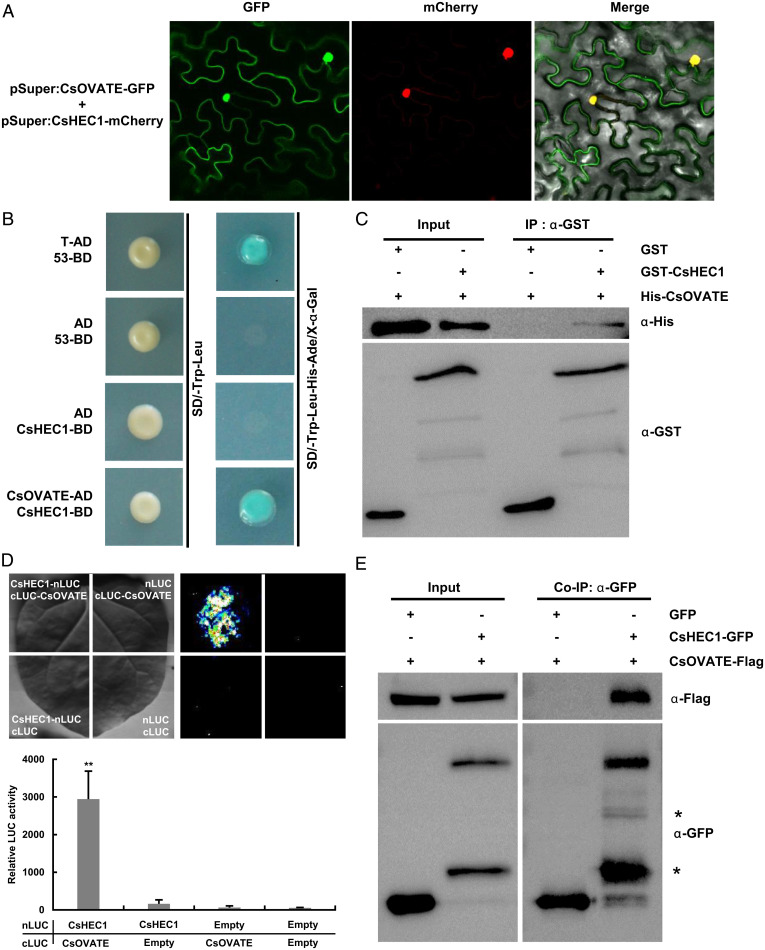
Given the opposite roles of CsHEC1 and CsOVATE during fruit neck elongation (Figs. 1, 2, and 4), we next explored any interactions between CsHEC1 and CsOVATE. Expression analyses were performed in the counterpart’s transgenic lines. The expression of CsOVATE was not altered in Cshec1 mutants or CsHEC1-OE lines (SI Appendix, Figs. S5C and S6B). Likewise, the CsHEC1 expression was unaffected in the Csovate mutants (SI Appendix, Fig. S10C), indicating no transcriptional regulation between CsHEC1 and CsOVATE in cucumber. Tomato OVATE was known to be localized in the cytoplasm of Nicotiana benthamiana leaf epidermal cells (26). However, CsOVATE-GFP (green fluorescent protein) had a localization signal both in the cytoplasm and in the nucleus, and CsOVATE colocalized with CsHEC1 in the nucleus (Fig. 5A). Yeast two-hybrid assay showed that CsHEC1 interacted with CsOVATE at the protein level (Fig. 5B). Pull-down experiment indicated that GST (glutathione S-transferase)-CsHEC1 was able to bind to His-CsOVATE (Fig. 5C). The firefly luciferase complementation imaging (LCI) assay in N. benthamiana showed that CsOVATE interacted with CsHEC1 but not with CsHEC2 (Fig. 5D and SI Appendix, Fig. S11). The specific interaction between CsOVATE and CsHEC1 was further verified by coimmunoprecipitation (Co-IP) analysis (Fig. 5E). Together, these results suggested that CsOVATE directly interacted with CsHEC1 at the protein level both in vitro and in vivo.
Fig. 5.
CsHEC1 directly interacts with CsOVATE at the protein level. (A) CsOVATE colocalization with the CsHEC1 protein in N. benthamiana leaves. The yellow signal in the merged field represents the colocalization signal in the nucleus. (B) Interaction of CsHEC1 and CsOVATE in the yeast two-hybrid system. The combination of T-AD and 53-BD was used as a positive control. (C) CsHEC1 interacts with CsOVATE in vitro tested by a GST pull-down assay. The combination of GST and His-CsOVATE was used as a control. (D) Firefly luciferase complementation imaging analysis. CsHEC1-nLUC and cLUC-CsOVATE were transiently coexpressed in N. benthamiana, and the remaining combinations were used as controls. Representative pictures (Top) and the relative luciferase activity value are shown (Bottom). Values are means ± SD (n = 6). Two-tailed Student’s t test (**P < 0.01). (E) Co-IP assay showing that CsHEC1 interacted with CsOVATE in vivo. The total and precipitated proteins were detected by immunoblotting using anti-GFP antibody or anti-Flag antibody. The asterisks indicate nonspecific bands.
Genetic Interaction of CsOVATE with CsHEC1 in Cucumber.
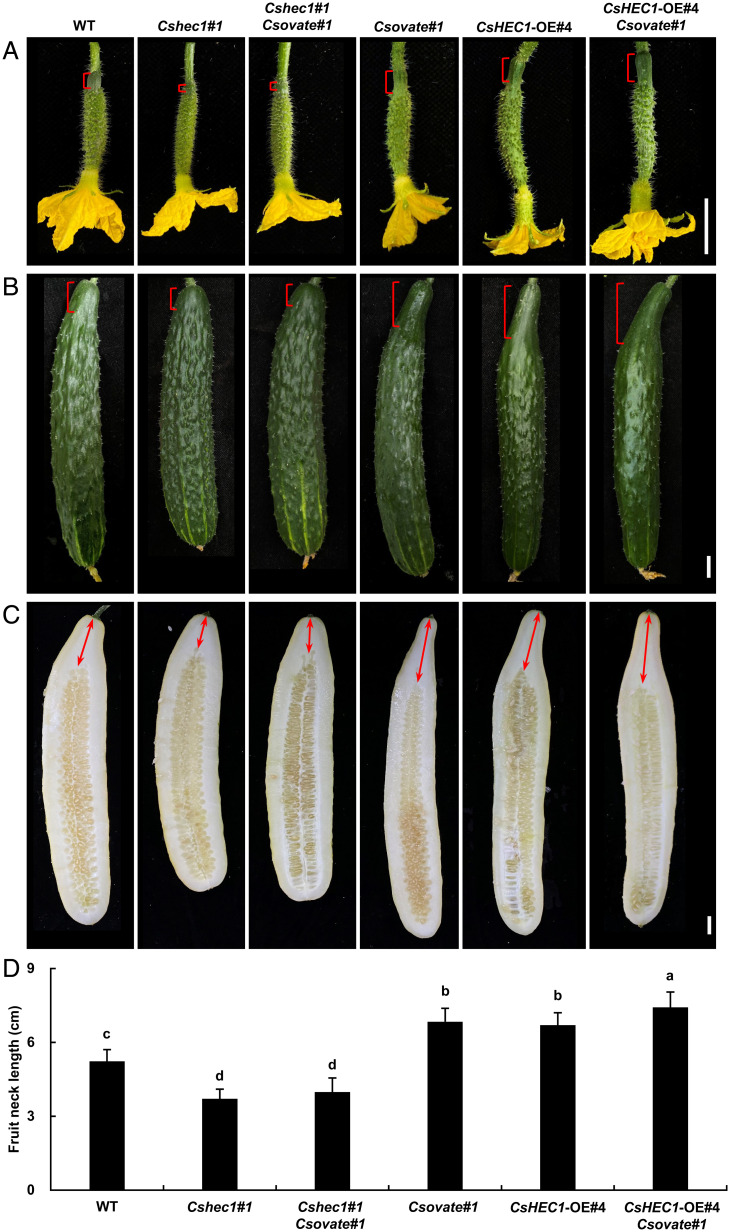
To determine the genetic relationships between CsHEC1 and CsOVATE, we generated a double mutant by crossing Csovate#1 with Cshec1#1. The FNLs of the double mutant Csovate#1 Cshec1#1 were indistinguishable from those of Cshec1#1 but markedly shorter than those of Csovate#1 and WT (Fig. 6 A–D), suggesting that the function of CsOVATE is dependent on CsHEC1 during fruit neck elongation. Meanwhile, plants overexpressing CsHEC1 (Pro35S:CsHEC1-Flag, OE#4) in the Csovate#1 mutant were obtained and exhibited further elongated fruit neck and increased FNL/FL ratio at anthesis and maturity compared with CsHEC1-OE#4 (Fig. 6 and SI Appendix, Fig. S12). Immunoblot analysis showed that the corresponding CsHEC1 protein was overexpressed in the respective plants with similar expression levels (SI Appendix, Fig. S13). Together, these data suggested that CsOVATE genetically interacted with CsHEC1 to inhibit CsHEC1 function in regulating fruit neck elongation in cucumber.
Fig. 6.
Genetic interaction of CsOVATE and CsHEC1 during fruit neck elongation in cucumber. (A–C) Fruit morphology of WT, Cshec1#1, Cshec1#1 Csovate#1, Csovate#1, CsHEC1-OE#4, and CsHEC1-OE#4/Csovate#1 at 0 DPP (A), 10 DPP (B), and 40 DPP (C). The brackets indicate the fruit neck and the double arrows represent the measured FNL. (Scale bars, 2 cm.) (D) Quantification of FNLs in the indicated lines at maturity. Values are means ± SD (n = 7). The different lowercase letters indicate significant differences (P < 0.05) by one-way ANOVA analysis with Duncan’s test.
CsOVATE Attenuates CsHEC1-Mediated CsYUC4 Activation and Auxin Accumulation.
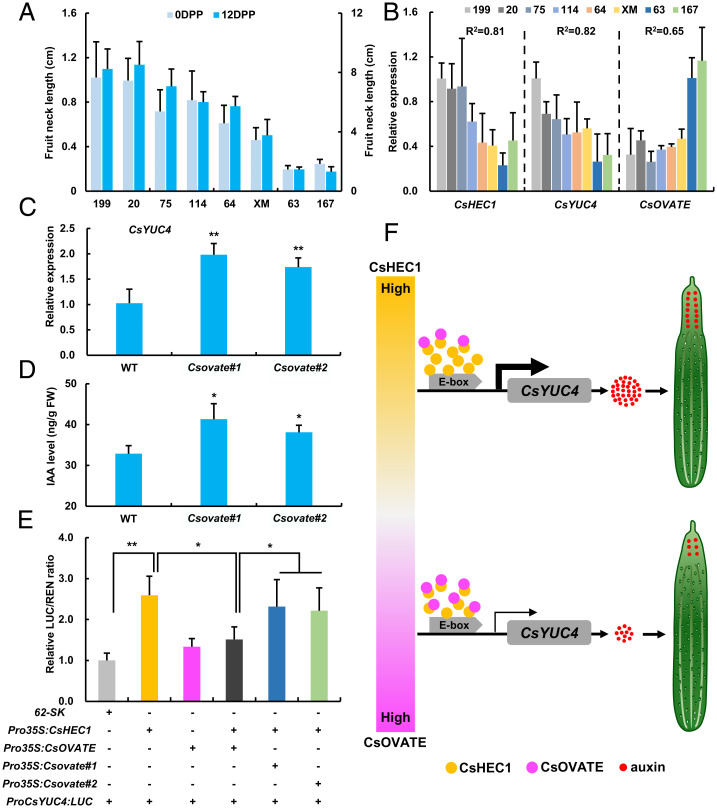
To further dissect the causal relationship between CsHEC1 and CsOVATE in the natural cucumber population, the expression of CsHEC1, CsYUC4, and CsOVATE was analyzed in eight cucumber inbred lines with different FNLs. Our data showed that the expression of CsHEC1 and CsYUC4 was positively correlated with FNL, with the correlation coefficient R2 0.81 for CsHEC1 and 0.82 for CsYUC4, whereas the expression of CsOVATE displayed a negative correlation (Fig. 7 A and B). Both CsYUC4 expression and auxin level were significantly increased in the fruit neck of Csovate knockout lines (Fig. 7 C and D), suggesting that CsOVATE may inhibit fruit neck elongation via decreasing the CsYUC4-mediated auxin biosynthesis. To further explore the relationship between CsOVATE and CsYUC4, a DLR assay was performed. Unlike the direct transcriptional activation of CsYUC4 by CsHEC1, the CsYUC4 expression was unaffected upon coexpression of CsOVATE. However, CsYUC4 transcription was significantly decreased upon coexpression of both CsOVATE and CsHEC1, while mutations of CsOVATE were unable to execute the attenuating effects (Fig. 7E), suggesting that CsOVATE antagonizes with CsHEC1 to modulate CsYUC4 transcriptional activation and auxin biosynthesis during fruit neck development in cucumber.
Fig. 7.
CsOVATE antagonizes CsHEC1-mediated CsYUC4 activation during fruit neck elongation in cucumber. (A) Cucumber inbred lines with different FNL at 0 and 10 DPP. (B) Expression analysis of CsHEC1, CsYUC4, and CsOVATE in different cucumber inbred lines. (C) Expression analysis of CsYUC4 in WT and Csovate mutants. (D) IAA content in the fruit necks of WT and Csovate mutants. Values are means ± SD (n = 3 in C and D). (E) Firefly luciferase and renilla reiformis luciferase activity assay in N. benthamiana leaves by coexpression of Pro35S:CsHEC1 and/or Pro35S:CsOVATE or Pro35S:Csovate#1 or Pro35S:Csovate#2 with ProCsYUC4:LUC. Values are means ± SD (n = 5). Significance analysis was calculated by Student’s t test (*P < 0.05, **P < 0.01). (F) The working model of CsHEC1 and CsOVATE regulating fruit neck elongation in cucumber. CsHEC1 promotes fruit neck elongation in cucumber by directly activating the expression of CsYUC4 and thus increasing auxin accumulation in the fruit neck. CsOVATE functions as a negative regulator for fruit neck elongation via interaction with CsHEC1 to attenuate the CsHEC1-mediated CsYUC4 activation, thus reducing auxin biosynthesis in the fruit neck.
Discussion
Unique Role of CsHEC1 during Fruit Neck Elongation in Cucumber.
The HEC genes encoding bHLH transcription factors play important roles in gynoecium development and shoot apical meristem maintenance (22, 24, 46). Here, we found that CsHEC1 was highly expressed in the fruit neck and promoted fruit neck elongation in cucumber (Figs. 1 and 2 and SI Appendix, Figs. S5–S7). Gene function is usually closely related to the gene expression pattern, modulated by upstream regulatory proteins and cis-regulatory elements (5, 6, 47–50). Variations in regulatory regions or expression domains have been found to be the main causes for the remarkable diversity in organ morphology during natural evolution and the domestication process of crops (5, 47, 49). In Arabidopsis, HEC1/2/3 are expressed in the female reproductive tissues, including stigma, style, septum, and transmitting tract. Accordingly, hec mutants display defects in the development of the stigma, transmitting tract, and septum, resulting in reduced fertility (22). In cucumber, CsHEC1 was highly expressed in the fruit neck and its mutation led to a shortened fruit neck, while CsHEC2 was expressed in the spine and tubercule and functions in wart density control (34), suggesting the functional diversification of HECs in the fleshy pepo fruit. Interestingly, CsHEC1 was highly expressed in the developing carpels as well while no aberrant carpel phenotype was observed in the Cshec1 mutant, which may be due to overlapping expression of the paralogous genes CsHEC2 and CsHEC3 in carpel primordium and thus the exertion of redundant functions during carpel development in cucumber (33) (Fig. 1 E and F and SI Appendix, Fig. S1).
Gene duplication event is another important cause for neofunctionalization in homologous genes (51). Differentiation of the HEC1/2 clade and HEC3/INDEHISCENT (IND) clade resulted from an early duplication event in angiosperms (33, 52). Within Solanaceae, the HEC1 clade and HEC2 clade have undergone further duplications producing SlHEC1 and SlHEC1-1, and SlHEC2 and SlHEC2-1 (SI Appendix, Fig. S1), which generally are not expressed in the gynoecium of dry dehiscent or fleshy fruits in this family (52). Unlike the apparent functional redundancy of HEC1/2/3 in Arabidopsis, CsHEC1/2 contribute to fruit development, while CsHEC3 regulates organ morphology and downy mildew resistance in cucumber (22, 33, 34). Thus, functional variations of HECs can be explained by their different expression patterns or duplication events in plant species with different fruit types.
CsHEC1 Regulates Auxin Biosynthesis to Promote Fruit Neck Elongation.
The phytohormone auxin plays a crucial role in multiple developmental processes including organ morphogenesis and plant architecture determination (38, 49, 53, 54). The IND in Capsella rubella exerts its function on the heart-shaped fruit by directly stimulating the expression of auxin biosynthesis genes TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS 1 (CrTAA1) and CrYUC9 to facilitate auxin maxima in the fruit shoulder (49). Auxin transporters PIN1/7 and PIN3 have been shown to mediate fruit elongation and shoot branching in cucumber, respectively (3, 54). In Arabidopsis, HEC1 functions in stigma formation and carpel fusion by directly activating the expression of PIN1/3 (24). Although sporadic results of gene expression or genetic data suggest the possibility of regulation between HECs and YUCs (24), the direct relationship between HECs and YUCs has not been established.
Here, we showed that CsHEC1 stimulates auxin accumulation in the fruit neck in cucumber (Figs. 1Q and 2J and SI Appendix, Fig. S7I). Among the CsYUC family, CsYUC4 is the only one displaying enriched expression in the fruit neck (SI Appendix, Fig. S8). Our data showed that CsHEC1 directly bound to the promoter of CsYUC4 to activate its expression. Enhanced expression of CsYUC4 driven by the CsHEC1 promoter resulted in an elongated fruit neck and increased auxin level in cucumber (Fig. 3). Csyuc4 mutants displayed shortened fruit neck and reduced fruit length (SI Appendix, Fig. S9), similar to that of Cshec1 mutants. In cucumber inbred lines with different FNLs, the expression of CsHEC1 and CsYUC4 displayed positive correlation with FNL (Fig. 7 A and B). These data suggested that CsHEC1 functions as an activator for FNL through direct regulation of CsYUC4-mediated auxin biosynthesis in cucumber (Fig. 7F).
CsOVATE Combats CsHEC1 during Fruit Neck Elongation in Cucumber.
Similar to the tomato fruit changing from round- to pear-shaped upon mutation in the OVATE gene (1, 27), Csovate mutants displayed increased FNL in cucumber (Fig. 4), suggesting that the function of OVATE appeared to be conserved in different species. The pear-shaped fruit caused by OVATE mutation in tomato was primarily due to an increase of the proximal cell numbers, resembling the effect of auxin application on fruit shape (40). In Csovate mutants, the longitudinal cell length was comparable to WT (Fig. 4 K and L), implying that the elongated fruit neck was due to an increase in cell numbers. The expression of CsYUC4 and IAA level was greatly increased in the Csovate knockout lines (Fig. 7 C and D). However, no direct regulation of CsYUC4 expression was detected by CsOVATE (Fig. 7E). Instead, CsOVATE directly interacts with CsHEC1 at the protein level (Fig. 5), and such interaction inhibits the CsHEC1-mediated transcriptional activation of CsYUC4 (Fig. 7E). Genetic analysis showed that plants overexpressing CsHEC1 in the Csovate#1 mutant background exhibited further elongation of fruit neck than that of CsHEC1-OE#4 alone, while the FNL of the double mutant Csovate Cshec1 resembles that of Cshec1 (Fig. 6). In the natural cucumber population, CsOVATE expression is negatively correlated with FNL and CsYUC4 transcription (Fig. 7 A and B). These data indicated that CsOVATE functions as a negative regulator for fruit neck elongation through direct interaction with CsHEC1 to attenuate the CsYUC4-mediated auxin biosynthesis. Thus, CsHEC1 and CsOVATE form a regulatory module to regulate local auxin content during controlling fruit neck elongation in cucumber (Fig. 7F). These findings provide a strategy to minimize the undesirable FNL by adjusting the levels of CsHEC1 and/or CsOVATE transcription to decrease auxin accumulation in fruit neck during cucumber breeding.
Further studies are needed to dissect the promoter differences and the underlying regulatory mechanism of CsHEC1 and CsOVATE expression in cucumber germplasms with various FNLs. Notably, similar to multiple regulators underlying fruit length variation in cucumber (3, 9, 55, 56), loss-of-function Cshec1 mutants showed only 21 to 28% decreases in FNL (Fig. 1M), suggesting that additional genes such as CsFnl7.1 (19) and other unidentified players may be involved in fruit neck development in cucumber. It would be of significant importance to dissect the gene network controlling fruit neck variation so as to accelerate genetic improvement of fruit shape in cucumber.
Materials and Methods
The details and procedures of plant materials and growth conditions, phenotypic characterization, phylogenetic analysis, generation of transgenic cucumber plants, subcellular localization, RNA extraction and expression analysis (qRT-PCR, in situ hybridization, and GUS staining), DNA–protein interaction assays (yeast one-hybrid, DLR, and ChIP-PCR), protein–protein interaction assays (yeast two-hybrid, LCI, pull-down, and Co-IP), and quantification of endogenous auxin and histology observation are provided in SI Appendix, Materials and Methods. Accession numbers used in this study are listed in SI Appendix, Table S1. Primers used in this study are provided in SI Appendix, Table S4.
Supplementary Material
Acknowledgments
We thank Prof. Sanwen Huang (Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences) for giving CRISPR-Cas9 vectors, and Dr. Li Yang for guidance in the cucumber transformation experiment. We are grateful to Prof. Xuexian Li (China Agricultural University) for critical comments. This work is supported by grants from the National Natural Science Foundation of China (32025033, 31930097, and 32102387), National Key Research and Development Program (2018YFD1000800), Construction of Beijing Science and Technology Innovation and Service Capacity in Top Subjects (CEFFPXM2019_014207_000032), and 111 Project (B17043).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. A.R. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2209717119/-/DCSupplemental.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Rodríguez G. R., et al. , Distribution of SUN, OVATE, LC, and FAS in the tomato germplasm and the relationship to fruit shape diversity. Plant Physiol. 156, 275–285 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Qi J., et al. , A genomic variation map provides insights into the genetic basis of cucumber domestication and diversity. Nat. Genet. 45, 1510–1515 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Zhao J., et al. , A functional allele of CsFUL1 regulates fruit length through repressing CsSUP and inhibiting auxin transport in cucumber. Plant Cell 31, 1289–1307 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xiao H., Jiang N., Schaffner E., Stockinger E. J., van der Knaap E., A retrotransposon-mediated gene duplication underlies morphological variation of tomato fruit. Science 319, 1527–1530 (2008). [DOI] [PubMed] [Google Scholar]
- 5.Cong B., Barrero L. S., Tanksley S. D., Regulatory change in YABBY-like transcription factor led to evolution of extreme fruit size during tomato domestication. Nat. Genet. 40, 800–804 (2008). [DOI] [PubMed] [Google Scholar]
- 6.Muños S., et al. , Increase in tomato locule number is controlled by two single-nucleotide polymorphisms located near WUSCHEL. Plant Physiol. 156, 2244–2254 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chu Y. H., Jang J. C., Huang Z., van der Knaap E., Tomato locule number and fruit size controlled by natural alleles of lc and fas. Plant Direct 3, e00142 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu C., et al. , A cascade of arabinosyltransferases controls shoot meristem size in tomato. Nat. Genet. 47, 784–792 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Weng Y., et al. , QTL mapping in multiple populations and development stages reveals dynamic quantitative trait loci for fruit size in cucumbers of different market classes. Theor. Appl. Genet. 128, 1747–1763 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Che G., Zhang X., Molecular basis of cucumber fruit domestication. Curr. Opin. Plant Biol. 47, 38–46 (2019). [DOI] [PubMed] [Google Scholar]
- 11.Roeder A. H., Yanofsky M. F., Fruit development in Arabidopsis. Arabidopsis Book 4, e0075 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao J., et al. , Phloem transcriptome signatures underpin the physiological differentiation of the pedicel, stalk and fruit of cucumber (Cucumis sativus L.). Plant Cell Physiol. 57, 19–34 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Sui X., et al. , Transcriptomic and functional analysis of cucumber (Cucumis sativus L.) fruit phloem during early development. Plant J. 96, 982–996 (2018). [DOI] [PubMed] [Google Scholar]
- 14.Fanourakis N. E., Tzifaki E. E., Correlated inheritance of fruit neck with fruit length and linkage relations with 10 other characteristics of cucumber. Euphytica 65, 71–77 (1992). [Google Scholar]
- 15.Gu X., Fang X., Han X., Preliminary study of fruit neck length inheritance in cucumber [in Chinese]. China Vegetables 2, 33–34 (1994). [Google Scholar]
- 16.Ma D., Lu S., Shen W., Huo Z., Li S., Combining ability analysis of cucumber quality characteristics [in Chinese]. Acta Agriculturae Boreali–Sinica 9, 65–68 (1994). [Google Scholar]
- 17.Wang G., Qin Z., Zhou X., Zhao Z., Mapping quantitative trait loci influencing cucumber carpopodium length using simple sequence repeat markers [in Chinese]. Acta Horticulturae Sinica 35, 543–546 (2008). [Google Scholar]
- 18.Wang M., et al. , Quantitative trait loci associated with fruit length and stalk length in cucumber using RIL population [in Chinese]. Acta Bot. Boreali–Occidential Sinica 34, 1764–1770 (2014). [Google Scholar]
- 19.Xu X., et al. , The major-effect quantitative trait locus Fnl7.1 encodes a late embryogenesis abundant protein associated with fruit neck length in cucumber. Plant Biotechnol. J. 18, 1598–1609 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sessions R. A., Zambryski P. C., Arabidopsis gynoecium structure in the wild and in ettin mutants. Development 121, 1519–1532 (1995). [DOI] [PubMed] [Google Scholar]
- 21.Marsch-Martínez N., de Folter S., Hormonal control of the development of the gynoecium. Curr. Opin. Plant Biol. 29, 104–114 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Gremski K., Ditta G., Yanofsky M. F., The HECATE genes regulate female reproductive tract development in Arabidopsis thaliana. Development 134, 3593–3601 (2007). [DOI] [PubMed] [Google Scholar]
- 23.Okada K., Ueda J., Komaki M. K., Bell C. J., Shimura Y., Requirement of the auxin polar transport system in early stages of Arabidopsis floral bud formation. Plant Cell 3, 677–684 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schuster C., Gaillochet C., Lohmann J. U., Arabidopsis HECATE genes function in phytohormone control during gynoecium development. Development 142, 3343–3350 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Snouffer A., Kraus C., van der Knaap E., The shape of things to come: Ovate family proteins regulate plant organ shape. Curr. Opin. Plant Biol. 53, 98–105 (2020). [DOI] [PubMed] [Google Scholar]
- 26.Wu S., et al. , A common genetic mechanism underlies morphological diversity in fruits and other plant organs. Nat. Commun. 9, 4734 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu J., Van Eck J., Cong B., Tanksley S. D., A new class of regulatory genes underlying the cause of pear-shaped tomato fruit. Proc. Natl. Acad. Sci. U.S.A. 99, 13302–13306 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu D., et al. , Phylogenetic analyses provide the first insights into the evolution of OVATE family proteins in land plants. Ann. Bot. 113, 1219–1233 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang S., Chang Y., Guo J., Chen J. G., Arabidopsis Ovate family protein 1 is a transcriptional repressor that suppresses cell elongation. Plant J. 50, 858–872 (2007). [DOI] [PubMed] [Google Scholar]
- 30.Zhou H., et al. , A 1.7-Mb chromosomal inversion downstream of a PpOFP1 gene is responsible for flat fruit shape in peach. Plant Biotechnol. J. 19, 192–205 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pan Y., et al. , Genetic architecture of fruit size and shape variation in cucurbits: A comparative perspective. Theor. Appl. Genet. 133, 1–21 (2020). [DOI] [PubMed] [Google Scholar]
- 32.Ma J., et al. , CmFSI8/CmOFP13 encoding an OVATE family protein controls fruit shape in melon. J. Exp. Bot. 73, 1370–1384 (2022). [DOI] [PubMed] [Google Scholar]
- 33.Yan S., et al. , CsIVP functions in vasculature development and downy mildew resistance in cucumber. PLoS Biol. 18, e3000671 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Z., et al. , HECATE2 acts with GLABROUS3 and Tu to boost cytokinin biosynthesis and regulate cucumber fruit wart formation. Plant Physiol. 187, 1619–1635 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heim M. A., et al. , The basic helix-loop-helix transcription factor family in plants: A genome-wide study of protein structure and functional diversity. Mol. Biol. Evol. 20, 735–747 (2003). [DOI] [PubMed] [Google Scholar]
- 36.Xu Y., et al. , A zinc finger protein BBX19 interacts with ABF3 to affect drought tolerance negatively in chrysanthemum. Plant J. 103, 1783–1795 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bai S. L., et al. , Developmental analyses reveal early arrests of the spore-bearing parts of reproductive organs in unisexual flowers of cucumber (Cucumis sativus L.). Planta 220, 230–240 (2004). [DOI] [PubMed] [Google Scholar]
- 38.Nemhauser J. L., Feldman L. J., Zambryski P. C., Auxin and ETTIN in Arabidopsis gynoecium morphogenesis. Development 127, 3877–3888 (2000). [DOI] [PubMed] [Google Scholar]
- 39.Sohlberg J. J., et al. , STY1 regulates auxin homeostasis and affects apical-basal patterning of the Arabidopsis gynoecium. Plant J. 47, 112–123 (2006). [DOI] [PubMed] [Google Scholar]
- 40.Wang Y., et al. , A comparison of sun, ovate, fs8.1 and auxin application on tomato fruit shape and gene expression. Plant Cell Physiol. 60, 1067–1081 (2019). [DOI] [PubMed] [Google Scholar]
- 41.Mashiguchi K., et al. , The main auxin biosynthesis pathway in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 108, 18512–18517 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Won C., et al. , Conversion of tryptophan to indole-3-acetic acid by TRYPTOPHAN AMINOTRANSFERASES OF ARABIDOPSIS and YUCCAs in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 108, 18518–18523 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yan S., et al. , Different cucumber CsYUC genes regulate response to abiotic stresses and flower development. Sci. Rep. 6, 20760 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Massari M. E., Murre C., Helix-loop-helix proteins: Regulators of transcription in eucaryotic organisms. Mol. Cell. Biol. 20, 429–440 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Toledo-Ortiz G., Huq E., Quail P. H., The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15, 1749–1770 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schuster C., et al. , A regulatory framework for shoot stem cell control integrating metabolic, transcriptional, and phytohormone signals. Dev. Cell 28, 438–449 (2014). [DOI] [PubMed] [Google Scholar]
- 47.Konishi S., et al. , An SNP caused loss of seed shattering during rice domestication. Science 312, 1392–1396 (2006). [DOI] [PubMed] [Google Scholar]
- 48.Arnaud N., Lawrenson T., Østergaard L., Sablowski R., The same regulatory point mutation changed seed-dispersal structures in evolution and domestication. Curr. Biol. 21, 1215–1219 (2011). [DOI] [PubMed] [Google Scholar]
- 49.Dong Y., et al. , Regulatory diversification of INDEHISCENT in the Capsella genus directs variation in fruit morphology. Curr. Biol. 29, 1038–1046.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rodríguez-Leal D., Lemmon Z. H., Man J., Bartlett M. E., Lippman Z. B., Engineering quantitative trait variation for crop improvement by genome editing. Cell 171, 470–480.e8 (2017). [DOI] [PubMed] [Google Scholar]
- 51.Panchy N., Lehti-Shiu M., Shiu S. H., Evolution of gene duplication in plants. Plant Physiol. 171, 2294–2316 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ortiz-Ramírez C. I., Plata-Arboleda S., Pabón-Mora N., Evolution of genes associated with gynoecium patterning and fruit development in Solanaceae. Ann. Bot. 121, 1211–1230 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cheng Y., Dai X., Zhao Y., Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev. 20, 1790–1799 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shen J., et al. , CsBRC1 inhibits axillary bud outgrowth by directly repressing the auxin efflux carrier CsPIN3 in cucumber. Proc. Natl. Acad. Sci. U.S.A. 116, 17105–17114 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xin T., et al. , Genetic regulation of ethylene dosage for cucumber fruit elongation. Plant Cell 31, 1063–1076 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang Z., et al. , Genome-wide target mapping shows histone deacetylase complex1 regulates cell proliferation in cucumber fruit. Plant Physiol. 182, 167–184 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.