Significance
Expression of androgen receptor variants (AR-Vs) is implicated in the development of castration-resistant prostate cancer (PCa). Others have shown that androgen depletion or antiandrogen treatment induces AR-V expression in PCa cell lines, xenografts, and patient samples, although the underlying mechanism remains unclear. Our findings reveal that hormonal therapy–induced CYCLIN K down-regulation represents a key mechanism that drives intronic polyadenylation (IPA) usage in the AR gene and AR-V expression and castration resistance in PCa, and that this mechanism of action can be therapeutically targeted by the PARP inhibitor.
Keywords: CDK12, CYCLIN K, AR, AR variants, prostate cancer
Abstract
Androgen receptor (AR) messenger RNA (mRNA) alternative splicing variants (AR-Vs) are implicated in castration-resistant progression of prostate cancer (PCa), although the molecular mechanism underlying the genesis of AR-Vs remains poorly understood. The CDK12 gene is often deleted or mutated in PCa and CDK12 deficiency is known to cause homologous recombination repair gene alteration or BRCAness via alternative polyadenylation (APA). Here, we demonstrate that pharmacological inhibition or genetic inactivation of CDK12 induces AR gene intronic (intron 3) polyadenylation (IPA) usage, AR-V expression, and PCa cell resistance to the antiandrogen enzalutamide (ENZ). We further show that AR binds to the CCNK gene promoter and up-regulates CYCLIN K expression. In contrast, ENZ decreases AR occupancy at the CCNK gene promoter and suppresses CYCLIN K expression. Similar to the effect of the CDK12 inhibitor, CYCLIN K degrader or ENZ treatment promotes AR gene IPA usage, AR-V expression, and ENZ-resistant growth of PCa cells. Importantly, we show that targeting BRCAness induced by CYCLIN K down-regulation with the PARP inhibitor overcomes ENZ resistance. Our findings identify CYCLIN K down-regulation as a key driver of IPA usage, hormonal therapy–induced AR-V expression, and castration resistance in PCa. These results suggest that hormonal therapy–induced AR-V expression and therapy resistance are vulnerable to PARP inhibitor treatment.
Prostate cancer (PCa) is the second-leading cause of cancer death in American men and androgen deprivation therapy (ADT) is the mainstay treatment of advanced/metastatic PCa. However, most prostate cancers become castration-resistant (CRPC). Androgen receptor variants (AR-Vs) have been implicated in the development of CRPC. AR-Vs are derived from alternative splicing of the AR gene and encode various truncated forms lacking a functional/intact ligand-binding domain in the C terminus. These AR-Vs become constitutively active in the absence of androgens, thus promoting PCa progression (1, 2). Increasing evidence suggests that AR-Vs contribute to antiandrogen therapy resistance and correlate with inferior clinical outcomes (3, 4). However, the precise mechanism underlying the genesis of AR-Vs remains poorly understood.
In eukaryotes, pre-mRNA cleavage and polyadenylation are essential for production of mature messenger RNA (mRNA) (5, 6). Most mammalian genes contain multiple cleavage and polyadenylation sites (PASs) in intronic (7–9) or 3′ untranslated regions (UTRs) (7, 10), and different mRNA isoforms of the same gene can be generated by the usage of alternative polyadenylation (APA) sites, including intronic polyadenylation (IPA) sites. These isoforms vary depending on cell types (11–13) and cellular conditions such as cell proliferation (14), differentiation (15, 16), developmental stage (17), oncogenesis (18, 19), and drug resistance (20, 21).
The Cyclin-dependent kinases (CDKs) are a group of serine/threonine kinases that regulate essential cellular processes. These kinases can be categorized into two major subclasses: 1) cell cycle–driving CDKs (e.g., CDK1, CDK2, CDK4, and CDK6), and 2) transcription-related CDKs (e.g., CDK7, CDK9, CDK12, and CDK13) (22). The CDK12 gene is implicated in PCa as it has been found deleted or mutated in metastatic CRPC (mCRPC). CDK12-deficient tumors often possess unique phenotypical features such as genomic tandem duplications and neoantigen expression (23–25). CDK12 deficiency also affects transcript elongation by promoting defects in RNA polymerase II serine 2 phosphorylation (26), 3′-end formation of pre-mRNA (27, 28), and DNA repair (29–31). CDK12-deficient prostate tumors display aggressive clinical behaviors including shorter time to biochemical recurrence and emergence of mCRPC (32). Additionally, CDK12 deficiency increases DNA damage and genomic instability by causing APA usage and shortening and down-regulation of homologous recombination (HR) repair genes such as ATM, FANCD2, and WRN, thereby creating a cellular condition mimicking the defects in BRCA1 and BRCA2 genes (termed homologous recombination deficiency, or BRCAness). However, the impact of CDK12 deficiency on other cancer-relevant signaling pathways such as AR APA usage and AR-V expression has not yet been explored.
In the present study, we demonstrate that CDK12 deficiency (mutation and/or deletion) or down-regulation of CYCLIN K, the regulatory subunit of the CYCLIN K–CDK12 complex induced by AR inhibition, promotes IPA usage in the AR gene, expression of AR-Vs, and endocrine therapy resistance in PCa.
Results
CDK12 Inhibition Confers AR Antagonist Resistance in PCa Cells.
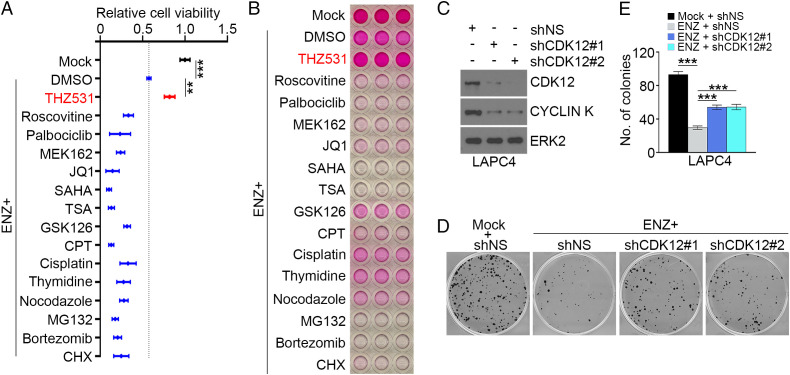
Enzalutamide (ENZ) is a Food and Drug Administration–approved second-generation antiandrogen for treatment of metastatic castration–sensitive and both nonmetastatic and metastatic CRPC (33, 34). While mCRPC tumors favorably respond to ENZ initially, patients survive a median of 16 mo. We therefore sought to identify signaling pathways that regulate ENZ resistance in PCa. To this end, we surveyed the sensitivity of AR-expressing LAPC4 PCa cells, which were originally derived from the lymph node metastasis of a male patient with CRPC, to an array of signaling pathway inhibitors following treatment with ENZ. In cell-viability assays, we demonstrated that LAPC4 cells responded in various degrees to the treatment of different drugs/chemicals we tested in the absence of ENZ; however, the effect of the CDK12 inhibitor THZ531 was opposite (SI Appendix, Fig. S1A). Further, treatment of LAPC4 cells with all the drugs except THZ531 substantially enhanced ENZ-induced cell-growth inhibition (Fig. 1A). Similar results were obtained in LAPC4 cells using the independent sulforhodamine B (SRB) assay which measured cell density and cellular protein content (Fig. 1B). To confirm this phenomenon, we employed a genetic approach to generate CDK12 knockdown–stable LAPC4 cells using two independent CDK12 gene–specific small hairpin RNAs (shRNAs). We then performed colony-formation assays using both control and CDK12-knockdown LAPC4 cells. CDK12 depletion decreased expression of its partner protein CYCLIN K (Fig. 1C), which is consistent with previous reports (29, 35). Importantly, we demonstrated that CDK12 knockdown conferred ENZ-resistant growth on LAPC4 cells (Fig. 1 D and E). Thus, data obtained from both pharmacological and genetic approaches consistently showed that CDK12 inactivation confers ENZ resistance in PCa cells.
Fig. 1.
CDK12 inhibition confers AR antagonist resistance in LAPC4 PCa cells. (A) LAPC4 cells cultured in regular medium (containing FBS without charcoal stripping) were treated with vehicle (mock) or ENZ in the presence or absence of the indicated drugs for 72 h and followed by MTS assay. Three biological replicates were analyzed. Data shown as means SD. **P < 0.01, ***P < 0.001. CHX, cycloheximide; CPT, camptothecin; DMSO, dimethyl sulfoxide; SAHA, suberoylanilide hydroxamic acid (Vorinostat); TSA, trichostatin A. (B) LAPC4 cells were treated with ENZ in combination with or without the indicated drugs for 1 wk and subjected to SRB assay. Similar results were obtained from two independent experiments. (C–E) LAPC4 cells stably infected with lentivirus expressing nonspecific control (shNS) or CDK12-specific shRNAs (shCDK12 #1 and #2) were subjected to Western blot (WB) analysis (C) for colony-formation assay in the presence or absence of ENZ treatment (5 µM) for 11 d. (D and E) Representative images showing colonies of the indicated cells are shown (D) as are quantitative data (E). ERK2 is a loading control for WB. Three biological replicates were analyzed. ***P < 0.001.
CDK12 Deficiency Induces Expression of AR-Vs in PCa Cells.
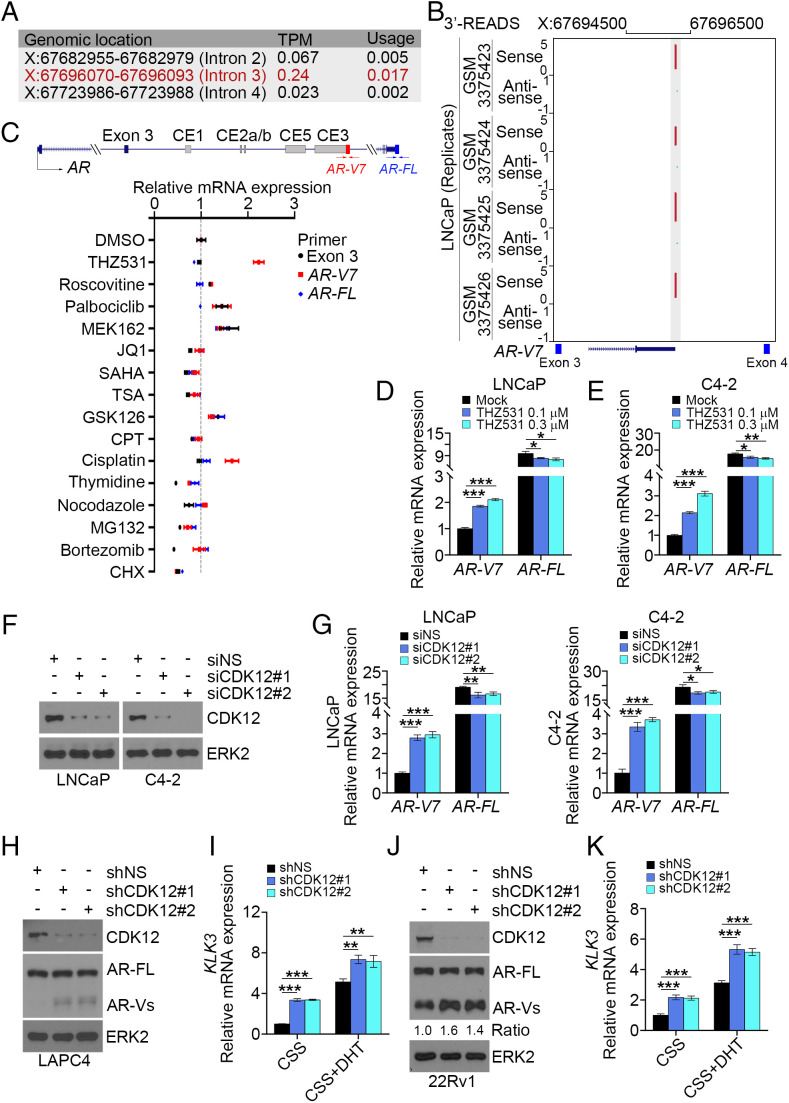
It is known that APA gives rise to mRNA variants with different coding regions or 3′ UTRs. It has been reported recently that CDK12 deficiency increases the incidence of APAs (30, 31). Moreover, increasing evidence suggests that expression of AR-Vs is one of the important mechanisms that contributes to ENZ resistance in PCa cells (3). We therefore sought to determine whether inactivation of CDK12 causes expression of AR-Vs in PCa cells. We observed that IPA usage was detectable in intron 3 of the AR gene in several normal tissues including prostate, testis, uterus, and liver, although the level was much lower compared with the usage of the distal PAS (SI Appendix, Fig. S1B). We also performed meta-analysis of the mRNA 3′-region extraction and deep sequencing (3′-READS) dataset generated from LNCaP cells, a cell line which predominantly expresses the full-length AR (AR-FL) (7). We uncovered three putative AR IPA sites in AR introns 2, 3, and 4 (Fig. 2A). Notably, the second peak (X: 67696070 to 67696093), located between exons 3 and 4, exhibited the highest transcripts per million (TPM) value and usage rate (Fig. 2 A and B). We further designed two pairs of primers unique for intronic PAS (within the third intron) and distal PAS sites, which correspond to AR-V7 and AR-FL, respectively (Fig. 2 C, Top), to define the IPA usage in LNCaP cells after treatment with various pathway inhibitors (Fig. 2 C, Bottom). Among the drugs/compounds we tested, THZ531 induced the highest IPA usage in intron 3 of AR but had limited effect on distal PAS usage. Accordingly, treatment with THZ531 or CR8, a CYCLIN K inhibitor (36), increased AR-V7 mRNA expression in LNCaP and its castration-resistant derivative cell line C4-2 (Fig. 2 D and E and SI Appendix, Fig. S2). Consistent with the effect of the CDK12 inhibitor, we demonstrated that knockdown of CDK12 by two independent small interference RNAs (siRNAs) increased IPA usage in AR intron 3 in both cell lines (Fig. 2 F and G). Similarly, stable depletion of CDK12 by gene-specific shRNAs also increased expression of AR-Vs and KLK3 (prostate-specific antigen [PSA]), a well-known AR target gene in LAPC4 and 22Rv1, a cell line that constitutively expresses AR-Vs (Fig. 2 H–K). Together, our data reveal that pharmacological inhibition or genetic depletion of CDK12 induces expression of AR variants in various PCa cell lines.
Fig. 2.
CDK12 alteration promotes AR IPA usage and AR-V7 expression. (A) The frequency of the usage of PASs between exons 2 and 5 of AR. X, chromosome X. (B) Analysis of 3′-READS shows an APA peak in the AR intron 3 region. (C) qRT-PCR analysis of AR-V7 and AR-FL mRNA using the indicated transcript-specific primers in LNCaP cells cultured in regular medium and treated with vehicle (DMSO) or the indicated drugs. Three biological replicates were analyzed. (D and E) LNCaP and C4-2 cells were treated with vehicle or THZ531 for 18 h and subjected to qRT-PCR analysis with the indicated primers. Three biological replicates were analyzed. *P < 0.05, **P < 0.01, ***P < 0.001. (F and G) LNCaP and C4-2 cells were transfected with the indicated siRNAs for 72 h and subjected to WB (F) and qRT-PCR analysis using the indicated primers (G). ERK2 is a loading control for WB. Three biological replicates were analyzed. *P < 0.05, **P < 0.01, ***P < 0.001. (H–K) 22Rv1 and LAPC4 cells stably infected with lentivirus expressing the indicated shRNAs were subjected to WB (H and J) or cultured in medium containing CSS in the presence or absence of DHT (1 nM) for 6 h followed by qRT-PCR analysis using KLK3 gene primers (I and K). Three biological replicates were analyzed. **P < 0.01, ***P < 0.001.
PCa-Associated CDK12 Mutations Enhance AR IPA Usage, AR Activity, and PCa Cell Growth.
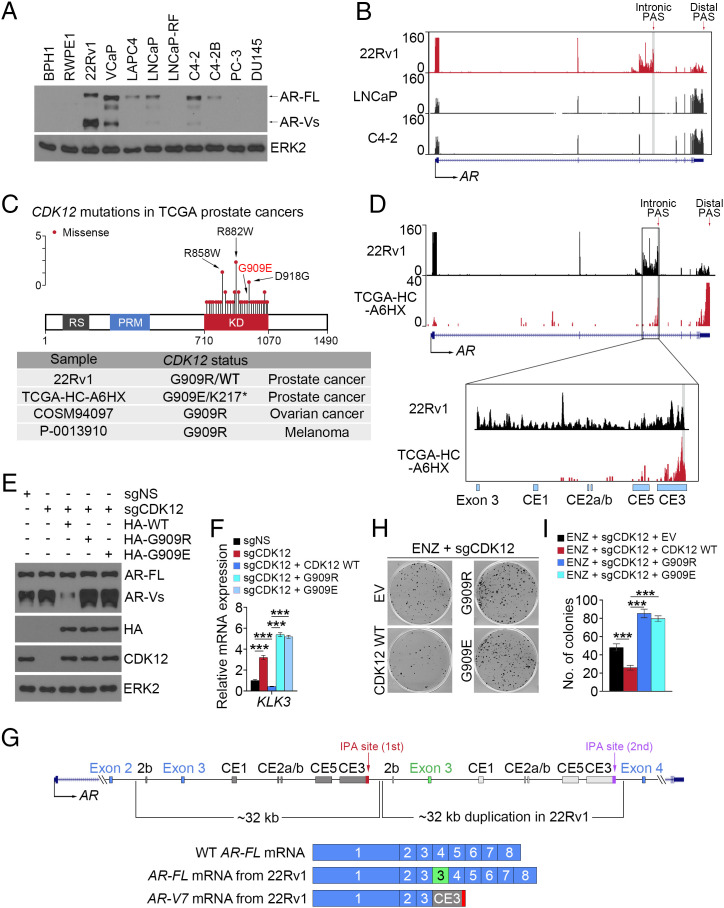
Among the six AR-expressing PCa cell lines examined, we noticed that expression of AR-Vs was much higher in 22Rv1 compared with VCaP, LNCaP, and C4-2 and that AR variant expression was almost undetectable in LAPC4 and C4-2B (Fig. 3A). In agreement with these results, meta-analysis of RNA-sequencing (RNA-seq) data showed that 22Rv1 cells had much higher IPA usage in intron 3 of the AR gene compared with the LNCaP and C4-2 cell lines (Fig. 3B). Further analysis showed that similar to the situation in The Cancer Genome Atlas (TCGA) PCa patient samples, there is a missense mutation (G909R) in the CDK12 gene in the 22Rv1 cell line (Fig. 3C and SI Appendix, Fig. S3 A and B). The same mutation was also detected in patient samples of other cancer types (Fig. 3 C, Bottom). Notably, a missense mutation (G909E, another mutation substituted with a charged amino acid) was also observed in the TCGA PCa patient sample TCGA-HC-A6HX (Fig. 3 C, Bottom). Analysis of RNA-seq data revealed that the IPA signaling in AR intron 3 was highly detectable in both the 22Rv1 cell line and this patient’s tumor (Fig. 3D and SI Appendix, Fig. S3C). Similarly, much higher IPA usage in DNA repair genes such as ATM and FANCD2 was also observed in CDK12-deficient patient samples compared with CDK12 wild-type (WT) counterparts (SI Appendix, Fig. S3D).
Fig. 3.
CDK12 alteration promotes AR IPA and ENZ resistance. (A) WB analysis of AR protein expression in the indicated PCa cell lines. ERK2 is a loading control for WB. (B) UCSC screenshot of RNA-seq in the AR locus in the indicated PCa cell lines. (C, Top) Spectrum of CDK12 gene mutations detected in TCGA PCa patient samples. (C, Bottom) Mutations in the CDK12 G909 residue in the 22Rv1 cell line and patient samples of the indicated cancer types. (D) UCSC screenshot of RNA-seq in the AR locus in the 22Rv1 cell line and the TCGA-HC-A6HX patient sample. (E and F) WB (E) and qRT-PCR analysis (F) in the stable 22Rv1 cell line with or without CDK12 knockout and/or rescued with WT or mutant CDK12. Three biological replicates were analyzed. ***P < 0.001. (G) Schematic diagram showing the genomic duplication in the region between exons 2 and 4 of the AR gene and the transcripts of AR-FL and AR-V7 in 22Rv1 cells. (H and I) CDK12-knockout and rescued 22Rv1 cells were treated with ENZ and subjected to colony-formation assay. At 11 d after treatment, cell colonies were photographed (H) and quantified (I). Three biological replicates were analyzed. ***P < 0.001.
We also examined the functional impact of these CDK12 mutations. We knocked out endogenous CDK12 in 22Rv1 cells and rescued with WT CDK12 or G909R or G909E mutants. Restored expression of mutant, but not WT, CDK12 enhanced expression of AR variants and KLK3 mRNA, a surrogate of AR activation in 22Rv1 cells (Fig. 3 E and F). Furthermore, we noticed that the molecular mass of AR-FL in 22Rv1 cells is larger (lower mobility by polyacrylamide gel) than the WT AR-FL in the VCaP, LNCaP, and C4-2 cell lines (Fig. 3A). This is the result of an additional zinc finger in the DNA-binding domain (DBD) caused by splicing of the extra copy of exon 3 (Fig. 3G). However, similar to a previous report (37), the additional zinc finger encoded by the extra exon 3 does not exist in the AR-Vs in 22Rv1 such that they migrate similar to those in the VCaP, LNCaP, and C4-2 cell lines (Fig. 3A). A plausible explanation is that independent IPA sites are present in the first intron 3 and the duplicated intron 3 of the AR gene; however, the pre-mRNA of the duplicated intron 3 should not be transcribed when the first IPA site was used in 22Rv1 cells (Fig. 3 G, Top) given that CDK12 is inactivated by the gene mutation. As a result, only the first exon 3, but not the additional exon 3, is expressed along with one copy of those cryptic exons in AR variants (Fig. 3 G, Bottom). Finally, we examined the effect of CDK12 mutations on AR antagonist resistance. We demonstrated that CDK12-knockout 22Rv1 cells with restored expression of G909R and G909E mutant CDK12 grew much faster than cells rescued with WT CDK12 when treated with ENZ (Fig. 3 H and I). Collectively, our data indicate that CDK12 mutations enhance AR IPA usage, AR activity, and ENZ-resistant growth of PCa cells.
CDK12 Deficiency Is Associated with Increased IPA Usage, AR Signaling Activation, and Poor Survival of PCa Patients.
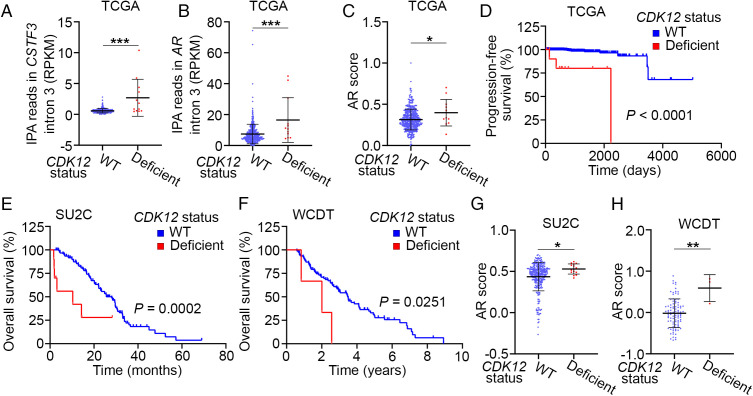
To determine the potential impact of CDK12 deficiency on AR signaling and overall survival in patients, we analyzed the PCa patient data of the TCGA cohort. The IPA usage in intron 3 of the CSTF3 gene is a well-studied APA indicator (38). We observed that patients with CDK12 defects (mutations and/or deletions) in prostate tumors had much higher CSTF3 IPA usage than patients with WT CDK12 (Fig. 4A), implying enhanced overall APA activities in the CDK12-deficient patient samples. Moreover, we found that the IPA usage in intron 3 of the AR gene locus was also higher in CDK12-deficient samples compared with CDK12 WT counterparts (Fig. 4B). We further showed that CDK12-deficient primary PCa in the TCGA cohort had higher AR activity than control samples (Fig. 4C), indicating the aberrant activation of the AR signaling in CDK12-deficient samples. It is worth noting that although the impact on IPA usage and AR score can be detected in CDK12-deficient tumors, the number of those cases (12 out of 499 cases, 2.4%) is very small in the TCGA cohort. This observation is consistent with the report that AR-V7 expression in primary PCa is minimal. Additionally, we found that CDK12 defects correlate with shorter progression-free survival of these patients (Fig. 4D). Most importantly, we demonstrated that CDK12 defects also correlate with inferior overall survival of mCRPC patients in both SU2C and West Coast Dream Team (WCDT) datasets (Fig. 4 E and F), consistent with a previous report (32). CDK12-deficient patients also had higher AR scores compared with CDK12 WT patients in these two cohorts (Fig. 4 G and H). Together, these data stress that AR signaling is hyperactivated in PCa harboring CDK12 gene deletions and/or mutations.
Fig. 4.
CDK12 alteration associates with hyperactivation of the AR signaling pathway and IPA usage in the AR gene locus in patient samples. (A and B) Comparison of RNA-seq reads in the IPA sites in intron 3 of the CSTF3 (A) and AR (B) genes between CDK12 WT (n = 487) and deficient (n = 12) PCa patient samples in the TCGA cohort. RPKM, reads per kilobase of transcript, per million mapped reads. ***P < 0.001. (C) Comparison of AR score (Materials and Methods) between CDK12 WT (n = 487) and deficient (n = 12) PCa patient samples from the TCGA cohort. *P < 0.05. (D) Progression-free survival (PFS) analysis between CDK12 WT (n = 487) and deficient (n = 12) PCa patient samples from the TCGA cohort. (E and F) OS analysis between CDK12 WT and deficient patient samples of the SU2C (E; n = 310 versus 15) or WCDT cohort (F; n = 95 versus 3). (G and H) Comparison of AR score (Materials and Methods) between CDK12 WT and deficient patient samples of the SU2C (G; n = 310 versus 15) or WCDT cohort (H; n = 95 versus 3). *P < 0.05, **P < 0.01.
Androgen Depletion and Antiandrogen Treatment Represses CCNK Gene Expression.
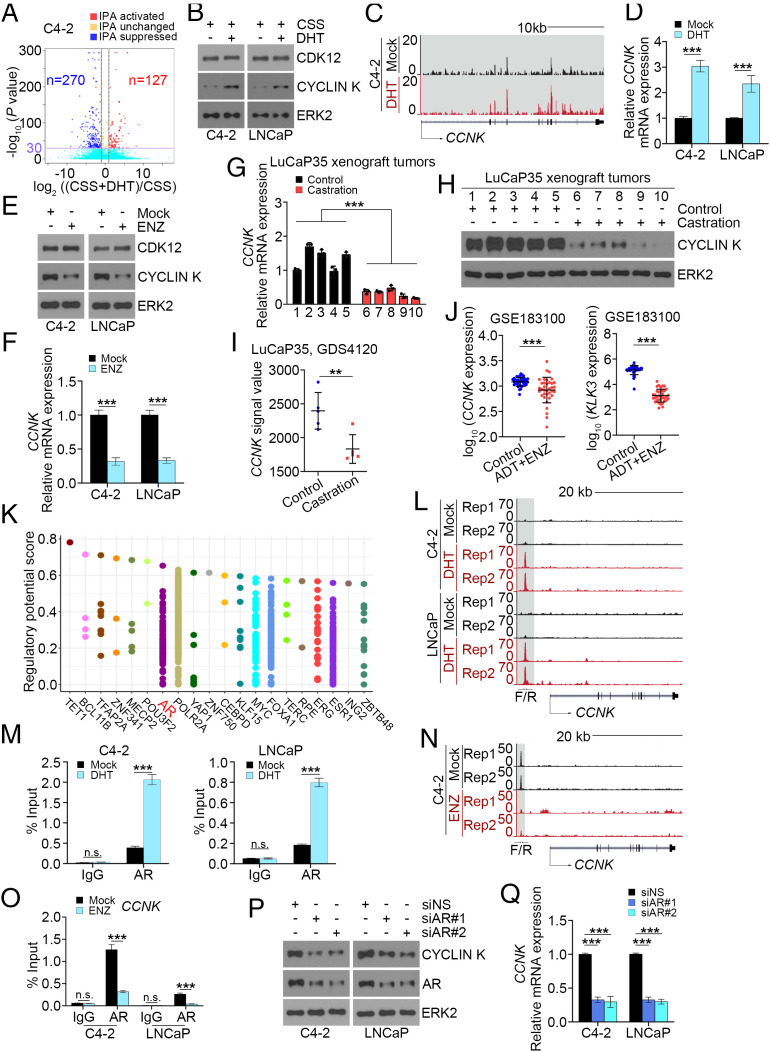
As mentioned above, ADT is the mainstay treatment of advanced/metastatic PCa. However, most progress to CRPC. It has been shown that castration induces increased expression of AR-Vs in both VCaP xenograft and LuCaP35 patient–derived xenograft (PDX) tumors in mice; however, this effect was completely reversed by testosterone replacement (39). We therefore examined whether androgen manipulation affects IPA usage globally and at the AR gene locus to induce AR-V expression. To this end, we employed APAlyzer software (40) to analyze RNA-seq data generated from C4-2 cells cultured in charcoal-stripped medium supplemented with or without dihydrotestosterone (DHT). We demonstrated that DHT treatment suppressed IPA peaks more than twofold versus activated peaks (Fig. 5A and SI Appendix, Fig. S4A), suggesting that DHT treatment induces net suppression of IPA usage. Because our data (Fig. 3) indicate that CDK12 acts as an upstream effector regulating AR-V genesis, we examined whether DHT treatment affects CDK12 expression. We showed that DHT had little or no effect on CDK12 protein expression in C4-2 cells (Fig. 5B). In contrast, we demonstrated that DHT treatment of C4-2 cells substantially increased the expression of CYCLIN K, the regulatory subunit of the CDK12–CYCLIN K kinase complex (Fig. 5B). Similar results were obtained at the mRNA level as revealed by RNA-seq data in C4-2 cells and qRT-PCR analysis in C4-2 and LNCaP cells (Fig. 5 C and D). Moreover, we showed that while ENZ treatment had no obvious effect on CDK12 expression, it substantially suppressed CYCLIN K expression at both protein and mRNA levels and increased AR-V7 expression (Fig. 5 E and F and SI Appendix, Fig. S4B). We also examined CCNK mRNA and CYCLIN K protein expression in LuCaP35 PDX tumors grown in mice with or without castration. We demonstrated that castration decreased expression of CCNK mRNA and CYCLIN K protein in LuCaP35 tumors (Fig. 5 G and H). These observations are consistent with microarray data in LuCaP35 PDX tumors (41) showing that the CCNK mRNA level was much lower in tumors in castrated mice compared with intact counterparts (Fig. 5I). Furthermore, we performed meta-analysis of RNA-seq data from 36 paired PCa patient samples before and after ADT and ENZ treatments. We observed that similar to the effect of KLK3 mRNA, ADT and ENZ treatment substantially decreased CCNK mRNA expression in these patient samples (Fig. 5J).
Fig. 5.
ADT promotes global IPA usage including AR IPA usage through repressing CCNK transcription. (A) Measurement of IPA usage in C4-2 cells cultured in CSS medium supplemented with or without DHT (10 nM) by analyzing RNA-seq data (GSM2432781 versus GSM2432783). (B) WB analysis of the indicated proteins in C4-2 and LNCaP cells cultured in CSS medium supplemented with or without DHT (10 nM) for 48 h. ERK2 is a loading control for WB. (C) UCSC screenshot of RNA-seq data in the CCNK locus in C4-2 cells cultured in CSS medium supplemented with vehicle (mock) or DHT (10 nM) for 24 h. (D) qRT-PCR analysis of CCNK mRNA expression in C4-2 and LNCaP cells cultured in CSS medium supplemented with vehicle (mock) or DHT (10 nM) for 48 h. Three biological replicates were analyzed. ***P < 0.001. (E and F) WB (E) and qRT-PCR analysis (F) in C4-2 and LNCaP cells cultured in regular medium treated with vehicle (mock) or ENZ (10 µM) for 72 h. Three biological replicates were analyzed. ***P < 0.001. (G and H) qRT-PCR (G) and WB (H) analysis of CYCLIN K in LuCaP35 xenograft tumors from male mice treated with sham castration (control) or castration for 1 wk. ***P < 0.001. (I) Analysis of microarray data (GDS4120) for CCNK mRNA expression in LuCaP35 xenograft tumors in mice with or without castration (control versus castration). **P < 0.01. (J) Analysis of RNA-seq for the expression of CCNK mRNA in paired PCa tissues from 36 patients before and after ADT and ENZ treatment. ***P < 0.001. (K) Meta-analysis of ChIP data of transcription factors using Cistrome software (http://dbtoolkit.cistrome.org) to define major possible transcription factors that could bind to the CCNK gene locus. (L) UCSC screenshot of AR ChIP-seq data in the CCNK gene locus in C4-2 and LNCaP cells cultured in CSS medium supplemented with vehicle or DHT (10 nM) for 24 h. (M) ChIP-qPCR analysis of AR occupancy in the CCNK gene promoter in C4-2 and LNCaP cells cultured in CSS medium supplemented with vehicle (mock) or DHT (10 nM) for 24 h. Three biological replicates were analyzed. ***P < 0.001; n.s., not significant. (N) UCSC screenshot of AR ChIP-seq data in the CCNK gene locus in C4-2 cells treated with vehicle (mock) or ENZ (10 µM) for 24 h. (O) ChIP-qPCR analysis of AR occupancy in the CCNK gene promoter in C4-2 and LNCaP cells treated with vehicle or ENZ (10 µM) for 24 h. Three biological replicates were analyzed. ***P < 0.001; n.s., not significant. (P and Q) WB (P) and qRT-PCR analysis (Q) in C4-2 and LNCaP cells transfected with control or AR-specific siRNAs for 72 h. Three biological replicates were analyzed. ***P < 0.001.
To determine the molecular mechanism underlying androgen regulation of CCNK expression, we performed meta-analysis of chromatin immunoprecipitation sequencing (ChIP-seq) data of transcription factors including those that are highly relevant in PCa such as AR, ERG, and FOXA1 and examined the enrichment of these factors in the CCNK gene locus. We demonstrated that AR is one of the top 10 transcript factors highly enriched in the CCNK locus (Fig. 5K), suggesting that AR might regulate CCNK gene expression by binding to this genomic region. Indeed, analysis of AR ChIP-seq data in the C4-2 and LNCaP cell lines (42) showed that DHT treatment largely increased AR occupancy in the proximity of the CCNK gene promoter (Fig. 5L). These results were further confirmed by AR ChIP-qPCR analysis (Fig. 5M). In contrast, ChIP-seq data showed that ENZ treatment inhibited AR occupancy at the CCNK gene locus in C4-2 cells, and these results were further confirmed by ChIP-qPCR analysis in both the C4-2 and LNCaP cell lines (Fig. 5 N and O). In agreement with these observations, we showed that AR knockdown by two independent siRNAs decreased CYCLIN K expression at both mRNA and protein levels in the C4-2 and LNCaP cell lines (Fig. 5 P and Q). We further showed that AR mRNA level was positively correlated with CCNK mRNA expression in PCa specimens in the TCGA cohort (SI Appendix, Fig. S4C). Together, these data indicate that CCNK expression is positively regulated by androgens but negatively regulated by androgen deprivation and antiandrogens in PCa cells in culture, PDX tumors, and patient samples.
Antiandrogen Induces Synthetic Lethality in PCa Cells upon PARP Inhibition.
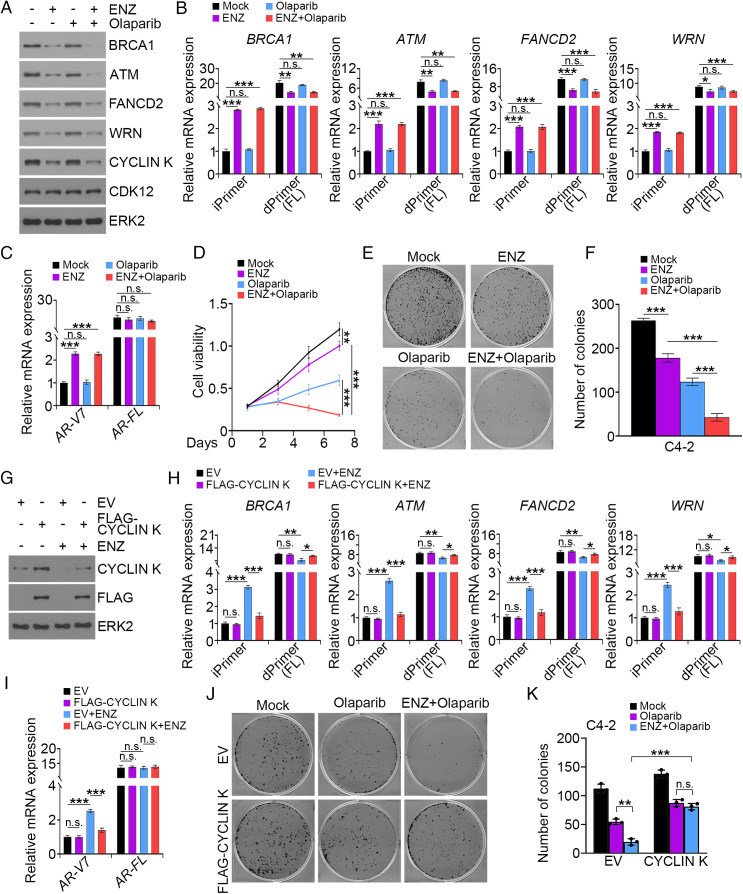
It has been shown recently that CDK12 inactivation induces IPA usage in the HR repair genes and down-regulation of the full-length forms of these genes (a condition mimicking BRCAness) and increases genomic instability (30). We therefore hypothesized that transcriptional suppression of CCNK by antiandrogen treatment induces IPA usage and down-regulation of HR repair genes and thereby promotes synthetic lethality in PCa cells upon PARP inhibition. To test this hypothesis, we treated C4-2 cells with ENZ and the PARP inhibitor olaparib alone or together. As expected, ENZ treatment decreased expression of HR regulators such as BRCA1, ATM, FANCD2, and WRN at both mRNA and protein levels (Fig. 6 A and B). We further confirmed that ENZ treatment induced IPA usage in introns of AR and HR genes (Fig. 6 B and C). Importantly, MTS assays showed that cotreatment of C4-2 cells with ENZ and olaparib resulted in much greater cell-growth inhibition compared with each agent treatment alone (Fig. 6D). These results were further confirmed by colony-formation assays (Fig. 6 E and F). Importantly, rescue experiments showed that forced expression of CYCLIN K not only abolished ENZ-induced IPA usage in both AR and HR repair gene loci but also prevented ENZ-mediated sensitization of PCa cells to olaparib (Fig. 6 G–K). Collectively, these data indicate that antiandrogen induces synthetic lethality in PCa cells upon PARP inhibition and that this process is mediated by down-regulation of CYCLIN K.
Fig. 6.
PARP inhibitor induces synthetic lethality in PCa cells treated with ENZ. (A–C) WB (A) and qRT-PCR analysis (B and C) in C4-2 cells treated with the indicated drugs for 3 d. ERK2 is a loading control for WB. Three biological replicates were analyzed. *P < 0.05, **P < 0.01, ***P < 0.001; n.s., not significant. (D) MTS assay in C4-2 cells treated with the indicated drugs. Three biological replicates were analyzed. **P < 0.01, ***P < 0.001. (E and F) Colony-formation assay in C4-2 cells treated with the indicated drugs. At 11 d after treatment, colonies were photographed (E) and quantified (F). Three biological replicates were analyzed. ***P < 0.001. (G–I) WB (G) and qRT-PCR analysis (H and I) in C4-2 cells transfected with the indicated plasmids and treated with vehicle or ENZ. iPrimer and dPrimer represent the pair of primers for amplification of transcripts generated by intronic PAS and distal PAS usage, respectively. Three biological replicates were analyzed. *P < 0.05, **P < 0.01, ***P < 0.001; n.s., not significant. (J and K) Colony-formation assay in C4-2 cells transfected with the indicated plasmids and treated with vehicle or indicated drugs. At 11 d after treatment, colonies were photographed (J) and quantified (K). EV, empty vector. Three biological replicates were analyzed. **P < 0.01, ***P < 0.001; n.s., not significant.
Discussion
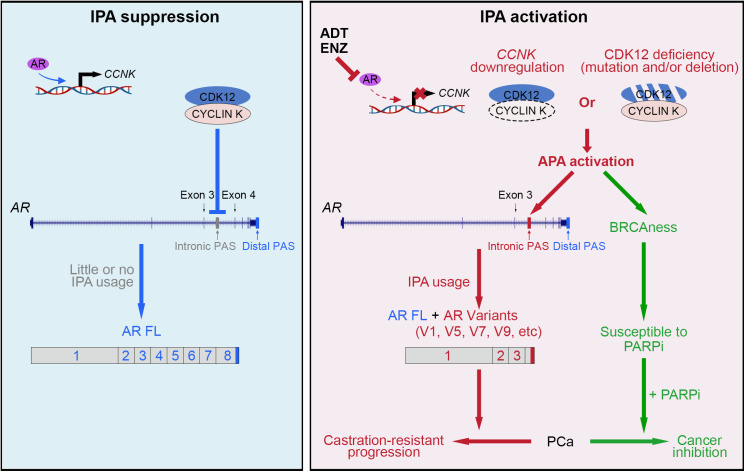
PCa often becomes therapy-resistant after ADT or antiandrogen treatment. Increasing evidence suggests that expression of AR-Vs is one of the critical mechanisms that confer hormone therapy resistance in advanced PCa (43). Importantly, it has been shown previously that androgen deprivation induces AR-V expression in human PCa xenografts in mice although the underlying mechanism remains to be elucidated (39). Additionally, APA has been implicated in the genesis of AR-Vs in PCa cells although the underlying mechanisms are largely unclear (44). In the present study, we identify defects in the CYCLIN K–CDK12 complex including CDK12 deletion and/or mutations and CYCLIN K transcriptional down-regulation as a mechanism that drives AR gene intron 3 IPA usage, thereby leading to AR-V production and hormonal therapy resistance in CRPC (Fig. 7, Right).
Fig. 7.
Hypothetical working model. (Left) Activation of the AR by androgens leads to transcriptional up-regulation of CCNK mRNA, which in turn results in the activation of the CYCLIN K–CDK12 complex and subsequent suppression of IPA usage in the AR gene locus and inhibition of AR variant expression. (Right) Inactivation of the CYCLIN K–CDK12 complex due to deletion or mutation of the CDK12 gene or down-regulation of CCNK caused by ADT or ENZ treatment leads to APA activation in the AR and HR repair gene loci (BRCAness), which promotes IPA usage in the AR locus and AR variant expression, thereby contributing to endocrine therapy resistance and castration-resistant progression of PCa. Most importantly, CYCLIN K–CDK12 inactivation also makes cancer cells susceptible to the PARP inhibitor.
We provide evidence showing that pharmacological inhibition of CDK12 promotes IPA usage at the AR gene intron 3 region, increases expression of AR-Vs, and augments AR activity. We further show that the CDK12 gene harbors a G909R missense mutation in the kinase domain in 22Rv1 cells and a similar charge-substitution mutation, G909E, in PCa patients. Most importantly, both mutants produce loss of function that promotes APA in AR intron 3 and expression of AR-Vs and AR target genes. Thus, using both pharmacological and genetic approaches, we convincingly show that inactivation of CDK12 promotes IPA usage in the AR gene locus and subsequently AR-V expression. Our finding that CDK12 loss increases AR-V expression also provides a mechanistic explanation for a previous report showing that CDK12-mutated PCa patients exhibit shorter time to PSA progression, development of CRPC, and metastasis compared with CDK12-WT patients (32). Notably, CDK12 appears to be a viable therapeutic target in triple-negative breast cancer (45), hepatocellular carcinoma (46), and osteosarcoma (47); however, because of their roles in induction of AR-V expression, CDK12 inhibitors might be used in a context-dependent manner for the treatment of PCa, especially those that are AR-positive.
It has been shown previously that AR-V expression increases acutely in response to androgen withdrawal in mice and that this process can be reversed by administration of testosterone, although the underlying mechanism remains to be determined (39, 48). We show that DHT treatment induces AR occupancy in a region close to the CCNK gene promoter and that AR occupancy can be completely inhibited by ENZ. Accordingly, we show that DHT induces and ENZ inhibits CCNK expression. Most importantly, we provide evidence showing that pharmacological inhibition of CYCLIN K induces IPA usage in the AR gene locus. Thus, similar to the effect of CDK12, a functional CYCLIN K is equivalently critical for the prevention of aberrant IPA usage; however, ADT or antiandrogen treatment abolishes this “safeguard” mechanism, thereby inducing AR-V expression and castration-resistant progression of PCa through aberrant IPA usage (Fig. 7, Right).
Others have observed that inactivation of CDK12 induces down-regulation of full-length HR genes including BRCA1 (30), implying that pharmacological inhibition of CDK12 may induce BRCAness and sensitize cancer cells to PARP inhibition. This notion is supported by the finding from a clinical trial showing the median overall survival (OS) (95% CI) for patients with CDK12-alteration CRPC tumors was 14.1 mo with olaparib but only 11.5 mo with placebo (49). Moreover, it has been shown previously that ENZ can decrease HR genes and sensitize to the PARP inhibitor although the underlying mechanism remains largely unclear (50–52). Similar to the effect of CDK12 inhibitors, our data convincingly show that ENZ also induces the BRCAness phenomenon by inactivating the CYCLIN K–CDK12 complex and this effect is mediated through ENZ-induced down-regulation of CYCLIN K. Most importantly, our data support the notion that while ADT induces aberrant IPA usage and AR-V expression, it also creates a synthetic lethal therapeutic scenario to use the PARP inhibitor to inhibit ENZ-resistant growth of CRPC (Fig. 7, Right). Thus, our finding provides a mechanistic explanation as to how second-generation AR pathway inhibitors such as abiraterone or ENZ in combination with the PARP inhibitor could improve the clinical outcome of CRPC patients (53, 54).
In summary, we identify the CYCLIN K–CDK12 complex as a key regulator of IPA usage in the AR gene locus (Fig. 7, Left). We further show that genetic alterations in the CDK12 gene, ADT, or antiandrogen treatment induce IPA usage in the AR gene locus and AR-V expression, thereby representing an intrinsic mechanism driving AR pathway inhibitor resistance during progression of CRPC (Fig. 7, Right). Our findings also suggest that defects in the CYCLIN K–CDK12 complex due to gene mutation or hormonal therapy also induce a BRCAness state, which generates a vulnerability for the effective treatment of hormonal therapy–resistant CRPC by the combination of ADT or ENZ with the PARP inhibitor (Fig. 7, Right).
Materials and Methods
Cell Culture and Chemicals.
LNCaP, 22Rv1, RWPE1, DU145, VCaP, PC-3, LAPC4, and 293T cell lines were purchased from the American Type Culture Collection (ATCC). The C4-2 cell line was purchased from Uro. The C4-2B cell line was obtained from ViroMed Laboratories. BPH1 cells were provided by S. Hayward from Northshore Research Institute, Chicago. LNCaP, C4-2, C4-2B, PC-3, DU145, BPH1, and 22Rv1 cell lines were cultured in RPMI 1640 cell-culture medium (Corning) containing 10% fetal bovine serum (FBS), together with 100 µg/mL streptomycin and 100 U/mL penicillin. VCaP and 293T cells were cultured in Dulbecco’s modified Eagle’s cell-culture medium (Corning) containing 10% FBS, together with 100 µg/mL streptomycin and 100 U/mL penicillin. RWPE1 cells were maintained in keratinocyte serum-free medium. LAPC4 cells were maintained in Iscove’s modified Dulbecco’s medium (Life Technologies) containing 10% FBS, together with 100 µg/mL streptomycin and 100 U/mL penicillin. LNCaP-RF cells were established by long-term culture of LNCaP cells (approximately >10 wk) in RPMI 1640 containing 10% charcoal-stripped serum (CSS). All cell lines were authenticated by short tandem repeat (STR) DNA profiling and cultured in an incubator at 37 °C with 5% CO2. Details of chemicals used in this study are provided in SI Appendix, Table S1.
Stable Cell-Line Generation.
The lentivirus transduction system was utilized to generate stable cell lines with specific gene knockdown and knockout as described previously (55). Polyethylenimine was used to transfect shRNA or single-guide RNA (sgRNA) plasmids together with lentivirus package plasmids (PSPAX2 and PMD2.G) into 293T cells. Forty-eight hours after transfection, supernatant containing viruses was collected, filtered, and utilized to infect the indicated cells. Polybrene (8 µg/mL) was added to the viral supernatant to increase the infection efficiency. Forty-eight hours after infection, culture medium was replaced with fresh medium, and puromycin (1 µg/mL) was administered for cell selection. Sequence information for shRNA and sgRNA is provided in SI Appendix, Table S2.
MTS Cell-Proliferation Assay.
Cell proliferation was measured utilizing the MTS assay (Promega) according to the manufacturer’s instructions (55). Briefly, cells were seeded in a 96-well plate with a density of 2,000 cells per well. At the indicated time points, 10 µL CellTiter 96R Aqueous One Solution reagent (Promega) was added to the cells. After incubation in a 37 °C incubator for 3 h, cell growth was measured on a microplate reader with absorbance at 490 nm.
SRB Assay.
Cells were resuspended in fresh culture medium, seeded in triplicate with a density of 2,000 cells per well in 96-well plates, and cultured with the indicated drugs for 1 wk. Colonies were fixed by adding 10% trichloroacetic acid (TCA) buffer for 1 h. The colonies were further stained by 0.4% (weight/volume) sulforhodamine B (SRB) solution in 1% acetic acid for 5 min, and then washed with 1% acetic acid twice to remove the SRB. Then, Tris base (pH 10.5) was added to solubilize the bound dye.
Colony-Formation Assay.
Cells were resuspended in fresh culture medium, seeded in triplicate with a density of 2,000 cells per well in 6-well plates, and cultured with the indicated drugs for various numbers of days depending on the cell lines used. Colonies were washed three times in phosphate-buffered saline (PBS) and fixed with 4% paraformaldehyde for 0.5 h. Colonies were further stained by 0.5% crystal violet for 2 h, and then washed with water twice to remove the crystal violet. The number of colonies in each well was counted.
Reverse Transcription and Real-Time PCR (qRT-PCR).
Total RNA was extracted from the cells by utilizing TRIzol reagent (Ambion) and reverse-transcribed into complementary DNA by utilizing the GoScript Kit (Promega). SYBR Green Master Mix (Bio-Rad) and the CFX96 Real-Time System (Bio-Rad) were utilized to conduct the real-time PCR according to the manufacturer’s instructions. Expression of the β-actin housekeeping gene was used as an inner control, and data are presented as mean ± SD. Sequence information for primers used for qPCR is provided in SI Appendix, Table S3.
Antibodies and Immunoblotting.
For immunoblotting analysis, cells were lysed in modified RIPA buffer (50 mM Tris⋅HCl, pH 7.4, 1% Nonidet P-40, 0.25% sodium deoxycholate, 150 mM NaCl, 0.1% sodium dodecyl sulfate [SDS], and 1 mM ethylenediaminetetraacetate) supplemented with 1% protease inhibitor mixture. Protein concentration was determined using DC protein assay reagent (Bio-Rad). Cell extracts were supplemented with 10% dithiothreitol (Thermo Fisher Scientific) and boiled at 95 °C for 3 min. Samples were subjected to SDS-polyacrylamide gel (Bio-Rad) separation, and the gels were further transferred to nitrocellulose (NC) membranes (Thermo Fisher Scientific). After transfer, the NC membranes were blocked in 5% nonfat milk (Bio-Rad) for 1 h at room temperature and incubated with the indicated primary antibodies at 4 °C overnight. The next day, the NC membranes were washed with 1× TBST for 10 min three times and incubated with matched secondary antibody for 1 h at room temperature. The membranes were washed with 1× TBST for 10 min three times. Lastly, the signals were developed with SuperSignal West Pico Luminal Enhancer Solution (Thermo Fisher Scientific) on autoradiography films (HyBlot). Detailed information for primary antibodies is provided in SI Appendix, Table S4.
ChIP-qPCR.
ChIP experiments were performed as described previously (56). In brief, chromatin was cross-linked for 10 min at room temperature with 11% formaldehyde/PBS solution added to the cell-culture medium. Cross-linked chromatin was then sonicated, diluted, and immunoprecipitated with Protein G Plus agarose beads (Bio-Rad) prebound with antibody at 4 °C overnight. Precipitated protein–DNA complexes were eluted, and cross-linking was reversed at 65 °C for 16 h. DNA fragments were purified and analyzed by qPCR. ChIP-qPCR data were analyzed as % input after normalizing each ChIP DNA fraction’s Ct value to the input DNA fraction’s Ct value.
AR Activity Score.
AR activity score was calculated based on the 20 AR target genes as described previously (57) including ABCC4, ACSL3, ADAM7, C1ORF116, CENPN, EAF2, ELL2, FKBP5, GNMT, HERC3, KLK2, KLK3, MAF, MED28, MPHOSPH9, NKX3.1, NNMT, PMEPA1, PTGER4, and ZBTB10. In brief, gene expression values [log2 (fragments per kilo base per million mapped reads (FPKM))] of each sample were converted to a Z score by Z = (x − µ)/σ, where µ is the average log2 (FPKM) across all samples of a gene and σ is the standard derivation (SD) of the log2 (FPKM). The Z scores were then summed across all genes for each sample.
Quantification and Statistical Analysis.
Analysis of RNA-seq for the expression of CCNK mRNA in paired PCa tissues from 36 patients was done before and after ADT and ENZ treatment. Data from RNA-seq (Gene Expression Omnibus accession no. GSE183100) were downloaded and used for analysis in this study.
GraphPad Prism 7 was used for statistical analyses of results from qRT-PCR, ChIP-qPCR, and cell-proliferation assays. The Student’s t test was used to compare data between two groups. Survival curves were obtained using the Kaplan–Meier method, and the log-rank test was used to test the difference in survival curves. P value < 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
We thank Jacob J. Orme, MD, PhD for critical reading of the manuscript. This work was supported in part by the NIH (R01 CA130908, R01 CA203849, and R01 CA271486 to H.H.) and the Mayo Clinic Foundation (H.H.).
Footnotes
Competing interest statement: R.S. and H.H. are listed as inventors on a provisional patent application filed by Mayo Clinic based on the findings in this report. All other authors declare no competing interests.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2205509119/-/DCSupplemental.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Dehm S. M., Tindall D. J., Alternatively spliced androgen receptor variants. Endocr. Relat. Cancer 18, R183–R196 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.He Y., et al. , Androgen receptor splice variants bind to constitutively open chromatin and promote abiraterone-resistant growth of prostate cancer. Nucleic Acids Res. 46, 1895–1911 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Antonarakis E. S., et al. , AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N. Engl. J. Med. 371, 1028–1038 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scher H. I., et al. , Association of AR-V7 on circulating tumor cells as a treatment-specific biomarker with outcomes and survival in castration-resistant prostate cancer. JAMA Oncol. 2, 1441–1449 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Proudfoot N. J., Transcriptional termination in mammals: Stopping the RNA polymerase II juggernaut. Science 352, aad9926 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shi Y., Manley J. L., The end of the message: Multiple protein-RNA interactions define the mRNA polyadenylation site. Genes Dev. 29, 889–897 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoque M., et al. , Analysis of alternative cleavage and polyadenylation by 3′ region extraction and deep sequencing. Nat. Methods 10, 133–139 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tian B., Pan Z., Lee J. Y., Widespread mRNA polyadenylation events in introns indicate dynamic interplay between polyadenylation and splicing. Genome Res. 17, 156–165 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Singh I., et al. , Widespread intronic polyadenylation diversifies immune cell transcriptomes. Nat. Commun. 9, 1716 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Derti A., et al. , A quantitative atlas of polyadenylation in five mammals. Genome Res. 22, 1173–1183 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lianoglou S., Garg V., Yang J. L., Leslie C. S., Mayr C., Ubiquitously transcribed genes use alternative polyadenylation to achieve tissue-specific expression. Genes Dev. 27, 2380–2396 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang E. T., et al. , Alternative isoform regulation in human tissue transcriptomes. Nature 456, 470–476 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang H., Lee J. Y., Tian B., Biased alternative polyadenylation in human tissues. Genome Biol. 6, R100 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sandberg R., Neilson J. R., Sarma A., Sharp P. A., Burge C. B., Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science 320, 1643–1647 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ji Z., Tian B., Reprogramming of 3′ untranslated regions of mRNAs by alternative polyadenylation in generation of pluripotent stem cells from different cell types. PLoS One 4, e8419 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shepard P. J., et al. , Complex and dynamic landscape of RNA polyadenylation revealed by PAS-seq. RNA 17, 761–772 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ji Z., Lee J. Y., Pan Z., Jiang B., Tian B., Progressive lengthening of 3′ untranslated regions of mRNAs by alternative polyadenylation during mouse embryonic development. Proc. Natl. Acad. Sci. U.S.A. 106, 7028–7033 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Masamha C. P., et al. , CFIm25 links alternative polyadenylation to glioblastoma tumour suppression. Nature 510, 412–416 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fu Y., et al. , Differential genome-wide profiling of tandem 3′ UTRs among human breast cancer and normal cells by high-throughput sequencing. Genome Res. 21, 741–747 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kunisky A. K., Anyaeche V. I., Herron R. S., Park C. Y., Hwang H. W., Shift in MSL1 alternative polyadenylation in response to DNA damage protects cancer cells from chemotherapeutic agent-induced apoptosis. Cell Rep. 37, 109815 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang J., et al. , A PolH transcript with a short 3′UTR enhances PolH expression and mediates cisplatin resistance. Cancer Res. 79, 3714–3724 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chou J., Quigley D. A., Robinson T. M., Feng F. Y., Ashworth A., Transcription-associated cyclin-dependent kinases as targets and biomarkers for cancer therapy. Cancer Discov. 10, 351–370 (2020). [DOI] [PubMed] [Google Scholar]
- 23.Quigley D. A., et al. , Genomic hallmarks and structural variation in metastatic prostate cancer. Cell 174, 758–769.e9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Viswanathan S. R., et al. ; PCF/SU2C International Prostate Cancer Dream Team, Structural alterations driving castration-resistant prostate cancer revealed by linked-read genome sequencing. Cell 174, 433–447.e19 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu Y. M., et al. , Inactivation of CDK12 delineates a distinct immunogenic class of advanced prostate cancer. Cell 173, 1770–1782.e14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bartkowiak B., et al. , CDK12 is a transcription elongation-associated CTD kinase, the metazoan ortholog of yeast Ctk1. Genes Dev. 24, 2303–2316 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davidson L., Muniz L., West S., 3′ end formation of pre-mRNA and phosphorylation of Ser2 on the RNA polymerase II CTD are reciprocally coupled in human cells. Genes Dev. 28, 342–356 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ketley A., et al. , CDK12 inhibition reduces abnormalities in cells from patients with myotonic dystrophy and in a mouse model. Sci. Transl. Med. 12, eaaz2415 (2020). [DOI] [PubMed] [Google Scholar]
- 29.Blazek D., et al. , The cyclin K/Cdk12 complex maintains genomic stability via regulation of expression of DNA damage response genes. Genes Dev. 25, 2158–2172 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dubbury S. J., Boutz P. L., Sharp P. A., CDK12 regulates DNA repair genes by suppressing intronic polyadenylation. Nature 564, 141–145 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krajewska M., et al. , CDK12 loss in cancer cells affects DNA damage response genes through premature cleavage and polyadenylation. Nat. Commun. 10, 1757 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reimers M. A., et al. , Clinical outcomes in cyclin-dependent kinase 12 mutant advanced prostate cancer. Eur. Urol. 77, 333–341 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tran C., et al. , Development of a second-generation antiandrogen for treatment of advanced prostate cancer. Science 324, 787–790 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Orme J. J., Pagliaro L. C., Quevedo J. F., Park S. S., Costello B. A., Rational second-generation antiandrogen use in prostate cancer. Oncologist 27, 110–124 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Choi S. H., et al. , CDK12 phosphorylates 4E-BP1 to enable mTORC1-dependent translation and mitotic genome stability. Genes Dev. 33, 418–435 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Słabicki M., et al. , The CDK inhibitor CR8 acts as a molecular glue degrader that depletes cyclin K. Nature 585, 293–297 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kohli M., et al. , Androgen receptor variant AR-V9 is coexpressed with AR-V7 in prostate cancer metastases and predicts abiraterone resistance. Clin. Cancer Res. 23, 4704–4715 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luo W., et al. , The conserved intronic cleavage and polyadenylation site of CstF-77 gene imparts control of 3′ end processing activity through feedback autoregulation and by U1 snRNP. PLoS Genet. 9, e1003613 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Watson P. A., et al. , Constitutively active androgen receptor splice variants expressed in castration-resistant prostate cancer require full-length androgen receptor. Proc. Natl. Acad. Sci. U.S.A. 107, 16759–16765 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang R., Tian B., APAlyzer: A bioinformatics package for analysis of alternative polyadenylation isoforms. Bioinformatics 36, 3907–3909 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun Y., et al. , Androgen deprivation causes epithelial-mesenchymal transition in the prostate: Implications for androgen-deprivation therapy. Cancer Res. 72, 527–536 (2012). [DOI] [PubMed] [Google Scholar]
- 42.Zhao Y., et al. , Activation of P-TEFb by androgen receptor-regulated enhancer RNAs in castration-resistant prostate cancer. Cell Rep. 15, 599–610 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Watson P. A., Arora V. K., Sawyers C. L., Emerging mechanisms of resistance to androgen receptor inhibitors in prostate cancer. Nat. Rev. Cancer 15, 701–711 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Van Etten J. L., et al. , Targeting a single alternative polyadenylation site coordinately blocks expression of androgen receptor mRNA splice variants in prostate cancer. Cancer Res. 77, 5228–5235 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Quereda V., et al. , Therapeutic targeting of CDK12/CDK13 in triple-negative breast cancer. Cancer Cell 36, 545–558.e7 (2019). [DOI] [PubMed] [Google Scholar]
- 46.Wang C., et al. , CDK12 inhibition mediates DNA damage and is synergistic with sorafenib treatment in hepatocellular carcinoma. Gut 69, 727–736 (2020). [DOI] [PubMed] [Google Scholar]
- 47.Bayles I., et al. , Ex vivo screen identifies CDK12 as a metastatic vulnerability in osteosarcoma. J. Clin. Invest. 129, 4377–4392 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sharp A., et al. , Androgen receptor splice variant-7 expression emerges with castration resistance in prostate cancer. J. Clin. Invest. 129, 192–208 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hussain M., et al. ; PROfound Trial Investigators, Survival with olaparib in metastatic castration-resistant prostate cancer. N. Engl. J. Med. 383, 2345–2357 (2020). [DOI] [PubMed] [Google Scholar]
- 50.Li L., et al. , Androgen receptor inhibitor-induced “BRCAness” and PARP inhibition are synthetically lethal for castration-resistant prostate cancer. Sci. Signal. 10, eaam7479 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Asim M., et al. , Synthetic lethality between androgen receptor signalling and the PARP pathway in prostate cancer. Nat. Commun. 8, 374 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Polkinghorn W. R., et al. , Androgen receptor signaling regulates DNA repair in prostate cancers. Cancer Discov. 3, 1245–1253 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Clarke N., et al. , Olaparib combined with abiraterone in patients with metastatic castration-resistant prostate cancer: A randomised, double-blind, placebo-controlled, phase 2 trial. Lancet Oncol. 19, 975–986 (2018). [DOI] [PubMed] [Google Scholar]
- 54.Thiery-Vuillemin A., et al. , Pain and health-related quality of life with olaparib versus physician’s choice of next-generation hormonal drug in patients with metastatic castration-resistant prostate cancer with homologous recombination repair gene alterations (PROfound): An open-label, randomised, phase 3 trial. Lancet Oncol. 23, 393–405 (2022). [DOI] [PubMed] [Google Scholar]
- 55.Hong Z., et al. , DNA damage promotes TMPRSS2-ERG oncoprotein destruction and prostate cancer suppression via signaling converged by GSK3β and WEE1. Mol. Cell 79, 1008–1023.e4 (2020). [DOI] [PubMed] [Google Scholar]
- 56.Zhang P., et al. , Intrinsic BET inhibitor resistance in SPOP-mutated prostate cancer is mediated by BET protein stabilization and AKT-mTORC1 activation. Nat. Med. 23, 1055–1062 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hieronymus H., et al. , Gene expression signature-based chemical genomic prediction identifies a novel class of HSP90 pathway modulators. Cancer Cell 10, 321–330 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.