Abstract
The extent of parallel evolution at the genotypic level is quantitatively linked to the distribution of beneficial fitness effects (DBFE) of mutations. The standard view, based on light-tailed distributions (i.e., distributions with finite moments), is that the probability of parallel evolution in duplicate populations is inversely proportional to the number of available mutations and, moreover, that the DBFE is sufficient to determine the probability when the number of available mutations is large. Here, we show that when the DBFE is heavy-tailed, as found in several recent experiments, these expectations are defied. The probability of parallel evolution decays anomalously slowly in the number of mutations or even becomes independent of it, implying higher repeatability of evolution. At the same time, the probability of parallel evolution is non-self-averaging—that is, it does not converge to its mean value, even when a large number of mutations are involved. This behavior arises because the evolutionary process is dominated by only a few mutations of high weight. Consequently, the probability varies widely across systems with the same DBFE. Contrary to the standard view, the DBFE is no longer sufficient to determine the extent of parallel evolution, making it much less predictable. We illustrate these ideas theoretically and through analysis of empirical data on antibiotic-resistance evolution.
Keywords: parallel evolution, distribution of fitness effects, predictability of evolution, antibiotic resistance
The repeatability and predictability of evolution are important questions in the field of evolutionary biology. In 1990, Stephen Jay Gould famously mused about “replaying life’s tape” (1). In subsequent years, the topic of parallel evolution has become a major subject of empirical research (2–4), and theoretical questions concerning the probability of parallel evolution within the mathematical theory of population genetics have also attracted substantial attention (5–7). Here, the questions are focused mostly on changes at the level of genetic sequences. According to a common definition (6, 7), parallel evolution is said to occur when the exact same mutation is substituted in replicate populations. It is in this strong sense that we shall use the term “parallel evolution” here. The computation of the probability of parallel evolution is often set in a simplified scenario (5–7), where an asexual population evolves by strong selection and weak mutation (SSWM), which applies for moderately large population size and low mutation rate (more details are in SI Appendix). The evolutionary process starts with a homogeneous population and n possible beneficial mutations that can occur. Denoting by ri the substitution rate of the i-th mutation, , the probability that the i-th mutation will be the first to fix is . Therefore, the probability that k replicate populations will all fix the same mutation is given by . Although we focus on the repeatability of the first substitution event, this measure can be generalized to address evolutionary trajectories with several mutations (see ref. 2 and further discussion in SI Appendix).
Within the SSWM approximation, the substitution rate of a mutation is the product of its mutation rate and fixation probability, , and the repeatability measure is affected equally by heterogeneities in μi and πi (4). However, because empirical information on mutation-rate heterogeneity is sparse, theoretical studies have commonly assumed that all mutations occur at the same rate. If, in addition, the selection coefficients are small, , it follows that , and the probability of parallel evolution is given by
| [1] |
We will adopt this simplification throughout, but emphasize that our theoretical results hold equally well for the full expression of Pk with ri in place of si, provided that the distribution of the substitution rates ri is heavy-tailed in the sense specified below.
The selection coefficients follow a distribution denoted by , which we refer to as the distribution of beneficial fitness effects (DBFE) (8). The mean probability of parallel evolution is , where denotes the average with respect to the DBFE. The extreme-value theory (EVT) hypothesis of fitness effects (see SI Appendix for details) predicts that the DBFE belongs to one of three classes of distributions: The Weibull and Gumbel classes contain distributions with finite moments, whereas the Fréchet class contains distributions with power-law tails (and therefore diverging moments). In the last case, the asymptotic form of the DBFE is
| [2] |
with a scale parameter and the tail exponent . The cases of the Gumbel and Weibull extreme-value distributions have been explored in some detail for k = 2 (5–7). The Fréchet EVT class had been conjectured to be relatively unimportant biologically (7), but several subsequent studies (9–11) have uncovered signatures of heavy-tailed distributions of fitness effects (see SI Appendix for further details on tails of empirical DBFEs). In the realistic case of a large number of available beneficial mutations n, the statistics of Pk for heavy-tailed distributions of the form Eq. 2 are markedly different from those of light-tailed distributions, as will be shown below.
Results
The number n of beneficial mutations that can occur in a population varies widely across organisms and environments, but it is likely to be large. For bacterial populations, one may conservatively estimate that there are several thousand beneficial mutations (SI Appendix). We therefore focus on Pk in the large-n regime. The simplest computation is in the limiting case of neutral variation, where all selection coefficients are identical. Since all mutations are equally likely to be the first to fix, (which is exact for all n). Specifically, the probability of parallel evolution in two replicates is . Using this observation, one can define, for any system, the quantity as the effective number of mutations that dominate the dynamics of fixation (further comments in SI Appendix). It is similar to the notion of the effective number of reproducing lineages studied in ref. 12 in the context of family size distributions.
The most commonly studied class of DBFEs is where all the moments are finite. In this case, the numerator and denominator in each term in Eq. 1 become uncorrelated as and the distribution of becomes sharply centered around the moment of . Therefore, for large n, we have
| [3] |
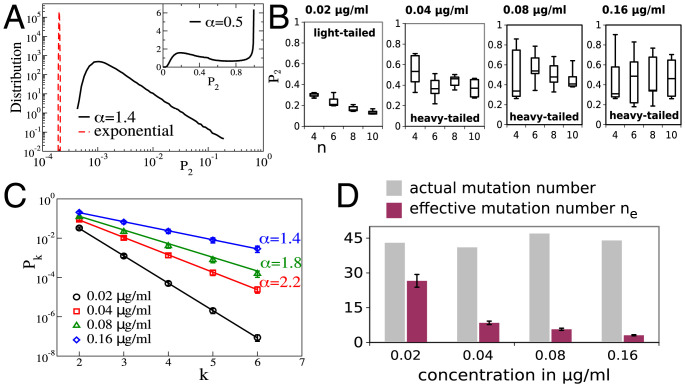
Notice that, so far, we have omitted the angular brackets around Pk since it converges to the mean value in the limit of large n, as shown by the highly localized distribution of Pk in Fig. 1A (red dashed curve). Such quantities are described as “self-averaging.” For the particular case k = 2, we have , which is the characteristic decay in self-averaging systems. It was shown in ref. 6 that, for an exponential distribution, (see SI Appendix for a general expression for ). Our focus here, however, is on heavy-tailed distributions with tails of the form Eq. 2. When , Eq. 3 continues to hold. Particularly for , P2 still decays as (see SI Appendix, Fig S1A). However, when , the moment diverges, and Eq. 3 no longer holds. We can now break the analysis down into two cases.
Fig. 1.
(A) The black curve is the numerically sampled distribution of P2 for , and Inset shows the same for ; we used 106 realizations and mutations. The dashed red curve is the distribution of P2 for an exponential distribution of selection coefficients and . (B) This and the following panels analyze data from the study based on mutant screening reported in ref. 9, which determined the selection coefficients for several resistance-conferring mutations in TEM-1 β-lactamase. Here, we numerically estimate the distribution of P2 from the selection coefficients reported in ref. 9. The dataset at each cefotaxime concentration was randomly split into subsets of size n in order to obtain distributions of P2 as a function of n. The box plots show median, quartiles, and extreme values. (C) The Pk were obtained from the entire dataset at each concentration, and Eq. 5 was used to infer α. (D) The effective mutation number has been computed and compared with the actual number of mutations in the available dataset at each concentration.
Case I.
The moderately heavy-tailed case occurs when ; in this case, is finite, but higher moments corresponding to diverge. For , the asymptotic behavior of is
| [4] |
where the constant . Note that decays with an exponent less than k – 1; therefore, the mean probability of parallel evolution is asymptotically much larger than in the case of light-tailed DBFEs. The scaling in Eq. 4 was first reported in ref. 9 and recently derived independently in ref. 12 in a different context. In particular, we see that when decays anomalously—i.e., with an exponent < 1—in contrast to , as in the light-tailed case (SI Appendix, Fig S1A).
It is important to point out that Pk does not become sharply centered around its mean value when , which can be shown as follows. The m-th moment is given by (SI Appendix). The value of can be read off from Eq. 4 by replacing k by km. Thus, all moments are of the same order . In particular, we notice that for . For self-averaging systems (which obey Eq. 3 for all k), this ratio goes asymptotically to one, and the SD vanishes relative to the mean (as seen in the red dashed curve in Fig. 1A). In contrast, here, we see that the SD diverges relative to the mean, implying a broad distribution for P2, as illustrated in Fig. 1A. This non-self-averaging effect arises because the sum is dominated by the largest weight (12, 13). According to EVT, the largest selection coefficient scales as , implying that . Therefore, the scale of typical Pk is
| [5] |
for , which is asymptotically smaller than , as given by Eq. 4. In fact, most of the weight is concentrated near the typical value, and the much higher mean is obtained from values of Pk that are much rarer, but have much higher magnitude.
Case II.
For the severely heavy-tailed case , all integer moments of s diverge. It was shown in ref. 13 that for a power-law distribution with ,
| [6] |
in the limit of large n. Specifically, the average probability of parallel evolution in two replicates is . Note that the asymptotic form in Eq. 6 is independent of n, and, thus, we have the striking result that the probability of parallel evolution remains finite, even in the limit of an infinite number of available alternative mutations. In the present case, all moments of P2 are of O(1), and, therefore, P2 is non-self-averaging. This is visible in the wide distribution of P2, as shown in the numerically sampled plot in Fig. 1 A, Inset. Similar non-self-averaging effects are familiar in the physics of disordered systems (ref. 13 and references therein) and in probability theory (14).
While the moderately () and severely () heavy-tailed cases display somewhat different behavior, we note that both Eqs. 4 and 6 give rise to the recursion relation
| [7] |
which, therefore, holds for the entire range . The result is independent of n and of all features of the underlying distribution, except the tail exponent α. It is, therefore, suitable for extracting α from empirical data; however, the disadvantage is that the averages require large datasets. Eq. 7 easily yields an approximate solution for large k, , shown in SI Appendix, Fig. S1B. The slow decay of with k contrasts with the exponential decay of the typical Pk, as given by Eq. 5.
The theoretical results discussed so far are valid in the limit of large n. Nonetheless, we will show that signatures of non-self-averaging effects can be discovered in limited empirical datasets. For this purpose, we use data on selection coefficients associated with antibiotic-resistance evolution reported in ref. 9. In this study, the fitness effects of 48 beneficial mutations in the resistance enzyme TEM-1 β-lactamase were reported for Escherichia coli growing at four different concentrations of the antibiotic cefotaxime (see SI Appendix for further details of the experiment). An analysis based on EVT indicated that the DBFE is light-tailed for the lowest concentration and heavy-tailed for the three higher concentrations, although large uncertainties were associated with the exponents in the latter case (9). We evaluate the statistics of P2 for the four different concentrations (see SI Appendix for further details). Fig. 1B shows P2 as a function of n. For the lowest concentration, P2 is seen to be small with a small dispersion, and it decreases with n, consistent with our expectation. For the three higher concentrations, the values of P2 are larger and have a large dispersion, which is consistent with heavy-tailed distributions. There is no discernible decrease with n. However, due to the relatively small values of n and the modest size of the datasets, it is not possible to distinguish this from a slow decrease with n. In Fig. 1C, we have plotted Pk as a function of k. Note that the distinction between the typical and mean values (12) has important implications here. Due to the limited size of the data, we have not used the recursion relation Eq. 7 to infer α. Instead, for each concentration, we have used the entire set of selection coefficients to create a single sample value of Pk, which is expected to be of the typical scale given by Eq. 5. Using this, we estimate the exponent α, which is seen to progressively decrease with increasing concentration, indicating an increasingly heavy-tailed distribution. Thus, stronger selection pressures amplify the differences between fitness effects of beneficial mutations, leading to a broader distribution. Nonetheless, we should mention that inferred power-law exponents should be treated with some caution, since these can be sensitive to experimental errors or methods of analysis (9, 11). What is clear from Fig. 1C, however, is that the behavior of Pk is at least a good qualitative indicator of the dispersion of selection coefficients. We have also computed and plotted the effective mutation number ne in Fig. 1D. The trend is, again, seen to be as predicted by theory. At the lowest concentration, ne is relatively large and close to (where n is the actual number of mutations), consistent with an exponential distribution of selection coefficients. The effective mutation number decreases progressively with increasing concentration and indicates a slower-than-exponential tail.
Conclusions
Parallel phenotypic evolution often proceeds through distinct genotypic pathways. Here, we have shown that heavy-tailed DBFEs can substantially enhance the probability of parallel evolution even at the genotypic level. However, we also find that this probability varies widely across large, independent samples generated from the same heavy-tailed DBFE. This makes it harder to generalize the degree of repeatability from one model system to other, closely related ones. On the flip side, the evolutionary process is dominated by a few mutations of high weight, making evolution of the system more repeatable and, therefore, more predictable. For example, in the study (15) on antibiotic-resistance evolution, the authors found that the mutation of highest effect (which also features in the heavy-tailed distributions reported in ref. 9 and discussed above) occurred in the majority of multiple-replicate experiments. The full implications of these ideas in the context of natural populations remain to be elucidated.
Supplementary Material
Acknowledgments
S.G.D. and J.K. acknowledge support by the Deutsche Forschungsgemeinschaft (German Research Foundation) within Collaborative Research Centre 1310 Predictability in Evolution. We thank Arjan de Visser for helpful comments on the manuscript.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2209373119/-/DCSupplemental.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix. The codes for numerical simulations are available on GitHub (https://github.com/meetsumand/Parallel-evolution) (16).
References
- 1.Gould S. J., Wonderful Life: The Burgess Shale and the Nature of History (WW Norton & Company, New York, 1990). [Google Scholar]
- 2.de Visser J. A. G. M., Krug J., Empirical fitness landscapes and the predictability of evolution. Nat. Rev. Genet. 15, 480–490 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Storz J. F., Causes of molecular convergence and parallelism in protein evolution. Nat. Rev. Genet. 17, 239–250 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bailey S. F., Blanquart F., Bataillon T., Kassen R., What drives parallel evolution? How population size and mutational variation contribute to repeated evolution. BioEssays 39, 1–9 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Gillespie J. H., A simple stochastic gene substitution model. Theor. Popul. Biol. 23, 202–215 (1983). [DOI] [PubMed] [Google Scholar]
- 6.Orr H. A., The probability of parallel evolution. Evolution 59, 216–220 (2005). [PubMed] [Google Scholar]
- 7.Joyce P., Rokyta D. R., Beisel C. J., Orr H. A., A general extreme value theory model for the adaptation of DNA sequences under strong selection and weak mutation. Genetics 180, 1627–1643 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bataillon T., Bailey S. F., Effects of new mutations on fitness: Insights from models and data. Ann. N. Y. Acad. Sci. 1320, 76–92 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schenk M. F., Szendro I. G., Krug J., de Visser J. A. G. M.. Quantifying the adaptive potential of an antibiotic resistance enzyme. PLoS Genet. 8, e1002783 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bank C., Hietpas R. T., Wong A., Bolon D. N., Jensen J. D., A Bayesian MCMC approach to assess the complete distribution of fitness effects of new mutations: Uncovering the potential for adaptive walks in challenging environments. Genetics 196, 841–852 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Foll M., et al., Influenza virus drug resistance: A time-sampled population genetics perspective. PLoS Genet. 10, e1004185 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Niwa H.-S., Reciprocal symmetry breaking in Pareto sampling. arXiv [Preprint] (2022). https://arxiv.org/abs/2202.04865 (Accessed 10 February 2022).
- 13.Derrida B., From random walks to spin glasses. Physica D 107, 186–198 (1997). [Google Scholar]
- 14.Pitman J., Yor M., The two-parameter Poisson-Dirichlet distribution derived from a stable subordinator. Ann. Probab. 25, 855–900 (1997). [Google Scholar]
- 15.van Dijk T., Hwang S., Krug J., de Visser J. A. G. M., Zwart M. P., Mutation supply and the repeatability of selection for antibiotic resistance. Phys. Biol. 14, 055005 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Das S. G., Codes to generate numerical data in “Unpredictable repeatability in molecular evolution.” GitHub. https://github.com/meetsumand/Parallel-evolution. Deposited 3 September 2022.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix. The codes for numerical simulations are available on GitHub (https://github.com/meetsumand/Parallel-evolution) (16).