Abstract
The heterofermentative lactic acid bacterium Lactobacillus brevis transports galactose and the nonmetabolizable galactose analogue thiomethyl-β-galactoside (TMG) by a permease-catalyzed sugar:H+ symport mechanism. Addition of glucose to L. brevis cells loaded with [14C]TMG promotes efflux and prevents accumulation of the galactoside, probably by converting the proton symporter into a uniporter. Such a process manifests itself physiologically in phenomena termed inducer expulsion and exclusion. Previous evidence suggested a direct allosteric mechanism whereby the phosphocarrier protein, HPr, phosphorylated at serine-46 [HPr(Ser-P)], binds to the galactose:H+ symporter to uncouple sugar transport from proton symport. To elucidate the molecular mechanism of inducer control in L. brevis, we have cloned the genes encoding the HPr(Ser) kinase, HPr, enzyme I, and the galactose:H+ symporter. The sequences of these genes were determined, and the relevant phylogenetic trees are presented. Mutant HPr derivatives in which the regulatory serine was changed to either alanine or aspartate were constructed. The cloned galP gene was integrated into the chromosome of Bacillus subtilis, and synthesis of the mutant HPr proteins in this organism was shown to promote regulation of GalP, as expected for a direct allosteric mechanism. We have thus reconstituted inducer control in an organism that does not otherwise exhibit this phenomenon. These results are consistent with the conclusion that inducer exclusion and expulsion in L. brevis operates via a multicomponent signal transduction mechanism wherein the presence of glycolytic intermediates such as fructose 1,6-bisphosphate (the intracellular effector), derived from exogenous glucose (the extracellular effector), activates HPr(Ser) kinase (the sensor) to phosphorylate HPr on Ser-46 (the messenger), which binds to the galactose:H+ symporter (the target), resulting in uncoupling of sugar transport from proton symport (the response). This cascade allows bacteria to quickly respond to changes in external sugar concentrations. Understanding the molecular mechanism of inducer control advances our knowledge of the link between metabolic and transport processes in bacteria.
The phosphoenolpyruvate: sugar phosphotransferase system (PTS) plays a major role in the regulation of carbon and energy metabolism in many diverse prokaryotes (26). However, different PTS proteins are involved, and they function by entirely different mechanisms in the various organisms (34). In gram-positive bacteria, glucose represses synthesis of PTS and non-PTS carbohydrate catabolic enzymes (catabolite repression), inhibits the uptake of PTS and non-PTS sugars (inducer exclusion), and stimulates dephosphorylation of intracellular sugar phosphates and/or elicits efflux of free sugars (inducer expulsion) (35, 40). A principal role of a metabolite-activated ATP-dependent protein kinase which phosphorylates seryl residue 46 (Ser-46) in HPr in the regulation of enzyme synthesis, inducer exclusion, and inducer expulsion via an allosteric mechanism that acts on several different target proteins has been suggested (4, 5, 30, 34, 36, 44).
Homofermentative lactic acid bacteria transport most sugars via the PTS. In contrast, heterofermentative lactobacilli such as Lactobacillus brevis transport glucose and galactose as well as their nonmetabolizable analogues 2-deoxyglucose and thiomethyl-β-galactoside (TMG), respectively, by active transport energized by the proton motive force (33). Previous biochemical studies have shown that L. brevis possesses a galactose:H+ symport permease, here designated GalP, which transports and accumulates cytoplasmic TMG. An ATP-dependent HPr(Ser) kinase was also identified, and evidence was presented suggesting that it participates in GalP regulation. A complex mechanism was proposed that led to metabolite-activated vectorial sugar exclusion and expulsion (30, 44). When provided with an exogenous energy source such as arginine, galactose-grown cells of L. brevis were shown to transport [14C]TMG and accumulate the free sugar analogue in the cytoplasm to a concentration at least 20-fold higher than that in the medium. However, addition of exogenous glucose, gluconate, or glucosamine to cells loaded with [14C]TMG resulted in efflux of the intracellular galactoside, giving rise to a reduced cytoplasmic concentration of the sugar. Counterflow experiments suggested that metabolism of glucose by intact L. brevis cells results in conversion of the active sugar:H+ transporter (symporter) into a facilitated diffusion system (uniporter [33]).
When right-side-out vesicles of L. brevis were loaded with free [14C]TMG and electroporated with purified HPr of Bacillus subtilis, rapid efflux of the galactoside was observed upon addition of glucose (44). Glucose could be replaced by any one of several glycolytic phosphorylated metabolites, such as fructose 1,6-bisphosphate, gluconate-6-phosphate, and 2-phosphoglycerate, when the compounds were electroporated into the vesicles. Allosteric activating effects of these compounds on the HPr(Ser) kinase in vitro were also observed (30). In the absence of glucose or phosphorylated metabolites, intravesicular HPr(S46D), a structural analogue of the seryl-phosphorylated form of HPr, promoted expulsion of accumulated free [14C]TMG. The noncharged HPr analogue HPr(S46A) did not promote expulsion of the accumulated galactoside, clearly implying a critical role for phosphorylation of HPr at Ser-46 by the HPr(Ser) kinase in inducer expulsion. In addition, uptake of lactose, glucose, mannose, and ribose was inhibited by wild-type HPr in the presence of fructose 1,6-bisphosphate or by HPr(S46D) alone, indicating that the metabolite-activated, ATP-dependent HPr(Ser) kinase regulates sugar permeases in L. brevis by a feedback-controlled process. Direct binding of 125I-labeled HPr(S46D) to L. brevis membranes containing high levels of the galactose permease and inhibition of the binding by nonradioactive HPr(S46D) were demonstrated (45).
Based on the extensive biochemical evidence summarized above, an allosteric model was proposed for the observed vectorial sugar expulsion in L. brevis (Fig. 1). To elucidate the details of the molecular mechanism of this process in vivo and to eventually determine the physiological significance of this process to growth and enzyme induction via the control of inducer levels, we have cloned, sequenced, and expressed the relevant L. brevis genes in B. subtilis. In this organism, which does not normally exhibit the phenomenon of PTS-mediated inducer control, we could observe regulation of the L. brevis GalP activity by mutant derivatives of the L. brevis HPr. We have therefore reconstituted the regulatory system in a bacterium that is readily amenable to genetic manipulation. This system should allow detailed studies of the type necessary to establish the mechanistic details of inducer control.
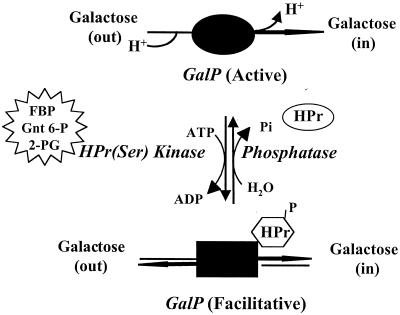
FIG. 1.
Proposed model for PTS-mediated regulation of vectorial sugar expulsion in L. brevis. Inducer expulsion in L. brevis is envisioned as a multicomponent signal transduction process. The galactose:H+ symporter GalP cotransports sugar and H+. H+ cotransport allows coupling of galactose uptake to the proton motive force. Upon addition of glucose (the extracellular effector), accumulation of glycolytic intermediates such as fructose 1,6-bisphosphate (FBP), gluconate-6-phosphate (Gnt 6-P), and 2-phosphoglycerate (2-PG) (the intracellular effectors) activate HPr(Ser) kinase (the sensor) to phosphorylate HPr on Ser-46 (the messenger), which binds to GalP (the target), resulting in uncoupling of sugar transport from H+ symport (the response). Consequently, the permease is converted from an active symporter to a facilitative diffusion uniporter that can no longer accumulate sugar substrates against a concentration gradient (inducer exclusion-expulsion).
MATERIALS AND METHODS
Bacterial strains and plasmids.
The strains used in this study were L. brevis ATCC 367, B. subtilis M168 (wild type), its isogenic ptsK mutant (32), Escherichia coli TOP10F′ (Invitrogen), E. coli XL10Gold, and E. coli TG1 (Stratagene). Growth conditions and media used to propagate the strains are described in the relevant sections. The plasmids used in this study were pCR2.1 (Invitrogen), PCRScript (Stratagene), pLA6 (6), pER82 (K. Pogliano, University of California at San Diego [UCSD]), pAG58 (15), and pMK4 (41). pCR2.1 and pCRScript were used for subcloning in E. coli. The pLA6, pAG58, and pMK4 plasmids were used for construction of expression vectors harboring L. brevis galP, wild-type HPr, HPr(S46A), and HPr(S46D). The pER82 plasmid was used to integrate L. brevis galP into the B. subtilis chromosome.
Cloning of the ptsK gene encoding the HPr(Ser) kinase from L. brevis.
To clone the HPr(Ser) kinase from L. brevis, we designed degenerate oligonucleotide primers based on the available multiple sequence alignment for PtsK proteins (32). Two of the primers, corresponding to the conserved amino acyl sequences HGVLV(l,m)D(e)V(i)Y(f)G (forward primer, alignment positions 143 to 151 [32]) and EI(v)RGL(v)GIIN (reverse primer, alignment positions 208 to 216) were used in a PCR (capital letters are conserved and lowercase letters are less well conserved residues). The actual sequences of the degenerate oligonucleotides were 5′CAiGGiGTiiTiiTiGAiiTiTAiGG3′ (forward, where i is inosine) and 5′TTDATDATiCCiAViCCiCKiAYYTC3′ (reverse, where D is T, A, or G; V is A, G, or C; K is G or T; and Y is C or T). A PCR DNA product of 217 bp was amplified from L. brevis ATCC 367 chromosomal DNA by Taq polymerase (Roche Molecular Biochemicals) under the following reaction conditions: denaturation of the template (8 min at 94°C), followed by 40 cycles of 1 min at 94°C (denaturation), 1 min at 32°C (annealing), 2 min at 72°C (extension), and 1 final cycle of 10 min at 72°C. The amplified product was subsequently cloned using the TA cloning kit (vector pCR2.1; Invitrogen). The 217-bp PCR product was sequenced, and high similarity to the corresponding region present in several ptsK genes (32) was detected.
The 217-bp DNA fragment was nonradioactively labeled with digoxigenin (DIG; High Prime DNA labeling system, Roche Molecular Biochemicals) and used as a probe in Southern hybridization against L. brevis chromosomal DNA fragments obtained by restriction with multiple enzyme combinations. A positive hybridization signal corresponded to the PvuII-SspI DNA fragment (approx. 2.9 kb). The mixture of DNA bands corresponding to the positive hybridization signal was cut from the gel, purified (gel extraction kit, Qiagen), and cloned into plasmid pUC18 digested with SmaI and dephosphorylated (shrimp alkaline phosphatase; USB). The ligation mixture was transformed into E. coli TOP10F′ (Invitrogen). Transformants were recovered, and individual colonies were inoculated into the wells of microtiter plates with Luria-Bertani (LB) broth (0.2 ml) and ampicillin (50 μg/ml). Following overnight growth, 0.1-ml aliquots of culture from 10 wells were pooled, and DNA was isolated (Spin kit 50; Qiagen). Pools that contained the cloned L. brevis ptsK gene gave positive signals in Southern hybridization and were further refined (individual DNA minipreps) to detect cultures containing the cloned gene. Of approximately 1,000 individual colonies that were screened, 2 contained the L. brevis ptsK gene cloned into pUC18. Sequencing of one of the clones, designated pK3, revealed that the ptsK gene was cloned on a 2,832-bp DNA fragment.
Cloning of the galP gene encoding the galactose permease of L. brevis.
To clone the gene encoding the galactose permease (GalP) of L. brevis, we designed several degenerate oligonucleotide primers based on the multiple sequence alignment of representative members of the LacS subfamily of the galactoside-pentoside-hexuronide (GPH) family of sugar transporters (25, 39). Two of the primers, 5′AAYGAYCARYTiHiTgg3′ (forward, where R is A or G and H is A, T, or C) and 5′TTCMRYTgiCCRTAYTCiAY3′ (reverse, where M is C or A), corresponding to the conserved amino acid regions NDQLM(l)W and V(i)EYGQW(l)K, respectively, were used in PCRs with L. brevis ATCC 367 chromosomal DNA (template) and Taq polymerase (Roche Molecular Biochemicals). The PCR product, a DNA fragment of 377 bp, was obtained under the following reaction conditions: one cycle of 8 min at 94°C, followed by 30 cycles of 1 min at 94°C (denaturation), 1 min at 37°C (annealing), 2 min at 72°C (extension), and one final cycle of 10 min at 72°C. The 377-bp DNA fragment was cloned into the pCR2.1 vector (Invitrogen) and sequenced. Sequence determination revealed similarity to internal genomic regions of several sugar permeases of the LacS subfamily. The 377-bp fragment was nonradioactively labeled with DIG and used as a probe in Southern hybridization against L. brevis chromosomal DNA digested with several restriction enzyme combinations. A strong hybridization signal corresponding to a 4.5-kb PvuII DNA fragment was detected. The mixture of DNA bands coinciding in size to this positive signal was isolated, and an attempt was made to clone it into the pUC18 vector. We had used this procedure previously to clone the L. brevis ptsK gene. Several cloning efforts were conducted with no success. Only deleterious or rearranged plasmids were recovered in the E. coli host. As an alternative, a mixture of PvuII DNA fragments corresponding to a positive hybridization signal was recovered from an agarose gel and digested with several restriction enzymes, including HindII, HpaI, DraI, FspI, and StuI. Following restriction, blunt-ended DNA fragments were self-ligated and used as templates in the inverse PCRs. PCR primers were designed based on the DNA sequence of the 377-bp fragment used as a probe in hybridization. The DNA sequences of the two primers were 5′CCAAGACTGGGAACAGAGGAACCG3′ (forward) and 5′CGTAAAGGTATCTACGTTGGCGG3′ (reverse). Pwo DNA polymerase (Roche Molecular Biochemicals) was used in the PCR with an initial denaturation (10 min at 94°C), 30 cycles of denaturation (1 min at 94°C), annealing (1 min at 50°C), extension (3 min at 72°C), and one cycle of final extension (10 min at 72°C). Initial rounds of inverse PCR amplified an HpaI DNA fragment of about 1.2 kb. This fragment was subcloned in the pCRScript vector (Stratagene). The DNA sequence was determined (1,197 bp), and it was used to design primers for the next round of inverse PCRs (nested PCRs). The DNA sequences of the two primers were 5′CATTCCGCACCCATTCTTCC3′ (forward) and 5′CGTCACCTTAGCGGTTCGTCC3′ (reverse). The 4.5-kb PvuII fragment mixture was then cut with EaeI; fragments were self-ligated, and inverse PCR was performed as described above. An additional 650 bp of new DNA sequence was determined, assembled with the previously determined 1,197-bp fragment, and used to design a new set of PCR primers to further extend the sequence. The DNA sequences of the new PCR primers were 5′GATAACAACAACCATTGTGATAACG3′ (forward) and 5′GCGTTTGATCCGATGATTGGTGGG3′ (reverse). For this purpose, L. brevis chromosomal DNA was digested with EcoRV, self-ligated, and used as a template for the inverse PCR. Finally, this approach led to subcloning of a 2,062-bp DNA fragment which encoded the complete galP gene.
Cloning and sequence of the ptsHI operon encoding HPr and EI of the PTS of L. brevis.
To clone the genes encoding HPr and enzyme I (EI), we designed several degenerate oligonucleotide primers based on multiple alignments of HPrs and EIs identified in several gram-positive and gram-negative bacteria. Two sets of primers, corresponding to the four conserved amino acid regions, were derived. The first set of primers, 5′GGiATHCAYGCiMGiCCiGCiAC3′ (forward) and 5′CCiARiSWCTAiACiCCCTADAT3′ (reverse, where S is G or C and W is A or T), corresponded to the amino acid regions GIHARPAT and IMGVMSLG, respectively. The second set of primers, 5′AARGARGTiGAYTTYTT3′ (forward) and 5′CATiSWRAAYTCRTC3′ (reverse), corresponded to the amino acid regions KEVDFF and DEFSM, respectively. These primers were used in PCRs with L. brevis chromosomal DNA (template) and Taq polymerase (Roche Molecular Biochemicals) to amplify DNA fragments corresponding to internal regions of genes coding for HPr (ptsH) and EI (ptsI), respectively. The PCR product corresponding to an internal region (129 bp) of the L. brevis ptsH gene was amplified under the following reaction conditions: one cycle of 8 min at 94°C, followed by 30 cycles of 1 min at 94°C (denaturation), 1 min at 42°C (annealing), 1 min at 72°C (extension), and one final cycle of 10 min at 72°C. The amplified product was cloned into the pCR2.1 vector (Invitrogen) and sequenced. The 129-bp fragment was nonradioactively labeled and used as a probe in Southern hybridization against L. brevis chromosomal DNA restricted with several enzymes, including StyI and SspI. DNA fragments giving positive hybridization signals were recovered from the agarose gel, self-ligated, and used as templates in inverse PCRs. PCR primers were designed based on the DNA sequence of the 129-bp fragment used as a probe for hybridization. The DNA sequences of the two primers were 5′AGCAGCCTGTACTAATAACG3′ (forward) and 5′CAGAAGTTAGCTTGCAATACC3′ (reverse). Pwo DNA polymerase (Roche Molecular Biochemicals) was used in the PCR where conditions were initial denaturation (10 min at 94°C), 30 cycles of denaturation (1 min at 94°C), annealing (1 min at 45°C), extension (2 min at 72°C), and one cycle of final extension (10 min at 72°C). An initial round of inverse PCR amplified a StyI DNA fragment of about 2 kb. This fragment was subcloned in the pCRScript vector (Stratagene), and the DNA sequence (1,877 bp) was determined. The entire ptsH gene was encoded on the 1,877-bp DNA fragment and could be directly amplified from L. brevis chromosomal DNA using PCR primers 5′TGTAGAGGTGAATGGTTGC3′ (forward) and 5′CTCATTAGTCAGATAAACCC3′ (reverse). To determine the complete sequence of ptsI, L. brevis genomic DNA was digested with XhoII and RcaI, self-ligated, and used as template in a set of two inverse PCRs with 5′TGACACTCCCTGTTGTCC3′ (forward for XhoII), 5′GTGAACCCACTGCTGCG3′ (reverse for XhoII), 5′TACGGCTGATAGAGATAGGC3′ (forward for RcaI), and 5′AGTGGATCGCATCGTTCGC3′ (reverse for RcaI). This approach led to subcloning of a 3,214-bp DNA fragment encoding the complete ptsH-ptsI operon.
Site-directed mutagenesis of L. brevis ptsH gene encoding HPr.
To create two site-directed mutants of L. brevis HPr, HPr(S46A), expected to mimic the unphosphorylated form of HPr, and HPr(S46D), expected to mimic the seryl-phosphorylated form of HPr, the experimental method described before (18) was employed. A two-step PCR approach resulted in replacement of serine residue 46 (S46) in the wild-type L. brevis HPr with either alanine (S46A) or aspartate (S46D). Three oligonucleotide primers were designed: 5′TGAAGTGCTATCATGGGCGTTATGTCATTAGG3′ (forward, for the S46A mutant, with G replacing T in the wild-type HPr), 5′TGAAGTGATATCATGGGCGTTATGTCATTAGG3′ (forward, for the S46D mutant, with GA replacing TC in the wild-type HPr), and 5′CTCTCATTAGTCAGATAAACCC3′ (reverse primer). In the first round of PCRs, two forward primers were combined with a reverse primer in a reaction in which the L. brevis wild-type HPr served as a template. Taq DNA polymerase (Roche Molecular Biochemicals) was used, and reaction conditions were as follows: one cycle of initial denaturation (8 min, 94°C), 30 cycles of denaturation (1 min, 94°C), annealing (1 min, 43°C), and extension (1 min, 72°C), and one cycle of final extension (10 min, 72°C). A unique 130-bp PCR product was amplified in each reaction, purified, and used as a primer in a second round of PCRs with the reverse primer described above. Similar reaction conditions were used as for the first round of PCRs except that annealing was done at 50°C. Amplified PCR products were cloned into the pCRScript vector (Stratagene). Replacement of the serine residue at position 46 in wild-type HPr by either alanine (S46A) or aspartate (S46D) was confirmed by DNA sequencing.
Chromosomal introduction of L. brevis galP gene in B. subtilis.
The L. brevis galP gene was amplified by PCR using the Pwo polymerase and L. brevis chromosomal DNA as the template. To express galP in B. subtilis, this gene was fused to a consensus B. subtilis Shine-Dalgarno (SD) sequence in a two-step PCR. Two different forward PCR primers were used, one for each reaction, together with a unique reverse PCR primer. The DNA sequences of the two forward primers were 5′TAGGAGGAGATAAAAATGGTTAAGCAATATTTATCATACGC C3′ (SD sequence in bold and 5′ end of galP underlined) and 5′AAAACTGCAGTTAGGAGGAGATAAAAATGGTTAAGC3′ (PstI restriction site in italics). The sequence of the reverse primer was 5′AAAATCGCGACATCATAGTCGCTTTATGCGACCG3′ (NruI restriction site in italics). The PstI-NruI-amplified galP gene was fused to shuttle promoter P6 (6) encoded on plasmid pLA6 restricted with Cfr10I (blunted) and PstI. P6 is a constitutive promoter functional in E. coli, B. subtilis, and numerous lactic acid bacteria (7). The P6/galP recombinant cassette was subcloned into the B. subtilis integration vector pER82 (kindly provided to us by K. Pogliano, UCSD). The P6/galP cassette was cloned into the amylase (amyE) gene on pER82 (restricted with BamHI and EcoRI [blunted]) next to the kanamycin resistance marker. Plasmid pEXP was assembled, propagated, linearized (PvuII), and transformed into B. subtilis M168 (wild type), and the isogenic strain bearing the inactivated gene encoding HPr(Ser) kinase (ptsK, Emr [32]). Following transformation, this cassette was integrated into the chromosome in the two isogenic bacteria via homologous recombination involving the amyE gene present on pEXP and a nonessential amyE gene product on the B. subtilis chromosome. Selection for kanamycin resistance (in wild-type B. subtilis) and for kanamycin and erythromycin resistance (in the B. subtilis ptsK mutant) resulted in the recovery of transformants containing the P6/L. brevis galP cassette integrated into the B. subtilis chromosome in both isogenic strains. The presence of the P6/galP cassette on the B. subtilis chromosome in both strains was confirmed by PCR.
Expression of L. brevis genes encoding HPr, HPr(S46A), and HPr(S46D) in B. subtilis.
To express L. brevis HPr (wild type), HPr(S46A), and HPr(S46D) in B. subtilis, L. brevis chromosomal DNA and the two previously constructed plasmids encoding HPr(S46A) and HPr(S46D) were used as templates in the two-step PCR. The three genes were simultaneously amplified by PCR and combined with the B. subtilis consensus SD sequence. Two different forward PCR primers were used, one in each reaction, combined with the unique reverse PCR primer. DNA sequences for the two forward primers were 5′AAGGAGGAGATCAATTATGGAAAAACGC3′ (SD sequence in bold and 5′ end of galP underlined) and 5′ATATGTCGACAAGGAGGAGATCAATTATGG3′ (SalI restriction site in italics). The sequence of the reverse primer was 5′ATATGGATCCCTCATTAGTCAGATAAACCC3′ (BamHI restriction site in italics). The SalI/-BamHI-amplified HPr (wild type) and the two mutant derivatives (S46A and S46D) were fused to the B. subtilis promoter Pspac present on plasmid pAG58 (15). The three HPr variants, each combined with Pspac, were further subcloned (as EcoRI-BamHI DNA fragments) into the E. coli–B. subtilis shuttle vector pMK4, which encodes the chloramphenicol resistance marker (41). Recombinant plasmids were originally recovered in E. coli XL10Gold (Stratagene) and propagated in E. coli TG1 (recA+; Stratagene). Plasmid DNA was transformed in B. subtilis M168 (wild type) and the M168 ptsK mutant, both harboring the P6/galP cassette encoding the L. brevis galactose permease integrated into the B. subtilis chromosome. B. subtilis was transformed using its natural competence system. The presence of the recombinant plasmids encoding L. brevis wild-type HPr, HPr(S46A), or HPr(S46D) was confirmed by restriction analysis and by PCR.
[3H]TMG uptake and accumulation in L. brevis.
L. brevis cultures were grown for 16 h at 30°C in complex medium (MRS) supplemented with 1% galactose. Cells were harvested in midlogarithmic phase and washed twice before resuspension in 50 mM Tris maleate–5 mM MgCl2 (TM) buffer (pH 7.0) to a cell density of 2 mg/ml (dry weight). The [3H]TMG (final concentration, 0.1 mM; 50 mCi/mmol), the energy source (20 mM arginine), and sugar (20 mM glucose) were added to a 0.5-ml cell suspension at the times indicated in the legend to Fig. 7. Transport assays were conducted at 30°C. Samples (0.1 ml) were removed at appropriate times, filtered (25-mm membrane filters, 45-μm pore size), washed three times with cold TM buffer, dried, and then transferred to vials containing 5 ml of scintillation fluid for determination of radioactivity. Values reported represent the averages of three independent assays.
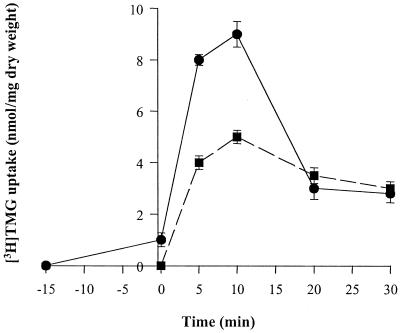
FIG. 7.
Effect of glucose on [3H]TMG uptake and accumulation in L. brevis. The experiments were conducted as described under Materials and Methods. L. brevis cultures were concentrated in TM buffer (pH 7.0) to a cell density of 2 mg/ml. [3H]TMG accumulation was measured at 30°C. In one set of experiments (●), 20 mM arginine was added to energize the cells, [3H]TMG (100 μM) was added at time −15 min, and 20 mM glucose was added at 0 min to induce [3H]TMG expulsion. In a separate set of experiments (■), glucose (20 mM, added at −1 min) was used as the sole energy source. [3H]TMG was added at 0 min.
[3H]TMG and [14C]glucose uptake and accumulation in B. subtilis.
The B. subtilis wild-type and ptsK mutant cultures harboring the L. brevis galP gene expressed from the constitutive promoter P6 were grown for 16 h at 30°C in complex medium (LB) with 1% glucose or 1% gluconate to midlogarithmic phase. Cells were washed twice and resuspended in 50 mM Tris maleate–5 mM MgCl2 buffer (pH 7.0) at a cell density of 1 mg/ml (dry weight). The energy source (20 mM arginine), the sugar (glucose or gluconate, each at 10 mM), and [3H]TMG (final concentration, 20 μM; 50 mCi/mmol) were added to a 0.1-ml cell suspension at −2, −1, and 0 min, respectively. Transport assays were conducted at 30°C. Samples (0.02 ml) were removed at appropriate times, filtered (25-mm membrane filters, 45-μm pore size), washed three times in cold TM buffer, dried, and then transferred to vials containing 5 ml of scintillation fluid for determination of radioactivity. Values reported represent the averages of three independent assays. Uptake of [14C]glucose was conducted using the conditions described in the legend to Fig. 9.
FIG. 9.
Time courses for uptake of [14C]glucose. The B. subtilis cells used in the experiment shown in Fig. 8 were grown in LB with 1% glucose (A) or LB with 1% gluconate (B). Arginine (20 mM) provided energy in both uptake experiments. The concentration of [14C]glucose was 20 μM.
Computer methods.
To determine the relatedness of L. brevis proteins to their protein homologues from other gram-positive and gram-negative bacteria, multiple sequence alignments and phylogenetic trees were generated with both the TREE program (9) and the ClustalX program (12).
Nucleotide sequence accession numbers.
The nucleotide sequence data reported in this paper were submitted to the GenBank nucleotide sequence database under accession numbers AF343443 (ptsK), AF343444 (ptsHI), and AF343445 (galP).
RESULTS
PtsK of L. brevis.
ptsK genes encoding HPr(Ser) kinases from B. subtilis (10, 32), Enterococcus faecalis (17), Streptococcus salivarius (2), Staphylococcus xylosus (14), and Lactobacillus casei (8) have been cloned. The ptsK gene of L. brevis was cloned and sequenced as described under Materials and Methods. A total of about 2.8 kb of L. brevis DNA was sequenced with the ptsK gene centrally located in this fragment. The gene encodes a protein of 312 amino acyl residues, with a calculated isoelectric point (pI) of 5.62 and molecular mass of 34.4 kDa. A total of 583 bp of DNA upstream of ptsK were sequenced, but no upstream open reading frame (ORF) within this region was detected. Instead, we identified: a typical putative −35, −10 promoter sequence for gram-positive bacteria and a consensus ribosome-binding site 6 bp upstream of the structural gene. The 1,314 bp of sequence downstream of ptsK were also determined. A second ORF encoding prolipoprotein diacylglyceryl transferase (functionally characterized homologue in Staphylococcus aureus, accession no. spP52282) was identified. This gene is homologous to the second gene in the ptsK operon of B. subtilis (32). The significance of the apposition of the two genes in these two evolutionarily divergent gram-positive bacterial species is not known.
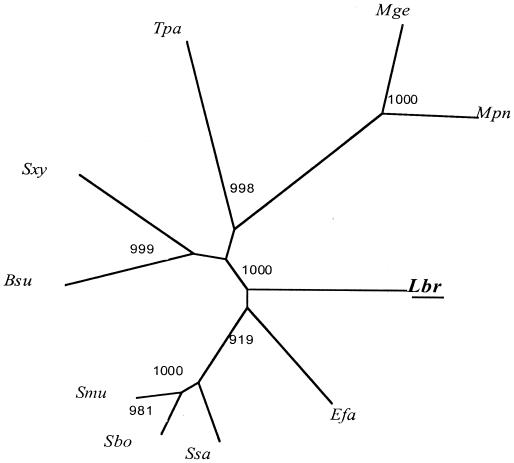
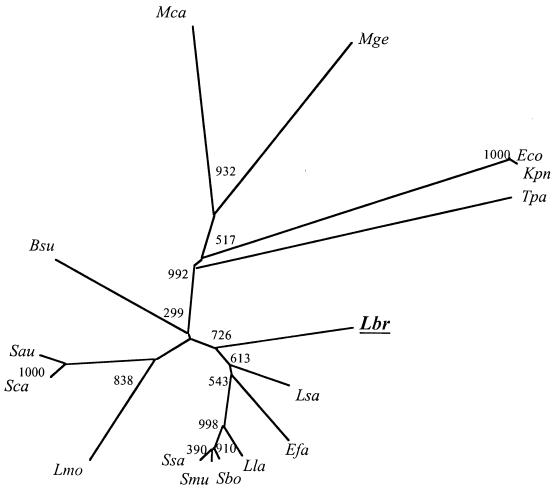
Figure 2 shows the phylogenetic tree of PtsK homologues generated with the ClustalX program. The L. brevis protein is distant from all other homologues. Of particular relevance to the work performed here is the great distance of this kinase from the B. subtilis homologue. This fact undoubtedly explains the very poor enzymatic cross-reactivity observed between these two enzymes (32) (see below).
FIG. 2.
Phylogenetic tree of L. brevis PtsK with several of its homologues from other bacteria. The phylogenetic tree was derived using the ClustalX program. Bootstrapping was applied, and bootstrap values (the relative probabilities for occurrence of internal nodes for the tree), based on 1,000 random runs, are provided. Abbreviations: Lbr, Lactobacillus brevis; Efa, Enterococcus faecalis; Sbo, Streptococcus bovis; Smu, Streptococcus mutans; Ssa, Streptococcus salivarius; Bsu, Bacillus subtilis; Sxy, Staphylococcus xylosus; Tpa, Treponema pallidum; Mge, Mycoplasma genitalium; Mpn, Mycoplasma pneumoniae. A very similar tree was generated with the TREE program (data not shown).
ptsHI operon of L. brevis.
Early biochemical evidence suggested that L. brevis possesses one of the two general PTS proteins, HPr, but that EI was missing in this organism (30). However, anaerobic growth of L. brevis in the presence of fructose resulted in the induction of a complete fructose-specific PTS (37). The ptsHI operon encoding HPr and EI was cloned as described under Materials and Methods. A DNA fragment of 3,214 bp was sequenced. This clone includes the complete ptsH and ptsI genes as well as 946 bp upstream and 263 bp downstream of the operon. The ptsH gene encodes a protein of 89 amino acyl residues (HPr), with a calculated pI of 4.7 and mass of 9.5 kDa. The ptsI gene is located immediately downstream to ptsH and encodes a protein of 576 residues, with a calculated pI of 5.4 and mass of 62.3 kDa. A putative promoter was identified upstream of ptsH. Putative ribosome-binding sites were identified 11 nucleotides in front of the ATG start codon of ptsH and 2 nucleotides in front of ptsI. A putative terminator downstream of ptsI was also identified. A truncated homologue of the Lactobacillus sake and Lactobacillus lactis clpE genes encoding a putative ATP-dependent protease was found upstream of ptsH. The L. brevis clpE gene proved to be transcribed in the opposite direction with respect to ptsHI.
The phylogenetic tree for several HPr proteins is shown in Fig. 3. The L. brevis protein clusters in the order indicated with the homologues from species of Lactobacillus > Enterococcus > Lactococcus > Streptococcus, as expected.
FIG. 3.
Phylogenetic tree for representative HPr proteins. The tree was generated with the ClustalX program. Bootstrapping was applied, and the bootstrap values based on 1,000 random runs are provided. Abbreviations: Bsu, Bacillus subtilis; Efa, Enterococcus faecalis; Eco, Escherichia coli; Kpn, Klebsiella pneumoniae; Lla, Lactococcus lactis; Lbr, Lactobacillus brevis; Lmo, Lysteria monocytogenes; Lsa, Lactobacillus sake; Mca, Mycoplasma capricolum; Mge, Mycoplasma genitalium; Sbo, Streptococcus bovis; Smu, Streptococcus mutans; Ssa, Streptococcus salivarius; Sau, Staphylococcus aureus; Sca, Staphylococcus carnosus; Tpa, Treponema pallidum.
We have published a multiple alignment of several relevant HPr proteins as well as the results of analyses of numerous site-specific mutants of E. coli HPr (16). The reported results lead to predictions for potential sites of interaction of HPr with PtsK and of HPr(Ser) with GalP. Although the L. brevis HPr was not included within the published analyses, we have updated that report (unpublished work) and summarize the results here.
The updated multiple alignment including the L. brevis HPr revealed that all of the residues shown in the sequence presented in Fig. 4 are fully conserved in the gram-positive bacterial HPrs that are known to be phosphorylated by a gram-positive bacterial PtsK. Note that enzymatic cross-reactivity between various gram-positive bacterial HPr kinases and various HPr proteins has been demonstrated (29), but that very considerable differences in phosphorylation rates are observed. For example, the B. subtilis PtsK barely phosphorylated L. brevis HPr.
FIG. 4.
Fully conserved residues in gram-positive bacterial HPrs.
In the diagrammed sequence (Fig. 4), nonconserved residues in the gram-positive bacterial proteins are indicated by dashes, underlined residues are conserved in all HPrs included in our alignment, overlined residues are conserved in all gram-positive HPrs but not in the E. coli HPr [which is known not to be a substrate of gram-positive HPr(Ser) kinases], and asterisks show the active-site histidine (H15*) and regulatory serine (S46*). Twenty-one residues are common to the gram-positive HPrs that are known to be phosphorylated by PtsKs but different from those in E. coli HPr, which is not phosphorylated (29). Of these 21 residues, 10 immediately surround Ser46 without interruption. These residues form the flexible loop connecting β-strand 3 and α-helix 2 in the open-faced sandwich structure of HPr. They also comprise the part of the protein that undergoes the largest conformational change in response to serine-46 phosphorylation (28). Phosphorylation of this residue causes helix 2 to extend in length, thereby decreasing the amount of random coil structure comprising in part the connecting loop.
The other residues conserved in HPrs of gram-positive bacteria but not in E. coli HPr are scattered, with six residues dispersed in the region near His15 to the left of the Ser46 conserved loop and three to the far right of this loop. It seems likely that the primary HPr site of interaction with PtsK (and possibly with GalP) is the region surrounding Ser46. These residues are reasonable candidates for mutagenesis.
GalP of L. brevis.
Although operons including genes for the transport and utilization of galactose via the Leloir pathway have been identified in several lactic acid bacteria (11), only two non-PTS galactose or galactoside permeases were identified, a galactose permease of Lactococcus lactis MG1363 (B. Grossiord, E. Vaughan, E. Luesink, and W. M. de Vos, Proc. 5th Symp. Lactic Acid Bacteria, 1996, abstr. H48) and a putative lactose permease of L. lactis ATCC 7962 (19). In this report we describe cloning of the L. brevis galP gene, encoding a protein of 475 amino acyl residues. The galP gene is followed by a galactokinase (galK) gene on the L. brevis chromosome. The calculated pI and mass of the GalP protein are 9.1 and 51.8 kDa, respectively. GalP is a member of the GPH family (TC 2.A.2 [39]). In contrast to the homologues from Streptococcus thermophilus, Lactobacillus bulgaricus, and Leuconostoc lactis, there is no IIAGlc-like domain C-terminal to the permease domain in L. brevis GalP. This fact immediately suggests (in accordance with the published results) that the L. brevis GalP permease is regulated by a mechanism that is distinct from that which has been reported for the lactose permeases of S. thermophilus and Leuconostoc lactis (22–24, 42).
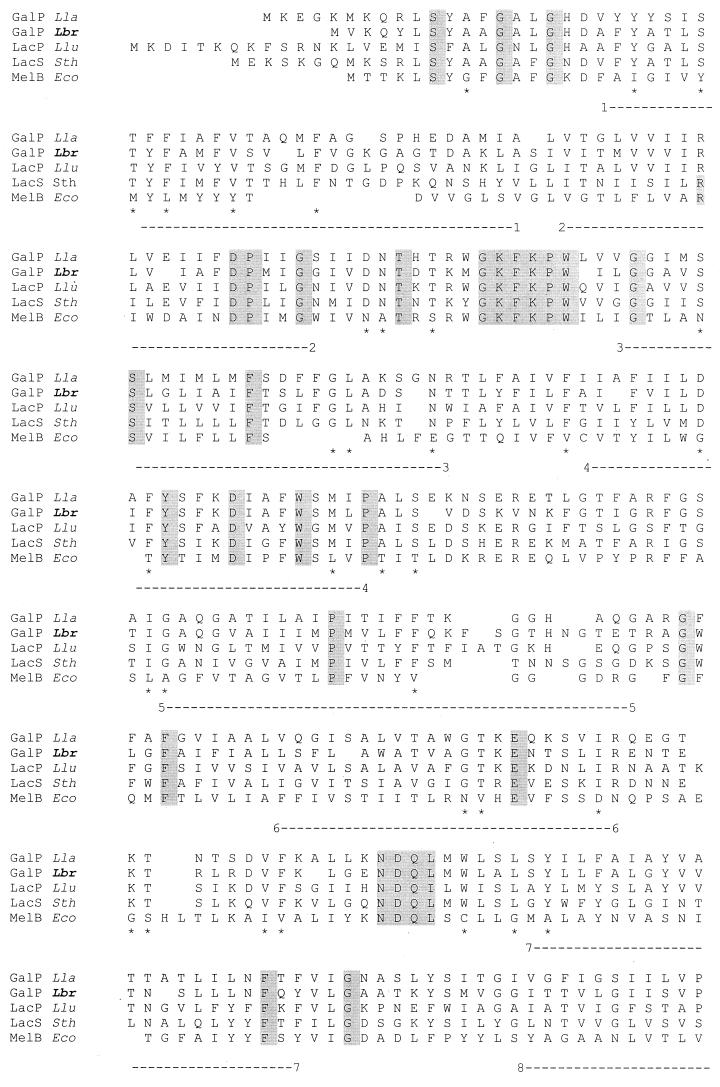
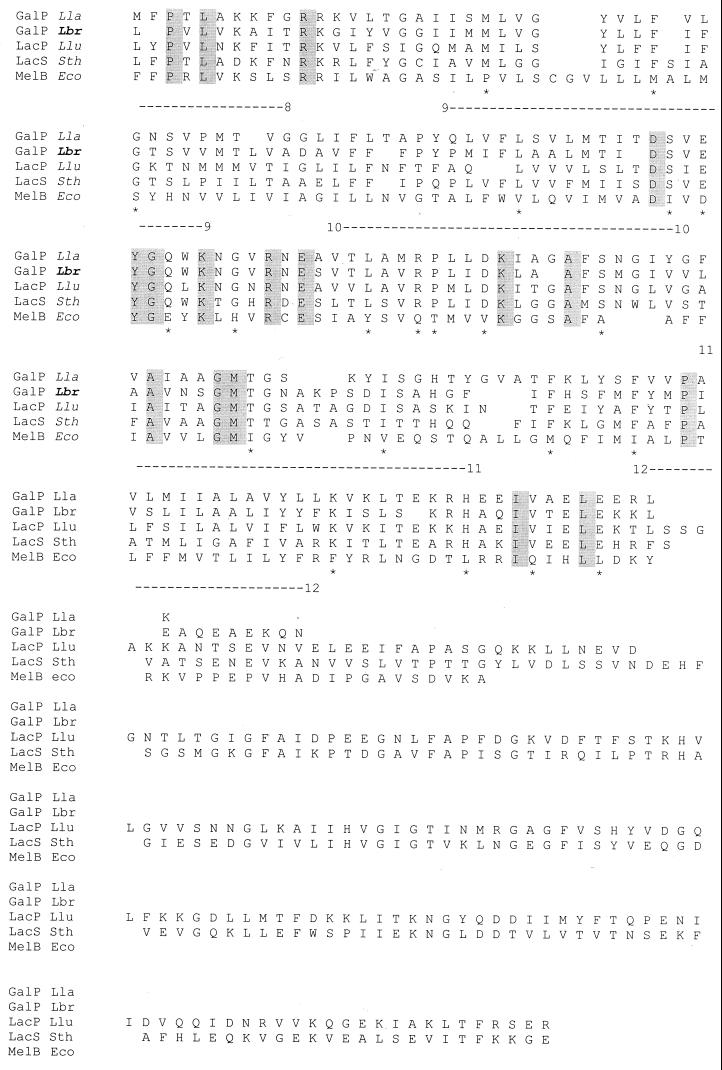
The multiple alignment of the L. brevis GalP protein with four related galactose and galactoside permeases is shown in Fig. 5. The deduced protein sequence of L. brevis GalP reveals strong similarity of this protein to the galactose permease of Lactococcus lactis and lactose permeases of S. thermophilus and Leuconostoc lactis (45 to 50% identity and 65 to 68% similarity for all four proteins). Fully conserved residues among the five proteins presented in the multiple alignment in Fig. 5 are shaded. Particularly worthy of note are the fully conserved GKFKPW and NDQL motifs in the first half of the protein between transmembrane segments (TMSs) 2 and 3 and in the central loop between TMSs 6 and 7, respectively. The former motif corresponds in position to the well-conserved motifs found between TMSs 2 and 3 in major facilitator superfamily (MFS) permeases (20). The GPH family is believed to be distantly related to the MFS (38). Several fully conserved residues were identified previously among lactic acid bacterial members of the GPH family (42), including E-67 and D-71 (numbers correspond to residues in the S. thermophilus LacS protein) and the conserved motif K-X-X-H-X-X-E, required for lactose and H+ symport. In L. brevis GalP, D-69 in TMS 2 corresponds to the conserved D-71 in TMS2 in the S. thermophilus LacS protein. No conserved residue corresponding to the E-67 in LacS could be identified in GalP. The conserved K-X-X-V-X-X-E motif, characteristic of members of the GPH family, was identified in the region between TMS 10 and TMS 11 in L. brevis GalP (the conserved H in this motif is replaced by V in GalP). The multiple alignment shown in Fig. 5 reveals that the C termini of the L. brevis and Lactococcus lactis GalP proteins are strongly hydrophilic, with 11 of 23 and 11 of 16 charged residues in equivalent regions of the two proteins, respectively. These regions correspond to the hydrophilic Q linkers in the S. thermophilus, L. bulgaricus, and Leuconostoc lactis proteins, which connect the permease domain to the regulatory IIAGlc-like domain.
FIG. 5.
Alignment of L. brevis GalP with representative members of the LacS and MelB subfamilies of the GPH family of sugar transporters. The multiple alignment was derived using the TREE program (9). Abbreviations: Lla, Lactococcus lactis; Lbr, Lactobacillus brevis; Llu, Leuconostoc lactis; Sth, Streptococcus thermophilus; Eco, Escherichia coli; GalP, galactose permease; LacP and LacS, lactose permease; MelB, melibiose permease. Complete protein sequences are shown. Completely conserved residues are shaded. Residues conserved only within the LacS subfamily are indicated by ∗. The 12 TMSs are indicated by numbered dashed lines under the alignment.
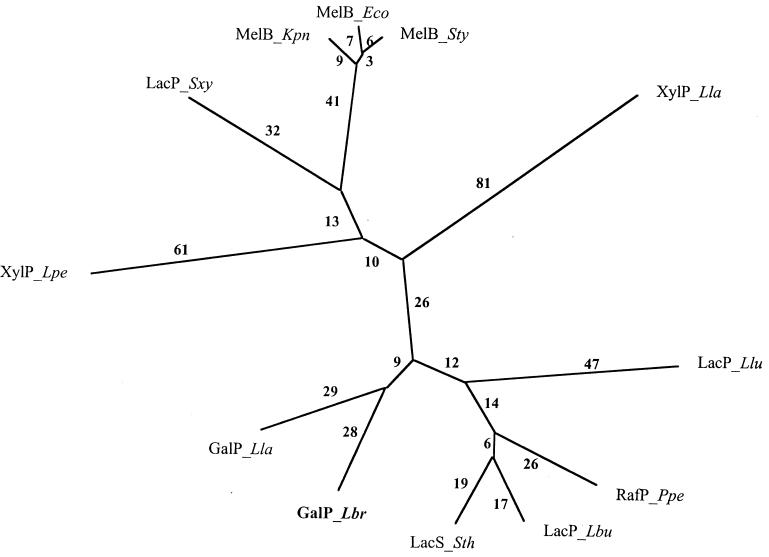
A phylogenetic tree for these proteins as well as for homologous xyloside, melibiose, and lactose permeases is shown in Fig. 6. The MelB proteins of gram-negative bacteria cluster with the LacP protein of S. xylosus, suggesting that the S. xylosus protein is in fact a melibiose permease. The two xyloside-transporting XylP proteins branch from the center of the tree and are not closely related. All of the lactose and galactose permeases are located at the bottom of the tree, and the two GalP proteins cluster together. Despite the strong overall sequence similarity between the L. brevis GalP, the S. thermophilus LacS, and the Leuconostoc lactis LacP, the L. brevis GalP protein clusters primarily with the Lactococcus lactis GalP, the only other protein besides the L. brevis GalP which is missing a regulatory C-terminal IIAGlc-like domain. It seems reasonable to suggest that the two GalP proteins may be subject to the same type of HPr(Ser)-mediated allosteric regulation. However, the presence of a lactose/galactose PTS in Lactococcus lactis will render this postulate difficult to test.
FIG. 6.
Phylogenetic tree for representative proteins of the GPH family. The phylogenetic tree was derived using the TREE program. Phylogenetic distance is approximately proportional to branch length. Abbreviations: Lla, Lactococcus lactis; Lbr, Lactobacillus brevis, Lpe, Lactobacillus pentosus; Lbu, Lactobacillus bulgaricus; Llu, Leuconostoc lactis; Ppe, Pediococcus pentosaceus; Sth, Streptococcus thermophilus; Sxy, Staphylococcus xylosus; Eco, Escherichia coli; Kpn, Klebsiella pneumoniae; Sty, Salmonella typhimurium; GalP, galactose permease; LacP or LacS, lactose permease; XylP, xyloside permease; RafP, raffinose permease; MelB, melibiose permease.
In view of this possibility, residues conserved between these two proteins but not the other homologues portrayed in Fig. 5 were identified. A surprisingly small number of such residues were found. Of particular note was a well-conserved LSY(I/1)LFA(I/l)(a/G)YV sequence, where the S--LFA motif is specific to the two GalP permeases. This motif is adjacent and C-terminal to the NDQL motif noted above. This region may prove to be important for function or regulation of the two GalP proteins.
Confirmation of inducer expulsion phenomenon in L. brevis.
In the presence of 20 mM arginine, addition of glucose to cells preloaded with [3H]TMG resulted in a transient increase in the rate of galactoside uptake, followed by rapid expulsion (Fig. 7). A similar phenomenon was observed when glucose was used as the energy source (added 1 min before the addition of [3H]TMG) or when arginine served as the initial energy source for TMG uptake and glucose was added 15 min after the addition of [3H]TMG (Fig. 7). According to the proposed model of inducer expulsion in L. brevis, the initial metabolism of glucose energizes active transport of TMG, but its continued metabolism, which results in the accumulation of fructose 1,6-bisphosphate, activates the HPr(Ser) kinase, phosphorylates HPr, and converts the GalP:H+ symporter into a facilitative, fully or partially uncoupled uniporter. Rapid efflux of the free sugar results.
Regulation of L. brevis GalP expressed in B. subtilis.
B. subtilis encodes HPr and HPr(Ser) kinase but does not exhibit the phenomena of inducer expulsion and inducer exclusion. To study the role of GalP in inducer expulsion, we integrated the L. brevis galP gene into the genome of B. subtilis, an organism that lacks the capacity to take up TMG. The galP gene was fused to a constitutive shuttle promoter (the P6/galP cassette) and expressed in both wild-type B. subtilis and the ptsK mutant strain.
The nucleotide sequences of the P6 promoter and of L. brevis galP were examined for the presence of putative catabolite-reponsive element sites. No such sequence was found, suggesting that the P6/galP cassette cannot be subject to catabolite repression by the PTS-dependent mechanism.
Plasmids encoding L. brevis wild-type HPr, HPr(S46A), and HPr(S46D) were introduced in the two B. subtilis strains. In this readily manipulatable organism, it was possible to study GalP regulation by mutant derivatives of L. brevis HPr.
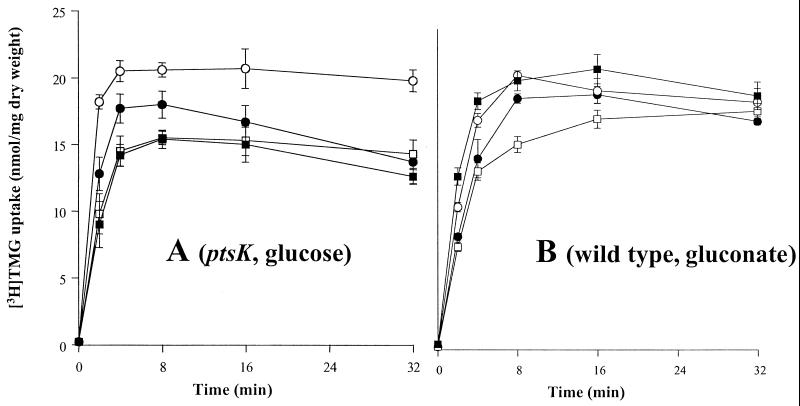
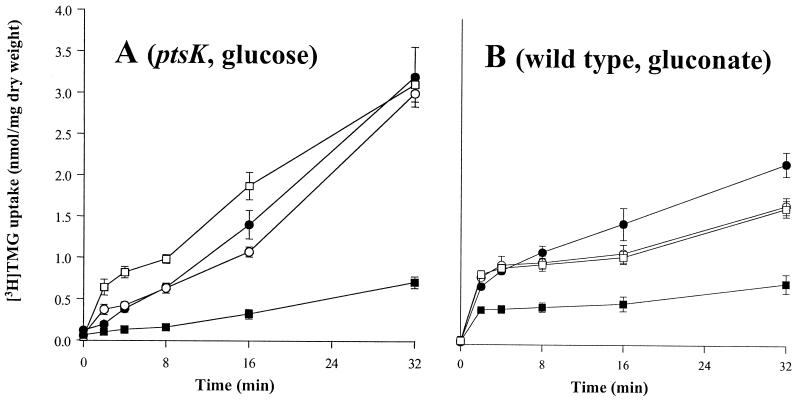
Although the B. subtilis genome includes a gene that is homologous to the L. brevis galP, we found that the capacity to take up TMG was lacking under the conditions of our assay. In the absence of the L. brevis galP gene, B. subtilis took up less then 0.05 nmol of [3H]TMG per mg (dry weight) of cells grown in LB supplemented with the appropriate carbohydrate (glucose or gluconate) and provided with an energy source, such as arginine plus glucose or gluconate. In the presence of the expressed L. brevis galP gene integrated into the B. subtilis chromosome, galactoside transport increased over 50-fold in both the B. subtilis wild-type strain and the ptsK mutant when arginine (20 mM) and either glucose or gluconate (10 mM) were added to cells at 2 min and 1 min, respectively, before the addition of [3H]TMG (0 min). However, in contrast to what was observed when the experiment was conducted in L. brevis, sugar efflux from B. subtilis did not follow the initial increase in sugar uptake (Fig. 8A and B). When the same assay was conducted with a B. subtilis wild-type or ptsK mutant strain expressing the P6/galP cassette and bearing the plasmid-encoded L. brevis wild-type HPr, HPr(S46A), or HPr(S46D), strong inhibition of [3H]TMG uptake was observed in the presence of HPr(S46D) relative to that observed when HPr or HPr(S46A) was present (Fig. 8A and B). The same pattern of inhibition of [3H]TMG uptake by L. brevis HPr(S46D) in wild-type B. subtilis and in the ptsK mutant was observed when the two strains were grown in the presence of either glucose or gluconate (data not shown). HPr(S46D) did not inhibit uptake of [14C]glucose under the same conditions (Fig. 9A and B). Inhibition of TMG uptake therefore could not be explained by a decrease in energy availability. Because HPr(S46D) resembles the phosphorylated form of HPr (31), these results confirm our earlier suggestion that GalP is subject to direct allosteric regulation by HPr(Ser). Although we could not demonstrate inducer expulsion in B. subtilis, we demonstrated for the first time in vivo inducer exclusion in this bacterium.
FIG. 8.
Time courses for uptake of [3H]TMG into B. subtilis cells expressing chromosomally encoded L. brevis galP gene. Cell cultures used either lacked (A) or possessed (B) a functional B. subtilis ptsK gene. Plasmid pMK4 with the inserted L. brevis ptsH genes encoding wild-type HPr (□), S46A HPr (○), or S46D HPr (■) is also present. The control strain (●) did not contain plasmid pMK4. Conditions were as described under Materials and Methods, with a [3H]TMG concentration of 20 μM. (A) Cells were grown in LB with 1% glucose, and 10 mM glucose and 20 mM arginine were added as energy sources for the transport studies. (B) Cells were grown in LB with 1% gluconate, and 10 mM gluconate and 20 mM arginine were added for the transport experiment as described under Materials and Methods. The differences between panels A and B reflect the conditions used rather than strain differences. Thus, when the two experiments (A and B) were conducted with both wild-type and ptsK mutant strains, the results were essentially the same for each of the two growth conditions.
DISCUSSION
The work reported in this communication sets the stage for detailed molecular genetic, biochemical, and biophysical experiments aimed at understanding in molecular detail the mechanism of inducer exclusion and expulsion in L. brevis. All of the genes which are known to be directly involved in this novel type of regulation (ptsK, ptsH, and galP) as well as one that may prove to be indirectly involved (ptsI) have now been cloned. Moreover, the essence of the regulatory system has been reconstituted in B. subtilis, an organism that does not normally exhibit the phenomenon of PTS-mediated inducer control. Thus, not only are the requisite genes available for study, but two distinct systems, the native L. brevis system and the reconstituted B. subtilis model system, can be exploited.
Availability of the L. brevis genes opens the door to several experimental approaches. These genes can be knocked out on the L. brevis chromosome to determine the physiological consequences to the cell of the presence of their gene products. The complete regulatory system [including the L. brevis HPr(Ser) kinase] can be reconstituted in B. subtilis to establish that these proteins comprise the entire regulatory cascade. We anticipate that it will also be possible to reconstitute the system in E. coli, an organism that lacks PtsK altogether. All of the plasmids constructed for expression in B. subtilis should be suitable for studies in E. coli, and mutants with pts gene deletions are available.
The cloned L. brevis ptsK gene will allow us to overproduce PtsK for in vitro biochemical analyses. To date, five HPr(Ser) kinases, those from B. subtilis, E. faecalis, S. salivarius, S. xylosus, and L. casei, have been purified and partially characterized. However, extensive evidence suggests that different kinases exhibit very different properties (2, 8, 10, 13, 14, 17, 32). Analyses of the cloned ptsK gene reported in this communication have provided revealing information that explains some earlier observations. For example, the great degree of sequence divergence between the PtsK proteins of B. subtilis and L. brevis (as well as the HPr proteins of these two organisms [Fig. 2 and 3]) explains the exceptionally poor enzymatic cross-reactivity between these proteins, as reported previously (30, 32) and confirmed here. Moreover, the phylogenetic trees shown in Fig. 2 and 3 are fully consistent with the interpretation that both PtsK and PtsH are orthologues in the organisms represented in both trees (L. brevis, B. subtilis, E. faecalis, S. salivarius, Streptococcus bovis, Streptococcus mutans, Mycoplasma genitalium, and Treponema pallidum). As the completely sequenced genomes of B. subtilis, Mycoplasma genitalium, M. pneumoniae, E. faecalis, and Treponema pallidum reveal only a single ptsK gene and a single ptsH gene (excluding the crh gene in B. subtilis, which lacks catalytic activity), we predict that all of these PtsK homologues and all of the PtsH homologues in gram-positive bacteria and perhaps in many gram-negative bacteria as well (i.e., T. pallidum and Neisseria gonorrhoeae) will prove to have been present in the common ancestor. Such a prediction leads in turn to the suggestion that PtsK-mediated regulatory mechanisms may be very old, dating back to a time that predates the divergence of gram-positive bacteria from each other and possibly also before the time when gram-positive and gram-negative bacteria diverged.
It has been reported recently that phosphorylation of HPr by the HPr(Ser) kinase of L. casei controls catabolite repression and inducer exclusion but not inducer expulsion (8). Involvement of HPr kinase in inducer expulsion in L. brevis was extensively documented in vitro (44, 45). To date, we have not been able to inactivate the ptsK gene in L. brevis. Therefore, in vivo confirmation of the role of HPr(Ser) kinase in this organism is not presently possible.
Availability of the galP clone will allow its protein product to be overproduced for reconstitution studies in defined liposome preparations. Only when the regulatory system is reconstituted in such an artificial system with purified components will it be possible to establish unequivocally the protein constituents involved and the details of the signal transduction pathway. The multiple alignment of GalP with its homologues (Fig. 5) provides clear clues as to possible residues involved in HPr(Ser) binding. Mutagenic approaches with GalP should be useful for defining catalytic residues as well as those concerned with allosteric regulation of the activity of this protein. Since the topology of GalP is probably similar to that of the E. coli melibiose permease (1, 27), one can narrow down the possibilities for binding sites. As for the interaction sites established for the E. coli lactose, maltose and melibiose permeases with the regulatory IIAGlc protein of the PTS (see references 3 and 35 for reviews), the allosteric sites of interaction between GalP and HPr(Ser) must be in the N and C termini and/or in the cytoplasmic loops.
Published evidence suggests that the L. brevis galactose permease is regulated by a mechanism that is entirely different from the mechanism that controls lactose permease activities in L. bulgaricus, Leuconostoc lactis, and S. thermophilus (22–24, 34, 36). These three proteins all possess a C-terminal IIAGlc-like domain that influences permease activity. This domain is lacking in L. brevis, clearly pointing to a distinct type of regulation. However, the sequence similarity observed for the GalP proteins from L. brevis and Lactococcus lactis (Fig. 5 and 6) suggests (in the absence of direct experimental evidence) that these permeases may be regulated by a common mechanism.
We have constructed site-specific mutations in L. brevis HPr, altered at the regulatory phosphorylation site. However, mutations at other sites are likely to reveal the sites of interaction with PtsK and GalP. It has been reported recently that replacement of isoleucine at position 47 with threonine in HPr of L. casei results in a less pronounced lag phase during diauxic growth in a mixture of PTS sugars (glucose and lactose) and that carbon catabolite repression was completely abolished when the organism was grown in the presence of non-PTS sugars maltose and ribose (43). Replacement of methionine at position 48 with valine in Streptococcus salivarius has been reported to abolish diauxy by allowing the cells to catabolize non-PTS sugars in the presence of glucose (21). In addition, the growth rate of the S. salivarius M48V mutant on galactose decreased rapidly over time, while growth on another non-PTS sugar (lactose or melibiose) was not affected. Both Ile-47 and Met-48 are conserved in HPr of L. brevis, a heterofermentative bacterium which has been shown to possess fructose PTS-inducible activity under anaerobic conditions (37). It would be most interesting to examine whether replacement of either of the two conserved residues, Ile-47 or Met-48, interferes with catabolite repression in L. brevis, and especially whether the M48V substitution interferes with growth on galactose (which is transported by GalP). Corresponding mutagenic approaches with HPr and GalP should be useful for defining catalytic residues as well as those concerned with allosteric regulation of the activities of these two proteins.
The GalP protein of L. brevis is the first transporter known to be involved in inducer control to have its gene introduced into the B. subtilis chromosome. By introducing the L. brevis GalP and derivatives of L. brevis HPr into B. subtilis, we confirmed directly, for the first time, the role of phosphorylation of HPr at Ser-46 in inducer control. However, under the conditions described in this work, we could not demonstrate the inducer expulsion phenomenon in B. subtilis. The extent to which the L. brevis GalP mechanism of inducer control is pertinent to other permeases in L. brevis and to the numerous proton:sugar symporters in other bacteria is unknown. However, extensive evidence suggests that several permeases, including the glucose permease in L. brevis, are controlled by this HPr(Ser-P)-mediated allosteric mechanism (45, 46). We anticipate that this mechanism will prove to be widespread in low-G+C-content gram-positive bacteria.
ACKNOWLEDGMENTS
Work in the authors' laboratory was supported by NIH grant 9RO1 GM55434 from the National Institute of General Medical Sciences.
We thank R. Bruckner, Universität Tübingen, Tübingen, Germany, for helpful suggestions during the initial efforts to clone the HPr kinase gene of L. brevis. We are grateful to Milda Simonaitis, Donna Yun, and Monica Mistry for assistance in the preparation of the manuscript. Don Jack provided useful assistance with the computer analyses.
REFERENCES
- 1.Botfield M C, Naguchi K, Tsuchiya T, Wilson T H. Membrane topology of the melibiose carrier of Escherichia coli. J Biol Chem. 1992;267:1818–1822. [PubMed] [Google Scholar]
- 2.Brochu D, Vadeboncoeur C. The HPr(Ser) kinase of Streptococcus salivarius: purification, properties, and cloning of the hprK gene. J Bacteriol. 1999;181:709–717. doi: 10.1128/jb.181.3.709-717.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dean D A, Reizer J, Nikaido H, Saier M H., Jr Regulation of the maltose transport system of Escherichia coli by the glucose-specific enzyme III of the PTS: characterization of inducer exclusion-resistant mutants and reconstitution of inducer exclusion in proteoliposomes. J Biol Chem. 1990;265:21005–21010. [PubMed] [Google Scholar]
- 4.Deutscher J, Saier M H., Jr ATP-dependent protein kinase-catalyzed phosphorylation of a seryl residue in HPr, the phosphoryl carrier protein of the phosphotransferase system in Streptococcus pyogenes. Proc Natl Acad Sci USA. 1983;80:6790–6795. doi: 10.1073/pnas.80.22.6790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deutscher J, Reizer J, Fischer C, Galinier A, Saier M H, Jr, Steinmetz M. Loss of protein kinase-catalyzed phosphorylation of HPr, a phosphocarrier protein of the phosphotransferase system, by mutation of the ptsH gene confers catabolite repression resistance to several catabolic genes of Bacillus subtilis. J Bacteriol. 1994;176:3336–3344. doi: 10.1128/jb.176.11.3336-3344.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Djordjevic G M, Bojovic B, Miladinov N, Topisirovic L. Cloning and molecular analysis of promoter sequences from the chromosomal DNA of Lactobacillus acidophillus ATCC 4356. Can J Microbiol. 1997;43:61–69. doi: 10.1139/m97-009. [DOI] [PubMed] [Google Scholar]
- 7.Djordjevic G M, Klaenhammer T R. Positive selection cloning vectors for Gram-positive bacteria based on a restriction endonuclease cassette. Plasmid. 1996;35:37–45. doi: 10.1006/plas.1996.0004. [DOI] [PubMed] [Google Scholar]
- 8.Dossonnet V, Monedero V, Zagorec M, Galinier A, Perez-Martinez G, Deutscher J. Phosphorylation of HPr by the bifunctional HPr kinase/P-Ser-phosphatase from Lactobacillus casei controls catabolite repression and inducer exclusion but not inducer expulsion. J Bacteriol. 2000;182:2582–2590. doi: 10.1128/jb.182.9.2582-2590.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feng D-F, Doolittle R F. Progressive alignment and phylogenetic tree construction of protein sequences. Methods Enzymol. 1990;183:375–387. doi: 10.1016/0076-6879(90)83025-5. [DOI] [PubMed] [Google Scholar]
- 10.Galinier A, Kravanja M, Engelmann R, Hengstenberg W, Kilhoffer M-C, Deutscher J, Haiech J. New protein kinase and protein phosphatase families mediate signal transduction in bacterial catabolite repression. Proc Natl Acad Sci USA. 1998;95:1823–1828. doi: 10.1073/pnas.95.4.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grossiord B, Vaughan E, Luesink E, de Vos W M. Genetics of galactose utilization via the Leloir pathway in lactic acid bacteria. Lait. 1998;78:77–84. [Google Scholar]
- 12.Heringa J. Two strategies for sequence comparison: profile-preprocessed and secondary structure-induced multiple alignment. Comput Chem. 1999;23:341–364. doi: 10.1016/s0097-8485(99)00012-1. [DOI] [PubMed] [Google Scholar]
- 13.Hoischen C, Reizer J, Dijkstra A, Rottem S, Saier M H., Jr Presence of protein constituents of the gram-positive bacterial phosphotransferase regulatory system in Acholeplasma laidlawii. J Bacteriol. 1993;175:6599–6604. doi: 10.1128/jb.175.20.6599-6604.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huynh P L, Jankovic I, Schnell N F, Bruckner R. Characterization of an HPr kinase mutant of Staphylococcus xylosus. J Bacteriol. 2000;182:1895–1902. doi: 10.1128/jb.182.7.1895-1902.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jaacks K J, Healy J, Losick R, Grossman A D. Identification and characterization of genes controlled by the sporulation-regulatory gene spo0H in Bacillus subtilis. J Bacteriol. 1989;171:4121–4129. doi: 10.1128/jb.171.8.4121-4129.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koch S, Sutrina S L, Wu L-F, Reizer J, Schnetz K, Rak B, Saier M H., Jr Identification of a site in the phosphocarrier protein, HPr, which influences its interactions with sugar permeases of the bacterial phosphotransferase system: kinetic analyses employing site directed mutants. J Bacteriol. 1996;178:1126–1133. doi: 10.1128/jb.178.4.1126-1133.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kravanja M, Engelmann R, Dossonnet V, Bluggel M, Meyer H E, Frank R, Galinier A, Deutscher J, Schnell N, Hengstenberg W. The hprK gene of Enterococcus faecalis encodes a novel bifunctional enzyme: the HPr kinase/phosphatase. Mol Microbiol. 1999;31:59–66. doi: 10.1046/j.1365-2958.1999.01146.x. [DOI] [PubMed] [Google Scholar]
- 18.Kuipers O P, Boot H J, de Vos W M. Improved site-directed mutagenesis method using PCR. Nucleic Acids Res. 1991;19:4558. doi: 10.1093/nar/19.16.4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee J M, Chung D K, Park J H, Lee W K, Chang H C, Kim J H, Lee H J. Cloning and nucleotide sequence of β-galactosidase gene from Lactococcus lactis ssp. lactis ATCC 7962. Biotechnol Lett. 1997;19:179–183. [Google Scholar]
- 20.Pao S S, Paulsen I T, Saier M H., Jr The major facilitator superfamily. Microbiol Mol Biol Rev. 1998;62:1–32. doi: 10.1128/mmbr.62.1.1-34.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Plamondon P, Brochu D, Thomas S, Fradette J, Gauthier L, Vaillancourt K, Buckley N, Frenette M, Vadeboncoeur C. Phenotypic consequences resulting from a methionine-to-valine substitution at position 48 in the HPr protein of Streptococcus salivarius. J Bacteriol. 1999;181:6914–6921. doi: 10.1128/jb.181.22.6914-6921.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poolman B, Royer T J, Mainzer S E, Schmidt B F. Lactose transport system of Streptococcus thermophilus: a hybrid protein with homology to the melibiose carrier and enzyme III of phosphoenolpyruvate-dependent phosphotransferase systems. J Bacteriol. 1989;171:244–253. doi: 10.1128/jb.171.1.244-253.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poolman B, Modderman R, Reizer J. Lactose transport system of Streptococcus thermophilus. J Biol Chem. 1992;267:9150–9157. [PubMed] [Google Scholar]
- 24.Poolman B, Knol J, Mollet B, Nieuwenhuis B, Sulter G. Regulation of bacterial sugar-H+ symport by phosphoenolpyruvate-dependent enzyme I/HPr-mediated phosphorylation. Proc Natl Acad Sci USA. 1995;92:778–782. doi: 10.1073/pnas.92.3.778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poolman B, Knol J, van der Does C, Henderson P J F, Liang W-J, Leblanc G, Pourcher T, Mus-Veteau I. Cation and sugar selectivity determinants in a novel family of transport proteins. Mol Microbiol. 1996;19:911–922. doi: 10.1046/j.1365-2958.1996.397949.x. [DOI] [PubMed] [Google Scholar]
- 26.Postma P W, Lengeler J W, Jacobson G R. Phosphoenolpyruvate: carbohydrate phosphotransferase systems. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: molecular and cellular biology. Washington, D.C.: ASM Press; 1996. pp. 1149–1174. [Google Scholar]
- 27.Pourcher T, Bibi E, Kaback H R, Leblanc G. Membrane topology of the melibiose permease of Escherichia coli studied by melB-phoA fusions analysis. Biochemistry. 1996;35:4161–4168. doi: 10.1021/bi9527496. [DOI] [PubMed] [Google Scholar]
- 28.Pullen K, Rajagopal P, Branchini B R, Huffine M E, Reizer J, Saier M H, Jr, Scholtz M, Klevit R E. Effects of phosphorylation of a serine onthe solution structure and stability of histidine-containing protein, a bacterial phosphotranfer protein. Protein Sci. 1996;4:2478–2486. doi: 10.1002/pro.5560041204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reizer J, Novotny M J, Hengstenberg W, Saier M H., Jr Properties of ATP-dependent protein kinase from Streptococcus pyogenes that phosphorylates a seryl residue in HPr, a phosphocarrier of the phosphotransferase system. J Bacteriol. 1984;160:333–3340. doi: 10.1128/jb.160.1.333-340.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reizer J, Peterkofsky A, Romano A H. Evidence of the presence of HPr and ATP-dependent HPr kinase in heterofermentative lactobacilli lacking a phosphoenolpyruvate:glucose phosphotransferase activity. Proc Natl Acad Sci USA. 1988;85:2041–2045. doi: 10.1073/pnas.85.7.2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reizer J, Sutrina S L, Saier M H, Jr, Stewart G C, Peterkofsky A, Reddy P. Mechanistic and physiological consequences of HPr(ser) phosphorylation on the activities of the phosphoenolpyruvate:phosphotransferase system in Gram-positive bacteria:studies with site-specific mutants of HPr. EMBO J. 1989;8:2111–2120. doi: 10.1002/j.1460-2075.1989.tb03620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reizer J, Hoischen C, Titgemeyer F, Rivolta C, Rabus R, Stulke J, Karamata D, Saier M H, Jr, Hillen W. A novel protein kinase that controls catabolite repression in bacteria. Mol Microbiol. 1998;27:1157–1169. doi: 10.1046/j.1365-2958.1998.00747.x. [DOI] [PubMed] [Google Scholar]
- 33.Romano A H, Brino G, Peterkofsky A, Reizer, J. J. Regulation of β-galactoside transport and accumulation in heterofermentative lactic acid bacteria. J Bacteriol. 1987;169:5589–5596. doi: 10.1128/jb.169.12.5589-5596.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saier M H, Jr, Chauvaux S, Deutscher J, Reizer J, Ye J J. Protein phosphorylation and regulation of carbon metabolism in Gram-negative versus Gram-positive bacteria. Trends Biol Sci. 1995;20:267–271. doi: 10.1016/s0968-0004(00)89041-6. [DOI] [PubMed] [Google Scholar]
- 35.Saier M H, Jr, Crasnier M. Inducer exclusion and the regulation of sugar transport. Res Microbiol. 1996;147:482–489. doi: 10.1016/s0923-2508(96)90150-3. [DOI] [PubMed] [Google Scholar]
- 36.Saier M H, Jr, Chauvaux S, Cook G M, Deutscher J, Paulsen I T, Reizer J, Ye J J. Catabolite repression and inducer control in Gram-positive bacteria. Microbiology. 1996;142:217–230. doi: 10.1099/13500872-142-2-217. [DOI] [PubMed] [Google Scholar]
- 37.Saier M H, Jr, Ye J J, Klinke S, Nino E. Identification of an anaerobically induced phosphoenolpyruvate-dependent fructose-specific phosphotransferase system and evidence for the Embden-Meyerhof glycolytic pathway in the heterofermentative bacterium Lactobacillus brevis. J Bacteriol. 1996;178:314–316. doi: 10.1128/jb.178.1.314-316.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saier M H, Jr, Beatty J T, Goffeau A, Harley K T, Heijne W H M, Huang S-C, Jack D L, Jahn P S, Lew K, Liu J, Pao S S, Paulsen I T, Tseng T-T, Virk P S. The major facilitator superfamily. J Mol Microbiol Biotechnol. 1999;1:257–279. [PubMed] [Google Scholar]
- 39.Saier M H., Jr A functional-phylogenetic classification system for transmembrane solute transporters. Microbiol Mol Biol Rev. 2000;64:254–411. doi: 10.1128/mmbr.64.2.354-411.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stülke J, Hillen W. Carbon catabolite repression in bacteria. Curr Opin Microbiol. 1999;2:195–201. doi: 10.1016/S1369-5274(99)80034-4. [DOI] [PubMed] [Google Scholar]
- 41.Sullivan M A, Yasbin R E, Young F E. New shuttle vectors for Bacillus subtilis and Escherichia coli, which allow rapid detection of inserted fragments. Gene. 1984;29:21–26. doi: 10.1016/0378-1119(84)90161-6. [DOI] [PubMed] [Google Scholar]
- 42.Vaughan E, David S, de Vos W M. The lactose transporter in Leuconostoc lactis is a new member of the LacS subfamily of galactoside-pentose-hexuronide translocators. Appl Environ Microbiol. 1996;62:1574–1582. doi: 10.1128/aem.62.5.1574-1582.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Viana R, Monedero V, Dossonnet V, Vadeboncoeur C, Perez-Martinez G, Deutscher J. Enzyme I and HPr from Lactobacillus casei: their role in sugar transport, carbon catabolite repression and inducer exclusion. Mol Microbiol. 2000;36:570–584. doi: 10.1046/j.1365-2958.2000.01862.x. [DOI] [PubMed] [Google Scholar]
- 44.Ye J J, Reizer J, Cui X, Saier M H., Jr ATP-dependent phosphorylation of serine in HPr regulates lactose:H+ symport in Lactobacillus brevis. Proc Natl Acad Sci USA. 1994;91:3102–3106. doi: 10.1073/pnas.91.8.3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ye J J, Saier M H., Jr Cooperative binding of lactose and HPr(Ser-P) to the lactose:H+ permease of Lactobacillus brevis. Proc Natl Acad Sci USA. 1995;92:417–421. doi: 10.1073/pnas.92.2.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ye J-J, Saier M H., Jr Allosteric regulation of the glucose:H+ symporter of Lactobacillus brevis: cooperative binding of glucose and HPr(Ser-P) J Bacteriol. 1995;177:1900–1902. doi: 10.1128/jb.177.7.1900-1902.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]