Significance
C-to-U RNA editing in plant organelles is carried out by a PPR-DYW protein or a protein complex that harnesses site recognition and deaminase activity. E-type PPR proteins can specify the editing site but lack the deaminase activity. This study shows that E-type PPR proteins specifically recruit trans deaminase MEF8/MEF8S, while GRP23, MORF1, and MORF8 facilitate this recruitment. In addition, some PPR-DYW proteins also need GRP23, presumably to recruit a second deaminase. This study reveals the functions of GRP23, MEF8/MEF8S, MORF1, and MORF8 and furthers our understanding of the RNA-editing mechanism in plant organelles.
Keywords: GRP23, editosome, Arabidopsis, mitochondrion, MORF
Abstract
Identifying the PPR-E+-NUWA-DYW2 editosome improves our understanding of the C-to-U RNA editing in plant organelles. However, the mechanism of RNA editing remains to be elucidated. Here, we report that GLUTAMINE-RICH PROTEIN23 (GRP23), a previously identified nuclear transcription regulator, plays an essential role in mitochondrial RNA editing through interacting with MORF (multiple organellar RNA-editing factor) proteins and atypical DYW-type pentatricopeptide repeat (PPR) proteins. GRP23 is targeted to mitochondria, plastids, and nuclei. Analysis of the grp23 mutants rescued by embryo-specific complementation shows decreased editing efficiency at 352 sites in mitochondria and 6 sites in plastids, with a predominant specificity for sites edited by the PPR-E and PPR-DYW proteins. GRP23 interacts with atypical PPR-DYW proteins (MEF8, MEF8S, DYW2, and DYW4) and MORF proteins (MORF1 and MORF8), whereas the four PPR-DYWs interact with the two MORFs. These interactions may increase the stability of the GRP23-MORF–atypical PPR-DYW complex. Furthermore, analysis of mef8N△64aamef8s double mutants shows that MEF8/MEF8S are required for the editing of the PPR-E protein–targeted sites in mitochondria. GRP23 could enhance the interaction between PPR-E and MEF8/MEF8S and form a homodimer or heterodimer with NUWA. Genetic complementation analysis shows that the C-terminal domains of GRP23 and NUWA possess a similar function, probably in the interaction with the MORFs. NUWA also interacts with atypical PPR-DYWs in yeast. Both GRP23 and NUWA interact with the atypical PPR-DYWs, suggesting that the PPR-E proteins recruit MEF8/MEF8S, whereas the PPR-E+ proteins specifically recruit DYW2 as the trans deaminase, and then GRP23, NUWA, and MORFs facilitate and/or stabilize the E or E+-type editosome formation.
Posttranscriptional C-to-U RNA editing alters the genetic information of DNA at the RNA level in plant chloroplasts and mitochondria (1, 2). In vascular plants, 30 to 40 editing sites are found in chloroplasts, whereas over 600 sites are in mitochondria (3, 4). RNA editing is considered a correction mechanism to reverse T-to-C mutations in the genomes to restore conserved residues that are usually essential for protein function (1, 2). Although RNA editing was first reported in plant mitochondria and plastids 30 y ago (5), the mechanism is not fully understood.
RNA editing is carried out by editing factors, which recognize the upstream sequences of the target cytidine (C) and then convert it to uridine (U) (6). Since 2005, many pentatricopeptide repeat (PPR) proteins have been found to be required for editing in mitochondria and chloroplasts (1, 2). PPR proteins contain a degenerate 35–amino acid (aa) repeat motif arrayed in tandem (7), which binds specifically to RNA sequences (8–12). PPR proteins are classified into P and PLS subfamilies according to the motifs. Based on the C-terminal domains, PLS subfamily PPRs are further divided into E, E+, and DYW subgroups (13, 14). The DYW domain was proven to possess the cytidine deamination activity (15, 16), as a single PPR-DYW protein could carry out the RNA editing in Physcomitrella patens. However, genetic and molecular evidence shows that RNA editing requires other factors in higher plants.
In higher-plant organelles, some PPR-E/E+ proteins can recruit a DYW-containing protein in trans to perform the deamination reaction. For example, the PPR-E protein CRR4 can recruit DYW1, a unique protein containing a mere DYW domain, to expedite editing at the ndhD-1 site in chloroplasts (17). Later, a general model for the E+-type PPR editosome in mitochondria and chloroplasts was proposed, in which a P-type PPR protein, NUWA, interacts with PPR-E+ proteins, such as SLO2 and CLB19, and an atypical PPR-DYW protein, DYW2, and enhances the interaction between PPR-E+s and DYW2 (18, 19). However, it is still unclear how PPR-E and PPR-DYW participate in RNA editing.
Besides PPRs, multiple organellar RNA-editing factors (MORFs) or RNA-editing interacting proteins were postulated to be a component of the RNA-editing complex (20, 21). The molecular function of MORF proteins is not entirely clear. MORF9 has been shown to increase the affinity of PPR motifs to their target RNAs in chloroplasts (22, 23). MORF proteins are proposed to bring PPR-E+s and DYW2 together (2), a similar function as NUWA. Although some progress has been made, the precise role of MORF proteins still needs to be deciphered.
GLUTAMINE-RICH PROTEIN23 (GRP23), a protein with 12 PPR motifs and a WQQ domain at its C terminus, was initially identified as a potential nuclear transcriptional regulator that interacts physically with RNA polymerase II subunit III (24). Loss of function of GRP23 arrests embryo development before the 16-cell dermatogen stage in Arabidopsis (24). Here, we show that GRP23 is localized in mitochondria, plastids, and nuclei and is required for the C-to-U editing at 358 sites, mostly in mitochondria. These sites are predominantly targeted by the E- and DYW-type PPR editing factors. Furthermore, we show that GRP23 interacts with atypical PPR-DYWs (DYW2, MEF8, MEF8S, and DYW4), MORF1, and MORF8. MORF1/MORF8 enhances the interaction between GRP23 and the atypical PPR-DYWs, and GRP23 enhances the interaction between PPR-Es and MEF8/MEF8S. These results indicate that GRP23 functions in RNA editing as a core member of the E-type editosomes in mitochondria. The E-type PPR editosome also contains MORF1 and MORF8.
Results
GRP23 Is Localized in Chloroplasts, Mitochondria, and Nuclei.
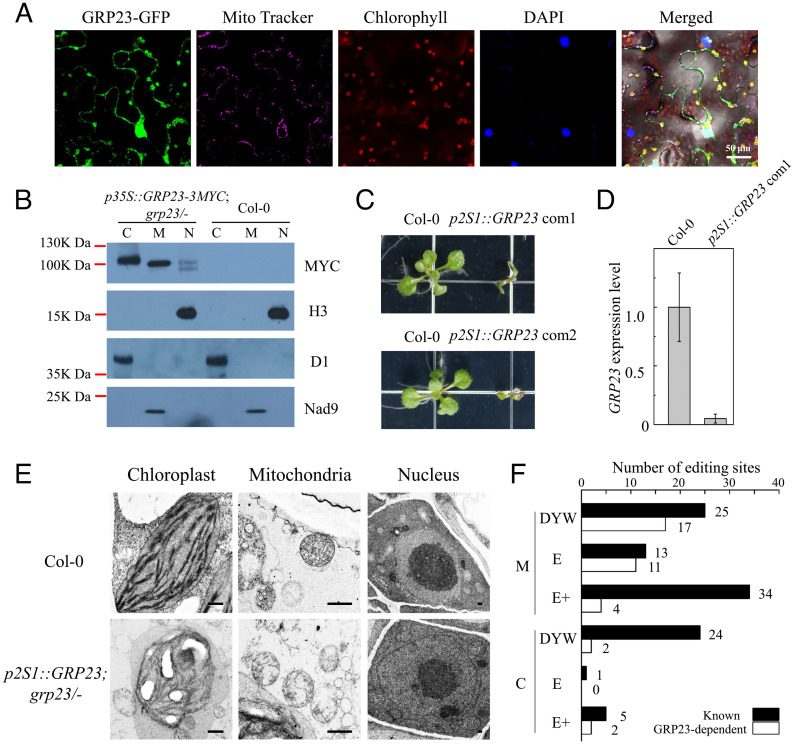
To localize GRP23, we fused GRP23 with green fluorescent protein (GFP) and then transiently expressed the fusion in tobacco leaves. Confocal microscopy revealed that GRP23-GFP signals were localized in chloroplasts, mitochondria, and nuclei (Fig. 1A). The subcellular localization of GRP23 was independently verified by immunoblotting. To do so, GRP23-3MYC driven by the CaMV 35S promoter was transformed into heterozygous grp23/+ plants (SALK_128329) of Arabidopsis. The expression of GRP23-3MYC rescued the grp23 embryo-lethal phenotype, and the rescued plants exhibited normal growth and development compared with the wild type (SI Appendix, Fig. S1), indicating that GRP23-3MYC has the GRP23 function. Then, the chloroplasts, mitochondria, and nuclei were fractionated from seedlings of the complemented mutants and the wild type, and the proteins were extracted and hybridized with MYC antibody. GRP23-3MYC was detected in the chloroplast, mitochondrial, and nuclear fractions but not in these fractions of the wild type (Fig. 1B). These results indicate that GRP23 is localized in chloroplasts, mitochondria, and nuclei. In addition, we found that the sizes of GRP23-3MYC were different in the three fractions. GRP23-3MYC appeared to be ∼100 kDa in chloroplasts but slightly smaller in mitochondria. In the nucleus, two sizes of GRP23-3MYC were detected, one similar to the mitochondrial one but the other marginally smaller (Fig. 1B). The different sizes of GRP23-3MYCs are probably caused by the cleavage of the transit and signal peptides.
Fig. 1.
GRP23 is required for the editing of the E- and DYW-type PPR-targeted sites in mitochondria. (A) Subcellular localization of the GRP23-GFP fusion in tobacco leaves. Young leaves of 4-wk-old tobacco were infiltrated with the cultures. At 22 to 24 h after infiltration, small pieces of infiltrated leaves were excised, soaked with phosphate-buffered saline containing MitoTracker Red and DAPI for 1 h, and observed under a Zeiss LSM 880 confocal microscope. In the merged picture, green plus purple: gray for mitochondrion; green plus red: yellow for chloroplast; purple plus blue: cyan for nucleus. (B) Determination of GRP23 localization by immunoblot assay. Protein fractions of mitochondria (M), chloroplasts (C), and nuclei (N) from the transgenic and the wild-type Arabidopsis were detected with antibody against MYC. The purity of the different fractions was tested with antibodies against the nuclear histone H3, the chloroplast D1, and the mitochondrial Nad9. (C) Phenotypes of the wild type and the grp23 mutants complemented by seed-specific expression of GRP23. Images of 2-wk-old seedlings grown on MS media were taken. (D) The expression of GRP23 in 2-wk-old seedlings of the wild type and the grp23 mutants complemented by seed-specific expression of GRP23. Data are mean ± SD of biological triplicates. (E) Cellular ultrastructure of the chloroplast, mitochondria, and nucleus of 2-wk-old seedlings from the wild type and the seed-specific complemented grp23 mutants. (Scale bars, 2 μm.) (F) RNA-editing analysis of the wild type and the seed-specific complemented grp23 mutants. The editing analysis of 2-wk-old seedlings of the wild type and two independent lines of seed-specific complemented grp23 mutants is shown. The editing status of the wild type and two independent lines of seed-specific complemented grp23 mutants was analyzed by STS-PCRseq and Sanger sequencing of PCR products. If the editing extent of one site in complemented grp23 mutants is decreased over 20% of that in the wild type, this site is considered as depending on GRP23. The known PPRs associated with editing sites are listed in Dataset S1A according to the literature (18, 19).
Depriving GRP23 Impairs Mitochondrial and Chloroplast Structures.
The arrest of embryos before the 16-cell dermatogen stage in grp23 hinders the use of the mutant to dissect the GRP23 function (24). To circumvent this obstacle, we used the At2S1 seed-specific promoter to express GRP23 specifically in the seeds. The At2S1 promoter confers a high level of expression at the torpedo stage but a low level at the cotyledon stage (25, 26). This partial complementation allows the formation of rescued grp23 seeds but deprives GRP23 after germination. Expression of p2S1::GRP23 rescued the grp23 seeds, but the grp23 seedlings only developed cotyledons and died in a few weeks (Fig. 1C and SI Appendix, Fig. S2). qRT-PCR analysis showed that the expression of GRP23 in the rescued seedlings is less than 10% of that in the wild type (Fig. 1D). Transmission electron microscopy analysis of 2-wk-old seedlings showed that the chloroplasts in the wild type developed an extensive thylakoid membrane system and contained small starch grains, whereas the chloroplasts in the rescued grp23 accumulated large starch grains, leading to abnormal chloroplast structure (Fig. 1E). The mitochondria in the rescued grp23 lacked distinctive cristae structures, in contrast to the mitochondria in the wild type that developed clear cristae structures and inner-membrane systems (Fig. 1E). No differences were detected in the nuclear morphology between the wild type and the rescued grp23 (Fig. 1E). These results suggest that the loss of GRP23 affects the structure and integrity of mitochondria and chloroplasts.
GRP23 Is Involved in the Editing of the E- and DYW-Type PPR-Targeted Sites.
As GRP23 shares a significant similarity with NUWA (19, 24), we first tested whether GRP23 is involved in RNA editing. STS-PCRseq (strand- and transcript-specific RT-PCRseq) (3) was used to analyze the editing status in the wild-type and the rescued grp23 seedlings. The RT-PCR amplicons of 34 mitochondrial transcripts and 17 chloroplast transcripts were quantified and mixed in an equimolar ratio. The complementary DNA (cDNA) mix was sheared by sonication and then used as a template for creating a TruSeq Nano DNA library. The sequencing of the wild type identified 587 editing sites in mitochondria and 33 sites in chloroplasts in Arabidopsis. Among these, 352 mitochondrial and 6 chloroplast sites showed an over 20% reduction of editing extent in the rescued grp23 mutants compared with the wild type (Dataset S1A). Sequencing the RT-PCR products of mitochondrial transcripts amplified from another rescued line showed similar results (SI Appendix, Fig. S3). Analysis against known PPR editing factors showed that the PPR-E– and PPR-DYW–targeted sites are enriched in the GRP23-mediated sites. In mitochondria, 11 of the 13 sites that are edited by the PPR-E proteins require GRP23, 17 of the 25 PPR-DYW–mediated sites are also dependent on GRP23, but only 4 of the 34 PPR-E+–mediated sites need GRP23 (Fig. 1F and Dataset S1A). In chloroplasts, 2 of the 24 PPR-DYW–dependent sites are mediated by GRP23, 2 of the 5 PPR-E+–mediated sites require GRP23, and no PPR-E–dependent site needs GRP23 (Fig. 1F). These results indicate that GRP23 is mainly required for the editing of PPR-E– and PPR-DYW–targeted sites in mitochondria. We also detected the splicing efficiency of mitochondrial introns in the wild type and the rescued mutants and found that the splicing efficiency of several introns is slightly decreased in the mutants (SI Appendix, Fig. S4). However, the splicing defect is not enough to cause the seedling-lethal phenotype of the rescued mutants and is probably a secondary effect of editing deficiency.
Identification of GRP23-Interacting Proteins by TurboID In Vivo.
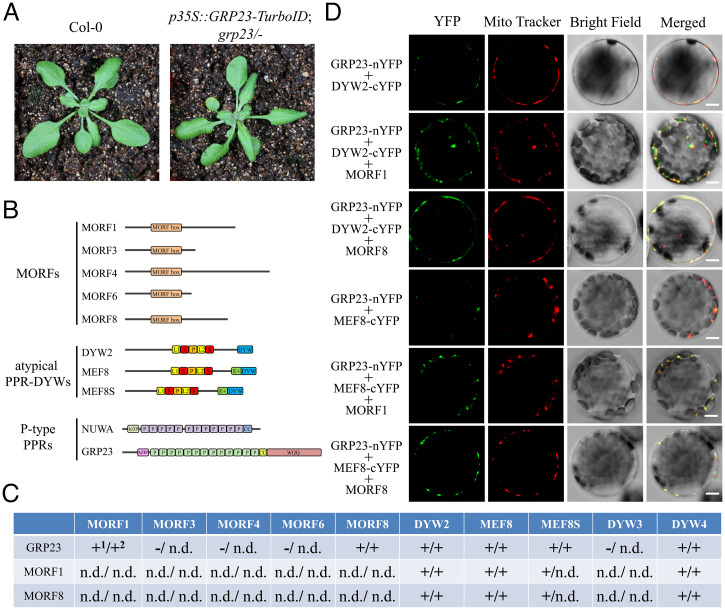
To explore the mechanism by which GRP23 participates in RNA editing, we performed TurboID-mediated proximity labeling analysis to capture the interacting partners of GRP23 (27). The full-length GRP23 cDNA was fused to a TurboID, and the fusion was expressed under the control of the CaMV 35S promoter. The expression of the 35S::GRP23-TurboID transgene completely rescued the embryo-lethal phenotype of grp23 (Fig. 2A and SI Appendix, Fig. S5), indicating that the GRP23-TurboID is functional. The biotinylated proteins from mitochondria of the complemented plants were extracted by streptavidin beads and identified by mass spectrometry. Ten potentially GRP23-interacting proteins were identified, including three atypical PPR-DYW proteins (MEF8, MEF8S, and DYW2), five MORF proteins (MORF1, MORF3, MORF4, MORF6, and MORF8), and two P-type PPR proteins (NUWA and GRP23) (Fig. 2B and Dataset S2).
Fig. 2.
Protein interactions between GRP23 and MORF1, MORF8, DYW2, MEF8, MEF8S, and DYW4. (A) Phenotypes of the wild type and the grp23 mutant complemented by p35S::GRP23-TurboID. Images of 4-wk-old plants were taken. (B) Scheme of the primary structure of GRP23-interacting proteins identified by TurboID-mediated proximity labeling. (C) Summary of the protein interactions between GRP23 and MORF1, MORF3, MORF4, MORF6, MORF8, DYW2, MEF8, MEF8S, and DYW4 in mitochondria detected by Y2H1 and BiFC2 assays shown in SI Appendix, Figs. S6 and S9. “+” represents interaction; “-” represents no interaction; n.d., not detected. (D) MORF1 and MORF8 increase the interactions between GRP23 and two atypical PPR-DYW proteins DYW2 and MEF8 in mitochondria. Arabidopsis protoplasts transformed with GRP23-nYFP together with DYW2-cYFP or MEF8-cYFP, in the absence or presence of MORF1 or MORF8, were observed by confocal microscopy 24 h after transformation. All images were obtained using the same setting and are representative of at least three independent experiments. (Scale bars, 5 μm.)
GRP23 Interacts with DYW2, MEF8, MEF8S, and DYW4.
Five atypical PPR-DYW proteins are present in Arabidopsis, including DYW2, DYW3, DYW4, MEF8, and MEF8S (19, 28). The identification of three atypical PPR-DYW proteins in the proximity labeling analysis prompted us to explore the interactions between GRP23 and all the atypical PPR-DYWs. We used yeast two-hybrid (Y2H) and bimolecular fluorescence complementation (BiFC) analyses to test the interactions. Y2H analysis indicated that GRP23 could interact with DYW2, MEF8, MEF8S, and DYW4 but not with DYW3 (Fig. 2C and SI Appendix, Fig. S6A). Delineation analysis showed that the N-terminal region and the PPR motif of GRP23 could mediate its interaction with atypical PPR-DYWs (SI Appendix, Fig. S6 B and C). DYW3 appears to be a pseudogene as its expressed sequence tags cannot be found, and RT-PCR failed to detect its expression in major tissues (SI Appendix, Fig. S7). BiFC analysis showed that GRP23 could interact with DYW2, MEF8, MEF8S, and DYW4 (Fig. 2C and SI Appendix, Fig. S6D). Coexpression of GRP23-nYFP (GRP23 fused with the N-terminal half of yellow fluorescent protein [YFP]) and DYW2-cYFP (DYW2 fused with the C-terminal half of YFP), MEF8-cYFP, MEF8S-cYFP, or DYW4-cYFP, respectively, reconstituted a functional YFP in mitochondria of protoplasts (SI Appendix, Fig. S6D).
GRP23 Interacts with MORF1 and MORF8.
Next, we tested the interactions between GRP23 and the MORF proteins. The Arabidopsis genome contains seven mitochondrial MORF proteins, MORF1, 3, 4, 5, 6, 7, and 8 (20, 21). All of these proteins were tested using the Y2H analysis. In yeast, GRP23 showed weak interactions with MORF1 and MORF8 but no interactions with MORF3, 4, 5, 6, and 7 (Fig. 2C and SI Appendix, Fig. S8A). The difference between the proximity labeling and the Y2H results suggests a possibly unstable interaction between GRP23 and MORF proteins. Proximity labeling is able to detect relatively weak and transient interactions (27). Additionally, the interactions may be missed in the Y2H assay due to improper folding of proteins in this heterologous system (29). We further analyzed the interactions between GRP23 with MORF1 and MORF8 by BiFC. As shown in SI Appendix, Fig. S8B, coexpression of GRP23-nYFP with MORF1-cYFP, MORF1-nYFP with GRP23-cYFP, GRP23-nYFP with MORF8-cYFP, or MORF8-nYFP with GRP23-cYFP reconstituted a functional YFP in mitochondria of protoplasts, confirming the interactions.
MORF1 and MORF8 Interact with Atypical PPR-DYWs.
GRP23 interacts with both atypical PPR-DYW and MORF proteins, and thus we speculated that atypical PPR-DYWs likely interact with MORF proteins. Y2H results showed that MORF1 and MORF8 interact with atypical PPR-DYWs, including DYW2, MEF8, MEF8S, and DYW4 (Fig. 2C and SI Appendix, Fig. S9A). The interactions between MORF proteins MORF1 and MORF8 and atypical PPR-DYWs were further confirmed by BiFC (Fig. 2C and SI Appendix, Fig. S9B). These results suggest that GRP23, MORF proteins, and atypical PPR-DYWs interact with each other.
GRP23, MORFs, and Atypical PPR-DYWs Are Probably Present in a Protein Complex.
The interactions of GRP23, MORF proteins, and atypical PPR-DYWs suggest that these proteins are probably present in a protein complex. We performed BiFC to test this hypothesis. When GRP23 was coexpressed with either DYW2 or MEF8 in Arabidopsis protoplasts, relatively weak YFP signals were detected in the mitochondria of protoplasts (Fig. 2D). When MORF1 or MORF8 was expressed in these protoplasts, however, the YFP signals in mitochondria were enhanced in parallel experiments (Fig. 2D). These results indicate that GRP23, MORF1 or MORF8, and DYW2 or MEF8 are likely to be present in a protein complex and suggest that these protein interactions probably increase the stability of GRP23-MORF-atypical PPR-DYW complexes.
MEF8 and MEF8S Are Involved in the Editing of the E-Type PPR-Targeted Sites.
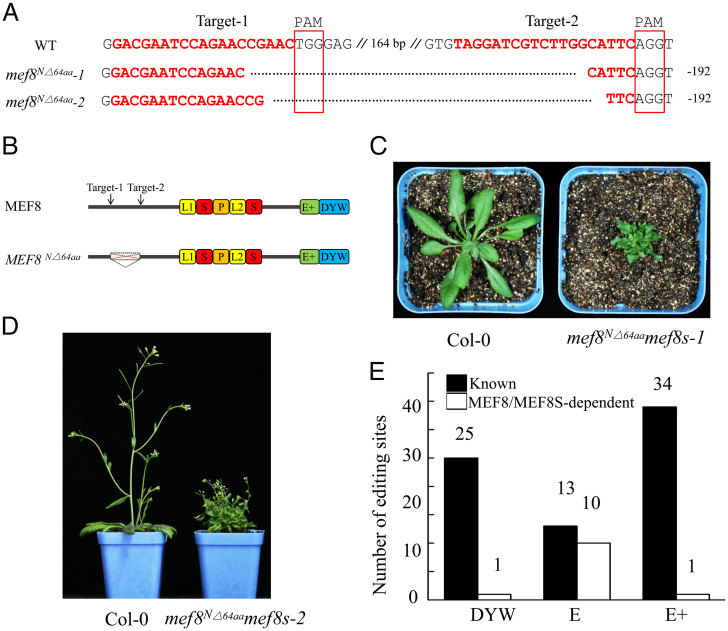
The functions of MEF8 and MEF8S in RNA editing have been analyzed in single mutants, which are viable (28, 30). However, the mef8mef8s double mutants are embryo-lethal, suggesting that these two proteins are functionally redundant (28). Thus, the function of MEF8/MEF8S remains unclear. To identify the function, we created mef8-knockout mutants using CRISPR-Cas9 in the mef8s-null allele background (SALK_047005C) and meanwhile partially rescued the double mutants by expressing MEF8S under the control of the seed-specific At2S1 promoter. However, the partially complemented plants could not be isolated after extensive attempts. Instead, we isolated two weak mutants of MEF8 in the mef8s mutant background, which contained deletions of the sequences between the two CRISPR target sites (Fig. 3A), resulting in the deletion of 64 aa at the N terminus of MEF8 (Fig. 3B). The mutants were named mef8N△64aamef8s. The mef8N△64aamef8s mutants exhibited curled leaf and sterile flower phenotypes (Fig. 3 C and D). Editing analysis of the mef8N△64aamef8s-1 mutants by STS-PCRseq revealed that the editing at 151 sites in mitochondria was impaired (Dataset S1B). Sanger sequencing of the RT-PCR products of the mitochondrial genes from the mef8N△64aamef8s-2 mutants confirmed the STS-PCRseq result (SI Appendix, Fig. S10). Further analysis indicated that most of the affected sites in the mef8N△64aamef8s mutants are targeted by the PPR-E proteins (Fig. 3E and Dataset S1B), suggesting that the PPR-E proteins probably recruit MEF8/MEF8S as the trans deaminase.
Fig. 3.
MEF8 and MEF8S are required for the editing of the E-type PPR-targeted sites in mitochondria. (A) Alignment of sequences of mutated alleles generated by CRISPR-Cas9. The fragments between two target sites were deleted. PAM, protospacer adjacent motif; WT, wild type. (B) Schematic illustration of MEF8 protein. The deleted region is indicated. (C and D) Morphology of the wild type and the mef8N△64aamef8s mutants. The seeds were germinated on MS media, and 1-wk-old seedlings were transferred to soil and grown in the greenhouse under long-day conditions (16 h light/8 h darkness). Images of a 4-wk-old wild-type plant and 6-wk-old mef8N△64aamef8s-1 mutant (C) and 6-wk-old wild-type plant and 10-wk-old mef8N△64aamef8s-2 mutant (D) were taken. The mef8N△64aamef8s mutants were slow-growing and produced sterile flowers. (E) RNA-editing analysis of the wild type and the mef8N△64aamef8s mutants. The editing analysis of 4-wk-old seedlings of the wild type and two independent lines of mef8N△64aamef8s mutants is shown. The editing status of the wild type and two independent lines of mef8N△64aamef8s mutants in mitochondria was analyzed by STS-PCRseq and Sanger sequencing of PCR products, respectively. If the editing extent of one site in the mef8N△64aamef8s mutant is decreased over 20% of that in the wild type, this site is considered as depending on MEF8/MEF8S. The known PPRs associated with editing sites are listed in Dataset S1B according to the literature (18, 19).
GRP23, NUWA, and MEF8/MEF8S Form Dimers.
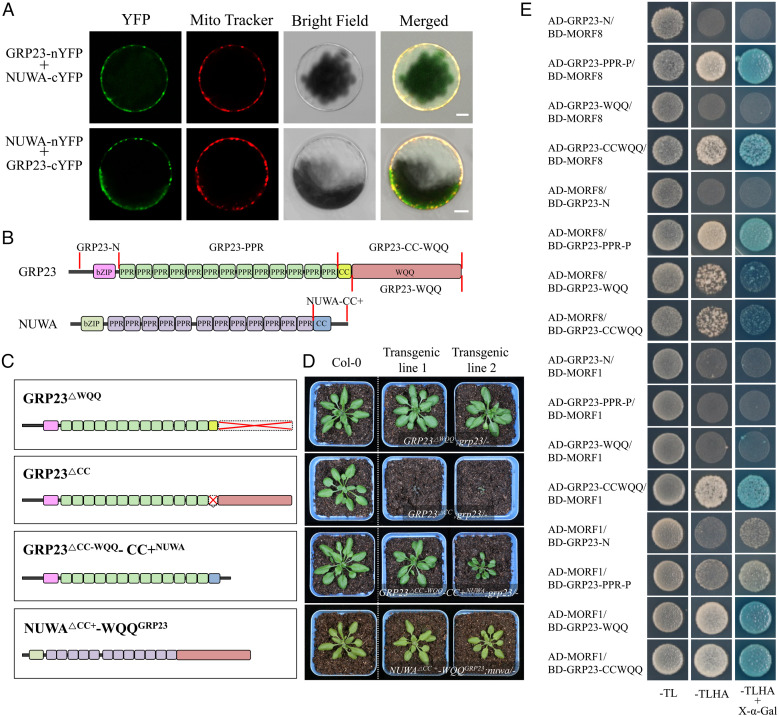
The proximity labeling result suggests that GRP23 may form homodimers and heterodimers with NUWA (Dataset S2). We tested the interactions by BiFC assay. The results showed that GRP23 could interact with itself and NUWA in mitochondria (Fig. 4A and SI Appendix, Fig. S11A). GRP23 homodimerization was also observed in the luciferase complementation assay (SI Appendix, Fig. S11B). MEF8 and MEF8S showed the capacity to form homodimers and heterodimers in mitochondria (SI Appendix, Fig. S11A).
Fig. 4.
CC and WQQ domains of GRP23 and the C terminus of NUWA have similar function and probably interact with MORF1 and MORF8. (A) Detection of the interaction between GRP23 and NUWA in Arabidopsis protoplasts. (Scale bars, 5 μm.) (B) Schematic illustrations of GRP23 and NUWA. (C) Chimeric proteins between GRP23 and NUWA were created to complement the grp23 or nuwa mutant. (D) Morphology of the wild type and grp23 or nuwa mutant complemented by chimeric proteins. The seeds were geminated on MS media, and 1-wk-old seedlings were transferred to soil and grown in the greenhouse under long-day conditions (16 h light/8 h darkness). Images of 4-wk-old plants of the wild type and the complemented mutants were taken. (E) The interactions between the truncations of GRP23 with MORF1 and MORF8. −TL, −TLHA, and −TLHA+X-α-Gal indicate SD/−Trp−Leu, SD/−Trp−Leu−His−Ade, and SD/−Trp−Leu−His−Ade containing X-α-Gal dropout plates, respectively. Images were taken after 3 d of incubation at 30 °C.
The CC and WQQ Domains of GRP23 and the C Terminus of NUWA Have Similar Functions and Probably Interact with MORF1 and MORF8.
The coiled-coil (CC) and WQQ domains are proposed to mediate protein–protein interactions (24, 31). The CC domain of NUWA (CCNUWA) is essential for plant survival in Arabidopsis (32), indicating an important role of CCNUWA for NUWA function. Compared with the conserved CC domain, CCNUWA is more conserved than the CC domain in GRP23 (CCGRP23). It is possible that the CC, WQQ, or CC-WQQ domains of GRP23 function similar to CCNUWA. To address that question, we tested whether CC-WQQGRP23, CCGRP23, or WQQGRP23 is required for survival. Three deletion variants, GRP23△CC, GRP23△WQQ, and GRP23△CC-WQQ, were created and used to complement the grp23 mutant genetically (Fig. 4C). Expression of GRP23△CC-WQQ cannot rescue grp23, as no rescued grp23 mutants were identified in a screen of 120 T2 transformants of GRP23△CC-WQQ, indicating an essential function of the CC-WQQ domain. However, the expression of GRP23△CC or GRP23△WQQ recused the grp23 mutant phenotype. The grp23 mutants complemented by GRP23△WQQ grew relatively normally (Fig. 4D). However, the mutants complemented by GRP23△CC grew rather slowly (Fig. 4D). These results suggest that both CCGRP23 and WQQGRP23 are not essential to the GRP23 functions, but CCGRP23 is functionally more crucial than WQQGRP23. To further explore the functional relationship between CCNUWA, CCGRP23, and WQQGRP23, we created chimeric proteins by exchanging the C-terminal region between GRP23 and NUWA and tested the complementation of grp23 or nuwa. GRP23△CC-WQQ-CC+NUWA is GRP23 with its CC-WQQ region replaced by the entire C-terminal regions (CC+) of NUWA (CC+NUWA; Fig. 4C). NUWA△CC+-WQQGRP23 is NUWA with its C terminus replaced by WQQGRP23. The nuwa mutants were complemented by expressing NUWA△CC+-WQQGRP23, and the grp23 mutants were complemented by expressing GRP23△CC-WQQ-CC+NUWA, evidenced by the relatively normal growth of the plants (Fig. 4D). This result suggests that CC+NUWA, CCGRP23, and WQQGRP23 likely have a similar function.
As the N terminus of GRP23 interacts with atypical PPR-DYWs, we speculated that the C terminus of GRP23 might interact with MORF proteins. In yeast cells, both WQQGRP23 and CC-WQQGRP23 domains showed interaction with MORF1 and MORF8, respectively (Fig. 4E). The PPR motifs also interacted with MORF1 and MORF8 (Fig. 4E).
GRP23 Assists the Interaction between MEF8 and PPR-E Proteins OTP71 and OTP87.
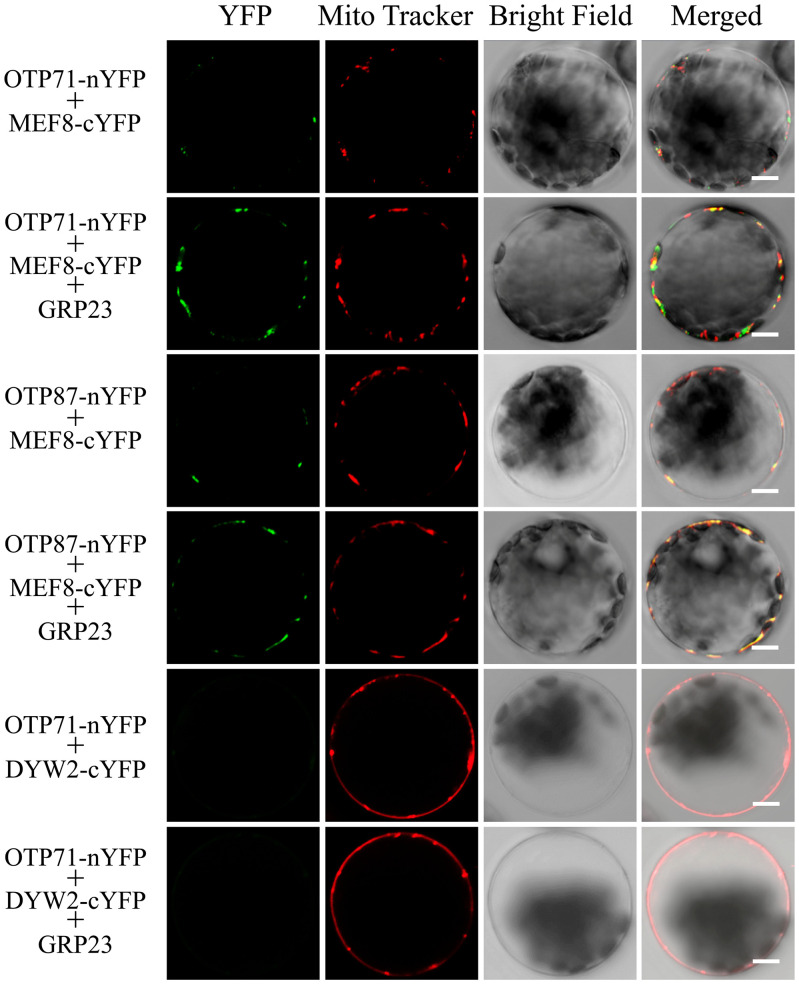
P-type PPR protein NUWA participates in E+-type PPR editing by assisting the interaction between PPR-E+ and DYW2 (18, 19). Therefore, we propose that GRP23 may enhance the interaction between PPR-E and MEF8/MEF8S. The editing at the ccmFN2-176 and atp1-1178 sites requires PPR-E proteins OTP71 and OTP87 (33, 34), respectively, and GRP23 and MEF8/MEF8S (Dataset S1 A and B). The interactions between these three proteins were analyzed by BiFC analysis. OTP71-nYFP and MEF8-cYFP, or OTP87-nYFP and MEF8-cYFP, showed a rather weak interaction in the mitochondria of Arabidopsis protoplasts, as indicated by the signals (Fig. 5). However, coexpression of wild-type GRP23 substantially increased the signals (Fig. 5), suggesting GRP23 enhances the interaction between MEF8 and the two OTP-PPR proteins. In contrast, OTP71 did not show any interaction with DYW2 in the absence or presence of GRP23 (Fig. 5).
Fig. 5.
GRP23 enhances the interactions between MEF8 and PPR-E proteins OTP71 and OTP87 in mitochondria. Arabidopsis protoplasts transformed with OTP71-nYFP or OTP87-nYFP together with MEF8-cYFP, in the absence or presence of GRP23, were observed by confocal microscopy 24 h after transformation. The PPR-E protein OTP71 is not DYW2-dependent and the interaction between OTP71 and DYW2 was used as a negative control. All images were obtained using the same setting and are representative of at least three independent experiments. (Scale bars, 5 μm.)
NUWA Interacts with Atypical PPR-DYWs.
Excluding predicted signal peptide and/or transit peptide regions, the remaining N termini of GRP23 and NUWA share 54% identity (SI Appendix, Fig. S12). As GRP23 interacts with atypical PPR-DYWs via its N-terminal region, we propose that NUWA also likely interacts with atypical PPR-DYWs. Y2H results showed that NUWA interacts with atypical PPR-DYWs, including DYW2, MEF8/MEF8S, and DYW4 (SI Appendix, Fig. S13).
Discussion
A single PPR-DYW protein can catalyze the C-to-U editing in P. patens mitochondria (15, 16), which is probably the simplest model of RNA editing. However, during the rapid expansion of PPR proteins in flowering plants, PPR proteins have diversified. Some PPR-DYW proteins lost the DYW domain to form PPR-E+ subclass proteins, and some lost the E+-DYW domain, forming PPR-E subclass proteins. Because the DYW domain confers the cytidine deaminase activity, the PPR-E and PPR-E+ subclasses of proteins need to recruit a cytidine deaminase in trans to catalyze the deamination reaction. PPR-E+ proteins have been found to recruit DYW2 as the trans deaminase (18, 19). The interaction between PPR-E+ proteins and DYW2 is weak. Thus, a unique P-type PPR protein, NUWA, is recruited to enhance this interaction (18, 19). So far, it is unclear how PPR-E proteins recruit the deaminase and whether PPR-DYW proteins require additional deaminase. This work demonstrates that MEF8/MEF8S provide the deaminase activity for the PPR-E proteins via protein interaction, and GRP23 facilitates this interaction by interacting with both proteins. GRP23 is also involved in the editing of PPR-DYW–targeted sites, possibly providing an additional deaminase. MORF proteins, MORF1 and MORF8, increase the stability of the E-type editosome.
The nature of editosomes in plant organelles is to form a protein complex that harnesses the target site recognition function and the deaminase activity. The structural features of PPR-E and PPR-E+ proteins may dictate which trans deaminase they recruit (35). PPR-E proteins lack the E+ and DYW domains, thus recruiting MEF8/MEF8S to supply the E+ and DYW domains (SI Appendix, Fig. S14). In contrast, PPR-E+ proteins lack the DYW domain, which can be provided by DYW2 (18, 19). These interactions result in the reconstruction of an editosome that possesses site specification and deaminase activity. The specific recruitment of DYW2 or MEF8/MEF8S may also be dictated by the gating domain that is essential to the deaminase activity (36). PPR-E+ proteins contain the first half (α-helix) of the gating domain, but PPR-E proteins do not. On the partner proteins, DYW2 has the second half of the gating domain (two β-strands), but MEF8/MEF8S have a complete gating domain. Thus, the gating domain may be supplied in trans through specific interactions.
The decrease in editing extent in the grp23 mutant never reaches complete obliteration. This raises the question of whether there is another protein with a redundant function to GRP23 or it is due to a residual expression of GRP23 in the rescued seedlings. The closest homolog of GRP23 in Arabidopsis is NUWA, sharing 34% aa identity (19). If GRP23 is redundant to NUWA, the knockout mutant of each may not show an embryo-lethal phenotype. Additionally, the functions of GRP23 and NUWA have significant differences (Fig. 1F) (18, 19). Therefore, the possibility of the existence of a redundant protein is ruled out. We detected a residual expression of GRP23 in the rescued seedlings (Fig. 1D). The residual transcripts can maintain the rescued plants in the seedling stage for a few weeks before dying.
The reduced RNA-editing efficiency at some sites in the mef8N△64aamef8s or grp23 mutants is likely to be a secondary effect. It is reported that the reduction of an editing event leads to a lack of downstream editing, as reduced RNA editing within the PPR protein-binding sequence would affect the editing efficiency of the downstream editing sites (37–39). A similar scenario is likely to happen to the mef8N△64aamef8s or grp23 mutant. Some RNA-editing defects in the mef8N△64aamef8s or grp23 mutant could be due to the secondary effect of the defect in the PPR-E protein target sequences. However, we think this secondary effect is not the cause of the editing deficiency of some PPR-DYW–targeted sites in the grp23 mutants. First, our data showed that the editing at 17 of the 25 PPR-DYW–targeted sites requires GRP23 (Fig. 1F). Second, we checked the cis-element sequences of the 17 PPR-DYW–targeted sites, and 13 sequences are not affected by editing. Therefore, based on our current data, we think GRP23 is involved in the editing of PPR-DYW–targeted sites.
In the mef8N△64aamef8s mutant, the N-terminal region of MEF8 from 62 to 125 aa is deleted in the mef8s mutant background (Fig. 3B), impairing the function of MEF8. This region is rich in glutamine residues (SI Appendix, Fig. S15). Interestingly, both DYW2 and MEF8S contain glutamine-rich regions as well (SI Appendix, Fig. S15). As the glutamine-rich region has been linked with protein–protein interactions (40), this region in DYW2 and MEF8/MEF8S may mediate the interaction with other editing factors, such as PPRs, GRP23, and NUWA.
Thirty-eight mitochondrial sites exhibited a reduction of editing extent in the single mef8 mutant (30). Strangely, none of them are targeted by known PPR-E proteins (30). One possibility is that some of these sites are indeed targeted by the PPR-E proteins, but these factors have not been identified. This explanation raises the question of why MEF8S could complement the function of MEF8 in the mef8 mutant by providing deaminase for those known PPR-E proteins but not for these unknown ones. The protein length difference between MEF8 and MEF8S at the N terminus suggests MEF8 may have a unique function (Fig. 2B), probably in interacting with those unidentified PPR-E proteins. It is also possible that these 38 sites are not targeted by PPR-E proteins. In this scenario, MEF8 has another function in addition to providing deaminase for PPR-E proteins. The alignment of the upstream sequences of the main targeted sites of MEF8 reveals some conserved nucleotides, suggesting that MEF8 might still recognize and bind its RNA targets (30).
The editing of some sites is increased in the mef8N△64aamef8s mutant (Dataset S1B). The increase was also observed in the single mef8 mutant (30). It is proposed that MEF8 also has an inhibitory effect on editing by occupying the upstream sequences of editing sites, which should be bound by PPR proteins (30). The DYW domain of MEF8 does not play a role in this process (30).
Our results reveal that MORF family proteins MORF1 and/or MORF8 are components of the E-type editosome. Although MORF proteins have been shown to be involved in RNA editing in plant organelles, the precise functions of those proteins are still poorly understood. A previous study shows that a chloroplast MORF protein, MORF9, increases the RNA-binding activity of PPR-PLS protein with substrate RNA (22, 23). We found that MORF1 and MORF8 stabilize the RNA editosomes through interacting with GRP23 and atypical PPR-DYWs (Fig. 2 C and D). Interestingly, the interactions between GRP23 and MORFs are weak and transient. On the one hand, the weak interaction is conducive to the formation of the editosome; on the other hand, it may promote the disassembly of the editosome. The MORF proteins interact with both GRP23 and PPR-E/E+ proteins. When the deamination reaction is in an inactive state, the interaction promotes the stability of the editosome. Upon activation, the DYW gating domain (E+ domain) changes its conformation (36). As MORF proteins bind to PPR-E and PPR-E+ through the E domain and the E domain is close to the E+ domain (41), the conformational change may destroy the interaction between MORF proteins and PPR-E or PPR-E+, promoting the disassembly of the editosome after the deamination process.
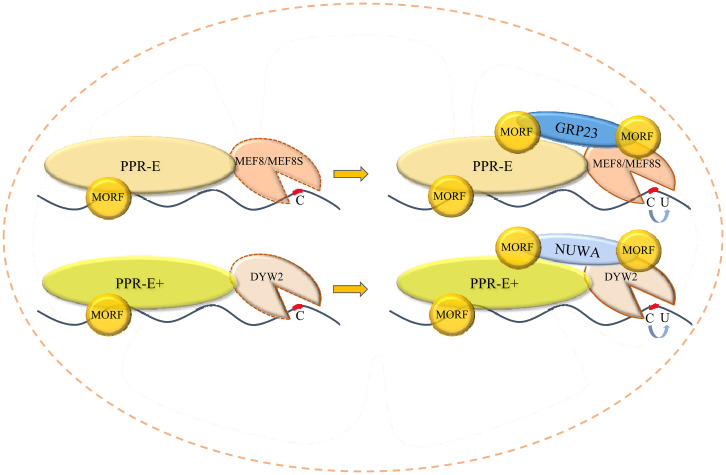
The organellar editosome is probably assembled in order (Fig. 6). GRP23 and NUWA can interact with MEF8, MEF8S, and DYW2 (Fig. 2C). However, PPR-Es specifically recruit MEF8/MEF8S (Fig. 5), and PPR-E+s recruit DYW2 (18, 19). If GRP23 or NUWA first interacts with atypical PPR-DYWs, the specific composition of the editosome is disrupted. Therefore, the combination of either PPR-E with MEF8/MEF8S or PPR-E+ with DYW2 is probably the first step in the assembly of the editosome. Then GRP23 or NUWA, possibly with a homo- or heterodimer form (Fig. 4A and SI Appendix, Fig. S11), binds to at least two positions with PPR-E-MEF8/MEF8S or PPR-E+-DYW2. The N terminus of GRP23 connects with MEF8/MEF8S (SI Appendix, Fig. S6C), and the CC-WQQ domain of GRP23 may connect with PPR-E. As the N-terminal sequences of NUWA and GRP23 share high similarity and both the CC and WQQ domains of GRP23 and the C-terminal domain of NUWA have the same function, NUWA likely interacts with PPR-E+-DYW2 in the same way as GRP23 interacts with PPR-E-MEF8/MEF8S. At least three MORF proteins are probably involved in the editosome. One increases the binding of PPR protein and RNA substrate (22); one interacts with both GRP23 and PPR protein; and one connects with both GRP23 and atypical PPR-DYWs.
Fig. 6.
Models of the assembly of E- and E+-type editosomes in mitochondria.
According to the RNA-editing analysis of the rescued grp23 mutant, most PPR-DYW proteins require the assistance of GRP23, suggesting PPR-DYWs likely need additional deaminase. However, the editing analysis results show that most PPR-DYW–dependent sites are not DYW2- and MEF8/MEF8S-dependent (Dataset S1B) (18, 19). We propose three possibilities to explain this. One possibility is that DYW2 and MEF8/MEF8S are redundant in providing deaminases for PPR-DYW. The interactions between GRP23 and DYW2 and MEF8/MEF8S offer a basis for this possibility. In this scenario, PPR-DYW forms a heterodimer with atypical PPR-DYWs. Indeed, DYW2 (18) and MEF8/MEF8S likely provide deaminase activity in dimer form (SI Appendix, Fig. S11A). Another possible explanation is that DYW4 is the trans deaminase for PPR-DYWs. The function of DYW4 has not been identified, and further studies are necessary to elucidate its molecular function. It is also possible that the interaction between GRP23 and PPR-DYW changes the conformation of the latter, making it easier to connect the target C without the need for an additional deaminase.
Materials and Methods
Plant Materials and Growth Conditions.
The grp23 (SALK_128329) and mef8s (SALK_047005C) mutants were obtained from the Arabidopsis Biological Resource Center. Arabidopsis seeds were sown on Murashige and Skoog (MS) medium containing 3% sucrose and 2 g/L Gelzan, and plants were grown at 22 °C under 16 h light/8 h darkness.
BiFC.
Vectors were cotransformed into Arabidopsis protoplasts as previously described (42). The protoplasts were incubated under weak light for 18 to 22 h, and the YFP fluorescence was observed using a confocal microscope under the same setting. For details, see SI Appendix, Materials and Methods.
Y2H Assay.
The sequences of genes of interest encoding the mature proteins without signal/transit peptides were ligated into the bait (pGBKT7) and prey (pGADT7) vectors, and the resulting constructs were cotransformed into the yeast strain Y2H Gold. The transformants were selected on Synthetic Dropout (SD)/−Trp/−Leu/−His/−Ade and SD/−Trp/−Leu/−His/−Ade/X-α-Gal plates. For details, see SI Appendix, Materials and Methods.
RNA-Editing Assay.
The editing efficiency was analyzed by STS-PCRseq and direct sequencing of the gene-specific RT-PCR products. For details, see SI Appendix, Materials and Methods.
TurboID-Mediated Proximity Labeling.
The TurboID sequence was amplified from the pENTR_L1-YFP-Turbo-L2 plasmid (Addgene). The 2-wk-old seedlings of grp23 mutants complemented by 35S-GRP23-TurboID were treated with 50 μM biotin. Biotin-labeled proteins were enriched using Dynabeads M-280 streptavidin and then analyzed by tandem mass spectrometry. For details, see SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
This research was supported by the National Natural Science Foundation of China (Project Nos. 32072126 and 31630053) and the Shandong Provincial Natural Science Foundation (Project No. ZR2019MC005).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2210978119/-/DCSupplemental.
Data, Materials, and Software Availability
The data reported in this paper have been deposited in the Sequence Read Archive database, https://www.ncbi.nlm.nih.gov/sra [accession no. PRJNA849404 (43)].
All study data are included in the article and/or supporting information.
References
- 1.Barkan A., Small I., Pentatricopeptide repeat proteins in plants. Annu. Rev. Plant Biol. 65, 415–442 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Small I. D., Schallenberg-Rüdinger M., Takenaka M., Mireau H., Ostersetzer-Biran O., Plant organellar RNA editing: What 30 years of research has revealed. Plant J. 101, 1040–1056 (2020). [DOI] [PubMed] [Google Scholar]
- 3.Bentolila S., Oh J., Hanson M. R., Bukowski R., Comprehensive high-resolution analysis of the role of an Arabidopsis gene family in RNA editing. PLoS Genet. 9, e1003584 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stern D. B., Goldschmidt-Clermont M., Hanson M. R., Chloroplast RNA metabolism. Annu. Rev. Plant Biol. 61, 125–155 (2010). [DOI] [PubMed] [Google Scholar]
- 5.Covello P. S., Gray M. W., RNA editing in plant mitochondria. Nature 341, 662–666 (1989). [DOI] [PubMed] [Google Scholar]
- 6.Farré J. C., Leon G., Jordana X., Araya A., cis recognition elements in plant mitochondrion RNA editing. Mol. Cell. Biol. 21, 6731–6737 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Small I. D., Peeters N., The PPR motif—A TPR-related motif prevalent in plant organellar proteins. Trends Biochem. Sci. 25, 46–47 (2000). [DOI] [PubMed] [Google Scholar]
- 8.Ke J., et al. , Structural basis for RNA recognition by a dimeric PPR-protein complex. Nat. Struct. Mol. Biol. 20, 1377–1382 (2013). [DOI] [PubMed] [Google Scholar]
- 9.Barkan A., et al. , A combinatorial amino acid code for RNA recognition by pentatricopeptide repeat proteins. PLoS Genet. 8, e1002910 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takenaka M., Zehrmann A., Brennicke A., Graichen K., Improved computational target site prediction for pentatricopeptide repeat RNA editing factors. PLoS One 8, e65343 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yagi Y., Hayashi S., Kobayashi K., Hirayama T., Nakamura T., Elucidation of the RNA recognition code for pentatricopeptide repeat proteins involved in organelle RNA editing in plants. PLoS One 8, e57286 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yin P., et al. , Structural basis for the modular recognition of single-stranded RNA by PPR proteins. Nature 504, 168–171 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Lurin C., et al. , Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 16, 2089–2103 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng S., et al. , Redefining the structural motifs that determine RNA binding and RNA editing by pentatricopeptide repeat proteins in land plants. Plant J. 85, 532–547 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Oldenkott B., Yang Y., Lesch E., Knoop V., Schallenberg-Rüdinger M., Plant-type pentatricopeptide repeat proteins with a DYW domain drive C-to-U RNA editing in Escherichia coli. Commun. Biol. 2, 85 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hayes M. L., Santibanez P. I., A plant pentatricopeptide repeat protein with a DYW-deaminase domain is sufficient for catalyzing C-to-U RNA editing in vitro. J. Biol. Chem. 295, 3497–3505 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boussardon C., et al. , Two interacting proteins are necessary for the editing of the NdhD-1 site in Arabidopsis plastids. Plant Cell 24, 3684–3694 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andrés-Colás N., et al. , Multiple PPR protein interactions are involved in the RNA editing system in Arabidopsis mitochondria and plastids. Proc. Natl. Acad. Sci. U.S.A. 114, 8883–8888 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guillaumot D., et al. , Two interacting PPR proteins are major Arabidopsis editing factors in plastid and mitochondria. Proc. Natl. Acad. Sci. U.S.A. 114, 8877–8882 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takenaka M., et al. , Multiple organellar RNA editing factor (MORF) family proteins are required for RNA editing in mitochondria and plastids of plants. Proc. Natl. Acad. Sci. U.S.A. 109, 5104–5109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bentolila S., et al. , RIP1, a member of an Arabidopsis protein family, interacts with the protein RARE1 and broadly affects RNA editing. Proc. Natl. Acad. Sci. U.S.A. 109, E1453–E1461 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yan J., et al. , MORF9 increases the RNA-binding activity of PLS-type pentatricopeptide repeat protein in plastid RNA editing. Nat. Plants 3, 17037 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Royan S., et al. , A synthetic RNA editing factor edits its target site in chloroplasts and bacteria. Commun. Biol. 4, 545 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ding Y. H., Liu N. Y., Tang Z. S., Liu J., Yang W. C., Arabidopsis GLUTAMINE-RICH PROTEIN23 is essential for early embryogenesis and encodes a novel nuclear PPR motif protein that interacts with RNA polymerase II subunit III. Plant Cell 18, 815–830 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De Clercq A., et al. , Expression and processing of an Arabidopsis 2s albumin in transgenic tobacco. Plant Physiol. 92, 899–907 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Devic M., Albert S., Delseny M., Induction and expression of seed-specific promoters in Arabidopsis embryo-defective mutants. Plant J. 9, 205–215 (1996). [DOI] [PubMed] [Google Scholar]
- 27.Branon T. C., et al. , Efficient proximity labeling in living cells and organisms with TurboID. Nat. Biotechnol. 36, 880–887 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Verbitskiy D., Zehrmann A., Härtel B., Brennicke A., Takenaka M., Two related RNA-editing proteins target the same sites in mitochondria of Arabidopsis thaliana. J. Biol. Chem. 287, 38064–38072 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fossum E., et al. , Evolutionarily conserved herpesviral protein interaction networks. PLoS Pathog. 5, e1000570 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Diaz M. F., Bentolila S., Hayes M. L., Hanson M. R., Mulligan R. M., A protein with an unusually short PPR domain, MEF8, affects editing at over 60 Arabidopsis mitochondrial C targets of RNA editing. Plant J. 92, 638–649 (2017). [DOI] [PubMed] [Google Scholar]
- 31.Kodama Y., Suetsugu N., Kong S. G., Wada M., Two interacting coiled-coil proteins, WEB1 and PMI2, maintain the chloroplast photorelocation movement velocity in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 107, 19591–19596 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.He S., et al. , A novel imprinted gene NUWA controls mitochondrial function in early seed development in Arabidopsis. PLoS Genet. 13, e1006553 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chateigner-Boutin A. L., et al. , The E domains of pentatricopeptide repeat proteins from different organelles are not functionally equivalent for RNA editing. Plant J. 74, 935–945 (2013). [DOI] [PubMed] [Google Scholar]
- 34.Hammani K., et al. , The pentatricopeptide repeat protein OTP87 is essential for RNA editing of nad7 and atp1 transcripts in Arabidopsis mitochondria. J. Biol. Chem. 286, 21361–21371 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gutmann B., Royan S., Small I., Protein complexes implicated in RNA editing in plant organelles. Mol. Plant 10, 1255–1257 (2017). [DOI] [PubMed] [Google Scholar]
- 36.Takenaka M., et al. , DYW domain structures imply an unusual regulation principle in plant organellar RNA editing catalysis. Nat. Catal. 4, 510–522 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Malbert B., et al. , The analysis of the editing defects in the dyw2 mutant provides new clues for the prediction of RNA targets of Arabidopsis E+-class PPR proteins. Plants 9, 280 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tasaki E., Hattori M., Sugita M., The moss pentatricopeptide repeat protein with a DYW domain is responsible for RNA editing of mitochondrial ccmFc transcript. Plant J. 62, 560–570 (2010). [DOI] [PubMed] [Google Scholar]
- 39.Ichinose M., Sugita C., Yagi Y., Nakamura T., Sugita M., Two DYW subclass PPR proteins are involved in RNA editing of ccmFc and atp9 transcripts in the moss Physcomitrella patens: First complete set of PPR editing factors in plant mitochondria. Plant Cell Physiol. 54, 1907–1916 (2013). [DOI] [PubMed] [Google Scholar]
- 40.Colgan J., Ashali H., Manley J. L., A direct interaction between a glutamine-rich activator and the N terminus of TFIIB can mediate transcriptional activation in vivo. Mol. Cell. Biol. 15, 2311–2320 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bayer-Császár E., et al. , The conserved domain in MORF proteins has distinct affinities to the PPR and E elements in PPR RNA editing factors. Biochim. Biophys. Acta. Gene Regul. Mech. 1860, 813–828 (2017). [DOI] [PubMed] [Google Scholar]
- 42.Yoo S. D., Cho Y. H., Sheen J., Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572 (2007). [DOI] [PubMed] [Google Scholar]
- 43.Yang Y. Z., Tan B. C., RT-PCT product sequencing of Arabidopsis. NCBI. https://www.ncbi.nlm.nih.gov/sra/?term=PRJNA849404. Accessed 15 June 2022.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data reported in this paper have been deposited in the Sequence Read Archive database, https://www.ncbi.nlm.nih.gov/sra [accession no. PRJNA849404 (43)].
All study data are included in the article and/or supporting information.