Significance
Atherosclerosis underpins the progression of stroke and myocardial infarction, both leading causes of death globally. Gene therapy is an emerging therapeutic approach for atherosclerosis, but the delivery of gene cargoes to the atherosclerotic plaque remains inefficient. In this study, we bypass this gene delivery obstacle by assembling a three-dimensional spherical nucleic acid nanostructure with therapeutic oligonucleotides that inhibit proinflammatory genes related to atherogenesis. The resultant nanostructure not only naturally targets the scavenger receptor on plaque macrophages and endothelial cells, but also reduces and stabilizes plaques. Our work offers insights into the translation of nucleic acid nanotechnology for treating and understanding cardiovascular diseases.
Keywords: nanomedicine, atherosclerosis, nucleic acid nanotechnology, gene therapy, cardiovascular disease
Abstract
Atherosclerosis treatments by gene regulation are garnering attention, yet delivery of gene cargoes to atherosclerotic plaques remains inefficient. Here, we demonstrate that assembly of therapeutic oligonucleotides into a three-dimensional spherical nucleic acid nanostructure improves their systemic delivery to the plaque and the treatment of atherosclerosis. This noncationic nanoparticle contains a shell of microRNA-146a oligonucleotides, which regulate the NF-κB pathway, for achieving transfection-free cellular entry. Upon an intravenous injection into apolipoprotein E knockout mice fed with a high-cholesterol diet, this nanoparticle naturally targets class A scavenger receptor on plaque macrophages and endothelial cells, contributing to elevated delivery to the plaques (∼1.2% of the injected dose). Repeated injections of the nanoparticle modulate genes related to immune response and vascular inflammation, leading to reduced and stabilized plaques but without inducing severe toxicity. Our nanoparticle offers a safe and effective treatment of atherosclerosis and reveals the promise of nucleic acid nanotechnology for cardiovascular disease.
Atherosclerosis is the chronic inflammation and remodeling of the vascular wall induced by hyperlipidemia, which underpins cardiovascular diseases (CVDs), the leading causes of death globally (1). Vascular inflammation in the context of atherosclerosis involves many cell types and their interactions, including endothelial cells (ECs), smooth muscle cells, and immune cells, mainly macrophages and T cells (2). Conventional interventions for atherosclerosis involve stent and balloon angioplasty (3, 4) that commonly cause recurring stent restenosis. Meanwhile, administration of statins to treat hyperlipidemia (5, 6) only slows down disease progression.
Bionanomaterials hold immense potential for managing atherosclerosis; they not only can be engineered to encapsulate therapeutic (7) or imaging agents (8), but also can be constructed with different types of biomolecular building blocks to achieve specific delivery to cell types of the plaque (9), e.g., lipoproteins that are naturally home to macrophages (10), nanoparticles (NPs) with peptides that target adhesion molecules of ECs (11), and sugar-based NPs that engage scavenger receptors (SRs) (12) or CD44 receptor (13) of macrophages in the plaque. Intriguingly, noncationic nucleic acid-based nanostructures derived from the assembly of plasmids or oligonucleotides are rarely used for constructing atherosclerosis nanomedicines despite their biocompatibility, abundant cellular entry without the aid of transfection agents (14), and convenient loading of functional nucleic acids for regulating disease pathways by hybridization (15). Past atherosclerosis nanomedicines that adopted a gene therapy approach mostly employed cationic carriers for electrostatically complexing gene cargoes and displaying targeting ligands for promoting plaque delivery. Yet, with genes, ligands, and the cationic carrier all included, this tricomponent NP often becomes bulky (>100 nm; prone to clearance by the liver and spleen that hampers systemic delivery to the plaque (16)) and remains cationic (leading to cytotoxicity that may limit clinical translation (17)).
We present a noncationic nucleic acid-based nanostructure to overcome these gene delivery obstacles to atherosclerotic plaques, with nucleic acid serving as a dual targeting agent for engaging plaque-related receptors and gene regulation agent for blocking biological pathways linked to atherogenesis. The core is a polyethylene glycol (PEG)-coated superparamagnetic iron oxide NP (SPION); both SPION and PEG are biocompatible and often used as a carrier and NP stabilizing agent, respectively, in cardiovascular drug delivery applications (18, 19). The outer shell contains a layer of phosphorothioate (PS)-modified microRNA-146a (miR-146a) oligonucleotides attached to the PEG-coated SPION core (PEG-SPION). MiR-146a, a key inhibitor of vascular inflammation, suppresses the NF-κB signaling pathway in macrophages and ECs (20, 21), two major cell types of the plaque. As NF-κB signaling mediates proinflammatory responses during atherogenesis, we believe that miR-146a is a suitable candidate for pharmacological intervention of atherosclerosis (22). The PS backbone endows the microRNA (miRNA) with protection against extra and intracellular nuclease degradation (23). Overall, miR-146a-functionalized SPION (miR-146a-SPION) is a spherical nucleic acid (SNA) nanostructure that abundantly enters both cell types without the aid of cationic or lipophilic transfection agents (24). Previously, we reported that modifying NPs with a shell of noncoding DNA oligonucleotides promotes their cellular uptake via class A scavenger receptors (SR-A) in vitro and delivery to atherosclerotic plaques in vivo (25). Here, we demonstrate that the miR-146a-SPIONs accumulate in the atherosclerotic plaques of apolipoprotein E knockout (ApoE−/−) mice fed on a high-cholesterol diet (HCD), an established model of atherosclerosis (26), upon an intravenous (i.v.) injection; notably, miR-146a-SPIONs naturally target SR-A on plaque macrophages and ECs in vivo. Aortic accumulation of miR-146a-SPIONs reach ∼1% of the injected dose (ID; 1.2% ID or 8.9% ID/g; SI Appendix, Fig. S1), a high-delivery efficiency when benchmarked against past reported atherosclerosis nanomedicines (SI Appendix, Table S1). Repeated injections of miR-146a-SPIONs reduce and stabilize the plaque by down-regulating genes linked to immune responses and vascular inflammation without inducing severe systemic toxicity.
Results
Synthesis and Characterization of miR-146a-SPIONs.
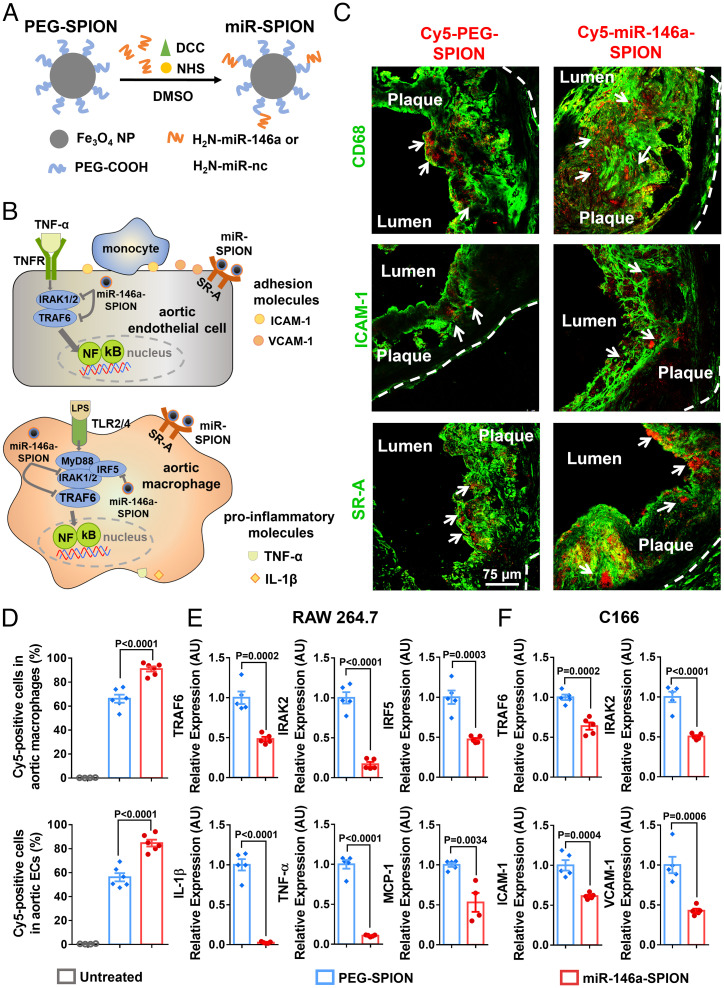
To prepare miR-SPIONs, we synthesized carboxy-terminated PEG-SPIONs with a core size of ∼20 nm (SI Appendix, Fig. S2) and covalently conjugated amine-terminated miR-146a oligonucleotides (sequence information in SI Appendix, Table S2) with the PEG via carbodiimide cross-linker chemistry (Fig. 1A) (25). The PEG-SPIONs have a mean hydrodynamic size of 59.3 nm and ζ-potential of –8.2 mV, and the miR-146a-SPIONs have a larger mean hydrodynamic size of 72.7 nm and a more negative ζ-potential of –21.8 mV. For all SPION types tested, the size distribution is homogeneous (SI Appendix, Table S3). Agarose gel electrophoresis of the SPIONs data show that the miRNA-conjugated SPIONs migrated closer to the positive electrode than PEG-SPIONs (SI Appendix, Fig. S3), matching the zeta potential measurements. The average loading of miR-146a-SPIONs is 275 strands per SPION. Denaturing polyacrylamide gel electrophoresis (PAGE) confirms the covalent loading of miRNA to the PEG-SPIONs. Whereas no free miRNA was detected inside the PAGE gel for the miRNA-conjugated SPIONs, distinct free miRNA bands were found for a physical mixture of PEG-SPIONs and free miR-146a strands (SI Appendix, Fig. S4). For ex vivo and in vivo distribution studies, we prepared Cy5-miR-SPIONs by covalently attaching Cyanine 5 (Cy5)- and amine-terminated miR-146a oligonucleotides to carboxy-terminated PEG-SPIONs. To reveal the effect of the miR shell on plaque-NP interactions, we prepared Cy5-PEG-SPIONs by covalently labeling amine-terminated Cy5 to carboxylated PEG-SPIONs. The Cy5-labeled PEG-SPIONs and miR-146a-SPIONs were 66.6 nm and 79.3 nm in mean hydrodynamic size, respectively. Attachment of Cy5 did not significantly alter their hydrodynamic sizes, zeta potentials, serum stability, cytotoxicity, and cellular uptake (SI Appendix, Table S3 and Figs. S3–S6).
Fig. 1.
Ex vivo binding of miR-146a-SPIONs to endothelial cells and macrophages in atherosclerotic plaque and in vitro down-regulation of genes related to NF-κB pathway by miR-146a-SPIONs in cell lines. (A) Preparation of miR-SPIONs. (B) The NF-κB pathway and miR-146a target in aortic macrophage and EC. (C) Representative confocal images showing the ex vivo association of Cy5-labeled PEG-SPIONs and miR-146a-SPIONs (red; with white arrows) to macrophages, ECs and SR-A in atherosclerotic plaques from aortic root sections. Green pseudo color: CD68 (macrophages), ICAM-1 (ECs) or SR-A. The white dotted lines indicate the border of the vascular wall from aortic roots. (D) Flow cytometric analysis shows the association of Cy5-labeled PEG-SPIONs (blue) and miR-146a-SPIONs (red) to total macrophages and total ECs in the aorta of ApoE−/− mice after 2 h of ex vivo incubation (n = 4–6, across two independent experiments). Statistical significance was calculated by one-way ANOVA with Tukey's test for post hoc analysis. (E and F) In vitro down-regulation of genes in RAW264.7 macrophages and C166 ECs related to the NF-κB proinflammatory pathway 24 h following incubation of miR-146a-SPIONs (red) or PEG-SPION (blue) (n = 5, across two independent experiments). Statistical significance was calculated by Student’s t test. All the data are presented as mean ± SEM.
We briefly characterized the SPIONs in serum-containing medium. Compared with serum-free conditions, incubation of PEG-SPIONs and miR-146a-SPIONs in serum-containing medium for 24 h led to an increase in their hydrodynamic diameters by 5–13 nm, indicating serum adsorption to the SPIONs but without inducing evident aggregation (SI Appendix, Table S3). Also, the serum protein corona did not drastically affect cellular uptake or receptor targeting of miR-146a-SPIONs in vitro. Compared with serum-free conditions, the cellular uptake of miR-146a-SPIONs by RAW 264.7 macrophages in serum-containing medium was only attenuated by 21.8%. Pretreatment with fucoidan (a ligand of SR-A) for 1 h drastically reduced cellular uptake of miR-146a-SPIONs by 68.4% and 84.7% under serum-containing and serum-free conditions, respectively (SI Appendix, Fig. S7).
Ex Vivo Delivery to Macrophages and ECs.
Initially, we evaluated the association of miR-SPIONs to the macrophages and ECs of atherosclerotic plaques ex vivo. Before our immunofluorescence (IF) studies, we verified that the aortic root tissue sections collected from uninjected plaque-bearing HCD-fed ApoE−/− mice do not emit detectable tissue autofluorescence in the Alexa Fluor 532, Cy3 (both dyes for labeling the secondary antibodies), or Cy5 wavelength (dye for labeling all SPION types tested) (SI Appendix, Fig. S8A). We also could not detect fluorescence when tissue sections from uninjected mice were stained with fluorescently labeled secondary antibodies only as negative control (SI Appendix, Fig. S8 B and C), confirming the specificity of our IF data below. By using confocal IF to track the tissue-level distribution of the SPIONs in the aortic root, we captured more intense Cy5 fluorescence in both the macrophage-rich region (CD68-positive) and the EC-rich region (ICAM-1-positive) for the miR-146a-SPION sample than the PEG-SPION sample (Fig. 1C, with additional nuclear counterstaining shown in SI Appendix, Figs. S9 and S10). Also, miR-146a-SPIONs bind to intraplaque SR-A-expressing cells more abundantly than PEG-SPIONs (Fig. 1C), proof of the utility of the miRNA shell in facilitating SR-A targeting ex vivo. To eliminate potential bias in data interpretation due to tissue autofluorescence, we further employed immunohistochemistry (IHC) to stain the aortic root sections with the same primary antibody against ICAM-1 as used for the IF images. IHC and IF of two adjacent aortic root sections revealed similar distribution of ICAM-1 expression (SI Appendix, Fig. S11). To remove potential bias due to antibody specificity, we also verified in aortic root tissue sections collected from nonatherosclerotic untreated C57BL/6 mice that no ICAM1 or CD68-positive regions were detected by both primary and secondary antibodies because normal blood vessels should not express ICAM1 or contain CD68-expressing macrophages (SI Appendix, Figs. S12 and S13).
To measure the cellular-level distribution of the SPIONs in the aorta, we analyzed the uptake of SPIONs by different cell types from the aorta by flow cytometry. We detected a significantly higher fraction of Cy5-positive plaque macrophages for the former group (90.9% vs. 66.2%) and a significantly higher fraction of Cy5-positive plaque ECs for the miR-146a-SPION sample (84.7%) than the PEG-SPION group (56.2%) (Fig. 1D). These data highlight the enhanced ex vivo binding of SPIONs to the plaque due to the miRNA shell. Because nearly all aortic ECs (84.7%) and macrophages (90.9%) were Cy5 positive for the miR-146a-SPION group due to the direct soaking of the aorta with SPIONs, it was challenging to ascertain the ex vivo association of SPIONs to SR-A-rich aortic ECs and macrophages. Therefore, we deferred our assessment of the association of miR-146-SPIONs to SR-A-rich aortic macrophages and ECs to in vivo studies (Fig. 2).
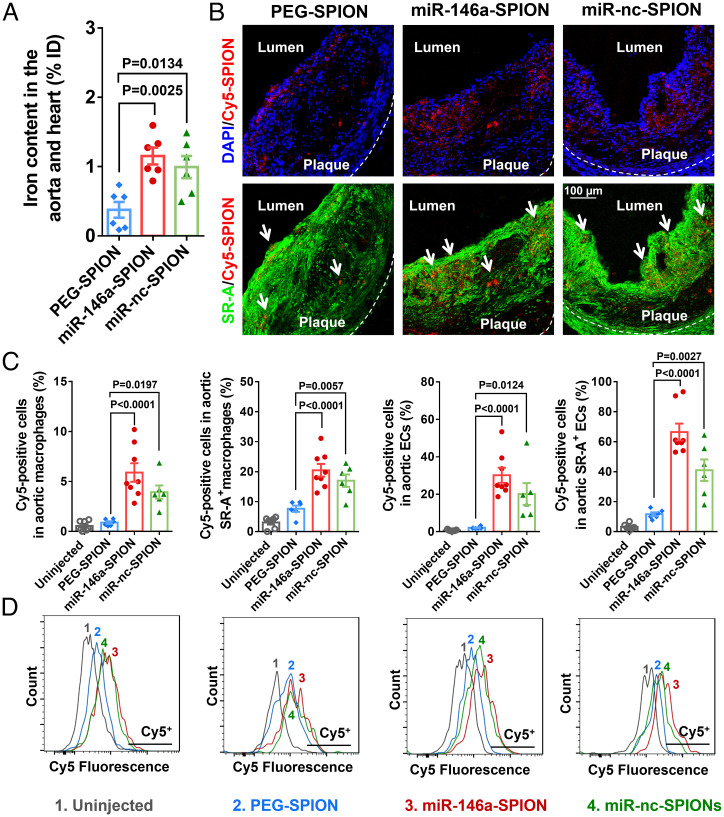
Fig. 2.
Accumulation of miR-146a-SPIONs inside atherosclerotic plaques. (A) ICP-MS measurements of the iron content in the aorta and heart (sites of plaques for ApoE−/− mice) show more abundant accumulation of miR-146a-SPIONs (red) and miR-nc-SPIONs (green) than PEG-SPIONs (blue) 2 h postinjection (n = 6 mice per group, across two independent experiments). (B) Representative confocal images of the aortic root confirm stronger colocalization of Cy5-labeled miR-146a-SPIONs and Cy5-labeled miR-nc-SPIONs than Cy5-PEG-SPIONs (red) with aortic SR-A-rich macrophages or ECs 2 h postinjection. Green pseudo color: SR-A. Blue: DAPI. (C) Flow cytometric analysis of the cells in the aorta reveal stronger association of miR-146a-SPIONs (red) and miR-nc-SPIONs (green) to the aortic cell types than PEG-SPIONs (blue), including total macrophages, SR-A+ macrophages, total ECs, and SR-A+ ECs 2 h postinjection (n = 6–8 mice per group, across two independent experiments). MFI: mean fluorescence intensity. (D) Representative flow cytometry histograms confirm that all types of aortic cells uptake Cy5-labeled miR-146a-SPIONs (red) and Cy5-labeled miR-nc-SPIONs (green) more readily than Cy5-labled PEG-SPIONs (blue) 2 h postinjection. Data are presented as means ± SEM. Statistical significance was calculated by one-way ANOVA with Tukey's test for post hoc analysis.
In Vitro Gene Regulation in Macrophages and ECs.
Next, we evaluated the ability of the miR-146a-SPIONs to down-regulate genes in vitro, using RAW264.7 mouse macrophage and C166 mouse EC cell lines to show whether the miR-146a-SPION effectively inhibits target gene expression. Representative transmission electron microscope images portray the intracellular accumulation of PEG-SPIONs and miR-146a-SPIONs after 2 h of incubation (SI Appendix, Fig. S14), proof of their in vitro cellular entry. We incubated RAW 264.7 macrophages with miR-146a-SPIONs or PEG-SPIONs for 24 h and treated the cells with lipopolysaccharide (LPS) for another 24 h to induce inflammatory responses (27). qRT-PCR measurements showed significant inhibition of several previously reported targets of miR-146a (28, 29) (Fig. 1B), including components of innate immune signaling (e.g., tumor necrosis factor [TNF] receptor-associated factor 6 [TRAF6] and interleukin 1 receptor-associated kinase 2 [IRAK2], and interferon regulatory factor 5), as well as some effector molecules (e.g., chemokine monocyte chemoattractant protein-1, cytokines such as interleukin-1β [IL-1β], and TNF-α) for miR-146a-SPION-treated cells compared with PEG-SPION-treated cells (Fig. 1E). Also, we detected a significant increase in miR-146a expression levels upon incubating the LPS-treated macrophages with miR-146a-SPIONs for 24 h (SI Appendix, Fig. S15A), verifying the efficient intracellular delivery of miR-146a. Then, we transfected C166 ECs with miR-146a-SPIONs or PEG-SPIONs for 24 h and treated the cells with TNF-α for another 24 h to induce inflammation (30). We observed similar inhibition of TRAF6 and IRAK2, and proinflammatory endothelial adhesion molecules, including ICAM-1 and vascular cell adhesion protein (VCAM)-1 in miR-146a-SPION-treated cells compared with PEG-SPION-treated cells (Fig. 1F). These data suggest that the effective delivery of miR-146a by the SNA nanostructure enhanced the suppression of marker genes of the NF-κB pathway in both cell types. These data also motivate us to evaluate the ability of miR-146a-SPIONs in inhibiting these genes in atherosclerotic plaque-bearing mice.
Blood Pharmacokinetics and Distribution to the Aorta.
We monitored the pharmacokinetics of miR-146a-SPIONs upon an i.v. injection into plaque-bearing HCD-fed ApoE−/− mice by sampling the blood plasma, because the presence of erythrocytes may bias the measurement of iron content, which was used to quantify the SPION concentration. Inductively coupled plasma mass spectrometry (ICP-MS) analysis revealed that PEG-SPIONs have a slightly longer half-life than miR-146a-SPIONs (2.67 vs. 1.89 h) (SI Appendix, Fig. S16). For our in vivo efficacy studies, we prepared miR-nc-SPIONs that contained scrambled miRNA sequences in the outer shell. The miR-nc-SPIONs had a hydrodynamic size, zeta potential, miRNA loading (263 strands per SPION), serum stability, cellular uptake properties, and plasma half-life (2.07 h) similar to the miR-146a-SPIONs (SI Appendix, Table S3 and Figs. S3–S6 and S16). To clarify the effect of surface charge on in vivo distribution, we further prepared abasic oligonucleotide-conjugated SPIONs (abasic-SPIONs) as another miRNA-free, negatively charged control NP by covalently attaching amine-terminated abasic oligonucleotides to carboxylated PEG-SPIONs. For this abasic oligonucleotide (the same length as miR-146a), more than half of the bases contained the negatively charged phosphate and ribose but without the nucleobase (SI Appendix, Table S2). The abasic-SPIONs had a hydrodynamic size, zeta potential, and serum stability similar to the miR-146a-SPIONs, but with a slightly higher oligonucleotide loading (SI Appendix, Table S3). The abasic sites were situated at the periphery of the SNA nanostructure, enabling us to explore the effect of negative charge on plaque delivery and SR-A targeting.
We chose 2 h postinjection as the time point for in vivo distribution studies. Because the blood circulation half-life of miR-146a SPIONs is ∼2 h, it is desirable for them to reach the plaques before predominant clearance from circulation. To bolster this claim, we showed by ICP-MS measurements that miR-146a-SPIONs accumulated in the phosphate buffered saline (PBS)-perfused aorta and heart most abundantly 2 h postinjection compared with other time points tested (0.5, 8, and 24 h) (SI Appendix, Fig. S17A), matching the distribution profile of DNA-coated SPIONs (25). Specifically, 2 h postinjection, the iron contents of the aorta and heart in the miR-146a-SPION (1.2% ID) and miR-nc-SPION groups (1.0% ID) were significantly higher than the PEG-SPION and abasic-SPION groups (both ∼0.4% ID) (Fig. 2A and SI Appendix, Fig. S18), indicating that a negative surface charge alone does not guarantee improved plaque delivery. Between miR-146a-SPIONs and PEG-SPIONs, we did not detect a significant difference in their organ-level distribution at other time points (SI Appendix, Fig. S17 B–D); both SPION types primarily accumulate in the liver (20–25% ID) and spleen (10–15% ID), with limited accumulation in the kidneys (<2% ID). These data support that 2 h is a sufficient duration for targeting and accumulation in the plaques. Ex vivo fluorescence imaging of the aorta excised from ApoE−/− mice injected with Cy5-labeled miR-146a-SPIONs or PEG-SPIONs revealed intense Cy5 fluorescence hot spots that indicate NP localization only in the miR-146a-SPION group but not the PEG-SPION group 2 h postinjection (SI Appendix, Fig. S19A). These data further confirm that the miRNA shell promotes the delivery of SPIONs to the aortic plaque, consistent with the aortic accumulation of DNA-SPIONs (1.25% ID) (25). As a negative control, we i.v. injected free Cy5 dyes into ApoE−/− mice but did not detect fluorescence hot spots in the aorta by ex vivo imaging (SI Appendix, Fig. S20). These distinct distribution patterns suggest that the Cy5 dyes are stably connected to miR-146a-SPION and that free dyes do not target plaques. Moreover, we measured the distribution of miR-146a-SPIONs upon injection into healthy C57BL/6 mice. ICP-MS measurements revealed similarly low iron contents in the aorta and heart for both miR-146a-SPION (0.3% ID) and PEG-SPION groups (0.2% ID) (SI Appendix, Fig. S21), and ex vivo fluorescence imaging of the aorta showed no obvious Cy5 fluorescence hot spots for both types of Cy5-labeled SPIONs (SI Appendix, Fig. S19). These data indicate selective delivery of miR-146a-SPIONs to plaques. At 24 h postinjection, the accumulation of miR-146a-SPIONs in the aorta was ∼0.5% ID, indicating considerable retention of SPIONs in the plaque (SI Appendix, Fig. S22). Therefore, we kept the consecutive injections 2–3 d apart in our efficacy studies (Figs. 3 and 4).
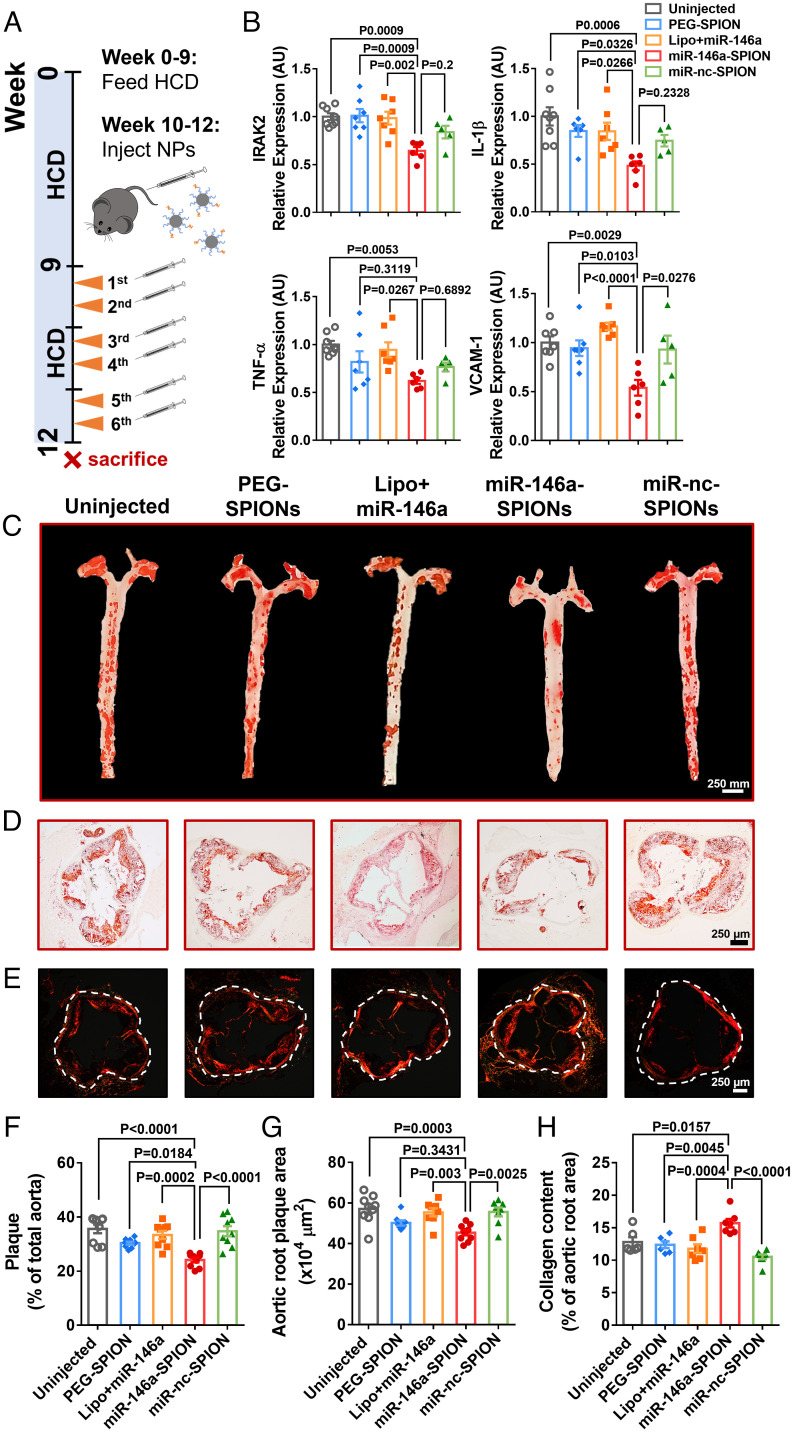
Fig. 3.
miR-146a-SPIONs reduced and stabilized atherosclerotic plaques and inhibited genes linked to atherogenesis. (A) After 9 wk of HCD, the ApoE−/− mice received two weekly i.v. injections from weeks 10–12 while continuing the HCD. (B) qRT-PCR analysis of aortic RNA reveals significant down-regulation of genes targeted by miR-146a (IRAK2) and genes downstream of the NF-κB pathway (IL-1β, TNF-α, and VCAM-1) by miR-146a-SPIONs. For the miR-146a-SPION group, representative images of (C) Oil Red O (ORO)-stained intact aortas and (D) ORO-stained sections of aortic roots indicate most substantial reduction in the plaque fat content (red), and (E) Sirius Red-stained aortic roots reveal most abundant accumulation of collagen (red). For the miR-146a-SPION group, the (F) fractional plaque area in total aorta (n = 8–10 mice per group, across three independent experiments) is the lowest, (G) plaque area in the aortic root is the lowest, and (H) collagen content in aortic root is the highest. Data of aortic root sections were chosen from n = 6 images per mouse from n = 6–8 mice per group. Data are presented as means ± SEM. Statistical significance was calculated by one-way ANOVA with Tukey's test for post hoc analysis.
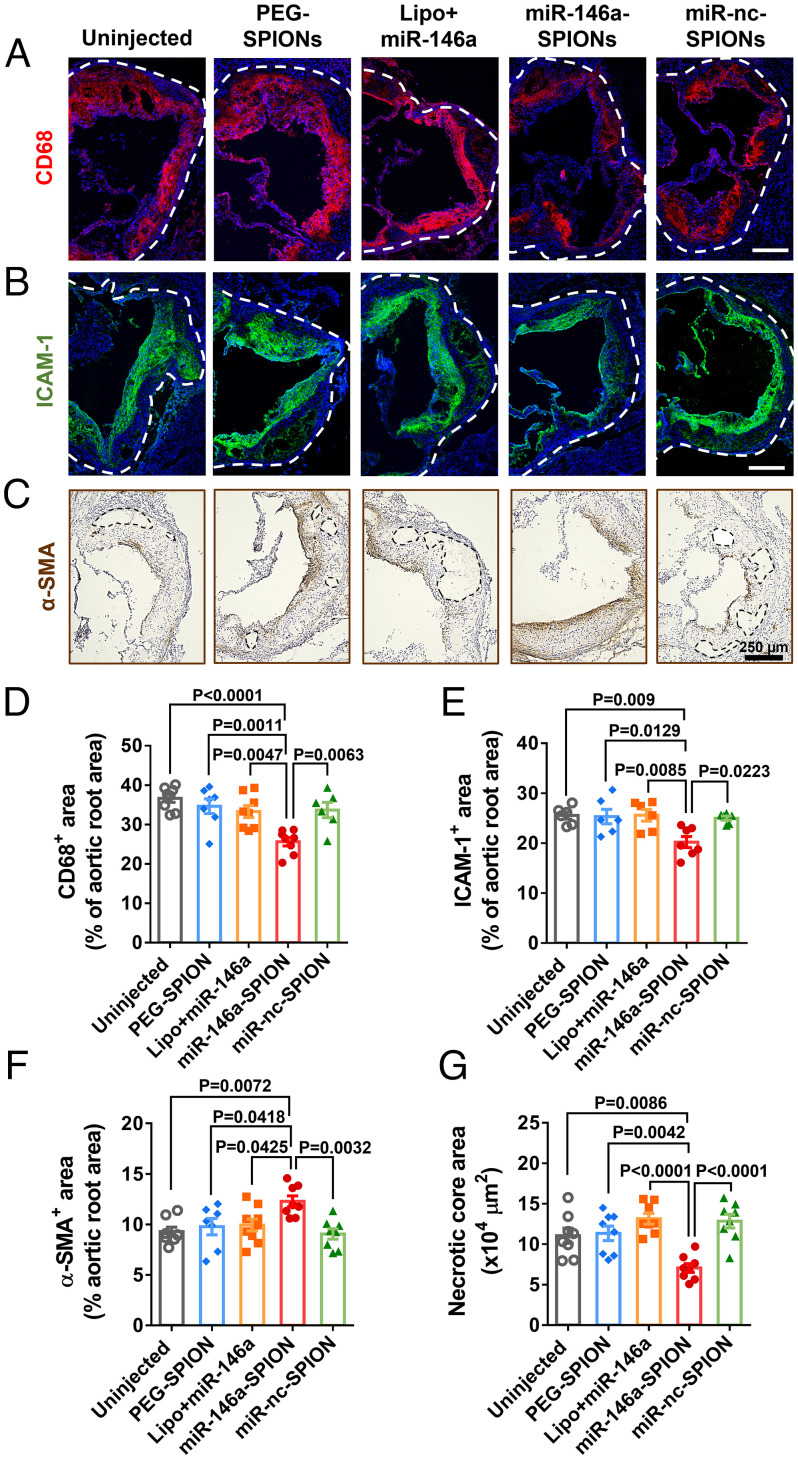
Fig. 4.
miR-146a-SPIONs inhibited the morphological markers of atherosclerotic plaques. Representative confocal images of the sections of aortic roots reveal most apparent reduction in (A) CD68 (marker of monocytes and macrophages; red pseudo color) for the miR-146a-SPION group and (B) ICAM-1 (marker of ECs; green pseudo color). Blue: DAPI. (C) Representative IHC images of the sections of aortic roots reveal most apparent enhancement in of α-SMA (highly expressed by smooth muscle cells; brown) for the miR-146a-SPION group. The dotted region indicates the necrotic core of the plaque. For the miR-146a-SPION group, (D) CD68-positive area in the aortic root is the lowest, (E) ICAM-1-positive area in the aortic root is the lowest, (F) α-SMA-positive area in the aortic root is the highest, and (G) area of necrotic core in the aortic root is the lowest. Data of aortic root sections were from an average of six images per mouse, from n = 6–8 mice per group (across three independent experiments). Data are presented as means ± SEM. Statistical significance was calculated by one-way ANOVA with Tukey's test for post hoc analysis.
To detect the intraplaque tissue-level distribution of SPIONs, we i.v. injected plaque-bearing mice with Cy5-labeled SPIONs and harvested the tissues 2 h postinjection. Representative confocal images revealed stronger Cy5 fluorescence signals in the aortic roots from the miR-146a-SPION and miR-nc-SPION groups than the PEG-SPION group, indicating more abundant accumulation of miRNA-coated SPIONs in the plaque (Fig. 2B). Notably, by jointly examining Cy5 fluorescence with SR-A IF, we further showed that both miR-SPION types colocalize more with SR-A than PEG-SPIONs, suggesting stronger binding of the SPIONs to SR-A due to the miRNA shell (Fig. 2B). Then, we quantified the intraplaque cellular-level distribution of SPIONs to the aorta by measuring Cy5 fluorescence of different aortic cell types by flow cytometry. Total macrophages and ECs from single-cell suspensions of the aorta were identified according to surface markers (see gating strategy in SI Appendix, Fig. S23). Data were expressed in terms of the fraction of Cy5-labeled SPION-positive cells in SPION-injected mice compared with uninjected mice for setting the negative gate (Fig. 2D). The mean fractions of Cy5-positive total macrophages for the miR-146a-SPION (5.9%) and miR-nc-SPION (3.9%) groups were ∼4–6 times of that of Cy5-PEG-SPION group (0.9%). Likewise, the mean fractions of Cy5-positive total ECs for the miR-146a-SPION (30.2%) and miR-nc-SPION (20.1%) groups were ∼10–15 times that of the PEG-SPION group (2.1%) (Fig. 2C). As control studies, we confirmed that the Cy5 fluorescence detected originated from Cy5-labeled NPs but not free Cy5 molecules and that miR-146a-SPIONs more efficiently delivered miRNA to aortic macrophages and aortic ECs than Lipofectamine, a commercial transfection agent (SI Appendix, Fig. S24). Our in vivo data indicate preferential uptake of miRNA-coated SPIONs by the two major aortic cell types over PEG-SPIONs, echoing the ex vivo binding to aortic macrophages and ECs of miR-146a-SPIONs (Fig. 1).
Furthermore, we probed the role of SR-A in mediating the intraplaque cellular-level distribution of SPIONs. By using unstained cells to set the negative gate (Fig. 2D), we verified that i.v. injection of PEG-SPIONs or miR-SPIONs did not significantly alter the expression of SR-A by aortic macrophages and ECs (SI Appendix, Fig. S25). The mean fractions of Cy5-positive SR-A-rich aortic macrophages for the Cy5-miR-146a-SPION (20.5%) and Cy5-miR-nc-SPION (17.0%) groups were approximately twofold that of the Cy5-PEG-SPION group (7.6%). Also, the mean fractions of Cy5-positive SR-A-rich aortic ECs for the Cy5-miR-146a-SPION (66.4%) and Cy5-miR-nc-SPION (41.0%) groups were four- to sixfold that of the Cy5-PEG-SPION group (11.6%) (Fig. 2C). The ∼2.5-fold larger fraction of Cy5-positive SR-A-rich macrophages (20.5%) than total macrophages (5.9%) together with the ∼1.2-fold larger fraction of Cy5-positive SR-A-aortic ECs (66.4%) than total aortic ECs (30.2%) collectively indicate the preferential uptake of miR-146a-SPIONs by SR-A-rich macrophages and ECs, matching our confocal IF data in the aortic root (Fig. 2B). By contrast, abasic-SPIONs did not associate with SR-A-rich macrophages and SR-A-rich ECs in the aorta as preferentially as miR-146a-SPIONs. The mean fractions of Cy5-positive SR-A-rich aortic macrophages (23.0%) and ECs (75.4%) were only ∼15–40% larger than those of Cy5-positive total aortic macrophages (16.2%) and total aortic ECs (66.0%) (SI Appendix, Fig. S26). These data suggest that negative surface charge is not a critical factor for SNAs to target SR-A. Finally, because ∼30% ID of the injected SPIONs accumulate in the liver (SI Appendix, Fig. S27), we investigated the intrahepatic cellular-level distribution of SPIONs (see gating strategy in SI Appendix, Fig. S28). The mean fractions of Cy5-positive liver SR-A-rich macrophages for the Cy5-miR-146a-SPION (60.4%) and Cy5-miR-nc-SPION (50.0%) groups were three- to fourfold higher than that of the Cy5-PEG-SPION group (16.1%) (SI Appendix, Figs. S29 and S30). The cellular-level distribution in the liver of ApoE−/− mice match the distribution to aortic macrophages, underscoring that the miRNA shell promotes delivery to SR-A-rich macrophages in vivo.
It is challenging to ascertain the effect of protein corona on SR-A targeting of i.v.-injected NPs in vivo because serum proteins are abundant in blood. To gain some insights, we analyzed the protein corona of PEG-SPIONs and miR-146a-SPIONs derived from 2 h of incubation in blood plasma collected from plaque-bearing ApoE−/− mice. PAGE analysis of the desorbed proteins revealed similar protein band patterns for both SPION types but darker bands for miR-146a-SPIONs (SI Appendix, Fig. S31A). Corroborating bicinchoninic acid assay showed approximately twofold more proteins adsorbed to miR-146a-SPIONs (SI Appendix, Fig. S31B). Downstream liquid chromatography-tandem mass spectrometry analysis confirmed the similar composition of their protein coronas. The most abundant proteins bound were all different isoforms of fibrinogen (∼55 kDa, which matched the darkest band of our PAGE data), albumin, beta globulin, and apolipoprotein A (SI Appendix, Fig. S31C). As protein adsorption moderately decreases cellular uptake in vitro (SI Appendix, Fig. S7A), we believe that the superior SR-A targeting by miR-146a-SPIONs compared with PEG-SPIONs is due to the miRNA shell rather than any additional amounts or different types of plasma proteins bound.
In Vivo Efficacy of miR-146a-SPIONs on Plaque Development and Morphology.
We then examined whether miR-146a-SPIONs exhibit anti-atherosclerosis efficacy by i.v. injecting them into ApoE−/− mice that were fed with HCD for the first 9 wk, a duration required for advanced plaques to be developed (26). Besides uninjected mice as negative control, our treatment groups included miR-146a-SPIONs, miR-nc-SPIONs, PEG-SPIONs, and miR-146a complexed with Lipofectamine (Lipo+miR-146a). Our injections took place twice a week from weeks 10–12 (Fig. 3A), keeping the amounts of miR strands and iron oxide injected constant across all groups. All mice were killed 24 h after the final dose. In our first set of studies, we extracted the RNA from intact aortas and examined the expression of target genes of miR-146a that were related to the NF-κB pathway by qRT-PCR. Injections of miR-146a-SPIONs significantly reduced the aortic expression of IRAK2 (direct target gene of miR-146a), IL-1β, VCAM-1, and TNF-α (downstream genes of the NF-κB pathway) compared with uninjected or Lipo+miR-146a; such treatments also significantly inhibited the first three marker genes in the aorta compared with PEG-SPIONs (Fig. 3B). Moreover, we detected ∼9.5-fold higher miR-146a expression in the aorta excised from mice that underwent a full treatment of miR-146a-SPIONs (SI Appendix, Fig. S15B), confirming the delivery of miR-146a to the plaques via SNAs. These data indicate the in vivo gene regulation affordable by miR-146a-SPIONs and match our in vitro data on the inhibited proinflammatory genes in RAW 264.7 macrophages and C166 ECs upon treatment of miR-146a-SPIONs (Fig. 1 E and F).
Next, we evaluated the effect of miR-146a-SPIONs on plaque size by staining whole aortas and aortic root sections with Oil Red O (Fig. 3 C and D and SI Appendix, Fig. S32). The lesion areas in the aortas of mice from the miR-146a-SPION group were significantly smaller than those for all other groups, with ∼30% reduction for miR-146a-SPION-treated mice than uninjected mice (Fig. 3F). The lesion areas in the aortic roots of mice from the miR-146a-SPION group were significantly smaller than those from the uninjected, miR-nc-SPION, and Lipo+miR-146a groups, with ∼20% smaller lesion areas for miR-146a-SPION-treated mice than uninjected mice (Fig. 3G). Furthermore, we assessed the effect of miR-146a-SPIONs on plaque stabilization because advanced plaques with low collagen contents are prone to rupture, inducing thrombosis and subsequent severe vascular events (31). Sirius Red staining of the aortic root sections showed significantly thicker collagen cap around the plaques for the miR-146a-SPION group than other groups (Fig. 3 E and H). These data suggest the efficacy of miR-146a-SPIONs in ameliorating atherosclerosis.
We addressed the plaque contents and inflammatory cell types in the aortic root in various treatment groups. Our IF data reveal smaller CD68-positive regions (an indicator of macrophage contents) and ICAM-1-positive regions (an indicator of endothelial inflammation) of the miR-146a-SPION treatment group than all other treatment groups (Fig. 4 A and B). Analysis of the fluorescence signals of the plaque area showed that the aortic CD68- and ICAM-1-positive areas for the miR-146a-SPION group were ∼30% and ∼20% smaller than other groups (Fig. 4 D and E), respectively. Corroborating IHC images showed a similar reduction (23.5%) in ICAM-1 expression due to miR-146a-SPION treatment (SI Appendix, Fig. S33). Besides intense ICAM-1-positive signals in the intimal layer of activated ECs, we also detected moderate signals in the plaque interior that contains smooth muscle cells, immune cells, and possibly ECs, in agreement with previous studies (32–34). In addition to inflamed ECs, ICAM-1 is also expressed by immune cells, e.g., infiltrating T cells (35). Moreover, IHC showed higher expression of α-SMA, a marker of the contents of vascular smooth muscle cells contributing to collagen production and fibrous cap formation (36), in the miR-146a-SPION treatment group than in all other treatment groups (Fig. 4C). The aortic α-SMA-positive area for the miR-146a-SPION group is ∼20% higher than other groups (Fig. 4F), indicating plaque stabilization and matching the Sirius Red staining data (Fig. 3E). Furthermore, treatment with miR-146a-SPIONs significantly reduced the size of the necrotic core, a key contributor to plaque vulnerability and rupture (37), compared with other treatments; the area of the necrotic core for the miR-146a-SPION group is ∼40% smaller than other groups (Fig. 4 C and G). Repeated injections of miR-146a-SPIONs did not change the body weight, weights, and histological structure of major organs, liver function, or blood cell composition of mice 24 h after full treatment (SI Appendix, Figs. S34–S36) and up to 4 wk after full treatment (SI Appendix, Figs. S37–S39). There was pronounced clearance of SPIONs from major internal organs 4 wk after full treatment (SI Appendix, Fig. S40) compared with 24 h after single injection (SI Appendix, Fig. S22). In short, miR-146a-SPIONs regressed and stabilized atherosclerotic plaques without causing severe toxicity.
Mechanism of Anti-Atherosclerosis Effect of miR-146a-SPIONs.
To gain insights into the anti-atherosclerosis efficacy of miR-146a-SPIONs, we extracted the intact aortas from plaque-bearing mice that received treatments of “miR-146a-SPIONs,” “miR-nc-SPIONs,” “PEG-SPIONs,” or “Untreated” mice (n = 3), followed by isolating the whole RNA for RNA-seq analysis. For pairwise comparisons, we identified differentially expressed transcripts (DETs) with statistical significance of Q < 0.05.
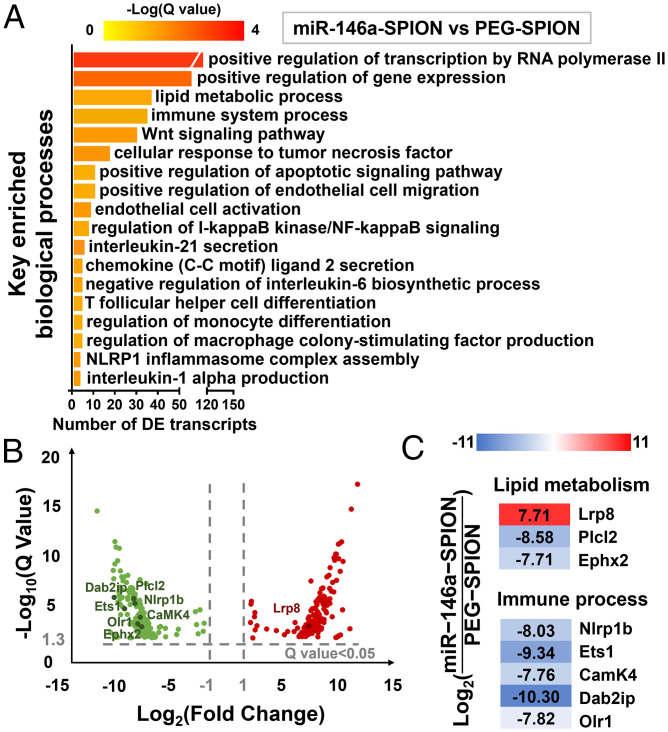
Initially, we analyzed the pairwise comparison between the “miR-146a-SPION” and “PEG-SPION” groups to evaluate the efficacy of the miRNA shell. Statistically enriched gene ontology (GO) terms most related to atherosclerosis are “lipid metabolic process” and “immune system process” (Fig. 5A and SI Appendix, Table S4). On lipid metabolic process, miR-146a-SPION treatment regulated several DETs, including the up-regulation of Lrp8, encoding ApoER2, whose deficiency accelerates atherosclerotic lesion necrosis formation (38); the down-regulation of Ephx2, which is an epoxide hydrolase associated with atherosclerosis and other CVDs (39, 40) and Plcl2, a susceptibility loci for atherosclerotic stroke and myocardial infarction (41, 42). On immune system process, miR-146a-SPION treatment down-regulated Olr1, Nlrp1b, Ets1, CaMK4, and Dab2ip (Fig. 5 B and C). Olr1, also known as oxidized low-density lipoprotein receptor-1 (LOX-1), is a SR for oxidized low-density lipoprotein (43). Nlrp1b is involved in inflammatory disorders and secretion of IL-1β (44, 45). Ets1 is a transcription factor for endothelial inflammation (46). CaMK4 regulates Ca2+ signaling in atherosclerosis and induces macrophage inflammation (47, 48). Dab2ip inhibits the DAB2IP-ASK1 pathway for alleviating atherosclerosis (49). Finally, the GO term of “regulation of I-κB kinase/NF-κB signaling” was also significantly enriched, echoing our in vitro (Fig. 1 E and F) and in vivo (Fig. 3B) qRT-PCR data on the regulation of the NF-κB signaling pathway affordable by miR-146a-SPIONs. Collectively, these data suggest that the 3D shell of miR-146a suppresses several biological processes related to the development of atherosclerosis (particularly inflammatory responses) and possibly lipid handling and entry to plaque cells, both contributing to plaque formation (Fig. 5).
Fig. 5.
miR-146a-SPIONs modulated the expression of genes related to immune response pathways and vascular inflammation in comparison to PEG-SPIONs. (A) Key enriched biological processes found by comparing the “miR-146a-SPION” group (n = 3) to the “PEG-SPION” group (n = 3). (B) Volcano plot showing the distribution of DETs by comparing the “miR-146a-SPION” group to the “PEG-SPION” group (absolute fold change > 2). (C) Representative DETs found in the comparison between “miR-146a-SPION” and “PEG-SPION” groups indicate the effect of miR-146a-SPION treatment on gene expression.
Finally, we analyzed the pairwise comparison between the “miR-146a-SPION” and “Untreated” groups to evaluate the efficacy of the overall NP. Statistically enriched GO terms most related to atherosclerosis are “immune system process” and “Wnt signaling pathway” (SI Appendix, Fig. S41A and Table S5). miR-146a-SPION treatment regulated several DETs related to “immune system process,” including the up-regulation of Otud7b (50) and Mapk7 (51) and down-regulation of Traf1 (52) (SI Appendix, Fig. S41B). miR-146a-SPION treatment also up-regulated Fzd5 (53, 54) and down-regulated Ndrg2 (55), which are DETs related to “Wnt signaling pathway” (SI Appendix, Fig. S41B). Wnt pathway components are abundantly detected in atherosclerotic plaque (56). From both comparisons, we conclude the efficacy of miR-146a-SPIONs for atherosclerosis stems from inhibiting EC activation, modulating immune responses, cholesterol transportation and processing, and vascular remodeling.
Discussion
Our miR-146a-SPIONs are safe and effective plaque targeting, gene delivery, and therapeutic nanostructures. First, the outer miRNA shell promotes the delivery to plaque macrophages and ECs by naturally targeting SR-A in vivo, so that this SNA nanostructure may overcome a major gene delivery hurdle to atherosclerotic plaques without the need for adding peptides, proteins, or aptamers for targeting other plaque components. Second, miR-146a-SPIONs are effective anti-atherosclerosis nanostructures that inhibit multiple proinflammatory genes related to both macrophages and ECs along the NF-κB pathway in the aorta. Researchers can now take advantage of miR-SPIONs to regulate other gene targets or biological pathways linked to atherosclerosis, inflammation, or CVDs in general by adjusting the oligonucleotide sequence. Finally, miR-146a-SPIONs do not induce severe toxicity, reinforcing the merit of using noncationic bionanomaterials in cardiovascular applications.
Further translation of miR-SPIONs as anti-atherosclerosis nanomedicines will require several limitations to be addressed. Our miR-SPIONs exhibit a blood half-life of ∼2 h, on par with noncationic nucleic acid-based nanostructures used in nonatherosclerosis applications (e.g., DNA nanogel [t1/2 = 1.14 h]) (57), DNA tetrahedron (t1/2 = 24.2 min) (58), and siRNA-protein octamer (t1/2 ∼ several hours) (59), as well as cationic lipid-nucleic acid nanocomplexes used in atherosclerosis applications (e.g., miRNA-loaded micelles [t1/2 < 2 h] (60), siRNA-loaded lipid NPs [t1/2 = 1.2 h] (61), and antagomir-loaded liposomes [t1/2 = 190.8 s] (30)). While the blood pharmacokinetics of miR-146a-SPIONs are sufficient for plaque delivery, their negative surface charge may contribute to clearance by the liver and spleen (17, 62). To make plaque delivery more efficient, there is still considerable room for improving the pharmacological properties of atherosclerosis nanomedicines that adopt a gene regulation approach, compared with those that exploit other therapeutic mechanisms (e.g., statin-loaded reconstituted HDL [t1/2 = 21.9 h] (63) and mucic acid NPs that modulate SR expression [t1/2 = 28 h] (12)). Moreover, as the central premise of our delivery strategy to atherosclerotic plaques lies in the ability of SNAs to naturally target SR-A, other types of nucleic acid-based nanostructures (64, 65) that enter cells via SR may also be gene carriers for atherosclerosis. For instance, DNA octahedral nanocages enter cells via LOX-1, another member of the SR family (66). The uncertainty is that whereas many past studies documented profuse SR-A expression in human atherosclerotic macrophages (67, 68), reports on the SR-A expression in human ECs remain scarce. Clear understanding in the cardiovascular biology of SR in humans will enhance the translational potential of nucleic acid nanotechnology.
On the role of miR-146a in atherogenesis, an established understanding is that miR-146a endows atheroprotective properties to the blood vessel wall and negatively regulates the NF-κB pathway linked to inflammation and atherosclerosis (22). Interestingly, deletion of miR-146a in bone marrow (BM)-derived cells may alleviate atherosclerosis. Cheng et al. prepared two groups of atherosclerosis mouse models with genetic knockout of the low-density lipoprotein receptor (Ldlr−/−) by providing the same HCD for 12 wk (69). The first group of Ldlr−/− mice received transplantation of BM collected from wildtype mice, whereas the second group received transplantation of BM collected from mice with genetic knockout of miR-146a (miR-146a−/−). The authors observed higher levels of circulating proinflammatory markers (e.g., soluble ICAM-1 and TNF-α), but paradoxically, smaller plaques in the aortic arch for the second group. Furthermore, the authors detected fewer circulating proatherogenic leukocytes (which led to reduced atherosclerosis) and larger spleen sizes in the second group. It turns out that miR-146a in BM-derived cells protects against HCD-induced exhaustion of hematopoietic stem cells in the BM (70) and prevents splenomegaly (71). However, the effect of miR-146a deficiency in hematopoietic cells on atherosclerosis remains inconsistent because Del Monte et al. later reported a limited difference in plaque sizes in the aortic arch and aortic root between these two mice groups that were fed with an HCD for 20 wk instead (72). A follow-up report even documented the presence of more large neutrophil extracellular traps in the atherosclerotic plaques and adhered to the vascular wall for the second group of mice (73). In our work, a full systemic treatment of miR-146a-SPIONs reduced proinflammatory cytokines in the aorta (Fig. 3B) without changing spleen size (SI Appendix, Fig. S37B) or population of leukocytes (SI Appendix, Fig. S38A) in plaque-bearing ApoE−/− mice 4 wk after treatment. As our treatment entails the delivery of miR-146a to the plaque rather than its depletion in the BM, the mechanism of action of miR-146a-SPIONs likely stems from regulation of the NF-κB pathway rather than exhaustion of hematopoietic stem cells in the BM.
Materials and Methods
For detailed Materials and Methods, please see SI Appendix.
Preparation of miR-146a-SPIONs.
Amine-modified, PS miR-146a strands (miR-NH2) were covalently attached to carboxylate-terminated PEG-SPIONs, following literature precedent on the attachment of DNA oligonucleotides to carboxylate-terminated PEG-SPIONs (25). The sequence of miR-146a is 5′ UGAGAACUGAAUUCCAUGGGUU 3′ (Idobio). See SI Appendix for methods of preparing PEG-SPIONs.
In Vivo Efficacy Studies.
ApoE−/− mice fed with an HCD for 9 wk were randomly divided into various treatment groups of nine animals each. Mice were i.v. injected with either PEG-SPIONs (10 mg/kg Fe), miR-146a-SPIONs (10 mg/kg Fe and 1.5 mg/kg miRNA), miR-nc-SPIONs (10 mg/kg Fe and 1.5 mg/kg miRNA), miR-146a (1.5 mg/kg) complexed with Lipofectamine 3000 (60 µg/kg), or PBS twice a week from weeks 10–12, with HCD maintained throughout the treatment period. The mice were killed on week 12 for further analysis. The size of treatment group was estimated based on Dunnett’s formalism and power calculation (α = 0.05, power = 0.80). Briefly, μ = √N δ/σ, where μ is the correlation coefficient that depends on the size of each group, N. With four different treatment groups, μ is 4.46 (74). If the superior treatment group gives an outcome (δ) of 1.5 SD (σ) better than the control group, the required size of N is (4.46 ÷ 1.5)2 = 8.84, or equivalently nine mice per group.
Supplementary Material
Acknowledgments
We thank Randy Cheung, Josie Lai, and Anny Cheung from the School of Biomedical Sciences at The Chinese University of Hong Kong (CUHK) for guidance in sectioning and staining, Prof. James Y. W. Lau from the Department of Surgery at CUHK for providing the ApoE−/− mouse model, and Prof. Dennis K. P. Ng from the Department of Chemistry at CUHK for supporting the near-infrared fluorescence imaging studies. Q.B. thanks Yao Liu and Lok Wai Cola Ho for guidance in IF staining. This work was supported in part by General Research Funds (Project No.14300120, No.14109519, and No.14105321) from the Research Grants Council, the Chow Yuk Ho Technology Centre for Innovative Medicine, a Vice Chancellor Discretionary Fund at CUHK, and the National Nature Science Foundation of China (No.81922078). C.H.J.C. acknowledges a Croucher Innovation Award from the Croucher Foundation.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. E.S.D. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2201443119/-/DCSupplemental.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Andreou I., Sun X., Stone P. H., Edelman E. R., Feinberg M. W., miRNAs in atherosclerotic plaque initiation, progression, and rupture. Trends Mol. Med. 21, 307–318 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hansson G. K., Libby P., The immune response in atherosclerosis: A double-edged sword. Nat. Rev. Immunol. 6, 508–519 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Liu M. W., et al. , Trapidil in preventing restenosis after balloon angioplasty in the atherosclerotic rabbit. Circulation 81, 1089–1093 (1990). [DOI] [PubMed] [Google Scholar]
- 4.Sarembock I. J., et al. , Influence of inflation pressure and balloon size on the development of intimal hyperplasia after balloon angioplasty. A study in the atherosclerotic rabbit. Circulation 80, 1029–1040 (1989). [DOI] [PubMed] [Google Scholar]
- 5.Fleg J. L., et al. , Effect of statins alone versus statins plus ezetimibe on carotid atherosclerosis in type 2 diabetes: The SANDS (Stop Atherosclerosis in Native Diabetics Study) trial. J. Am. Coll. Cardiol. 52, 2198–2205 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tang J., et al. , Inhibiting macrophage proliferation suppresses atherosclerotic plaque inflammation. Sci. Adv. 1, 1400223 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan C. K. W., et al. , Recent advances in managing atherosclerosis via nanomedicine. Small 14, 1702793 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Cormode D. P., et al. , Atherosclerotic plaque composition: Analysis with multicolor CT and targeted gold nanoparticles. Radiology 256, 774–782 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim B. Y. S., Rutka J. T., Chan W. C. W., Nanomedicine. N. Engl. J. Med. 363, 2434–2443 (2010). [DOI] [PubMed] [Google Scholar]
- 10.Kuai R., Li D., Chen Y. E., Moon J. J., Schwendeman A., High-density lipoproteins: Nature’s multifunctional nanoparticles. ACS Nano 10, 3015–3041 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nahrendorf M., et al. , Noninvasive vascular cell adhesion molecule-1 imaging identifies inflammatory activation of cells in atherosclerosis. Circulation 114, 1504–1511 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Lewis D. R., et al. , Sugar-based amphiphilic nanoparticles arrest atherosclerosis in vivo. Proc. Natl. Acad. Sci. U.S.A. 112, 2693–2698 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beldman T. J., et al. , Hyaluronan nanoparticles selectively target plaque-associated macrophages and improve plaque stability in atherosclerosis. ACS Nano 11, 5785–5799 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chiu Y. T. E., Li H., Choi C. H. J., Progress toward understanding the interactions between DNA nanostructures and the cell. Small 15, e1805416 (2019). [DOI] [PubMed] [Google Scholar]
- 15.Yamankurt G., et al. , The effector mechanism of siRNA spherical nucleic acids. Proc. Natl. Acad. Sci. U.S.A. 117, 1312–1320 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang J., et al. , Immune cell screening of a nanoparticle library improves atherosclerosis therapy. Proc. Natl. Acad. Sci. U.S.A. 113, E6731–E6740 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ho L. W. C., Liu Y., Han R., Bai Q., Choi C. H. J., Nano-cell interactions of non-cationic bionanomaterials. Acc. Chem. Res. 52, 1519–1530 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Thakor A. S., et al. , Clinically approved nanoparticle imaging agents. J. Nucl. Med. 57, 1833–1837 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ye M., et al. , SR-A-targeted phase-transition nanoparticles for the detection and treatment of atherosclerotic vulnerable plaques. ACS Appl. Mater. Interfaces 11, 9702–9715 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Li K., Ching D., Luk F. S., Raffai R. L., Apolipoprotein E enhances microRNA-146a in monocytes and macrophages to suppress nuclear factor-κB-driven inflammation and atherosclerosis. Circ. Res. 117, e1–e11 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma S., et al. , E-selectin-targeting delivery of microRNAs by microparticles ameliorates endothelial inflammation and atherosclerosis. Sci. Rep. 6, 22910 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feinberg M. W., Moore K. J., MicroRNA regulation of atherosclerosis. Circ. Res. 118, 703–720 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu L. S., et al. , α-L-threose nucleic acids as biocompatible antisense oligonucleotides for suppressing gene expression in living cells. ACS Appl. Mater. Interfaces 10, 9736–9743 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Choi C. H. J., Hao L., Narayan S. P., Auyeung E., Mirkin C. A., Mechanism for the endocytosis of spherical nucleic acid nanoparticle conjugates. Proc. Natl. Acad. Sci. U.S.A. 110, 7625–7630 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang L., et al. , Promoting the delivery of nanoparticles to atherosclerotic plaques by DNA coating. ACS Appl. Mater. Interfaces 11, 13888–13904 (2019). [DOI] [PubMed] [Google Scholar]
- 26.Getz G. S., Reardon C. A., Diet and murine atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 26, 242–249 (2006). [DOI] [PubMed] [Google Scholar]
- 27.Kim H., Kumar S., Kang D.-W., Jo H., Park J.-H., Affinity-driven design of cargo-switching nanoparticles to leverage a cholesterol-rich microenvironment for atherosclerosis therapy. ACS Nano 14, 6519–6531 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taganov K. D., Boldin M. P., Chang K.-J., Baltimore D., NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. U.S.A. 103, 12481–12486 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheng H. S., et al. , MicroRNA-146 represses endothelial activation by inhibiting pro-inflammatory pathways. EMBO Mol. Med. 5, 1017–1034 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kheirolomoom A., et al. , Multifunctional nanoparticles facilitate molecular targeting and miRNA delivery to inhibit atherosclerosis in ApoE−/− mice. ACS Nano 9, 8885–8897 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rekhter M. D., Collagen synthesis in atherosclerosis: Too much and not enough. Cardiovasc. Res. 41, 376–384 (1999). [DOI] [PubMed] [Google Scholar]
- 32.Kuehn C., et al. , Suppression of proatherogenic leukocyte interactions by MCS-18--Impact on advanced atherosclerosis in ApoE-deficient mice. Atherosclerosis 245, 101–110 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Roth Flach R. J., et al. , Endothelial protein kinase MAP4K4 promotes vascular inflammation and atherosclerosis. Nat. Commun. 6, 8995 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Budatha M., et al. , Inhibiting integrin α5 cytoplasmic domain signaling reduces atherosclerosis and promotes arteriogenesis. J. Am. Heart Assoc. 7, e007501 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stanciu L. A., Djukanovic R., The role of ICAM-1 on T-cells in the pathogenesis of asthma. Eur. Respir. J. 11, 949–957 (1998). [DOI] [PubMed] [Google Scholar]
- 36.Basatemur G. L., Jørgensen H. F., Clarke M. C. H., Bennett M. R., Mallat Z., Vascular smooth muscle cells in atherosclerosis. Nat. Rev. Cardiol. 16, 727–744 (2019). [DOI] [PubMed] [Google Scholar]
- 37.Bentzon J. F., Otsuka F., Virmani R., Falk E., Mechanisms of plaque formation and rupture. Circ. Res. 114, 1852–1866 (2014). [DOI] [PubMed] [Google Scholar]
- 38.Komaravolu R. K., Waltmann M. D., Konaniah E., Jaeschke A., Hui D. Y., ApoER2 (Apolipoprotein E receptor-2) deficiency accelerates smooth muscle cell senescence via cytokinesis impairment and promotes fibrotic neointima after vascular injury. Arterioscler. Thromb. Vasc. Biol. 39, 2132–2144 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee C. R., et al. , Genetic variation in soluble epoxide hydrolase (EPHX2) and risk of coronary heart disease: The atherosclerosis risk in communities (ARIC) study. Hum. Mol. Genet. 15, 1640–1649 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fornage M., et al. , Polymorphism of the soluble epoxide hydrolase is associated with coronary artery calcification in African-American subjects: The coronary artery risk development in young adults (CARDIA) study. Circulation 109, 335–339 (2004). [DOI] [PubMed] [Google Scholar]
- 41.Huang X., et al. , Two novel SNPs in the PLCL2 gene associated with large artery atherosclerotic stroke identified by fine-mapping. J. Mol. Neurosci. 70, 496–503 (2020). [DOI] [PubMed] [Google Scholar]
- 42.Hirokawa M., et al. , A genome-wide association study identifies PLCL2 and AP3D1-DOT1L-SF3A2 as new susceptibility loci for myocardial infarction in Japanese. Eur. J. Hum. Genet. 23, 374–380 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mehta J. L., Chen J., Hermonat P. L., Romeo F., Novelli G., Lectin-like, oxidized low-density lipoprotein receptor-1 (LOX-1): A critical player in the development of atherosclerosis and related disorders. Cardiovasc. Res. 69, 36–45 (2006). [DOI] [PubMed] [Google Scholar]
- 44.Yan Y., et al. , Omega-3 fatty acids prevent inflammation and metabolic disorder through inhibition of NLRP3 inflammasome activation. Immunity 38, 1154–1163 (2013). [DOI] [PubMed] [Google Scholar]
- 45.Greaney A. J., et al. , Frontline science: Anthrax lethal toxin-induced, NLRP1-mediated IL-1β release is a neutrophil and PAD4-dependent event. J. Leukoc. Biol. 108, 773–786 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sato Y., Role of ETS family transcription factors in vascular development and angiogenesis. Cell Struct. Funct. 26, 19–24 (2001). [DOI] [PubMed] [Google Scholar]
- 47.Zhang X., et al. , Calcium/calmodulin-dependent protein kinase (CaMK) IV mediates nucleocytoplasmic shuttling and release of HMGB1 during lipopolysaccharide stimulation of macrophages. J. Immunol. 181, 5015–5023 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Junho C. V. C., Caio-Silva W., Trentin-Sonoda M., Carneiro-Ramos M. S., An overview of the role of calcium/calmodulin-dependent protein kinase in cardiorenal syndrome. Front. Physiol. 11, 735 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zheng J., et al. , Polygonum multiflorum and Codonopsis pilosula granule alleviates atherosclerosis by inhibiting the expression of DAB2IP-ASK1 pathway in vascular endothelial cells. Vasc. Investig. Ther. 3, 6–14 (2020). [Google Scholar]
- 50.Luong A., et al. , Cezanne regulates inflammatory responses to hypoxia in endothelial cells by targeting TRAF6 for deubiquitination. Circ. Res. 112, 1583–1591 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Le N.-T., et al. , A crucial role for p90RSK-mediated reduction of ERK5 transcriptional activity in endothelial dysfunction and atherosclerosis. Circulation 127, 486–499 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Missiou A., et al. , Tumor necrosis factor receptor-associated factor 1 (TRAF1) deficiency attenuates atherosclerosis in mice by impairing monocyte recruitment to the vessel wall. Circulation 121, 2033–2044 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ackers I., Szymanski C., Silver M. J., Malgor R., Oxidized low-density lipoprotein induces WNT5A signaling activation in THP-1 derived macrophages and a human aortic vascular smooth muscle cell line. Front. Cardiovasc. Med. 7, 567837 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brandt M. M., et al. , Endothelial loss of Fzd5 stimulates PKC/Ets1-mediated transcription of Angpt2 and Flt1. Angiogenesis 21, 805–821 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu S., et al. , NDRG2 induced by oxidized LDL in macrophages antagonizes growth factor productions via selectively inhibiting ERK activation. Biochim. Biophys. Acta 1801, 106–113 (2010). [DOI] [PubMed] [Google Scholar]
- 56.Matthijs Blankesteijn W., Hermans K. C. M., Wnt signaling in atherosclerosis. Eur. J. Pharmacol. 763 (Pt A), 122–130 (2015). [DOI] [PubMed] [Google Scholar]
- 57.Ding F., et al. , A crosslinked nucleic acid nanogel for effective siRNA delivery and antitumor therapy. Angew. Chem. Int. Ed. Engl. 57, 3064–3068 (2018). [DOI] [PubMed] [Google Scholar]
- 58.Lee H., et al. , Molecularly self-assembled nucleic acid nanoparticles for targeted in vivo siRNA delivery. Nat. Nanotechnol. 7, 389–393 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tai W., Li J., Corey E., Gao X., A ribonucleoprotein octamer for targeted siRNA delivery. Nat. Biomed. Eng. 2, 326–337 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chin D. D., et al. , miR-145 micelles mitigate atherosclerosis by modulating vascular smooth muscle cell phenotype. Biomaterials 273, 120810 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tao W., et al. , siRNA nanoparticles targeting CaMKIIγ in lesional macrophages improve atherosclerotic plaque stability in mice. Sci. Transl. Med. 12, eaay1063 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Han S., Mahato R. I., Sung Y. K., Kim S. W., Development of biomaterials for gene therapy. Mol. Ther. 2, 302–317 (2000). [DOI] [PubMed] [Google Scholar]
- 63.Duivenvoorden R., et al. , A statin-loaded reconstituted high-density lipoprotein nanoparticle inhibits atherosclerotic plaque inflammation. Nat. Commun. 5, 3065 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang P., et al. , Visualization of the cellular uptake and trafficking of DNA origami nanostructures in cancer cells. J. Am. Chem. Soc. 140, 2478–2484 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Peng X., et al. , DNA nanostructure-programmed cell entry via corner angle-mediated molecular interaction with membrane receptors. Nano Lett. 21, 6946–6951 (2021). [DOI] [PubMed] [Google Scholar]
- 66.Vindigni G., et al. , Receptor-mediated entry of pristine octahedral DNA nanocages in mammalian cells. ACS Nano 10, 5971–5979 (2016). [DOI] [PubMed] [Google Scholar]
- 67.Gough P. J., et al. , Analysis of macrophage scavenger receptor (SR-A) expression in human aortic atherosclerotic lesions. Arterioscler. Thromb. Vasc. Biol. 19, 461–471 (1999). [DOI] [PubMed] [Google Scholar]
- 68.de Winther M. P. J., van Dijk K. W., Havekes L. M., Hofker M. H., Macrophage scavenger receptor class A: A multifunctional receptor in atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 20, 290–297 (2000). [DOI] [PubMed] [Google Scholar]
- 69.Cheng H. S., et al. , Paradoxical suppression of atherosclerosis in the absence of microRNA 146a. Circ. Res. 121, 354–367 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhao J. L., Rao D. S., O’Connell R. M., Garcia-Flores Y., Baltimore D., MicroRNA-146a acts as a guardian of the quality and longevity of hematopoietic stem cells in mice. eLife 2, e00537 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Boldin M. P., et al. , miR-146a is a significant brake on autoimmunity, myeloproliferation, and cancer in mice. J. Exp. Med. 208, 1189–1201 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Del Monte A., et al. , miR-146a deficiency in hematopoietic cells is not involved in the development of atherosclerosis. PLoS One 13, e0198932 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Arroyo A. B., et al. , miR-146a is a pivotal regulator of neutrophil extracellular trap formation promoting thrombosis. Haematologica 106, 1636–1646 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dunnett C. W., A multiple comparison procedure for comparing several treatments with a control. J. Am. Stat. Assoc. 50, 1096–1121 (1955). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.